Abstract
RNA sequencing (RNA-seq) has become a standard procedure to investigate transcriptional changes between conditions and is routinely used in research and clinics. While standard differential expression (DE) analysis between two conditions has been extensively studied, and improved over the past decades, RNA-seq time course (TC) DE analysis algorithms are still in their early stages. In this study, we compare, for the first time, existing TC RNA-seq tools on an extensive simulation data set and validated the best performing tools on published data. Surprisingly, TC tools were outperformed by the classical pairwise comparison approach on short time series (<8 time points) in terms of overall performance and robustness to noise, mostly because of high number of false positives, with the exception of ImpulseDE2. Overlapping of candidate lists between tools improved this shortcoming, as the majority of false-positive, but not true-positive, candidates were unique for each method. On longer time series, pairwise approach was less efficient on the overall performance compared with splineTC and maSigPro, which did not identify any false-positive candidate.
Keywords: time course RNA-seq, differential expression, tools comparison, simulated and biological data sets
Introduction
Since its invention [1], RNA sequencing (RNA-seq) protocols have been continuously improved. Nowadays, this technique is used for many applications including the identification of regulatory elements such as enhancer RNAs or long noncoding RNAs, biomarkers, responses to a signal, as well as to capture and model whole biological processes by time course (TC) data [2–6].
RNA-seq data are represented by counts for each gene, which have been obtained by mapping short reverse transcribed RNA fragments to a reference genome/transcriptome. Differences between two conditions are considered significant if the parameters used to model the gene counts do not originate from the same underlying statistical distribution. Negative binomial (NB) distribution is the established gold standard, because of its ability to accurately model RNA-seq data with a low number of available replicates [7]. A plethora of differential expression (DE) tools exist, varying on their assumptions on how to model the variance of the NB distribution.
Additionally to standard differential gene expression, RNA-seq experiments encompass a vast number of other features such as: alternative splicing [8], detection of fusion transcripts [9], circular RNAs [10] and gene regulatory network (GRN) modeling [11]. Finally, it is expected that novel insights will be gained by sequencing full-length transcripts [12] or creating algorithms and standards for analyzing single cells’ transcriptomes [13], novel transcript identification [14] as well as by unraveling the code of RNA modifications [15].
Whereas standard transcriptomic differential gene expression analysis tools have been benchmarked and are now integrated regularly with other omics data [16], TC expression analysis has no established standards. Specific methods are needed to account for the temporal correlation between time points and easier candidate gene identification. Most of the available software perform a pairwise comparison of each time point to the first one, or to the same time point of a second time series/treatment [17], thereby neglecting temporal dependencies and information that could increase predictive power.
In the recent years, many tools have been implemented to characterize longitudinal data sets such as gene set [18] and pathway analysis [19], GRN identification [20], inference of perturbation times [21, 22] or clustering [23]. Here, we focus our analysis specifically on the comparison of tools developed for TC data DE analysis. First, we generated simulated data sets to benchmark available tools (Table 1, ‘Material and methods’ section) for the analysis of differentially expressed gene (DEG) of TC RNA-seq. Subsequently, we compared their performance with standard pairwise comparison strategies. Finally, after this assessment, we tested best performing tools on a published biological data set [33].
Table 1.
Properties of available TC analysis tools
| Method | Normalization method | Model | DEG test | Uneven sampling allowed | Isoforms | Clustering | Time | Citation |
|---|---|---|---|---|---|---|---|---|
| DyNB | Variance estimation+ scaling factors on GP | NBa+GPb | MLc BY MCMCd | Yes | No | No | Days | [24] |
| EBSeq-HMM | Median/quantile | Beta NB+AR-HMMe | EBf | Yes | Yes | Yes | Minutes | [25] |
| FunPat | – | γ/logNorm/Weibull | Bounded Area | No | No | Yes | Seconds | [26] |
| ImpulseDE2 | – | NB+impulse model | LLRg | Yes | No | No | Minutes | [27] |
| lmms | – | lmmsh | LLR | Yes | No | Yes | Minutes | [28] |
| Next maSigPro | – | NB+PRi | LLR | No | No | Yes | Minutes | [29] |
| nsgp | – | Nonstationary/stati GP | ML by gradj/ HMC-NUTSk | No | No | No | Days | [30] |
| splineTC | – | Spline regression | Moderate F-statistic | No | No | No | Seconds | [31] |
| timeSeq | Via edgeR | NBMMl | Kullback–Leibler distance ratio | Yes | Exon level | No | Days | [32] |
aNB model,
bGP,
cML,
dMCMC,
eAR-HMM,
fempirical Bayesian method,
glog likelihood ratio,
hlinear mixed model splines,
ipolynomial regression,
jgradient descent,
kHastings-Monte-Carlo no U-turn sampling,
lnegative binomial mixed model. If a tool has several normalization methods, the standard method is underlined.
Material and methods
RNA-seq time course tools
Time course DE analysis tools used in this study are summarized in Table 1 and have already been partially reviewed [17]. Main features of each tool are highlighted here:
AdaptiveGP (nsgp) is a Gaussian process (GP) regression method implemented in MATLAB. This implementation models noise variance, signal variance and length scale as nonstationary (latent) functions (nonstationary Gaussian process—nsgp), which are inferred by one of two gradient-based techniques: maximum a posteriori estimation or by Hamiltonian Monte Carlo (HMC) sampling. DEG classification is performed by calculating a Bayes factor (BF) of the ratio of marginal likelihoods (MLs) of separated and combined data sets.
DyNB is another MATLAB implementation that uses NB distribution and GP to model RNA-seq counts and their temporal correlations. Normalization and variance estimation are performed as in DESeq2, with the difference that steps are performed on the GP instead of the discrete read counts. Markov chain Monte Carlo (MCMC) sampling is performed to obtain an ML to classify DEG as described before for nsgp. DyNB allows for irregular sampling and is further able to detect delays in replicates or the whole time series.
EBSeqHMM is an extension of the EBSeq R package. It applies an empirical Bayes autoregressive hidden Markov model (AR-HMM) to identify dynamic genes in two steps. First, parameters are estimated using a negative binomial (NB) model, and then in a second step categorize genes at each time point by a Markov-switching autoregressive model and classify genes into expression paths (upregulated/downregulated or constant for each time point). The package comprises visualization and clustering methods and further allows the analysis to be performed at the isoform level. EBSeqHMM requires a minimum of 3 time points.
edgeR/DESeq2 (pairwise) are established R packages for DE analysis. While having different methods for estimating the dispersion, both tools are based on a NB model and are considered as gold standards in the DE analysis field [34]. While traditional comparison does not consider TC analysis, complex designs and combination of pairwise comparisons allow naïve investigation of TC data. Generalized linear models (GLM) as well as a likelihood ratio test with a full and reduced formula in edgeR and DESeq, respectively, were used as gold standard for DEG.
FunPat is an R package allowing DE analysis by comparing the enclosed area between two expression profiles. P-values are assigned by testing the bounded area for significance against a null hypothesis area that is computed by sampling from a best fit distribution (gamma, log-normal or Weibull), which has been created using the mean/variance of the supplied replicates. Additional features included are the functional annotation and extraction of genes, which share annotation and temporal patterns.
ImpulseDE2 is an R package that performs DE analysis of single or case/control TC data in a three-step workflow. First, parameters are estimated, and second, data are fitted against a constant, an optional sigmoid and an impulse model. The impulse model denots the divergence between samples of the transition from a steady state to an intermediate state and back to a steady state. Third, DE is assessed by a log likelihood ratio test. ImpulseDE2 also offers the possibility to consider batch effect factors in the model fitting process.
lmms is an R package modeling time series data via serial fitting of linear mixed model whose goodness of fit is assessed by an analysis of variance (ANOVA) log likelihood ratio test. Differential expression is assessed either over time, between groups or a combination of both. Model fits are tested against a null hypothesis with the extended linear model parameters set to zero and compared using an ANOVA log likelihood ratio test. lmms further offers functions for quality control, filtering and clustering.
Next maSigPro is an update of the maSigPro R package, enabling users to analyze RNA-seq count data by adding GLMs. MaSigPro models count data with a NB distribution to subsequently perform polynomial regression and a log likelihood ratio test to fit genes and detect DE, respectively. The regression is performed in two steps, first selecting non-flat expression profiles, and second, finding the best model by goodness of fit according to a user specified cutoff.
splineTimeR (splineTC) is an R package, which fits natural cubic splines (functions defined by piecewise polynomials) between time points and subsequently applies empirical Bayes moderate F-statistics on the coefficients of the spline regression model between two groups. The package offers downstream features, such as visualization, pathway enrichment and gene association network reconstruction functions.
TimeSeq is accounting for read counts using a NB mixed effect model and a bivariate function of time and treatment that are fitted by a penalized maximum likelihood. P-values of significant genes are computed via the Kullback–Leibler distance ratio and permutation of time point labels. TimeSeq further differentiates between parallel and nonparallel expression profile (control and treatment) DEGs and offers gene set testing for nonparallel DEGs. TimeSeq requires at least one replicate for the first time point.
Detailed formulas used for data modeling and DE testing of each tool are supplied in Supplementary File S1.
Data simulation
To simulate a realistic data set with biological characteristics, a mean/dispersion index was extracted from expression data sets comprising 41 immortalized B-cell samples [35]. Processed samples were obtained from the ReCount project [36]. Next, we randomly sampled 30 000 genes from the mean index, and created for each gene a new NB distribution using corresponding dispersion parameters. The NB distribution was subsequently used to sample time points and replicates on a gene-specific basis. To prevent abnormal expression distributions between replicates, read counts were drawn repeatedly from the NB distribution according to n = min(500, mean/4) times and averaged to resemble biological replicates. Libraries were adjusted by multiplying with the corresponding library size factor or time pattern vectors. The subsequent expression table was subsampled for 20 000 genes and nonexpressed genes were removed, resulting in a final number of 18 503 expressed genes (all count tables are supplied online). For data sets with >4 time points, the increase in time points was seen as increase in the sampling rate to keep the possible expression patterns compact. Therefore, a spline was fitted to the existing expression pattern using lmmspline and predicts functions of the lmms R package, of which new mean values at the desired time points were sampled. In a second step, replicates were sampled from the initial mean/dispersion dictionary as described before.
Processing of biological data
For the biological data set, we used TC RNA-seq data from a recent publication by Kiselev and colleagues [33]. Raw reads of data set (GSE69822) were downloaded from the GEO database [37], quality controlled using fastQC [38], mapped to the ensemble GRCh38.83 genome annotation using the STAR aligner [39] and quantified with featureCounts [38].
Computational resources
All software was run on a MacBook Pro 2.4 Ghz Intel Core i7 with 16 GB of RAM. To accelerate Matlab tools and the timeSeq R package, data sets were split into 100 or 10 subsets for the Matlab and R implementations, respectively. Each subset was run on a single node of the ETH cluster with 16 cores and 16 GB of RAM.
Parameters
Differential expression analysis for all tools was performed using a P-value of 0.01 (if applicable). Additional filtering approaches and variation of parameters are noted in Supplementary Table S6.
Results
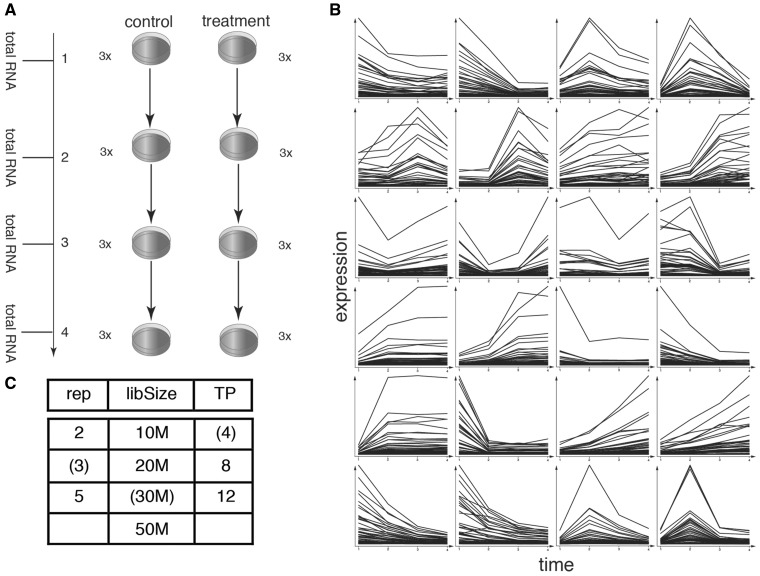
The study designed comprises a standard layout of a control and treatment TC. The standard parameters were selected according to realistic biological conditions and contain 4 time points and three biological replicates per time point (Figure 1A). To simulate DE, only the treatment samples were modified, and the control was considered to be constant (having a baseline biological variation). In total, 24 pattern categories were simulated, each spanning either 2 or 3 consecutive time points (Supplementary Table S1). Categories consisted of 50 genes each, summing up to 1200 DEGs with randomly sampled expression levels (see ‘Material and methods’ section). Thereby, each pattern mainly contained weakly expressed and only few highly expressed genes (Figure 1B), as it is the case in a biological data set [40]. While in various biological scenarios often the control TC changes over time as well, we again choose the simplest case to limit the extent of the study. To characterize the selected TC tools, several scenarios were tested, assessing the behavior of the tools on different library sizes, number of replicates or number of time points (Figure 1C).
Figure 1.
The standard experimental design (A) consisted of two TCs having 4 time points with three replicates each. A single value for each gene was sampled from a negative binomial distribution using mean/dispersion value pairs of a biological data set. Time points and replicates were then drawn from the same distribution and expression patterns applied by multiplying with the pattern vector. Of the total 24 patterns that were simulated, each consisted of 50 genes, resulting in the simulation of 1200 DEGs in total (B). As genes were drawn from a negative binomial distribution, each pattern mostly consists of lowly expressed genes and a few highly expressed genes. (C) Other experimental designs were tested by increasing or reducing the library size, replicates or time points (standard parameters in parenthesis).
Regular pairwise comparison outperformed most TC tools
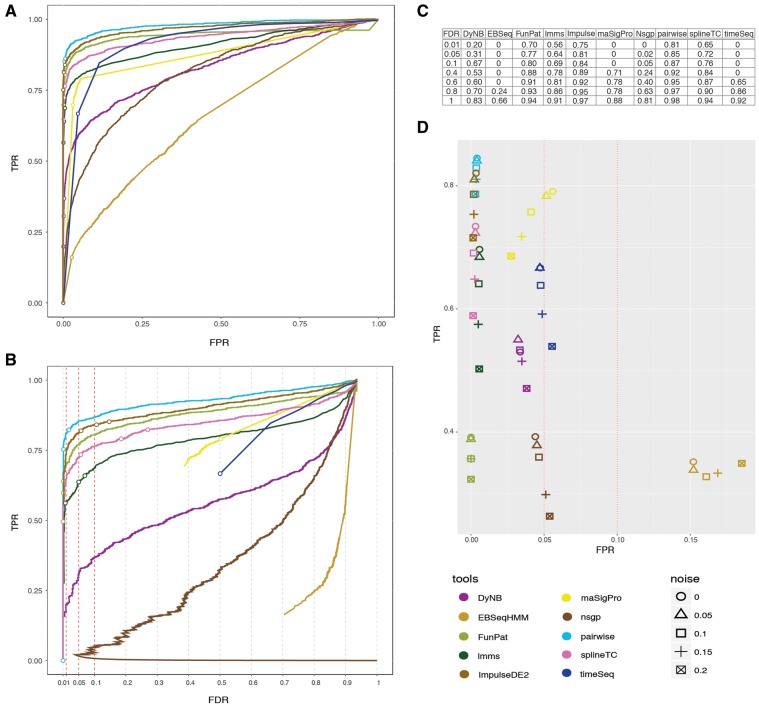
Results of each method were evaluated using a stringent P-value cutoff of 0.01 and summarized by standard classification terms (Table 2). While all methods were highly accurate, half of the methods suffered from a low sensitivity (correctly identifying DEGs). Consequently, these methods had high false discovery rates (FDRs) and low precision as reflected by the overall measure of the F1 score (combined score of precision and sensitivity) (Table 2). In more detail, EBSeqHMM [25] identified more false positives (FPs) than true positives (TPs). MaSigPro [29] and timeSeq [32] identified the majority of all candidates, but suffered from a high number of FPs. As both DyNB [24] and nsgp [30] only report a BF, we selected genes with the top 1200 absolute log BFs. This is equivalent to the total number of DEGs and should not arbitrarily increase FPs. Nevertheless, both DyNB and nsgp did not properly control their FDR. FunPat [26] identified only a fraction of TPs correctly, resulting in a low F1 score, though its FDR was outstanding, as it did not produce a single FP. The pairwise comparison by edgeR [41], ImpulseDE2 [27] and splineTC [31] identified almost the exact number of DEGs. The same was true for lmms [28], with exception that the aforementioned tools were able to keep their FDR <10%. The performance of all tools is illustrated in Figure 2 with receiver operating curve (ROC) (Figure 2A), true-positive rate (TPR)/FDR (Figure 2B) plots and by the calculated area under the curve (AUC) for different FDR thresholds of the ROC (Figure 2C).
Table 2.
Test summary statistics of the standard scenario
| Tools | TP | FP | FN | TN | sen | spec | prec | FNR | FDR | acc | F1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DyNB | 675 | 525 | 525 | 16 778 | 0.56 | 0.97 | 0.56 | 0.44 | 0.44 | 0.94 | 0.56 |
| EBSeqHMM | 527 | 3889 | 673 | 13 414 | 0.44 | 0.78 | 0.12 | 0.56 | 0.88 | 0.75 | 0.19 |
| FunPat | 470 | 0 | 730 | 17 303 | 0.39 | 1.00 | 1.00 | 0.61 | 0.00 | 0.96 | 0.56 |
| ImpulseDE2 | 980 | 57 | 220 | 17 246 | 0.82 | 1.00 | 0.95 | 0.18 | 0.05 | 0.99 | 0.88 |
| lmms | 836 | 99 | 364 | 17 303 | 0.70 | 0.99 | 0.89 | 0.30 | 0.11 | 0.97 | 0.78 |
| maSigPro | 947 | 699 | 253 | 16 604 | 079 | 0.96 | 0.58 | 0.21 | 0.42 | 0.95 | 0.67 |
| nsgp | 482 | 718 | 718 | 16 585 | 0.40 | 0.96 | 0.40 | 0.60 | 0.60 | 0.92 | 0.40 |
| pairwise | 1014 | 70 | 186 | 17 233 | 0.85 | 1.00 | 0.94 | 0.16 | 0.06 | 0.97 | 0.89 |
| splineTC | 881 | 53 | 319 | 17 250 | 0.73 | 1.00 | 0.94 | 0.27 | 0.06 | 0.98 | 0.83 |
| timeSeq | 801 | 802 | 399 | 16 501 | 0.67 | 0.95 | 0.50 | 0.33 | 0.50 | 0.94 | 0.57 |
FN, false negatives; TN, true negatives; sen(sitivity), correctly identify TP; spec(ificity), correctly identify FP; prec(ision), ratio of correctly identified candidates; FNR (false-negative rate), ratio of falsely refused candidates; FDR, ratio of falsely identified candidates; acc(uracy), ratio of correctly identified TP and FP; F1, weighted harmonic mean of precision and sensitivity.
Figure 2.
Results of standard simulation scenario. (A) ROC showing the TPR and FPR on the x and y axis, respectively. FDR thresholds of 0.01, 0.05 and 0.1 are indicated by rings on each curve. (B) TPR/FDR curves with ring indicated adjusted P-value thresholds of 0.01, 0.05 and 0.1. (C) AUC fraction (ranging from 0/worst to 1/best) calculated for the ROC on several FDR thresholds. (D) Performance of TC tools on noisy data ranging from 0.05 to 0.2 white noise added to the samples.
Identification of late and small pattern changes was limited
For a better characterization of the performance of tools on specific expression patterns, we further grouped the simulated patterns into two classes and several categories each. Classes addressed pattern types (Supplementary Figure S1A) or timing of expression changes (Supplementary Figure S1B) and were visualized using the iCOBRA R package [42]. The overall performance of all methods was decreased when the expression pattern change was small or occurred at later time points. ImpulseDE2, splineTC, maSigPro, lmms and the pairwise comparison achieved the highest score in all categories. In more detail, DyNB, the pairwise approach, maSigPro, lmms and splineTC more likely detected gradual and mixed patterns. nsgp performed well on fast/abruptly changing and mixed patterns. FunPat and EBSeq-HMM were more appropriate for gradual but not abrupt changing patterns. TimeSeq and ImpulseDE2 had an almost stable performance with the exception for mixed and low/late patterns.
Proper FDR control decreased susceptibility to noise
To further assess the robustness of methods as well as to ensure that simulated data were not too artificial, we introduced white noise ranging from 5 to 20% of the signal. DyNB and nsgp showed reduced TPRs and increased false-positive rates (FPRs) (chance of falsely classifying a gene as DE). EBSeq-HMM and timeSeq showed rather constant TPR with increases in FPR only. In this aspect, MaSigPro was a positive exception, as both the FPR and TPR were decreasing. On the contrary, well-controlled FDR methods only had reduced TPR power while keeping their FPR stable. Comparing tools with a stable FPR, the pairwise approach had the smallest spread, followed by ImpulseDE2, FunPat and splineTC (Figure 2D).
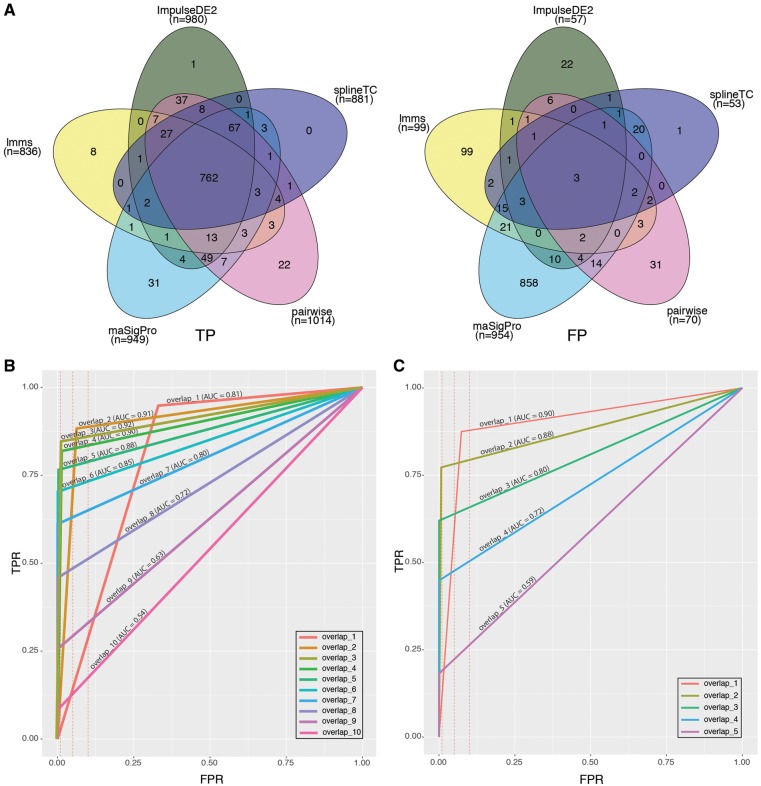
True-positive but not false-positive genes overlapped
Comparison of separately overlapping TP and FP genes between methods (Supplementary Tables S2 and S3) allowed us to make two observations. First, on average only ∼2% of TP candidates identified by the individual tools were unique. Second, 37% of FP candidates were unique to each of the tools (Figure 3A, see Supplementary Tables S2a and S3a for all tools). Subsequently, we investigated the effect of overlapping the candidate lists of individual tools on the overall performance, by computing the AUC using the R package ROCR [43]. Comparisons were performed for DEGs identified by at least 1 and up to 10 tools (Figure 3B). The best TPR/FPR ratio was achieved by using genes found by at least three methods. Moreover, further filtering by increasing the number of minimum overlaps did neither increase the TPR nor reduce the FPR (Figure 3B). The analysis was repeated using the top five scoring tools only, and similar results were obtained with the difference that the minimum required overlap by two methods resulted in the best TPR/FPR ratio (Figure 3C).
Figure 3.
Results of overlapping candidate lists. (A) Overlaps of true-positive (left) and false-positive (right) candidates of top five tools. (B) ROC curve with computed AUC for overlaps of candidate lists. The number of the overlap indicates the minimum number of lists sharing candidates. (C) ROC curve with computed AUC for top five tools. FPR thresholds of 0.01, 0.05 and 0.1 are indicated by dashed red lines.
Increases in replicates or time points improved statistical power
As previously observed for conventional RNA-seq [44], increasing sequencing depth only marginally increases power, whereas replicates have a major impact on the overall performance of the analysis (Supplementary Table S4, Supplementary Data S1). Addition of time points boosted the performances of maSigPro by 30%, not reporting any FP at all, and splineTC by 10%, outperforming the pairwise approach (Supplementary Table S4). Other tools were discarded, as they either used up too much memory (EBSeq-HMM) or took too long to run (DyNB, nsgp, timeSeq), even when splitting the data set.
Performance on biological data
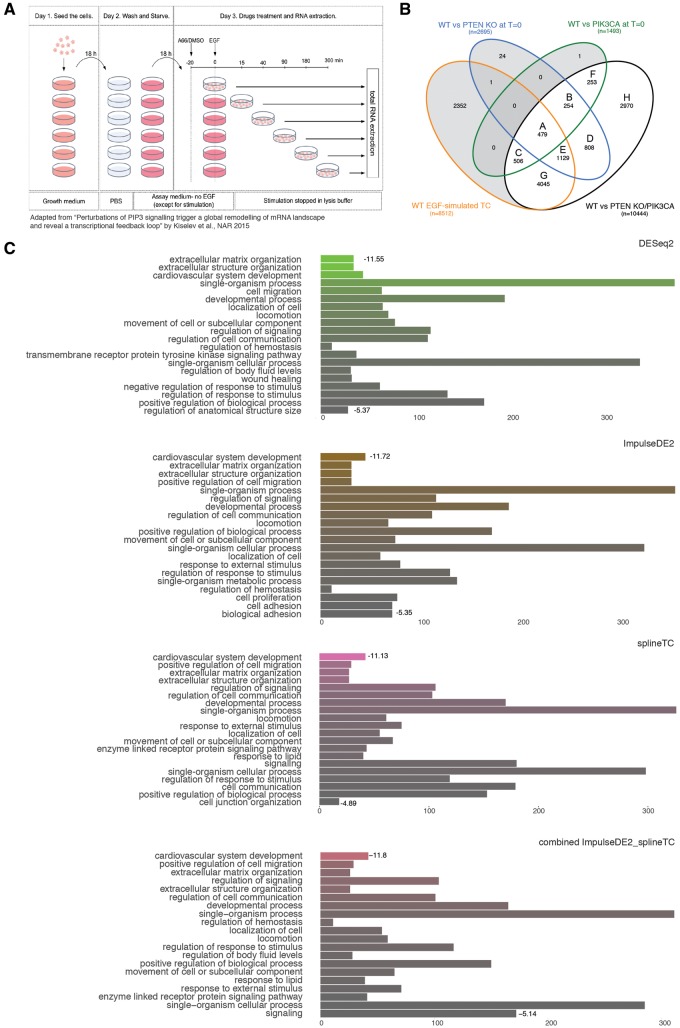
To evaluate the performances on actual biological data, we selected a published data set (accession number GSE69822) [24]. The aim of the study was to identify transient and chronic perturbations of phosphatidylinositol-trisphosphate (PIP3) signaling. To identify such perturbations, four TCs under epidermal growth factor (EGF) stimulation were performed using: wild-type cells, PTEN knockout and PIK3CA knockin cells (both mutations leading to chronic EGF stimulation) as well as cells treated with a transient EGF inhibitor (Figure 4A). All TC consisted of 6 time points, each having three replicates. For simplicity, we focused our analysis on the comparison of the perturbations compared with the control group over time and at time point 0 (Figure 4B). DEGs were further categorized into subgroups A–H to identify chronically/induced and baseline-activated candidate genes. A step log likelihood ratio test of DESeq2 [45] was used to identify DEGs. As DESeq2 performed equally well as edgeR on the simulated data set (Supplementary Table S5, Supplementary Figure S2), we used DESeq2 as the pairwise comparison approach for this analysis. Moreover, to identify the potential of a combinational approach of top-performing tools, overlapping candidates of ImpulseDE2, splineTC and maSigPro were investigated as well. MaSigPro was selected instead of lmms, as lmms did not perform well on longer time series. In addition, splineTC already represented a spline fitting approach.
Figure 4.
Experimental design and results of published data on PIP3 signaling perturbations. (A) Experimental design and processing steps of samples. (B) DESeq2 overlaps of DEGs between TCs and T0 for further categorization and GO analysis. (C) GO enrichment for Class A DEGs for each method and the combined approach. The length of the bar depicts the number of enriched genes in each term. Log10 P-value is indicated by color (increasing from colored to gray), and is shown for the first and last term to indicate the range.
Time course tools reproduced pairwise comparison candidates
As described in the study, DEG candidates of the perturbation TCs were merged and overlapped with DEGs of the control TC (Figure 4B). Overlaps of wild type (WT) and perturbation TC candidates were highest with about 59% for DESeq2, 53% for splineTC, 52% for ImpulseDE2 and clearly reduced with only 37% for maSigPro (Supplementary Figure S3A). Considering each TC separately, the overlap between the four methods was extensive, containing at least 30% of each tool’s candidates (Supplementary Figure S3B and C). Comparing the overlaps between perturbation and WT TCs, all approaches identified the same core set of candidates (Supplementary Figure S3D), whereas maSigPro missed a second set of genes that were reported by the other tools. Therefore, we concluded that ImpulseDE2 and splineTC were able to reproduce the findings of the pairwise DESeq2 approach.
Functional annotations between tools were highly similar
To recapture more functional groups, TC overlaps were categorized as described in the original study [24] (Figure 4B and Supplementary Figure S4) Functional annotation was performed via the topGO function of the FGNet [46] R package, and subsequently gene ontology (GO) terms were collapsed by the REVIGO tool [47]. Exemplary results for genes overlapping in all categories (Class A) are shown as bar plots (Figure 4C) containing the (up to) 20 most enriched GO terms for each comparison. Following up on the combinatorial approach of the simulation study, we also tested an intersection of ImpulseDE2 and splineTC candidates. MaSigPro was excluded as it failed to identify a second gene set reported by the other tools (Supplementary Figure S4) and because of the fact that best results on simulated data were obtained by overlapping results of two tools (Figure 3C). Terms and enrichments were highly similar between methods, though the ranking and enrichment scores of categories varied between tools and were reordered correspondingly for the combined approach (Figure 4C and Supplementary Data S2).
Discussion
In this manuscript, we have evaluated the performance of nine RNA-seq TC DE tools (Table 1) and standard pairwise comparison. Performance was first assessed on simulated data (Figure 2, Table 2 and Supplementary Table S3), and top-performing tools were applied to a published biological data set (Figures 3 and 4).
ImpulseDE2 was the overall best performing tool, achieving results comparable with the classical pairwise comparison approach, with a higher number of replicates even outperforming edgeR. With increasing time points, ImpulseDE2 performance dropped, caused by increased numbers of FPs. Most likely, the data modeling approach facilitating an impulse model is not the best fit for the stretched patterns simulated in this study. Moreover, authors state that performance and runtime are linearly correlated to the number of time points.
SplineTC was the best performing tool on long time series (8+ time points) simulation data, excelled in running time (a few seconds) and achieved good control of the FDR. The major pitfalls are the vulnerability to noisy data and the requirement of log count data as input. Log transformation is standard for microarray data and smoothens the signal for better spline fitting. However, this might not be describing RNA-seq count data optimally, thereby providing a possible explanation for the proneness to noise.
lmms reached the fourth place in terms of F1 score. Performance was highly similar to the second spline approach on the standard experimental setup. Nevertheless, it performed slightly worse than splineTC and could not properly account for the increased number of time points. lmms was broadly designed for omics data; therefore, RNA-seq-specific normalization and data transformation might increase performance.
MaSigPro’s performance is placed in the upper section compared with other tools. The main strength of this tool is that no preference for any patterns was observed. Interestingly, maSigPro had decreased FPRs with increasing noise, which might be attributed to the model selection step. Nonetheless, it did not control its FDR as good as the pairwise or splineTC approaches. This was highly dependent on the goodness-of-fit threshold, whose modifications could lead to a better performance. Increasing the number of time points dramatically improved the performance of maSigPro up to 30%, without increasing the number of FPs. Finally, positive features of maSigPro include the running time of only several minutes.
TimeSeq was robust to noise, despite the long running times when computing P-values and the fact that it identified as many FP as TP candidates. Further, no information on how to include replicates and how the tool handles them is supplied in the original study [36]. Therefore, replicates had to be supplied as separate gene entries. TimeSeq offers the additional feature of gene set-level analysis, which is thought to increase performance of DEG identification, but was not tested in this simulation study.
FunPat’s performance was only average, as it only identified a third of all DEGs of the simulated data set. Nevertheless, considering that it did not identify a single FP, this tool might be appealing for biologists to select candidates for downstream validation, to gain confidence for certain candidates, with the trade-off to potentially loose TP candidates. FunPat requires only a single time point to be replicated to extract the variance parameters and has an extremely short running time. Nevertheless, several drawbacks for this method were identified, including the need for data transformation to account for replicates, a poor performance on minor pattern changes, only moderate robustness to noise and the aforementioned conservative candidate selection, especially when increasing the number of time points.
The performance of DyNB and nsgp ranked them in the middle section of the tested tools. Several drawbacks and aspects might be considered before using these tools. Among them is the long running time, the vulnerability to noise, as well as the output format of BFs making the interpretation of the results more demanding. Furthermore, these two tools are Matlab implementations, which require a commercial license. A potential way to improve these tools could be to select genes by plotting the distribution of BF and filtering candidates by setting a threshold at the drop-off the BF distribution plots. Owing to the underlying machine learning algorithms, a higher number of replicates and time points are likely to improve the performance but also profoundly the runtime.
The purpose of EBSeq-HMM is to identify dynamic patterns and clustering them by setting their expression behavior to be either up/down or constant. The main issue observed was that no constant components were observed, because of the fact that it is unlikely that 2 time points have exactly equal expression values. Another drawback was that no P-values of DE but posterior probabilities belonging to an expression path (see ‘Material and methods’ section) are reported. Additionally, it was not possible to test for DE between two TCs, as the first time point of the individual TC instead of a control TC was taken as reference. While performing robustly on noisy data, the overall performance of EBSeq-HMM was rather poor. Improvements might be achieved by changing the normalization method as recently described [48].
Other insights gained from this study are in agreement with previous publications on pairwise RNA-seq data sets analysis [44]. Tools performed poorly on small and late pattern changes. However, the late category should not affect the pairwise comparison; therefore, this category might not be properly simulated or biased by too high variance. Further, addition of replicates or time points increased statistical power to a greater extent compared with an increasing sequencing depth.
Overlapping candidate lists from different tools is a standard approach to increase confidence and reduce the number of candidate genes. Here, we showed that TPs in contrast to FPs are highly overlapping between tools (Figure 3A). Stringent filtering was not improving the FPR but only decreasing the sensitivity and thereby limiting the ability to find TPs. Consequently, the minimal overlap of three and two methods had the best TPR/FPR ratio, considering all or the top five tools, respectively (Figure 3B and C).
To confirm insights gained from the simulation studies, best performing tools and their combination were applied to a biological data set. The tested tools gave similar and specific enrichment results, whereas the combined approach seemed to filter out more general GO terms and re-rank essential ones in similar classes (e.g. Classes B and E) but negatively dominated nonsimilar classes, as most genes were discarded (e.g. Classes C and H). While all tools identify the same core set of genes, intersection of all tools might have eliminated a second bigger set of genes identified by ImpulseDE2, splineTC and DESeq2 only. This pointed out that conservative or tools that identify different sets of genes might dominate final results by excluding large fraction of candidates shared by other tools. Therefore, we suggest only combining result lists of well-performing tools yielding similar results.
Conclusions
Overall, we concluded that except for ImpulseDE2, splineTC and maSigPro, TC RNA-seq tools are only partially able to account for the temporal character of data sets. Unexpectedly, pairwise comparison of time points was the most robust and accurate approach on the standard experimental setup. The only exception was ImpulseDE2 that performed almost equally, but was more prone to noise. While increasing the number of replicates improved performances of all tools, increasing the number of time points boosted maSigPro and splineTC performance only. MaSigPro did not report any additional FP, and splineTC outperformed the pairwise approach. Other tools proved impractical on longer time series because of their computational demands. Therefore, TC tools only outperform classical approaches on time series with a higher number of time points, implying greater costs because of a greater number of samples that have to be sequenced.
Finally, we conclude that combining candidates of several methods is the most reliable and cost-effective trade-off to increasing replicates or time points. Nevertheless, possible domination of candidate selection by conservative tools or tools that identify different set of genes has to be considered. Candidates from this first-level analysis can be further reduced using, e.g., clustering and enrichment techniques as well as integration of other omics data fitting the hypothesis for downstream analysis and candidate selection for validation.
Future perspectives
Improving technologies and more powerful computing devices enable scientists to apply algorithms that have been so far too computational demanding. These newly available algorithms, shorter runtimes as well as the active research in RNA-seq TC field will enable better and easier analysis of time series data. Existing tools allow only the analysis on a single or on two TCs, while future tools might allow multiple group TC DE analysis. Further, integration of other data types such as chromatin immunoprecipitation (ChIP)/assay for transposase-accessible chromatin with high throughput sequencing (ATAC-seq), variant information, adenylation or microbial data, will allow for better DE, dissection of underlying regulatory mechanisms and to test more specific hypotheses. Taken together, future RNA-seq TC analysis will enable scientists to better elucidate general and specific temporal biological processes, their dependencies and help to understand the bigger picture of, e.g., cancer types or disease progression.
Key Points
Time course analysis tools are outperformed by the classical pairwise comparison approach on short time series, except of ImpulseDE2.
Overlapping of candidate lists between similar tools reduced FPs to a greater extent than TPs.
splineTC and maSigPro have best overall performance on long time series data.
Availability
All data simulated are supplied as Supplementary Files (Supplementary Data S1 and S2), scripts are accessible at GitHub (https://github.com/daniel-spies/rna-seq_tcComp) and biological data are freely available from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE69822.
Supplementary Material
Acknowledgements
The authors thank the Ciaudo laboratory for the critical reading of the manuscript and fruitful discussions. The authors deeply acknowledge Drs Manfred Claassen, Christian von Mering, Mark Robinson, Uwe Ohler and Charlotte Soneson for critical comments and discussions.
Funding
This work was supported by a core grant from ETH-Z (supported by Roche). D.S. is supported by the Peter und Traudl Engelhorn foundation and P.F.R. is supported by a PhD fellowship from the ETH-Z foundation (grant number ETH-05 14-3).
Biographies
Daniel Spies is a PhD candidate at ETH. His research focuses on RNA-seq time course analysis.
Peter Renz is a PhD candidate at ETH. His research focuses on regulatory mechanisms of pluripotency in human embryonic stem cells.
Tobias Beyer, PhD, has been a senior scientist and junior group leader at ETH since 2014. His research focuses on regulatory mechanisms of pluripotency in human embryonic stem cells.
Constance Ciaudo, PhD, has been an assistant professor at the Institute of Molecular Health Science at ETH since 2013. Her research group focuses on RNAi and Genome Integrity
References
- 1. Nagalakshmi U, Wang Z, Waern K. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008;320:1341–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Acerbi E, Viganò E, Poidinger M. Continuous time Bayesian networks identify Prdm1 as a negative regulator of TH17 cell differentiation in humans. Sci Rep 2016;6:23128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. do Amaral MN, Arge LW, Benitez LC. Comparative transcriptomics of rice plants under cold, iron, and salt stresses. Funct Integr Genomics 2016;16:567–79. [DOI] [PubMed] [Google Scholar]
- 4. Giannopoulou EG, Elemento O, Ivashkiv LB.. Use of RNA sequencing to evaluate rheumatic disease patients. Arthritis Res Ther 2015;17:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sudmant PH, Alexis MS, Burge CB.. Meta-analysis of RNA-seq expression data across species, tissues and studies. Genome Biol 2015;16:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Z, Gerstein M, Snyder M.. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Korpelainen E, Tuimala J, Somervuo P. Differential expression analysis In: RNA-Seq Data Analysis. 2014, 147–80. [Google Scholar]
- 8. Eswaran J, Horvath A, Godbole S. RNA sequencing of cancer reveals novel splicing alterations. Sci Rep 2013;3:1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar S, Vo AD, Qin F. Comparative assessment of methods for the fusion transcripts detection from RNA-Seq data. Sci Rep 2016;6:21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Su H, Lin F, Deng X. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med 2016;14:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schulze S, Henkel SG, Driesch D. Computational prediction of molecular pathogen-host interactions based on dual transcriptome data. Front Microbiol 2015;6:783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tilgner H, Jahanbani F, Blauwkamp T. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat Biotechnol 2015;33:736–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaakkola MK, Seyednasrollah F, Mehmood A. Comparison of methods to detect differentially expressed genes between single-cell populations. Brief Bioinform 2016. pii: bbw057. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pertea M, Kim D, Pertea GM. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 2016;11:1650–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gilbert WV, Bell TA, Schaening C.. Messenger RNA modifications: form, distribution, and function. Science 2016;352:1408–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Conesa A, Madrigal P, Tarazona S. A survey of best practices for RNA-seq data analysis. Genome Biol 2016;17:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spies D, Ciaudo C.. Dynamics in transcriptomics: advancements in RNA-seq time course and downstream analysis. Comput Struct Biotechnol J 2015;13:469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hejblum BP, Skinner J, Thiebaut R.. Time-course gene set analysis for longitudinal gene expression data. PLoS Comput Biol 2015;11:e1004310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kayano M, Matsui H, Yamaguchi R. Gene set differential analysis of time course expression profiles via sparse estimation in functional logistic model with application to time-dependent biomarker detection. Biostatistics 2015;17:235–48. [DOI] [PubMed] [Google Scholar]
- 20. Iglesias-Martinez LF, Kolch W, Santra T.. BGRMI: a method for inferring gene regulatory networks from time-course gene expression data and its application in breast cancer research. Sci Rep 2016;6:37140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wise A, Bar-Joseph Z.. SMARTS: reconstructing disease response networks from multiple individuals using time series gene expression data. Bioinformatics 2014;31:1250–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang J, Penfold CA, Grant MR. Inferring the perturbation time from biological time course data. Bioinformatics 2016;32:2956–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hensman J, Rattray M, Lawrence ND.. Fast nonparametric clustering of structured time-series. IEEE Trans Pattern Anal Mach Intell 2014;37:383–93. [DOI] [PubMed] [Google Scholar]
- 24. Äijö T, Butty V, Chen Z. Methods for time series analysis of RNA-seq data with application to human Th17 cell differentiation. Bioinformatics 2014;30:i113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leng N, Li Y, Mcintosh BE. EBSeq-HMM: a Bayesian approach for identifying gene-expression changes in ordered RNA-seq experiments. Bioinformatics 2015;31:2614–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sanavia T, Finotello F, Di Camillo B.. FunPat: function-based pattern analysis on RNA-seq time series data. BMC Genomics 2015;16:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fischer DS, Theis FJ, Yosef N.. Impulse model-based differential expression analysis of time course sequencing data. bioRxiv 2017: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Straube J, Gorse AD; PROOF Centre of Excellence Team, A linear mixed model spline framework for analysing time course ‘Omics’ data. PLoS One 2015;10:e0134540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nueda MJ, Tarazona S, Conesa A.. Next maSigPro: updating maSigPro bioconductor package for RNA-seq time series. Bioinformatics 2014;30:2598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heinonen M, Mannerström H, Rousu J. Non-Stationary Gaussian Process Regression with Hamiltonian Monte Carlo. In: Proceedings of the 19th International Conference on Artificial Intelligence and Statistics (AISTATS), Cadiz, Spain, 2016. Vol. 41, pp. 732–40. JMLR: W&CP. [Google Scholar]
- 31. Michna A, Braselmann H, Selmansberger M. Natural cubic spline regression modeling followed by dynamic network reconstruction for the identification of radiation-sensitivity gene association networks from time-course transcriptome data. PLoS One 2016;11:e0160791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun X, Dalpiaz D, Wu D. Statistical inference for time course RNA-Seq data using a negative binomial mixed-effect model. BMC Bioinformatics 2016;17:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kiselev VY, Juvin V, Malek M. Perturbations of PIP3 signalling trigger a global remodelling of mRNA landscape and reveal a transcriptional feedback loop. Nucleic Acids Res 2015;43:9663–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Finotello F, Di Camillo B.. Measuring differential gene expression with RNA-seq: challenges and strategies for data analysis. Brief Funct Genomics 2015;14:130–42. [DOI] [PubMed] [Google Scholar]
- 35. Cheung VG, Nayak RR, Wang IX. Polymorphic cis- and trans-regulation of human gene expression. PLoS Biol 2010;8:e1000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Frazee AC, Langmead B, Leek JT.. ReCount: a multi-experiment resource of analysis-ready RNA-seq gene count datasets. BMC Bioinformatics 2011;12:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barrett T, Wilhite SE, Ledoux P. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 2013;41:D991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liao Y, Smyth GK, Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 39. Dobin A, Davis CA, Schlesinger F. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson MD, Oshlack A.. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Robinson MD, McCarthy DJ, Smyth GK.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Soneson C, Robinson MD.. iCOBRA: open, reproducible, standardized and live method benchmarking. Nat Methods 2016;13:283. [DOI] [PubMed] [Google Scholar]
- 43. Sing T, Sander O, Beerenwinkel N. ROCR: visualizing classifier performance in R. Bioinformatics 2005;21:3940–1. [DOI] [PubMed] [Google Scholar]
- 44. Liu Y, Zhou J, White KP.. RNA-seq differential expression studies: more sequence or more replication? Bioinformatics 2014;30:301–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aibar S, Fontanillo C, Droste C. Functional gene networks: R/Bioc package to generate and analyse gene networks derived from functional enrichment and clustering. Bioinformatics 2015;31:1686–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Supek F, Bošnjak M, Škunca N. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 2011;6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lin Y, Golovnina K, Chen ZX. Comparison of normalization and differential expression analyses using RNA-Seq data from 726 individual Drosophila melanogaster. BMC Genomics 2016; 17:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.