Abstract
Modification of protein cysteine residues by disulfide formation with glutathione (glutathionylation) is a reversible posttranslational modification of critical importance in controlling cell signaling events following oxidative and/or nitrosative stress. Here, we show that human sulfiredoxin, a small redox protein conserved in eukaryotes, can act as a novel regulator of the redox-activated thiol switch in cells by catalyzing deglutathionylation of a number of distinct proteins in response to oxidative and/or nitrosative stress. Actin and protein tyrosine phosphatase 1B were identified in vitro as targets of sulfiredoxin 1 (Srxl)-dependent deglutathionylation and confirmed in vivo by two-dimensional gel electrophoresis analysis. In addition, we show that Srxl-dependent degluta-thionylation is functionally relevant through restoration of phosphatase activity. Human sulfiredoxin contains one cysteine residue (Cys99) that is conserved in all family members. Mutation of the cysteine residue inhibits deglutathionylation but did not affect its capacity to bind intracellular proteins. Furthermore, sulfiredoxin is not an acceptor molecule for the GS− moiety during the reaction process. Using two-dimensional gel electrophoresis, we identified multiple protein targets in vivo that are deglutathionylated by sulfiredoxin following oxidative and/or nitrosative stress. This novel deglutathiony-lation function of sulfiredoxin suggests it has a central role in redox control with potential implications in cell signaling.
Introduction
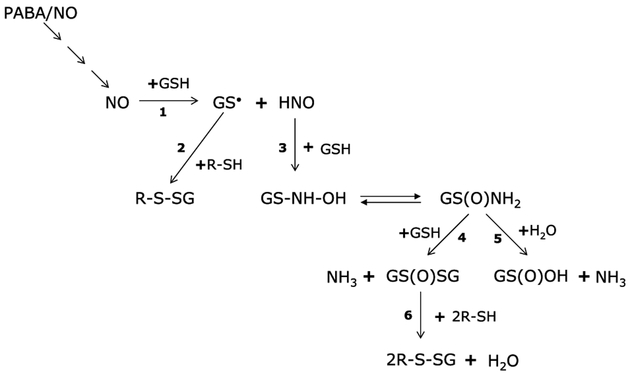
The redox state of the cell is distinguished as the ratio of oxidized to reduced redox molecules, a major component of which is the tripeptide (Gly-Glu-Cys) glutathione (GSH). GSH is present in cells at millimolar concentrations (1), and the ratio of the reduced pool to glutathione disulfide (GSSG) is critical to cellular redox balance. In models of oxidative stress, transient shifts in the GSH/GSSG ratio from 100 to 10 or even 1 have been described and found to correlate with the amount of protein mixed disulfide formation (2). Glutathionylation is a novel posttranslational modification of low pKa cysteine residues in proteins by addition of GS−. Oxidative or nitrosative stress can cause this reversible modification on multiple proteins through intermediary formation of radicals (Fig. 1). In brief, a reduced protein thiol (R-SH) can react with an activated rGSH radical (GS), or an activated protein thiol (R-S−) can react with GSH. Figure 1 illustrates a subset of reactions that can generate radicals purported to result in glutathionylation.
Figure 1.
PABA/NO–induced radical formation leading to glutathionylation. PABA/NO releases NO (4), which can react with GSH in the cell (1) to form a glutathionyl radical GS ˙ and nitroxyl (HNO), which in turn reacts with GSH (3) to give N-hydroxysulfenamide (GS-NH-OH), which can rearrange to generate a sulfinamide [GS(O)NH2] Reaction of GS(O)NH2 with GSH (4) forms glutathione disulfide-S-oxide [GS(O)SG] and NH3 and with H2O (5) to form sulfinic acid [GS(O)OH] and NH3. The key intermediates leading to the synthesis of GS(O)SG are the sulfinamides GS(O)NH2 and GS(O)-NH-SG. The reaction of GS(O)SG (6) or GS● (2) with a reduced protein thiol (R-SH) leads to the formation of mixed disulfide (6).
Diazeniumdiolates are a family of compounds, also called NONOates, that have proven useful for treating an increasing diversity of medical disorders in relevant animal models (for a review, see ref. 3). PABA/NO belongs to this family, and upon activation by the glutathione S-transferase π (GSTπ)-catalyzed conjugation to GSH, the drug releases the NONOate moiety that can further break down to nitric oxide (NO; ref. 4). Subsequent intracellular reactions of NO, outlined in Fig. 1, can produce the proximal donor for the glutathionylation reaction. For example, NO can react with GSH to form a GSH radical (GS·) and nitroxyl (HNO). The GS· can react directly with a protein thiol (R-SH) to form a mixed disulfide. Alternatively, HNO can react with GSH to give N-hydroxysulfenamide (GS-NH-OH), which rearranges to form sulfinamide [GS(O)-NH2; ref. 5]. The glutathione thiosulfinamide can subsequently react with GSH to form GS(O)SG, which is purported to be the most likely candidate driving the formation of mixed disulfides (6). Glutathionylation can act as a protective cellular mechanism preventing proteins from terminal modifications as a consequence of exposure to reactive oxygen and nitrogen species, or perhaps more importantly can contribute to redox-mediated cell signaling. There is a growing list of proteins that are potential targets for glutathionylation (reviewed in ref. 7). Of consequence to our studies, the list includes protein tyrosine phosphatase 1B (PTP1B; ref. 8), c-jun (9), GST (10), glutaredoxin, thioredoxin (7), H-Ras (11), and mitogen-activated protein kinase kinase kinase 1 (12). It is apparent that the entire scope of glutathionylation and its implications for cellular signaling has yet to be fully explored. The identification of enzymes directly involved in the regulation of glutathionylation and deglutathionylation will be critical in understanding the process of redox switching. Glutaredoxin was the first protein identified as a specific glutathionyl-mixed disulfide oxidoreductase (13). Interestingly, glutaredoxin is able to catalyze both the glutathionylation and deglutathionylation of specific proteins (for a review, see ref. 14).
Recently, a small redox protein, sulfiredoxin, involved in redox chemistry through its cysteine residues was identified in the budding yeast Saccharomyces cerevisiae (15). Sulfiredoxin is redox regulated and contains one critical cysteine residue that is conserved in mammals and yeast. Our studies with the human homologue show the redox function of sulfiredoxin is conserved, and importantly, that the conserved cysteine residue within the active site is critical for these events. The present studies show a novel role for human sulfiredoxin (Srx1) in redox-regulated events in response to oxidative and/or nitrosative stress. Specifically, it seems that Srx1 can participate in the catalytic reversal of NO-induced protein glutathionylation both in vitro and in vivo. The conserved cysteine is critical for the deglutathionylating activity of Srx1. Moreover, the deglutathionylating activity of Srx1 is of physiologic significance to the cell, as a restoration of PTP1B phosphatase activity is observed upon deglutathionylation by Srx1.
Materials and Methods
Materials and cell lines.
HEK293 cells were maintained in DMEM containing 10% FCS, 100 Ag/mL streptomycin, 100 units/mL penicillin, and 2 mmol/L L-glutamine at 5% CO2 and 37°C.
Cloning and site-directed mutagenesis.
Human SRX1 was amplified using PCR from a cDNA prepared from HEK293 cells. After PCR amplification the fragments were cloned into the Xba l-Bam HI recognition sites of pcDNA3.1/his myc for expression of Srx1 in a mammalian system or into the NdeI-BamHI recognition sites of pET28b for expression of Srx1 in a bacterial system. Clones were sequenced to ensure the integrity of the insert. For site-directed mutagenesis experiments, pcDNA3.1/his myc/SRX1 and pET28/SRX1 were then mutated using gene-specific primers by site-directed mutagenesis using the kit supplied by Stratagene (La Jolla, CA).
Transfected cell lines.
HEK293 cells were transfected with pcDNA3.1/hismyc plasmid vector (Invitrogen, Carlsbad, CA) containing either human SRX1 or the sulfiredoxin mutant SRX1/C99S. HEK293 cells were then transfected with pcDNA3.1/his myc/SRX1 (HEK/SRX1) or pcDNA3.1/his myc/SRX1C99S (HEK/C99S). Mock transfectants (HEK/pc) were made by transfecting HEK293 cells with pcDNA3.1/his myc containing no insert. After 48 hours, cells were transferred to 10-cm plates and allowed to grow in drug-free medium for 24 hours before colony selection in medium containing 400 Ag/mL G418 sulfate (Mediatech, Herndon,VA). Several colonies were chosen and characterized for each of the transfected cell lines made. Cells were maintained in the medium and under the conditions described above.
Cytotoxicity analysis.
Cells were plated onto 96-well plates at a density of 5,000 per well in 100 AL of medium. After 24 hours, increasing concentrations of drug were added, and the cells were maintained in drug for a further 72 hours. After this period of drug exposure, cell survival was determined using standard Sulforhodamine B staining procedures (16). Cell survival values were expressed and plotted as a fraction of vehicle-treated controls.
Protein purification.
For expression of recombinant proteins, pET28/SRX or pET28/C99S were transformed into BL21(DE3) pLysS-competent Escherichia coli cells and grown in Luria-Bertani media (supplemented with 50 Ag/mL kanamycin at 37 °C) to an A600 nm of 0.6. At this time, they were induced with 1 mmol/L isopropyl-l-thio-B-d-galactopyranoside for 2 hours. Cells were spun at 4°C for 30 minutes, and pellets were resuspended in 1x bacterial lysis buffer [10 mmol/L sodium phosphate buffer (pH 7.2), 0.5 mol/L NaCl, 10% glycerol, 1× protease inhibitor cocktail (Sigma, St. Louis, MO)] and stored at −80°C until needed. Cell pellets were thawed at room temperature and sonicated on ice for 4 minutes and then centrifuged at 4°C for 30 minutes at 5,000 × g. Imidazole was added to the supernatant to a final concentration of 10 mmol/L and loaded onto a Ni2+ column pre-equilibrated to 10 mmol/L imidazole in 10 mmol/L potassium phosphate buffer. Supernatant was allowed to drip through by gravity flow followed by 10 mL each of 10, 50, and 100 mmol/L imidazole wash buffers. Proteins were eluted with a 200 mmol/L imidazole wash, and a >95% protein purity was confirmed by SDS-PAGE of eluate fractions; 200 mmol/L wash was dialyzed at 4°C overnight in dialysis buffer [25 mmol/L HEPES (pH 7.6), 100 mmol/L NaCl, 1 mmol/L EDTA, and 1 mmol/L phenylmethylsulfonyl fluoride], and concentration was determined by the extinction coefficient.
Immunoblot analysis.
For all of the experimental approaches requiring Western blot analysis, cells were collected after treatment and washed twice in ice-cold PBS. Pellets were resuspended in lysis buffer [20 mmol/L Tris-HCl (pH 7.5), 15 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 1% Triton X-100, 2.5 mmol/L sodium pyrophosphate, and 1 mmol/L h-glycerophosphate] and incubated for 30 minutes on ice. Phosphatase inhibitors (5 mmol/L NaF and 1 mmol/L Na3VO4) and 1× protease inhibitor cocktail (Sigma) were added fresh to the lysis buffer. Lysates were vortexed vigorously and centrifuged for 30 minutes at 10,000 × g at 4°C, and subsequently, protein concentrations were assayed with the Bradford reagent (Bio-Rad Laboratories, Hercules, CA). Equal amounts of protein were separated on 12% SDS polyacrylamide gels and transferred at room temperature for 1 hour onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories) for immunodetection using the requisite primary and secondary antibodies. Antibodies were purchased from the following sources: phospho-specific c-Jun NH2-terminal kinase (JNK; Promega, Madison, WI), JNK1/2 (BD PharMingen, San Diego, CA), actin (Calbiochem, San Diego, CA), PTP1B (R&D Biosystems, Minneapolis, MN). Glutathiony-lation was determined using monoclonal anti-GSS antibody from Virogen (Watertown, MA).
Pull-down assays.
Where possible, protein/protein interactions were implicated; cells were harvested and lysates were prepared as described above using the following lysis buffer [50 mmol/L NaH2PO4, 300 mmol/L NaCl, 10 mmol/L Imidazole, 0.05% Tween 20]; 6× His myc epitope-tagged wild type and mutant Srx1 were purified from 1 mg of lysate using Ni2+ charged agarose (Amersham, Arlington Heights, IL). Proteins were separated by electrophoresis on a nonreducing 15% SDS-polyacrylamide gel and then transferred to PVDF (see above). Actin was detected using anti-actin primary antibody followed by horseradish peroxidase-conjugated anti-mouse IgG secondary antibody, and proteins were visualized as described above.
Human Srx1 antibody production.
Polyclonal Srx1 antibodies were made by standard protocols at the Medical University of South Carolina Polyclonal Antibody Production Facility. Rabbits were challenged with purified recombinant Srx1 protein as the antigen. Blood was drawn at monthly intervals to check the titer, and serum was eventually harvested after 5 months. Specificity of reactivity was initially tested using SDS-PAGE and recombinant protein preparations.
In vitro glutathionylation/deglutathionylation.
Actin (34 μmol/L; Sigma) or PTP1B (R&D Biosystems) was incubated at 37°C in 50 mmol/L potassium phosphate buffer (pH 7.2), 10 mmol/L GSH, and 100 Amol/L PABA/NO in the presence or absence of purified recombinant wild-type (sulfiredoxin) or mutant (C99S) sulfiredoxin proteins in 100 μL total volume. After 1, 5, 10, 15, and 30 minutes, 20 AL samples were removed and placed on ice. Samples were subsequently run on 15% SDS-PAGE gels, transferred, and probed with the appropriate antibodies as described in the respective figure legends.
HEK293 cells were treated with 10 Amol/L PABA/NO for 1 hour at room temperature. After treatment, cells were collected, washed twice in ice cold PBS, and lysed as described above using a different lysis buffer [150 mmol/L NaCl, 1% NP40, 50 mmol/L Tris-Cl (pH 8)]. Equal amounts of protein lysate were incubated at 37°C with buffer (control), 2.75 Amol/L sulfiredoxin, or C99S protein in 20 μL total volume. After 30 minutes, samples were placed on ice and subsequently run on a 15% SDS-PAGE gel, transferred, and probed with the appropriate antibodies.
Phosphatase assay.
To determine the effect of glutathionylation on target proteins, phosphatase activity of purified PTP1B was measured using the Malachite Green Phosphate detection kit (R&D Biosystems) according to the manufacturer’s instructions. PTP1B was preincubated with GSH, PABA/NO, or various combinations thereof in the presence or absence of purified sulfiredoxin protein for 15 minutes at 37°C before phosphatase activity was measured. Purified sulfiredoxin protein used in the phosphatase assay was prepared using Tris buffer in the absence of any phosphate ions that could interfere with the reaction.
Two-dimensional SDS-PAGE analysis.
For the first dimension, 100 Ag of whole cell protein was resuspended in 8 mol/L urea with 0.5% carrier ampholytes and run on immobilized pH gradients (IPG) covering the exponential pH range 3 to 10. For the second dimension, the IPG strips were separated on 10% polyacrylamide gels. All buffers were maintained under nonreducing conditions to facilitate separation of the glutathiony-lated products (17).
Globular actin/filamentous actin assay.
To show a direct biological importance for glutathionylation, filamentous actin (F-actin) and free globular actin (G-actin) contents in HEK293 cells were measured using an assay previously described (18) with the following adaptations. Cells were plated onto six-well plates at a density of 0.5 × 106 per well in 2 mL of medium. After 24 hours, cells were treated with DMSO (control) or 10 Amol/L PABA/NO for 1 hour. After this period of drug exposure, cells were washed once with 1× PBS. Subsequently, 400 μL 1% Triton X-100 was added per well for 2 minutes, after which the detergent soluble fraction was collected and centrifuged at 3,000 × g, and the supernatant was kept as the soluble fraction containing the G-actin. The pellet was washed in 1× PBS and contained the F-actin. The soluble fraction was mixed 1:1, and the insoluble pellet was resuspended in Laemlli buffer (Bio-Rad Laboratories). Ten microliters of each fraction were separated on a 10% SDS-PAGE gel, transferred, and probed with an antibody specific for actin as described above.
Phalloidin staining.
For imaging microfilaments, HEK293/pc and HEK293/SRX cells were seeded onto sterile 18-mm diameter coverslips precoated with 5 Ag/mL fibronectin (Fisher Scientific, Pittsburgh, PA), allowed to attach overnight and then washed twice with 1× PBS to remove unattached cells. Cells were then left untreated or treated with 10 Amol/L PABA/NO. After 1 hour, cells were washed twice with 1× PBS then fixed with 3.7% formaldehyde and permeabilized with 0.1% Triton X-100. F-actin distribution was then examined by phalloidin staining as per manufacturer instructions (Molecular Probes, Eugene, OR) using a Nikon Eclipse E800 microscope with Plan Fluor DIC optics and a Photometrics Coolsnap ES camera.
Statistical analysis.
Where immunoblots are shown, each is a representative of at least three essentially identical results. Other quantitative data are reported as means ± SD for at least three experiments. Student’s paired t test was used to determine the significance of differences between the means of control and experimental/treated groups, and P < 0.05 was taken as significant (although in some cases a more significant value is given).
Results
Human Srx1 protects cells from oxidative stress by H2O2. To assess the importance of human Srx1 in response to oxidative stress, we used a human embryonic kidney (HEK293) cell line that was stably transfected with Srx1 (HEK/SRX). We observed an increase in resistance to H2O2 in HEK/SRX (IC50 = 46.7 ± 4.85 μmol/L) when compared with the mock-transfected parental control (HEK/pc; IC50 = 23.5 ± 1.21 μmol/L). This result confirms that human Srx1 with only the one conserved cysteine residue functions in a manner similar to the yeast protein (15) in protecting against peroxidative stress products.
Srx1 is involved in the pathways leading to deglutathiony-lation.
Human Srx1 has been shown to have a role in the reduction of sulfinic to sulfenic acid in the typical 2-Cys peroxiredoxin family members (19, 20). The sulfinic acid is unstable and easily oxidized to the irreversible sulfonic acid. One of the possible biological functions of glutathionylation is its capacity to protect target proteins from undergoing this irreversible modification. In effect, this could serve to prevent the protein from degradation. We hypothesized that Srx1 may be involved in the “reduction” of glutathionylated proteins back to the reduced thiol form. Studies in our lab identified PABA/NO as a potent inducer of glutathionylation (21). Figure 2 shows that PABA/NO induces glutathionylation in HEK/pc cells in a time-dependent manner. However, in HEK/SRX cells, the total levels of glutathionylation are less, and the modification rate is delayed compared with nontransfected cells. This would suggest that Srx1 is involved in pathways leading to deglutathionylation. It is unlikely that Srx1 is simply acting as an oxidant scavenger byremoving any toxic metabolites of PABA/NO because PABA/NO is equally cytotoxic in both HEK/pc and HEK/SRX cells (IC50 = 20.4 ± 3.3 and 20.2 ± 0.5 μmol/L, respectively).
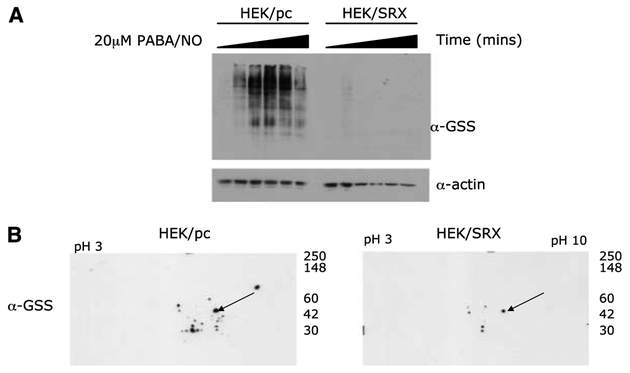
Figure 2.
Srx1 deglutathionylates multiple cellular proteins. A, HEK293 cells stably transfected with pcDNA containing no insert (HEK/pc) or wild-type Srx (HEK/SRX) were treated with 20 Amol/L PABA/NO for 0, 15, 30, 60, 120, and 180 minutes. Transferred membranes were probed with anti-GSS (α-GSS) antibody to detect protein glutathionylation and anti-actin (α-actin) to show equivalent protein loading. Representative Western blot of at least three independent experiments. B, HEK/pc and HEK/SRX cells were treated with 30 μmol/L PABA/NO for 2 hours. Proteins were separated by two-dimensional SDS-PAGE, and transferred membranes were probed with anti-GSS (α-GSS) antibody to detect protein glutathionylation. Coomassie-stained membranes were used to confirm equivalent loading (data not shown). Actin was identified by Western blot (arrow).
Sulfiredoxin deglutathionylates PTP1B and actin in vitro.
To investigate further the role of Srxl in the glutathionylation pathway, we used recombinant actin and PTP1B, proteins that are known to be glutathionylation targets in vitro. Figure 3 shows that in the presence of PABA/NO and GSH, both actin and PTP1B are glutathionylated in a time-dependent manner. However, incubation with Srx1 inhibits the formation of this modification. The inhibition seems to be catalytic in nature because heat-inactivated protein did not reverse the effect (Fig. 3B). Similar results were observed for both actin and PTP1B (Fig. 3A). Incubation of PTP1B with PABA/NO and GSH led to a time-dependent increase in glutathionylation. As with actin, incubation with Srx1 inhibited this modification, suggesting that Srx1 may have a general (rather than protein specific) role in the reversal, or prevention, of protein glutathionylation.
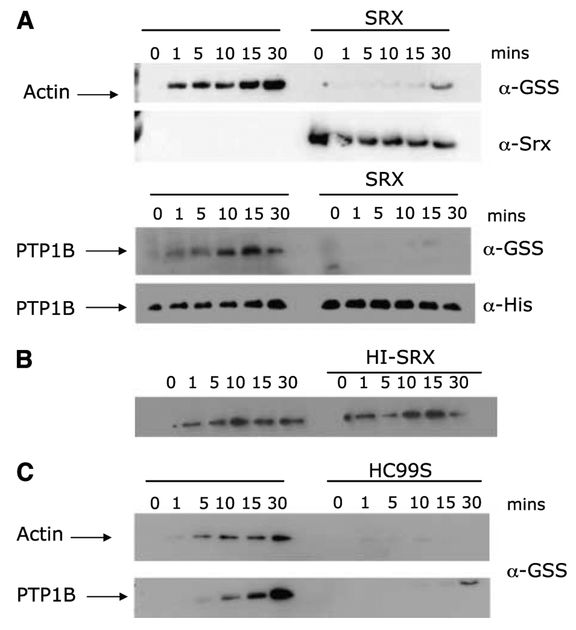
Figure 3.
Actin and PTP1B are in vitro targets of Srx1 deglutathionylation. Actin (34 Amol/L) or his-tagged PTP1B were incubated with 10 mmol/L GSH and 100 Amol/L PABA/NO for the indicated times at 37°C in the presence or absence of purified human wild-type (A), heat-inactivated (HI-SRX; B), or mutant (HC99S) (C) Srx1. Samples were run on 15% SDS-PAGE gels, and transferred membranes were probed with anti-GSS (α-GSS) antibody to detect protein glutathionylation, anti-Srx (α-Srx) to show the presence of purified Srx protein, and anti-His (α-His) to show the presence of his-tagged PTP1B.
Two-dimensional SDS-PAGE analysis confirms intracellular targets of sulfiredoxin.
In vitro analysis with purified protein suggests a role for sulfiredoxin in the deglutathionylation of both actin and PTP1B. Through two-dimensional SDS-PAGE analysis, we sought to elucidate the potential in vivo targets of degluta-thionylation by Srxl. HEK/SRX and HEK/pc cells were treated for 2 hours with 30 Amol/L PABA/NO, and total glutathionylated proteins was evaluated using two-dimensional SDS-PAGE. Gluta-thionylated proteins were at undetectable levels in untreated cells (data not shown). Figure 2B shows that a number of distinct glutathionylated proteins were observed in HEK/pc cells. However, the corresponding quantity and intensity of the spots was reduced or diminished in HEK/SRX cells (Fig. 2B). Coomassie-stained membranes show equivalent protein loading. These data suggest that Srxl nonspecifically deglutathionylates a range of proteins that have been modified by nitrosative stress.
Where diminution of immunoreactive intensity is apparent in the presence of Srxl, those proteins can be identified as substrates for deglutathionylation. We have previously identified, using matrix-assisted laser desorption ionization mass spectrometer time-of-flight, proteins that are glutathionylated in vivo in response to PABA/NO. These proteins include actin, elongation factor 2, chaperonin, β-lactate dehydrogenase, ATP synthase β-subunit, Rho GDP dissociation inhibitor β, nucleophosmin-1, protein disulfide isomerase, protein tyrosine phosphatase 1B, and glucosidase II (21). In the present study, we confirm that actin is a specific substrate of Srxl. Membranes from the two-dimensional SDS-PAGE were stripped and probed with an anti-actin antibody. The immunoblot of actin corresponds to the glutathionylated protein indicated by the arrow (Fig. 2B). Although complete deglutathio-nylation of actin was not observed within 2 hours, its level was significantly diminished in HEK/SRX cells.
Cys99 is essential for Srx1 deglutathionylating activity.
Srxl contains a “catalytic” cysteine residue located at the COOH terminus of the protein that is conserved in yeast, mouse, and human. To investigate the importance of this residue in the deglutathionylation function of this protein, we did site-directed mutagenesis to create a cysteine to serine mutant (C99S). Incubation of actin or PTP1B with recombinant C99S (HC99S) prevented glutathionylation, initially bringing into question the importance of the cysteine residue to the deglutathionylating activity of the protein (Fig. 3C). However, treatment of HEK293 cells with 10 Amol/L PABA/NO for 1 hour followed by in vitro incubation with recombinant Srx1 revealed that wild-type, but not mutant, sulfiredoxin was able to reverse glutathionylation compared with lysates treated with buffer only (Fig. 4A). Taken together, these results suggest that the mutant protein was not able to remove the GS− moiety from actin but instead bound to the protein with consequent steric inhibition of the glutathionylation site. This hypothesis was further supported by the coimmunoprecipitation of actin with either the wild-type or mutant-tagged Srxl as shown in Fig. 4B.
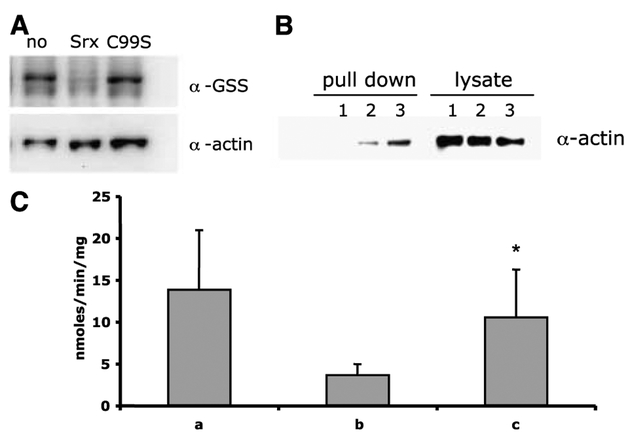
Figure 4.
Srx deglutathionylates actin through the conserved cysteine residue and restores functionality to PTP1B. A, drug-treated HEK293 cell protein lysates were incubated with buffer (no), wild-type (Srx), or mutant (C99S) Srx at 37°C for 30 minutes. Samples were run on 15% SDS-PAGE gels, and transferred membranes were probed with anti-GSS (α-GSS) antibody to detect protein glutathionylation. Stripped blots were then probed with anti-actin (α-actin) to confirm equivalent loading. B, HEK293 cells were stably transfected (see Materials and Methods) with an empty plasmid containing a 6xhis myc tag [HEK/pc (1)] or the same plasmid containing wild-type Srx [HEK/SRX (2)] or mutant Srx [HEK/C99S (3)]. Pull down of his-tagged proteins was achieved by incubating with Ni2+-charged agarose followed by 12% SDS-PAGE gels. Transferred membranes were probed with anti-actin (α-actin) to show that actin binds to wild-type and mutant-tagged Srx1. Lysates (50 μg) were run as control. C, amount of phosphate liberated from a known PTP1B-specific phosphopeptide substrate DADEY(PO3)LIPQQG when incubated at 30°C for 30 minutes with purified his-tagged PTP1B alone (A), PTP1B preincubated with 10 mmol/L GSH and 100 Amol/L PABA/NO (B), and PTP1B preincubated with 10 mmol/L GSH and 100 Amol/L PABA/NO and purified human Srx1 (C). Amount of phosphate liberated from phosphopeptide substrate by PTP1B over time (nmol/min/mg). Columns, means of three independent experiments; bars, SD. *, P < 0.05, (C) compared with (B).
Deglutathionylation of PTP1B correlates with restoration of phosphatase activity.
Preincubation of recombinant PTP1B with PABA/NO in the presence of GSH resulted in a loss of phosphatase activity (Fig. 4C). Immunoblot analysis showed this loss of activity was coincident with glutathionylation of PTP1B. The addition of purified Srx1 resulted in deglutathionylation and restored phosphatase activity to PTP1B. The fact that phosphatase activity was not fully restored by Srx1 is perhaps a consequence of PABA/NO-induced nitration of susceptible amino acids of PTP1B by Srxl (data not shown). These data are consistent with a role for Srx1 in regulation of phosphorylation/dephosphorylation pathways by restoring functionality to the phosphatase through deglutathionylation.
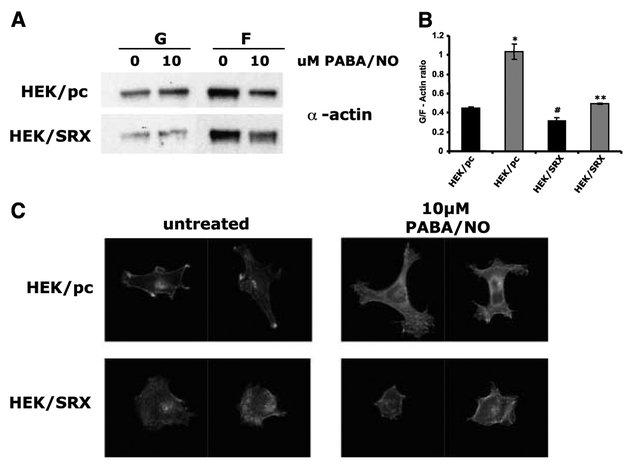
Effect of sulfiredoxin overexpression and PABA/NO treatment on G-actin and F-actin distribution.
It has been reported previously that glutathionylation of actin can inhibit the formation of actin filaments and alter the relative ratio of G-actin and F-actin within the cell (22). To investigate the effect of sulfiredoxin on this process, the relative amounts of G-actin and F-actin were measured in HEK293 cells. Parental HEK293 (HEK/pc) cells contained a higher level of G-actin and a lower level of F-actin when compared with sulfiredoxin-overexpressing cells (HEK/SRX; Fig. 5A). The ratio of G-actin/F-actin was calculated as 0.448 ± 0.016 for HEK/pc when compared with a value of 0.313 ± 0.034 for HEK/SRX cells, suggesting that higher levels of sulfiredoxin expression result in a larger proportion of polymerized (or filamentous) actin. The addition of PABA/NO to HEK/pc cells produced an increase in G-actin and a decrease in F-actin, with a concurrent increase in the ratio of G-actin/F-actin from 0.448 ± 0.016 to 1.032 ± 0.079 (Fig. 5B). This showed that PABA/NO treatment caused depolymerization of F-actin, leading to a relative increase in the amount of G-actin. This increase in G-actin corresponded to an increase in the amount of glutathionylated actin in the soluble fraction (immunoblot data not shown). Although a change in G-actin and F-actin was also observed in the HEK/SRX cells, the increase in the ratio of G-actin/F-actin was less pronounced (0.313 ± 0.034 compared with 0.495 ± 0.009). Because these cells also have a lower amount of glutathionylated actin, the results suggest that the lower levels of actin glutathionylation mediated by increased expression of sulfiredoxin leads to a less intense change in the actin distribution.
Figure 5.
Quantification of G-actin/F-actin ratio and phalloidin staining of F-actin in HEK293/pc and HEK293/SRX cells. A, representative Western blot of G-actin and F-actin from parental (HEK/pc) and Srx-overexpressing (HEK/SRX) cells either untreated (0) or treated (10) with 10 μmol/L PABA/NO for 1 hour. B, grouped densitometric data for three separate experiments expressed as the ratio of G-actin to F-actin. Black columns, untreated samples; gray columns, samples treated with 10 μmol/L PABA/NO for 1 hour. Significance difference in G-actin/F-actin ratio of HEK/pc to HEK/SRX cells (*, P < 0.01), untreated to treated HEK/pc cells (#, P < 0.05), untreated to treated HEK/SRX cells (**, P < 0.02; paired t tests). C, confocal microscope images of phalloidin-stained parental (HEK/pc) or Srx-overexpressing (HEK/SRX) cells either untreated or treated with 10 μmol/L PABA/NO for 1 hour.
Cell morphology and sulfiredoxin.
Figure 5 indicates how sulfiredoxin expression can influence actin microfilament organization and cell morphology. Sulfiredoxin overexpression resulted in a more rounded cell morphology (HEK/SRX untreated) when compared with the parental control, with no distinct protrusions or peripheral microfilament bundles (Fig. 5C). Treatment with PABA/NO resulted in the formation of lamellipodial extensions in the HEK/pc cells, and these were absent or significantly less pronounced in the HEK/SRX cells (Fig. 5C). Recent studies have illustrated the importance of a lateral flow of actin depolymerization and polymerization in the formation of lamellipodial extensions (reviewed in ref. 23). Thus, our present data are consistent with the conclusion that high levels of sulfiredoxin influence glutathionylation and inhibit actin depolymerization/polymerization flow.
Discussion
Reversible and irreversible posttranslational modifications of proteins are central to the regulation of multiple intracellular signaling pathways that may regulate cellular stress response, cell cycle, differentiation, or apoptosis. Phosphorylation is one of the most studied of the reversible posttranslational modifications and is regulated through the action of kinases and phosphatases. Glutathionylation is a reversible posttranslational modification that has only recently received attention. This has been due, in part, to the lack of available reagents sensitive enough to permit detection of this modification. We have enhanced the detection limits by using a novel GST-activated NO-releasing drug (3) that causes potent glutathionylation through one or more of its metabolites (21). The breakdown products that emerge from the metabolism of PABA/NO are obviously strong inducers of protein glutathionylation (Fig. 1).
Under conditions of oxidative/nitrosative stress, cysteine residues within proteins are primary targets for oxidation. The presence of millimolar levels of GSH within the cell (1) helps to maintain an equilibrium favorable for the formation of possible glutathionylation precursors (see Fig. 1). Glutathionylation can be protective, preventing the sequential oxidation of thiol groups to sulfonic acid, a product that leads to inevitable proteosomal degradation. In addition, glutathionylation has been shown to regulate the structure/function of a quite diverse range of cellular proteins. As such, reversibility of glutathionylation provides a viable switch of some importance in controlling cellular response to changes in redox conditions. Perhaps because of this, there is a benefit from a degree of functional redundancy in regulation of this reversible reaction. Proteins involved in the reduction of oxidized cysteine residues include thioredoxin and glutaredoxin. Thioredoxin can reduce protein disulfides and protein sulfenic acid intermediates. Glutaredoxin not only enzymatically deglutathionylates certain proteins (13) but has also been shown to catalyze the oxidative modification (i.e., glutathionylation) of several proteins in the presence of a GS− radical-generating system (24). Hence, glutaredoxin is capable of catalyzing both glutathionylation and deglutathionylation of proteins via distinct mechanisms. Glutare-doxin contains a conserved two-cysteine residue motif (CXXC). Both cysteines are required for its reductive deglutathionylation function via a dithiol mechanism of disulfide exchange. However, studies with mutant glutaredoxin containing only the NH2-terminal cysteine residue within the conserved motif (CXXS) showed that glutaredoxin can function as an oxidase through a monothiol mechanism (25). Studies have also shown that glutaredoxin can form a mixed disulfide (glutaredoxin-SG) as a consequence of both its oxidative and reductive mechanisms. Human Srx1 contains only one cysteine residue within its sequence, and our present results suggest that it is involved specifically in the reductive deglutathio-nylation of proteins. Although the conserved cysteine residue in Srx1 (Cys99) is essential for the deglutathionylation reaction, our data suggest that it is not a direct acceptor for the GSH moiety, as Srx1 does not form a mixed disulfide with GSH during the deglutathionylation reaction. In addition, the recombinant sulfir-edoxin protein is not glutathionylated in vitro after treatment with PABA/NO. In yeast, five different glutaredoxins have been described. Grx1 and Grx2 are classic dithiolic glutaredoxins containing both conserved cysteine residues (26). Interestingly, Grx3, Grx4, and Grx5 contain only one conserved NH2-terminal cysteine residue and function through a monothiol mechanism (27). However, Grx5 contains an additional nonconserved COOH-terminal cysteine residue, which studies have shown is involved in deglutathionylation of mitochondrial proteins through a monothiol mechanism (28). This “nonconserved” COOH-terminal cysteine residue aligns with the Cys99 residue in human Srx1 (Fig. 6). The similarity of human Srx1 to Grx5 in yeast suggests a potential monothiol mechanism of disulfide exchange reaction, although a glutathiony-lated Srx1 intermediate was not observed. The crystal structure of human Srx1 (29) identifies the active site motif as FGGCHR, along with detail of H bonding with residues surrounding the active site cysteine. However, the importance of this residue to the degluta-thionylation reaction has yet to be established.
Figure 6.
Sequence alignment of yeast Grx5 and human Srx1. S. cerevisiae Grx5 (accession no. Q02784) and human Srx1 (accession no. Q9BYN0) aligned using MAFFT version 5.667 (multiple sequence alignment at GenomeNet). The aligned cysteine residues are highlighted in bold. *, identical residues; double/single dots, similar residues. Srx1 is 21% identical and 53% similar to yeast Grx5.
As well as catalyzing deglutathionylation of proteins, Srx1 also seems to have the capacity to act as a ligand-binding protein and inhibit glutathionylation. This is based on the lack of glutathio-nylation of either actin or PTP1B when preincubated with Srx1, before the addition of PABA/NO and GSH. A plausible explanation would be that protein/protein interactions cause steric hindrance of glutathionylation. This observation lends further weight to the idea that Srx1 is involved specifically in the reversal of glutathionylation. Furthermore, although the cysteine residue is essential for deglutathionylation, the ligand binding or protein/protein interaction properties of Srx1 are apparently independent of its conserved cysteine residue because both inhibition of glutathionylation and immunoprecipitation of actin are observed in the presence of mutant Srx1. Moreover, our data imply that the mutant protein may be more effective at preventing glutathionylation than the wild type. This is presumably a consequence of enhanced binding affinity of the mutant for the target protein (in this case, actin; cf. Fig. 4).
Previous studies by our group observed an in vivo induction of protein nitration by PABA/NO (30). Furthermore, a critical ratio of GSH:PABA/NO is necessary to induce glutathionylation versus nitration in vitro. We were able to observe a strong induction of glutathionylation with a ratio of 100:1 (GSH:PABA/NO). No detectable modification was observed with a 1:1 ratio, and a ratio of 1:10 resulted specifically in nitration (data not shown). Therefore, it seems that high levels of GSH are critical to the formation of the radical species required for the formation of glutathionylated proteins. Although Fig. 1 details a number of plausible reactive proximal donors, recent studies suggest that glutathione disulfide monoxide may be the critical species (6).
Functional consequences of glutathionylation have been documented. For example, when actin is glutathionylated, it has a decreased capacity to polymerize compared with the native form, mediated through inhibition of filament elongation (22). Fibroblasts of patients with Friedreich’s ataxia (FRDA) disease have an increase in actin glutathionylation. When FRDA fibroblasts are treated with GSH, a complete rescue of cytoskeletal abnormalities and cell viability is observed. Oxidative stress-induced glutathionylation of actin leads to an impairment of cytoskeletal function in FRDA fibroblasts (31). We observe an Srxl-dependent reversal of actin glutathionylation in vitro that was confirmed in vivo by twodimensional gel analysis. In addition, sulfiredoxin levels influenced the ratio of G-actin to F-actin and produced changes in cell morphology that would be of importance to cell structure/function relationships. Such changes were further defined in the presence of nitrosative stress. Taken together, these results imply potential therapeutic relevance for Srx1 in diseases with phenotypes associated with dysfunctional protein glutathionylation. Other diseases associated with aberrant protein glutathionylation include diabetes (type I and II), HIV infections, hyperlipidemia, and renal cell carcinoma (for a review, see ref. 32). It is salient to note that interference with phosphorylation/dephosphorylation is a recognized strategy for the design of drugs in the treatment of cancer. In some regards, Srx1 could be viewed as analogous to the phosphatases involved in such cascades. Indeed, at least one such phosphatase (PTP1B) has a secondary level of regulation that involves deglutathionylation. PTP1B is a tyrosine phosphatase with a primary role in insulin signaling regulation ( for a review, see ref. 33). Others have shown and we now confirm that glutathionylation inhibits and deglutathionylation restores the phosphatase activity of PTP1B (8, 34). Srxl-dependent deglutathionylation of PTP1B in parallel with a functional restoration of phosphatase activity suggests a role for Srx1 in the regulation of kinase signaling in response to an oxidative and/or nitrosative stress. The identification of a novel role for Srx1 as a protein specifically involved in the deglutathionylation of proteins is a discovery that has implications for regulation in response to oxidative and/or nitrosative stress. With the growing number of disease states that are associated with aberrant protein glutathionylation, design and testing of agonists/antagonists of Srx1 could prove to be a fruitful field for future therapeutic application.
Acknowledgments
Received 2/7/2006; revised 3/30/2006; accepted 5/4/2006.
Grant support: National Cancer Institute grant CA R01 CA 08660 and the South Carolina Centers of Excellence award (K.D. Tew).
The costs of publication of this article were defrayed in part bythe payment ofpage charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
We thank Drs. L. Keefer (Laboratory of Comparative Carcinogenesis, NCI, Frederick, MD) and J.E. Saavedra (Basic Science Research Program, SAIC, Frederick, MD) for the supply of PABA/NO.
References
- 1.Tew KD. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res 1994;54:4313–20. [PubMed] [Google Scholar]
- 2.Gilbert HF. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol 1995;251: 8–28. [DOI] [PubMed] [Google Scholar]
- 3.Keefer LK. Progress toward clinical application of the nitric oxide-releasing diazeniumdiolates. Annu Rev Pharmacol Toxicol 2003;43:585–607. [DOI] [PubMed] [Google Scholar]
- 4.Saavedra JE, Billiar TR, Williams DL, Kim YM, Watkins SC, Keefer LK. Targeting nitric oxide (NO) delivery in vivo. Design of a liver-selective NO donor prodrug that blocks tumor necrosis factor-alpha-induced apoptosis and toxicity in the liver. J Med Chem 1997;40:1947–54. [DOI] [PubMed] [Google Scholar]
- 5.Singh SP, Wishnok JS, Keshive M, Deen WM, Tannenbaum SR. The chemistry of the S-nitrosogluta-thione/glutathione system. Proc Natl Acad Sci U S A 1996;93:14428–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang KP, Huang FL. Glutathionylation of proteins by glutathione disulfide S-oxide. Biochem Pharmacol 2002; 64:1049–56. [DOI] [PubMed] [Google Scholar]
- 7.Klatt P, Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitro-sative stress. Eur J Biochem 2000;267:4928–44. [DOI] [PubMed] [Google Scholar]
- 8.Barrett WC, DeGnore JP, Konig S, et al. Regulation of PTP1B via glutathionylation of the active site cysteine 215. Biochemistry 1999;38:6699–705. [DOI] [PubMed] [Google Scholar]
- 9.Klatt P, Lamas S. c-Jun regulation by S-glutathionylation. Methods Enzymol 2002;348:157–74. [DOI] [PubMed] [Google Scholar]
- 10.Dafre AL, Sies H, Akerboom T. Protein S-thiolation and regulation of microsomal glutathione transferase activity by the glutathione redox couple. Arch Biochem Biophys 1996;332:288–94. [DOI] [PubMed] [Google Scholar]
- 11.Ji Y, Akerboom TP, Sies H, Thomas JA. S-nitrosylation and S-glutathiolation of protein sulfhydryls by S-nitroso glutathione. Arch Biochem Biophys 1999;362:67–78. [DOI] [PubMed] [Google Scholar]
- 12.Cross JV, Templeton DJ. Oxidative stress inhibits MEKK1 by site-specific glutathionylation in the ATP-binding domain. Biochem J 2004;381:675–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gravina SA, Mieyal JJ. Thioltransferase is a specific glutathionyl mixed disulfide oxidoreductase. Biochemistry 1993;32:3368–76. [DOI] [PubMed] [Google Scholar]
- 14.Shelton MD, Chock PB, Mieyal JJ. Glutaredoxin: role in reversible protein S-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid Redox Signal 2005;7:348–66. [DOI] [PubMed] [Google Scholar]
- 15.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 2003;425:980–4. [DOI] [PubMed] [Google Scholar]
- 16.Skehan P, Storeng R, Scudiero D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst 1990;82:1107–12. [DOI] [PubMed] [Google Scholar]
- 17.Righetti PG, Gianazza E, Bianchi-Bosisio A, Sinha P, Kottgen E. Isoelectric focusing in immobilized pH gradients: applications in clinical chemistry and forensic analysis. J Chromatogr 1991;569:197–228. [DOI] [PubMed] [Google Scholar]
- 18.Ke X, Terashima M, Nariai Y, Nakashima Y, Nabika T, Tanigawa Y. Nitric oxide regulates actin reorganization through cGMP and Ca(2+)/calmodulin in RAW 264.7 cells. Biochim Biophys Acta 2001;1539:101–13. [DOI] [PubMed] [Google Scholar]
- 19.Chang TS, Jeong W, Woo HA, Lee SM, Park S, Rhee SG. Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J Biol Chem 2004;279:50994–1001. [DOI] [PubMed] [Google Scholar]
- 20.Woo HA, Jeong W, Chang TS, et al. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. J Biol Chem 2005;280:3125–8. [DOI] [PubMed] [Google Scholar]
- 21.Townsend DM, Findlay VJ, Fazilev F, et al. A glutathione S-transferase pi-activated prodrug causes kinase activation concurrent with S-glutathionylation of proteins. Mol Pharmacol 2006;69:501–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalle-Donne I, Scaloni A, Giustarini D, et al. Proteins as biomarkers of oxidative/nitrosative stress in diseases: the contribution of redox proteomics. Mass Spectrom Rev 2005;24:55–99. [DOI] [PubMed] [Google Scholar]
- 23.Small JV, Resch GP. The comings and goings of actin: coupling protrusion and retraction in cell motility. Curr Opin Cell Biol 2005;17:517–23. [DOI] [PubMed] [Google Scholar]
- 24.Starke DW, Chock PB, Mieyal JJ. Glutathione-thiyl radical scavenging and transferase properties of human glutaredoxin (thioltransferase). Potential role in redox signal transduction. J Biol Chem 2003;278:14607–13. [DOI] [PubMed] [Google Scholar]
- 25.Xiao R, Lundstrom-Ljung J, Holmgren A, Gilbert HF. Catalysis of thiol/disulfide exchange. Glutaredoxin 1 and protein-disulfide isomerase use different mechanisms to enhance oxidase and reductase activities. J Biol Chem 2005;280:21099–106. [DOI] [PubMed] [Google Scholar]
- 26.Luikenhuis S, Perrone G, Dawes IW, Grant CM. The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol Biol Cell 1998;9: 1081–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez-Manzaneque MT, Ros J, Cabiscol E, Sorribas A, Herrero E. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol Cell Biol 1999;19: 8180–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamarit J, Belli G, Cabiscol E, Herrero E, Ros J. Biochemical characterization of yeast mitochondrial Grx5 monothiol glutaredoxin. J Biol Chem 2003;278: 25745–51. [DOI] [PubMed] [Google Scholar]
- 29.Jonsson TJ, Murray MS, Johnson LC, Poole LB, Lowther WT. Structural basis for the retroreduction of inactivated peroxiredoxins by human sulfiredoxin. Biochemistry 2005;44:8634–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Findlay VJ, Townsend DM, Saavedra JE, et al. Tumor cell responses to a novel glutathione S-transferase-activated nitric oxide-releasing prodrug. Mol Pharmacol 2004;65:1070–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pastore A, Tozzi G, Gaeta LM, et al. Actin glutathionylation increases in fibroblasts of patients with Friedreich’s ataxia: a potential role in the pathogenesis of the disease. J Biol Chem 2003;278: 42588–95. [DOI] [PubMed] [Google Scholar]
- 32.Giustarini D, Rossi R, Milzani A, Colombo R, Dalle-Donne I. S-glutathionylation: from redox regulation of protein functions to human diseases. J Cell Mol Med 2004;8:201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asante-Appiah E, Kennedy BP. Protein tyrosine phosphatases: the quest for negative regulators of insulin action. Am J Physiol Endocrinol Metab 2003;284: E663–70. [DOI] [PubMed] [Google Scholar]
- 34.Barrett WC, DeGnore JP, Keng YF, Zhang ZY, Yim MB, Chock PB. Roles of superoxide radical anion in signal transduction mediated by reversible regulation of protein-tyrosine phosphatase 1B. J Biol Chem 1999; 274:34543–6. [DOI] [PubMed] [Google Scholar]