See Calafato and Bramon (doi:10.1093/brain/awy345) for a scientific commentary on this article.
Cognitive deficits accompany schizophrenia and are associated with genetic risk but the direction of causality is unclear. Using causal modelling, Toulopoulou et al. show that cognitive deficits partially mediate the effects of cumulative genetic risk for schizophrenia. Impaired cognition is an intermediate phenotype on the causal path to the disorder with implications for diagnosis and intervention.
Keywords: schizophrenia, cognition, polygenic risk scores, intermediate phenotypes, endophenotypes
Abstract
Cognitive deficit is thought to represent, at least in part, genetic mechanisms of risk for schizophrenia, with recent evidence from statistical modelling of twin data suggesting direct causality from the former to the latter. However, earlier evidence was based on inferences from twin not molecular genetic data and it is unclear how much genetic influence ‘passes through’ cognition on the way to diagnosis. Thus, we included direct measurements of genetic risk (e.g. schizophrenia polygenic risk scores) in causation models to assess the extent to which cognitive deficit mediates some of the effect of polygenic risk scores on the disorder. Causal models of family data tested relationships among key variables and allowed parsing of genetic variance components. Polygenic risk scores were calculated from summary statistics from the current largest genome-wide association study of schizophrenia and were represented as a latent trait. Cognition was also modelled as a latent trait. Participants were 1313 members of 1078 families: 416 patients with schizophrenia, 290 unaffected siblings, and 607 controls. Modelling supported earlier findings that cognitive deficit has a putatively causal role in schizophrenia. In total, polygenic risk score explained 8.07% [confidence interval (CI) 5.45–10.74%] of schizophrenia risk in our sample. Of this, more than a third (2.71%, CI 2.41–3.85%) of the polygenic risk score influence was mediated through cognition paths, exceeding the direct influence of polygenic risk score on schizophrenia risk (1.43%, CI 0.46–3.08%). The remainder of the polygenic risk score influence (3.93%, CI 2.37–4.48%) reflected reciprocal causation between schizophrenia liability and cognition (e.g. mutual influences in a cyclical manner). Analysis of genetic variance components of schizophrenia liability indicated that 26.87% (CI 21.45–32.57%) was associated with cognition-related pathways not captured by polygenic risk score. The remaining variance in schizophrenia was through pathways other than cognition-related and polygenic risk score. Although our results are based on inference through statistical modelling and do not provide an absolute proof of causality, we find that cognition pathways mediate a significant part of the influence of cumulative genetic risk on schizophrenia. We estimate from our model that 33.51% (CI 27.34–43.82%) of overall genetic risk is mediated through influences on cognition, but this requires further studies and analyses as the genetics of schizophrenia becomes better characterized.
See Calafato and Bramon (doi:10.1093/brain/awy345) for a scientific commentary on this article.
Introduction
A wealth of evidence from adoption, family, and twin studies as well as from linkage and association studies confirms genetics contribute significantly to risk for schizophrenia (Sullivan et al., 2003b; Lichtenstein et al., 2009; Purcell et al., 2009; Ripke et al., 2013). While earlier association studies resulted in few replicated findings, recent large-scale genome-wide association studies (GWAS), which allow for testing of millions of single nucleotide polymorphisms (SNPs) in the genome, have produced statistically robust results implicating over 100 independent risk loci across the genome (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014; Gandal et al., 2016). Common SNPs appear to contribute the majority of interindividual variation in schizophrenia risk (Purcell et al., 2009; Bergen and Petryshen, 2012; Ripke et al., 2013), while copy number variants (CNVs), including rare de novo and inherited CNVs, contribute small increments in population risk variation despite larger effect sizes (Manolio et al., 2009; Malhotra and Sebat, 2012; Kotlar et al., 2015; Genovese et al., 2016). The cumulative sum of risk associated alleles at common variants across the genome derived from the recent Psychiatric Genomic Consortium (PGC) genome-wide association study, a so-called polygenic risk score (PRS), has been shown to account for ∼7% of the variance in disease risk (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014).
One strategy that has been used to understand how risk-associated SNPs might affect brain function is to examine their relationship with measures of brain structure and function that are consistently altered in schizophrenia, and which may represent inherited biological mechanisms of risk (e.g. endophenotypes or intermediate phenotypes) (Flint et al., 2014; Glahn et al., 2014; Blokland et al., 2018). In this regard, numerous patient, family, twin, prospective, and high-risk studies have shown that schizophrenia is associated with deviations in cognition, in brain neurophysiology, and to a less consistent degree, structure (Friston and Frith, 1995; Callicott et al., 2000; Pantelis et al., 2003; Bramon et al., 2004; Ragland et al., 2009; Olincy et al., 2010; van den Heuvel et al., 2013; Crossley et al., 2016). To the extent that these deviations are found in unaffected relatives, they have been considered as representing inherited neurobiological risk rather than effects of the disease state (Goldberg et al., 1995; Gur et al., 2007a, b; Wood et al., 2008; Waters-Metenier and Toulopoulou, 2010, 2011a, b; Owens et al., 2012; Millard et al., 2016).
One of the most consistently and robustly implicated intermediate phenotypes in schizophrenia is cognitive deficit (Schaefer et al., 2013; Mark and Toulopoulou, 2016). Cognitive deficits associated with schizophrenia have been shown to be: (i) heritable and aggregate amongst unaffected family members of individuals with schizophrenia including parents and healthy siblings (Egan et al., 2001a; Toulopoulou et al., 2003a, b; Gur et al., 2007b; Calkins et al., 2013; Zhang et al., 2016); (ii) are more concordant among identical than non-identical schizophrenia twins who are discordant for the clinical disorder (Cannon et al., 2000b; Toulopoulou et al., 2007); and (iii) are observed in children and adults who later develop schizophrenia (Cannon et al., 2000a; Reichenberg et al., 2010; Seidman et al., 2013; Meier et al., 2014; Agnew-Blais et al., 2015). Statistical modelling of twin and family data have further suggested a shared genetic link between cognition and schizophrenia risk, which has been extended by recent genetic association studies of schizophrenia PRSs and cognitive performance (Toulopoulou et al., 2007, 2010b, 2015; Owens et al., 2011a, b; Fowler et al., 2012; McIntosh et al., 2013; Walters et al., 2013; Lencz et al., 2014; Hatzimanolis et al., 2015; Kauppi et al., 2015; Germine et al., 2016; Hagenaars et al., 2016; Hubbard et al., 2016; Liebers et al., 2016; Alloza et al., 2017; Cosgrove et al., 2017; Rampino et al., 2017; Ranlund et al., 2018). In summary, this extensive literature suggests that cognitive deficit is reliably linked with inherited risk for schizophrenia and that it often predates the diagnosis.
Studies of genetic associations between schizophrenia and cognition, either based on molecular data or statistical modeming of twin and family samples, provide evidence of shared genetic influences but typically do not address causation. However, in a recent paper in a pan-European sample of twins with schizophrenia, Toulopoulou et al. (2015) sought to identify through statistical inference the direction of causation between the liability to schizophrenia and several candidate intermediate phenotypes. Using novel reciprocal causation models, we reported that cognitive deficits lay upstream of schizophrenia liability, with about a quarter of the genetic variation in schizophrenia risk mediated through genetic variation in cognition (Toulopoulou et al., 2015). This finding was directionally specific for cognition, suggesting that cognitive deficit contributed causally to schizophrenia, but not vice versa.
While the aforementioned study examined the potentially causal relationship between cognitive deficit and schizophrenia liability, the models were based on inferences from twin rather than molecular genetic data. Here we extended the earlier causal modelling to the molecular genetic level, seeking to quantify the extent to which cognitive deficit may mediate some of the effect of cumulative genetic risk as measured by the PRS for schizophrenia. Specifically, we used trivariate causal modelling to test our hypothesis that cognition mediated the effect of PRSs on schizophrenia risk against alternative hypothetical models (e.g. that schizophrenia mediated the polygenic score effect on cognition). We report support for our hypothesis that cognitive deficit mediates some of the effects of PRS on schizophrenia, adding substantial evidence that cognitive deficit is an intermediate phenotype on the causal path to diagnosis with possible implications for diagnosis, prediction, and intervention.
Materials and methods
Participants
Participants were 1313 Caucasian individuals with European ancestry from 1078 families including 416 schizophrenia patients, 290 unaffected siblings, and 607 unrelated controls. All participants were assessed as part of the Clinical Brain Disorders Branch (CBDB) Sibling Study, one of the largest and most comprehensively-phenotyped studies of patients with schizophrenia and their healthy siblings, which has been described in detail before (Egan et al., 2001b). There were 227 families with a schizophrenia patient and an unaffected sibling, 189 additional families with one schizophrenia patient and no additional family members, and 55 families with one or more unaffected siblings. All participants were able to provide informed consent after the procedures had been fully explained for a protocol approved by the NIH IRB.
Measurement
Clinical assessment
All participants were medically screened and completed separate DSM-IV diagnostic interviews (First et al., 1994) with two research psychiatrists. Individuals were included in the schizophrenia group if they had schizophrenia, schizoaffective disorder, psychosis not otherwise specified, or schizoid personality disorder (Dickinson et al., 2011). Schizoid personality diagnosis was included as part of the schizophrenia spectrum, consistent with prior work in the PGC and with evidence of strong genetic overlap with schizophrenia. Siblings were allowed to have a history of mood/anxiety or personality disorder but no schizophrenia spectrum disorder history. Controls were not included in the study if they had first-degree relatives with schizophrenia spectrum disorders or Axis I or Axis II diagnosis history, or if they were currently on psychotropic medication. Controls were determined to be unrelated based on genome-wide identity-by-state estimation in PLINK. If identity-by-state of paired samples had PH_HAT > 0.2, one of the samples was excluded. Exclusion criteria for all participants included history of head trauma with extended loss of consciousness, alcohol, or drug abuse within the past 6 months, IQ < 70, or evidence of learning disability. Schizophrenia participants were stable and receiving neuroleptic medications at the time of the study.
Polygenic risk score
DNA was extracted from blood using standard procedures, and genotyping was done using various Illumina Bead Chips including 510K/610K/660K/2.5M. We divided samples into two groups according to genotyping chips: one group included samples genotyped with low-resolution BeadChips (510K/610K/660K), and the other included samples genotyped with high-resolution BeadChips (2.5 M). For each type of chip, both cases and controls were genotyped. There are many reasons that the different chips are unlikely to influence our results—these include: very high genotyping and imputation accuracies; we used only common SNPs for the analyses; and the fact that we ran several samples on most chips and observed genotype concordance rates > 99.5%. Imputation was performed separately for these two groups. To control for the use of two different imputations, we included genotyping batch label as a covariate in the statistical analysis. Quality control was performed before imputation using PLINK (version 1.07; http://pngu.mgh.harvard.edu/purcell/plink/), as reported by the PGC (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). The quality control parameters for retaining SNPs and subjects were: SNP missingness < 0.05 (before sample removal); subject missingness < 0.02; autosomal heterozygosity deviation (|Fhet | < 0.2); SNP missingness < 0.02 (after sample removal); difference in SNP missingness between cases and controls < 0.02; and SNP Hardy-Weinberg equilibrium (P > 10−6 in controls or P > 10−10 in cases). Pre-phasing was done before imputation with SHAPEIT, and imputation was done with IMPUTE2 using 1000 Genome Phase 1 as reference panel. SNPs were clumped using PLINK. In each step of clumping, if two SNPs are within 500 kb and with r2 ≥ 0.1, the more significant SNP was kept. To control for population stratification in the association analysis, the first 10 principal components of the whole genome data were calculated using EIGENSOFT v5.01 (EIGENSOFT, http://www.hsph.harvard.edu/alkes-price/software/).
To assess cumulative polygenic risk at the genomic level, we used the results from the 2014 PGC schizophrenia meta-analysis (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), after excluding the present sample, to construct PRS in our sample using R scripts. Thus, the data comprised 36 573 cases and 112 468 controls. Polygenic risk was estimated in each individual separately by adding up the number of risk alleles (0, 1, 2) of each SNP that was found to be associated with schizophrenia in the PGC sample multiplied by the logarithm of the SNP’s odds ratio as described before (Vassos et al., 2017). Ten different PRS levels were calculated based on the SNPs associated with schizophrenia at different P-value thresholds (PT < 5 × 10−8, 10−6, 10−4, 10−3, 0.01, 0.05, 0.1, 0.2, 0.5, 1). PRS was standardized using the mean and variance of the control group for all subsequent analysis.
Neuropsychological assessment and cognitive factors
All participants were administered a comprehensive neuropsychological battery of assessments, previously reported to be consistently and commensurately impaired in schizophrenia (Dickinson et al., 2011). Guided by principal components analysis, composites were created reflecting six broad cognitive domains: verbal memory [cognitive factor (CF) 1], n-back (CF 2), visual memory (CF 3), processing speed (CF 4), card sorting (CF 5) and digit span (CF 6). All cognitive composites were standardized using the mean and variance of the control group. Using similar methodology, a single composite measure, g, was also created as one estimate of general cognitive ability (Dickinson et al., 2011) (used in correlation analyses, described below). However, in the current study, we focused on the six domain-specific cognitive factors as the key indices of the cognitive effects of schizophrenia, and modelled general cognitive ability as a latent variable underlying performance in these domains [see ‘Bivariate non-causal and causal models (Cholesky)’ section below]. The main advantage of using several cognitive domains is the ability to model a latent variable that is free of measurement error. Using a measurement model when attempting to infer direction of causation ensures parameter estimates will be unbiased, rather than being attenuated by unknown varying degrees of measurement error (Duffy and Martin, 1994).
Correlations
Prior to model fitting we ran correlational analysis to illustrate the relationship among key variables. Pearson correlations between two continuous variables (e.g. g and PRS) were estimated using SPSS 23.0 (Corp, 2015). The biserial correlation was estimated between a dichotomous and a continuous variable (e.g. schizophrenia and PRS; schizophrenia and g) using OpenMx (Boker et al., 2011). While the cases include family members, ignoring non-independence among data points does not introduce bias to the estimates of correlation coefficients, but will produce standard errors and P-values that are too small. We therefore randomly selected one member from each family to run such analysis.
Model fitting
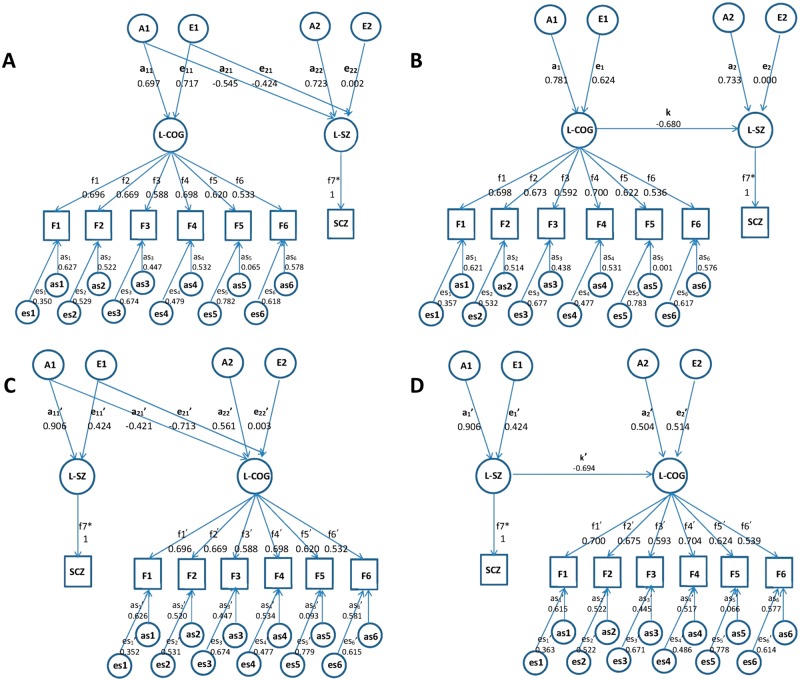
We used causal modelling of family and control data to explore the underlying relationships between schizophrenia, cognitive deficit and PRS. These complex models are based on the following assumptions. Both genetic (A) and environmental (E) components contribute to the variance of the phenotypes (e.g. schizophrenia or cognition), while half of the genetic variance contributes to the covariance between siblings. Specifically, the relatedness between patients and their siblings is incorporated into the standard error of the mean (SEM) through the correlations between their additive components (set to be 0.5 as appropriate for first degree relatives). This is equivalent to a random effects model with pre-specified correlational structure. As in twin studies, the reciprocal causal relationships between two phenotypes can be estimated from sibling data, assuming that each phenotype has specific A and E components (Duffy and Martin, 1994). Because patients were recruited based on their clinical status, our subjects do not represent a random sample of the population. Since parameter estimates (e.g. heritability) obtained from non-random data would be misleading unless ascertainment is correctly modelled, we do not attempt to estimate the model parameters for schizophrenia but assume values supported by the literature, an approach adopted in previous studies (Toulopoulou et al., 2007, 2010a, 2015). Thus, we assume a liability threshold model for schizophrenia, where the liability is normally distributed with mean 0, variance 1, with a threshold that corresponds to a lifetime population prevalence of 1% (Sullivan et al., 2003a), and components of variance A = 0.82 and E = 0.18 (Cardno et al., 1999). This is a standard procedure used for analysing samples ascertained to contain affected family members. Figure 1 shows the causal paths between schizophrenia and cognition. The observed phenotype, e.g. schizophrenia diagnosis labelled as SCZ, is denoted by a square, while the latent variable L-SZ in the circle represents schizophrenia liability and is continuous. The arrow pointing from L-SZ to SCZ is fixed to unity, which denotes that SCZ is obtained directly from L-SZ through a liability-threshold model.
Figure 1.
Testing causal paths between schizophrenia and cognition. Bivariate non-causal and causal models (Cholesky) with observed variables: SCZ = schizophrenia and F1–6 = cognitive factors 1–6 (F1: verbal memory; F2: N back; F3: visual memory; F4: processing speed; F5: card sorting; F6: digit span), and latent variables: L-SZ = schizophrenia liability; L-COG = general cognitive factor; A = genetic component; E = environmental component; for each observed cognitive factor, variance could be explained by both shared (L-COG) and specific components, including genetic (as1–6) and environmental (es1–6); the path coefficients amn in lowercase is the path coefficient from the mth genetic factor to the nth latent variable; the path coefficients from L-COG to F1–6 are shown as f1–6; path coefficients f1 and f1' from L-COG to F1 are set to unity to identify the model; all path coefficients are standardized after model fitting; parameters labelled with an asterisk have been fixed to 1. The overall genetic variance to L-SZ is constrained to 0.82 (see ‘Materials and methods’ section). Models A and C are equivalent Cholesky models with different ordering of L-SZ and L-COG; Models B and D are both sub-models (nested models) of Cholesky model, representing different directions of causation between L-SZ and L-COG; the parameters estimates obtained are such that in Model Ba1 and e1 multiplied by k are approximately equal to a21 and e21, whereas in Model Da1′ and e1′ are far from proportional to a21′ and e21′; thus, one would expect Model D to be rejected, as is indeed the case.
The genetic components of cognition are estimated from the data. Even though the data were ascertained from families with schizophrenia, we have previously shown (by theory and simulation studies) that unbiased estimates can be obtained when the parameters of the phenotype responsible for ascertainment (i.e. schizophrenia in our study) are correctly specified (Rijsdijk et al., 2005).
Bivariate non-causal and causal models (Cholesky)
To build the causal model, and before incorporating molecular genetic data, we first checked that the data are consistent with our earlier work that showed genetic overlap between schizophrenia and cognition due to a potentially causal relationship between the two phenotypes. Specifically, we employed bivariate models (Neale and Cardon, 1992) to explore the covariance between the two phenotypes, schizophrenia and cognition, a necessary feature to infer causation. These models consider every variance and covariance between all pairs of variables in the sibling data, and provide the baseline for causation models. In the Cholesky decomposition, amn is the path coefficient from the mth genetic factor to the nth latent phenotype (Fig. 1A and C). When modelling the relationship between schizophrenia and cognition (six cognitive factors), two latent (unobserved, statistically-defined) variables are employed to build the bivariate model. As shown in Fig. 1, one latent variable is L-SZ, representing schizophrenia liability in a liability threshold model, and the other is L-COG, reflecting shared influences on the six broad cognitive domains mentioned above, F1–6.
Models A and C in Fig. 1 are assumed to be statistically equivalent while testing different ordering of the variables cognition and schizophrenia; the order of the variables (here L-SZ and L-COG) being arbitrary in the Cholesky decomposition. In total, 29 parameters are estimated in the full bivariate model: three genetic parameters (a11, a21, a22), three environmental parameters (e11, e21, e22), five factor loadings f2–6 from L-COG to F2–6 (f1 is set to unity as customary), six genetic and six environmental residual parameters (as1–6, es1–6), and six means of cognitive factors (MF1–6) (MF1–6 not shown in Fig. 1). All the path coefficients are standardized such that all variables have unit variance after model fitting.
Causal model
The models above describe the covariance between two phenotypes but cannot provide information on the direction of causation. The causal model attempts to explore their causal relationship. The full reciprocal causal models, represented in Fig. 1B and C include two opposite arrows between two latent variables, the causal path k pointing from L-COG→L-SZ and k′ pointing from L-SZ→L-COG. The full reciprocal causal model with two causal paths between two latent phenotypes would estimate all the variance and covariance between them, which is in principle the same as the Cholesky model, and therefore their model fit would be the same. The significance of causal paths k and k′ can be tested by examining sub-models, representing different directions of causation between L-SZ and L-COG, shown in Fig. 1B and D dropping one causal path at a time, then comparing the outcome with the full reciprocal causal model (see ‘Results’ section).
The models with one causal path are considered as nested models of the Cholesky model, because the causal arrow imposes a certain constraint in the genetic and environmental components. In the Cholesky model, genetic contribution to the covariance between L-SZ and L-COG is expressed as a11 × a21, and environmental contribution is e11 × e21, and the proportion of A and E component is freely estimated, expressed as (a11/e11) × (a21/e21). In the sub-model with causal path k (Fig. 1B), the proportion of genetic and environmental covariance is related to (a1/e1)2, with genetic contribution a1 × a1 × k, and environmental contribution e1 × e1 × k. If the sub-model could describe the genetic architecture of variables, the value of a1 × k and e1 × k, representing A1 and E1 contribution to the second latent variables L-SZ, should be close to a21 and e21 in the Cholesky model, respectively. On the other hand, they could be discrepant with each other, which would mean that the causation model is not suitable to describe the data.
The genetic and environmental components of schizophrenia liability should be set to a specific value, here 0.82 and 0.18 as described above. In this scenario, more than one genetic or environmental single-headed arrow is pointing to L-SZ. As shown in Fig. 1, in the Cholesky model, both a21 and a22 (e21 and e22) contribute to L-SZ’s genetic (environmental) variance, the genetic and environmental parts are constrained as a212 + a222 = 0.82, e212 + e222 = 0.18. While in the causation model, both k and a2 (k and e2) contribute to A (E) components, so the genetic and environmental parts are constrained as (a1 × k)2 × a22 = 0.82 and (e1 × k)2 × e22 = 0.18. In this situation, all of the genetic and environmental parameters (ep) are freely estimated, and the observed statistics (os) would decrease by 2 because of the constraints, so the degrees of freedom (df) is calculated as df = os − ep.
In the other scenario, when testing the causation model with causal path k′ (i.e. schizophrenia is causal of cognitive deficit) (Fig, 1D), only one genetic (environmental) path a1′ (e1′) is pointing to L-SZ, and the comparable full model (the Cholesky model) shown in the Fig. 1C, also has one single-headed arrow pointing to L-SZ. Therefore, only parameters a11′ and e11′ (a1′ and e1′) contribute to the genetic and environmental components of L-SZ, and a11′2 and e11′2 (a1′2 and e1′2) could be directly set to 0.82 and 0.18. In this situation, the number of ep could decrease by 2, so df is the same as the first scenario. Though the Cholesky model in Fig. 1A and C has different constraint methods and different order of the variables in the figures, they are identical with the same −2LL (minus 2 log likelihood) and df (degree of freedom). Therefore, they would be the full models and provide the baseline for the causal models with different causal direction.
If the causal model is not significantly worse than the Cholesky model, it would mean that the bivariate causal model fits the data well and with fewer parameters, meaning that it would represent the best model to describe the relationship between schizophrenia liability and cognition. Otherwise, the Cholesky model would be the more appropriate, which does not implicate a specific order of variables.
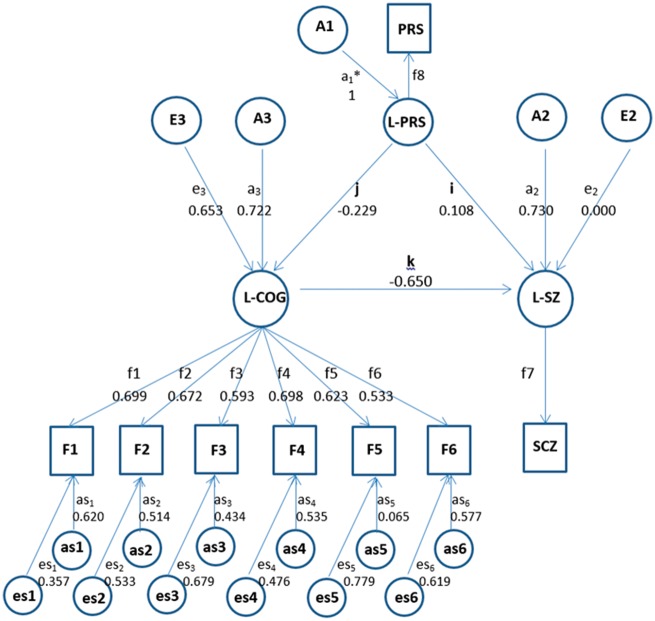
Trivariate causal model
The trivariate model incorporates molecular genetic data, and forms a more detailed description of the full dataset, compared to the bivariate models, to explore the underlying causal relationships between schizophrenia, cognition and PRS. Figure 2 shows the causal relationships between key variables. Latent PRS (L-PRS) determines the observed phenotype [i.e. PRS threshold (PT < 0.05)] while L-COG and L-SZ reflect latent variables as described before. In our model, the PRS is treated as a phenotype (just as schizophrenia and cognition). However, PRS is special in that it is calculated from individuals’ genotypes in an additive fashion, and is therefore guaranteed to have a heritability of 1 and a correlation of 0.5 between siblings (and other types of first-degree relatives). As shown in Fig. 2, latent variables were modelled to be influenced through causal paths i, j, k and k′ (k′ from L-SZ→L-COG not shown in Fig. 2), and residual genetic and environmental components of latent variables, As1–6 and Es1–6, which incorporated measurement error. Reciprocal causal models typically use two opposite single-headed arrows between each pair of latent variables to represent these potential causal relationships. Genetic causation assumes that PRS can only cause phenotypes and genotypes cannot be influenced by the phenotypes, so there is no reciprocal causation involving L-PRS. As with L-COG, the path loadings for L-SZ and L-PRS from the latent variables to the observed phenotypes were constrained to 1.
Figure 2.
Model of causal relationships among polygenic risk scores, schizophrenia liability and cognition. Trivariate causal model with observed variables: SCZ = schizophrenia, F1–6 = cognitive factors 1–6, and PRS = polygenic risk score (P-value threshold 0.05); and latent variables: L-SZ = schizophrenia liability, L-COG = general cognitive factor, L-PRS = polygenic risk score; A = genetic component, E = environmental component; for each observed cognitive factor, variance could be explained by both shared (L-COG) and specific components, including genetic (as1–6) and environmental (es1–6); the path coefficients am in lowercase is the path coefficient from the mth genetic factor to the latent variable; the path coefficients from L-COG to F1–6 are shown as f1–6; path coefficient f1 from L-COG to F1 is set to unity to identify the model; i, j, k represent causal paths: causal path i, L-PRS to L-SZ; causal path j, L-PRS to L-COG; causal path k, L-COG to L-SZ; all path coefficients are standardized after model fitting; parameters labelled with an asterisk have been fixed to 1; the overall genetic variance to L-SZ is constrained to 0.82. Note that L-PRS influences L-SZ both directly and indirectly (through L-COG).
In this analysis, the full trivariate model has four causal paths at the latent variable level. The significance of each causal path would be tested by comparing nested models dropping one causal path then comparing to the full model, using the likelihood ratio chi-squared statistic. Thus, there would be four sub-models dropping single-headed arrows i, j, k and k′ sequentially. Again, the model with the fewest variables is deemed the best fitting model for the data.
Multiple testing and sensitivity analysis
A number of bivariate models, some nested within others, were fitted to test the two reciprocal paths between L-SZ and L-COG. It would be reasonable and appropriate to adjust for testing two hypotheses. We regarded 0.025 as the critical P-value for statistical significance rather than the usual 0.05. The purpose of the trivariate model was to estimate how much of the genetic contribution of the polygenic score to schizophrenia liability is mediated through cognitive impairment. This is a single specific question and does not involve multiple testing.
In the model fitting, the genetic and environmental contributions to schizophrenia liability are assumed to be 0.82 and 0.18, respectively. Many researchers suggest that the heritability of schizophrenia ranges at 0.7–0.9 (Farmer et al., 1987; Sullivan et al., 2003b). To explore whether the value of heritability affects the model fitting results, we also fitted the data fixing schizophrenia heritability (a2) ranging from 0.7 to 0.9.
Data availability
The data that support the findings of this study are available from the corresponding author, upon request.
Results
Mean comparisons
Demographics, means, standard deviations (SDs) and P-values for group comparisons on cognitive factors, PRSs, and the g composite are given in Table 1. As expected, participants with schizophrenia performed significantly worse than siblings and healthy controls, and siblings scored lower than controls (all P < 0.001). Patients with schizophrenia and siblings have significantly higher PRS than controls (all P < 0.001).
Table 1.
Demographic, cognitive factor and PRSs
| Control | Siblings | Schizophrenia | Schizophrenia and Control group | Sibling and Control group | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | 607 | 290 | 416 | - | - | ||||||
| Male, n | 285 | 122 | 316 | - | - | ||||||
| Age, mean (SD) | 31.05 (9.78) | 35.58* (9.64) | 34.38* (9.89) | - | - | ||||||
| n | Mean | SD | n | Mean | SD | n | Mean | SD | P-value | P-value | |
| PRS | 607 | 0 | 1 | 286 | 0.31 | 0.94 | 416 | 0.78 | 1.05 | 4.10 × 10−31 | 1.37 × 10−5 |
| Verbal memory (F1) | 598 | 0 | 1 | 284 | −0.38 | 1.08 | 415 | −2.00 | 1.33 | 7.97 × 10−114 | 4.97 × 10−7 |
| n-back (F2) | 561 | 0 | 1 | 216 | −0.57 | 1.31 | 282 | −1.82 | 1.53 | 4.38 × 10−62 | 8.51 × 10−9 |
| Visual memory (F3) | 345 | 0 | 1 | 280 | −0.10 | 1.36 | 405 | −1.92 | 2.32 | 3.96 × 10−45 | 0.33 |
| Processing speed (F4) | 603 | 0 | 1 | 283 | −0.53 | 1.06 | 416 | −1.98 | 1.1 | 2.36 × 10−137 | 1.20 × 10−12 |
| Card sorting (F5) | 583 | 0 | 1 | 279 | −0.47 | 1.24 | 391 | −1.80 | 1.65 | 8.14 × 10−71 | 3.16 × 10−8 |
| Span (F6) | 523 | 0 | 1 | 286 | −0.27* | 1.04 | 414 | −1.15*,^ | 1.13 | 3.95 × 10−54 | 2.71 × 10−4 |
| g | 594 | 0 | 1 | 277 | −0.60* | 1.16 | 412 | −2.96*,^ | 1.72 | 3.90 × 10−151 | 2.86 × 10−13 |
PRS = PRS with P-value cut-off at 0.05. Data on PRS and cognitive variables have been standardized according to the mean and standard deviation of the control group. *Indicates P < −0.001 when compared to controls; ^indicates P < 0.001 when compared to siblings.
Polygenic risk score analysis
The means and standard deviations for PRS in schizophrenia, siblings and control groups calculated for the different thresholds are given in Supplementary Table 1. The numbers of SNPs for the different thresholds are shown in Supplementary Table 2. We conducted logistic regression to estimate the proportion of schizophrenia variation in case/control status that was explained by each PRS and found that the P-value threshold 0.05 accounted for more variance in our sample than PRS for other thresholds, about 9% of the variance in schizophrenia liability (Supplementary Table 2). This PRS threshold showing greatest risk prediction is consistent with results of the original PGC report (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Thus, we chose this PRS threshold (PT < 0.05), which included 24 694 SNPs, for all our subsequent analysis.
Correlational analysis
Supplementary Fig. 1 shows the correlation coefficients between each pair of variables. Schizophrenia correlated with g at −0.461 (P-value < 0.001), and PRS at 0.142 (P-value < 0.001). The Pearson correlation between g and PRS was −0.297 (P-value < 0.001).
Bivariate non-causal and causal models (Cholesky)
The bivariate model fitting results are shown in the first part of Table 2. Model 1 is the baseline Cholesky model, which tests for covariation between schizophrenia liability and cognition. Model 2 (full reciprocal causal model) with two causal paths has an identical fit, as expected, as the two models are created to be equivalent (see above). Model 3, a nested model, which drops the causal path k′ (L-SZ→L-COG), was not significantly worse (P-value = 0.18) than Models 1 and 2. While Model 4, which drops path k (L-COG→L-SZ), was significantly worse (P-value < 0.01) compared with Models 1 and 2, suggesting that the causal path k was important and could not be dropped. Therefore, Model 3 fitted the data well and with fewer parameters, supporting our earlier work in a pan-European twin with schizophrenia sample that found cognition to be upstream of schizophrenia liability (Toulopoulou et al., 2015).
Table 2.
Model fitting results
| ep | −2LL | df | AIC | Δ−2LL | Δdf | P | Comparison model | |
|---|---|---|---|---|---|---|---|---|
| Bivariate model | ||||||||
| 1: Cholesky | 29 | 26 072.78 | 8446 | 9179.78 | – | – | – | – |
| 2: L-COG↔L-SZ | 29 | 26 072.78 | 8446 | 9179.78 | – | – | – | – |
| 3: Dropping L-SZ→L-COG | 28 | 26 074.57 | 8447 | 9179.57 | 1.79 | 1 | 0.18 | 1 and 2 |
| 4: Dropping L-COG→L-SZ | 28 | 26 092.64 | 8447 | 9197.64 | 19.85 | 1 | <0.01 | 1 and 2 |
| Trivariate model | ||||||||
| 1: Full | 32 | 29 736.61 | 9752 | 10 232.61 | – | – | – | – |
| 2: Dropping PRs→L-COG (j) | 31 | 29 759.54 | 9753 | 10 253.54 | 22.94 | 1 | <0.01 | 1 |
| 3: Dropping PRS→L-SZ (i) | 31 | 29 751.26 | 9753 | 10 245.26 | 14.65 | 1 | <0.01 | 1 |
| 4: Dropping L-SZ→L-COG (k′) | 31 | 29 737.85 | 9753 | 10 231.85 | 1.24 | 1 | 0.26 | 1 |
| 5: Dropping L-COG→L-SZ (k) | 31 | 29 758.10 | 9753 | 10 252.10 | 21.49 | 1 | <0.01 | 1 |
Δ − 2LL = the difference of minus 2 log likelihood between two models; Δdf = the difference of the degrees of freedom; −2LL = minus 2 log likelihood; AIC = Akaike information criterion; ep = estimate parameter; df = degree of freedom; P = P-value, when P-value < 0.05 (P-value < 0.025 for bivariate), the model is significantly worse than the comparison model.
The overall genetic variance to L-SZ is constrained to 0.82.
The same model fitting was performed assuming different heritability levels, from 0.7 to 0.9, and the model fitting results are presented in Supplementary Table 3. When heritability, a2 is 0.7 to 0.8, Model 4, which drops path L-COG→L-SZ, deteriorates statistically significantly from baseline models, while Model 3, which drops path L-SZ→L-COG, is not significantly worse and thus the model with the best fit for the data. When a2 is 0.9, both Models 3 and 4 are significantly worse than the baseline model (Model 2), and Model 3 is still chosen due to the smaller Akaike information criterion (AIC). Thus, the model fitting results do not change with different heritability levels.
According to the chosen Model 3 (Fig. 1B), A contributes 61% (0.7812 = 0.61 × 100% = 61%; CI 0.748–0.814%) to the variance in cognition, and it is moderately heritable. The causal path k from L-COG→L-SZ is −0.680, which suggests that 46% [(−0.680)2 = 0.46 × 100% = 46%; CI −0.711 to −0.674] of variance in schizophrenia liability is explained by variation in cognition.
Figure 1B and D illustrate the two nested models with opposite direction of causation, and based on the two Cholesky models (Fig. 1A and C), which are equivalent. The parameter estimation in the nested model that describes the relationship best would be closer to the equivalent Cholesky. In Fig. 1, the upper part illustrates the nested model (Fig. 1B) from cognition to schizophrenia liability (L-COG→L-SZ), and its comparable Cholesky model (Fig. 1A). A1 would affect both of the phenotypes simultaneously, and its contribution on the variance of the second phenotype L-SZ is a212 (0.552) in the Cholesky model (Fig. 1A), and (a1 × k)2 [(0.781 × 0.680)2 = 0.532] in the causal model (Fig. 1B). Figure 1 also illustrates the causal model L-SZ→L-COG (Fig. 1D), and its comparable Cholesky model (Fig. 1C). The contribution of A1 on variance of L-COG is a21′2 (0.422) in the Cholesky model and (a1′ × k′)2 [(0.906 × 0.694)2 = 0.632] in the nested model. The path estimates are closer between a21 (0.55) and a1 × k (0.53) [e21 (0.42) and e1 × k (0.42)] (Fig. 1A and B), comparing with the path estimates between a21′ (0.42) and a1′ × k′ (0.63) [e21′ (0.71) and e1′ × k′ (0.29)] (Fig. 1C and D). Thus, the value of A1 and E1 contribution on the second phenotype is closer between the Cholesky model and nested model L-COG→L-SZ. This result is concordant with our assumption, and duplicates and verifies the results of the model fitting comparison (above), which supports a direction of causation from cognition to schizophrenia liability. The details of parameter comparison, such as the value of a21 and a1 × k, at different schizophrenia heritability levels, and the corresponding confidence intervals, can be found in Supplementary Table 4.
Trivariate causal model
The model fitting comparison results are listed in the second part of Table 2. Model 1 is the full model with four causal paths, i, j, k and k′ among three latent variables. When the causal path k′ is dropped (k′ = 0), the model (Model 4) is not significantly different from the full model (Model 1). In contrast, when we drop the causal path k, which indicates that the L-SZ is accounted for by variation in L-COG (L-COG→L-SZ) (Model 5), the model fit deteriorated significantly compared to its base model (Model 1). Models 2 and 3 show that causal paths j and i could not be dropped, thus L-PRS contributes significantly to both L-SZ and L-COG.
The model fitting results at different schizophrenia heritability levels (0.7–0.9) are presented in Supplementary Table 3. When the heritability of schizophrenia is set to 0.7 and 0.8, the model fitting results are similar and model selections are the same as the results when the heritability of schizophrenia as 0.82. When it is fixed to 0.9, all nested models (Models 2–5) are significantly worse than Model 1, and could not select the model according to P-value; however, Model 4 still has the smallest AIC among them, and fit the data best.
Figure 2 shows the parameter estimates of the best fitting model (Model 5) with three significant causal paths (i, j, k) at heritability of schizophrenia as 0.82. Table 3 shows the corresponding genetic variance components of schizophrenia liability including those that ‘pass through’ cognition. In total, L-PRS explained 8.07% (CI 5.45–10.74%) of the genetic variance components of schizophrenia liability, directly and indirectly through L-COG. L-PRS affected L-SZ directly by causal path i (Fig. 2), with 1.43% (CI 0.46–3.08%) of genetic variance in schizophrenia liability explained by L-PRS. L-PRS also explained 2.71% (CI 2.41–3.85%) of genetic variance of schizophrenia liability indirectly through L-COG. The remainder 3.93% (CI 2.37–4.48%) reflected correlated variation in which variation in L-SZ was attributed to variation from cognition and vice versa. Of the remaining genetic variance components of schizophrenia liability 26.87% (CI 21.45–32.57%) was accounted for by L-COG-relevant pathways not captured by L-PRS and 65.06% (CI 59.96–70.95%) was through paths other than L-COG and L-PRS (Table 3). The parameter estimates of the other models, based on different heritability assumptions are shown in the Supplementary material.
Table 3.
Genetic variance components of schizophrenia liability
| Expression | Variance component | Estimate, % | CIs, % | |
|---|---|---|---|---|
| L-PRS contributed directly | 1.43 | 0.46–3.08 | ||
| L-PRS through L-COG | 2.71 | 2.41–3.85 | ||
| Covariance between L-PRS and L-COG | 3.93 | 2.37–4.48 | ||
| From L-COG excluded L-PRS | 26.87 | 21.45–32.57 | ||
| L-SZ independently | 65.06 | 59.96–70.95 |
% = percentage of variance in liability to schizophrenia explained by additive genetic differences; latent variables: L-SZ = schizophrenia liability; L-COG = general cognitive factor; L-PRS = polygenic risk score; i, j, k represent causal paths: causal path i, L-PRS to L-SZ; causal path j, L-PRS to L-COG; causal path k, L-COG to L-SZ, causal path k′, L-SZ to L-COG; am2 is the path coefficient from the mth genetic factor to the latent variable in Fig. 2. Genetic variance to L-SZ is constrained to 0.82. Confidence intervals that do not include 0 are significant.
The parameter estimates of Model 4 with a2 of schizophrenia fixed to 0.7, 0.8 and 0.9 are shown in Supplementary Fig. 2. The corresponding amounts and percentages of genetic variance components of schizophrenia liability contributed by L-PRS at different schizophrenia heritability levels are provided in Supplementary Table 5. As shown in Supplementary Fig. 2 when schizophrenia heritability a2 increases, genetic parameters (a2 and a3) increase and environmental parameters (e2 and e3) decrease. Accordingly, when heritability is set at a2 = 0.7 the genetic (a3) and environmental (e3) path loadings for L-COG are a3 = 0.662 and e3 = 0.748 respectively, while at a2 = 0.9, the equivalent path loadings change to a3 = 0.843 and e3 = 0.488. Because the total variance of schizophrenia is fixed, including genetic and environmental part, the environmental path loading of L-SZ (e2) decreases with increasing a2, and becomes nearly 0 when a2 is at 0.82. Thus, when heritability of schizophrenia is fixed to 0.82 and above, the environmental variance is totally through the environmental component of L-COG. As shown in Supplementary Table 5 the amount of every component of schizophrenia genetic variance accounted by L-PRS is similar at different a2 levels, with percentages decreasing as a2 increases: 9.3% of schizophrenia genetic variance is explained by L-PRS at a2 = 0.7, and 7.4% at a2 = 0.9. The genetic variance components of schizophrenia liability related L-COG at different schizophrenia heritability levels are shown in Supplementary Table 6. The total variance related to L-COG increases as a2 increases from 0.7 to 0.9. Thus at a2 = 0.7, 31.68% (CI 23.20–42.60%) of overall genetic risk is mediated through influences on cognition, and at a2 = 0.9, 39.23% (CI 36.35–50.90%).
Discussion
Recent studies have shown genetic overlap between schizophrenia and cognition; however, the direction of causation remains unclear. We used causal modelling to address this question. A central aim of the current work was to determine whether and to what extent cognitive deficit mediates the influence of common genetic variants on schizophrenia. Results of modelling incorporating molecular genetic data, in the form of PRSs, were consistent with earlier statistical modelling in twin data—both approaches suggest that genetics, in part, move through cognition to exert an effect on schizophrenia risk (Toulopoulou et al., 2015). More specifically, current analyses indicated that, out of 8.07% of the variation in schizophrenia explained by PRS, more than one-third of that variation, i.e. 2.71% was mediated through cognitive deficit. We found that this model fit the data better than one with opposite directionality, which represented schizophrenia as mediating the relationship between PRS and cognitive deficit. Further parsing of genetics variance components based on family data (e.g. siblings) suggested that cognitive deficit mediated an even greater part of the genetic influences on schizophrenia, beyond what is accounted for by the PRS (i.e. 26.87% of the inherited liability to schizophrenia not captured in the modelling by PRS).
The findings have implications for the question of whether many genes could act through a constrained set of pathways (Geschwind and Flint, 2015). Specifically, results suggested that as much as 33.51% of the overall heritable liability to schizophrenia may be mediated through cognitive operations. Only a modest portion of this is captured by current PRS. It is not clear that rare variant or epigenetic influences can be reflected in a PRS type scheme. Nevertheless, results were robust, and changes in the value of schizophrenia’s heritability did not appreciably affect model fitting. Further study of the molecular and cellular genetic basis of variation in cognition, which involves the basic mechanisms of brain development and function (Birnbaum and Weinberger, 2017), will likely provide insights about the mechanisms by which risk genes bias the brain toward inefficient cognition in schizophrenia.
Interpreting the polygenic effects of risk variants in terms of disease mechanisms will require integration of genetic, biological, and circuit levels of analysis (Gandal et al., 2016), but recent attempts have highlighted the role of histone methylation, dendritic spines, calcium signalling, glutamatergic transmission, plasticity, neurogenesis, synaptic pruning, and immunity (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014; The Network and Pathway Analysis Subgroup of the Psychiatric Genomics Consortium, 2015; Gandal et al., 2016). Most of the risk variants are regulatory, exerting their influence through modifications in gene expression (e.g. in the context of gene environment interactions) (Birnbaum and Weinberger, 2017; Ursini et al., 2018). A possibility highlighted by our findings is that some of the aforementioned processes first influence alterations in cognition and have later effects contributing to the more acute symptoms of schizophrenia. At the same time, we found that about 65% of the genetic influences on schizophrenia were not related to genetic influences on cognition and PRS, highlighting the challenges to the current approach to identifying intermediate phenotypes.
One finding consistent with current genome wide heritability estimates (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) is that most of the heritable influence on schizophrenia is independent of current PRS. Even though polygenic risk was calculated based on the large PGC dataset (36 573 schizophrenia cases and 112 468 controls), PRS is still an incomplete measure of risk. As mentioned earlier, PRS derived from the current state of the art GWAS and accounts for part of the variation in schizophrenia liability (∼7%), less than the estimated additive heritability from twin studies (Wray et al., 2014).
We modelled general cognition as a latent trait, reflecting the covariance of six broad cognitive domains that are consistently altered in schizophrenia. Similar latent cognition variables have been widely used in genetic studies (Dickinson et al., 2014; Trampush et al., 2017). Latent cognition correlated robustly with PRS and had the added advantage of facilitating model fitting by reducing the number of variables for analysis. When cognitive factors were modelled separately in exploratory bivariate models, individual analyses were consistent with the main findings, but results were generally weaker. These observations are in line with earlier reports from statistical modelling of twin and sibling data that suggested a stronger link between schizophrenia and general cognition than between schizophrenia and individual cognitive abilities (Toulopoulou et al., 2007, 2010b, 2015; Owens et al., 2011a, 2012).
Findings from the current study suggest that cognitive deficit mediates (or causes) part of the observed association between schizophrenia and genetic factors. Although it is unlikely that cognitive deficit can ‘cause’ psychotic symptoms in a straightforward mechanistic sense, it is possible that poor cognition might leave an individual with fewer resources to combat psychotic symptoms. Alternatively, poor cognition can be viewed as a necessary but not sufficient component of an altered developmental trajectory that may involve psychosis-related neural functions as a later emerging component (Birnbaum and Weinberger, 2017). Thus, some individuals may have cognitive compromise but not sufficient developmental deviation to manifest psychosis, or alternatively, some individuals with mild psychotic symptoms might not develop clinical schizophrenia unless their cognition is also impaired. By either scenario, cognitive deficit would be in the causative chain from genetic risk to schizophrenia, in combination with other factors.
It has been a long-standing assumption that cognitive deficit and schizophrenia both reflect abnormal neurodevelopment. Indeed, recent findings that several of the risk variants predict expression in brain of genes that are more likely to be expressed prenatally, are consistent with evidence of an early developmental contribution to risk (Birnbaum et al., 2015). Literature addressing the antecedents of schizophrenia suggest that some developmental programming connected to schizophrenia occurs early in life and affects cognitive development in childhood, well before the onset of acute psychotic symptoms. Other developmental programming, in adolescence, may further hamper cognitive development, and also generate psychotic phenomena. The current modelling cannot address these intertwined developmental hypotheses but offer clearer support for an aetiological trajectory through cognitive deficit toward schizophrenia.
Our results should be viewed in light of several limitations. First, while the cumulative effect of the genome-wide schizophrenia-associated common risk loci identified in the latest studies provides an optimal starting point for exploring the role of cognition as an intermediate disease mechanism, the risk scores, as mentioned earlier, represent only part of the heritable variation that contributes to schizophrenia. Second, the PRS includes SNPs that may not be causative variants. As discussed by others (Wray et al., 2014) schizophrenia-associated SNPs correlate with many other variants, which could be the true risk-conferring ones. Third, we constructed PRSs based on the standard methodology. The methodology is robust; however, other approaches, such as empirical Bayes, which applies an automatic PRS weighting, might capture risk better (So and Sham, 2017). Fourth, our estimates are limited to the cognitive assessments used, participant characteristics, and study design. Other assessments, participants, or study designs could yield different results. It would be of interest to model both the more upstream (i.e. earlier in the sequence of cognitive operations) and downstream cognitive processes that recent research has highlighted as important in schizophrenia (e.g. source memory, prediction error, motivational salience, and social cognition). Fifth, the models are limited by the validity of the assumptions we made. With three variables (schizophrenia, cognition, and PRS) in our causative models, we effectively assumed that there was no other reason for these three variables to correlate than each other, which is likely an oversimplification of reality. For example, in our models, we assumed that genes cause increases in liability for schizophrenia and reductions in cognitive function. These assumptions are appropriate, but there might be other reasons that cognition and schizophrenia correlate with each other, other than in relation to the PRS. Sixth, cognition was defined as a latent variable with continuous indicators, while schizophrenia as a binary outcome variable. This difference in measurement approaches could lead to differences in statistical power to detect the two reciprocal causal paths. Indeed, in the bivariate model where both paths are estimated, the path from L-SZ to L-COG has a wider 95% CI than the path from L-COG to L-SZ. Thus, while our results support a causal relationship from L-COG to L-SZ, we cannot exclude the possibility of type 2 error in relation to the path from L-SZ to L-COG. Seventh, we estimated the E component in our models, as studies do not attribute significance to shared environment (C) for both schizophrenia (Sullivan et al., 2003a) and cognition (Bouchard, 2013). However, the study design cannot differentiate between C and E (for this, we would need twin data); thus, E should be better interpreted as both common environment and unique environment. Eighth, to assess PRS we used the results from the latest PGC schizophrenia meta-analysis. As the PGC schizophrenia meta-analysis did not stratify on environmental risk (e.g. obstetrical complications) we cannot know the extent to which risk variants may be dependent on the environment. Ninth, causal modelling evaluates consistency of various models to the data, and does not provide absolute proof of causality. A longitudinal design, starting before illness onset, would be more informative and definitive. Alternatively, Mendelian randomization may also be a powerful approach. However, to perform a robust Mendelian randomization, it is preferable to use individual SNPs as multiple instruments (as opposed to PRSs applied here), and to control for some of these SNPs being pleiotropic (Davey Smith and Hemani, 2014; Hemani et al., 2018; Verbanck et al., 2018). Finally, when PGC sample size is further enlarged to produce a PRS that captures a greater proportion of the variance in schizophrenia liability, it is uncertain whether the proportion of variance mediated through cognitive impairment will remain unchanged.
In conclusion, the underlying biology of polygenic risk in schizophrenia is poorly understood. One strategy to translate polygenic burden into brain mechanisms of disease is to examine the causal relationships between schizophrenia, PRS, and proposed biological associations of risk, i.e. intermediate phenotypes that presumably lie on the chain of causation from gene to phenotype (e.g. cognitive deficit). We showed that cognitive deficit partially mediates the relationship between PRS and the disorder. Modelling sibling data suggested an even greater role for cognition in transmitting genetic influences on schizophrenia risk. Other genetic influences on diagnosis are more independent of cognition. Further discovery and analysis will be needed to understand more fully the degree to which genetic risk for schizophrenia is mediated through cognition.
Funding
X.Z. was funded by T.T. and P.S. through State Key Laboratory of Brain and Cognitive Sciences funds and an National Institutes of Health (NIH) subcontract (NIH-260850043) awarded to T.T. The work was supported by the National Institute of Mental Health (NIMH) Division of Intramural Research Programs through funding to the Clinical Brain Disorders Branch (D.R.W., P.I.) and, later, the Clinical and Translational Neuroscience Branch (K.F.B., P.I. NCT00001486, ZIAMH002712), and by direct funding from the Lieber Institute for Brain Development and the Maltz Research Laboratories.
Competing interests
The authors report no competing interests.
Supplementary Material
Glossary
Abbreviations
- L-COG
cognition, latent
- L-PRS
polygenic risk scores, latent
- L-SZ
schizophrenia liability
- PGC
Psychiatric Genomics Consortium
- PRS
polygenic risk score
- SNP
single nucleotide polymorphism
References
- Agnew-Blais JC, Buka SL, Fitzmaurice GM, Smoller JW, Goldstein JM, Seidman LJ. Early childhood IQ trajectories in individuals later developing schizophrenia and affective psychoses in the New England family studies. Schizophr Bull 2015; 41: 817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alloza C, Bastin ME, Cox SR, Gibson J, Duff B, Semple SI et al. Central and Non-central networks, cognition, clinical symptoms, and polygenic risk scores in schizophrenia. Hum Brain Mapp 2017; 38: 5919–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen SE, Petryshen TL. Genome-wide association studies of schizophrenia: does bigger lead to better results? Curr Opin Psychiatry 2012; 25: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum R, Jaffe AE, Chen Q, Hyde TM, Kleinman JE, Weinberger DR. Investigation of the prenatal expression patterns of 108 Schizophrenia-Associated genetic loci. Biol Psychiatry 2015; 77: e43–51. [DOI] [PubMed] [Google Scholar]
- Birnbaum R, Weinberger DR. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci 2017; 18: 727. [DOI] [PubMed] [Google Scholar]
- Blokland GAM, del Re EC, Mesholam-Gately RI, Jovicich J, Trampush JW, Keshavan MS et al. The genetics of endophenotypes of neurofunction to understand schizophrenia (GENUS) consortium: a collaborative cognitive and neuroimaging genetics project. Schizophr Res 2018; 195: 306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boker S, Neale M, Maes H, Wilde M, Spiegel M, Brick T et al. OpenMx: an open source extended structural equation modeling framework. Psychometrika 2011; 76: 306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard TJ. The wilson effect: the increase in heritability of IQ with age. Twin Res Hum Genet 2013; 16: 923–30. [DOI] [PubMed] [Google Scholar]
- Bramon E, Rabe-Hesketh S, Sham P, Murray RM, Frangou S. Meta-analysis of the P300 and P50 waveforms in schizophrenia. Schizophr Res 2004; 70: 315–29. [DOI] [PubMed] [Google Scholar]
- Calkins ME, Ray A, Gur RC, Freedman R, Green MF, Greenwood TA et al. Sex differences in familiality effects on neurocognitive performance in schizophrenia. Biol Psychiatry 2013; 73: 976–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callicott JH, Bertolino A, Mattay VS, Langheim FJP, Duyn J, Coppola R et al. Physiological dysfunction of the dorsolateral prefrontal cortex in schizophrenia revisited. Cereb Cortex 2000; 10: 1078–92. [DOI] [PubMed] [Google Scholar]
- Cannon TD, Bearden CE, Hollister JM, Rosso IM, Sanchez LE, Hadley T. Childhood cognitive functioning in schizophrenia patients and their unaffected siblings: a prospective cohort study. Schizophr Bull 2000a; 26: 379–93. [DOI] [PubMed] [Google Scholar]
- Cannon TD, Huttunen MO, Lonnqvist J, Tuulio-Henriksson A, Pirkola T, Glahn D et al. The inheritance of neuropsychological dysfunction in twins discordant for schizophrenia. Am J Hum Genet 2000b; 67: 369–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardno AG, Marshall E, Coid B, Macdonald AM, Ribchester TR, Davies NJ et al. Heritability estimates for psychotic disorders: the maudsley twin psychosis series. Arch Gen Psychiatry 1999; 56: 162–8. [DOI] [PubMed] [Google Scholar]
- Corp I. IBM SPSS Statistics for Windows. 23.0 ed. Armonk, NY: IBM Corp; 2015. [Google Scholar]
- Cosgrove D, Harold D, Mothersill O, Anney R, Hill MJ, Bray NJ et al. MiR-137-derived polygenic risk: effects on cognitive performance in patients with schizophrenia and controls. Transl Psychiatr 2017; 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley NA, Mechelli A, Ginestet C, Rubinov M, Bullmore ET, McGuire P. Altered hub functioning and compensatory activations in the connectome: a meta-analysis of functional neuroimaging studies in schizophrenia. Schizophr Bull 2016; 42: 434–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014; 23: R89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D, Goldberg TE, Gold JM, Elvevag B, Weinberger DR. Cognitive factor structure and invariance in people with schizophrenia, their unaffected siblings, and controls. Schizophr Bull 2011; 37: 1157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D, Straub RE, Trampush JW, Gao Y, Feng N, Xie B et al. Differential effects of common variants in scn2a on general cognitive ability, brain physiology, and messenger RNA expression in schizophrenia cases and control individuals. JAMA Psychiatry 2014; 71: 647–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DL, Martin NG. Inferring the direction of causation in cross-sectional twin data: theoretical and empirical considerations. Genet Epidemiol 1994; 11: 483–502. [DOI] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Gscheidle T, Weirich M, Rawlings R, Hyde TM et al. Relative risk for cognitive impairments in siblings of patients with schizophrenia. Biol Psychiatry 2001a; 50: 98–107. [DOI] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA 2001b; 98: 6917–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer AE, McGuffin P, Gottesman II. Twin concordance for DSM-III schizophrenia. Scrutinizing the validity of the definition. Arch Gen Psychiatry 1987; 44: 634–41. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer R, Gibbon M, Williams JB. Structured clinical interview for axis I DSM-IV. New York: Biometrics Research Department, New State Psychiatric Institute; 1994. [Google Scholar]
- Flint J, Timpson N, Munafò M. Assessing the utility of intermediate phenotypes for genetic mapping of psychiatric disease. Trends Neurosci 2014; 37: 733–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler T, Zammit S, Owen MJ, Rasmussen F. A population-based study of shared genetic variation between premorbid iq and psychosis among male twin pairs and sibling pairs from sweden. Arch Gen Psychiatry 2012; 69: 460–6. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Frith CD. Schizophrenia - a disconnection syndrome. Clin Neurosci 1995; 3: 89–97. [PubMed] [Google Scholar]
- Gandal MJ, Leppa V, Won HJ, Parikshak NN, Geschwind DH. The road to precision psychiatry: translating genetics into disease mechanisms. Nat Neurosci 2016; 19: 1397–9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landen M et al. Increased burden of ultra-rare protein-altering variants among 4 877 individuals with schizophrenia. Nat Neurosci 2016; 19: 1433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germine L, Robinson EB, Smoller JW, Calkins ME, Moore TM, Hakonarson H et al. Association between polygenic risk for schizophrenia, neurocognition and social cognition across development. Transl Psychiatr 2016; 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science 2015; 349: 1489–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Knowles EE, McKay DR, Sprooten E, Raventos H, Blangero J et al. Arguments for the sake of endophenotypes: examining common misconceptions about the use of endophenotypes in psychiatric genetics. Am J Med Genet B Neuropsychiatr Genet 2014; 165B: 122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg TE, Torrey EF, Gold JM, Bigelow LB, Ragland RD, Taylor E et al. Genetic risk of neuropsychological impairment in schizophrenia: a study of monozygotic twins discordant and concordant for the disorder. Schizophr Res 1995; 17: 77–84. [DOI] [PubMed] [Google Scholar]
- Gur RE, Calkins ME, Gur RC, Horan WP, Nuechterlein KH, Seidman LJ et al. The consortium on the genetics of schizophrenia: neurocognitive endophenotypes. Schizophr Bull 2007a; 33: 49–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur RE, Nirngaonkar VL, Almasy L, Calkins ME, Ragland JD, Pogue-Geile MF et al. Neurocognitive endophenotypes in a multiplex multigenerational family study of schizophrenia. Am J Psychiatry 2007b; 164: 813–19. [DOI] [PubMed] [Google Scholar]
- Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DCM, Ritchie SJ et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N = 112151) and 24 GWAS consortia. Mol Psychiatr 2016; 21: 1624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzimanolis A, Bhatnagar P, Moes A, Wang R, Roussos P, Bitsios P et al. Common genetic variation and schizophrenia polygenic risk influence neurocognitive performance in young adulthood. Am J Med Genet B Neuropsychiatr Genet 2015; 168: 392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Zheng J, Elsworth B, Wade K, Haberland V, Baird D et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018; 7: e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard L, Tansey KE, Rai D, Jones P, Ripke S, Chambert KD et al. Evidence of common genetic overlap between schizophrenia and cognition. Schizophr Bull 2016; 42: 832–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi K, Westlye LT, Tesli M, Bettella F, Brandt CL, Mattingsdal M et al. Polygenic risk for schizophrenia associated with working Memory-related prefrontal brain activation in patients with schizophrenia and healthy controls. Schizophr Bull 2015; 41: 736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlar AV, Mercer KB, Zwick ME, Mulle JG. New discoveries in schizophrenia genetics reveal neurobiological pathways: a review of recent findings. Eur J Med Genet 2015; 58: 704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Knowles E, Davies G, Guha S, Liewald DC, Starr JM et al. Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: a report from the Cognitive Genomics consorTium (COGENT). Mol Psychiatr 2014; 19: 168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 2009; 373: 234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebers DT, Pirooznia M, Seiffudin F, Musliner KL, Zandi PP, Goes FS. Polygenic risk of schizophrenia and cognition in a Population-Based survey of older adults. Schizophr Bull 2016; 42: 984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 2012; 148: 1223–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ et al. Finding the missing heritability of complex diseases. Nature 2009; 461: 747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark W, Toulopoulou T. Cognitive intermediate phenotype and genetic risk for psychosis. Curr Opin Neurobiol 2016; 36: 23–30. [DOI] [PubMed] [Google Scholar]
- McIntosh AM, Gow A, Luciano M, Davies G, Liewald DC, Harris SE et al. Polygenic risk for schizophrenia is associated with cognitive change between childhood and old age. Biol Psychiatry 2013; 73: 938–43. [DOI] [PubMed] [Google Scholar]
- Meier MH, Caspi A, Reichenberg A, Keefe RSE, Fisher H, Harrington H et al. Neuropsychological decline in schizophrenia from the premorbid to the postonset period: evidence from a population-representative longitudinal study. Am J Psychiatry 2014; 171: 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard SP, Shofer J, Braff D, Calkins M, Cadenhead K, Freedman R et al. Prioritizing schizophrenia endophenotypes for future genetic studies: an example using data from the COGS-1 family study. Schizophr Res 2016; 174: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MC, Cardon LR. Data summary. Methodology for genetic studies of twins and families. Dordrecht: Springer Netherlands; 1992. p. 35–53. [Google Scholar]
- Olincy A, Braff DL, Adler LE, Cadenhead KS, Calkins ME, Dobie DJ et al. Inhibition of the P50 cerebral evoked response to repeated auditory stimuli: results from the Consortium on Genetics of Schizophrenia. Schizophr Res 2010; 119: 175–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens SF, Picchioni MM, Ettinger U, McDonald C, Walshe M, Schmechtig A et al. Prefrontal deviations in function but not volume are putative endophenotypes for schizophrenia. Brain 2012; 135: 2231–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens SF, Picchioni MM, Rijsdijk FV, Stahl D, Vassos E, Rodger AK et al. Genetic overlap between episodic memory deficits and schizophrenia: results from the maudsley twin study. Psychol Med 2011a; 41: 521–32. [DOI] [PubMed] [Google Scholar]
- Owens SF, Rijsdijk F, Picchioni MM, Stahl D, Nenadic I, Murray RM et al. Genetic overlap between schizophrenia and selective components of executive function. Schizophr Res 2011b; 127: 181–7. [DOI] [PubMed] [Google Scholar]
- Pantelis C, Velakoulis D, McGorry PD, Wood SJ, Suckling J, Phillips LJ et al. Neuroanatomical abnormalities before and after onset of psychosis: a cross-sectional and longitudinal MRI comparison. Lancet 2003; 361: 281–8. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland JD, Laird AR, Ranganath C, Blumenfeld RS, Gonzales SM, Glahn DC. Prefrontal activation deficits during episodic memory in schizophrenia. Am J Psychiatry 2009; 166: 863–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampino A, Taurisano P, Fanelli G, Attrotto M, Torretta S, Antonucci LA et al. A polygenic risk score of glutamatergic SNPs associated with schizophrenia predicts attentional behavior and related brain activity in healthy humans. Eur Neuropsychopharmacol 2017; 27: 928–39. [DOI] [PubMed] [Google Scholar]
- Ranlund S, Calafato S, Thygesen JH, Lin K, Cahn W, Crespo-Facorro B et al. A polygenic risk score analysis of psychosis endophenotypes across brain functional, structural, and cognitive domains. Am J Med Genet B 2018; 177: 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenberg A, Caspi A, Harrington H, Houts R, Keefe RS, Murray RM et al. Static and dynamic cognitive deficits in childhood preceding adult schizophrenia: a 30-year study. Am J Psychiatry 2010; 167: 160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijsdijk F, Haren NEM, Picchioni M, McDonald C, Toulopoulou T, Pol H et al. Brain MRI abnormalities in schizophrenia: same genes or same environment? Psychol Med 2005; 35: 1399–409. [DOI] [PubMed] [Google Scholar]
- Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 2013; 45: 1150–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer J, Giangrande E, Weinberger DR, Dickinson D. The global cognitive impairment in schizophrenia: consistent over decades and around the world. Schizophr Res 2013; 150: 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman LJ, Cherkerzian S, Goldstein JM, Agnew-Blais J, Tsuang MT, Buka SL. Neuropsychological performance and family history in children at age 7 who develop adult schizophrenia or bipolar psychosis in the New England Family Studies. Psychol Med 2013; 43: 119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So HC, Sham PC. Improving polygenic risk prediction from summary statistics by an empirical Bayes approach. Sci Rep 2017; 7: 41262. doi: 10.1038/srep41262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 2003a; 60: 1187–92. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 2003b; 60: 1187–92. [DOI] [PubMed] [Google Scholar]
- The Network and Pathway Analysis Subgroup of the Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 2015; 18: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulopoulou T, Goldberg TE, Mesa IR, Picchioni M, Rijsdijk F, Stahl D et al. Impaired intellect and memory: a missing link between genetic risk and schizophrenia? Arch Gen Psychiatry 2010a; 67: 905–13. [DOI] [PubMed] [Google Scholar]
- Toulopoulou T, Goldberg TE, Mesa IR, Picchioni M, Rijsdijk F, Stahl D et al. Impaired intellect and memory a missing link between genetic risk and schizophrenia? Arch Gen Psychiatry 2010b; 67: 905–13. [DOI] [PubMed] [Google Scholar]
- Toulopoulou T, Morris RG, Rabe-Hesketh S, Murray RM. Selectivity of verbal memory deficit in schizophrenic patients and their relatives. Am J Med Genet B 2003a; 116B: 1–7. [DOI] [PubMed] [Google Scholar]
- Toulopoulou T, Picchioni M, Rijsdijk F, Hua-Hall M, Ettinger U, Sham P et al. Substantial genetic overlap between neurocognition and schizophrenia—genetic modelling in twin samples. Arch Gen Psychiatry 2007; 64: 1348–55. [DOI] [PubMed] [Google Scholar]
- Toulopoulou T, Rabe-Hesketh S, King H, Murray RM, Morris RG. Episodic memory in schizophrenic patients and their relatives. Schizophr Res 2003b; 63: 261–71. [DOI] [PubMed] [Google Scholar]
- Toulopoulou T, van Haren N, Zhang X, Sham PC, Cherny SS, Campbell DD et al. Reciprocal causation models of cognitive vs volumetric cerebral intermediate phenotypes for schizophrenia in a pan-European twin cohort. Mol Psychiatr 2015; 20: 1386–96. [DOI] [PubMed] [Google Scholar]
- Trampush JW, Yang MLZ, Yu J, Knowles E, Davies G, Liewald DC et al. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol Psychiatr 2017; 22: 336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini G, Punzi G, Chen Q, Marenco S, Robinson JF, Porcelli A et al. Convergence of placenta biology and genetic risk for schizophrenia. Nat Med 2018; 24: 792–801. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O, Collin G, Scheewe T, Mandl RCW, Cahn W et al. Abnormal rich club organization and functional brain dynamics in schizophrenia. JAMA Psychiatry 2013; 70: 783–92. [DOI] [PubMed] [Google Scholar]
- Vassos E, Di Forti M, Coleman J, Iyegbe C, Prata D, Euesden J et al. An examination of polygenic score risk prediction in individuals with First-Episode psychosis. Biol Psychiatry 2017; 81: 470–7. [DOI] [PubMed] [Google Scholar]
- Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018; 50: 693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters JTR, Rujescu D, Franke B, Giegling I, Vasquez AA, Hargreaves A et al. The role of the major histocompatibility complex region in cognition and brain structure: a schizophrenia GWAS follow-up. Am J Psychiatry 2013; 170: 877–85. [DOI] [PubMed] [Google Scholar]
- Waters-Metenier S, Toulopoulou T. Putative structural neuroimaging endophenotypes in schizophrenia: a comprehensive review of the current evidence. Future Neurol 2011a; 6: 679–715. [Google Scholar]
- Waters-Metenier SL, Toulopoulou T. Qualifying brain functional MRI parameters as endophenotypes in schizophrenia. Future Neurol 2010; 5: 817–38. [Google Scholar]
- Waters-Metenier SL, Toulopoulou T. Putative diffusion tensor neuroimaging endophenotypes in schizophrenia: a review of the early evidence. Future Neurol 2011b; 6: 415–33. [Google Scholar]
- Wood SJ, Pantelis C, Velakoulis D, Yücel M, Fornito A, McGorry PD. Progressive changes in the development toward schizophrenia: studies in subjects at increased symptomatic risk. Schizophr Bull 2008; 34: 322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F, Middeldorp CM. Research Review: polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry 2014; 55: 1068–87. [DOI] [PubMed] [Google Scholar]
- Zhang R, Picchioni M, Allen P, Toulopoulou T. Working memory in unaffected relatives of patients with schizophrenia: a meta-analysis of functional magnetic resonance imaging studies. Schizophr Bull 2016; 42: 1068–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon request.