Phenotypic drug susceptibility testing (DST) for the two first-line tuberculosis drugs ethambutol and pyrazinamide is known to yield unreliable and inaccurate results. In this prospective study, we propose a diagnostic algorithm combining phenotypic DST with Sanger sequencing to inform clinical decision-making for drug-resistant Mycobacterium tuberculosis complex isolates.
KEYWORDS: Mycobacterium tuberculosis, ethambutol, pyrazinamide
ABSTRACT
Phenotypic drug susceptibility testing (DST) for the two first-line tuberculosis drugs ethambutol and pyrazinamide is known to yield unreliable and inaccurate results. In this prospective study, we propose a diagnostic algorithm combining phenotypic DST with Sanger sequencing to inform clinical decision-making for drug-resistant Mycobacterium tuberculosis complex isolates. Sequencing results were validated using whole-genome sequencing (WGS) of the isolates. Resistance-conferring mutations obtained by pncA sequencing correlated well with phenotypic DST results for pyrazinamide. Phenotypic resistance to ethambutol was only partly explained by mutations in the embB 306 codon. Additional resistance-conferring mutations were found in the embB gene at codons 354, 406, and 497. In several isolates that tested ethambutol susceptibility by phenotypic DST, well-known resistance-conferring embB mutations were determined. Thus, targeted Sanger sequencing beyond the embB 306 codon or WGS together with phenotypic DST should be employed to ensure reliable ethambutol drug susceptibility testing, as a basis for the rational design of multidrug-resistant tuberculosis regimens with or without ethambutol.
INTRODUCTION
Isoniazid (INH), rifampin (RMP), ethambutol (EMB), and pyrazinamide (PZA) are first-line drugs used to treat tuberculosis (TB) caused by the fully susceptible Mycobacterium tuberculosis complex (MTBC) (1). In contrast, multidrug-resistant TB (MDR-TB) caused by MTBC resistant to both RMP and INH requires prolonged treatment with less efficacious and more toxic second-line drugs. Drug susceptibility testing (DST) is crucial to detect resistance, as it informs the choice of effective treatment. While phenotypic DST (pDST) provides reliable and reproducible results for INH and RMP, it yields less accurate results for EMB and PZA (2). Potential explanations include an inaccurate current critical concentration splitting the upper end of the wild-type distribution as well as methodological variations within diagnostic laboratories (3).
In the context of MDR-TB, EMB and PZA are recommended as “add-on agents” regardless of pDST results (4). Side effects caused by PZA and EMB, such as nausea, vomiting, hepatoxicity, and optic neuropathy, often accrete the multiple side effects caused by second-line drugs (5, 6). If reliable and reproducible DST for EMB and PZA was available, unnecessary exposure and harmful side effects could be avoided. With PZA being an integral part of the World Health Organization (WHO) recommended short-course MDR regimen, the reliable detection of PZA resistance is important for countries considering a roll-out of this regimen (4).
Molecular-based methods detecting resistance-conferring mutations proffer alternatives to pDST for select drugs. EMB targets the mycobacterial cell wall by inhibiting the arabinosyl transferases encoded by the embCAB operon, an ∼10-kb region comprising the three adjacent genes embC, embA, and embB (7). Its effect is mainly exerted upon the polymerization steps in the biosynthesis of the arabinan component of cell wall arabinogalactan (8). The pncA-encoded nicotinamidase/pyrazinamidase converts the prodrug PZA to pyrazinoic acid, which disrupts membrane permeability and transport. In accordance with these mechanisms, resistance to EMB and PZA are mainly associated with mutations in the embCAB operon, notably codon 306 of embB, and the pncA gene (9–12).
The present study aimed to investigate an algorithm combining phenotypic and genotypic (pncA and embB306, Sanger sequencing) approaches to determine PZA and EMB susceptibility in RMP-resistant TB isolates for routine use in a laboratory.
RESULTS
A total of 85 unrelated clinical isolates, including 7 RMP-monoresistant and 78 MDR isolates were included. These isolates comprised all four main lineages of M. tuberculosis. A total of 43 isolates belonged to lineage 2 “Beijing” (50.6%), 10 to lineage 3 “Delhi-CAS” (11.8%), 2 to lineage 1 “East-African-Indian” (2.4%), and 30 to lineage 4 “Euro-American” (35.3%) (see Fig. S1 in the supplemental material). For a detailed list of all detected mutations known to be implicated in EMB and PZA resistance as well as the excluded phylogenetic single nucleotide polymorphisms (SNPs), see Table S1 in the supplemental material.
Sequencing and phenotypic drug susceptibility.
pDST classified 52 isolates (61.2%) as PZA resistant. There was 100% concordance between pDST and pncA sequencing results for 49 PZA-resistant and 33 PZA-susceptible isolates. No PCR results could be obtained for three resistant isolates. Next-generation sequencing (NGS) revealed large pncA deletions for two isolates and a pncA T47A mutation for one of these isolates. A total of 28 different mutations, including one double mutation, were identified in the pncA gene. pncA D8G, L4S, and T76P represented the most common variants (Table S1).
For EMB, 42 (49.4%) isolates tested resistant and 43 (50.6%) susceptible by pDST. Overall, 60 of the 85 isolates (70.6%) showed concordance between pDST and embB306 sequencing.
Ethambutol-resistant isolates.
In less than two-thirds of the 42 EMB-resistant isolates (24 of 42, 57.14%), phenotypic resistance was confirmed by identifying a mutation of the embB codon 306 by Sanger sequencing. The remaining 18 isolates were phenotypically resistant to EMB without evidence of embB306 mutations. We repeated pDST for isolates with discordant results. Of 18 phenotypically resistant isolates with embB306 wild type, one isolate tested susceptible on repeat testing. For 16 of the remaining 17 strains, NGS analysis revealed nonsynonymous, nonphylogenetic mutations in the embCAB operon up- or downstream from the embB306 codon. The mutation embB Q497R was present in 7 isolates (once in combination with embB A453A), and embB G406A was found in 5 isolates (once in combination with embB D1024N). The following SNPs were identified in only one isolate each: embB S297A together with embB D1024N, embB D354A with embB D1024N, and embB Q497K. The mutations embBY319C and embB S297A were present only in pDST-resistant strains. Phenotypic resistance to EMB occurred at a significantly higher frequency in isolates that carried mutations at embB codon 306 or 497 (chi-square, P < 0.0001).
Ethambutol-susceptible isolates.
In total, 43 out of 85 isolates (50.6%) tested phenotypically susceptible. Among these isolates, 7 (16.3%) displayed a codon 306 (M306I) mutation in the embB gene, while the majority were found to be embB306 wild type (36 out of 43 susceptible isolates [83.7%]). Of these 7 phenotypically susceptible isolates with an embB306 mutation, 2 isolates tested resistant in a repeated pDST. Of the remaining 5 phenotypically susceptible isolates with an embB306 mutation, 4 displayed elevated MICs (Table 1). Aside from variants in the embB306 codon, 18 susceptible isolates had at least one mutation within embCAB. The mutations embBD354A, embBG406A, and embBQ497K occurred in both pDST-resistant and -susceptible isolates (Table 1; Fig. 1). Increased MICs were detected in 4 of the 5 susceptible isolates with these mutations. One isolate with an embBD354A and an additional embBD1024N mutation was tested resistant when repeated. Of the remaining 31 susceptible isolates, with and without the embCAB mutation, 4 were tested resistant when repeated and 3 had a double mutation in embB (2 times Y334H + embA -12c/T and once embB D328G + embB L74R).
TABLE 1.
embCAB mutations and their corresponding MICs in the selected clinical isolates
| pDST |
||
|---|---|---|
| Mutation(s) | Susceptible (n = 43) (MICs, μg/mla) | Resistant (n = 42) |
| embA -12c/t | 2 (2.5, 3.75) | |
| embA -11c/a | 1 (2.5) | |
| embA P838L | 1 (≤1.25) | |
| embB N296H | 1 (3.75) | |
| embB S297A + embB D1024N | 1 | |
| embB M306I | 6 (3.75 [n = 3], 5 [n = 1], ≥5b [n = 2]) | 3 |
| embB M306I + embB N296H | 1 | |
| embB M306I + embA -16c/a | 1 | |
| embB M306V | 1 (2.5) | 17 |
| embB M306V + Q497P | 1 | |
| embB M306V + embA -11c/t | 1 | |
| embB Y319C | 1 | |
| embB D328G + embB L74R | 1 (≥5b) | |
| embB Y334H + embA -12c/t | 2 (≥5b, ≥5b) | |
| embB D354A | 1 (5) | |
| embB D354A + embB D1024N | 1 (≥5b) | 1 |
| embB T393A | 1 (≤1.25) | |
| embB G406A | 1 (5) | 4 |
| embB G406A + embB D1024N | 1 | |
| embB G406D | 2 (≤1.25, 3.75) | |
| embB Q497K | 2 (5, 5) | 1 |
| embB Q497R | 4 | |
| embB Q497R + embB Q453A | 1 | |
| embB Q497R + embC A387V | 1 | |
| embB Q497R + embA -8c/a | 1 | |
| embB D1024N | 1 (3.75) | |
| embC P707L | 1 (2.5) | |
| No mutation | 18 (c) | 2d |
MICs were tested for susceptible strains only.
Tested resistant when repeated.
≤1.25 (n = 11), 2.5 (n = 6), ≥5 (n = 1) (gspI -240 c/T and Rv3785 c/G at position 4,243,217).
One was tested susceptible, when repeated (MIC, ≤1.25).
FIG 1.
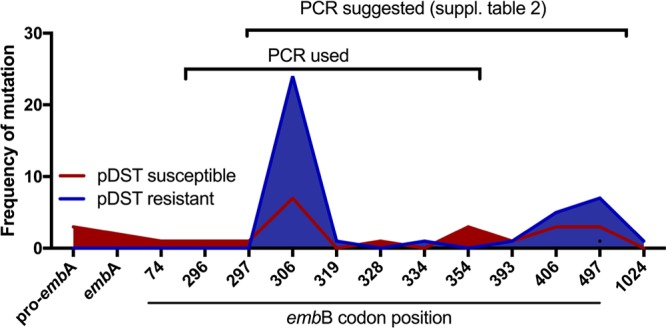
Distribution of mutations across the embB codons. Frequencies of mutations at their respective codon positions are shown. The region covered by the PCR primers currently used and the validated PCR primers suggested to be used (Table S2) are indicated by black brackets.
No embR mutation was detected in our data set, while variants in ubiA and aftA were each found in three cases representing both susceptible and resistant isolates. The two EMB-resistant isolates with a variant in ubiA and one EMB-resistant isolate with a mutation in aftA also harbored mutations in the embCAB operon. All mutations found in EMB-resistant and -susceptible isolates within the embCAB operon and in other candidate genes and the MICs of EMB-susceptible strains are listed in Table S1 and Table 1.
DISCUSSION
This study showed 100% concordance between pDST and pncA Sanger sequencing for PZA. However, our study affirms that pDST and embB306 sequencing alone is not sufficient for a reliable determination of EMB resistance (13). Concordance with phenotypic resistance was high for some mutations identified within the embCAB operon, notably embB M306V and embBQ497R. Additional mutations that were present in both phenotypically susceptible and resistant isolates in our collection and were associated with elevated MICs comprise embBQ497K, embBD354A, and embBG406A. This is in agreement with previously published MIC data showing an association between these codons and high MICs (9, 14–16). The fact that some of these isolates were initially classified as susceptible by pDST is explained by isolate MICs close to the EMB critical concentration. Our data suggest that an isolate should be regarded as EMB resistant if any of these mutations are present and an elevated MIC is observed, regardless of the initial pDST result. Yet, because of the relatively small sample size, no conclusions can be drawn for other mutations detected in only one or two isolates.
Earlier work suggests that ubiA (Rv3806c) and notably nonsynonymous mutations in codons 237 and 240 may confer EMB resistance (17). We found three ubiA mutations in our data set (V55G and two times E149D) in both phenotypically resistant and susceptible isolates. aftA, another enzyme-encoding gene implicated in cell wall synthesis has also been found to be associated with EMB resistance (18). Our data set revealed mutations at codon 575 in a susceptible isolate and mutation T611M in a resistant and susceptible isolate.
pDST in combination with Sanger sequencing of codon 306 in embB correctly identified 57.1% (24/42) of EMB-resistant isolates. An additional 16 isolates would have been detected as EMB resistant if the region covered by Sanger sequencing was widened to include codons embB297 and embB497, as previously proposed (13, 19, 20). The sensitivity to predict phenotypic EMB resistance of the embB codon 306 mutations was 0.57 (specificity 0.83) which could be increased to 0.9 (specificity 0.67) when including embB codons 306, 354, 406, and 497. With all embCAB mutations identified among our study isolates, the sensitivity to predict EMB resistance was 0.95 (specificity 0.42). Primers for an extended PCR spanning these codons have been validated as part of this study (see Table S2 in the supplemental material). No PCR product could be obtained for three pDST-resistant isolates. The phenotypic resistance can be explained by larger deletions in two isolates and a T47A mutation in one isolate. The mutation T47A has been described to lead to increased MICs close to the resistance breakpoints (21).
In contrast to previous publications, we report 100% concordance between molecular and phenotypic PZA DST (22). NGS confirmed resistant pDST results in three additional isolates with noninterpretable Sanger sequencing results. Two of these isolates had large deletions in the pncA gene. A recent study involving six national reference laboratories investigating 1,142 MDR strains showed that 10% of isolates with pncA mutations tested phenotypically susceptible. The majority of pncA variants (85%) in this study were high-confidence mutations known to be associated with PZA resistance (11). In a comprehensive mutational screening approach, the PZA-resistance phenotype of 977 pncA nonsynonymous SNPs was assessed. One-third of the mutations (n = 301) were resistance conferring while another one-third (n = 310) were not associated with phenotypic resistance (23). Of the 28 different pncA mutations identified in our study, 18 belonged to the resistance-conferring group as per Yadon et al. (23); none were in the group not associated with resistance.
Recommendation for patient management.
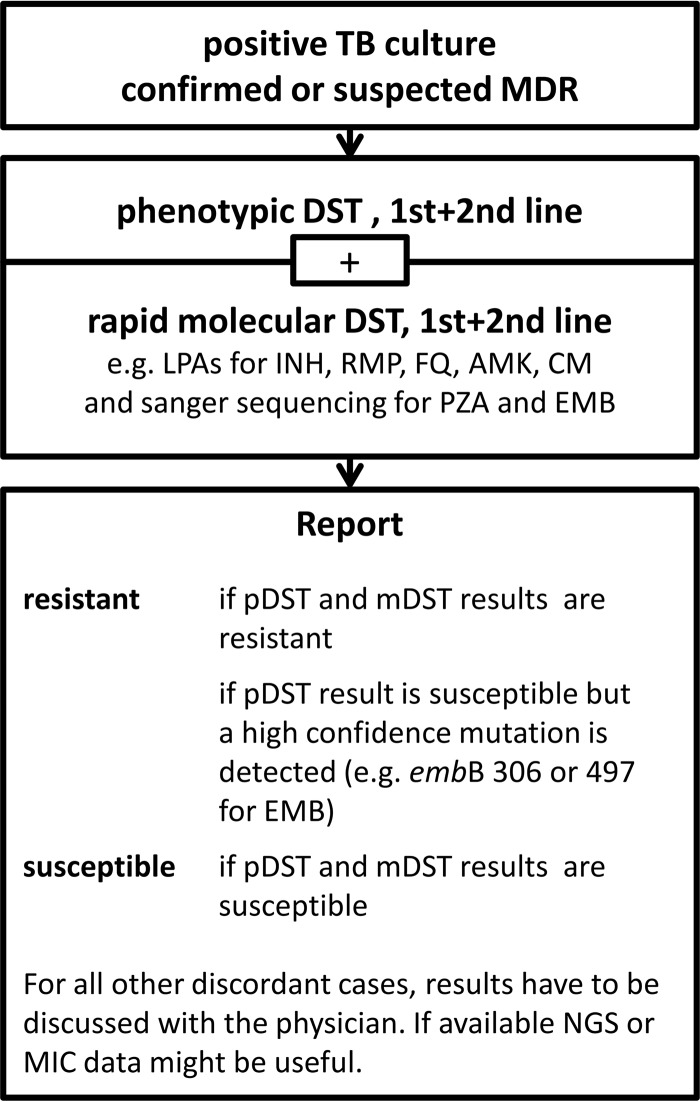
One method alone, i.e., pDST, Sanger sequencing around codon 306 of embB, or NGS, is insufficient to reliably detect EMB resistance. Thus, we propose a diagnostic algorithm using phenotypic and genotypic methods in parallel for all clinical isolates with RMP resistance detected by rapid molecular (GeneXpert or line probe assays) or phenotypic tests (Fig. 2). If NGS is not available or only available as a research tool, Sanger sequencing between codon 297 and 497 of embB should be considered (see Table S2 for a validated primer sequence spanning these embB codons). Interpretation of DST results is straightforward if phenotypic and genotypic test results are in agreement. Discordances between phenotypic and genotypic results may need to be further investigated. However, isolates with mutations in the embB codons 306, 354, 406, and 497 should be assumed EMB resistant regardless of the phenotypic DST result (9, 14–16). MICs should be determined for all phenotypically susceptible isolates with mutations in the genes/operon aftA, ubiA, and embCAB to elucidate putative additive effects of linked mutations (e.g., embB S297A and embB D1024N) and the relevance of debatable mutations (e.g., embB Y319C and N296H) on the EMB-resistance level. If MIC testing is not feasible, repeated pDST at the critical concentrations should be performed. For those isolates, the microbiologist or biomedical scientist should relay the uncertainty about the effectiveness of EMB to the clinician who may or may not consider EMB as an add-on agent.
FIG 2.
Algorithm proposed for ethambutol resistance determination in a mycobacterial laboratory in a low incidence-setting.
Implementation of the proposed algorithm is likely to be feasible and relevant in most low TB incidence, high-resource settings where pDST is routinely performed and access to NGS is widely available. The majority of MDR-TB cases in Western European countries are diagnosed among migrants from high TB and/or MDR-TB burden countries. In Germany, a high proportion of MDR-TB cases are from former Soviet Union and Eastern Europe countries (24). While the prevalence of EMB resistance among MDR-TB cases is not systematically reported, the studies reporting EMB resistance show a consistently high prevalence of >50% (25, 26).
The feasibility of implementing this algorithm in high MDR-TB burden middle or low-income settings depends on the availability of phenotypic DST and sequencing. This, in turn, requires technical skills and expertise, infrastructure, appropriate biosafety measures, and bioinformatics. Over the past years the capacity to perform phenotypic DSTs has greatly increased in many middle or low-income settings. There is great interest and enthusiasm to rolling out sequencing in low-resource settings, but wide implementation has not yet happened (27; https://unitaid.org/call-for-proposal/seeking-projects-to-fight-tuberculosis-and-its-drug-resistant-strains/#en). With sequencing becoming more widely available, algorithms such as the one proposed in this study will be implemented in high MDR-TB burden settings, where they are most needed.
Conclusions.
At present, pDST for PZA and EMB cannot be replaced by any commercially available molecular diagnostic. The one line-probe assay aimed at detecting EMB resistance covers a limited range of resistance-conferring mutations and does not differentiate between resistance-conferring or silent variants. Thus, Sanger sequencing or NGS together with pDST should be employed to ensure reliable EMB DST results, enabling clinicians to decide whether to include EMB as part of an MDR-TB regimen.
MATERIALS AND METHODS
M. tuberculosis isolates.
We included all RMP-resistant clinical isolates referred to the German National Reference Laboratory for Mycobacteria, Borstel (NRL), between January 2016 and March 2017.
Phenotypic and Sanger-based drug susceptibility testing.
pncA and embB306 Sanger sequencing and pDST for first and second-line drugs were done in parallel as part of the diagnostic service. pDST was performed using the MGIT960 system according to the manufacturer’s instructions (Becton, Dickinson, Sparks, MD). Processing of isolates and DNA extraction were performed as previously described (28). MGIT tubes were prepared with 0.8 ml MGIT960 SIRE supplement and, with exception of the drug-free growth control tubes, 0.1 ml drug solution. The following drug concentrations were tested: PZA at 100 and EMB at 5.0 μg/ml. Bacterial suspensions of 0.5 ml were added to the test tubes and the growth-control tubes.
For molecular DST, pncA and embB306 sequencing were performed as previously described (9, 29).
MIC testing for ethambutol.
For EMB MIC determination, pDST-susceptible strains were subsequently tested at 5.0, 3.75, 2.5, and 1.25 μg/ml. The provided drug of the Bactec MGIT 960 SIRE kit was reconstituted into sterile distilled/deionized water as described in the package insert. For the test concentrations 3.75, 2.5, and 1.25 μl/ml, the stock solution was diluted 3:1, 1:1, and 1:3 with sterile distilled/deionized water before 100 μl were added to the respective MGIT tubes. The interpretation was done with the EpiCenter TBeXiST software according to Springer et al. (30).
Next-generation sequencing and phylogenomic analyses.
All isolates underwent next-generation sequencing (NGS) to confirm mutations and investigate relatedness of isolates. From extracted genomic DNA, sequencing libraries were constructed using the Nextera XT kit and run on the NextSeq (2 × 150 bp) sequencing platform (Illumina, San Diego, CA, USA). Reads were mapped to the reference genome M. tuberculosis H37Rv (GenBank accession number NC_000962.3) with the alignment program BWA. Reads were refined using the Genome Analysis Toolkit (GATK) and SAMtools. For variant calling in the mapped reads, we used SAMtools and custom perl scripts with minimum thresholds of four reads in both forward and reverse orientation and 75% allele frequency. Variants in repetitive regions or genes were masked. Single nucleotide polymorphism (SNP) positions with a clear base call in all isolates were concatenated to a sequence alignment. A maximum likelihood tree was inferred using FastTree with 1,000 resamples (31). The consensus tree was midpoint rooted in FigTree, and nodes were arranged in increasing order.
Accession number(s).
The sequence read sets were deposited in the European Nucleotide Archive under the BioProject accession number PRJEB27354.
Supplementary Material
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01798-18.
REFERENCES
- 1.WHO. Guidelines for treatment of drug-susceptible tuberculosis and patient care (2007 update). 2017. WHO, Geneva, Switzerland. [Google Scholar]
- 2.WHO. Implementing tuberculosis diagnostics: a policy framework. 2015. WHO, Geneva, Switzerland. [Google Scholar]
- 3.Schön T, Juréen P, Giske CG, Chryssanthou E, Sturegård E, Werngren J, Kahlmeter G, Hoffner SE, Angeby KA. 2009. Evaluation of wild-type MIC distributions as a tool for determination of clinical breakpoints for Mycobacterium tuberculosis. J Antimicrob Chemother 64:786–793. doi: 10.1093/jac/dkp262. [DOI] [PubMed] [Google Scholar]
- 4.WHO. 2016. Treatment guidelines for drug-resistant tuberculosis, 2016 update. WHO, Geneva. [PubMed] [Google Scholar]
- 5.Elsevier Ltd. 2008. Pyrazinamide. Tuberculosis 88:141–144. doi: 10.1016/S1472-9792(08)70021-0. [DOI] [PubMed] [Google Scholar]
- 6.Elsevier Ltd. 2008. Ethambutol. Tuberculosis 88:102–105. doi: 10.1016/S1472-9792(08)70008-8. [DOI] [PubMed] [Google Scholar]
- 7.Belanger AE, Besra GS, Ford ME, Mikusová K, Belisle JT, Brennan PJ, Inamine JM. 1996. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc Natl Acad Sci U S A 93:11919–11924. doi: 10.1073/pnas.93.21.11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mikusová K, Slayden RA, Besra GS, Brennan PJ. 1995. Biogenesis of the mycobacterial cell wall and the site of action of ethambutol. Antimicrob Agents Chemother 39:2484–2489. doi: 10.1128/AAC.39.11.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plinke C, Cox HS, Zarkua N, Karimovich HA, Braker K, Diel R, Rüsch-Gerdes S, Feuerriegel S, Niemann S. 2010. embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J Antimicrob Chemother 65:1359–1367. doi: 10.1093/jac/dkq120. [DOI] [PubMed] [Google Scholar]
- 10.Zignol M, Dean AS, Alikhanova N, Andres S, Cabibbe AM, Cirillo DM, Dadu A, Dreyer A, Driesen M, Gilpin C, Hasan R, Hasan Z, Hoffner S, Husain A, Hussain A, Ismail N, Kamal M, Mansjö M, Mvusi L, Niemann S, Omar SV, Qadeer E, Rigouts L, Ruesch-Gerdes S, Schito M, Seyfaddinova M, Skrahina A, Tahseen S, Wells WA, Mukadi YD, Kimerling M, Floyd K, Weyer K, Raviglione MC. 2016. Population-based resistance of Mycobacterium tuberculosis isolates to pyrazinamide and fluoroquinolones: results from a multicountry surveillance project. Lancet Infect Dis 16:1185–1192. doi: 10.1016/S1473-3099(16)30190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miotto P, Cabibbe AM, Feuerriegel S, Casali N, Drobniewski F, Rodionova Y, Bakonyte D, Stakenas P, Pimkina E, Augustynowicz-Kopeć E, Degano M, Ambrosi A, Hoffner S, Mansjö M, Werngren J, Rüsch-Gerdes S, Niemann S, Cirillo DM. 2014. Mycobacterium tuberculosis pyrazinamide resistance determinants: a multicenter study. mBio 5:e01819-14. doi: 10.1128/mBio.01819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Telenti A, Philipp WJ, Sreevatsan S, Bernasconi C, Stockbauer KE, Wieles B, Musser JM, Jacobs WR. 1997. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat Med 3:567–570. doi: 10.1038/nm0597-567. [DOI] [PubMed] [Google Scholar]
- 13.Brossier F, Sougakoff W, Bernard C, Petrou M, Adeyema K, Pham A, Amy de la Breteque D, Vallet M, Jarlier V, Sola C, Veziris N. 2015. Molecular analysis of the embCAB locus and embR gene involved in ethambutol resistance in clinical isolates of Mycobacterium tuberculosis in France. Antimicrob Agents Chemother 59:4800–4808. doi: 10.1128/AAC.00150-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park YK, Ryoo SW, Lee SH, Jnawali HN, Kim C-K, Kim HJ, Kim SJ. 2012. Correlation of the phenotypic ethambutol susceptibility of Mycobacterium tuberculosis with embB gene mutations in Korea. J Med Microbiol 61:529–534. doi: 10.1099/jmm.0.037614-0. [DOI] [PubMed] [Google Scholar]
- 15.Sekiguchi J, Miyoshi-Akiyama T, Augustynowicz-Kopeć E, Zwolska Z, Kirikae F, Toyota E, Kobayashi I, Morita K, Kudo K, Kato S, Kuratsuji T, Mori T, Kirikae T. 2007. Detection of multidrug resistance in Mycobacterium tuberculosis. J Clin Microbiol 45:179–192. doi: 10.1128/JCM.00750-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, Kontsevaya I, Corander J, Bryant J, Parkhill J, Nejentsev S, Horstmann RD, Brown T, Drobniewski F. 2014. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet 46:279–286. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Safi H, Lingaraju S, Amin A, Kim S, Jones M, Holmes M, McNeil M, Peterson SN, Chatterjee D, Fleischmann R, Alland D. 2013. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-D-arabinose biosynthetic and utilization pathway genes. Nat Genet 45:1190–1197. doi: 10.1038/ng.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alderwick LJ, Seidel M, Sahm H, Besra GS, Eggeling L. 2006. Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J Biol Chem 281:15653–15661. doi: 10.1074/jbc.M600045200. [DOI] [PubMed] [Google Scholar]
- 19.Zhao L-L, Sun Q, Liu H-C, Wu X-C, Xiao T-Y, Zhao X-Q, Li G-L, Jiang Y, Zeng C-Y, Wan K-L. 2015. Analysis of embCAB mutations associated with ethambutol resistance in multidrug-resistant mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother 59:2045–2050. doi: 10.1128/AAC.04933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Q, Xiao T-Y, Liu H-C, Zhao X-Q, Liu Z-G, Li Y-N, Zeng H, Zhao L-L, Wan K-L. 2018. Mutations within embCAB are associated with variable level of ethambutol resistance in Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother 62:e01279-17. doi: 10.1128/AAC.01279-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werngren J, Sturegård E, Juréen P, Ängeby K, Hoffner S, Schön T. 2012. Reevaluation of the critical concentration for drug susceptibility testing of Mycobacterium tuberculosis against pyrazinamide using wild-type MIC distributions and pncA gene sequencing. Antimicrob Agents Chemother 56:1253–1257. doi: 10.1128/AAC.05894-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramirez-Busby SM, Valafar F. 2015. Systematic review of mutations in pyrazinamidase associated with pyrazinamide resistance in Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother 59:5267–5277. doi: 10.1128/AAC.00204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yadon AN, Maharaj K, Adamson JH, Lai Y-P, Sacchettini JC, Ioerger TR, Rubin EJ, Pym AS. 2017. A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide. Nat Commun 8:588. doi: 10.1038/s41467-017-00721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robert Koch-Institut. 2016. Bericht zur Epidemiologie der Tuberkulose in Deutschland für 2015. Robert Koch-Institut, Berlin, Germany. [Google Scholar]
- 25.Stagg HR, Brown J, Ibraim E, Riekstiņa V, Viiklepp P, Cīrule A, Cocei H, Danilovitš M, Dravniece G, Jackson C, White PJ. 2015. Drug susceptibility patterns in MDR-TB patients: challenges for future regimen design. A cross-sectional study. PLoS One 10:e0142425. doi: 10.1371/journal.pone.0142425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Günther G, van Leth F, Alexandru S, Altet N, Avsar K, Bang D, Barbuta R, Bothamley G, Ciobanu A, Crudu V, Davilovits M, Dedicoat M, Duarte R, Gualano G, Kunst H, de Lange W, Leimane V, Magis-Escurra C, McLaughlin A-M, Muylle I, Polcová V, Pontali E, Popa C, Rumetshofer R, Skrahina A, Solodovnikova V, Spinu V, Tiberi S, Viiklepp P, Lange C, TBNET. 2015. Multidrug-resistant tuberculosis in Europe, 2010–2011. Emerg Infect Dis 21:409–416. doi: 10.3201/eid2103.141343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinmann P, Auer C, Clary B, Guinot D, Bosch-Capblanch X. 2011. Narrowing the gap—expanding and accelerating access to diagnostics for patients at risk of multi-drug resistant tuberculosis (MDR-TB). Swiss Centre for International Health, Basel, Switzerland. https://docplayer.net/89075841-Mid-term-review-expand-tb.html.
- 28.Hillemann D, Rüsch-Gerdes S, Richter E. 2007. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol 45:2635–2640. doi: 10.1128/JCM.00521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feuerriegel S, Köser CU, Richter E, Niemann S. 2013. Mycobacterium canettii is intrinsically resistant to both pyrazinamide and pyrazinoic acid. J Antimicrob Chemother 68:1439–1440. doi: 10.1093/jac/dkt042. [DOI] [PubMed] [Google Scholar]
- 30.Springer B, Lucke K, Calligaris-Maibach R, Ritter C, Böttger EC. 2009. Quantitative drug susceptibility testing of Mycobacterium tuberculosis by use of MGIT 960 and EpiCenter instrumentation. J Clin Microbiol 47:1773–1780. doi: 10.1128/JCM.02501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.