SUMMARY
Recent observations showed that nascent RNA polymerase II transcripts, pre-mRNAs, and noncoding RNAs are highly susceptible to premature 3′-end cleavage and polyadenylation (PCPA) from numerous intronic cryptic polyadenylation signals (PASs). The importance of this in gene regulation was not previously appreciated as PASs, despite their prevalence, were thought to be active in terminal exons at gene ends. Unexpectedly, antisense oligonucleotide interference with U1 snRNA base-pairing to 5′ splice sites, which is necessary for U1 snRNP’s (U1) function in splicing, caused widespread PCPA in metazoans. This uncovered U1’s PCPA suppression activity, termed telescripting, as crucial for full-length transcription in thousands of vertebrate genes, providing a general role in transcription elongation control. Progressive intron-size expansion in metazoan evolution greatly increased PCPA vulnerability and dependence on U1 telescripting. We describe how these observations unfolded and discuss U1 telescripting’s role in shaping the transcriptome.
1. INTRODUCTION
Studies on the survival of motor neuron (SMN) complex have shown that it is an assembly chaperone for RNA–protein complexes (RNPs) best characterized for outfitting uridine-rich, noncoding small nuclear RNAs (snRNAs) with seven-membered Sm protein rings (Sm cores) (Fischer et al. 1997; Liu et al. 1997; Yong et al. 2004b; Cauchi 2010; Fischer et al. 2011; So et al. 2017). The assembly of Sm cores is a rate-limiting step in the biogenesis of small nuclear ribonucleo proteins (snRNPs), designated U1, U2, U4, U5, U11, U12, U4atac, which are well-characterized for their functions in splicing (Guthrie and Patterson 1988; Patel and Steitz 2003; Yong et al. 2004a; Battle et al. 2006; Wahl et al. 2009; Cauchi 2010; Yong et al. 2010; Fischer et al. 2011; Matera and Wang 2014). However, SMN deficiency, which causes spinal muscular atrophy (SMA), changes snRNP levels nonuniformly; rather than reducing snRNPs proportionally, SMA cells and cells engineered to have low SMN have an altered snRNP repertoire (Gabanella et al. 2007; Zhang et al. 2008; Workman et al. 2009). It had been noted earlier that snRNPs’ abundances vary despite their 1:1 stoichiometry in spliceosomes (Baserga and Steitz 1993). For example, at around 1,000,000 copies per human (HeLa) cell, U1 snRNP (U1) is several-fold more abundant than U4 and U6. However, the potential role of snRNP abundance in pre-messenger RNA (mRNA) processing was unknown. As we had also observed numerous splicing abnormalities in SMA (Zhang et al. 2008), we set out to determine what effects altering snRNP repertoire might have on the transcriptome. Both the snRNP repertoire and splicing changes were complex and varied in SMA mouse tissues (Zhang et al. 2008, 2013), suggesting a role for cell-specific factors and making it difficult to recapitulate in an experimental cell system. To circumvent this, we chose to systematically inhibit individual snRNPs one at a time.
Decreasing individual snRNPs levels to varying degrees by targeted RNA interference (RNAi) with shRNA or short interfering RNAs (siRNAs) did not seem suitable, as it takes more than 48 hours and is difficult to control. Instead, we chose to systematically inhibit individual snRNPs’ splicing activity using antisense morpholino oligonucleotides (AMOs) to mask the sequences in their snRNAs that are necessary for base-pairing with pre-mRNA (Kaida et al. 2010). Transfection of 25-mer AMOs to U2 and U12, tested on select introns, had been previously shown by Harald Konig and colleagues to inhibit splicing (Matter and Konig 2005). Based on this, we designed a series of snRNA targeting AMOs, including U1, U2, U5, U6, U11, and U6atac. RT-PCR on select introns validated their efficacy and guided optimization of transfection conditions and selection of doses and times.
Several features of AMOs make them an advantageous tool for probing function and interactions of specific RNA sequences. Their small size facilitates cell penetration, ensuring transfection of all cells in the experimental population. They are fast-acting, binding target RNAs nearly instantaneously, which minimizes the likelihood of indirect effects, and they are nondestructive to the RNP target (unlike antisense deoxy oligonucleotides that elicit RNase H-mediated RNA degradation), thereby preventing changes in the bound versus free RNP proteins pools. The morpholino chemistry also increases base-pairing avidity for RNA targets, and they are more RNase resistant compared with RNA antisense oligonucleotides (Summerton 1999; Heasman 2002), allowing the effects of a single transfection to be measured 24–72 hours later. Importantly, AMO dose can be readily varied over a large range and nontargeting AMOs can be used as controls.
2. U1 snRNP SUPPRESSES PASs IN NASCENT TRANSCRIPTS
U1 AMO, which emerged as the key probe in this study, illustrates the principles that guided the design of splicing-interfering AMOs. U1 is a ubiquitous RNP in eukaryotes, comprising U1 snRNA (164 nucleotides in human) and 11 proteins, including seven Sm proteins and three U1-specific proteins: U1-70K, U1A, and U1C. U1-70K and U1A bind stem-loops 1 and 2, designated in 5′ to 3′ order, out of U1 snRNA’s four stem-loops. In addition to its potential to advance understanding of SMA pathogenesis, U1 was a target of interest in the context of snRNPs’ stoichiometry because it is the most abundant small noncoding RNA in vertebrates, which was unexplained. Although U1, along with U2 snRNA, had been detected in the 1960s (Hodnett and Busch 1968; Weinberg and Penman 1968), they received little attention and there was no biological framework in which to consider them. That changed with a series of landmark discoveries by Joan A. Steitz’s group, beginning in 1979 (Lerner and Steitz 1979), of a new class of RNPs—the snRNPs, which contained U1 and U2 snRNAs as well as additional snRNPs they discovered, U4, U5, and U6. The same group then put forward the transformative insight (Lerner et al. 1980), confirmed shortly thereafter, by direct evidence (Mount et al. 1983; Padgett et al. 1983), that U1 snRNP recognizes the 5′ splice site (SS) by RNA:RNA base-pairing between U1 snRNA’s 5′-end and the pre-mRNA, thus identifying the first factors and initiating step in the removal of introns. Based on this, to inhibit splicing, we have designed U1 AMO, a 25-nt AMO complementary to U1 snRNA’s 5′-end, thereby providing a functional knockdown of U1’s known function.
Several assays were performed to verify that the transfected U1 AMO bound U1’s 5′-end, determine the dose–response profile and identify the dose required to mask all, or nearly all, of U1 in these cells (Kaida et al. 2010). An RNase H protection assay was performed on extracts from cells transfected with various U1 AMO doses. This assay, in which RNase H and an antisense DNA oligonucleotide probe complementary to U1 snRNA’s 5′-sequence was added to the extract, cleaves U1 snRNA that has this sequence accessible. The results showed a U1 AMO dose-dependent decrease in U1 snRNA cleavage, indicating that U1 AMO prevented the antisense DNA oligonucleotide probe from binding. This determined the U1 AMO dose at which complete or near complete interference with U1 snRNA 5′-base-pairing in cells was achieved (hereafter, high U1 AMO). The same dose–response was observed in a fluorescent in situ hybridization assay using a fluorescent labeled locked nucleic acid (LNA) probe complementary to U1 snRNA’s 5′ sequence on cells transfected with various U1 AMO doses. In addition, U1 AMO inhibited splicing of several test introns in vitro. Thus, U1 AMO functionally inactivated U1 snRNP both in vivo and in vitro (Kaida et al. 2010).
As these initial experiments (ca. 2008/2009) predated high-throughput RNA sequencing (RNA-seq), we used high density genomic tiling arrays, consisting of glass slide-immobilized oligonucleotides covering three human chromosomes harboring more than 3600 annotated genes, the method of choice at the time for surveying transcriptome differences between samples. For this, cDNA was prepared from total RNA from HeLa cells 8-hours posttransfection with U1 AMO at a dose that masks all, or nearly all, of U1’s 5′-end, and from cells transfected with nontargeting AMO control in the same experiment. Hybridization of the cDNAs to the tiling arrays identified differential RNA expression at each chromosomal location.
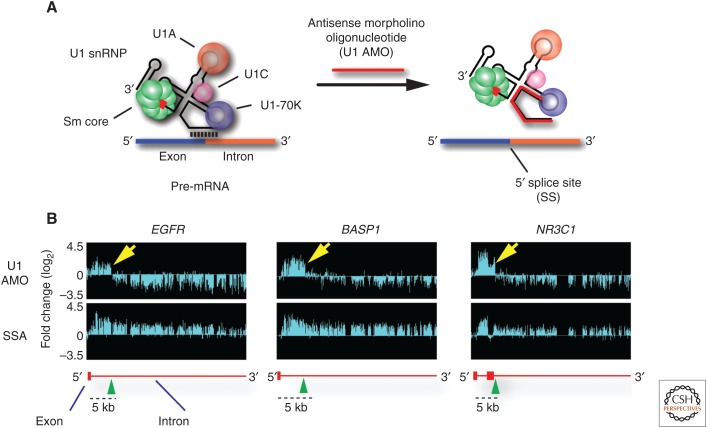
Genome browser visualization of the first series of experiments produced a surprising result, which was challenging to explain (Fig. 1). Expecting U1 AMO to inhibit splicing, we performed in parallel the same analysis on cells treated with spliceostatin A (SSA), a small molecule splicing inhibitor that targets SF3B1 (Kaida et al. 2007; Kotake et al. 2007), and with U2 AMO. Like AMOs, SSA is fast-acting, and it produced patterns consistent with splicing inhibition, including accumulation of signals covering entire introns. However, in the majority of genes, U1 AMO produced a remarkably different pattern, consisting of reads extending several kilobases from the transcription start site (TSS) into an intron were they abruptly ended, followed by a precipitous drop over the rest of the gene. Confirming U1’s expected activity, widespread splicing inhibition was evident by the accumulation of unspliced introns upstream of the point where the transcripts precipitously ended. U2 AMO produced a similar pattern to SSA, indicating that the phenomenon reflected U1-specific activity.
Figure 1.
Genomic tiling arrays detect, in U1 antisense morpholino oligonucleotide (AMO) transfected cells, transcripts extending from the transcription start site (TSS) of the genes EGFR, BASP1, and NR3C1 into the first part of the intron in which they abruptly end. (A) AMO (25-mer) to U1 snRNA’s 5′-end (U1 AMO) interferes with U1 snRNP (U1) base-pairing with 5′ splice sites, necessary for splicing of introns, and with other sequences on nascent transcripts. (B) Transcriptome profiling with genomic tiling arrays show the fold change in RNA signals compared with control. The top panel shows that, in addition to inhibiting splicing, U1 AMO also induces premature 3′-end cleavage and polyadenylation (PCPA) as RNA reads downstream from these end points are strongly decreased. In contrast (bottom panels), splicing inhibition with spliceostatin A (SSA) shows RNA reads increasing over the full length of introns, as expected for unspliced pre-mRNAs. Schematic gene structures (based on RefSeq, hg19) are depicted in red, with horizontal lines indicating introns and boxes indicating exons.
However, we could not discern from the pattern what the mechanism might be. Possibilities included that U1 is a transcription factor, particularly for elongation; that RNA polymerases (Pol II) stalled at road blocks that required U1 base-pairing to overcome (namely, termination did not occur; polymerases remained engaged but stopped moving); that transcription termination occurred early; or that transcription was in fact full-length, but nascent transcripts were being rapidly degraded leaving only 5′-side RNAs. It was puzzling why such a dramatic effect had not been previously detected or reported considering that U1 has been one of the most intensively investigated RNAs over decades and in many organisms. The likelihood of a technical artifact seemed remote, as the same result was observed in three separate biological experiments, but it could not be ruled out.
Significant progress in this respect came from application of another tool, which we used to define the 3′-end points of the U1 AMO-induced transcripts (Kaida et al. 2010). For this, we performed 3′ rapid amplification of cDNA ends (3′-RACE) (Scotto-Lavino et al. 2006) on RNAs using primers to produce cDNAs for 3′-end location of several genes. Surprisingly, sequencing of these revealed that they had 3′-end nongenomic poly(A) sequences ∼20–60 nt downstream from a consensus polyadenylation signal, typically AAUAAA and its variants (Proudfoot and Brownlee 1976; Magana-Mora et al. 2017), indistinguishable from canonical PASs that induce cleavage and polyadenylation (CPA) (Shi and Manley 2015; Tian and Manley 2017) at genes’ ends. This suggested that they were produced by premature cleavage and polyadenylation (PCPA) from cryptic polyadenylation signals (PASs) in introns, counter to the prevailing view that PASs marked the end of genes. Mutational inactivation of intronic PASs from which PCPA is elicited with U1 AMO (actionable PASs, used hereafter to indicate locations that not only have consensus PAS hexamer, but also show CPA activity) showed the PAS-dependent nature of this phenomenon, confirming that these intronic signals function like PASs at the canonical 3′-end of genes’ full-length transcripts. Interestingly, PAS inactivation did not necessarily prevent PCPA; instead, PCPA occurred from downstream PASs in the same intron or pre-mRNA. Having the same 5′ to 3′ polarity as transcription suggested that PCPA and its suppression by U1 is a directional, cotranscriptional mechanism (Berg et al. 2012).
These experiments revealed that, in addition to its splicing role, U1 is also a PCPA suppressor. PCPA is not a secondary effect of splicing inhibition as it frequently occurs in introns at great distances before transcription reaches the 3′SS. Early, promoter proximal PCPA is the default state for the majority of human genes, and full-length transcription requires U1. For brevity, we refer to this activity as telescripting because it is necessary for long-distance, full-length synthesis.
As there was nothing known about this new process, much more information was needed to understand its generality, potential role in biology and regulation. However, genomic tiling arrays were a significant bottleneck in terms of throughput, cost, labor, resolution, and availability of arrays for probing diverse organisms. RNA-seq became more widely available, but sequencing capacity, access to instruments and informatics expertise were major constraints. The expense of transcriptome sequencing to a depth required to identify differences between experimental samples also limited its application more broadly. To overcome these limitations, we devised a strategy for rapid transcriptome profiling that greatly accelerated the pace of our research by applying high-throughput RNA-seq only to differentially expressed transcripts (HIDE-seq; Berg et al. 2012). Reasoning that the most relevant information we sought was about what changed on U1 snRNP inhibition, we used subtractive hybridization (Diatchenko et al. 1996; Gurskaya et al. 1996) on cDNA fragments prepared from poly(A) RNA from control and U1 AMO-treated samples to eliminate identical sequences and thus only PCR amplify differentially expressed fragments for sequencing. In developing HIDE-seq we have introduced several technical improvements to subtractive hybridization, including digestion of the cDNAs with two or more 4-cutter restriction enzymes, which refined resolution, and ligation of bar-coded adaptors. These adaptors allowed samples to be combined and sequenced together, drastically reducing cost and controlling for sample-to-sample variation. By physically eliminating sequences that did not change, HIDE-seq increased sequencing depth of differential effects and greatly simplified data analysis as the number of reads directly reflected transcriptome change at any location. Algorithms developed to process the HIDE-seq datasets helped classify transcriptome changes, detect PCPA locations and visualize them on genome browsers. It confirmed the finding from genomic tiling arrays and provided a wealth of new information.
Applying HIDE-seq to human, mouse, and fruit fly (using U1 AMO matching the U1 snRNA 5′-sequence of each organism), we showed that U1 telescripting is evolutionarily conserved in metazoans (Berg et al. 2012). Genome-wide maps of PCPA locations at high U1 AMO doses further showed that it generally occurs in one of the first introns, frequently within 1 kb from the intron’s 5′SS. Assuming that U1 base-paired at that 5′SS for the purpose of splicing also provided telescripting, then effective U1 telescripting could be estimated to have a range of around 1 kb. However, in many genes, U1 AMO-induced PCPA occurred at much greater distances from the intron’s 5′SS, raising the possibility that U1 bound to 5′SS alone may be insufficient to ensure telescripting of introns longer than 1 kb. Mutating a 5′SS to a sequence that is incompatible with splicing in cases in which PCPA occurred from a PAS <1 kb downstream, caused constitutive PCPA from that PAS. However, U1 AMO increased that PCPA several-fold, suggesting that the PAS received additional suppression from U1 base-paired elsewhere. Furthermore, transfection of synthetic U1 snRNA with 5′-end complementary to the mutated 5′SS restored PAS suppression. This separated U1 splicing and telescripting requirements and indicated that U1 base-paired to any sequence could potentially function in telescripting, even if it is nonfunctional in splicing. U1 base-pairing to 5′SS is highly degenerate and depends on various pre-mRNA binding proteins, which likely facilitates its binding to cryptic 5′SS and many other RNA sequences. An important conclusion from these experiments was that U1 snRNP bound at 5′SS alone would likely be insufficient to protect long introns (>1 kb), which are common in complex organisms, requiring additional U1 base-paired in the intron to fully suppress PASs within them.
The genomic HIDE-seq maps revealed that telescripting is a physiological process, as many PCPA sites coincide with 3′-poly(A) sequence tags from diverse biological specimens, including normal human tissues. Therefore, while telescripting occurs naturally the PCPA elicited artificially in our experiments with U1 AMO enhance PCPA detection of actionable PASs that are widespread nature. Thus, serendipitously, U1 AMO uncovered telescripting and its ability to recapitulate it in a controlled experimental setting makes it a useful research tool for this process.
Testing the effect of various U1 AMO doses showed an interesting and dose-dependent effect on PCPA locations. While high U1 AMO triggered drastic TSS-proximal PCPA, generally in the first quarter of the gene, at low U1 AMO doses (masking <15% of U1) PCPA occurred at greater distances downstream, resulting in widespread 3′ untranslated regions (3′ UTRs) shortening. These PCPAs corresponded to an alternative 3′-end processing and polyadenylation (APA) shift to usage of a more proximal PAS in 3′ UTRs that contain tandem PASs. Such 3′ UTR shortening, which maintains the full-length coding sequence (CDS) but could de-regulate mRNA translation, stability, and localization, is associated with stimulated immune cells and neurons, cell proliferation, and cancer (Niibori et al. 2007; Flavell et al. 2008; Sandberg et al. 2008; Mayr and Bartel 2009; Lianoglou et al. 2013). Thus, U1 AMO recapitulated 3′ UTR APA. Knockdown of several 3′-end CPA factors (CPAFs), CFIm25/CPSF5, CFIm68/CFSF6, or PABPN1, also shifts APA to proximal PASs in the terminal exon, but apparently not in introns (Gruber et al. 2012, 2014; Jenal et al. 2012; Elkon et al. 2013; Masamha et al. 2014; Li et al. 2015; Zhu et al. 2018).
In addition, in many genes low U1 AMO induced a shift to usage of a PAS in an intron, resulting in shorter mRNA isoforms that lack the carboxy-terminal portion of the full-length CDS. These events are frequently attributed to alternative splicing that gives rise to an alternative last exon that has a PAS. However, as it emerged that potentially PAS usage anywhere in the transcript is under U1 control we considered a scenario whereby the primary event is PCPA in an intron. Mutational inactivation of the intronic PAS supported the PCPA-first scenario as U1 AMO no longer induced the splicing of the alternative last exon. An example of such a case, homer-1, illustrates the biological importance of this type of U1-controlled PCPA. Homer-1 encodes a scaffold protein critical for synaptogenesis and synapse strengthening in neurons. Neuronal stimulation, which can be experimentally mimicked in mouse neuronal-type PC12 cells, rapidly shifts synthesis from full-length (homer-ll) to a shorter isoform lacking the protein’s carboxy-terminal domain (CTD) (homer-1s). homer-1s antagonizes homer-ll, which is necessary to prevent overstimulation causing epilepsy (Niibori et al. 2007). Interestingly, low U1 AMO recapitulated homer-1l to homer-1s shift by PCPA in the intron downstream from homer-1s’ alternative last exon (Berg et al. 2012). Using several low U1 AMO concentrations showed a dose-dependent increase in the fraction of homer-1s and a reciprocal decrease in homer-1l.
3. U1 AMO PREMATURELY TERMINATES ELONGATING Pol II IN GENE BODIES
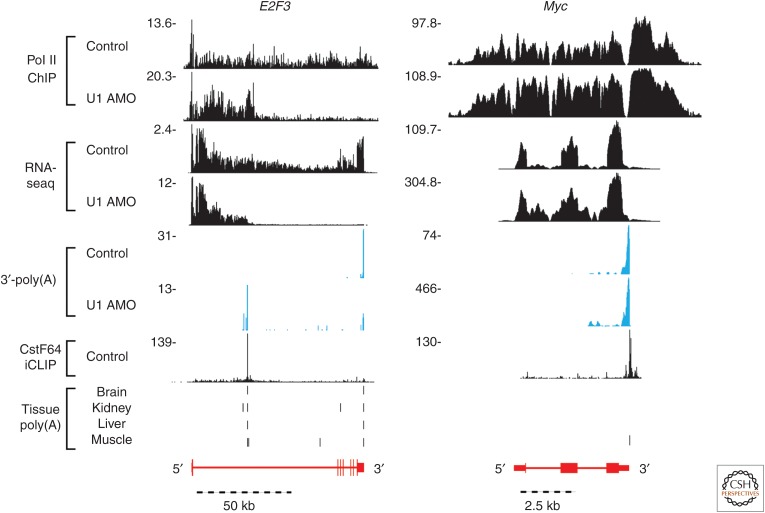
We used chromatin immunoprecipitation-sequencing (ChIP-seq) (Gilmour and Lis 1984) with antibodies to RNA Pol II to investigate the relationship between PCPA and Pol II transcription (Oh et al. 2017). ChIP-seq maps of Pol II chromosomal locations in controls extended from the TSS to the canonical 3′-end of genes, from which point it progressively declined to background level within a few kb downstream from the PAS (Fig. 2). The gradual decline in the post-3′-end section, described as the termination zone (TZ) (Fong et al. 2015), is consistent with the torpedo termination model in which XRN2 exonuclease degrades, in the 5′ to 3′ direction, the unprotected 5′-end of the transcript (because of a lack of a 5′-methylated G cap) trailing from Pol II before it catches up and causes the polymerase to release (Connelly and Manley 1988; Fong et al. 2015; Proudfoot 2016). In PCPAed genes, Pol II patterns were the same, including a TZ, except that the PCPA point marked the 3′-end. Pol II occupancy downstream, through the rest of the gene, was sharply reduced or eliminated. These observations indicated that PCPA causes premature transcription termination, supporting a cotranscriptional U1 telescripting model as opposed to an alternative mechanism whereby PCPA products are processed from full-length transcripts, posttranscriptionally. The Pol II ChIP-seq patterns, which tracked with nascent RNAs also indicated that transcription initiation was not inhibited despite PCPA; rather, transcription continued to flow into genes.
Figure 2.
Genome browser images of a medium/large, PCPAed gene, E2F3, and a small, up-regulated gene, Myc, in control and U1 antisense morpholino oligonucleotide (AMO)-transfected cells. The distinct RNA-seq read changes in these two genes show the gene-size-dependent transcriptome changes after U1 AMO treatment. Further evidence for these peaks being premature 3′-end cleavage and polyadenylation (PCPA) is shown with the overlap between nongenomically encoded 3′-poly(A) reads from our own poly(A) selected RNA-seq samples shown in blue, known iCLIP binding sites for the cleavage and polyadenylation (CPA) factor CstF64 (Yao et al. 2012) shown in black, and poly(A) sites found in various human tissues (brain, kidney, liver, and muscle; Derti et al. 2012) shown as vertical black bars. Schematic gene structures (based on RefSeq, hg19) are depicted in red, with horizontal lines indicating introns and boxes indicating exons.
4. SELECTIVE TELESCRIPTING DEPENDENCE OF LONG GENES
Improvements in RNA-seq instruments and greater availability made next-generation RNA-seq a tool of choice for whole-transcriptome sequencing, which we used to obtain more comprehensive definition of U1 AMO’s effect at nucleotide resolution (Fig. 2). Knowing that PCPA and telescripting are cotranscriptional, we used a brief (5–30 min at 3.5 to 7.5 h post-AMO transfection) metabolic labeling with 4-thiouridine and selected thiol-labeled RNAs for RNA-seq, which enhanced detection of nascent transcripts, short-lived PCPA products and newly spliced mRNAs (Dolken et al. 2008; Younis et al. 2013; Oh et al. 2017). Of note, in terms of technologies, HIDE-seq remains a powerful tool detecting segmental differences, especially rare ones, among RNAs in multiple samples. Although dedicated 3′-poly(A)-seq is more suitable for pinpointing locations of this specific processing step, it does not inform about any other transcriptome changes.
PCPA calling algorithms developed to capture characteristic RNA-seq features of PCPAed transcripts, consisting of increased reads in the 5′-portions of introns and decreased reads in downstream exons, detected PCPA in transcripts of thousands of genes in high U1 AMO-treated human (HeLa) cells. Multiple 3′-poly(A) reads were frequently detected in the same gene along with clusters of closely spaced 3′-poly(A) reads in the same intron, suggesting that full-length transcription in some genes requires multiple actionable PASs to be suppressed. Actionable PASs in introns and throughout pre-mRNAs are therefore transcription elongation checkpoints that depend on U1 and other factors to clear. The higher number of PASs in large genes, that arose stochastically, makes them more dependent on telescripting, which makes PCPA a gene regulation mechanism based on gene size, which to our knowledge is unprecedented.
Many U1 AMO-induced PCPAs coincided with 3′-poly(A) reads in normal (control) cells and tissues (Derti et al. 2012), indicating that, despite U1’s abundance, telescripting is insufficient to permanently suppress all PASs (Fig. 2). The resulting loss of polymerases in midgenes is a process we call transcription attrition. The extent of this unanticipated phenomenon is probably much greater than the 3′-poly(A) reads suggest because PCPAed transcripts are generally unstable and rapidly cleared away by the exosome (Lubas et al. 2015; Ogami et al. 2018). Although it seems wasteful, transcription attrition likely has a biological purpose.
However, around 1000 genes showed no evidence of PCPA under the same U1 AMO conditions (e.g., Myc; Fig. 2). A clear difference between this group and the PCPAed genes was gene size, which in higher eukaryotes is derived almost entirely from intron size; as PCPAed genes had a median size of 39 kb versus 14.2 kb for non-PCPAed genes (median size of all expressed genes was 22.8 kb). This showed that U1 telescripting is selectively required for full-length transcription of large genes. Remarkably, many of the PCPA-resistant genes were up-regulated (median 6.8 kb), producing full-length, spliced mRNAs, in the same environment where widespread splicing inhibition was evident and large genes were PCPAed (Oh et al. 2017).
The ability of acute stimuli to elicit PCPA, including in 3′ UTRs, could be the result of a transient U1 shortage caused by transcription up-regulation that draws U1 to an increasing number of nascent transcripts. Increasing U1 levels to keep up with greater demand would inevitably be slower because it entails an elaborate assembly of U1 snRNA with Sm proteins by the SMN complex in the cytoplasm and additional processing and transport steps. Alternatively, cell stimulation might change the balance between PCPA and telescripting in some other way. Regardless of its mechanism, the shift to shorter transcripts suggests that transient U1 telescripting insufficiency is a built-in aspect of immediate/early response.
The U1 AMO transfections have also tested the effect of U1 base-pairing interference on splicing in human cells. As expected, high U1 AMO strongly reduced splicing, but not uniformly. Intron retention, a reflection of splicing inhibition, was apparent in small and intermediate size introns. In large genes, the number of splicing events was sharply lower, however much of it was secondary to PCPA, which eliminated the opportunity of splicing from downstream introns. However, splicing in small genes that were up-regulated with U1 AMO was surprisingly robust in the same environment. How might this be explained? One potential explanation is a residual amount of uninhibited U1 remained and is sufficient for splicing in these genes, typically in all their introns. Another possibility is that these introns can use U1 that has U1 AMO bound, without U1 snRNA base-pairing. U1’s ability to interact with nascent transcripts independent of base-pairing has been described (Spiluttini et al. 2010). A third potential explanation is that these introns could splice even without U1 at all. Previous studies have shown that splicing in U1-depleted extracts can be restored by the addition of excess of SR proteins, a group of splicing-activating hnRNP proteins (Crispino et al. 1994; Tarn and Steitz 1994; Fukumura et al. 2009). It is plausible that by decreasing the number of introns that compete for the same factors, PCPA creates similar conditions of excess SR proteins in the nuclei of cells.
For this model to make sense, sustaining or increasing mRNA productivity from small genes that are up-regulated with U1 AMO must be critically important, or, more generally, up-regulation of these genes and simultaneous PCPA in large genes must be beneficial. Several arguments support this proposition. The small genes that are up-regulated with U1 AMO are ubiquitously expressed and enriched in primary response genes that are induced during acute cell stimulation. They include genes such as Myc, Cyr61, and GADD45B whose functions are necessary to adapt to adverse environmental changes and stressors. In contrast, these functions are underrepresented in large genes, which encode more diverse functions generally expressed in differentiated tissues. For example, neuronal and developmental genes are among the largest (Bertagnolli et al. 2013; Gabel et al. 2015), which makes them highly susceptible to PCPA (Fig. 3). Under circumstances that threaten cell survival PCPA of large genes transiently might be a strategic sacrifice to enhance expression of cell survival genes.
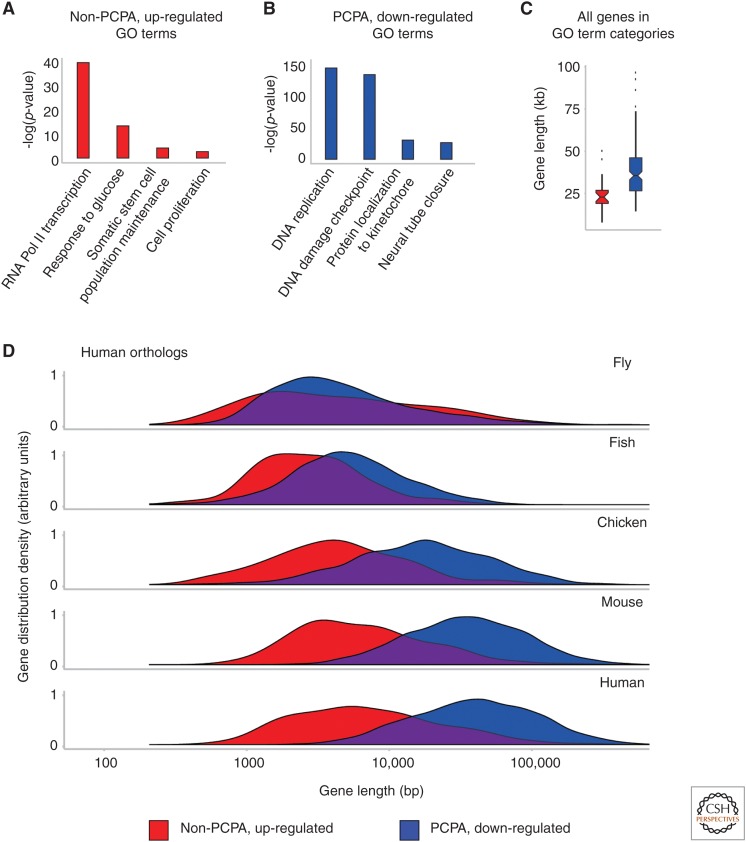
Figure 3.
Gene ontology (GO) term enrichment and gene size expansion in premature 3′-end cleavage and polyadenylation (PCPA) sensitive genes compared with non-PCPAed, up-regulated genes. The enrichment of the GO terms (Supek et al. 2011; Fang et al. 2016) for the top 50% of genes, ranked by fold change, in either non-PCPAed, up-regulated (A) or PCPAed, down-regulated (B) genes are shown. The gene size for all human genes in these functional categories, not just expressed in our samples, are shown as box plots (C) that were found enriched in either PCPAed genes (blue, median size 38 kb) or non-PCPAed, up-regulated genes (red, median size 23 kb). Density plots showing the gene size (D) of human gene orthologs among non-PCPAed, up-regulated genes and PCPAed genes in five metazoans. The sizes of non-PCPAed and up-regulated genes did not change across Drosophila melanogaster (median 3.5 kb), Takifugu rubripes (median 2.5 kb), Gallus gallus (median 4.3 kb), Mus musculus (median 5.9 kb), and Homo sapiens (median 6.8 kb); but PCPAed and down-regulated genes expanded significantly in these same species (medians 4.4 kb, 5.2 kb, 18.4 kb, 38 kb, and 45.0 kb, respectively).
The architecture of large and small genes seems well-suited for such a model. Small genes take less time to transcribe and their small size reduces susceptibility to PCPA. Interestingly, comparison of orthologous genes in several organisms showed that the size of PCPAed genes increased in vertebrates, especially in mammals, whereas small genes that are up-regulated with U1 AMO maintained a relatively small size (Fig. 3). Striking intron size expansion occurred in the course of evolution of vertebrates, especially in mammals (Catania and Lynch 2008; Gelfman et al. 2012; Rogozin et al. 2012). The significance of this expansion and its impact on gene expression are unknown. However, our studies suggest that intron size expansion was a strategic process, rather than random. It was selected against in essential acute response and survival genes but permissible in other genes. The selective intron size expansion stratified genes according to size and function, made progressively more polarized in vertebrate evolution. Its adoption suggests that it was beneficial.
We suggest that an important benefit of large introns is that they increase the opportunity to use PCPA to rapidly decrease competition from large genes for both transcription and pre-mRNA processing factors to boost expression from small, PCPA-insensitive genes. Many studies suggest that competition among pre-mRNAs for various binding factors can have strong effects on mRNA synthesis of many other genes (Timchenko et al. 1996; Miller et al. 2000; Cooper et al. 2009; Berg et al. 2012; Elkon et al. 2013; Munding et al. 2013). Another interesting and unexplained aspect of intron expansion is its positional bias for introns in the 5′ portion of genes. In humans, the first or second introns are frequently the largest (Bradnam and Korf 2008), and it is there that PCPA typically occurs. As large genes tend to have more exons, PCPA in the first or second introns eliminates multiple downstream exons and introns. We speculate that this architecture was selected for because it leverages PCPA to maximize the loss of competition for splicing resources and thus the potential gain for non-PCPAed genes.
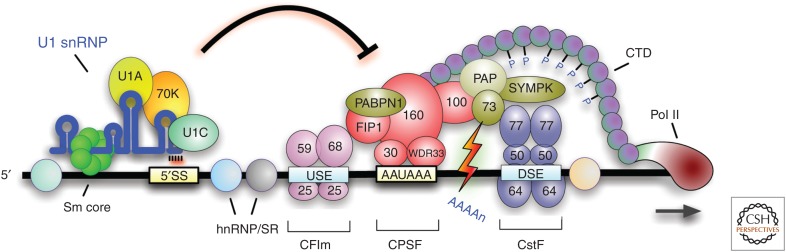
A potential advantage of a polarized genome with genes stratified by size and function is that provides a layer of regulation and an ability to shift gene expression priorities nearly instantly, without a need for de novo synthesis of transcription and pre-mRNA processing factors. It is difficult to imagine that metazoan genome evolution could have taken such a course without U1 telescripting. Figure 4 presents a schematic overview of the role of U1 telescripting in gene expression regulation.
Figure 4.
A schematic representing the role of U1 telescripting, suppression of premature transcription termination, by 3′ cleavage and polyadenylation (CPA) from polyadenylation signals (PASs) scattered throughout genes’ nascent transcripts. Shown are the common components known to carry out 3′ cleavage and polyadenylation processing at the canonical terminal PAS in the last exon; it is unknown which of these play a role in premature 3′-end cleavage and polyadenylation (PCPA) in introns. The precise location of U1 small nuclear ribonucleo protein (snRNP) binding relative to PASs is unknown but is likely to be generally within 1 kb. Transcription length depends on the balance between available U1 and CPA factors, and on the density of potential U1 base-pairing sites, which include 5′SS, cryptic 5′SS, and other sequences that U1 can bind, as well as PASs.
5. BEYOND PROTEIN-CODING GENES: TELESCRIPTING’S ROLE IN SHAPING THE TRANSCRIPTOME
As the shift to shorter mRNA and protein isoforms, represented by homer-1, illustrated, PCPA is not necessarily destructive and is also potentially useful for increasing proteome diversity (Berg et al. 2012). An additional way in which PCPA in introns contributes to protein isoform diversity is by the translation of an open reading frame that extends into the intron. Cartegni’s group described such cases for the EGF receptor and related receptor protein kinases (Vorlova et al. 2011). Like in homer-1, the short protein isoform lacks or has a different CTD and is antagonistic to the full-length protein.
Adding an important dimension, several studies from Phillip Sharp’s and Torben Jensen’s groups discovered that Pol II upstream antisense RNAs (uaRNAs) (Flynn et al. 2011, also known as PROMPTs; Preker et al. 2008) generally receive insufficient U1 telescripting relative to their prevalence of PASs (referred to in that study as U1-PAS axis), resulting in their rapid elimination by the nuclear exosome targeting complex, thereby reinforcing transcription in the sense direction (Almada et al. 2013; Andersen et al. 2013; Ntini et al. 2013; Lubas et al. 2015). This highlighted U1’s important role in transcription regulation, placing it as a central and bidirectional regulator of transcription elongation from divergent promoters (Almada et al. 2013; Ntini et al. 2013). This same study noted the evolutionary pressure to maintain and reinforce a U1 to PAS ratio in the sense direction and a low or inverse ratio in the antisense direction.
6. TELESCRIPTING MECHANISM
The molecular mechanism by which U1 telescripting suppresses actionable PASs particularly in introns remains unknown. CPAFs bind directly to nascent transcripts’ PASs cotranscriptionally with the transcription elongation complex and are dependent on Pol II CTD phosphorylation state (Buratowski 2009; Perales and Bentley 2009; Eick and Geyer 2013; Bentley 2014). We have noted that ultraviolet (UV) cross-linking studies from other groups (Martin et al. 2012; Yao et al. 2012; Chan et al. 2014; Schonemann et al. 2014) show several CPAFs binding in the vicinity of prominent PCPA locations in introns in control cells (e.g., Fig. 2) (Oh et al. 2017), indicating that intronic actionable PASs are recognized by at least some of the same or very similar factors that cleave and polyadenylate canonical 3′-end PASs. This makes it unlikely that U1 shields PASs. However, there is lack of critical information on where U1 binds such that it suppresses a PAS. Eric Lander’s group (Engreitz et al. 2014) has identified U1 base-pairing at 5′SS and in introns in mouse embryonic stem cells, but the dataset is not sufficiently deep and is from a different organism and cell type (our studies have focused on human cells) to address this question. It is therefore not possible to distinguish among various general models, including whether U1 telescripting is mediated by direct U1 interaction with CPAFs.
Previous studies showing CPA-inhibitory activity of two U1 proteins, U1A and U1-70K albeit in specific contexts, may be relevant. U1A as a U1 snRNP-free protein dimer inhibits CPA from an adjacent PAS in the terminal exon of both its own pre-mRNA and a few others that contain tandem stem-loops highly similar to U1 snRNA SL2 (Boelens et al. 1993; Gunderson et al. 1994, 1997; Klein Gunnewiek et al. 2000; Phillips and Gunderson 2003; Workman et al. 2014). In vitro studies showed that this inhibition is mediated by U1A binding to poly(A) polymerase (PAP). U1-70K can also inhibit PAP bound to U1 using the similar PAP-inhibitory motifs in U1A (Gunderson et al. 1998). As a U1-free protein, U1-70K can also associate with the PAS’ upstream element (USE)-binding CFIm subunit (Ruegsegger et al. 1996; Awasthi and Alwine 2003). It remains to be determined whether U1A and U1-70K also have such CPA-inhibitory activities in the context of U1 snRNP and, if so, whether such activities applies to PASs in introns. In specific cases, U1 base-paired to a 5′SS-like sequence adjacent to the PAS in a late-phase transcript in bovine papilloma virus (BPV) and 5′LTRs in HIV-1 and foamy viruses or tethered by engineering extensive base-pairing in proximity of a PAS in the terminal exon can inhibit usage of that PAS (Gunderson et al. 1998; Ashe et al. 2000; Vagner et al. 2000; Fortes et al. 2003; Schrom et al. 2013).
U1 base-paired upstream and in the vicinity of the PAS (<1 kb) could either prevent formation of an active CPA complex or inhibit its activity. CPA occurs at the first actionable PAS that is not suppressed by U1. U1 shortage thus increases the likelihood of PCPA. Like U1 AMO, knockdowns of CFIm25/CPSF5, CFIm68/CFSF6 and PABPN1 can elicit proximal PAS usage in tandem PASs in 3′ UTRs (Gruber et al. 2012; Jenal et al. 2012; Elkon et al. 2013; Masamha et al. 2014; Li et al. 2015). It will be interesting to determine whether these knockdowns and U1 AMO induce the same APA changes, which will help determine whether these CPAFs act in concert with U1 in the last exon.
Few PCPAs were detected in internal exons in our studies. It is possible that exons are bound by factors that shield PASs and that their small size and relative GC-richness (Amit et al. 2012) decreased the likelihood of having AU-rich PAS hexanucleotides. Small genes and introns are less susceptible both because their small introns decrease the opportunity for PASs to arise and because kinetic competition from splicing removes introns more quickly.
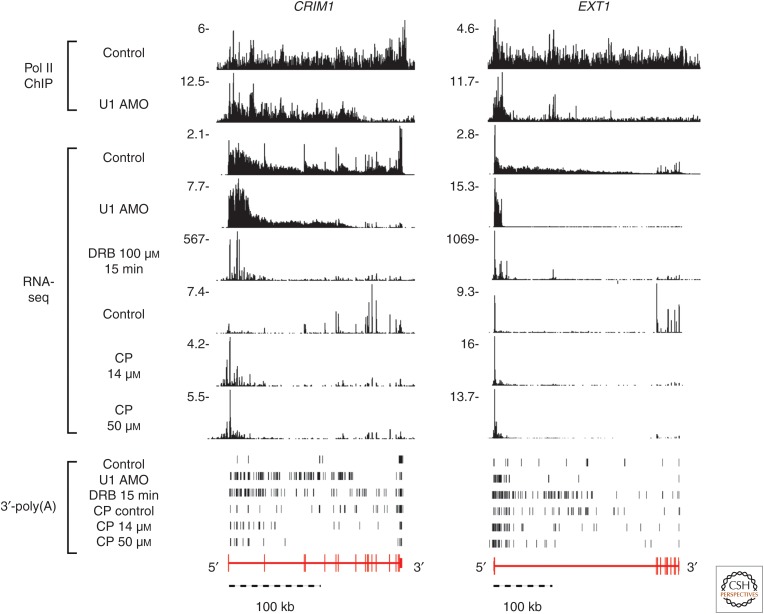
As PCPA is cotranscriptional and PASs induce Pol II pausing (Rigo et al. 2005; Davidson et al. 2014; Kaida 2016), we investigated the effect of small molecule reagents, 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) (Chodosh et al. 1989) and camptothecin (CP) (Pommier 2006), which slow down Pol II transcription on PCPA. DRB inhibits CDK9 and related kinases that phosphorylate the Pol II CTD, which is required to release Pol II pausing at promoter proximal pause sites (PPP; within 100 nt from the TSS) and at PASs (Guo and Price 2013; Kwak and Lis 2013; Laitem et al. 2015). Immediately following DRB washout, as PPP is released, polymerases rapidly flow into genes and are likely slowed down as the leading polymerases encounter a barrier or checkpoint, forcing lagging polymerases to also slow. CP slows transcription through inhibition of topoisomerase I, which relieves transcription-obstructing DNA supercoiling. Despite different chemistries and mechanisms of action, both DRB and CP caused PCPA at similar locations as U1 AMO (Fig. 5), suggesting that slower transcription and/or pausing increases the propensity for PCPA. Interestingly, CP and DNA damage, which create transcription roadblocks, have been shown previously to selectively decrease expression from large genes (McKay et al. 2004; Andrade-Lima et al. 2015). PCPA could provide checkpoints for orderly removal of polymerases to prevent overcrowding and transcription errors.
Figure 5.
Genome browser views showing that, in the example genes CRIM1 and EXT1, DRB and CP induce premature 3′-end cleavage and polyadenylation (PCPA), suggesting that slower Pol II elongation speed or pausing increases the propensity for PCPA. PCPA locations induced by DRB and CP overlap with U1 antisense morpholino oligonucleotide (AMO)-induced PCPAs and their nature is confirmed by nongenomic 3′-poly(A) reads from samples, shown as vertical black bars, indicating that these termination sites are true PCPAs, and not just random nuclease cleavage sites. RNAs produced during the 15-min DRB washout phase and 30-min CP treatment were metabolically labeled, selected, and sequenced (RNA-seq). The signal beyond the PCPA point seen in the Pol II ChIP-seq sample represents a transcription termination zone. Pol II occupancy beyond that site was strongly decreased.
A recent study from Sharp’s group showed that PCPA frequently occurs at the first stable nucleosomes relative to the TSS (Chiu et al. 2018). This study showed that U1 AMO-induced PCPA sites tend to cluster at the edge of CpG islands at the first stable nucleosomes downstream from the TSS. Global run-on sequencing and use of flavopiridol (a CDK9 inhibitor, similar to DRB) linked PCPAs to Pol II pausing in these locations, thus showing the influence of transcription elongation speed on telescripting. These first stable nucleosomes act as speed bumps, slowing Pol II and allowing more time for the CPAF machinery to act on a PAS. Chromatin structure, through its effect on transcription elongation speed and resulting changes in Pol II CTD phosphorylation, could therefore also play a role in the balance between telescripting and PCPA.
7. HOW CELLS MAKE A GREAT ABUNDANCE OF U1
The demand for high U1 abundance to use for telescripting required cells of higher organisms to produce more U1 than other spliceosomal snRNPs (with the possible exception of U2, which is also very abundant). To some extent, this is helped by the greater number (potentially >100) of U1 genes, although it is unknown how many of those are transcriptionally active (O’Reilly et al. 2013). However, pre-snRNA transcription may not be a limiting factor for U1 synthesis. The rate-limiting step in snRNP biogenesis is likely the assembly of the Sm core. Interestingly, recent studies revealed that the U1-specific RBP, U1-70K can hijack the SMN–Gemin2–Sm pentamer complex, a key intermediate of the Sm core assembly, and promote access of U1 snRNA to the SMN complex over other snRNAs (So et al. 2016). This reveals an additional and a U1-exclusive Sm core assembly pathway at the expense of other snRNPs, which explains at least in part how U1 abundance is achieved in cells.
8. CONCLUDING REMARKS
The discovery that U1 snRNA 5′-end base-pairing (to pre-mRNAs’ 5′SS) provides crucial recognition for the removal of introns was a conceptual breakthrough later generalized to other trans-acting small RNPs. It is a marvel of nature’s ingenuity that U1, through the same U1 snRNA 9-nucleotide sequence, is also crucial for ensuring that transcription continues far enough to produce introns. By suspending termination, U1 increases its own chances to find a 3′SS, and thereby remove itself from the pre-mRNA. The much earlier discovery of U1’s role in splicing influences a more splicing-centric perspective, but an evolutionary perspective invites the question of which came first: splicing or telescripting? Although the answer is unknown and U1’s origin is uncertain, this abundant noncoding small RNP has emerged as a central regulator of the Pol II transcriptome.
Technological advances, application of new tools to study RNA and RNPs have made possible the discoveries on telescripting. Understanding the mechanism by which U1 telescripting suppresses PASs to avert premature transcription termination now presents exciting new challenges. Additional tools will be necessary to dissect the biochemical and structural basis of U1 telescripting, including U1 interactions with components of the transcription and CPA machineries, and to precisely determine its binding locations on nascent transcripts and visualize PCPA in cells in real time, and identify modulators by high-throughput screening.
Although physiological circumstances in which telescripting is drastically inhibited, as occurs with high U1 AMO doses, have not been described, it can be reasonably predicted that some viruses have evolved mechanisms to inhibit telescripting, including by decoying U1, to benefit from resources that are tied up by long-distance transcription.
ACKNOWLEDGMENTS
We thank our laboratory colleagues, present and past, whose shared curiosity to understand unexpected observations and team work contributed to this research. This work was supported by a grant from the National Institutes of Health (R01GM112923 to G.D.). G.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Editors: Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. 2013. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit M, Donyo M, Hollander D, Goren A, Kim E, Gelfman S, Lev-Maor G, Burstein D, Schwartz S, Postolsky B, et al. 2012. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep 1: 543–556. [DOI] [PubMed] [Google Scholar]
- Andersen PR, Domanski M, Kristiansen MS, Storvall H, Ntini E, Verheggen C, Schein A, Bunkenborg J, Poser I, Hallais M, et al. 2013. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat Struct Mol Biol 20: 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade-Lima LC, Veloso A, Paulsen MT, Menck CF, Ljungman M. 2015. DNA repair and recovery of RNA synthesis following exposure to ultraviolet light are delayed in long genes. Nucleic Acids Res 43: 2744–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe MP, Furger A, Proudfoot NJ. 2000. Stem-loop 1 of the U1 snRNP plays a critical role in the suppression of HIV-1 polyadenylation. RNA 6: 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi S, Alwine JC. 2003. Association of polyadenylation cleavage factor I with U1 snRNP. RNA 9: 1400–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga SJ, Steitz JA. 1993. The diverse world of small ribonucleoproteins. In The RNA world (ed. Gesteland RF, Atkins JF), pp. 359–382. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. 2006. The SMN complex: An assembly machine for RNPs. Cold Spring Harb Symp Quant Biol 71: 313–320. [DOI] [PubMed] [Google Scholar]
- Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. 2012. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertagnolli NM, Drake JA, Tennessen JM, Alter O. 2013. SVD identifies transcript length distribution functions from DNA microarray data and reveals evolutionary forces globally affecting GBM metabolism. PLoS ONE 8: e78913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens WC, Jansen EJ, van Venrooij WJ, Stripecke R, Mattaj IW, Gunderson SI. 1993. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell 72: 881–892. [DOI] [PubMed] [Google Scholar]
- Bradnam KR, Korf I. 2008. Longer first introns are a general property of eukaryotic gene structure. PLoS ONE 3: e3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania F, Lynch M. 2008. Where do introns come from? PLoS Biol 6: e283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchi RJ. 2010. SMN and Gemins: “We are family” … or are we? Insights into the partnership between Gemins and the spinal muscular atrophy disease protein SMN. Bioessays 32: 1077–1089. [DOI] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR III, Ule J, Manley JL, Shi Y. 2014. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev 28: 2370–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu AC, Suzuki HI, Wu X, Mahat DB, Kriz AJ, Sharp PA. 2018. Transcriptional pause sites delineate stable nucleosome-associated premature polyadenylation suppressed by U1 snRNP. Mol Cell 69: 648–663.e647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh LA, Fire A, Samuels M, Sharp PA. 1989. 5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J Biol Chem 264: 2250–2257. [PubMed] [Google Scholar]
- Connelly S, Manley JL. 1988. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev 2: 440–452. [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. 2009. RNA and disease. Cell 136: 777–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino JD, Blencowe BJ, Sharp PA. 1994. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 265: 1866–1869. [DOI] [PubMed] [Google Scholar]
- Davidson L, Muniz L, West S. 2014. 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev 28: 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. 2012. A quantitative atlas of polyadenylation in five mammals. Genome Res 22: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, et al. 1996. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci 93: 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, et al. 2008. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14: 1959–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D, Geyer M. 2013. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev 113: 8456–8490. [DOI] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. 2013. Alternative cleavage and polyadenylation: Extent, regulation and function. Nat Rev Genet 14: 496–506. [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. 2014. RNA–RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell 159: 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Knezevic B, Burnham KL, Knight JC. 2016. XGR software for enhanced interpretation of genomic summary data, illustrated by application to immunological traits. Genome Med 8: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Liu Q, Dreyfuss G. 1997. The SMN–SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 90: 1023–1029. [DOI] [PubMed] [Google Scholar]
- Fischer U, Englbrecht C, Chari A. 2011. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip Rev RNA 2: 718–731. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. 2008. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Almada AE, Zamudio JR, Sharp PA. 2011. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci 108: 10460–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Brannan K, Erickson B, Kim H, Cortazar MA, Sheridan RM, Nguyen T, Karp S, Bentley DL. 2015. Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA polymerase II termination suggest widespread kinetic competition. Mol Cell 60: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Cuevas Y, Guan F, Liu P, Pentlicky S, Jung SP, Martinez-Chantar ML, Prieto J, Rowe D, Gunderson SI. 2003. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc Natl Acad Sci 100: 8264–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumura K, Taniguchi I, Sakamoto H, Ohno M, Inoue K. 2009. U1-independent pre-mRNA splicing contributes to the regulation of alternative splicing. Nucleic Acids Res 37: 1907–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. 2007. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS ONE 2: e921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, Hemberg M, Ebert DH, Greenberg ME. 2015. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfman S, Burstein D, Penn O, Savchenko A, Amit M, Schwartz S, Pupko T, Ast G. 2012. Changes in exon–intron structure during vertebrate evolution affect the splicing pattern of exons. Genome Res 22: 35–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. 1984. Detecting protein–DNA interactions in vivo: Distribution of RNA polymerase on specific bacterial genes. Proc Natl Acad Sci 81: 4275–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Keller W, Zavolan M. 2012. Cleavage factor Im is a key regulator of 3′ UTR length. RNA Biol 9: 1405–1412. [DOI] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Keller W, Zavolan M. 2014. Means to an end: Mechanisms of alternative polyadenylation of messenger RNA precursors. Wiley Interdiscip Rev RNA 5: 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Beyer K, Martin G, Keller W, Boelens WC, Mattaj LW. 1994. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell 76: 531–541. [DOI] [PubMed] [Google Scholar]
- Gunderson SI, Vagner S, Polycarpou-Schwarz M, Mattaj IW. 1997. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev 11: 761–773. [DOI] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell 1: 255–264. [DOI] [PubMed] [Google Scholar]
- Guo J, Price DH. 2013. RNA polymerase II transcription elongation control. Chem Rev 113: 8583–8603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya NG, Diatchenko L, Chenchik A, Siebert PD, Khaspekov GL, Lukyanov KA, Vagner LL, Ermolaeva OD, Lukyanov SA, Sverdlov ED. 1996. Equalizing cDNA subtraction based on selective suppression of polymerase chain reaction: Cloning of Jurkat cell transcripts induced by phytohemaglutinin and phorbol 12-myristate 13-acetate. Anal Biochem 240: 90–97. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Patterson B. 1988. Spliceosomal snRNAs. Annu Rev Genet 22: 387–419. [DOI] [PubMed] [Google Scholar]
- Heasman J. 2002. Morpholino oligos: Making sense of antisense? Dev Biol 243: 209–214. [DOI] [PubMed] [Google Scholar]
- Hodnett JL, Busch H. 1968. Isolation and characterization of uridylic acid-rich 7 S ribonucleic acid of rat liver nuclei. J Biol Chem 243: 6334–6342. [PubMed] [Google Scholar]
- Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, Vrielink JA, Bos AJ, Drost J, Rooijers K, et al. 2012. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149: 538–553. [DOI] [PubMed] [Google Scholar]
- Kaida D. 2016. The reciprocal regulation between splicing and 3′-end processing. Wiley Interdiscip Rev RNA 7: 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. 2007. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3: 576–583. [DOI] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Gunnewiek JM, Hussein RI, van Aarssen Y, Palacios D, de Jong R, van Venrooij WJ, Gunderson SI. 2000. Fourteen residues of the U1 snRNP-specific U1A protein are required for homodimerization, cooperative RNA binding, and inhibition of polyadenylation. Mol Cell Biol 20: 2209–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. 2007. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol 3: 570–575. [DOI] [PubMed] [Google Scholar]
- Kwak H, Lis JT. 2013. Control of transcriptional elongation. Annu Rev Genet 47: 483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitem C, Zaborowska J, Isa NF, Kufs J, Dienstbier M, Murphy S. 2015. CDK9 inhibitors define elongation checkpoints at both ends of RNA polymerase II-transcribed genes. Nat Struct Mol Biol 22: 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Steitz JA. 1979. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci 76: 5495–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. 1980. Are snRNPs involved in splicing? Nature 283: 220–224. [DOI] [PubMed] [Google Scholar]
- Li W, You B, Hoque M, Zheng D, Luo W, Ji Z, Park JY, Gunderson SI, Kalsotra A, Manley JL, et al. 2015. Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet 11: e1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. 2013. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev 27: 2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fischer U, Wang F, Dreyfuss G. 1997. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 90: 1013–1021. [DOI] [PubMed] [Google Scholar]
- Lubas M, Andersen PR, Schein A, Dziembowski A, Kudla G, Jensen TH. 2015. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep 10: 178–192. [DOI] [PubMed] [Google Scholar]
- Magana-Mora A, Kalkatawi M, Bajic VB. 2017. Omni-PolyA: A method and tool for accurate recognition of poly(A) signals in human genomic DNA. BMC Genomics 18: 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Gruber AR, Keller W, Zavolan M. 2012. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep 1: 753–763. [DOI] [PubMed] [Google Scholar]
- Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, Wagner EJ. 2014. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510: 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Wang Z. 2014. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter N, Konig H. 2005. Targeted “knockdown” of spliceosome function in mammalian cells. Nucleic Acids Res 33: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay BC, Stubbert LJ, Fowler CC, Smith JM, Cardamore RA, Spronck JC. 2004. Regulation of ultraviolet light–induced gene expression by gene size. Proc Natl Acad 101: 6582–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. 2000. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J 19: 4439–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount SM, Pettersson I, Hinterberger M, Karmas A, Steitz JA. 1983. The U1 small nuclear RNA–protein complex selectively binds a 5′ splice site in vitro. Cell 33: 509–518. [DOI] [PubMed] [Google Scholar]
- Munding EM, Shiue L, Katzman S, Donohue JP, Ares M Jr. 2013. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol Cell 51: 338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niibori Y, Hayashi F, Hirai K, Matsui M, Inokuchi K. 2007. Alternative poly(A) site-selection regulates the production of alternatively spliced vesl-1/homer1 isoforms that encode postsynaptic scaffolding proteins. Neurosci Res 57: 399–410. [DOI] [PubMed] [Google Scholar]
- Ntini E, Jarvelin AI, Bornholdt J, Chen Y, Boyd M, Jorgensen M, Andersson R, Hoof I, Schein A, Andersen PR, et al. 2013. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol 20: 923–928. [DOI] [PubMed] [Google Scholar]
- Ogami K, Chen Y, Manley JL. 2018. RNA surveillance by the nuclear RNA exosome: Mechanisms and significance. Noncoding RNA 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JM, Di C, Venters CC, Guo J, Arai C, So BR, Pinto AM, Zhang Z, Wan L, Younis I, et al. 2017. U1 snRNP telescripting regulates a size-function-stratified human genome. Nat Struct Mol Biol 24: 993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly D, Dienstbier M, Cowley SA, Vazquez P, Drozdz M, Taylor S, James WS, Murphy S. 2013. Differentially expressed, variant U1 snRNAs regulate gene expression in human cells. Genome Res 23: 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett RA, Mount SM, Steitz JA, Sharp PA. 1983. Splicing of messenger RNA precursors is inhibited by antisera to small nuclear ribonucleoprotein. Cell 35: 101–107. [DOI] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. 2003. Splicing double: Insights from the second spliceosome. Nat Rev Mol Cell Biol 4: 960–970. [DOI] [PubMed] [Google Scholar]
- Perales R, Bentley D. 2009. “Cotranscriptionality”: The transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C, Gunderson S. 2003. Sequences adjacent to the 5′ splice site control U1A binding upstream of the IgM heavy chain secretory poly(A) site. J Biol Chem 278: 22102–22111. [DOI] [PubMed] [Google Scholar]
- Pommier Y. 2006. Topoisomerase I inhibitors: Camptothecins and beyond. Nat Rev Cancer 6: 789–802. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ. 2016. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science 352: aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Brownlee GG. 1976. 3′ Non-coding region sequences in eukaryotic messenger RNA. Nature 263: 211–214. [DOI] [PubMed] [Google Scholar]
- Rigo F, Kazerouninia A, Nag A, Martinson HG. 2005. The RNA tether from the poly(A) signal to the polymerase mediates coupling of transcription to cleavage and polyadenylation. Mol Cell 20: 733–745. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Carmel L, Csuros M, Koonin EV. 2012. Origin and evolution of spliceosomal introns. Biol Direct 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegsegger U, Beyer K, Keller W. 1996. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem 271: 6107–6113. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonemann L, Kuhn U, Martin G, Schafer P, Gruber AR, Keller W, Zavolan M, Wahle E. 2014. Reconstitution of CPSF active in polyadenylation: Recognition of the polyadenylation signal by WDR33. Genes Dev 28: 2381–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrom EM, Moschall R, Hartl MJ, Weitner H, Fecher D, Langemeier J, Bohne J, Wohrl BM, Bodem J. 2013. U1snRNP-mediated suppression of polyadenylation in conjunction with the RNA structure controls poly(A) site selection in foamy viruses. Retrovirology 10: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto-Lavino E, Du G, Frohman MA. 2006. 3′ end cDNA amplification using classic RACE. Nat Protoc 1: 2742–2745. [DOI] [PubMed] [Google Scholar]
- Shi Y, Manley JL. 2015. The end of the message: Multiple protein–RNA interactions define the mRNA polyadenylation site. Genes Dev 29: 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So BR, Wan L, Zhang Z, Li P, Babiash E, Duan J, Younis I, Dreyfuss G. 2016. A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat Struct Mol Biol 23: 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So BR, Zhang Z, Dreyfuss G. 2017. The function of survival motor neuron complex and its role in spinal muscular atrophy pathogenesis. In Spinal muscular atrophy: Disease mechanisms and therapy (ed. Sumner CJ, Paushkin S, Ko C-P), pp. 99–111. Academic, San Diego. [Google Scholar]
- Spiluttini B, Gu B, Belagal P, Smirnova AS, Nguyen VT, Hebert C, Schmidt U, Bertrand E, Darzacq X, Bensaude O. 2010. Splicing-independent recruitment of U1 snRNP to a transcription unit in living cells. J Cell Sci 123: 2085–2093. [DOI] [PubMed] [Google Scholar]
- Summerton J. 1999. Morpholino antisense oligomers: The case for an RNase H-independent structural type. Biochim Biophys Acta 1489: 141–158. [DOI] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. 1994. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev 8: 2704–2717. [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. 1996. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res 24: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Ruegsegger U, Gunderson SI, Keller W, Mattaj IW. 2000. Position-dependent inhibition of the cleavage step of pre-mRNA 3′-end processing by U1 snRNP. RNA 6: 178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlova S, Rocco G, Lefave CV, Jodelka FM, Hess K, Hastings ML, Henke E, Cartegni L. 2011. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol Cell 43: 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Weinberg RA, Penman S. 1968. Small molecular weight monodisperse nuclear RNA. J Mol Biol 38: 289–304. [DOI] [PubMed] [Google Scholar]
- Workman E, Saieva L, Carrel TL, Crawford TO, Liu D, Lutz C, Beattie CE, Pellizzoni L, Burghes AH. 2009. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum Mol Genet 18: 2215–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman E, Veith A, Battle DJ. 2014. U1A regulates 3′ processing of the survival motor neuron mRNA. J Biol Chem 289: 3703–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Biesinger J, Wan J, Weng L, Xing Y, Xie X, Shi Y. 2012. Transcriptome-wide analyses of CstF64–RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci 109: 18773–18778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Golembe TJ, Battle DJ, Pellizzoni L, Dreyfuss G. 2004a. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol Cell Biol 24: 2747–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Wan L, Dreyfuss G. 2004b. Why do cells need an assembly machine for RNA–protein complexes? Trends Cell Biol 14: 226–232. [DOI] [PubMed] [Google Scholar]
- Yong J, Kasim M, Bachorik JL, Wan L, Dreyfuss G. 2010. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol Cell 38: 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis I, Dittmar K, Wang W, Foley SW, Berg MG, Hu KY, Wei Z, Wan L, Dreyfuss G. 2013. Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. eLife 2: e00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. 2008. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133: 585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Pinto AM, Wan L, Wang W, Berg MG, Oliva I, Singh LN, Dengler C, Wei Z, Dreyfuss G. 2013. Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc Natl Acad Sci 110: 19348–19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Wang X, Forouzmand E, Jeong J, Qiao F, Sowd GA, Engelman AN, Xie X, Hertel KJ, Shi Y. 2018. Molecular mechanisms for CFIm-mediated regulation of mRNA alternative polyadenylation. Mol Cell 69: 62–74.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]