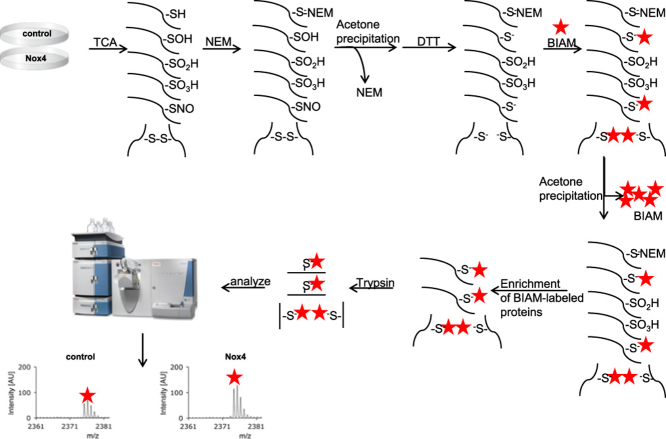
Abstract
Aim
NADPH oxidase (Nox) -derived reactive oxygen species have been implicated in redox signaling via cysteine oxidation in target proteins. Although the importance of oxidation of target proteins is well known, the specificity of such events is often debated. Only a limited number of Nox-oxidized proteins have been identified thus far; especially little is known concerning redox-targets of the constitutively active NADPH oxidase Nox4.
In this study, HEK293 cells with tetracycline-inducible Nox4 overexpression (HEK-tet-Nox4), as well as podocytes of WT and Nox4-/- mice, were utilized to identify Nox4-dependent redox-modified proteins.
Results
TGFβ1 induced an elevation in Nox4 expression in podocytes from WT but not Nox4-/- mice. Using BIAM based redox switch assay in combination with mass spectrometry and western blot analysis, 142 proteins were identified as differentially oxidized in podocytes from wild type vs. Nox4-/- mice and 131 proteins were differentially oxidized in HEK-tet-Nox4 cells upon Nox4 overexpression. A predominant overlap was found for peroxiredoxins and thioredoxins, as expected. More interestingly, the GRB2-associated-binding protein 1 (Gab1) was identified as being differentially oxidized in both approaches. Further analysis using mass spectrometry-coupled BIAM switch assay and site directed mutagenesis, revealed Cys374 and Cys405 as the major Nox4 targeted oxidation sites in Gab1.
Innovation & conclusion
BIAM switch assay coupled to mass spectrometry is a powerful and versatile tool to identify differentially oxidized proteins in a global untargeted way. Nox4, as a source of hydrogen peroxide, changes the redox-state of numerous proteins. Of those, we identified Gab1 as a novel redox target of Nox4.
Abbreviations: Nox, NADPH oxidase; PMA, Phorbol Myristate Acetate; ROS, Reactive Oxygen Species; BIAM, biotinylated Iodoacetamide; Tet, tetracycline; Prx, Peroxiredoxin; Trx, Thioredoxin; AMS, 4-Acetamido-4′-Maleimidylstilbene-2,2′-Disulfonic Acid
Keywords: NADPH oxidase, Nox4, BIAM switch assay, Reactive oxygen species, Mass spectrometry
Graphical abstract
Highlights
-
•
BIAM switch assay coupled to MS is a powerful tool to identify oxidized proteins.
-
•
Stable Nox4 overexpression oxidized Prx and Trx in presence of Auranofin only.
-
•
Gab1 was identified as novel redox-target of Nox4.
-
•
Cys374 and Cys405 were identified as the major oxidized cysteines in Gab1.
1. Introduction
NADPH oxidases of the Nox family are important sources of reactive oxygen species (ROS). In humans, several Nox homologues are expressed which can be classified by their mode of activation. Nox1, Nox2 and Nox3 are mainly activated by an assembly of cytosolic proteins, whereas the activities of Nox5, DUOX1 and DUOX2 are calcium dependent. Nox4 is a unique NADPH oxidase family member - owing to the fact that it is constitutively active and independent from cytosolic subunits [1]. The sole function of Nox4 is to produce hydrogen peroxide (H2O2) [2], [3]. ROS, particularly H2O2 in high doses, may induce oxidative stress, apoptosis, age-related pathologies and death [4], [5], [6]. In contrast ROS elicit protective effects [7] and functions as second messengers [8], [9]. Signaling by H2O2 is mainly accomplished via oxidation of cysteines in target proteins.
Four oxidation states of cysteines are considered: disulfide (-S-S-), sulfenic acid (-SOH), sulfinic acid (SO2H) and sulfonic acid (SO3H). The formation of the latter two requires an excess of H2O2 and the reaction is considered to be irreversible under physiological conditions [10]. However, oxidation towards disulfide or sulfenic acid is reversible with the aid of two major cellular reducing systems; peroxiredoxin/thioredoxin and glutathione [11], [12], [13], [14]. Therefore, these states represent molecular redox switches, which alter protein function. Most published methods for identification of the redox status of proteins focus on indirect versus direct detection of oxidative cysteine modifications in targeted approaches, in order to identify oxidative modifications of a defined protein [15], [16], [17], [18], [19]. A major tool for detection of redox modifications is the BIAM (biotinylated iodoacetamide) switch assay, which was initially developed as a method to detect S-nitrosylated proteins [20]. Briefly, the BIAM switch assay follows a 3-step-protocol with (1) Blocking of reduced thiols and (2) Reduction of reversible oxidized thiols. The reduction step (2) allows the option to refine the specificity of the assay. Specific agents reducing S-glutathionylations include ascorbate for S-nitrosylations and glutaredoxin. Global reduction of all proteins can be achieved with DTT or TCEP, which are able to reduce disulfide bonds and sulfenic acid. The last step (3) is labeling of the nascent reduced thiols with biotinylated iodoacetamide. Biotinylated proteins are then concentrated with the aid of streptavidin labeled antibodies and eventually analyzed by western blot or mass spectrometry.
Some known redox-targets are phosphatases, SERCA and small GTPases. (1) In protein phosphatases, oxidation results in transient inactivation [21], [22], [23]. (2) The oxidation of sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) prevents its S-glutathionylation and subsequently NO formation [24], [25] and eventually (3) oxidation of Ras and RhoGTPases increases their activity [26]. However, the majority of direct redox targets oxidized by Nox4 or any other NADPH oxidase remain unidentified.
To screen for novel redox-targets of the constitutive active NADPH oxidase 4 in an untargeted approach, we established a BIAM switch assay coupled to mass spectrometry with DTT as a global reductant. We utilized HEK293 cells that overexpress Nox4 in a tetracycline-inducible manner (HEK-tet-Nox4), as well as primary murine podocytes from WT and Nox4-/- mice. With this work, we present a method to screen for redox-targets of Nox4 in an unbiased fashion.
2. Results
2.1. External H2O2 and stable overexpression of Nox5, but not Nox4 results in robust oxidation of peroxiredoxins and thioredoxins
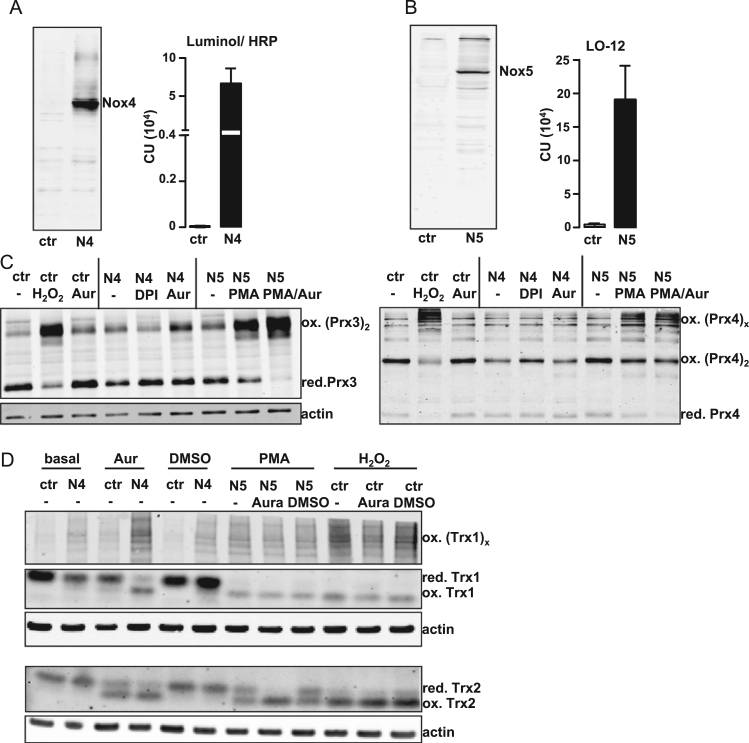
Individual ROS producing systems may elicit both specific and unspecific effects. Major differences can be expected when comparing H2O2 from Nox4 and •O2ˉ from Nox5. Accordingly, we first established HEK293 cells with stable overexpression of Nox4 and Nox5 (Fig. 1A, B). Overexpression of Nox4 results in a 2000-fold increase in H2O2 formation, as measured with Luminol/ HRP chemiluminescence in intact cells. Surprisingly, overexpression of Nox4 only resulted in a minor increase in oxidation of Prx3, Prx4 and Trx1, compared to the oxidation of these proteins in cells treated with 100 mM H2O2. Additional treatment of the cells overexpressing Nox4 with the thioredoxin reductase inhibitor auranofin [27] unmasked a Nox4-dependent oxidation of Trx2 and forced the oxidation of Prx3, Prx4 and Trx1.
Fig. 1.
Characterisation and redox-western for HEK293 cells with stable overexpression of Nox4 and Nox5. A Representative Western blot for Nox4 in HEK293 cells stably overexpressing Nox4 (N4) and H2O2 measurements assessed by chemiluminescence with luminol (100 μM)/HRP (1 U/ml). N = 3. B Representative Western blot for Nox5 in HEK293 cells overexpressing Nox5 (N5) and O2•− measurements after PMA activation (100 nM, 40 min) assessed by chemiluminescence with L-012(100 μM). N = 4. C,D Redox-Western for Prx3, Prx4. Cells were treated with either 100 mM H2O2, 3 μM DPI (3 h), PMA (100 nM, 40 min) or auranofin (3 μM, 40 min). Cells were blocked with NEM (100 mM), washed with PBS-NEM (100 mM). Cells were scraped in alkylation buffer (40 mM Hepes, 50 mM NaCl, 1 mM EGTA, Inhibitors, Catalase, 100 mM NEM) and 1% CHAPS for solubilization. For Redox-Western Trx1 and Trx2 the cells were scraped in 20% TCA followed by acetonewashing. Proteins were dissolved and incubated with 15 mM AMS for 3 h.
In contrast to Nox4, Nox5 requires activation; which was achieved by treatment of the cells with PMA (phorbol myristate acetate). PMA treatment and subsequent Nox5 activation in Nox5 overexpressing cells increased the formation of •O2ˉ 100 fold as measured with L-012 chemiluminescence in intact cells. Acute activation of Nox5 in these cells resulted in a strong oxidation of peroxiredoxins and thioredoxins, which was further increased upon treatment with auranofin (Fig. 1C, D). Upon oxidation, Prx and Trx can multimerize [28], [29]. Mitochondrial Prx3 (oxidized to its dimeric form) and cytosolic/ endoplasmic reticulum-located Prx4 multimeric oxidation products were highly abundant after Nox5 activation and external H2O2 treatment. With Nox4 overexpression, such effects were only observed upon inhibition of the reductases. A similar observation was made with thioredoxins: intramolecular oxidation of the mitochondrial Trx2 and multimeric oxidation of the cytosolic Trx1 were much more abundant in cells overexpressing active Nox5 and treated with external H2O2 than in Nox4 overexpressing cells without auranofin treatment.
These findings indicate that in an overexpression system, Nox4-derived H2O2 is less potent at oxidizing Prx and Trx than both Nox5-derived •O2ˉ and treatment with 100 mM H2O2. Therefore, oxidation by H2O2 through Nox4 allows a faster and more robust reversion of Prx and Trx oxidation by endogenous reductases.
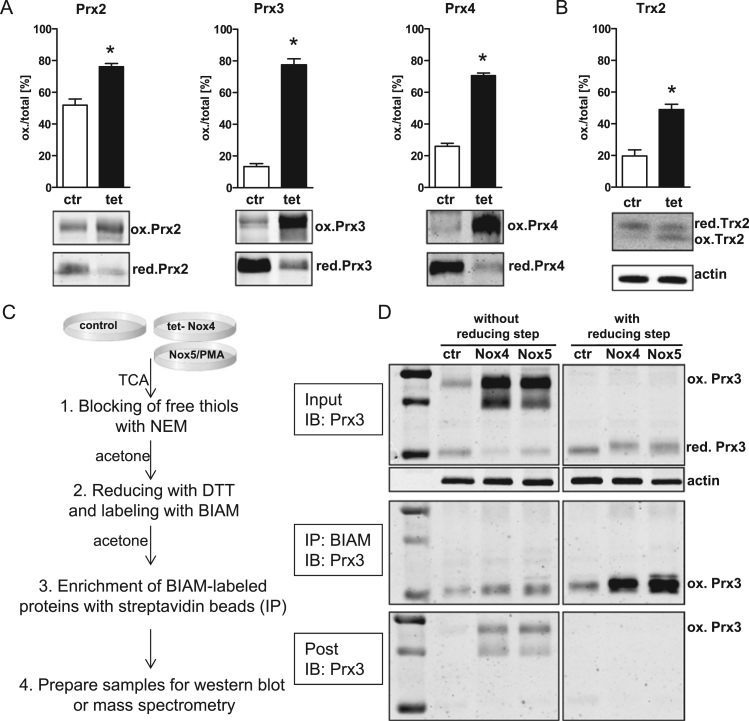
2.2. Tetracyline induced overexpression of Nox4 results in oxidation of peroxiredoxins and thioredoxins
The above finding led us to conclude that the adaptation of a cell to permanent overexpression of Nox4 might not be ideal when searching for its redox targets. Accordingly, we continued using tetracyline (tet) inducible overexpression of Nox4 in HEK293 cells which actually results in a more physiological localization of Nox4. [30]. Tet-induced overexpression of Nox4 (24 h, 1 µg/ml) resulted in strong H2O2 formation, while basal H2O2 was not detectable [31] (Suppl. S1). Redox western blots revealed 80% oxidation for Prx2, Prx3 and Prx4 and 50% oxidation for Trx2, whereas under basal conditions most of these proteins were present in a reduced state (Fig. 2A, B). In subsequent experiments with the BIAM switch assay, Prx3 was used as a bench mark protein to monitor the success of the assay and functional Nox4 overexpression. In tetracyline treated HEK-tet-Nox4 cells and in Nox5-overexpressing HEK293 cells activated with PMA, Prx3 was oxidized and present as homodimer. Following the established BIAM labeling protocol, (1) free thiols were blocked with N-ethylmaleimid, then (2) samples were reduced using DTT and eventually (3) nascent free thiols were labeled/alkylated with BIAM. BIAM- labeled proteins were precipitated with streptavidin coupled beads (Fig. 2C) and immunoblotted for Prx3. Due to the reduction of the sample in step (2), oxidized Prx3 should appear as a monomeric BIAM-labeled band at the height of the formerly reduced Prx3 band. If the reduction step is omitted, no BIAM labeling should be present. Indeed, without reduction, no notable band was detected; while reducing the samples allowed the BIAM to label -SH groups, resulting in a monomeric band with Prx3 immunodetection (Fig. 2D). Remarkably, in HEK-tet-Nox4 and PMA-activated Nox5 overexpressing HEK293 cells, Prx3 oxidation was induced. This was not the case for the control HEK293 cells. Considering these data, tet- inducible Nox4 overexpression in HEK293 cells represents a strong system for establishing the BIAM switch assay in the identification of Nox4 redox targets.
Fig. 2.
Redox-western for tetracycline induced (24 h, 1 μg/ml) Nox4 overexpression in HEK-tet-Nox4 cells and principle of BIAM switch assay. A Redox-Western for Prx2, Prx3, Prx4. Cells were blocked with NEM (50 mM), washed with PBS-NEM (50 mM). Cells were scraped in alkylation buffer (40 mM Hepes, 50 mM NaCl, 1 mM EGTA, Inhibitors, Catalase, 100 mM NEM) and 1% CHAPS for solubilization. B Redox-Western for Trx2. Cells were scraped in 20% TCA followed by acetone-washing. Proteins were dissolved and incubated with 15 mM AMS for 3 h. C Principle of BIAM switch assay. D Western blot for Prx3 after BIAM switch assay in tetracyline induced HEK-tet-Nox4 cells (Nox4) in non-induced HEKtet-Nox4 cells (ctr) and PMA-activated Nox5 overexpressing HEK 293 cells (Nox5). Left: without DTT reduction; Right: with DTT reduction. Cells were blocked with NEM. Reduction was performed with 4 mM DTT if indicated. 100 μg of protein were used for the pull down of BIAM-labeled proteins with streptavidin agarose beads. SDS-Page gel electrophoresis under non-reducing conditions.
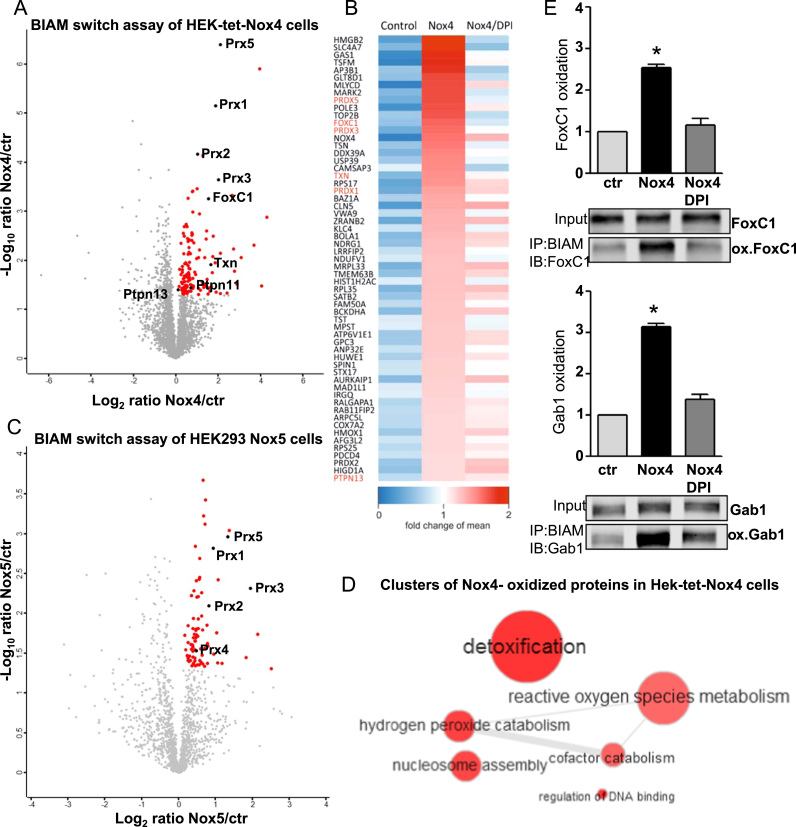
2.3. Overexpression/activation of Nox4 and Nox5 results in differential oxidation of several target proteins
In order to identify novel redox-targets for Nox4 in a global and untargeted manner, BIAM switch assay was coupled to mass spectrometry. In HEK-tet-Nox4 cells tetracycline induced Nox4 overexpression caused 131 proteins to be significantly oxidized (Suppl. Table 1). Importantly, the flavoprotein inhibitor DPI was used as a control, to ensure, that the proteins identified were true redox targets. Indeed, co-treatment with DPI prevented the oxidation of most proteins upon Nox4 overexpression (Fig. 3A, B). Nox5 overexpression and PMA-stimulated •O2ˉ production caused significantly higher oxidation of 85 proteins (Fig. 3C, Suppl. Table 2). Here, normal HEK293 cells with PMA stimulation served as a control. Proteins identified in DPI or PMA treated HEK293 cells were considered as false positives and were excluded from further analysis. Within the remaining fraction, peroxiredoxins and thioredoxin were verified as redox-targets of Nox4 and Nox5. Further analysis of the data clustered the Nox4 oxidized proteins in different functional groups such as proteins involved in detoxification of the cell (ROS metabolism), proteins responsible for regulation of DNA binding or involved in nucleosome assembly (Fig. 3D). The same analysis performed in the Nox5-dependent oxidized proteins clustered them into proteins involved in oxidation-reduction process, cell redox-homeostasis or proteins responsible for DNA replication initiation (Suppl. S2). There was an overlap between proteins involved in antioxidant defense for both cluster analyses (Fig. 3D, Suppl. S2). This indicates an unspecific response of the cells upon overexpression/ -activation of any ROS source. Beside many others, two interesting Nox4 redox-targets, GRB2-associated-binding protein 1 (Gab1) and the transcription factor FoxC1, were not oxidized by Nox5. Gab1 and FoxC1 showed a 3-fold increase in oxidation in tet-treated HEK-tet-Nox4 cells when compared to base level. Treatment of the Nox4 overexpressing cells with DPI restored the basal level (Fig. 3E).
Fig. 3.
BIAM switch assay for tetracycline induced (24 h, 1 μg/ml) Nox4 overexpression in HEK-tet-Nox4 cells for identification of oxidized proteins. A Mass spectrometry coupled BIAM switch assay for tetracycline induced (24 h, 1 μg/ml) Nox4 overexpression in HEKtet-Nox4 cells compared to non-induced HEK-tet-Nox4 cells. Significant differentially oxidized proteins marked as red and black dots (p-value < 0.05; fold-change >1). B Top 25 differentially oxidized proteins in HEK-tet-Nox4 cells in response to DPI (3 h, 3 μM). C Mass spectrometry coupled BIAM switch assay for HEK293 cells stable overexpressing Nox5 and activated with PMA (24 h, 1 μg/ml) compared to PMA-activated HEK293 cells (ctr). Significant differentially oxidized proteins marked as red and black dots (p-value < 0.05; fold-change >1). D Clustering of oxidized proteins in Hek-tet-Nox4 cells according to their involvment in different cellular processes. E Western blot based BIAM switch assay for FoxC1, Gab1 and Prx3 in tetracycline induced (24 h, 1 μg/ml) Nox4 overexpression in HEK293-tet-Nox4 cells compared to non-induced HEK293-tet-Nox4 cells (ctr) and tetracycline induced (24 h, 1 μg/ml) Nox4 overexpression in HEK293-tet-Nox4 co-treated with DPI (3 h, 3 μM).
In conclusion, Nox4 and Nox5 oxidize many proteins. However, although we analyzed the effects of both NADPH oxidases in overexpressing systems, several differentially oxidized proteins were identified, due to the differences in the nature of the ROS produced and potentially the localization of both enzymes.
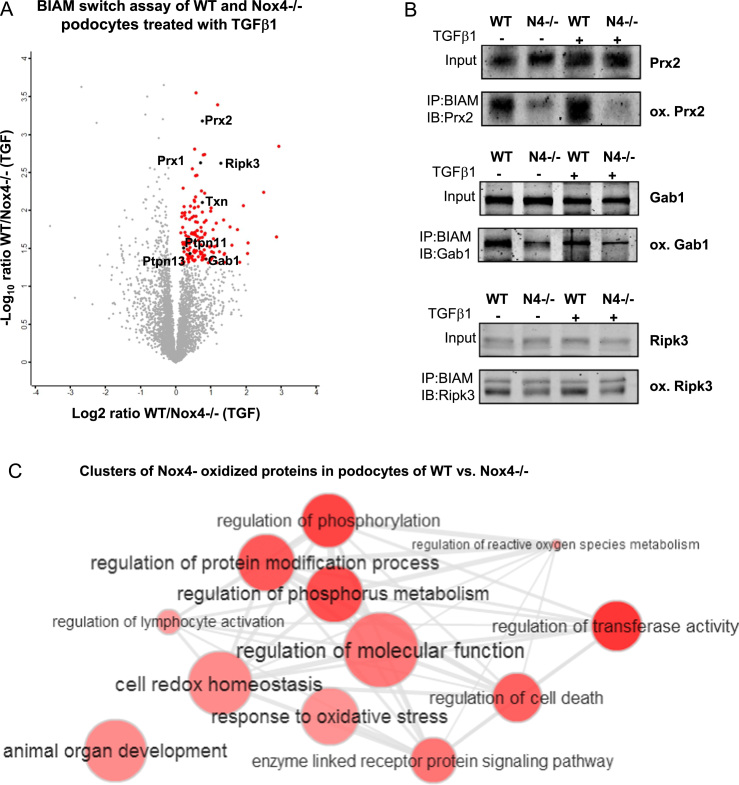
2.4. BIAM switch assay in primary murine podocytes of WT vs. Nox4-/- identified 142 potential redox targets
Overexpression often has many unwanted secondary effects and should serve only as a model to obtain a first impression. Accordingly, we used primary murine podocytes as cells with high endogenous Nox4 expression. This provided a more relevant analysis of differentially oxidized proteins in vivo. Primary podocytes were isolated from wild type and Nox4-/- mice. As a model to analyze the effect of Nox4 in these cells we treated wild type and Nox4 deficient cells with TGFβ1. This cytokine induces a 2-fold increase in Nox4 mRNA- expression and a 4-fold rise in H2O2 production in wild type but not in Nox4-/- cells (Suppl. S3). Additionally, treatment with TGFβ1 increased the oxidation for Prx2 in podocytes from wild type but not from Nox4-/- mice (Suppl. S3). In response to TGFβ1, oxidation of 142 proteins increased in podocytes from wild type but not Nox4-/- mice (Fig. 4A, Suppl. Table 3). Again, peroxiredoxins and thioredoxin as redox- markers could be identified as being oxidized by Nox4. Identified proteins clustered in many different functional groups such as proteins involved in cellular redox homeostasis, regulation of cell death and regulation of phosphorylation and molecular functions (Fig. 4C). According to the finding in the overexpression system, we found Gab1 to be oxidized to a lesser extent in Nox4-/- cells with and without TGFβ1-treatment. Additionally, Prx2 and receptor interacting serin/threonine kinase (Ripk3) were substantially more oxidized in podocytes of wild type mice compared to Nox4-/-. Taken together, Gab1 represents a novel redox target differentially oxidized by Nox4.
Fig. 4.
BIAM switch assay of primary murine podocytes of WT and Nox4-/- mice treated with TGFβ1 (24 h, 10 ng/ml) for identification of oxidized proteins. A Mass spectrometry coupled BIAM switch assay of podocytes of WT and Nox4-/- mice treated with TGFβ1 (24 h,10 ng/ml). Significant differentially oxidized proteins marked as red and black dots (p-value < 0.05; fold-change >1). B Western blot based BIAM switch assay for Prx2, Gab1 and Ripk3 in podocytes of WT and Nox4-/- mice treated with TGFβ1 (24 h, 10 ng/ml). C Clustering of Nox4-oxidized proteins in podocytes of WT mice according to their involvement in different cellular processes.
2.5. Gab1 Cys374 and Cys405 as major oxidation targets
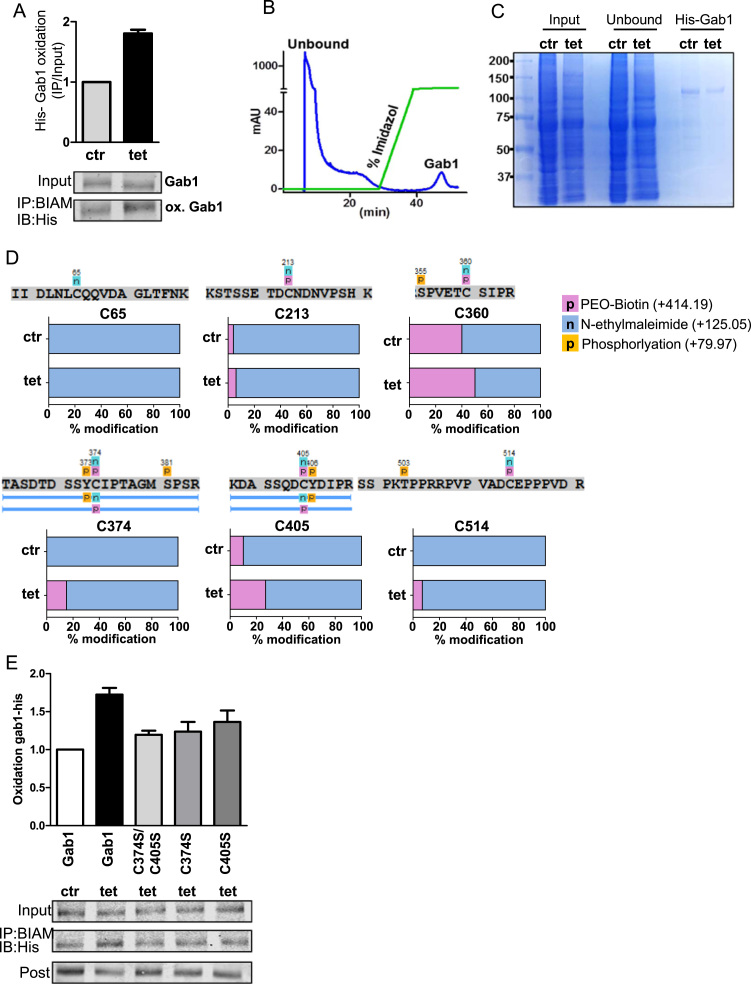
To identify the cysteines in Gab1 that were most susceptible to oxidation, His-Gab1 was transiently overexpressed in HEK-tet-Nox4 cells. Induction of Nox4 expression by tet lead to an increase in oxidation of His-Gab1 by 80% (Fig. 5A). BIAM modified His-Gab1 was purified by HisTrap columns and loaded onto a SDS page gel (Fig. 5B,C). The His-Gab1 bands were analyzed by mass spectrometry (Gab1 protein sequence coverage of 92%, Data not shown). We identified 6 cysteines and, of those, Cys374 and Cys405 were differentially oxidized in cells overexpressing Nox4 (Fig. 5D, Suppl. Table 4). Single mutations of C374S and C405S and the double mutant C374S/ C405S lead to a loss in redox modification of His-Gab1 upon Nox4 overexpression (Fig. 5E). These results indicate that, Cys374 and Cys405 in Gab1 are major redox targets of Nox4.
Fig. 5.
Identification of oxidized cysteines in GAB1 in tetracycline induced (24 h, 1 μg/ml) Nox4 overexpressing HEK-tet-Nox4 cells. A Western Blot based BIAM switch assay for His-Gab in tet-induced (24 h, 1 μg/ml) HEK-tet-Nox4 cells and in non-induced HEK-tet-Nox4 cells (ctr). BIAM switch assay was performed as described. B Chromatogram of affinity purification of His -Gab1 after performing the BIAM switch assay. 2 mg of total protein was used for His-trap affinity purification. Elution was performed with an imidazole gradient (20–500 mM) over 20 min with a flow rate of 1 ml/min. C SDS-PAGE coomassie gel of different fractions obtained during affinity purification. D Identified oxidized Cysteines of Gab1 and corresponding peptide sequences obtained from mass spectrometry. His- Gab1 coomassie gel bands from ctr and tet-induced Hek-tet-Nox4 cells were cut out and analyzed by mass spectrometry. Percentage of modification (PEO-Biotin=oxidized and N-ethylmaleimide=reduced) was obtained by spectral counting. E Western Blot based BIAM switch assay for His-Gab1 and His-Gab1 mutants in tet-induced (24 h, 1 μg/ml) HEK-tet-Nox4 cells and in noninduced HEK-tet-Nox4 cells (ctr).
Gab1 is an adaptor protein and plays a pivotal role in many signaling pathways [32]. If phosphorylated at tyrosine residues, Gab1 provides binding sites for multiple effector proteins, such as Src homology-2 (SH2)-containing protein tyrosine phosphatase 2 (SHP2) and phosphatidylinositol 3-kinase (PI3K) regulatory subunit p85. Gab1 is important for receptor tyrosine kinase -mediated signals into pathways with diverse biological functions [33]. We hypothesized that oxidation of Gab1 interferes with phosphorylation of the protein.
Indeed, overexpression of Nox4 in HEK-tet-Nox4 cells shows a complete loss of phosphorylation within the peptides identified if the adjacent cysteine is oxidized (PEO-Biotin). Tyr373 and Tyr406, which are in the direct vicinity of Cys374 and C405, are of special interest. These cysteines of Gab1 are major oxidation targets of Nox4, which might serve as a redox-phospho switch (Fig. 5D).
Binding partners of Gab1 include the regulatory subunit p85 of the PI3K, phospholipase Cγ (PLC-γ), CRK and the protein tyrosine phosphatase SHP2 (PTPN11). SHP2, identified in our study as differentially oxidized as well, under basal conditions is auto-inhibited by intramolecular interaction of its N-terminal SH2 domain and its PTP domain. Downstream of EGFR, SHP2 can be activated by binding to phosphorylated tyrosine residues of Gab1. Mass spectrometry showed that Gab1 phosphorylation of tyrosine residues in direct vicinity of H2O2 sensitive cysteins was absent upon oxidation, whereas the phosphorylation was present if cysteines were reduced. This therfore represents a potential redox-phospho switch mechanism. Accordingly, overexpression of Nox4 and therby an increase in H2O2, should reduce the ability of Gab1 to interact with SHP2. As shown in Supplementary Fig. S4, Nox4 overexpression indeed, reduces the Gab1- SHP2 interaction upon EGF stimulation. We conclude that Nox4 derived H2O2 interferes with Gab1s tyrosine phoshorylation which inhibits the binding of Gab1 and SHP2.
3. Discussion
Identification of redox-targets has so far been largely restricted to targeted approaches where proteins of interest were either used in overexpression systems [23] or as recombinant proteins [34], [35]. Untargeted approaches, such as OxICAT or modified ICAT-BIAM switch assays identified several oxidized proteins [36], [37] often without naming the source of the ROS. In this study, we describe a BIAM switch assay coupled to mass spectrometry as a novel screening method to identify oxidized proteins in a global and untargeted manner. In doing so, we concentrated on Nox4 and Nox5 as two NADPH oxidases with one producing H2O2 and the other •O2ˉ, respectively. Thus, identified proteins may represent novel redox-targets of Nox4 and Nox5. For the newly identified redox-target of Nox4, Gab1, we discovered Cys374 and Cys405 as the two major cysteines oxidized by Nox4.
Nox4 produced H2O2 in a constitutive manner, as shown in HEK293 cells stably overexpressing Nox4. However, no strong oxidation of peroxiredoxins or thioredoxins was observed upon stable overexpression of Nox4 unless unmasked by auranofin. Auranofin selectively blocks antioxidant enzymes like thioredoxin reductase (TrxR) and glutathione peroxidase (GPx) [38], [39]. These enzymes are responsible for maintenance of the redox homeostasis in the cell. Overall these findings suggest an adaptation of the cellular antioxidant defense machinery towards the long-term massive production of H2O2 by the stable overexpression of Nox4 in HEK293 cells [40].
Acute induction of Nox4 overexpression in HEK-tet-Nox4 cells identified 131 differentially oxidized proteins, while in murine podocytes 142 proteins were identified as differentially oxidized by Nox4. These proteins mainly cluster in proteins involved in cellular redox homeostasis, in regulation of cell death or in regulation of phosphorylation and molecular functions. Besides peroxiredoxins and thioredoxins two tyrosine-protein phosphatases PTPN11 (SHP2) and PTPN13 which are generally accepted and often described as oxidation targets [41], [42] were found to be differentially oxidized in both Nox4 systems. In contrast, Nox5 dependent redox targets clustered in proteins involved in oxidation-reduction process, cell redox-homeostasis or proteins responsible for DNA replication initiation. An overlap between proteins oxidized by Nox4 and oxidized by PMA-activated HEK293 cells overexpressing Nox5 was found predominantly for the peroxiredoxins. This result supports the enormous relevance of the peroxiredoxins as anti-oxidant enzymes [43] as they can modulate the concentration of H2O2 and •O2ˉ.
With Gab1, we identified a novel redox target of Nox4. Gab1 is a multifunctional docking protein that plays a central role in receptor tyrosine kinase signaling, i.e. is crucial for many cellular processes, including growth, cell cycle progression and apoptosis. Upon interaction with activated growth factor and cytokine receptors Gab1 is phosphorylated at multiple sites, such as serine, threonine and tyrosine residues. Phosphorylated Gab1 represents a perfect binding “platform” for various SH2 domain- containing proteins and therefore mediates receptor tyrosine kinase signaling into diverse downstream signaling events
We discovered Cys374 and Cys405 as the two major cysteines in murine Gab1, oxidized by Nox4. Furthermore the mass spectrometry data show that if the cysteine was oxidized (labeled with BIAM), the phosphorylation within the related peptide was completely absent. Conversely, if cysteines were reduced, phosphorylation was present.
Gab1 has several tyrosine phosphorylation sites and serves as a docking protein for multiple effector proteins such as Src homology-2 (SH2)-containing protein tyrosine phosphatase 2 (SHP2) and phosphatidylinositol 3-kinase (PI3K) regulatory subunit p85 [32], [44].
Six tyrosine residues in Gab1 (Y242, Y259, Y307, Y317, Y373 and Y406) are located in Tyr-X-X-Pro (YXXP) motifs, which serve as binding sites for the adaptor protein Crk [45] and for phospholipase Cγ (PLCγ) that binds to Y307, Y373 and Y406 respectively [46]. Interestingly, the two identified major oxidized cysteines Cys374 and Cys405 are located within or in direct proximity of an YXXP motif with non-phosphorylated Y373 and Y406 if the cysteines were oxidized. This observation indicates interference between the oxidation of cysteines and the phosphorylation of tyrosines in immediate vicinity, which may represent a newly identified “redox-phospho” switch mechanism. This Nox4-dependent redox-switch mechanism might change the interaction between Gab1 and its aforementioned interacting partners as shown for the reduced Gab1-SHP2 interaction leading to potential various altered downstream signaling effects.
Overall, in the present study we provide evidence that an untargeted BIAM switch assay coupled to mass spectrometry represents a robust and reliable screening method to identify redox targets. Peroxiredoxins are differentially oxidized by Nox4 and Nox5. Further, Gab1 is a newly discovered redox- target of Nox4, with Cys374 and Cys405 as the two major oxidized cysteines.
4. Innovation
With the aid of mass spectrometry coupled BIAM switch assay, we present a robust screening tool to identify redox-targets of the NADPH oxidase, Nox4. In total, we identified 142 proteins, which were differentially oxidized by Nox4 in podocytes of WT but not in Nox4-/- mice and therefore potential novel redox-targets of Nox4. Out of those, we selected Gab1 for a more intense analysis and identified Cys374 and Cys405 a two major cysteines oxidized by Nox4. Importantly, oxidation of these may elicit inhibition of phosphorylation of Gab1 and thereby interfere with the function of this adaptor protein.
5. Material and methods
5.1. Overexpression system using HEK293 cells
Human embryonic kidney 293 cells (HEK293) obtained from ATCC (Manassas, VA, USA) were stably transfected with a plasmid coding for Nox4 or Nox5. ROS formation in HEK293 cells stably overexpressing Nox5 were activated with 100 nM PMA (from SigmaAldrich) for 40 min.
HEK293 expressing human Nox4 with a tetracycline inducible system were kindly provided by K.H. Krause. Overexpression of Nox4 in HEK-tet-Nox4 cells was induced by 1 µg/ml tetracycline (from Cayman Chemicals) for 24 h in culture medium.
HEK293 cells were cultured in Minimum Essential Medium (MEM, Gibco) supplemented with 8% fetal bovine serum, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate and 50 µg/ml gentamycin, maintained at 37 °C with 5% CO2.
5.2. Isolation of murine podocytes
Primary murine podocytes were isolated as described [47], [48]. Briefly, mice were perfused with 8×107 beads (Dynabeads M-450 Tosylactivated, Thermo Scientific) in 20 ml podocyte growth medium (RPMI 1640 (Gibco), 8% FCS, 5 mm HEPES, 0.1% non-essential amino acids, 0.1 mm sodium pyruvate, 0.01 mg/ml of insulin/transferrin/sodium selenite solution, penicillin/streptomycin). Minced kidneys were digested in 1 mg/ml collagenase A (from Roche) for 30 min at 37 °C. Kidney suspension was sieved through 100 µm filters twice and was centrifuged for 4 min at 1200 rpm. The pellet was resuspended and the glomeruli bound on the Dynabeads were collected with the aid of a magnetic stand. Glomeruli were resuspended in 1 ml podocyte medium and seeded onto a 6 cm plate coated with collagen. Podocytes were cultured in podocyte medium in a humidified atmosphere (5% CO2, 37 °C). For experiments primary murine podocytes of WT and Nox4-/- mice were treated with or without 10 ng/ml TGFβ1 (from PromoCell) for 24 h, if indicated.
5.3. ROS measurements with chemiluminescence
ROS production was assessed in intact cells with 100 µmol/L luminol (from SigmaAldrich) / 1 U/ml horseradish peroxidase (HRP, from SigmaAldrich) for H2O2 or with 200 µmol/L L-012 (from Wako Chemicals) for •O2ˉ in a Berthold TriStar2 microplate reader (LB942, Berthold, Wildbad, Germany). All measurements were performed in HEPES-Tyrode buffer containing in mmol/L: 137 NaCl, 2.7 KCl, 0.5 MgCl2, 1.8 CaCl2, 5 glucose, 0.36 NaH2PO4, 10 HEPES.
5.4. Redox-western blot and antibodies
For Prx2, Prx3 and Prx4 cells were incubated with NEM (50 mM) and washed with PBS- NEM (100 mM). Cells were scraped in alkylation buffer (40 mM Hepes, 50 mM NaCl, 1 mM EGTA, Inhibitors, Catalase, 100 mM NEM) and 1% CHAPS (from Applichem) for solubilization. For Trx1 and Trx2 cells were scraped in 20% TCA followed by two acetone washing steps. Proteins were dissolved in EB buffer (10% SDS, 150 mM NaCl, 50 mM Hepes) and incubated with 15 mM AMS (from Life Technologies) for 3 h. Protein amount was determined by Lowry protein assay. Samples were substituted with sample buffer (8.5% glycerin, 2% SDS, 6.25% TRIS/HCl pH 6.8, 0.013% bromphenol blue) and separated on a non-reducing SDS-PAGE gel, followed by Western blot analysis and detection by antibodies. Primary antibodies against Prx2 (#LF-PA0091), Prx3 (#LF-PA0030), Prx4 (#LF-PA0009), Trx1 (#LF-PA0187) and Trx2 (#LF-PA0012) were diluted 1:1000 and were purchased from AbFrontier. The antibody against Nox4 was a gift from Ajay Shah from Kings College London and was diluted 1:1000. After incubation with first antibodies, membranes were analyzed with an infrared-based detection system, using fluorescent-dye-conjugated secondary antibodies from LI-COR biosciences.
5.5. BIAM switch assay for identification of oxidized proteins
For blocking of free thiols living cells were incubated with N-ethylmaleimide (NEM, 50 mM, SigmaAldrich) for 5 min and then washed gently with PBS-NEM (100 mM). Cells were denatured and scraped from the plate in 1 ml of cold (4 °C) 20% TCA followed by two washing steps (TCA 10%, TCA 5%) and centrifugation for 30 min, 10,000 g, 4 °C. The pellet was resolved in 200 µl NEM-resolution buffer (50 mM NEM; 50 mM Tris-HCl, pH 8,5; 8 M Urea; 5 mM EDTA; 20% SDS) on a shaker for 1 h at 37 °C, 1000 rpm. Reaction was stopped by adding 1 ml of ice- cold (-20 °C) acetone. Excess of NEM was removed by centrifugation for 30 min, 10,000 g, 4 °C and washing the pellet twice with 1 ml of ice-cold acetone.
Initial reduction of oxidized thiols was performed with 200 µl of 4 mM DTT solution (from Applichem) for 5 min directly followed by incubation with 200 µl BIAM (EZ-Link™ Iodoacetyl-PEG2-Biotin, Thermo Scientific) -resolution buffer (20 mg/ml BIAM; 50 mM Tris-HCl, pH 8,5; 8 M Urea; 5 mM EDTA; 1% SDS) on a shaker for 1 h at 37 °C, 1000 rpm. Excess of BIAM was removed by adding 1 ml of ice-cold acetone and centrifugation for 30 min, 10,000 g, 4 °C and washing twice with 1 ml of ice-cold acetone. Proteins were resolved in 200 µl resolving buffer (50 mM Tris-HCl, pH 8,5; 5 mM EDTA; 20% SDS; 10% Triton). 100–500 µg of protein were used for the pull down of BIAM-labeled proteins with 25–100 µl streptavidin agarose beads (from Thermo Scientific) per sample. For Western blot analysis, beads were boiled in 50 µl sample buffer (8.5% glycerin, 2% SDS, 6.25% Tris/HCl, pH 6.8, 20 mM DTT, 0.013% bromphenol blue). For mass spectrometry proteins were eluted with 8 M guanidium-HCl and boiled at 95 °C for 5 min. Samples were digested for 16 h with 1 µg trypsin (sequencing grade, Promega) at 37 °C in 50 mM ABC and 1 mM CaCl2. Purification and fractionation of peptides was performed with C18 stage-tips and SCX stage-tips [49] respectively. Peptides were dried and resolved in 1% acetonitrile and 0.5% formic acid. Samples were analyzed by mass spectrometry.
5.6. Purification of His- Gab1 for cysteine identification
His-tagged murine Gab1 was overexpressed in HEK-tet-Nox4 cells. BIAM switch assay was performed as described. BIAM labeled proteins were loaded on HisTrap™ FF crude columns packed with Ni-Sepharose (from GE Healthcare) and purified using the Äkta FPLC from GE Healthcare. Purified protein was eluted with an 20–500 mM imidazole (from SigmaAldrich) gradient over 20 min with a flow rate of 1 ml/min. Collected fractions were combined as indicated and proteins were separated on a SDS-page and stained with coomassie. Gab1 coomassie band was cut out of the gel.
The gel pieces were de-stained in 60% Methanol, 50 mM ammoniumbicarbonate (ABC) and washed in 50 mM ABC. Samples were digested for 16 h with trypsin or GluC (sequencing grade, Promega) at 37 °C in 50 mM ABC, 0.01% Protease Max (Promega) and 1 mM CaCl2. Peptides were eluted in 30% acetonitrile and 3% formic acid, centrifuged into a fresh 96 well plate, dried in a speed vac and resolved in 1% acetonitrile and 0.5% formic acid. Samples were analyzed by mass spectrometry.
5.7. Mass spectrometry
Liquid chromatography/mass spectrometry (LC/MS) was performed with the aid of a Thermo Scientific™ Q Exactive Plus equipped with an ultra-high performance liquid chromatography unit (Thermo Scientific Dionex Ultimate 3000) and a Nanospray Flex Ion-Source (Thermo Scientific). Peptides were loaded onto a C18 reversed-phase pre-column (Thermo Scientific), followed by separation on a picotip emitter tip (diameter 100 µm, 15 cm from New Objectives) in-house packed with 2.4 µm Reprosil C18 resin (Dr. Maisch GmbH). Elution of peptides from the BIAM switch assay performed in HEK293 cells (Nox4 and Nox5), was performed with the aid of a gradient from 100% eluent A (4% acetonitrile, 0.1% formic acid) to 30% eluent B (80% acetonitrile, 0.1% formic acid) for 60 min and a second gradient to 60% B for 30 min. The gradient for the fractions from BIAM switch assay performed with murine podocytes was from 100% eluent A to 30% B for 20 min followed by a second gradient to 60% B for 10 min. The gradient for the cysteine identification of His-Gab1 was from 100% eluent A to 30% B for 90 min and the second gradient to 60% B for 15 min.
MS data were recorded by data dependent acquisition. The full MS scan ranged from 300 to 2000 m/z, with resolution of 70.000 and an automatic gain control (AGC) value of 3 * 106 total ion counts, with a maximal ion injection time of 160 ms. Only higher charged ions (2+) were selected for MS/MS scans with a resolution of 17500, an isolation window of 2 m/z and an automatic gain control value set to 105 ions with a maximal ion injection time of 150 ms. MS1 data were acquired in profile mode.
5.8. BIAM switch assay data analysis
MaxQuant (v1.5.3.30, podocyte dataset; v1.5.2.8, HEK cell datasets (Nox4 and Nox5)) [50], Perseus 1.5.2.6 [51] and Excel (Microsoft Office 2013) were used. N-terminal acetylation (+42.01) and oxidation of methionine (+15.99), biotinylated iodoacetamide on cyteines (414.19) and N-ethylmaleimide (125.05) on cysteines were selected as variable modifications. The human reference proteome set (Uniprot, 4/2015, 68511 entries for HEK cell datasets) and the mouse reference proteome set (Uniprot, 2/2016, 79950 entries for podocyte dataset) were used to identify peptides and proteins with a false discovery rate (FDR) of less than 1%. Minimal ratio count for label-free quantification (LFQ) was 1. Reverse identifications, only identified by site and common contaminants were removed and the data-set was reduced to proteins that were quantified in at least 4 of 6 samples for the podocytes dataset, 4 of 5 sample for the HEK-tet-Nox4 cells, or 3 of 4 for the HEK293 Nox5 cells in one experimental group. Missing LFQ values were replaced by random background values. Significant interacting proteins were determined by Students t-test.
5.9. Data analysis for identification of modified amino acids
MS Data were analyzed by Peaks7. Proteins were identified using mouse reference proteome database UniProtKB with 70947 entries, released in 12/2016 with a false discovery rate of 1%. The enzyme specificity was set to trypsin or Gluc (bicarbonate). Acetylation (+42.01) at N-terminus, oxidation of methionine (+15.99), deamidation at asparagine and glutamine, N-ethylmaleimide on cysteines (+125.05), biotinylated iodoacetamide on cysteines (414.19) and phosphorylation on serine, threonine and tyrosine (+79.97) were variable modifications. For quantification of modified cysteines in tet- induced HEK-tet-Nox4 cells (tet) and in non-induced HEK-tet-Nox4 cells (ctr) spectral counting of peptides with modified cysteines was performed.
5.10. RT-qPCR
Total mRNA from murine podocytes or HEK-tet-Nox4 cells was isolated with a RNA-Mini-kit (Bio&Sell, Feucht, Germany) according to the manufacturers’ protocol. Random hexamer primers (Promega, Madison,WI, USA) and Superscript III Reverse Transcriptase (Invitrogen, Darmstadt, Germany) were used for cDNA synthesis. Semi-quantitative real-time PCR was performed with AriaMX qPCR system (Agilent Technologie, Santa Clara, CA, USA) using iQ™ SYBR® Green Supermix (BioRad, Hercules, CA, USA) with appropriate primers. Relative expression of target genes was normalized to GAPDH and analyzed by the delta-delta-ct method. Primer sequences were for murirne GAPDH (fwd: 5′-GTGTGAACGGATTTGGCCGTATTG-3′, rev: 5′-ACCAGTAGACTCCACGACATACTC-3′), human GAPDH (fwd: 5′-TGCACCACCAACTGCTTAGC-3′, rev: 5′-GGCATGGACTGTGGTCATGAG-3′), murine Nox4 (fwd: 5′-TGTTGGGCCTAGGATTGTGTT-3′, rev: 5′-AGGGACCTTCTGTGATCCTCG-3′) and human Nox4 (fwd: 5′-TCCGGAGCAATAAGCCAGTC-3′, rev: 5′-CCATTCGGATTTCCATGACAT-3′).
5.11. Cluster analysis and statistics
Cluster analysis was performed with gorilla [52] and the mode of two unranked lists and a p-value threshold of 10−3. Clusters were visualized by using REVIGO [53].
Unless otherwise indicated, data are given as means ± standard error of mean (SEM). Calculations were performed with Prism 5.0. Individual statistics of unpaired samples was performed by t-test and if not normal distributed by Mann-Whitney test. A p-value of < 0.05 was considered as significant. All experiments were performed at least three time. n indicates the number of individual experiments or animals.
Acknowledgement
We are grateful for excellent technical assistance of Katalin Palfi and Maria Walter.
Acknowledgments
Source of funding
This research was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) (to KS SCHR1241/1-1, SFB815/TP1, SFB834/TPA2) and the Excellence Cluster Cardio-Pulmonary Institute (CPI), EXC 2026, Project ID: 390649896, the Faculty of Medicine, Goethe-Universität, Frankfurt am Main, Germany, the DZHK, Partner Site RheinMain, Frankfurt.
Disclosures
The authors declare that they have no relevant financial, personal or professional relationships to disclose which could be perceived as a conflict of interest or as potentially influencing or biasing the authors’ work.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2019.101125.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Bedard K., Krause K.-H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 2.Takac I., Schröder K., Zhang L., Lardy B., Anilkumar N., Lambeth J.D., Shah A.M., Morel F., Brandes R.P. The E-loop is involved in hydrogen peroxide formation by the NADPH oxidase Nox4. J. Biol. Chem. 2011;286:13304–13313. doi: 10.1074/jbc.M110.192138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandes R.P., Weissmann N., Schröder K. Redox-mediated signal transduction by cardiovascular Nox NADPH oxidases. J. Mol. Cell. Cardiol. 2014;73:70–79. doi: 10.1016/j.yjmcc.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Simon H.-U., Haj-Yehia A., Levi-Schaffer F. Apoptosis. 2000;5:415–418. doi: 10.1023/a:1009616228304. [DOI] [PubMed] [Google Scholar]
- 5.Finkel T., Holbrook N.J. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 6.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24:R453–62. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schröder K., Zhang M., Benkhoff S., Mieth A., Pliquett R., Kosowski J., Kruse C., Luedike P., Michaelis U.R., Weissmann N., Dimmeler S., Shah A.M., Brandes R.P. Nox4 is a protective reactive oxygen species generating vascular NADPH oxidase. Circ. Res. 2012;110:1217–1225. doi: 10.1161/CIRCRESAHA.112.267054. [DOI] [PubMed] [Google Scholar]
- 8.Reth M. Hydrogen peroxide as second messenger in lymphocyte activation. Nat. Immunol. 2002;3:1129–1134. doi: 10.1038/ni1202-1129. [DOI] [PubMed] [Google Scholar]
- 9.Finkel T. Reactive oxygen species and signal transduction. IUBMB Life. 2001;52:3–6. doi: 10.1080/15216540252774694. [DOI] [PubMed] [Google Scholar]
- 10.Lim J.C., Choi H.-I., Park Y.S., Nam H.W., Woo H.A., Kwon K.-S., Kim Y.S., Rhee S.G., Kim K., Chae H.Z. Irreversible oxidation of the active-site cysteine of peroxiredoxin to cysteine sulfonic acid for enhanced molecular chaperone activity. J. Biol. Chem. 2008;283:28873–28880. doi: 10.1074/jbc.M804087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berndt C., Lillig C.H., Holmgren A. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H1227–36. doi: 10.1152/ajpheart.01162.2006. [DOI] [PubMed] [Google Scholar]
- 12.Casagrande S., Bonetto V., Fratelli M., Gianazza E., Eberini I., Massignan T., Salmona M., Chang G., Holmgren A., Ghezzi P. Glutathionylation of human thioredoxin: a possible crosstalk between the glutathione and thioredoxin systems. Proc. Natl. Acad. Sci. USA. 2002;99:9745–9749. doi: 10.1073/pnas.152168599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das K.C., White C.W. Redox systems of the cell: possible links and implications. Proc. Natl. Acad. Sci. USA. 2002;99:9617–9618. doi: 10.1073/pnas.162369199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallogly M.M., Mieyal J.J. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Cadenas E., Packer L., editors. Thiol Redox Transitions in Cell Signaling: Part A: Chemistry and Biochemistry of Low Molecular Weight and Protein Thiols. Elsevier/Academic; Amsterdam, Netherlands, Boston, Mass: 2010. [Google Scholar]
- 16.Chiappetta G., Ndiaye S., Igbaria A., Kumar C., Vinh J., Toledano M.B. Proteome screens for cys residues oxidation. In: Cadenas E., Packer L., editors. Thiol Redox Transitions in Cell Signaling: Part A: Chemistry and Biochemistry of Low Molecular Weight and Protein Thiols. Elsevier/Academic; Amsterdam, Netherlands, Boston, Mass: 2010. pp. 199–216. [Google Scholar]
- 17.Gupta V., Carroll K.S. Sulfenic acid chemistry, detection and cellular lifetime. Biochim. Biophys. Acta. 2014;1840:847–875. doi: 10.1016/j.bbagen.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonard S.E., Carroll K.S. Chemical 'omics' approaches for understanding protein cysteine oxidation in biology. Curr. Opin. Chem. Biol. 2011;15:88–102. doi: 10.1016/j.cbpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Ying J., Clavreul N., Sethuraman M., Adachi T., Cohen R.A. Thiol oxidation in signaling and response to stress: detection and quantification of physiological and pathophysiological thiol modifications. Free Radic. Biol. Med. 2007;43:1099–1108. doi: 10.1016/j.freeradbiomed.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffrey S.R., Snyder S.H. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 21.Lou Y.-W., Chen Y.-Y., Hsu S.-F., Chen R.-K., Lee C.-L., Khoo K.-H., Tonks N.K., Meng T.-C. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. FEBS J. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- 22.Meng T.-C., Fukada T., Tonks N.K. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell. 2002;9:387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 23.Chen K., Kirber M.T., Xiao H., Yang Y., Keaney J.F. Regulation of ROS signal transduction by NADPH oxidase 4 localization. J. Cell Biol. 2008;181:1129–1139. doi: 10.1083/jcb.200709049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong X., Schröder K. NADPH oxidases are responsible for the failure of nitric oxide to inhibit migration of smooth muscle cells exposed to high glucose. Free Radic. Biol. Med. 2009;47:1578–1583. doi: 10.1016/j.freeradbiomed.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong X., Hou X., Jourd'heuil D., Weisbrod R.M., Cohen R.A. Upregulation of Nox4 by TGF{beta}1 oxidizes SERCA and inhibits NO in arterial smooth muscle of the prediabetic Zucker rat. Circ. Res. 2010;107:975–983. doi: 10.1161/CIRCRESAHA.110.221242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell L., Hobbs G.A., Aghajanian A., Campbell S.L. Redox regulation of Ras and Rho GTPases: mechanism and function. Antioxid. Redox Signal. 2013;18:250–258. doi: 10.1089/ars.2012.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gromer S., Arscott L.D., Williams C.H., Schirmer R.H., Becker K. Human placenta thioredoxin reductase. J. Biol. Chem. 1998;273:20096–20101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- 28.Cao Z., Tavender T.J., Roszak A.W., Cogdell R.J., Bulleid N.J. Crystal structure of reduced and of oxidized peroxiredoxin IV enzyme reveals a stable oxidized decamer and a non-disulfide-bonded intermediate in the catalytic cycle. J. Biol. Chem. 2011;286:42257–42266. doi: 10.1074/jbc.M111.298810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tavender T.J., Sheppard A.M., Bulleid N.J. Peroxiredoxin IV is an endoplasmic reticulum-localized enzyme forming oligomeric complexes in human cells. Biochem. J. 2008;411:191–199. doi: 10.1042/BJ20071428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prior K.-K., Leisegang M.S., Josipovic I., Löwe O., Shah A.M., Weissmann N., Schröder K., Brandes R.P. CRISPR/Cas9-mediated knockout of p22phox leads to loss of Nox1 and Nox4, but not Nox5 activity. Redox Biol. 2016;9:287–295. doi: 10.1016/j.redox.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serrander L., Cartier L., Bedard K., Banfi B., Lardy B., Plastre O., Sienkiewicz A., Fórró L., Schlegel W., Krause K.-H. NOX4 activity is determined by mRNA levels and reveals a unique pattern of ROS generation. Biochem. J. 2007;406:105–114. doi: 10.1042/BJ20061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meng S., Chen Z., Munoz-Antonia T., Wu J. Participation of both Gab1 and Gab2 in the activation of the ERK/MAPK pathway by epidermal growth factor. Biochem. J. 2005;391:143–151. doi: 10.1042/BJ20050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlessinger J., Lemmon M.A. SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE. 2003;2003:RE12. doi: 10.1126/stke.2003.191.re12. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y.-Y., Chu H.-M., Pan K.-T., Teng C.-H., Wang D.-L., Wang A.H.-J., Khoo K.-H., Meng T.-C. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J. Biol. Chem. 2008;283:35265–35272. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter J., Ilbert M., Graf P.C.F., Ozcelik D., Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leichert L.I., Gehrke F., Gudiseva H.V., Blackwell T., Ilbert M., Walker A.K., Strahler J.R., Andrews P.C., Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. USA. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.García-Santamarina S., Boronat S., Domènech A., Ayté J., Molina H., Hidalgo E. Monitoring in vivo reversible cysteine oxidation in proteins using ICAT and mass spectrometry. Nat. Protoc. 2014;9:1131–1145. doi: 10.1038/nprot.2014.065. [DOI] [PubMed] [Google Scholar]
- 38.Marzano C., Gandin V., Folda A., Scutari G., Bindoli A., Rigobello M.P. Inhibition of thioredoxin reductase by auranofin induces apoptosis in cisplatin-resistant human ovarian cancer cells. Free Radic. Biol. Med. 2007;42:872–881. doi: 10.1016/j.freeradbiomed.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 39.Kudin A.P., Augustynek B., Lehmann A.K., Kovács R., Kunz W.S. The contribution of thioredoxin-2 reductase and glutathione peroxidase to H(2)O(2) detoxification of rat brain mitochondria. Biochim. Biophys. Acta. 2012;1817:1901–1906. doi: 10.1016/j.bbabio.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 40.Pickering A.M., Vojtovich L., Tower J., Davies K.J.A. Oxidative stress adaptation with acute, chronic, and repeated stress. Free Radic. Biol. Med. 2013;55:109–118. doi: 10.1016/j.freeradbiomed.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groen A., Lemeer S., van der Wijk T., Overvoorde J., Heck A.J.R., Ostman A., Barford D., Slijper M., den Hertog J. Differential oxidation of protein-tyrosine phosphatases. J. Biol. Chem. 2005;280:10298–10304. doi: 10.1074/jbc.M412424200. [DOI] [PubMed] [Google Scholar]
- 42.Tsutsumi R., Harizanova J., Stockert R., Schröder K., Bastiaens P.I.H., Neel B.G. Assay to visualize specific protein oxidation reveals spatio-temporal regulation of SHP2. Nat. Commun. 2017;8:466. doi: 10.1038/s41467-017-00503-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knoops B., Loumaye E., van der Eecken V. Evolution of the peroxiredoxins. Subcell. Biochem. 2007;44:27–40. doi: 10.1007/978-1-4020-6051-9_2. [DOI] [PubMed] [Google Scholar]
- 44.Chan P.-C., Sudhakar J.N., Lai C.-C., Chen H.-C. Differential phosphorylation of the docking protein Gab1 by c-Src and the hepatocyte growth factor receptor regulates different aspects of cell functions. Oncogene. 2010;29:698–710. doi: 10.1038/onc.2009.363. [DOI] [PubMed] [Google Scholar]
- 45.Lamorte L., Kamikura D.M., Park M. A switch from p130Cas/Crk to Gab1/Crk signaling correlates with anchorage independent growth and JNK activation in cells transformed by the Met receptor oncoprotein. Oncogene. 2000;19:5973–5981. doi: 10.1038/sj.onc.1203977. [DOI] [PubMed] [Google Scholar]
- 46.Gual P., Giordano S., Williams T.A., Rocchi S., van Obberghen E., Comoglio P.M. Sustained recruitment of phospholipase C-gamma to Gab1 is required for HGF-induced branching tubulogenesis. Oncogene. 2000;19:1509–1518. doi: 10.1038/sj.onc.1203514. [DOI] [PubMed] [Google Scholar]
- 47.Babelova A., Jansen F., Sander K., Löhn M., Schäfer L., Fork C., Ruetten H., Plettenburg O., Stark H., Daniel C., Amann K., Pavenstädt H., Jung O., Brandes R.P. Activation of Rac-1 and RhoA contributes to podocyte injury in chronic kidney disease. PLoS One. 2013;8:e80328. doi: 10.1371/journal.pone.0080328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takemoto M., Asker N., Gerhardt H., Lundkvist A., Johansson B.R., Saito Y., Betsholtz C. A new method for large scale isolation of kidney glomeruli from mice. Am. J. Pathol. 2002;161:799–805. doi: 10.1016/S0002-9440(10)64239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 50.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 51.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 52.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material