Abstract
Phosphoinositide membrane signaling is critical for normal physiology, playing well-known roles in diverse human pathologies. The basic mechanisms governing phosphoinositide signaling within the nucleus, however, have remained deeply enigmatic owing to their presence outside the nuclear membranes. Over 40% of nuclear phosphoinositides can exist in this non-membrane state, held soluble in the nucleoplasm by nuclear proteins that remain largely unidentified. Recently, two nuclear proteins responsible for solubilizing phosphoinositides were identified, steroidogenic factor-1 (SF-1; NR5A1) and liver receptor homolog-1 (LRH-1; NR5A2), along with two enzymes that directly remodel these phosphoinositide/protein complexes, phosphatase and tensin homolog (PTEN; MMAC) and inositol polyphosphate multikinase (IPMK; ipk2). These new footholds now permit the assignment of physiological functions for nuclear phosphoinositides in human diseases, such as endometriosis, nonalcoholic fatty liver disease/steatohepatitis, glioblastoma, and hepatocellular carcinoma. The unique nature of nuclear phosphoinositide signaling affords extraordinary clinical opportunities for new biomarkers, diagnostics, and therapeutics. Thus, phosphoinositide biology within the nucleus may represent the next generation of low-hanging fruit for new drugs, not unlike what has occurred for membrane phosphatidylinositol 3-kinase drug development. This review connects recent basic science discoveries in nuclear phosphoinositide signaling to clinical pathologies, with the hope of inspiring development of new therapies.
Keywords: nuclear lipid signaling; phosphatidylinositol (3,4,5) triphosphate; glioblastoma; endometriosis; diabetes; hepatocellular carcinoma; inositol phosphate multikinase
As we learn more about the unique ways phosphoinositide signaling works in the nucleus (1), we can connect these basic science observations with unexplained mechanisms of human pathologies that could lead to new diagnostics, biomarkers, or therapies (2). From a clinical perspective, it is easy to wonder why phosphoinositide signaling in the nucleus has not realized the meteoric rise in therapeutic development that plasma membrane phosphoinositide, phosphatidylinositol 3-kinase (PI3-kinase), pharmacology has enjoyed over the last 20 years (3–7). Indeed, at the time this article was written, clinicaltrials.gov contained well over 200 ongoing trials assessing PI3-kinase inhibitors at various stages of development. The lag in clinical translation of nuclear phosphoinositide signaling stems from a lag in our basic understanding of how these signaling processes operate at the molecular level (8). This uncertainty is centered around one specific mystery that has clouded nuclear phosphoinositide biology for the last 40 years: that nuclear phosphoinositides exist outside membranes (9, 10).
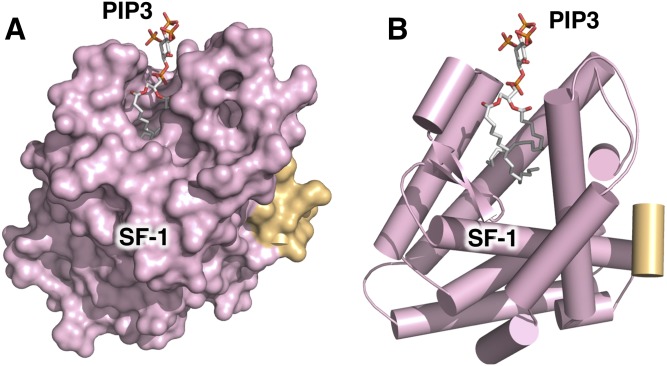
The biophysical basis for this phenomenon had been speculated on for decades (11, 12), but only recently has X-ray crystallography provided concrete data to reveal the structural format of non-membrane nuclear phosphoinositides (13, 14). These studies showed that certain non-membrane nuclear proteins can bury the acyl chains of phosphoinositides deep in the hydrophobic core of the protein (Fig. 1A), while the phosphoinositide headgroups remain solvent-exposed on the surface of the protein (Fig. 1B). This structural format “presents” the phosphoinositide headgroup as a unique signaling platform, granting extraordinary signaling capacity to the limited number of these complexes that have been characterized (1). Even with just this handful of validated examples, the mechanism represents a fundamentally new way that phosphoinositide signaling can work outside membranes.
Fig. 1.
Nuclear receptor SF-1 binds PI(3,4,5)P3 (PIP3) with acyl chains buried in a deep hydrophobic pocket, with the PIP3 headgroup on the surface of the SF-1 protein. A: Surface representation of the 2.4 Å crystal structure of the ligand binding domain of SF-1 (colored pink) bound by dipalmitoyl (di-C16) PIP3 (represented as atom colored sticks) (13). The lower right contains the integrated surface representation of the cocrystallized nuclear receptor coactivator LXXLL peptide, PGC1α (colored gold), which functions to regulate SF-1 transcriptional activity in chromatin. This helical bundle architecture is conserved throughout the ligand binding domains of the nuclear receptor superfamily of transcription factors, most of which bind hydrophobic cholesterol-, fatty acid- or phospholipid-based ligands (54, 195). Note the absence of the DNA-binding domain from this structure; the full-length structures of SF-1 and LRH-1 are currently unknown. B: Cartoon representation of A, with identical coloring scheme, α helices represented as cylinders, highlighting the positioning of the dipalmitoyl acyl chains deep in the hydrophobic core of the SF-1 helical bundle. These structures exemplify how non-membrane nuclear phosphoinositides can exist outside membrane systems (8).
The clinical significance of these basic science discoveries is only now being explored, remaining quite hypothetical and, thus, completely untapped by the pharmaceutical industry. The infancy of the field is perhaps reflected in the fact that non-membrane phosphoinositide signaling has only been confirmed to be mediated by inositol polyphosphate multikinase (IPMK; ipk2) and the classic phosphatidylinositol (3, 4, 5) trisphosphate [PI(3,4,5)P3] phosphatase and tensin homolog (PTEN; MMAC). However, several other candidate mediators can be readily identified in the literature that might operate similarly, such as StAR-PAP (15), SHIP and B53/nucleophosmin (16), PI3Kβ (17), TAF3 (18), ING2 (19), PPARα (20, 21), and PPARγ (22–25). These proteins and enzymes have accumulated circumstantial evidence over the past two decades suggesting that they too could participate in similar non-membrane phosphoinositide signaling mechanisms, consistent with non-membrane pathways being as ubiquitous as classic membrane pathways, and potentially far more penetrant (1, 13, 14, 26, 27). While many of these candidates have demonstrated pathological impact in the clinic that suggest connections between nuclear phosphoinositide signaling and human disease, a more honest assessment would be that we have yet to scratch the surface of the clinical repercussions these signaling mechanisms may have. Still, it is hard to ignore the potential opportunities for new diagnostics, biomarkers, and therapeutics that non-membrane nuclear phosphoinositide signaling could provide (28), linking basic science to better clinical outcomes for patients suffering from several pathologies, including endometriosis, obesity, nonalcoholic fatty liver disease (NAFLD), nonalcoholic steatohepatitis (NASH), and several varieties of PTEN-dependent cancers. This review presents the evidence linking these diseases to nuclear phosphoinositides, at the infancy of our understanding of these basic signaling mechanisms.
NON-MEMBRANE NUCLEAR PHOSPHOINOSITIDES
History
Many biochemical studies in the 1950s, 1960s, and 1970s were consistent with the presence of non-membrane phospholipids bound to chromatin and other non-membrane bound nuclear substructures (29–36). These results were rightfully critiqued as potential biochemical artifacts due to the tendency of phospholipids to interact nonspecifically with almost every biomolecule, as well laboratory glass- and plasticware. However, breakthroughs at the dawn of phosphoinositide signaling in the 1980s and 1990s spearheaded by Robin Irvine, Nullin Divecha, and Lucio Cocco showed clear differences in membrane versus non-membrane nuclear phosphoinositide metabolism by multiple approaches (12, 33, 37–41), which have been extensively reviewed elsewhere (8–10, 39, 42, 43). These studies collectively showed functional differences in cytoplasmic versus nuclear phosphoinositide metabolism and signaling, presumably due to the non-membrane nature of nuclear phosphoinositides. Later work by Peter Downes (44) showed further convincing functional evidence that nuclear pools of PI(3,4,5)P3 are metabolically distinct from membrane PI(3,4,5)P3 in certain human cell lines, but still provided very little mechanistic information about how membrane and non-membrane pathways could be different. Later seminal studies by Nullin Divecha (37, 45–47), Richard Andersen (15, 48), Or Gozani (19), Pascale Zimmerman (49, 50), Aurelia Lewis (51), and several others (18, 52, 53) produced data that were clearly consistent with the existence of non-membrane nuclear phosphoinositides and provided functional connections to basic cellular processes, but again provided little data explaining how nuclear phosphoinositides could operate outside membranes and even less information on the physicochemical structural format of the endonuclear phosphoinositides (8). Thus by 2010, it remained possible that all non-membrane nuclear phosphoinositides were simply just biochemical artifacts (9). What was needed to address the question was concrete structural biology that could definitively explain how nuclear phosphoinositides existed outside membranes, perhaps providing clues as to how they executed functions in the nucleus (8). That structural biology came from studies examining the nuclear receptor, steroidogenic factor-1 (SF-1: NR5A1), and the enzyme action of IPMK (ipk2, Arg82p, ArgR) and PTEN on phosphoinositides bound to this nuclear receptor (1, 13, 54). These studies provided the first structural and mechanistic enzymology required to prove that non-membrane nuclear phosphoinositides must exist, at least in the handful of human cell lines used in these studies, and explained how and why non-membrane nuclear phosphoinositide signaling was inherently different from membrane signaling (1, 26, 55–59).
Nuclear receptor mammalian physiology
Before delving into the interesting signaling properties of these nuclear receptors, it is worthwhile to introduce the mammalian physiology controlled by this class of transcription factors. SF-1 (NR5A1) and the highly homologous liver receptor homolog-1 (LRH-1; NR5A2) are both members of the exclusively metazoan nuclear hormone receptor superfamily of ligand-regulated transcription factors (54, 60, 61), each encoded by separate genes. SF-1 and LRH-1 activate very similar subsets of genes in human cell lines and bind almost identical response elements in vitro and in cells (62) to generally activate expression of genes encoding steroidogenic enzymes (63–66); however, the tissue expression patterns of these nuclear receptors are very distinct in humans (67). SF-1 expression is highly restricted to the gonads, adrenals, and a small portion of the hypothalamus (the ventral medial) (60, 67), while LRH-1 is found in liver, pancreas, gut, and brain (68). SF-1 and LRH-1 share orthologs in all animals with various expression patterns, including nhr-25 in worms (26), Ftz-F1/HR39 in flies (69, 70), and lrh-1/Ftz-F1/nr5a1a (sf-1a) in fish (71).
Global Sf-1 knockout mice die quickly after birth from a lack of steroids, due to complete developmental agenesis of the adrenals (64). Postpartum rescue of Sf-1 knockout mice with appropriate steroids allows these sterile animals to attain normal lifespan (72). Global Lrh-1 knockout mice are embryonic lethal with defects in gastrulation (73, 74). Recent studies using viruses to acutely rescue Lrh-1 in liver-specific knockout mice have physiologically linked Lrh-1 with arachidonic acid metabolism and several other aspects of lipid homeostasis in the liver (75). Both Sf-1 and Lrh-1 are downregulated by posttranslational modification with the small ubiquitin-like modifier (SUMO), and several elegant knock-in studies, which generated “SUMO-less” Sf-1 and Lrh-1 mice, have shown that SUMO physiologically regulates the activities of both Sf-1 (76) and Lrh-1 (77–80). It remains unclear how SUMO regulates SF-1 and LRH-1 at the molecular level, because it is unknown whether SUMO allosterically regulates NR5A structure or acts as a docking site to alter recruitment of transcriptional regulatory proteins. Both SF-1 (13) and LRH-1 (55) bind the signaling phosphoinositides, PI(4,5)P2 and PI(3,4,5)P3, with nanomolar affinity (14, 57).
Nuclear receptors hide phosphoinositide acyl chains
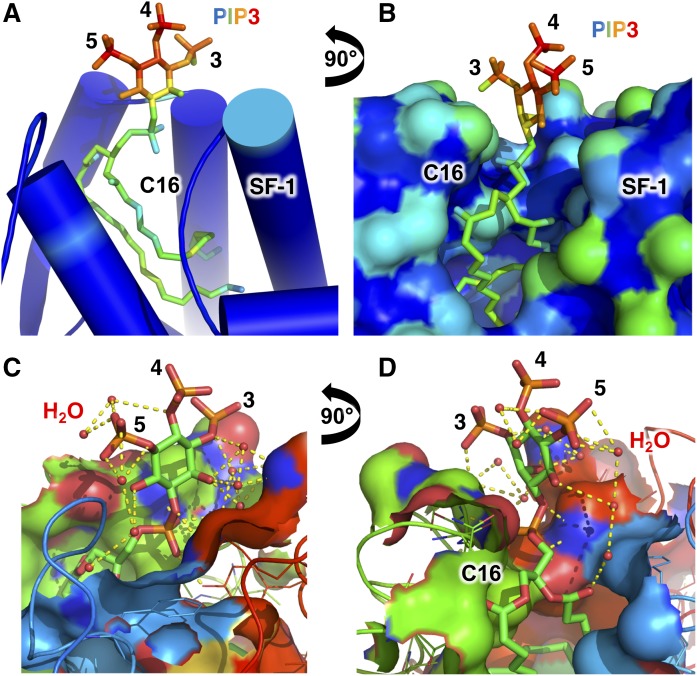
Perhaps the most obvious mechanistic breakthrough came with the first crystal structures that definitively showed how phosphoinositides exist outside of membranes in the nucleus. High resolution X-ray crystal structures of PI(4,5)P2 and PI(3,4,5)P3 phosphoinositide species bound to the phospholipid-binding domain of human SF-1 revealed that the acyl chains are buried deep within the hydrophobic core of the globular SF-1 protein (Fig. 2A), while the phosphoinositide headgroups remain highly exposed to solvent (Fig. 2B) (13). The PI(3,4,5)P3 headgroup also participates in an extensive water-mediated hydrogen bonding network with the SF-1 protein (Fig. 2C, D). These structures revealed a definitive physicochemical format for non-membrane nuclear phosphoinositides by providing a conclusive structural explanation for how hydrophobic phosphoinositides can exist outside membranes, with stoichiometry and biological ramifications consistent with a highly specific signaling function for non-membrane nuclear phosphoinositides. The crystal structure of LRH-1 bound to PI(3,4,5)P3 confirmed the same type of format (55) and also raised questions as to whether structural genomics could be used to identify novel nuclear proteins that bind the phosphoinositide acyl chains through a similar mechanism (55). The current state of structural genomics (81–83) is not yet capable of predicting whether the hydrophobic core of a protein can accommodate the acyl chains of a phospholipid with the level of detail and confidence required to be predictively useful. However, with some a priori structural information, a candidate protein could be computationally modeled around a phosphoinositide using ROSETTA, integrated with biophysically derived experimental restraints from nuclear magnetic resonance spectroscopy (84), small angle X-ray scattering (85), or double electron-electron paramagnetic resonance spectroscopy (86). These types of integrated structural analyses represent one of the few feasible paths forward that could identify how nuclear phosphoinositides interact with nuclear binding proteins, short of full-scale crystallographic or other biochemical efforts. Several crystal structures were able to show how bacterial lipids (87) and phosphatidylcholines (88) can exist outside membranes, however the structures of SF-1 and LRH-1 bound by signaling phosphoinositides (13, 14) permitted the generation of new signaling hypotheses that this structural platform makes possible.
Fig. 2.
Solvent accessibility of the SF-1 bound PI(3,4,5)P3 headgroup is high and participates in an extensive water-mediated hydrogen bonding network. A: Cartoon representation of the crystal structure of SF-1 bound to dipalmitoyl PI(3,4,5)P3 (13) colored as a spectrum to solvent accessibility, where the least solvent accessible is blue<green<yellow<orange<red is most solvent accessible. Note the very high solvent accessibility of the PI(3,4,5)P3 headgoup phosphorylations on the indicated 3, 4, and 5 positions, and the low solvent accessibility of the di-C16 palmitoyl acyl chains. B: Surface representation and orthogonal view of A with identical coloring scheme, again showing high solvent accessibility of the PI(3,4,5)P3 phosphorylations even relative to hydrophilic surface residues on the SF-1 protein. C, D: Orthogonal ligand-sites surface cutaway views of the crystal structure of SF-1 bound to dipalmitoyl PI(3,4,5)P3, showing the extensive water-mediated hydrogen bonding network the 3, 4, and 5 phosphorylations participate in. Water molecules are shown as small red spheres. The crystal structure of SF-1 bound to PI(4,5)P2 is very similar in terms of the solvent accessibility of the phosphoinositide headgroup [Protein Data Bank (PDB): 4QK4]. The high solvent accessibility of PIP3 bound to SF-1 highlights that the headgroup in nuclear phosphoinositide signaling complexes can be readily accessible for catalysis and other signaling events.
Nuclear phosphoinositide signaling and catalysis
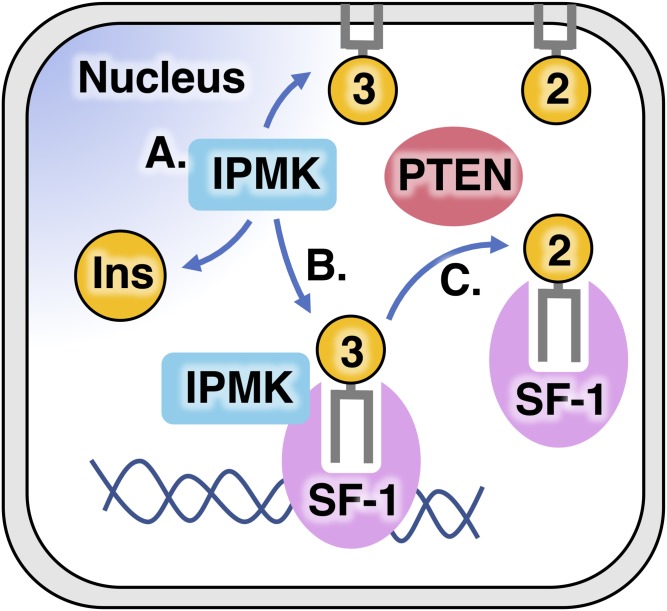
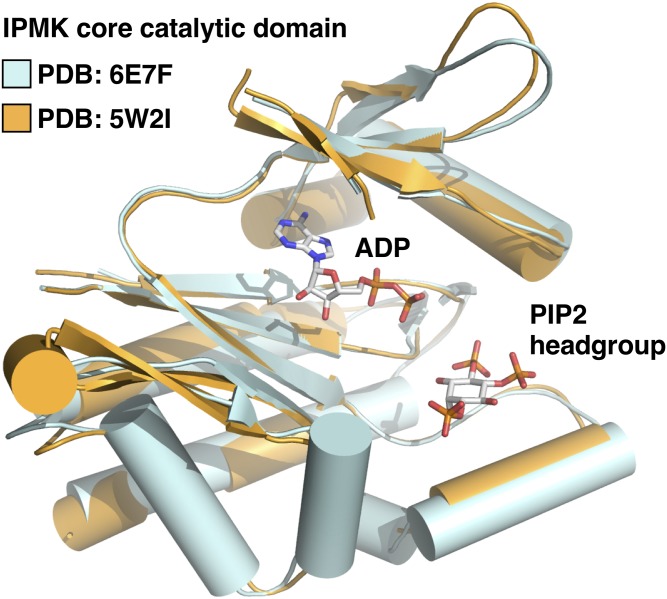
We showed that the complex of SF-1 bound by phosphoinositides is a direct substrate for the phosphoinositide signaling enzymes, PTEN and IPMK (Fig. 3) (1). The tumor suppressor and PI(3,4,5)P3-phosphatase PTEN have been extensively reviewed elsewhere (89), but IPMK is a far less well-studied inositol kinase that localizes to the nucleus and bears membrane PI3-kinase activity (90) along with the ability to phosphorylate several soluble inositol species (91). IPMK is ubiquitously expressed in all eukaryotes from yeast (92, 93) to human (94), including plants (95, 96), protists (97), and fungi (43), although the plant IPMK lacks any detectable PI3-kinase activity (1, 96). Recent human IPMK crystal structures of the core catalytic domain of IPMK (C. D. Seacrist and R. D. Blind, unpublished observations) revealed the atomic-resolution mechanism IPMK uses to interact with several ligands, including ATP and small molecule enzyme substrates of IPMK (Fig. 4) (99). These high-resolution crystal structures showed how IPMK coordinates each ligand, giving insight into how IPMK discriminates between its many substrates (99). IPMK kinase activity activates SF-1 gene expression programs in human cells, while PTEN activity inhibits SF-1 transcriptional activity in human cells (1). These signaling activities are dependent on the ability of SF-1 to bind phosphoinositides, as mutants of SF-1 that do not bind any phospholipids become decoupled from any regulation by IPMK and PTEN (1). Further in vitro studies (described below) conclusively demonstrated that the complex of SF-1 bound to phosphoinositides is a bona fide direct substrate of IPMK and PTEN.
Fig. 3.
Model of PTEN- and IPMK-mediated nuclear phosphoinositide signaling through SF-1. A: IPMK is a nuclear protein conserved in all eukaryotes (144) that was first characterized functionally as a transcriptional coregulator (196, 197) before the inositol kinase enzyme activity was discovered (92). B: Shortly thereafter, Adam Resnick in Solomon Synder’s laboratory discovered that IPMK also has a PI3-kinase enzyme activity, with the ability to phosphorylate PI(4,5)P2 in membrane systems (90), confirmed to occur in vivo in mouse models (103). IPMK is a close structural homolog to the IP3-kinase superfamily and protein kinase A (99), but is structurally unrelated to the class 1 PI3-kinases. C: IPMK was then shown to bind and directly phosphorylate PI(4,5,)P2 bound to the nuclear receptor SF-1, generating PI(3,4,5)P3 bound to SF-1 resulting in robust activation of SF-1 transcriptional programs in human cell lines (1). Nuclear PTEN opposes this function of IPMK, dephosphorylating PIP3 bound to SF-1 and downregulating SF-1 transcriptional activity (1). This nuclear phosphoinositide signaling pathway is the first described in which the structural biology and mechanistic enzymology fully describe the mechanism of the signal transduction, which is extremely important in drug design efforts that could impact the clinic.
Fig. 4.
Superposition of two recently solved crystal structures of IPMK, a nuclear PI(4,5)P2 kinase. Cartoon views of recently published human IPMK catalytic core PDB 6C8A (apo) and PDB 5W2I (bound to ADP and di-C4 PI(4,5,)P2) (99, C. D. Seacrist and R. D. Blind, unpublished observations). These extensive crystallographic and kinetic studies revealed novel kinetic properties of IPMK and how IPMK interacts with ligands, ATP and PI(4,5)P2. However, because crystallization requires removal of the disordered domains of IPMK, the structure of these domains (one disordered domain on the N terminus and one which interrupts the core kinase domain) remains undescribed. Note that the PI(4,5)P2 glycerol backbone and the di-C4 acyl chains were not ordered (99).
Catalytically, IPMK directly phosphorylates PI(4,5)P2 while PI(4,5)P2 is bound to SF-1, with the best catalytic efficiency (kcat/KM) of any IPMK kinase substrate ever tested (1, 100). Further, PTEN dephosphorylates PI(3,4,5)P3 while PI(3,4,5)P3 is bound to SF-1, again with better catalytic efficiency than any other reported substrate for PTEN (1, 101). The better catalytic efficiency is driven by lower KM numbers for these enzymes when the phosphoinositide substrate is bound to SF-1, suggesting that there are direct protein-protein contacts between SF-1 and the enzymes, which has also been observed in cells and in vitro (1). The enzymology providing these catalytic details is crucial for our understanding of how non-membrane signaling differs from membrane signaling. What the numbers show in this regard is that IPMK and PTEN enzymatically prefer to act on phosphoinositides bound to SF-1, providing in vitro evidence that these important enzymes could be more active in the nucleus than the same enzymes at the plasma membrane (102). It is important to note that membrane signaling also relies on protein-protein interactions, which are usually not accounted for in enzyme kinetic experiments. Further, it is currently unknown if any enhanced catalysis in non-membrane phosphoinositide signaling is also true in living cells, or in any physiologically relevant animal tissue (103). It was shown that IPMK has PIP2-kinase activity on pure SF-1 immunoprecipitated from HEK cells (1), suggesting that SF-1 is bound by PIP2 in human cell lines. It is also clear that PTEN functionally downregulates SF-1 transcriptional activity while IPMK functionally upregulates SF-1 activity in human cell lines, and that both IPMK and PTEN activity are dependent on the ability of SF-1 to bind to phosphoinositides (1). These studies revealed a new way nuclear phosphoinositides directly control transcriptional activation of a phosphoinositide-binding nuclear receptor (42).
Nuclear phosphoinositide effector mechanisms
The X-ray crystal structures of SF-1 bound to PI(3,4,5)P3 and PI(4,5)P2 showed how these phosphoinositides are solubilized by SF-1 (Fig. 1B), revealing how SF-1 coordinates the PI(3,4,5)P3 and PI(4,5)P2 headgroups (Fig. 3C, D). These studies also suggested that PI(3,4,5)P3 can act as “molecular glue” between SF-1 and potential coregulator proteins (55, 57), used as the basis for studies by Michael Sheetz’s group (104). Together, these basic science studies provided a structural model explaining how non-membrane phosphoinositides exist (Fig. 1A) and identified transcription as a cellular function regulated by non-membrane nuclear phosphoinositides (Fig. 3), while determining a structural mechanism explaining how nuclear phosphoinositides regulate their cognate receptor. However, because SF-1 is restricted only to very limited metazoan tissues, SF-1 cannot be the only factor responsible for all eukaryotic non-membrane phosphoinositides, as non-membrane nuclear phosphoinositides have been observed in many mammalian cell lines and tissues that do not express detectable levels of either SF-1 or LRH-1. Thus, the identity of the other nuclear phosphoinositide binding proteins that solubilize these phosphoinositides awaits discovery. The potential clinical ramifications of these nuclear phosphoinositide signaling pathways in specific pathologies are highlighted below.
ENDOMETRIOSIS
Endometriosis is a very painful endocrine disorder afflicting six million women in the United States alone (105, 106), with some studies estimating that 1 in 10 women will be afflicted with this disease (105, 107), making the impact in the hundreds of millions of women worldwide. Endometriosis is defined as the ectopic presence of steroidogenic uterine endometrial tissue in either the pelvic peritoneum or on the ovaries (108), which can cause severe pain, damage to surrounding organs, sterility, and can threaten life in severe cases (109). Endometriotic tissue often overexpresses SF-1 (110, 111), which through the activation of genes encoding steroidogenic enzymes, such as CYP11A1, CYP17A1, CYP19A1, and 17BHSD, drives estradiol production that is required to maintain the endometriotic tissue in the ectopic locations (61, 112). The current standard of care for endometriosis is analgesics, birth control hormone contraceptives (113), and the androgen nuclear receptor antagonist, Danazol (114), which carries some significant side effects (115). Aromatase inhibitors, such as aminoglutethimide, can be effective in postmenopausal women (116, 117) by decreasing the global production of estradiol, effectively decreasing endometriotic tissue growth (112, 117). However, aromatase inhibitors also inhibit normal physiological hormone production from the adrenals, thyroid (118), and gonads (119–121). Global aromatase inhibitors thus have serious side effects on bone metabolism (122), adrenal insufficiency (123), and kidney failure (121). For these reasons, a more targeted therapy specifically inhibiting autocrine production of steroids by endometriotic tissue has been oft-predicted as a potentially efficacious route for therapy (124). The role of SF-1 in steroidogenesis, when taken with its prominent overexpression, makes SF-1 a viable therapeutic target for endometriosis (61), but specifically targeting SF-1 in the endometriotic tissue, while leaving adrenal and gonadal SF-1 unperturbed to execute normal adrenal and gonadal physiology, has prevented development of a targeted therapy for endometriosis.
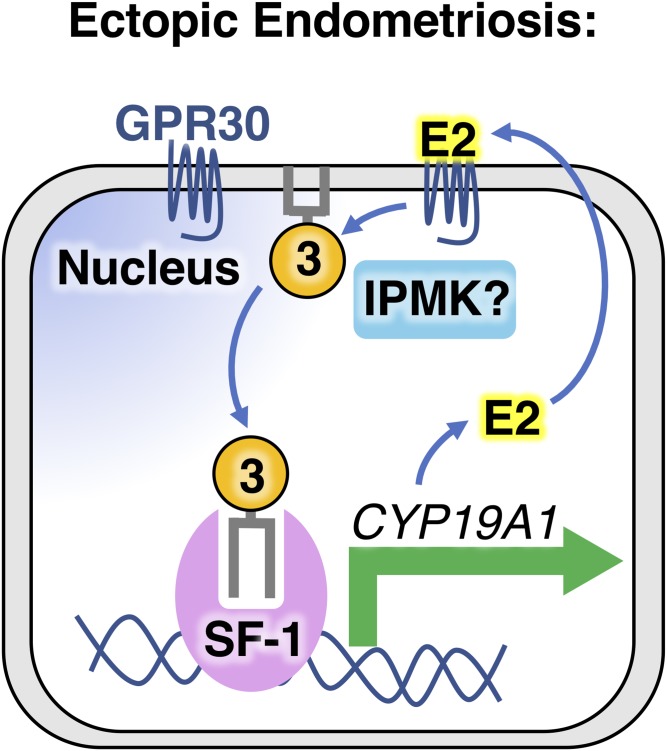
Prior to elucidation of the details of SF-1 regulation by nuclear phosphoinositides, we were able to show that estradiol activates steroidogenic gene expression programs in primary human endometriotic H38 cells by increasing levels of PI(3,4,5)P3 (Fig. 5) (112). The PI(3,4,5)P3 produced by this pathway is regulated by activation of the G protein-coupled receptor 30 (GPR30, GPER1). GPR30 activation by estradiol stimulates PI(3,4,5)P3 production, activating SF-1 and initiating an autocrine feed-forward loop in endometriotic cells. Continuous SF-1 activation of steroidogenic gene programs results in overproduction of estradiol, perpetuating endometriotic steroidogenesis (112), which maintains the endometriotic tissue in the ectopic location, preventing atrophy and clearance. In terms of therapeutic targeting possibilities, GPR30 remains high on the list of hopefuls for endometriosis (112). Although it is unknown if GPR30 can specifically stimulate accumulation of nuclear PI(3,4,5)P3, GPR30 is often observed on the nuclear membrane, where its signaling role remains unexplored (125). Regardless, inhibitors targeting GPR30 would be predicted to block nuclear SF-1 activation by decreasing PI(3,4,5)P3, effectively interfering with the feed-forward autocrine stimulation of ectopic endometriotic tissue (112). Global GPR30 developmental knockout animals have no differences in fertility, body weight, estrous cycle, or breeding rates when compared with wild-type (126), suggesting that compounds inhibiting GPR30 might have minimal adverse side effects (127, 128). Further, because SF-1-regulated gene expression is only sensitive to IPMK activity, and not class I PI3-kinases (1), one might predict that IPMK is the source of the PI(3,4,5)P3 that stimulates SF-1 in endometriosis. Indeed, SF-1 target RNAs that are important in steroidogenesis are downregulated in HEK cells when IPMK is knocked down using small interfering RNAs (1). Taken together, these data suggest that an antagonist of IPMK kinase activity, combined with a GPR30 antagonist, could pharmacologically converge on SF-1 in endometriotic tissue (112). A better understanding of how nuclear phosphoinositides are regulated by GPR30 and IPMK would greatly enhance any effort attempting to develop treatment options for women suffering from endometriosis.
Fig. 5.
GRP30 could be a good drug target in nuclear phosphoinositide signaling and endometriosis. Endometriotic uterine endometrial cells growing in the pelvic peritoneum or on the ovaries require estradiol to maintain growth in these ectopic locations. Ectopic endometriotic tissue itself is often steroidogenic, overexpressing SF-1. Estradiol or synthetic agonists can activate the G protein-coupled receptor, GPR30 (GPER1), present on the plasma membrane and nuclear envelope of endometriotic cells, resulting in accumulation of global cell levels of PIP3 (112). The mechanism GPR30 uses to increase PIP3 is unknown and it is further unknown whether IPMK might play a role in endometriosis. GPR30-mediated increases in PIP3 correlate with activation of SF-1, leading to increased gene expression of steroidogenic genes involved in biosynthesis of estradiol, such as aromatase (CYP19A1). Increased expression of these genes leads to increased production of steroids, which can then reactivate membrane-bound GPR30, resulting in a feed-forward activation loop that perpetuates ectopic growth of the endometrial tissue (112). For these reasons, GPR30 could be an excellent candidate for drug development efforts targeting endometriosis.
NEUROENDOCRINOLGY OF OBESITY
In addition to being expressed in the gonads and adrenals (129), SF-1 defines neurons located in the ventral medial hypothalamus (VMH) (63), a portion of the human brain well-known to control satiety, sexual activity, and fear (130). Genetic studies in mice indicate that an SF-1 agonist targeting the VMH would be predicted to have high therapeutic value in obesity (131) and in enhancing metabolic responses to exercise (132). Well-executed tissue-specific knockout studies have shown that SF-1 VMH neurons control insulin-mediated protection from diet-induced leptin resistance, weight gain, adiposity, and impaired glucose tolerance (133–136); and SF-1 neurons are well-known to be critical in mediating diet-induced obesity (72). Tissue-specific knockout studies of SF-1 in the VMH have demonstrated SF-1 as an anorexigenic factor regulated by nutritional status (137), and have shown that SF-1 is required for the development of the complete functional hypothalamic-pituitary-gonadal axis, as well as the hypothalamic-pituitary-adrenal axis (64, 134, 138). These studies are very important clinically because nuclear receptors like SF-1 are some of the best targets for agonist drug design, if molecular details on how the receptor is activated are known (139–141). The molecular and structural detail describing the SF-1/IPMK nuclear phosphoinositide signaling axis represents an ideal candidate for such an effort. Although global IPMK knockout mice die at embryonic day 9.5 (92), data generated from Ipmkflx/flx mice link IPMK to obesity, type 2 diabetes, and metformin action (142). Mice with viral-CRE-mediated knockout of IPMK in the hypothalamic region have altered glucose-mediated AMPK regulation (143). Murine embryonic fibroblasts with viral-CRE-mediated knockout of IPMK from Ipmkflx/flx mice cannot induce AMPK activation in response to metformin (142), indicating that IPMK is required for full response to metformin. These data are consistent with a model where IPMK inhibitors have therapeutic value in type 2 diabetic insulin resistance (142). Identifying biomarkers that will indicate sensitivity to IPMK inhibitors will help any sort of clinically relevant IPMK-drug design effort, and thus should begin to have higher priority within the nuclear phosphoinositide signaling field (144). It is certainly worth mentioning that several genetic knockout studies of PIK3CA class 1 PI3-kinase p110α in SF-1 neurons of the VMH have been executed (132, 133, 136, 145), showing that these animals have increased sensitivity to high-fat diet-induced obesity due to decreased energy expenditure (133). More recent studies have shown an estrogen-dependent sexually dimorphic effect of PIK3CA in decreasing energy expenditure (146), which, when coupled with recent SF-1 studies (147), shows that phosphoinositides within the VMH could be an important aspect of sexually dimorphic phenotypes in mammals. While these PIK3CA genetic studies have shown that all aspects of phosphoinositide signaling are clearly important in the VMH (148), it remains unclear what fraction, if any, of the phenotypes from the PIK3CA knockout studies could be attributed to nuclear pathways.
NAFLD AND NASH
The American Liver Foundation estimates that 100 million Americans today have NAFLD (149). Pten liver-specific knockout mice driven by albumin CRE develop fatty liver (150), which progresses to NASH (151) though a mechanism that is incompletely understood (152). What is clear is that this phenotype is very different from liver-specific knockout of the tyrosine phosphatase, Ptp-1 (150), despite both Pten and Ptp-1 acting as common negative downstream regulators of insulin membrane signaling, suggesting that either PTEN or Ptp-1 is acting through nonclassical roles (150). Data suggesting nonclassical roles for PTEN in the liver come from studies showing that high-fat diet in Pten liver-specific knockout mice elicits liver damage, but independently of AKT (153), and that loss of specifically nuclear PTEN associates with hepatitis C virus-infected liver inflammation in primary hepatocytes from human patients (154). These data suggest that nuclear PTEN may have a role in NAFLD and NASH progression, but the mechanism PTEN could use to mediate these effects remains unclear.
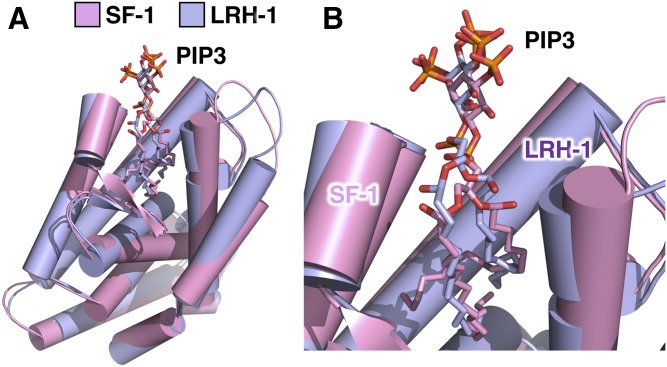
The nuclear receptor, LRH-1 (68), is a close structural and functional homolog of SF-1, and is highly expressed in liver where it controls lipid homeostasis (68, 155, 156). Because LRH-1 can bind PI(3,4,5)P3 in a structural format very similar to SF-1 (Fig. 6A) (55), it is worth mentioning that LRH-1 could be regulated similarly to SF-1, where PTEN might remodel PI(3,4,5)P3 bound to LRH-1 to regulate LRH-1 activity in lipid metabolism (157). This is certainly possible from a structural biology perspective (Fig. 6B) (55, 57). Several liver-specific knock-in studies that generated SUMO-less LRH-1 mutants in the mouse liver lead to hyper-activation of LRH-1 (80), with phenotypes that closely phenocopy PTEN loss in the mouse liver (150). These data are consistent with PTEN negatively regulating a potential mouse liver signaling pathway that is also controlled by LRH-1. Another study showed that the plant phospholipid, dilauryl phosphatidylcholine (DLPC), activates LRH-1 physiologically in the mouse liver leading to complete resolution of diet-induced fatty liver (155, 158), again demonstrating that activation of LRH-1 phenocopies PTEN inactivation in the mouse liver. These circumstantial data still require a great deal of experimental work to determine how or if nuclear PTEN and LRH-1 might act in the same nuclear phosphoinositide signaling pathway in the mammalian liver, but are consistent with a model in which PTEN downregulates nuclear LRH-1 transcriptional activity in a pathway capable of generating NAFLD and NASH in the mouse liver. Given that IPMK counteracts the effects of PTEN in the nucleus of HEK cells (1), and that IPMK is highly expressed in the human liver (94), it stands to reason that an inhibitor of IPMK could impact liver physiology (142, 143, 159). Understanding which targets are important in NAFLD and NASH, and how those targets operate at the molecular level, can lead to more clinically favorable outcomes by funneling efforts toward new lower-hanging pharmacological targets. Indeed, nuclear phosphoinositide signaling pathways have yet to be targeted in NAFLD or NASH by any effort from the pharmaceutical industry.
Fig. 6.
Human LRH-1 and SF-1 both bind nuclear PIP3 in very similar ways. A: Superposition of the 1.8 Å crystal structure of human LRH-1 ligand binding domain (14) (represented as purple cartoon) and the crystal structure of human SF-1 ligand binding domain (13) (represented as pink cartoon), each bound individually to one dipalmitoyl PI(3,4,5)P3 molecule. The two superposed structures have a root mean square deviation of less than 1 Å (0.898 Å) that, while totally identical, shows the high degree of homology between SF-1 and LRH-1. B: Close-up of A showing similar solvent accessibility of PI(3,4,5)P3 headgroups when the phosphoinositide is bound to either human SF-1 or human LRH-1. This structural comparison suggests that the same structural format of PI(3,4,5)P3 that grants special signaling capacity to SF-1 is conserved within LRH-1. It remains to be determined whether IPMK or PTEN can directly remodel phosphoinositides bound to LRH-1.
GLIOBLASTOMA AND HEPATOCELLULAR CARCINOMA
Glioblastoma multiforme is a devastating tumor, with one of the worst 5 year survival rates of any human cancer (160) and no targeted therapies that have been approved (161–163). Resected glioblastoma tumors (164) and glioblastoma cell lines often have reduced or null expression of the PTEN tumor suppressor (165). The loss of PTEN is part of the rationale for current efforts in class 1 PI3-kinase inhibitor trials determining efficacy against glioblastoma (166), as these inhibitors in part counteract loss of PTEN to decrease PI(3,4,5)P3 levels in tumors (167). Although several studies over the past decade have revealed important enzyme-independent functions for nuclear PTEN (164, 168–172), these discoveries are difficult to act on in the clinic pharmacologically, as protein-protein interactions are difficult to interfere with using small molecules (173, 174). Kinases and other enzymes are far easier drug targets, with a very druggable ligand-binding pocket for ATP, which can be relatively easily screened using large compound libraries (7, 174–178). Further, high-resolution structural studies can provide information to rationally improve lead compounds (179), further inspiring investment of resources from the pharmaceutical industry. Indeed, the discovery that IPMK counteracts the phosphatase activity of nuclear PTEN instantly raised interest in IPMK as a potential new drug target in any tumor that has lost PTEN (1, 180). Still, PTEN action on SF-1/PI(3,4,5)P3 remains the only known phosphatase-dependent function of PTEN in the nucleus where the mechanistic details have been well-characterized by structural biology and enzymology. Those types of details remain unknown for all other phosphatase-dependent studies of nuclear PTEN (164, 168–172), leaving open the possibility that nuclear PTEN action on non-membrane phosphoinositide/nuclear protein complexes may penetrate beyond SF-1. Still, what role the PI(3,4,5)P3-phosphatase enzyme activity of PTEN has in the nucleus remains a severely understudied area of PTEN tumor biology (181).
One study that revealed specific functions for nuclear PTEN phosphatase activity came from Alfred Yung’s group, which has studied glioblastoma for almost 40 years. This work used the PTEN-null human U251MG glioblastoma cell line to show that PTEN functionally regulates cell proliferation and growth in soft agar (165). These growth phenotypes could only be rescued by complementation with exclusively nuclear phosphatase-active PTEN, whereas complementation with cytoplasmic or phosphatase-dead PTEN had no effect on any aspect of the growth of these human glioblastoma cells (165, 171). Neither cytoplasmic nor nuclear PTEN had any effect on cell migration or invasiveness in this cell line, suggesting that PTEN regulation is limited to growth phenotypes in this cell line (165). These data suggested that nuclear PTEN phosphatase has specific functions in the nucleus that are distinct from the cytoplasm, which can control human glioblastoma cell growth. Several studies have also linked IPMK to growth control of glioblastoma cell lines (90, 103), suggesting that PTEN and IPMK could functionally oppose each other in the nucleus of glioblastoma cells. However, many, if not most, glioblastomas and PTEN-null glioblastoma cell lines are sensitive to both cytoplasmic and nuclear PTEN, suggesting that nuclear PTEN action does not solely drive growth in all glioblastoma tumors (165). Still, the potential to use nuclear PTEN staining as a biomarker in glioblastoma clinical trials (3, 28), coupled with the new links to IPMK as the counteracting kinase in the nucleus (1), could provide enough incentive for pharmaceutical development of an IPMK inhibitor designed to counteract loss of nuclear PTEN in glioblastoma patients (28). Given that glioblastoma multiforme currently has no approved targeted therapeutics, the clinical need is overwhelming.
Liver cancer also represents potentially fertile ground to develop new therapeutics within classically underserved populations. Although incidence rates have been falling for almost all cancers since 1999, the incidence rates of liver cancer in African Americans and Hispanics have continued to rise (182–184). Many hepatocellular carcinoma and cholangiocellular carcinoma liver tumors and many liver cancer cell lines have lost PTEN (150, 152, 185). Cancers of the liver may be an appealing potential therapeutic target for nuclear phosphoinositide intervention for several reasons. These tumors have excellent therapeutic potential as they have a poor prognosis (186), sorafenib (Nexavar) is the only currently approved therapeutic (187–189), and loss of specifically nuclear PTEN associates with increased progression to liver cancer in human primary hepatocytes infected with hepatitis C virus, one of the most common causes of liver cancers (190). Pten liver-specific knockout generates liver tumors in mice that are models for human hepatocellular and cholangiocellular carcinomas, which are dependent on loss of PTEN (150). Further, IPMK expression is highest in the human liver (94) and IPMK expression is associated with hyper-proliferation of liver tissue and cells (94, 191), providing an ideal potential kinase target for new drug design efforts (28). Several studies have linked prolonged metformin use in diabetic patients to decreased incidence of hepatocellular carcinoma (142, 192–194). IPMK knockout from homozygous IPMK floxed mouse embryonic fibroblasts using virally mediated CRE-recombinase decreases AMPK phosphorylation and activation mediated by metformin treatment, suggesting that IPMK may be required for full responsiveness to metformin treatment (142, 143). Clearly, the role of nuclear phosphoinositide signaling in glioblastoma and hepatocellular carcinoma will require much more investigation, but PTEN-null glioblastoma and hepatocellular carcinoma represent excellent first model systems to pursue clinical applications.
CONCLUSIONS
Non-membrane nuclear phosphoinositide signaling represents a novel unexplored opportunity to pharmacologically target several human pathologies that have been historically recalcitrant to clinical intervention. Some of the diseases most likely to be treatable with therapeutics targeting nuclear phosphoinositide signaling are endometriosis, NAFLD, NASH, glioblastomas, and hepatocellular carcinomas. The evidence linking nuclear phosphoinositide signaling to the molecular underpinnings of these diseases suggests that further exploration and validation in preclinical models is warranted. The more we learn about nuclear phosphoinositide signaling, the more likely that we can harness their unique signaling potential to reprogram the pathological state in these human diseases.
Footnotes
Abbreviations:
- GPR30
- G protein-coupled receptor 30 (GPER1)
- IPMK
- inositol polyphosphate multikinase (ipk2)
- LRH-1
- liver receptor homolog-1 (NR5A2)
- NAFLD
- nonalcoholic fatty liver disease
- NASH
- nonalcoholic steatohepatitis
- PDB
- Protein Data Bank
- PI3-kinase
- phosphatidylinositol 3-kinase (p110s)
- PI(3
- 4,5)P3, phosphatidylinositol (3,4,5) trisphosphate (PIP3)
- PTEN
- phosphatase and tensin homolog (MMAC)
- SF-1
- steroidogenic factor-1 (NR5A1)
- SUMO
- small ubiquitin-like modifier
- VMH
- ventral medial hypothalamus
This work was supported by American Cancer Society Grant RSG-17-063-01, V Foundation for Cancer Research Grant V2016-015, a Vanderbilt Diabetes Center Discovery Award from Vanderbilt University Medical Center, and an Ambassador’s Discovery Award from the Vanderbilt-Ingram Cancer Center. No author has an actual or perceived conflict of interest with the contents of this article.
REFERENCES
- 1.Blind R. D., Suzawa M., and Ingraham H. A.. 2012. Direct modification and activation of a nuclear receptor-PIP2 complex by the inositol lipid kinase IPMK. Sci. Signal. 5: ra44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Persidis A. 1998. Signal transduction as a drug-discovery platform. Nat. Biotechnol. 16: 1082–1083. [DOI] [PubMed] [Google Scholar]
- 3.Janku F., Hong D. S., Fu S., Piha-Paul S. A., Naing A., Falchook G. S., Tsimberidou A. M., Stepanek V. M., Moulder S. L., Lee J. J., et al. . 2014. Assessing PIK3CA and PTEN in early-phase trials with PI3K/AKT/mTOR inhibitors. Cell Reports. 6: 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thorpe L. M., Yuzugullu H., and Zhao J. J.. 2015. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer. 15: 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tzenaki N., and Papakonstanti E. A.. 2013. p110δ PI3 kinase pathway: emerging roles in cancer. Front. Oncol. 3: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toker A., and Cantley L. C.. 1997. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. 387: 673–676. [DOI] [PubMed] [Google Scholar]
- 7.Wong K. K., Engelman J. A., and Cantley L. C.. 2010. Targeting the PI3K signaling pathway in cancer. Curr. Opin. Genet. Dev. 20: 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barlow C. A., Laishram R. S., and Anderson R. A.. 2010. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol. 20: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irvine R. F. 2003. Nuclear lipid signalling. Nat. Rev. Mol. Cell Biol. 4: 349–360. [DOI] [PubMed] [Google Scholar]
- 10.Irvine R. F. 2006. Nuclear inositide signalling–expansion, structures and clarification. Biochim. Biophys. Acta. 1761: 505–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irvine R. F., and Divecha N.. 1992. Phospholipids in the nucleus–metabolism and possible functions. Semin. Cell Biol. 3: 225–235. [DOI] [PubMed] [Google Scholar]
- 12.Cocco L., Martelli A. M., Gilmour R. S., Ognibene A., Manzoli F. A., and Irvine R. F.. 1988. Rapid changes in phospholipid metabolism in the nuclei of Swiss 3T3 cells induced by treatment of the cells with insulin-like growth factor I. Biochem. Biophys. Res. Commun. 154: 1266–1272. [DOI] [PubMed] [Google Scholar]
- 13.Blind R. D., Sablin E. P., Kuchenbecker K. M., Chiu H-J., Deacon A. M., Das D., Fletterick R. J., and Ingraham H. A.. 2014. The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1. Proc. Natl. Acad. Sci. USA. 111: 15054–15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sablin E. P., Blind R. D., Uthayaruban R., Chiu H-J., Deacon A. M., Das D., Ingraham H. A., and Fletterick R. J.. 2015. Structure of liver receptor homolog-1 (NR5A2) with PIP3 hormone bound in the ligand binding pocket. J. Struct. Biol. 192: 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mellman D. L., Gonzales M. L., Song C., Barlow C. A., Wang P., Kendziorski C., and Anderson R. A.. 2008. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 451: 1013–1017. [DOI] [PubMed] [Google Scholar]
- 16.Ahn J-Y., Liu X., Cheng D., Peng J., Chan P-K., Wade P. A., and Ye K.. 2005. Nucleophosmin/B23, a nuclear PI(3,4,5)P(3) receptor, mediates the antiapoptotic actions of NGF by inhibiting CAD. Mol. Cell. 18: 435–445. [DOI] [PubMed] [Google Scholar]
- 17.Kumar A., Redondo-Munoz J., Perez-Garcia V., Cortes I., Chagoyen M., and Carrera A. C.. 2011. Nuclear but not cytosolic phosphoinositide 3-kinase beta has an essential function in cell survival. Mol. Cell. Biol. 31: 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stijf-Bultsma Y., Sommer L., Tauber M., Baalbaki M., Giardoglou P., Jones D. R., Gelato K. A., van Pelt J., Shah Z., Rahnamoun H., et al. . 2015. The basal transcription complex component TAF3 transduces changes in nuclear phosphoinositides into transcriptional output. Mol. Cell. 58: 453–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gozani O., Karuman P., Jones D. R., Ivanov D., Cha J., Lugovskoy A. A., Baird C. L., Zhu H., Field S. J., Lessnick S. L., et al. . 2003. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 114: 99–111. [DOI] [PubMed] [Google Scholar]
- 20.Chakravarthy M. V., Lodhi I. J., Yin L., Malapaka R. R. V., Xu H. E., Turk J., and Semenkovich C. F.. 2009. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 138: 476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H., Shi W., Tontonoz P., Wang S., Subbanagounder G., Hedrick C. C., Hama S., Borromeo C., Evans R. M., Berliner J. A., et al. . 2000. Role for peroxisome proliferator-activated receptor alpha in oxidized phospholipid-induced synthesis of monocyte chemotactic protein-1 and interleukin-8 by endothelial cells. Circ. Res. 87: 516–521. [DOI] [PubMed] [Google Scholar]
- 22.McIntyre T. M., Pontsler A. V., Silva A. R., St Hilaire A., Xu Y., Hinshaw J. C., Zimmerman G. A., Hama K., Aoki J., Arai H., et al. . 2003. Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARgamma agonist. Proc. Natl. Acad. Sci. USA. 100: 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oishi-Tanaka Y., and Glass C. K.. 2010. A new role for cyclic phosphatidic acid as a PPARgamma antagonist. Cell Metab. 12: 207–208. [DOI] [PubMed] [Google Scholar]
- 24.Delerive P., Furman C., Teissier E., Fruchart J., Duriez P., and Staels B.. 2000. Oxidized phospholipids activate PPARalpha in a phospholipase A2-dependent manner. FEBS Lett. 471: 34–38. [DOI] [PubMed] [Google Scholar]
- 25.Tsukahara T., Tsukahara R., Fujiwara Y., Yue J., Cheng Y., Guo H., Bolen A., Zhang C., Balazs L., Re F., et al. . 2010. Phospholipase D2-dependent inhibition of the nuclear hormone receptor PPARgamma by cyclic phosphatidic acid. Mol. Cell. 39: 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullaney B. C. C., Blind R. D. D., Lemieux G. A. A., Perez C. L. L., Elle I. C. C., Faergeman N. J. J., Van Gilst M. R. R., Ingraham H. A. A., and Ashrafi K.. 2010. Regulation of C. elegans fat uptake and storage by acyl-CoA synthase-3 is dependent on NR5A family nuclear hormone receptor nhr-25. Cell Metab. 12: 398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blind R. D. 2014. Disentangling biological signaling networks by dynamic coupling of signaling lipids to modifying enzymes. Adv. Biol. Regul. 54: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith A. D., Roda D., and Yap T. A.. 2014. Strategies for modern biomarker and drug development in oncology. J. Hematol. Oncol. 7: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chayen J., Gahan P. B., and La Cour L. F.. 1959. The masked lipids of nuclei. Q. J. Microsc. Sci. 100: 325–337. [Google Scholar]
- 30.Rose H. G., and Frenster J. H.. 1965. Composition and metabolism of lipids within repressed and active chromatin of interphase lymphocytes. Biochim. Biophys. Acta. 106: 577–591. [DOI] [PubMed] [Google Scholar]
- 31.Berezney R., and Coffey D. S.. 1974. Identification of a nuclear protein matrix. Biochem. Biophys. Res. Commun. 60: 1410–1417. [DOI] [PubMed] [Google Scholar]
- 32.Cocco L., Rubbini S., Manzoli L., Billi A. M., Faenza I., Peruzzi D., Matteucci A., Artico M., Gilmour R. S., and Rhee S. G.. 1999. Inositides in the nucleus: presence and characterisation of the isozymes of phospholipase beta family in NIH 3T3 cells. Biochim. Biophys. Acta. 1438: 295–299. [DOI] [PubMed] [Google Scholar]
- 33.Manzoli F. A., Maraldi N. M., Cocco L., Capitani S., and Facchini A.. 1977. Chromatin phospholipids in normal and chronic lymphocytic leukemia lymphocytes. Cancer Res. 37: 843–849. [PubMed] [Google Scholar]
- 34.Zilversmit D. B., and Hughes M. E.. 1977. Extensive exchange of rat liver microsomal phospholipids. Biochim. Biophys. Acta. 469: 99–110. [DOI] [PubMed] [Google Scholar]
- 35.Smith C. D., and Wells W. W.. 1983. Phosphorylation of rat liver nuclear envelopes. I. Characterization of in vitro protein phosphorylation. J. Biol. Chem. 258: 9360–9367. [PubMed] [Google Scholar]
- 36.Smith C. D., and Wells W. W.. 1983. Phosphorylation of rat liver nuclear envelopes. II. Characterization of in vitro lipid phosphorylation. J. Biol. Chem. 258: 9368–9373. [PubMed] [Google Scholar]
- 37.Divecha N., Banfić H., and Irvine R. F.. 1991. The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-I) in the plasma membrane, and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase C to the nucleus. EMBO J. 10: 3207–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Divecha N., Rhee S. G., Letcher A. J., and Irvine R. F.. 1993. Phosphoinositide signalling enzymes in rat liver nuclei: phosphoinositidase C isoform beta 1 is specifically, but not predominantly, located in the nucleus. Biochem. J. 289: 617–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manzoli F. A., Martelli A. M., Capitani S., Maraldi N. M., Rizzoli R., Barnabei O., and Cocco L.. 1989. Nuclear polyphosphoinositides during cell growth and differentiation. Adv. Enzyme Regul. 28: 25–34. [DOI] [PubMed] [Google Scholar]
- 40.Cocco L., Gilmour R. S., Ognibene A., Letcher A. J., Manzoli F. A., and Irvine R. F.. 1987. Synthesis of polyphosphoinositides in nuclei of Friend cells. Evidence for polyphosphoinositide metabolism inside the nucleus which changes with cell differentiation. Biochem. J. 248: 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vann L. R., Wooding F. B., Irvine R. F., and Divecha N.. 1997. Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. Biochem. J. 327: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamann B. L. B. L., and Blind R. D. R. D.. 2018. Nuclear phosphoinositide regulation of chromatin. J. Cell. Physiol. 233: 107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malabanan M. M., and Blind R. D.. 2016. Inositol polyphosphate multikinase (IPMK) in transcriptional regulation and nuclear inositide metabolism. Biochem. Soc. Trans. 44: 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindsay Y., McCoull D., Davidson L., Leslie N. R., Fairservice A., Gray A., Lucocq J., and Downes C. P.. 2006. Localization of agonist-sensitive PtdIns(3,4,5)P3 reveals a nuclear pool that is insensitive to PTEN expression. J. Cell Sci. 119: 5160–5168. [DOI] [PubMed] [Google Scholar]
- 45.Shah Z. H., Jones D. R., Sommer L., Foulger R., Bultsma Y., D’Santos C., and Divecha N.. 2013. Nuclear phosphoinositides and their impact on nuclear functions. FEBS J. 280: 6295–6310. [DOI] [PubMed] [Google Scholar]
- 46.Jones D. R., Bultsma Y., Keune W-J., Halstead J. R., Elouarrat D., Mohammed S., Heck A. J., D’Santos C. S., and Divecha N.. 2006. Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol. Cell. 23: 685–695. [DOI] [PubMed] [Google Scholar]
- 47.Vann L. R., Wooding F. B., Irvine R. F., and Divecha N.. 1997. Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. Biochem. J. 327: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boronenkov I. V., Loijens J. C., Umeda M., and Anderson R. A.. 1998. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell. 9: 3547–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mortier E., Wuytens G., Leenaerts I., Hannes F., Heung M. Y., Degeest G., David G., and Zimmermann P.. 2005. Nuclear speckles and nucleoli targeting by PIP2-PDZ domain interactions. EMBO J. 24: 2556–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Egea-Jimenez A. L., Gallardo R., Garcia-Pino A., Ivarsson Y., Wawrzyniak A. M., Kashyap R., Loris R., Schymkowitz J., Rousseau F., and Zimmermann P.. 2016. Frizzled 7 and PIP2 binding by syntenin PDZ2 domain supports Frizzled 7 trafficking and signalling. Nat. Commun. 7: 12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis A. E., Sommer L., Arntzen M. Ø., Strahm Y., Morrice N. A., Divecha N., and D’Santos C. S.. 2011. Identification of nuclear phosphatidylinositol 4,5-bisphosphate-interacting proteins by neomycin extraction. Mol. Cell. Proteomics. 10: M110.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao S., Lyons L. S., Fahrenholtz C. D., Wu F., Farooq A., Balkan W., and Burnstein K. L.. 2012. A novel nuclear role for the Vav3 nucleotide exchange factor in androgen receptor coactivation in prostate cancer. Oncogene. 31: 716–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osborne S. L., Thomas C. L., Gschmeissner S., and Schiavo G.. 2001. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J. Cell Sci. 114: 2501–2511. [DOI] [PubMed] [Google Scholar]
- 54.Crowder M. K., Seacrist C. D., and Blind R. D.. Phospholipid regulation of the nuclear receptor superfamily. Adv. Biol. Regul. 63: 6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.
- 56.Whitby R. J., Stec J., Blind R. D., Dixon S., Leesnitzer L. M., Orband-Miller L. A., Williams S. P., Willson T. M., Xu R., Zuercher W. J., et al. . 2011. Small molecule agonists of the orphan nuclear receptors steroidogenic factor-1 (SF-1, NR5A1) and liver receptor homologue-1 (LRH-1, NR5A2). J. Med. Chem. 54: 2266–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blind R. D., Sablin E. P., Kuchenbecker K. M., Chiu H. J., Deacon A. M., Das D., Fletterick R. J., and Ingraham H. A.. 2014. The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1. Proc. Natl. Acad. Sci. USA. 111: 15054–15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sablin E. P., Blind R. D., Krylova I. N., Ingraham J. G., Cai F., Williams J. D., Fletterick R. J., and Ingraham H. A.. 2009. Structure of SF-1 bound by different phospholipids: evidence for regulatory ligands. Mol. Endocrinol. 23: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blind R. D. D., Pineda-Torra I., Xu Y., Xu H. E. E., and Garabedian M. J. J.. 2012. Ligand structural motifs can decouple glucocorticoid receptor transcriptional activation from target promoter occupancy. Biochem. Biophys. Res. Commun. 420: 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ikeda Y. 1996. SF-1: a key regulator of development and function in the mammalian reproductive system. Acta Paediatr. Jpn. 38: 412–419. [DOI] [PubMed] [Google Scholar]
- 61.Ferraz-de-Souza B., Lin L., and Achermann J. C.. 2011. Steroidogenic factor-1 (SF-1, NR5A1) and human disease. Mol. Cell. Endocrinol. 336: 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohno C. K., Ueda H., and Petkovich M.. 1994. The Drosophila nuclear receptors FTZ-F1 alpha and FTZ-F1 beta compete as monomers for binding to a site in the fushi tarazu gene. Mol. Cell. Biol. 14: 3166–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanley N. A., Ikeda Y., Luo X., and Parker K. L.. 2000. Steroidogenic factor 1 (SF-1) is essential for ovarian development and function. Mol. Cell. Endocrinol. 163: 27–32. [DOI] [PubMed] [Google Scholar]
- 64.Luo X., Ikeda Y., and Parker K. L.. 1994. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 77: 481–490. [DOI] [PubMed] [Google Scholar]
- 65.Luo X., Ikeda Y., Schlosser D. A., and Parker K. L.. 1995. Steroidogenic factor 1 is the essential transcript of the mouse Ftz-F1 gene. Mol. Endocrinol. 9: 1233–1239. [DOI] [PubMed] [Google Scholar]
- 66.Ikeda Y., Shen W. H., Ingraham H. A., and Parker K. L.. 1994. Developmental expression of mouse steroidogenic factor-1, an essential regulator of the steroid hydroxylases. Mol. Endocrinol. 8: 654–662. [DOI] [PubMed] [Google Scholar]
- 67.Abd-Elaziz M., Moriya T., Akahira J., Nakamura Y., Suzuki T., and Sasano H.. 2005. Immunolocalization of nuclear transcription factors, DAX-1 and Ad4BP/SF-1, in human common epithelial ovarian tumors: correlations with StAR and steroidogenic enzymes in epithelial ovarian carcinoma. Int. J. Gynecol. Pathol. 24: 153–163. [DOI] [PubMed] [Google Scholar]
- 68.Fayard E., Auwerx J., and Schoonjans K.. 2004. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 14: 250–260. [DOI] [PubMed] [Google Scholar]
- 69.Allen A. K., and Spradling A. C.. 2008. The Sf1-related nuclear hormone receptor Hr39 regulates Drosophila female reproductive tract development and function. Development. 135: 311–321. [DOI] [PubMed] [Google Scholar]
- 70.Boulanger A., Clouet-Redt C., Farge M., Flandre A., Guignard T., Fernando C., Juge F., and Dura J-M.. 2011. ftz-f1 and Hr39 opposing roles on EcR expression during Drosophila mushroom body neuron remodeling. Nat. Neurosci. 14: 37–44. [DOI] [PubMed] [Google Scholar]
- 71.Lin W., Wang H. W., Sum C., Liu D., Hew C. L., and Chung B.. 2000. Zebrafish ftz-f1 gene has two promoters, is alternatively spliced, and is expressed in digestive organs. Biochem. J. 348: 439–446. [PMC free article] [PubMed] [Google Scholar]
- 72.Majdic G., Young M., Gomez-Sanchez E., Anderson P., Szczepaniak L. S., Dobbins R. L., McGarry J. D., and Parker K. L.. 2002. Knockout mice lacking steroidogenic factor 1 are a novel genetic model of hypothalamic obesity. Endocrinology. 143: 607–614. [DOI] [PubMed] [Google Scholar]
- 73.Labelle-Dumais C., Jacob-Wagner M., Paré J-F., Bélanger L., and Dufort D.. 2006. Nuclear receptor NR5A2 is required for proper primitive streak morphogenesis. Dev. Dyn. 235: 3359–3369. [DOI] [PubMed] [Google Scholar]
- 74.Heng J-C. D., Feng B., Han J., Jiang J., Kraus P., Ng J-H., Orlov Y. L., Huss M., Yang L., Lufkin T., et al. . 2010. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 6: 167–174. [DOI] [PubMed] [Google Scholar]
- 75.Miranda D. A., Krause W. C., Cazenave-Gassiot A., Suzawa M., Escusa H., Foo J. C., Shihadih D. S., Stahl A., Fitch M., Nyangau E., et al. . 2018. LRH-1 regulates hepatic lipid homeostasis and maintains arachidonoyl phospholipid pools critical for phospholipid diversity. JCI Insight. 3: 96151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee F. Y., Faivre E. J., Suzawa M., Lontok E., Ebert D., Cai F., Belsham D. D., and Ingraham H. A.. 2011. Eliminating SF-1 (NR5A1) sumoylation in vivo results in ectopic hedgehog signaling and disruption of endocrine development. Dev. Cell. 21: 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stein S., and Schoonjans K.. 2015. Molecular basis for the regulation of the nuclear receptor LRH-1. Curr. Opin. Cell Biol. 33: 26–34. [DOI] [PubMed] [Google Scholar]
- 78.Stein S., Oosterveer M. H., Mataki C., Xu P., Lemos V., Havinga R., Dittner C., Ryu D., Menzies K. J., Wang X., et al. . 2014. SUMOylation-dependent LRH-1/PROX1 interaction promotes atherosclerosis by decreasing hepatic reverse cholesterol transport. Cell Metab. 20: 603–613. [DOI] [PubMed] [Google Scholar]
- 79.Mataki C., Magnier B. C., Houten S. M., Annicotte J-S., Argmann C., Thomas C., Overmars H., Kulik W., Metzger D., Auwerx J., et al. . 2007. Compromised intestinal lipid absorption in mice with a liver-specific deficiency of liver receptor homolog 1. Mol. Cell. Biol. 27: 8330–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stein S., Lemos V., Xu P., Demagny H., Wang X., Ryu D., Jimenez V., Bosch F., Lüscher T. F., Oosterveer M. H., et al. . 2017. Impaired SUMOylation of nuclear receptor LRH-1 promotes nonalcoholic fatty liver disease. J. Clin. Invest. 127: 583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dyer K. N., Hammel M., Rambo R. P., Tsutakawa S. E., Rodic I., Classen S., Tainer J. A., and Hura G. L.. 2014. High-throughput SAXS for the characterization of biomolecules in solution: a practical approach. Methods Mol. Biol. 1091: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bradley A. R., Echalier A., Fairhead M., Strain-Damerell C., Brennan P., Bullock A. N., Burgess-Brown N. A., Carpenter E. P., Gileadi O., Marsden B. D., et al. . 2017. The SGC beyond structural genomics: redefining the role of 3D structures by coupling genomic stratification with fragment-based discovery. Essays Biochem. 61: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grabowski M., Niedzialkowska E., Zimmerman M. D., and Minor W.. 2016. The impact of structural genomics: the first quindecennial. J. Struct. Funct. Genomics. 17: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weiner B. E., Alexander N., Akin L. R., Woetzel N., Karakas M., and Meiler J.. 2014. BCL:Fold–protein topology determination from limited NMR restraints. Proteins. 82: 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Putnam D. K., Weiner B. E., Woetzel N., Lowe E. W., and Meiler J.. 2015. BCL:SAXS: GPU accelerated Debye method for computation of small angle X-ray scattering profiles. Proteins. 83: 1500–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krug U., Alexander N. S., Stein R. A., Keim A., Mchaourab H. S., Sträter N., and Meiler J.. 2016. Characterization of the domain orientations of E. coli 5′-nucleotidase by fitting an ensemble of conformers to DEER distance distributions. Structure. 24: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krylova I. N., Sablin E. P., Moore J., Xu R. X., Waitt G. M., MacKay J. A., Juzumiene D., Bynum J. M., Madauss K., Montana V., et al. . 2005. Structural analyses reveal phosphatidyl inositols as ligands for the NR5A orphan receptors SF-1 and LRH-1. Cell. 120: 343–355. [DOI] [PubMed] [Google Scholar]
- 88.Sablin E. P., Blind R. D., Krylova I. N., Ingraham J. G., Cai F., Williams J. D., Fletterick R. J., and Ingraham H. A.. 2009. Structure of SF-1 bound by different phospholipids: Evidence for regulatory ligands. Mol. Endocrinol. 23: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee Y-R., Chen M., and Pandolfi P. P.. 2018. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat. Rev. Mol. Cell Biol. 19: 547–562. [DOI] [PubMed] [Google Scholar]
- 90.Resnick A. C., Snowman A. M., Kang B. N., Hurt K. J., Snyder S. H., and Saiardi A.. 2005. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc. Natl. Acad. Sci. USA. 102: 12783–12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Odom A. R., Stahlberg A., Wente S. R., and York J. D.. 2000. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 287: 2026–2029. [DOI] [PubMed] [Google Scholar]
- 92.Frederick J. P., Mattiske D., Wofford J. A., Megosh L. C., Drake L. Y., Chiou S. T., Hogan B. L., and York J. D.. 2005. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc. Natl. Acad. Sci. USA. 102: 8454–8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qiu H. F., Dubois E., Broën P., Messenguy F., Broen P., and Messenguy F.. 1990. Functional analysis of ARGRI and ARGRIII regulatory proteins involved in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Gen. Genet. 222: 192–200. [DOI] [PubMed] [Google Scholar]
- 94.Nalaskowski M. M., Deschermeier C., Fanick W., and Mayr G. W.. 2002. The human homologue of yeast ArgRIII protein is an inositol phosphate multikinase with predominantly nuclear localization. Biochem. J. 366: 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chattaway J. A., Drobak B. K., Watkins P. A. C., Dawson A. P., Letcher A. J., Stephens L. R., and Irvine R. F.. 1992. An inositol 1,4,5-trisphosphate-6-kinase activity in pea roots. Planta. 187: 542–545. [DOI] [PubMed] [Google Scholar]
- 96.Endo-Streeter S., Tsui M. K., Odom A. R., Block J., and York J. D.. 2012. Structural studies and protein engineering of inositol phosphate multikinase. J. Biol. Chem. 287: 35360–35369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cestari I., Haas P., Moretti N. S., Schenkman S., and Stuart K.. 2016. Chemogenetic characterization of inositol phosphate metabolic pathway reveals druggable enzymes for targeting kinetoplastid parasites. Cell Chem. Biol. 23: 608–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.
- 99.Wang H., and Shears S. B.. 2017. Structural features of human inositol phosphate multikinase rationalize its inositol phosphate kinase and phosphoinositide 3-kinase activities. J. Biol. Chem. 292: 18192–18202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mayr G. W., Windhorst S., and Hillemeier K.. 2005. Antiproliferative plant and synthetic polyphenolics are specific inhibitors of vertebrate inositol-1,4,5-trisphosphate 3-kinases and inositol polyphosphate multikinase. J. Biol. Chem. 280: 13229–13240. [DOI] [PubMed] [Google Scholar]
- 101.McConnachie G., Pass I., Walker S. M., and Downes C. P.. 2003. Interfacial kinetic analysis of the tumour suppressor phosphatase, PTEN: evidence for activation by anionic phospholipids. Biochem. J. 371: 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blind R. D. 2014. Disentangling biological signaling networks by dynamic coupling of signaling lipids to modifying enzymes. Adv. Biol. Regul. 54: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maag D., Maxwell M. J., Hardesty D. A., Boucher K. L., Choudhari N., Hanno A. G., Ma J. F., Snowman A. S., Pietropaoli J. W., Xu R., et al. . 2011. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc. Natl. Acad. Sci. USA. 108: 1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Y-H., Hariharan A., Bastianello G., Toyama Y., Shivashankar G. V., Foiani M., and Sheetz M. P.. 2017. DNA damage causes rapid accumulation of phosphoinositides for ATR signaling. Nat. Commun. 8: 2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Parasar P., Ozcan P., and Terry K. L.. 2017. Endometriosis: epidemiology, diagnosis and clinical management. Curr. Obstet. Gynecol. Rep. 6: 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Greene A. D., Lang S. A., Kendziorski J. A., Sroga-Rios J. M., Herzog T. J., and Burns K. A.. 2016. Endometriosis: where are we and where are we going? Reproduction. 152: R63–R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fuldeore M. J., and Soliman A. M.. 2017. Prevalence and symptomatic burden of diagnosed endometriosis in the United States: national estimates from a cross-sectional survey of 59,411 women. Gynecol. Obstet. Invest. 82: 453–461. [DOI] [PubMed] [Google Scholar]
- 108.Foti P. V., Farina R., Palmucci S., Vizzini I. A. A., Libertini N., Coronella M., Spadola S., Caltabiano R., Iraci M., Basile A., et al. . 2018. Endometriosis: clinical features, MR imaging findings and pathologic correlation. Insights Imaging. 9: 149–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Canis M., Bouchet P., Botchorichvili R., Bourdel N., Pouly J-L., and Slim K.. 2015. Long-term evaluation of clinical results and quality of life for deeply infiltrating pelvic endometriosis. J. Minim. Invasive Gynecol. 22: S172. [DOI] [PubMed] [Google Scholar]
- 110.Bulun S. E., Utsunomiya H., Lin Z., Yin P., Cheng Y-H., Pavone M. E., Tokunaga H., Trukhacheva E., Attar E., Gurates B., et al. . 2009. Steroidogenic factor-1 and endometriosis. Mol. Cell. Endocrinol. 300: 104–108. [DOI] [PubMed] [Google Scholar]
- 111.Tian Y., Kong B., Zhu W., Su S., and Kan Y.. 2009. Expression of steroidogenic factor 1 (SF-1) and steroidogenic acute regulatory protein (StAR) in endometriosis is associated with endometriosis severity. J. Int. Med. Res. 37: 1389–1395. [DOI] [PubMed] [Google Scholar]
- 112.Lin B. C. C., Suzawa M., Blind R. D. D., Tobias S. C. C., Bulun S. E. E., Scanlan T. S. S., and Ingraham H. A. A.. 2009. Stimulating the GPR30 estrogen receptor with a novel tamoxifen analogue activates NR5A and promotes endometrial cell proliferation. Cancer Res. 69: 5415–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vercellini P., Trespidi L., Colombo A., Vendola N., Marchini M., and Crosignani P. G.. 1993. A gonadotropin-releasing hormone agonist versus a low-dose oral contraceptive for pelvic pain associated with endometriosis. Fertil. Steril. 60: 75–79. [PubMed] [Google Scholar]
- 114.Dmowski W. P., and Cohen M. R.. 1975. Treatment of endometriosis with an antigonadotropin, Danazol. A laparoscopic and histologic evaluation. Obstet. Gynecol. 46: 147–154. [PubMed] [Google Scholar]
- 115.Nagata Y., Nakamura G., and Kusuda M.. 1982. Therapeutic effect and side effects of danazol in endometriosis. Asia Oceania J. Obstet. Gynaecol. 8: 229–236. [DOI] [PubMed] [Google Scholar]
- 116.Abu Hashim H. 2014. Potential role of aromatase inhibitors in the treatment of endometriosis. Int. J. Womens Health. 6: 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pavone M. E., and Bulun S. E.. 2012. Aromatase inhibitors for the treatment of endometriosis. Fertil. Steril. 98: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dowsett M., Mehta A., Cantwell B. M., and Harris A. L.. 1991. Low-dose aminoglutethimide in postmenopausal breast cancer: effects on adrenal and thyroid hormone secretion. Eur. J. Cancer. 27: 846–849. [DOI] [PubMed] [Google Scholar]
- 119.Casper R. F. 2007. Aromatase inhibitors in ovarian stimulation. J. Steroid Biochem. Mol. Biol. 106: 71–75. [DOI] [PubMed] [Google Scholar]
- 120.Scaglia H. E., Carrere C. A., Mariani V. A., Zylbersztein C. C., Rey-Valzacchi G. J., Kelly E. E., and Aquilano D. R.. 1991. Altered testicular hormone production in infertile patients with idiopathic oligoasthenospermia. J. Androl. 12: 273–280. [PubMed] [Google Scholar]
- 121.Brodie A. M., Garrett W. M., Hendrickson J. R., Tsai-Morris C. H., and Williams J. G.. 1983. 1. Estrogen antagonists. Aromatase inhibitors, their pharmacology and application. J. Steroid Biochem. 19: 53–58. [PubMed] [Google Scholar]
- 122.Folkestad L., Bjarnason N. H., Bjerregaard J. K., and Brixen K.. 2009. The effect of aromatase inhibitors on bone metabolism. Basic Clin. Pharmacol. Toxicol. 104: 3–10. [DOI] [PubMed] [Google Scholar]
- 123.Harvey P. W., and Everett D. J.. 2003. The adrenal cortex and steroidogenesis as cellular and molecular targets for toxicity: critical omissions from regulatory endocrine disrupter screening strategies for human health? J. Appl. Toxicol. 23: 81–87. [DOI] [PubMed] [Google Scholar]
- 124.Garmendia J. V., and De Sanctis J. B.. 2012. Perspectives of new therapies for endometriosis. Recent Pat. Endocr. Metab. Immune Drug Discov. 6: 218–223. [DOI] [PubMed] [Google Scholar]
- 125.Cheng S-B., Graeber C. T., Quinn J. A., and Filardo E. J.. 2011. Retrograde transport of the transmembrane estrogen receptor, G-protein-coupled-receptor-30 (GPR30/GPER) from the plasma membrane towards the nucleus. Steroids. 76: 892–896. [DOI] [PubMed] [Google Scholar]
- 126.Otto C., Fuchs I., Kauselmann G., Kern H., Zevnik B., Andreasen P., Schwarz G., Altmann H., Klewer M., Schoor M., et al. . 2009. GPR30 does not mediate estrogenic responses in reproductive organs in mice. Biol. Reprod. 80: 34–41. [DOI] [PubMed] [Google Scholar]
- 127.Dennis M. K., Burai R., Ramesh C., Petrie W. K., Alcon S. N., Nayak T. K., Bologa C. G., Leitao A., Brailoiu E., Deliu E., et al. . 2009. In vivo effects of a GPR30 antagonist. Nat. Chem. Biol. 5: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chourasia T. K., Pang Y., and Thomas P.. 2015. The catecholestrogen, 2-hydroxyestradiol-17beta, acts as a G protein-coupled estrogen receptor 1 (GPER/GPR30) antagonist to promote the resumption of meiosis in zebrafish oocytes1. Biol. Reprod. 92: 69. [DOI] [PubMed] [Google Scholar]
- 129.Sadovsky Y., Crawford P. A., Woodson K. G., Polish J. A., Clements M. A., Tourtellotte L. M., Simburger K., and Milbrandt J.. 1995. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc. Natl. Acad. Sci. USA. 92: 10939–10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Choi Y-H., Fujikawa T., Lee J., Reuter A., and Kim K. W.. 2013. Revisiting the ventral medial nucleus of the hypothalamus: the roles of SF-1 neurons in energy homeostasis. Front. Neurosci. 7: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.. [Google Scholar]
- 132.Fujikawa T., Castorena C. M., Pearson M., Kusminski C. M., Ahmed N., Battiprolu P. K., Kim K. W., Lee S., Hill J. A., Scherer P. E., et al. . 2016. SF-1 expression in the hypothalamus is required for beneficial metabolic effects of exercise. eLife. 5: e18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Klöckener T., Hess S., Belgardt B. F., Paeger L., Verhagen L. A. W., Husch A., Sohn J-W., Hampel B., Dhillon H., Zigman J. M., et al. . 2011. High-fat feeding promotes obesity via insulin receptor/PI3K-dependent inhibition of SF-1 VMH neurons. Nat. Neurosci. 14: 911–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shinoda K., Lei H., Yoshii H., Nomura M., Nagano M., Shiba H., Sasaki H., Osawa Y., Ninomiya Y., Niwa O., et al. . 1995. Developmental defects of the ventromedial hypothalamic nucleus and pituitary gonadotroph in the Ftz-F1 disrupted mice. Dev. Dyn. 204: 22–29. [DOI] [PubMed] [Google Scholar]
- 135.Ikeda Y., Luo X., Abbud R., Nilson J. H., and Parker K. L.. 1995. The nuclear receptor steroidogenic factor 1 is essential for the formation of the ventromedial hypothalamic nucleus. Mol. Endocrinol. 9: 478–486. [DOI] [PubMed] [Google Scholar]
- 136.Kim K. W., Zhao L., Donato J., Kohno D., Xu Y., Elias C. F., Lee C., Parker K. L., and Elmquist J. K.. 2011. Steroidogenic factor 1 directs programs regulating diet-induced thermogenesis and leptin action in the ventral medial hypothalamic nucleus. Proc. Natl. Acad. Sci. USA. 108: 10673–10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fabelo C., Hernandez J., Chang R., Seng S., Alicea N., Tian S., Conde K., and Wagner E. J.. 2018. Endocannabinoid signaling at hypothalamic steroidogenic factor-1/proopiomelanocortin synapses is sex- and diet-sensitive. Front. Mol. Neurosci. 11: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Morohashi K. I., and Omura T.. 1996. Ad4BP/SF-1, a transcription factor essential for the transcription of steroidogenic cytochrome P450 genes and for the establishment of the reproductive function. FASEB J. 10: 1569–1577. [DOI] [PubMed] [Google Scholar]
- 139.Gronemeyer H., Gustafsson J-Å., and Laudet V.. 2004. Principles for modulation of the nuclear receptor superfamily. Nat. Rev. Drug Discov. 3: 950–964. [DOI] [PubMed] [Google Scholar]
- 140.Shi Y. 2006. Orphan nuclear receptors, excellent targets of drug discovery. Comb. Chem. High Throughput Screen. 9: 683–689. [DOI] [PubMed] [Google Scholar]
- 141.Shi Y. 2007. Orphan nuclear receptors in drug discovery. Drug Discov. Today. 12: 440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bang S., Chen Y., Ahima R. S., and Kim S. F.. 2014. Convergence of IPMK and LKB1-AMPK signaling pathways on metformin action. Mol. Endocrinol. 28: 1186–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bang S., Kim S., Dailey M. J., Chen Y., Moran T. H., Snyder S. H., and Kim S. F.. 2012. AMP-activated protein kinase is physiologically regulated by inositol polyphosphate multikinase. Proc. Natl. Acad. Sci. USA. 109: 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.
- 145.Kim K. W., Li S., Zhao H., Peng B., Tobet S. A., Elmquist J. K., Parker K. L., and Zhao L.. 2010. CNS-specific ablation of steroidogenic factor 1 results in impaired female reproductive function. Mol. Endocrinol. 24: 1240–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Saito K., He Y., Yang Y., Zhu L., Wang C., Xu P., Hinton A. O., Yan X., Zhao J., Fukuda M., et al. . 2016. PI3K in the ventromedial hypothalamic nucleus mediates estrogenic actions on energy expenditure in female mice. Sci. Rep. 6: 23459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Santiago A. M., Clegg D. J., and Routh V. H.. 2016. Estrogens modulate ventrolateral ventromedial hypothalamic glucose-inhibited neurons. Mol. Metab. 5: 823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kim K. W., Sohn J-W., Kohno D., Xu Y., Williams K., and Elmquist J. K.. 2011. SF-1 in the ventral medial hypothalamic nucleus: A key regulator of homeostasis. Mol. Cell. Endocrinol. 336: 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Snyder H. S., Sakaan S. A., March K. L., Siddique O., Cholankeril R., Cummings C. D., Gadiparthi C., Satapathy S. K., Ahmed A., and Cholankeril G.. 2018. Non-alcoholic fatty liver disease: a review of anti-diabetic pharmacologic therapies. J. Clin. Transl. Hepatol. 6: 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stiles B., Wang Y., Stahl A., Bassilian S., Lee W. P., Kim Y. J., Sherwin R., Devaskar S., Lesche R., Magnuson M. A., et al. . 2004. Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity [corrected]. Proc. Natl. Acad. Sci. USA. 101: 2082–2087. [Erratum. 2004. Proc. Natl. Acad. Sci. USA. 101: 5180.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen W. T., Zhu G., Pfaffenbach K., Kanel G., Stiles B., and Lee A. S.. 2014. GRP78 as a regulator of liver steatosis and cancer progression mediated by loss of the tumor suppressor PTEN. Oncogene. 33: 4997–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Peyrou M., Bourgoin L., and Foti M.. 2010. PTEN in liver diseases and cancer. World J. Gastroenterol. 16: 4627–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shearn C. T., Mercer K. E., Orlicky D. J., Hennings L., Smathers-McCullough R. L., Stiles B. L., Ronis M. J. J., and Petersen D. R.. 2014. Short term feeding of a high fat diet exerts an additive effect on hepatocellular damage and steatosis in liver-specific PTEN knockout mice. PLoS One. 9: e96553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bao W., Florea L., Wu N., Wang Z., Banaudha K., Qian J., Houzet L., Kumar R., and Kumar A.. 2014. Loss of nuclear PTEN in HCV-infected human hepatocytes. Infect. Agent. Cancer. 9: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Musille P. M., Pathak M., Lauer J. L., Hudson W. H., Griffin P. R., and Ortlund E. A.. 2012. Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation. Nat. Struct. Mol. Biol. 19: 532-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Moore D. D. 2012. Nuclear receptors reverse McGarry’s vicious cycle to insulin resistance. Cell Metab. 15: 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.
- 158.Lee J. M., Lee Y. K., Mamrosh J. L., Busby S. A., Griffin P. R., Pathak M. C., Ortlund E. A., and Moore D. D.. 2011. A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature. 474: 506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.de Mestier L., Pasmant E., Fleury C., Brixi H., Sohier P., Féron T., Diebold M. D., Clauser E., and Cadiot G.; Groupe d’Étude des Tumeurs Endocrines. 2017. Familial small-intestine carcinoids: chromosomal alterations and germline inositol polyphosphate multikinase sequencing. Dig. Liver Dis. 49: 98–102. [DOI] [PubMed] [Google Scholar]
- 160.Tykocki T., and Eltayeb M.. 2018. Ten-year survival in glioblastoma. A systematic review. J. Clin. Neurosci. 54: 7–13. [DOI] [PubMed] [Google Scholar]
- 161.Jones C., Perryman L., and Hargrave D.. 2012. Paediatric and adult malignant glioma: close relatives or distant cousins? Nat. Rev. Clin. Oncol. 9: 400–413. [DOI] [PubMed] [Google Scholar]
- 162.Vinci M., Burford A., Molinari V., Kessler K., Popov S., Clarke M., Taylor K. R., Pemberton H. N., Lord C. J., Gutteridge A., et al. . 2018. Functional diversity and cooperativity between subclonal populations of pediatric glioblastoma and diffuse intrinsic pontine glioma cells. Nat. Med. 24: 1204–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cai X., and Sughrue M. E.. 2017. Glioblastoma: new therapeutic strategies to address cellular and genomic complexity. Oncotarget. 9: 9540–9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bassi C., Ho J., Srikumar T., Dowling R. J. O., Gorrini C., Miller S. J., Mak T. W., Neel B. G., Raught B., and Stambolic V.. 2013. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 341: 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Liu J-L. L. J-L., Sheng X., Hortobagyi Z. K., Mao Z., Gallick G. E., and Yung W. K. A.. 2005. Nuclear PTEN-mediated growth suppression is independent of Akt down-regulation. Mol. Cell. Biol. 25: 6211–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.DeGraffenried L. A., Fulcher L., Friedrichs W. E., Grünwald V., Ray R. B., and Hidalgo M.. 2004. Reduced PTEN expression in breast cancer cells confers susceptibility to inhibitors of the PI3 kinase/Akt pathway. Ann. Oncol. 15: 1510–1516. [DOI] [PubMed] [Google Scholar]
- 167.Maehama T., and Dixon J. E.. 1998. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273: 13375–13378. [DOI] [PubMed] [Google Scholar]
- 168.Trotman L. C., Wang X., Alimonti A., Chen Z., Teruya-Feldstein J., Yang H., Pavletich N. P., Carver B. S., Cordon-Cardo C., Erdjument-Bromage H., et al. . 2007. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 128: 141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gong L., Govan J. M., Evans E. B., Dai H., Wang E., Lee S-W., Lin H-K., Lazar A. J., Mills G. B., and Lin S-Y.. 2015. Nuclear PTEN tumor-suppressor functions through maintaining heterochromatin structure. Cell Cycle. 14: 2323–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fan C., He L., Kapoor A., Rybak A. P., De Melo J., Cutz J. C., and Tang D.. 2009. PTEN inhibits BMI1 function independently of its phosphatase activity. Mol. Cancer. 8: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.. [Google Scholar]
- 172.Song M. S., Carracedo A., Salmena L., Song S. J., Egia A., Malumbres M., and Pandolfi P. P.. 2011. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 144: 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rask-Andersen M., Almén M. S., and Schiöth H. B.. 2011. Trends in the exploitation of novel drug targets. Nat. Rev. Drug Discov. 10: 579–590. [DOI] [PubMed] [Google Scholar]
- 174.Bull S. C., and Doig A. J.. 2015. Properties of protein drug target classes. PLoS One. 10: e0117955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gross S., Rahal R., Stransky N., Lengauer C., and Hoeflich K. P.. 2015. Targeting cancer with kinase inhibitors. J. Clin. Invest. 125: 1780–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sawyers C. L. 2002. Rational therapeutic intervention in cancer: kinases as drug targets. Curr. Opin. Genet. Dev. 12: 111–115. [DOI] [PubMed] [Google Scholar]
- 177.Wesche H., Xiao S-H., and Young S. W.. 2005. High throughput screening for protein kinase inhibitors. Comb. Chem. High Throughput Screen. 8: 181–195. [DOI] [PubMed] [Google Scholar]
- 178.Stephens L., Williams R., and Hawkins P.. 2005. Phosphoinositide 3-kinases as drug targets in cancer. Curr. Opin. Pharmacol. 5: 357–365. [DOI] [PubMed] [Google Scholar]
- 179.Erlanson D. A., Fesik S. W., Hubbard R. E., Jahnke W., and Jhoti H.. 2016. Twenty years on: the impact of fragments on drug discovery. Nat. Rev. Drug Discov. 15: 605–619. [DOI] [PubMed] [Google Scholar]
- 180.Kim E., Beon J., Lee S., Park J., and Kim S.. 2016. IPMK: A versatile regulator of nuclear signaling events. Adv. Biol. Regul. 61: 25–32. [DOI] [PubMed] [Google Scholar]
- 181.Baker S. J. 2007. PTEN enters the nuclear age. Cell. 128: 25–28. [DOI] [PubMed] [Google Scholar]
- 182.Perez A., Anzaldua M., McCormick J., and Fisher-Hoch S.. 2004. High frequency of chronic end-stage liver disease and hepatocellular carcinoma in a Hispanic population. J. Gastroenterol. Hepatol. 19: 289–295. [DOI] [PubMed] [Google Scholar]
- 183.El-Serag H. B., Lau M., Eschbach K., Davila J., and Goodwin J.. 2007. Epidemiology of hepatocellular carcinoma in Hispanics in the United States. Arch. Intern. Med. 167: 1983–1989. [DOI] [PubMed] [Google Scholar]
- 184.Lazo M., Bilal U., and Perez-Escamilla R.. 2015. Epidemiology of NAFLD and type 2 diabetes: health disparities among persons of Hispanic origin. Curr. Diab. Rep. 15: 116. [DOI] [PubMed] [Google Scholar]
- 185.Horie Y., Suzuki A., Kataoka E., Sasaki T., Hamada K., Sasaki J., Mizuno K., Hasegawa G., Kishimoto H., Iizuka M., et al. . 2004. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J. Clin. Invest. 113: 1774–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cristea C. G., Gheonea I. A., Sandulescu L. D., Gheonea D. I., Ciurea T., and Purcarea M. R.. 2015. Considerations regarding current diagnosis and prognosis of hepatocellular carcinoma. J. Med. Life. 8: 120–128. [PMC free article] [PubMed] [Google Scholar]
- 187.Ha T. Y., Hwang S., Moon K. M., Won Y. J., Song G. W., Kim N., Tak E., Ryoo B. Y., and Hong H. N.. 2015. Sorafenib inhibits migration and invasion of hepatocellular carcinoma cells through suppression of matrix metalloproteinase expression. Anticancer Res. 35: 1967–1976. [PubMed] [Google Scholar]
- 188.Ganten M. K., Schuessler M., Bruckner T., Ganten T. M., and Koschny R.. 2015. Pancreatic atrophy in hepatocellular carcinoma patients receiving long-term treatment with sorafenib. Oncology. 89: 88–94. [DOI] [PubMed] [Google Scholar]
- 189.Abdel-Rahman O., and Fouad M.. 2014. Sorafenib-based combination as a first line treatment for advanced hepatocellular carcinoma: a systematic review of the literature. Crit. Rev. Oncol. Hematol. 91: 1–8. [DOI] [PubMed] [Google Scholar]
- 190.
- 191.Lee Y. K., and Moore D. D.. 2008. Liver receptor homolog-1, an emerging metabolic modulator. Front. Biosci. 13: 5950–5958. [DOI] [PubMed] [Google Scholar]
- 192.Chan K-M., Kuo C-F., Hsu J-T., Chiou M-J., Wang Y-C., Wu T-H., Lee C-F., Wu T-J., Chou H-S., and Lee W-C.. 2017. Metformin confers risk reduction for developing hepatocellular carcinoma recurrence after liver resection. Liver Int. 37: 434–441. [DOI] [PubMed] [Google Scholar]
- 193.Zhang Q., Kong J., Dong S., Xu W., and Sun W.. 2017. Metformin exhibits the anti-proliferation and anti-invasion effects in hepatocellular carcinoma cells after insufficient radiofrequency ablation. Cancer Cell Int. 17: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tseng C-H. Metformin and risk of hepatocellular carcinoma in patients with type 2 diabetes. Liver Int. Epub ahead of print. June 29, 2018; doi:10.1111/liv.13872. [DOI] [PubMed] [Google Scholar]
- 195.Musille P. M., Kohn J. A., and Ortlund E. A.. 2013. Phospholipid–driven gene regulation. FEBS Lett. 587: 1238–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Dubois E., Bercy J., and Messenguy F.. 1987. Characterization of two genes, ARGRI and ARGRIII required for specific regulation of arginine metabolism in yeast. Mol. Gen. Genet. 207: 142–148. [DOI] [PubMed] [Google Scholar]
- 197.Bercy J., Dubois E., and Messenguy F.. 1987. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene. 55: 277–285. [DOI] [PubMed] [Google Scholar]