Knowledge of lncRNAs in multiple myeloma (MM) is continuously increasing.1,2 We and others have recently provided evidence by microarray3 or RNA sequencing (RNA-seq) analyses2,4 that distinct lncRNA transcriptional signatures distinguished myeloma cells from their normal counterparts, were specifically associated with molecular subgroups or progressive disease phases, and could be independently related to MM clinical outcome.4 Among the most expressed lncRNAs, we identified the nuclear paraspeckle assembly transcript 1 (NEAT1),2 already reported to be over-expressed in many types of solid tumors, raising the hypothesis that it may play a critical oncogenic role and facilitate tumorigenesis.5 Herein, we demonstrate that MM cells significantly over-express NEAT1 and its deregulation is unrelated to patients’ prognosis. However, the putative NEAT1 involvement in cellular stress response makes it an attractive candidate for targeted therapy in the disease.
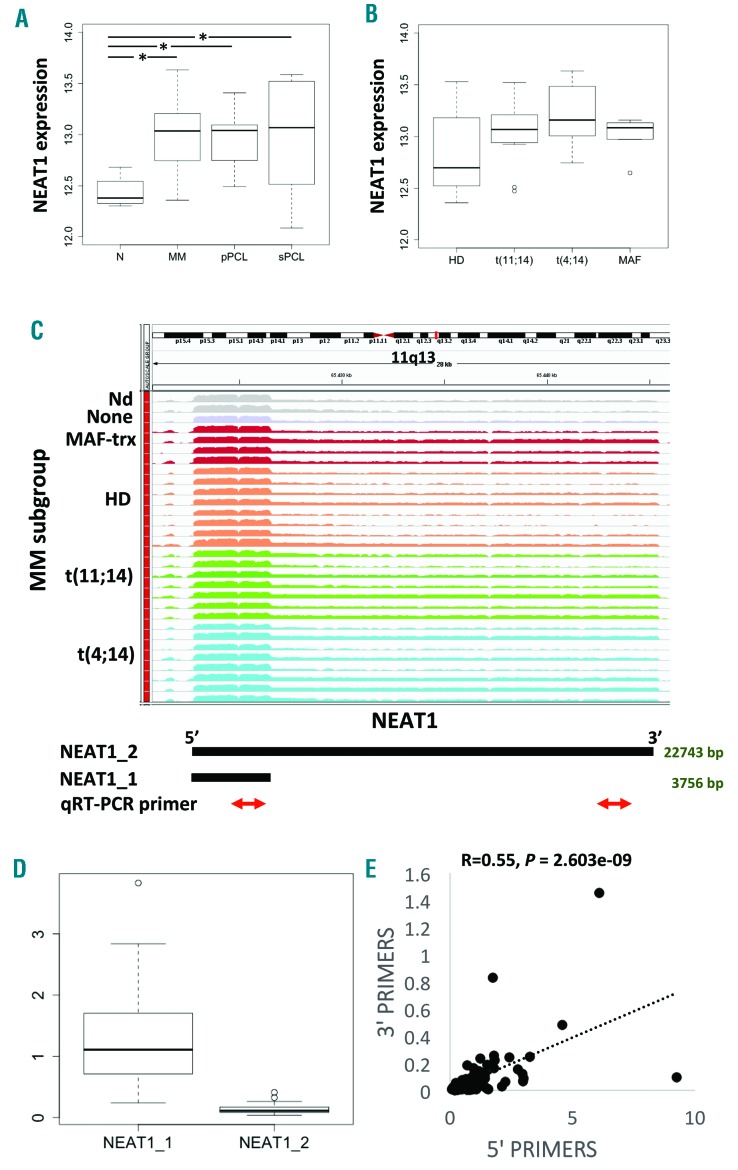
The expression profile of NEAT1 has been primarily investigated by GeneChip® Human Gene 2.0 ST array in a cohort of 50 MMs representative of the major molecular characteristics of the disease (Online Supplementary Table S1), in 9 primary plasma cell leukemia (pPCL) and 6 secondary PCL (sPCL) patients, and in 4 purified bone marrow (BM) PCs from healthy controls (N). Almost 19,000 coding genes and 10,138 lncRNAs have been annotated based on the GENCODE encyclopedia, as previously described.3 NEAT1 belonged to a short list of 17 lncRNAs markedly expressed in MM patients (Online Supplementary Table S2). NEAT1 presents two isoforms: a canonically polyadenylated short transcript of 3.7 kb (NEAT1_1) and a longer non-polyadenylated transcript (NEAT1_2) of about 23 kb. The analysis indicated that NEAT1 showed significant overexpression in pathological MM and both pPCL and sPCL samples as compared with healthy donors (Figure 1A). Notably, MM patients over-expressed NEAT1 irrespectively of their molecular characteristic, i.e. the presence of chromosomal translocations, hyperdiploidy, 13q and 17p13 deletion, gain of 1q arm, mutations of the MAPK-pathway (BRAF, NRAS, or KRAS), or FAM46C and DIS3 genes (Figure 1B and Online Supplementary Figure S1).
Figure 1.
NEAT1 expression by GeneChip® Human Gene 2.0 ST arrays. (A) Boxplot of NEAT1 expression shows a significant upregulation in pathological multiple myeloma (MM), primary plasma cell leukemia (pPCL) and secondary PCL (sPCL) samples as compared with healthy control based on Dunn test; *P<0.01. (B) Boxplot of NEAT1 expression does not show any significant correlation with group membership defined by t(11;14), t(4;14), MAF translocation, and hyperdiploid status. (C) NEAT1 expression by RNA sequencing. Visualization of RNA-seq data: zoomed view of the NEAT1 lncRNA region; the coverage bigWig files generated using bamCoverage function in deeptools (http://deeptools.readthedocs.io/en/latest/content/tools/bamCoverage.html) and the human genome annotation file (GENCODE v.25) were loaded into the Integrated Genome Viewer (IGV) (http://www.broadinstitute.org/igv/). The y-axis shows the scaled number of reads mapping to each location of the genome in the NEAT1 region (x-axis) schematically reported below together with qRT-PCR primers. Each lane represents a MM patient: different colors refer to the sample molecular characteristic indicated at the left. In order to compare samples, coverage values from all patients were group-scaled. (D) Boxplot of NEAT1_1 and NEAT1_2 expression by RNA-seq showing that the shorter transcript is expressed approximately 10-fold higher than the longer isoform. Transcript abundance was estimated as Fragments Per Kilobase per Million mapped reads (FPKM). (E) Pearson analysis correlating NEAT1 total expression (x-axis) with the long transcript expression (y-axis). Correlation coefficient R and significant P-value are reported. HD: hyperdiploid.
Subsequently, we evaluated the expression of NEAT1 in 30 of the 50 MM patients for whom RNA-seq data were available2 (Figure 1C). RNA-seq allowed estimation of NEAT1_1 and NEAT1_2 isoforms abundance based on the presence of unambiguously mapped reads. Importantly, we found that NEAT1_1 isoform was much more abundant than the longer NEAT1_2 isoform, being approximately 90% of total NEAT1 abundance (Figure 1D). The results on these 30 patients, representative of the major genetic/prognostic lesions, confirmed that NEAT1 is not differentially expressed in the diverse MM molecular subtypes.2
NEAT1 expression levels were therefore validated by quantitative real-time PCR (qRT-PCR) (Online Supplementary Table S3) in 60 (46 MM and 14 PCL) of the pathological samples examined by arrays and in 46 additional patients including 36 MM, 5 smoldering MM, 5 PCL. NEAT1 global expression levels were first obtained by targeting the 5’-region, to amplify both transcripts. The expression pattern observed was consistent with the array results (Online Supplementary Figure S2). Furthermore, we used a second primer configuration that targeted the 3’-region, to amplify exclusively the NEAT1_2 isoform. As such, we confirmed the estimated proportion of NEAT1_2 in the overall NEAT1 expression as shown by RNA-seq and proved that the long isoform NEAT1_2 was positively correlated with total NEAT1 (Figure 1E). No differential expression was found in the small subset of smoldering MM (Online Supplementary Figure S3), in line with previous observations that indicated no significant differences between pathological specimens. Next, we validated these findings in two publicly available array-based datasets from the University of Arkansas for Medical Science (UAMS), including 22 healthy donors, 12 monoclonal gammopathy of undetermined significance (MGUS), 44 smoldering MM (#GSE5900 series on NCBI GEO repository) and the large array-profiled UAMS TT2/TT3 trials cohort encompassing more than 550 patients (#GSE2658 and #GSE24080). Underexpression of NEAT1 was confirmed in healthy donors; NEAT1 levels were also significantly reduced in MGUS patients compared to MM (Online Supplementary Figure S4). However, for the sake of clarity, it is worth specifying that these datasets, given the related array configuration, investigated preferentially the short polyadenylated NEAT1_1 isoform.
To gain insights into the possible role of deregulated NEAT1 in MM, we investigated the protein-coding genes concurrently detected by Gene 2.0 ST array. NEAT1 is an indispensable structural component of paraspeckles (PSs), peculiar lncRNA-directed nuclear bodies potentially involved in stress response. Although their exact function still has to be fully elucidated, PSs may affect gene expression by regulating the transcription and pre-mRNA splicing events and holding nuclear mRNA for editing. NEAT1 could control these events by modulating the functions of PSs upon exposure to specific stress events.6–8 Notably, in our cohort, NEAT1 expression is significantly correlated with that of NONO and SFPQ (Pearson correlation R>0.3, P<0.01) (Online Supplementary Figure S5), both encoding for proteins essential to the formation of minimal ribonucleoprotein particles.9 These data suggest that NEAT1 overexpression in MM may be associated with increased PSs formation.
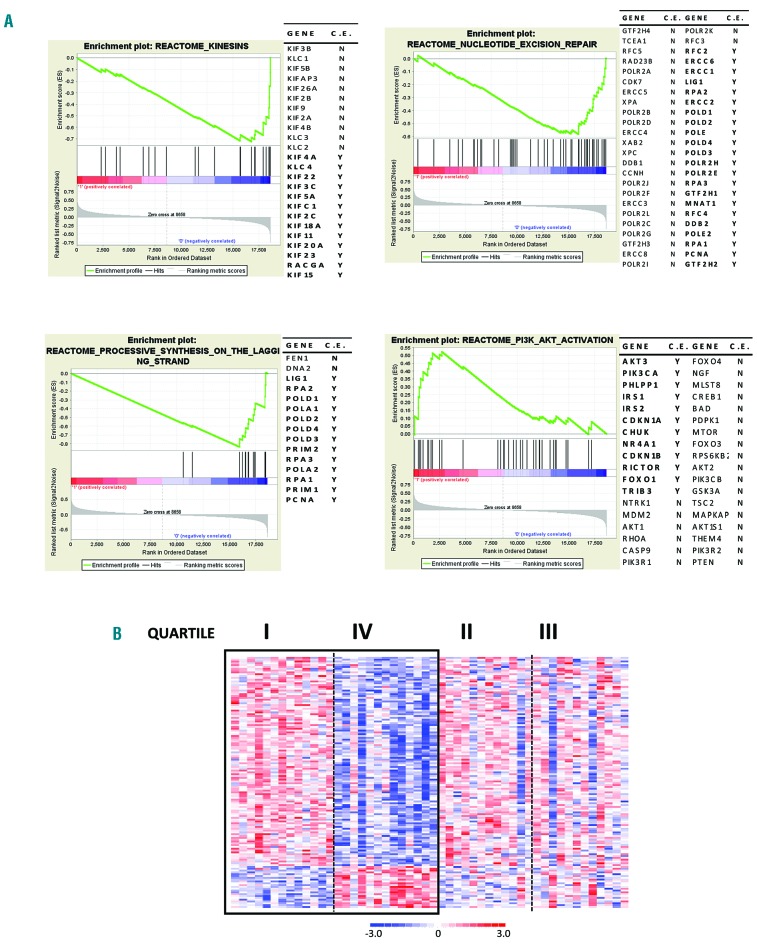
With NEAT1 being expressed at a high level in all tumor samples but presenting low variance across the whole dataset, we focused on those patients at the extremes of the expression distribution to unravel putative biological effects associated with its modulation. Namely, we compared the transcriptional profiles of patients showing the highest and the lowest quartile of NEAT1 expression levels, obtaining 138 differentially expressed genes (Online Supplementary Table S4). Functional annotation analysis of this signature, aimed at identifying highly significant represented categories, interestingly revealed enrichment in the unfolded protein response (UPR) category, which is the cellular response to the endoplasmic reticulum stress due to the accumulation of unfolded or misfolded proteins (Online Supplementary Table S5). Specifically, among the 7 genes included in this gene set (BAG3, EXOSC2, TUBB2A, DDX10, PREB, NFYA, GEMIN4), MM with the highest NEAT1 transcript levels under-expressed all of them but BAG3. Since persistent activation of UPR in MM leads to apoptosis,10 the downmodulation of UPR pathway genes might suggest a survival attempt for myeloma cells. Furthermore, the functional enrichment analysis indicated that higher NEAT1 expression was associated with lower expression of DYNLL1, essential for p53 nuclear trafficking, with consequent transcriptional activation of genes involved in growth arrest and apoptosis in response to DNA damage.11 Consistently with a reduced amount of p53 dynein-dependent nuclear translocation, enrichment analysis using gene set enrichment analysis (GSEA) showed that higher NEAT1 expression was correlated with weakened DNA repair-associated pathways. GSEA identified also enrichment in pathways associated with DNA synthesis and repair in MM with lower NEAT1 levels, and PI3K/AKT activation pathway in patients with high NEAT1 expression (Figure 2), in agreement with recent data describing NEAT1 as a crucial player in suppressing transformation in response to oncogenic signals.12 For completeness, GSEA analysis was run in the #GSE2658 series, which confirmed significance in PI3KT/AKT activation pathway (Online Supplementary Table S6).
Figure 2.
Functional annotation of NEAT1 signature in multiple myeloma (MM). (A) Enrichment plots of four selected gene sets detected by gene set enrichment analysis. The green curves show the enrichment score and reflect the degree to which each gene (black vertical lines) is represented at the bottom of the ranked gene list. Genes contributing to the core enrichment (CE) in the gene set are indicated in bold with Y. (B) Heatmap of the 138 differentially expressed genes identified by comparing the first versus fourth quartile of 50 MM patients stratified into four groups based on NEAT1 expression level.
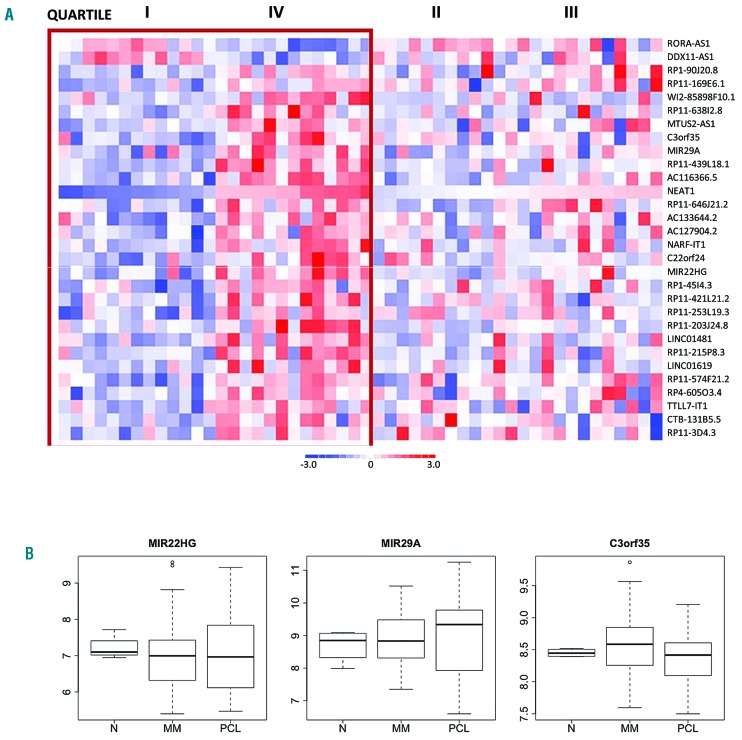
Subsequently, we found that high NEAT1 expression was associated with 27 over-expressed and 2 under-expressed lncRNA transcripts (Figure 3 and Online Supplementary Table S7). Although most of these are virtually uncharacterized, three over-expressed ncRNAs, namely the two miRNA precursors MIR22HG and MIR29A, and C3orf35 (Figure 3), have already been reported in the context of MM. In particular, MIR22HG located at 17p13 was under-expressed in different solid tumors13 and in MM.4 It produces the mature miR-22, positively associated with progression-free survival (PFS) in pPCL.14 In our cohort of patients, no differences in MIR22HG expression could be observed among healthy donors, MM and PCL samples. MIR29A, mapped at 7q32, was over-expressed in MM showing the highest NEAT1 level. From MIR29A originate 2 miRNAs, miR-29a and miR-29b-1, whose tumor suppressor activities have been well documented in MM.15 With regard to the expression levels of lncRNA, no significant differences were found between MIR29A expression in healthy and pathological samples, the latter showing a very heterogeneous expression pattern. Finally, C3orf35, mapped at 3p22, was recently described as over-expressed in MM versus normal control,4 although these data were not confirmed in our microarray dataset.
Figure 3.
lncRNAs differentially expressed based on NEAT1 expression. (A) Heatmap of the 30 differentially expressed lncRNAs identified by comparing the first versus fourth quartile of 50 multiple myeloma (MM) patients stratified into four groups based on NEAT1 expression level. (B) Boxplot of MIR22HG, MIR29A, and C3orf35 expression levels in 4 healthy controls (N), 50 MM and 15 plasma cell leukemia (PCL) samples profiled on GeneChip® Human Gene 2.0 ST arrays.
To evaluate the potential prognostic value of NEAT1 in MM, we analyzed a retrospectively collected proprietary dataset including 55 MM at diagnosis for whom clinical data were available, over a median follow up of 54 months. In particular, we assessed the expression of both global NEAT1 gene and the longer NEAT1_2 transcript separately by qRT-PCR, neither of them are correlated with overall survival nor the time-to-next-treatment (Online Supplementary Table S8). Furthermore, we extended the qRT-PCR analysis to 12 additional MM patients (7 of them included in the 55-sample dataset) for whom we had serial samples at the onset and relapse; but even in this case no significant variation in NEAT1 expression was observed (Online Supplementary Figure S6). Finally, we evaluated the NEAT1 prognostic significance in the TT2/TT3 trials cohort, showing no significant correlation between NEAT1_1 isoform expression and overall survival (Online Supplementary Figure S7).
In conclusion, MM significantly over-expresses NEAT1, although, based on the available data, its deregulation does not appear to directly affect patients’ prognosis. However, the putative NEAT1 involvement in different mechanisms of cellular stress response, such as the UPR and p53 pathways, makes it a confident candidate for further studies in a perspective of targeted therapy in the disease.
Supplementary Material
Footnotes
Funding: this work was financially supported by grants from Associazione Italiana Ricerca sul Cancro (AIRC) to Antonino Neri (IG16722, IG10136, and the “Special Program Molecular Clinical Oncology-5 per mille” #9980, 2010/15); ET was supported by a fellowship (#19370) from Fondazione Italiana Ricerca sul cancro (FIRC); KT was supported by a fellowship from Fondazione Umberto Veronesi.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Nobili L, Ronchetti D, Agnelli L, Taiana E, Vinci C, Neri A. Long Non-Coding RNAs in Multiple Myeloma. Genes (Basel). 2018;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ronchetti D, Agnelli L, Pietrelli A, et al. A compendium of long non-coding RNAs transcriptional fingerprint in multiple myeloma. Sci Rep. 2018;8(1):6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronchetti D, Agnelli L, Taiana E, et al. Distinct lncRNA transcriptional fingerprints characterize progressive stages of multiple myeloma. Oncotarget. 2016;7(12):14814–14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samur MK, Minvielle S, Gulla A, et al. Long intergenic non-coding RNAs have an independent impact on survival in multiple myeloma. Leukemia. 2018;32(12):2626–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu X, Li Z, Zheng H, Chan MT, Wu WK. NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif. 2017;50(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adriaens C, Standaert L, Barra J, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22(8):861–868. [DOI] [PubMed] [Google Scholar]
- 7.Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10(3):456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3’-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31(20): 4020–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox AH, Nakagawa S, Hirose T, Bond CS. Paraspeckles: Where Long Noncoding RNA Meets Phase Separation. Trends Biochem Sci. 2018;43(2):124–135. [DOI] [PubMed] [Google Scholar]
- 10.Woo CW, Cui D, Arellano J, et al. Adaptive suppression of the ATF4-CHOP branch of the unfolded protein response by toll-like receptor signalling. Nat Cell Biol. 2009;11(12):1473–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo KW, Kan HM, Chan LN, et al. The 8-kDa dynein light chain binds to p53-binding protein 1 and mediates DNA damage-induced p53 nuclear accumulation. J Biol Chem. 2005;280(9):8172–8179. [DOI] [PubMed] [Google Scholar]
- 12.Mello SS, Sinow C, Raj N, et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31(11): 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui Z, An X, Li J, Liu Q, Liu W. LncRNA MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression and inhibits endometrial carcinoma cells proliferation. Biomed Pharmacother. 2018;104:223–228. [DOI] [PubMed] [Google Scholar]
- 14.Lionetti M, Musto P, Di Martino MT, et al. Biological and clinical relevance of miRNA expression signatures in primary plasma cell leukemia. Clin Cancer Res. 2013;19(12):3130–3142. [DOI] [PubMed] [Google Scholar]
- 15.Stamato MA, Juli G, Romeo E, et al. Inhibition of EZH2 triggers the tumor suppressive miR-29b network in multiple myeloma. Oncotarget. 2017;8(63):106527–106537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.