Abstract
Parvoviruses are highly attractive templates for the engineering of safe, efficient, and specific gene therapy vectors, as best exemplified by adeno-associated virus (AAV). Another candidate that currently garners increasing attention is human bocavirus 1 (HBoV1). Notably, HBoV1 capsids can cross-package recombinant (r)AAV2 genomes, yielding rAAV2/HBoV1 chimeras that specifically transduce polarized human airway epithelia (pHAEs). Here, we largely expanded the repertoire of rAAV/BoV chimeras, by assembling packaging plasmids encoding the capsid genes of four additional primate bocaviruses, HBoV2–4 and GBoV (Gorilla BoV). Capsid protein expression and efficient rAAV cross-packaging were validated by immunoblotting and qPCR, respectively. Interestingly, not only HBoV1 but also HBoV4 and GBoV transduced pHAEs as well as primary human lung organoids. Flow cytometry analysis of pHAEs revealed distinct cellular specificities between the BoV isolates, with HBoV1 targeting ciliated, club, and KRT5+ basal cells, whereas HBoV4 showed a preference for KRT5+ basal cells. Surprisingly, primary human hepatocytes, skeletal muscle cells, and T cells were also highly amenable to rAAV/BoV transduction. Finally, we adapted our pipeline for AAV capsid gene shuffling to all five BoV isolates. Collectively, our chimeric rAAV/BoV vectors and bocaviral capsid library represent valuable new resources to dissect BoV biology and to breed unique gene therapy vectors.
Keywords: adeno-associated virus, AAV, bocavirus, BoV, capsid engineering
Introduction
For several decades, parvoviruses have been developed as safe, efficient, and versatile DNA delivery vectors for human gene therapy applications. In particular, recombinant adeno-associated viruses (rAAVs) have emerged as leading candidates due to their amenability to genetic modification and to their excellent safety profile, culminating in the authorization of Glybera1 and, most recently, Luxturna.2 Still, there remains room for improvement considering their limited cargo capacity of 4.7 kb, which hampers delivery of larger expression cassettes, such as those encoding Streptococcus pyogenes (Sp) Cas93 or cystic fibrosis transmembrane conductance regulator (CFTR).4 Additionally, the high pre-existing anti-AAV immunity in humans and the insufficient cell specificity of natural AAVs represent further clinical translation hurdles.
To date, numerous efforts have been made to overcome the limitations of AAV vectors. AAV variants derived by random5 or rational capsid engineering6 that exhibit lower antibody reactivity and/or higher cell specificity are particularly promising. Additionally, the size restriction of AAV vectors was partly alleviated by the use of mini- or micro-genes7 or, alternatively, by distributing large inserts over two or three distinct AAV genomes.8 Another solution is to harness other parvoviruses displaying clinically relevant properties. For example, autonomous parvoviruses such as H1, minute virus of mice (MVM), and LuIII are attractive for cancer therapy due to the propensity of the wild-type viruses to specifically replicate in, and lyse, proliferating cells, including tumor cells.9, 10 Akin to AAVs, they were also engineered as gutless vectors devoid of viral genes,11 albeit, in most cases, the non-structural coding region was retained to maintain selectivity for cells in S phase. The resulting replication-competent oncolytic viruses showed great potential for the treatment of glioblastoma, and they are presently under clinical investigation.12
Yet another promising mean to overcome the limitations of naturally occurring AAVs is the generation and use of pseudotyped vectors that originate from cross-packaging of a genome of one parvovirus into the capsid of another.11, 13, 14, 15 The resulting chimeric vector will ideally combine the best of both worlds, e.g., the low genotoxicity of an episomal rAAV genome with the specific tropism and/or larger cargo capacity of the heterologous second parvovirus. In one of the earliest reports, Ponnazhagan et al.13 packaged rAAV genomes into the B19 capsid, which allowed efficient and specific targeting of erythroid cells. Additionally, a chimeric H1/AAV vector, consisting of the tumor-specific replication gene and promoters of the autonomous parvovirus H1, was packaged into an AAV2 capsid, and it was designed to selectively kill tumor cells in vitro.16 Finally, the most recent report of a chimeric parvovirus vector was published in 2013.17 In this study, Yan et al.17 packaged an rAAV genome into the capsid of human bocavirus 1 (HBoV1), yielding rAAV/HBoV1 chimeras that specifically and potently transduced polarized human airway epithelia (pHAEs). This approach is very intriguing due to the larger size of the wild-type (WT) HBoV1 genome of 5.5 kb, as opposed to 4.7 kb for most AAV serotypes, implying a higher transgene packaging capacity. Indeed, Yan et al.17 were able to experimentally verify this predicted benefit of rAAV/HBoV1 hybrid vectors, by demonstrating efficient packaging of oversized (with respect to AAV capsids) single-stranded (ss)AAV2 vector genomes of up to 5.5 kb into HBoV1 capsids. Further increasing the attractiveness of this latest chimeric parvoviral vector is the availability of numerous BoV serotypes in primates and non-primates with as-of-yet unknown tropisms, which can be harnessed and may be useful for therapeutic gene transfer.
Here we further explored and extended the rAAV/HBoV1 system, by exploiting four additional primate BoVs—three from humans (HBoV2, 3, and 4) and one from Gorilla (GBoV)—that have not been studied as vectors before. To this end, we first created new helper plasmids expressing the capsid open reading frames (ORFs) of these BoVs, by swapping the respective HBoV1 ORF in the previously reported HBoV1 helper construct,17 similar to AAV vector pseudotyping. We then used our new helpers to produce reporter-encoding rAAV/BoV vectors, and we assessed them in various cell lines and primary cells, including organoids. Finally, we leveraged our technology for AAV capsid evolution18 by shuffling all five BoVs, providing the first proof of concept for the feasibility to create highly diverse libraries of chimeric BoV capsids that will help to study WT BoV biology and to isolate new and superior gene therapy vectors.
Results
Development of a Streamlined Protocol for Bocaviral Vector Production Based on Iodixanol Purification and Triple-Plasmid Transfection
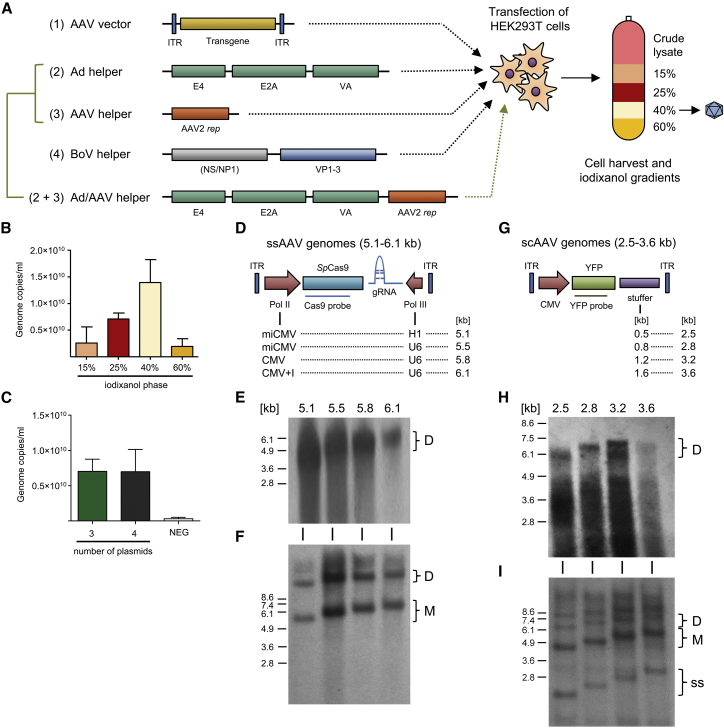
In the original report by Yan et al.17 that motivated this work, hybrid vectors derived from rAAV2 genomes and HBoV1 capsids were produced through co-transfection of four plasmids (Figure 1A, plasmids 1–4), followed by the purification of full viral particles from cell lysates by two rounds of cesium chloride (CsCl) density ultracentrifugation.17, 19 As the latter is laborious and requires 2–3 days, we tested whether rAAV/HBoV1 particles can also be purified akin to rAAVs using a much faster (2.5-h), discontinuous iodixanol density gradient centrifugation (Figure 1A, right). Indeed, we found that the majority (up to 70%) of chimeric rAAV/HBoV1 viral particles accumulated in the 40% iodixanol phase, identical to rAAVs, as detected by qPCR analysis of the different iodixanol fractions (Figure 1B).
Figure 1.
Recombinant HBoV1 Vector Production and Analysis of Packaging Capacity
(A) Plasmids for rAAV/HBoV1 production using either the 4-plasmid (constructs 1–4) or 3-plasmid (constructs 1, 2+3, and 4) transfection protocols. In both systems, the plasmids are co-transfected into HEK293T cells, which are then harvested 72 h later. To release the viral particles, cells are subjected to five freeze-thaw cycles, before free plasmid DNA is digested with benzonase. The resulting crude lysate is purified using iodixanol gradient centrifugation, and the vector-containing 40% phase is collected. (B) Purification of scAAV-YFP/HBoV1 via iodixanol gradient centrifugation. Shown is the distribution of benzonase-resistant particles in the different indicated iodixanol fractions. Data are mean (±SD) genome copies per milliliter (n = 3), as determined by TaqMan RT-PCR. (C) Production of scAAV-YFP/HBoV1 using the 3- or 4-plasmid transfection protocols. Data are mean (±SD) genome copies per milliliter (n = 3), as determined by TaqMan real-time PCR. (D) Oversized ssAAV-CRISPR constructs used in this work. SpCas9 and gRNA cassettes are expressed from different RNA polymerase II (Pol II) (first column) or Pol III (second column) promoters, respectively. Total genome sizes are shown in the third column. (E) Southern blot analysis of the ssAAV-CRISPR genomes from (D), which were packaged into and isolated from HBoV1 particles and then resolved on an alkaline agarose gel. The number above each lane indicates the size of the packaged genome. AAV vector genomes were detected with a probe against SpCas9. (F) Low-molecular-weight (Hirt) extracts of the indicated constructs followed by Southern blot analysis. Brackets indicate monomeric (M) and dimeric (D) AAV replicative forms. (G) Oversized scAAV genomes used in this work. Stuffer sequences from lacZ with the indicated lengths (first column) were inserted to increase the total genome size (second column). (H) Southern blot analysis of the scAAV-YFP genomes from (G), which were packaged into and isolated from HBoV1 particles and then resolved on an alkaline agarose gel. The number above each lane indicates the size of the packaged genome. AAV vector genomes were labeled with a probe against yfp. (I) Low-molecular-weight (Hirt) extracts of the indicated constructs followed by Southern blot analysis. Brackets indicate monomeric (M) and dimeric (D) AAV replicative forms. ss, single-stranded; sc, self-complementary.
Of the four plasmids needed for rAAV/HBoV production, two are routinely also used for rAAV generation, namely, the rAAV vector and the adenoviral (Ad) helper plasmid (plasmids 1 and 2 in Figure 1A). A difference between the two vector types is that only one helper plasmid encoding rep, cap, and aap is needed for rAAV vector production. In contrast, two separate plasmids are used for chimeric rAAV/HBoV1 production, one expressing AAV rep and the other HBoV1 ns, np1, and cap17 (plasmids 3 and 4 in Figure 1A, respectively). Notably, the Kleinschmidt lab has previously reported a series of plasmids that co-express AAV and Ad helper functions from a single backbone. This includes pDG that encodes the complete AAV2 genome (minus the encapsidation elements [ITRs, inverted terminal repeats])20 or pDGΔVP that was derived from pDG by deleting the cap gene21 (Ad/AAV helper in Figure 1A). Consequently, we tested whether the latter could replace the two separate AAV rep and Ad helper plasmids used for rAAV/HBoV1 vector production. To this end, we produced rAAV/HBoV1 vectors encoding a yfp (yellow fluorescent protein) expression cassette, using either the two individual helpers or pDGΔVP to supply AAV rep and Ad functions, and then we measured particle yields after iodixanol purification by qPCR. As shown in Figure 1C, both approaches yielded largely comparable rAAV/HBoV1 vector amounts in a range of 5 × 109–1 × 1010 vector genomes/mL from five 15-cm plates of HEK293T cells. These numbers are in line with previous data showing that the original four-plasmid protocol typically yields particle amounts reaching up to 10% of standard AAV vectors.17
Notably, we encountered no difficulties in propagating the pDGΔVP helper plasmid in regular DH10B bacteria, and we obtained similar yields as for the two separate, smaller helper plasmids (data not shown). Therefore, and in view of the reduced costs, time, and workload for preparing only three instead of four plasmids, all further rAAV/BoV vector preparations in this work were performed using the newly established triple-transfection protocol.
Analysis of rAAV/HBoV1 Packaging Capacity Using Single-Stranded or Self-Complementary Vector Genomes
As noted, Yan et al.17 have previously demonstrated the ability of hybrid rAAV/HBoV1 vectors to encapsidate large ssAAV vector genomes of up to 5.5 kb. Here, we independently confirmed and extended these results, by first generating a series of ssAAV vector genomes encoding the two components of the gene-editing tool CRISPR, i.e., the endonuclease SpCas9 and a guide RNA (gRNA) expression cassette. The total size of the resulting rAAV genomes, including the AAV2 ITRs, ranged from 5.1 to 6.1 kb (corresponding to 92%–110% of the 5.5-kb WT HBoV1 genome or 109%–130% of the 4.7-kb WT AAV2 genome), with gradual increases between 300 and 400 bp (Figure 1D). This was achieved by cloning and using different RNA polymerase II or III promoters for the expression of Cas9 or gRNA, respectively (Figure 1D). Generally, such all-in-one CRISPR constructs are attractive tools, but they present a challenge for standard AAV vectors due to their limited packaging capacity.3, 22
We first tested whether AAV2 can package these oversized CRISPR constructs, by visualizing encapsidated viral genomes using alkaline gel electrophoresis and Southern blot analysis (Figure S1A). The maximum size for efficient packaging into AAV2 was found to be around 5.1 kb, consistent with previous reports.22, 23, 24 The larger oversized ssAAV genomes did not result in distinct bands at the expected heights, but they produced a smear below 4.9 kb that most likely represents truncated genomes, also congruent with prior observations.22, 23
In contrast to AAV2 capsids, we could verify packaging of intact, full-length AAV2 vector genomes into the HBoV1 capsid over the entire size range (Figure 1E). Accordingly, HBoV1 can encapsidate up to 1 kb more DNA than AAV2 (6.1 versus 5.1 kb, respectively). Moreover, analysis of low-molecular-weight DNA isolated from vector-producing cells demonstrated proper rescue and replication of the AAV genomes (Figure 1F).
While the original data of Yan et al.17 as well as our own findings consistently showed the capacity of the HBoV1 capsid to accommodate oversized ssAAV vectors, the question remained whether this would also apply to self-complementary (sc)AAV genomes, and, if so, up to what size. For AAV capsids, the upper size limit for efficient encapsidation of scAAV vectors is typically reported to be around 2.5 kb, consistent with the fact that these genomes are packaged as a dimer of two inverted transgene copies, flanked by two ITRs and separated by a third, truncated ITR.25
To assess the corresponding capacity of the HBoV1 capsid, we cloned and packaged a set of oversized scAAV vector genomes akin to the ssAAV constructs, ranging from 2.5 to 3.6 kb (corresponding to 91%–131% of the WT HBoV1 genome or 106%–153% of the WT AAV2 genome), in intermediate steps of 300 to 400 bp (Figure 1G). The encapsidated AAV genomes were extracted from the viral particles and visualized by alkaline agarose gel electrophoresis and Southern blot analysis. In agreement with the analysis of ssAAV genomes (Figures 1D–1F), we found that scAAV genomes of up to 3.2 kb in size (migrating as ssDNA in alkaline gels at a height of around 6.4 kb) can be packaged efficiently into the HBoV1 capsid (Figure 1H). Moreover, we confirmed proper rescue and replication of all oversized scAAV genomes (Figure 1I). In contrast to HBoV1, we found that scAAV genomes larger than 2.8 kb cannot be packaged into the AAV2 capsid (Figure S1B). Thus, HBoV1 can encapsidate roughly 0.5 kb more scAAV DNA than AAV2, which is congruent with our conclusion for the ssAAV genomes (1 kb difference, see above). Notably, as subsequently discussed, intriguing data from the Srivastava lab26 indicated that the absolute packaging limit may be pushed even further, by optimizing scAAV genome sequence and structure.
Together, these results verify the ability of our modified rAAV/HBoV1 production scheme to replicate and package oversized AAV genomes of both types, sc or ss, into HBoV1 capsids.
Creation of New Helper Plasmids for rAAV/BoV Vector Production Based on Four Additional BoV Serotypes
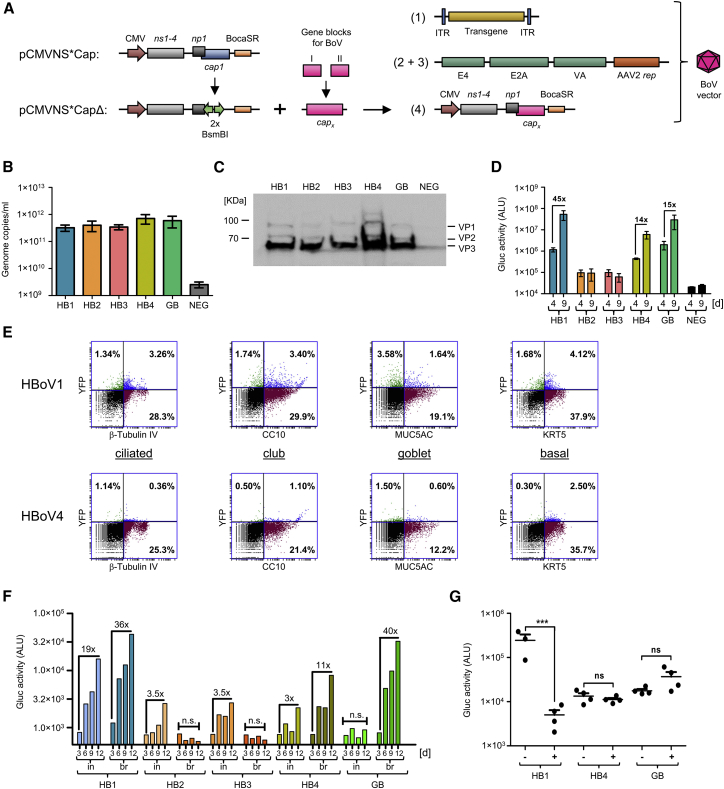
Hybrid rAAV/HBoV1 vectors are highly efficient at transducing human polarized (p)HAEs17 as well as ferret lungs.19 Together with the ability to package large DNAs, this may enable encapsidation of a full-length CFTR gene and its delivery and expression in lungs of cystic fibrosis patients. These exciting prospects inspired us to begin to also explore the potential of other reported bocaviral isolates for transgene delivery into different cells and tissues. Specifically, we aimed to expand the repertoire of BoV-derived vectors by investigating four additional primate BoVs that are commonly detected in stool,27, 28 three from humans (HBoV2, 3, and 4) and one from Gorilla (GBoV). To this end, we assembled the corresponding cap ORFs based on published sequences, and we cloned them individually into the HBoV1 helper plasmid (pCMVNS*CapΔ in Figure 2A) in place of the HBoV1 cap ORF.
Figure 2.
Pseudotyping of rAAV Genomes with Capsids Derived from Four Additional Bocavirus Serotypes
(A) BoV helper plasmid (pCMVNS*Cap1) for chimeric rAAV/HBoV1 production and acceptor plasmid (pCMVNS*CapΔ) derived thereof for cloning of the different BoV cap ORFs. Each cap sequence was ordered as two gene blocks, assembled to a full-length cap ORF (capx, where x = HBoV2–4 or GBoV) and subsequently cloned into the acceptor plasmid using a Golden Gate reaction. BocaSR, BoV-transcribed small non-coding RNA. Numbers in brackets refer to the construct labels in Figure 1A. (B) Production and iodixanol purification of chimeric HBoV1-4 and GBoV vectors encoding Gluc. The amount of genome copies per milliliter was determined with TaqMan RT-PCR. Shown are averages (±SEM) of four independent productions. (C) Western blot analysis of the indicated iodixanol-purified BoV stocks. Detected are the three BoV capsid proteins VP1, VP2, and VP3. NEG, iodixanol gradient from untransfected cells. (D) Transduction of pHAEs with the indicated scAAV-Gluc/BoV variants at an MOI of 2 × 104. Gluc activity in the medium was measured at 4 and 9 days post-transduction as arbitrary light units (ALU). Data are the mean Gluc expression (±SEM, n = 3). Numbers above the columns depict fold increases in expression. (E) Flow cytometry analysis of pHAEs transduced with scAAV-YFP/HBoV1 or scAAV-YFP/HBoV4. scAAV-Gluc/HBoV1 was used as a control (both at an MOI of 5 × 104, n = 4 independent transwells). Cells were co-stained for YFP and a cell type-specific marker: β-Tubulin IV (ciliated cells), MUC5AC (goblet cells), CC10 (club cells), or KRT5 (basal cells). Percentages of double-positive cells are shown in the upper right quarter. (F) Transduction of primary lung organoids with the indicated scAAV-Gluc/BoV variants. Several organoids per donor were either mechanically broken (br) and incubated with a total of 5 × 109 viral genomes, or they were individually microinjected (in) with 5 × 108–1 × 109 viral particles. Total Gluc activity in the medium was measured 3–12 days post-transduction and plotted on the y axis as ALU. Numbers above the columns depict fold increases in expression. (G) Transduction of pHAEs with the indicated scAAV-Gluc/BoV variants at an MOI of 1 × 104 in the presence (+) or absence (−) of IVIg. Gluc activity in the medium was measured 5 days post-transduction as ALU. Shown is the mean Gluc activity (+SEM, n = 4, except for HB1 [–] n = 3). For pHAEs transduction, LLnL and Doxorubicin were added at concentrations of 40 and 5 μM, respectively. Transductions of primary lung organoids were performed in the presence of 1 μM Doxorubicin. For statistical analysis, one-way ANOVA was used. Significance at p < 0.001 is indicated by a triple asterisk. ns, non-significant.
To test whether the resulting bocaviral helper plasmids can produce DNA-containing viral particles, HEK293T cells were transfected with (1) one of the new HBoV helpers (HBoV2–4 and GBoV), (2) an scAAV-YFP vector, and (3) pDGΔVP. Based on our finding that iodixanol density gradients permit rAAV/HBoV1 purification (Figure 1B), as well as on prior data showing that the accumulation of DNA-containing AAV particles in the 40% phase is serotype independent,29 we used iodixanol to purify all five bocaviral vector variants. Indeed, qPCR analyses of the 40% phase showed the presence of DNaseI-resistant particles for all five BoV serotypes (Figure 2B; Table S1). Importantly, the comparable particle quantities imply that changing the HBoV1 cap ORF had not impaired the ability of our helper plasmids to support robust capsid assembly and rAAV packaging for any of the tested bocaviral serotypes. Moreover, correct expression of VP1, VP2, and VP3 proteins was confirmed by Western blot analyses of purified vectors (Figure 2C).
Next, we compared the results obtained with the iodixanol protocol with purification by CsCl density centrifugation. Regardless of serotype, the rAAV/BoV vectors banded at densities from 1.43 to 1.48 g/mL, as detected by DNA dot blot analysis (Figure S2). Following dialysis and concentration (see the Materials and Methods for details), the amount of viral particles obtained ranged from 1.5 to 5.4 × 103 viral genomes/cell (equivalent to 5.3 × 1011–1.9 × 1012 viral genomes/mL), which is comparable to the yields obtained from the iodixanol gradients (Table S1).
In view of the same efficiency of both methodologies and of the fact that iodixanol purification is not only faster and easier than CsCl purification but also that the inert and non-toxic properties of iodixanol allow us to use the purified viruses directly on cells without dialysis or purification steps, iodixanol gradient centrifugation was favored in this work for vector purification.
Assessment of Multiple BoV Vectors in Primary Airway Epithelial Cells
Despite a global prevalence of human BoVs,30 little is known about their tropism and pathogenicity, mainly due to the restricted animal models and the lack of convenient cell culture systems that support viral replication. In turn, this limited knowledge prevented us from rationally selecting specific cells for functional validation of the new BoV vectors. Instead, we took the following indirect approach to predict cells that may be susceptible to rAAV/BoV transduction.
In essence, we harnessed findings that the so-called hypervariable regions that reside within the cap ORF determine host and tissue specificity31 of both non-primate BoVs and other parvoviruses. Therefore, we performed a phylogenetic analysis of this region to predict the closest BoV relatives within the animal kingdom. To this end, we first aligned the DNA sequence of the entire cap ORF of the primate BoV variants as well as representative non-primate BoVs with known and unknown tropism. This revealed a low DNA sequence homology of below 45% between primate and non-primate BoVs, whereas all primate BoVs share over 75% homology. Next, the alignment was used to perform a phylogenetic analysis (Figure S3A), which showed a close relationship of all five BoV isolates studied here to (1) Canine minute virus (GenBank: AB518884), a BoV known to spread through the dog placenta to infect the lung and intestine of fetuses;32 and (2) Canine BoV3 (GenBank: KC580640), a distinct canine BoV variant detected in the liver.33 Also, HBoV1/GBoV and HBoV3/HBoV4 clustered together in clades, respectively. From this, we concluded that cells from the lung, liver, or gastrointestinal tract might be promising targets for HBoV2–4 or GBoV vectors.
To test this prediction, rAAV vectors encoding either Gaussia luciferase (Gluc) or YFP were packaged into the BoV capsids or into AAV2 as a control. The use of these two reporters allowed for direct visualization (YFP) or monitoring (Gluc) of transgene expression over time. As it was shown that the proteasome inhibitors N-acetyl-L-leucyl-L-leucyl-L-norleucinal (LLnL) and Doxorubicin boost the transduction of HBoV1 and of selected AAV serotypes in pHAEs,17, 34 transductions were always performed in the presence of one or both of these reagents.
We first evaluated the transduction ability of the different BoV serotypes in pHAEs, a known target of HBoV1.35 Therefore, cells from different donors were grown as air-liquid-interface cultures and polarized on transwells as a pseudostratified heterogenous epithelium. Transduction efficiency was tested by first inoculating the cells exclusively from the apical side with equal amounts of viral particles. YFP expression became detectable at 48–72 h post-transduction (data not shown), and it was sustained until the end of the experiment (day 14; Figure S3B). Interestingly, we found that not only HBoV1 but also HBoV4 and GBoV transduce pHAEs to different extents, whereas HBoV serotypes 2 and 3 remained at background level (Figure 2D; Figure S3B; Table S2). Since Gluc is a secreted protein, transgene expression could be monitored over time by taking aliquots from the basolateral media. We observed an increase in luciferase expression from day 4 to 9 (Figure 2D), which reached a plateau after 12–14 days in culture (data not shown). We could further confirm these findings in polarized CuFi-8 cells, a cell line derived from a cystic fibrosis patient and previously reported to be permissive to HBoV1 infection, albeit less than pHAEs17 (Figure S3C).
Previous studies have shown that HBoV1 displays a polarity bias for the apical side of pHAEs.17 To study whether the other BoV variants also favor one side of cells over the other, we transduced pHAEs from the basolateral or apical surface with equal amounts of viral particles, and then we compared transgene expression (Figure S3D). In accordance with prior observations,17 HBoV1 displayed a high polarity of transduction (apical > basolateral, 100-fold). Similarly, GBoV was 10-fold more efficient at transducing from the apical than from the basolateral side. Overall, transduction could not be enhanced for any of the chimeric vectors via inoculation from the basolateral side. Of note, in contrast to earlier data,17 we detected a higher transduction with AAV2 than with HBoV1 in pHAEs, both from the apical and basolateral sides. As the transduction kinetics of BoV are poorly understood, this discrepancy might be explained by differences in the experimental settings, e.g., transduction for 1 h here versus 16 h in the prior study, or differential formation of cell junctions, which may have permitted concomitant access to both sides. Moreover, since proteasome inhibitors mostly increase transduction from the apical side,34 their use may have particularly boosted AAV2, which normally prefers the basolateral side.36
The use of scAAV-YFP vectors furthermore enabled flow cytometry analysis of transduced pHAEs. This revealed an overall transduction efficiency from 2% to 15% (data not shown), with the maximum observed for HBoV1, in line with its known lung tropism.17, 19 Co-staining with antibodies against YFP and cell-specific markers allowed us to determine the susceptibility of different cell populations within the heterogeneous pHAEs culture to BoV transduction. In detail, we stained with antibodies against KRT5, β-Tubulin IV, CC10, and MUC5AC, which are specific markers for basal, ciliated, club, and goblet cells, respectively. For rAAV/HBoV1, this showed that basal, ciliated, and club cells were transduced to comparable levels, while secretory MUC5AC-positive goblet cells were less susceptible (Figure 2E). Interestingly, while a similar result was obtained with GBoV (Figure S4A), we observed a strikingly different cellular distribution profile for HBoV4, which preferentially transduced basal cells and was least effective in ciliated cells (Figure 2E).
Recently, 3D primary human lung organoid cultures have been established,37 which reflect in vivo conditions better than conventional pHAEs grown in transwells. Therefore, we attempted to recapitulate our data in such a primary organoid culture system (Figure 2F). Since BoVs need to access the apical surface (Figure S3D), which is located inside the organoids, we used two distinct methods for transduction with scAAV-Gluc/BoV: (1) direct microinjection into the organoids (in); and (2) mechanical breaking of the organoids (br), followed by incubation with viral particles. Both methods resulted in a time-dependent increase in Gluc expression for HBoV1, HBoV4, and GBoV, which is in line with the trend that we had noted before in the pHAE cultures (Figure 2D). Intriguingly, we observed striking differences between the two application routes (Figure 2F; Table S2). In particular, HBoV2 and HBoV3 only transduced following direct microinjection, whereas the opposite was observed for GBoV, which required mechanical breaking. The other two serotypes, HBoV1 and HBoV4, tolerated both routes, but they showed a slightly better transduction of sheared organoids.
Collectively, the successful transduction of pHAEs and lung organoids provided a first experimental evidence for the functionality of all four new viral vectors, in particular HBoV4 and GBoV. This motivated us to further characterize our bocaviral vector collection, including measurements of its reactivity with human antibodies and of its activity in additional cell types.
Differential Effect of Purified Human Immunoglobulins on pHAEs Transduction with rAAV/BoV
Prior data show that a majority of human serum samples are positive for neutralizing antibodies (NAbs) against AAV2 (∼70%)38 or HBoV1 (∼58%).39 Depending on the target tissue and route of administration, NAbs will eventually neutralize the viral capsid and, hence, negatively affect vector transduction efficiency. Thus, we used pHAEs to test whether HBoV4, which is the least seroprevalent variant of human BoVs (∼2%),39 and GBoV displayed less reactivity than HBoV1 with intravenous immunoglobulin (IVIg). Therefore, we adapted a previously described in vitro virus neutralization assay.40 Briefly, scAAV-Gluc/BoV vectors were incubated with 25 mg/dL IVIg solution (representing a 1:40 dilution of the normal average IgG concentration in adults) or PBS as a control for 1 h at 37°C. Next, the mixtures were added to the apical surface of pHAEs, and luciferase activity was measured after 5 days in culture. As expected, HBoV1 displayed a high loss of activity after IVIg exposure (50-fold). In stark contrast, the transduction by HBoV4 and GBoV remained entirely unaffected by IVIg under our experimental conditions (Figure 2G).
High Susceptibility of Primary Hepatocytes to Primate BoV Infection
In dogs, the occurrence of animal BoVs in liver tissue has been documented repeatedly. For example, a strain of the Canine minute virus (GenBank: KT241026) was found in intranuclear inclusion bodies in hepatocytes and associated with hepatitis.41 Also, Canine BoV 3 (GenBank: KC580640), a genetically distinct canine BoV, was found in a concatemerized form in the liver.33 In contrast, only a few reports so far have documented the presence of HBoV in the human liver42 or have associated BoV liver infection with hepatitis.43, 44
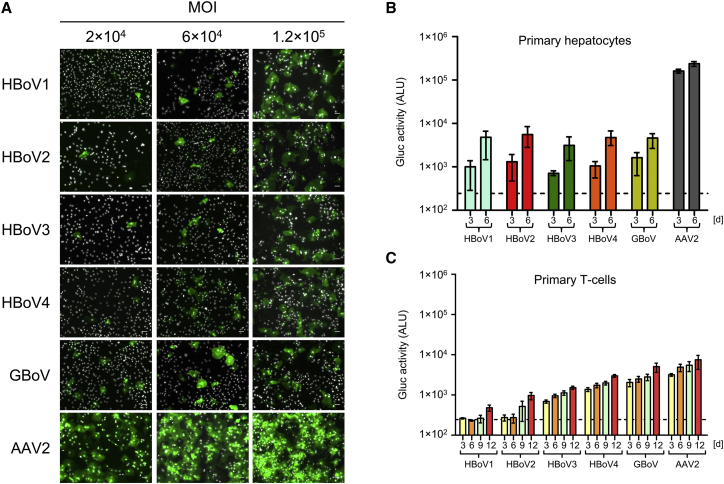
Therefore, we tested whether human hepatocytes can be transduced with our different rAAV/BoV vectors. To this end, monolayers of primary human hepatocytes were transduced with equal amounts of scAAV-YFP/BoV vector at three MOIs (2 × 104, 6 × 104, and 1.2 × 105 genome copies/cell). YFP expression could be detected at all MOIs, starting at 2 days post-transduction (Figure 3A). Using Gluc as an alternative reporter, we likewise measured an increase of transgene expression over time (Figure 3B; Table S2), akin to our data in pHAEs (Figure 2D). Notably, all five BoV serotypes performed comparably well in hepatocytes, including HBoV2 and HBoV3. The latter is particularly interesting as these two serotypes were inferior to the other three in pHAEs and lung organoids (Figures 2D and 2F), which further exemplifies the distinct cellular tropisms of the five bocaviral isolates that we studied here.
Figure 3.
Transduction of Primary Human Hepatocytes and T Cells with rAAV/BoV Vectors
(A) YFP expression 5 days after transduction of primary human hepatocytes (pHeps) with scAAV-YFP/BoV vectors at the indicated MOIs (n = 3 donors). Expression first became detectable at day 2 (not shown) and then increased over time. Cells were fixed and stained with an FITC-coupled anti-GFP antibody, and nuclei were stained with Hoechst. Scale bar, 50 μm. (B) Gluc activity at 3 and 6 days post-transduction of pHeps with scAAV-Gluc/BoV at an MOI of 1 × 104. Data represent the mean and range of two independent measurements (n = 2 donors). (C) Transduction of primary human T cells with scAAV-Gluc/BoV (MOI = 1 × 104). Gluc activity in the medium was measured 3–12 days post-transduction. Plotted are means ±SEM of three independent experiments (n = 3 donors). For all transductions, Doxorubicin was added to a final concentration of 1 μM/well. Dashed lines indicate the assay background.
Robust Transduction of Human CD4+ T Cells with Primate BoV Vectors
Numerous viruses, such as smallpox, HIV, and measles, use the hematopoietic system to spread throughout the body and to reach their target cells. Based on this as well as on evidence for HBoV1 viremia in healthy and diseased individuals,45 we tested the ability of chimeric rAAV/BoV vectors to target different types of blood cells. Measurements of Gluc activity over time implied that macrophages are not susceptible to BoV infection, while PBMCs (peripheral blood mononuclear cells) displayed a low level of transduction (Figures S4B and S4C). Interestingly, we found human CD4+ T cells to be highly permissive to BoV transduction (Figure 3C; Table S2), showing robust Gluc expression that gradually increased over the 12 days in culture. Moreover, using chimeric scAAV-YFP/BoV vectors, we also detected YFP-positive cells with all vectors. Particularly notable is the performance of HBoV4 and GBoV, which clearly outperformed the HBoV1 prototype with both reporter genes and nearly matched the efficiency of AAV2 (Figures 3C and S4D).
Detection of a Broad Cell Tropism of Primate BoV Vectors In Vitro
Motivated by our identification of two novel primary cell types that support BoV transduction, next to the previously reported (for HBoV1)17 pHAE cultures, we decided to further extend our screen for target cells. Consequently, we evaluated all vectors in additional primary cell types, namely, (1) skeletal myoblasts or tubes and cardiomyocytes, (2) pulmonary fibroblasts, and (3) endothelial cells that line blood and lymphatic vessels; furthermore, as BoV infection has been correlated to gastrointestinal disease,30, 46 we included both (4) intestinal and colon primary human mini-gut cultures.
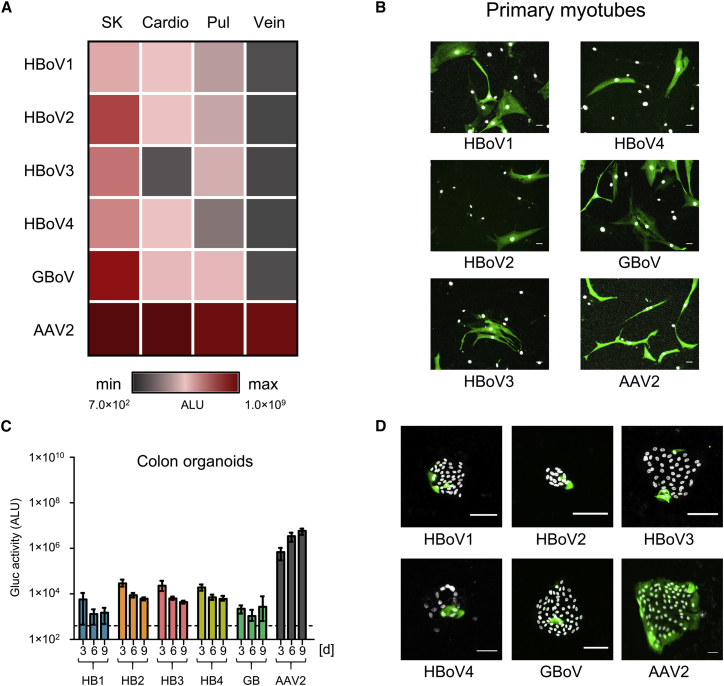
Interestingly, unlike pHAEs, lung fibroblasts showed a preference for transduction with GBoV (7- to 13-fold higher than all other BoV serotypes) (Figure 4A; Table S2). Although HBoV has never been associated with myopathies, we found that undifferentiated muscle cells (myoblasts) can be transduced with all the different BoV serotypes (column SK in Figure 4A; Table S2). Of particular note is that, akin to other viruses such as influenza virus,47 differentiated muscle cells (myotubes) were highly susceptible to scAAV-YFP/BoV vectors of all five serotypes, comparable with AAV2 (Figure 4B). In contrast to skeletal muscle and its diseases, bocaviral DNA was often detected in the heart and BoV was linked to myocarditis.42, 48 In our in vitro transduction assays using YFP (data not shown) or Gluc (Figure 4A; Table S2), we noted that all BoV variants were able to transduce primary cardiomyocytes but less efficiently (10- to 200-fold) than skeletal muscle cells. Finally, transduction of human vein endothelial cells resulted in low (HBoV1 and GBoV) or no (HBoV2–4) Gluc expression (Figure 4A; Table S2).
Figure 4.
Transduction of Primary Human Cells with rAAV/BoV Vectors
(A) Heatmap showing the results of transduction of the indicated primary cells with the different scAAV-Gluc/BoV vectors at an MOI of 5 × 104. Plotted is the Gluc activity measured in the medium 9 days post-transduction. (B) Transduction of primary myotubes with 1 × 109 genomes of scAAV-YFP/BoV vectors or AAV2 as control. (C and D) Transduction of differentiated primary human colon organoids with 5 × 109 viral genomes of scAAV-Gluc/BoV (C) or 1 × 1011 viral genomes of scAAV-YFP/BoV (D). Gluc activity shown in (C) was measured at the indicated time points. Data represent the mean and range of two independent experiments (n = 2 donors). Images in (D) were taken 9 days post-transduction. The exposure was reduced 3-fold for AAV2 to avoid saturation of signal. All transductions were performed in the presence of 1 μM Doxorubicin. For all images, cells were fixed and stained with an FITC-coupled anti-GFP antibody. Scale bar, 50 μm. Nuclei were stained with Hoechst. SK, skeletal muscle cells; cardio, cardiac myocytes; pul, pulmonary fibroblasts; vein, saphenous vein endothelial cells. Dashed lines indicate the assay background.
To study the transduction of intestinal organoids, mini-guts were removed from Matrigel, trypsinized, and seeded in 2D to allow for viral transduction through the apical membrane. These 2D primary intestinal cultures were maintained in basal or differentiation media. The resulting organoids were inoculated with equal amounts of viral particles, and Gluc expression was monitored over time. We detected low luciferase expression in undifferentiated ileum (Figure S5A), as well as in differentiated ileum (data not shown), while a more robust and sustainable expression was observed in differentiated primary colon organoids (Figures 4C and 4D). Moreover, in contrast to our data in lung organoids, HBoV2–4 showed a higher activity than HBoV1 or GBoV in colon organoids (Table S2). These experiments were further extended by transducing T84 cells, a human colon cancer cell line that we have previously found to be susceptible to WT HBoV1 infection.49 In these cells, HBoV2–4 also showed a higher activity than HBoV1, while the best results were obtained with GBoV (Figure S5B; Table S2).
Transduction of Human and Mouse Cell Lines with BoV Vectors
The finding that T84 cells, an immortalized intestinal cell line, are readily transduced with all five BoV vectors is notable, as Caco-2 cells, another intestinal cell line, do not support productive infection.50 Moreover, HAEs or CuFi-8 cells, the only other two cell culture systems that are currently available for ex vivo BoV research, are considerably more difficult to obtain, culture, and polarize. Thus, we tested six additional immortalized cell lines derived from different tissues for their susceptibility to BoV infection (Figure 5).
Figure 5.
Transduction of Cell Lines with rAAV/BoV Vectors
The indicated cell lines were transduced with scAAV-YFP/BoV or scAAV-YFP/AAV2 as a control at an MOI of 2 × 105. Cells were fixed 5 days post-transduction and stained with an FITC-coupled anti-GFP antibody. Cell nuclei were stained with Hoechst. Scale bar, 50 μm.
As expected from our data in primary hepatocytes, two human liver-derived cell lines, Huh7 (hepatocytes) and LX-2 (stellate cells), could be transduced as well, albeit at lower efficiency than the primary cells. In contrast, PancI (a pancreatic cell line) and RAW264.7 (a mouse macrophage cell line) were largely resistant to BoV transduction. Notably, transduction of the mouse cells could have been hampered by the high species specificity of primate BoVs.28 Surprisingly, the cell lines MCF-7 (breast cancer) and HeLa (cervix carcinoma) showed robust YFP expression, especially after transduction with HBoV4 and GBoV. The efficiency in HeLa cells is of note in view of an ongoing debate whether WT BoV infection may be associated with miscarriage, based on sporadic detection of HBoV DNA in placenta and aborted tissue.51
Proof of Concept for the Feasibility to Create Highly Complex BoV Cap Libraries via DNA Shuffling
Previously, we and others have comprehensively exemplified the power of directed molecular evolution to breed synthethic AAV capsids with unique assets for gene transfer and gene therapy.52 One of the key technologies is DNA family shuffling, i.e., a process whereby cap genes of multiple AAV serotypes are first fragmented and then reassembled based on their partial homologies, resulting in libraries of chimeric AAV capsids that can be subjected to various positive and/or negative selection pressures.
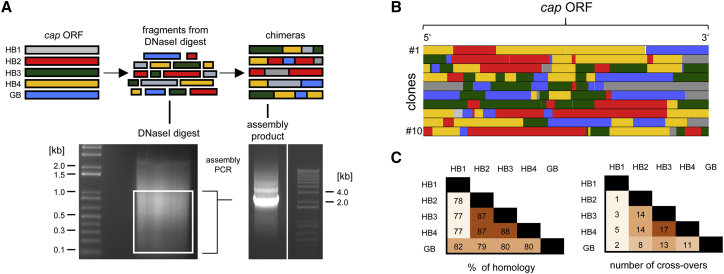
Here we aimed to establish the first proof of concept that DNA family shuffling can also be applied to the primate BoVs used in this work. Thus, we first PCR-amplified the cap ORFs of HBoV1–4 and GBoV, and then we fragmented the PCR products using DNaseI (Figure 6A). Following gel electrophoresis and excision of a DNA range between 0.1 and 1 kb, the fragments were used as a template for the assembly of full chimeric cap sequences (Figures 6A and 6B). Finally, this sequence pool was cloned into a BoV helper plasmid comprising AAV2 ITRs, resulting in the plasmid library pAAVCMVNS*Capchim (see the Materials and Methods for details). Based on bacterial colony counts after transformation, this library had a high diversity of 5 × 107 capsid variants. Moreover, we used our in-house Shuffling Alignment Analysis Tool (SALANTO)53, 54 to quantitatively and qualitatively analyze library composition and cap crossover frequency. While we observed a similar distribution of HBoV2, 3, 4, and GBoV (23%, 21%, 28%, and 18%, respectively), HBoV1 was slightly underrepresented (8%). Congruent with our recent data from AAV cap shuffling,54 the frequency of recombination events between different serotypes correlated with their degree of homology and was thus highest among HBoV2–4, which share 87%–88% homology (Figure 6C).
Figure 6.
DNA Family Shuffling of Primate BoVs
(A) The cap ORFs from HBoV1–4 and GBoV were PCR-amplified and subjected to a controlled DNaseI digest. The white box in the first gel indicates the range of excised fragments between 1 and 0.1 kb. These cap fragments served as a template in two subsequent PCRs (see the Materials and Methods for details) that resulted in the ∼2.2-kb amplicon shown in the second gel, which represents a mixture of full-length chimeric cap ORFs. (B) Complete cap ORFs of 10 clones from a library of chimeric bocaviral plasmids. Analysis was done using SALANTO v.3,53 which assigns each sequence to the corresponding parental serotypes (shown in A). White lines, mutations to the parental sequences; light gray, ambiguous sequences that can be assigned to more than one parental serotype. (C) The first table shows the DNA homology of the different bocaviral cap ORFs used in this work. The second table shows the number of crossover events between pairs of BoV measured in a representative shuffled library.
As the last step, we produced a viral library by co-transfection of the plasmid pool together with pDGΔVP into HEK293T cells. After CsCl purification, virus-containing fractions were identified via DNA dot blot, which showed a peak at a density of 1.43 g/mL. Moreover, real-time qPCR revealed a good titer of the final stock of 5 × 109 genome copies/mL.
Discussion
This work was inspired by a seminal study from 2013, in which Yan et al.17 described a chimeric rAAV2/HBoV1 vector that packaged the complete CFTR gene and potently transduced human pHAEs. As a novel gene delivery vector, chimeric BoVs possess a variety of interesting features and assets, several of which were validated or newly identified in this study.
A first notable advantage of rAAV/BoV vectors is their expanded packaging capacity as compared to vectors based on AAV capsids. The relative gain of up to 1 kb observed here for ssAAV genomes or 0.5 kb for scAAV is crucial, as it allows for the encapsidation of larger transgenes or regulatory elements, such as promoters, enhancers, and others. In fact, we were able to efficiently package ssAAV cassettes with sizes of up to 5.8 kb (105% of the WT HBoV1 genome) into the HBoV1 capsid. Encapsidation of 6.1-kb-long rAAV genomes (110% of WT HBoV1) was also possible, albeit with lower efficiency. These findings are congruent with and extend prior notions that at least 5.5 kb foreign DNA can be packaged into HBoV1.17 Our observation that scAAV genomes of up to 3.2 kb (115% of WT HBoV1, if 100% is considered to be 2.75 kb, i.e., 50% of 5.5 kb) can also be packaged into the HBoV1 capsid, but not into AAV, further corroborates this conclusion.
Moreover, important to point out is that the absolute limit of scAAV vector DNA that can be encapsidated is likely also governed by a variety of additional parameters, such as Guanine-Cytosine (GC) content, secondary structure, presence of cryptic terminal resolution sites, or levels of Rep proteins expressed from the helper plasmid, as implied by Wu and colleagues.26 Importantly, the ss or scAAV vector plasmids were kept constant between our AAV and BoV productions, ensuring that the relative increases in packaging capacity that we noted for BoV were exclusively due to the capsid. Further of note, all AAV helper plasmids used and compared here express lower levels of Rep78/68 than conventional helper constructs, due to a shift (pWHC2) or replacement of the AAV2 p5 promoter (pDGΔVP, with the weaker mouse mammary tumor virus [MMTV] promoter). This was found by the Srivastava lab26 to favor encapsidation of intact scAAV vector genomes, when comparing an AAV helper plasmid (pACG-2) with a weak ACG start codon for Rep78/68 to a traditional construct. This can readily explain why we also noted a dominant packaging of full-length scAAV vector genomes and thus represents another advantage of using pDGΔVP for BoV vector production.
Notably, while the original data were obtained with the full-length CFTR gene,17 they were independently validated here with another therapeutically relevant expression cassette, encoding all components of the CRISPR machinery, i.e., SpCas9 and gRNA. Considering the difficulties in encapsidating SpCas9—the largest (4.2 kb) of all currently studied Cas9 orthologs—into AAV capsids, the ability to readily package all-in-one CRISPR cassettes into HBoV1 is remarkable. Thus, we have already created constructs expressing gRNAs directed against the dF508 mutation in the CFTR gene, one of the leading causes of cystic fibrosis (CF), and we will soon begin to evaluate these in CF organoids. Moreover, by using smaller orthologs such as SaCas9 (3.1 kb), it should even be possible to co-package Cas9 and multiple or concatemerized gRNA expression cassettes plus a DNA template for homologous recombination (if desired) into a single BoV particle.
It must be pointed out, however, that mere DNA encapsidation does not always translate into efficient transduction, based on data with AAV. Hence, a crucial task for future work remains to assess the infectivity of BoV particles containing oversized ss or sc genomes of various lengths. A complication that prevented us from performing these analyses is the strict dependency of the current BoV vector generation on proteasome inhibitors (reported for HBoV117 and also seen here for all other serotypes; data not shown). Also, others have documented a correlation of increasing AAV genome size and more effective proteasomal degradation, and they have shown that the use of proteasome inhibitors can restore transduction by oversized vectors to nearly WT levels.22 Here, this caused the conundrum that it would be hard to disentangle general effects of proteasome inhibitors on the BoV capsid and its degradation from a specific impact on particles carrying oversized genomes. Thus, a unanimous conclusion on the infectivity of BoV particles containing large genomes has to await the next generation of vectors that is less or no longer dependent on proteasome inhibitors, which, as subsequently discussed, may well be in sight.
A second benefit of BoV vectors is our finding that, akin to AAVs, they are readily amenable to pseudotyping, i.e., cross-packaging of a vector genome into various capsids. Our respective studies were fueled by the recent discovery of additional primate BoV serotypes beyond HBoV1, such as HBoV2–4 in humans55, 56 and GBoV in Gorilla.28 We were further motivated by the interchangeability of AAV ITRs as well as rep and cap genes, which has greatly facilitated the development of different AAV serotypes as viral vectors.57 In particular, it enabled AAV pseudotyping by simply replacing the cap ORF between helper plasmids, and thus it paved the way for AAV to become one of the most widely used viral vectors in gene therapy so far.
Since the non-structural proteins of primate BoVs are highly homologous (over 73% for NS and NP1), we postulated that the BoV genome may offer a similar flexibility as AAV. Therefore, we generated new BoV helper plasmids by replacing the HBoV1 cap ORF with that of HBoV2–4 or GBoV while maintaining the non-structural ORFs. Indeed, we found that all helper plasmids gave comparable expression of the three capsid proteins and that all pseudotyped AAV/BoV vectors encapsidated AAV genomes at efficiencies equal to or even exceeding an HBoV1 helper (up to 2.8 × 1012 genomes/mL). This is intriguing especially in the case of GBoV, which was shown to carry a unique ns ORF that encodes an extended version of the NS protein.28
The successful generation of new BoV helpers for vector pseudotyping laid an essential foundation for our next major goal, which was to study the functionality of the different BoV serotypes and to compare their cellular tropisms. In all these experiments, we included AAV2 as a positive control based on the known wide tropism and high efficiency of this AAV serotype in cultured cells. Our major rationale was to validate that our experimental conditions, including cell vitality, permitted transduction, which was especially important in the case of macrophages where none of the bocaviral vectors were efficient. At the same time, we acknowledge that other AAV serotypes may perform even better than AAV2 in specific cultured cell types, such as AAV3 in hepatocytes;58 AAV5, 6, or 9 in lung cells;59 or AAV6 in T cells,60 and we will thus include these in future, more quantitative comparisons (also in vivo, see below).
Based on the sporadic detection of HBoV2 DNA in lungs of patients61 and on our own phylogenetic analyses (Figure S3A), we started with the pHAEs model. This already provided the first evidence for the functionality of two of our new vectors, those based on GBoV and HBoV4. In addition, we screened a panel of primary cells, organoids, and cell lines, and we found a wide range of them to be efficiently transduced by different BoVs, including human hepatocytes, T cells, and skeletal muscle cells. In particular, the potency in hepatocytes is clinically relevant, as it implies that (1) all five BoV serotypes are intriguing candidates for liver gene transfer, and that, vice versa, (2) the liver should be considered as an off-target in case of another on-target. Also notable is the efficiency of HBoV4 and GBoV in primary human T cells, which, combined with the high packaging capacity, makes these two isolates very interesting for the transfer of large gene expression cassettes to T cells. A particularly intriguing application could be delivery of all-in-one CRISPR cassettes, with the aim to excise and thus eradicate latent genomes of the HIV. Finally, we are intrigued by the distinct cell specificities that we observed within pHAEs, where HBoV4 was 10-fold less efficient at transducing ciliated cells as compared to HBoV1. While these data need independent validation with additional pHAEs preparations, they may already imply that HBoV4 is a preferred vector for selective gene transfer to non-ciliated cells in the lung, such as club or basal cells.
Altogether, our various screens revealed that BoVs have a much wider tropism in vitro than previously anticipated. This particularly pertains to GBoV that has the broadest target range and was frequently the most efficient in primary human cells, making it a preferred candidate for further development that may complement or even replace the current HBoV1 vector prototype. Along these lines, we are currently assembling a comprehensive library of DNA-barcoded, next-generation sequencing-compatible AAV and BoV vectors, whose biodistribution we will then analyze in non-human primates to study bocaviral (and AAV) tropism in a large animal species in vivo and compare the results to our present in vitro data.
In addition, we obtained the first evidence that pseudotyped rAAV/BoV vectors not only differ in their cell specificity but also in their reactivity to pooled human antibodies (IVIg). In particular, HBoV4 and GBoV appear more capable of escaping neutralization than HBoV1, which is in line with data that HBoV4 is the least prevalent BoV variant in the human population (∼2%).39 These initial data, combined with the good efficiency of HBoV4 and GBoV, are very encouraging from a clinical perspective, as they imply the possibility of using these two serotypes as alternatives to HBoV1, which has the highest seroprevalence in adults (∼59%).39 Also, identical to strategies used in the AAV field, it should enable vector re-dosing in patients after capsid swapping, which is otherwise impeded by neutralizing antibodies that emerge after the first administration. Thus, it will be interesting and important for follow-up work to validate and extend these findings, by testing more IVIg concentrations or patient sera and by determining virus-neutralizing titers.
In this context, we note our recent findings that natural HBoV1 variants differing in single amino acids exhibit distinct transduction efficiencies and IVIg reactivities (J.F., unpublished data). This provides additional optimism that it will be possible to breed superior BoV variants with minimal antibody reactivity, e.g., via rational engineering of critical surface residues or by using IVIg as negative selection pressure during capsid library evolution.
Despite these promising prospects, it is clear that the rAAV/BoV system requires further improvements on multiple levels to ultimately facilitate the breadth of applications that is already possible with AAV vectors. On the one hand, the original protocols for vector production were more complicated, as they required four instead of the three or two plasmids that are commonly used in the AAV field. Importantly, in this work, we have already demonstrated the ability to also produce rAAV/BoV vectors via triple transfection, owing to our use of a combined helper plasmid that co-expresses all adenoviral and AAV helper functions. Furthermore, we showed that rAAV/BoV vectors of all five serotypes can be purified by iodixanol gradient density centrifugation, which is much easier and faster than the originally proposed CsCl protocol.17 In this context, we are excited by the most recent report of an alternative BoV production system by Yan et al.,62 which is completely devoid of the BoV non-structural proteins and likewise uses three instead of four plasmids. Interestingly, it resembles the AAV vector production protocol in its sole dependency on AAV2 Rep proteins and adenoviral helper functions.2 Also, the authors noted a 4-fold increase in virus titers as compared to the conventional HBoV1 helper plasmid that was also used here. It will thus be very interesting to test whether this optional production system will further increase the yields of the other serotypes reported in this work.
On the other hand, regardless of the production scheme, the current first generation of BoV vectors is limited by its strict dependency on proteasome inhibitors for efficient transduction. This is best exemplified by prior data with rAAV/HBoV1 vectors in pHAEs, where transduction was more than 1,000-fold improved upon proteasome inhibition.17 The same inhibitor agents are also commonly used to facilitate transduction with genuine AAV vectors, especially in the airway epithelia.34 We note that we have routinely used these inhibitors for all AAV2 controls in this work to remain consistent with the BoV samples and that AAV2 transduction in the absence of these inhibitors was typically weaker (data not shown). Possible targets of proteasome inhibitors include a post-entry block to viral transduction,63 since AAV is thought to be ubiquitinated once it reaches the cytoplasm and to thereby become a substrate for proteasome-dependent degradation.64 Hence, interfering with this pivotal step in viral trafficking increases the amount of rAAV particles that can reach the nucleus and release their cargo.63
Luckily, there are multiple ways to alleviate this specific block in rAAV/BoV transduction. First, one option to circumvent proteasomal degradation is to mutate exposed tyrosine residues on the BoV capsid surface, which are likely targets for phosphorylation and ensuing ubiquitination, to phenylalanines.65, 66, 67, 68 For multiple AAV serotypes, this was shown to result in an up to 30-fold increase in transgene expression. Owing to the new high-resolution structures of HBoV1–4 that were recently reported by the Agbandje-McKenna lab,69 it is now indeed feasible to map and modify surface tyrosines in the different BoV capsids in such a rational manner.
Second, new options may arise as our understanding of WT BoV biology continues to improve. Already it is known that HBoV1 interferes with different cellular pathways to escape defense mechanisms and to accomplish infection, which distinguishes it from AAVs that cause only a mild innate immune response in vitro70, 71 and in vivo.72, 73 This raises the question whether special regulatory functions present in the WT BoV genome are impaired in the vector context. For example, the nucleoprotein protein NP1 negatively regulates interferon (IFN) signaling by downregulating IFN-beta promoter activity. Consequently, NP1 was suggested to be an early gene in the viral life cycle, whose expression fosters infection by suppression of the IFN response.74 Another interesting and, so far, unique observation for HBoV1 is that the VP2 capsid protein interacts with RNF125, which in turn inhibits the ubiquitination and hence proteasome-mediated degradation of RNF125 itself and of RIG-I. Both proteins are negative regulators of the IFN response; thus, it was speculated that VP2 might act as a late gene during the establishment of BoV latency.75 Moreover, NS1 was shown to play a role in immune evasion by antagonizing the nuclear factor κB (NF-κB)-signaling pathway.76 While a dissection of these possibilities was beyond our present scope, one can readily anticipate that a deeper understanding of fundamental BoV biology, including intracellular trafficking, capsid uncoating, or immune responses, will inform and benefit the design of enhanced, second-generation bocaviral vectors.
Third, until such rational design approaches have been enabled, directed molecular capsid evolution represents a powerful alternative to increase viral fitness through iterative selection, as extensively documented for AAV capsids and vectors.52 Here, we exemplified this potential by leveraging, for the first time, DNA family shuffling77, 78 to create a combinatorial BoV library with a high diversity of 5 × 107 clonal variants. As a full selection scheme typically requires at least three and often up to five time-consuming rounds of (1) target cell infection, (2) PCR rescue, and (3) cloning of a secondary library for re-infection, we cannot yet present a fully evolved, synthetic BoV akin to AAV-DJ or AAV-LK03.5, 79, 80, 81 Still, we note that a single round of selection on pHAEs has already led to the emergence of chimeric BoV capsids that yield higher titers than the WT BoV isolates (data not shown), indicative of an effective ongoing molecular evolution process. This fuels our optimism that this approach will indeed allow for the selection of next-generation, designer BoV vectors with desirable features not occurring in nature, such as new tropism, greater efficiency, and/or lower immunoreactivity. At the same time, as with all directed molecular evolution strategies, it will be key to carefully screen lead candidates for possible de novo emergence of adverse properties, such as additional T cell epitopes.
Concurrently, studies of the evolved capsids will provide further insights into the biology of BoV infection. To this end, it should be rewarding to also include other BoV variants, such as those reported in chimpanzees,82 pigs,83 and dogs.33, 41 Importantly, our data with five different BoVs in this work suggest that our helper plasmids can be easily modified to accommodate and express other BoV variants as well. Furthermore, it may even be possible to perform a pan-parvoviral shuffling between BoV and AAV variants (e.g., AAV3 for selection on human hepatocytes), provided there are sequence stretches that correspond to comparable regions in the assembled capsid and these share over 50% homology, which is a prerequisite for efficient DNA shuffling.54 Finally, we have already started to create and evaluate alternative library designs, including one where the shuffled bocaviral capsid genes are flanked by the authentic HBoV1 terminal repeats rather than the AAV2 ITRs, which makes this library capable of autonomous replication. In turn, this could streamline subsequent selection schemes, as it should alleviate the necessity of time-consuming PCR rescues, which is an intriguing possibility that we will study in the future.
In conclusion, as the field of BoV gene therapy vectors remains in its early days, we believe that the data and strategies presented here and elsewhere already offer a first glimpse into its great potential and that, given the inspiring experience with AAV over the last five decades, there is every reason to be excited over this rapidly emerging gene delivery system.
Materials and Methods
Cloning Procedures
To generate the different bocaviral helper plasmids that express HBoV2–4 and GBoV VPs, the entire 2-kb HBoV1 cap ORF was deleted in a previously described62, 84 HBoV1 helper plasmid (pCMVNS*Cap) and replaced by two inverted BsmBI sites (at nucleotide position 646, np1 numbering), to allow for subsequent seamless insertion of alternative cap sequences. To this end, an overlap extension (OE-)PCR was performed using the two flanking primers 1/4 (all primer sequences are shown in Table S3) and two overlapping primers 2/3. The resulting 1,918-bp fragment was cloned into pCMVNS*Cap using HindIII/NotI restriction sites (also present in primer 1 or 4, respectively). This resulted in the cap acceptor plasmid pCMVNS*ΔVP-2xBsmBI.
Each BoV cap ORF (GenBank: HBoV2 FJ170278, HBoV3 EU918736, HBoV4 FJ973561, GBoV HM145750) was ordered as two gene blocks (each 1,000 bp) from Integrated DNA Technologies (Leuven, Belgium). For HBoV3 and HBoV4, one PCR reaction was performed to amplify each gene block, using primers 5–8 (HBoV3) and 9–12 (HBoV4). For the first and second gene block of HBoV2 and GBoV, respectively, no PCR product could be obtained, possibly due to secondary structures in the sequence. This was solved by performing three separate PCR reactions for each bocaviral isolate. Accordingly, gene block I of GBoV was amplified using primers 13 and 14, while gene block II was amplified in two separate reactions using primers 15–18. Likewise, gene block I of HBoV2 was amplified in two separate reactions using primers 19–22, and gene block II was amplified using primers 23 and 24. All PCR reactions were gel-purified using the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany), following the manufacturer’s instructions. For each bocaviral cap ORF, the fragments were assembled and cloned via a Golden Gate reaction into an empty pBSII-KS(+) plasmid (Agilent, Waldbronn, Germany), using NotI and ClaI restriction sites. Correct assembly and integrity of the resulting 2,004- to 2,016-bp-long cap ORFs were validated by Sanger sequencing (GATC, Konstanz, Germany). Next, the complete cap ORFs were amplified using primers 25–32 and subcloned into pCMVNS*ΔVP-2xBsmBI using Golden Gate cloning, resulting in helper plasmids pCMVNS*HBoV2-4Cap and pCMVNS*GBoVCap.
The shuffling acceptor pCMVNS*ΔVP-1xBsmBI was derived from pCMVNS*Cap by first deleting the cap ORF and inserting a BsmBI site at nucleotide position 592 (np1 numbering) using OE-PCR with overlapping primers 33/35 and flanking primers 34/1. Next, the complete pCMVNS*ΔVP-1xBsmBI sequence was PCR-amplified using primers 36/37. The PCR product was digested with EagI/PacI and cloned into a derivative of pSSV9,85 i.e., a conventional AAV vector construct harboring two AAV2 ITRs. This resulted in the final shuffled-capsid acceptor pAAVCMVNS*ΔVP-1xBsmBI.
To obtain oversized scAAV constructs, portions of the lacZ cDNA ranging from 500 to 1,600 bp were PCR-amplified from pTRUFlacZ (a plasmid harboring the full-length lacZ),86 using forward primer 41 in combination with reverse primer 42 (yielding a 500-bp fragment), 43 (800 bp), 44 (1,200 bp), or 45 (1,600 bp). All PCR products were digested with EcoRI, which enabled cloning into an EcoRI site downstream of the yfp ORF in pAAV-CMV-YFP,87 a plasmid carrying a yfp transgene driven by the CMV (Cytomegalovirus) promoter. In this backbone, two AAV ITRs, one of which carries a mutation of the terminal resolution site, flank the expression cassettes and allow for their packaging as scAAV genomes.88
Oversized ssAAV vector constructs expressing Cas9 from Streptococcus pyogenes (Sp) were derived from our previously reported Cas9 expression plasmid3 containing the SpCas9 cDNA from the Church lab (Addgene plasmid 41815). This variant was now replaced with the SpCas9 cDNA from the Zhang lab, by PCR-amplifying this cDNA from former Addgene plasmid 49535 (in the meantime replaced by Addgene by plasmid 52961) using primers 46 and 47 and by cloning the NheI/ClaI-digested PCR product into our appropriately cleaved original AAV/SpCas9 plasmid,3 resulting in pAAV-FZSpCas9. The full-length and minimal CMV promoters (all promoter sequences are shown in Table S4) were directly cloned into the pAAV-FZSpCas9 acceptor by PacI/NheI digest of the source and acceptor plasmids. CMV + I, a CMV promoter with an SV40 intron, was PCR-amplified from pAAV-CMV-YFP using primers 48 and 49 and by cloning the digested PacI/NheI fragment into pAAV-FZSpCas9. U6 and H1 promoters, together with their gRNA expression scaffold89 carrying two inverted BbsI restriction sites, were directly cloned as AscI/NotI fragments.
Cell Culture
All standard cell lines (HeLa, Huh7, LX-2, Panc-I, MCF-7, and RAW264.7) were maintained at 37°C with 5% CO2 incubation. Cells were maintained in DMEM with GlutaMAX (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL penicillin-streptomycin (both Sigma-Aldrich). T84 human colon carcinoma cells (ATCC CCL-248) were cultured in a 50:50 mixture of DMEM and F12 (Thermo Fisher Scientific) supplemented with 10% FBS and 1% penicillin-streptomycin (Thermo Fisher Scientific). CuFi-8 cells were cultured and differentiated as described previously.90
The following primary cells were purchased from PromoCell (Heidelberg, Germany) and were grown as monolayers: Saphenous vein endothelial cells (C-12231), skeletal muscle cells (C-12530), pulmonary fibroblasts (C-12360), and cardiac myocytes (C-12810). Primary human hepatocytes were purchased from Cytes Biotechnologies (Barcelona, Spain). All primary cells were maintained following the supplier’s instructions.
pHAE cells were kindly provided by the Thoraxklinik (Heidelberg University Hospital, Heidelberg, Germany). Resected bronchial tissue was used to generate pHAEs. Primary epithelial cells were cultivated in DMEM-Ham’s F-12 medium (Thermo Fisher Scientific) supplemented with Airway Epithelial Cell Growth Medium Supplement Pack (PromoCell) and ROCK-inhibitor Y-27632 (STEMCELL Technologies, Vancouver, BC, Canada). After an extension phase, cells were transferred onto ThinCerts (Greiner Bio-One, Kremsmünster, Austria) and differentiated at air-liquid interface in PneumaCult ALI Basal Medium supplemented with PneumaCult ALI 10xSupplement (all STEMCELL Technologies). After 3 weeks, established pHAEs were used for experiments.
PBMCs, primary T cells, and macrophages were isolated and purified at the Department of Virology of the Heidelberg University Hospital (Germany). Macrophages were maintained in RPMI 1640 with GlutaMAX medium (Thermo Fisher Scientific) supplemented with 10% FBS and 100 U/mL penicillin-streptomycin. T cells and PBMCs were maintained under the same conditions but additionally supplemented with Phytohemagglutinin (PHA, 2 μg/mL; Merck, Darmstadt, Germany) and interleukin-2 (IL-2, 10 μg/mL; Biomol, Hamburg, Germany).
Human lung organoids were expanded as previously described.91 Differentiation was induced 5 days prior to viral vector microinjection through media change, as described previously.91
Primary intestinal organoids were cultured as previously described.92, 93 Human tissue was received from colon and small intestine resection from the Heidelberg University Hospital. Stem cells containing crypts were isolated following 2 mM EDTA dissociation of tissue sample for 1 h at 4°C. Crypts were spun and washed in ice-cold PBS. Fractions enriched in crypts were filtered with 70-μM filters, and the fractions were observed under a light microscope. Fractions containing the highest number of crypts were pooled and spun again. The supernatant was removed and crypts were re-suspended in Matrigel (Corning, Wiesbaden, Germany). Crypts were trypsinized, seeded onto collagen-coated plates, and maintained in a basal or differentiated state for 5 days in differentiation culture media (see Table S5 for medium components).
Transductions
For transduction of cell lines and primary cells in monolayers, cells were seeded into 96-well plates (Greiner Bio-One) 1 day prior to transduction at densities of 5 × 103 or 1 × 104 cells/well. The next day, the different vectors were added at MOIs of 2 × 104–2 × 105 (scAAV-YFP/BoV) or 1 × 104–5 × 104 (scAAV-Gluc/BoV). The same MOIs were used for the AAV2 controls carrying the same expression cassettes. Transductions were always performed in the presence of 0.5–1 μM Doxorubicin (Santa Cruz Biotechnology, Dallas, TX, USA; 25316-40-9), and the viruses were left on the cells overnight. On the next day, the medium was replaced and the cells were further incubated for 48 h (cell lines) or up to 12 days (primary cells). Transgene expression was assessed via fluorescence microscopy or Gaussia luciferase assay.
For the transduction of pHAEs, viral particles (BoV or AAV2 as control) were added to the apical side at MOIs ranging from 2 × 104 to 1 × 105 (as indicated for each experiment), and they were incubated overnight in the presence of two proteasome inhibitors, Doxorubicin (Santa Cruz Biotechnology, 5 μM) and Calpain inhibitor 1 ALLN (G-Biosciences, St. Louis, MO, USA; 786-057, 40 μM).
For basolateral infection, the transwell inserts were flipped, and scAAV-Gluc/BoV was directly added to the surface of the filter at an MOI of 2 × 104. After 1 h at 37°C, the virus was removed and inserts were inverted again into the medium. The same procedure was applied for the infection from the apical side. Cells were further incubated with the two proteasome inhibitors overnight. The next day, the medium was replaced on the basolateral side with fresh medium without any inhibitors, and transgene expression was assessed over the following 9 days.
For the transduction of lung organoids, luminal access was provided to the viral vectors. To this end, organoids were sheared gently with a flame-narrowed glass pipette and resuspended in 1 vol culture medium. Next, 5 × 109 scAAV-Gluc/BoV viral particles and 3 vol Cultrex growth factor-reduced BME type 2 (Trevigen, Gaithersburg, MD, USA, 3533-010-02) were added to the culture medium and mixed by pipetting. After solidifying for 30 min, 1 mL differentiation medium91 was added, and organoids were assessed microscopically for YFP expression every day for the following 2 weeks.
To transduce intestinal organoids, 5 × 109 scAAV-Gluc/BoV particles were added to each well and were incubated overnight in the presence of 0.5 μM Doxorubicin. The next day, the media were changed, and aliquots were taken at various time points for luciferase activity measurements.
Virus Production, Purification, and Titration
HEK293T cells were grown in 15-cm dishes to about 70% confluency. For the AAV2 control productions, 14 μg of each, AAV2 helper plasmid pWHC2 (encoding AAV2 cap and rep), adenoviral helper (pAdh), and the AAV vector plasmid (pAAV) containing the transgene of interest, was used. For the production of rAAV/BoV chimeric vectors, two methods were employed. The first was previously described by Yan et al.17 and uses a standard four-component protocol that includes co-transfection of pAAV2-rep, pAdh, the HBoV1 helper plasmid pCMVNS*NP1BoV1Cap, and pAAV at a 1.5:3:3:1 molar ratio. Here, to produce the different BoV serotypes, we used our corresponding helper constructs pCMVNS*NP1BoVxCap, where x denotes the specific bocaviral serotype. The second, modified protocol involves a triple-transfection approach comprising three components, i.e., pCMVNS*NP1BoVxCap, pDGΔVP,21 and pAAV at a 3.5:2:1 ratio. For the production of chimeric bocavirus libraries, two plasmids are needed: pAAVCMVNS*ΔVP-Capchim (pool of plasmids with chimeric cap sequences) and pDGΔVP (24 μg each/plate).
In all cases, the DNA was mixed with 0.8 mL 300 mM NaCl and 0.8 mL H2O (per 15-cm dish). Next, a mixture composed of 0.8 mL 300 mM NaCl, 0.4 mL Polyethylenimine (PEI; Polysciences Europe, Eppelheim, Germany) and 0.4 mL H2O was added to each DNA mixture. After a 10-min incubation at room temperature (RT), the DNA-PEI mixtures were added dropwise to the cells. Cells were harvested 72 h post-transfection by scraping and centrifugation at 800 × g. After Benzonase (Merck) digestion for 1 h at 37°C to remove free DNA, rAAV/BoV or rAAV particles were purified from the crude cell lysate as previously described for rAAV vectors, using two rounds of CsCl17, 19 or iodixanol density gradient ultracentrifugation.29 Titers of viral stocks were determined by TaqMan real-time PCR analysis using probes against the transgene (for yfp), the promoter (for Gluc), or np190 for chimeric bocavirus libraries (Table S6).
SDS-PAGE and Western Blotting
Western blot analysis was performed as previously described.94 Briefly, 10 μL from each iodixanol-purified virus stock was mixed with an equal volume of 2× SDS sample loading buffer and boiled for 5 min at 95°C. Then, the samples were separated on 8% SDS-PAGE gels and transferred to a nitrocellulose membrane (NeoLab, Heidelberg, Germany) via semi-dry transfer. Membranes were blocked with 5% milk (Roth, Karlsruhe, Germany) for 1 h at RT and incubated overnight with an anti-VP polyclonal primary rabbit antibody (1:1,000 dilution) recognizing the three bocaviral capsid proteins VP1, VP2, and VP3 (kind gift from Maria Söderlund-Venermo, University of Helsinki). For detection, a horseradish peroxidase-conjugated secondary donkey anti-rabbit antibody (GE Healthcare, Chicago, IL, USA; NA934V) was used at a 1:10,000 dilution. To visualize protein bands, Western Lightning Plus-ECL reagent (PerkinElmer, Waltham, MA, USA) was used, and the emitted signal was detected with a chemiluminescence imager (Intas ChemoStar, Göttingen, Germany).
Dot Blot Analysis
Per CsCl gradient fraction, 15 μL was mixed with 189 μL 50 mM Tris-HCl (pH 8), 1 μL 1 M MgCl2, 1 μL 1 M CaCl2, 2 μL 1 mg/mL DNaseI (Roche, Basel, Switzerland), and 2 μL 1 mg/mL RNase A (QIAGEN), and the entire mixture was incubated for 30 min at 37°C. DNase I was inactivated by adding 2 μL 0.5 M EDTA, 4 μL 0.25 M EGTA, and 10 μL 10% sarcosine, and by then heating the samples to 70°C for 20 min. Next, 10 μL 20 mg/mL proteinase K (Roche) was added to the samples and incubated overnight at 55°C. The next day, each sample was mixed with 40 μL 5 M NaOH, 20 μL 0.5 M EDTA, and 225 μL H2O and incubated at 55°C for 10 min before loading on a Bio-Dot Microfiltration Apparatus (Bio-Rad, Hercules, CA, USA). A nylon membrane (Hybond-N+, GE Healthcare) was cut to size, soaked in 6× saline-sodium citrate (SSC) for 10 min, and assembled into the dot blot device, according to the manufacturer’s protocol. The samples were then applied in two aliquots, followed by a final wash with 200 μL 2× SSC per well. Membranes were placed on a Whatman paper soaked in 10× SSC, and the DNA was crosslinked with a UV-crosslinker for 2 min. A probe binding in the yfp transgene was generated using the DIG Starter Kit II (Roche), according to the manufacturer’s instructions. The blots were pre-hybridized and incubated with probe (roughly 25 ng/mL) overnight. Blots were exposed to X-ray films (GE Healthcare) and developed in an X-OMAT 2000 X-ray film processor (Kodak, Rochester, NY, USA). After background subtraction, dot blots were analyzed using Fiji software.95
Southern Blot Analysis of rAAV Vector Genomes
HEK293T cells were seeded in 15-cm dishes at a density of 4 × 106 cells/dish. After 48 h, cells were transfected for BoV or AAV2 production as described above. 3 days later, cells were harvested in 1 mL 50 mM Tris-HCl and subjected to five freeze-thaw cycles. Then, 540 μg DNase I (Roche), 5 μL 1 M MgCl2, and 5 μL 1 M CaCl2 were added, and the cell lysate was further incubated for 3 h at 37°C. To inactivate the DNase I, 10 μL 0.5 M EDTA and 20 μL 0.25 M EGTA were added, and the cell lysate was incubated at 70°C for 20 min. Viral DNA was then purified using the QIAGEN Blood and Tissue Kit, following the manufacturer’s instructions. Viral DNA was additionally digested with DpnI for 1 h prior to loading on 0.7% alkaline agarose gels (50 mM NaOH and 1 mM EDTA), followed by Southern blotting as previously described.96
Hirt DNA extracts from rAAV- or rAAV/HBoV1-transfected cells were also prepared as previously described.97 Southern blots were probed with DIG-labeled YFP or SpCas9 probes (∼25 ng/mL) and further processed according to the manufacturer’s instructions (DIG starter Kit II, Roche). Blots were exposed to X-ray films (GE Healthcare) and developed in an X-OMAT 2000 X-ray film processor (Kodak).
Measurement of Gaussia Luciferase Reporter Expression
Gaussia luciferase activity in the cell medium was determined by using a GloMax96 microplate luminometer equipped with an automatic injector (Promega, Madison, WI, USA). 20 μL medium was incubated with 100 μL Luciferase assay buffer (1.1 M NaCl, 2.2 mM Na2EDTA, 0.22 M potassium phosphate buffer [pH 5.1], 0.44 mg/mL BSA, and 1.3 mM NaN3) supplied with Coelenterazine (PJK, Kleinblittersdorf, Germany) at a final dilution of 11.7 μM.
Fluorescence-Activated Cell Sorting Analysis
The apical surface of pHAE cells was washed with PBS for 20 min at 37°C to remove cell-associated mucus. Cells were then detached by adding 0.25% Trypsin-EDTA (Thermo Fisher Scientific) to the apical side for 10–15 min at 37°C. Trypsinization was stopped by adding 1% BSA in PBS, and four transwells (from different donors) were pooled and centrifuged at 400 × g for 15 min. Next, cells were washed twice with PBS and resuspended in PBS to a final concentration of 1 × 106 cells/mL. Live-dead cell marker (Fixable Aqua Dead Cell Stain Kit, Thermo Fisher Scientific) was added to the cell suspension (1 μL for 1 × 106 cells) for 30 min in the dark at 4°C. Cells were washed twice with PBS and then fixed with 4% paraformaldehyde (PFA) for 15 min at RT, followed by permeabilization with 1% Triton X-100 for 20 min. Then, cells were washed twice with 0.5% BSA in PBS, and a fluorescein isothiocyanate (FITC)-coupled antibody against GFP/YFP (Novus Biologicals, Littleton, CO, USA; NB100-1771) was added to all samples (including the negative controls) to enhance the signal in YFP-expressing cells.
To stain the different cell types in the airway epithelia, the following primary antibodies were used: (1) goblet cell marker MUC5A/C (Abcam, Cambridge, UK; ab3649) diluted 1:100, ciliated cell marker β-Tubulin IV (Merck, T7941) diluted 1:100, basal cell marker Cytokeratin5 (Santa Cruz Biotechnology, RCK103) diluted 1:50, or Clara (club) cell marker CC16 (BioVendor, Heidelberg, Germany; RD181022220-01) diluted 1:200. The primary antibodies were incubated for 1 h at 4°C, followed by treatment with secondary anti-mouse (AF-647 goat anti-mouse A21235, Thermo Fisher Scientific) or secondary anti-rabbit (AF-645 donkey anti-rabbit 711-605-152; Dianova, Hamburg, Germany) antibodies for 30 min at RT. Cells were measured on a FACSVerse (BD Biosciences, Franklin Lakes, NJ, USA), and analysis was performed using Flowing Software (version 2.5.1; Turku Centre for Biotechnology, Turku, Finland). Only living cells were used for the analysis, and gates were placed according to the following control conditions: (1) cells transduced with scAAV-Gluc/HBoV1 and then stained with FITC-coupled anti-GFP antibody, the different cell-specific primary antibody, and the respective secondary antibody; (2) cells transduced with scAAV-YFP/HBoV1 and then stained with each secondary antibody alone and FITC-coupled anti-GFP antibody; or (3) cells transduced with scAAV-YFP/HBoV1 and only stained with FITC-coupled anti-GFP antibody.
Immunostainings
Cells transduced with recombinant viruses were fixed with 4% PFA for 15 min at various time points post-transduction (as indicated in the Results). The cells were then washed three times with PBS and permeabilized with 0.1% Triton X-100 for 15 min. Next, 3% BSA in PBS was added to the cells, and the plates were blocked for 1 h in the dark. To enhance the YFP signal, 100–200 μL 1:1,000 diluted primary FITC-coupled anti-GFP antibody (Novus Biologicals) was added to each well. After overnight incubation at 4°C, plates were washed three times with PBS before a secondary anti-goat IgG antibody (Alexa Fluor 488-labeled, Thermo Fisher Scientific) was added for 1 h at RT. Plates were washed again three times with PBS, and Hoechst was added at a 1:3,000 dilution to the PBS from the last wash. Finally, plates were stored with 200 μL PBS/well in the dark at 4°C until microscopy analysis. Microscopy pictures were taken with an Olympus inverted fluorescence microscope IX-81 and were processed using Fiji.
Shuffling of Bocavirus Capsids and Generation of Libraries
BoV cap ORFs (spanning vp1–3) were PCR-amplified with HotStar HiFidelity Polymerase (QIAGEN) using primers 38 and 39, which bind in regions flanking the cap ORF that are shared among all BoV helpers. The forward primer 38 binds upstream in the ns1 ORF, and the reverse primer 39 binds in a genomic region downstream of the cap ORF. The resulting 2,631-bp PCR products were separated on agarose gels and purified using the QIAquick Gel Extraction Kit (QIAGEN). Then, the PCR products were mixed in equimolar ratios to a total amount of 4 μg. DNA fragmentation through DNase I digestion was performed as previously described for AAV.5 The reactions were resolved by agarose gel electrophoresis, and fragments between 100 and 1,000 bp were excised and purified using the QIAquick Gel Extraction Kit (QIAGEN). Reassembly of the capsid genes was achieved in a primer-less reaction as previously described.5, 18, 80 Briefly, 500 ng DNase I-digested fragments was amplified with Phusion HS II polymerase (Thermo Fisher Scientific), following the manufacturer’s instructions. PCR conditions were 98°C for 30 s; 40 cycles of 98°C for 10 s, 52°C for 30 s, and 72°C for 45 s + 1 s/cycle; and final elongation at 72°C for 10 min. Elongation time was increased by 1 s in each cycle.80
Afterward, 1–2 μL of this assembly reaction was used to PCR-amplify and enrich complete capsid genes using Phusion HS II polymerase and primers 34/40 that bind to regions flanking all capsid genes. Conditions for the second PCR were 98°C for 2 min; 40 cycles of 98°C for 10 s, 65°C for 15 s, and 72°C for 90 s; and final elongation at 72°C for 1 min.
To verify successful reassembly and amplification, an aliquot of the second PCR was visualized on a 1% agarose gel (expected size was about 2.2 kb). Next, the second PCR was scaled up to 16 reactions of 50 μL each. Products were then purified using multiple QIAquick PCR Purification columns (QIAGEN) for subsequent cloning of a capsid library. To this end, purified capsid genes were digested with Esp3I (BsmBI) in Tango Buffer (both Thermo Fisher Scientific) overnight at 37°C. The digest was then separated on a gel and purified using the QIAquick Gel Extraction Kit. The shuffled capsid genes were ligated overnight with 1 μg purified BsmBI-digested acceptor vector using a 3:1 insert:vector ratio.
Next, the ligation reaction was dialyzed using MF-Millipore Membrane Filter (Merck) and transformed into E. cloni SUPREME electrocompetent cells (Lucigen, Madison, WI, USA), according to the manufacturer’s instructions. After 45 min recovery in the supplied medium, the reactions were pooled and 100 μL of this suspension (undiluted as well as 1:10 and 1:100 dilutions) were plated on Luria-Bertani broth (LB)-ampicillin agar plates to estimate the number of clones in the library. The rest of the transformation suspension was grown overnight in 400 mL LB-ampicillin to amplify the plasmid library. DNA maxi preps (Macherey-Nagel, Düren, Germany) were performed to obtain a plasmid library for viral library production.
Statistical Methods
The statistical analysis in Figure 2G was performed in GraphPad Prism v.5.0 (https://www.graphpad.com). The experiment was performed in at least three biological replicates (i.e., transwells). A one-way ANOVA with Tukey’s multiple comparison test was used for statistical analysis, as indicated in the figure legend.
Ethical Approval
This study was carried out in accordance with the recommendations of the Heidelberg University Hospital, with written informed consent from all subjects in accordance with the Declaration of Helsinki. All samples were received and maintained in an anonymized manner. The protocols were approved by the ethics commission at Heidelberg University Hospital under the protocols S-270/2001 (collection of surgical material for lung research) or S-443/2017 (collection of colon and intestinal material to study infectious diseases).
PBMCs were obtained as leftover material from anonymous blood donations at the blood bank at Heidelberg University. As this material could not be used otherwise in the blood bank and as the samples have been fully anonymized, the ethics committee at Heidelberg University Hospital does not require specific approval for the use of this material.
Written informed consent for the derivation and usage of human lung organoid lines was obtained from patients of the St. Antonius Hospital Nieuwegein (protocol Z-12.55). In all cases, patients can withdraw their consent at any time, leading to the prompt disposal of their tissue and any derived material.
Author Contributions
J.F. and D.G. conceived and designed the experiments. J.F. generated constructs and performed the majority of experiments. M.A.S. and M.M. established the pHAEs culture. S.H. and J.E.A. designed and performed DNA family shuffling. Y.V. generated oversized CRISPR cassettes. J.P., M.S., H.C., S.B., and J.F. conceived and performed experiments in primary organoid and crypt cultures. V.S. and O.S. performed transductions of the CuFi-8 cell line. Z.Y. and J.Q. provided practical advice. J.F. and D.G. wrote the manuscript. All authors read the manuscript and approved the final version.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
J.F. and D.G. are grateful for funding from the Cystic Fibrosis Foundation (CFF, grant GRIMM15XX0), the German Research Foundation (DFG, Cluster of Excellence CellNetworks, EXC81), as well as from the Heidelberg Biosciences International Graduate School HBIGS at Heidelberg University. D.G. acknowledges additional funding by the German Center for Infection Research (DZIF, BMBF; TTU-HIV 04.803). M.A.S. and M.M. acknowledge funding by the German Center for Lung Research (DZL, BMBF; 82DZL00402). J.E.A. and D.G. are grateful for support through the MYOCURE project. MYOCURE has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement 667751. S.B. and D.G. appreciate support from the Collaborative Research Center SFB1129 (German Research Foundation, DFG; TP14 to S.B. and TP2 to D.G.). S.B. was supported by a research grant from the Chica and Heinz Schaller Foundation. V.S. and O.S. are grateful to the Beatrix-Lichtken-Stiftung Cologne for funding. Z.Y. and J.Q. appreciate funding through grant AI139572 from the National Institute of Allergy and Infectious Diseases, NIH, USA. This work was further supported by the gravitation program NOCI: 024.003.001 from the Netherlands Organisation for Scientific Research (NWO). Finally, we thank Janina Haar for sharing various primary cells including human hepatocytes and cells shown in Figure 4A; Manuela Nickl for providing T cells; as well as Andrea Imle, Kathleen Börner, and David Bejarano (all Department of Infectious Diseases/Virology, Heidelberg University Hospital, Heidelberg, Germany) for preparing and providing PBMCs and macrophages.
Footnotes
Supplemental Information includes five figures and six tables and can be found with this article online at https://doi.org/10.1016/j.omtm.2019.01.003.
Supplemental Information
References
- 1.Carpentier A.C., Frisch F., Labbé S.M., Gagnon R., de Wal J., Greentree S., Petry H., Twisk J., Brisson D., Gaudet D. Effect of alipogene tiparvovec (AAV1-LPL(S447X)) on postprandial chylomicron metabolism in lipoprotein lipase-deficient patients. J. Clin. Endocrinol. Metab. 2012;97:1635–1644. doi: 10.1210/jc.2011-3002. [DOI] [PubMed] [Google Scholar]
- 2.Bennett J., Wellman J., Marshall K.A., McCague S., Ashtari M., DiStefano-Pappas J., Elci O.U., Chung D.C., Sun J., Wright J.F. Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: a follow-on phase 1 trial. Lancet. 2016;388:661–672. doi: 10.1016/S0140-6736(16)30371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Senís E., Fatouros C., Große S., Wiedtke E., Niopek D., Mueller A.K., Börner K., Grimm D. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol. J. 2014;9:1402–1412. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- 4.Flotte T.R., Afione S.A., Solow R., Drumm M.L., Markakis D., Guggino W.B., Zeitlin P.L., Carter B.J. Expression of the cystic fibrosis transmembrane conductance regulator from a novel adeno-associated virus promoter. J. Biol. Chem. 1993;268:3781–3790. [PubMed] [Google Scholar]
- 5.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Münch R.C., Muth A., Muik A., Friedel T., Schmatz J., Dreier B., Trkola A., Plückthun A., Büning H., Buchholz C.J. Off-target-free gene delivery by affinity-purified receptor-targeted viral vectors. Nat. Commun. 2015;6:6246. doi: 10.1038/ncomms7246. [DOI] [PubMed] [Google Scholar]
- 7.Hakim C.H., Wasala N.B., Pan X., Kodippili K., Yue Y., Zhang K., Yao G., Haffner B., Duan S.X., Ramos J. A Five-Repeat Micro-Dystrophin Gene Ameliorated Dystrophic Phenotype in the Severe DBA/2J-mdx Model of Duchenne Muscular Dystrophy. Mol. Ther. Methods Clin. Dev. 2017;6:216–230. doi: 10.1016/j.omtm.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan D., Yue Y., Engelhardt J.F. Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Mol. Ther. 2001;4:383–391. doi: 10.1006/mthe.2001.0456. [DOI] [PubMed] [Google Scholar]
- 9.Angelova A.L., Geletneky K., Nüesch J.P., Rommelaere J. Tumor Selectivity of Oncolytic Parvoviruses: From in vitro and Animal Models to Cancer Patients. Front. Bioeng. Biotechnol. 2015;3:55. doi: 10.3389/fbioe.2015.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paglino J.C., Ozduman K., van den Pol A.N. LuIII parvovirus selectively and efficiently targets, replicates in, and kills human glioma cells. J. Virol. 2012;86:7280–7291. doi: 10.1128/JVI.00227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maxwell I.H., Maxwell F., Rhode S.L., 3rd, Corsini J., Carlson J.O. Recombinant LuIII autonomous parvovirus as a transient transducing vector for human cells. Hum. Gene Ther. 1993;4:441–450. doi: 10.1089/hum.1993.4.4-441. [DOI] [PubMed] [Google Scholar]
- 12.Geletneky K., Hajda J., Angelova A.L., Leuchs B., Capper D., Bartsch A.J., Neumann J.O., Schöning T., Hüsing J., Beelte B. Oncolytic H-1 Parvovirus Shows Safety and Signs of Immunogenic Activity in a First Phase I/IIa Glioblastoma Trial. Mol. Ther. 2017;25:2620–2634. doi: 10.1016/j.ymthe.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponnazhagan S., Weigel K.A., Raikwar S.P., Mukherjee P., Yoder M.C., Srivastava A. Recombinant human parvovirus B19 vectors: erythroid cell-specific delivery and expression of transduced genes. J. Virol. 1998;72:5224–5230. doi: 10.1128/jvi.72.6.5224-5230.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivastava C.H., Samulski R.J., Lu L., Larsen S.H., Srivastava A. Construction of a recombinant human parvovirus B19: adeno-associated virus 2 (AAV) DNA inverted terminal repeats are functional in an AAV-B19 hybrid virus. Proc. Natl. Acad. Sci. USA. 1989;86:8078–8082. doi: 10.1073/pnas.86.20.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spitzer A.L., Maxwell F., Corsini J., Maxwell I.H. Species specificity for transduction of cultured cells by a recombinant LuIII rodent parvovirus genome encapsidated by canine parvovirus or feline panleukopenia virus. J. Gen. Virol. 1996;77:1787–1792. doi: 10.1099/0022-1317-77-8-1787. [DOI] [PubMed] [Google Scholar]
- 16.Krüger L., Eskerski H., Dinsart C., Cornelis J., Rommelaere J., Haberkorn U., Kleinschmidt J.A. Augmented transgene expression in transformed cells using a parvoviral hybrid vector. Cancer Gene Ther. 2008;15:252–267. doi: 10.1038/sj.cgt.7701113. [DOI] [PubMed] [Google Scholar]
- 17.Yan Z., Keiser N.W., Song Y., Deng X., Cheng F., Qiu J., Engelhardt J.F. A novel chimeric adenoassociated virus 2/human bocavirus 1 parvovirus vector efficiently transduces human airway epithelia. Mol. Ther. 2013;21:2181–2194. doi: 10.1038/mt.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kienle E., Senís E., Börner K., Niopek D., Wiedtke E., Grosse S., Grimm D. Engineering and evolution of synthetic adeno-associated virus (AAV) gene therapy vectors via DNA family shuffling. J. Vis. Exp. 2012;(62):3819. doi: 10.3791/3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan Z., Feng Z., Sun X., Zhang Y., Zou W., Wang Z., Jensen-Cody C., Liang B., Park S.Y., Qiu J., Engelhardt J.F. Human Bocavirus Type-1 Capsid Facilitates the Transduction of Ferret Airways by Adeno-Associated Virus Genomes. Hum. Gene Ther. 2017;28:612–625. doi: 10.1089/hum.2017.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm D., Kern A., Rittner K., Kleinschmidt J.A. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 1998;9:2745–2760. doi: 10.1089/hum.1998.9.18-2745. [DOI] [PubMed] [Google Scholar]
- 21.Dubielzig R., King J.A., Weger S., Kern A., Kleinschmidt J.A. Adeno-associated virus type 2 protein interactions: formation of pre-encapsidation complexes. J. Virol. 1999;73:8989–8998. doi: 10.1128/jvi.73.11.8989-8998.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grieger J.C., Samulski R.J. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J. Virol. 2005;79:9933–9944. doi: 10.1128/JVI.79.15.9933-9944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Z., Yang H., Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong J.Y., Fan P.D., Frizzell R.A. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum. Gene Ther. 1996;7:2101–2112. doi: 10.1089/hum.1996.7.17-2101. [DOI] [PubMed] [Google Scholar]
- 25.McCarty D.M., Monahan P.E., Samulski R.J. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8:1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- 26.Wu J., Zhao W., Zhong L., Han Z., Li B., Ma W., Weigel-Kelley K.A., Warrington K.H., Srivastava A. Self-complementary recombinant adeno-associated viral vectors: packaging capacity and the role of rep proteins in vector purity. Hum. Gene Ther. 2007;18:171–182. doi: 10.1089/hum.2006.088. [DOI] [PubMed] [Google Scholar]
- 27.Guido M., Tumolo M.R., Verri T., Romano A., Serio F., De Giorgi M., De Donno A., Bagordo F., Zizza A. Human bocavirus: Current knowledge and future challenges. World J. Gastroenterol. 2016;22:8684–8697. doi: 10.3748/wjg.v22.i39.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapoor A., Mehta N., Esper F., Poljsak-Prijatelj M., Quan P.L., Qaisar N., Delwart E., Lipkin W.I. Identification and characterization of a new bocavirus species in gorillas. PLoS ONE. 2010;5:e11948. doi: 10.1371/journal.pone.0011948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zolotukhin S., Byrne B.J., Mason E., Zolotukhin I., Potter M., Chesnut K., Summerford C., Samulski R.J., Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 30.Jartti T., Hedman K., Jartti L., Ruuskanen O., Allander T., Söderlund-Venermo M. Human bocavirus-the first 5 years. Rev. Med. Virol. 2012;22:46–64. doi: 10.1002/rmv.720. [DOI] [PubMed] [Google Scholar]
- 31.Hueffer K., Parrish C.R. Parvovirus host range, cell tropism and evolution. Curr. Opin. Microbiol. 2003;6:392–398. doi: 10.1016/s1369-5274(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 32.Manteufel J., Truyen U. Animal bocaviruses: a brief review. Intervirology. 2008;51:328–334. doi: 10.1159/000173734. [DOI] [PubMed] [Google Scholar]
- 33.Li L., Pesavento P.A., Leutenegger C.M., Estrada M., Coffey L.L., Naccache S.N., Samayoa E., Chiu C., Qiu J., Wang C. A novel bocavirus in canine liver. Virol. J. 2013;10:54. doi: 10.1186/1743-422X-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan Z., Zak R., Zhang Y., Ding W., Godwin S., Munson K., Peluso R., Engelhardt J.F. Distinct classes of proteasome-modulating agents cooperatively augment recombinant adeno-associated virus type 2 and type 5-mediated transduction from the apical surfaces of human airway epithelia. J. Virol. 2004;78:2863–2874. doi: 10.1128/JVI.78.6.2863-2874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng X., Yan Z., Luo Y., Xu J., Cheng F., Li Y., Engelhardt J.F., Qiu J. In vitro modeling of human bocavirus 1 infection of polarized primary human airway epithelia. J. Virol. 2013;87:4097–4102. doi: 10.1128/JVI.03132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duan D., Yue Y., Yan Z., Yang J., Engelhardt J.F. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J. Clin. Invest. 2000;105:1573–1587. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barkauskas C.E., Chung M.I., Fioret B., Gao X., Katsura H., Hogan B.L. Lung organoids: current uses and future promise. Development. 2017;144:986–997. doi: 10.1242/dev.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 39.Kantola K., Hedman L., Arthur J., Alibeto A., Delwart E., Jartti T., Ruuskanen O., Hedman K., Söderlund-Venermo M. Seroepidemiology of human bocaviruses 1-4. J. Infect. Dis. 2011;204:1403–1412. doi: 10.1093/infdis/jir525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meliani A., Leborgne C., Triffault S., Jeanson-Leh L., Veron P., Mingozzi F. Determination of anti-adeno-associated virus vector neutralizing antibody titer with an in vitro reporter system. Hum. Gene Ther. Methods. 2015;26:45–53. doi: 10.1089/hgtb.2015.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi J.W., Jung J.Y., Lee J.I., Lee K.K., Oem J.K. Molecular characteristics of a novel strain of canine minute virus associated with hepatitis in a dog. Arch. Virol. 2016;161:2299–2304. doi: 10.1007/s00705-016-2895-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brebion A., Vanlieferinghen P., Déchelotte P., Boutry M., Peigue-Lafeuille H., Henquell C. Fatal subacute myocarditis associated with human bocavirus 2 in a 13-month-old child. J. Clin. Microbiol. 2014;52:1006–1008. doi: 10.1128/JCM.03013-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kainulainen L., Waris M., Söderlund-Venermo M., Allander T., Hedman K., Ruuskanen O. Hepatitis and human bocavirus primary infection in a child with T-cell deficiency. J. Clin. Microbiol. 2008;46:4104–4105. doi: 10.1128/JCM.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haytoğlu Z., Canan O. Bocavirus Viremia and Hepatitis in an Immunocompetent Child. Balkan Med. J. 2017;34:281–283. doi: 10.4274/balkanmedj.2015.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonvicini F., Manaresi E., Gentilomi G.A., Di Furio F., Zerbini M., Musiani M., Gallinella G. Evidence of human bocavirus viremia in healthy blood donors. Diagn. Microbiol. Infect. Dis. 2011;71:460–462. doi: 10.1016/j.diagmicrobio.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 46.Kapoor A., Simmonds P., Slikas E., Li L., Bodhidatta L., Sethabutr O., Triki H., Bahri O., Oderinde B.S., Baba M.M. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis. 2010;201:1633–1643. doi: 10.1086/652416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desdouits M., Munier S., Prevost M.C., Jeannin P., Butler-Browne G., Ozden S., Gessain A., Van Der Werf S., Naffakh N., Ceccaldi P.E. Productive infection of human skeletal muscle cells by pandemic and seasonal influenza A(H1N1) viruses. PLoS ONE. 2013;8:e79628. doi: 10.1371/journal.pone.0079628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuethe F., Lindner J., Matschke K., Wenzel J.J., Norja P., Ploetze K., Schaal S., Kamvissi V., Bornstein S.R., Schwanebeck U., Modrow S. Prevalence of parvovirus B19 and human bocavirus DNA in the heart of patients with no evidence of dilated cardiomyopathy or myocarditis. Clin. Infect. Dis. 2009;49:1660–1666. doi: 10.1086/648074. [DOI] [PubMed] [Google Scholar]
- 49.Schildgen V., Longo Y., Pieper M., Schildgen O. T84 air-liquid interface cultures enable isolation of human bocavirus. Influenza Other Respir. Viruses. 2018;12:667–668. doi: 10.1111/irv.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghietto L.M., Toigo D’Angelo A.P., Viale F.A., Adamo M.P. Human bocavirus 1 infection of CACO-2 cell line cultures. Virology. 2017;510:273–280. doi: 10.1016/j.virol.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansen M., Brockmann M., Schildgen V., Schildgen O. Human bocavirus is detected in human placenta and aborted tissues. Influenza Other Respir. Viruses. 2019;13:106–109. doi: 10.1111/irv.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grimm D., Zolotukhin S. E Pluribus Unum: 50 Years of Research, Millions of Viruses, and One Goal--Tailored Acceleration of AAV Evolution. Mol. Ther. 2015;23:1819–1831. doi: 10.1038/mt.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schürmann N., Trabuco L.G., Bender C., Russell R.B., Grimm D. Molecular dissection of human Argonaute proteins by DNA shuffling. Nat. Struct. Mol. Biol. 2013;20:818–826. doi: 10.1038/nsmb.2607. [DOI] [PubMed] [Google Scholar]
- 54.Herrmann A.K., Bender C., Kienle E., Grosse S., El Andari J., Botta J., Schürmann N., Wiedtke E., Niopek D., Grimm D. A robust and all-inclusive pipeline for shuffling of Adeno-associated viruses (AAV) ACS Synth. Biol. 2019;8:194–206. doi: 10.1021/acssynbio.8b00373. [DOI] [PubMed] [Google Scholar]
- 55.Arthur J.L., Higgins G.D., Davidson G.P., Givney R.C., Ratcliff R.M. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog. 2009;5:e1000391. doi: 10.1371/journal.ppat.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kapoor A., Slikas E., Simmonds P., Chieochansin T., Naeem A., Shaukat S., Alam M.M., Sharif S., Angez M., Zaidi S., Delwart E. A newly identified bocavirus species in human stool. J. Infect. Dis. 2009;199:196–200. doi: 10.1086/595831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grimm D., Kay M.A., Kleinschmidt J.A. Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol. Ther. 2003;7:839–850. doi: 10.1016/s1525-0016(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 58.Li S., Ling C., Zhong L., Li M., Su Q., He R., Tang Q., Greiner D.L., Shultz L.D., Brehm M.A. Efficient and Targeted Transduction of Nonhuman Primate Liver With Systemically Delivered Optimized AAV3B Vectors. Mol. Ther. 2015;23:1867–1876. doi: 10.1038/mt.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Limberis M.P., Vandenberghe L.H., Zhang L., Pickles R.J., Wilson J.M. Transduction efficiencies of novel AAV vectors in mouse airway epithelium in vivo and human ciliated airway epithelium in vitro. Mol. Ther. 2009;17:294–301. doi: 10.1038/mt.2008.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J., DeClercq J.J., Hayward S.B., Li P.W., Shivak D.A., Gregory P.D., Lee G., Holmes M.C. Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic Acids Res. 2016;44:e30. doi: 10.1093/nar/gkv1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song J.R., Jin Y., Xie Z.P., Gao H.C., Xiao N.G., Chen W.X., Xu Z.Q., Yan K.L., Zhao Y., Hou Y.D., Duan Z.J. Novel human bocavirus in children with acute respiratory tract infection. Emerg. Infect. Dis. 2010;16:324–327. doi: 10.3201/eid1602.090553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan Z., Zou W., Feng Z., Shen W., Park S.Y., Deng X., Qiu J., Engelhardt J.F. Establishment of a High Yield rAAV/HBoV Vector Production System Independent of Bocavirus Non-structural Proteins. Hum. Gene Ther. 2018 doi: 10.1089/hum.2018.173. Published online November 6, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Douar A.M., Poulard K., Stockholm D., Danos O. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J. Virol. 2001;75:1824–1833. doi: 10.1128/JVI.75.4.1824-1833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan Z., Zak R., Luxton G.W., Ritchie T.C., Bantel-Schaal U., Engelhardt J.F. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 2002;76:2043–2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhong L., Li B., Mah C.S., Govindasamy L., Agbandje-McKenna M., Cooper M., Herzog R.W., Zolotukhin I., Warrington K.H., Jr., Weigel-Van Aken K.A. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. USA. 2008;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Markusic D.M., Herzog R.W., Aslanidi G.V., Hoffman B.E., Li B., Li M., Jayandharan G.R., Ling C., Zolotukhin I., Ma W. High-efficiency transduction and correction of murine hemophilia B using AAV2 vectors devoid of multiple surface-exposed tyrosines. Mol. Ther. 2010;18:2048–2056. doi: 10.1038/mt.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petrs-Silva H., Dinculescu A., Li Q., Deng W.T., Pang J.J., Min S.H., Chiodo V., Neeley A.W., Govindasamy L., Bennett A. Novel properties of tyrosine-mutant AAV2 vectors in the mouse retina. Mol. Ther. 2011;19:293–301. doi: 10.1038/mt.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mao Y., Wang X., Yan R., Hu W., Li A., Wang S., Li H. Single point mutation in adeno-associated viral vectors -DJ capsid leads to improvement for gene delivery in vivo. BMC Biotechnol. 2016;16:1. doi: 10.1186/s12896-015-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mietzsch M., Kailasan S., Garrison J., Ilyas M., Chipman P., Kantola K., Janssen M.E., Spear J., Sousa D., McKenna R. Structural Insights into Human Bocaparvoviruses. J. Virol. 2017;91:e00261-17. doi: 10.1128/JVI.00261-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zaiss A.K., Liu Q., Bowen G.P., Wong N.C., Bartlett J.S., Muruve D.A. Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J. Virol. 2002;76:4580–4590. doi: 10.1128/JVI.76.9.4580-4590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laredj L.N., Beard P. Adeno-associated virus activates an innate immune response in normal human cells but not in osteosarcoma cells. J. Virol. 2011;85:13133–13143. doi: 10.1128/JVI.05407-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vandendriessche T., Thorrez L., Acosta-Sanchez A., Petrus I., Wang L., Ma L., DE Waele L., Iwasaki Y., Gillijns V., Wilson J.M. Efficacy and safety of adeno-associated viral vectors based on serotype 8 and 9 vs. lentiviral vectors for hemophilia B gene therapy. J. Thromb. Haemost. 2007;5:16–24. doi: 10.1111/j.1538-7836.2006.02220.x. [DOI] [PubMed] [Google Scholar]
- 73.Mingozzi F., High K.A. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood. 2013;122:23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Z., Zheng Z., Luo H., Meng J., Li H., Li Q., Zhang X., Ke X., Bai B., Mao P. Human bocavirus NP1 inhibits IFN-β production by blocking association of IFN regulatory factor 3 with IFNB promoter. J. Immunol. 2012;189:1144–1153. doi: 10.4049/jimmunol.1200096. [DOI] [PubMed] [Google Scholar]
- 75.Luo H., Zhang Z., Zheng Z., Ke X., Zhang X., Li Q., Liu Y., Bai B., Mao P., Hu Q., Wang H. Human bocavirus VP2 upregulates IFN-β pathway by inhibiting ring finger protein 125-mediated ubiquitination of retinoic acid-inducible gene-I. J. Immunol. 2013;191:660–669. doi: 10.4049/jimmunol.1202933. [DOI] [PubMed] [Google Scholar]
- 76.Liu Q., Zhang Z., Zheng Z., Zheng C., Liu Y., Hu Q., Ke X., Wang H. Human Bocavirus NS1 and NS1-70 Proteins Inhibit TNF-α-Mediated Activation of NF-κB by targeting p65. Sci. Rep. 2016;6:28481. doi: 10.1038/srep28481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stemmer W.P. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurtzman A.L., Govindarajan S., Vahle K., Jones J.T., Heinrichs V., Patten P.A. Advances in directed protein evolution by recursive genetic recombination: applications to therapeutic proteins. Curr. Opin. Biotechnol. 2001;12:361–370. doi: 10.1016/s0958-1669(00)00228-7. [DOI] [PubMed] [Google Scholar]
- 79.Excoffon K.J., Koerber J.T., Dickey D.D., Murtha M., Keshavjee S., Kaspar B.K., Zabner J., Schaffer D.V. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc. Natl. Acad. Sci. USA. 2009;106:3865–3870. doi: 10.1073/pnas.0813365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koerber J.T., Jang J.H., Schaffer D.V. DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol. Ther. 2008;16:1703–1709. doi: 10.1038/mt.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lisowski L., Dane A.P., Chu K., Zhang Y., Cunningham S.C., Wilson E.M., Nygaard S., Grompe M., Alexander I.E., Kay M.A. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brožová K., Hrazdilová K., Slaninková E., Modrý D., Černý J., Celer V. Genetic and phylogenetic characterization of novel bocaparvovirus infecting chimpanzee. Infect. Genet. Evol. 2016;37:231–236. doi: 10.1016/j.meegid.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 83.Zhou F., Sun H., Wang Y. Porcine bocavirus: achievements in the past five years. Viruses. 2014;6:4946–4960. doi: 10.3390/v6124946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zou W., Cheng F., Shen W., Engelhardt J.F., Yan Z., Qiu J. Nonstructural Protein NP1 of Human Bocavirus 1 Plays a Critical Role in the Expression of Viral Capsid Proteins. J. Virol. 2016;90:4658–4669. doi: 10.1128/JVI.02964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Samulski R.J., Berns K.I., Tan M., Muzyczka N. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc. Natl. Acad. Sci. USA. 1982;79:2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grimm D., Kern A., Pawlita M., Ferrari F., Samulski R., Kleinschmidt J. Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 1999;6:1322–1330. doi: 10.1038/sj.gt.3300946. [DOI] [PubMed] [Google Scholar]
- 87.Kunze C., Börner K., Kienle E., Orschmann T., Rusha E., Schneider M., Radivojkov-Blagojevic M., Drukker M., Desbordes S., Grimm D., Brack-Werner R. Synthetic AAV/CRISPR vectors for blocking HIV-1 expression in persistently infected astrocytes. Glia. 2018;66:413–427. doi: 10.1002/glia.23254. [DOI] [PubMed] [Google Scholar]
- 88.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 89.Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang Q., Deng X., Yan Z., Cheng F., Luo Y., Shen W., Lei-Butters D.C., Chen A.Y., Li Y., Tang L. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog. 2012;8:e1002899. doi: 10.1371/journal.ppat.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sachs N., Zomer-van Ommen D.D., Papaspyropoulos A., Heo I., Bottinger L., Klay D., Weeber F., Huelsz-Prince G., Iakobachvili N., Viveen M.C. Long-term expanding human airway organoids for disease modelling. bioRxiv. 2018 doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 93.Pervolaraki K., Stanifer M.L., Münchau S., Renn L.A., Albrecht D., Kurzhals S., Senís E., Grimm D., Schröder-Braunstein J., Rabin R.L., Boulant S. Type I and Type III Interferons Display Different Dependency on Mitogen-Activated Protein Kinases to Mount an Antiviral State in the Human Gut. Front. Immunol. 2017;8:459. doi: 10.3389/fimmu.2017.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grosse S., Penaud-Budloo M., Herrmann A.K., Börner K., Fakhiri J., Laketa V., Krämer C., Wiedtke E., Gunkel M., Ménard L. Relevance of Assembly-Activating Protein for Adeno-associated Virus Vector Production and Capsid Protein Stability in Mammalian and Insect Cells. J. Virol. 2017;91:e01198-17. doi: 10.1128/JVI.01198-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sambrook J., Russell D.W. Alkaline agarose gel electrophoresis. CSH Protoc. 2006;2006 doi: 10.1101/pdb.prot4027. pdb.prot4027. [DOI] [PubMed] [Google Scholar]
- 97.Bardelli M., Zarate-Perez F., Agundez L., Jolinon N., Michael Linden R., Escalante C.R., Henckaerts E. Analysis of Replicative Intermediates of Adeno-associated Virus through Hirt Extraction and Southern Blotting. Bio Protoc. 2017;7:e2271. doi: 10.21769/BioProtoc.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.