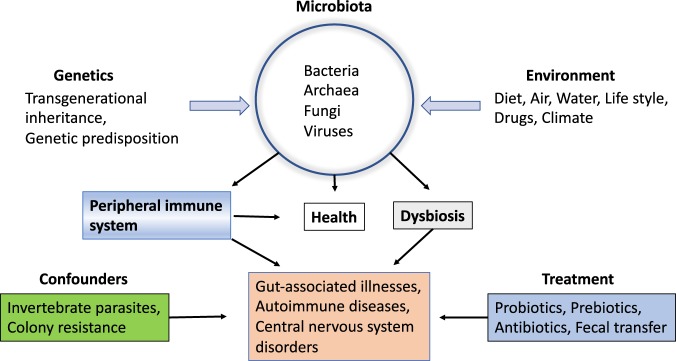
Elie Metchnikoff’s lecture illustrated that ‘men were free from microbes at birth and within tenth and seventeenth hour of birth the surface of skin, mucous membranes, respiratory passages, and more abundantly the digestive organs became peopled with microbes derived from the air or water’.1 It is interesting to note that the revised ratio of human cells to the number of bacteria in the body is 1:1, which is further tipped towards the human cell mass following each defecation.2 Based on metagenomic sequencing of microbial genes in the fecal samples of 124 European individuals, the MetaHIT Consortium reported 3.3 million non-redundant bacterial genes, 1000–1150 bacterial species, and 160 species per individual among which 57 species are common to >90% of individuals.3 Although subsequent studies indicated that microbes play an important role in human health and disease, a daunting task is to decipher whether ethnicity (genetics) or environment (diet, lifestyle, drugs, and dwelling conditions) has the most impact on the fecal microbiome (Fig. 1).
Fig. 1.
Interrelationships between genetic and environmental factors and their impact on health and disease
In a recent study, Deschasaux et al.4 have used the second-generation sequencing to analyze the V4 hypervariable region of the bacterial 16S ribosomal RNA (rRNA) gene in the fecal matter of 2084 participants consisting of the Dutch and immigrant populations—Ghanaians, Moroccans, African Surinamese, South-Asian Surinamese, and Turkish.4 Principal coordinate analysis indicated that ethnicity significantly contributed to the inter-individual dissimilarities (β-diversity) at the operational taxonomic unit (OTU) level. Similarly, the richness and evenness of bacterial composition and α-diversity (Shannon diversity index) also differed dramatically among various ethnicities. The strongest determinant of α- and β-diversity remained ethnicity and none of the other factors including dietary patterns, age, gender, education, body mass index, lifestyle, area of residence, sample collection, and sequencing run appeared to have an effect on microbial taxa distribution in analyzed samples. Since a previous study suggested an association between fecal microbiota and metabolic syndrome, type 2 diabetes,5 a restricted number of participants without this metabolic syndrome were also analyzed, which reiterated similar ethnic patterns in α- and β-diversity.4 Notably, the Dutch displayed the highest α-diversity in contrast to the ethnic groups who immigrated from non-Western countries. Several bacterial families were highly represented in the Dutch which were depleted in South-Asian Surinamese. Overall, distinct OTUs were mutually associated with one or other ethnicities, indicating that the ethnic origin of individuals could be a marker for differences in the fecal microbial composition. This is consistent with a previous report of ethnic-specific fecal microbial taxa distribution in Chinese patients with ulcerative colitis.6
Many studies indicated that although a proportion of the microbial taxa is heritable, they are subject to modulation by environmental cues. A recent investigation of a highly homogeneous ancestry (99% Han Chinese) allowed the isolation of the effect of geography without ethnicity as a confounding factor.7 In this study, the 16S rRNA gene V4 hypervariable region was analyzed on stool samples from 7009 individuals from 14 districts within one province of China. Geographic location dominated the variation among the ‘healthy’ microbial taxa found in controls. It also exceeded the common metabolic diseases such as type 2 diabetes, obesity, and fatty liver. Importantly, extrapolation of healthy reference baselines of fecal microbiota derived from one location to others led to incorrect healthy status predictions even in a population of highly homogeneous ancestry. Thus, these data are in support of the influence of the environment on the fecal microbial composition. Further support for this notion comes from the work of Rothschild et al.8 who analyzed 1046 healthy Israeli individuals of different ancestral origins—Ashkenazi, North African, Middle Eastern, Sephardi, Yemenite, and admixed ancestries who shared a common environment. Although a previous study indicated that 42 single-nucleotide polymorphisms (SNPs) in the peripheral blood-derived DNA could be associated with 10% of the β-diversity variance in the fecal matter,9 similar analysis by Rothschild et al.8 did not result in a statistically significant outcome.8 Principal coordinate analysis indicated no significant correlation between any of the top five host genetic groups and any of top five microbiome β-diversity. No significant differences between ancestries in microbiome composition, Shannon diversity index (α-diversity), or abundance of specific taxa and evenness were observed at the phylum level. Significant similarity among microbiomes of genetically unrelated individuals who shared a household but not among the relatives who lived in different dwellings was evident. Correlation between phenotypes based on metabolic parameters such as obesity and blood glucose levels and microbial taxa found in the stool reinforced the idea that host genetics and environment are independent of each other.
Previously, heritable bacteria were identified based on co-occurrence among family members or associations between specific SNPs and bacterial taxa.9 Rothschild et al.8 concurred with the inheritance of a small number of bacterial groups in their subjects (such as Bifidobacterium associated with two loci close to the LCT gene, which encodes lactase enzyme enabling lactose consumption) and the requirement of larger sample sizes to expand the numbers of heritable bacterial taxa and SNP associations. Although Deschasaux et al.4 did not analyze SNPs to evaluate the inheritability of bacterial taxa, they found that Firmicutes was the most abundant phylum in all ethnic groups analyzed, followed by Bacteroidetes, Actinobacteria, and Proteobacteria. In the Dutch, Firmicutes and Bacteroidetes were respectively enriched and depleted, whereas Actinobacteria were abundant in South-Asian Surinamese.4 The preponderance of Prevotella OTU was observed in Moroccans, Turkish, and Ghanaians. On the other hand, Bacteroides OTU was prevalent in Surinamese, and Clostridiales in the Dutch. Interestingly, a high level of Proteobacteria was reported in the Han Chinese population.7 Inasmuch as these studies were performed on healthy individuals representing each ethnicity, the dominance of various bacterial taxa found in these multi-ethnic groups suggests that the characteristics of ‘healthier gut microbiome’ vary among different ethnic populations. Nevertheless, these data are consistent with the contention that the enterotypes inherited from ancestors are subject to further environmental influences including diet, residence, drugs, lifestyle, and health status.
Since the recognition that ‘meat and vegetable diets stimulated the development of special bacterial forms and about forty-five species of microbes inhabited the large intestine1’, our understanding of the impact of genetic and environmental factors on gut microbiota has advanced considerably (Fig. 1). Transgenerational inheritance and genetic predisposition play an essential role in the establishment of the intestinal microbial patterns in newborns. The inherited microbial taxa are subject to modulation by environmental factors including diet, water, and drugs. Intestinal microbes play a crucial role in the maturation of the peripheral immune system, essential for the maintenance of the health status. Deviation from the healthy state, dysbiosis, directly or indirectly via the adaptive immune system, can lead to the onset of several gut-associated illnesses, autoimmune diseases, and central nervous system disorders, among others. The usefulness of helminth parasites, probiotics, prebiotics, and antibiotics for the treatment of a variety of diseases remains contentious. Colony resistance influenced by host genetics may account for the nebulous beneficial effects afforded by substitution with exogenous bacteria under certain pathological conditions.
A thorough understanding of the impact of intestinal microbiota on health and disease is a prerequisite for the implementation of microbe-based treatments for a myriad of ailments. However, many hurdles remain to be overcome in order to realize this goal fully. Recently, several ‘pitfalls’ of microbiome studies have been noted.10 These include the omission of fungi, viruses, and parasites from microbiome studies, improper attention to microbial taxonomy, failure to quantify the total numbers of bacteria, not distinguishing the cause or outcome of microbial changes, discordance between experimental and clinical studies, lack of thorough understanding of the influence of environmental and other factors on gut microbiota, and uncertain benefits and safety of probiotics, prebiotics, antibiotics, and fecal microbial transfer for disease treatment.10
Many other shortcomings that could contribute to discrepancies of results obtained in various studies deserve consideration and are briefly discussed below. Currently, direct comparison of microbiome data from various investigations is not possible due to a number of variations in experimental conditions including sample collection, storage prior to DNA extraction, use of different sequencing platforms, as well as the lack of standard data processing and analysis methods. Some of these variables could account for the vast differences in fecal microbiome between control subjects and specific patient populations ranging from no effect to varying representation of microbes and Archaea. Similarly, the lack of concordance in fecal microbiota of inbred strains of mice following uniform manipulation could also be attributed to experimental and analytical variables. The fact that patient-derived fecal matter or certain specific microbial groups have been shown respectively to induce or repress autoimmune diseases in animal models requires replication and thorough evaluation prior to their clinical application. Although 16S rRNA is the target of choice, the presence of variable copy numbers in bacterial genomes and sequence variation within closely related taxa or within a genome limit their use in bacterial ecology.11 The choice of primers that target different hypervariable regions of the 16S rRNA gene plays a critical role in determining the profile of mostly uncultivated, complex microbial community generated by DNA sequencing.12 Whereas primers spanning the V1–V3 and V7–V9 hypervariable regions provided greater depth of coverage than Sanger sequencing,13 these and V4 primers have been used variably in different studies, which do not permit direct comparison of data. Sequence identity clusters or OTUs provide an imperfect representation of bacterial taxa of a certain phylogenetic rank.14 Development of bacterial genomics can help to make bacterial abundance estimates more accurate. The possibility exists that sequencing bias could account for almost 100% representation of certain bacterial genera reported in some studies. Finally, some environmental factors may have different effects depending on other confounders. One such example is the lack of influence of acidified drinking water, known to deplete intestinal microbes, on the development of autoimmune diabetes and experimental encephalomyelitis in genetically susceptible non-obese diabetic (NOD) mice, whereas the same maneuver had different effects, attributed to other confounders.15 Taken together, these data emphasize the requirement for careful consideration of these and other aspects discussed earlier10 to device optimal bacteria-based treatment modalities for a number of pathological conditions.
Acknowledgements
Arathi Jayaraman is acknowledged for critical comments on the manuscript.
Competing interests
The author declares no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dr. Metchnikoff on microbes and the human body. Nature 63, 621–622 (1901).
- 2.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deschasaux M, et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat. Med. 2018;24:1526–1531. doi: 10.1038/s41591-018-0160-1. [DOI] [PubMed] [Google Scholar]
- 5.Bouter KE, van Raalte DH, Groen AK, Nieuwdorp M. Role of the gut microbiome in the pathogenesis of obesity and obesity-related metabolic dysfunction. Gastroenterology. 2017;152:1671–1678. doi: 10.1053/j.gastro.2016.12.048. [DOI] [PubMed] [Google Scholar]
- 6.Prideaux L, et al. Impact of ethnicity, geography, and disease on the microbiota in health and inflammatory bowel disease. Inflamm. Bowel Dis. 2013;19:2906–2918. doi: 10.1097/01.MIB.0000435759.05577.12. [DOI] [PubMed] [Google Scholar]
- 7.He Y, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat. Med. 2018;24:1532–1535. doi: 10.1038/s41591-018-0164-x. [DOI] [PubMed] [Google Scholar]
- 8.Rothschild D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 2016;48:1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park AM, Omura S, Fujita M, Sato F, Tsunoda I. Helicobacter pylori and gut microbiota in multiple sclerosis versus Alzheimer’s disease: 10 pitfalls of microbiome studies. Clin. Exp. Neuroimmunol. 2017;8:215–232. doi: 10.1111/cen3.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouhy F, Clooney AG, Stanton C, Claesson MJ, Cotter PD. 16S rRNA gene sequencing of mock microbial populations- impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016;16:123. doi: 10.1186/s12866-016-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker AW, et al. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome. 2015;3:26. doi: 10.1186/s40168-015-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar PS, Brooker MR, Dowd SE, Camerlengo T. Target region selection is a critical determinant of community fingerprints generated by 16S pyrosequencing. PLoS One. 2011;6:e20956. doi: 10.1371/journal.pone.0020956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One. 2013;8:e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayaraman S, Jayaraman A. Long-term provision of acidified drinking water fails to influence autoimmune diabetes and encephalomyelitis. J. Diabetes Res. 2018;2018:3424691. doi: 10.1155/2018/3424691. [DOI] [PMC free article] [PubMed] [Google Scholar]