Abstract
Identification of fungal organisms often poses a problem for pathologists because the histomorphology of some fungal organisms is not specific, fresh tissues may not be available, and isolation and identification in culture may take a long time. The purpose of this study was to validate the use of panfungal PCR to identify fungal organisms from formalin-fixed paraffin-embedded (FFPE) tissues. FFPE curls were tested from 128 blocks containing canine, feline, equine, and bovine tissues with cutaneous, nasal, pulmonary, and systemic fungal infections, identified by the presence of fungi in histologic sections. Quantitative scoring of histologic sections identified rare (11.9%), occasional (17.5%), moderate (17.5%) or abundant (53%) fungal organisms. DNA was isolated from FFPE tissues and PCR was performed targeting the Internal Transcribed Spacer 2 (ITS-2) region, a segment of non-coding DNA found in all eukaryotes. PCR products were sequenced and identified at ≥97% identity match using the basic local alignment search tool (BLAST) and the NCBI database of ITS sequences. Of the 128 blocks, 117 (91.4%) yielded PCR products and high quality sequences were derived from 89 (69.5%). Sequence and histological identifications matched in 79 blocks (61.7%). This assay was capable of providing genus- and species-level identification when histopathology could not and thus is a beneficial complementary tool for diagnosis of fungal diseases.
Keywords: Panfungal Polymerase Chain Reaction, Formal-fixed paraffin-embedded, Fungi, mycoses, mycology, pathology, Internal Transcribed Spacer, lyticase, DNA isolation
An exponential growth in molecular diagnostics for infectious diseases has produced opportunities for confirming culture results, and can be combined with other methods, such as histopathology, to confirm a diagnosis when culture is not possible.1,5,17 DNA-based assays have been particularly helpful in diagnosis of fungal infections due to the speed of these tests compared to fungal culture, which can take two to four weeks.12 Many fungi are not readily cultivable in the laboratory and others require biosafety level 3 precautions. In addition to culture, fungal identification has relied on histopathology and cytology, although inherent challenges exist due to the subtle nuances in distinguishing fungal morphology.52
Polymerase chain reaction (PCR) assays have been developed to target and quantify specific genera and species of fungi from clinical samples.13,15 These assays, however, require prior suspicion of the etiologic agent, and are not available for every fungal organism including emerging pathogens. The greatest utility has come from panfungal PCR which amplifies conserved genes that are present in all fungi such as the ribosomal RNA (rRNA) region including the large (28S) and small subunits (18S), 5.8S, internal transcribed spacer (ITS-1 and - 2), and intergenic spacer (IGS) regions. Primers targeting these conserved rRNA sequences were originally developed for phylogenetic studies.60 The resulting PCR products have regions that are hypervariable amongst fungi allowing for identification by comparison of their sequences to a reference fungal database. The ITS regions are currently considered the superior region for species resolution.55 Panfungal PCR can potentially identify any fungal agent without prior knowledge of its suspected identity, which can greatly reduce the time and cost spent on other molecular diagnostics.
Panfungal PCR on formalin-fixed paraffin-embedded (FFPE) tissues is useful when fresh tissue was not or could not be collected. It has been successfully applied in different human tissues and for a variety of fungal infections,38,45,49 however, less work has been performed in veterinary cases. Two of the human studies38,45 have included small subsets of animal tissues (6 and 19, respectively), but were lacking in breadth of animal species and tissue types. Recently, there have been more extensive applications of this assay in cats,3,9 but there is still a need for validation in other animals. Furthermore, there does not appear to be a general consensus on DNA extraction methods for FFPE tissues45,50 and many of the recommended kits are expensive or the protocols are time-intensive.
Improving fungal diagnostics in veterinary medicine has the potential to not only improve animal healthcare but also enable proper selection of anti-fungal therapeutics, thereby reducing the development of resistance that has already been documented in poultry,7 dogs,46 and other animals.14 Additionally, improvement in the accuracy and specificity of identifying fungi to the genus and species level would allow for better epidemiological studies of current and emerging fungal pathogens. The purpose of this study was to validate the use of panfungal PCR on FFPE specimens in a variety of veterinary cases from birds, cats, camels, cattle, dogs, horses, and sheep. Furthermore, we aimed to test and optimize a cost-effective DNA extraction kit for amplification of fungal DNA from FFPE animal tissues.
Materials and methods
Case selection
From the pathology archives at the Texas A&M College of Veterinary Medicine, the Dermatopathology Specialty Service, and the Texas A&M Veterinary Medical Diagnostic Laboratory, an exhaustive query was performed, and 128 paraffin blocks from 96 animals were selected for inclusion in this study. The initial selection criteria included the presence of fungi observed histologically (Supplemental Figure 1), a diagnosis of primary fungal infection, and an attempt to select representative numbers of animal species, tissue types, and fungal etiological agents. Once the blocks were selected, a single pathologist reviewed all archived slides to confirm the histological diagnosis and to categorize the cases based on the amount of fungal organisms. Separate guidelines were used for yeast and hyphal morphologies since hyphae are generally more prolific, and cases were classified according to the number of organisms seen in 20 20X fields as either rare (<3 yeast or <8 hyphae), occasional (3–10 yeast or 8–20 hyphae), moderate (10–15 yeast or 20–50 hyphae), or abundant (>15 yeast or >50 hyphae). The cases included dogs (66), cats (25), horses (19), cattle (7), birds (6), camels (3), sheep (1) or wild cats (1) tissues. The tissue types included oral cavity, respiratory tract, gastrointestinal tract, urinary tract, skin, brain, heart, lymph node, and bone. Previous ancillary testing included 69 cases with special stains, 26 with culture, 4 with cytology, 3 with PCR performed at other institutions, and one with immunohistochemistry. Most cases without ancillary testing showed strong fungal identification based on histopathology, and additional tests were deemed unnecessary due to costs or unavailability of fresh samples.
Twenty-four blocks were used to compare two commercially available kits for extraction of fungal DNA from FFPE tissues, another 52 blocks were used to further optimize the superior kit’s protocol, and the remaining 52 blocks were processed following the optimized protocol (Supplemental Figure 1). From each block, sections totalling 50 μm depth were cut in five μm scrolls with a microtome and the blade was cleaned with absolute alcohol between blocks. The initial scroll for each block was discarded to minimize environmental and carry-over contamination. The scrolls were stored in DNase-, DNA-, and RNA-free microcentrifuge tubes until DNA was extracted.
ITS-2 and IRBP polymerase chain reactions and sequence analysis
Fungal DNA was amplified by PCR targeting the internal transcribed spacer-2 (ITS-2) region, located between the coding regions for the 5.8S and small ribosomal RNA subunits, using the following panfungal primers: ITS3-F (5’-GCATCGATGAAGAACGCAGC-3’) and ITS4-R (5’-TCCTCCGCTTATTGATATGC-3’).60 In order to evaluate quality of host DNA in each sample, a PCR targeting the Canis familiaris interphotoreceptor retinoid-binding protein (IRBP) gene, which is highly conserved in a large number of mammalian species, was performed using the following primers: IRBP-F (5’-TCCAACACCACCACTGAGATCTGGAC-3’) and IRBP-R (5’-GTGAGGAAGAAATCGGACTGGCC-3’).21 For both PCRs, the total reaction volume was 25μl containing 1ul of DNA extract, 0.5 μl 10uM forward primer, 0.5 μl 10uM reverse primer, 10.5 μl Ultrapure nuclease free water (Invitrogen, Carlsbad, CA), and 12.5 μl Accustart II Toughmix (Hot-start Taq Polymerase, Quanta Biosciences, Gaithersburg, MD). Thermal cycler conditions were identical for both PCRs and consisted of 5 minutes at 95°C; 40 cycles of 95°C for 30 seconds, 57°C for 30 seconds, and 68°C for 30 seconds; and a final extension step at 68°C for 5 minutes. The amplification positive control included with every group of ITS-2 reactions was one microliter of purified genomic DNA from Cryptococcus neoformans (ATTC 208821, strain H99). The positive control included with IRBP reactions was one microliter of DNA extracted from a block containing an assortment of animal skin biopsies that were unremarkable. A negative control was included with both PCRs by adding one microliter of UltraPure water (Invitrogen, Carlsbad, CA) instead of DNA. Samples that tested negative for ITS-2 and IRBP were repeated and spiked with one microliter of positive control DNA to determine if false negative results were due to PCR inhibitors, likely from the formalin fixation.
All PCR products were separated on a 2% agarose gel made with 1X Tris boric acid ethylene-diamine-tetra-aceitc acid (TBE) buffer, and GelRed ® (Biotium, Inc., Hayward, CA). The sample was separated by electrophoresis for one hour at 120V and visualized with a UV trans-illuminator (GelDoc EZ™ system, Bio-rad Laboratories, Inc., Hercules, CA). Every ITS-2 band was excised from the gel, and the DNA was isolated using the E.Z.N.A.® gel extraction kit (Omega Bio-Tek, Inc., Norcross, GA). This purified DNA was then submitted for sequencing (Eton Bioscience, Inc., San Diego, CA) using the ITS3-F and ITS4-R primers. Sequences were trimmed for quality by removal of the beginning and end of sequences, joined as contigs using Sequencher®, and then queried against the GenBank database using the Basic Local Alignment Search Tool (BLAST, NCBI). Published GenBank accession matches were given precedence over unpublished ones. Sequences were assessed both quantitatively with the quality score given in Sequencher® and qualitatively by visual inspection of the electropherogram. Sequences were identified as ‘poor quality’ for the remaining analyses when the overall quality score was less than 70%, where no or little sequence was detected, and when a majority of peaks overlapped for each base. A fungal identification was made when a sequence matched the reference at ≥ 97% identity (for genus and species levels). When the sequence matched to more than one taxon at ≥ 97%, the one with the highest identity score was chosen. When there were multiple matches at the highest identity score, all matches were reported. Also, when there were multiple matches from different bands, all matches were reported. IRBP bands were considered positive or negative based on the presence or absence of a band at the same level as positive controls, and were not submitted for sequencing.
Comparison of DNA extraction from paraffin blocks using two commercially-available kits
Two commercially available kits, Qiagen QIAamp® DNA FFPE Tissue Kit with Deparaffinization Solution (Valencia, CA) and MoBio BiOstic® FFPE Tissue DNA Isolation Kit (Carlsbad, CA), were compared through assessment of DNA extract quality (calculated as the number of IRBP-positive samples divided by the total number of samples),45 PCR efficiency (calculated as the number of ITS-2 PCR-positive samples divided by the number of IRBP- positive samples),45 and ITS-2 sequence quality (calculated as the number of high quality sequences divided by the total number of samples). The 24 blocks included in this experiment included roughly equal numbers of blocks with either yeast or hyphae, to evaluate the efficiency of DNA extraction from yeast capsules as well as hyphal mats and chitin-containing hyphae. The kit protocols (including an initial deparaffinization step) were followed according to manufacturer’s directions, except the combined thickness of the scrolls included in the first step was standardized at 50 micrometers, and the incubation time with proteinase K was standardized at two hours for both protocols, the minimum recommended incubation time period. All extracted DNA was stored at −20°C prior to PCR amplification. For every block, fungal DNA was amplified through ITS-2 PCR and host DNA was amplified through IRBP PCR.
Optimization of the MoBio BiOstic® kit protocol
The MoBio BiOstic® kit protocol was further optimized to determine whether addition of lyticase would improve PCR efficiency. The DNA was extracted from scrolls of the 52 blocks following the manufacturer’s directions, except for a two hour incubation time with proteinase K. Following the proteinase K digestion, the lysate was equally divided into two microcentrifuge tubes. One of these tubes was then processed following the manufacturer’s directions, but 2U/100ul of lyticase (L2524, Sigma-Aldrich, St Louis, MO) was added to the other tube and incubated at 37°C for 45 minutes.45 After lyticase incubation, these samples were processed following the manufacturer’s directions. From each block, fungal DNA was amplified using the ITS-2 PCR and host DNA was amplified using the IRBP PCR. DNA quality, PCR efficiency, and sequence quality was calculated for all extractions.
Phylogenetic analysis of Aspergillus spp.
All Aspergillus spp. sequences (n=20) from this study were aligned with eight reference sequences for A. flavus, A. fumigatus, A. niger, A. terreus, A. ustus, A. nidulans, A. felis, and Phialosimplex caninus (NCBI accession numbers: AF138287, AF138288, AF138904, AF138290, AF157507, AF138289,30 KF558318.1,6 and GQ169311.1,28 respectively) using molecular evolutionary genetic analysis 758 (MEGA 7) and multiple sequence comparison by log-expectation (MUSCLE) algorithm.20 The alignment was chopped at the 3’ end to exclude the ITS-1 region found in the reference sequences. The alignment was then imported into Geneious35 and using the Tamura-Nei model to calculate the distances, neighbor joining was performed, with F. solani (NCBI accession: HQ026747.1) selected as outgroup. Bootstrap analysis was performed with 1000 replications and the consensus trees were reported including the bootstrap support values at branching points.
Statistical analysis
Contingency analysis was performed in JMP® Pro12 (SAS Institute, Inc., Cary, NC) to determine whether an association existed between the number of organisms observed histologically and ITS-2 PCR results, and with the quality of sequences derived from the PCR products. First the correlation between variables was assessed with a Chi square test. When a significant correlation was identified for variables with multiple categories, the samples were then scored (Yes/No) for the category in each variable that was suspected to contribute to the correlation. A second Chi square test was then performed between one category from each variable, and significant p-values were reported.
Results
Comparison of two DNA extraction kits
DNA extraction using the Qiagen QIAamp® DNA FFPE kit and MoBio BiOstic® FFPE Tissue DNA Isolation kit were compared using 24 blocks of FFPE tissue from animals with suspected histoplasmosis (7), Malassezia infection (5), cryptococcosis (1), dermatophytosis (5), and phaeohyphomycosis (6) (Table 1). To assess the ability of the kit to remove inhibitors and preserve DNA quality we calculated the number of IRBP-positive samples divided by the total number of samples tested (DNA quality) and found that the kits were equally successful at 100%. To test the ability of the kit to extract fungal DNA from the block, we calculated the number of ITS-2 PCR-positive samples divided by the number of IRBP-positive samples (PCR Efficiency) and found that the kits were equally successful at 83%. The percentage of high quality sequences derived from ITS-2 PCR products that yielded an identification (ITS-2 Sequence Qualtity) were the same for each kit at 71%. Based on DNA quality, PCR efficiency, and the quality of ITS-2 sequences for these twenty four blocks, we determined that the MoBio BiOstic® kit was comparable to the Qiagen QIAamp® kit (with deparaffinization solution), but the MoBio BiOstic® kit had practicality advantages including a lower price and shorter protocol completion time. Therefore we decided to further optimize the MoBio BiOstic kit’s protocol for the extraction of fungal DNA from FFPE animal tissues.
Table 1.
Comparison of two DNA-extraction kits, and effect of adding lyticase, with respect to DNA Quality, PCR Efficiency, and Sequence Quality.
| Kit Experiment | lyticase Experiment | |||
|---|---|---|---|---|
| Measures | Qiagen | MoBio | MoBio +lyticase | MoBio -lyticase |
| Total number of blocks (n) | 24 | 24 | 51 | 51 |
| IRBP PCR Positives (n) | 24 | 24 | 49 | 49 |
| ITS-2 PCR Positives (n) | 17 | 19 | 32 | 44 |
| ITS-2 PCR Faint Bands (n) | 3 | 1 | 11 | 2 |
| ITS-2 Sequence Identification (n) | 17 | 17 | 35 | 41 |
| DNA Quality | 100% | 100% | 94.2% | 96.1% |
| PCR Efficiency | 83.3% | 83.3% | 87.8% | 93.9% |
| Sequence Identification | 70.8% | 70.8% | 67.3% | 80.4% |
| Correlation with histology | 70.8% | 70.8% | 57.7% | 68.6% |
DNA Quality=#IRBP Positive/Total number; PCR Efficiency=(ITS-2 Positive+ITS-2 Faint)/IRBP Positive; Sequence Identification=Sequence Identification/Total number.
Qiagen: Qiagen QIAamp® DNA FFPE Tissue Kit; MoBio: MoBio BiOstic® FFPE Tissue DNA Isolation Kit
lyticase experiment: lyticase was added or not added, prior to DNA extraction using the MoBio kit.
Optimization of the MoBio BiOstic® FFPE DNA extraction kit
The addition of lyticase digestion to the Qiagen QIAamp® kit protocol has been shown to improve PCR efficiency3,45 so we wanted to test whether this would also be true for the MoBio BiOstic® kit. A set of 52 blocks of FFPE tissue (different from those mentioned above) were included in this experiment. The calculated PCR efficiency for samples digested with lyticase was 87.8%, and for samples without lyticase was 93.9%. Furthermore, only 67.3% of the samples digested with lyticase produced a high quality sequence for which identification was possible (ITS-2 sequence quality), and only 30/51 (57.7%) of the samples yielded a sequence which correlated with the histologic diagnosis. In comparison, 80.4% of samples without lyticase produced high quality sequences and 35/51 (68.6%) of samples yielded a sequence that correlated with the histology. We found that lyticase did not improve amplification of fungal DNA extracted from FFPE blocks using the MoBio BiOstic® kit, and therefore was not necessary to be included in the optimized protocol.
Sequence Analysis of Archived Case Material
An additional 52 blocks were processed with the MoBio BiOstic® kit according to the manufacturer’s directions, except proteinase K incubation time was two hours. findings of For all processed samples (n=128; using the MoBio kit without lyticase; Supplemental Table S1), 117/128 (91.4%) were ITS-2 PCR positive, 9/128 (7.0%) were negative, and 1.5% (2/128) were smeared on an agarose gel (Supplemental Table S1). Of the blocks that were ITS-2 PCR-positive, 37/117 (31%) demonstrated multiple PCR product bands on electrophoresis, 17/117 (14.5%) had faint band(s), and the remaining (63/117) showed a single strongly positive band. Most PCR-positives (89/117) yielded high quality sequences, although 28/117 (23.9%) yielded a poor quality sequence. Roughly equal numbers of the poor quality sequences came from faint bands (13/28) as from strongly positive bands. None of the poor quality sequences were queried against the database. In 6 samples, non-fungal eukaryotic DNA was amplified including the ITS-2 sequences of plants and mites.
To calculate sensitivity relative to histologic examination, the number of cases for which genus-level identification of fungi was possible through both histology and the panfungal PCR (n=66) was divided by the number of cases for which identification was possible through histology (n=95). The diagnostic assay had a sensitivity of 69.5% relative to histologic examination (Table 2).
Table 2.
Number of fungal identifications made through histopathology or panfungal PCR for calculation of sensitivity.
| Histopathology | |||
|---|---|---|---|
| PCR | + | − | Total |
| + | 66 | 18 | 84 |
| − | 29 | 15 | 44 |
| Total | 95 | 33 | 128 |
(+) indicates fungal identification was made to at least the genus level.
A fungal identification was made through ITS-2 PCR and sequencing for 83/128 (65%) of blocks tested. The vast majority of these sequence-derived identities (96%, 79/83) corresponded to those made histologically, and only a small number (4%) identified as an organism that did not match the histological descriptions. A genus- or species-level identification was made by BLAST analysis of sequences in 48/83 (60%) of cases, whereas histological evaluation was unable to do so. This included 24 samples for which sequencing provided a genus level resolution, and 24 samples a species level resolution. Three representative cases were selected to demonstrate the benefit of using panfungal PCR where histological fungal morphology could not provide a genus level identification (Figs. 1 to 3).
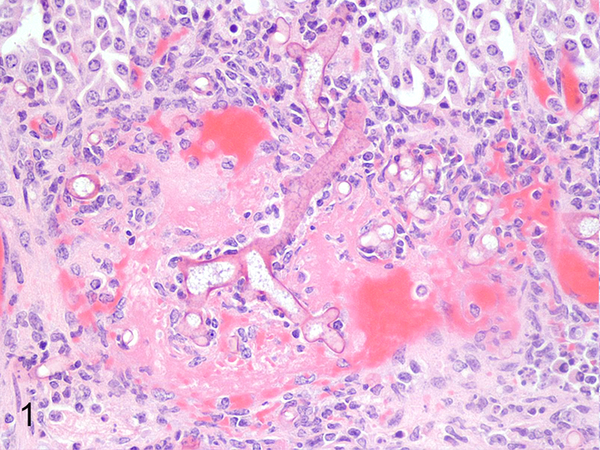
Fig. 1. Various mycotic infections in domestic species that were classified using panfungal PCR.
Abomasum, cow, case 118. Large, irregular-walled fungal hyphae identified as Rhizopus microsporus are within a thrombosed vessel. HE.
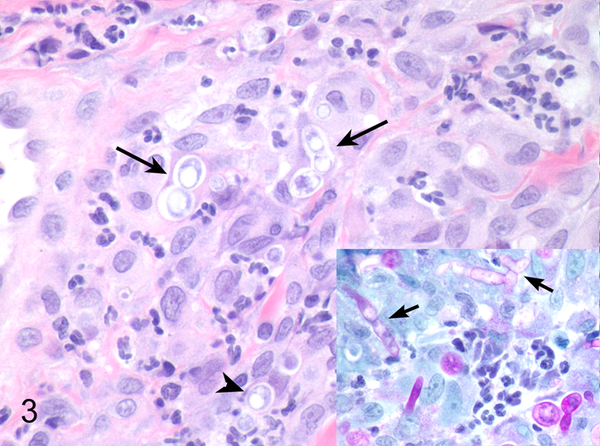
Fig. 3. Various mycotic infections in domestic species that were classified using panfungal PCR.
Dermis, dog, case 63. Curvularia spicifera showing varied fungal morphology including yeasts (arrowhead) and pseudohyphae (long arrows). HE. Inset: PAS with yeasts, pseudohyphae, and hyphae (short arrows).
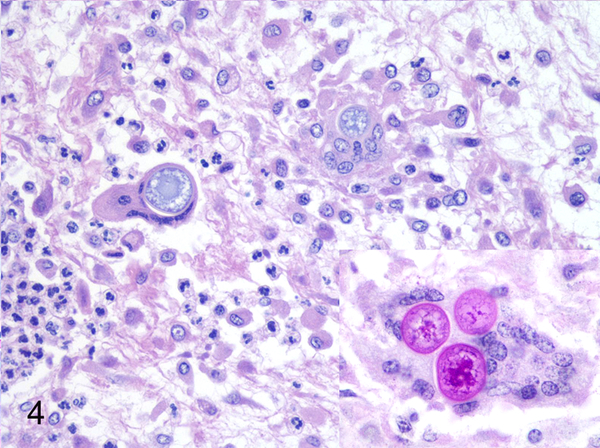
Some of the cases that were included from the archives also had ancillary testing performed which enabled further comparison with culture results. Fungal culture had been successfully performed on 25 cases. Panfungal PCR and sequencing results correlated with culture in 12/25 (48%) of cases, while 7/25 (28%) did not. Aspergillus was sequenced from three of the blocks, and from the tissues contained within these blocks an isolate of Mucor had been cultured. The histopathologic findings for two of the cases were consistent with aspergillosis, agreeing with the sequencing results, and in the third the hyphae could not be further identified. Lastly, six of the cases that had positive fungal culture results were PCR-positive, but the resultant sequences were of poor quality. To highlight the utility of this panfungal PCR assay when culture could pose a threat to the clinical microbiology personnel, several cases of dimorphic fungal infections were included in this study, and one of these cases of coccidioidomycosis with rare organisms is presented in Figure 4.
Fig. 4. Various mycotic infections in domestic species that were classified using panfungal PCR.
Subcutis, horse, case 48. Multinucleated giant cells contain large Coccidioides posadasii fungal spherules. HE. Inset: PAS.
Due to the high prevalence of Aspergillus cases in the present study, and the previously proposed ability of the ITS-2 region to provide speciation within the genus, additional phylogenetic analysis was performed on Aspergillus sequences to confirm BLAST results, and further validate this assay. From the 20 Aspergillus sequences included, 19/20 (95%) demonstrated correlation between the BLAST result and the phylogenetic clusters into which it was placed using neighbor joining analysis (Table 3). The one sequence that did not fall into a cluster, Equ1, matched to A. fumigatus using BLAST, and fell into the A. fumigatus cluster without repeat bootstrapping. Upon increasing the bootstrap replicates to 1000, Equ1 fell out of the A. fumigatus cluster in the consensus tree. Boot strap support values indicate high confidence (>90) for the separation of major clusters by species except for the branch points leading to the A. fumigatus (75) and A. felis (83) clusters (Supplemental Figure 2). Furthermore, the majority (17/20) of culture results also correlated with the BLAST sequencing results, and the phylogenetic clade placement. There were three samples from which Mucor sp. was cultured, but the sequencing either matched to A. fumigatus or A. felis.
Table 3.
Aspergillus sequences comparing BLAST, phylogenetic and ancillary testing results.
| SeqID | Case | BLAST Results | Phylogenetic Clade | Ancillary Testing |
|---|---|---|---|---|
| Avi1 | 24 | A. flavus, A. oryzae | A. flavus | Aspergillus |
| Avi2 | 31 | A. flavus, A. oryzae | A. flavus | Aspergillus |
| Avi3 | 32 | A. terreus | A. terreus | Aspergillus terreus |
| Avi4 | 33 | A. terreus | A. terreus | Aspergillus terreus |
| Bov1 | 27 | A. fumigatus | A. fumigatus | - |
| Bov2 | 28 | A. fumigatus | A. fumigatus | - |
| Can1 | 23 | P. caninus | P. caninus | Aspergillus |
| Can2 | 40 | A. terreus | A. terreus | Aspergillus |
| Can3 | 25 | A. flavus, A. oryzae | A. flavus | Aspergillus |
| Can4 | 34 | A. terreus | A. terreus | Aspergillus |
| Can5 | 38 | A. terreus | A. terreus | Aspergillus |
| Can6 | 35 | A. terreus | A. terreus | - |
| Can7 | 37 | A. terreus | A. terreus | Aspergillus terreus |
| Can8 | 36 | A. terreus | A. terreus | Aspergillus |
| Can9 | 39 | A. terreus | A. terreus | Aspergillus |
| Equ1 | 29 | A. fumigatus | -* | Mucor spp.* |
| Equ2 | 30 | A. nidulans, A. quadrilineata | A. nidulans | - |
| Equ3 | 26 | A. flavus, A. oryzae | A. flavus | Aspergillus |
| Fel1 | 41 | A. aureolus, N. fischeri, A. felis | A. felis | Mucor spp.* |
| Fel2 | 42 | A. aureoles, N. fischeri, A. felis | A. felis | Mucor spp.* |
Avi, avian; Bov, bovine; Can, canine; Equ, equine; Fel, feline. Results that do not correlate with other categories are identified by an asterisk (*). Blocks with no phylogenetic clade placement or no culture results are marked with a dash (-).
Statistical analysis revealed no influence of host, tissue, or fungal type on PCR results (positive/negative). Respiratory tract samples in this study were significantly associated with increased likelihood of having more than one band on gel electrophoresis (P=0.015), and skin samples were significantly associated with having poor quality sequences (P=0.002). Chi square analysis further demonstrated that the abundance of fungal organisms observed histologically was correlated with the PCR result (positive/faint/negative), number of bands, and quality of sequence. Rare organisms observed histologically were associated with an increased likelihood of having faint bands (P=0.0054), and poor quality sequences (P=0.0045). Tissues with abundant organisms were associated with an increased likelihood of having a strongly positive (P=0.0367), single band (P=0.0061). Overall, poor quality sequences were more likely obtained from blocks that demonstrated multiple PCR product bands on electrophoresis (P=0.0053), and from blocks with faint bands (P<0.0001).
Discussion
The panfungal PCR diagnostic assay presented here had a sensitivity of 69.5%, calculated as the percentage of cases for which ITS-2 sequencing provided a fungal identification. This sensitivity correlates well with previous studies, which also amplified the ITS-2 region from DNA extracts of FFPE tissues, and showed sensitivities of 53.8%4 and 64.3%.38 These two studies with similar sensitivities both targeted the ITS-2 regions, but used other methods of DNA extraction. Our work demonstrated this assay was successful in a wide variety of animal hosts, tissues, and fungi and thus is a valuable test when used in conjunction with histopathology for identification of fungal infections in animals. The results presented here also show that the MoBio BiOstic® FFPE tissue DNA isolation kit was successful at isolating high quality fungal DNA from FFPE animal tissues without requiring the use of lyticase. The time required for assay completion is approximately three days. The assay has the ability to identify fungi in FFPE specimens regardless of their transport temperature, and thus in some cases may be superior to culture, although culture remains the gold standard.
Many fungi have characteristic histologic features, which can be used to identify fungi in typical presentations and representative sections. However, there are the cases of atypical morphology, stemming from genotypic variation.29 For example, some Cryptococcus cases have varying amounts of polysaccharide capsule and thus resemble yeasts of similar size such as Candida or Blastomyces, or one of the smaller yeasts such as Histoplasma or Candida.26 Also, Candida has the ability to produce pseudohyphae and may even appear to be branching, and thus could be confused with hyphae.42 The present assay enabled sequencing of a variety of yeast from FFPE tissues including Blastomyces (3), Coccidioides (7), Cryptococcus (8), Histoplasma (16), and Malassezia (1). For one of the yeast cases, the histological diagnosis could not be identified further than “yeast with narrow-based budding,” but sequencing was able to confirm the etiologic agent as Histoplasma sp.
There appears to be greater challenges in identifying hyphae compared to yeast, especially the hyaline septate molds that are indistinguishable from one another morphologically, including Fusarium and Scedosporium.29 Even for Aspergillus species that have a characteristic branching angle, reports of misidentification include one study with 122 cases of Aspergillus identified based upon histological morphology, where only 83% had positive culture results for Aspergillus and the remaining culture results were positive for Scedosporium, Fusarium, Pseudallescheria, Phialophora, and Trichophyton.39 Although pigmented fungi may appear to be a straightforward identification on HE-stained sections, there exists variability in the amount of melanin produced by a genotypic variant and thus could be confused with other hyaline hyphae, and these fungi cannot be identified to the genus level.29,36 The present assay was able to sequence a variety of hyphae including Arthroderma (3), Aspergillus (20), Chaetomium (1), Curvularia (4), Curvularia/Bipolaris (8), Penicillium (1), Pythium (1), Rhizopus (3), and Scedosporium/Pseudallescheria (2). Eleven of these could not be identified further than hyphae based on histopathology alone. Overall there were three cases in this study where the fungi observed histologically were not pigmented, but the fungal sequence identified a phaeohyphomycoses, further supporting the variability in melanin production in these pigmented fungi (Fig. 3).
Statistical analyses demonstrated this assay is equally successful for both yeast and hyphae. There was not any influence of the type of tissue from which the DNA was isolated on PCR results (positive/negative), however, lung samples were more likely to have multiple PCR product bands, and skin samples were more likely to yield poor quality sequences. The skin and respiratory tracts are known to have resident microbiota, which might influence PCR performed on these samples. It is also possible that the method of sample collection, or post-mortem handling of the tissues prior to embedding, could have introduced contamination. Another limitation of this assay is that sensitivity decreases when organisms are rarely observed. More sensitive sequencing techniques, such as next-generation sequencing, might have greater success in amplifying fungal DNA when there are rare organisms in the embedded tissue. However, these techniques are costly and not yet suitable for diagnostics.
There was variable ability of the assay to derive genus and species level resolution. The sequence analysis provided genus- and species-level resolution in 36% of all cases where they could not be distinguished based only on histologic morphology. For some genera, such as those belonging to the phaeohyphomycoses, the ITS-2 region was unable to distinguish between genera such as Curvularia, Bipolaris, and Alternaria. On the contrary, the ITS-2 sequence was sufficient to derive a genus-level identification for fungi such as Coccidioides, Cryptococcus, Penicillium, Rhizopus, Scedosporium, and the fungal-like Pythium. In other cases, species level resolution was possible for Aspergillus, Blastomyces, Histoplasma, and Microsporum species. The ability to distinguish between as many as six different Aspergillus species prompted further phylogenetic analysis to confirm these results derived through BLAST. Bioinformatics literature discusses the limitations of BLAST to distinguish between taxa, and promotes the superior ability of neighbor-joining analysis to do so.43 Agreement of the BLAST results, neighbor-joining analysis, and culture (Table 3) demonstrate that this assay is indeed capable of reliably speciating Aspergillus. The ability to differentiate between species of fungal organisms has clinical relevance due to the documented differences in susceptibility to antifungal agents between fungal genera, species,27,37 and even subspecies.56 Taken in conjunction with the documented rise of fungal resistance to antifungal agents specifically in Aspergillus,7 this tool may be extremely valuable for pathologists and clinicians treating these systemic mycoses.
Interestingly, 28/128 (21.9%) of all FFPE blocks returned sequences that were of poor quality in spite of being purified from a strong band on the gel. Similar reports of ambiguous or poor quality sequences were reported in one of the original applications of this assay,38 but has not been described in another more recent work.45 Studies have shown through cloning that ITS sequence heterogeneity exists in some fungal genera including Fusarium47 and even in strains of Rhizopus microsporus.61 Most likely there has not been an exhaustive investigation into which fungal genera, species, or strains possess this ITS sequence heterogeneity. We hypothesize that at least some of the ‘poor quality’ sequences in this study were the result of ITS sequence heterogeneity present in the genomes of fungi infecting those animal tissues. Statistical analysis revealed that poor quality sequences were more common when there were rare organisms on the slide, multiple bands present, or the bands were faint. These findings elucidate some of the limitations of this assay, and further molecular work such as cloning and sequencing and/or high-throughput sequencing might be used to identify fungi observed histologically when panfungal PCR with second generation sequencing fails to identify an etiologic agent.
Another rarely reported finding38 that was encountered in the present study was the presence of multiple bands in a sample identified with gel electrophoresis. Use of the IRBP PCR enabled confirmation of high quality extracted DNA for these samples, and thus the presence of multiple bands is not likely due to fragmentation of the DNA. It is possible that these additional sequences were amplified from commensal or contaminating fungi present in the tissues. At least for the skin,44 gastrointestinal tract,22 and respiratory tract19 tissues, recent next generation sequencing studies have identified an enormous diversity of commensal fungi inhabiting these tissues. Although these commensal fungi are rarely seen histologically, their DNA could still be present in the tissues and amplified with this assay, possibly producing a false positive result. An additional possibility that could explain the presence of multiple bands is variation in the copy number of the ITS coding region,11,40 which is present as tandem repeats in some species of fungi.31,34 Another recent application of this assay on tropical mycoses24 found that some of the amplified fungal ITS-2 sequences were not consistent with the histological findings on those blocks. That study suggested that the amplified fungal DNA came from environmental contamination of the blocks. While this is possible, it is also possible that commensal fungal DNA was amplified, as hypothesized here. This limitation indicates that the PCR and sequencing results provided by this assay need to be interpreted in the context of fungal morphology observed histologically.
In summary, this assay has been improved through reduction in cost and time, and it has been validated for a variety of animal hosts and fungal pathogens that commonly infect animals. Although some limitations still exist for the assay, it could provide a diagnosis when morphology based on histological examination alone could not, and when fresh tissues were not available for fungal culture. Having a quick and accurate diagnosis enables the correct selection of antifungal therapeutics. Accurate identification of fungal infections in animals will not only enhance epidemiological studies of current and emerging fungal threats to animals, but to humans as well. Further development of genus- and species-specific PCR could help to resolve fungal identification for instances when the ITS region possesses sequence heterogeneity within species, or when the ITS region is homogenous across genera.
Supplementary Material
Flowchart explaining selection criteria for blocks and assignment to experiments. Blocks were never used in more than one experiment, and each block was only represented once in the final analyses that are summarized in Supplemental Table 1. The chronology of the experiments followed from testing two kits (1), to assessing addition of lyticase (2), and lastly performance of optimized protocol on remaining blocks (3). An approximately equal and representative number of yeast or hyphal cases were chosen to be included in each experiment.
Phylogenetic analysis of Aspergillus sequences amplified from FFPE animal tissues. The phylogenetic relationship of all Aspergillus sequences amplified from FFPE animal tissues in this study, along with reference Aspergillus sp. sequences, was calculated using Tamura-nei nearest neighbor analysis, and bootstrap (n=1000) values located at branch points. Sequences were named according to the host animal to show the host adaptability of various Aspergillus sp.
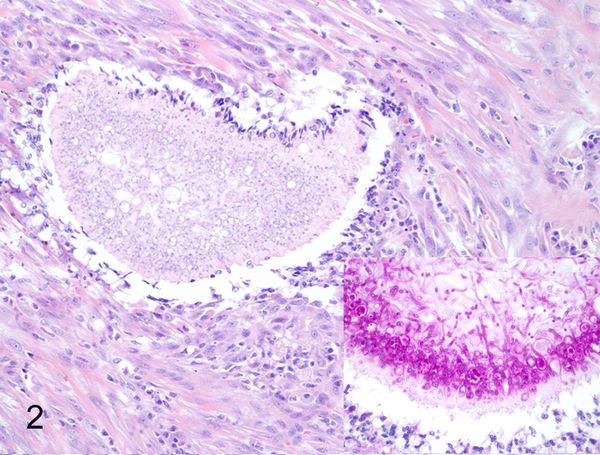
Fig. 2. Various mycotic infections in domestic species that were classified using panfungal PCR.
Urinary bladder, dog, case 125. Hyphae form large fungal mats and were classified into the Scedosporium spp./Pseudallescheria spp. family. HE. Inset: PAS.
Acknowledgements:
Dr. Xiaorong Lin, Department of Biology, Texas A&M University, for providing purified fungal genomic DNA, and Anna Blick, Department of Veterinary Pathobiology, Texas A&M University, for assistance with phylogenetic analysis.
Contributor Information
Courtney Meason-Smith, Department of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, TX, USA.
Erin E. Edwards, Department of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, TX, USA
Caitlin E. Older, Department of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, TX, USA
Mackenzie Branco, Department of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, TX, USA.
Laura K. Bryan, Department of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, TX, USA
Sara D. Lawhon, Department of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, TX, USA
Jan S. Suchodolski, Department of Small Animal Clinical Sciences, College of Veterinary Medicine, Texas A&M University, College Station, TX, USA
Gabriel Gomez, Texas A&M Veterinary Medical Diagnostic Laboratories, College Station, TX, USA.
Joanne Mansell, Department of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, TX, USA.
Aline Rodrigues Hoffmann, Department of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, TX, USA.
References
- 1.Alexander BD, Pfaller MA: Contemporary tools for the diagnosis and management of invasive mycoses. Clinical Infectious Diseases 2006:43:S15–S27. [Google Scholar]
- 2.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol 1990:215(3):403–410. [DOI] [PubMed] [Google Scholar]
- 3.Arunmozhi Balajee S, Hurst SF, Chang LS, Miles M, Beeler E, Hale C, et al. : Multilocus sequence typing of Histoplasma capsulatum in formalin-fixed paraffin-embedded tissues from cats living in non-endemic regions reveals a new phylogenetic clade. Med Mycol 2013:51(4):345–351. [DOI] [PubMed] [Google Scholar]
- 4.Babouee Flury B, Weisser M, Prince SS, Bubendorf L, Battegay M, Frei R, et al. : Performances of two different panfungal PCRs to detect mould DNA in formalin-fixed paraffin-embedded tissue: what are the limiting factors? BMC Infect Dis 2014:14:692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balajee SA, Sigler L, Brandt ME: DNA and the classical way: identification of medically important molds in the 21st century. Med Mycol 2007:45(6):475–490. [DOI] [PubMed] [Google Scholar]
- 6.Barrs VR, van Doorn TM, Houbraken J, Kidd SE, Martin P, Pinheiro MD, et al. : Aspergillus felis sp. nov., an emerging agent of invasive aspergillosis in humans, cats, and dogs. PLoS One 2013:8(6):e64871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beernaert LA, Pasmans F, Van Waeyenberghe L, Dorrestein GM, Verstappen F, Vercammen F, et al. : Avian Aspergillus fumigatus strains resistant to both itraconazole and voriconazole. Antimicrob Agents Chemother 2009:53(5):2199–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger L, Speare R, Daszak P, Green DE, Cunningham AA, Goggin CL, et al. : Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci U S A 1998:95(15):9031–9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernhardt A, von Bomhard W, Antweiler E, Tintelnot K: Molecular identification of fungal pathogens in nodular skin lesions of cats. Med Mycol 2015:53(2):132–144. [DOI] [PubMed] [Google Scholar]
- 10.Bieganska M, Dardzinska W, Dworecka-Kaszak B: Fungal colonization - an additional risk factor for diseased dogs and cats? Ann Parasitol 2014:60(3):139–146. [PubMed] [Google Scholar]
- 11.Black J, Dean T, Byfield G, Foarde K, Menetrez M: Determining fungi rRNA copy number by PCR. J Biomol Tech 2013:24(1):32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosshard PP: Incubation of fungal cultures: how long is long enough? Mycoses 2011:54(5):e539–545. [DOI] [PubMed] [Google Scholar]
- 13.Boyle DG, Boyle DB, Olsen V, Morgan JA, Hyatt AD: Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis Aquat Organ 2004:60(2):141–148. [DOI] [PubMed] [Google Scholar]
- 14.Brilhante RS, de Jesus Santos Rodrigues T, de Souza Collares Maia Castelo-Branco D, Teixeira CE, de Brito Macedo R, Bandeira SP, et al. : Antifungal susceptibility and virulence attributes of animal-derived isolates of Candida parapsilosis complex. J Med Microbiol 2014:63(Pt 11):1568–1572. [DOI] [PubMed] [Google Scholar]
- 15.Brinkman NE, Haugland RA, Wymer LJ, Byappanahalli M, Whitman RL, Vesper SJ: Evaluation of a rapid, quantitative real-time PCR method for enumeration of pathogenic Candida cells in water. Appl Environ Microbiol 2003:69(3):1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC: Hidden killers: human fungal infections. Sci Transl Med 2012:4(165):165rv113. [DOI] [PubMed] [Google Scholar]
- 17.Chen SC, Halliday CL, Meyer W: A review of nucleic acid-based diagnostic tests for systemic mycoses with an emphasis on polymerase chain reaction-based assays. Med Mycol 2002:40(4):333–357. [DOI] [PubMed] [Google Scholar]
- 18.Cheng TL, Rovito SM, Wake DB, Vredenburg VT: Coincident mass extirpation of neotropical amphibians with the emergence of the infectious fungal pathogen Batrachochytrium dendrobatidis. Proc Natl Acad Sci U S A 2011:108(23):9502–9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delhaes L, Monchy S, Frealle E, Hubans C, Salleron J, Leroy S, et al. : The airway microbiota in cystic fibrosis: a complex fungal and bacterial community--implications for therapeutic management. PLoS One 2012:7(4):e36313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC: MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004:32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira EC, Gontijo CM, Cruz I, Melo MN, Silva AM: Alternative PCR protocol using a single primer set for assessing DNA quality in several tissues from a large variety of mammalian species living in areas endemic for leishmaniasis. Mem Inst Oswaldo Cruz 2010:105(7):895–898. [DOI] [PubMed] [Google Scholar]
- 22.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, et al. : Topographic diversity of fungal and bacterial communities in human skin. Nature 2013:498(7454):367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frick WF, Pollock JF, Hicks AC, Langwig KE, Reynolds DS, Turner GG, et al. : An emerging disease causes regional population collapse of a common North American bat species. Science 2010:329(5992):679–682. [DOI] [PubMed] [Google Scholar]
- 24.Frickmann H, Loderstaedt U, Racz P, Tenner-Racz K, Eggert P, Haeupler A, et al. : Detection of tropical fungi in formalin-fixed, paraffin-embedded tissue: still an indication for microscopy in times of sequence-based diagnosis? Biomed Res Int 2015:2015:938721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gargas A, Trest MT, Christensen M, Volk TJ, Blehert DS: Geomyces destructans sp nov associated with bat white-nose syndrome. Mycotaxon 2009:108:147–154. [Google Scholar]
- 26.Gazzoni AF, Severo CB, Salles EF, Severo LC: Histopathology, serology and cultures in the diagnosis of cryptococcosis. Rev Inst Med Trop Sao Paulo 2009:51(5):255–259. [DOI] [PubMed] [Google Scholar]
- 27.Goncalves SS, Stchigel AM, Cano J, Guarro J, Colombo AL: In vitro antifungal susceptibility of clinically relevant species belonging to Aspergillus section Flavi. Antimicrob Agents Chemother 2013:57(4):1944–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greiner K, Persoh D, Weig A, Rambold G: Phialosimplex salinarum, a new species of Eurotiomycetes from a hypersaline habitat. IMA Fungus 2014:5(2):161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guarner J, Brandt ME: Histopathologic diagnosis of fungal infections in the 21st century. Clin Microbiol Rev 2011:24(2):247–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henry T, Iwen PC, Hinrichs SH: Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J Clin Microbiol 2000:38(4):1510–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrera ML, Vallor AC, Gelfond JA, Patterson TF, Wickes BL: Strain-dependent variation in 18S ribosomal DNA Copy numbers in Aspergillus fumigatus. J Clin Microbiol 2009:47(5):1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hove MG, Woods GL: Duration of fungal culture incubation in an area endemic for Histoplasma capsulatum. Diagn Microbiol Infect Dis 1997:28(1):41–43. [DOI] [PubMed] [Google Scholar]
- 33.Isenberg HD: 8.4: Processing specimens for fungal culture In: Isenberg HD, ed. Clinical microbiology procedures handbook. Second ed.: American Society for Microbiology Press; 2004. [Google Scholar]
- 34.Iwen PC, Hinrichs SH, Rupp ME: Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med Mycol 2002:40(1):87–109. [DOI] [PubMed] [Google Scholar]
- 35.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. : Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012:28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura M, McGinnis MR: Fontana-Masson--stained tissue from culture-proven mycoses. Arch Pathol Lab Med 1998:122(12):1107–1111. [PubMed] [Google Scholar]
- 37.Lass-Florl C, Alastruey-Izquierdo A, Cuenca-Estrella M, Perkhofer S, Rodriguez-Tudela JL: In vitro activities of various antifungal drugs against Aspergillus terreus: Global assessment using the methodology of the European committee on antimicrobial susceptibility testing. Antimicrob Agents Chemother 2009:53(2):794–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lau A, Chen S, Sorrell T, Carter D, Malik R, Martin P, et al. : Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J Clin Microbiol 2007:45(2):380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S, Yun NR, Kim KH, Jeon JH, Kim EC, Chung DH, et al. : Discrepancy between histology and culture in filamentous fungal infections. Med Mycol 2010:48(6):886–888. [DOI] [PubMed] [Google Scholar]
- 40.Longo AV, Rodriguez D, da Silva Leite D, Toledo LF, Mendoza Almeralla C, Burrowes PA, et al. : ITS1 copy number varies among Batrachochytrium dendrobatidis strains: implications for qPCR estimates of infection intensity from field-collected amphibian skin swabs. PLoS One 2013:8(3):e59499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorch JM, Meteyer CU, Behr MJ, Boyles JG, Cryan PM, Hicks AC, et al. : Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature 2011:480(7377):376–378. [DOI] [PubMed] [Google Scholar]
- 42.Luna M: Candidiasis Pathology of infectious diseases. First ed.; 1997. [Google Scholar]
- 43.Matsen FA, Kodner RB, Armbrust EV: pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 2010:11:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meason-Smith C, Diesel A, Patterson AP, Older CE, Mansell JM, Suchodolski JS, et al. : What is living on your dog’s skin? Characterization of the canine cutaneous mycobiota and fungal dysbiosis in canine allergic dermatitis. FEMS Microbiol Ecol 2015:91(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munoz-Cadavid C, Rudd S, Zaki SR, Patel M, Moser SA, Brandt ME, et al. : Improving molecular detection of fungal DNA in formalin-fixed paraffin-embedded tissues: comparison of five tissue DNA extraction methods using panfungal PCR. J Clin Microbiol 2010:48(6):2147–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nijima M, Kano R, Nagata M, Hasegawa A, Kamata H: An azole-resistant isolate of Malassezia pachydermatis. Vet Microbiol 2011:149(1–2):288–290. [DOI] [PubMed] [Google Scholar]
- 47.O’Donnell K, Cigelnik E: Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 1997:7(1):103–116. [DOI] [PubMed] [Google Scholar]
- 48.Patel AJ, Gattuso P, Reddy VB: Diagnosis of blastomycosis in surgical pathology and cytopathology: correlation with microbiologic culture. Am J Surg Pathol 2010:34(2):256–261. [DOI] [PubMed] [Google Scholar]
- 49.Paterson PJ, Seaton S, McLaughlin J, Kibbler CC: Development of molecular methods for the identification of aspergillus and emerging moulds in paraffin wax embedded tissue sections. Mol Pathol 2003:56(6):368–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rabelo-Goncalves E, Roesler B, Guardia AC, Milan A, Hara N, Escanhoela C, et al. : Evaluation of five DNA extraction methods for detection of H. pylori in formalin-fixed paraffin-embedded (FFPE) liver tissue from patients with hepatocellular carcinoma. Pathol Res Pract 2014:210(3):142–146. [DOI] [PubMed] [Google Scholar]
- 51.Ribes JA, Vanover-Sams CL, Baker DJ: Zygomycetes in human disease. Clin Microbiol Rev 2000:13(2):236–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sangoi AR, Rogers WM, Longacre TA, Montoya JG, Baron EJ, Banaei N: Challenges and pitfalls of morphologic identification of fungal infections in histologic and cytologic specimens: a ten-year retrospective review at a single institution. Am J Clin Pathol 2009:131(3):364–375. [DOI] [PubMed] [Google Scholar]
- 53.Sarmiento-Ramirez JM, Abella E, Martin MP, Telleria MT, Lopez-Jurado LF, Marco A, et al. : Fusarium solani is responsible for mass mortalities in nests of loggerhead sea turtle, Caretta caretta, in Boavista, Cape Verde. Fems Microbiology Letters 2010:312(2):192–200. [DOI] [PubMed] [Google Scholar]
- 54.Saubolle MA: Laboratory aspects in the diagnosis of coccidioidomycosis. Ann N YAcad Sci 2007:1111:301–314. [DOI] [PubMed] [Google Scholar]
- 55.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. : Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 2012:109(16):6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singer LM, Meyer W, Firacative C, Thompson GR, 3rd, Samitz E, Sykes JE: Antifungal drug susceptibility and phylogenetic diversity among Cryptococcus isolates from dogs and cats in North America. J Clin Microbiol 2014:52(6):2061–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Songer JG, Post KW: Veterinary microbiology : bacterial and fungal agents of animal disease. St. Louis, Mo.: Elsevier Saunders, 2005. [Google Scholar]
- 58.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S: MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013:30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson DW, Kaplan W, Phillips BJ: The effect of freezing and the influence of isolation medium on the recovery of pathogenic fungi from sputum. Mycopathologia 1977:61(2):105–109. [DOI] [PubMed] [Google Scholar]
- 60.White TBT, Taylor JW, Taylor JW: Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Academic Press 1990. [Google Scholar]
- 61.Woo PC, Leung SY, To KK, Chan JF, Ngan AH, Cheng VC, et al. : Internal transcribed spacer region sequence heterogeneity in Rhizopus microsporus: implications for molecular diagnosis in clinical microbiology laboratories. J Clin Microbiol 2010:48(1):208–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flowchart explaining selection criteria for blocks and assignment to experiments. Blocks were never used in more than one experiment, and each block was only represented once in the final analyses that are summarized in Supplemental Table 1. The chronology of the experiments followed from testing two kits (1), to assessing addition of lyticase (2), and lastly performance of optimized protocol on remaining blocks (3). An approximately equal and representative number of yeast or hyphal cases were chosen to be included in each experiment.
Phylogenetic analysis of Aspergillus sequences amplified from FFPE animal tissues. The phylogenetic relationship of all Aspergillus sequences amplified from FFPE animal tissues in this study, along with reference Aspergillus sp. sequences, was calculated using Tamura-nei nearest neighbor analysis, and bootstrap (n=1000) values located at branch points. Sequences were named according to the host animal to show the host adaptability of various Aspergillus sp.