Abstract
Background
Cyclin-dependent protein kinase 9 (CDK9) has been shown to play an important role in the pathogenesis of malignant tumors. However, the expression and function of CDK9 remain unknown in osteosarcomas. The purpose of this study is to assess the expression, function and clinical prognostic relationship of CDK9 in osteosarcomas.
Methods
A tissue microarray of 70 patient specimens was analyzed by immunohistochemistry to measure CDK9 expression, which was further investigated for correlation with patient clinical characteristics. CDK9 expression in osteosarcoma cell lines and patient tissues was also evaluated by Western blotting. CDK9-specific siRNA and the CDK9 inhibitor were applied to determine the effect of CDK9 inhibition on osteosarcoma cell proliferation and anti-apoptotic activity. The clonogenicity and migration activity were also examined using clonogenic and wound healing assays. A 3D cell culture model was performed to mimic the in vivo osteosarcoma environment to further validate the effect of CDK9 inhibition on osteosarcoma cells.
Findings
We demonstrated that higher CDK9-expression is associated with significantly shortened patient survival by immunohistochemistry. Expression of CDK9 is inversely correlated to the percent of tumor necrosis post-neoadjuvant chemotherapy, which is the most important predictive factor of disease outcome for osteosarcoma patients. Knockdown of CDK9 with siRNA and inhibition of CDK9 activity with inhibitor decreased cell proliferation and induced apoptosis in osteosarcoma.
Interpretation
High expression of CDK9 is an independent predictor of poor prognosis in osteosarcoma patients. Our results suggest that CDK9 is a novel prognostic marker and a promising therapeutic target for osteosarcomas.
Keywords: CDK9, Osteosarcoma, Prognostic marker, Therapeutic target, Apoptosis
Research in context.
Evidence before this study
CDK9 has also emerged as an important therapeutic target in cancer, due to its crucial role in RNA transcription, elongation and other cellular processes. In complex with regulatory cyclins, CDK9 drives the transcription of global (non-ribosomal) genes, including genes that are regulated by super enhancers, a large cluster of DNA regulatory element enhancers. As a consequence, CDK9 is involved in cell cycle progression, growth, proliferation, differentiation, and apoptosis. Dysregulation of CDK9 has been observed in a number of human solid tumors, including prostate cancer, neuroblastoma, hepatocellular carcinoma and lymphoma, but remains to be investigated in osteosarcoma.
Added value of this study
In this study, we demonstrate that osteosarcoma patients with high CDK9 tumor-expression levels have significantly shorter survival than patients with low CDK9 expression. In addition to this, the expression of CDK9 is inversely correlated to the percent of tumor necrosis that occurs post-neoadjuvant chemotherapy. Inhibition of CDK9 either by siRNA or an inhibitor, can abrogate osteosarcoma cell growth or induce apoptosis both in vitro and mimic in vivo. Collectively, these findings suggest that CDK9 is a critical component of osteosarcoma cell growth, proliferation and migration. Therefore, CDK9 is an attractive and promising molecular biomarker and therapeutic target for patients with osteosarcoma.
Implications of all the available evidence
Osteosarcomas are currently treated using chemotherapy regimens established over 30 years ago. Although chemotherapy has dramatically improved their 5-year survival, recurrent and metastatic osteosarcoma still have a high mortality rate. The development of novel therapeutic strategies for the treatment of osteosarcoma remains an important and unmet clinical need. Our study demonstrates that the expression of CDK9 is associated with the clinical prognosis of patients with osteosarcoma. CDK9 plays an important role in osteosarcoma cell growth and proliferation. A recent study has demonstrated that the treatment of CDK9 overexpressing patient-derived tumor xenograft (PDTX) osteosarcoma models, with the CDK inhibitor AT7519, resulted in decreased tumor growth. Similarly, another novel highly selective CDK9 inhibitor, MC180295, has demonstrated broad anti-cancer activity in vitro and is effective in in vivo cancer models. These findings suggest that CDK9 is a promising molecular target in osteosarcoma.
Alt-text: Unlabelled Box
1. Introduction
Osteosarcoma is the most common malignant tumor that affects children, adolescents, and young adults [1]. It is responsible for 20% of all primary bone sarcomas [2]. Before 1970, treatment for osteosarcoma primarily involved surgical resection. Chemotherapy has dramatically improved 5-year survival for patients with localized osteosarcoma from <20% to over 65% following the advent of multiagent regimens [3]. However, recurrent and metastatic osteosarcoma have retained a high mortality rate, with patient survival usually less than one year [1,4,5]. In the last 30 years, the treatment and survival rates of osteosarcoma patients have shown very little improvement. Therefore, the development of novel therapeutic strategies for the treatment of osteosarcoma remains an important and unmet clinical need.
Cyclin-dependent kinases (CDKs) are members of a complex family of heterodimeric serine/threonine protein kinases and are involved in critical cellular processes, including in cellular DNA transcription and cell-cycle progression, among others [6]. Mammalian cells contain at least 20 different CDKs, but only a few subsets of CDK–Cyclin complexes are directly associated with cell-cycle progression. Previous studies have demonstrated that many CDKs are associated with tumorigenesis and progression of different cancers, including osteosarcoma [[7], [8], [9], [10], [11], [12], [13]]. Therefore, pharmacological inhibition of CDKs has been considered as an attractive option for treating a number of human malignancies. Palbociclib (IBRANCE®), a dual CDK4/6 inhibitor, has already received U.S FDA approval for the treatment of breast cancer [14,15]. Palbociclib has also demonstrated promising antitumor potential both as a monotherapy and in combination in many preclinical studies and clinical trials for a number of other cancer types [[16], [17], [18]].
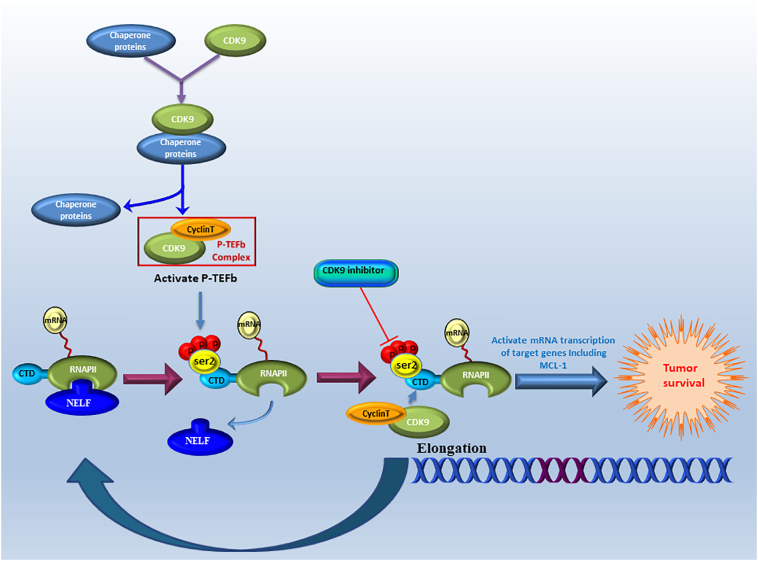
Recently, cyclin-dependent protein kinase 9 (CDK9) has been shown to play an essential role in acute myeloid leukemia, breast cancer, melanoma, prostate cancer and lung cancer [12,[19], [20], [21], [22], [23], [24], [25]]. CDK9 and cyclin T complex, which is a component of the positive transcription elongation factor b (P-TEFb), promotes release of paused RNA polymerase II (RNAPII) into elongation process [26]. CDK9 is expressed in two isoforms, a lighter 42 kDa isoform and a heavier 55 kDa isoform, the latter is translated from the same mRNA but at an upstream transcriptional start site of the 42 kDa protein [27]. Compared with the lighter isoform, the 55 kDa protein has an additional 117 amino acids at the N-terminus. These two isoforms of CDK9 have mostly been attributed to the regulation of transcription but not cell-cycle progression [27,28]. Both isoforms have been shown to be expressed in human cancer cell lines and in normal tissues. CDK9 has been reported to regulate RNAPII-associated transcription by phosphorylating the large subunit of RNAPII, at the C-terminal domain (CTD) [19,29]. RNAPII suppressed by CDK9 inhibition has been shown to block transcriptional elongation leading to oppression of short-living anti-apoptotic proteins, such as MCL-1, thereby promoting the apoptosis of tumor cells [30]. Accordingly, targeting CDK9, or blocking its pathway of transcription, offers a potentially effective therapy for malignant tumors (Supplementary Fig. S1). However, the relationship between CDK9 expression and clinical prognosis, and the therapeutic potential of targeting CDK9 in osteosarcoma patients remains to be elucidated. This prompted us to evaluate the role of CDK9 in osteosarcoma.
This is the first study to examine the expression of CDK9 in osteosarcoma patient specimens and correlate this to post-neoadjuvant chemotherapy tumor necrosis as well as the clinical prognosis of the patients. We also investigated the role of CDK9 in cell proliferation, colonization and migration in osteosarcoma cells.
2. Materials and methods
2.1. Osteosarcoma sample collection and tissue microarrays (TMA)
A total of 70 osteosarcoma specimens with formalin fixed paraffin-embedded (FFPE) blocks and 8 fresh tissue samples were obtained from patients who were diagnosed with osteosarcoma and who had received preoperative chemotherapy and surgical treatment at the Orthopaedic Department of Massachusetts General Hospital (MGH) between 1993 and 2010. Clinical information was also collected, including age, gender, tumor location, disease status, and follow-up time. The samples included 46 (65.7%) males and 24 (34.3%) females, with the mean age of 34.1 years old (range: 6–77 years old). All osteosarcoma patients were followed up for a mean period of 81.1 months after surgery (range: 1–273 months). The TMA of osteosarcoma FFPE blocks were provided by the Sarcoma Tissue Bank in MGH. To ensure that the selection included the core of the tumor, three sites of each FFPE block were selected for assembling the recipient master block. The TMA was constructed by the Tissue Microarray and Imaging Core at the Dana-Farber/Harvard Cancer Center. Representative triplicate 0.5 mm-diameter core biopsies of each tissue block were obtained through the pathology reports and reading of corresponding Hematoxylin and eosin (HE)-stained slides by a pathologist. The study was approved by the Partners Human Research Committee (#: 2007P-002464). All patients signed a consent form for their clinical information to be used for this research.
2.2. Immunohistochemistry (IHC)
The expression of CDK9 was determined using IHC assays according to the manufacturer's instructions (Cell Signaling Technology, Beverly, MA, USA). In brief, the paraffin-embedded slides were baked for 1 h at 60 °C before xylene deparaffinization and subsequent rehydration through graded ethanol (100% and 95%). 3% hydrogen peroxide was used to quench endogenous peroxidase activity after heated epitope retrieval. Following this, the slide was blocked for 1 h with normal goat serum, and then incubated with polyclonal rabbit antibody to human CDK9 (Cell Signaling Technology, catalog #2316S, 1:50 dilution, in 1% bovine serum albumin PBS) overnight in a humidified chamber set at 4 °C. SignalStain® Boost Detection Reagent (Cell Signaling Technology) and SignalStain® DAB (Cell Signaling Technology) were then utilized to detect the bound antibody. A hematoxylin QS (Vector Laboratories, Burlingame, CA) counterstain was used to obtain clearer images of the osteosarcoma cell nuclei before final long-term preservation using VectaMount AQ (Vector Laboratories) section mounting. Even in the absence of CDK4 antibody binding, the TMA slides were stained to reveal any nonspecific secondary antibody reactions.
2.3. Analysis of immunohistochemistry staining
Two independent pathologists, who were blinded to patient data and tumor characteristics, viewed and scored the immunostained slides. CDK9 expression was subsequently divided up into 6 groups based on the percentage of cells showing positive nuclear staining: 0, no nuclear staining; 1+, <10% of positive cells; 2+, 10%–25% of positive cells; 3+, 26%–50% of positive cells; 4+, 51%–75% of positive cells; 5+, >75% of positive cells. The low-CDK9 expression subset included groups 0; 1+ and 2+, while the high-CDK9 expression subset included groups 3+; 4+ and 5+. The tumor necrosis data was collected from the clinical data, and grouped according to the percent of tumor tissue necrosis of pathological specimens. Data was divided into two groups; good response: ≥ 90% necrosis; poor response: <90% necrosis. CDK9 staining images were obtained using a Nikon Eclipse Ti—U fluorescence microscope (Diagnostic Instruments Inc., NY, USA) with a SPOT RT™ digital camera (Diagnostic Instruments Inc.).
2.4. Human osteoblast and osteosarcoma cell lines and cell culture
The human osteoblast cell line HOB-c was purchased from PromoCell GmbH (Heidelberg, Germany). Osteoblast cell lines were cultured in osteoblast growth medium (PromoCell). Human osteosarcoma cell lines U2OS, MG63, MNNG/HOS, Saos-2, and 143B were obtained from the American Type Culture Collection (Rockville, MD). Osteosarcoma cell lines KHOS, KHOSR2, and U2OSR2 were provided by Dr. Efstathios Gonos (Institute of Biological Research & Biotechnology, Athens, Greece). All osteosarcoma cell lines were cultured at 37 °C in a humidified 5% CO2 atmosphere in RPMI 1640 (GE Healthcare Life Sciences, Logan, Utah, USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, MO, USA) and 1% penicillin/streptomycin.
2.5. Protein preparation and western blotting
1× RIPA lysis buffer (Sigma-Aldrich) and protease inhibitor cocktail tablets (Roche Applied Science, IN, USA) were used to extract protein lysates from the cells or tissues. The protein lysate concentrations were then determined using the DC™ protein assay reagents (BIORAD, Hercules, CA, USA) and a spectrophotometer SPECTRA max 340PC (Molecular Devices, Inc., CA, USA). Western blotting was performed using similar methods to those previously described [13]. In short, an SDS-PAGE gel was used to run the denatured proteins before they were transferred to nitrocellulose membranes. The membranes were incubated with monoclonal rabbit antibodies to human RNAPII ser2 (1:1000 dilution, Abcam, San Francisco, California, USA), CDK9, MCL-1, BIRC5 (1:1000 dilution, Cell Signaling Technology), and monoclonal mouse antibodies to human RNAPII total (1:1000 dilution, Abcam), Tubulin (1:1000 dilution, Cell Signaling Technology) at 4 °C overnight after they were blocked in 5% nonfat milk for 1 h. Following incubation with the primary antibody, TBST was then used as a membrane wash (3 times, 5 min, room temperature). Next, goat anti-rabbit IRDye 800CW (926–32,211, 1:5000 dilution) or goat anti-mouse IRDye 680LT secondary antibody (926–68,020, 1:15000 dilution) (Li-COR Biosciences, NE, USA) was applied for 2 h at room temperature followed by another TBST membrane wash (3 times, 5 min, room temperature). Bands were detected using an Odyssey Infrared Fluorescent Western Blots Imaging System from Li-COR Bioscience (Lincoln, NE, USA) and Odyssey software 3.0 was used to quantify the bands.
2.6. Immunofluorescence assay
The cultured osteosarcoma cell lines were used in the immunofluorescence assays. The cells were grown for three days in 6-well plates and fixed with 4% paraformaldehyde for 15 min before being permeabilized with ice-cold methanol and blocked with 1% BSA. A CDK9 primary antibody (1:200 dilution, Cell Signaling Technology), and β-Actin (1:500 dilution, Sigma-Aldrich) were incubated with the cells at 4 °C overnight. The next day cells were incubated for an additional 1 h with Alexa Fluor 488 (Green) conjugated goat anti-rabbit antibody or Alexa Fluor 594 (red) goat anti-mouse antibody (Invitrogen, NY, USA). Cells were imaged using a Nikon Eclipse Ti—U fluorescence microscope (Diagnostic Instruments Inc., NY, USA) equipped with a SPOT RT™ digital camera.
2.7. Knockdown of CDK9 by siRNA Transfection and MTT Assay
Knockdown of CDK9 in osteosarcoma cells was performed by a specific siRNA transfection. In brief, U2OS and KHOS cells were grown at a density of 2 × 103 cells/well in 96-well plates or at a density of 4 × 104 cells/well in 12-well plates and transfected with increasing concentrations (0, 10, 30, 60 nM) of synthesized CDK9 siRNA (5’-GCUGCUAAUGUGCUUAUCA-3′) (Sigma-Aldrich) using the Lipofectamine RNAiMax reagent (Invitrogen) according to the manufacturer's instructions. Nonspecific siRNA (60 nM) was used as a negative control. Three or five days after transfection of the CDK9 siRNA, the proteins of U2OS and KHOS cell were extracted for protein measurement with Western blotting or assessment of cellular proliferation by MTT assays. At the end of the 5-day cell treatment, 20 μL of MTT (5 mg/mL, Sigma-Aldrich) was added to each well of the 96-well plates. After incubating at 37 °C in a humidified 5% CO2 atmosphere for 4 h, the resulting formazan product was solubilized with 100 μL of acid isopropanol and the absorbance was measured at a wavelength of 490 nm on the SpectraMax Microplate® Spectrophotometer (Molecular Devices LLC, Sunnyvale, CA).
2.8. CDK9 suppression by LDC000067 inhibitor treatment and MTT assay
The novel highly selective CDK9 inhibitor, LDC000067 (abbreviated as LDC067; Sigma-Aldrich), has been verified at a concentration of 10 μM to inhibit the effects of CDK9 in several cancer cell lines [31]. Here, U2OS and KHOS cells were seeded into 96-well plates at a density of 4 × 103 cells/well or 6-well plates at a density of 6 × 105 cells/well and incubated with increasing concentrations (0, 2.5, 5, 10, 20 μM) of LDC067 for 2, 3, or 5 days prior to subsequent experiments. After LDC067 treatment for 5 days, the cell proliferation of U2OS and KHOS was investigated using the MTT assay (see above mentioned experimental procedure). Meanwhile, in order to detect the morphological changes of U2OS and KHOS cells, a Nikon microscope (Diagnostic Instruments Inc., NY, USA) was used after 3 days of LDC067 treatment.
2.9. Clonogenic assays
The clonogenic assay is a well-established in vitro method for evaluating the viability and proliferation capabilities of cells. Osteosarcoma cells, U2OS and KHOS, were seeded at 150 cells/well in the 12-well plates and treated with the CDK9 inhibitor, LDC067, at different concentrations (0, 5, 10 μM), and incubated at 37 °C for 12 days. Colonies were subsequently fixed with methanol for 10 min, and then washed three times with PBS before staining with 10% Giemsa stain (Sigma-Aldrich) for 20 min. The cells were washed with flowing water and allowed to dry. Pictures of the stained colonies were captured using digital camera (Olympus, Tokyo, Japan).
2.10. Cell wounding healing migration assays
Wound healing assays were applied to test cell migration activities. U2OS and KHOS cells were seeded into 6-well plates at a density of 4 × 105 cells per well and incubated overnight. A sterile 10 μL tip was then used to scrape two parallel lines within the adherent cell layer. Next, 10 μM of LDC067 was added and left to incubate for 72 h in a low-serum medium containing 2% FBS. Wounds were photographed using a Nikon microscope (Diagnostic Instruments Inc., NY, USA) with Zen Imaging software after 0, 24, 48, and 72 h of the LDC067 treatment. The distance between the two edges of the scratch, at five distinct sites of each image, was used to measure the width of the wounds. The following formula was applied to measure the cell migration distance: (wound width at the 0 h time point - wound width at the observed time point) / 2.
2.11. Three-dimension(3D) cell culture
A 3D cell culture is an improved method and a more accurate model that can mimic the in vivo environment to evaluate the behavior of cancer cell growth [32]. Spheroid formation of the osteosarcoma cell lines, U2OS and KHOS, was established in 24-well VitroGel™ 3D cell culture plates with a density of 2 × 105 cells/well, according to the manufacturer's protocol (TheWell Bioscience Inc., NJ, USA). Immediately following this, 10 μM of LDC067 was added into the cell medium. Osteosarcoma cells without treatment were considered as the blank control. The plates were incubated at 37 °C in a humidified 5% CO2 atmosphere. Medium was changed every 24–48 h to provide enough nutrients for the cells and to prevent an osmolality shift in the medium. The spheroids were photographed under a Nikon microscope (Diagnostic Instruments Inc.) equipped with the Zen Imaging software every 2 days. At the point of 12 days, the spheroids were harvested from the bottom side of the plate by gently pipetting 100 μL PBS into each well. After 15 min of incubation with 0.25 μM Calcein AM (Invitrogen™, Oregon, USA), the spheroids were imaged on the Nikon Eclipse Ti—U fluorescence microscope (Diagnostic Instruments Inc.) equipped with a SPOT RT™ digital camera. The diameter of spheroids was measured three times using software ImageJ.
2.12. Statistical analysis
GraphPad Prism 7 software was used for statistical analyses. Independent two-tailed Student t-tests were performed for independent data to determine the statistical significance. One-way ANOVA tests were performed for multiple comparisons. Comparison of differences in survival curves was analyzed by Kaplan-Meier plots and log-rank tests. The χ2 test was used to evaluate the relationship between CDK9 expression and osteosarcoma clinical-pathological parameters. Prognostic factors associated with overall survival were analyzed through the Cox proportional hazards regression model, in a stepwise manner. Only those factors that were statistically significant (P < .05) in the univariate survival analysis were involved in the multivariate analysis. The correlations analysis was investigated by a Spearman's rank correlation. All results were presented as mean ± SD, and P values <.05 was deemed statistically significant.
3. Results
3.1. CDK9 expression is associated with the clinicopathological features and clinical prognosis of osteosarcoma patients
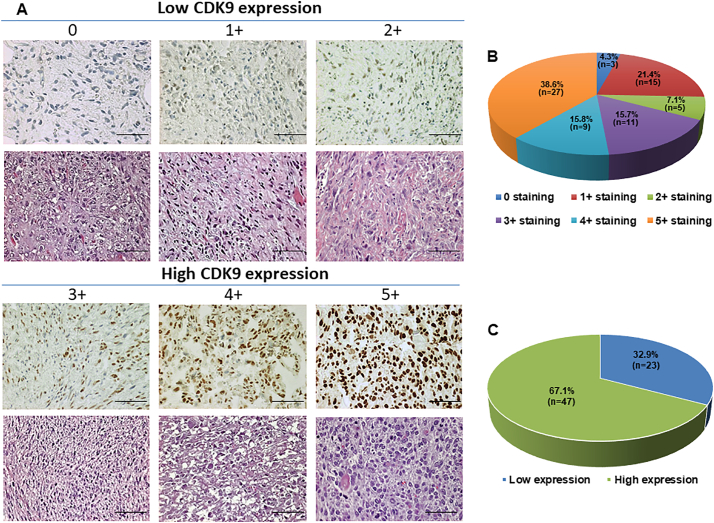
We first evaluated the CDK9 expression of osteosarcoma patient samples and analyzed if this could be correlated to their pathological characteristics, as well as the patient clinical prognosis. Of the available patient samples, 67 of 70 (95.7%) tissues exhibited CDK9 immunostaining in the cell nucleus, ranging from 0 staining (3 of 70, 4.3%); 1+ staining (15 of 70, 21.4%), 2+ staining (5 of 70, 7.1%), and 3+ staining (11 of 70, 15.7%), 4+ staining (9 of 70, 12.8%), 5+ staining (27 of 70, 38.6%) (Fig. 1A and B, Supplementary Table S1). The stained specimens were subdivided into two categories: ≤2+ were defined as low CDK9 expression (32.9%) and ≥3+ were defined as high CDK9 expression (67.1%) (Fig. 1A and C, Table 1).
Fig. 1.
Evaluation of CDK9 expression and staining in an osteosarcoma TMA by immunohistochemistry. (A) Representative images of different immunohistochemistry staining intensities of CDK9 and HE staining are shown in osteosarcoma tissues. According to the CDK9 staining in the tumor samples, the staining patterns were divided into 6 groups: no positive staining (0); <10% positive cells (1+); 10–25% positive cells (2+); 26–50% positive cells (3+); 51–75% positive cells (4+); >75% positive cells (5+). (Original magnification, 400×; Scale bar, 50 μm). (B) Pie chart representing relative frequency of different CDK9 expression levels in osteosarcoma tissue microarrays. (C) Tumors with the staining score of ≤2+ were defined as low CDK9 expression group (blue), ≥3+ were defined as the high CDK9 expression group (green). Pie chart representing relative frequency of the two groups in osteosarcoma tissue microarrays.
Table 1.
The relationship between CDK9 expression and clinicopathological features of osteosarcoma patients.
| Clinicopathological features | Number of cases |
CDK9 Expression low |
CDK9 expression high |
P value |
|---|---|---|---|---|
| (n, %) | (n, %) | (n, %) | ||
| All patients | 70(100) | 23 (32.9) | 47(67.1) | |
| Age, year | ||||
| ≤18 | 20(28.6) | 5 (25.0) | 15(75.0) | 0.1719 |
| >18 | 50(71.4) | 18 (36.0) | 32(64.0) | |
| Gender | ||||
| Male | 46(65.7) | 15(32.6) | 31(67.4) | 0.6799 |
| Female | 24(34.3) | 8(33.3) | 16(66.7) | |
| Tumor site | ||||
| Femur | 33(47.1) | 10(30.3) | 23(69.7) | 0.9945 |
| Tibia | 13(18.6) | 4(30.8) | 9(69.2) | |
| Humeral bone | 8 (11.4) | 3(37.5) | 5(62.5) | |
| Other | 16(22.9) | 6(37.5) | 10(62.5) | |
| Metastasis | ||||
| Absent | 13(18.6) | 8(61.5) | 5(38.5) | 0.0064a |
| Present | 57(81.4) | 14(24.6) | 43(75.4) | |
| Recurrence | ||||
| Absent | 47(67.1) | 16(34.0) | 31(66.0) | 0.9899 |
| Present | 23(32.9) | 7(30.4) | 16(69.6) |
Statistically significant.
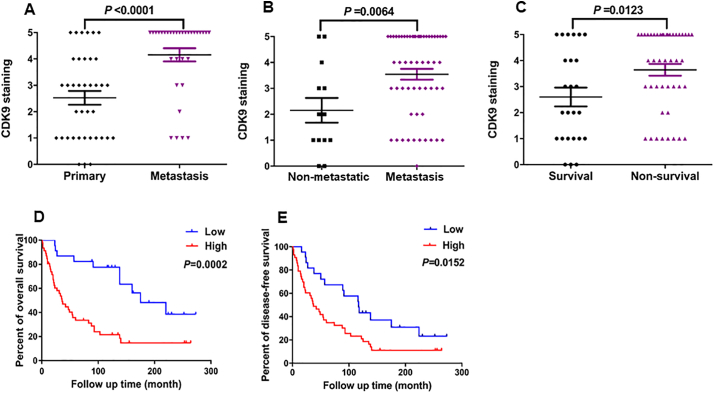
We also compared patient prognosis with the expression of CDK9. There was a significant difference in CDK9 expression between primary tumor tissues (patients without metastatic/recurrent disease) and tissues taken from patients with metastatic disease (P < .0001, based on the Independent two-tailed Student t-test) (Fig. 2A). Based on the patient follow-up data, the CDK9 expression was significantly higher in the osteosarcoma tissues from the patients who developed metastatic disease compared with those patients who did not (P = .0064, based on the Independent two-tailed Student t-test) (Fig. 2B). We also found that the expression of CDK9 in patients who did not survive (non-survival) was significantly higher than those who did survive (survival) (P = .0123, based on the Independent two-tailed Student t-test) (Fig. 2C). Further analysis showed that CDK9 expression had no relationship to other clinical pathological features of the patients, including age (P = .1719, based on the χ2 test), gender (P = .6799, based on the χ2 test), and tumor site (P = .9945, based on the χ2 test) (Table 1).
Fig. 2.
The correlation between CDK9 expression and clinicopathological characteristics and prognosis of osteosarcoma patients. (A) Distribution of CDK9 immunostaining scores in primary tumor tissues (patients without metastasis/recurrence of disease) and tissues from patients with metastatic disease. (B) Comparison of CDK9 immunohistochemistry staining scores between osteosarcoma tissues of patients with metastatic (including patients with primary metastatic disease and patients who developed metastatic disease) and non-metastatic disease (based on the disease status of patients at the end of follow-up time). (C) Comparison of CDK9 immunohistochemistry staining scores between survivor and non-survivor osteosarcoma tissues. (D) Kaplan-Meier overall-survival curve of patients with osteosarcoma were sub-grouped as either CDK9 low-expression group (staining score ≤ 2+) or high-expression group (staining score ≥ 3+). Compared with the low-expression group, the patients with high CDK9 staining had a shorter overall survival. (E) Kaplan-Meier disease-free survival curves of patients with osteosarcoma that were sub-grouped as either CDK9 low expression group (staining score ≤ 2+) or high expression group (staining score ≥ 3+). Comparison between the two groups, patients with high CDK9 expression had worse disease-free survival.
Next, we further evaluated the association between CDK9 expression and other clinical outcomes of osteosarcoma patients, including overall patient survival and disease-free survival. The Kaplan-Meier analysis revealed that patients with high CDK9 expressing tumors had significantly worse overall survival rates compared to patients with low CDK9 expressing tumors (P = .0002, based on the log-rank test) (Fig. 2D). Similarly, patients with high CDK9 expression had significantly worse disease-free survival rates than the patients with low CDK9 expression (P = .0152, based on the log-rank test) (Fig. 2E). Furthermore, to confirm whether CDK9 expression is an independent prognostic factor for osteosarcoma patients, we applied a Cox regression analysis. In the univariate Cox regression analysis, overexpression of CDK9, presence of metastatic disease, and the response to preoperative chemotherapy were all associated with a shorter survival rate in osteosarcoma patients. However, other clinicopathological features showed no prognostic correlations (Table 2). Notably, the analysis of multivariate Cox regression indicated that high CDK9 expression was an independent predictor for survival of osteosarcoma patients (P = .004, based on the Cox proportional hazards regression model) (Table 2). These results demonstrated that CDK9 expression independently predicts osteosarcoma patient outcomes.
Table 2.
Univariate and multivariate survival analysis of prognostic factors in osteosarcoma.
| Variable | Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | P value | Hazard Ratio | 95% CI | P value | |
| All patients | ||||||
| Age, year | 0.996 | 0.979–1.014 | 0.680 | |||
| ≤18 | ||||||
| >18 | ||||||
| Gender | 0.932 | 0.520–1.663 | 0.811 | |||
| Male | ||||||
| Female | ||||||
| Tumor site | 1.008 | 0.353–2.715 | 0.422 | |||
| Femur | ||||||
| Tibia | ||||||
| Humeral bone | ||||||
| Other | ||||||
| Metastasis | 7.819 | 4.068–15.030 | <0.001a | 4.893 | 2.036-13.162 | <0.001a |
| Present | ||||||
| Absent | ||||||
| Recurrence | 1.496 | 0.822–2.720 | 0.187 | |||
| Present | ||||||
| Absent | ||||||
| CDK9 expression | 3.411 | 1.924–6.045 | <0.001a | 3.568 | 1.512-8.417 | 0.004a |
| High | ||||||
| Low | ||||||
| Response to pre-operative chemotherapy | 0.240 | 0.115–0.499 | 0.002a | 0.128 | 0.026-0.651 | 0.008a |
| Good | ||||||
| Poor | ||||||
Statistically significant.
3.2. CDK9 expression is inversely correlated to percent of tumor necrosis post-neoadjuvant chemotherapy in osteosarcoma patients
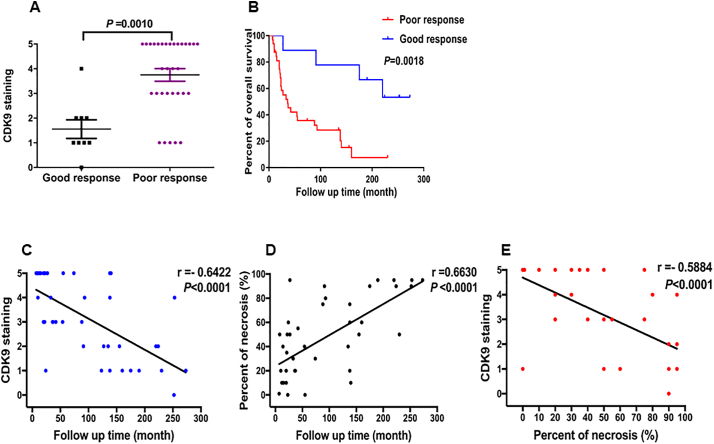
It is well known that the percent of necrosis after neoadjuvant chemotherapy is the most important predictive factor of disease outcome for osteosarcoma patients [[33], [34], [35]]. Significant necrosis is associated with better disease-free survival and overall survival. First, we evaluated whether CDK9 expression is associated with the degree of necrosis as determined by the percent of tumor necrosis on histological examination of osteosarcoma tumor specimens. Among the 70 patient samples, the percent of tumor necrosis post-neoadjuvant chemotherapy data was available for 41 specimens. Eight tumor tissues demonstrated ≥90% necrosis (good responders) and 33 patients showed <90% necrosis (poor responders) after neoadjuvant chemotherapy. Patients with a poor response had significantly higher CDK9 expression compared with those who had a good response (P = .0010, based on the Independent two-tailed Student t-test) (Fig. 3A and Supplementary Table S2). In addition, there was a significant difference in the overall survival rate between good responders and poor responders (P = .0018, based on the log-rank test) (Fig. 3B). We further compared the relationship between CDK9 expression, the percent of tumor necrosis and overall survival in these osteosarcoma patients. The data revealed the overall survival of patients is inversely correlated to CDK9 expression levels (P < .0001, r = −0.6422, based on the Spearman's rank correlation) (Fig. 3C). In contrast, as anticipated, the overall survival of patients is directly correlated with the increase of the percent of tumor necrosis (P < .0001, r = 0.6630, based on the Spearman's rank correlation) (Fig. 3D) We also evaluated if high CDK9 expression is related to a decrease in the necrosis of osteosarcoma tissues. Indeed, linear regression analysis demonstrated that the expression of CDK9 is inversely proportional to the percent of tumor necrosis in osteosarcoma tissues (P < .0001, r = −0.5884, based on the Spearman's rank correlation) (Fig. 3E). These results indicate that CDK9 expression can act as an indirect indicator of the percent of tumor necrosis post-neoadjuvant therapy in osteosarcomas.
Fig. 3.
Association of CDK9 expression with the response of preoperative chemotherapy and tumor necrosis of osteosarcomas patients. (A) Comparison of CDK9 immunostaining scores between good responders and poor responders osteosarcoma tissues based on the histological necrosis percentage after preoperative chemotherapy. (B) Kaplan-Meier overall survival curves of patients with osteosarcoma that were sub-grouped as either good or poor responders to preoperative chemotherapy. The overall survival of good responders was better than poor responders. (C) Inverse correlation between the expression of CDK9 and overall survival of osteosarcoma patients (P < .0001, r = −0.6422, based on the Spearman's rank correlation). (D) Positive correlation between percent tumor necrosis and overall survival in osteosarcoma patients (P < .0001, r = 0.6630, based on the Spearman's rank correlation). (E) Inverse correlation between CDK9 expression and overall survival of osteosarcoma patients (P < .0001, r = −0.5884, based on the Spearman's rank correlation).
3.3. CDK9 is highly expressed in human osteosarcoma cell lines and tissues
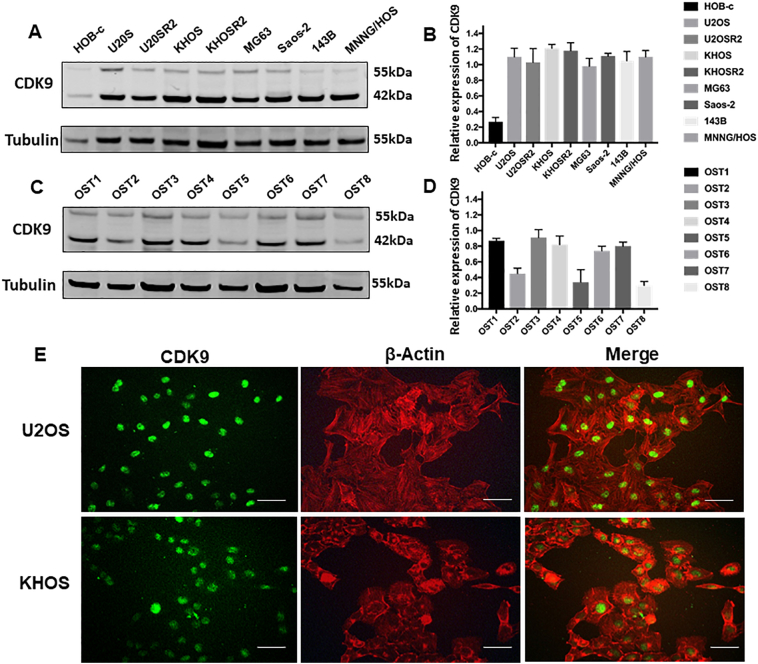
To determine the potential role of CDK9 in human osteosarcoma cells, we first examined the expression of CDK9 in human osteosarcoma cell lines. Western blotting analysis showed that CDK9 is highly expressed in osteosarcoma cell lines (U2OS, U2OSR2, KHOS, KHOSR2, MG63, Saos-2, 143B and MNNG/HOS), whereas the expression of CDK9 is markedly lower in the normal osteoblast cell line (HOB-c) (Fig. 4A and B). In order to preclude the possibility that CDK9 expression is an artifact induced by in vitro propagation, we further evaluated the expression of CDK9 in eight osteosarcoma fresh specimens. The results demonstrated that CDK9 is highly expressed in the majority of osteosarcoma tissues (Fig. 4C and D). There are 2 known isoforms of the CDK9 protein, our study found both CDK9 42 kDa and CDK9 55 kDa isomers had variable expression in osteosarcoma cell lines and osteosarcoma tissues. The 42 kDa form is predominantly overexpressed, thus the 42 kDa band was used for the quantification of CDK9 expression (Fig. 4B and D). We also explored the expression of CDK9 and confirmed its cellular localization in osteosarcoma cells by immunofluorescence in U2OS and KHOS cell lines. We found that the CDK9 protein was mainly localized to the cell nucleus (Fig. 4E). Collectively, these results are consistent with the TMA data showing marked CDK9 expression in osteosarcoma samples (Fig. 1A).
Fig. 4.
Expression of CDK9 in osteosarcoma cell lines and osteosarcoma patient fresh tissues. (A) Expression levels of CDK9 in osteosarcoma cell lines (U2OS, U2OSR2, KHOS, KHOSR2, MG63, Saos-2, 143B and MNNG/HOS) were stronger than the expression of CDK9 in the normal osteoblast cell line (HOB-c) measured by Western blotting. (B) Densitometry quantification of the Western blots of CDK9 from Fig. 4A, presented as relative to tubulin expression. The data represent the mean ± SE of the experiment carried out in triplicate. (C) CDK9 expression in eight human osteosarcoma fresh tissues measured by Western blotting. (D) Densitometry quantification of the Western blots of CDK9 from Fig. 4C, presented as relative to tubulin expression. The data represent the mean ± SE of the experiment carried out in triplicate. (E) Confirmation of CDK9 expression in osteosarcoma cell lines by immunofluorescence with antibodies to CDK9 (green) and Actin (red). Green fluorescence of CDK9 protein was mainly localized in the nucleus of osteosarcoma cells (Scale bar, 50 μm).
3.4. CDK9 knockdown by siRNA decreased human osteosarcoma cell proliferation
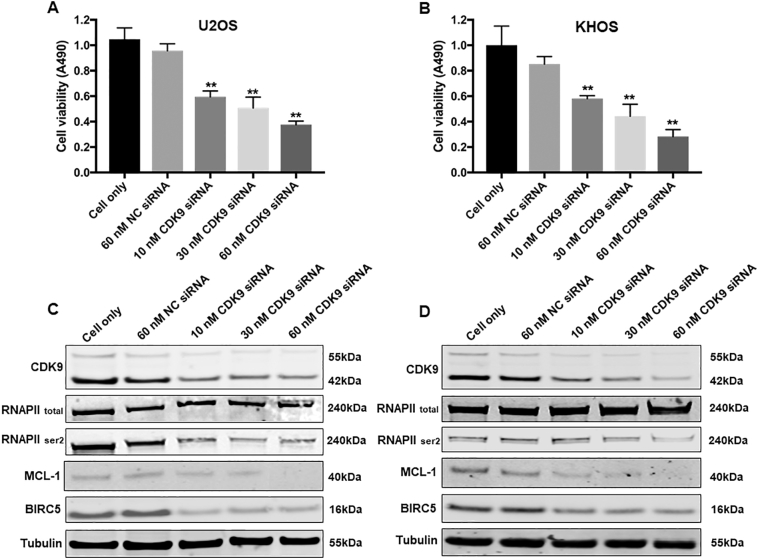
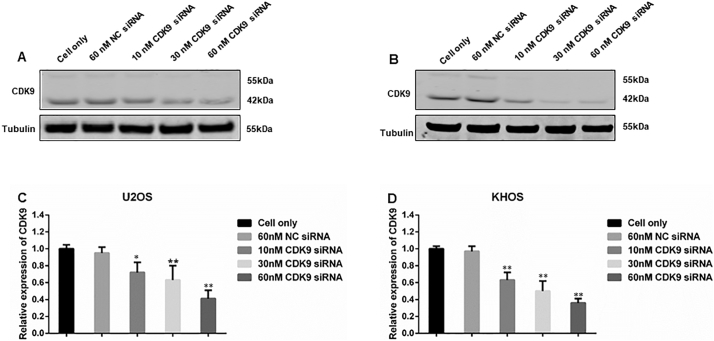
In order to validate the role of CDK9 in osteosarcoma cell proliferation and growth, we knocked down CDK9 expression with a CDK9-specific siRNA and investigated the change in osteosarcoma cell viability and cell signaling pathways. After transfection with increasing concentrations of CDK9 siRNA for 3 days, there was a dose-dependent decrease in the cell viability of both U2OS and KHOS cells, which was not present in the nonspecific siRNA transfected cells and the cell only control (Fig. 5A and B). In addition to this, CDK9 siRNA transfection significantly inhibited CDK9 expression as determined by Western blotting and immunofluorescence assays (Supplementary Figs. S2 and S3A and B). Silencing of CDK9 with siRNA inhibited the CDK9 signaling pathway in a dose-dependent manner, as indicated by the reduced expression of phosphorylated RNAPII ser2 as well as MCL-1 and BIRC5, while total RNAPII expression did not markedly change (Fig. 5C and D).
Fig. 5.
CDK9 inhibition by siRNA decreased osteosarcoma cell proliferation by suppression of the RNAPII phosphorylation pathway. Cell viability of (A) U2OS and (B) KHOS, determined by MTT assays measured at 490 nm after CDK9 siRNA negative control (NC) siRNA transfection. The data is mean ± SE of the 2 experiments carried out in triplicate. The expression of respective proteins in CDK9-associated signaling pathway was measured by Western blotting in the osteosarcoma cell lines (C) U2OS, and (D) KHOS after 72 h of siRNA transfection. (** indicates P < .01).
3.5. Activity of CDK9 is suppressed by the CDK9 inhibitor LDC067 in osteosarcomas
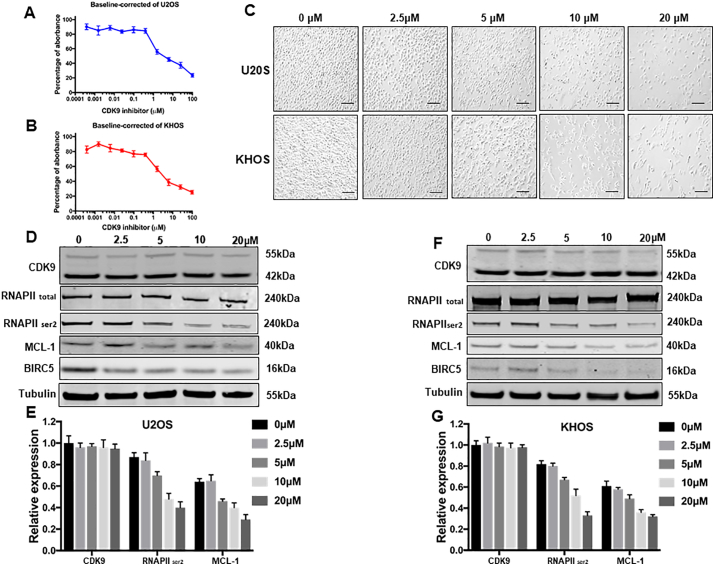
Kinase inhibitors are the backbone for many current targeted cancer therapies. However, due to the heterogeneity of the disease, kinase based therapeutic options have not yet been established for osteosarcoma treatment. After validating the expression and clinical significance of CDK9 in patients osteosarcoma samples, we further assessed the effect of CDK9 inhibition on the proliferation of osteosarcoma cells. Cell viability was decreased in a dose-dependent manner in both osteosarcoma cell lines U2OS and KHOS, with the IC50 values for LDC067 at 5.80 μM and 6.50 μM, respectively. The effect of treatment with the increasing concentrations of LDC067 for 5 days was analyzed (Fig. 6A and B). Similarly, the morphologic changes and a reduction of the viable cell number were observed with an increase in the concentration of LDC067 in osteosarcoma cell lines, when treated for 72 h (Fig. 6C).
Fig. 6.
Effects of the CDK9 inhibitor LDC067 on the expression of CDK9 and cell growth in osteosarcoma cell lines. LDC067, at the indicated concentrations, inhibited osteosarcoma cell proliferation in (A) U2OS, and (B) KHOS cell lines, which was determined by MTT assays. The data represent the mean ± SE of 2 experiments carried out in triplicate. (C) Microscopy images of morphologic changes and a reduction in cells number after 72 h of LDC067 treatment (Scale bar, 100 μm). (D) The expression of proteins involved in the CDK9-signaling pathway in the U2OS osteosarcoma cell line was examined by Western blotting after 48 h of LDC067 treatment. (E) Semiquantitative analysis of Fig. 6D densitometry relative to tubulin. The data represent the mean ± SE of the experiment carried out in triplicate. (F) The expression of proteins involved in the CDK9-signaling pathway in the KHOS osteosarcoma cell line was examined by Western blotting after 48 h of LDC067 treatment. (G) Semiquantitative analysis of Fig. 6F densitometry relative to tubulin. The data are mean ± SE of the experiment carried out in triplicate.
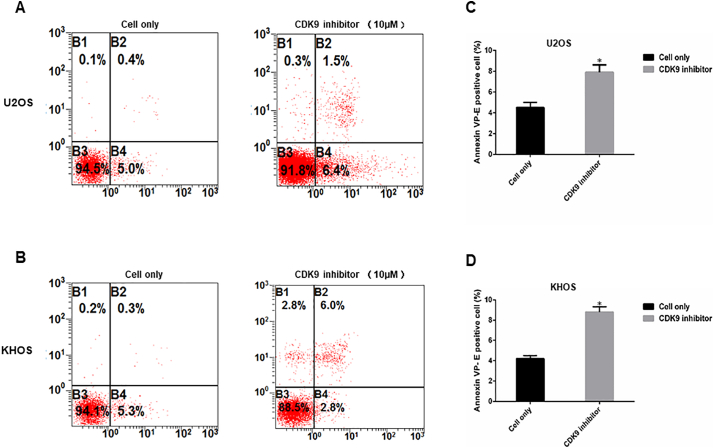
To investigate the CDK9 cell signaling pathway, we measured the expression of several CDK9 downstream target proteins following LDC067 treatment. After incubation of the cell lines with 2.5, 5, 10, and 20 μM LDC067 for 48 h, expression of RNAPII ser2, the major downstream target of CDK9, and the expression of anti-apoptotic proteins, MCL-1 and BIRC5, were significantly decreased in a dose-dependent manner, whereas the expression of total RNAPII did not significantly change (Fig. 6D, E, F, and G). Importantly, LDC067 only inhibits CDK9 activity, but not its expression, as demonstrated by Western blotting (Fig. 6D, E, F, and G). Furthermore, a flow cytometry analysis was performed to evaluate cellular apoptosis induced by inhibiting CDK9 activity. Compared with the untreated cells, the apoptosis rates of U2OS and KHOS cells, after 10 μM LDC067 treatment, were significantly increased (Supplementary Fig. S4).
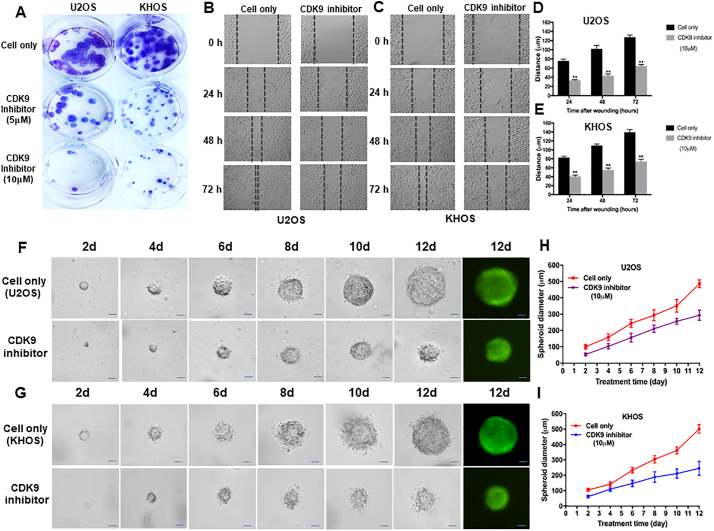
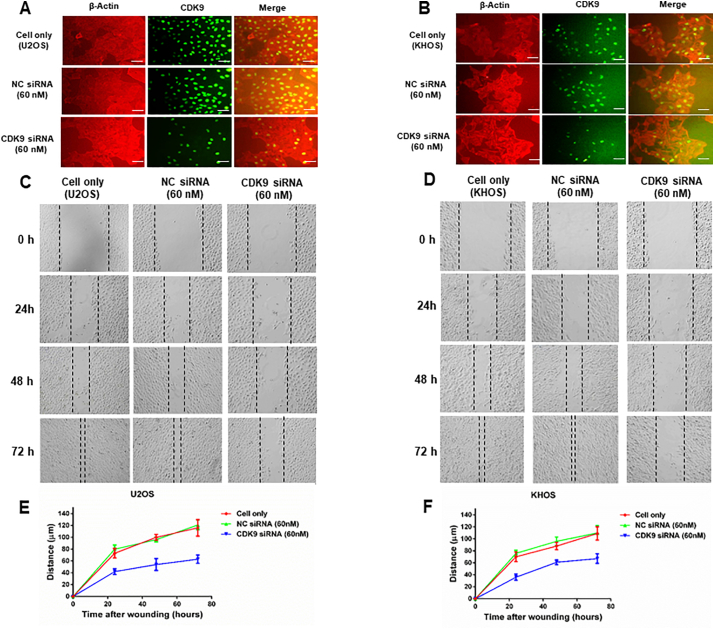
We next assessed the effect of LDC067 on the colony forming ability of osteosarcoma cells using a clonogenic assay. The clonogenicity of U2OS and KHOS, when treated with LDC067, was reduced in a dose-dependent manner, compared with untreated cells (Fig. 7A). Cell migration and invasion are the crucial steps in the event of cancer metastasis. As TMA results showed that the expression of CDK9 was significantly correlated to metastatic disease in osteosarcoma patients, we further evaluated the role of CDK9 in osteosarcoma cell migration in vitro, after exposure to 10 μM of LDC067 for 24, 48, and 72 h. Cell migration was significantly suppressed both in U2OS and KHOS cells in a time-dependent manner (Fig. 7B, C, D, and E).
Fig. 7.
CDK9 inhibition reduced osteosarcoma cell clonogenicity and migration in vitro and decreased the spheroid diameter of osteosarcoma cell lines in a 3D cell culture. (A) Representative results of colony formation in U2OS and KHOS. The numbers of colonies and their sizes were markedly decreased in cells treated with LDC067. Representative images of (B) U2OS, and (C) KHOS cell migration after LDC067 treatment for 24 h, 48 h, and 72 h. Cell migration distance of osteosarcoma cell lines (D) U2OS, and (E) KHOS were measured after 10 μM of LDC067 treatment. Spheroid formation of (F) U2OS, and (G) KHOS were significantly smaller than untreated cells at all observation points. Cells fluorescence images of spheroid formation were taken after 12 days of cultivation. The curves of relative spheroid diameter of (H) U2OS, and (I) KHOS treated with CDK9 inhibitor compared with untreated cells at all observation points. The data are mean ± SE of the 2 experiments carried out in triplicate (Scale bar, 100 μm). (** indicates P < .01).
Additionally, to evaluate whether suppression of CDK9 could alter the tumorigenicity of osteosarcoma in a simulated in vivo environment, a three-dimension(3D) cell culture was used to test the effect. Observations of spheroid size were recorded at different time points. The spheroid diameters in CDK9 inhibitor-treated U2OS and KHOS cells were significantly smaller than the untreated cells (Fig. 7F and G). After 12 days of 10 μM CDK9 inhibitor treatment, the spheroid diameter of U2OS cell was 57.4% of the untreated U2OS cell (P < .001, based on the Independent two-tailed Student t-test) (Fig. 7H). Similar results were also found in the KHOS cell line, with the diameter of the KHOS spheroid at 48.8% of the untreated KHOS cell (P < .001, based on the Independent two-tailed Student t-test) (Fig. 7I).
4. Discussion
Previous studies have demonstrated that CDK9 expression is a poor prognostic factor for the clinical outcomes of many tumors [20,21,24]. In our study, we analyzed the expression of CDK9 in 70 osteosarcoma tissue specimens and, for the first time, we have identified a close relationship between CDK9 expression and poor patient clinical outcomes. TMA of osteosarcoma tissue samples showed that 95.7% expressed CDK9. Notably, 27 of 32 (84.4%) samples from patients with metastatic disease revealed a high expression of CDK9 (staining score ≥ 3+), which was significantly higher than tumor tissues of patients without metastatic disease. Additionally, our study revealed that osteosarcoma patients with high CDK9 expression had a worse overall survival rate and disease-free survival rate than those patients with low CDK9 expression.
Although the histological response to preoperative chemotherapy has generally been accepted as one of the most important prognostic factors of the clinical outcomes of osteosarcoma patients, and is frequently used to guide the choice of postoperative chemotherapy [[33], [34], [35], [36]], few studies have focused on the correlation between the percent tumor necrosis post-neoadjuvant therapy and associated biomarker in patients with osteosarcoma. Patients with a histological response to preoperative chemotherapy, i.e. ≥90% necrosis (good responders), have been shown to have a much better survival rate than those with <90% necrosis (poor responders) [33,[36], [37], [38]]. Therefore, patients with a tumor necrosis rate of ≥90% should be continued on a similar chemotherapy to the preoperative regimen for a likely successful treatment. However, patients with tumor necrosis rates <90% are at a much higher risk of metastasis and recurrence even after complete surgical resection of the primary tumor. Although usually a new chemotherapy regimen that differs from the preoperative regimen by combining or replacing new chemotherapeutic agents is applied in order to improve the outcome of poor responders, the efficacy of these changed regimens has shown limited benefit for patients [[39], [40], [41], [42]]. Furthermore, the degree of necrosis is usually only known after 8 to 10 weeks of preoperative chemotherapy. In this period, the tumor cells may develop resistance or metastasize to the lungs if the preoperative chemotherapy is ineffective. Thus, it is necessary to identify the patients who are likely to not respond to the standard preoperative treatment at the time of initial diagnosis. In this study, we have demonstrated that the expression of CDK9 is inversely correlated to the degree of tumor necrosis that arises post-neoadjuvant chemotherapy, and, importantly, this relates to the clinical prognosis of osteosarcoma patients. These data indicate that CDK9 can serve as an alternative predictor of neoadjuvant therapy outcomes (tumor necrosis) and as a prognostic marker in osteosarcoma.
To date, over-expression of CDK9 has been reported in a number of cancer types [20,43]. Here, we find that CDK9 is also highly expressed in both osteosarcoma cell lines and in most of the fresh osteosarcoma tissues, which was consistent with the CDK9 TMA data. We also explored if the expression of CDK9 played a critical role in osteosarcoma growth and proliferation. CDK9 is a transcription factor reported to regulate the transcription of RNAPII, the key enzyme involved in the synthesis of mRNA [19,29]. To verify the biological role of CDK9 in osteosarcoma cell growth and proliferation, we conducted a knockdown analysis using a specific siRNA to CDK9. The results demonstrated a reduction in the cell growth and viability of U2OS and KHOS osteosarcoma cells when CDK9 was silenced.
As CDK9 inhibitors have recently been assessed in preclinical studies for the treatment of hematologic cancers [44,45], we also performed in vitro CDK9 loss-of-function studies to assess cell proliferation and growth of osteosarcoma cells. We found that the inhibition of CDK9 activity also decreased osteosarcoma cell growth and proliferation in dose-dependent manner. The transcriptional function of CDK9 requires phosphorylation on the ser2 of the CTD of RNAPII during elongation [46,47]. Moreover, LDC067 has been confirmed to target the ser2 of the RNAPII CTD, preventing phosphorylation and inducing apoptosis [31]. To further characterize the function of CDK9 in osteosarcoma cell survival and proliferation, we also investigated how interrupting the CDK9-signaling pathway impacts the target genes RNAPII ser2 in osteosarcoma cells. We found that siRNA and LDC067 both reduced the downstream phosphorylation of RNAPII ser2 and decreased expression of the anti-apoptotic proteins, BIRC5 and MCL-1. As anticipated, we also confirmed that LDC067 did not alter the expression of CDK9. Inhibition of CDK9 has been shown to be an effective anti-cancer strategy by blocking transcriptional elongation, thereby suppressing the expression of anti-apoptotic proteins, such as MCL-1, resulting in the induction of apoptosis in cancer cells [48]. Previous reports have demonstrated that chronic lymphocytic leukemia cells treated with a CDK9 inhibitor also suppressed expression of MCL-1 [31,49]. Therefore, our results in osteosarcomas are compatible with leukemia cells, in which CDK9 inhibition led to a decrease in the expression of anti-apoptotic proteins and induction of apoptosis.
Clonogenic assays are an in vitro cell survival assay based on the ability of a single cell to grow colonies [50,51]. The size and number of colonies in osteosarcoma cells were reduced in a dose-dependent manner with LDC067 treatment. While newer treatments have improved osteosarcoma patient survival, the high incidence of lung metastasis means the mortality rate still remains very high [1,52]. CDK9 has been confirmed as a target gene, suppressing tumor cell proliferation, migration and invasion [53,54]. As CDK9 was expressed highly in 84.4% of the tissues taken from osteosarcoma patients with metastatic disease, we further investigated the role of CDK9 in osteosarcoma cell migration in vitro by treating with LDC067. The cell migration activities were significantly suppressed both in U2OS and KHOS cells after LDC067 treatment in a time-dependent manner. Previous studies have shown that in triple negative breast cancer cell lines treatment with a CDK2/9 inhibitor resulted in decreased cell migration through a reduction in the phosphorylation of CDK-mediated Smad3 [55]. Accordingly, it is possible that the mechanism by which inhibition of CDK9 leads to decreased cell migration in osteosarcomas may also be through the suppression of Smad3 phosphorylation, which warrants further investigation. A more recent study has revealed that treatment of MYC-amplified and CDK9 overexpressing patient-derived tumor xenografts (PDTX) osteosarcoma mouse models, with a multi-CDK inhibitor, resulted in decreased tumor growth [56]. Similar results have been demonstrated with the multi-CDK inhibitor, Flavopiridol, targeting CDK1, 2, 6, 7 and 9, which decreases metastasis and cell survival in osteosarcomas [57]. Recently, another novel highly selective CDK9 inhibitor, MC180295, shown broad anti-cancer activity in vitro and demonstrated efficacy against lung cancer in vivo [25]. As 3D cell models have been authenticated to be more realistic for translating in vitro study results for in vivo application [32,58], we further validated the effect of CDK9 inhibition on cell proliferation using 3D cell cultures to simulate the in vivo tumor environment. Furthermore, we also found that the spheroid diameter of cells treated by the CDK9 inhibitor was significantly decreased compared with the untreated cells. Collectively, these results indicate that CDK9 plays a crucial role in the growth and proliferation of osteosarcoma cells.
In conclusion, our study demonstrated that the level of CDK9 expression is associated with the clinical prognosis of patients with osteosarcoma. CDK9 inhibition decreases osteosarcoma cell growth and proliferation by preventing RNAPII phosphorylation. These findings suggest that CDK9 may be a potential novel molecular biomarker for the early detection of disease and also may offer to be an effective therapeutic target in osteosarcoma.
The following are the supplementary data related to this article.
Fig. S1.
Schematic demonstrating the role of CDK9 in the transcription elongation process involved in tumor cell survival. CDK9 and cyclin T form the P-TEFb complex that regulates gene transcription elongation and mRNA maturation. CDK9 is involved in the phosphorylation on the ser2 of the CTD of RNAPII during elongation when the negative elongation factor (NELF) is released from the RNAPII, allowing transcriptional elongation and expression of anti-apoptotic genes including MCL-1, which together promote tumor cell survival.
Fig. S2.
Dose-dependent Knockdown of CDK9 by siRNA Transfection. The expression of both the 55 kDa and 42 kDa CDK9 was measured by Western blotting in the osteosarcoma cell lines (A) U2OS, and (B) KHOS after 72 h of siRNA transfection. (C) and (D) Western blotting densitometry quantification of the 42 kDa isoform of CDK9 from Fig. S2A and B, respectively. Densitometry is presented as relative to tubulin expression. The data represent the mean ± SE of the experiment carried out in triplicate (* indicates P < .05, ** indicates P < .01).
Fig. S3.
CDK9 inhibition by siRNA transfection decreases osteosarcoma cell proliferation and migration. CDK9 expression and osteosarcoma cell proliferation of (A) U2OS and (B) KHOS were determined by immunofluorescence after an incubation with a CDK9 antibody (green) and β-Actin (red). Representative images of (C) U2OS, and (D) KHOS cell migration after CDK9 siRNA and negative control (NC) siRNA transfection for 0 h, 24 h, 48 h, and 72 h. The distance of cell migration of the osteosarcoma cell lines (E) U2OS, and (F) KHOS were measured after siRNA transfection. (Scale bar, 50 μm).
Fig. S4.
CDK9 inhibition induced osteosarcoma cell apoptosis measured by flow cytometry analysis. Representative images of cell apoptosis in osteosarcoma cell lines (A) U2OS and (B) KHOS treated with the 10 μM of CDK9 inhibitor. (C and D) Representative bar chart showed apoptosis rate (reflect by the Annexin VP-E positive cells) of cell lines U2OS and KHOS, respectively, after CDK9 inhibitor treatment compared with untreated cells. The data are mean ± SE of the 2 experiments carried out in triplicate (* indicates P < .05).
Supplementary material
Funding sources
This project was partially supported by grants from the Stephan L. Harris Fund, and the Gattegno and Wechsler funds. Dr. Duan is supported, in part, through a grant from the Sarcoma Foundation of America (SFA), a grant from the National Cancer Institute (NCI)/ National Institutes of Health (NIH), UO1, CA151452–01, a pilot grant from Sarcoma SPORE/NIH, and support from UCLA Orthopedic Surgery.
Declaration of interests
The authors have no financial conflicts to declare.
Author contributions
Conception and design: ZD.
Experiments: HM, NS.
Acquisition of data: HM, NS and ZD.
Analysis and interpretation of data: HM, NS, FH and ZD.
Writing, review, and/or revision of the manuscript: HM, NS, FH, and ZD,
References
- 1.Moore D.D., Luu H.H. Osteosarcoma. Cancer Treat. Res. 2014;162:65–92. doi: 10.1007/978-3-319-07323-1_4. [DOI] [PubMed] [Google Scholar]
- 2.Harrison D.J., Geller D.S., Gill J.D., Lewis V.O., Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev. Anticancer Ther. 2018;18(1):39–50. doi: 10.1080/14737140.2018.1413939. [DOI] [PubMed] [Google Scholar]
- 3.Lamora A., Talbot J., Bougras G. Overexpression of smad7 blocks primary tumor growth and lung metastasis development in osteosarcoma. Clin. Cancer Res. 2014;20(19):5097–5112. doi: 10.1158/1078-0432.CCR-13-3191. [DOI] [PubMed] [Google Scholar]
- 4.Meyers P.A. Muramyl tripeptide (mifamurtide) for the treatment of osteosarcoma. Expert Rev. Anticancer Ther. 2009;9(8):1035–1049. doi: 10.1586/era.09.69. [DOI] [PubMed] [Google Scholar]
- 5.He J.P., Hao Y., Wang X.L. Review of the molecular pathogenesis of osteosarcoma. Asian Pac. J. Cancer Prev. 2014;15(15):5967–5976. doi: 10.7314/apjcp.2014.15.15.5967. [DOI] [PubMed] [Google Scholar]
- 6.Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15(6):122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y., Shen J.K., Yu Z., Hornicek F.J., Kan Q., Duan Z. Expression and therapeutic implications of cyclin-dependent kinase 4 (CDK4) in osteosarcoma. Biochim. Biophys. Acta. 2018;1864(5 Pt A):1573–1582. doi: 10.1016/j.bbadis.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Alexander K., Yang H.S., Hinds P.W. Cellular senescence requires CDK5 repression of Rac1 activity. Mol. Cell Biol. 2004;24(7):2808–2819. doi: 10.1128/MCB.24.7.2808-2819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asghar U., Witkiewicz A.K., Turner N.C., Knudsen E.S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 2015;14(2):130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malumbres M., Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 11.Satyanarayana A., Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28(33):2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 12.Abdullah C., Wang X., Becker D. Expression analysis and molecular targeting of cyclin-dependent kinases in advanced melanoma. Cell Cycle (Georgetown, Tex) 2011;10(6):977–988. doi: 10.4161/cc.10.6.15079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan Z., Zhang J., Choy E. Systematic kinome shRNA screening identifies CDK11 (PITSLRE) kinase expression is critical for osteosarcoma cell growth and proliferation. Clin Cancer Res. 2012;18(17):4580–4588. doi: 10.1158/1078-0432.CCR-12-1157. [DOI] [PubMed] [Google Scholar]
- 14.Turner N.C., Ro J., Andre F. Palbociclib in hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 2015;373(3):209–219. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 15.Finn R.S., Crown J.P., Ettl J. Efficacy and safety of palbociclib in combination with letrozole as first-line treatment of ER-positive, HER2-negative, advanced breast cancer: expanded analyses of subgroups from the randomized pivotal trial PALOMA-1/TRIO-18. Breast Cancer Res. 2016;18(1):67. doi: 10.1186/s13058-016-0721-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thangavel C., Boopathi E., Liu Y. Therapeutic challenge with a CDK 4/6 inhibitor induces an RB-dependent SMAC-mediated apoptotic response in non-small cell lung cancer. Clin. Cancer Res. 2018;24(6):1402–1414. doi: 10.1158/1078-0432.CCR-17-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin C.A., Cullinane C., Kirby L. Palbociclib synergizes with BRAF and MEK inhibitors in treatment naive melanoma but not after the development of BRAF inhibitor resistance. Int J Cancer. 2018;142(10):2139–2152. doi: 10.1002/ijc.31220. [DOI] [PubMed] [Google Scholar]
- 18.Mills C.C., Kolb E.A., Sampson V.B. Recent advances of cell-cycle inhibitor therapies for pediatric cancer. Cancer Res. 2017;77(23):6489–6498. doi: 10.1158/0008-5472.CAN-17-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romano G., Giordano A. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle. 2008;7(23):3664–3668. doi: 10.4161/cc.7.23.7122. [DOI] [PubMed] [Google Scholar]
- 20.Sengupta S., Biarnes M.C., Jordan V.C. Cyclin dependent kinase-9 mediated transcriptional de-regulation of cMYC as a critical determinant of endocrine-therapy resistance in breast cancers. Breast Cancer Res Treat. 2014;143(1):113–124. doi: 10.1007/s10549-013-2789-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boffo S., Damato A., Alfano L., Giordano A. CDK9 inhibitors in acute myeloid leukemia. J. Exp. Clin. Cancer Res. 2018;37(1):36. doi: 10.1186/s13046-018-0704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan B., Zhuo Y., Chin D., Morris C.A., Morris G.F., Lasky J.A. Cyclin-dependent kinase 9 is required for tumor necrosis factor-alpha-stimulated matrix metalloproteinase-9 expression in human lung adenocarcinoma cells. J. Biol. Chem. 2005;280(2):1103–1111. doi: 10.1074/jbc.M406293200. [DOI] [PubMed] [Google Scholar]
- 23.Whittaker S.R., Barlow C., Martin M.P. Molecular profiling and combinatorial activity of CCT068127: a potent CDK2 and CDK9 inhibitor. Mol. Oncol. 2018;12(3):287–304. doi: 10.1002/1878-0261.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahaman M.H., Kumarasiri M., Mekonnen L.B. Targeting CDK9: a promising therapeutic opportunity in prostate cancer. Endocr. Relat. Cancer. 2016;23(12):T211–T226. doi: 10.1530/ERC-16-0299. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H., Pandey S., Travers M. Targeting CDK9 reactivates epigenetically silenced genes in cancer. Cell. 2018;175(5):1244–1258. doi: 10.1016/j.cell.2018.09.051. (e26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanso M., Levin R.S., Lipp J.J. P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev. 2016;30(1):117–131. doi: 10.1101/gad.269589.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H., Herrmann C.H. Differential localization and expression of the Cdk9 42k and 55k isoforms. J. Cell Physiol. 2005;203(1):251–260. doi: 10.1002/jcp.20224. [DOI] [PubMed] [Google Scholar]
- 28.Shore S.M., Byers S.A., Dent P., Price D.H. Characterization of Cdk9(55) and differential regulation of two Cdk9 isoforms. Gene. 2005;350(1):51–58. doi: 10.1016/j.gene.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Morales F., Giordano A. Overview of CDK9 as a target in cancer research. Cell Cycle. 2016;15(4):519–527. doi: 10.1080/15384101.2016.1138186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregory G.P., Hogg S.J., Kats L.M. CDK9 inhibition by dinaciclib potently suppresses Mcl-1 to induce durable apoptotic responses in aggressive MYC-driven B-cell lymphoma in vivo. Leukemia. 2015;29(6):1437–1441. doi: 10.1038/leu.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albert T.K., Rigault C., Eickhoff J. Characterization of molecular and cellular functions of the cyclin-dependent kinase CDK9 using a novel specific inhibitor. Br J Pharmacol. 2014;171(1):55–68. doi: 10.1111/bph.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravi M., Paramesh V., Kaviya S.R., Anuradha E., Solomon F.D. 3D cell culture systems: advantages and applications. J. Cell Physiol. 2015;230(1):16–26. doi: 10.1002/jcp.24683. [DOI] [PubMed] [Google Scholar]
- 33.Khoury J.F., Ben-Arush M.W., Weintraub M. Alkaline phosphatase level change in patients with osteosarcoma: its role as a predictive factor of tumor necrosis and clinical outcome. Isr. Med. Assoc. J. 2014;16(1):26–32. [PubMed] [Google Scholar]
- 34.Man T.K., Chintagumpala M., Visvanathan J. Expression profiles of osteosarcoma that can predict response to chemotherapy. Cancer Res. 2005;65(18):8142–8150. doi: 10.1158/0008-5472.CAN-05-0985. [DOI] [PubMed] [Google Scholar]
- 35.Bacci G., Longhi A., Versari M., Mercuri M., Briccoli A., Picci P. Prognostic factors for osteosarcoma of the extremity treated with neoadjuvant chemotherapy: 15-year experience in 789 patients treated at a single institution. Cancer. 2006;106(5):1154–1161. doi: 10.1002/cncr.21724. [DOI] [PubMed] [Google Scholar]
- 36.Im H.J., Kim T.S., Park S.Y. Prediction of tumour necrosis fractions using metabolic and volumetric 18F-FDG PET/CT indices, after one course and at the completion of neoadjuvant chemotherapy, in children and young adults with osteosarcoma. Eur. J. Nucl. Med. Mol. Imaging. 2012;39(1):39–49. doi: 10.1007/s00259-011-1936-4. [DOI] [PubMed] [Google Scholar]
- 37.Ferrari S., Mercuri M., Bacci G. Comment on "Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant Cooperative Osteosarcoma Study Group protocols". J. Clin. Oncol. 2002;20(12) doi: 10.1200/JCO.2002.20.12.2910. (2910; author reply −1) [DOI] [PubMed] [Google Scholar]
- 38.Bielack S.S., Kempf-Bielack B., Delling G. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002;20(3):776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 39.Meyers P.A., Heller G., Healey J. Chemotherapy for nonmetastatic osteogenic sarcoma: the Memorial Sloan-Kettering experience. J. Clin. Oncol. 1992;10(1):5–15. doi: 10.1200/JCO.1992.10.1.5. [DOI] [PubMed] [Google Scholar]
- 40.Ferrari S., Serra M. An update on chemotherapy for osteosarcoma. Expert Opin. Pharmacother. 2015;16(18):2727–2736. doi: 10.1517/14656566.2015.1102226. [DOI] [PubMed] [Google Scholar]
- 41.DeVita V.T., Jr., Chu E. A history of cancer chemotherapy. Cancer Res. 2008;68(21):8643–8653. doi: 10.1158/0008-5472.CAN-07-6611. [DOI] [PubMed] [Google Scholar]
- 42.Jaffe N. Historical perspective on the introduction and use of chemotherapy for the treatment of osteosarcoma. Adv. Exp. Med. Biol. 2014;804:1–30. doi: 10.1007/978-3-319-04843-7_1. [DOI] [PubMed] [Google Scholar]
- 43.Kretz A.L., Schaum M., Richter J. CDK9 is a prognostic marker and therapeutic target in pancreatic cancer. Tumour Biol. 2017;39(2) doi: 10.1177/1010428317694304. (1010428317694304) [DOI] [PubMed] [Google Scholar]
- 44.Narita T., Ishida T., Ito A. Cyclin-dependent kinase 9 is a novel specific molecular target in adult T-cell leukemia/lymphoma. Blood. 2017;130(9):1114–1124. doi: 10.1182/blood-2016-09-741983. [DOI] [PubMed] [Google Scholar]
- 45.Parry D., Guzi T., Shanahan F. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol. Cancer Ther. 2010;9(8):2344–2353. doi: 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- 46.Fuda N.J., Ardehali M.B., Lis J.T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461(7261):186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Q., Li T., Price D.H. RNA polymerase II elongation control. Annu. Rev. Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S., Fischer P.M. Cyclin-dependent kinase 9: a key transcriptional regulator and potential drug target in oncology, virology and cardiology. Trends Pharmacol. Sci. 2008;29(6):302–313. doi: 10.1016/j.tips.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Chen R., Wierda W.G., Chubb S. Mechanism of action of SNS-032, a novel cyclin-dependent kinase inhibitor, in chronic lymphocytic leukemia. Blood. 2009;113(19):4637–4645. doi: 10.1182/blood-2008-12-190256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franken N.A., Rodermond H.M., Stap J., Haveman J., van Bree C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006;1(5):2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 51.Fedr R., Pernicova Z., Slabakova E. Automatic cell cloning assay for determining the clonogenic capacity of cancer and cancer stem-like cells. Cytometry A. 2013;83(5):472–482. doi: 10.1002/cyto.a.22273. [DOI] [PubMed] [Google Scholar]
- 52.Liao D., Zhong L., Duan T. Aspirin Suppresses the growth and Metastasis of Osteosarcoma through the NF-kappaB Pathway. Clin. Cancer Res. 2015;21(23):5349–5359. doi: 10.1158/1078-0432.CCR-15-0198. [DOI] [PubMed] [Google Scholar]
- 53.Lu Y., Tang L., Zhang Q., Zhang Z., Wei W. MicroRNA-613 inhibits the progression of gastric cancer by targeting CDK9. Artif. Cells Nanomed. Biotechnol. 2018;46(5):980–984. doi: 10.1080/21691401.2017.1351983. [DOI] [PubMed] [Google Scholar]
- 54.Tang W., Wang W., Zhao Y., Zhao Z. MicroRNA-874 inhibits cell proliferation and invasion by targeting cyclin-dependent kinase 9 in osteosarcoma. Oncol. Lett. 2018;15(5):7649–7654. doi: 10.3892/ol.2018.8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas A.L., Lind H., Hong A. Inhibition of CDK-mediated Smad3 phosphorylation reduces the Pin1-Smad3 interaction and aggressiveness of triple negative breast cancer cells. Cell Cycle. 2017;16(15):1453–1464. doi: 10.1080/15384101.2017.1338988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sayles L.C., Breese M.R., Koehne A.L. Genome-informed targeted therapy for osteosarcoma. Cancer Discov. 2018 Sep 28 doi: 10.1158/2159-8290.CD-17-1152. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zocchi L., Wu S.C., Wu J., Hayama K.L., Benavente C.A. The cyclin-dependent kinase inhibitor flavopiridol (alvocidib) inhibits metastasis of human osteosarcoma cells. Oncotarget. 2018;9(34):23505–23518. doi: 10.18632/oncotarget.25239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Duinen V., Trietsch S.J., Joore J., Vulto P., Hankemeier T. Microfluidic 3D cell culture: from tools to tissue models. Curr. Opin. Biotechnol. 2015;35:118–126. doi: 10.1016/j.copbio.2015.05.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material