Abstract
Phosphoinositides are key regulators of a large number of diverse cellular processes that include membrane trafficking, plasma membrane receptor signaling, cell proliferation, and transcription. How a small number of chemically distinct phosphoinositide signals are functionally amplified to exert specific control over such a diverse set of biological outcomes remains incompletely understood. To this end, a novel mechanism is now taking shape, and it involves phosphatidylinositol (PtdIns) transfer proteins (PITPs). The concept that PITPs exert instructive regulation of PtdIns 4-OH kinase activities and thereby channel phosphoinositide production to specific biological outcomes, identifies PITPs as central factors in the diversification of phosphoinositide signaling. There are two evolutionarily distinct families of PITPs: the Sec14-like and the StAR-related lipid transfer domain (START)-like families. Of these two families, the START-like PITPs are the least understood. Herein, we review recent insights into the biochemical, cellular, and physiological function of both PITP families with greater emphasis on the START-like PITPs, and we discuss the underlying mechanisms through which these proteins regulate phosphoinositide signaling and how these actions translate to human health and disease.
Keywords: lipid signaling, lipid and membrane trafficking, cell signaling, diseases, lipids • membranes
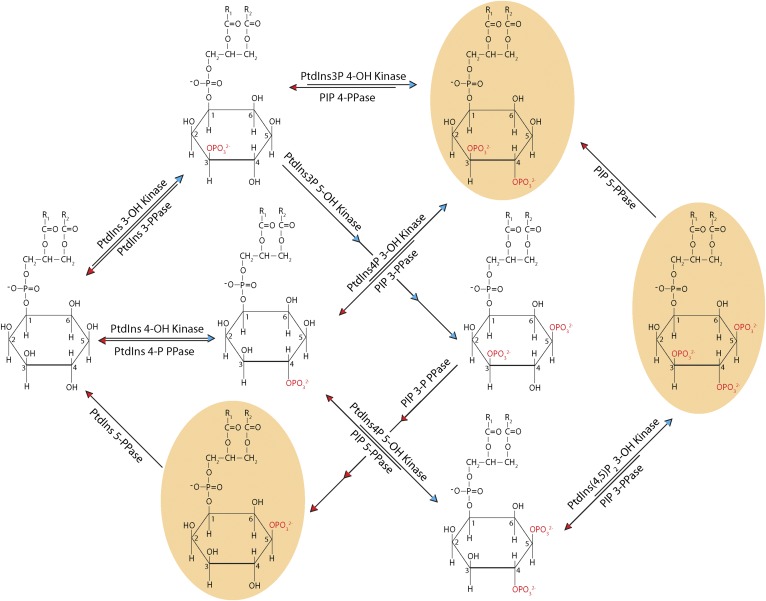
Phosphoinositides are a set of molecules derived from phosphatidylinositol (PtdIns), which define a chemical code that facilitates the conversion of intracellular membrane surfaces into high-definition lipid signaling screens (Fig. 1). Mammalian cells synthesize seven chemically distinct phosphoinositides: PtdIns 3-phosphate (PtdIns3P), PtdIns 4-phosphate (PtdIns4P), PtdIns 5-phosphate (PtdIns5P), PtdIns 3,5-bisphosphate [PtdIns(3,5)P2], PtdIns 4,5-bisphosphate [PtdIns(4,5)P2], PtdIns 3,4-bisphosphate [PtdIns(3,4)P2], and PtdIns 3,4,5-triphosphate [PtdIns(3,4,5)P3]. Yeasts (Saccharomyces) synthesize all of these except PtdIns(3,4)P2 and PtdIns(3,4,5)P3. This cohort of molecules is produced by a series of reversible phosphorylation and dephosphorylation reactions catalyzed by phosphoinositide kinases (PIKs) and phosphatases, respectively, that target the inositol headgroup at positions C3, C4, and C5 (1, 2). These chemically distinct lipids, although small in number, control an impressively large set of intracellular activities. How is such diversification of biological function achieved? Current views for how PtdIns and phosphoinositide signaling are diversified focus on regulatory mechanisms that include: the regulation and localization of the enzymes that produce, degrade, or sequester phosphoinositides; the identities of the effector proteins that recognize these molecules with the associated coincidence detection mechanisms that accompany productive phosphoinositide/effector engagement; and the physical properties of the membranes in which these lipids reside (3, 4). In more recent progress, PtdIns transfer proteins (PITPs) are increasingly recognized as central components in mechanisms for diversification of phosphoinositide biological outcomes.
Fig. 1.
Phosphoinositide metabolism. The synthesis and degradation of phosphoinositides by kinases and phosphatases (denoted as “PPase”), respectively, is depicted in schematic form. PLs in tan bubbles exist in higher eukaryotes but not S. cerevisiae.
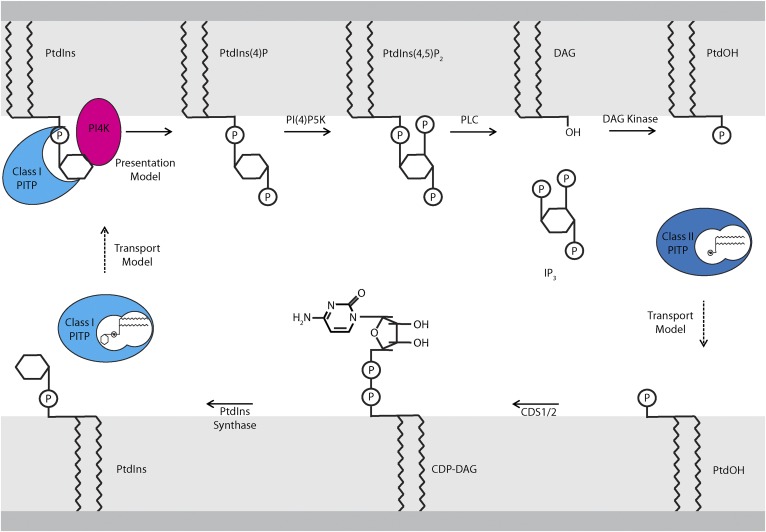
PITPs were historically interpreted as “lipid carriers” that ferry PtdIns from sites of synthesis (i.e., the ER) to membranes depleted of PtdIns as a consequence of phosphoinositide-driven signaling reactions. The genesis of this view came from pioneering studies demonstrating that PtdIns turnover and phospholipase C (PLC)-catalyzed inositol phosphate headgroup cleavage rapidly increase following stimulation of professional secretory tissues with carbachol (5–8). Those observations ultimately led to the discovery that PtdIns(4,5)P2 is the metabolic precursor for the second messengers, inositol triphosphate (IP3) and diacylglycerol (DAG), via the action of plasma-membrane resident PLC (9–17). IP3 promotes Ca2+ release from ER stores, and DAG activates downstream protein kinase signaling. The various data prompted Robert Michell (18) to raise the question: how are phosphoinositides replenished at the plasma membrane after consumption by PLC? That question highlighted the conundrum that PtdIns synthesis is presumably restricted to the ER, whereas the phosphoinositide cycle runs in a physically distinct subcellular compartment. Michell hypothesized that plasma membrane PtdIns pools are resupplied from the ER in a lipid-trafficking cycle where soluble lipid-carrier proteins ferry PtdIns from the ER to the plasma membrane, and that a reciprocal lipid carrier-mediated pathway returns phosphatidic acid (PtdOH) [produced by DAG kinases using plasma membrane DAG substrates generated by PLC-mediated hydrolysis of PtdIns(4,5)P2] back to the ER to refuel PtdIns synthesis (Fig. 2). The Michell hypothesis launched a search for such lipid transfer proteins and defined a presumptive mechanism of action for proteins so identified that dominates the field to this day.
Fig. 2.
Models of PITP function in the PIP cycle. PtdIns is synthesized at the ER by the sequential action of the enzymes, CDS1/2 and PtdIns synthase, which convert PtdOH to CDP-DAG and CDP-DAG to PtdIns, respectively. “Transport models” for describing PITP function postulate that class I PITPs transfer PtdIns from the ER to signaling membranes (i.e., the plasma membrane). “Presentation models” describe PITPs as noncatalytic factors that present PtdIns to PI4K, thereby directly stimulating PtdIns4P synthesis. In the presentation model, PITP utilizes PtdIns at the signaling membrane and does not catalyze PtdIns transport from the ER. PtdIns4P synthesized at the plasma membrane can be converted to PtdIns(4,5)P2, which is then hydrolyzed by agonist-stimulated PLC activity to produce DAG and IP3. DAG is converted to PtdOH by DAG kinase. PtdOH must then be replenished at the ER to restore the PtdIns biosynthetic cycle. Class II PITPs, which bind PtdOH, are proposed to fulfill this role in a transport mechanism.
A major obstacle to progress on understanding PITPs (and lipid transfer proteins in general) was the dearth of genuine functional assays from which to analyze their mechanism(s) of function in a reliable and physiologically relevant way. Recognition that Sec14 is the major PITP of yeast broke this barrier, and other studies subsequently followed (19, 20). Building evidence now reports that PITPs are central factors in determining the biological outcomes of phosphoinositide signaling and that these do so via a novel mechanism where at least some PITPs are not lipid carriers at all. Rather they function as nanoreactors that utilize their lipid-exchange cycle to present PtdIns to otherwise biologically insufficient PtdIns kinases, thereby making these kinases better enzymes on demand (21–23). The biological functions of PITPs and how these proteins diversify phosphoinositide signaling is the central focus of this review.
PHOSPHOINOSITIDE SIGNALING IS A MAJOR REGULATOR OF MEMBRANE TRAFFICKING
The discovery that Sec14 is the major yeast PITP (19), and subsequent studies demonstrating that Sec14 executes an essential coordination of multiple aspects of lipid metabolism to support membrane trafficking from the yeast trans-Golgi network (TGN) (20, 24), revealed for the first time that lipid signaling is a core feature of constitutive membrane trafficking in eukaryotic cells, particularly as it relates to the Golgi system. It is now well-appreciated that lipid signaling in TGN/endosomal membranes defines their functionality in terms of controlling vesicular trafficking into and from these compartments (19, 20, 25–27). As a growing body of work indicates that PITPs generally function in the regulation of membrane trafficking at the level of the Golgi/endosomal systems, these involvements will be a significant point of discussion in this review. Thus, we first give a brief overview of the TGN/endosomal system.
The TGN and endosomal networks define a continuum of maturing compartments that serve as intracellular regulatory hubs in which lipid signaling events are integrated with cargo sorting and membrane trafficking. The TGN/endosomal system not only orchestrates shuttling of Golgi-derived cargo to specific cellular locations but also receives important endocytic cargo (e.g., signaling receptors) recycled from the plasma membrane (28, 29). Endosomes link the plasma membrane with the Golgi and the lysosome, and are comprised of three morphologically and biochemically distinct compartments: the early, late, and recycling endosomes (30). The early endosome is the first compartment to accept and sort internalized cargo from the plasma membrane (31). Cargo destined for recycling to the cell surface can be transported by at least two routes: a fast route involving membrane tubulation or a slow route involving transport by the recycling endosome (32–34). Endosomes containing cargo destined for degradation mature into late endosomes, a process marked by an exchange of the surface Rab GTPase population (35, 36). This is accompanied by endosome acidification and internalization of intraluminal vesicles containing cargo marked for degradation, giving rise to a multivesicular endosome (or body) that fuses with the lysosome, facilitating cargo degradation (37–41).
PtdIns4P AND MEMBRANE TRAFFICKING IN EUKARYOTES
As the bulk of the evidence to date connects PITP function with PtdIns4P synthesis/signaling, we focus on PtdIns4P and the 4-OH phosphoinositide signaling in this review. The roles of PtdIns3P and PtdIns(3,5)P2 in the endosomal/lysosomal compartments are primary themes of other outstanding reviews (42, 43). PtdIns4P pools are detected on the TGN and early endosomal membranes, on the plasma membrane, and on late endosomal and lysosomal membranes (44–49). Direct evidence that PtdIns4P regulates Golgi trafficking was first described in yeast through the identification of secretory pathway defects associated with hypomorphic PtdIns 4-OH kinase (PI4K) mutants compromised for PtdIns4P synthesis (50, 51). A number of PtdIns4P effectors have since been discovered, which function on Golgi membranes to regulate vesicle budding (52, 53), maintenance of Golgi structure (54), and perhaps nonvesicular lipid transport (55, 56).
The most immediate control levels for PtdIns4P signaling come from the PI4Ks that generate PtdIns4P by phosphorylating PtdIns on the 4-OH position of the inositol ring (Fig. 1). Mammals produce two classes of structurally distinct PI4Ks: type II (PI4KIIα and PI4KIIβ) and type III (PI4KIIIα and PI4KIIIβ) PI4Ks (57). The type II enzymes are orthologs of the yeast Lsb6 PI4K and primarily localize to the Golgi and endosomal membranes (57, 58), although there are reports that PI4KIIα chiefly localizes to the TGN with a minor pool present at the ER (59, 60). PI4KIIβ is found on both trafficking vesicles and the plasma membrane (61, 62). The mammalian type PI4KIIIα and PI4KIIIβ enzymes are orthologs of the yeast plasma membrane Stt4 and the Golgi Pik1 PI4Ks, respectively (63–66). The mammalian type III PI4Ks are variously reported to localize to the cytosol (67, 68), nucleolus (69, 70), cis-Golgi (71), and ER (72). As is the case in yeast, PI4KIIIα synthesizes PtdIns4P at the plasma membrane (48).
PI4K activity provides the next control point for regulation of PtdIns4P production, and much effort is devoted to identifying such regulators of kinase activity. Calcium and the Arf1 GTPase regulate yeast and mammalian Pik1/PI4KIIIβ. Yeast Pik1 forms a complex with a small noncatalytic integral membrane Ca2+-binding protein subunit (frequenin) required for enzymatic activity (74–79). Mammals present a similar scenario as the PI4KIIIβ complexes with a frequenin ortholog [neuronal calcium sensor-1 (NCS-1)], and PI4KIIIβ recruitment to the Golgi is promoted by activated Arf1 (63, 79).
PtdIns4P signals are interpreted by the direct binding of effector proteins to the lipid, and these effectors include activated small GTPases of the Arf and Rab families (3). Arf1-GTP cooperates with PtdIns4P to recruit vesicle biogenesis machinery that nucleates vesicle formation. These factors include components of clathrin adaptor protein complex 1 (AP-1), Rab GTPases and Rab-guanine nucleotide exchange factors, oxysterol binding proteins (OSBPs), and effector proteins, such as the Gga Arf-GTPase-binding proteins that regulate trafficking between the TGN and lysosomes (81–83). Four phosphate adaptor proteins (FAPPs) also bind both ARF GTPases and PtdIns4P at the Golgi, and FAPP membrane recruitment induces membrane tubulation and formation of transport carriers (53, 84).
The functions of discrete PtdIns4P pools can be further specified by the local PtdIns4P concentration, such as in the activation of yeast Sec4, a ras-like G-protein required for consumption of post-Golgi secretory vesicles at the plasma membrane. A critical aspect of the Sec4 cycle involves the association of Sec4-GTP with Sec15, an essential subunit of the exocyst complex that tethers secretory vesicles to the plasma membrane (85, 86). Sec2, a Sec4-GEF, temporally controls Sec4 activation via coincidence detection of PtdIns4P and the Rab GTPase, Ypt31, which itself regulates TGN exocytic vesicle biogenesis (87, 88). Whether Sec2 associates with Ypt31 for its activation, or with Sec15, is determined by local PtdIns4P availability. Basal PtdIns4P (as is presumably the prevailing condition on post-Golgi membranes) facilitates Sec2 binding to Sec15 for downstream activation of Sec4, whereas elevated PtdIns4P promotes Sec2 binding to Golgi Ypt31 (89).
HIGHER ORDER PHOSPHOINOSITIDES
PtdIns4P further serves as a metabolic precursor for the generation of higher-order phosphoinositides, including PtdIns(4,5)P2 in all eukaryotes and PtdIns(3,4)P2 and PtdIns(3,4,5)P3 in higher organisms. The synthesis of PtdIns(4,5)P2 is dependent not only on the kinases that directly produce it but also on the PI4Ks and PI5Ks that serve to produce metabolic precursors for the reaction. Yeasts express a single PtdIns4P 5-OH kinase (PtdIns4P5K), Mss4, that catalyzes synthesis of plasma membrane PtdIns(4,5)P2 from Pik1- or Stt4-generated PtdIns4P (Fig. 1). The specific PtdIns4P pool utilized by Mss4 likely dictates downstream PtdIns(4,5)P2 signaling outcome, suggesting the existence of at least two distinct functional pools of yeast plasma membrane PtdIns(4,5)P2 (65, 90).
As in the case of PtdIns4P, the cohort of PtdIns(4,5)P2-producing lipid kinases is larger in mammals, consistent with an expanded capacity for PtdIns(4,5)P2 pool specification. Mammals express three type I PtdIns4P5K isoforms: PIPKIα, PIPKIβ, and PIPKIγ (91). Mammalian type II PIKs are encoded by three genes designated as α, β, and γ, and these enzymes produce PtdIns(4,5)P2 by phosphorylating PtdIns5P on the 4-OH position on the inositol ring. However, as PtdIns5P is a quantitatively a trace phosphoinositide, it remains an open question as to why cells use this pathway to produce PtdIns(4,5)P2 at all when, by far, the dominant channel for producing PtdIns(4,5)P2 is through the action of PtdIns4P5Ks (92–94).
PtdIns(4,5)P2 is also a substrate for PI3K in a reaction that produces PtdIns(3,4,5)P3 (95, 96). This phosphoinositide class specifies localization and activation of various effectors through direct (e.g., PDK1) and indirect interactions (e.g., AKT), and these protein::PtdIns(3,4,5)P3 interactions are typically mediated through pleckstrin homology (PH) domains that have evolved as phosphoinositide recognition units (97–102).
PtdIns(4,5)P2 SIGNALING AND MEMBRANE TRAFFICKING
PtdIns(4,5)P2 functions in anterograde trafficking to the plasma membrane in regulated exocytic pathways, such as those involving plasma membrane targeting and fusion of dense core secretory granules in neuroendocrine cells and synaptic vesicles in neurons (103, 104). In these cases, this phosphoinositide modulates activities of PtdIns(4,5)P2-binding proteins, such as calcium-activated protein for secretion (CAPS) (105), the synaptotagmin Syt1 isoform (106, 107), rabphilin (108, 109), synaptotagmin (110), and secretory carrier membrane protein 2 (SCAMP2) (111).
PtdIns(4,5)P2 has a dual role in growth factor receptor signaling, as it is essential for the downregulation of the very signaling reactions it propagates. For example, PtdIns(4,5)P2 recruits clathrin coat adaptors to the plasma membrane by a direct-binding mechanism (112–114). These recruitment events drive internalization of activated receptors, as evidenced by the fact that depletion of plasma membrane PtdIns(4,5)P2 blocks clathrin coat adaptor recruitment to the mammalian plasma membrane and retards internalization of activated-receptors (115, 116). Following internalization, disassembly of the clathrin coat on endocytic vesicles requires PtdIns(4,5)P2 degradation by the synaptojanin family of phosphoinositide phosphatases (117–119). This is a highly conserved process. Clathrin-dependent endocytosis is a PtdIns(4,5)P2-mediated process in yeast as well (120).
THE INTRINSIC BIOLOGICAL INSUFFICIENCY OF PI4Ks
The discussion of PITPs is introduced by a simple, yet underappreciated, concept. Namely, that PI4Ks are biologically insufficient enzymes that, on their own, even as wild-type enzymes, cannot produce sufficient PtdIns4P to execute meaningful signaling in cells (22, 23, 121). This is not to say that these enzymes fail to produce PtdIns4P in cells on their own. They do. However, the levels produced are insufficient to overcome the action of antagonists of PtdIns4P signaling, such as phosphoinositide phosphatases and PtdIns4P-binding proteins that sequester the phosphoinositide from its effectors. From an alternative perspective, if PI4K is viewed as a writer for PtdIns4P signaling, its intrinsic signature is too weak to overcome the action of erasers of PtdIns4P signaling. It is here where PITPs enter the picture. These proteins boost PtdIns4P production by PI4Ks so that the pool produced is sufficient to overcome the action of erasers and support PtdIns4P signaling. Obviously, such a mechanism also lends itself not only to exquisite control of PI4K activity, and therefore PtdIns4P signaling, but also to diversifying the outcomes for PtdIns4P signaling.
PITPs AND MECHANISMS FOR DIVERSIFYING PtdIns4P SIGNALING
PITPs are ancient molecules conserved from single-celled eukaryotes to man. These fall into two distinct families: the Sec14-PITPs and the StAR-related lipid transfer domain (START)-like PITPs (19, 20, 24). Though structurally unrelated, the two families share the ability to transfer PtdIns between membranes in vitro. There are several compelling reasons to study PITP reaction mechanisms. First, the biochemistry of phospholipid (PL) exchange by PITPs describes a unique interfacial reaction that operates in the absence of energy. That biochemistry alone begs mechanistic understanding, and it is clear that PITP function in cells employs lipid exchange as a core functional property. Second, and in that regard, the lipid exchange properties of PITPs are associated with diverse biology across the Eukaryota, including a number of mammalian disease conditions (122). Third, the biological derangements associated with individual PITP insufficiencies report novel PITP-dependent mechanisms for regulating PtdIns kinase activities, and that PITPs are central participants in mechanisms for diversifying the biological outcomes of phosphoinositide signaling (121, 123). As Sec14 represents the best understood PITP in terms of its biochemistry, structural biology, and biological activity, we first describe the Sec14 knowledge base and then apply it to a conceptual framework for discussing START-like PITPs.
STRUCTURAL INSIGHTS INTO Sec14 FUNCTION: THE CURIOUS ENGINEERING OF THE Sec14-FOLD
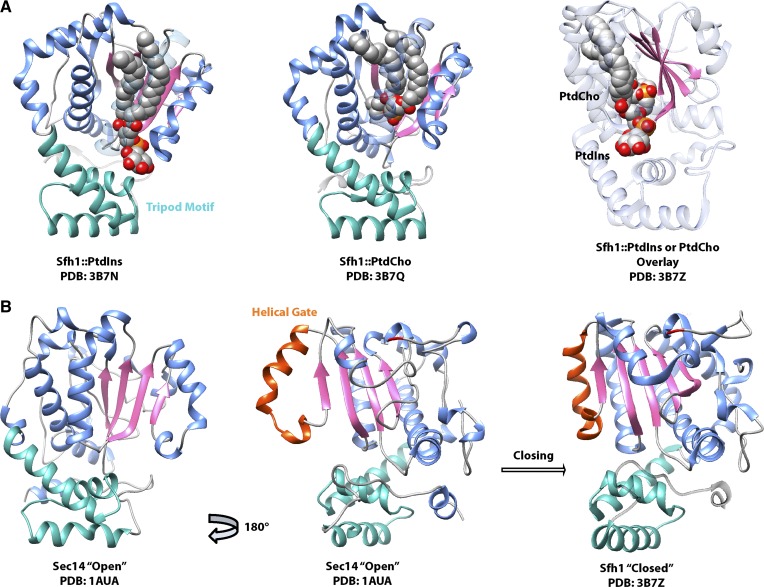
Solution structures of several Sec14 and Sec14-like PITP conformers provide insights into how this PITP is engineered for biological function, as well as a conceptual model for how Sec14-like domains work. The Sec14-fold is comprised of around 280 amino acid residues folded into a two-lobed globular structure containing a large hydrophobic cavity (Fig. 3) (22, 123, 124). This cavity comprises the PL binding pocket where either PtdIns or phosphatidylcholine (PtdCho) bind. Because the pocket is only large enough to accommodate one PL at a time, Sec14 binds PtdIns or PtdCho in a mutually exclusive manner (Fig. 3A). The lipid-free apo-Sec14 structure is considered to approximate the “open” conformer state that Sec14 transiently assumes on the membrane surface during the lipid exchange reaction (Fig. 3B). As the hydrophobic cavity presents a chemical environment whose hydrophobicity gradient is nearly identical to that of a membrane leaflet (125, 126), lipid exchange into and out of the pocket occurs by simple chemical partitioning, thereby accounting for the ATP-independence of the exchange reaction. What marks a PL for exchange by Sec14 is not yet clear, although a localized perturbation of the membrane surface induced upon Sec14 binding is an attractive possibility. Access to the Sec14 hydrophobic pocket is controlled by a short helical gate, which is flipped open in the open conformer and shut in the “closed” lipid-bound conformers. These helical gate dynamics involve large conformational transitions (∼18 Å) governed by a small, but highly-conserved, conformational switch motif termed the “G (gating)-module” (Fig. 3B) (127, 128). That the G-module is a hotspot for disease missense mutations in mammalian Sec14-like proteins/domains not only emphasizes its functional importance across the Sec14 superfamily but also suggests that Sec14-like domains share similar properties in terms of their conformational dynamics (127, 128).
Fig. 3.
The Sec14 structure. Crystal structures of Sec14 and the Sec14-like PITP Sfh1 are shown, with the tripod motif (green) oriented as if toward the membrane. The lipid binding β-sheet floor is rendered in pink, and the helices that line the binding pocket are in blue. A: Crystal structures of the Sec14-like PITP Sfh1 reveal that PtdIns and PtdCho both bind in the lipid binding pocket, with the PtdCho headgroup buried deep within the pocket and the PtdIns headgroup facing outwards toward the protein surface. Overlay of Sfh1 bound to either lipid illustrates that PtdIns and PtdCho acyl chain binding regions overlap, and Sfh1 cannot fully accommodate both lipids at the same time. B: The crystal structure of open lipid-free Sec14 is compared with Sfh1 bound to PtdCho (lipid not shown for clarity). The helical gate (orange) closes around the lipid binding pocket, with a key residue of the G-module (G266) highlighted in red.
An unexpected feature of the Sec14 molecule is its remarkable physical segregation of the substructures organizing binding of the PtdIns and PtdCho headgroups (22, 23). The PtdIns headgroup is coordinated by an extensive H-bond network near the protein surface, while the PtdCho headgroup is sequestered deep within the interior of the hydrophobic pocket where it is stabilized by cation-π interactions with the side chains of a three-walled tyrosine cage (22). The primary sequence motifs that converge in three dimensions to form the phosphoinositol and phosphocholine coordinating units constitute what we term the PtdIns- and PtdCho-binding barcodes, respectively. Interestingly, the PtdIns-binding barcode is conserved across the Sec14 superfamily (forecasting that Sec14 domains generally represent inositol lipid-binding modules), whereas the PtdCho-binding barcode is not. These signatures forecast that the noncanonical Sec14-like PITPs (i.e., bind PtdIns but not PtdCho) greatly outnumber the canonical Sec14-like PITPs (i.e., the PtdIns/PtdCho transfer proteins) in the eukaryotic proteome (22, 122).
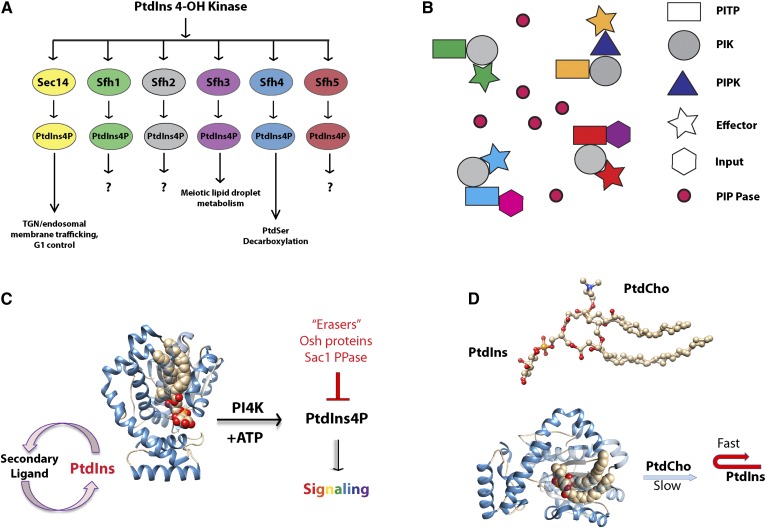
The Sec14-fold is a versatile module, as evidenced by the impressive expansion of Sec14 proteins and domains throughout the eukaryotic kingdom. Even the simple unicellular eukaryote, Saccharomyces cerevisiae, expresses five Sec14 homology (Sfh) proteins in addition to Sec14 (129). Significantly, it is these PITPs and not the PI4K itself that specify (i.e., diversify) the various biological outcomes of PtdIns4P signaling (Fig. 4A). The mammalian repertoire of stand-alone Sec14 domain proteins includes α-tocopherol transfer protein (αTTP), Caytaxin, and cellular retinaldehyde binding protein (CRALBP). Mutations in these proteins lead to human diseases, such as ataxia with vitamin E deficiency, Cayman-type cerebellar ataxia, and retinal degeneration, respectively (122). In higher eukaryotes, however, the Sec14 module is most frequently represented as a functional unit of multi-domain proteins housing other biochemical activities including: Rho-guanine nucleotide exchange factors (e.g., three of the four splice variants of Dbl, six of eleven Kalirin/Duo proteins, Dbs, Trio), Rho-GTPase activating proteins (e.g., CDC42GAP, p50RhoGAP), the neurofibromin-1 Ras-GTPase activating protein, and the MEG2/PTPN9 protein-tyrosine phosphatase (122, 130). A similar case also applies to plants. Higher plants such as Arabidopsis express some 25 Sec14-like proteins of which most exhibit demonstrable PITP activities. Of these, approximately half are two-domain PITPs that link a Sec14-domain to a coiled-coil domain unique to plants (the nodulin domain) that in certain cases constitutes a PtdIns(4,5)P2-binding motif (131–134).
Fig. 4.
The Sec14 PtdIns presentation mechanism. A: Sec14-like PITPs diversify the biological outcomes of PI4K in cells by specifying unique PtdIns4P pools that promote unique cellular processes. B: Transient complexes that bring together an individual PITP with a PI4K and a set of PtdIns4P effectors, either as individual proteins or in PITP-multidomain arrangements, generate a signaling pixel. The identities of the PITPs in the complex, the specific metabolic input that these sense in the form of the second ligands they bind for priming PtdIns presentation to the PI4K, and the PtdIns4P effectors determine distinct biological outcomes. The pixel boundary is the molecular space of each PITP/PI4K/effector complex. Populating interstitial areas of the membrane with PtdIns4P phosphatases sharpens pixel boundaries and enables PtdIns4P signaling at essentially point resolution. C: Sec14-like PITPs exchange a second ligand for PtdIns, and present PtdIns to PI4K, which generates PtdIns4P used for signaling reactions. The forward reaction is antagonized by PtdIns4P “erasers,” or negative regulators, such as Osh proteins or Sac1 phosphatase. D: PtdIns and PtdCho occupy overlapping positions in the Sec14 lipid-binding pocket. The slow egress of PtdCho from the Sec14 pocket frustrates entry of incoming PtdIns, resulting in an abortive exchange that exposes (presents) the frustrated PtdIns to the PI4K.
Based on these lines of evidence, we propose the concept of a signaling “pixel”: a PtdIns-presentation subunit (the PITP) engaged with a PI4K that itself interacts with a defined set of PtdIns4P effectors. The signaling pixel facilitates the engineering of phosphoinositide signaling with essentially point resolution. The proposed signaling pixel arrangement allows functionally distinct PITP/PI4K/PtdIns4P effector complexes, dedicated to distinct biological outcomes, to be physically segregated on a membrane surface, even though these pixels might be positioned adjacent to each other on that same surface. Phosphoinositide phosphatases are posited to sharpen pixel boundaries by degrading any phosphoinositides that escape pixel boundaries, thereby specifying functional compartmentation of lipid signaling on a membrane surface with high definition (Fig. 4B).
KEY PREDICTIONS OF INTER-COMPARTMENTAL LIPID TRANSFER MODELS
As described above, the existence of PITPs as cytosolic carriers that ferry PtdIns from the ER to distal compartments that consume PtdIns in phosphoinositide signaling cascades was predicted by Michell (1). This hypothesis guides broad extrapolations of the in vitro lipid transfer activities of proteins to in vivo function, circular though such arguments may be. Lipid transfer models for PITP function postulate that the soluble PITP::PtdIns complex is the mobile intermediate in a PtdIns transport step between two distinct membranes (Fig. 2). The PITP loads with a PtdIns molecule in the ER, and this preferential loading is governed by the higher affinity of PITPs for PtdIns over other lipids (e.g., PtdCho). Specific targeting of the soluble complex to the acceptor membrane (e.g., the plasma membrane) is also a key principle of transfer mechanisms. At the acceptor membrane, the PtdIns is unloaded and the PITP reloads with a counter-ligand (i.e., a lipid that is not PtdIns, classically, and in the case of Sec14, PtdCho). In this model, PITP loading and unloading is governed by an “accessible” or “free” PtdIns concentration gradient. The acceptor compartment is PtdIns-deficient relative to the ER, and the mass excess of the counter-ligand governs the altered specificity of lipid loading at this organelle.
Transfer models assume that the ER is the sole compartment of PtdIns synthesis, the veracity of which is not clear (135–139). Transfer models make several key predictions, but these are surprisingly difficult to interrogate. For example, the acid test of lipid transfer models is a direct demonstration that a PITP engages in monomeric PtdIns transport from the ER to the acceptor organelle. However, in vivo assays claiming to test this functional mode are themselves indirect and subject to multiple interpretations. While in vitro PtdIns exchange assays are consistent with inter-compartmental transport activities, these assays are set up as simple equilibration systems (140). As such, these too are subject to multiple interpretations. Thus, direct evidence in favor of lipid transfer mechanisms for PITP function is presently missing.
Transfer models demand vectorial PtdIns transfer from the ER to distal organelle systems. This vectorial transfer is predicted to derive from the greater affinity of PITPs for PtdIns relative to other lipid counter-ligands, and this concept rests on measurements that PtdIns is a relatively minor PL in mammalian cells (5 mol% of total glycerol-PL). Thus, even relatively subtle alterations in this PITP lipid affinity balance are expected to strongly compromise phosphoinositide signaling. Again, while this issue has not been addressed experimentally in mammalian systems, it has in yeast and with surprising results (see below).
There is also the question of scale. While the magnitude of the flux required for PITP-dependent PtdIns transport to sustain phosphoinositide signaling is currently unknown, a PtdIns supply model would suggest that it should be high in professional high-demand signaling systems. Yet, monomer transport of PtdIns via cytosolic carriers seems to be an inefficient mechanism when vesicle-based strategies, or potentially membrane contact-site mechanisms (see below), provide higher capacity systems for PtdIns supply. Given the expectation that PITP-dependent PtdIns-transport thresholds must reflect high flux systems in cells, even modestly reduced PITP expression should be detrimental to sustained phosphoinositide signaling. If the requisite flux is low, however, then a clean experimental test of transfer models becomes ever more difficult as the experimentalist encounters confounding “limit of detection” barriers. A low flux requirement is also difficult to reconcile with a general PtdIns supply function. Thus, we argue that the direct evidentiary case for PITPs serving as trans-organellar PtdIns carriers is presently weak.
THE Sec14 NANOREACTOR AND ALTERNATIVES TO LIPID TRANSFER MECHANISMS
As the major yeast PITP, Sec14 executes functions essential for yeast viability. It localizes to TGN/endosomal membranes, regulating anterograde trafficking from this compartment to the plasma membrane and retrograde trafficking from later endosomal compartments to earlier stages of the TGN/endosomal system (141, 142). Yeast cells acutely compromised for Sec14 activity do not efficiently form post-Golgi secretory vesicles from the TGN and endosomal compartments (19, 141, 143). Because there is an unambiguous physiological coupling of Sec14 activity to cell viability and membrane trafficking through the yeast TGN/endosomal system, Sec14 provides the first reliable system for experimentally dissecting mechanisms for PITP function. It is in this context that several lines of evidence converge on the idea that Sec14 functional mechanisms do not involve lipid transfer between membranes in vivo. We highlight the following:
First, yeasts differ from mammalian cells in that PtdIns is second only to PtdCho as the most abundant cellular PL, comprising 20–25 mol% of total glycerophospholipid in yeast membranes versus ∼5 mol% in mammals (144). It is unlikely that monomer PtdIns transfer would play a significant biological PtdIns supply role under these normal physiological conditions of PtdIns surfeit. Remarkably, even when yeasts are genetically manipulated so that PtdIns constitutes the most abundant PL in yeast membranes, the cellular Sec14 requirement for viability is not relieved (20). The biological threshold for Sec14 activity is also surprisingly low, lower than would be expected if this major yeast PITP functioned as a bulk monomeric PtdIns carrier. Furthermore, PtdIns synthesizing activities (e.g., CDP-DAG synthase, PtdIns synthase, PtdIns kinase) are detected in secretory vesicles generated at the Golgi, suggesting that the plasma membrane has the capacity to synthesize its own PtdIns (137). In mammalian cells, an ER-derived organelle containing PtdIns synthase shuttles rapidly to various intracellular compartments, thereby mobilizing the capacity to synthesize PtdIns away from the ER (135).
Second, the normally essential Sec14 requirement for cell viability is “bypassed” by loss-of-function mutations in specific and biologically nonessential lipid biosynthetic genes. The biochemical identities of the “bypass Sec14” mutations are not easily reconciled with lipid transfer models (20, 24, 145–149). In particular, the finding that functional ablation of the DAG-consuming CDP-choline pathway for PtdCho biosynthesis effects bypass Sec14, even under conditions where this pathway makes no net contribution to PtdCho biosynthesis (i.e., functions as a salvage pathway only), is not at all congruent with transfer models for Sec14 function (24). Rather, this evidence demonstrates that the essential function of Sec14 is to properly integrate PtdIns, PtdCho, PtdIns4P, and DAG metabolism in order to establish a TGN/endosomal lipid-signaling environment conducive to formation of secretory vesicles (23).
Third, transfer models rely on vectorial transfer of lipids driven by concentration gradients and, for PITPs, this is attributed to a decreasing gradient of accessible or free PtdIns from its site of synthesis in the ER, through the Golgi, and to the plasma membrane. Thus, the relative affinity for PtdIns versus PtdCho is predicted to be an essential PITP feature for appropriate response to these PtdIns gradients. However, a mutant Sec14 with defects in PtdIns-binding (such that the PITP preferably binds PtdCho) nevertheless retains considerable biological activity (64, 150).
Taken together, these findings demand fresh ideas regarding Sec14 function specifically and PITP function in general. A promising alternative perspective is described by “nanoreactor” models of PITP action (22, 23). That is, Sec14 “presents” PtdIns to the Pik1 and Stt4 PI4Ks (this PITP does stimulate the activity of both of these lipid kinases in vivo), with the result that PtdIns4P synthesis is stimulated (Fig. 4C). This presentation function obligatorily requires the ability of a single Sec14 protein to exchange bound PtdCho for PtdIns, and we posit that Sec14 binding PtdIns and PtdCho at distinct, but overlapping, sites is an essential design component of the presentation mechanism. Detailed discussions of the nanoreactor model are published elsewhere (21, 23, 125). But, the basic concept is that Sec14 PITPs present PtdIns to the kinase during heterotypic exchange reactions where a slowly egressing PtdCho is being exchanged for PtdIns (Fig. 4D). Slow egress of PtdCho from the Sec14 lipid-binding pocket results in a series of abortive PtdIns exchange events, which leave PtdIns in a transitional state between the membrane surface and the Sec14 lipid binding pocket. It is this transitional PtdIns molecule that defines the preferred PI4K substrate “pool”. That is, an abortive exchange intermediate neither fully bilayer-incorporated nor incorporated in the Sec14 lipid binding pocket provides a superior substrate for the otherwise biologically inadequate interfacial PI4K enzyme (22). In this scenario, PtdIns molecules that successfully load into the Sec14 pocket and can be transferred between membranes are not eligible to be kinase substrates. Moreover, in this scenario, PtdCho acts as a “coincidence-detector” whose binding primes PtdIns presentation (and potentiation of phosphoinositide synthesis) in response to PtdCho metabolic cues.
This “PtdCho-sensing” model raises an interesting question: how can the most abundant PL in yeast cell membranes possibly effectively function as a coincidence-detector? But, strong in vivo data indicate that this is so. A large body of genetic and cell biological data demonstrate that Sec14 regulates the critical interface between PtdCho synthesis and PtdIns4P signaling in vivo in the context of TGN/endosomal membrane trafficking. The specificity of this interface yields a clue, however, as it is the activity of the CDP-choline pathway for PtdCho biosynthesis that is physiologically relevant (22–24). The data are consistent with the PtdCho-sensing activity of Sec14 constituting a mechanism for sensing metabolic flux through the CDP-choline pathway and coordinating the metabolism of two pro-trafficking lipids, DAG and PtdIns4P (23). This view implies that Sec14 has functional access to a restricted pool of PtdCho and forecasts a more intimate and intricate organization of Sec14, CDP-choline biosynthetic enzymes, and PI4K than presently appreciated.
Nanoreactor mechanisms postulate that Sec14 is active only when it is engaged in multiple cycles of heterotypic PtdCho/PtdIns exchange while transiently associated with membrane surfaces, and that “activation” of PtdIns molecules as lipid kinase substrates does not involve complete sequestration of PtdIns inside the lipid-binding pocket. Thus, the soluble PtdIns-bound protein is not a productive intermediate in some inter-membrane lipid transfer reactions. Rather, the cytosolic Sec14::PtdIns and Sec14::PtdCho complexes represent functionally quiescent pools that define the energy-zero “noise” that accompanies the presentation regime and bears no energy cost to the cell.
Nanoreactor models are also attractive, as these proffer new perspectives for viewing the expanded Sec14 superfamily, and the prevalence of Sec14-domains in multi-domain arrangements. We propose that these domains are modules that stimulate specific phosphoinositide synthesis in the immediate locale of other domains whose activities are modulated by that phosphoinositide. The high conservation of the PtdIns barcode among Sec14-like proteins, in the absence of conserved PtdCho barcodes, forecasts that the expansion of the Sec14-superfamily reflects expansion of the cohort of other lipophilic priming ligands that Sec14-like modules sense for coupling to a PtdIns or PIK for stimulated phosphoinositide production, or potentially a phosphoinositide phosphatase for stimulated degradation. This concept was initially supported by the observation that the most commonly inherited missense alleles of αTTP associated with ataxia with vitamin E deficiency and of CRALBP associated with macular degeneration directly compromise the PtdIns barcode (122). It was proposed that αTTP and CRALBP operate along Sec14-like nanoreactor principles by binding inositol lipids, as well as vitamin E and retinaldehyde, as part of their functional mechanisms. The solution of the crystal structure of αTTP complexed with PtdIns(4,5)P2 confirms these basic predictions (152).
THE ANTAGONISTIC RELATIONSHIP OF Sec14 WITH LIPID EXCHANGE PROTEINS OF THE OSBP FAMILY
A remarkable development that has come from the Sec14 studies is that lipid transfer proteins function as an antagonistic pair in coupling PtdIns4P signaling with TGN/endosomal membrane trafficking. That is, the action of the pro-trafficking Sec14 is countered by the action of the Kes1/Osh4, a PtdIns4P/ergosterol exchange protein that functions as a trafficking brake in this system (23, 146, 149, 153–156). Moreover, the antagonistic actions of Kes1 and Sec14 in TGN/endosomal membrane trafficking extend to cell cycle contexts associated with cellular commitment to a new round of cell division. That is, Kes1 is a negative regulator of progression through the G1 stage of the cell cycle in the face of nutrient-deprived environments, and it also antagonizes Sec14 activity in regulating the timing of the G1 phase of the yeast cell cycle (156). Although Kes1 is one of seven yeast oxysterol binding-related protein (ORP) homologs, it is unique among yeast Kes1-like proteins in its role as antagonist to Sec14-dependent PtdIns4P signaling in membrane trafficking and cell-cycle contexts (146, 149, 155, 156).
The biological function of ORPs is a matter of strong contemporary interest, as these proteins are highly conserved in eukaryotes. ORPs remain functionally enigmatic, however, because of the general lack of information regarding their biological activities. It is for this reason that this topic is discussed briefly here, as it affords an opportunity to relate the Sec14/Kes1 functional antagonism to current controversies associated with the physiological activities of ORPs.
There are proposals that yeast and mammalian ORPs function as lipid carriers that promote nonvesicular lipid trafficking between intracellular membranes (154, 157), and other studies that argue that these proteins play no significant role in mobilizing sterols between intracellular membranes in vivo (155, 158, 159). The most detailed intermembrane transfer proposal, one that has gathered significant momentum in the literature, is a countercurrent hypothesis. This model posits that ORPs (including Kes1) transfer sterol from the ER to TGN membranes and subsequently execute the reciprocal transfer of PtdIns4P from the TGN to the ER where hydrolysis of PtdIns4P by the Sac1 phosphatase provides the energy to drive the cycle (157, 160). The evidence in support of this proposal comes largely from elegant in vitro reconstitutions of sterol/PtdIns4P vectorial transfer systems. Of particular relevance to this discussion are several key predictions of the countercurrent hypothesis. First, that sterol binding is an essential functional property of Kes1. Second, that PtdIns4P binding is an essential functional property of Kes1. Third, that the Sac1 PtdIns4P phosphatase is a central player in the sterol/PtdIns4P countercurrent cycle. A brief examination of the veracity of these predictions using authentic functional readouts based on the Sec14/Kes1 functional antagonism identifies compelling in vivo data that are inconsistent with the countercurrent proposal.
Whereas PtdIns4P-binding is indeed an essential in vivo activity of Kes1 (149, 154, 156), and Sac1 deficits interfere with Kes1 function in cells (149), genetic and functional data demonstrate that it is the PtdIns4P pool produced by the Pik1, and not the Stt4, PI4K, that is the Kes1-relevant pool in cells. This issue of the Pik1 versus Stt4 pool is of critical importance because the Sac1 phosphatase accesses the Stt4 pool exclusively in cells (65, 161). Those data argue that Sac1 is not directly involved in Kes1 biological function and are therefore contrary to a key tenet of the countercurrent hypothesis. Indeed, PtdIns4P accumulation in the ER in cells lacking Sac1 induces Kes1 mislocalization from Golgi membranes to ER, thereby resulting in diminished Kes1 activity on Golgi membranes (149). This is the proposed basis for how Sac1 deficiencies restore Golgi function to cells lacking Sec14. In fact, elevated Kes1 expression rescues Kes1 biological function in cells lacking Sac1. With regard to sterol binding, loss of sterol binding strongly enhances Kes1 activity in vivo, as assessed by membrane trafficking or cell cycle readouts (155, 156). These data are fundamentally at odds with the base predictions of the countercurrent hypothesis. One would expect a bona fide sterol transporter to be inactivated by loss of sterol binding. The hypothesis most consistent with the available data describes a Kes1 sterol/PtdIns4P exchange cycle on TGN/endosomal membranes that senses available sterol, which, upon displacement of the PtdIns4P bound to Kes1, releases the sterol-bound Kes1 from the membrane. In this manner, sterol availability controls the amplitude of the Kes1-mediated trafficking brake by tuning the ability of Kes1 to sequester PtdIns4P from pro-trafficking effectors (155). It remains to be determined whether mammalian ORPs will conform closely to the Kes1 precedent. But, we argue that the concepts gleaned from Kes1 biology provide new perspectives from which to view potential ORP functions in cells. Indeed, the evidence identifies Kes1 PtdIns4P binding activity as a key nonhistone target for the highly conserved NuA4 lysine acetyltransferase (156).
START-LIKE PITPs
The START-like PITPs bear no primary sequence homology at all to Sec14-like PITPs and, as discussed below, this lack of homology corresponds to the fact that these PITPs assume different protein folds. Initially, START-like PITPs were to represent higher eukaryotic versions, as these are absent from S. cerevisiae and other fungi, and were not initially found in plant genomes. However, with the advent of high throughput genome sequencing, it is now apparent that candidate START-like PITPs are present in the genomes of ancient eukaryotes, including members of the Alveolata (a protist subgroup) and the Diplomonadida (flagellates including Giardia lamblia). So, START-like PITPs represent ancient constructions.
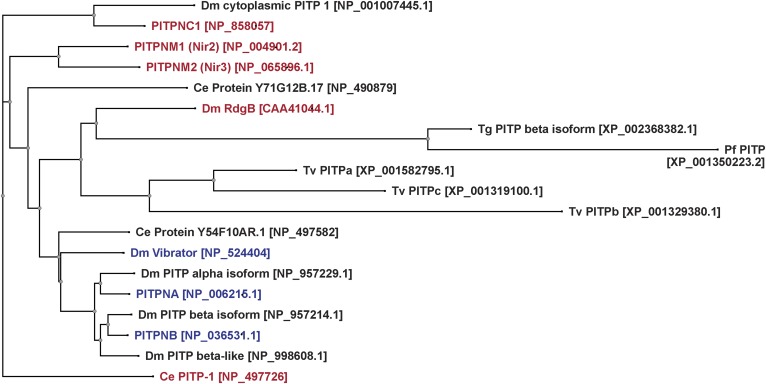
In terms of classification, the START PITP family is subdivided into two structural classes: class I PITPs with high sequence homology to mammalian PITPα and PITPβ (the first established mammalian PITPs) and class II PITPs with high homology to Drosophila retinal degeneration B (RdgB) (including human proteins PITPnm1 or RdgBα1, PITPnm2 or RdgBα2, and PITPnc1 or RdgBβ) (Fig. 5). The PITP domains of class I and class II proteins share ∼40% primary sequence identity. Unfortunately, an alternative nomenclature also exists within the literature, which defines class I and class II PITPs by whether they are single- or multi-domain proteins, respectively. In that classification system, PITPnc1 is assigned to class I (162). In this review, we use the former definition and so describe PITPnc1 as a class II protein. Class II PITPs are also subject to yet another standard of nomenclature, which we will not use but describe, for purpose of reference, based on their association with the N-terminal domain of protein kinase PYK2 (where PITPnm1 is Nir2, PITPnm2 is Nir3, and PITPnc1 has no designation) (163). The primary sequence divergence between class I and class II START-like PITPs reflects biochemical distinctions as well. Whereas the class I proteins are PtdIns/PtdCho transfer proteins, the class II PITPs are primarily PtdIns/PtdOH transfer proteins (164–166).
Fig. 5.
The START PITP family. START PITPs are aligned by primary sequence using VectorNTI (Life Technologies), and the alignment is visualized as a cladogram. Proteins discussed in this review are highlighted in color. Accession numbers for the specific amino acid sequences used are in brackets. Class I PITPs are designated in blue, and class II PITPs in red. PITPs from ancient eukaryotes, including Toxoplasma and Trichomonas do not fall into either category.
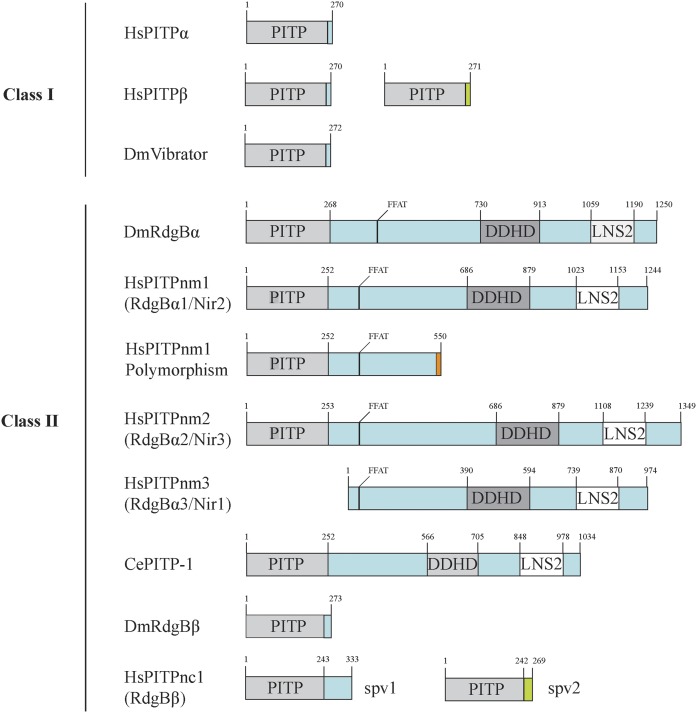
One of the more striking differences between the various START-like PITPs is the length and composition of their C termini (Fig. 6). Mammalian PITPβ is expressed as either a “canonical” or an “alternative” spliceoform, and these spliceoforms differ in their extreme C termini (167). This pattern is recapitulated in the transcript processing of PITPγ, a PITPβ-like class I PITP unique to zebrafish (168). PITPnc1 is also alternatively spliced into two variants that differ in their C-terminal sequences. The longer isoform contains a proline, glutamic acid, serine, threonine (PEST) sequence that mediates binding to 14-3-3 proteins and promotes turnover of the protein. This PEST motif is absent from the short PITPnc1 splice variant, however (169), suggesting that the two splice variants may not be functionally identical. The remaining class II PITPs are distinguished by multiple domains C-terminal to the PITP domain, as discussed below.
Fig. 6.
START PITP domain architecture. The class I and class II PITPs discussed in this review are depicted in schematic form with specific domains indicated. Two transcriptional variants each for PITPβ and PITPnc1 are indicated. PITPnm1 is depicted in its full-length wild-type form, as well as the premature truncation found in a subset of the human population [Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/about)].
THE START PITP DOMAIN
Crystal structures for mouse, human, and rat PITPα are available in both lipid-free as well as in PtdIns- or PtdCho-bound conformers (170–172) (Fig. 7). The structure for rat PITPβ bound to PtdCho has also been solved (173). Although there are no crystal structures of the PITP domains of class II PITPs at present, the high primary sequence conservation between the two PITP classes forecasts that class II PITP structures will closely resemble those of the class I PITPs.
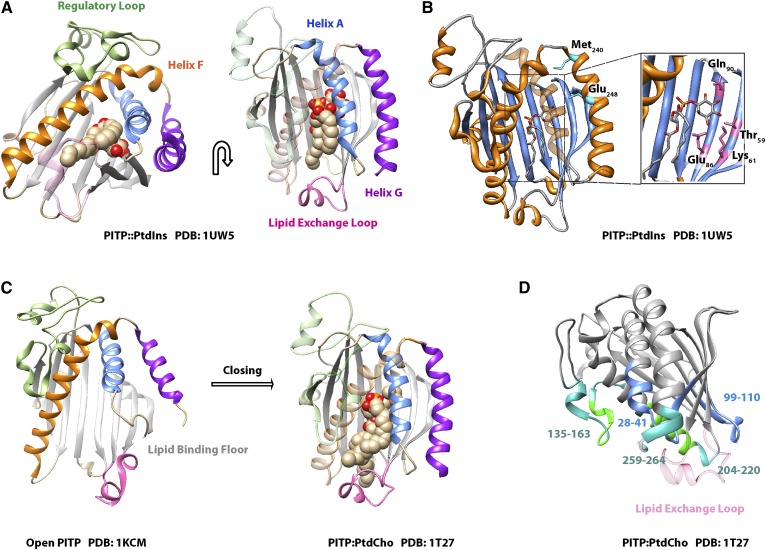
Fig. 7.
START PITP structure. A: The crystal structure of PITPα bound to PtdIns is shown in two orientations as indicated by the arrow. The β-sheet floor of the lipid binding pocket (gray), the regulatory loop (green), helix F (orange), helix A (blue), helix G (purple), and the lipid exchange loop (pink) are highlighted. B: The PtdIns headgroup-binding motif of PITPα is illustrated and the coordinating residues within the lipid binding pocket (inset: Thr59, Lys61, Glu86, Gln90) are highlighted. Residues on helix G that do not bind the headgroup directly but modulate the conformational dynamics that specifically influence PtdIns-binding are also highlighted (cyan: Met240, Glu248). C: The crystal structures of open lipid-free PITPα and PtdCho-bound PITPα are shown to illustrate the conformational dynamics associated with gating of the hydrophobic lipid-binding pocket during the exchange cycle. The lipid exchange loop controls access to the pocket and helix G and helix A fold over the lipid binding floor. Coloring scheme is as in A. D: The crystal structure of PITPα bound to PtdCho is shown with membrane-association regions. Hydrophobic interactions with the membrane are rendered in green and involve residues 135-163 and 259-264, while electrostatic interactions with the membrane involve residues 28-41 and 99-110 and are highlighted in blue. Residues that interact with the membrane only when PtdIns is present in the bilayer are indicated in bright green. The lipid exchange loop is made translucent for the purpose of clarity, and this substructure penetrates the bilayer surface.
The core START PITP consists of four domains: 1) the lipid binding pocket composed of an eight-stranded β sheet and bounded by two α helices; 2) an unstructured region called the “regulatory loop”; 3) the C-terminal helix G; and 4) the “lipid exchange loop” that gates entry to the lipid-binding pocket (Fig. 7A). The START PITP PtdIns binding barcode consists of four residues on the cavity β-sheet floor that coordinate the PtdIns headgroup. For PITPα, these residues are Thr59, Lys61, Glu86, and Asn90 (Fig. 7B) (172, 174). In contrast to Sec14, PtdIns and PtdCho bind in nearly identical orientations in the START-like PITP lipid-binding pocket (Fig. 7A, C). Both headgroups are sequestered within the cavity. A random mutagenesis screen also identified Glu248 on the C-terminal G-helix, a structure far removed from the headgroup-coordinating region of the lipid-binding pocket, as being specifically involved in PtdIns-binding and transfer (Fig. 7A, B) (174). The G-helix is conformationally dynamic during PITP association with membranes (175). Molecular dynamics simulations indicate that the G-helix undergoes a localized unfolding around Glu248 upon PtdIns binding, which changes its hydrogen bonding network in such a way that interaction of the G-helix with the A-helix is altered and promotes closure of the gate over the lipid binding pocket (176). This is interesting from the standpoint that the transition is required for stabilizing PtdIns but not PtdCho binding in the lipid binding pocket, suggesting that the trajectory of these lipids during lipid exchange may not be the same. Moreover, certain PITPα substructures associate with the bilayer only when PtdIns is present (residues 147-153 and 208-219) (Fig. 7D). Thus, the PITPα configuration may differ when the protein is engaged in heterotypic PtdIns/PtdCho versus homotypic PtdCho/PtdCho exchange reactions (176).
Whereas functional analysis of START-like PITPs has lagged behind those of Sec14-like PITPs, the START-like PITP system is leading the way in terms of defining the details of the lipid-exchange cycle that is central to PITP biological function. All-atom molecular dynamics simulations reveal that PITPα engages membranes primarily though the lipid exchange loop, which inserts into the bilayer upon membrane binding and induces a conformational change to a more helical structure (176). Other regions involved in membrane association include surface residues that promote membrane interaction via hydrophobic interactions (residues 135-163, 204-220, and 259-264) and those that engage the membrane in electrostatic interactions (residues 28-41 and 99-110) (Fig. 7D). Interestingly, such simulations not only draw a plausible trajectory for PL movement into and out of the PITPα hydrophobic pocket but also indicate that PITPα interactions with bound PL (i.e., the exchange substrate) dramatically lower the energy barrier for transition of PL ligand between PITP and the membrane bilayer so that it is within the bounds of thermal energy (176). These findings rationalize the ATP-independence of PITP-mediated PL exchange/transfer.
START-LIKE PITP FUNCTIONS FROM THE PERSPECTIVE OF PERMEABILIZED CELL SYSTEMS
A number of biochemical reconstitution studies using permeabilized cell systems implicated membrane trafficking activities for START-like PITPs. These include the pioneering work of Tom Martin and colleagues (177, 178) who reconstituted regulated dense core vesicle exocytosis in permeabilized neuroendocrine cells, vesicle budding from the TGN and other mammalian Golgi compartments (179–181), and the scission of coatomer-coated transport vesicles (182). Other permeabilized cell models reconstituting PLC-coupled receptor (i.e., epidermal growth factor receptor) signaling at the plasma membrane also identified PITPα as an essential cytosolic factor required for signaling (183–185). In those cases, PITPα was purified from cytosol as a factor promoting the corresponding reactions. Interestingly, in all the cases where the experiment was done, recombinant yeast Sec14 also fulfilled the PITP requirement in the reconstitution (185, 186). With the benefit of hindsight, it is likely that these semi-intact cell systems suffered from phosphoinositide degradation (i.e., run-down) during cell permeabilization and fractionation, and that phosphoinositide pools needed to be restored for the systems to function. Thus, whereas permeabilized cell systems faithfully reconstituted phosphoinositide requirements for the corresponding reactions, it remains unclear whether the projected roles for PITPs in the reconstituted processes are physiologically relevant.
IN VIVO CLASS I PITP FUNCTIONS FROM THE PERSPECTIVE OF CULTURED CELL MODELS
Localization studies report that PITPα localizes to the nucleus and the cytoplasm, whereas PITPβ localizes predominantly to the Golgi complex (168, 187, 188). Interestingly, mutant PITPα defective for PtdIns is excluded from the nucleus, whereas nuclear pools are readily detected for wild-type PITPα and PITPα mutants defective in both PtdIns and PtdCho binding (189). While the idea that PITPα is somehow involved in regulating nuclear phosphoinositide metabolism is an attractive one, it remains an open question. Pulse-chase experiments indicate that PITPα does not play a significant role in bulk PtdIns import into the nuclear matrix (189). There are conflicting assignments as to which Golgi compartment PITPβ targets: one study localizes this class I PITP to the TGN, whereas another claims a cis-Golgi localization (168, 188).
Initial studies of class I PITP functions in cellular systems involved overexpression and siRNA approaches. Overexpression strategies suggested that PITPα promotes secretion of mitogenic and survival factors. NIH3T3 fibroblasts engineered for high-level PITPα expression displayed increased survival upon in vitro induction of apoptosis (190, 191). Transfer of conditioned medium from those cells to naive NIH3T3 cells increased proliferation rates and conferred protection against UV radiation and TNFα treatment (192). Fractionation studies identified the survival factor as a COX-2-dependent endocannabinoid that signals through a cannabinoid 1-like receptor (192, 193). Media from cells overexpressing PITPα were also neuroprotective for primary spinal cord-derived motor neurons in terms of serum deprivation-induced cell death, and this effect was traced to production of arachidonic acid metabolites (191).
An RNA interference approach suggested that PITPβ is required for cargo transport from the Golgi to the ER in HeLa cells (188). Transient knockdown of PITPβ resulted in Golgi compaction, changes in nuclear morphology, a reduction in cellular PtdIns4P levels, and strong defects in COPI-mediated Golgi to ER retrograde trafficking in the face of unaffected anterograde membrane trafficking. A lipid-transfer mechanism was proposed where PITPβ delivers PtdIns from the ER to cis-Golgi membranes to make substrate available to PI4KIIB, which produces PtdIns4P and ultimately affects the actin dynamics that mobilize COPI-coated vesicles (188). This model has its curious features, however. Because retrograde trafficking is an essential pathway for recharging the ER with the v-SNARE molecules required for targeting of ER-derived vesicles to the Golgi, it remains unclear how PITPβ-depleted cells would sustain robust anterograde trafficking under conditions of strongly compromised retrograde Golgi to ER trafficking. With the advent of CRISPR/Cas9 technology, strategies for engineering cleaner cellular models for class I PITP function are available for independent address of these issues.
IN VIVO CLASS I PITP FUNCTIONS FROM THE PERSPECTIVE OF VERTEBRATE MODELS
Both mammalian class I PITPs are ubiquitously expressed in all tissues, but PITPα is more abundant in the brain while PITPβ is more abundant in the liver and neutrophils (187, 194–196). Mice genetically ablated for PITPα are born alive at the expected Mendelian frequencies, but suffer neonatal/perinatal death as a result of spinocerebellar disease, intestinal and hepatic steatosis, and hypoglycemia (197, 198). Further studies demonstrated that PtdIns-binding is an essential biological activity for PITPα and, interestingly, that neurons ablated for PITPα activity are nonetheless uncompromised relative to wild-type when subjected to intense trains of neuronal stimulation (199). Thus, PITPα does not function as a PtdIns carrier to the neuronal plasma membrane, at least not as drawn by the Michell hypothesis. However stark the phenotypes in the PITPα-null mouse may be, cells derived from the mouse and cultured ex vivo, including neurons, show no obvious phenotypes.
The severe effects of PITPβ depletion on the Golgi reported by Carvou et al. (188) suggested an important, and perhaps essential, role for PITPβ in mammalian development. While initial attempts to generate PITPβ-deficient mice and embryonic stem cells were unsuccessful (200), it is now clear that PITPβ-null mice are viable and show no obvious phenotypes (201). Furthermore, the available data indicate that a level of functional redundancy exists between PITPα and PITPβ in the developing mouse. As described below, this understanding is driving pioneering studies of class I PITP function in mammalian neural stem cells.
Class I PITP studies in zebrafish are also informative. Unlike the case in mammals where PITPα deficiencies exerted postnatal consequences, morpholino-mediated knockdown of the zebrafish PITPα resulted in early embryonic lethality (168). The zebrafish PITPβ ortholog is expressed as two spliceoforms and these are collectively not required for development, but knockdown of both PITPβ spliceoforms in this organism causes dramatic defects in photoreceptor outer segment biogenesis and maintenance in double cone photoreceptor cells (168). This phenotype is consistent with the tissue expression profile of zebrafish PITPβ, as expression of this protein is restricted to double cone cells of the fish retina (168). Given that the vertebrate photoreceptor outer segment defines a context of extremely active membrane biogenesis and turnover, a role for PITPβ in photoreceptor cell membrane trafficking remains a plausible functional mechanism. Zebrafish express yet another PITPβ-like PITP, designated PITPγ, but the biological functions of this isoform and any potential functional redundancies with PITPβ remain to be interrogated (168).
IN VIVO CLASS I PITPs AND EMBRYONIC DEVELOPMENT OF THE MAMMALIAN NEOCORTEX
PITPα and PITPβ are both expressed in the ventricular zone of embryonic mouse brain, indicative of their coexpression in the neural stem cells from which the mammalian neocortex is ultimately derived. A recent study took advantage of this fact to investigate PITPα and PITPβ function in neural stem cells and the consequences of loss of function of these proteins for development of the mammalian forebrain during embryogenesis. Neural stem cell-specific eviction of both PITPα and PITPβ structural genes in murine neural stem cells gave rise to progeny that were born alive but which lacked the forebrain. A single copy of either class I PITP gene was sufficient to support development of an anatomically normal brain (201).
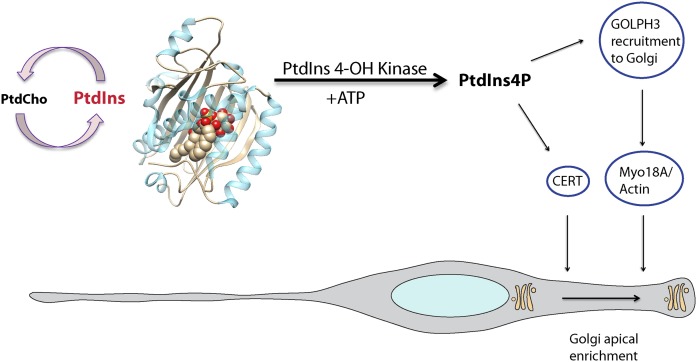
Forebrain loss in the neural stem cell double knockout mouse was the consequence of loss of neural stem cell polarity and subsequent rampant apoptosis throughout the developing forebrain that erased the compromised tissue by late gestation. When cell autonomous defects were examined by in utero electroporation, the earliest event was loss of polarity of the Golgi system. That is, repositioning of the Golgi system from the apical process of the neural stem cell to a perinuclear location coupled to its morphological collapse from an extended ribbon to a more compact distribution (201). A pathway was written where the class I PITPs potentiate formation of a Golgi PtdIns4P pool that recruits the PtdIns4P-binding protein, Golgi phosphoprotein 3 (GOLPH3) (ortholog of yeast Vps74), and the nonconventional myosin, Myo18A, to Golgi membranes, thereby linking the system to the F-actin cytoskeleton that is itself highly enriched in the neural stem cell apical process. GOLPH3 had previously been reported to bind PtdIns4P and to cooperate with Myo18A and F-actin to drive release of secretory vesicles from the Golgi (Fig. 8) (54, 202, 203). Contrary to published reports from cultured cell models (54, 204), class I PITP- and GOLPH3-deficient neural stem cell Golgi were not compromised for bulk membrane trafficking and gave rise to differentiated progeny that were capable of correct directional migration over large distances in the developing neocortex (201).
Fig. 8.
Class I START PITP function in apical loading of the Golgi system in neural stem cells. Class I START PITPs exchange PtdCho and PtdIns, thereby stimulating PI4K activity on late Golgi membranes. The PtdIns4P pool recruits GOLPH3 and CERT to Golgi membranes with GOLPH3 subsequently engaging the apically directed actin machinery via the nonconventional myosin, Myo18A. This interaction promotes loading of the Golgi system to the neural stem cell apical process, thereby setting up an asymmetry critical for neural establishment/maintenance of neural stem cell polarity.
Interestingly, while both PITPα PtdIns- and PtdCho-binding activities were required for rescue of the neural stem cell Golgi defects, expression of a fully functional copy of the yeast Sec14 was not at all able to rescue class I PITP deficiencies (201). Given that class I PITPs and Sec14 are very similar in their PtdIns- and PtdCho-binding and transfer properties, and these proteins are very similar in their differential affinities for PtdIns versus PtdCho, these data argue strongly that class I PITPs do not operate as lipid carriers whose vectorial transport of PtdIns is driven by simple ER-Golgi PtdIns and PtdCho gradients. If that were the case, Sec14 should have scored as an effective surrogate.
Are the activities of class I PITPs in neural stem cells conserved processes? It seems so. The sole Drosophila class I PITP [vibrator (Vib)] was identified in a genetic screen for mutants with dysregulated neuroblast homeostasis and self-renewal (205). Indeed, Vib regulates asymmetric division of neuroblasts (i.e., fly orthologs of neural stem cells) by anchoring the nonmuscle myosin II regulatory light chain Spaghetti-squash (Sph) to the cell cortex during cell division, thereby promoting the asymmetric partitioning of cellular components involved in cell fate determination as well as cytokinesis (206, 207). Vib is inferred to cooperate with PI4KIIIa to maintain a PtdIns4P pool on the neuroblast cell cortex for Sph recruitment (205).
CLASS I PITPs AND RECEPTOR TYROSINE KINASE SIGNALING: THE CASE FOR THE EGF RECEPTOR
PITPα was reported as an essential component of EGFR signaling in permeabilized cell reconstitution experiments in which addition of PITPα to cytosol-depleted cells recovered receptor-stimulated PLCγ activity (183). These data were interpreted in the context of the Michell hypothesis, wherein PITPα delivers PtdIns to the plasma membrane for PtdIns4P and PtdIns(4,5)P2 production by PI4K and PI4P5K, respectively (Fig. 2). This provides PtdIns(4,5P)2 for subsequent metabolism by PLC and PI3K. Studies with genetically engineered mice do not provide obvious support for such a link, however. The waved mouse is deficient in EGF signaling due to a hypomorphic EGFR mutation that compromises receptor activation. This mutant mouse presents defective hair follicle development, while EGFR nullizygosity is embryonic lethal in some backgrounds, but not in others (208). EGFR-deficient mice that survive through the first postnatal week exhibit defective eyelid formation (are born with eyes open), hair follicle differentiation (wavy coats), and a shortened intestine with fewer villi (209). EGFR-deficient mice also have defects in lung development as evidenced by immature alveoli and respiratory failure (210). None of these defects are observed in PITPα-null mice (198). Furthermore, mice defective in EGFR signaling do not exhibit pathologies associated with PITPα deficiency, such as neurodegeneration, liver steatosis, and chylomicron retention disease (199, 208, 210, 211). At any rate, embryonic stem cells genetically depleted of PITPα can still form tumors in nude mice as efficiently as wild-type cells, a process that requires competitive growth factor scavenging from the environment (200).
Recent cancer biology studies indicate that the idea of some functional linkage between PITPα and EGFR signaling cannot yet be discounted, however. Analysis of human tumor samples treated with therapeutics targeting EGF signaling suggest an interesting link between PITPα and the EGF pathway. PITPα was identified as one of 28 genes highly predictive for human EGFR (HER2)-positive metastatic breast cancer resistance to trastuzumab-docetaxel therapy (212). Trastuzumab (trade name Herceptin) is a humanized monoclonal antibody that binds an extracellular domain of HER2 and inhibits signaling by promoting receptor dephosphorylation and/or blocking dimerization. The result is an anti-proliferative response through diminished signaling through the PI3K/Akt pathway (213, 214) and induction of the cyclin-dependent kinase (CDK) inhibitor, p27kip1 (215). Whether the correlation of elevated PITPα expression with trastuzumab-docetaxel resistance is related to a function for PITPα in HER2 signaling remains to be determined and identifies an interesting future direction for investigation.
CLASS I PITPs AND RECEPTOR TYROSINE KINASE SIGNALING: THE CASE FOR THE NETRIN RECEPTOR
PITPα has also been reported as essential for signaling by deleted in colorectal cancer (DCC), a netrin receptor (216). Netrins are secreted factors that regulate brain and spinal cord development by acting as neuronal guidance cues for axon elongation and pathfinding (217, 218). As for the EGFR pathway, PITPα is posited to physically interact with netrin-activated DCC, thereby supplying the plasma membrane with PtdIns as substrate during signaling. Several lines of evidence support the case (216). First, pull-down assays report that PITPα binds the DCC cytosolic tail. Second, the kanga mouse, which presents a natural deletion of the region of the DCC cytosolic tail that binds PITPα in vitro, phenocopies netrin-deficient mice. Third, some data are presented to suggest axon-guidance deficits in the PITPα hypomorphic Vib mouse (216). Again, the neuronal phenotypes associated with PITPα-null mice are not congruent for an essential role for PITPα in netrin signaling. While netrin-deficient and DCC-deficient (e.g., kanga) animals exhibit major defects in brain structure, especially in the anterior and hippocampal commissures and corpus callosum (218), PITPα-null mice do not (198, 199). Moreover, PITPα-null mice do not exhibit an obvious thinning of the ventral plate of the spine (198, 199), a structure whose formation requires netrin-dependent axon guidance of spinal commisural neurons (218).
CLASS I PITPs AS GENETIC MODIFIERS OF DISEASE
Recent work suggests that PITPα is involved in control of PtdIns(3,4,5)P3/Akt signaling, although perhaps not via its propagation. Specifically, PITPα is a genetic modifier for Duchenne muscular dystrophy (DMD). DMD results from the absence of the dystrophin protein in muscle and presents as a progressive muscle wasting disease that ultimately leads to cardio-respiratory failure. Expression profiling of DMD model dogs (termed the Brazilian golden retriever muscular dystrophy or GRMD) reported that dogs with less severe GRMD phenotypes and slowed disease progression present significantly lower levels of PITPα expression (219). That this correlation is of functional relevance is indicated by several independent lines of evidence. First, partial knockdown of PITPα expression in the dystrophin-deficient sapje zebrafish model for DMD improved muscle structure and swim test performance in affected fish, as well as improving overall survival. Second, lentiviral knockdown of PITPα expression in human muscle cells derived from DMD patients improved the fusion index, a sign of healthy myoblasts. In all three models, disease severity also correlated directly with reduced levels of AKT phosphorylation on Ser473, as well as increased PTEN levels. As loss of PITPα expression in those models consistently increased phospho-AKT levels and reduced PTEN levels (219), the DMD models may thus represent an interesting case where PITPα acts as a negative regulator of PtdIns(3,4,5)P3 and AKT signaling via modulation of PtdIns(3,4,5)P3 degradation by PTEN.
CLASS II START-LIKE PITPs
Though all PITPs analyzed to date bind PtdIns, the prevailing view is that the cellular function of PITPs requires the binding of an additional ligand, presumably a lipid. It is in this property that class II PITPs diverge biochemically from class I PITPs. Whereas all class I PITPs analyzed to date bind PtdIns and PtdCho, the class II PITPs bind PtdIns and PtdOH (164–166, 220). As such, it is the class II PITPs that adhere to the biochemical properties envisioned by Michell for soluble proteins that recharge PtdIns resynthesis and transport in cells undergoing agonist-stimulated phosphoinositide hydrolysis and PLC-signaling (Fig. 2). It remains unclear whether class II PITPs transport PtdIns and PtdOH between the plasma membrane and the ER to supply plasma membrane signaling reactions or to carry out a unique presentation function analogous to that of Sec14.
THE STAND-ALONE CLASS II START-LIKE PITP, PITPnc1
PITPnc1, the only single-domain mammalian homolog of the class II proteins, is expressed ubiquitously across many tissues, with highest mRNA expression in the heart, muscle, kidney, liver and peripheral blood, and lower levels in the brain, eye, spleen, small intestine, lungs, and leukocytes (221). Some studies suggest that PITPnc1 activity responds to DAG signaling. After prolonged treatment with the DAG analog, PMA, PITPnc1 redistributes to the Golgi system where it is proposed to interact with the angiotensin II receptor-associated protein (ATRAP) (169). ATRAP localizes to secretory organelles, including the ER, Golgi, and endocytic vesicles (222), and promotes angiotensin II type I receptor internalization in response to angiotensin II-mediated activation via a mechanism involving PLC inhibition (222–224). Upon angiotensin II stimulation, cellular DAG levels increase in a biphasic manner; the first peak occurs at 15 s post stimulation due to PtdIns(4,5)P2 hydrolysis, and the second peak occurs at 5 min due to PtdCho hydrolysis (225). As 16 h treatment of cells with PMA reflects neither condition, whether interaction of PITPnc1 with ATRAP represents a genuine cellular response to DAG signaling remains an open question.
PITPnc1 is gathering interest from the perspective of cancer biology as elevations in PITPnc1 expression correlate with metastatic progression in human metastatic breast, melanoma, and colon tumors, and targeted PITPnc1 overexpression in mice was sufficient to promote tumor metastasis (226). The PITPnc1 gene is also part of a microRNA (miRNA) regulon involved in cancer cell metastasis (227). Silencing of miR-126 in MDA-MB-231 is observed in several human cancers (228, 229), and its silencing in breast cancer cells promotes recruitment of endothelial cells and the induction of angiogenesis that facilitates metastatic colonization (227). Three miR-126-targeted genes encode components of two parallel signaling pathways that regulate endothelial cell recruitment by cancer cells: PITPnc1, the secreted insulin-like growth factor binding protein 2 (IGFBP2), and the c-Mer tyrosine kinase (MERTK) receptor. PITPnc1 is required for the IGFBP2-mediated increase in endothelial recruitment, tumor colonization, and angiogenesis, and reduction of PITPnc1 expression in breast cancer cells results in diminished IGFBP2 secretion (227).
Extended analyses show that PITPnc1 promotes the secretion of multiple factors (e.g., metalloproteases and pro-angiogenic growth factor) that likely contribute to metastasis (226). From the functional perspective, liposome binding experiments identify PITPnc1 as primarily a PtdIns4P-binding protein, with negligible PtdIns- and PtdOH-binding capacity, and mutation analysis argued that PtdIns4P binding is “conventional,” as it is mediated by the PtdIns-binding barcode of this protein. Halberg et al. (226) posit that PITPnc1 binds PtdIns4P on Golgi membranes through an unspecified mechanism involving association with the small GTPase, Rab1B. It is also proposed to stimulate Golgi PtdIns4P synthesis, thereby promoting GOLPH3 recruitment to Golgi membranes. GOLPH3 is subsequently envisioned to drive membrane tubulation and promotes vesical biogenesis (54, 202, 203, 226). These conclusions are not exactly in line with previous reports that define PtdIns and PtdOH as the primary PITPnc1 ligands (164, 220), although the results are not mutually exclusive. While available START-like PITP structures suggest that it unlikely that a phosphorylated inositol headgroup can be accommodated in the START-like PITP lipid-binding pocket, resolution of the question awaits structural characterization of a class II PITP. Finally, although cell culture work suggests important roles for PITPnc1 in membrane trafficking and cell motility, mice genetically ablated for PITPnc1 are born alive and do not exhibit obvious phenotypic deficits, even as adults (A. Grabon, V. A. Bankaitis, and M. I. McDermott, unpublished observations).
THE MULTI-DOMAIN CLASS II START-LIKE PITPs
The multi-domain class II PITPs include the Drosophila RdgBa, the founding member of the class, as well as human PITPnm1 and PITPnm2, and the Caenorhabditis elegans PITP-1 (Fig. 6). These proteins contain N-terminal PITP domains with ∼40% homology to class I PITPs (164, 221, 230, 231). In addition, they have several domains, including a hydrophobic region that associates with membranes (232), a DDHD domain that binds PtdIns4P in vitro (233–235), and a FFAT (EFFDAxE) motif that is proposed to anchor PITPnm1 to the ER (166). Class II PITPs contain a LNS2 domain found in lipins and lipin homologs in S. cerevisiae (Smp2) and Schizosaccharomyces pombe (Ned1) (236). Lipins are PtdOH phosphatases, and the LNS2 domain of PITPnm1 binds PtdOH but does not harbor PtdOH phosphatase activity (237, 238). The C termini of class II PITPs also interact with the protein tyrosine kinase, PYK2, a protein involved in signaling from cell surface receptors (163, 239), from which the alternative Nir nomenclature derives (discussed above). The physical properties of the multi-domain class II PITPs are consistent with these proteins functioning at membrane contact sites, and this mechanism has been proposed for Drosophila RdgB and human PITPnm1 in the context of a transport/supply function (165, 166, 240–242). However, as all of the available data are also equally compatible with PtdIns presentation models, we argue that the functional data available for multi-domain PITPs should be interpreted with an open mind.
THE Drosophila CLASS II PITP RdgBα IN THE PHOSPHOINOSITIDE CYCLE AT MCS
The sole class II PITP of Drosophila is RdgB and mutant flies lacking this activity altogether are fully viable, albeit blind. Mutants in the rdgB gene were identified in a screen for light-enhanced retinal degeneration (243). The Drosophila melanogaster compound eye consists of 750–800 unit eyes, or ommatidia, each of which contains eight photoreceptor neurons that project axons to the optic lobe (244). RdgB is expressed in these photoreceptor cells, where it localizes to a plasma membrane-adjacent subcompartment of the ER called the submicrovillar cisternae (245, 246). Mutants in rdgB exhibit abnormal electroretinogram patterns in response to light (243, 247). The rhabdomere membranes housing the photoreceptors are vesicularized and internalized, resulting in rhabdomere degeneration (243, 247–250). In addition, the cell bodies feature signs suggestive of autophagy, and the photoreceptor axon terminals exhibit a marked decrease in synaptic vesicles as well as in presynaptic structure (249, 250). The light dependence of rdgB mutant retinal degeneration is prevented either by calcium depletion or by inactivation of IP3 signaling components that occur downstream of rhodopsin activation, including PLC and PKC. That the screen for light-enhanced retinal degeneration that identified rdgB also identified a mutant in the fly DAG kinase (rdgA) lends further support to the notion that deficits in PLC activation, IP3 production, and DAG conversion to PtdOH promote light-induced photoreceptor degeneration (251, 252).
Light signaling through G protein-coupled receptors in the rhabdomere is phosphoinositide mediated, and it represents a high-demand phosphoinositide turnover/resynthesis circuit, particularly when challenged with intense light. As such, it is a relevant physiological system for studying the functional role of a class II PITPs. In that regard, RdgB localization to the submicrovillar cisternae is suited to its proposed role in transfer of PtdIns from the ER to the plasma membrane to propel the PtdIns(4,5)P2 cycle, as envisioned by Michell (Fig. 2) (253). Recent characterization of RdgB as a PtdOH-binding protein invites speculation that it may be involved in the reverse portion of the cycle as well, transferring PtdOH back to the ER (165). Indeed, RdgB loss-of-function mutants display an inability to restore plasma membrane PtdIns(4,5)P2 levels in photoreceptor cells following light stimulation, and heterologous cytosol reconstitution experiments measuring generation of inositol phosphates following stimulation of permeabilized mammalian cells suggest that the fly RdgB PITP domain is required for optimal PLC activity (165). Basal PtdIns(4,5)P2 levels in rdgB mutant retina are also lower compared with wild-type retina, and rdgB mutants also show elevated retinal PtdOH levels following light stimulation, suggesting that RdgB is involved in downstream PtdOH metabolic pathways (165).
However, the data are more complicated. For example, rhodopsin levels are also downregulated in rdgB-null flies, possibly as a mechanism to quench a hyperactive PLC signaling cascade in these mutants. Indeed, rdgB mutants fail to terminate signaling in single light pulse stimulation experiments (247, 250). Mutations in a simple PtdIns supply activity would be predicted to show signal failure due to PtdIns(4,5)P2 rundown, rather than a deficit in signal termination. Thus, the data do not exclude the possibility that PtdIns(4,5)P2 deficits to some extent reflect extended PLC activity profiles following light stimulation. In that scenario, RdgB quenches the plasma membrane signal rather than, or in addition to, restoring signaling capacity.
It also bears emphasis that it is the extended domain architecture of class II PITPs that encourages interpretation of these proteins as PtdIns transporters that operate at membrane contact sites, given their presumed abilities to participate in ER/plasma membrane bridges. As the PITP domain of the multi-domain class II proteins represents only some 20–25% of the total primary sequence and does not contain any of the other domains involved in such functions (e.g., PtdIns4P-binding, ER-anchoring, etc.), one would expect that the class II PITP domain alone would be insufficient to complement defects in the full-length protein. Yet, in the fly, a context of unambiguous physiological relevance, expression of the RdgB PITP domain alone fully restores all RdgB functions to rdgB-null flies as measured by corrected retinal degeneration and by electrophysiological criteria, even under conditions of intense light stimulation (247, 250). This is an altogether remarkable (and largely ignored) result that does not easily conform to prevailing (and broadly held) ideas for how class II PITPs function in mammalian cells (as reviewed below). Interestingly, expression of mammalian PITPα cannot rescue rdgB-null phenotypes in the fly (247), and this failure might reflect the fact that PITPα is a PtdIns/PtdCho-exchange protein, whereas the RdgB PITP domain has PtdIns/PtdOH exchange activity.
THE C. elegans CLASS II PITP-1 AND PIP/DAG SIGNALING IN SENSORY NEURONS
PITP-1 is the single worm ortholog of the class II START-like PITPs and, like RdgB in flies, it too does not execute functions essential for viability of the organism. Rather, loss-of-function alleles of the corresponding structural gene were identified in a genetic screen for C. elegans mutants defective in ASE right (ASER) gustatory neuron plasticity (254). ASER is a sensory neuron responsible for sensing salt under attraction and avoidance conditions, and C. elegans chemotaxis as a function of prior experience is dependent on ASER plasticity. For example, worms normally exhibit attractive behavior to NaCl but switch to NaCl avoidance if the animals had prior experience to NaCl-exposure in a starvation environment (255). This behavioral conditioning is reversible and is dependent on two opposing phosphoinositide signaling cascades: 1) PLC/DAG/PKC signaling for attractive cues, such that high DAG levels are associated with attraction and low levels with avoidance; and 2) PtdIns(3,4,5)P3/PI3K signaling for plasticity and repulsive chemotaxis. Genetic or pharmacological activation of DAG signaling pathways results in constitutive attraction to salt, and inactivation of the PI3K pathway eliminates plasticity (256, 257). The PITP-1 mutant identified in the screen, pitp-1(pe1209), exhibits reduced NaCl attraction and loss of plasticity in response to starvation conditioning (255).
DAG kinase mutations rescue attraction defects in pitp-1(pe1209) worms, but not plasticity defects, suggesting that PITP-1 mediates the interface between two distinct signaling pathways that determine attractive or repulsive responses to a specific cue: 1) a DAG-dependent pathway, possibly due to regulation of PtdIns(4,5)P2 pools; and 2) a pathway insensitive to DAG pools (at least those subject to regulation by DAG kinase), possibly through PtdIns(3,4,5)P3 signals mediating worm starvation responses. Though the data were interpreted from the perspective that PITP-1 provides ER PtdIns to the synaptic membrane during signaling, we suggest that the data fit more closely with the nanoreactor model. PITP-1 might modulate PtdIns availability to opposing pools of PtdIns/phosphoinositide-modifying enzymes (signaling pixels?) that compete for substrate, such as between PLC and PI3K. Whether PITP-1 has intrinsic PtdIns-transfer activity has not yet been tested. The nature of its second ligand is also unknown.
PITP-1 localizes to RAB-3-positive synaptic vesicles in the axon of presynaptic regions in ASER neurons, and is also localized to presynaptic regions of amphid interneuron B (AIB) interneurons. Unlike the Drosophila rdgB mutant, which manifests light-induced degeneration of photoreceptor cells, pitp-1 worms do not suffer ASER neuron degeneration when exposed to stimulatory cues (243, 254). Yet again, the PITP-1 PITP domain was necessary and, remarkably, sufficient to rescue the chemosensory attraction and plasticity defects of pitp-1 mutants (254).
THE HUMAN CLASS II PITP, PITPnm1, IN THE PHOSPHOINOSITIDE CYCLE AT MCS
PITPnm1 is the mammalian ortholog to fly RdgB. Analogous to the proposed function of RdgB at submicrovillar cisternae in Drosophila photoreceptor cells, PITPnm1 is identified as an ER-plasma membrane contact site component in human cells (166). HEK293T cells stably overexpressing angiotensin receptor (HEF293T-AT1 cells) displayed plasma membrane PtdIns(4,5)P2 cycle stimulation following addition of angiotensin II. As seen in rdgB mutant fly photoreceptor cells (165), unstimulated HEK293T-AT1 cells knocked down for PITPnm1 had lower basal PtdIns(4,5)P2 levels. Additionally, lower levels of PtdIns and PtdIns4P were observed, suggesting that PITPnm1 plays a role in maintaining signaling lipid pools under basal conditions (166). PITPnm1 was required for sustained PLC signaling, as measured by intracellular calcium and DAG and PtdOH FRET-based biosensors. Additionally, depletion of PITPnm1 with siRNA led to PtdOH accumulation, and these cells were unable to convert PtdOH to CDP-DAG and PtdIns. This effect was PITPnm1 specific in that siRNA-mediated PITPα and PITPβ depletion did not recapitulate these effects. One interpretation is that PITPnm1 binds PtdOH as a counter-ligand to PtdIns, while PITPα and PITPβ bind PtdCho and not PtdOH (211). However, counter-ligand identity alone cannot entirely account for the PITPnm1 specificity in these assays because PITPnc1 (the soluble human RdgB ortholog), a PtdOH transfer protein in vitro, also did not recapitulate the effect (164, 166, 220).
At steady-state, PITPnm1 localizes to the cytoplasm and the ER. Following stimulation of cells with angiotensin II, PITPnm1 translocates to ER-plasma membrane contact sites, as marked by the contact site marker, STIM1. PITPnm1 C-terminal regions were reported to be important for plasma membrane association, while the PITP domain was inhibitory (258). Considering that the PITPnm1 LNS2 domain binds PtdOH (238), perhaps this domain mediates plasma membrane interaction following PLC activation. Alternatively, the PtdOH-transfer ability of the PITPnm1 PITP-domain may potentiate propagation of the phosphoinositide cycle as forecast by Michell (1).
THE CLASS II PITPnm1 AND PIP/DAG SIGNALING AT THE GOLGI
Like Sec14 in yeast, PITPnm1 is also linked to Golgi trafficking and promotes membrane fission facilitating vesicular biogenesis from the TGN (259). Acute siRNA-mediated depletion of PITPnm1 in HeLa cells causes Golgi membranes to become dispersed and swollen, resulting in retention of secretory cargo in the TGN and a subsequent delay of cargo transport to the plasma membrane. Cargo retained in the TGN under PITPnm1-deficient conditions localizes to highly tubular membranes associated with the Golgi, and is accompanied by a marked decrease in intermediate carriers emerging from the Golgi.
Trafficking defects resulting from transient PITPnm1 knockdown stem from a reduction of DAG pools at the TGN (259). PITPnm1 is required for maintenance of the DAG pool used in the CDP-choline pathway for PtdCho biosynthesis (259). The specificity for sensing CDP-choline pathway activity is analogous to the case of yeast Sec14 for which the PtdCho counter-ligand is the end-product of the CDP-choline pathway activity (21, 23, 260). Interestingly, Litvak et al. (259) excluded Golgi PtdIns4P as a consequence of PITPnm1 depletion on the basis that Golgi-targeting of a PtdIns4P biosensor (the OSBP PH-domain) was indifferent to PITPnm1 knockdown. However, because there are multiple PtdIns4P pools at the Golgi, and PH-domains are sensitive to these differences, analysis of a single biosensor may be insufficient to arrive at firm conclusions regarding what effects PITPnm1 depletion has on Golgi PtdIns4P levels. Truncation studies show that the PITPnm1 PITP domain is required, but not sufficient, to restore cargo trafficking from the TGN under PITPnm1-silenced conditions (259).
CLASS II PITPs AND RECEPTOR TYROSINE KINASE SIGNALING: THE CASE FOR THE EGF RECEPTOR
PITPnm1 is reported to be required for propagation of EGF receptor signaling responses in HeLa cells (238). PITPnm1 localizes to the Golgi in this context, though a distinct report localized PITPnm1 to the ER in HEK293T cells (166). The subcellular localization of PITPnm1 may therefore be cell type dependent. In both cases, however, PITPnm1 translocates to the plasma membrane upon receptor stimulation (166, 238). Modulation of PITPnm1 expression affects signaling pathways downstream of EGFR consistent with a role in activation or propagation of the EGFR response. However, transient siRNA-mediated PITPnm1 knockdown also causes Golgi defects (259), raising the possibility that EGFR signaling defects under PITPnm1-depleted conditions result from inadequate forward trafficking of components required for plasma membrane signaling. The stable shRNA-expressing knockdown clones used in this study did not display Golgi defects, however (238). So, there remains some uncertainty surrounding the issue of PITPnm1 involvement in mammalian Golgi function.
Propagation of EGF signaling required both the PITPnm1 PITP- and LNS2-domains (238). LNS2 domains are characterized by lipid phosphate phosphatase (LPP) (or PtdOH phosphatase) activity. The multi-domain class II PITPs contain LNS2 domains, but the canonical DXDX(T/V) motifs involved in phosphatase function are altered such that the initial aspartate (D) residue is replaced with a serine. As this residue is required for LPP activity in other HAD superfamily members, it is unlikely that the PITPs possess LPP activity, although this remains to be tested. The PITPnm1 LNS2 domain associates with PtdOH-containing vesicles and to PtdIns4P-containing vesicles to a lesser extent. Addition of PtdOH to cells resulted in LNS2-domain-dependent PITPnm1 translocation to the plasma membrane, suggesting that PtdOH is the PITPnm1 recruitment signal following EGF stimulation. To test this possibility, the authors interfered with phospholipase D (PLD)-mediated PtdOH production using primary alcohols. Indeed, inhibition of EGF-stimulated PITPnm1 redistribution to the plasma membrane was observed. However, use of primary alcohols as inhibitors of PLD-mediated PtdOH production is not a rigorous approach (262–266). With the availability of isoform-specific PLD inhibitors, the issue can now be revisited in a more rigorous way (267–270).
PITPnm1 AND MAMMALIAN DISEASE
PITPnm1 exhibits a broad tissue expression profile (271–273). This fact, when coupled with reports of an essential role for PITPnm1 in Golgi trafficking and receptor tyrosine kinase signaling, suggest that this class II PITP is important for mammalian development. In that regard, PITPnm1 was initially reported as an essential gene in mice, as nullizygosity resulted in preimplantation lethality (274). However, a more recent report demonstrates that PITPnm1-null mice are viable and fertile with the only reported defect being decreased circulating cholesterol levels in male homozygotes (275). In that regard, 5% of the human population expresses a prematurely truncated PITPnm1 gene from which only the PITP domain is expressed [from the Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/about)] (276) (Fig. 6). To date, the available genomic data only identify heterozygous states for this allele, suggesting that either the homozygous condition is lethal in humans or that homozygous individuals exist but have not yet been identified in exome sequencing efforts. The best data presently suggest that, like its fly and worm orthologs, PITPnm1 is not an essential gene in mammals either.
Ironically, in terms of human disease, it is a Nir protein that is composed of the C-terminal domain arrangement that typifies the class II StART-like PITPs, but lacks the PITP domain entirely, that enters the picture. Defects in PITPnm3 result in an autosomal dominant cone-rod dystrophy 5 retinal dystrophy in humans. This disease manifests itself as an onset of blindness in early adulthood due to cone photoreceptor degeneration (277, 278).
SUMMARY
A chemically simple phosphoinositide repertoire is nonetheless responsible for regulating a diverse set of cellular responses in mammals. The mechanisms through which the phosphoinositide code is functionally diversified are critical to cellular function as well as being of direct relevance to human health and disease. PITPs are assuming their place on the center stage of the mechanisms by which phosphoinositide signaling is functionally channeled to diverse biological outcomes. The importance of PITPs is evidenced by the remarkable range of highly specific physiological functions these proteins play, from unicellular eukaryotes to multicellular organisms such as plants, Drosophila, vertebrates, and even mammals. Although, the collaboration between PITPs and PI4Ks in promoting PtdIns4P signaling has been featured, it remains an open question of whether other PITP-like activities play similar roles in regulating the activities of PI3K- and phosphoinositide-kinases.
Whereas the history of how PITPs were first identified supports a broad view of these proteins as inter-organellar carriers of PtdIns between membranes to support phosphoinositide signaling, an alternative view of these proteins as essential potentiators of PtdIns-kinase activity cells is emerging, and these presentation (or nanoreactor) models are conceptually powerful in terms of describing the roles of PITPs in instructive regulation of PtdIns kinase activities and how such regulation contributes to diversification of phosphoinositide signaling. Defining the ligand specificities of each PITP now assumes central importance, as PITPs act as “sensors” for such ligands and translate that sensing capacity to PtdIns4P signaling. The compositions of the signaling pixels for each PITP also represent critical parts of the functional puzzle. Regardless of whether one favors PtdIns-presentation or PtdIns-transfer models for PITPs (and this may prove to be a case-by-case basis), elucidation of the molecular dynamics of the lipid exchange cycle in atomistic detail is an important area of investigation, as that cycle is central to PITP function.
Finally, the genetic diversity of mammals will ultimately have its input into our understanding of PITP function. The identification of genetic modifier loci whose loss of function bypasses the cellular Sec14 requirement for viability in yeast provided invaluable and completely unexpected insights into the role of Sec14 as an integrator of membrane trafficking, PtdIns4P signaling, and PtdCho metabolism. It is in that same regard that new and exciting opportunities are becoming available for studying PITP, and particularly class I StART-like PITP, function in mammalian disease. These new opportunities are forecast by genetic studies that demonstrate the existence of powerful naturally occurring modifiers of PITPα deficiencies in mice. Several genetic modifiers of the hypomorphic Vib allele that reduces PITPα expression some 80% are now identified (211, 278). Homozygous Vib mice of the C57/B6 genetic background die 33–35 days after birth from an escalating neurodegenerative disease and an ascending motor paralysis. Yet, Vib homozygotes crossed onto the A/J genetic background live much longer, some to nearly 2 years. What is interesting is that the A/J Vib mice preserve the pathologies of B6 Vib mice, including severity and rate of onset of tremors and neurodegenerative disease (269). Those observations raise a number of questions regarding what the relationship is between neurodegeneration and morbidity: what is the actual cause of death in these mice and what are the mechanisms by which lifespan is extended in PITPα-deficient A/J mice? Identification of the responsible polymorphisms, or the epigenetic signatures associated with phenotype, is expected to promote dissection of both the pathologies of PITPα-deficiency diseases as well as the cellular mechanisms of PITPα function. It is clear that the PITP field is one rich for exploration from structural, biophysical, and biological perspectives. We argue that this essentially unplowed field offers outstanding opportunities for talented young scientists with new ideas to contribute to a major problem in contemporary cell biology.
Footnotes
Abbreviations:
- ASER
- ASE right
- ATRAP
- angiotensin II receptor-associated protein
- αTTP
- α-tocopherol transfer protein
- CRALBP
- cellular retinaldehyde binding protein
- DAG
- diacylglycerol
- DCC
- deleted in colorectal cancer
- DMD
- Duchenne muscular dystrophy
- G-module
- gating-module
- GOLPH3
- Golgi phosphoprotein 3
- IGFBP2
- insulin-like growth factor binding protein 2
- IP3
- inositol triphosphate
- LPP
- lipid phosphate phosphatase
- ORP
- oxysterol binding-related protein
- OSBP
- oxysterol binding protein
- PH
- pleckstrin homology
- PIK
- phosphoinositide kinase
- PI4K
- phosphatidylinositol 4-OH kinase
- PIP
- phosphoinositide
- PIP Pase
- phosphoinositide phosphatase
- PITP
- phosphatidylinositol transfer protein
- PL
- phospholipid
- PLC
- phospholipase C
- PLD
- phospholipase D
- PtdCho
- phosphatidylcholine
- PtdIns
- phosphatidylinositol
- PtdIns3P
- phosphatidylinositol 3-phosphate
- PtdIns4P
- phosphatidylinositol 4-phosphate
- PtdIns5P
- phosphatidylinositol 5-phosphate
- PtdIns(3,5)P2
- phosphatidylinositol 3,5-bisphosphate
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PtdIns(3,4)P2
- phosphatidylinositol 3,4-bisphosphate
- PtdIns(3,4,5)P3
- phosphatidylinositol 3,4,5-triphosphate
- PtdIns4P5K
- PtdIns4P 5-OH kinase
- PtdOH
- phosphatidic acid
- RdgB
- retinal degeneration B
- Sfh
- Sec14 homology
- START
- StAR related lipid transfer domain
- TGN
- trans-Golgi network
- Vib
- vibrator
This work was supported by National Institutes of Health Grants GM44530 and GM112591 and Welch Foundation Award BE-0017 to V.A.B. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Michell R. H. 2008. Inositol derivatives: evolution and functions. Nat. Rev. Mol. Cell Biol. 9: 151–161. [DOI] [PubMed] [Google Scholar]
- 2.Michell R. H. 2013. Inositol lipids: from an archaeal origin to phosphatidylinositol 3,5-bisphosphate faults in human disease. FEBS J. 280: 6281–6294. [DOI] [PubMed] [Google Scholar]
- 3.Behnia R., and Munro S.. 2005. Organelle identity and the signposts for membrane traffic. Nature. 438: 597–604. [DOI] [PubMed] [Google Scholar]
- 4.Kutateladze T. G. 2010. Translation of the phosphoinositide code by PI effectors. Nat. Chem. Biol. 6: 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hokin M. R., and Hokin L. E.. 1954. Effects of acetylcholine on phospholipides in the pancreas. J. Biol. Chem. 209: 549–558. [PubMed] [Google Scholar]
- 6.Hokin M. R., Benfey B. G., and Hokin L. E.. 1958. Phospholipides and adrenaline secretion in guinea pig adrenal medulla. J. Biol. Chem. 233: 814–817. [PubMed] [Google Scholar]
- 7.Hokin L. E. 1968. Dynamic aspects of phospholipids during protein secretion. Int. Rev. Cytol. 23: 187–208. [DOI] [PubMed] [Google Scholar]
- 8.Kemp P., Hübscher G., and Hawthorne J. N.. 1961. Phosphoinositides. 3. Enzymic hydrolysis of inositol-containing phospholipids. Biochem. J. 79: 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michell R. H., Kirk C. J., Jones L. M., Downes C. P., and Creba J. A.. 1981. The stimulation of inositol lipid metabolism that accompanies calcium mobilization in stimulated cells: defined characteristics and unanswered questions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 296: 123–138. [DOI] [PubMed] [Google Scholar]
- 10.Weiss S. J., McKinney J. S., and Putney J. W.. 1982. Receptor-mediated net breakdown of phosphatidylinositol 4,5-bisphosphate in parotid acinar cells. Biochem. J. 206: 555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berridge M. J. 1983. Rapid accumulation of inositol trisphosphate reveals that agonists hydrolyse polyphosphoinositides instead of phosphatidylinositol. Biochem. J. 212: 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creba J. A., Downes C. P., Hawkins P. T., Brewster G., Michell R. H., and Kirk C. J.. 1983. Rapid breakdown of phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate in rat hepatocytes stimulated by vasopressin and other Ca2+-mobilizing hormones. Biochem. J. 212: 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berridge M. J., and Irvine R. F.. 1989. Inositol phosphates and cell signalling. Nature. 341: 197–205. [DOI] [PubMed] [Google Scholar]
- 14.Nishizuka Y. 1984. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature. 308: 693–698. [DOI] [PubMed] [Google Scholar]
- 15.Nishizuka Y. 1986. Studies and perspectives of protein kinase C. Science. 233: 305–312. [DOI] [PubMed] [Google Scholar]
- 16.Nishizuka Y. 1992. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science. 258: 607–614. [DOI] [PubMed] [Google Scholar]
- 17.Michell R. H. 2009. First came the link between phosphoinositides and Ca2+ signalling, and then a deluge of other phosphoinositide functions. Cell Calcium. 45: 521–526. [DOI] [PubMed] [Google Scholar]
- 18.Michell R. H. 1975. Inositol phospholipids and cell surface receptor function. Biochim. Biophys. Acta. 415: 81–147. [DOI] [PubMed] [Google Scholar]
- 19.Bankaitis V. A., Aitken J. R., Cleves A. E., and Dowhan W.. 1990. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 347: 561–562. [DOI] [PubMed] [Google Scholar]
- 20.Cleves A., McGee T., and Bankaitis V. A.. 1991a. Phospholipid transfer proteins: a biological debut. Trends Cell Biol. 1: 30–34. [DOI] [PubMed] [Google Scholar]
- 21.Ile K. E., Schaaf G., and Bankaitis V. A.. 2006. Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nat. Chem. Biol. 2: 576–583. [DOI] [PubMed] [Google Scholar]
- 22.Schaaf G., Ortlund E. A., Tyeryar K. R., Mousley C. J., Ile K. E., Garrett T. A., Ren J., Woolls M. J., Raetz C. R., Redinbo M. R., et al. 2008. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol. Cell. 29: 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankaitis V. A., Mousley C. J., and Schaaf G.. 2010. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem. Sci. 35: 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cleves A. E., McGee T. P., Whitters E. A., Champlon K. M., Altken J. R., Dowhan W., Goebl M., and Bankaitis V. A.. 1991b. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 64: 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth M. G. 2004. Phosphoinositides in constitutive membrane traffic. Physiol. Rev. 84: 699–730. [DOI] [PubMed] [Google Scholar]
- 26.Bankaitis V. A., Garcia-Mata R., and Mousley C. J.. 2012. Golgi membrane dynamics and lipid metabolism. Curr. Biol. 22: R414–R424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balla T. 2013. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93: 1019–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glick B. S., and Nakano A.. 2009. Membrane traffic within the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 25: 113–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu F., Crump C. M., and Thomas G.. 2001. Trans-Golgi network sorting. Cell. Mol. Life Sci. 58: 1067–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellman I. 1996. Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 12: 575–625. [DOI] [PubMed] [Google Scholar]
- 31.Jovic M., Sharma M., Rahajeng J., and Caplan S.. 2010. The early endosome: a busy sorting station for proteins at the crossroads. Histol. Histopathol. 25: 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxfield F. R., and McGraw T. E.. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5: 121–132. [DOI] [PubMed] [Google Scholar]
- 33.Bonifacino J. S., and Rojas R.. 2006. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 7: 568–579. [DOI] [PubMed] [Google Scholar]
- 34.Deneka M., Neeft M., and Sluijs P. V. D.. 2003. Regulation of membrane transport by rab GTPases. Crit. Rev. Biochem. Mol. Biol. 38: 121–142. [DOI] [PubMed] [Google Scholar]
- 35.Rink J., Ghigo E., Kalaidzidis Y., and Zerial M.. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 122: 735–749. [DOI] [PubMed] [Google Scholar]
- 36.Poteryaev D., Datta S., Ackema K., Zerial M., and Spang A.. 2010. Identification of the switch in early-to-late endosome transition. Cell. 141: 497–508. [DOI] [PubMed] [Google Scholar]
- 37.Futter C. E., Pearse A., Hewlett L. J., and Hopkins C. R.. 1996. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell Biol. 132: 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luzio J. P., Rous B. A., Bright N. A., Pryor P. R., Mullock B. M., and Piper R. C.. 2000. Lysosome-endosome fusion and lysosome biogenesis. J. Cell Sci. 113: 1515–1524. [DOI] [PubMed] [Google Scholar]
- 39.Woodman P. G., and Futter C. E.. 2008. Multivesicular bodies: co-ordinated progression to maturity. Curr. Opin. Cell Biol. 20: 408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piper R. C., and Katzmann D. J.. 2007. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23: 519–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huotari J., and Helenius A.. 2011. Endosome maturation. EMBO J. 30: 3481–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marat A. L., and Haucke V.. 2016. Phosphatidylinositol 3-phosphates—at the interface between cell signalling and membrane traffic. EMBO J. 35: 561–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nascimbeni A. C., Codogno P., and Morel E.. 2017. Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J. 284: 1267–1278. [DOI] [PubMed] [Google Scholar]
- 44.Hammond G. R., Schiavo G., and Irvine R. F.. 2009. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2). Biochem. J. 422: 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hammond G. R., Machner M. P., and Balla T.. 2014. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol. 205: 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarkes D., and Rameh L. E.. 2010. A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem. J. 428: 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rong Y., Liu M., Ma L., Du W., Zhang H., Tian Y., Cao Z., Li Y., Ren H., Zhang C., et al. 2012. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat. Cell Biol. 14: 924–934. [DOI] [PubMed] [Google Scholar]
- 48.Nakatsu F., Baskin J. M., Chung J., Tanner L. B., Shui G., Lee S. Y., Pirruccello M., Hao M., Ingolia N. T., Wenk M. R., et al. 2012. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 199: 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sridhar S., Patel B., Aphkhazava D., Macian F., Santambrogio L., Shields D., and Cuervo A. M.. 2013. The lipid kinase PI4KIIIβ preserves lysosomal identity. EMBO J. 32: 324–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hama H., Schnieders E. A., Thorner J., Takemoto J. Y., and DeWald D. B.. 1999. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274: 34294–34300. [DOI] [PubMed] [Google Scholar]
- 51.Walch-Solimena C., and Novick P.. 1999. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1: 523–525. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y. J., Wang J., Sun H. Q., Martinez M., Sun Y. X., Macia E., Kirchhausen T., Albanesi J. P., Roth M. G., and Yin H. L.. 2003. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 114: 299–310. [DOI] [PubMed] [Google Scholar]
- 53.Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M., and De Matteis M. A.. 2004. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6: 393–404. [DOI] [PubMed] [Google Scholar]
- 54.Dippold H. C., Ng M. M., Farber-Katz S. E., Lee S. K., Kerr M. L., Peterman M. C., Sim R., Wiharto P. A., Galbraith K. A., Madhavarapu S., et al. 2009. GOLPH3 bridges phosphatidylinositol-4-phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 139: 337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tóth B., Balla A., Ma H., Knight Z. A., Shokat K. M., and Balla T.. 2006. Phosphatidylinositol 4-kinase IIIβ regulates the transport of ceramide between the endoplasmic reticulum and Golgi. J. Biol. Chem. 281: 36369–36377. [DOI] [PubMed] [Google Scholar]
- 56.D’Angelo G., Polishchuk E., Di Tullio G., Santoro M., Di Campli A., Godi A., West G., Bielawski J., Chuang C. C., van der Spoel A. C., et al. 2007. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 449: 62–67. [DOI] [PubMed] [Google Scholar]
- 57.Clayton E. L., Minogue S., and Waugh M. G.. 2013. Phosphatidylinositol 4-kinases and PI4P metabolism in the nervous system: roles in psychiatric and neurological diseases. Mol. Neurobiol. 47: 361–372. [DOI] [PubMed] [Google Scholar]
- 58.Salazar G., Craige B., Wainer B. H., Guo J., De Camilli P., and Faundez V.. 2005. Phosphatidylinositol-4-kinase type II α is a component of adaptor protein-3-derived vesicles. Mol. Biol. Cell. 16: 3692–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waugh M. G., Minogue S., Anderson J. S., Balinger A., Blumenkrantz D., Calnan D. P., Cramer R., and Hsuan J. J.. 2003. Localization of a highly active pool of type II phosphatidylinositol 4-kinase in a p97/valosin-containing-protein-rich fraction of the endoplasmic reticulum. Biochem. J. 373: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weixel K. M., Blumental-Perry A., Watkins S. C., Aridor M., and Weisz O. A.. 2005. Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J. Biol. Chem. 280: 10501–10508. [DOI] [PubMed] [Google Scholar]
- 61.Balla A., Tuymetova G., Barshishat M., Geiszt M., and Balla T.. 2002. Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J. Biol. Chem. 277: 20041–20050. [DOI] [PubMed] [Google Scholar]
- 62.Wei Y. J., Sun H. Q., Yamamoto M., Wlodarski P., Kunii K., Martinez M., Barylko B., Albanesi J. P., and Yin H. L.. 2002. Type II phosphatidylinositol 4-kinase β is a cytosolic and peripheral membrane protein that is recruited to the plasma membrane and activated by Rac-GTP. J. Biol. Chem. 277: 46586–46593. [DOI] [PubMed] [Google Scholar]
- 63.Godi A., Pertile P., Meyers R., Marra P., Di Tullio G., Iurisci C., Luini A., Corda D., and De Matteis M. A.. 1999. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1: 280–287. [DOI] [PubMed] [Google Scholar]
- 64.Huijbregts R. P., Topalof L., and Bankaitis V. A.. 2000. Lipid metabolism and regulation of membrane trafficking. Traffic. 1: 195–202. [DOI] [PubMed] [Google Scholar]
- 65.Audhya A., Foti M., and Emr S. D.. 2000. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell. 11: 2673–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baird D., Stefan C., Audhya A., Weys S., and Emr S. D.. 2008. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J. Cell Biol. 183: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Balla A., Vereb G., Gülkan H., Gehrmann T., Gergely P., Heilmeyer L. M. Jr., and Antal M.. 2000. Immunohistochemical localisation of two phosphatidylinositol 4-kinase isoforms, PI4K230 and PI4K92, in the central nervous system of rats. Exp. Brain Res. 134: 279–288. [DOI] [PubMed] [Google Scholar]
- 68.Balla A., and Balla T.. 2006. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 16: 351–361. [DOI] [PubMed] [Google Scholar]
- 69.Kakuk A., Friedländer E., Vereb G. Jr., Kása A., Balla A., Balla T., Heilmeyer L. M. Jr., Gergely P., and Vereb G.. 2006. Nucleolar localization of phosphatidylinositol 4-kinase PI4K230 in various mammalian cells. Cytometry A. 69: 1174–1183. [DOI] [PubMed] [Google Scholar]
- 70.Kakuk A., Friedländer E., Vereb G. Jr., Lisboa D., Bagossi P., Tóth G., Gergely P., and Vereb G.. 2008. Nuclear and nucleolar localization signals and their targeting function in phosphatidylinositol 4-kinase PI4K230. Exp. Cell Res. 314: 2376–2388. [DOI] [PubMed] [Google Scholar]
- 71.Dumaresq-Doiron K., Savard M. F., Akam S., Costantino S., and Lefrancois S.. 2010. The phosphatidylinositol 4-kinase PI4KIIIα is required for the recruitment of GBF1 to Golgi membranes. J. Cell Sci. 123: 2273–2280. [DOI] [PubMed] [Google Scholar]
- 72.Wong K., Meyers R., and Cantley L. C.. 1997. Subcellular localization of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 272: 13236–13241. [DOI] [PubMed] [Google Scholar]
- 73.Deleted in proof. [Google Scholar]
- 74.Hendricks K. B., Wang B. Q., Schnieders E. A., and Thorner J.. 1999. Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat. Cell Biol. 1: 234–241. [DOI] [PubMed] [Google Scholar]
- 75.Ames J. B., Hendricks K. B., Strahl T., Huttner I. G., Hamasaki N., and Thorner J.. 2000. Structure and calcium-binding properties of Frq1, a novel calcium sensor in the yeast Saccharomyces cerevisiae. Biochemistry. 39: 12149–12161. [DOI] [PubMed] [Google Scholar]
- 76.Zheng Q., Bobich J. A., Vidugiriene J., McFadden S. C., Thomas F., Roder J., and Jeromin A.. 2005. Neuronal calcium sensor-1 facilitates neuronal exocytosis through phosphatidylinositol 4-kinase. J. Neurochem. 92: 442–451. [DOI] [PubMed] [Google Scholar]
- 77.Bourne Y., Dannenberg J., Pollmann V., Marchot P., and Pongs O.. 2001. Immunocytochemical localization and crystal structure of human frequenin (neuronal calcium sensor 1). J. Biol. Chem. 276: 11949–11955. [DOI] [PubMed] [Google Scholar]
- 78.Kapp-Barnea Y., Melnikov S., Shefler I., Jeromin A., and Sagi-Eisenberg R.. 2003. Neuronal calcium sensor-1 and phosphatidylinositol 4-kinase β regulate IgE receptor-triggered exocytosis in cultured mast cells. J. Immunol. 171: 5320–5327. [DOI] [PubMed] [Google Scholar]
- 79.Haynes L. P., Thomas G. M., and Burgoyne R. D.. 2005. Interaction of neuronal calcium sensor-1 and ADP-ribosylation factor 1 allows bidirectional control of phosphatidylinositol 4-kinase beta and trans-Golgi network-plasma membrane traffic. J. Biol. Chem. 280: 6047–6054. [DOI] [PubMed] [Google Scholar]
- 80.Deleted in proof. [Google Scholar]
- 81.Heldwein E. E., Macia E., Wang J., Yin H. L., Kirchhausen T., and Harrison S. C.. 2004. Crystal structure of the clathrin adaptor protein 1 core. Proc. Natl. Acad. Sci. USA. 101: 14108–14113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang J., Sun H. Q., Macia E., Kirchhausen T., Watson H., Bonifacino J. S., and Yin H. L.. 2007. PI4P Promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol. Biol. Cell. 18: 2646–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Demmel L., Gravert M., Ercan E., Habermann B., Müller-Reichert T., Kukhtina V., Haucke V., Baust T., Sohrmann M., Kalaidzidis Y., et al. 2008. The clathrin adaptor Gga2p is a phosphatidylinositol 4-phosphate effector at the Golgi exit. Mol. Biol. Cell. 19: 1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cao X., Coskun U., Rössle M., Buschhorn S. B., Grzybek M., Dafforn T. R., Lenoir M., Overduin M., and Simons K.. 2009. Golgi protein FAPP2 tubulates membranes. Proc. Natl. Acad. Sci. USA. 106: 21121–21125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salminen A., and Novick P. J.. 1989. The Sec15 protein responds to the function of the GTP binding protein, Sec4, to control vesicular traffic in yeast. J. Cell Biol. 109: 1023–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Novick P., and Brennwald P.. 1993. Friends and family: the role of the Rab GTPases in vesicular traffic. Cell. 75: 597–601. [DOI] [PubMed] [Google Scholar]
- 87.Ortiz D., Medkova M., Walch-Solimena C., and Novick P.. 2002. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J. Cell Biol. 157: 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benli M., Döring F., Robinson D. G., Yang X., and Gallwitz D.. 1996. Two GTPase isoforms, Ypt31p and Ypt32p, are essential for Golgi function in yeast. EMBO J. 15: 6460–6475. [PMC free article] [PubMed] [Google Scholar]
- 89.Mizuno-Yamasaki E., Medkova M., Coleman J., and Novick P.. 2010. Phosphatidylinositol 4-phosphate controls both membrane recruitment and a regulatory switch of the Rab GEF Sec2p. Dev. Cell. 18: 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garrenton L. S., Stefan C. J., McMurray M. A., Emr S. D., and Thorner J.. 2010. Pheromone-induced anisotropy in yeast plasma membrane phosphatidylinositol-4,5-bisphosphate distribution is required for MAPK signaling. Proc. Natl. Acad. Sci. USA. 107: 11805–11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Doughman R. L., Firestone A. J., and Anderson R. A.. 2003. Phosphatidylinositol phosphate kinases put PI4,5P(2) in its place. J. Membr. Biol. 194: 77–89. [DOI] [PubMed] [Google Scholar]
- 92.Clarke J. H., and Irvine R. F.. 2013. Evolutionarily conserved structural changes in phosphatidylinositol 5-phosphate 4-kinase (PI5P4K) isoforms are responsible for differences in enzyme activity and localization. Biochem. J. 454: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boronenkov I. V., and Anderson R. A.. 1995. The sequence of phosphatidylinositol-4-phosphate 5-kinase defines a novel family of lipid kinases. J. Biol. Chem. 270: 2881–2884. [DOI] [PubMed] [Google Scholar]
- 94.Rameh L. E., Tolias K. F., Duckworth B. C., and Cantley L. C.. 1997. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 390: 192–196. [DOI] [PubMed] [Google Scholar]
- 95.Stephens L. R., Hughes K. T., and Irvine R. F.. 1991. Pathway of phosphatidylinositol (3,4,5)-trisphosphate synthesis in activated neutrophils. Nature. 351: 33–39. [DOI] [PubMed] [Google Scholar]
- 96.Hawkins P. T., Jackson T. R., and Stephens L. R.. 1992. Platelet-derived growth factor stimulates synthesis of PtdIns(3,4,5)P3 by activating a PtdIns(4,5)P2 3-OH kinase. Nature. 358: 157–159. [DOI] [PubMed] [Google Scholar]
- 97.Hemmings B. A. 1997. Akt signaling: linking membrane events to life and death decisions. Science. 275: 628–630. [DOI] [PubMed] [Google Scholar]
- 98.Stokoe D., Stephens L. R., Copeland T., Gaffney P. R., Reese C. B., Painter G. F., Holmes A. B., McCormick F., and Hawkins P. T.. 1997. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science. 277: 567–570. [DOI] [PubMed] [Google Scholar]
- 99.Cohen P., Alessi D. R., and Cross D. A.. 1997. PDK1, one of the missing links in insulin signal transduction? 1. FEBS Lett. 410: 3–10. [DOI] [PubMed] [Google Scholar]
- 100.Krugmann S., Anderson K. E., Ridley S. H., Risso N., McGregor A., Coadwell J., Davidson K., Eguinoa A., Ellson C. D., Lipp P., et al. 2002. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol. Cell. 9: 95–108. [DOI] [PubMed] [Google Scholar]
- 101.Vanhaesebroeck B., Leevers S. J., Ahmadi K., Timms J., Katso R., Driscoll P. C., Woscholski R., Parker P. J., and Waterfield M. D.. 2001. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 70: 535–602. [DOI] [PubMed] [Google Scholar]
- 102.Engelman J. A., Luo J., and Cantley L. C.. 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7: 606–619. [DOI] [PubMed] [Google Scholar]
- 103.Holz R. W., Hlubek M. D., Sorensen S. D., Fisher S. K., Balla T., Ozaki S., Prestwich G. D., Stuenkel E. L., and Bittner M. A.. 2000. A pleckstrin homology domain specific for phosphatidylinositol 4,5-bisphosphate (PtdIns-4,5-P2) and fused to green fluorescent protein identifies plasma membrane PtdIns-4,5-P2 as being important in exocytosis. J. Biol. Chem. 275: 17878–17885. [DOI] [PubMed] [Google Scholar]
- 104.Brown F. D., Rozelle A. L., Yin H. L., Balla T., and Donaldson J. G.. 2001. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 154: 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Loyet K. M., Kowalchyk J. A., Chaudhary A., Chen J., Prestwich G. D., and Martin T. F.. 1998. Specific binding of phosphatidylinositol 4,5-bisphosphate to calcium-dependent activator protein for secretion (CAPS), a potential phosphoinositide effector protein for regulated exocytosis. J. Biol. Chem. 273: 8337–8343. [DOI] [PubMed] [Google Scholar]
- 106.Chang C. L., Hsieh T. S., Yang T. T., Rothberg K. G., Azizoglu D. B., Volk E., Liao J. C., and Liou J.. 2013. Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Reports. 5: 813–825. [DOI] [PubMed] [Google Scholar]
- 107.Giordano F., Saheki Y., Idevall-Hagren O., Colombo S. F., Pirruccello M., Milosevic I., Gracheva E. O., Bagriantsev S. N., Borgese N., and De Camilli P.. 2013. PI(4,5)P2-dependent and Ca2+-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 153: 1494–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chung S. H., Song W. J., Kim K., Bednarski J. J., Chen J., Prestwich G. D., and Holz R. W.. 1998. The C2 domains of Rabphilin3A specifically bind phosphatidylinositol 4,5-bisphosphate containing vesicles in a Ca2+-dependent manner. In vitro characteristics and possible significance. J. Biol. Chem. 273: 10240–10248. [DOI] [PubMed] [Google Scholar]
- 109.Guillén J., Ferrer-Orta C., Buxaderas M., Pérez-Sánchez D., Guerrero-Valero M., Luengo-Gil G., Pous J., Guerra P., Gómez-Fernández J. C., Verdaguer N., et al. 2013. Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1. Proc. Natl. Acad. Sci. USA. 110: 20503–20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bai J., Tucker W. C., and Chapman E. R.. 2004. PIP 2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat. Struct. Mol. Biol. 11: 36–44. [DOI] [PubMed] [Google Scholar]
- 111.Liao H., Ellena J., Liu L., Szabo G., Cafiso D., and Castle D.. 2007. Secretory carrier membrane protein SCAMP2 and phosphatidylinositol 4,5-bisphosphate interactions in the regulation of dense core vesicle exocytosis. Biochemistry. 46: 10909–10920. [DOI] [PubMed] [Google Scholar]
- 112.Itoh T., Koshiba S., Kigawa T., Kikuchi A., Yokoyama S., and Takenawa T.. 2001. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 291: 1047–1051. [DOI] [PubMed] [Google Scholar]
- 113.Gaidarov I., and Keen J. H.. 1999. Phosphoinositide-AP-2 interactions required for targeting to plasma membrane clathrin-coated pits. J. Cell Biol. 146: 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rohde G., Wenzel D., and Haucke V.. 2002. A phosphatidylinositol (4,5)-bisphosphate binding site within μ2-adaptin regulates clathrin-mediated endocytosis. J. Cell Biol. 158: 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zoncu R., Perera R. M., Sebastian R., Nakatsu F., Chen H., Balla T., Ayala G., Toomre D., and De Camilli P. V.. 2007. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc. Natl. Acad. Sci. USA. 104: 3793–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Abe N., Inoue T., Galvez T., Klein L., and Meyer T.. 2008. Dissecting the role of PtdIns(4,5)P2 in endocytosis and recycling of the transferrin receptor. J. Cell Sci. 121: 1488–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cremona O., Di Paolo G., Wenk M. R., Lüthi A., Kim W. T., Takei K., Daniell L., Nemoto Y., Shears S. B., Flavell R. A., et al. 1999. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell. 99: 179–188. [DOI] [PubMed] [Google Scholar]
- 118.Haffner C., Di Paolo G., Rosenthal J. A., and De Camilli P.. 2000. Direct interaction of the 170 kDa isoform of synaptojanin 1 with clathrin and with the clathrin adaptor AP-2. Curr. Biol. 10: 471–474. [DOI] [PubMed] [Google Scholar]
- 119.Verstreken P., Koh T. W., Schulze K. L., Zhai R. G., Hiesinger P. R., Zhou Y., Mehta S. Q., Cao Y., Roos J., and Bellen H. J.. 2003. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 40: 733–748. [DOI] [PubMed] [Google Scholar]
- 120.Weinberg J., and Drubin D. G.. 2012. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 22: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Grabon A., Khan D., and Bankaitis V. A.. 2015. Phosphatidylinositol transfer proteins and instructive regulation of lipid kinase biology. Biochim. Biophys. Acta. 1851: 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nile A. H., Bankaitis V. A., and Grabon A.. 2010. Mammalian diseases of phosphatidylinositol transfer proteins and their homologs. Clin. Lipidol. 5: 867–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ren J., Pei-Chen Lin C., Pathak M. C., Temple B. R., Nile A. H., Mousley C. J., Duncan M. C., Eckert D. M., Leiker T. J., Ivanova P. T., et al. 2014. A phosphatidylinositol transfer protein integrates phosphoinositide signaling with lipid droplet metabolism to regulate a developmental program of nutrient stress–induced membrane biogenesis. Mol. Biol. Cell. 25: 712–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sha B., Phillips S. E., Bankaitis V. A., and Luo M.. 1998. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature. 391: 506–510. [DOI] [PubMed] [Google Scholar]
- 125.Bankaitis V. A., Ile K. E., Nile A. H., Ren J., Ghosh R., and Schaaf G.. 2012. Thoughts on Sec14-like nanoreactors and phosphoinositide signaling. Adv. Biol. Regul. 52: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Smirnova T. I., Chadwick T. G., Voinov M. A., Poluektov O., van Tol J., Ozarowski A., Schaaf G., Ryan M. M., and Bankaitis V. A.. 2007. Local polarity and hydrogen bonding inside the Sec14p phospholipid-binding cavity: high-field multi-frequency electron paramagnetic resonance studies. Biophys. J. 92: 3686–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ryan M. M., Temple B. R., Phillips S. E., and Bankaitis V. A.. 2007. Conformational dynamics of the major yeast phosphatidylinositol transfer protein sec14p: insight into the mechanisms of phospholipid exchange and diseases of sec14p-like protein deficiencies. Mol. Biol. Cell. 18: 1928–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schaaf G., Dynowski M., Mousley C. J., Shah S. D., Yuan P., Winklbauer E. M., de Campos M. K., Trettin K., Quinones M. C., Smirnova T. I., et al. 2011. Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution. Mol. Biol. Cell. 22: 892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bankaitis V. A. 2009. The Cirque du Soleil of Golgi membrane dynamics. J. Cell Biol. 186: 169–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ghosh R., and Bankaitis V. A.. 2011. Phosphatidylinositol transfer proteins: negotiating the regulatory interface between lipid metabolism and lipid signaling in diverse cellular processes. Biofactors. 37: 290–308. [DOI] [PubMed] [Google Scholar]
- 131.Vincent P., Chua M., Nogue F., Fairbrother A., Mekeel H., Xu Y., Allen N., Bibikova T. N., Gilroy S., and Bankaitis V. A.. 2005. A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J. Cell Biol. 168: 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ghosh R., de Campos M. K., Huang J., Huh S. K., Orlowski A., Yang Y., Tripathi A., Nile A., Lee H. C., Dynowski M., et al. 2015. Sec14-nodulin proteins and the patterning of phosphoinositide landmarks for developmental control of membrane morphogenesis. Mol. Biol. Cell. 26: 1764–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang J., Ghosh R., Tripathi A., Lönnfors M., Somerharju P., and Bankaitis V. A.. 2016. Two-ligand priming mechanism for potentiated phosphoinositide synthesis is an evolutionarily conserved feature of Sec14-like phosphatidylinositol and phosphatidylcholine exchange proteins. Mol. Biol. Cell. 27: 2317–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang J., Ghosh R., and Bankaitis V. A.. 2016. Sec14-like phosphatidylinositol transfer proteins and the biological landscape of phosphoinositide signaling in plants. Biochim. Biophys. Acta. 1861: 1352–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim Y. J., Guzman-Hernandez M. L., and Balla T.. 2011. A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev. Cell. 21: 813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Imai A., and Gershengorn M. C.. 1987. Independent phosphatidylinositol synthesis in pituitary plasma membrane and endoplasmic reticulum. Nature. 325: 726–728. [DOI] [PubMed] [Google Scholar]
- 137.Kinney A. J., and Carman G. M.. 1990. Enzymes of phosphoinositide synthesis in secretory vesicles destined for the plasma membrane in Saccharomyces cerevisiae. J. Bacteriol. 172: 4115–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Monaco M. E., and Adelson J. R.. 1991. Evidence for coupling of resynthesis to hydrolysis in the phosphoinositide cycle. Biochem. J. 279: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Santiago O. M., Rosenberg L. I., and Monaco M. E.. 1993. Organization of the phosphoinositide cycle. Assessment of inositol transferase activity in purified plasma membranes. Biochem. J. 290: 179–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Davison J. M., Bankaitis V. A., and Ghosh R.. 2012. Devising powerful genetics, biochemical and structural tools in the functional analysis of phosphatidylinositol transfer proteins (PITPs) across diverse species. Methods Cell Biol. 108: 249–302. [DOI] [PubMed] [Google Scholar]
- 141.Bankaitis V. A., Malehorn D. E., Emr S. D., and Greene R.. 1989. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J. Cell Biol. 108: 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mousley C. J., Tyeryar K., Ile K. E., Schaaf G., Brost R. L., Boone C., Guan X., Wenk M. R., and Bankaitis V. A.. 2008. Trans-Golgi network and endosome dynamics connect ceramide homeostasis with regulation of the unfolded protein response and TOR signaling in yeast. Mol. Biol. Cell. 19: 4785–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Novick P., Field C., and Schekman R.. 1980. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 21: 205–215. [DOI] [PubMed] [Google Scholar]
- 144.Strahl T., and Thorner J.. 2007. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1771: 353–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cleves A. E., Novick P. J., and Bankaitis V. A.. 1989. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J. Cell Biol. 109: 2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fang M., Kearns B. G., Gedvilaite A., Kagiwada S., Kearns M., Fung M. K., and Bankaitis V. A.. 1996. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 15: 6447–6459. [PMC free article] [PubMed] [Google Scholar]
- 147.Xie Z., Fang M., Rivas M. P., Faulkner A. J., Sternweis P. C., Engebrecht J., and Bankaitis V. A.. 1998. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc. Natl. Acad. Sci. USA. 95: 12346–12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rivas M. P., Kearns B. G., Xie Z., Guo S., Sekar M. C., Hosaka K., Kagiwada S., York J. D., and Bankaitis V. A.. 1999. Pleiotropic alterations in lipid metabolism in yeast sac1 mutants: relationship to “bypass Sec14p” and inositol auxotrophy. Mol. Biol. Cell. 10: 2235–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li X., Rivas M. P., Fang M., Marchena J., Mehrotra B., Chaudhary A., Feng L., Prestwich G. D., and Bankaitis V. A.. 2002. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 157: 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Phillips S. E., Sha B., Topalof L., Xie Z., Alb J. G., Klenchin V. A., Swigart P., Cockcroft S., Martin T. F., Luo M., et al. 1999. Yeast Sec14p deficient in phosphatidylinositol transfer activity is functional in vivo. Mol. Cell. 4: 187–197. [DOI] [PubMed] [Google Scholar]
- 151.Deleted in proof. [Google Scholar]
- 152.Kono N., Ohto U., Hiramatsu T., Urabe M., Uchida Y., Satow Y., and Arai H.. 2013. Impaired α-TTP-PIPs interaction underlies familial vitamin E deficiency. Science. 340: 1106–1110. [DOI] [PubMed] [Google Scholar]
- 153.Im Y. J., Raychaudhuri S., Prinz W. A., and Hurley J. H.. 2005. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 437: 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.de Saint-Jean M., Delfosse V., Douguet D., Chicanne G., Payrastre B., Bourguet W., Antonny B., and Drin G.. 2011. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 195: 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mousley C. J., Yuan P., Gaur N. A., Trettin K. D., Nile A. H., Deminoff S. J., Dewar B. J., Wolpert M., Macdonald J. M., Herman P. K., et al. 2012. A sterol-binding protein integrates endosomal lipid metabolism with TOR signaling and nitrogen sensing. Cell. 148: 702–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huang J., Mousley C. J., Dacquay L., Maitra N., Drin G., He C., Ridgway N. D., Tripathi A., Kennedy M., Kennedy B. K., et al. 2018. A lipid transfer protein signaling axis exerts dual control of cell-cycle and membrane trafficking systems. Dev. Cell. 44: 378–391.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schulz T. A., and Prinz W. A.. 2007. Sterol transport in yeast and the oxysterol binding protein homologue (OSH) family. Biochim. Biophys. Acta. 1771: 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Georgiev A. G., Sullivan D. P., Kersting M. C., Dittman J. S., Beh C. T., and Menon A. K.. 2011. Osh proteins regulate membrane sterol organization but are not required for sterol movement between the ER and PM. Traffic. 12: 1341–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Quon E., Sere Y. Y., Chauhan N., Johansen J., Sullivan D. P., Dittman J. S., Rice W. J., Chan R. B., Di Paolo G., Beh C. T., et al. 2018. Endoplasmic reticulum-plasma membrane contact sites integrate sterol and phospholipid regulation. PLoS Biol. 16: e2003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Antonny B., Bigay J., and Mesmin B.. 2018. The oxysterol-binding protein cycle: burning off PI(4)P to transport cholesterol. Annu. Rev. Biochem. 87: 809–837. [DOI] [PubMed] [Google Scholar]
- 161.Nemoto Y., Kearns B. G., Wenk M. R., Chen H., Mori K., Alb J. G., De Camilli P., and Bankaitis V. A.. 2000. Functional characterization of a mammalian Sac1 and mutants exhibiting substrate-specific defects in phosphoinositide phosphatase activity. J. Biol. Chem. 275: 34293–34305. [DOI] [PubMed] [Google Scholar]
- 162.McDermott M. I., and Mousley C. J.. 2016. Lipid transfer proteins and the tuning of compartmental identity in the Golgi apparatus. Chem. Phys. Lipids. 200: 42–61. [DOI] [PubMed] [Google Scholar]
- 163.Lev S., Hernandez J., Martinez R., Chen A., Plowman G., and Schlessinger J.. 1999. Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mol. Cell. Biol. 19: 2278–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Garner K., Hunt A. N., Koster G., Somerharju P., Groves E., Li M., Raghu P., Holic R., and Cockcroft S.. 2012. Phosphatidylinositol transfer protein, cytoplasmic 1 (PITPNC1) binds and transfers phosphatidic acid. J. Biol. Chem. 287: 32263–32276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yadav S., Garner K., Georgiev P., Li M., Gomez-Espinosa E., Panda A., Mathre S., Okkenhaug H., Cockcroft S., and Raghu P.. 2015. RDGBα, a PtdIns-PtdOH transfer protein, regulates G-protein-coupled PtdIns(4,5)P2 signalling during Drosophila phototransduction. J. Cell Sci. 128: 3330–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kim Y. J., Guzman-Hernandez M. L., Wisniewski E., and Balla T.. 2015. Phosphatidylinositol-phosphatidic acid exchange by Nir2 at ER-PM contact sites maintains phosphoinositide signaling competence. Dev. Cell. 33: 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Phillips S. E., Ile K. E., Boukhelifa M., Huijbregts R. P., and Bankaitis V. A.. 2006. Specific and nonspecific membrane-binding determinants cooperate in targeting phosphatidylinositol transfer protein β-isoform to the mammalian trans-Golgi network. Mol. Biol. Cell. 17: 2498–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ile K. E., Kassen S., Cao C., Vihtehlic T., Shah S. D., Mousley C. J., Alb J. G. Jr., Huijbregts R. P., Stearns G. W., Brockerhoff S. E., et al. 2010. Zebrafish class 1 phosphatidylinositol transfer proteins: PITPβ and double cone cell outer segment integrity in retina. Traffic. 11: 1151–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Garner K., Li M., Ugwuanya N., and Cockcroft S.. 2011. The phosphatidylinositol transfer protein RdgBβ binds 14–3-3 via its unstructured C-terminus, whereas its lipid-binding domain interacts with the integral membrane protein ATRAP (angiotensin II type I receptor-associated protein). Biochem. J. 439: 97–111. [DOI] [PubMed] [Google Scholar]
- 170.Schouten A., Agianian B., Westerman J., Kroon J., Wirtz K. W., and Gros P.. 2002. Structure of apo-phosphatidylinositol transfer protein α provides insight into membrane association. EMBO J. 21: 2117–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yoder M. D., Thomas L. M., Tremblay J. M., Oliver R. L., Yarbrough L. R., and Helmkamp J.. 2001. Structure of a multifunctional protein: mammalian phosphatidylinositol transfer protein complexed with phosphatidylcholine. J. Biol. Chem. 276: 9246–9252. [DOI] [PubMed] [Google Scholar]
- 172.Tilley S. J., Skippen A., Murray-Rust J., Swigart P. M., Stewart A., Morgan C. P., Cockcroft S., and McDonald N. Q.. 2004. Structure-function analysis of phosphatidylinositol transfer protein alpha bound to human phosphatidylinositol. Structure. 12: 317–326. [DOI] [PubMed] [Google Scholar]
- 173.Vordtriede P. B., Doan C. N., Tremblay J. M., Helmkamp G. M., and Yoder M. D.. 2005. Structure of PITPβ in complex with phosphatidylcholine: comparison of structure and lipid transfer to other PITP isoforms. Biochemistry. 44: 14760–14771. [DOI] [PubMed] [Google Scholar]
- 174.Alb J. G., Gedvilaite A., Cartee R. T., Skinner H. B., and Bankaitis V. A.. 1995. Mutant rat phosphatidylinositol/phosphatidylcholine transfer proteins specifically defective in phosphatidylinositol transfer: implications for the regulation of phospholipid transfer activity. Proc. Natl. Acad. Sci. USA. 92: 8826–8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tremblay J. M., Unruh J. R., Johnson C. K., and Yarbrough L. R.. 2005. Mechanism of interaction of PITPα with membranes: Conformational changes in the C-terminus associated with membrane binding. Arch. Biochem. Biophys. 444: 112–120. [DOI] [PubMed] [Google Scholar]
- 176.Grabon A., Orlowski A., Tripathi A., Vuorio J., Javanainen M., Róg T., Lönnfors M., McDermott M. I., Siebert G., Somerharju P., et al. 2017. Dynamics and energetics of the mammalian phosphatidylinositol transfer protein phospholipid exchange cycle. J. Biol. Chem. 292: 14438–14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hay J. C., and Martin T. F.. 1993. Phosphatidylinositol transfer protein required for ATP-dependent priming of Ca2+-activated secretion. Nature. 366: 572–575. [DOI] [PubMed] [Google Scholar]
- 178.Hay J. C., Fisette P. L., Jenkins G. H., Fukami K., Takenawa T., Anderson R. A., and Martin T. F.. 1995. ATP-dependent inositide phosphorylation required for Ca2+-activated secretion. Nature. 374: 173–177. [DOI] [PubMed] [Google Scholar]
- 179.Ohashi M., de Vries K. J., Frank R., Snoek G., Bankaitis V., Wirtz K., and Huttner W. B.. 1995. A role for phosphatidylinositol transfer protein in secretory vesicle formation. Nature. 377: 544–547. [DOI] [PubMed] [Google Scholar]
- 180.Jones S. M., Alb J. G., Phillips S. E., Bankaitis V. A., and Howell K. E.. 1998. A phosphatidylinositol 3-kinase and phosphatidylinositol transfer protein act synergistically in formation of constitutive transport vesicles from the trans-Golgi network. J. Biol. Chem. 273: 10349–10354. [DOI] [PubMed] [Google Scholar]
- 181.Paul K. S., Bogan A. A., and Waters M. G.. 1998. Phosphatidylinositol transfer protein (PITPα) stimulates in vitro intra-Golgi transport. FEBS Lett. 431: 91–96. [DOI] [PubMed] [Google Scholar]
- 182.Simon J. P., Morimoto T., Bankaitis V. A., Gottlieb T. A., Ivanov I. E., Adesnik M., and Sabatini D. D.. 1998. An essential role for the phosphatidylinositol transfer protein in the scission of coatomer-coated vesicles from the trans-Golgi network. Proc. Natl. Acad. Sci. USA. 95: 11181–11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kauffmann-Zeh A., Thomas G. M., Ball A., Prosser S., Cunningham E., Cockcroft S., and Hsuan J. J.. 1995. Requirement for phosphatidylinositol transfer protein in epidermal growth factor signaling. Science. 268: 1188–1190. [DOI] [PubMed] [Google Scholar]
- 184.Fensome A., Cunningham E., Prosser S., Tan S. K., Swigart P., Thomas G., Hsuan J., and Cockcroft S.. 1996. ARF and PITP restore GTPγS-stimulated protein secretion from cytosol-depleted HL60 cells by promoting PIP2 synthesis. Curr. Biol. 6: 730–738. [DOI] [PubMed] [Google Scholar]
- 185.Cunningham E., Tan S. K., Swigart P., Hsuan J., Bankaitis V., and Cockcroft S.. 1996. The yeast and mammalian isoforms of phosphatidylinositol transfer protein can all restore phospholipase C-mediated inositol lipid signaling in cytosol-depleted RBL-2H3 and HL-60 cells. Proc. Natl. Acad. Sci. USA. 93: 6589–6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cockcroft S., and Carvou N.. 2007;. Biochemical and biological functions of class I phosphatidylinositol transfer proteins. Biochim. Biophys. Acta. 1771: 677–691. [DOI] [PubMed] [Google Scholar]
- 187.De Vries K. J., Westerman J., Bastiaens P. I., Jovin T. M., Wirtz K. W., and Snoek G. T.. 1996. Fluorescently labeled phosphatidylinositol transfer protein isoforms (α and β), microinjected into fetal bovine heart endothelial cells, are targeted to distinct intracellular sites. Exp. Cell Res. 227: 33–39. [DOI] [PubMed] [Google Scholar]
- 188.Carvou N., Holic R., Li M., Futter C., Skippen A., and Cockcroft S.. 2010. Phosphatidylinositol-and phosphatidylcholine-transfer activity of PITPβ is essential for COPI-mediated retrograde transport from the Golgi to the endoplasmic reticulum. J. Cell Sci. 123: 1262–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tribble E. K., Ivanova P. T., Grabon A., Alb J. G., Faenza I., Cocco L., Brown H. A., and Bankaitis V. A.. 2016. Quantitative profiling of the endonuclear glycero-phospholipidome of murine embryonic fibroblasts. J. Lipid Res. 57: 1492–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Snoek G. T., Berrie C. P., Geijtenbeek T. B., van der Helm H. A., Cadeé J. A., Iurisci C., Corda D., and Wirtz K. W.. 1999. Overexpression of phosphatidylinositol transfer protein α in NIH3T3 cells activates a phospholipase A. J. Biol. Chem. 274: 35393–35399. [DOI] [PubMed] [Google Scholar]
- 191.Bunte H., Schenning M., Sodaar P., Bär D. P., Wirtz K. W., Van Muiswinkel F. L., and Snoek G. T.. 2006. A phosphatidylinositol transfer protein α-dependent survival factor protects cultured primary neurons against serum deprivation-induced cell death. J. Neurochem. 97: 707–715. [DOI] [PubMed] [Google Scholar]
- 192.Schenning M., van Tiel C. M., van Manen D., Stam J. C., Gadella B. M., Wirtz K. W., and Snoek G. T.. 2004. Phosphatidylinositol transfer protein α regulates growth and apoptosis of NIH3T3 cells involvement of a cannabinoid 1-like receptor. J. Lipid Res. 45: 1555–1564. [DOI] [PubMed] [Google Scholar]
- 193.Snoek G. T. 2004. Phosphatidylinositol transfer proteins: emerging roles in cell proliferation, cell death and survival. IUBMB Life. 56: 467–475. [DOI] [PubMed] [Google Scholar]
- 194.Venuti S. E., and Helmkamp G. M. Jr.. 1988. Tissue distribution, purification and characterization of rat phosphatidylinositol transfer protein. Biochim. Biophys. Acta. 946: 119–128. [DOI] [PubMed] [Google Scholar]
- 195.Utsunomiya A., Owada Y., Yoshimoto T., and Kondo H.. 1997. Localization of gene expression for phosphatidylinositol transfer protein in the brain of developing and mature rats. Brain Res. Mol. Brain Res. 45: 349–352. [DOI] [PubMed] [Google Scholar]
- 196.Cosker K. E., Shadan S., Van Diepen M., Morgan C., Li M., Allen-Baume V., Hobbs C., Doherty P., Cockcroft S., and Eickholt B. J.. 2008. Regulation of PI3K signalling by the phosphatidylinositol transfer protein PITPα during axonal extension in hippocampal neurons. J. Cell Sci. 121: 796–803. [DOI] [PubMed] [Google Scholar]
- 197.Hamilton B. A., Smith D. J., Mueller K. L., Kerrebrock A. W., Bronson R. T., van Berkel V., Daly M. J., Kruglyak L., Reeve M. P., Nemhauser J. L., et al. 1997. The vibrator mutation causes neurodegeneration via reduced expression of PITPα: positional complementation cloning and extragenic suppression. Neuron. 18: 711–722. [DOI] [PubMed] [Google Scholar]
- 198.Alb J. G., Cortese J. D., Phillips S. E., Albin R. L., Nagy T. R., Hamilton B. A., and Bankaitis V. A.. 2003. Mice lacking phosphatidylinositol transfer protein alpha exhibit spinocerebellar degeneration, intestinal and hepatic steatosis, and hypoglycemia. J. Biol. Chem. 278: 33501–33518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Alb J. G., Phillips S. E., Wilfley L. R., Philpot B. D., and Bankaitis V. A.. 2007. The pathologies associated with functional titration of phosphatidylinositol transfer protein α activity in mice. J. Lipid Res. 48: 1857–1872. [DOI] [PubMed] [Google Scholar]
- 200.Alb J. G. Jr., Phillips S. E., Rostand K., Cui X., Pinxteren J., Cotlin L., Manning T., Guo S., York J. D., Sontheimer H., et al. 2002. Genetic ablation of phosphatidylinositol transfer protein function in murine embryonic stem cells. Mol. Biol. Cell. 13: 739–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xie Z., Hur S. K., Zhao L., Abrams C. S., and Bankaitis V. A.. 2018. A Golgi lipid signaling pathway controls apical Golgi distribution and cell polarity during neurogenesis. Dev. Cell. 44: 725–740.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wood C. S., Schmitz K. R., Bessman N. J., Setty T. G., Ferguson K. M., and Burd C. G.. 2009. PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J. Cell Biol. 187: 967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Taft M. H., Behrmann E., Munske-Weidemann L. C., Thiel C., Raunser S., and Manstein D. J.. 2013. Functional characterization of human myosin-18A and its interaction with F-actin and GOLPH3. J. Biol. Chem. 288: 30029–30041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Xing M., Peterman M. C., Davis R. L., Oegema K., Shiau A. K., and Field S. J.. 2016. GOLPH3 drives cell migration by promoting Golgi reorientation and directional trafficking to the leading edge. Mol. Biol. Cell. 27: 3828–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Koe C. T., Tan Y. S., Lönnfors M., Hur S. K., Low C. S. L., Zhang Y., Kanchanawong P., Bankaitis V. A., and Wang H.. 2018. Vibrator and PI4KIIIα govern neuroblast polarity by anchoring non-muscle myosin II. eLife. 7: e33555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Karess R. E., Chang X. J., Edwards K. A., Kulkarni S., Aguilera I., and Kiehart D. P.. 1991. The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell. 65: 1177–1189. [DOI] [PubMed] [Google Scholar]
- 207.Barros C. S., Phelps C. B., and Brand A. H.. 2003. Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev. Cell. 5: 829–840. [DOI] [PubMed] [Google Scholar]
- 208.Luetteke N. C., Phillips H. K., Qiu T. H., Copeland N. G., Earp H. S., Jenkins N. A., and Lee D. C.. 1994. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 8: 399–413. [DOI] [PubMed] [Google Scholar]
- 209.Sibilia M., and Wagner E. F.. 1995. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 269: 234–238. [DOI] [PubMed] [Google Scholar]
- 210.Miettinen P. J., Berger J. E., Meneses J., Phung Y., Pedersen R. A., Werb Z., and Derynck R.. 1995. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature. 376: 337–341. [DOI] [PubMed] [Google Scholar]
- 211.Weimar W. R., Lane P. W., and Sidman R. L.. 1982. Vibrator (vb): a spinocerebellar system degeneration with autosomal recessive inheritance in mice. Brain Res. 251: 357–364. [DOI] [PubMed] [Google Scholar]
- 212.Végran F., Boidot R., Coudert B., Fumoleau P., Arnould L., Garnier J., Causeret S., Fraise J., Dembélé D., and Lizard-Nacol S.. 2009. Gene expression profile and response to trastuzumab–docetaxel-based treatment in breast carcinoma. Br. J. Cancer. 101: 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kute T., Lack C. M., Willingham M., Bishwokama B., Williams H., Barrett K., Mitchell T., and Vaughn J. P.. 2004. Development of Herceptin resistance in breast cancer cells. Cytometry A. 57: 86–93. [DOI] [PubMed] [Google Scholar]
- 214.Nagata Y., Lan K. H., Zhou X., Tan M., Esteva F. J., Sahin A. A., Klos K. S., Li P., Monia B. P., Nguyen N. T., et al. 2004. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 6: 117–127. [DOI] [PubMed] [Google Scholar]
- 215.Le X. F., Claret F. X., Lammayot A., Tian L., Deshpande D., LaPushin R., Tari A. M., and Bast R. C.. 2003. The role of cyclin-dependent kinase inhibitor p27Kip1 in anti-HER2 antibody-induced G1 cell cycle arrest and tumor growth inhibition. J. Biol. Chem. 278: 23441–23450. [DOI] [PubMed] [Google Scholar]
- 216.Xie Y., Ding Y. Q., Hong Y., Feng Z., Navarre S., Xi C. X., Zhu X. J., Wang C. L., Ackerman S. L., Kozlowski D., et al. 2005. Phosphatidylinositol transfer protein-α in netrin-1-induced PLC signalling and neurite outgrowth. Nat. Cell Biol. 7: 1124–1132. [DOI] [PubMed] [Google Scholar]
- 217.Keino-Masu K., Masu M., Hinck L., Leonardo E. D., Chan S. S. Y., Culotti J. G., and Tessier-Lavigne M.. 1996. Deleted in colorectal cancer (DCC) encodes a netrin receptor. Cell. 87: 175–185. [DOI] [PubMed] [Google Scholar]
- 218.Serafini T., Colamarino S. A., Leonardo E. D., Wang H., Beddington R., Skarnes W. C., and Tessier-Lavigne M.. 1996. Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell. 87: 1001–1014. [DOI] [PubMed] [Google Scholar]
- 219.Vieira N. M., Spinazzola J. M., Alexander M. S., Moreira Y. B., Kawahara G., Gibbs D. E., Mead L. C., Verjovski-Almeida S., Zatz M., and Kunkel L. M.. 2017. Repression of phosphatidylinositol transfer protein α ameliorates the pathology of Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA. 114: 6080–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cockcroft S., Garner K., Yadav S., Gomez-Espinoza E., and Raghu P.. 2016. RdgBα reciprocally transfers PA and PI at ER-PM contact sites to maintain PI(4,5)P2 homoeostasis during phospholipase C signalling in Drosophila photoreceptors. Biochem. Soc. Trans. 44: 286–292. [DOI] [PubMed] [Google Scholar]
- 221.Fullwood Y., dos Santos M., and Hsuan J. J.. 1999. Cloning and characterization of a novel human phosphatidylinositol transfer protein, rdgBβ. J. Biol. Chem. 274: 31553–31558. [DOI] [PubMed] [Google Scholar]
- 222.Lopez-Ilasaca M., Liu X., Tamura K., and Dzau V. J.. 2003. The angiotensin II type I receptor-associated protein, ATRAP, is a transmembrane protein and a modulator of angiotensin II signaling. Mol. Biol. Cell. 14: 5038–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Daviet L., Lehtonen J. Y., Tamura K., Griese D. P., Horiuchi M., and Dzau V. J.. 1999. Cloning and characterization of ATRAP, a novel protein that interacts with the angiotensin II type 1 receptor. J. Biol. Chem. 274: 17058–17062. [DOI] [PubMed] [Google Scholar]
- 224.Cui T., Nakagami H., Iwai M., Takeda Y., Shiuchi T., Tamura K., Daviet L., and Horiuchi M.. 2000. ATRAP, novel AT1 receptor associated protein, enhances internalization of AT1 receptor and inhibits vascular smooth muscle cell growth. Biochem. Biophys. Res. Commun. 279: 938–941. [DOI] [PubMed] [Google Scholar]
- 225.Kondo T., Konishi F., Inui H., and Inagami T.. 1992. Diacylglycerol formation from phosphatidylcholine in angiotensin II-stimulated vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 187: 1460–1465. [DOI] [PubMed] [Google Scholar]
- 226.Halberg N., Sengelaub C. A., Navrazhina K., Molina H., Uryu K., and Tavazoie S. F.. 2016. PITPNC1 recruits RAB1B to the Golgi network to drive malignant secretion. Cancer Cell. 29: 339–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Png K. J., Halberg N., Yoshida M., and Tavazoie S. F.. 2011. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 481: 190–194. [DOI] [PubMed] [Google Scholar]
- 228.Guo C., Sah J. F., Beard L., Willson J. K., Markowitz S. D., and Guda K.. 2008. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 47: 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Feng R., Chen X., Yu Y., Su L., Yu B., Li J., Cai Q., Yan M., Liu B., and Zhu Z.. 2010. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 298: 50–63. [DOI] [PubMed] [Google Scholar]
- 230.Chang J. T., Milligan S., Li Y., Chew C. E., Wiggs J., Copeland N. G., Jenkins N. A., Campochiaro P. A., Hyde D. R., and Zack D. J.. 1997. Mammalian homolog of Drosophila retinal degeneration B rescues the mutant fly phenotype. J. Neurosci. 17: 5881–5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lu C., Vihtelic T. S., Hyde D. R., and Li T.. 1999. A neuronal-specific mammalian homolog of the drosophila retinal degeneration B gene with expression restricted to the retina and dentate gyrus. J. Neurosci. 19: 7317–7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Elagin V. A., Elagina R. B., Doro C. J., Vihtelic T. S., and Hyde D. R.. 2000. Cloning and tissue localization of a novel zebrafish RdgB homolog that lacks a phospholipid transfer domain. Vis. Neurosci. 17: 303–311. [DOI] [PubMed] [Google Scholar]
- 233.Lev S. 2004. The role of the Nir/rdgB protein family in membrane trafficking and cytoskeleton remodeling. Exp. Cell Res. 297: 1–10. [DOI] [PubMed] [Google Scholar]
- 234.Klinkenberg D., Long K. R., Shome K., Watkins S. C., and Aridor M.. 2014. A cascade of ER exit site assembly that is regulated by p125A and lipid signals. J. Cell Sci. 127: 1765–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Inoue H., Baba T., Sato S., Ohtsuki R., Takemori A., Watanabe T., Tagaya M., and Tani K.. 2012. Roles of SAM and DDHD domains in mammalian intracellular phospholipase A1 KIAA0725p. Biochim. Biophys. Acta. 1823: 930–939. [DOI] [PubMed] [Google Scholar]
- 236.Tange Y., Hirata A., and Niwa O.. 2002. An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J. Cell Sci. 115: 4375–4385. [DOI] [PubMed] [Google Scholar]
- 237.Csaki L. S., Dwyer J. R., Fong L. G., Tontonoz P., Young S. G., and Reue K.. 2013. Lipins, lipinopathies, and the modulation of cellular lipid storage and signaling. Prog. Lipid Res. 52: 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kim S., Kedan A., Marom M., Gavert N., Keinan O., Selitrennik M., Laufman O., and Lev S.. 2013. The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling. EMBO Rep. 14: 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Verma N., Keinan O., Selitrennik M., Karn T., Filipits M., and Lev S.. 2015. PYK2 sustains endosomal-derived receptor signalling and enhances epithelial-to-mesenchymal transition. Nat. Commun. 6: 6064. [DOI] [PubMed] [Google Scholar]
- 240.Holthuis J. C., and Levine T. P.. 2005. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6: 209–220. [DOI] [PubMed] [Google Scholar]
- 241.Lev S. 2012. Nonvesicular lipid transfer from the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 4: a013300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Prinz W. A. 2014. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 205: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Harris W. A., and Stark W. S.. 1977. Hereditary retinal degeneration in Drosophila melanogaster. A mutant defect associated with the phototransduction process. J. Gen. Physiol. 69: 261–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Cagan R. 2009. Principles of Drosophila eye differentiation. Curr. Top. Dev. Biol. 89: 115–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Vihtelic T. S., Goebl M., Milligan S., O’tousa J. E., and Hyde D. R.. 1993. Localization of Drosophila retinal degeneration B, a membrane-associated phosphatidylinositol transfer protein. J. Cell Biol. 122: 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Suzuki E., and Hirosawa K.. 1994. Immunolocalization of a Drosophila phosphatidylinositol transfer protein (rdgB) in normal and rdgA mutant photoreceptor cells with special reference to the subrhabdomeric cisternae. J. Electron Microsc. (Tokyo). 43: 183–189. [PubMed] [Google Scholar]
- 247.Milligan S. C., Alb J. G., Elagina R. B., Bankaitis V. A., and Hyde D. R.. 1997. The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J. Cell Biol. 139: 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Hotta Y., and Benzer S.. 1969. Abnormal electroretinograms in visual mutants of Drosophila. Nature. 222: 354–356. [DOI] [PubMed] [Google Scholar]
- 249.Stark W. S., and Carlson S. D.. 1982. Ultrastructural pathology of the compound eye and optic neuropiles of the retinal degeneration mutant (w rdgB KS222) Drosophila melanogaster. Cell Tissue Res. 225: 11–22. [DOI] [PubMed] [Google Scholar]
- 250.Trivedi D., and Padinjat R.. 2007. RdgB proteins: functions in lipid homeostasis and signal transduction. Biochim. Biophys. Acta. 1771: 692–699. [DOI] [PubMed] [Google Scholar]
- 251.Masai I., Okazaki A., Hosoya T., and Hotta Y.. 1993. Drosophila retinal degeneration A gene encodes an eye-specific diacylglycerol kinase with cysteine-rich zinc-finger motifs and ankyrin repeats. Proc. Natl. Acad. Sci. USA. 90: 11157–11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Masai I., Suzuki E., Yoon C. S., Kohyama A., and Hotta Y.. 1997. Immunolocalization of Drosophila eye-specific diacylgylcerol kinase, rdgA, which is essential for the maintenance of the photoreceptor. J. Neurobiol. 32: 695–706. [DOI] [PubMed] [Google Scholar]
- 253.Vihtelic T. S., Hyde D. R., and O’Tousa J. E.. 1991. Isolation and characterization of the Drosophila retinal degeneration B (rdgB) gene. Genetics. 127: 761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Iwata R., Oda S., Kunitomo H., and Iino Y.. 2011. Roles for class IIA phosphatidylinositol transfer protein in neurotransmission and behavioral plasticity at the sensory neuron synapses of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 108: 7589–7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Saeki S., Yamamoto M., and Iino Y.. 2001. Plasticity of chemotaxis revealed by paired presentation of a chemoattractant and starvation in the nematode Caenorhabditis elegans. J. Exp. Biol. 204: 1757–1764. [DOI] [PubMed] [Google Scholar]
- 256.Tomioka M., Adachi T., Suzuki H., Kunitomo H., Schafer W. R., and Iino Y.. 2006. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron. 51: 613–625. [DOI] [PubMed] [Google Scholar]
- 257.Adachi T., Kunitomo H., Tomioka M., Ohno H., Okochi Y., Mori I., and Iino Y.. 2010. Reversal of salt preference is directed by the insulin/PI3K and Gq/PKC signaling in Caenorhabditis elegans. Genetics. 186: 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Kim Y. J., Guzman-Hernandez M. L., Wisniewski E., Echeverria N., and Balla T.. 2016. Phosphatidylinositol and phosphatidic acid transport between the ER and plasma membrane during PLC activation requires the Nir2 protein. Biochem. Soc. Trans. 44: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Litvak V., Dahan N., Ramachandran S., Sabanay H., and Lev S.. 2005. Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat. Cell Biol. 7: 225–234. [DOI] [PubMed] [Google Scholar]
- 260.Kearns B. G., McGee T. P., Mayinger P., Gedvilaite A., Phillips S. E., Kagiwada S., and Bankaitis V. A.. 1997. Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 387: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Deleted in proof. [Google Scholar]
- 262.Su W., Chen Q., and Frohman M. A.. 2009. Targeting phospholipase D with small-molecule inhibitors as a potential therapeutic approach for cancer metastasis. Future Oncol. 5: 1477–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Yanase Y., Carvou N., Frohman M. A., and Cockcroft S.. 2009. Reversible bleb formation in mast cells stimulated with antigen is Ca2+/calmodulin-dependent and bleb size is regulated by ARF6. Biochem. J. 425: 179–193. [DOI] [PubMed] [Google Scholar]
- 264.Kanaho Y., Sato T., Hongu T., and Funakoshi Y.. 2013. Molecular mechanisms of fMLP-induced superoxide generation and degranulation in mouse neutrophils. Adv. Biol. Regul. 53: 128–134. [DOI] [PubMed] [Google Scholar]
- 265.Sato T., Hongu T., Sakamoto M., Funakoshi Y., and Kanaho Y.. 2013. Molecular mechanisms of N-formyl-methionyl-leucyl-phenylalanine-induced superoxide generation and degranulation in mouse neutrophils: phospholipase D is dispensable. Mol. Cell. Biol. 33: 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Frohman M. A. 2015. The phospholipase D superfamily as therapeutic targets. Trends Pharmacol. Sci. 36: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Zheng Y., Rodrik V., Toschi A., Shi M., Hui L., Shen Y., and Foster D. A.. 2006. Phospholipase D couples survival and migration signals in stress response of human cancer cells. J. Biol. Chem. 281: 15862–15868. [DOI] [PubMed] [Google Scholar]
- 268.Monovich L., Mugrage B., Quadros E., Toscano K., Tommasi R., LaVoie S., Liu E., Du Z., LaSala D., Boyar W., et al. 2007. Optimization of halopemide for phospholipase D2 inhibition. Bioorg. Med. Chem. Lett. 17: 2310–2311. [DOI] [PubMed] [Google Scholar]
- 269.Scott S. A., Selvy P. E., Buck J. R., Cho H. P., Criswell T. L., Thomas A. L., Armstrong M. D., Arteaga C. L., Lindsley C. W., and Brown H. A.. 2009. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat. Chem. Biol. 5: 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lavieri R. R., Scott S. A., Selvy P. E., Kim K., Jadhav S., Morrison R. D., Daniels J. S., Brown H. A., and Lindsley C. W.. 2010. Design, synthesis, and biological evaluation of halogenated N-(2-(4-oxo-1-phenyl-1, 3, 8-triazaspiro [4.5] decan-8-yl) ethyl) benzamides: discovery of an isoform-selective small molecule phospholipase D2 inhibitor. J. Med. Chem. 53: 6706–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Uhlén M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. 2015. Tissue-based map of the human proteome. Science. 347: 1260419. [DOI] [PubMed] [Google Scholar]
- 272.Thul P. J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Blal H. A., Alm T., Asplund A., Björk L., Breckels L. M., et al. 2017. A subcellular map of the human proteome. Science. 356: eaal3321. [DOI] [PubMed] [Google Scholar]
- 273.Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., et al. 2017. A pathology atlas of the human cancer transcriptome. Science. 357: eaan2507. [DOI] [PubMed] [Google Scholar]
- 274.Lu C., Peng Y. W., Shang J., Pawlyk B. S., Yu F., and Li T.. 2001. The mammalian retinal degeneration B2 gene is not required for photoreceptor function and survival. Neuroscience. 107: 35–41. [DOI] [PubMed] [Google Scholar]
- 275.Carlisle F. A., Pearson S., Steel K. P., and Lewis M. A.. 2013. Pitpnm1 is expressed in hair cells during development but is not required for hearing. Neuroscience. 248: 620–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Lek M., Karczewski K. J., Minikel E.V., Samocha K. E., Banks E., Fennell T., O’Donnell-Luria A. H., Ware J. S., Hill A. J., Cummings B. B., and Tukiainen T.. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Köhn L., Kadzhaev K., Burstedt M. S., Haraldsson S., Hallberg B., Sandgren O., and Golovleva I.. 2007. Mutation in the PYK2-binding domain of PITPNM3 causes autosomal dominant cone dystrophy (CORD5) in two Swedish families. Eur. J. Hum. Genet. 15: 664–671. [DOI] [PubMed] [Google Scholar]
- 278.Reinis A., Golovleva I., Köhn L., and Sandgren O.. 2013. Ocular phenotype of CORD5, an autosomal dominant retinal dystrophy associated with PITPNM3 p. Q626H mutation. Acta Ophthalmol. 91: 259–266. [DOI] [PubMed] [Google Scholar]
- 279.Concepcion D., Johannes F., Lo Y. H., Yao J., Fong J., and Hamilton B. A.. 2011. Modifier genes for mouse phosphatidylinositol transfer protein α (vibrator) that bypass juvenile lethality. Genetics. 187: 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]