Abstract
Genomic imprinting leads to different expression levels of maternally and paternally derived alleles. Over the last years, major progress has been made in identifying novel imprinted candidate genes in plants, owing to affordable next-generation sequencing technologies. However, reports on sequencing the transcriptome of hybrid F1 seed tissues strongly disagree about how many and which genes are imprinted. This raises questions about the relative impact of biological, environmental, technical, and analytic differences or biases. Here, we adopt a statistical approach, frequently used in RNA-seq data analysis, which properly models count overdispersion and considers replicate information of reciprocal crosses. We show that our statistical pipeline outperforms other methods in identifying imprinted genes in simulated and real data. Accordingly, reanalysis of genome-wide imprinting studies in Arabidopsis and maize shows that, at least for Arabidopsis, an increased agreement across datasets could be observed. For maize, however, consistent reanalysis did not yield a larger overlap between the datasets. This suggests that the discrepancy across publications might be partially due to different analysis pipelines but that technical, biological, and environmental factors underlie much of the discrepancy between datasets. Finally, we show that the set of genes that can be characterized regarding allelic bias by all studies with minimal confidence is small (~8,000/27,416 genes for Arabidopsis and ~12,000/39,469 for maize). In conclusion, we propose to use biologically replicated reciprocal crosses, high sequence coverage, and a generalized linear model approach to identify differentially expressed alleles in developing seeds.
Introduction
In a diploid cell, the maternal and paternal alleles of a given gene usually share the same expression state in a specific tissue, meaning that they are either both expressed or both silent. Important exceptions to this rule are genes regulated by genomic imprinting, where the expression state depends on the parental origin of the alleles, and only one is expressed while the other remains silent or is weakly expressed. The two alleles do not differ in their sequence but rather carry parent-specific, epigenetic imprints that allow the cell to distinguish the two alleles1–8. Genomic imprinting evolved independently in mammals and flowering plants (angiosperms) (reviewed in9–15). In both groups, offspring develop within the mother and depend solely on her to supply nutrients for growth and development. This common reproductive strategy results in an intragenomic parental conflict over resource allocation, which likely underlies the evolution of genomic imprinting, at least for loci that control growth14,16,17. Accordingly, some imprinted genes in both, mammals and plants, have a role in controlling growth (e.g.18–26). Consistent with this function, many imprinted genes are preferentially expressed in the tissues that support embryonic growth, i.e. the placenta in mammals or the triploid endosperm in the seeds of flowering plants.
Over the last decade, the advent of Next-Generation Sequencing (NGS) allowed (nearly) genome-wide imprinting studies by sequencing the transcriptome of hybrid F1 seed tissues: Given exonic polymorphisms between the parents, reads overlapping heterozygous SNPs can be assigned to their parent-of-origin, and reciprocal crosses allow the discrimination between parent-of-origin-dependent and strain-specific genetic effects. Accordingly, a number of research groups performed genome-wide, allele-specific transcriptome profiling studies of hybrid seeds in Arabidopsis and maize to identify genes that are preferentially expressed from one parental allele27–38. As a result, the total number of imprinted genes increased from around 206 to over 900 potentially imprinted plant genes28–33,35,36,38.
However, comparisons of the identified imprinted candidate genes revealed little overlap between the studies30,34,39. In general, the analysis of RNA-sequencing (RNA-seq) data to identify allele-specific expression is prone to false positives due to both, biological and technical variation40–42. Thus, even studies with seemingly similar design heavily disagree on the number of imprinted genes in the mouse brain, e.g. ranging from less than 20040 to over a thousand43. To date, although guidelines for the analysis of allele-specific expression have recently become available42, many different methods have been applied to filter, normalize, and statistically assess allelic imbalance from RNA-seq data. For the analysis of allele-specific expression, several analysis methods and software42 have been developed, yet only very few are suitable for an analysis of imprinted expression. Moreover, no specialized method is available for statistical testing of imprinting in the triploid endosperm, where the expected allelic ratio is 2:1 because the mother contributes two genomes to this tissue. In plants, many authors have used count tests (such as Chi-Square, binomial, or Fisher’s exact tests), which heavily underestimate the count dispersion typically observed in RNA-seq data41,42,44, resulting in increased numbers of false positives particularly for large counts. Highly expressed transcripts may appear imprinted with high statistical significance, as count tests are sensitive to very small allelic imbalance at high counts, requiring additional filtering with somewhat arbitrary imbalance cut-offs.
Here, we present a new statistical approach to call imprinted genes from large allele-specific RNA-seq datasets from endosperm that outperforms other methods in simulated and real data. We propose a generally applicable approach using generalized linear models (GLM) implemented in edgeR45, which is based on the negative binomial distribution to deal with potential count overdispersion46 as it is typically seen in RNA-seq data. The presented pipeline outperforms other methods using simulated data. Furthermore, we reanalyze the raw data from seven studies to assess the relative importance of differences in data generation and data analysis. The consistent reanalysis by the proposed pipeline results in a larger overlap of imprinted candidate genes across Arabidopsis datasets, but showed little improvement across maize datasets. In conclusion, consistent data analysis can improve concordance between datasets but biological and technical variation in data generation contributes most to the differences among datasets.
Results
Comparison of Genome-wide Imprinting Studies in Plants: Biological, Technical, and Statistical Differences
To shed light onto discrepancies between published studies and potential biases, we compared the genome-wide, allele-specific transcriptome profiling studies of hybrid seeds in Arabidopsis and maize that were designed to identify imprinted candidate genes in the endosperm. All studies were based on reciprocal crosses of two polymorphic inbred strains, and all studies used manually dissected endosperm. For simplicity, we only compared studies using Ler and Col-0 accessions (Arabidopsis) or B73 and Mo17 inbreds (maize). First, we set out to identify the genes that can be evaluated for imprinting by all studies and reanalyzed the raw data from the seven published studies starting from the raw reads. We examined all reads overlapping with previously known exonic SNPs (see Methods), separated and counted maternal and paternal reads overlapping SNPs, and summed up informative (i.e. SNP containing) reads across transcripts. After discarding very lowly expressed genes (<10 counts per gene), median reads per transcript were 95–654 for the different datasets.
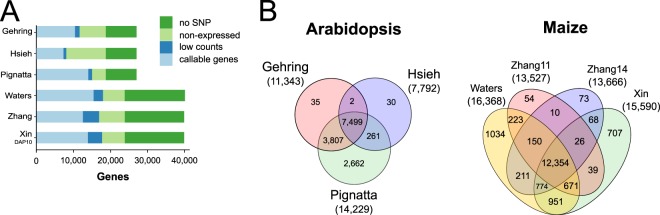
The power of allelic imbalance detection depends mainly on the degree of allelic bias, the sequencing read length and coverage (expression strength), as well as the divergence of the crossed strains (number of SNPs per transcript)47. Although the three Arabidopsis studies all use the same accessions, the number of callable genes ranges from 7,792 for the Hsieh dataset to 14,229 for the Pignatta dataset (Fig. 1A): Among the 27,416 protein-coding Arabidopsis genes, 31% do not overlap with any exonic SNP and their imprinting cannot be assessed. Another 17–39% of the genes are not or only very weakly expressed (<10 allelic counts overall), and were thus discarded from further analysis. Ultimately, only 28–52% of all predicted Arabidopsis genes can be assessed for genomic imprinting in the three different studies, because only those have a sufficient number of allele-specific reads to identify statistically significant biased expression (Fig. 1A). 7,499 genes (27% of all Arabidopsis genes) can be assessed for genomic imprinting in all three datasets (Fig. 1B).
Figure 1.
Power of detecting imprinted candidate genes in reanalyzed datasets. (A) Callable genes assessable for genomic imprinting were required to overlap with at least 1 exonic SNP and to have read counts of at least 10. Number of genes are shown as percentages of the total number of genes. (B) Venn diagrams showing the overlap of callable genes for Arabidopsis and maize datasets.
Among the 39,469 maize protein-coding genes, 33% do not overlap with any exonic SNP (Fig. 1A) between the B73 and Mo17 inbreds. Another 26–33% were not or very weakly expressed (<10 counts overall) and were discarded (Fig. 1A). In the end, between 34% (Zhang11) and 41% (Waters) of maize genes can be assessed for genomic imprinting per dataset and only 31% of all maize genes (12,354 genes) can be assessed in all datasets (Fig. 1A,B). In conclusion, even after consistent reanalysis, the set of genes that can be assessed for genomic imprinting differs considerably between the datasets. For all the following analyses, we considered only genes that could be evaluated for imprinting in all datasets.
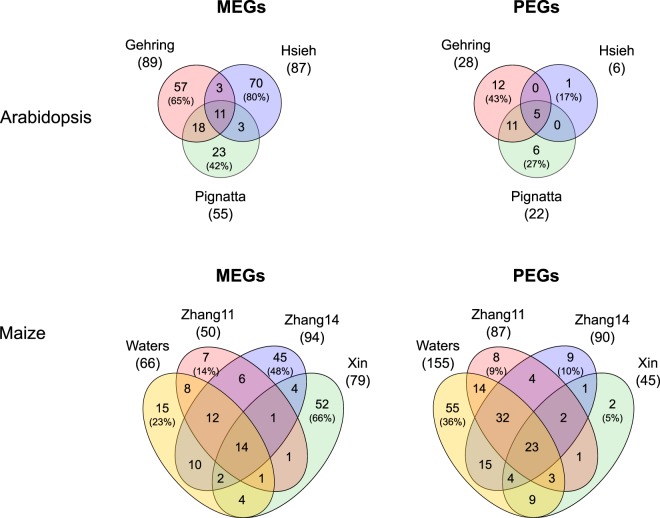
In Arabidopsis, a total of 185 genes that could be evaluated by all datasets were proposed to be maternally expressed imprinted genes (MEGs) in at least one study using the Landsberg erecta (Ler) and Columbia-0 (Col-0) accessions (Fig. 2). The three studies proposed between 55 and 89 MEGs28,29,38. The large majority of genes (81%) were unique to a single study, and only eleven MEGs were identified as imprinted in all three studies (6%; Fig. 2). Thirtyfive genes were proposed to be paternally expressed imprinted genes (PEGs) in at least one study with five PEGs being commonly identified in all three studies (14%; Fig. 2). In maize, the four available studies using the inbreds B73 and Mo1731,33,36,37 listed 182 different MEGs and 182 PEGs (Fig. 2). The majority of genes (65% and 41% for MEGs and PEGs, respectively) was proposed by a single study only. The overlap between studies was also small, with 14 MEGs (8%) and 23 PEGs (13%) being commonly identified by all four studies.
Figure 2.
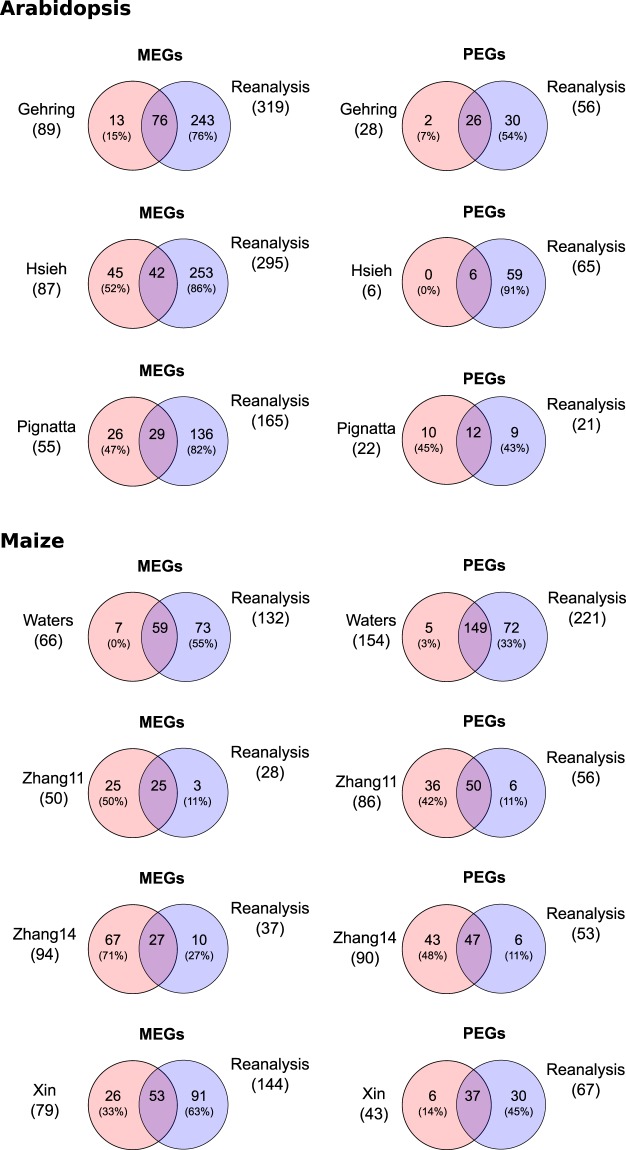
Venn diagrams showing the number of imprinted genes in hybrid endosperm reported by different studies in Arabidopsis Ler/Col accessions and in maize B73/Mo17 inbreds. Only genes were considered that could be evaluated for imprinting in all datasets. Numbers in brackets denote the percentage of non-shared genes relative to the full set reported. The “Xin” and “Zhang11” sets comprise genes identified at 10 days after pollination and “Zhang14” comprises genes at 12 days after pollination. Genes with an accession/inbred-specific bias in expression were excluded from the analysis.
The low concordance between studies could be due (i) to intrinsic, biological differences (e.g. developmental stage analyzed), (ii) to technical differences (particularly library preparation and complexity, sequencing depth and batch effects, reviewed in41,42, and/or (iii) to the varying bioinformatics/statistical analysis protocols applied. As summarized in Table 1, all studies differ in terms of developmental stage and some use different accessions creating considerable biological variation. Technical differences like library preparation, sequencing platform, read length, and single-end vs. paired-end sequencing introduce a further level of variation. Particularly, the observed differences in sequencing depth, expected read-mapping biases48, as well as in the completeness and quality of available SNP annotations, present likely technical sources of inconsistency (Table 1). Regarding statistical analysis, all studies applied count statistics (Table 1), which do not properly model count dispersion of RNA-Seq data, resulting in increased numbers of false positives particularly for large counts41,44. Furthermore, the studies apply very different criteria for filtering potentially imprinted genes according to the allelic bias (Table 1). The requirement to call a gene’s expression parentally biased differed tremendously between the studies and ranged from 90% of all reads that have to derive from one parent31, over 5 times more reads from one parent33, to simply assessing deviations from the expected 2:1 ratio in the endosperm28. Lastly, to deal with potential contamination from the seed coat, transcripts that were highly expressed in this maternal tissue were filtered out in silico in most studies (Table 1). The filtering conditions and the expression data used varied considerably across studies.
Table 1.
Characteristics of data generation and data analysis of published genome-wide imprinting datasets in Arabidopsis and maize.
| Wolff 2011 | Gehring 2011 | Hsieh 2011 | Pignatta 2014 | Waters 2011 | Zhang 2011 | Xin 2013 | Waters 2013 | Zhang 2014 | |
|---|---|---|---|---|---|---|---|---|---|
| Organism | Arabidopsis | Arabidopsis | Arabidopsis | Arabidopsis | Maize | Maize | Maize | Maize | Maize |
| Strains | Col-0, Bur-0 | Col-0, Ler | Col-0, Ler | Col-0, Ler, Cvi | B73, Mo17 | B73, Mo17 | B73, Mo17 | B73,Mo17,Ki11,Oh43 | B73, Mo17 |
| Starting Material | whole seeds | dissected endosperm/embryo | dissected endosperm/embryo | dissected endosperm/embryo | dissected endosperm/embryo | dissected endosperm/embryo | whole kernels (0,3,5 DAP)/dissected endosperm (7, 10,15 DAP) | dissected endosperm | dissected endosperm |
| Timepoint | 4 DAP | 6 – 7 DAP | 7- 8 DAP | 6 DAP | 14 DAP | 10 DAP | 0,3,5,7,10,15 DAP | 14 DAP | 12 DAP |
| Biological Replicates per cross | 1 | 1 | 2 | 3 | 1 | 1 | 1 | 1 | 1 |
| Sequencing Platform | Illumina GAII | Illumina GAII | Illumina GAII | Illumina HiSeq | Illumina GAII/HiSeq | Illumina HiSeq | Illumina HiSeq | Illumina HiSeq | Illumina HiSeq |
| Read Length | 36 bp | 50/36 bp | 76 bp | 40/80 bp | 2 × 76 bp | 2 × 100 bp | 2 × 90 bp | 2 × 100/2 × 50 bp | 2 × 100 |
| NCBI SRA study ID | SRP005700 | SRP007424 | SRP003799 | SRP033371 | SRP009313 | SRP011991 | SRP026399 | SRP031872 | SRP011991 |
| Genome annotation | TAIR 8 | TAIR 9 | TAIR 8 | TAIR 10 | B73 AGPv2 | n/a | B73 5b.60 | n/a | B73 (V2) |
| Total number of raw reads (hybrids) | 122 mio | 100 mio | 165 mio | 1,837 mio | 245 mio | 149 mio | 379 mio | 1,969 mio | 154 mio |
| Total Number of SNPs | 569,859 | 347,928 | 402,226 | 384,612 | 1.6 mio | 51,416 exonic | 6.5 mio | 28,195–142,033 exonic | 4.2 mio |
| Mapping | |||||||||
| Mapping software | vmatch | TopHat | Bowtie | Tophat v2.0.8 | GSNAP | bwa | TopHat2 | TopHat | TopHat |
| Number mismatches allowed | 2/36 | n/a | 3/76 | 1/40 | 2/36 | n/a | n/a | 2 | n/a |
| Allele-specific mapping bias | Alignment to Col and Bur pseudoreference (only SNPs) | n/a | Alignment to Col and Ler pseudoreference (only SNPs) | n/a | n/a | Mapping to SNP-masked genes | Mapping to both genomes (reference-guided assembly of Mo17) | Mapping to SNP-masked transcripts (filtered gene set v5b.60) | Mapping to SNP-masked genes |
| Counting and Statistics | |||||||||
| Minimal coverage (allelic reads) | ≥10 (≥30) | ≥15 | n/a | n/a | ≥10 reads assigned to one allele in both hybrids | ≥10 per cross | ≥40 (non-stringent: ≥5 per cross) | ≥10 per cross | n/a |
| Summing Reads across | gene | gene | individual SNPs | gene | gene | gene | |||
| Statistical Test | Binomial | Storer-Kim | Fisher’s exact test | Fisher’s exact test | Chi-Square | Chi-Square | Chi-Square | Chi-Square | Chi-Square |
| Multiple Testing Correction | yes (FDR 5%) | no (p < 0.01) | no (p < 0.001) | yes (FDR 1%) | no (p < 0.01) | no (p < 0.05) | unknown adjustment (p < 0.001) | no (p < 0.05 or p < 0.01) | no (p < 0.05) |
| Allelic bias filtering | n/a | only unique expression | MEGs: maternal score ≥ 4x paternal score; PEGs: paternal score ≥ 1.5x maternal score | MEGs: ≥ 85% maternal reads PEGs: ≥ 50% paternal reads | ≥90% reads from one parent in both reciprocal crosses | ≥83% reads from one parent | MEGs: ≥ 90% maternal reads PEGs: ≥ 70% paternal reads < 6 paternally derived reads at 0 DAP | Moderate MEGs: > 90% maternal reads PEGS: > 60% paternal reads. Strong MEGs/PEGs: > 90% maternal/paternal and p < 0.01 | ≥ 83% reads from one parent (reduced criteria: Chi-Square p < 0.05) |
| Filtering out potentially contaminating transcripts derived from the seed coat | Expression SLR in endosperm ≥ 3x seed coat and SLRs in vegetative tissues < 5 (less strigent criteria for low expressed genes) | MEGs expression in endosperm ≥ 2x higher than in the seed coat | Expression levels in endosperm < 4x than LCM-disected endosperm | Expression levels < 2x higher in the seed coat than embryo or endosperm | n/a | n/a | n/a | B73 expression atlas | n/a |
Abbreviations: DAP, days after pollination; FDR, false discovery rate; LCM, Laser Capture Microdissection; MEG, maternally expressed imprinted gene; PEG, paternally expressed imprinted gene; SNP, single-nucleotide polymorphism; SLR, signal log ratio; SRA, short read archive (http://www.ncbi.nlm.nih.gov/sra).
Analysis of Allelic Bias Using edgeR Outperforms other Methods
It was previously noticed that a large part of the differences between publications is owed to different statistical pipelines to call imprinted genes. When the two Arabidopsis datasets that analyzed the same accessions and a similar developmental stage and tissue (Gehring and Hsieh datasets) were analyzed in the same way, the overlap increased substantially (from 14 to 56 MEGs and from 6 to 18 PEGs28). Therefore, we created a statistical pipeline to identify genes with statistically significant allelic imbalance from the expected, endosperm-specific 2:1 ratio. Our pipeline is based on edgeR45 and analyzes counts by a generalized linear model (GLM), based on a negative binomial distribution in a paired design (parentals of the same cross) with two or more biological replicates (or reciprocal crosses). Importantly, edgeR models count overdispersion as shown exemplarily for the Pignatta dataset (Supplementary Fig. S1).
We then tested the performance of our statistical pipeline compared to other methods based on synthetic data, where we could control the settings and the true genomic imprinting status of each gene. We simulated counts using negative binomial distributions, with mean and dispersion parameters estimated from real data (see Material and Methods). Imprinted genes were introduced by adding 200 genes with a parent-of-origin-specific allelic bias: 50 MEGs each with strong (99% maternal reads) or moderate (85% maternal reads) allelic bias, as well as 50 PEGs each with strong (34% maternal reads) or moderate (48% maternal reads) allelic bias. Random allelic read sampling was modeled by sampling from a binomial distribution.
We evaluated the performance of three different methods: our edgeR-based pipeline, Fisher’s exact test, and Stouffer’s method. Fisher’s exact test is the analysis method used in most published plant studies where a Fisher’s exact test is performed with allelic counts of summed reciprocal crosses. From now on this method will be called “Fisher-summed”. Stouffer’s method49 is a method to combine p-values bearing the same null hypothesis. We combined the two p-values per gene calculated by Fisher’s exact tests comparing to expectations for each reciprocal cross separately, and calculated a combined adjusted p-value using Stouffer’s method. From now on this method will be called “Fisher-combined”.
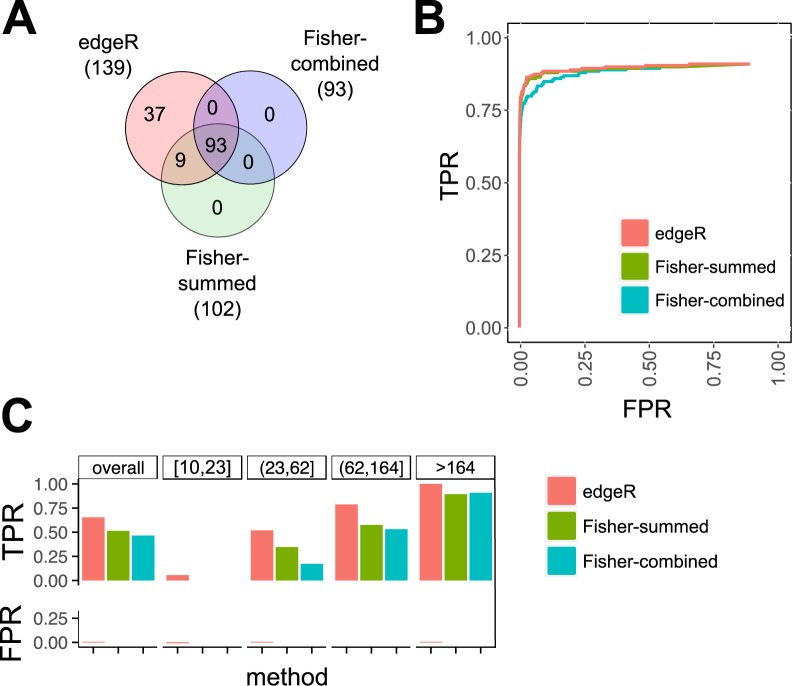
In the simulation setting, our pipeline (edgeR) identified the largest number of observed true positives (spiked-in MEGs and PEGs) (Fig. 3A), identifying 131 true positives (of 200, 66% true positive rate [TPR]) with 8 false positives (falsely detected as imprinted), whereas the second-best method, Fisher-summed, identified only 102 true positives (51% TPR) with 0 false positives (0% FPR). Fisher-combined was close third, identifying 93 true positives (47% TPR) with 0 false positives (0% FPR).
Figure 3.
Benchmarking of three tested methods to identify imprinted genes using simulated data. (A) Overlap of detected spike-in imprinted genes between different methods (FDR 5%). (B) ROC curves. (C) True positive rates (TPR) and false positive rates (FPR) across four equally sized categories of genes with increasing expression levels (number of counts) at FDR 5%.
The Venn diagram (Fig. 3A) shows that 93 genes were identified between all three methods, 9 were shared between edgeR and Fisher-summed, 37 genes were only identified by edgeR, and 61 true positive genes were not identified by any method. Considering the genes that were simulated to be imprinted as the true positive group, and the remaining genes as the true negative group, we computed the false positive rate and the true positive rate for all possible score thresholds and constructed a ROC (Receiver Operating Characteristic) curve for each method (Fig. 3B). edgeR and Fisher-summed had a small performance advantage in detecting the spike-in genes over Fisher-combined. We divided the assessed genes into four equally sized bins according to their number of counts. As expected, the true positive rate (TPR) increased with larger counts per gene for all methods (Fig. 3C; Supplementary Fig. S2). TPR was highest for edgeR in all categories with a large advantage in the two categories with the lowest counts, while the other two methods only passed the 50% TPR for the two quartiles of genes with the largest counts. True positive rates also depended on the degree of allelic imbalance: at a 5% False Discovery Rate (FDR) cut-off, edgeR achieved TPRs of 80% and 54%, respectively, for strong and weak MEGs, and TPRs of 70% and 58%, respectively, for strong and weak PEGs. In contrast, the second best method Fisher-summed achieved TPRs of 74% and 44%, respectively, for strong and weak MEGs, and TPRs of 58% and 28%, respectively, for strong and weak PEGs. Simulations with count distributions similar to the largest observed datasets showed that edgeR still performed better than the other methods (data not shown). EdgeR outperformed the other methods when 200 genes with an accession-specific bias in expression were added to the 200 simulated imprinted genes (data not shown). In addition, the performance of edgeR was robust against varying proportions of simulated strongly and moderately expressed imprinted genes (Supplementary Table S4) and against varying numbers of imprinted genes, as long as the total number did not drop below 50 genes (Supplementary Table S5).
Consistent Reanalysis of Published Imprinting Studies with edgeR Identifies More Common Imprinted Candidate Genes in Arabidopsis but not in Maize
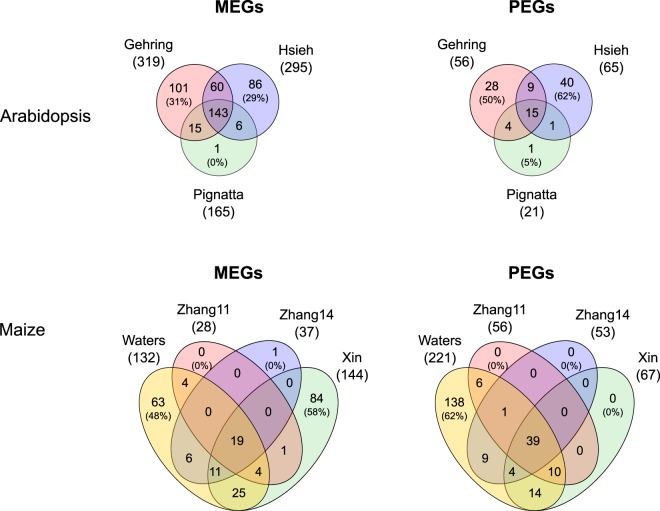
To test our statistical pipeline on real data, we identified genes with statistically significant allelic imbalance from the expected, endosperm-specific 2:1 ratio using edgeR45 with reanalyzed data starting from raw reads. We identified between 165–319 candidate MEGs and 21–65 candidate PEGs for the examined datasets using a 5% FDR cut-off (Fig. 4). Diagnostic plots exemplary for the Pignatta dataset display a good model fit (Supplementary Fig. S3). A list of the called imprinted candidate genes in Arabidopsis and maize can be found in Supplementary Tables S1 and S2, respectively.
Figure 4.
Venn diagrams showing the overlap between imprinted candidate genes across datasets when reanalyzing the raw data using the same standardized method with generalized linear models/edgeR at a FDR cut-off of 5%. Only genes were considered that could be evaluated for imprinting in all datasets. Numbers in brackets denote the percentage of non-shared genes relative to the full set detected in the dataset. The “Xin” and “Zhang11” sets comprise genes identified at 10 days after pollination and “Zhang14” comprises genes at 12 days after pollination.
In Arabidopsis, we identified a larger number of potentially imprinted genes with clearly increased overlaps: 143 common MEGs were found from all three datasets (87% of the smallest dataset [Pignatta], and 45% of the largest dataset [Gehring]) and, in addition, 60 MEGs (20% of the smaller dataset) were shared by the Gehring and Hsieh datasets (Fig. 4). Now only 31% and 29% of identified MEGs were unique to the Gehring and Hsieh datasets, respectively. In the Pignatta dataset, only a single MEG (1/165, 0%) and a single PEG (1/21, 5%) were not shared with any other dataset, potentially owing to the fact that (i) the Pignatta dataset had the largest sequencing depth and therefore the largest gene coverage, and (ii) three biological replicates per cross could be analyzed, allowing the identification of high-confidence candidates with a small number of false positives. We also identified an almost three-fold increased number of PEGs (98 instead of 35 genes as originally published), although with an increased proportion of non-shared PEGs in the Hsieh dataset (from 17% [1/6] to 62% [40/65]).
In maize, we identified a large number of imprinted candidate genes, 218 MEGs and 221 PEGs (Fig. 4). 70 MEGs (32%) and 83 PEGs (38%) were identified from at least two different datasets, leading to an overlap of MEGs similar to the one found in the originally published lists (Fig. 2). The number of candidate imprinted genes varied a lot between datasets, limiting the maximum number of genes shared by all four datasets. The proportion of non-shared MEGs and PEGs decreased for all datasets, except for Waters MEGs and PEGs, which increased from 22% to 48% and from 36% to 62%, respectively, likely due to the significant increase in the number of imprinted candidate genes (Fig. 4). In contrast, the Zhang datasets, as well as the PEGs identified from the Xin dataset, showed almost no exclusive candidate imprinted genes.
Furthermore, we identified the top50 imprinted candidate genes, which are statistically most significant for each dataset and compared the number of shared genes (Supplementary Fig. S4). Between 17–28 candidate genes were common to all compared datasets and the proportion of non-shared candidates decreased or was similar as in the originally published lists except for the Hsieh, Pignatta, and Xin PEGs.
In summary, a reanalysis of the datasets using our edgeR-based pipeline produced gene lists with a clearly larger overlap in Arabidopsis, despite identifying a larger number of imprinted candidate genes with less pronounced allelic imbalance. Thus, the Jaccard similarity indices between Arabidopsis datasets were higher for MEGs and identical for PEGs after reanalysis with edgeR compared to the originally published gene lists (Supplementary Table S6). For maize, we saw an increase in dataset concordance after reanalysis for both MEGs and PEGs.
Reanalysis Using the edgeR Analysis Pipeline Identifies Novel Imprinted Candidate Genes in Comparison to the Original Analysis
Having identified imprinted candidate genes using our new analysis pipeline, we compared the candidate genes pairwise with the lists from the original publications (Fig. 5). When comparing with our candidate MEGs and PEGs at a 5% FDR cut-off, 0–71% of previously published imprinted candidate genes were also identified by our pipeline.
Figure 5.
Pairwise comparison of imprinted genes between the originally published analysis and the reanalysis using generalized linear models and edgeR with a 5% FDR cut-off. Only genes were considered that could be evaluated for imprinting in all datasets. Numbers in brackets denote the percentage of non-shared genes relative to the full set detected in the dataset. For the “Xin” dataset only one timepoint (10 days after pollination) is shown. The “Zhang11” set comprises genes identified at 10 days after pollination and “Zhang14” comprises genes expressed at 12 days after pollination.
From the reanalysis we selected the genes with topmost significance to get the equivalent number of the previously published imprinted candidate genes and compared them to the original publications. Arabidopsis PEGs and maize MEGs and PEGs generally overlapped at 44–85% between the reanalysis and the original gene lists (Supplementary Fig. S5), much higher than for Arabidopsis MEGs, where often only negligible overlaps were observed.
In conclusion, the reanalysis with a standardized analytical pipeline identifies a large number of similar candidates but also additional novel candidate genes. Furthermore, it fails to identify previously called imprinted candidates, likely owing to the improved statistical analysis, which takes count overdispersion into account and neglects large allelic biases in transcripts covered by a low number of reads.
Consistent Reanalysis Using Fisher’s Exact Test Identifies Fewer Overlapping Imprinted Candidate Genes
We also reanalyzed the data from the seven studies using the “Fisher-summed” method in order to assess its performance with real data. We selected the genes with topmost significance to get for each dataset the equivalent number of the previously published imprinted candidate genes (Supplementary Fig. S6). A proportion of maternal reads of ≥85% and ≤50% was required for candidate MEGs and PEGs, respectively. In Arabidopsis, the number of genes identified from all three datasets (27 MEGs and 1 PEGs; Supplementary Fig. S6) was only a fraction of those identified after reanalysis with edgeR (41 MEGs and 5 PEGs; Supplementary Fig. S7). Also, the number of genes shared between at least two different datasets (63 MEGs and 10 PEGs; Supplementary Fig. S6) was smaller than after reanalysis with edgeR (70 MEGs and 19 PEGs; Supplementary Fig. S7). The proportion of non-shared genes was 29–68% per dataset, much higher than after reanalysis with edgeR, where proportions of non-shared genes were 4–32%. A reanalysis of the maize datasets using the “Fisher-summed” method produced a similar concordance between datasets as edgeR (Supplementary Figs S6 and S7). Accordingly, Jaccard similarity indices between datasets were markedly higher in Arabidopsis after reanalysis with edgeR for MEGs compared with the Fisher-summed reanalysis or the originally published gene lists (Supplementary Table S6). For PEGs, Jaccard similarity indices after reanalysis with edgeR were higher than after Fisher-summed reanalysis and identical with the originally published gene lists.
Power of Detecting Imprinted Genes Is Relatively Small due to Non-saturating Sequencing Depth
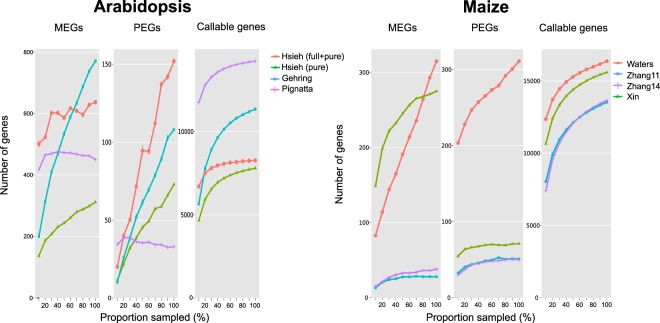
The power-sample size relationship in allelic imbalance detection is not well understood as it depends mainly on the degree of allelic bias, the sequencing read length and coverage (expression strength), but also on the detection procedure. We show that sequencing depth is far from being saturated, as shown by random subsampling of each dataset and detecting MEGs and PEGs by our edgeR-based pipeline (Fig. 6). For most datasets, the number of detected MEGs and PEGs do flatten to some extent with increasing sampling proportions. The low numbers of detectable genes in the Zhang 11 and Zhang 14 are likely due to low mapping rates and low sequencing depth. The relatively flat slopes for the total number of callable genes indicate that a huge increase in sequencing depth would be required to assess the remaining 4–10% genes that were expressed, but did not reach the minimal read coverage of 10 reads.
Figure 6.
Saturation curves showing numbers of detected MEGs and PEGs from various datasets. The curves were generated by randomly sampling increasing proportions of each dataset and identifying imprinted candidate genes using the same pipeline. Values are means (and standard errors) of 10 random subsamples.
Discussion
Reanalysis of Imprinting Studies Reveals that Biological and Technical Differences Strongly Contribute to Biases in Identifying Imprinted Genes
Published studies strongly disagree in the extent and composition of imprinted genes in the endosperm (Fig. 2). We reanalyzed both simulated and real data from seven genome-wide imprinting studies in Arabidopsis and maize, using a standardized bioinformatics pipeline based on generalized linear models. In Arabidopsis, our analysis identified an increased number of imprinted genes co-identified by at least two different datasets, particularly for MEGs where the overlap between datasets was strikingly larger (Fig. 4 and Supplementary Fig. S4). In maize, consistent reanalysis identified a slightly increased number of imprinted genes shared between studies both for MEGs and PEGs. For MEGs, it is conceivable that the increased concordance is partially due to the fact that we did not filter out potential seed coat contamination in the reanalysis. However, as it has previously been shown that most imprinted genes in the embryo are also expressed in the seed coat50, such a filter could also eliminate truly imprinted genes.
It was previously proposed that the discrepancies might at least in part stem from different data analysis pipelines to call imprinted genes and, in fact, using similar filtering conditions produced more overlap28,34. However, even after consistent reanalysis, a high number of imprinted candidate genes are unique to a single dataset, and reanalysis is able to increase the overlap across datasets only by a limited degree. This suggests that biological variation (such as different environmental growth condition, different developmental seed stage, stochastic allelic expression differences), technical differences (e.g., library preparation/complexity, batch effects41,42), and differences in sequencing depth inherent to the datasets contribute to these discrepancies and cannot be corrected for in silico.
Particularly in maize, reanalysis did not increase the concordance between datasets. Even though the developmental seed stages did vary to some extent (Table 1; 10–14 DAP), we cannot fully explain this finding by biological variation only. All original analyses used the same statistical test and highly similar conditions for allelic bias filtering. Possibly, the published original analyses of maize datasets used filtering conditions close to the optimum, such that edgeR-based reanalysis could not further improve concordance. Furthermore, the co-identification of imprinted genes is additionally hampered by unequal read coverage: 48 of 129 imprinted genes proposed by Zhang and colleagues33 had too few reads to characterize imprinted expression in the data of Waters and colleagues31. The relative importance of biological versus technical biases on the large discrepancy between the datasets from maize is currently difficult to assess.
A Statistical Approach that Uses Generalized Linear Models and Takes Replicate Information into Account Largely Increases Sensitivity
Our statistical approach involves modeling of count data and is quite different from the approach chosen by the authors in the original publications. Our approach relies on generalized linear models and edgeR, which is a commonly used, well-established and robust method for differential expression analysis of RNA-seq data. By interpreting allelic counts of a cross as separate samples and imprinting analysis as a differential gene expression problem, we benefit from the power and flexibility of generalized linear models based on edgeR. The method is highly flexible, i.e. also batch effects can be modeled. The general methodology used here is also applicable to imprinting analysis in other tissues, where a deviation from 1:1 is tested. A further advantage of our statistical approach is that it circumvents somewhat arbitrary minimum cut-offs of effect size (allelic imbalance), which would require an individual optimization for each dataset. Most genes are only partially imprinted and we believe that the current knowledge does not support the exclusion of genes with moderate allelic imbalance yet reaching statistical significance. Our approach also allows (nearly) genome-wide ranking of genes according to their likelihood of being imprinted, allowing downstream applications, such as gene set enrichment analysis, with increased statistical power (e.g. for Gene Ontology analysis).
Prior to read counting, many steps are required to map, filter, and count allelic reads. To assess the importance of the preprocessing/counting relative to statistics, we reanalyzed the original allelic counts from28 using our method. We found a large discrepancy for MEGs and PEGs with original gene lists, whereas MEGs and PEGs largely agreed with the gene lists obtained from complete reanalysis with our analysis pipeline (data not shown). Given the large discrepancy, the statistical approach seems to have a larger relative contribution than the earlier steps required to obtain the variant counts.
Simulation Reveals that edgR-based Analysis Outperforms Other Methods
The edgeR-based approach performed well in our simulation setting and clearly outperformed the other methods we tested. Notably, the other methods based on Fisher’s exact test seem overly conservative and missed most spike-in imprinted genes. edgeR was the only method to predict false positives due to a relatively poor FDR control for the lowest quartile of counts (10–23 counts per gene) where 4 of edgeR’s totally 8 false positive imprinted genes were identified. edgeR’s specificity could be further increased by selecting a larger minimal number of counts per gene (e.g., a cut-off of 20 allelic counts per gene). In our simulation setting, edgeR’s sensitivity was highest at a minimal cut-off of 10 or 20 reads with TPRs of 0.66 and 0.65, respectively, but dropped markedly at a cut-off of 50 reads with a TPR of 0.54 (Supplementary Table S7).
Importantly, our simulation did not aim to model biological variability predicting biological replicates. All biological systems have inherent biological variation and edgeR can account for it, whereas Fisher’s exact test completely ignores the within-condition variability, as it requires counts from replicates to be summed up for each condition. Therefore, we expect that edgeR would outperform methods using Fisher’s exact tests even more strikingly when biological replicates are available.
Higher Coverage, Replicate Samples, and edgeR-based Analysis Could Improve the Identification of Imprinted Genes
The first generation of genome-wide studies identified many new imprinted genes in plants, yet a considerable proportion of genes could not be characterized due to low sequencing depth or insufficient genetic heterogeneity between the parents. Future experiments in genomic imprinting will be performed using paired-end reads with high sequencing coverage. With the rapid development in NGS, higher coverage is now readily achievable, which will notably increase the statistical power to detect imprinted genes. In the studies reanalyzed here, low counts were observed for many genes close to the minimal coverage cut-off of 10, where the variance is large and a large allelic bias is required to reach significance. Importantly, a sufficient number of biological replicates (i.e., at least three per reciprocal cross51) would permit to reliably estimate the variability from the data, enabling the performance of a more robust differential expression analysis and a more reliable estimation of the total number of imprinted genes.
Biological Perspectives
When comparing datasets from different studies, we assume that the same set of genes is imprinted over the sampling time period. Partial or complete violations of the assumption also decrease the amount of overlap across datasets in addition to technical and analytic biases. Indeed, first studies showed dynamic expression of imprinted genes in maize36. Furthermore, we cannot solely rely on statistical significance in calling a gene imprinted or not. There are several ways to prioritize a gene list after statistically calling genes with an allelic bias in a given tissue. First, filtering the gene lists by fold difference between the two parental alleles could help to identify genes that can more easily be validated experimentally. Second, filtering the gene lists against tissue-specific expression data from seeds might identify genes relevant to the tissue of interest52,53. However, expression of a given gene in other (e.g. maternal seed coat) tissues does not exclude an allelic bias in the fertilization products50. Third, comparing the gene list against central cell and sperm cell expression data54–56 can inform to what extent the allelic bias is a result of expression in the fertilization products or might at least partially represent carry-over of gametic transcripts that were produced prior to fertilization.
An improved assessment including larger gene sets (by using different sets of strains), as well as imprinted genes with moderate allelic imbalance, will provide further insights into the extent and biological significance of genomic imprinting. Having the full catalog of imprinted genes in several plant species will also allow the tackling of evolutionary questions about genomic imprinting, including its origin and fixation, the conservation of imprinted genes, and their gain and loss of imprinting status on the phylogenetic tree.
Lastly, we have to stress that without further confirmation, gene lists are not representative with regard to the absolute number of imprinted candidate genes expressed in the endosperm. Considering this, it is indispensable to confirm bioinformatically identified imprinted candidate genes by alternative methods, such as allele-specific expression analysis using RT-PCR and Sanger sequencing, pyrosequencing, and/or reporter gene assays.
Conclusions
A new, effective data analysis pipeline is reported that allows for an improved analysis of RNA-seq data from reciprocal F1 crosses. The pipeline allows for the modelling of biological replicates and is applicable to any diploid or triploid tissue. Furthermore, our genome-wide ranking of Arabidopsis and maize imprinted candidate genes, which integrates all available datasets, provides a useful resource to inform future experiments focused on understanding genomic imprinting in plants.
Methods
Read Mapping and Counting
FASTQ-formatted raw reads were downloaded from the NCBI Short Read Archive (SRA) for endosperm experiments for Arabidopsis (GSM674847, GSM674848, GSM756822, GSM756824, GSM607727, GSM607728, GSM607732, GSM607735, GSM1276498, GSM1276500, GSM1276502, GSM1276504, GSM1276505, GSM1276508, GSM1276509, GSM1276512, GSM1276514, GSM1276515) and for maize (SRX105679, SRX105678, SRX114629, SRX114630, SRX047539, SRX047544, SRP031872, GSE48425). Hsieh samples obtained through laser capture microdissection were not included in the analysis as their inclusion reduced the overlap with other datasets (data not shown). Reads were quality-checked with the FastQC application (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). To reduce the bias in mapping reads towards reference alleles, we aligned the reads to a masked reference genome, in which bases at known polymorphic sites were replaced with “N”. Reads were mapped to the genome using STAR v2.3.0e57. Only reads that mapped to a unique position in the genome were considered for further analysis.
For Arabidopsis the genome annotation TAIR10 (http://www.arabidopsis.org/) was used, and for maize AGPv3 annotated genes were downloaded from Ensembl Plants Release 31 (http://plants.ensembl.org/)58. Arabidopsis SNP annotation files were obtained from the 1001 Genomes project59 http://1001genomes.org/data/MPI/MPISchneeberger2011/releases/2012_03_14/). Maize hapmap v3.2.1 SNP variants60 in VCF format (Release date 3/3/2016) were obtained from the Panzea database (http://cbsusrv04.tc.cornell.edu/users/panzea/download.aspx?filegroupid=15) and filtered for high-confidence (flag LLD) biallelic SNP variants, which were polymorphic between B73 and Mo17 and homozygous in both inbred strains. Allelic reads were counted at previously identified SNP positions between homozygous parentals, using Python v3.2 with pysam v0.8.461, while positions with low sequencing quality (phred quality <20) were excluded. No more than one SNP was counted per read to prevent pseudo-replication. Counts were summed up per gene and variants with less than 10 reads (summed across the two reciprocal crosses) were discarded.
Testing for Allele-specific Expression
To assess allele-specific expression, we used edgeR version 3.4.245. It uses an empirical Bayes estimation based on the negative binomial distribution. For library size normalization and to eliminate composition biases between libraries, we used the TMM (Trimmed Mean of M-values) method. TMM normalization keeps the ratio between maternal and paternal allelic reads in a cross at approx. 2 (data not shown). Experiments were analyzed using a generalized linear model with a paired design (the two allelic counts of the same cross treated as paired samples) and at least two biological replicates (the two reciprocal crosses). When biological replicates were available, they were included in the model. We used tagwise dispersion estimates. False Discovery Rate (FDR) was calculated according to Benjamini and Hochberg62. The R software version 3.0.363 was used for statistical analysis and for creating the graphs.
Simulation of genomic imprinting
To generate synthetic data we used the function makeExampleDESeqDataSet of the DESeq2 version 1.2.1046 R/Bioconductor package. Mean and dispersion parameters that were used in the simulation were estimated from real RNA-seq data (interceptMean = 2, interceptSD = 3). Two biological replicates were simulated to serve as the two reciprocal crosses. No outlier counts and differential expression were introduced. The total number of genes in each simulated dataset was 15,000, and their true proportion of maternal reads was set to 2/3. Then we randomly picked 200 genes and modified their imprinting status, 50 each of strong MEGs (99% maternal reads), weak MEGs (85% maternal reads), strong PEGs (34% maternal reads) and weak PEGs (48% maternal reads). Random allelic read sampling was modeled by sampling from a binomial distribution with the probability of success set to the true proportion of maternal reads.
We evaluated three methods for identifying genes with parent-of-origin-specific expression: edgeR, Fisher’s exact test, and Stouffer’s method. Fisher’s exact test does two separate statistical tests for the two reciprocal crosses, and the resulting two p-values per gene were combined with Stouffer’s method (calculated by sumz function of R package metap). Benchmarking was performed using Venn diagrams and Receiver Operating Characteristic (ROC) curves with iCOBRA64 https://github.com/markrobinsonuzh/iCOBRA.
Comparison with Published Gene Lists
Published gene lists were compiled from the Supplementary Data of the respective publications. For the Waters B73xMo17 comparison, the updated list from35 was used, not the original one31. If lists of imprinted genes were available at various stringencies, we used the least stringent list (e.g. moderately imprinted genes for the Waters dataset). Genes with an accession/inbred-specific bias in expression were omitted. The Zhang14 dataset comprised only the lists of 106 MEGs and 91 PEGs at 12DAP (Prof. Xiaomei Lai, personal communication) described in37 but not the endosperm samples at 10DAP which were already described in33 and contained in the Zhang11 dataset. Genes that could not be evaluated for imprinting in all datasets were filtered out.
Saturation Plots
In order to assess the degree of undersampling, we performed random subsampling on the count data and performed the same processing and statistical analysis steps as for the full data.
Electronic supplementary material
Acknowledgements
We thank Malgorzata Nowicka and Mark Robinson (Department of Molecular Life Sciences, University of Zurich), as well as Marc W. Schmid (Department of Plant and Microbial Biology, University of Zurich) for helpful discussions. This work was supported by the University Research Priority Program ‘Evolution in Action’ of the University of Zurich and, in part, by an Advanced Grant of the European Research Council (ERC AdG Nr. 243996) to U.G.
Author Contributions
S.W., M.T.R., and U.G. designed the study, U.G. raised money and supervised the study, S.W. performed the analyses, and all authors wrote the paper. All authors read and approved the final manuscript.
Data Availability
Scripts and data used in this manuscript are available on github (http://www.github.com/swyder/Reanalysis_plant_imprinting) and as supplementary files.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36768-4.
References
- 1.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 2.Grossniklaus, U. Genomic Imprinting in Plants: A Predominantly Maternal Affair in Plant Epigenetics (ed. Meyer, P.) 174–200 (Blackwell Publishing Ltd, 2007).
- 3.Bartolomei, M. S. & Ferguson-Smith, A. C. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 3, 10.1101/cshperspect.a002592 (2011). [DOI] [PMC free article] [PubMed]
- 4.Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- 5.Barlow DP. Genomic imprinting: a mammalian epigenetic discovery model. Annu Rev Genet. 2011;45:379–403. doi: 10.1146/annurev-genet-110410-132459. [DOI] [PubMed] [Google Scholar]
- 6.Raissig MT, Baroux C, Grossniklaus U. Regulation and flexibility of genomic imprinting during seed development. Plant Cell. 2011;23:16–26. doi: 10.1105/tpc.110.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang H, Köhler C. Evolution, function, and regulation of genomic imprinting in plant seed development. J Exp Bot. 2012;63:4713–4722. doi: 10.1093/jxb/ers145. [DOI] [PubMed] [Google Scholar]
- 8.Gehring M. Genomic imprinting: insights from plants. Annu Rev Genet. 2013;47:187–208. doi: 10.1146/annurev-genet-110711-155527. [DOI] [PubMed] [Google Scholar]
- 9.Messing J, Grossniklaus U. Genomic imprinting in plants. Results Probl Cell Differ. 1999;25:23–40. doi: 10.1007/978-3-540-69111-2_2. [DOI] [PubMed] [Google Scholar]
- 10.Baroux C, Spillane C, Grossniklaus U. Genomic imprinting during seed development. Adv Genet. 2002;46:165–214. doi: 10.1016/S0065-2660(02)46007-5. [DOI] [PubMed] [Google Scholar]
- 11.Feil R, Berger F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007;23:192–199. doi: 10.1016/j.tig.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Köhler C, Weinhofer-Molisch I. Mechanisms and evolution of genomic imprinting in plants. Heredity. 2010;105:57–63. doi: 10.1038/hdy.2009.176. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez-Marcos JF, Constância M, Burton GJ. Maternal to offspring resource allocation in plants and mammals. Placenta. 2012;33(Suppl. 2):e3–10. doi: 10.1016/j.placenta.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Pires ND, Grossniklaus U. Different yet similar: evolution of imprinting in flowering plants and mammals. F1000prime Rep. 2014;6:63. doi: 10.12703/P6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues JA, Zilberman D. Evolution and function of genomic imprinting in plants. Genes Dev. 2015;29:2517–2531. doi: 10.1101/gad.269902.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haig D, Westoby M. Parent-specific gene expression and the triploid endosperm. Am. Nat. 1989;134:147–155. doi: 10.1086/284971. [DOI] [Google Scholar]
- 17.Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 18.Lau MM, et al. Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev. 1994;8:2953–2963. doi: 10.1101/gad.8.24.2953. [DOI] [PubMed] [Google Scholar]
- 19.Ludwig T, et al. Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev Biol. 1996;177:517–535. doi: 10.1006/dbio.1996.0182. [DOI] [PubMed] [Google Scholar]
- 20.Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita T, Yadegari R, Harada JJ, Goldberg RB, Fischer RL. Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell. 1999;11:1945–1952. doi: 10.1105/tpc.11.10.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiyosue T, et al. Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:4186–4191. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA. 2000;97:10637–10642. doi: 10.1073/pnas.170292997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingouff M, Haseloff J, Berger F. Polycomb group genes control developmental timing of endosperm. Plant J. 2005;42:663–674. doi: 10.1111/j.1365-313X.2005.02404.x. [DOI] [PubMed] [Google Scholar]
- 25.Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol. 2002;192:245–258. doi: 10.1002/jcp.10129. [DOI] [PubMed] [Google Scholar]
- 26.Monk D. Genomic imprinting in the human placenta. Am. J. Obstet. Gynecol. 2015;213:S152–162. doi: 10.1016/j.ajog.2015.06.032. [DOI] [PubMed] [Google Scholar]
- 27.Autran D, et al. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell. 2011;145:707–719. doi: 10.1016/j.cell.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 28.Gehring M, Missirian V, Henikoff S. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One. 2011;6:e23687. doi: 10.1371/journal.pone.0023687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsieh T-F, et al. Inaugural Article: Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci USA. 2011;108:1755–1762. doi: 10.1073/pnas.1019273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKeown PC, et al. Identification of imprinted genes subject to parent-of-origin specific expression in Arabidopsis thaliana seeds. BMC Plant Biol. 2011;11:113. doi: 10.1186/1471-2229-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waters AJ, et al. Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell. 2011;23:4221–4233. doi: 10.1105/tpc.111.092668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolff P, et al. High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 2011;7:e1002126. doi: 10.1371/journal.pgen.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M, et al. Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc Natl Acad Sci USA. 2011;108:20042–20047. doi: 10.1073/pnas.1112186108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pignatta D, Gehring M. Imprinting meets genomics: new insights and new challenges. Curr Opin Plant Biol. 2012;15:530–535. doi: 10.1016/j.pbi.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Waters AJ, et al. Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc Natl Acad Sci USA. 2013;110:19639–19644. doi: 10.1073/pnas.1309182110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xin M, et al. Dynamic expression of imprinted genes associates with maternally controlled nutrient allocation during maize endosperm development. Plant Cell. 2013;25:3212–3227. doi: 10.1105/tpc.113.115592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang M, et al. Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize. Genome Res. 2014;24:167–176. doi: 10.1101/gr.155879.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pignatta D, et al. Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. eLife. 2014;3:e03198. doi: 10.7554/eLife.03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Köhler C, Wolff P, Spillane C. Epigenetic mechanisms underlying genomic imprinting in plants. Annu Rev Plant Biol. 2012;63:331–352. doi: 10.1146/annurev-arplant-042811-105514. [DOI] [PubMed] [Google Scholar]
- 40.DeVeale B, van der Kooy D, Babak T. Critical evaluation of imprinted gene expression by RNA-Seq: a new perspective. PLoS Genet. 2012;8:e1002600. doi: 10.1371/journal.pgen.1002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Clark AG. Using next-generation RNA sequencing to identify imprinted genes. Heredity. 2014;113:156–166. doi: 10.1038/hdy.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castel SE, Levy-Moonshine A, Mohammadi P, Banks E, Lappalainen T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2015;16:195. doi: 10.1186/s13059-015-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregg C, et al. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Y-H, Xia K, Wright FA. A powerful and flexible approach to the analysis of RNA sequence count data. Bioinformatics. 2011;27:2672–2678. doi: 10.1093/bioinformatics/btr449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stouffer, S. A. The American Soldier: Adjustment During Army Life. (Princeton University Press, 1949).
- 48.Fontanillas P, et al. Key considerations for measuring allelic expression on a genomic scale using high-throughput sequencing. Mol. Ecol. 2010;19(Suppl. 1):212–227. doi: 10.1111/j.1365-294X.2010.04472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Degner JF, et al. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25:3207–3212. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raissig MT, Bemer M, Baroux C, Grossniklaus U. Genomic imprinting in the Arabidopsis embryo is partly regulated by PRC2. PLoS Genet. 2013;9:e1003862. doi: 10.1371/journal.pgen.1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Zhou J, White KP. RNA-seq differential expression studies: more sequence or more replication? Bioinformatics. 2014;30:301–304. doi: 10.1093/bioinformatics/btt688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belmonte MF, et al. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA. 2013;110:E435–444. doi: 10.1073/pnas.1222061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schon MA, Nodine MD. Widespread Contamination of Arabidopsis Embryo and Endosperm Transcriptome Data Sets. Plant Cell. 2017;29:608–617. doi: 10.1105/tpc.16.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borges F, et al. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 2008;148:1168–1181. doi: 10.1104/pp.108.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wuest SE, et al. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol. 2010;20:506–512. doi: 10.1016/j.cub.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 56.Schmid MW, et al. A powerful method for transcriptional profiling of specific cell types in eukaryotes: laser-assisted microdissection and RNA sequencing. PLoS One. 2012;7:e29685. doi: 10.1371/journal.pone.0029685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monaco MK, et al. Gramene 2013: comparative plant genomics resources. Nucleic Acids Res. 2014;42:D1193–1199. doi: 10.1093/nar/gkt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 2011;43:956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 60.Bukowski, R. et al. Construction of the third generation Zea mays haplotype map. Preprint at, https://www.biorxiv.org/content/early/2015/09/16/026963 (2015). [DOI] [PMC free article] [PubMed]
- 61.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 63.R Core Team. R: A Language and Environment for Statistical Computing, http://www.r-project.org (2012).
- 64.Soneson C, Robinson MD. ICOBRA: open, reproducible, standardized and live method benchmarking. Nat. Methods. 2016;13:283. doi: 10.1038/nmeth.3805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Scripts and data used in this manuscript are available on github (http://www.github.com/swyder/Reanalysis_plant_imprinting) and as supplementary files.