Abstract
Phosphoinositides (PIs) are recognized as major signaling molecules in many different functions of eukaryotic cells. PIs can be dephosphorylated by multiple phosphatase activities at the 5-, 4-, and 3- positions. Human PI 5-phosphatases belong to a family of 10 members. Except for inositol polyphosphate 5-phosphatase A, they all catalyze the dephosphorylation of PI(4,5)P2 and/or PI(3,4,5)P3 at the 5- position. PI 5-phosphatases thus directly control the levels of PI(3,4,5)P3 and participate in the fine-tuning regulatory mechanisms of PI(3,4)P2 and PI(4,5)P2. Second messenger functions have been demonstrated for PI(3,4)P2 in invadopodium maturation and lamellipodia formation. PI 5-phosphatases can use several substrates on isolated enzymes, and it has been challenging to establish their real substrate in vivo. PI(4,5)P2 has multiple functions in signaling, including interacting with scaffold proteins, ion channels, and cytoskeleton proteins. PI 5-phosphatase isoenzymes have been individually implicated in human diseases, such as the oculocerebrorenal syndrome of Lowe, through mechanisms that include lipid control. Oncogenic and tumor-suppressive functions of PI 5-phosphatases have also been reported in different cell contexts. The mechanisms responsible for genetic diseases and for oncogenic or tumor-suppressive functions are not fully understood. The regulation of PI 5-phosphatases is thus crucial in understanding cell functions.
Keywords: cancer, hormones, growth factors, protein
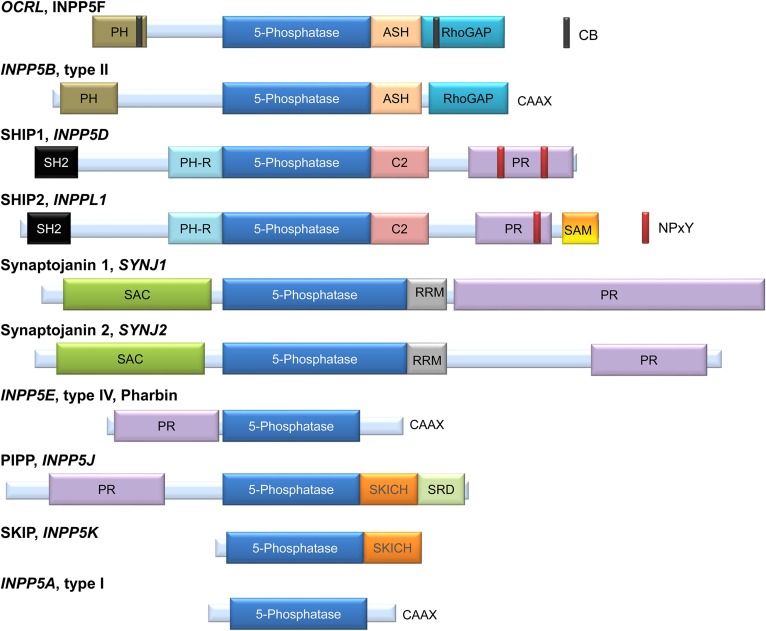
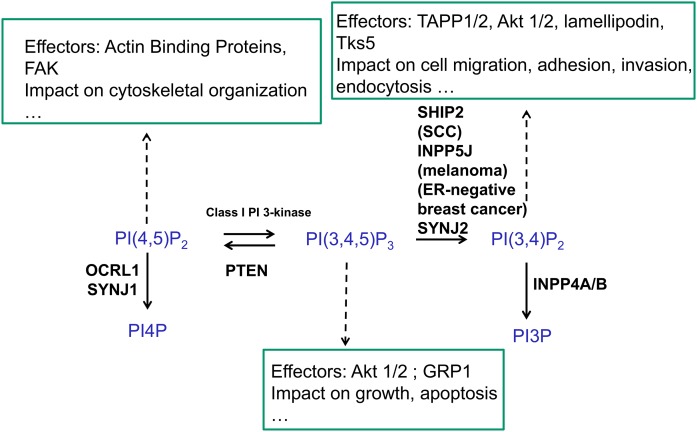
Phosphoinositides (PIs) can be dephosphorylated by multiple mechanisms involving the participation of 3-, 4-, and 5-phosphatase activities (1–3). Inositol polyphosphate or phosphoinositide 5-phosphatases (INPP5Ps) represent a large family of 10 different enzymes in the human genome (referred to as oculocerebrorenal syndrome of Lowe [OCRL]/INPP5F, INPP5B, SHIP1/2, synaptojanins (SYNJs) 1 and 2, INPP5E, INPP5J, INPP5K, and INPP5A) (4) (Fig. 1). They all share an inositol 5-phosphatase domain with a series of conserved amino acids and a general comparable structural organization (5). As seen for INPP5B, conserved sequence motifs support a common interaction mode with the PI in most PI 5-phosphatases (5). A similar part of these active enzyme sites are involved in membrane interactions, consistent with a model in which the catalytic domain inserts slightly into the membrane. Except for INPP5A, which recognizes only soluble inositol 1,4,5-trisphosphate (Ins(1,4,5)P3) and inositol 1,3,4,5-tetrakisphosphate (Ins(1,3,4,5)P4) as substrates (6), they all catalyze the dephosphorylation of essentially PI(3,4,5)P3, and PI(4,5)P2 (7, 8) at the 5-phosphate position of the inositol ring (Table 1). They therefore display a negative control on PI(3,4,5)P3, acting as potential tumor suppressors in tumor cells (9), as well as a positive control on PI(3,4)P2 formation (Fig. 2). Actually, PI(3,4)P2 can be rapidly dephosphorylated by the active PI 4-phosphatases INPP4A/B or PI 3-phosphatase PTEN (phosphatase and tensin homolog) (10). Both INPP4B and PTEN are important tumor suppressors, particularly in breast cancer cells, and can be absent in some tumors (11). The relative flux of 3- versus 5-dephosphorylation of PI(3,4,5)P3 is cell context dependent. There is indeed a large variability in the expression of PTEN or INPP4B in different tissues and cell types, as shown, for example, in triple-negative breast cancer cells (12).
Fig. 1.
Domain and motive organization of the mammalian PI 5-phosphatases. OCRL and INPP5B are very similar in terms of internal domain organization. Both present a PH domain at the N terminus and an ASH domain followed by an RhoGAP domain at the C terminus. OCRL also has two CB domains. INPP5B lacks the CB domains and has a CAAX domain at the C terminus (57, 88–90). SHIP1 and SHIP2 are also very similar in terms of organization; both contain an SH2 domain at the N terminus; a PH-R domain and C2 domain, upstream and downstream of the 5-phosphatase domain, respectively; and a PR domain with two NPxY sequence motifs for SHIP1 and one for SHIP2. SHIP2 also has a SAM domain in the C terminus, but this SAM domain is absent in SHIP1 (91, 92). The SHIP2 ubiquitin-interacting motif at amino acids 1117–1134 has also been reported for SHIP2 (17). SYNJ1 and SYNJ2 show the presence of a SAC at the N terminus, an RRM domain next to the PI 5-phosphatase domain; and a PR domain at the C terminus (93). INPP5E or pharbin is composed of a PR domain at the N terminus and a CAAX sequence at the C terminus (94). PIPP and SKIP have in a common a SKICH domain downstream of the 5-phosphatase domain. PIPP also has a PR domain at the N terminus and an SRD at the C terminus (95, 96). INPP5A or type I 5-phosphatase is the smallest from the family and is constituted, in addition to the 5-phosphatase domain, by a CAAX sequence at the C terminus (6). Figure is adapted from (1, 62). ASH, ASPM-SPD2-hydin; CB, clathrin binding; PH-R, PH-related; PIPP, proline-rich inositol polyphosphate 5-phosphatase; PR, proline-rich; RhoGAP, Rho GTPase-activating protein; RRM, RNA recognition motif; SAC, SAC1-like phosphatase domain; SH2, Src homology 2; SKICH, SKIP COOH terminal homology; SRD, serine-rich domain.
TABLE 1.
Substrate specificities of the PI 5-phosphatase
| PI 5-Phosphatase | Substrate(s) Identified on Isolated Enzyme | Intact Cell Substrate(s) | References |
| OCRL, INPP5F | PI(3,4,5)P3, PI(4,5)P2, PI(3,5)P2, Ins(1,3,4,5)P4, Ins(1,4,5)P3 | PI(4,5)P2 | (99) |
| INPP5B, type II | PI(3,4,5)P3, PI(4,5)P2, Ins(1,3,4,5)P4, Ins(1,4,5)P3 | PI(4,5)P2 | (100, 101) |
| SHIP1, INPP5D | PI(3,4,5)P3, Ins(1,3,4,5)P4 | PI(3,4,5)P3 | (26, 91) |
| SHIP2, INPPL1 | PI(3,4,5)P3, PI(4,5)P2, Ins(1,3,4,5)P4 | PI(3,4,5)P3, PI(4,5)P2 | (28, 31, 98, 102) |
| SYNJ1 | PI(4,5)P2, PI(3,4,5)P3 | PI(4,5)P2, PI(3,4,5)P3, | (93) |
| SYNJ2 | PI(4,5)P2, PI(3,4,5)P3 | PI(4,5)P2, PI(3,4,5)P3 | (103) |
| INPP5E, type IV, pharbin | PI(4,5)P2, PI(3,5)P2, PI(3,4,5)P3 | PI(4,5)P2, PI(3,4,5)P3 | (16, 94, 104, 105) |
| PIPP, INPP5J | PI(3,4,5)P3, PI(4,5)P2, Ins(1,3,4,5)P4, Ins(1,4,5)P3 | PI(3,4,5)P3 | (96, 106) |
| SKIP, INPP5K | PI(3,4,5)P3, PI(4,5)P2, Ins(1,3,4,5)P4, Ins(1,4,5)P3 | PI(3,4,5)P3, PI(4,5)P2 | (107–109) |
| INPP5A, type I | Ins(1,4,5)P3, Ins(1,3,4,5)P4 | Ins(1,4,5)P3, Ins(1,3,4,5)P4 | (28, 56, 91, 110) |
Substrates of the PI 5-phosphatases are indicated both on the isolated enzyme and in intact cells. It is possible that the intact cell substrate could vary depending on the cell context, particularly in cancer cells (48). In addition to a PI 5-phosphatase reaction, both SYNJs show the presence of a SAC domain that hydrolyzes phosphatidylinositol 3-phosphate, phosphatidylinositol 4-phosphate, and PI(3,5)P2 to PI (1). PIPP, proline-rich inositol polyphosphate 5-phosphatase.
Fig. 2.
The control of PI(3,4,5)P3 by PI 3- and 5-phosphatases. Although not indicated in this figure, the metabolism of PI(3,4)P2 can also occur by PTEN 3-phosphatase activity (10). Major effectors of the PIs and their impact are indicated. ER, estrogen receptor; FAK, focal adhesion kinase; GRP1, general receptor for phosphoinositide 1-associated scaffold protein; PI3P, phosphatidylinositol 3-phosphate; PI4P, phosphatidylinositol 4-phosphate; SCC, squamous cell carcinoma; Tks5, tyrosine kinase substrate with five Src homology 3 domains.
PI(4,5)P2 is the second major substrate of the PI 5-phosphatases to produce phosphatidylinositol 4-phosphate (13). PI(4,5)P2 can act as a signaling molecule in many biological mechanisms (e.g., endocytosis, exocytosis, migration, adhesion, ion channel control), and its content can be controlled by various members of the PI 5-phosphatase family. SHIP2, for example, can use PI(4,5)P2 in some glioblastoma cell lines, but in others it acts mainly on PI(3,4,5)P3 (14). The PI(4,5)P2 dephosphorylation pathway is tremendously important in some human genetic diseases such as Lowe syndrome, which results from OCRL mutations (15), or Joubert and MORM syndromes, which result from INPP5E mutations (16).
Moreover, PI 5-phosphatases interact with a very large number of proteins, thereby mediating noncatalytic properties or effectively targeting subcellular locations (i.e., plasma membrane, endosomes, lamellipodia, cilia, nucleus, etc.). Identifying these interactors has been very helpful in understanding the function of the different PI 5-phosphatases in cells (17). Focal adhesion intrinsic and associated proteins represent 232 human proteins (18): among the 10 different PI 5-phosphatases, only SHIP1 and SHIP2 have been reported as part of the focal adhesion intrinsic proteins or adhesome (see http://www.adhesome.org). Interestingly, most of the SHIP1/2 interactors such as filamin, Abl, Src, Cbl, or RhoA are also part of focal adhesion intrinsic proteins and participate in the control of cell migration, adhesion, and invasion (19–21).
SHIP1, which is very much expressed in immune cells, also influences downstream signaling via both phosphatase-dependent and -independent mechanisms (22). For example, in B cells, SHIP1 catalyzes an important production of PI(3,4)P2. Evidence has been provided that PI(3,4)P2-dependent processes may contribute to the therapeutic efficacy of PI 3-kinase inhibitors in B-cell malignancies (23). The role of SHIP1 is different in T cells, where it can have multiple functions [reviewed in (24)]. In the context of apoptosis, SHIP1 inhibits CD95/APO-1/Fas-induced apoptosis in primary T lymphocytes and T leukemic cells by promoting CD95 glycosylation (25). This activity of SHIP1 requires the SHIP1 Src homology 2 domain and probably occurs in the endoplasmic reticulum independently of its phosphatase activity. In this particular cell context, the inhibitory function for SHIP1 underscores the role of glycosylation in the regulation of CD95 signaling in T cells (25).
THE CONTROL OF PI(3,4,5)P3 BY PI 3- AND 5-PHOSPHATASES: THE IMPORTANCE AND IMPACT OF PTEN AND INPP4B
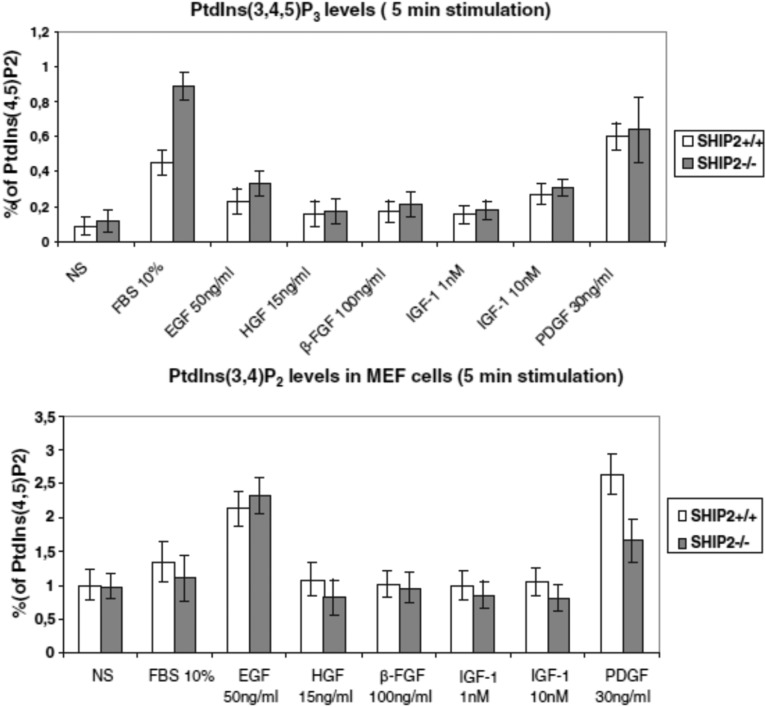
PI(3,4,5)P3 can be dephosphorylated in two very distinct pathways, PI 3-phosphatase PTEN and PI 5-phosphatases, to produce the very different signaling lipids PI(4,5)P2 and PI(3,4)P2, respectively (Fig. 2). Several PI 5-phosphatases can take PI(3,4,5)P3 as a substrate in an acellular assay of activity. This is the case for SHIP1/2, INPP5K, INPP5E, INPP5J, or SYNJ2 (Table 1). In intact cells, the flux of 5-dephosphorylation of PI(3,4,5)P3 is more complex and very much depends on precise enzyme localization and interaction with the membrane. This was very much studied for SHIP1/2. SHIP1 is a major phosphatase that controls the content of PI(3,4,5)P3 and PI(3,4)P2 in thrombin-stimulated platelets (26). In zebrafish, SHIP1 limits the motility of neutrophils and their recruitment to wounds. The effect appears to be mediated by PI(3,4,5)P3, which is located at the leading edge (27). By the use of a model of mouse embryonic fibroblasts (MEFs) deficient or not for SHIP2, PI(3,4,5)P3 was upregulated in SHIP2−/− cells compared with SHIP2+/+ cells (Fig. 3). This was shown in response to serum (10%) added for 5 or 10 min to the cells but not in starved cells (28). The data were obtained by labeling the PIs with phosphorus-32, lipid isolation, and deacylation. It is important to note that the PI(3,4)P2 measured in parallel in the same HPLC run was present but unchanged between +/+ and −/− cells at the same time points for serum stimulation (Fig. 3). This suggested that PI(3,4)P2 was rapidly dephosphorylated by PI 4-phosphatase INPP4A/B or PI 3-phosphatase PTEN. The only treatments that provided a significant increase of PI(3,4)P2 in MEFs were the addition of either platelet-derived growth factor (PDGF), epidermal growth factor (EGF), or H2O2 for 5–15 min to the cells (28, 29). This was due to the inactivation of both PTEN and INPP4A/B after the oxidation of the enzyme(s) at critical cysteine residues (30).
Fig. 3.
PI(3,4,5)P3 and PI(3,4)P2 levels in SHIP2+/+ and SHIP2−/− MEF cells. MEFs were stimulated for 5 min with 10% serum, 50 ng/ml EGF, 15 ng/ml HGF, 100 ng/ml β-FGF, 1 and 10 nM IGF-1, and 30 ng/ml PDGF. Data taken with permission from (28). HGF, hepatocyte growth factor; IGF-1, interleukin-like growth factor 1.
Through an siRNA approach directed against all PI phosphatases (INPP5B, INPP5E, INPP5J, INPP5K, SHIP1/2, SYNJ1/2, OCRL, and PTEN) expressed in Mcf10a cells, it appears that the only genetic manipulations that altered the PI(3,4,5)P3 response to EGF were knockdown or deletion of the PI phosphatases PTEN or SHIP2 (10). This was unexpected based on enzymatic activities determined on purified catalytic domains where SHIP2 had lower activity compared with INPP5B, OCRL, and SHIP2 (5). This suggests that the N- or C-terminal part of SHIP2, or both, are required for optimal activity. Again, the differences in PI(3,4,5)P3 content (measured by mass spectrometry) between siRNA-treated and wild-type cells were at best seen at short times of stimulation with EGF (1–5 min). This suggests that SHIP2 is a very potent PI(3,4,5)P3 5-phosphatase in this model either in being recruited to the EGF receptor (31) and/or displaying a very high Vmax/low Km compared with other PI 5-phosphatases. The effect is transient, suggesting that SHIP2 is only active at the plasma membrane for a short time, probably due to its localization upon stimulation to a fraction of the cell where PI(3,4,5)P3 is also present. This could be secondary to SHIP2 interaction with cytoskeletal proteins [actin-binding proteins such as filamin (19) or myosin 1c (32)] in plasma membrane-associated platforms and scaffold proteins (33). Furthermore, the stimulation of PI(3,4,5)P3 in response to EGF was very much amplified in cells in which both PTEN and SHIP2 were deleted (10). PI(3,4)P2 was also not very much different between wild-type and SHIP2/PTEN-deleted cells in response to EGF treatment. A minor and nonsignificant increase of PI(3,4)P2 was observed in SHIP2 knockdown cells compared with control cells. In contrast, PI(3,4)P2 was very much potentiated in response to EGF in both PTEN- and INPP4B-deleted cells compared with wild-type cells. The effect is synergistic between the two PI phosphatases and underscores the role of both enzymes in the control of PI(3,4)P2 when mutated together, particularly in cancer cells (10).
PI 5-PHOSPHATASES AND PI(3,4)P2, A NEW SIGNAL MOLECULE
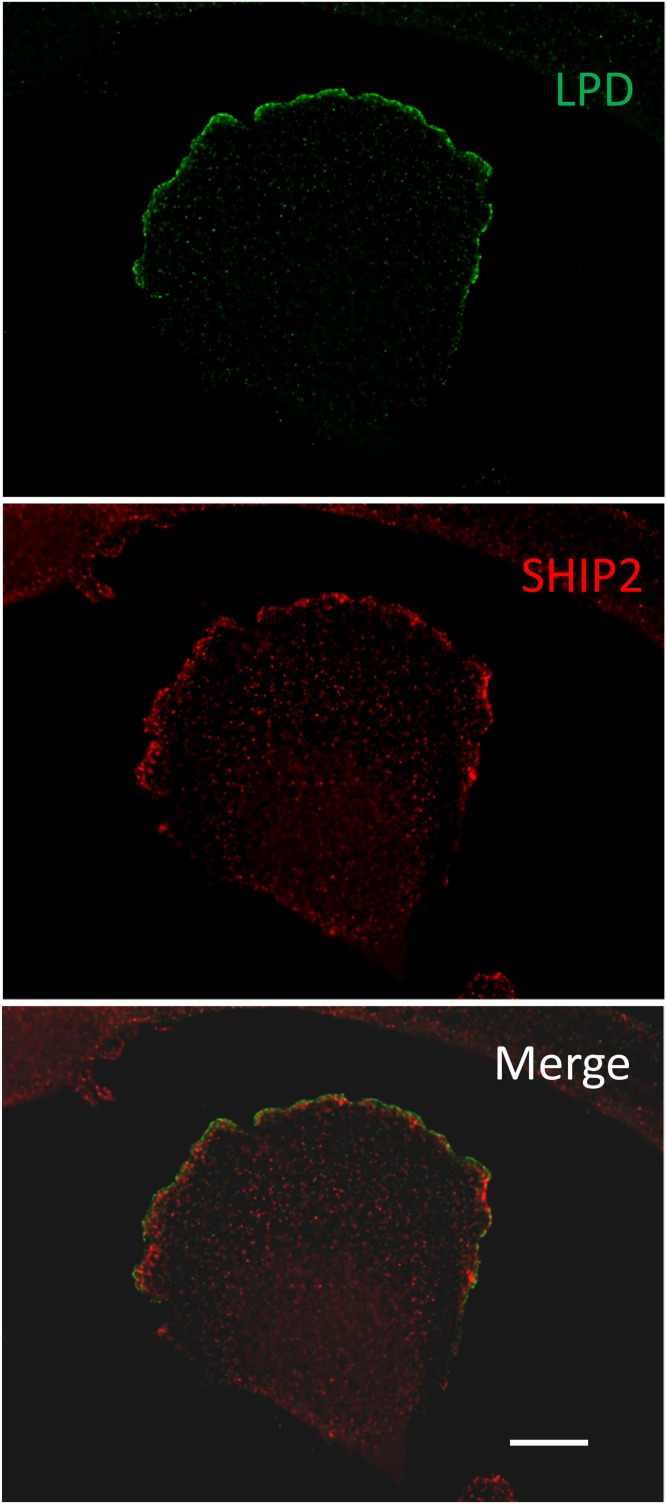
PI(3,4)P2 can be produced by PI 5-phosphatases such as SYNJ2, INPP5J, or SHIP1/2 (1–3). There is debate concerning whether PI(3,4)P2 contributes to protein kinase B (Akt) and downstream effector activation together with PI(3,4,5)P3. PI(3,4)P2 is able to interact with the pleckstrin homology (PH) domain of Akt as well as PI(3,4,5)P3 (34). Some proteins do interact specifically with PI(3,4)P2, such as tandem-PH-domain-containing proteins (TAPPs) 1 and 2 (35); PI(3,4)P2 also recruits lamellipodin (Ras association and PH domains 1) (36), tyrosine kinase substrate with five Src homology 3 domains at the invadopodium (37), or the sorting nexin 9 at late-stage endocytic intermediates (38). Based on evidence obtained in vivo, it has been proposed that the binding of TAPP1 and TAPP2 to PI(3,4)P2 provides a mechanism to downregulate the insulin-signaling and PI 3-kinase pathway (39). Interestingly, SHIP2 and lamellipodin partially colocalize in glioblastoma 1321 N1 cells, suggesting that both proteins could cooperate in PI(3,4)P2 signaling (C. Erneux, unpublished observations; Fig. 4). Moreover, diC8-PI(3,4)P2 addition to breast cancer MDA-MB-231 cells potentiated cell migration velocity and lamellipodia formation. This was not observed with its isomer diC8-PI(3,5)P2 (40). This attributes a signaling function to PI(3,4)P2 in cell migration.
Fig. 4.
SHIP2 and lamellipodin (LPD) colocalization in glioblastoma 1321 N1 cells. 1321 N1 cells were plated on coverslips and kept in culture in the presence of 10% serum for 24 h. The cells were fixed and stained with anti-lamellipodin in green (Alexa Fluor 488) and anti-SHIP2 (Novus) in red (Alexa Fluor 594). Images were obtained on Axioimager (Zeiss) at 100× 1.45 NA oil after deconvolution. Scale bar = 10 µm.
In the developing cerebral cortex, PI(3,4)P2, lamellipodin, and Ena/vasodilator-stimulated phosphoprotein regulate the dynamic morphology of multipolar migrating cells (41). In another study of neurite initiation, a positive role of PI(3,4)P2 in regulating actin aggregation and neuritogenesis has been proposed (42). Two enzymes, SHIP2 and class II PI 3-kinase α, are complementarily required for the production of PI(3,4)P2. Moreover, recently, novel second messenger functions of PI(3,4)P2 have been identified in the control of invadopodium precursor stabilization (43), feedback control of PI(3,4,5)P3 generation (44) in breast cancer cells, and basal mammalian target of rapamycin complex 1 activity in many different cells (45). Therefore, PI(3,4)P2 must be considered as a second messenger on its own not only in B cells and breast cancer cells but also in many other cells (11, 40). Its role could be particularly relevant in cancer cells in which INPP4B is mutated or absent, a situation that frequently occurs in aggressive hormone receptor-negative basal-like breast carcinomas (12).
CONSEQUENCES IN SIGNALING PATHWAYS: PI(3,4,5)P3 AND PI(4,5)P2 CONTENT, Akt AND Erk ACTIVITY, AND CELL SURVIVAL
PI(3,4,5)P3 and PI(4,5)P2 are major PI 5-phosphatase substrates that show signaling properties (Table 1, Fig. 2). Their content in membranes could be affected by PI phosphatase activities. OCRL is described as a PI(4,5)P2 5-phosphatase (Table 1), and PI(4,5)P2 levels have been shown to be very much increased in cell lines from kidney proximal tubules of a patient with Lowe syndrome compared with controls (46). OCRL localizes to the primary cilium and cilia from Lowe-patient fibroblasts, where it exhibits increased levels of PI(4,5)P2 (47). SHIP2 also controls plasma membrane PI(4,5)P2, thereby participating in the control of cell migration in 1321 N1 glioblastoma cells (48). PI(3,4,5)P3 is a positive effector of Akt activity (Fig. 2). In experiments performed in MEFs, Akt phosphorylation and activity were always upregulated in SHIP2-depleted cells compared with control cells when stimulated for a short period of time by serum (5–10 min; Fig. 5) (28, 49). This follows an upregulation of PI(3,4,5)P3 in MEFs at the same time points. Similar results on pAkt were also reported in human embryonic kidney cells in response to insulin-like growth factor 1 added for 5 min (50). pAkt upregulation in SHIP2-depleted cells is thus transient; it is not observed when cells are maintained in serum. In colorectal cancer cells, pAkt was rather decreased in SHIP2-depleted cells (51). This was observed as well in MDA-MB-231 breast cancer cells grown in serum alone (40).
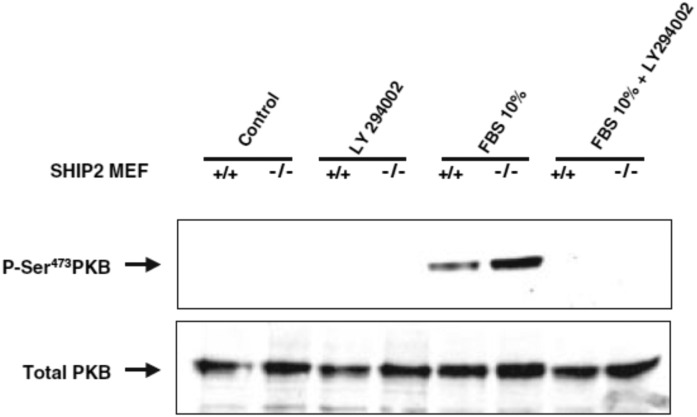
Fig. 5.
pAkt/PKB S473 level in SHIP2+/+ and SHIP2−/− MEF cells. MEF cells were stimulated in the presence of 10% serum for 5 min in the presence and absence of PI 3-kinase inhibitor LY-294002 (25 µM). Data were taken with permission from (28). PKB, protein kinase B.
Actually, many factors may influence the pAkt response and possible influence of both PI 5-phosphatases and PTEN activities. The existence of separate pools of PI 3-kinase-produced lipids in differentiating 3T3-L6 myoblasts has been proposed (52): a pool of nascent PI(3,4,5)P3 that is mainly dephosphorylated by PTEN and is able to activate Akt and a more stable pool that is dephosphorylated by SHIP2 that is unable to activate Akt. Knockdown of SHIP2 in this model did not affect pAkt but increased apoptotic cell death. The time course of stimulation by an agonist as discussed previously (e.g., in MEFs) may also play a role. In platelets, a striking correlation was observed between PI(3,4)P2 production and the tyrosine phosphorylation of SHIP1 upon thrombin stimulation (53). The data suggested that SHIP1 is only active for a short amount of time. The nature of stimuli added to cells and possible synergism between SHIP2 and PTEN or INPP4B that will increase PI(3,4,5)P3 and PI(3,4)P2 and pAkt as shown in Mcf10a cells may also be involved (10). In addition to Akt, pErk levels could also be modified in PI 5-phosphatase-deficient cells, e.g., for SHIP2 in 1321 N1 glioblastoma cells (49) or recently in a model of rat chondrosarcoma chondrocytes in fibroblast growth factor (FGF)-FGF receptor signaling (54). SHIP2 inhibition by the use of a specific competitive inhibitor diminishes the activation of both Akt- and Erk-signaling pathways in CD2-associated protein-deficient podocytes (55). This leads to an increase in apoptosis in podocytes with reduced expression of the CD2-associated protein (55). The depletion of SHIP2 in MDA-MB-231 cells or addition of a SHIP2 inhibitor in the same model resulted in a decrease in living cells (40). Therefore, changes in expression or activity of PI 5-phosphatases could affect cell survival in cells incubated in vitro or in xenograft mice in vivo (40).
INTERACTION BETWEEN PTEN OR PI 5-PHOSPHATASES AND THE PLASMA MEMBRANE
How do PI 5-phosphatases interact with their lipid substrate? None of the PI 5-phosphatases interact with the plasma membrane with only a PI substrate binding site. Some members of the family are attached to membranes by prenylation (INPP5B, INPP5E, or INPP5A; Fig. 1), a mechanism required to support activity (56). For other isoforms, the interaction also involves specific domains or sequences of the PI phosphatases. For OCRL and INPP5B (Fig. 1), a C-terminal Rho GTPase-activating protein-like domain participates with the 5-phosphatase catalytic domain in the interaction to form an arc below the membrane monolayer (57). For SHIP2, both the PI catalytic domain and a C2 domain interact with phosphatidylserine to facilitate the recognition of the substrate PI(3,4,5)P3 (58). The C2 domain also acts as an allosteric activator of PI phosphatase activity emanating from hydrophobic or polar interdomain interactions (58). Therefore, lipids at the plasma membrane could act as allosteric activators, a situation that also occurs for PI(4,5)P2 and the PI 3-phosphatase PTEN (59). In SHIP2, the addition of phosphatidylserine enhances catalytic activity, as measured with diC8-PI(3,4,5)P3 as a substrate (60). A similar effect has been reported for PTEN (61). Furthermore, the direct or indirect interaction of SHIP1/2 with immune or growth factor receptors (EphA2, FcγRIIb, EGF, or PDGF) has been reported and may also facilitate PI substrate recognition and phosphatase reaction (4, 17).
PI 5-PHOSPHATASE MUTATIONS ARGUE IN FAVOR OF SPECIFICITY OF PI 5-PHOSPHATASE ISOFORMS IN HUMANS
Could the distinct PI 5-phosphatase cooperate in metabolizing PIs, i.e., PI(4,5)P2 and PI(3,4,5)P3, or act separately in a given genetic or cellular context? Mutations of the PI 5-phosphatases have been reported in rare but very specific human genetic diseases (62). The INPPL1 gene that encodes SHIP2 has been found to be mutated in opsismodysplasia (MIM 258480), a rare autosomal-recessive disease characterized by growth-plate defects and delayed bone maturation (63, 64). Most of the mutations in INPPL1 lead to a premature stop codon at the N-terminal end or occur in the catalytic domain at positions that are crucial for the catalytic activity and therefore impair activity (65). One mutation was found in the C2 domain that controls activity (58). No mutations were found in the C-terminal proline-rich sequences and sterile α motif (SAM) domain (Fig. 6). Therefore, genetic evidence suggests that impaired SHIP2 catalytic activity, i.e., lack of dephosphorylation of PI(3,4,5)P3, plays a role in the phenotype. In humans, the loss of OCRL function results in the X-linked oculocerebrorenal syndrome of Lowe and type 2 Dent disease (MIM 309000) (66, 67). Mutations occurred in the catalytic domain of the protein. The same phenomenon occurs for skeletal muscle- and kidney-enriched inositol phosphatase (SKIP; also referred to as INPP5K) (Fig. 1): recessive mutations in INPP5K (MIM 607875) have been reported in a syndrome overlapping both the dystroglycanopathy and the Marinesco-Sjögren spectrum (68). Those rare mutations also impair catalytic activity. Moreover, in the case of the worm orthologue of INPP5K, the same mutations could also affect the fine control of the endoplasmic reticulum network organization (69). Thus, genetic evidence of very specific human diseases argues against the redundancy between PI 5-phosphatases and suggests that in a genetic context PI 5-phosphatase could have very specific and individual functions.
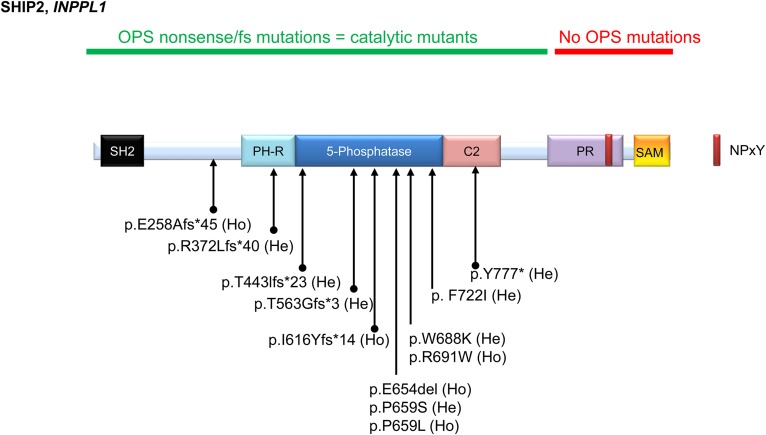
Fig. 6.
INPPL1 (gene that encodes SHIP2) mutated in OPS (MIM 258480). The INPPL1 sequence is accessible as NCBI reference sequence NM_001567.3. SHIP2 domains in the human sequence (1258 amino acids) are shown in Fig. 1. Homozygous or compound heterozygous mutations have been reported in p.E654del(Ho) (97); p.R691W(Ho), p.P659L(Ho), p.T563Gfs*(He), and p.E258Afs*45(Ho) (63); p.F722I(He), p.W688K(He), p.P659S(He), p.T443Ifs*23(He), and p.I616Yfs*14(Ho) (64); and p.Y777*(He) and p.R372Lfs*40(He) (98). So far, most mutations have been shown in the N-terminal and catalytic domain. No mutations have yet to be found in the C-terminal PR and SAM domain. He, heterozygous; Ho, homozygous; OPS, opsismodysplasia; PH-R, PH-related; PR, proline-rich; SH2, Src homology 2.
COMPENSATION BETWEEN DIFFERENT PI 5-PHOSPHATASES IN CELLS DEPENDS ON THE CELL CONTEXT
OCRL encodes a very active PI(4,5)P2 5-phosphatase (Table 1). In terms of structure, INPP5B is very close to OCRL, with approximately 45% sequence identity (62). Mice deficient for OCRL do not have the characteristics of a Lowe syndrome, but double-knockout mice for OCRL and INPP5B are embryonic lethal (70). Thus, in mice, OCRL and INPP5B must cooperate in function. In humans, this appears not to be the case. Recent studies have shown that PI 5-phosphatase compensation may occur in Mcf10a cells deficient for PTEN and SHIP2. The content of PI(3,4,5)P3 was particularly high in the absence of both PTEN and SHIP2 compared with wild-type, PTEN-knockout, or SHIP2-knockdown cells (10). Evidence of PI(3,4)P2 formation has been presented to suggest that another phosphatase (or phosphatases) must compensate for the loss of SHIP2 and PTEN in this particular model and dephosphorylate PI(3,4,5)P3 in EGF-stimulated cells (10). Whether this is applicable to other cells or depends only on some growth factors, or both, remain open questions. The data underscore the difference between measuring PI phosphatase activity on purified enzymes and its activity in intact cells in agonist-stimulated cells.
PI 5-PHOSPHATASES: TUMOR PROMOTORS OR TUMOR SUPPRESSORS?
A tumor promotor or suppressor role of the different PI 5-phosphatases in different cancer cells have been reported. For example, in breast cancer MDA-MB-231 cells as a model, SYNJ2 has been presented as an oncogene (71). The overexpression of SYNJ2 correlated with a shorter survival of breast cancer patients. In xenograft mice, the catalytic activity of SYNJ2 promotes both tumorigenic growth and metastatic spread. It was proposed that SYNJ2’s oncogenic activity relates to its ability to dephosphorylate PI(3,4,5)P3, producing PI(3,4)P2, which acts at key steps of both invadopodia and lamellipodia formation (71). In U87-MG glioblastoma cells, siRNA depletion of SYNJ2 also inhibits invasion with the involvement of Rac1 (72). In contrast to the data obtained with SYNJ2, the depletion of INPP5J in MDA-MB-231 cells increases breast cancer cell transformation (suggesting tumor-suppressive activity) but reduces cell migration, invasion, and metastasis by a mechanism that is driven by Akt1 (73).
In blood cancer cells expressing SHIP1, the addition of a SHIP1-specific inhibitor, 3AC, decreases pAkt and promotes apoptosis (74). When added to the cells pretreated with 3AC, PI(3,4)P2 increases cell survival, raising the possibility that the SHIP1 reaction product is part of a survival mechanism in this model. Altogether, the data suggest that SHIP1 inhibition and its impact on pAkt could be seen as a strategy to decrease the survival of hematologic malignancies. In esophageal squamous cell carcinoma, miR-508 suppresses multiple PI phosphatases, in particular INPP5J, leading to constitutive activation of PI 3-kinase/Akt and inducing an aggressive phenotype of those tumor cells (75). SHIP2 has both oncogenic and tumor-suppressive effects depending on the cell context. In glioblastoma and squamous cell carcinoma, it shows tumor-suppressive functions that are also mediated at least in part by Akt and the control of survival (76), whereas in some breast cancer cells it shows tumor-promoting activity (40, 77). Because high expression of SHIP2 was found in very aggressive human breast cancer samples such as triple-negative breast cancer or colorectal cancer, SHIP2 was proposed as oncogenic in these models (77–79). SHIP2 also played an essential oncogenic role for the maintenance of a subpopulation of breast cancer stem cells and tumorigenicity in vivo through c-Jun N-terminal kinase 1/vimentin activation (80).
INHIBITORS OF PI 5-PHOSPHATASES
PI 5-phosphatase inhibitors SYNJ1, OCRL, and SHIP1/2 have been identified by multiple screening strategies (81–83). They represent valuable tools for probing PI phosphatase function in cell models but also in vivo. SHIP1 inhibitors have been proven to be effective in treating human multiple myeloma and acute lymphoblastic leukemia in vivo using severe combined immunodeficiency xenogeneic cancer models (84). More recently, the use of a SHIP1-selective inhibitor in mice and in vivo, referred to as 3AC, increased both natural killer and T-cell responsiveness and reduced the growth of hematological and solid tumors (85). Because the effect is lost when the T-cell compartment lacks SHIP1, the effect appears to be SHIP1-specific or requires at least the in vivo interaction between SHIP1 and the drug. A SHIP2 inhibitor injected in mice was not efficient in providing the same protection, suggesting nonredundancy between SHIP1 and SHIP2 in the T cells of these mice and that selective SHIP1 inhibitors have to be used to promote antitumor properties. It is equally possible that SHIP1 is much more expressed or active in T cells compared with SHIP2 or other PI 5-phosphatases, as proposed in platelets (26). SHIP2 modulators could also be useful in the treatment of some cancers, e.g., in breast cancer and colorectal cancer in PTEN/INPP4B-deficient cells. Considering the role of SHIP2 in migration and invasion particularly, in some breast cancer cell types (86), or colorectal cancer (51), inhibitors of SHIP2 could be useful as an additional therapy in preventing or controlling cell migration and metastasis by inhibiting PI(3,4)P2 signaling (40). This could be particularly important in estrogen receptor-negative breast cancers, in which SHIP2 phosphatase activity has been suggested to modulate tumorigenicity in vivo (80). The same approach could be of interest in the treatment of other diseases. For example, recent studies have provided evidence that potent SHIP2 inhibitors would provide effective treatment options for Alzheimer’s disease (87). The substrate lipid binding site may thus provide alternatives for the synthesis of effective drugs targeting the PI 5-phosphatases.
Acknowledgments
The authors thank Matthias Krause for providing the anti-lamellipodin antibody.
Footnotes
Abbreviations:
- Akt
- protein kinase B
- EGF
- epidermal growth factor
- FGF
- fibroblast growth factor
- INPP5P
- phosphoinositide 5-phosphatase
- Ins(1,3,4,5)P4
- inositol 1,3,4,5-tetrakisphosphate
- Ins(1,4,5)P3
- inositol 1,4,5-trisphosphate
- MEF
- mouse embryonic fibroblast
- OCRL
- oculocerebrorenal syndrome of Lowe
- PDGF
- platelet-derived growth factor
- PH
- pleckstrin homology
- PI
- phosphoinositide
- PTEN
- phosphatase and tensin homolog
- SAM
- sterile α motif
- SKIP
- skeletal muscle- and kidney-enriched inositol phosphatase
- SYNJ
- synaptojanin
- TAPP
- tandem-pleckstrin-homology-domain-containing protein
This work was supported by Fonds de la Recherche Scientifique Médicale Grant J.0078.18, the Université Libre de Bruxelles, and Télévie. A.R.R. is supported by a grant from Télévie. S.G. is supported by fellowships from Fondation Rose et Jean Hoguet, Fonds Lekime-Ropsy, and Télévie.
REFERENCES
- 1.Balla T. 2013. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93: 1019–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edimo W. E., Janssens V., Waelkens E., and Erneux C.. 2012. Reversible Ser/Thr SHIP phosphorylation: a new paradigm in phosphoinositide signalling? BioEssays. 34: 634–642. [DOI] [PubMed] [Google Scholar]
- 3.Eramo M. J., and Mitchell C. A.. 2016. Regulation of PtdIns(3,4,5)P3/Akt signalling by inositol polyphosphate 5-phosphatases. Biochem. Soc. Trans. 44: 240–252. [DOI] [PubMed] [Google Scholar]
- 4.Blero D., Payrastre B., Schurmans S., and Erneux C.. 2007. Phosphoinositide phosphatases in a network of signalling reactions. Pflugers Arch. 455: 31–44. [DOI] [PubMed] [Google Scholar]
- 5.Trésaugues L., Silvander C., Flodin S., Welin M., Nyman T., Graslund S., Hammarstrom M., Berglund H., and Nordlund P.. 2014. Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases. Structure. 22: 744–755. [DOI] [PubMed] [Google Scholar]
- 6.Verjans B., De Smedt F., Lecocq R., Vanweyenberg V., Moreau C., and Erneux C.. 1994. Cloning and expression in Escherichia coli of a dog thyroid cDNA encoding a novel inositol 1,4,5-trisphosphate 5-phosphatase. Biochem. J. 300: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erneux C., Edimo W. E., Deneubourg L., and Pirson I.. 2011. SHIP2 multiple functions: a balance between a negative control of PtdIns(3,4,5)P3 level, a positive control of PtdIns(3,4)P2 production, and intrinsic docking properties. J. Cell. Biochem. 112: 2203–2209. [DOI] [PubMed] [Google Scholar]
- 8.Erneux C., Ghosh S., Ramos A. R., and Edimo W. E.. 2016. New functions of the inositol polyphosphate 5-phosphatases in cancer. Curr. Pharm. Des. 22: 2309–2314. [DOI] [PubMed] [Google Scholar]
- 9.Toker A., and Rameh L.. 2015. PIPPing on AKT1: How Many Phosphatases Does It Take to Turn off PI3K? Cancer Cell. 28: 143–145. [DOI] [PubMed] [Google Scholar]
- 10.Malek M., Kielkowska A., Chessa T., Anderson K. E., Barneda D., Pir P., Nakanishi H., Eguchi S., Koizumi A., Sasaki J., et al. . 2017. PTEN regulates PI(3,4)P2 signaling downstream of class I PI3K. Mol. Cell. 68: 566–580.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., and Marshall A. J.. 2015. Phosphatidylinositol (3,4) bisphosphate-specific phosphatases and effector proteins: A distinct branch of PI3K signaling. Cell. Signal. 27: 1789–1798. [DOI] [PubMed] [Google Scholar]
- 12.Fedele C. G., Ooms L. M., Ho M., Vieusseux J., O’Toole S. A., Millar E. K., Lopez-Knowles E., Sriratana A., Gurung R., Baglietto L., et al. . 2010. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc. Natl. Acad. Sci. USA. 107: 22231–22236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thapa N., Tan X., Choi S., Lambert P. F., Rapraeger A. C., and Anderson R. A.. 2016. The Hidden Conundrum of Phosphoinositide Signaling in Cancer. Trends Cancer. 2: 378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos A. R., Elong E. W., and Erneux C.. 2018. Phosphoinositide 5-phosphatase activities control cell motility in glioblastoma: Two phosphoinositides PI(4,5)P2 and PI(3,4)P2 are involved. Adv. Biol. Regul. 67: 40–48. [DOI] [PubMed] [Google Scholar]
- 15.De Matteis M. A., Staiano L., Emma F., and Devuyst O.. 2017. The 5-phosphatase OCRL in Lowe syndrome and Dent disease 2. Nat. Rev. Nephrol. 13: 455–470. [DOI] [PubMed] [Google Scholar]
- 16.Chávez M., Ena S., Van S. J., de Kerchove A., Schurmans S., and Schiffmann S. N.. 2015. Modulation of ciliary phosphoinositide content regulates trafficking and sonic hedgehog signaling output. Dev. Cell. 34: 338–350. [DOI] [PubMed] [Google Scholar]
- 17.Xie J., Erneux C., and Pirson I.. 2013. How does SHIP1/2 balance PtdIns(3,4)P2 and does it signal independently of its phosphatase activity? BioEssays. 35: 733–743. [DOI] [PubMed] [Google Scholar]
- 18.Winograd-Katz S. E., Fassler R., Geiger B., and Legate K. R.. 2014. The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 15: 273–288. [DOI] [PubMed] [Google Scholar]
- 19.Dyson J. M., O’Malley C. J., Becanovic J., Munday A. D., Berndt M. C., Coghill I. D., Nandurkar H. H., Ooms L. M., and Mitchell C. A.. 2001. The SH2-containing inositol polyphosphate 5-phosphatase, SHIP-2, binds filamin and regulates submembraneous actin. J. Cell Biol. 155: 1065–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato K., Yazawa T., Taki K., Mori K., Wang S., Nishioka T., Hamaguchi T., Itoh T., Takenawa T., Kataoka C., et al. . 2012. The inositol 5-phosphatase SHIP2 is an effector of RhoA and is involved in cell polarity and migration. Mol. Biol. Cell. 23: 2593–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandenbroere I., Paternotte N., Dumont J. E., Erneux C., and Pirson I.. 2003. The c-Cbl-associated protein and c-Cbl are two new partners of the SH2-containing inositol polyphosphate 5-phosphatase SHIP2. Biochem. Biophys. Res. Commun. 300: 494–500. [DOI] [PubMed] [Google Scholar]
- 22.Pauls S. D., and Marshall A. J.. 2017. Regulation of immune cell signaling by SHIP1: A phosphatase, scaffold protein, and potential therapeutic target. Eur. J. Immunol. 47: 932–945. [DOI] [PubMed] [Google Scholar]
- 23.Li H., Wu X., Hou S., Malek M., Kielkowska A., Noh E., Makondo K. J., Du Q., Wilkins J. A., Johnston J. B., et al. . 2016. Phosphatidylinositol-3,4-bisphosphate and its binding protein lamellipodin regulate chemotaxis of malignant B lymphocytes. J. Immunol. 196: 586–595. [DOI] [PubMed] [Google Scholar]
- 24.Srivastava N., Sudan R., and Kerr W. G.. 2013. Role of inositol poly-phosphatases and their targets in T cell biology. Front. Immunol. 4: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charlier E., Conde C., Zhang J., Deneubourg L., Di V. E., Rahmouni S., Chariot A., Agostinis P., Pang P. C., Haslam S. M., et al. . 2010. SHIP-1 inhibits CD95/APO-1/Fas-induced apoptosis in primary T lymphocytes and T leukemic cells by promoting CD95 glycosylation independently of its phosphatase activity. Leukemia. 24: 821–832. [DOI] [PubMed] [Google Scholar]
- 26.Giuriato S., Pesesse X., Bodin S., Sasaki T., Viala C., Marion E., Penninger J., Schurmans S., Erneux C., and Payrastre B.. 2003. SH2-containing inositol 5-phosphatases 1 and 2 in blood platelets: their interactions and roles in the control of phosphatidylinositol 3,4,5-trisphosphate levels. Biochem. J. 376: 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam P. Y., Yoo S. K., Green J. M., and Huttenlocher A.. 2012. The SH2-domain-containing inositol 5-phosphatase (SHIP) limits the motility of neutrophils and their recruitment to wounds in zebrafish. J. Cell Sci. 125: 4973–4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blero D., Zhang J., Pesesse X., Payrastre B., Dumont J. E., Schurmans S., and Erneux C.. 2005. Phosphatidylinositol 3,4,5-trisphosphate modulation in SHIP2-deficient mouse embryonic fibroblasts. FEBS J. 272: 2512–2522. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J., Liu Z., Rasschaert J., Blero D., Deneubourg L., Schurmans S., Erneux C., and Pesesse X.. 2007. SHIP2 controls PtdIns(3,4,5)P(3) levels and PKB activity in response to oxidative stress. Cell. Signal. 19: 2194–2200. [DOI] [PubMed] [Google Scholar]
- 30.Leslie N. R., Bennett D., Lindsay Y. E., Stewart H., Gray A., and Downes C. P.. 2003. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J. 22: 5501–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pesesse X., Dewaste V., De Smedt F., Laffargue M., Giuriato S., Moreau C., Payrastre B., and Erneux C.. 2001. The Src homology 2 domain containing inositol 5-phosphatase SHIP2 is recruited to the epidermal growth factor (EGF) receptor and dephosphorylates phosphatidylinositol 3,4,5-trisphosphate in EGF- stimulated COS-7 cells. J. Biol. Chem. 276: 28348–28355. [DOI] [PubMed] [Google Scholar]
- 32.Edimo W. E., Ramos A. R., Ghosh S., Vanderwinden J. M., and Erneux C.. 2016. The SHIP2 interactor Myo1c is required for cell migration in 1321 N1 glioblastoma cells. Biochem. Biophys. Res. Commun. 476: 508–514. [DOI] [PubMed] [Google Scholar]
- 33.Astro V., and I. de Curtis. 2015. Plasma membrane-associated platforms: dynamic scaffolds that organize membrane-associated events. Sci. Signal. 8: re1. [DOI] [PubMed] [Google Scholar]
- 34.Franke T. F., Kaplan D. R., Cantley L. C., and Toker A.. 1997. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 275: 665–668. [DOI] [PubMed] [Google Scholar]
- 35.Kimber W. A., Trinkle-Mulcahy L., Cheung P. C., Deak M., Marsden L. J., Kieloch A., Watt S., Javier R. T., Gray A., Downes C. P., et al. . 2002. Evidence that the tandem-pleckstrin-homology-domain-containing protein TAPP1 interacts with Ptd(3,4)P2 and the multi-PDZ-domain-containing protein MUPP1 in vivo. Biochem. J. 361: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krause M., Leslie J. D., Stewart M., Lafuente E. M., Valderrama F., Jagannathan R., Strasser G. A., Rubinson D. A., Liu H., Way M., et al. . 2004. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell. 7: 571–583. [DOI] [PubMed] [Google Scholar]
- 37.Sharma V. P., Eddy R., Entenberg D., Kai M., Gertler F. B., and Condeelis J.. 2013. Tks5 and SHIP2 regulate invadopodium maturation, but not initiation, in breast carcinoma cells. Curr. Biol. 23: 2079–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ketel K., Krauss M., Nicot A. S., Puchkov D., Wieffer M., Muller R., Subramanian D., Schultz C., Laporte J., and Haucke V.. 2016. A phosphoinositide conversion mechanism for exit from endosomes. Nature. 529: 408–412. [DOI] [PubMed] [Google Scholar]
- 39.Wullschleger S., Wasserman D. H., Gray A., Sakamoto K., and Alessi D. R.. 2011. Role of TAPP1 and TAPP2 adaptors binding to PtdIns(3,4)P2 in regulating insulin sensitivity defined by knock-in analysis. Biochem. J. 434: 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosh S., Scozzaro S., Ramos A. R., Delcambre S., Chevalier C., Krejci P., and Erneux C.. 2018. Inhibition of SHIP2 activity inhibits cell migration and could prevent metastasis in breast cancer cells. J. Cell Sci. 131(16). [DOI] [PubMed] [Google Scholar]
- 41.Yoshinaga S., Ohkubo T., Sasaki S., Nuriya M., Ogawa Y., Yasui M., Tabata H., and Nakajima K.. 2012. A phosphatidylinositol lipids system, lamellipodin, and Ena/VASP regulate dynamic morphology of multipolar migrating cells in the developing cerebral cortex. J. Neurosci. 32: 11643–11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang S. X., Duan L. H., He S. J., Zhuang G. F., and Yu X.. 2017. Phosphatidylinositol 3,4-bisphosphate regulates neurite initiation and dendrite morphogenesis via actin aggregation. Cell Res. 27: 253–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eddy R. J., Weidmann M. D., Sharma V. P., and Condeelis J. S.. 2017. Tumor cell invadopodia: invasive protrusions that orchestrate metastasis. Trends Cell Biol. 27: 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reed D. E., and Shokat K. M.. 2017. INPP4B and PTEN loss leads to PI-3,4–P2 accumulation and inhibition of PI3K in TNBC. Mol. Cancer Res. 15: 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marat A. L., Wallroth A., Lo W. T., Muller R., Norata G. D., Falasca M., Schultz C., and Haucke V.. 2017. mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science. 356: 968–972. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X., Hartz P. A., Philip E., Racusen L. C., and Majerus P. W.. 1998. Cell lines from kidney proximal tubules of a patient with Lowe syndrome lack OCRL inositol polyphosphate 5-phosphatase and accumulate phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 273: 1574–1582. [DOI] [PubMed] [Google Scholar]
- 47.Prosseda P. P., Luo N., Wang B., Alvarado J. A., Hu Y., and Sun Y.. 2017. Loss of OCRL increases ciliary PI(4,5)P2 in Lowe oculocerebrorenal syndrome. J. Cell Sci. 130: 3447–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elong Edimo W., Ghosh S., Derua R., Janssens V., Waelkens E., Vanderwinden J. M., Robe P., and Erneux C.. 2016. SHIP2 controls plasma membrane PI(4,5)P2 thereby participating in the control of cell migration in 1321 N1 glioblastoma. J. Cell Sci. 129: 1101–1114. [DOI] [PubMed] [Google Scholar]
- 49.Elong Edimo W., Derua R., Janssens V., Nakamura T., Vanderwinden J. M., Waelkens E., and Erneux C.. 2011. Evidence of SHIP2 S132 phosphorylation, its nuclear localization and stability. Biochem. J. 439: 391–401. [DOI] [PubMed] [Google Scholar]
- 50.Malik N., Macartney T., Hornberger A., Anderson K. E., Tovell H., Prescott A. R., and Alessi D. R.. 2018. Mechanism of activation of SGK3 by growth factors via the class 1 and class 3 PI3Ks. Biochem. J. 475: 117–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoekstra E., Das A. M., Willemsen M., Swets M., Kuppen P. J., van der Woude C. J., Bruno M. J., Shah J. P., Ten Hagen T. L., Chisholm J. D., et al. . 2016. Lipid phosphatase SHIP2 functions as oncogene in colorectal cancer by regulating PKB activation. Oncotarget. 7: 73525–73540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mandl A., Sarkes D., Carricaburu V., Jung V., and Rameh L.. 2007. Serum withdrawal-induced accumulation of phosphoinositide 3-kinase lipids in differentiating 3T3–L6 myoblasts: distinct roles for Ship2 and PTEN. Mol. Cell. Biol. 27: 8098–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giuriato S., Payrastre B., Drayer A. L., Plantavid M., Woscholski R., Parker P., Erneux C., and Chap H.. 1997. Tyrosine phosphorylation and relocation of SHIP are integrin-mediated in thrombin-stimulated human blood platelets. J. Biol. Chem. 272: 26857–26863. [DOI] [PubMed] [Google Scholar]
- 54.Fafilek B., Balek L., Kunova Bosakova M., Varecha M., Nita A., Gregor T., Gudemova I., Krenova J., Ghosh S., Piscacek M., et al. . 2018. Inositol phosphatase SHIP2 enables sustained MAP kinase activation by fibroblast growth factor via recruiting SRC kinases to the FGF-receptor signaling complex. Sci. Signal. 11(548). [DOI] [PubMed] [Google Scholar]
- 55.Saurus P., Tolvanen T. A., Lindfors S., Kuusela S., Holthofer H., Lehtonen E., and Lehtonen S.. 2017. Inhibition of SHIP2 in CD2AP-deficient podocytes ameliorates reactive oxygen species generation but aggravates apoptosis. Sci. Rep. 7: 10731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Smedt F., Missiaen L., Parys J. B., Vanweyenberg V., De Smedt H., and Erneux C.. 1997. Isoprenylated human brain type I inositol 1,4,5-trisphosphate 5- phosphatase controls Ca2+ oscillations induced by ATP in Chinese hamster ovary cells. J. Biol. Chem. 272: 17367–17375. [DOI] [PubMed] [Google Scholar]
- 57.Erdmann K. S., Mao Y., McCrea H. J., Zoncu R., Lee S., Paradise S., Modregger J., Biemesderfer D., Toomre D., and De Camilli C. P.. 2007. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev. Cell. 13: 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Le Coq J., Camacho-Artacho M., Velazquez J. V., Santiveri C. M., Gallego L. H., Campos-Olivas R., Dolker N., and Lietha D.. 2017. Structural basis for interdomain communication in SHIP2 providing high phosphatase activity. eLife. 6: e26640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker S. M., Leslie N. R., Perera N. M., Batty I. H., and Downes C. P.. 2004. The tumour-suppressor function of PTEN requires an N-terminal lipid-binding motif. Biochem. J. 379: 301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vandeput F., Backers K., Villeret V., Pesesse X., and Erneux C.. 2006. The influence of anionic lipids on SHIP2 phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase activity. Cell. Signal. 18: 2193–2199. [DOI] [PubMed] [Google Scholar]
- 61.McConnachie G., Pass I., Walker S. M., and Downes C. P.. 2003. Interfacial kinetic analysis of the tumour suppressor phosphatase, PTEN: evidence for activation by anionic phospholipids. Biochem. J. 371: 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pirruccello M., and De Camilli P.. 2012. Inositol 5-phosphatases: insights from the Lowe syndrome protein OCRL. Trends Biochem. Sci. 37: 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Below J. E., Earl D. L., Shively K. M., McMillin M. J., Smith J. D., Turner E. H., Stephan M. J., Al-Gazali L. I., Hertecant J. L., Chitayat D., et al. . 2013. Whole-genome analysis reveals that mutations in inositol polyphosphate phosphatase-like 1 cause opsismodysplasia. Am. J. Hum. Genet. 92: 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huber C., Faqeih E. A., Bartholdi D., Bole-Feysot C., Borochowitz Z., Cavalcanti D. P., Frigo A., Nitschke P., Roume J., Santos H. G., et al. . 2013. Exome sequencing identifies INPPL1 mutations as a cause of opsismodysplasia. Am. J. Hum. Genet. 92: 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fradet A., and Fitzgerald J.. 2017. INPPL1 gene mutations in opsismodysplasia. J. Hum. Genet. 62: 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Attree O., Olivos I. M., Okabe I., Bailey L. C., Nelson D. L., Lewis R. A., McInnes R. R., and Nussbaum R. L.. 1992. The Lowe’s oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature. 358: 239–242. [DOI] [PubMed] [Google Scholar]
- 67.Bökenkamp A., Bockenhauer D., Cheong H. I., Hoppe B., Tasic V., Unwin R., and Ludwig M.. 2009. Dent-2 disease: a mild variant of Lowe syndrome. J. Pediatr. 155: 94–99. [DOI] [PubMed] [Google Scholar]
- 68.Osborn D. P., Pond H. L., Mazaheri N., Dejardin J., Munn C. J., Mushref K., Cauley E. S., Moroni I., Pasanisi M. B., Sellars E. A., et al. . 2017. Mutations in INPP5K cause a form of congenital muscular dystrophy overlapping Marinesco-Sjogren syndrome and dystroglycanopathy. Am. J. Hum. Genet. 100: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong R., Zhu T., Benedetti L., Gowrishankar S., Deng H., Cai Y., Wang X., Shen K., and De Camilli C. P.. 2018. The inositol 5-phosphatase INPP5K participates in the fine control of ER organization. J. Cell Biol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Inoue K., Balkin D. M., Liu L., Nandez R., Wu Y., Tian X., Wang T., Nussbaum R., De C. P., and Ishibe S.. 2017. Kidney tubular ablation of Ocrl/Inpp5b phenocopies Lowe syndrome tubulopathy. J. Am. Soc. Nephrol. 28: 1399–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ben-Chetrit N., Chetrit D., Russell R., Korner C., Mancini M., Abdul-Hai A., Itkin T., Carvalho S., Cohen-Dvashi H., Koestler W. J., et al. . 2015. Synaptojanin 2 is a druggable mediator of metastasis and the gene is overexpressed and amplified in breast cancer. Sci. Signal. 8: ra7. [DOI] [PubMed] [Google Scholar]
- 72.Chuang Y. Y., Tran N. L., Rusk N., Nakada M., Berens M. E., and Symons M.. 2004. Role of synaptojanin 2 in glioma cell migration and invasion. Cancer Res. 64: 8271–8275. [DOI] [PubMed] [Google Scholar]
- 73.Ooms L. M., Binge L. C., Davies E. M., Rahman P., Conway J. R., Gurung R., Ferguson D. T., Papa A., Fedele C. G., Vieusseux J. L., et al. . 2015. The inositol polyphosphate 5-phosphatase PIPP regulates AKT1-dependent breast cancer growth and metastasis. Cancer Cell. 28: 155–169. [DOI] [PubMed] [Google Scholar]
- 74.Brooks R., Fuhler G. M., Iyer S., Smith M. J., Park M. Y., Paraiso K. H., Engelman R. W., and Kerr W. G.. 2010. SHIP1 inhibition increases immunoregulatory capacity and triggers apoptosis of hematopoietic cancer cells. J. Immunol. 184: 3582–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin C., Liu A., Zhu J., Zhang X., Wu G., Ren P., Wu J., Li M., Li J., and Song L.. 2014. miR-508 sustains phosphoinositide signalling and promotes aggressive phenotype of oesophageal squamous cell carcinoma. Nat. Commun. 5: 4620. [DOI] [PubMed] [Google Scholar]
- 76.Yu J., Ryan D. G., Getsios S., Oliveira-Fernandes M., Fatima A., and Lavker R. M.. 2008. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc. Natl. Acad. Sci. USA. 105: 19300–19305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prasad N. K., Tandon M., Badve S., Snyder P. W., and Nakshatri H.. 2008. Phosphoinositol phosphatase SHIP2 promotes cancer development and metastasis coupled with alterations in EGF receptor turnover. Carcinogenesis. 29: 25–34. [DOI] [PubMed] [Google Scholar]
- 78.Fuhler G. M., Brooks R., Toms B., Iyer S., Gengo E. A., Park M. Y., Gumbleton M., Viernes D. R., Chisholm J. D., and Kerr W. G.. 2012. Therapeutic potential of SH2 domain-containing inositol-5′-phosphatase 1 (SHIP1) and SHIP2 inhibition in cancer. Mol. Med. 18: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prasad N. K. 2009. SHIP2 phosphoinositol phosphatase positively regulates EGFR-Akt pathway, CXCR4 expression, and cell migration in MDA-MB-231 breast cancer cells. Int. J. Oncol. 34: 97–105. [PubMed] [Google Scholar]
- 80.Fu C. H., Lin R. J., Yu J., Chang W. W., Liao G. S., Chang W. Y., Tseng L. M., Tsai Y. F., Yu J. C., and Yu A. L.. 2014. A novel oncogenic role of inositol phosphatase SHIP2 in ER-negative breast cancer stem cells: involvement of JNK/vimentin activation. Stem Cells. 32: 2048–2060. [DOI] [PubMed] [Google Scholar]
- 81.Pirruccello M., Nandez R., Idevall-Hagren O., Alcazar-Roman A., Abriola L., Berwick S. A., Lucast L., Morel D., and De C. P.. 2014. Identification of inhibitors of inositol 5-phosphatases through multiple screening strategies. ACS Chem. Biol. 9: 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suwa A., Yamamoto T., Sawada A., Minoura K., Hosogai N., Tahara A., Kurama T., Shimokawa T., and Aramori I.. 2009. Discovery and functional characterization of a novel small molecule inhibitor of the intracellular phosphatase, SHIP2. Br. J. Pharmacol. 158: 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Viernes D. R., Choi L. B., Kerr W. G., and Chisholm J. D.. 2014. Discovery and development of small molecule SHIP phosphatase modulators. Med. Res. Rev. 34: 795–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Z., Shojaee S., Buchner M., Geng H., Lee J. W., Klemm L., Titz B., Graeber T. G., Park E., Tan Y. X., et al. . 2015. Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature. 521: 357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gumbleton M., Sudan R., Fernandes S., Engelman R. W., Russo C. M., Chisholm J. D., and Kerr W. G.. 2017. Dual enhancement of T and NK cell function by pulsatile inhibition of SHIP1 improves antitumor immunity and survival. Sci. Signal. 10: eaam5353. [DOI] [PubMed] [Google Scholar]
- 86.Rajadurai C. V., Havrylov S., Coelho P. P., Ratcliffe C. D., Zaoui K., Huang B. H., Monast A., Chughtai N., Sangwan V., Gertler F. B., et al. . 2016. 5′-Inositol phosphatase SHIP2 recruits Mena to stabilize invadopodia for cancer cell invasion. J. Cell Biol. 214: 719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lim J. W., Kim S. K., Choi S. Y., Kim D. H., Gadhe C. G., Lee H. N., Kim H. J., Kim J., Cho S. J., Hwang H., et al. . 2018. Identification of crizotinib derivatives as potent SHIP2 inhibitors for the treatment of Alzheimer’s disease. Eur. J. Med. Chem. 157: 405–422. [DOI] [PubMed] [Google Scholar]
- 88.Mao Y., Balkin D. M., Zoncu R., Erdmann K. S., Tomasini L., Hu F., Jin M. M., Hodsdon M. E., and De C. P.. 2009. A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism. EMBO J. 28: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mitchell C. A., Connolly T. M., and Majerus P. W.. 1989. Identification and isolation of a 75-kDa inositol polyphosphate-5- phosphatase from human platelets. J. Biol. Chem. 264: 8873–8877. [PubMed] [Google Scholar]
- 90.Ponting C. P. 2006. A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics. 22: 1031–1035. [DOI] [PubMed] [Google Scholar]
- 91.Damen J. E., Liu L., Rosten P., Humphries R. K., Jefferson A. B., Majerus P. W., and Krystal G.. 1996. The 145-kDa protein induced to associate with Shc by multiple cytokines is an inositol tetraphosphate and phosphatidylinositol 3,4,5- triphosphate 5-phosphatase. Proc. Natl. Acad. Sci. USA. 93: 1689–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pesesse X., Deleu S., De Smedt F., Drayer L., and Erneux C.. 1997. Identification of a second SH2-domain-containing protein closely related to the phosphatidylinositol polyphosphate 5-phosphatase SHIP. Biochem. Biophys. Res. Commun. 239: 697–700. [DOI] [PubMed] [Google Scholar]
- 93.McPherson P. S., Garcia E. P., Slepnev V. I., David C., Zhang X., Grabs D., Sossin W. S., Bauerfeind R., Nemoto Y., and De Camilli P.. 1996. A presynaptic inositol-5-phosphatase. Nature. 379: 353–357. [DOI] [PubMed] [Google Scholar]
- 94.Kong A. M., Speed C. J., O’Malley C. J., Layton M. J., Meehan T., Loveland K. L., Cheema S., Ooms L. M., and Mitchell C. A.. 2000. Cloning and characterization of a 72-kDa inositol-polyphosphate 5- phosphatase localized to the Golgi network. J. Biol. Chem. 275: 24052–24064. [DOI] [PubMed] [Google Scholar]
- 95.Gurung R., Tan A., Ooms L. M., McGrath M. J., Huysmans R. D., Munday A. D., Prescott M., Whisstock J. C., and Mitchell C. A.. 2003. Identification of a novel domain in two mammalian inositol-polyphosphate 5-phosphatases that mediates membrane ruffle localization - The inositol 5-phosphatase skip localizes to the endoplasmic reticulum and translocates to membrane ruffles following epidermal growth factor stimulation. J. Biol. Chem. 278: 11376–11385. [DOI] [PubMed] [Google Scholar]
- 96.Mochizuki Y., and Takenawa T.. 1999. Novel inositol polyphosphate 5-phosphatase localizes at membrane ruffles. J. Biol. Chem. 274: 36790–36795. [DOI] [PubMed] [Google Scholar]
- 97.Iida A., Okamoto N., Miyake N., Nishimura G., Minami S., Sugimoto T., Nakashima M., Tsurusaki Y., Saitsu H., Shiina M., et al. . 2013. Exome sequencing identifies a novel INPPL1 mutation in opsismodysplasia. J. Hum. Genet. 58: 391–394. [DOI] [PubMed] [Google Scholar]
- 98.Ghosh S., Huber C., Siour Q., Sousa S. B., Wright M., Cormier-Daire V., and Erneux C.. 2017. Fibroblasts derived from patients with opsismodysplasia display SHIP2-specific cell migration and adhesion defects. Hum. Mutat. 38: 1731–1739. [DOI] [PubMed] [Google Scholar]
- 99.Zhang X., Jefferson A. B., Auethavekiat V., and Majerus P. W.. 1995. The protein deficient in Lowe syndrome is a phosphatidylinositol-4,5- bisphosphate 5-phosphatase. Proc. Natl. Acad. Sci. USA. 92: 4853–4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jefferson A. B., and Majerus P. W.. 1995. Properties of type II inositol polyphosphate 5-phosphatase. J. Biol. Chem. 270: 9370–9377. [DOI] [PubMed] [Google Scholar]
- 101.Schmid A. C., Wise H. M., Mitchell C. A., Nussbaum R., and Woscholski R.. 2004. Type II phosphoinositide 5-phosphatases have unique sensitivities towards fatty acid composition and head group phosphorylation. FEBS Lett. 576: 9–13. [DOI] [PubMed] [Google Scholar]
- 102.Pesesse X., Moreau C., Drayer A. L., Woscholski R., Parker P., and Erneux C.. 1998. The SH2 domain containing inositol 5-phosphatase SHIP2 displays phosphatidylinositol 3,4,5-trisphosphate and inositol 1,3,4,5- tetrakisphosphate 5-phosphatase activity. FEBS Lett. 437: 301–303. [DOI] [PubMed] [Google Scholar]
- 103.Nemoto Y., Arribas M., Haffner C., and DeCamilli P.. 1997. Synaptojanin 2, a novel synaptojanin isoform with a distinct targeting domain and expression pattern. J. Biol. Chem. 272: 30817–30821. [DOI] [PubMed] [Google Scholar]
- 104.Kisseleva M. V., Wilson M. P., and Majerus P. W.. 2000. The isolation and characterization of a cDNA encoding phospholipid- specific inositol polyphosphate 5-phosphatase. J. Biol. Chem. 275: 20110–20116. [DOI] [PubMed] [Google Scholar]
- 105.Kisseleva M. V., Cao L., and Majerus P. W.. 2002. Phosphoinositide-specific inositol polyphosphate 5-phosphatase IV inhibits Akt/protein kinase B phosphorylation and leads to apoptotic cell death. J. Biol. Chem. 277: 6266–6272. [DOI] [PubMed] [Google Scholar]
- 106.Ooms L. M., Fedele C. G., Astle M. V., Ivetac I., Cheung V., Pearson R. B., Layton M. J., Forrai A., Nandurkar H. H., and Mitchell C. A.. 2006. The inositol polyphosphate 5-phosphatase, PIPP, Is a novel regulator of phosphoinositide 3-kinase-dependent neurite elongation. Mol. Biol. Cell. 17: 607–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Davies E. M., Kong A. M., Tan A., Gurung R., Sriratana A., Bukczynska P. E., Ooms L. M., McLean C. A., Tiganis T., and Mitchell C. A.. 2015. Differential SKIP expression in PTEN-deficient glioblastoma regulates cellular proliferation and migration. Oncogene. 34: 3711–3727. [DOI] [PubMed] [Google Scholar]
- 108.Ijuin T., Mochizuki Y., Fukami K., Funaki M., Asano T., and Takenawa T.. 2000. Identification and characterization of a novel inositol polyphosphate 5- phosphatase. J. Biol. Chem. 275: 10870–10875. [DOI] [PubMed] [Google Scholar]
- 109.Ijuin T., and Takenawa T.. 2012. Regulation of insulin signaling by the phosphatidylinositol 3,4,5-triphosphate phosphatase SKIP through the scaffolding function of Pak1. Mol. Cell. Biol. 32: 3570–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Verjans B., Erneux C., Raspe E., and Dumont J. E.. 1991. Kinetics of inositol 1,4,5-trisphosphate and inositol 1,3,4,5- tetrakisphosphate generation in dog-thyroid primary cultured cells stimulated by carbachol. Eur. J. Biochem. 196: 43–49. [DOI] [PubMed] [Google Scholar]