Abstract
Targeted monoclonal antibody therapy is a promising therapeutic strategy for cancer and antibody-dependent cell-mediated cytotoxicity (ADCC) represents a crucial mechanism underlying these approaches. The majority of patients have limited responses to monoclonal antibody therapy due to the development of resistance. Models of ADCC provide a system for uncovering immune resistance mechanisms. We continuously exposed epidermal growth factor receptor (EGFR+) A431 cells to KIR-deficient NK92-CD16V effector cells and the anti-EGFR cetuximab. Persistent ADCC exposure yielded ADCC-resistant cells (ADCCR1) that, compared with control ADCC-sensitive cells (ADCCS1), exhibited reduced EGFR expression, overexpression of histone- and interferon-related genes, and a failure to activate NK cells, without evidence of epithelial to mesenchymal transition. These properties gradually reversed following withdrawal of ADCC selection pressure. The development of resistance was associated with lower expression of multiple cell surface molecules that contribute to cell:cell interactions and immune synapse formation. Classic immune checkpoints did not modulate ADCC in this unique model system of immune resistance. We showed that the induction of ADCC resistance involves genetic and epigenetic changes that lead to a general loss of target cell adhesion properties that are required for the establishment of an immune synapse, killer cell activation, and target cell cytotoxicity.
Keywords: Antibody-dependent cell-mediated cytotoxicity, cetuximab, resistance, cell adhesion
Introduction
Antibody-dependent cell-mediated cytotoxicity (ADCC) was first described as a mechanism of action for monoclonal antibody therapy more than 30 years ago (1). Most efforts to modulate ADCC depend upon the incubation of potential effector cells with cytokines or chemokines that modify effector cell function. Relatively little is known about the mechanism by which tumor cells develop resistance to ADCC. Prior studies have examined only a restricted number of candidate genes/proteins (e.g. epidermal growth factor receptor [EGFR] network (2)) or receptor tyrosine kinases linked to PD-L1 expression (e.g. JAK1 and JAK2 (3)).
To further explore tumor cell-based mechanisms of ADCC resistance, we developed a model system for anti-EGFR ADCC. This model system included a high-EGFR expressing squamous cell carcinoma cell line (A431), an EGFR-targeting monoclonal antibody (cetuximab), and a KIR-deficient NK cell line transduced with CD16V to create the optimal conditions for ADCC (NK92-CD16V) (4). Binding of NK92-CD16V to A431 through cetuximab led to the activation of the NK cells, formation of immune synapses, and killing of target cells by the release of perforins and granzymes. Previously, we used this model system to study resistance to ADCC mediated by selected components of the EGFR signaling network (2). The use of NK92-CD16V cells allows for the study of resistance mechanisms unrelated to most known KIR-ligand interactions. Thus, this model system examines preferentially diverse target cell resistance mechanisms that may be seen in response to other immune effector mechanisms. We hypothesized that continuous ADCC exposure would create strong, durable selection pressure on A431 cells that recapitulates in vivo immune attack and induces distinct resistance mechanisms. This approach led to the development of several ADCC resistant cell lines, differing from the parental ADCC-sensitive A431 cell line in morphology, gene expression, proliferation, and sensitivity to other drugs.
We extensively characterized the ADCC resistant cell line termed ADCCR1 and found that this resistant cell line, which does not possess markers of epithelial-mesenchymal transition, had a distinctive transcriptional profile highlighted by overexpression of CD74, histone- and interferon-related genes, and significant reduction in HSPB1 and EGFR expression. Surface expression of cell adhesion molecules was dramatically altered, indicating that the developed ADCC resistance was specific to the formation and function of an immune synapse. Similar characteristics were observed in a second ADCC resistant line. This current work defines a molecular mechanism of resistance to ADCC involving the loss of molecules necessary for immune cell conjugation, and may inform on the mechanisms of resistance in other cases of immune synapse-mediated cytotoxicity.
Materials and Methods
Cell lines and cell culture
The A431 cell line was obtained from the Georgetown Lombardi Tissue Culture Shared Resource (TCSR) in 2010 and 2014, and its origin was verified by DNA fingerprinting by short tandem repeat (STR) analysis prior to utilization, as described (5). ADCCS1 and ADCCR1 were derived from cells obtained in 2010 and have been used during 2010-2018. Tissue culture growth conditions for this cell line were: High-glucose Dulbecco’s Modified Eagle Medium (DMEM, HyClone) supplemented with 10% fetal bovine serum (FBS, Omega Scientific) and 2 mM (1X) L-glutamine (Gibco). NK92-CD16V cells that express GFP due to transduction with pBMN-IRES-EGFP were kindly provided by Kerry S. Campbell from Fox Chase Cancer Center, Philadelphia PA. They were cultured in MEMα-modification (HyClone) supplemented with 10% FBS, 10% horse serum, 1 mM sodium pyruvate, and 1X non-essential amino acids (Gibco), as well as 0.1 mM β-mercaptoethanol (Sigma) as described (2,4,6). NK92-CD16V cells were maintained in suspension and passaged every 2-3 days by resuspending the cells in NK media (described above) at a concentration of 0.2 × 106 cells/mL and stimulated with 1% v/v of IL2 supernatant derived from J558L cells (4). All cell lines were maintained at 37°C in 5% CO2 and tested negative for mycoplasma. Cell counts were estimated by hemocytometer, and viable cells identified by trypan blue (Invitrogen) exclusion.
Inhibitors and treatment antibodies
Inhibitors of histone acetyltransferase C646 (Cat. No. S7152), DNA methyltransferase azacytidine (Cat. No. S1782), histone demethylase GSK J4 HCL (Cat. No. S7070), and HDAC panobinostat (Cat. No. S1030) were purchased from Selleck Chemicals. Inhibitors were solubilized in DMSO at 20 μM. Vehicle treatment (DMSO) was used at the highest equivalent v/v used in inhibitor treatments. 10,000 cells/well of both ADCCS1 and ADCCR1 were plated over-night in 96-well, clear-bottom white plates (Corning, Cat. No. 3903) then treated in the presence of the inhibitors at 0.01-10 μM concentrations for 2 hours prior to ADCC assay. Cetuximab (Bristol-Myers Squibb) and trastuzumab (Genentech) were purchased from the MedStar Georgetown University Hospital Pharmacy.
Flow cytometry
A431 cells were cultured for 3-6 passes then were dissociated using 0.25% trypsin, resuspended in DMEM plus 10% FBS and 1% L-glutamine. 0.5-1 × 106 cells were aliquoted into Eppendorf tubes, spun at 5000 rpm for 1 minute at 4°C, washed twice with HBSS (Fisher Scientific Cat. No. SH3058801), and resuspended in 100 μl of FACS buffer (PBS plus 1% BSA). All antibodies used are labeled antibodies, and no blocking step was performed. Labeled antibodies were then added at the manufacturer’s recommended concentrations and incubated at 4° C for 30 minutes, with vortexing at 15 minutes. For intracellular staining cells were resuspended in 50 μl of BD perm/wash (Cat. No.554723) for 20 minutes before proceeding to staining with antibody at 4°C for 30 minutes. Cells were then washed with FACS buffer twice and resuspended in FACS buffer or fixative (1% PFA in PBS). Flow antibodies were purchased from BioLegend: EGFR (Cat. No. 352904), CD74 (Cat. No. 326807), CD54/ICAM (Cat. No. 322713), CD142 (Cat. No. 365205), CD73 (Cat. No. 344021), ITGB4/CD104 (Cat. No. 343903) ALCAM/CD166 (Cat. No. 343903), CD95/Fas (Cat. No. 305611), CD138, (Cat. No. 352307), and APC-labelled IgG1 isotype control (Cat. No. 400121). CD107a (Cat. No. 641581), CD44 (Cat. No. 559942), HER2 (Cat. No. 340879), and PD-L1 (Cat. No. 557929) were purchased from BD Biosciences. PE-labelled IgG1 isotype control was purchased from eBioscience (Cat. No. 12-4714-81). Samples were run in the Georgetown Lombardi Comprehensive Cancer Center Flow Cytometry & Cell Sorting Shared Resource using BD LSRFortessa. Analyses were performed using FlowJo (v10.4.1).
Derivation of ADCC resistance
Initial derivation of ADCC resistance
A431 cells were seeded overnight in six-well plates (Greiner bio-one, Cat.No. 657160) at 150,000 cells per well. The following day, six different treatments groups were added for the initial ADCC challenge: 1) vehicle (media); 2) cetuximab (0.01 or 1 μg/mL); 3) 500,000 NK92-CD16V cells, cetuximab (0.01 μg/mL) plus 500,000 NK92-CD16V cells (low ADCC), or cetuximab (1 μg/mL) and 500,000 NK92-CD16V cells (high ADCC). Adding 500,000 NK92-CD16V cells under these culture conditions equates to ~ 2:1 effector-to-target (E:T) ratio at the time of treatment addition. Three or four days later, all wells were aspirated of treatments, washed, and the remaining adherent cells were collected by trypsinization. Viable cell density for each treatment was assessed by trypan blue exclusion. Identical conditions were employed for each subsequent ADCC challenge. Over six months, 34 consecutive, subsequent challenges were conducted. Viable cell density was used as a surrogate to assess for resistance in the treatment groups. After every fifth treatment cycle (Ch5, Ch10, Ch15, etc.) cells from each treatment were also expanded for one passage and cryopreserved.
Rederivation of ADCC resistance
A431 cells were seeded overnight in 5 T75 flasks (Greiner bio-one, Cat.No. 658175) at 500,000 cells per flask. The following day, the flasks were divided into 4 treatments groups: untreated (media only), cetuximab (1 μg/mL), 1× 106 NK cells (1:1 E:T), and ADCC (1 μg/mL cetuximab plus 1:1 E:T). Each of the control groups contained 1 flask, and the ADCC group was distributed into two flasks to allow for sufficient cells numbers when pooled to re-plate and expand for cryopreservation, Western blot, flow cytometry, and ADCC assays. Treatments were applied for 72 hours and then the flasks were aspirated, the cells were washed, and the remaining adherent cells were collected by trypsinization. Viable cell density for each treatment was assessed by trypan blue exclusion. Forty-nine additional challenges were conducted. Resistance in ADCC treatment groups was assessed by morphology, cell proliferation rate, and ADCC assay.
ADCC assay
ADCC assays were performed in 96-well, clear-bottom white plates (Corning, Cat. No. 3903) using the Cytotox-Glo Cytotoxicity assay (Promega, Cat. No. G291). ADCC assays were preformed using A431/ADCCS1/ADCCR1 as target cells and NK92-CD16V cells as the effector cells. Target cells are plated at 10,000 cells/well overnight (A431 cells double overnight) Specific lysis was assessed at 4 hours post exposure to NK92-CD16V cells (20,000 cells/well) at 1:1 E:T ratio in the presence or absence of cetuximab (1 μg/mL) or trastuzumab (5 µg/mL) as described (2). For assessment of specific lysis after blocking ICAM-1, cells were plated in medium containing the blocking antibody (10 µg/mL; BioLegend, Cat. No. 322703).
Western blot
Cells were lysed in boiling buffer with EDTA (Boston BioProducts) supplemented with 1X protease and 1% phosphatase inhibitor prepared following the manufacturer protocols (Sigma-aldrich, Cat.No. 11697498001 and P5726). Cleared lysate concentrations were obtained by a DC Protein Assay (BioRad). Lysates 30-40 μg were run on SDS-PAGE gels and transferred to nitrocellulose membranes (GE Healthcare). Western blots were conducted using: the Abcam antibodies to EGFR (Cat. No. 52892) and perforin (Cat. No. ab180773), and Cell Signaling Technology (CST) antibodies to GAPDH (Cat. No. 5174), JAK1 (Cat. No. 3332), STAT1 (Cat. No. 14994), p-STAT1 Y701 (Cat. No. 9167), PCAF/KA2B (Cat. No. 3378), Granzyme B (Cat. No. 4275), NFKB p65 (Cat. No. 82420), p-NFKB P65 (Cat. No. 3031S), and HSPB1 (Cat. No. 2402S). Goat anti-rabbit or donkey anti-mouse IgG HRP-conjugated secondary antibodies (GE Healthcare) were used with chemiluminescence substrates (Pierce). Densitometry was measured using ImageJ (v1.48).
NK cell activation assay
CD107α was used as a marker of NK cell degranulation and activation. ADCCS1 and ADCCR1 cells were seeded overnight in six-well plates at 500,000 and 700,000 cells, respectively. We looked at the effect of ADCCS1 and ADCCR1 cells on the activation of NK cells in the presence and absence of cetuximab. We also measured CD107α expression on unexposed NK cells, ADCCS1, and ADCCR1 cells to insure no auto-fluorescence and background. 1×106 NK cells and cetuximab for final 1µg/mL concentration were added to each well. The exposure time was two hours, after which, cells were collected and stained as described in the flow cytometry methods. Samples were run in the Georgetown Lombardi Comprehensive Cancer Center Flow Cytometry & Cell Sorting Shared Resource using BD LSRFortessa. Analyses were performed using FlowJo (v10.4.1).
NK cell conjugation assay
NK conjugation was assessed using a multi-well conjugation assay. Target cells (ADCCS1 or ADCCR1) were plated at a density of 10,000 cells per well on 96-well clear bottom black plates (Greiner, 655090) in FluoroBrite DMEM (Gibco) supplemented with 10% FBS and incubated overnight at 37°C, 5% CO2. NK92-CD16V cells at a density of 8 × 105 cells/mL in Dulbecco’s PBS were labeled with 5 µM carboxyfluorescein diacetate (Molecular Probes) for 20 minutes at 37°C, 5% CO2. The labelled NK92-CD16V cells were spun at 1500 rpm for 5 minutes and resuspended in NK medium (described above) and incubated for an additional 10 minutes at 37°C, 5% CO2. The labelled NK92-CD16V cells were spun again at 1500 rpm for 5 minutes and resuspended in the FluoroBrite DMEM to 8 × 105 cells/mL. NK cells (25 µl representing ~ 1:1 E:T) were added in sextuplets to target cells. Then, either 25 µL of medium or cetuximab (1 µg/mL) was added to target cells. As background, 50 µL of medium alone was added to a row of target cells. The plate was incubated at for 2 hours and then initial fluorescence was read using a PerkinElmer’s Envision 2104 Multilabel Reader set to 492/517 nm excitation/emission. Wells were emptied of non-adherent NK cells, washed twice with 200 µL of FluoroBrite DMEM, refilled with 150 µL FluoroBrite DMEM, and ending fluorescence was measured. Percentage of NK cells in conjugate was calculated as [(fluorescenceend-fluorescencebackground)/(fluorescenceinitial-fluorescencebackground)]*100. The mean of all replicates for each target cell line was then determined and SEM calculated.
Cell surface screen
The BD Lyoplate™ Human Cell Surface Marker Screening Panel (BD Biosciences, 560747) contains purified monoclonal antibodies to 242 cell surface markers. We compared ADCCS1 and ADCCR1 cell lines. Each cell line was screened twice. The cells were dissociated from flasks using BD Accutase™ (Cat. No.561527) and resuspended in BD Pharmingen™ Stain Buffer (FBS) (Cat. No. 554656) at 5 × 106 cells/mL. 100 μl/well (5 × 105 cells) were then dispensed into three 96-well round bottom plates (BD Falcon, 353910). The assay was conducted according to the manufacturer’s instructions. Samples were run in the Georgetown Lombardi Comprehensive Cancer Center Flow Cytometry & Cell Sorting Shared Resource using BD LSRFortessa. The flow cytometry analysis was done using FlowJo (v10.4.1).
Viability and proliferation assays
ADCCS1 and ADCCR1 cells were plated at 1000 cells/well and 2000 cells/well in 96 well plates (Fisher Scientific, Cat. No. 720089), respectively. Seven plates were prepared for each cell line to measure proliferation across seven days without treatment or with effector cell exposure. CellTiter-Blue (Promega) assays were conducted in 96-well format per manufacturer instructions on one plate per cell line for seven days to measure in vitro proliferation of ADCCS1 and ADCCR1. Prism Graphpad 5 was used to conduct two-tailed t tests and p value.
ELISA assays
Human IFNγ ELISA MAX™ Deluxe Kit (Biolegend, 430104) was used to measure IFNγ in the media four hours post ADCC exposure. ADCCS1 and ADCCR1 cells were plated in 96-well clear bottom plates (Corning, 3300) at 10,000 cells/well and incubated in culture conditions overnight at 37°C in 5% CO2. The control wells were then exposed to either media, cetuximab (1 μg/mL), or NK92-CD16V cells at the indicated E:T ratios in the absence of antibody. The ADCC wells all were incubated with cetuximab (1 μg/mL) and NK92-CD16V cells reflecting E:T ratios of 0:1, 1:1, 2:1 and 4:1 by adding 0; 20,000; 40,000; and 80,000 NK cells, respectively, to the wells. After 4 hours incubation, the plates were spun down at 1000 × g for 5 minutes, and the supernatant was collected and transferred into a fresh round bottom plate. IFNγ detection in supernatants was done using the ELISA Max Deluxe Kit (Biolegend, Cat. No. 430105) according to the manufacturer’s instructions.
In vivo tumor growth
Cohorts of ten 6-8 week-old female BALB/c nude mice were injected subcutaneously (s.c.) in the right flanks with 1 × 106 cells of ADCCS1 or 2 × 106 ADCCR1 cells suspended in 100 μL PBS. Tumor size was monitored twice weekly and measured using a caliper, and the volume was calculated using the following formula: Volume = (1/2)*length*width. Animals were euthanized when tumors reached 2 cm in the largest diameter or exhibited undue suffering. All animal experiments were carried out with Georgetown University Institutional Animal Care and Use Committee approval.
RNA isolation and gene expression analysis
Six pairs (12 total samples) of serially passaged vehicle-treated ADCCS1 cells and ADCCR1 cells from challenges 30-35 were passaged twice without treatments and collected by trypsinization. RNA was isolated using the PureLink RNA Mini Kit (Ambion). RNA quality was assessed for quality by Bioanalyzer (Agilent) for an RNA Integrity Number (RIN) >6. The direct hybridization assay method (as per manufacturer’s instructions) was used to generate biotin-labeled cRNA from 100 ng of RNA, which was then hybridized to the HumanHT-12 v4 Expression BeadChip, washed, and scanned per manufacturer’s instructions (Illumina). All data were obtained from a single BeadChip. Data has been submitted to GEO repository, GEO accession number GSE114545.
Data were preprocessed with log2 variance stabilization and quantile normalization using the R/Bioconductor package lumi (7) and subset to detected probes. Differential expression analysis was performed with the R/Bioconductor package LIMMA (8), using unpaired, empirical Bayes moderated t-tests to compare sensitive and resistant cells. Probes with FDR adjusted p-values below 0.01 were called statistically significant.
CoGAPS analysis (9) and PatternMarker statistics (10) was performed for time course analysis. Probes with less than one log fold-change between any two samples were filtered from analysis. Mean and standard deviation for probes annotated to the same gene were computed. Standard deviations were assigned to be the maximum of 10% of the mean gene expression value or standard deviation computed across all probes. These gene level data summaries were input to CoGAPS and the algorithm was run for a range of 2-8 patterns, with 5 found to be optimal fit based upon ClutrFree analysis (11). Three of the five patterns inferred changes in transcription across the passages and two stable changes between sensitive and resistant cells across all passage numbers, the latter of which were selected for further analysis. PatternMarker genes for the pattern upregulated in resistant cells were input to STRING (12) (version 6.2) to generate networks. Gene-level expression values were z-scored across all samples and visualized in the STRING network using the R package network.
Sample preparation for proteomics and phosphoproteomics
Cell pellets from ADCCS1 and ADCCR1 cells were resuspended in lysis buffer containing 50 mM Tris HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 5 mM EDTA, 1× Protease Inhibitor Cocktail (Roche, Cat. No. 04693132001)) and 1x Phosphatase Inhibitor Cocktail (Sigma, Cat. No. P5726). The suspension was sonicated using a probe-tip ultrasonic processor (Vibra Cell™; with the AMPL setting of 30%) 2 times for 10 seconds and spun down at 12,000 × g for 15 minutes. The supernatant was collected, with proteins extracted by methanol/chloroform precipitation (13). The precipitated proteins were then dissolved in 8 M urea and 50 mM triethylammonium bicarbonate, pH 8, with the protein concentration determined by the BCA assay (Thermo Fisher, Cat. No. 23225). Equal amounts (50 µg for proteomics and 300 µg for phosphoproteomics) of proteins from each sample were reduced with 10 mM DTT for 30 minutes at 37 °C and alkylated with 30 mM iodoacetamide for 30 minutes at room temperature in the dark, followed by quenching with 10 mM DTT for another 30 minutes. After decreasing the urea concentration with 50 mM triethylammonium bicarbonate to 1 M, sequencing-grade trypsin (Promega) was added and incubated overnight at 37 °C. After acidification with trifluoroacetic acid (final: 2%), tryptic digests were desalted with C18 spin columns (Nest Group) and dried with a SpeedVac. Each sample was then labeled with one isotopic reagent in a 6-plex iTRAQ labeling kit (Sciex) according to the manufacturer instructions. Differentially labeled peptides were then pooled and dried by vacuum centrifugation. Dried peptide mixtures were then fractionated with an Agilent 1260 Infinity HPLC system by using a C18 column (3.5 µm 2.1 × 100 mm XTerra MS; for proteomics) or another C18 column (5 µm 4.6 × 250 mm xBridge; for phosphoproteomics) with a 60-minute gradient of buffer A (20 mM ammonium formate in H2O, pH 10) and buffer B (20 mM ammonium formate in ACN, pH10). All the fractions were collected (one fraction for every 1 minute) and combined into 12 fractions with a concatenation method (14). Phosphoproteomic samples were processed with one more step: after being dried with a SpeedVac, phosphopeptides in each fraction were enriched with a Titansphere Phos-Tio Kit (GL Sciences), according to manufacturer instructions.
NanoUPLC-MS/MS
Dried peptides and phosphopeptides from each fraction were dissolved into 20 µL of 0.1% formic acid. Each sample (1 µL for proteomics and 10 µL for phosphoproteomics) was loaded onto a C18 Trap column (Waters Acquity UPLC Symmetry C18 NanoAcquity 10 K 2G V/M, 100 A, 5 µm, 180 µm × 20 mm) at 15 µL/minute for 4 minutes. Peptides were then separated with an analytical column (Waters Acquity UPLC M-Class, peptide BEH C18 column, 300 A, 1.7 µm, 75 µm × 150 mm), which was temperature controlled at 40°C. The flow rate was set at 400 nL/minute. A 90-minute gradient of buffer A (2% ACN, 0.1% formic acid) and buffer B (0.1% formic acid in ACN) was used for separation: 1% buffer B at 0 minute, 5% buffer B at 1 minute, 40% buffer B at 80 minutes, 99% buffer B at 85 minutes, 99% buffer B at 90 minutes. The gradient went back to 1% buffer B in 10 minutes, with the column equilibrated with 1% buffer B for 20 minutes. Data were acquired using an ion spray voltage of 2.3 kV, GS1 5 psi, GS2 0, CUR 30 psi and an interface heater temperature of 150°C. Mass spectra were recorded with Analyst TF 1.7 (AB SCIEX™) in the information-dependent acquisition (IDA) mode. Each cycle consisted of a full scan (m/z 400-1600) and fifty information dependent acquisitions (IDAs) (m/z 100-1800) in the high sensitivity mode with a 2+ to 5+ charge state. Rolling collision energy was used, with iTRAQ reagent collision energy adjustment on.
Proteomic and phosphoproteomic data analysis
Data files were submitted for simultaneous searches using Protein Pilot version 5.0 software (Sciex) utilizing the Paragon and Progroup algorithms (15) and the integrated false discovery rate (FDR) analysis function (16). MS/MS data was searched against the NCBI Homo Sapiens Proteome (UP000005640) of the UniProt-Sprot database containing 20,316 entries (Filtered by reviewed and downloaded on June 2, 2015). For proteomics, ‘Trypsin’ was selected as the enzyme, ‘Carbamido-methylation’ was set as a fixed modification on cysteine. Variable peptide modifications included methionine (M) oxidation and iTRAQ labeling of the N-terminal lysine (K) and tyrosine (Y). For phosphoproteomics, search parameters were set as: sample type [iTRAQ-8plex], cys alkylation (Iodoacetamide), digestion (Trypsin), instrument (TripleTOF 5600), special factors (phosphorylation emphasis), species (Homo Sapiens), ID Focus (Biological modifications), database (uniprot_sprot.fasta), search effort (Thorough), false discovery rate (FDR) analysis (Yes), and user-modified parameter files (No). The proteins were inferred based on the ProGroupTM algorithm associated with the ProteinPilot software. Peptides were defined as redundant if they had identical cleavage site(s), amino acid sequence, and modification. All peptides were filtered with confidence to 5% FDR, with the confidence of phosphorylation sites such as phospho-serine (p-Ser), phospho-threonine (p-Thr), and phospho-tyrosine (p-Tyr) automatically calculated. Quantitative phosphopeptide selection criteria are as follows: 1) The phosphopeptides without quantitative information were discarded. 2) The phosphor-peptides that were annotated with “auto-discordant peptide-type” and “auto-shared MS/MS” were excluded. For both data sets the detected protein threshold in the software was set to the value which corresponded to 1% FDR. Automatic normalization of quantitative data (bias correction) was performed to correct any experimental or systematic bias.
Statistics
Statistical analysis done in in vitro cell proliferation, in vivo tumor growth, specific lysis, Target:NK cells conjugation cell viability was two-tailed t tests conducted using prism GraphPad 5. Gene expression analysis was conducted via R/Bioconductor package lumi (7-8), and data time course analysis using CoGAPS analysis (9) and PatternMarker statistics (10). Proteomic and phosphor proteomic analysis was conducted using Paragon and Progroup algorithms (15) and the integrated false discovery rate (FDR) analysis function(16).
Measures of mRNA expression, proteomic and phosphoproteomic peptide counts were normalized by mean-centered scaling across sample groups (Z-score) using R to provide comparable distributions between assay types.
Results
Deriving resistance to ADCC
Previously, we have shown that A431 cells are sensitive to cetuximab-mediated ADCC (2), using a model system consisting of EGFR-overexpressing A431 cells, NK92-CD16V, and cetuximab (Fig. 1A). In order to explore mechanisms of resistance to ADCC, we continuously exposed A431 cells in vitro to ADCC conditions for 30~50 challenges, consisting of the addition of fresh NK cells and cetuximab every three days following the removal of exhausted media and non-adherent cells (Fig. 1B). A431 cell survival in response to ADCC conditions (Fig. 1C) showed the most target cell death (90%) at 24hrs. After 72hrs. of ADCC exposure, more than a 90% difference in cell survival between ADCC treated cells and untreated cells (0.6 × 106 and 9.3 × 106 cells, respectively) was still observed. A431 cell numbers had only recovered to 25% of their pretreatment baseline at 48 hours post challenge but were almost fully recovered at 72 hours, demonstrating that 72 hour cycles of ADCC conditions permitted sufficient target cell killing and recovery of residual viable A431 cells to generate conditions permissive for the emergence of ADCC-resistant cells.
Figure 1: ADCC model system and the derivation of ADCC resistance.
A, An in vitro NK cell-mediated ADCC model system consisting of an NK-like cell line (NK92-CD16V), an EGFR monoclonal antibody (cetuximab; red-filled circles), and EGFR-expressing A431 cells. The combination of target, effector, and antibody create optimal conditions for ADCC. B, Schematic of the work flow of the four conditions of continuous exposure. Untreated control, cetuximab (1 μg/ml)-treated contro,l and NK cell-mediated ADCC in absence and presence of cetuximab (1 μg/ml). C, Time course of A431 cell survival in response to ADCC exposure conditions. A431 cells were seeded and exposed to ADCC conditions for the indicated times, as described in Materials and Methods. ***, p < 0.001 by two-tailed t-test across all time points as indicated on graph. Error bars represent SEM. D, Specific lysis of 20,000 ADCCR1 cells and 20,000 ADCCS1 cells by NK92-CD16V cells in the presence of cetuximab (1 µg/mL) for 4 hours at a 1:1 E:T ratio. **, p < 0.01 by two-tailed t-test. Error bars represent SEM. E, In vitro proliferation of ADCCS1 cells and ADCCR1 cells in absence of ADCC conditions. ***, p < 0.001 and **, p < 0.01 by two-tailed t-test for day 2-6 and day 7, respectively. Error bars represent SEM. F, Growth of subcutaneous tumors derived from ADCCS1 and ADCCR1 cells in Balb/c nude mice. N = 10 in each group. p-value calculated by two-tailed t-test as indicated on graph. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars represent SEM. G, Influence of secreted factors by ADCCR1 cells on ADCC sensitivity of ADCCS1 cells. Bar graph shows ADCCR1 (R1) cells compared to mixed ADCCR1/ADCCS1 (S) cells at indicated percentages. Error bars represent SEM.
Properties of ADCCR1 cells
After 34 consecutive ADCC challenges, the surviving A431 cells (designated ADCCR1) demonstrated slower proliferation, morphologic changes, and an increased number of cells surviving the ADCC challenge. ADCC sensitivity was assessed and quantified by measuring specific lysis in ADCCR1 cells compared to contemporaneously cultured but untreated A431 cells (ADCCS1). There was a significant difference between ADCC-induced specific lysis in ADCCR1 and ADCCS1 cells (p<0.01 by two-tailed t-test) (Fig. 1D).
In comparison with ADCCS1 cells, ADCCR1 morphology was elongated with a ‘spindle-like’ appearance reminiscent of fibroblasts, with apparent contrast at cell margins. ADCCR1 cells displayed less distinct colony or clonal organization, with a tendency for reduced cell-cell contact (Supplementary Fig. S1). Both in vitro proliferation (Fig. 1E) and in vivo subcutaneous xenograft growth in nude mice (Fig. 1F) with ADCCR1 tumors was significantly slower compared to ADCCS1 tumors. ADCCR1 cell proliferation was reduced by 50% (***p<0.001 by two-tailed t test). Mice bearing ADCCS1 and ADCCR1 tumors had median survivals of 15 and 33 days, respectively.
We considered the possibility that ADCCR1 cells secrete factors that mediate ADCC resistance, and addressed this by admixing ADCCS1 and ADCCR1 cells at varying ratios, and also reciprocally substituting supernatants from ADCCS1 cells with media from ADCCR1 cells (Fig. 1G). Specific lysis correlated with the proportion of ADCCS1 cells added to ADCCR1 (p = 0.003, R2 = 0.904), whereas the media exchanges had no effects on cytotoxicity.
ADCCR1 and ADCCS1 cells possessed significantly different phosphoproteomic and proteomic profiles (Supplementary Fig. S2A-B). Amongst the phosphorylated proteins with statistically significantly altered phosphorylation in ADCCR1 vs. ADCCS1 cells, a general tendency towards hyper-phosphorylation of proteins in the ADCCR1 cells was seen (Supplementary Fig. S2B). Only five proteins were selectively hypo-phosphorylated in the ADCCR1 cells (Supplementary Fig. S2C). The protein, phosphoprotein, and mRNA of selected proteins in ADCCR1 cells compared to ADCCS1 cells showed a similar pattern across the data sets.
Relation of EGFR expression to ADCC resistance
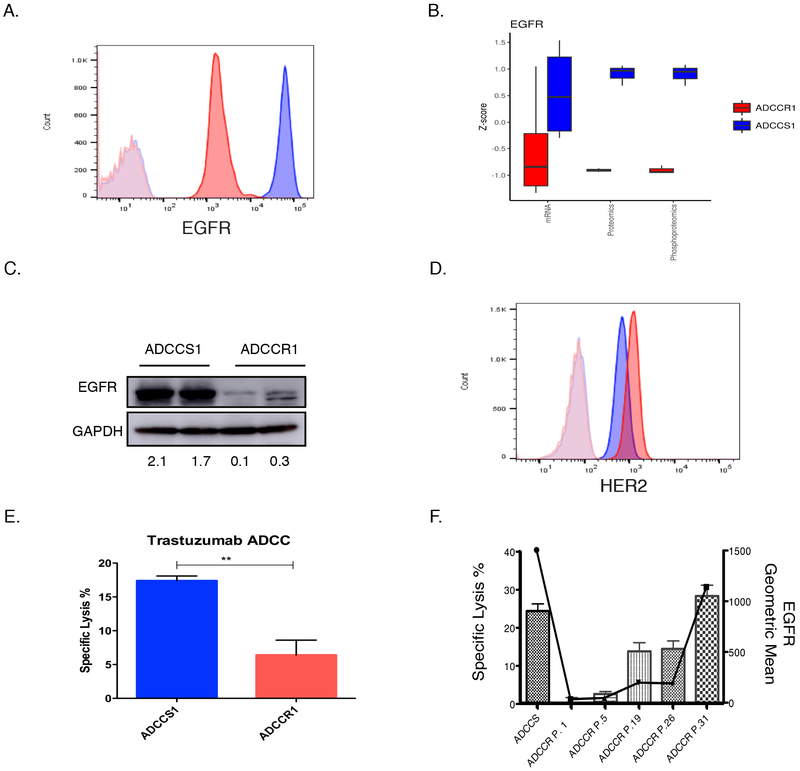
EGFR is the target of cetuximab. Therefore, we investigated the role of EGFR in the ADCCR1 cells to better understand the EGFR association with the ADCC resistance phenotype. EGFR was significantly reduced on the cell surface of ADCCR1 cells compared to ADCCS1 cells (Fig. 2A). EGFR protein had concordantly reduced gene expression in the ADCCR1 cells (Fig. 2B). Reduced EGFR protein expression was also found by proteomic analysis and western blot, and reduced EGFR phosphorylation was demonstrated by phosphoproteomic analysis (Fig. 2B-C).
Figure 2: EGFR and HER2 expression in ADCCS1 and ADCCR1 cells.
A, A representative flow cytometry analysis of EGFR cell surface expression in ADCCS1 (blue) cells and ADCCR1 (red) cells with isotype control expression for ADCCS1 (light blue) and ADCCR1 (light red). B, EGFR expression in ADCCS1 and ADCCR1 cells measured by mRNA, proteomic, and phosphoproteomic analysis. Measures of mRNA expression as well as proteomic and phosphoproteomic peptide counts were normalized by mean-centered scaling across sample groups (Z-score) to provide comparable distributions between assay types. Analysis were done on ADCCS1 and ADCCR1 cells post 33 challenges passaged for 3-4 times. C, Western blot for EGFR protein expression in ADCCS1 and ADCCR1 cells. ADCCS1 and ADCCR1 cells post 33 challenges passaged for 3-4 times were used. Densitometry values of expression relative to GAPDH indicated below the blot. D, Representative flow cytometry analysis of HER2 cell surface expression in ADCCS1 (blue) and ADCCR1 (red) cells with isotype control expression for ADCCS1 (light blue) and ADCCR1 (light red). E, Specific lysis of ADCCR1(target) and ADCCS1(target) cells by NK92-CD16V(effectors) cells at a 1:1 E:T ratio in the presence of trastuzumab (5 µg/mL) for 4 hours. **, p < 0.01 by two-tailed t-test. F, ADCC-induced specific lysis percentage (bars) and corresponding EGFR expression geometric mean by flow cytometry (solid line) in ADCCS1 cells and in ADCCR1 cells as a function of serial in vitro passaging (P. denotes passage number) following the cessation of ADCC exposure.
We next assessed whether the loss of EGFR was responsible for the ADCC-resistant phenotype exhibited by ADCCR1 cells. We previously demonstrated that EGFR knockdown in parental A431 cells results in a moderate reduction of sensitivity to ADCC (2). Although the EGFR surface expression, measured by flow cytometry, in the cells with EGFR knockdown were similar to what we observed in ADCCR1 cells, they did not exhibit the complete ADCC resistance displayed by ADCCR1 cells. This indicated that although loss of EGFR contributed to ADCC resistance in ADCCR1 cells, it was not the sole mediator of resistance. We next examined the ADCC sensitivity of ADCCR1 cells using a different antibody target. ADCCR1 and ADCCS1 cells expressed HER2 to similarly (Fig. 2D). However, ADCCR1 cells displayed resistance to ADCC mediated by trastuzumab (Fig. 2E). This suggested that additional ADCC resistance mechanisms, beyond EGFR loss, mediate the ADCCR1 phenotype.
ADCC resistance and the EGFR-loss phenotype were not durable in the absence of continued ADCC selection pressure. When ADCCR1 cells were cultured in the absence of cetuximab and NK92-CD16V cells, the expression of EGFR slowly returned to that of wild-type A431 cells over 31 passages (approximately three months; Fig. 2F). The restoration of ADCC sensitivity had some correlation with EGFR recovery, and ADCC sensitivity returned rapidly, even with minimal increases in EGFR surface expression.
Overexpression of interferon- and histone-associated genes in ADCCR1 cells
To investigate the difference between ADCC-resistant and -sensitive cells, we examined the gene expression profile of ADCCR1 and ADCCS1 cells using the Illumina HumanHT-12 v4 Expression BeadChip array. The cell lines showed distinct transcriptional profiles (Fig. 3A). EGFR and HSPB1 showed the most significant loss of gene expression in the ADCCR1 cells, whereas CD74 was the most enriched (Fig. 3B). Although HSPB1 loss was found consistently across data sets and validated by western blot (Supplementary Fig. S3A-B), overexpression of HSPB1 in ADCCR1 cells did not re-sensitize ADCCR1 cells to ADCC (Supplementary Fig. S3C-D). Elevation of CD74 in ADCCR1 cells compared to ADCCS1 cells was observed in the proteomic analysis, in addition to the gene expression analysis (Supplementary Fig. S3E). Total CD74 protein in the cell was significantly higher. However, CD74 was not present on the cell surface (Supplementary Fig. S3F). The cytosolic intracellular domain of CD74 is known to regulate the transcription of cell survival genes (17). When pro-survival molecules known to associate with CD74 were examined, we found a selective activation of RELA (NFκB p65) in ADCCR1 cells (Fig. 3C).
Figure 3: Association of interferon response and histone gene expression with ADCC resistance.
A, Heat map of gene expression assessed by whole genome Illumina bead arrays in ADCCS1 and ADCCR1 cells. Differential gene expression analysis was conducted for genes possessing at least 2-fold changes and adjusted false discovery rates (FDR) of p < 0.01. The heat map is based on hierarchical clustering of both samples (columns) and probes (rows) and contains 388 total probes for 334 unique genes. Reduced (blue) and increased (red) gene expression is shown based on z-score assessment across each probe (row). B, Volcano plot of differential gene expression in ADCCR1 compared to ADCCS1 cells. Differential gene expression analysis was conducted for genes possessing adjusted false discovery rates (FDR) of p < 0.01. The dotted line (vertical) indicates the p-value threshold 4 and −4 Log2 FC indicating significantly upregulated and downregulated genes respectively (red). C, Western blot of JAK1, STAT1, NFκB p65, and p-NFκB p65 in ADCCS1 and ADCCR1 cells. ADCCS1 and ADCCR1 cells post 33 challenges passaged for 3-4 times were used. Densitometry values of expression relative to GAPDH indicated below the blots. D, Diagram of 300 genes found to be upregulated in ADCCR1 cells compared to ADCCS1 cells by CoGAPS analysis. Overexpressed (red) and under-expressed (green) genes in ADCCR1 compared to ADCCS1 cells. The interferon-induced and histone-associated gene clusters are identified in the lower right portion of the diagram within hatched boxes.
Analysis of ADCCS1 and ADCCR1 cells from challenges 30 to 35 was performed with the Coordinated Gene Activity in Pattern Sets (CoGAPS) algorithm (9), using the time course analysis pipeline from Stein-O’Brien et al. (18). The PatternMarker statistic for CoGAPS (10) identified 300 genes with consistent upregulation and 450 genes with consistent downregulation in ADCCR1 cells compared to ADCCS1 cells across all challenges. The 300 genes upregulated in ADCCR1 cells contained clusters of interferon-associated and histone-associated genes (Fig. 3D). No observed upregulation in gene expression of any known marker for epithelial to mesenchymal transition was seen.
Ingenuity Pathway Analysis was used to analyze the expression pattern of genes upregulated in ADCCR1 cells. Interferon signaling, antigen presentation, and communication between innate and adaptive immune cells were the top canonical pathways identified (Supplementary Fig. S4A), and IFNγ was found to be a top upstream regulator of these cells with a p value of overlap 1.62 × 10−35 (Supplementary Fig. S4B). Although IFNγ itself was not significantly overexpressed in these cells, proteins downstream of IFNγ were overexpressed in ADCCR1 compared to ADCCS1 cells (JAK1, STAT1), further supporting activation of IFN signaling (Fig. 3C).
Upregulated histone-associated gene expression (Supplementary Table S1) pointed to a possible epigenetic mechanism driving ADCC resistance. KAT2B, a p300-associated histone acetyltransferase found within this histone cluster, was relatively overexpressed in ADCCR1 cells compared with ADCCS1 cells (Fig. 4A). ADCC resistance was partially reversed by inhibition of p300 using the histone acetyltransferase inhibitor C646 (Fig. 4B). No re-sensitization to ADCC in ADCCR1 cells was seen when using pan-HDAC, histone demethylase, or DNMT inhibitors (Fig. 4C).
Figure 4: Effects of pharmacological modification of histone-associated proteins identified by CoGAPS gene expression analysis on ADCC sensitivity.
A, Western blot of PCAF (KAT2B) in ADCCS1 and ADCCR1 cells. ADCCS1 and ADCCR1 cells post 33 challenges passaged for 3-4 times were used. Densitometry values of expression relative to GAPDH indicated below. B, Specific lysis of ADCCS1 (blue) and ADCCR1 (red) measured by ADCC assay at 4 hrs when pre-treated with increasing concentrations of histone acetyl transferase inhibitor (HATi) C646 for 2 hours. C, Specific lysis of ADCCS1 (blue) and ADCCR1 (red) measured by ADCC assay at 4 hrs when pre-treated with increasing concentrations of DNMT (DNMTi), pan-HDAC (pan-HDACi), and histone demethylase inhibitors.
ADCCR1 cells fail to activate or bind NK cells
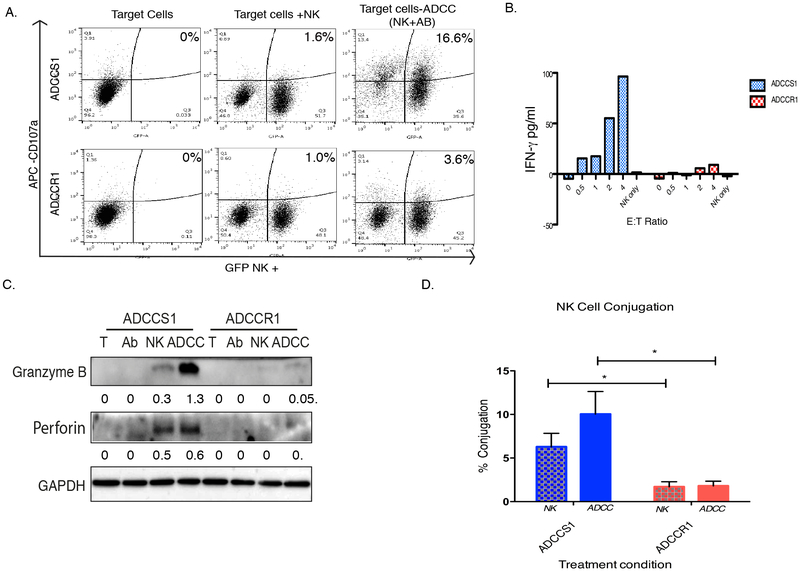
We examined whether the resistance to ADCC-mediated lysis in ADCCR1 cells was due to an intrinsic mechanism (resistance to perforin/granzyme or blocking apoptosis) or to defective cell:cell conjugation. To assess NK activity, expression of CD107α, a marker of NK degranulation and activation, were quantified in the NK92-CD16V cells two hours post exposure to target cells in the absence or presence of cetuximab (Fig. 5A). CD107αwas -exposed NK cells compared with ADCCR1-exposed NK cells (t test, p <0.0001). We also measured IFNγ secreted in the media post-ADCC (Fig. 5B). A correlation between the number of NK cells added and the amount of IFNγ released when using ADCCS1 cells was observed (p = 0.001, R2 = 0.982), whereas no IFNγ was released at any effector to target ratio with ADCCR1 cells. Taken together, these results indicated that ADCCR1 cells failed to activate NK cells even in ADCC conditions. This was further verified by the absence of perforin and granzyme B released upon exposure of ADCCR1 cells to NK cells for one hour in the absence (NK lane) or presence of cetuximab (1 μg/mL; ADCC lane) by Western blot (Fig. 5C). In contrast, exposure of ADCCS1 cells to NK cells resulted in detectable levels of granzyme B and perforin by Western blot, even when exposed to NK cells alone.
Figure 5: NK cell activation and conjugation to EGFR+ target cells under ADCC conditions.
A, Representative dot plots of NK activation measured by flow cytometry analysis using CD107α (APC) and GFP+ NK92-CD16V cells at a 1:1 E:T for 2 hours as described in Materials and Methods. ADCCS1 (top row) and ADCCR1 cells (bottom row) were incubated with NK92-CD16V cells in the absence (middle panels) or in the presence of cetuximab (1 µg/mL; right panel) for 2 hours. B, ELISA measuring IFNγ levels in the media of ADCCS1 (blue bars) and ADCCR1 (red bars) 4 hours post exposure to ADCC conditions (cetuximab [1 µg/mL] plus NK92-CD16V cells) at E:T ratios of 0-4:1 and NK92-CD16V cells in the absence of cetuximab. C, Western blot analysis of granzyme B and perforin protein expression in target cells two hours after exposure to T (media control), Ab (cetuximab only at 1µg/mL), NK (NK92-CD16V cells only at 1:1 E:T) and ADCC (NK92-CD16V cells at 1:1 E:T plus 1 µg/mL cetuximab). D, Percentage of NK cells conjugated to target was measured by a multi-well conjugation assay as described in Materials and Methods. ADCCS1 (blue) and ADCCR1 (red) were incubated with NK92-CD16V cells at a 1:1 E:T ratio in the absence (NK, checkered bar) or in the presence of cetuximab (1 µg/mL; ADCC, solid bar) for 2 hours. *, p < 0.05 by two tailed t test.
To further investigate ineffective NK cell activation by ADCCR1 cells, we examined whether effector:target cell conjugation was occurring after treatment with NK cells in the presence or absence of cetuximab (1 μg/mL) for four hours. ADCCS1 cells conjugated to NK92-CD16V cells effectively in the presence and absence of cetuximab, whereas ADCCR1 cells’ ability to conjugate was significantly less in both conditions (Fig. 5D). Therefore, ADCCR1 cells failed to activate NK cells and resisted ADCC-mediated lysis by avoiding NK cell conjugation.
ADCCR1 cells exhibit reduced expression of multiple cell surface proteins
We conducted a BD Lyoplate cell surface molecule screen to better understand the differences in conjugation of NK92-CD16V cells to ADCCS1 and ADCCR1 cells. Many ADCCR1 cell surface molecules were reduced compared to ADCCS1 cells, including cell adhesion molecules that play a role in the immune response, such as CD54 (ICAM-1), CD81 (TAPA-1), CD59, CD58, CD9, and HLA-A, B, and C (Fig. 6A-B).
Figure 6: Cell Surface Screen of ADCCS1 and ADCCR1 Cells.
A, Dot plot comparing geometric means of ADCCS1 and ADCCR1 cell surface molecule expression measured by the BD Lyoplate assay as described in Materials and Methods. Molecules with highest differential cell surface expression in ADCCS1 cells are shown in the box. BD lyoplate screen geometric means of ADCCS1 and ADCCR1 are in Supplementary Table S1. B, Geometric means of molecules with the highest differential expression (box in Fig. 6A) in ADCCS1 (blue) compared to ADCCR1 (red) cells. C, Representative histograms of selected cell surface molecules with reduced cell surface expression on ADCCR1 cells based on lower protein expression. Light gray histograms: negative control. Dark gray histograms: expression of total protein in permeabilized ADCCR1 cells. Open histograms: cell surface expression of indicated molecule in ADCCR1 cells. D, Representative histograms of selected cell surface molecules with reduced cell surface expression based on reduced transport to cell surface. Light gray histograms: negative control. Dark gray histograms: expression of total protein in permeabilized ADCCR1 cells. Open histograms: cell surface expression of indicated molecule in ADCCR1 cells.
ICAM-1, a known LFA-1 ligand, was significantly downregulated in ADCCR1 cells, and LFA-1/ICAM-1 interactions are essential for NK cell activation (19-23). Blocking LFA-1/ICAM-1 interactions significantly reduced ADCC sensitivity in ADCCS1 cells (Supplementary Fig. S5). CD81 and CD9 are tetraspanin proteins that play roles in adhesion and formation of the immune synapse (24,25). Tetraspanins are known to associate at the immune synapse with receptors and integrins, including ICAM-1 and LFA-1 (26-28). No upregulation of known immune checkpoints, including PD-L1, in ADCCR1 cells was observed (Supplementary Fig. S6).
We found that the reduced presence of select molecules on the cell surface did not necessarily correspond to a reduction in mRNA expression in ADCCR1 cells, with the exception of EGFR. Although some adhesion molecules with reduced cell surface expression had concomitant reductions in protein expression (Fig. 6C), several molecules found to be downregulated in ADCCR1 cells on the cell surface did not have reduced protein expression (Fig. 6D), suggesting a failure of transport to the cell surface. Total BD Lyoplate geometric mean values of ADCCS1 and ADCCR1 are in Supplementary Table S2.
Rederivation of ADCC resistance
To shed light on the sequence of events as ADCC resistance develops, we re-derived ADCC resistance from parental A431 cells, monitoring specific lysis under ADCC conditions, cell surface EGFR expression, proliferation, and cellular morphology. Changes in morphology towards the appearance of ADCCR1 cells were first observed at challenge 27, whereas no significant changes in EGFR cell surface expression or specific lysis was seen (Supplementary Fig. S7A). ADCC resistance was first observed at challenge 39, with accompanying changes in cell proliferation, morphology, significantly reduced specific lysis, but no significant changes in cell surface EGFR expression, as was seen in ADCCR1 cells. Despite an additional ten ADCC challenges, these ADCC-resistant cells (ADCCR2). EGFR cell surface expression was reduced in ADCCR2 by only 45% as compared to the 70% in ADCCR1 (Supplementary Fig. S7B). Hence, the ADCC resistance phenotype in these cells was unrelated to changes in cell surface EGFR expression. Even without a significant loss of antibody target, the ADCCR2 line failed to activate NK cells (Supplementary Fig. S7C) and displayed reduced NK cell conjugation (Supplementary Fig. S7D). ADCCR2 cells also exhibited reduced ICAM-1 expression similar to ADCCR1 cells (Supplementary Fig. S7E), suggesting that ADCC resistance in both lines can be attributed to evasion of NK cell binding.
Discussion
This paper represents a comprehensive analysis of tumor cell-based resistance mechanisms to ADCC, which serves as a model for the induction of resistance to continuous immune attack. Hence, some of the mechanisms that emerge may be anticipated to occur in response to other mechanisms of immune attack such as cytotoxic T-cell attack through the formation of immune synapses. To assure consistency, we employed the NK92-CD16V cell line as effectors, which we and others have used to explore various facets of ADCC (2,4,29-31). This model system permitted us to explore mechanisms of ADCC resistance unrelated to known KIR molecule–related regulatory NK cell mechanisms, as we have previously demonstrated (2). Resistance mechanisms identified in the current study, thus, may be relevant to other forms of immune attack, such as T-cell receptor-mediated cytolysis. Future studies will address these possibilities.
We have isolated two distinct ADCC-resistant cell lines from A431 cells, termed ADCCR1 and ADCCR2. In comparison with parental ADCC-sensitive A431 cells, ADCCR1 is characterized by reduced cell surface expression of EGFR and other molecules associated with cell adhesion and immune synapse formation, reduced expression of HSPB1 (a known chaperone of EGFR), and increased expression of CD74 (a known MHC II chaperone and regulator of antigen presentation) (32). ADCCR1 cells had a distinct transcriptional profile characterized by upregulation of genes associated with interferon response and histone function. The ADCCR1 resistance phenotype was partially reversed by inhibition of the histone acetyltransferase p300. ADCCR1 cells proliferated more slowly than A431 cells, and ADCCR1 tumors also grew more slowly in nude mouse xenografts. The ADCCR2 cell line, which was induced using a similar ADCC selection pressure strategy, did not exhibit reduced cell surface EGFR expression.
In this model system, we demonstrated a loss of numerous cell surface molecules associated with adhesion and immune synapse formation. NK cell activation is a dynamic process mediated by multiple factors, many of which promote adhesion. ADCCR1 and ADCCR2 cells exhibited significant loss of ICAM-1, a known LFA-1 ligand that mediates the tight adhesion between target cells and cytotoxic lymphocytes required for cytotoxic activity of T-cells and NK cells (33,34). This was evident by the inhibitory effect of blocking LFA-1/ICAM-1 interactions on ADCC and NK cell natural cytotoxicity that we and others have observed (35-38). NFκB p65 activity has been inversely associated with ICAM-1 expression and consequently ICAM-1/LFA-1 binding and NK cytotoxicity (39-41). ADCCR1 cells overexpressed CD74 in the cytoplasm but not on the cell surface. This finding was of some interest, as the cytoplasmic tail of CD74 regulates NFκB activity (17,42,43).
In this model system, ADCC resistance was not associated with PD-L1 expression, in contrast to the findings described by others implicating IFNγ in the upregulation of PD-L1 and resistance (44,45). Our findings indicated that, in contrast to establishment of immune blockade, ADCCR1 and ADCCR2 cells achieved resistance to immune attack by lowering cell surface expression of molecules known to mediate cell adhesion and formation of the immune synapse in order to prevent immune cell conjugation. The distinct EGFR expression in ADCCR1 and ADCCR2 cells demonstrated that altered levels of EGFR expression are independent of changes in cell adhesion molecule expression and sensitivity to ADCC. Ultimately, reduced NK cell conjugation, activation, and degranulation are likely the result of a combination of factors, including but not limited to loss of cell adhesion receptors and/or EGFR. We speculate that the ADCC resistance phenotype described here represents an adaptive mechanism to shield cells from immune attack. Future studies in ADCCR2 cells and isolation of ADCC-resistant lines from other parental cell lines will determine whether this phenotype may have broader relevance.
The reversibility of the resistance phenotype, coupled with a histone-related gene signature in the ADCCR1 cells suggested this was an epigenetic phenomenon, linked to interferon response genes and CD74 upregulation, the induction of NFκB p65, and then modulation of cell surface receptor expression to reduce the conjugation of effector cells. Future work will shed light on the relationship between the epigenetic changes and cell surface receptor modulation. Although ADCC resistance is likely the result of multiple cellular changes, the modification of the immune synapse function is of particular interest. We speculate that similar phenotypes could be induced by powerful immune selection through ADCC or similar mechanisms that involve the formation of immune synapses. This hypothesis is supported by our findings that the resistance phenotype reverted back to the ADCC-sensitive phenotype after continuous culture in non-ADCC conditions.
These findings address important questions related to the induction of cellular resistance to ADCC and possibly other immune therapies. It has long been assumed that therapy-imposed selection pressures would induce genetic or epigenetic changes to permit targeted cells to escape immune control. Epigenetic modifications are established tumorigenic mechanisms (46). Earlier studies have linked anti-cancer drug resistance to epigenetic modification leading to transcriptional silencing of genes necessary for drug activation (47). However, its role in cancer immunopathology and immunotherapy is poorly understood. The IFNγ pathway has been linked to primary, adaptive, and acquired resistance to checkpoint blockade therapy (48-50). Prolonged exposure to IFN-γ, can lead to immune escape due to cell desensitization and immune editing (51,52). The immune system can be hindered by epigenetic changes within the target cell, which prevent the recruitment or activation of effector cells. A study has demonstrated that epigenetic suppression of TH1 chemokines suppresses cell trafficking to the tumor microenvironment (53). Multiple studies have shown that epigenetic modification by HDAC inhibitors alone (54,55) or in combination with DNMT inhibitors (56) can enhance immunotherapy.
It should be noted that tumor cell resistance to T-cell attack has been known to involve defective antigen presentation, depriving killer cells of their targets. Similarly, target antigen modulation is a known mechanism of resistance to monoclonal antibody therapy. Here, we showed that the induction of ADCC resistance could lead to the more general loss of target cell adhesion properties required for the establishment of an immune synapse, killer cell activation, and target cell cytotoxicity. In contrast to models of cellular cytotoxicity resistance that invoke the establishment of immune checkpoints, this work demonstrates that target cells can evade conjugation by rendering the cells invisible to the cytotoxic apparatus.
Supplementary Material
Acknowledgments
Grant Support
DSA was supported by a scholarship from the Government of Saudi Arabia and King Saud bin Abdulaziz University for Health Sciences. JCM was supported by an individual pre-doctoral NRSA fellowship, NCI F30-CA165474. LMW was supported by NCI grants R01 CA050633 and P30 CA051008. The Tissue Culture, Proteomics and Metabolomics, Flow Cytometry & Cell Sorting, Genomics & Epigenomics, and Microscopy & Imaging Shared Resources were supported by Lombardi Comprehensive Cancer Center Support Grant, NCI P30-CA051008. EJF was supported by NCI R01 CA177669, P30 CA006973, and the Johns Hopkins University Award.
Footnotes
Conflicts of Interest: Disclosed in forms submitted by Author
References
- 1.Herlyn M, Sears HF, Steplewski Z, Koprowski H. Monoclonal antibody detection of a circulating tumor-associated antigen. I. Presence of antigen in sera of patients with colorectal, gastric, and pancreatic carcinoma. J Clin Immunol. 1982;2:135–40. [DOI] [PubMed] [Google Scholar]
- 2.Murray JC, aldeghaither D, Wang S, Nasto RE, Jablonski SA, Tang Y, et al. c-Abl modulates tumor cell sensitivity to antibody-dependent cellular cytotoxicity. Cancer Immunol Res. American Association for Cancer Research; 2014;2:1186–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellucci R, Martin A, Bommarito D, Wang K, Hansen SH, Freeman GJ, et al. Interferon-γ-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression oncoimmunology. Taylor & Francis; 2015;4:e1008824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binyamin L, Alpaugh RK, Hughes TL, Lutz CT, Campbell KS, Weiner LM. Blocking NK cell inhibitory self-recognition promotes antibody-dependent cellular cytotoxicity in a model of anti-lymphoma therapy. J Immunol. NIH Public Access; 2008;180:6392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 melanoma cells--a loss for breast cancer, but a boon for melanoma research Breast Cancer Res Treat. 3rd ed. Kluwer Academic Publishers-Plenum Publishers; 2007;104:13–9. [DOI] [PubMed] [Google Scholar]
- 6.Weiner LM, Surana R, Wang S. Monoclonal antibodies: versatile platforms for cancer immunotherapy. Nat Rev Immunol. 2010;10:317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. [DOI] [PubMed] [Google Scholar]
- 8.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fertig EJ, Ding J, Favorov AV, Parmigiani G, Ochs MF. CoGAPS: an R/C++ package to identify patterns and biological process activity in transcriptomic data. Bioinformatics. 2010;26:2792–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein-O’Brien GL, Carey JL, Lee WS, Considine M, Favorov AV, Flam E, et al. PatternMarkers & GWCoGAPS for novel data-driven biomarkers via whole transcriptome NMF. Bioinformatics. 2017;33:1892–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bidaut G, Ochs MF. ClutrFree: cluster tree visualization and interpretation. Bioinformatics. 2004;20:2869–71. [DOI] [PubMed] [Google Scholar]
- 12.Mering von C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins Nucleic Acids Res. Oxford University Press; 2003;31:258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–3. [DOI] [PubMed] [Google Scholar]
- 14.Ma J, Hart GW. Analysis of Protein O-GlcNAcylation by Mass Spectrometry Curr Protoc Protein Sci. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2017;87:24.10.1–24.10.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, et al. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–55. [DOI] [PubMed] [Google Scholar]
- 16.Tang WH, Shilov IV, Seymour SL. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J Proteome Res. 2008;7:3661–7. [DOI] [PubMed] [Google Scholar]
- 17.Gil-Yarom N, Radomir L, Sever L, Kramer MP, Lewinsky H, Bornstein C, et al. CD74 is a novel transcription regulator. PNAS. 2017;114:562–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein-O’Brien G, Kagohara LT, Li S, Thakar M, Ranaweera R, Ozawa H, et al. Integrated time course omics analysis distinguishes immediate therapeutic response from acquired resistance. 2018;10(37):1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urlaub D, Höfer K, Müller M-L, Watzl C. LFA-1 Activation in NK Cells and Their Subsets: Influence of Receptors, Maturation, and Cytokine Stimulation. J Immunol. American Association of Immunologists; 2017;198:1944–51. [DOI] [PubMed] [Google Scholar]
- 20.Gross CC, Brzostowski JA, Liu D, Long EO. Tethering of intercellular adhesion molecule on target cells is required for LFA-1-dependent NK cell adhesion and granule polarization. J Immunol. American Association of Immunologists; 2010;185:2918–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryceson YT, Ljunggren H-G, Long EO. Minimal requirement for induction of natural cytotoxicity and intersection of activation signals by inhibitory receptors. Blood. 2009;114:2657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barber DF, Faure M, Long EO. LFA-1 contributes an early signal for NK cell cytotoxicity. J Immunol. 2004;173:3653–9. [DOI] [PubMed] [Google Scholar]
- 23.Bryceson YT, March ME, Barber DF, Ljunggren H-G, Long EO. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells J Exp Med. Rockefeller University Press; 2005;202:1001–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rocha-Perugini V, González-Granado JM, Tejera E, López-Martín S, Yáñez-Mó M, Sánchez-Madrid F. Tetraspanins CD9 and CD151 at the immune synapse support T-cell integrin signaling. Eur J Immunol. 2014;44:1967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rocha-Perugini V, Zamai M, González-Granado JM, Barreiro O, Tejera E, Yañez-Mó M, et al. CD81 controls sustained T cell activation signaling and defines the maturation stages of cognate immunological synapses. Mol Cell Biol. 2013;33:3644–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tardif MR, Tremblay MJ. Tetraspanin CD81 provides a costimulatory signal resulting in increased human immunodeficiency virus type 1 gene expression in primary CD4+ T lymphocytes through NF-kappaB, NFAT, and AP-1 transduction pathways. J Virol. American Society for Microbiology; 2005;79:4316–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rocha-Perugini V, Sánchez-Madrid F, Martínez Del Hoyo G. Function and Dynamics of Tetraspanins during Antigen Recognition and Immunological Synapse Formation. Front Immunol. 2015;6:653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittelbrunn M, Yáñez-Mó M, Sancho D, Ursa A, Sánchez-Madrid F. Cutting edge: dynamic redistribution of tetraspanin CD81 at the central zone of the immune synapse in both T lymphocytes and APC. J Immunol. 2002;169:6691–5. [DOI] [PubMed] [Google Scholar]
- 29.Paolino M, Choidas A, Wallner S, Pranjic B, Uribesalgo I, Loeser S, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells Nature. Nature Publishing Group; 2015;507:508–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tonn T, Becker S, Esser R, Schwabe D, Seifried E. Cellular immunotherapy of malignancies using the clonal natural killer cell line NK-92. J Hematother Stem Cell Res. 2001;10:535–44. [DOI] [PubMed] [Google Scholar]
- 31.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. [DOI] [PubMed] [Google Scholar]
- 32.Stumptner-Cuvelette P, Benaroch P. Multiple roles of the invariant chain in MHC class II function. Biochim Biophys Acta. 2002;1542:1–13. [DOI] [PubMed] [Google Scholar]
- 33.Marlin SD, Springer TA. Purified intercellular adhesion molecule-1 (ICAM-1) is a ligand for lymphocyte function-associated antigen 1 (LFA-1). Cell. 1987;51:813–9. [DOI] [PubMed] [Google Scholar]
- 34.Helander T, Timonen T, Kalliomäki P, Schröder J. Recognition of chromosome 6-associated target structures by human lymphokine-activated killer cells. J Immunol. 1991;147:2063–7. [PubMed] [Google Scholar]
- 35.Miedema F, Tetteroo PA, Hesselink WG, Werner G, Spits H, Melief CJ. Both Fc receptors and lymphocyte-function-associated antigen 1 on human T gamma lymphocytes are required for antibody-dependent cellular cytotoxicity (killer cell activity). Eur J Immunol. 1984;14:518–23. [DOI] [PubMed] [Google Scholar]
- 36.Balasa B, Yun R, Belmar NA, Fox M, Chao DT, Robbins MD, et al. Elotuzumab enhances natural killer cell activation and myeloma cell killing through interleukin-2 and TNF-α pathways Cancer Immunol Immunother. Springer Berlin Heidelberg; 2015;64:61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarahian M, Watzl C, Issa Y, Altevogt P, Momburg F. Blockade of natural killer cell-mediated lysis by NCAM140 expressed on tumor cells Int J Cancer. Wiley Subscription Services, Inc., A Wiley Company; 2007;120:2625–34. [DOI] [PubMed] [Google Scholar]
- 38.Anikeeva N, Steblyanko M, Fayngerts S, Kopylova N, Marshall DJ, Powers GD, et al. Integrin receptors on tumor cells facilitate NK cell-mediated antibody-dependent cytotoxicity. Eur J Immunol. 2014;44:2331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jewett A, Wang M-Y, Teruel A, Poupak Z, Bostanian Z, Park N-H. Cytokine dependent inverse regulation of CD54 (ICAM1) and major histocompatibility complex class I antigens by nuclear factor kappaB in HEp2 tumor cell line: effect on the function of natural killer cells. Hum Immunol. 2003;64:505–20. [DOI] [PubMed] [Google Scholar]
- 40.Ledebur HC, Parks TP. Transcriptional regulation of the intercellular adhesion molecule-1 gene by inflammatory cytokines in human endothelial cells. Essential roles of a variant NF-kappa B site and p65 homodimers. J Biol Chem. 1995;270:933–43. [DOI] [PubMed] [Google Scholar]
- 41.Rahman A, Anwar KN, True AL, Malik AB. Thrombin-induced p65 homodimer binding to downstream NF-kappa B site of the promoter mediates endothelial ICAM-1 expression and neutrophil adhesion. J Immunol. 1999;162:5466–76. [PubMed] [Google Scholar]
- 42.Starlets D, Gore Y, Binsky I, Haran M, Harpaz N, Shvidel L, et al. Cell-surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood. American Society of Hematology; 2006;107:4807–16. [DOI] [PubMed] [Google Scholar]
- 43.Gore Y, Starlets D, Maharshak N, Becker-Herman S, Kaneyuki U, Leng L, et al. Macrophage migration inhibitory factor induces B cell survival by activation of a CD74-CD44 receptor complex. J Biol Chem. 2008;283:2784–92. [DOI] [PubMed] [Google Scholar]
- 44.Abiko K, Matsumura N, Hamanishi J, Horikawa N, Murakami R, Yamaguchi K, et al. IFN-γ from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer Br J Cancer. Nature Publishing Group; 2015;112:1501–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang SC, Latchman YE, Buhlmann JE, Tomczak MF, Horwitz BH, Freeman GJ, et al. Regulation of PD-1, PD-L1, and PD-L2 expression during normal and autoimmune responses Eur J Immunol. WILEY‐VCH Verlag; 2003;33:2706–16. [DOI] [PubMed] [Google Scholar]
- 46.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host nature reviews cancer. Nature Publishing Group; 2013;13:497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nyce J, Leonard S, Canupp D, Schulz S, Wong S. Epigenetic mechanisms of drug resistance: drug-induced DNA hypermethylation and drug resistance. PNAS. National Academy of Sciences; 1993;90:2960–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-γ Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell. 2016;167:397–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discovery. American Association for Cancer Research; 2017;7:188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. Massachusetts Medical Society; 2016;375:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell. 2016;167:1540–1554.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity Nature. Nature Publishing Group; 2001;410:1107–11. [DOI] [PubMed] [Google Scholar]
- 53.Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy Nature. Nature Publishing Group; 2015;527:249–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terranova-Barberio M, Thomas S, Ali N, Pawlowska N, Park J, Krings G, et al. HDAC inhibition potentiates immunotherapy in triple negative breast cancer. Oncotarget. Impact Journals; 2017;8:114156–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pili R, Quinn DI, Hammers HJ, Monk P, George S, Dorff TB, et al. Immunomodulation by Entinostat in Renal Cell Carcinoma Patients Receiving High-Dose Interleukin 2: A Multicenter, Single-Arm, Phase I/II Trial (NCI-CTEP#7870). Clin Cancer Res. American Association for Cancer Research; 2017;23:7199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Topper MJ, Vaz M, Chiappinelli KB, DeStefano Shields CE, Niknafs N, Yen R-WC, et al. Epigenetic Therapy Ties MYC Depletion to Reversing Immune Evasion and Treating Lung Cancer. Cell. 2017;171:1284–1300.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.