Abstract
Piperlongumine is a natural product found in the plant species Piper longum and this compound exhibits potent anticancer activity in multiple tumor types and has been characterized as an inducer of reactive oxygen species (ROS). Treatment of Panc1 and L3.6pL pancreatic, A549 lung, 786-O kidney, and SKBR3 breast cancer cell lines with 5-15 μM piperlongumine inhibited cell proliferation and induced apoptosis and ROS, and these responses were attenuated after cotreatment with the antioxidant glutathione. Piperlongumine also downregulated expression of Sp1, Sp3, Sp4 and several pro-oncogenic Sp-regulated genes including cyclin D1, survivin, cMyc, epidermal growth factor receptor (EGFR) and hepatocyte growth factor receptor (cMet), and these responses were also attenuated after cotreatment with glutathione. Mechanistic studies in Panc1 cells showed that piperlongumine-induced ROS decreased expression of cMyc via an epigenetic pathway and this resulted in downregulation of cMyc-regulated microRNAs (miRs)-27a, miR-20a and miR-17 and induction of the transcriptional repressors ZBTB10 and ZBTB4. These repressors target GC-rich Sp binding sites to decrease transactivation. This pathway observed for piperlongumine in Panc1 cells has previously been observed for other ROS-inducing anticancer agents and shows that an important underlying mechanism of action of piperlongumine is due to downregulation of Sp1, Sp3, Sp4 and pro-oncogenic Sp-regulated genes.
Keywords: piperlongumine, ROS, Sp transcription factors, cMyc
Introduction
Piperlongumine is an alkaloid natural product found in the plant species Piper longum Linn that exhibits a broad spectrum of biological effects (1-5) including antitumorigenic activities in cancer cell lines and animal models (6-19). Raj and coworkers identified piperlongumine in a high throughput screening assay and demonstrated the highly selective killing of cancer cell lines compared to normal untransformed cells. Their studies also demonstrated in vivo antitumor activity in both mouse and rat models and they also reported that piperlongumine induced ROS in several cancer cell lines (6). It was concluded that piperlongumine was a potent inducer of oxidative stress-dependent cell killing and this was due, in part, to depletion of glutathione and other thiol-containing proteins involved in maintaining cellular redox homeostasis (6,10). Several subsequent studies have confirmed the anticancer activities of piperlongumine and these include pathways/genes that are ROS-dependent (6-13) and other pathways in which the role of ROS was not determined (14-19).
Studies in this laboratory have investigated the anticancer activities and mechanism of action of several ROS-inducing anticancer agents including curcumin, a nitro-aspirin derivative, betulinic acid, methyl 2-cyano-3,12-dioxooleana-1,9-dien-28-oate (CDDO-Me), histone deacetylase (HDAC) inhibitors, phenethylisothiocyanate (PEITC), celastrol, penfluridol and benzylisothiocyanate (BITC) (20-28). For some of these drugs such as curcumin and betulinic acid, their induction of ROS was cell context-dependent; however, the induction of ROS by these compounds was functionally important since compound-dependent inhibition of cancer cell proliferation and survival were reversed after cotreatment with antioxidants. Drug-induced ROS via alkylation of GSH and redox genes or by direct effects on mitochondria also leads to oxidative stress-induced endoplasmic reticulum (ER) stress and increased apoptosis (29). Studies in this laboratory have demonstrated that ROS inducers (20-28) and also hydrogen peroxide and t-butyl hydroperoxide (30,31) decrease expression of specificity protein 1 (Sp1), Sp3 and Sp4 transcription factors (TFs) and also several pro-oncogenic Sp-regulated genes and non-coding RNAs (32). The mechanism of ROS-dependent downregulation of Sp TFs involves initial ROS-induced repression of cMyc, decreased expression of cMyc-regulated microRNAs (miRs), miR-27a and miR-20a/miR-17-5p which results in the induction of miR-regulated ZBTB10 (ZBTB34) and ZBTB4 (25,27,28,32). ZBTBs are transcriptional repressors that competitively bind GC-rich cis-elements and displace Sp TFs resulting in decreased Sp-regulated gene expression (33,34).
ROS-dependent targeting of Sp TFs represents an important pathway that contributes to the anticancer activity of ROS-inducers since this results in downregulation of pro-oncogenic Sp-regulated genes including survivin, cyclin D1, vascular endothelial growth factor (VEGF) and its receptors, epidermal growth factor receptor (EGFR), and other receptor tyrosine kinases (32,35). In this study, we show that piperlongumine induces ROS, inhibits cell growth, and induces apoptosis in several cancer cell lines and cotreatment with glutathione reverses these responses. Piperlongumine also induces ROS-dependent downregulation of Sp1, Sp3, Sp4 and pro-oncogenic Sp-regulated genes, demonstrating that the anticancer activity of this compound is also due, in part, to targeting of Sp TFs.
Materials and Methods
Cell lines, antibodies, and reagents
Pancreatic cancer cells (Panc1, L3.6pL), kidney (786-O), lung (A549), and breast (SKBR3) cancer cell lines were purchased from American Type Culture Collection (Manassas, VA), and the SKBR3, L3.6pL and Panc1 cells were authenticated in 2016 by Biosynthesis (Lewisville, TX). Cells were grown and maintained at 37°C in the presence of 5% CO2 in Dulbecco's modified Eagle's medium/Ham's F-12 medium supplemented with 10% fetal bovine serum or RPMI-1640 Medium with 10% fetal bovine serum (FBS). DMEM, RPMI 1640-Medium, FBS, formaldehyde, trypsin and glutathione (98% pure) were purchased from Sigma-Aldrich (St. Louis, MO). cMyc, survivin, cleaved poly (ADP-ribose) polymerase (cPARP) and cMet antibodies were from Cell Signaling (Boston, MA); ZBTB4 and Sp1 antibodies were from Abcam (Cambridge, MA); and ZBTB10 antibody from Bethyl Laboratories Inc. (Montgomery, TX). Chemiluminescence reagents (Immobilon Western) for western blot imaging were purchased from Millipore (Billerica, MA), and piperlongumine (PL) was purchased from INDOFINE Chemical Company, Inc. (Hillsborough, NJ). The Apoptotic, Necrotic, and Healthy Cells Quantification Kit was purchased from Biotium (Hayward, CA). The ROS Determination Kit was purchased from Invitrogen (Grand Island, NY); Chromatin Immunoprecipitation Kit was purchased from Active Motif (Carlsbad, CA); and XTT cell viability kit purchased from Cell Signaling (Boston, MA)
Cell viability assay
Cells were plated in 96 well plate at a density of 3000 per well with DMEM containing 10% charcoal-stripped FBS. After 24 hr, cells were treated with DMSO and different concentrations of piperlongumine with DMEM containing 2.5% charcoal-stripped FBS for 0 to 48 hr. After treatment with piperlongumine, 25 μL (XTT with 1% of electron coupling solution) were added to each well and incubated for 4 hr as described in the manufacturer's instruction (Cell Signaling, Boston, MA). After incubation, absorbance was measured at wavelength of 450 nm in a 96 well plate reader.
Measurement of ROS
Cell permeable probe CM-H2DCFDA [5-(and 6)-chloromethyl-2,7-dichlorodihydrofluorescein diacetate acetyl ester] as described in manufacturer's instructions (Life Technologies, Carlsbad, CA) was used to measure ROS level in cancer cells. Cells were seeded at density of 1.5×105 per ml in 6 well plates, allowed to attach for 24 hr, pretreated with GSH for 30 min, and treated with vehicle (DMSO), piperlongumine alone or with GSH for 30 mins to 9 hr. ROS levels were measured by flow cytometry as previously described (27,28).
Measurement of apoptosis (Annexin V staining)
Cells were seeded at density of 1.5×105 per ml in 6 well plates and allowed to attach for 24 hr, pretreated with GSH for 30 min, treated with either vehicle or piperlongumine or combination with GSH for 24 hr. Cells were then stained and analyzed by flow cytometry using the Vybrant apoptosis assay kit according to the manufacturer's protocol (Biotium, CA).
Western blot analysis
Panc1, L3.6pL, SKBR3, 786-O, and A549 cells were seeded at a density of 1.5×105 per ml in 6 well plates and allowed to attach for 24 hr. Cells were treated with various concentration of piperlongumine alone or in combination with GSH, and whole cell proteins were extracted using RIPA lysis buffer containing 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 (w/v), 0.5% sodium deoxycholate and 0.1% SDS with protease and phosphatase inhibitor cocktail. Protein concentrations were measured using Lowry's method and equal amounts of protein were separated in 10% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. Membranes were incubated with primary antibodies overnight at 4°C and incubated with corresponding HRP-conjugated secondary IgG antibodies and immuno-reacted proteins were detected with chemiluminescence reagent.
Chromatin immunoprecipitation (ChIP) assay
Panc1 cells were seeded at a density of 5×106 and allowed to attach for 24 hr. Cells were treated with piperlongumine for 3 hr and subjected to ChIP analysis using the ChIP-IT Express magnetic chromatin immunoprecipitation kit (Active Motif, Carlsbad, CA) according to the manufacturer's protocol using 1% formaldehyde for crosslinking. The sonicated chromatin was immunoprecipitated with normal IgG (Santa Cruz Biotech (Santa Cruz, CA), and antibodies for RNA polymerase II (pol II; GeneTex, Irving, CA), H3K27me3 (Abcam, Cambridge, MA), H3K4me3 (Abcam), H4K16Ac (Active Motif), Sp1 (Abcam), Sp3 and Sp4 (SantaCruz) incubated with protein A-conjugated magnetic beads at 4°C for overnight. Magnetic beads were extensively washed, and crosslinked protein-DNA was reversed and eluted. DNA was extracted from the immunoprecipitates and PCR was performed using following primers. The primers for detection of the c-Myc promoter region were 5′-GCC CTT TCC CCA GCC TTA GC-3′ (sense) and 5′-AAC CGC ATC CTT GTC CTG TGA GTA-3′ (antisense); the primers for the detection of the β-actin (ACTB) promoter region were 5′-CTC CCT CCTCCT CTT CCT CA-3′ (sense) and 5′-TCG AGC CAT AAA AGGCAA CTT-3′ (antisense); the primers for detection of the Sp1 promoter region were 5′-CTA ACT CCA ATC ATA ACG TTC C-3′ (sense) and 5′-GAG CTG GAG ATG ATT GGC TTG-3′ (antisense). PCR products were resolved on a 2% agarose gel in the presence of ethidium bromide (EtBr) (Denville Scientific Inc., Holliston, MA).
Real-time polymerase chain reaction (RT-PCR)
Expression of miR-17, miR-20a and miR-27a after treatment with piperlongumine alone or in combination with GSH was measured using RT-PCR. Panc1 cells were plated at density of 4×105 in 60 mm dish and were allowed to attach for 24 hr. Cells were treated with piperlongumine alone or in combination with GSH for 0 to 24 hr. Total RNA was extracted using the mirVana miRNA isolation kit (Ambion, Austin, TX) according to the manufacturer's instruction. TaqMan microNRA assays (Life Technologies, Carlsbad, CA) were used to quantify expression of miR-17, miR-20a and miR-27a, and RNU6 was used as a control to determine relative miRNA expression.
Xenograft Study
Female athymic nu/nu mice (4-6 weeks old) were purchased from Harlan Laboratories (Houston, TX). L3.6pL cells (1×106) were harvested in 100 μL of DMEM and suspended in ice-cold Matrigel (1:1 ratio) and s.c. injected to either side of the flank area of the mice. After one week of tumor cell inoculation, mice were divided in to two groups of 5 animals each. The first group received 100 μL of vehicle (corn oil), and second group of animal received an injection of 30 mg/kg/day of piperlongumine in 100 μL volume of corn oil by i.p. for three weeks. All mice were weighed once a week over the course of treatment to monitor changes in body weight. Tumor volumes could not be determined over the period of treatment because xenografted tumors were relatively deep. After three weeks of treatment, mice were sacrificed and tumor weights were determined. All animal studies were carried out according to the procedures approved by the Texas A&M University Institutional Animal Care and Use Committee.
Statistical analysis
Student's t-test was used to determine the statistical significance between two groups. The experiments for treatment group were performed at least three independent times and results were expressed as means ± SEM. P-values less than 0.05 were considered to be statistically significant.
Results
1. Piperlongumine induces ROS-dependent growth inhibition and apoptosis
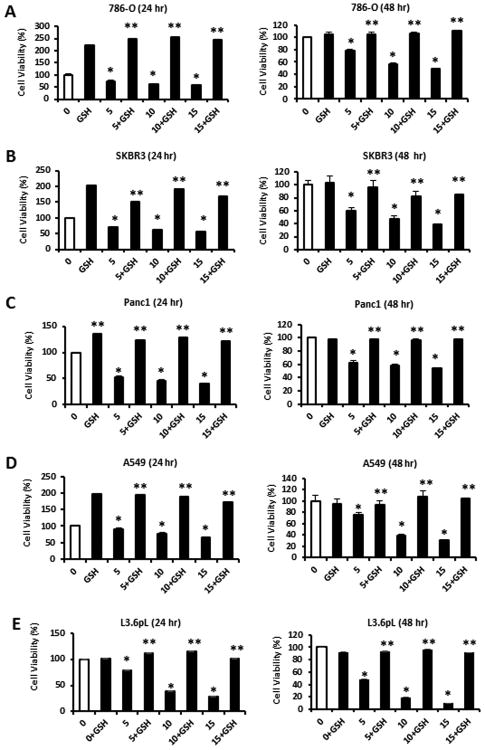
In this study, we initially used 786-O kidney, SKBR3 breast, Panc1 and L3.6pL pancreatic, and A549 lung cancer cell lines to investigate the growth inhibitory effects of piperlongumine. Treatment of 786-O cells with 5, 10 and 15 μM piperlongumine for 24 and 48 hr significantly decreased cell proliferation at all concentrations (Fig. 1A) and these effects were blocked after cotreatment with 5 mM GSH. Interestingly, we observed that GSH alone enhanced proliferation of these cells at the 24 hr but not the 48 hr time point, suggesting that endogenous ROS may have been decreasing cell proliferation at the former time point. The same experimental protocol was used for SKBR3 (Fig. 1B), Panc1 (Fig. 1C), A549 (Fig. 1D) and L3.6pL (Fig. 1E) cells and the results confirmed that for this panel of cancer cell lines that piperlongumine-induced ROS was a major factor in the growth inhibitory effects observed for this compound.
Figure 1.
Piperlongumine inhibits cancer cell proliferation. 786-O (A), SKBR3 (B), Panc1 (C), A549 (D) and L3.6pL (E) cancer cell lines were treated with different concentrations of piperlongumine or 5 mM GSH alone or in combination for 24 and 48 hr and cell numbers were determined as outlined in the Materials and Methods. Results are means ± SEM for at least 3 replicate determinations and significant (p<0.05) growth inhibition by piperlongumine (*), growth induction or reversal of piperlongumine-dependent growth inhibition by GSH (**) are indicated.
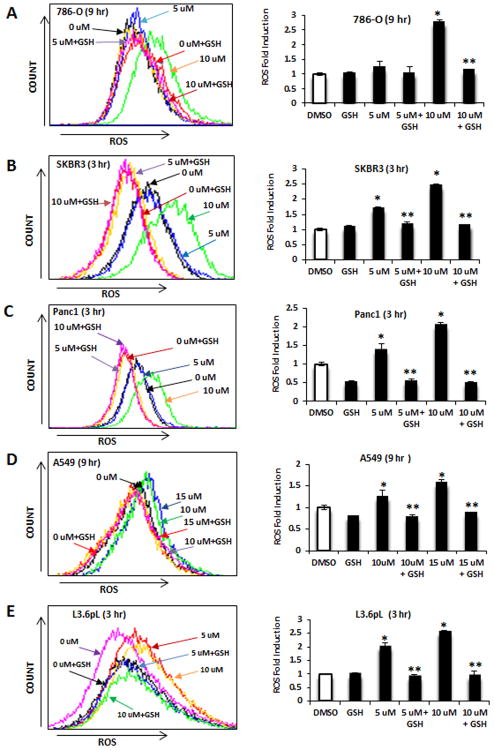
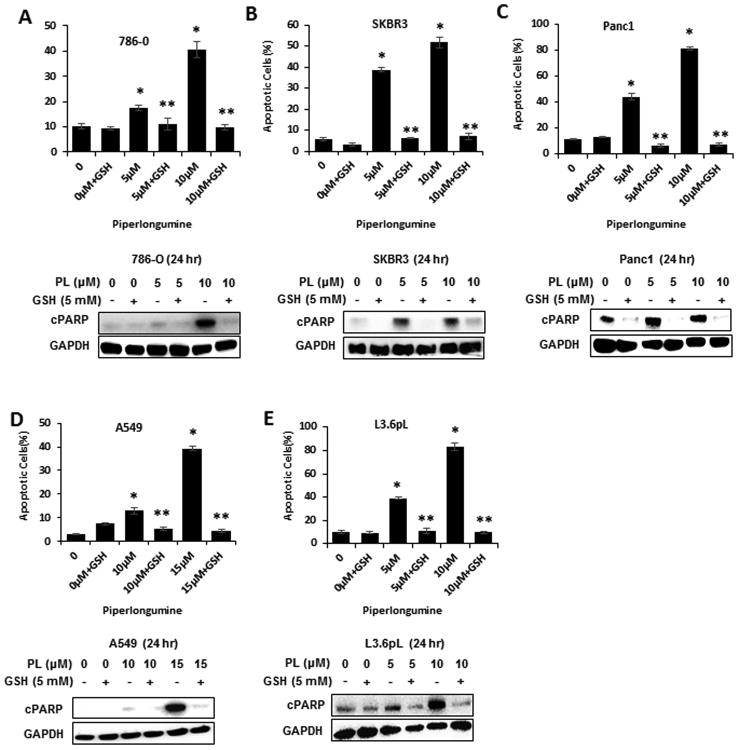
We also investigated induction of ROS by piperlongumine by FACS analysis using the cell permeable CM-H2DFCDA dye. Piperlongumine induced ROS in 786-O, SKBR3, Panc1, A549 and L3.6pL cells (Figs. 2A-2E); this response was attenuated in cells cotreated with GSH and these results are consistent with previous studies showing the piperlongumine induces ROS (6). Treatment of these cells with piperlongumine also induced Annexin V staining and PARP cleavage which are markers of apoptosis (Figs. 3A-3E) and cotreatment with GSH attenuated these responses, demonstrating that piperlongumine induces ROS which in turn inhibits cell growth and induces apoptosis. Supplemental Figure S1 illustrates the flow cytometric analysis of the piperlongumine-induced Annexin V staining.
Figure 2.
Piperlongumine induces ROS in cancer cell lines. 786-O (A), SKBR3 (B), Panc1 (C), A549 (D) and L3.6pL (E) cells were treated with piperlongumine or 5 mM GSH alone or in combination and ROS was determined by FACS analysis of the cell permeant dye. CM-H2DFCDA as outlined in the Materials and Methods. Results are expressed as means ± SEM for at least 3 replicate determinations and significant (p<0.05) induction of ROS by piperlongumine (*) and inhibition by GSH (**) are indicated.
Figure 3.
Piperlongumine induces apoptosis in cancer cells. 786-O (A), SKBR3 (B), Panc1 (C), A549 (D) and L3.6pL (E) cells were treated with 5 mM GSH and different concentration of piperlongumine alone and in combination and after 24 hr, effects on Annexin V staining and PARP cleavage were determined fluorimetrically or by western blot analyses of whole cell lysates, respectively, as outlined in the Materials and Methods. Results of Annexin V staining are expressed as means ± SEM of at least 3 replicate determinations and significant (p<0.05) induction of Annexin V (*) and inhibition by GSH (**) are indicated.
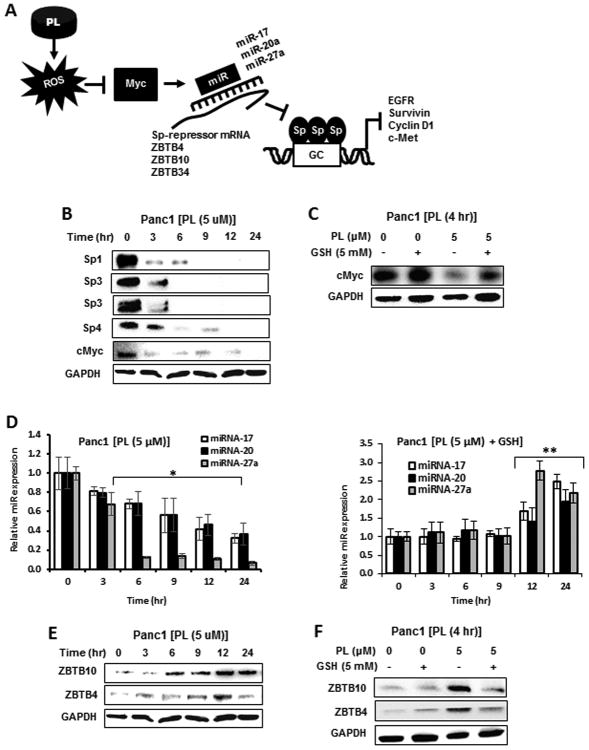
2. Piperlongumine downregulates Sp1, Sp3, Sp4 and Sp-regulated genes
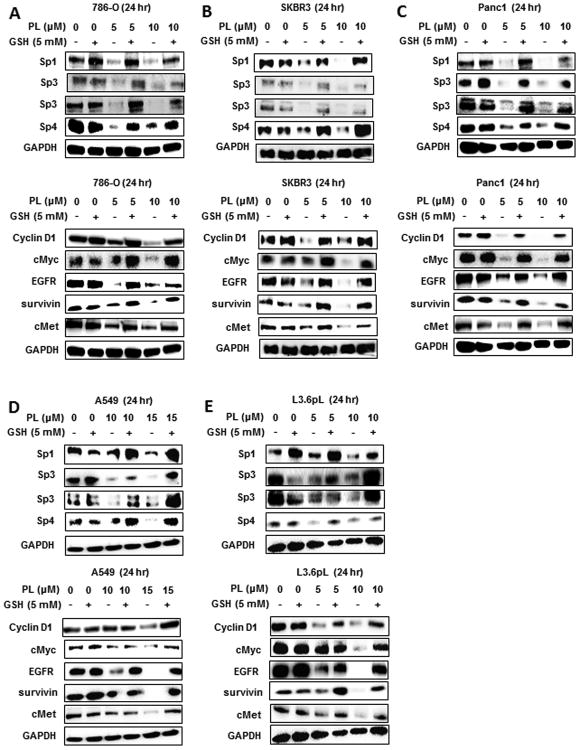
The effects of piperlongumine on downregulation of Sp1, Sp3, Sp4 and Sp-regulated genes including cyclin D1, EGFR, hepatocyte growth factor receptor (cMET), and survivin were also investigated in the 5 cancer cell lines. Treatment of 786-O cells with 5 or 10 μM piperlongumine decreased expression of Sp1, Sp3 and Sp4 and after cotreatment with GSH, these responses were blocked and similar results were observed for effects of piperlongumine on Sp-regulated cMyc, EGFR, survivin and cMET (Fig. 4A). This same approach was used to investigate the effects of piperlongumine alone or in combination with GSH on Sp TFs and Sp-regulated genes in SKBR3 (Fig. 4B), Panc1 (Fig. 4C), A549 (Fig. 4D), and L3.6pL (Fig. 4E) cells. The higher concentration of piperlongumine (15 μM for A549 cells and 10 μM for the other cell lines) decreased expression of Sp1, Sp3, Sp4 and Sp-regulated genes and this response was attenuated after cotreatment with GSH. We also observed that 5 μM piperlongumine was effective in reducing one or more Sp proteins and Sp-regulated genes in 786-O, SKBR3, Panc1 and L3.6pL cells and 10 μM piperlongumine in A549 cells which was the most piperlongumine-resistant cell line after treatment for 24 hr.
Figure 4.
Piperlongumine downregulates Sp1, Sp3, Sp4 and Sp-regulated genes: effects of GSH. 786-O (A), SKBR3 (B), Panc1 (C), A549 (D), and L3.6pL (E) cells were treated with 5 mM GSH or different concentrations of piperlongumine alone and in combination for 24 hr, and whole cell lysates were analyzed by western blots. Effects on Sp proteins and Sp-regulated gene expression and PARP cleavage (Fig. 3) were all obtained in the same experiment and have the same GAPDH loading control. Similar results were observed in duplicate experiments.
Piperlongumine modulates expression of or inhibits redox enzymes and the conjugated en-one structure alkylates thiol-containing molecules (7,15) and we therefore further investigate the effects of the non-thiol-containing reductant Tiron on piperlongumine-dependent Sp downregulation (Suppl. Fig. S2). The results were similar to that observed after treatment with piperlongumine ± GSH (Fig. 4); piperlongumine decreased Sp1, Sp3, Sp4 and Sp-regulated genes in 786-O, SKBR3, Panc1, A549 and L3.6pL cells and cotreatment with 5 mM Tiron blocked the effects of piperlongumine in all but SKBR3 cells where some responses were decreased by 10 μM Tiron (Suppl. Fig. S2). These data confirm the piperlongumine-induced ROS results in downregulation of Sp TFs and pro-oncogenic Sp-regulated genes and this was similar to the effects observed for other ROS-inducing anticancer agents (20-28).
3. Mechanism of PL-induced downregulation of Sp TFs and in vivo studies
Figure 5A outlines the mechanism of ROS-induced downregulation of Sp TFs by initially targeting cMyc which results in downregulation of cMyc-regulated miRs and induction of miR suppressed ZBTB transcriptional repressors (32). Using Panc1 cells as a model, Figure 5B shows that 5 μM piperlongumine decreases cMyc expression within 3 hr after treatment and similar results were observed for Sp1, Sp3 and Sp4. Piperlongumine-dependent downregulation of cMyc was blocked after cotreatment with GSH (Fig. 5C) and piperlongumine-induced downregulation of miR-27a and miR-17/miR-20 (Fig. 5D) was also inhibited by cotreatment with GSH (Fig. 5E) and at longer time points (12 and 24 hr), GSH enhanced miR expression. We also observed that 5 μM piperlongumine induced expression of ZBTB10 and ZBTB4 proteins (Fig. 5E) and cotreatment with GSH attenuated this response (Fig. 5F) and these effects are consistent with the pathway illustrated in Figure 5A.
Figure 5.
Mechanism of piperlongumine induced Sp downregulation. (A) Proposed mechanism of piperlongumine-induced Sp downregulation by initial induction of ROS. (B) Panc1 cells were treated with 5 μM piperlongumine for up to 24 hr, and whole cell lysates were analyzed by western blots. (C) Panc1 cells were treated with 5 μM piperlongumine or 5 mM GSH alone for 4 hr, and whole cell lysates were analyzed by western blots. Panc1 cells with 5 μM piperlongumine or 5 mM GSH alone and in combination for up to 24 hr, and the extracted RNA (D) or protein (E) was analyzed by real time PCR or western blots, respectively. Results in (D) are means ± SEM for at least 3 replicates and significant (p<0.05) miR downregulation by piperlongumine (*) and reversal by cotreatment with GSH (**) are indicated.
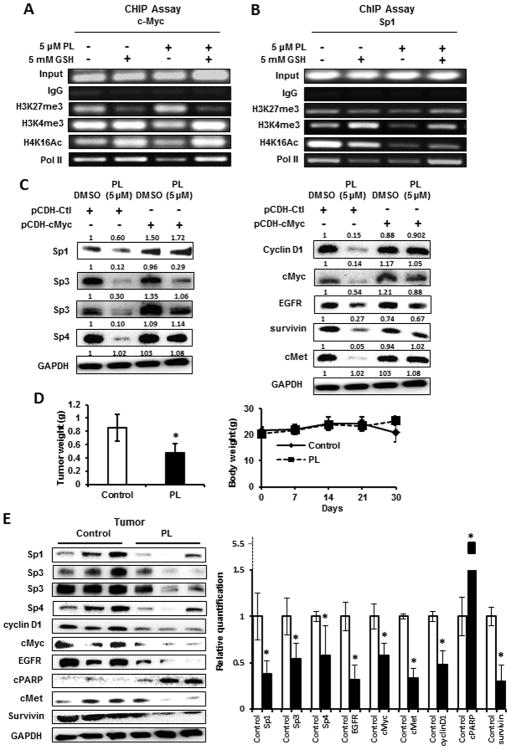
ROS induces rapid shifts of chromatin-modifying complexes from non-GC-rich to GC-rich sequences (36), and ChIP analysis of the cMyc promoter showed that piperlongumine increased the gene inactivation mark H3K27 and slightly decreased the activation marks H3K4me3 and H4K16Ac and pol II (Fig. 6A). GSH reversed the piperlongumine-induced interactions with the cMyc promoter and GSH alone enhanced H4K16Ac. In contrast, the major piperlongumine-dependent changes on the GC-rich region of the Sp1 promoter were decreased in the H3K4me3 and H4K16Ac histone marks (Fig. 6B) which is consistent with the decreased expression of cMyc (Fig. 5B). We further confirmed the critical role of cMyc in ROS-dependent downregulation of Sp TFs by showing that piperlongumine-induced decreases in Sp1, Sp3, Sp4 and Sp-regulated gene products were rescued by overexpression of cMyc (Fig. 6C). We also observed that piperlongumine (30 mg/kg/d) decreased tumor weight but not body weight in athymic nude mice bearing L3.6pL cells as a xenograft (Fig. 6D) and this was accompanied by significant downregulation of Sp1, Sp3, Sp4 and pro-oncogenic Sp-regulated gene products and induction of PARP cleavage in tumors from piperlongumine-treated mice compared to the vehicle controls. Thus, like other ROS-inducing anticancer agents, an important underlying mechanism of action is due to targeting of Sp transcription factors (Fig. 5A).
Figure 6.
Piperlongumine-dependent Sp downregulation is cMyc-dependent and in vivo studies. Panc1 cells were treated with 5 μM piperlongumine or 5 mM GSH alone or in combination for 3 hr, and interactions with the cMyc (A) and Sp1 (B) promoters were determined in ChIP assays. Quantitation of the bands was carried out by quantitative PCR and results are illustrated in Supplemental Figure S3. (C) Panc1 cells were treated with DMSO or 5 μM piperlongumine alone or after transfection with a cMyc expression plasmid and after 3 hr, whole cell lysates were analyzed by western blots. Athymic nude mice bearing L3.6pL cells as xenografts were treated with piperlongumine (30 mg/kg/d), and effects on tumor weights and body weights (D) and expression of various gene products (E) in tumors from control (corn oil) and piperlongumine-treated mice were determined by western blot analysis of tumor lysates. Expression levels of various proteins in control vs. piperlongumine-treated mice were determined (normalized to GAPDH). Significant (p<0.05) changes in protein levels in tumors from piperlongumine-treated mice compared to controls (*) are indicated.
Discussion
Sp1, Sp3 and Sp4 transcription factors are overexpressed in pancreatic cancer lines (20-28,30-32) and Sp1 is a negative prognostic factor for patient survival (37,38), and similar results have been reported for other tumors (32). Results of RNA interference studies demonstrate that individual knockdown of Sp1, Sp3 and Sp4 inhibits growth and migration and induces apoptosis in 785-O, SKBR3, Panc1, A549 and L3.6pL cells and other cell lines (24,35). The responses observed after Sp knockdown are due to the parallel decrease in genes that regulate cancer cell growth, survival and migration/invasion and these include multiple receptor tyrosine kinase, angiogenic factor and pro-survival genes such as bcl2 and surviving (35). The results suggest that Sp transcription factors are non-oncogene addiction genes and are therefore important drug targets for cancer chemotherapy.
Studies in this laboratory have focused on identifying anticancer agents that target Sp proteins and these include several ROS-inducing agents such as BITC, PEITC, curcumin, betulinic acid and HDAC inhibitors (20-28). Initial studies on the broad spectrum anticancer activity of piperlongumine showed that this compound also induced ROS (6) and this was confirmed in the 5 cancer cell lines used in this study (Fig. 2). Like other ROS-inducing agents, we also observed that piperlongumine decreased Sp1, Sp3, Sp4 and pro-oncogenic Sp-regulated genes in vitro and in vivo (Figs. 4, 5 and Suppl. Fig. S2). Since GSH also reversed the growth inhibitory and pro-apoptotic effects of piperlongumine, we conclude that an important underlying mechanism of action of piperlongumine as an anticancer agent is due to ROS-dependent Sp downregulation; however, we did not further investigate the specific ROS species induced by piperlongumine. Several reports show that piperlongumine also induces many other effects in cancer cell lines (1-19); however, some of the specific piperlongumine-induced downregulated gene products in these studies include NFκB, bcl-2, cMyc, cyclin D1, VEGF, surviving (7,14,19) which are also Sp-regulated genes (32).
It was initially reported by O'Hagan and coworkers that hydrogen peroxide induced genome-wide shifts of chromatin-modifying complexes from non-GC-rich to GC-rich promoters and this resulted in decreased expression of cMyc (36). This represents a novel epigenetic pathway for ROS-mediated gene repression; moreover, studies in this laboratory have also observed these effects in cancer cells treated with other ROS inducers including PEITC, celastrol, HDAC inhibitors, BITC and penfluridol (20-28). Induction of ROS by these agents was accompanied by decreased expression of cMyc-regulated miRs (27a, 17 and 20), resulting in induction of miR-repressed ZBTB10 and ZBTB4 as illustrated in Figure 5A. Piperlongumine also rapidly decrease cMyc expression in Panc1 cells and this was accompanied by ROS-dependent downregulation of miR-27a and miR-17/20a (part of the miR-17-92 cluster) and induction of ZBTB10 and ZBTB4 (Figs. 5C-5F). Piperlongumine also decreased interactions of pol II, slightly increased H3K27me3, and decreased H3K4me3/H4K16Ac interactions with the GC-rich cMyc promoter (Fig. 6A) and these results were similar to that observed for other ROS inducers (25,27,28). Examination of the GC-rich region of the Sp1 promoter in a ChIP assay also showed decreased interactions with pol II and the H3K4me3 and H4K16Ac activation markers (Fig. 6B), consistent with the rapid downregulation of Sp protein (Fig. 5B).
In summary, results of this study demonstrate that in important underlying mechanism of action of piperlongumine is due to the ROS-dependent downregulation of cMyc and a cMyc-regulated pathway (Fig. 5A), resulting in downregulation of Sp1, Sp3, Sp4 and pro-oncogenic Sp-regulated genes. This observation is consistent with previous studies on ROS-inducing anticancer agents including CDDO-Me, celastrol, PEITC, BITC, HDAC inhibitors and penfluridol (20-28). Many ROS-inducing anticancer agents induce important ROS-independent and -dependent responses that contribute to the overall compound efficacy. Recognition of the ROS-Sp downregulation pathway could be important for designing drug-drug and drug-radiation combination therapies since many treatment-related drug resistance genes (e.g. survivin) are Sp-regulated.
Supplementary Material
Acknowledgments
Funding: The financial assistance of the National Institutes of Health (P30-ES023512, S. Safe), Texas AgriLife Research and Sid Kyle endowment, is gratefully acknowledged.
Footnotes
Conflict of Interest: The authors declare that there are no competing financial interests.
Author Contributions: Participated in research design: K.K., S.S.
Conducted experiments: K.K., R.K., U.H.J., E.H.
Contributed new reagents or analytic tools: S.S.
Performed data analysis: K.K., R.K.
Wrote or contributed to the writing of the manuscript: S.S., K.K.
Literature Cited
- 1.Yang YC, Lee SG, Lee HK, Kim MK, Lee SH, Lee HS. A piperidine amide extracted from Piper longum L. fruit shows activity against Aedes aegypti mosquito larvae. J Agric Food Chem. 2002;50:3765–7. doi: 10.1021/jf011708f. [DOI] [PubMed] [Google Scholar]
- 2.Son DJ, Kim SY, Han SS, Kim CW, Kumar S, Park BS, et al. Piperlongumine inhibits atherosclerotic plaque formation and vascular smooth muscle cell proliferation by suppressing PDGF receptor signaling. Biochem Biophys Res Commun. 2012;427:349–54. doi: 10.1016/j.bbrc.2012.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cicero Bezerra Felipe F, Trajano Sousa Filho J, de Oliveira Souza LE, Alexandre Silveira J, Esdras de Andrade Uchoa D, Rocha Silveira E, et al. Piplartine, an amide alkaloid from Piper tuberculatum, presents anxiolytic and antidepressant effects in mice. Phytomedicine. 2007;14:605–12. doi: 10.1016/j.phymed.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 4.Lee SA, Hong SS, Han XH, Hwang JS, Oh GJ, Lee KS, et al. Piperine from the fruits of Piper longum with inhibitory effect on monoamine oxidase and antidepressant-like activity. Chem Pharm Bull (Tokyo) 2005;53:832–5. doi: 10.1248/cpb.53.832. [DOI] [PubMed] [Google Scholar]
- 5.Fontenele JB, Leal LK, Silveira ER, Felix FH, Bezerra Felipe CF, Viana GS. Antiplatelet effects of piplartine, an alkamide isolated from Piper tuberculatum: possible involvement of cyclooxygenase blockade and antioxidant activity. J Pharm Pharmacol. 2009;61:511–5. doi: 10.1211/jpp/61.04.0014. [DOI] [PubMed] [Google Scholar]
- 6.Raj L, Ide T, Gurkar AU, Foley M, Schenone M, Li X, et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature. 2011;475:231–4. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Adams DJ, Dai M, Pellegrino G, Wagner BK, Stern AM, Shamji AF, et al. Synthesis, cellular evaluation, and mechanism of action of piperlongumine analogs. Proc Natl Acad Sci U S A. 2012;109:15115–20. doi: 10.1073/pnas.1212802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin HO, Lee YH, Park JA, Lee HN, Kim JH, Kim JY, et al. Piperlongumine induces cell death through ROS-mediated CHOP activation and potentiates TRAIL-induced cell death in breast cancer cells. J Cancer Res Clin Oncol. 2014;140:2039–46. doi: 10.1007/s00432-014-1777-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu JM, Pan F, Li L, Liu QR, Chen Y, Xiong XX, et al. Piperlongumine selectively kills glioblastoma multiforme cells via reactive oxygen species accumulation dependent JNK and p38 activation. Biochem Biophys Res Commun. 2013;437:87–93. doi: 10.1016/j.bbrc.2013.06.042. [DOI] [PubMed] [Google Scholar]
- 10.Zou P, Xia Y, Ji J, Chen W, Zhang J, Chen X, et al. Piperlongumine as a direct TrxR1 inhibitor with suppressive activity against gastric cancer. Cancer Lett. 2016;375:114–26. doi: 10.1016/j.canlet.2016.02.058. [DOI] [PubMed] [Google Scholar]
- 11.Roh JL, Kim EH, Park JY, Kim JW, Kwon M, Lee BH. Piperlongumine selectively kills cancer cells and increases cisplatin antitumor activity in head and neck cancer. Oncotarget. 2014;5:9227–38. doi: 10.18632/oncotarget.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginzburg S, Golovine KV, Makhov PB, Uzzo RG, Kutikov A, Kolenko VM. Piperlongumine inhibits NF-κB activity and attenuates aggressive growth characteristics of prostate cancer cells. Prostate. 2014;74:177–86. doi: 10.1002/pros.22739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Wang JW, Xiao X, Shan Y, Xue B, Jiang G, et al. Piperlongumine induces autophagy by targeting p38 signaling. Cell Death Dis. 2013;4:e824. doi: 10.1038/cddis.2013.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng J, Son DJ, Gu SM, Woo JR, Ham YW, Lee HP, et al. Piperlongumine inhibits lung tumor growth via inhibition of nuclear factor kappa B signaling pathway. Sci Rep. 2016;6:26357. doi: 10.1038/srep26357. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Han JG, Gupta SC, Prasad S, Aggarwal BB. Piperlongumine chemosensitizes tumor cells through interaction with cysteine 179 of IkappaBalpha kinase, leading to suppression of NF-kappaB-regulated gene products. Mol Cancer Ther. 2014;13:2422–35. doi: 10.1158/1535-7163.MCT-14-0171. [DOI] [PubMed] [Google Scholar]
- 16.Shrivastava S, Kulkarni P, Thummuri D, Jeengar MK, Naidu VG, Alvala M, et al. Piperlongumine, an alkaloid causes inhibition of PI3 K/Akt/mTOR signaling axis to induce caspase-dependent apoptosis in human triple-negative breast cancer cells. Apoptosis. 2014;19:1148–64. doi: 10.1007/s10495-014-0991-2. [DOI] [PubMed] [Google Scholar]
- 17.Bharadwaj U, Eckols TK, Kolosov M, Kasembeli MM, Adam A, Torres D, et al. Drug-repositioning screening identified piperlongumine as a direct STAT3 inhibitor with potent activity against breast cancer. Oncogene. 2015;34:1341–53. doi: 10.1038/onc.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randhawa H, Kibble K, Zeng H, Moyer MP, Reindl KM. Activation of ERK signaling and induction of colon cancer cell death by piperlongumine. Toxicol In Vitro. 2013;27:1626–33. doi: 10.1016/j.tiv.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han SS, Son DJ, Yun H, Kamberos NL, Janz S. Piperlongumine inhibits proliferation and survival of Burkitt lymphoma in vitro. Leuk Res. 2013;37:146–54. doi: 10.1016/j.leukres.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jutooru I, Chadalapaka G, Abdelrahim M, Basha MR, Samudio I, Konopleva M, et al. Methyl 2-cyano-3,12-dioxooleana-1,9-dien-28-oate decreases specificity protein transcription factors and inhibits pancreatic tumor growth: role of microRNA-27a. Mol Pharmacol. 2010;78:226–36. doi: 10.1124/mol.110.064451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chintharlapalli S, Papineni S, Lei P, Pathi S, Safe S. Betulinic acid inhibits colon cancer cell and tumor growth and induces proteasome-dependent and - independent downregulation of specificity proteins (Sp) transcription factors. BMC Cancer. 2011;11:371. doi: 10.1186/1471-2407-11-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathi SS, Jutooru I, Chadalapaka G, Sreevalsan S, Anand S, Thatcher GR, et al. GT-094, a NO-NSAID, inhibits colon cancer cell growth by activation of a reactive oxygen species-microRNA-27a: ZBTB10-specificity protein pathway. Mol Cancer Res. 2011;9:195–202. doi: 10.1158/1541-7786.MCR-10-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chadalapaka G, Jutooru I, Safe S. Celastrol decreases specificity proteins (Sp) and fibroblast growth factor receptor-3 (FGFR3) in bladder cancer cells. Carcinogenesis. 2012;33:886–94. doi: 10.1093/carcin/bgs102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedrick E, Crose L, Linardic CM, Safe S. Histone deacetylase inhibitors inhibit rhabdomyosarcoma by reactive oxygen species-dependent targeting of specificity protein transcription factors. Mol Cancer Ther. 2015;14:2143–53. doi: 10.1158/1535-7163.MCT-15-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jutooru I, Guthrie AS, Chadalapaka G, Pathi S, Kim K, Burghardt R, et al. Mechanism of action of phenethylisothiocyanate and other reactive oxygen species-inducing anticancer agents. Mol Cell Biol. 2014;34:2382–95. doi: 10.1128/MCB.01602-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gandhy SU, Kim K, Larsen L, Rosengren RJ, Safe S. Curcumin and synthetic analogs induce reactive oxygen species and decreases specificity protein (Sp) transcription factors by targeting microRNAs. BMC Cancer. 2012;12:564. doi: 10.1186/1471-2407-12-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hedrick E, Li X, Safe S. Penfluridol represses integrin expression in breast cancer through induction of reactive oxygen species and downregulation of Sp transcription factors. Mol Cancer Ther. 2017;16:205–16. doi: 10.1158/1535-7163.MCT-16-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasiappan R, Jutooru I, Karki K, Hedrick E, Safe S. Benzyl isothiocyanate (BITC) induces reactive oxygen species-dependent repression of STAT3 protein by down-regulation of specificity proteins in pancreatic cancer. J Biol Chem. 2016;291:27122–33. doi: 10.1074/jbc.M116.746339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–52. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Jutooru I, Chadalapaka G, Sreevalsan S, Lei P, Barhoumi R, Burghardt R, et al. Arsenic trioxide downregulates specificity protein (Sp) transcription factors and inhibits bladder cancer cell and tumor growth. Exp Cell Res. 2010;316:2174–88. doi: 10.1016/j.yexcr.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jutooru I, Chadalapaka G, Lei P, Safe S. Inhibition of NFκB and pancreatic cancer cell and tumor growth by curcumin is dependent on specificity protein down-regulation. J Biol Chem. 2010;285:25332–44. doi: 10.1074/jbc.M109.095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Safe S, Imanirad P, Sreevalsan S, Nair V, Jutooru I. Transcription factor Sp1, also known as specificity protein 1 as a therapeutic target. Expert Opin Ther Targets. 2014;18:759–69. doi: 10.1517/14728222.2014.914173. [DOI] [PubMed] [Google Scholar]
- 33.Kim K, Chadalapaka G, Lee SO, Yamada D, Sastre-Garau X, Defossez PA, et al. Identification of oncogenic microRNA-17-92/ZBTB4/specificity protein axis in breast cancer. Oncogene. 2012;31:1034–44. doi: 10.1038/onc.2011.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28:1194–202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 35.Hedrick E, Cheng Y, Jin UH, Kim K, Safe S. Specificity protein (Sp) transcription factors Sp1, Sp3 and Sp4 are non-oncogene addiction genes in cancer cells. Oncotarget. 2016;7:22245–56. doi: 10.18632/oncotarget.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–19. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang NY, Woda BA, Banner BF, Whalen GF, Dresser KA, Lu D. Sp1, a new biomarker that identifies a subset of aggressive pancreatic ductal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:1648–52. doi: 10.1158/1055-9965.EPI-07-2791. [DOI] [PubMed] [Google Scholar]
- 38.Hu J, Hu H, Hang JJ, Yang HY, Wang ZY, Wang L, et al. Simultaneous high expression of PLD1 and Sp1 predicts a poor prognosis for pancreatic ductal adenocarcinoma patients. Oncotarget. 2016;7:78557–65. doi: 10.18632/oncotarget.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.