Abstract
Extracellular adenosine 5′‐triphosphate (eATP) is an essential signaling molecule that mediates different cellular processes through its interaction with membrane‐associated receptor proteins in animals and plants. eATP regulates plant growth, development, and responses to biotic and abiotic stresses. Its accumulation in the apoplast induces ROS production and cytoplasmic calcium increase mediating a defense response to invading microbes. We show here that perception of extracellular nucleotides, such as eATP, is important in plant–fungus interactions and that during colonization by the beneficial root endophyte Serendipita indica eATP accumulates in the apoplast at early symbiotic stages. Using liquid chromatography–tandem mass spectrometry, and cytological and functional analysis, we show that S. indica secrets SiE5′NT, an enzymatically active ecto‐5′‐nucleotidase capable of hydrolyzing nucleotides in the apoplast. Arabidopsis thaliana lines producing extracellular SiE5′NT are significantly better colonized, have reduced eATP levels, and altered responses to biotic stresses, indicating that SiE5′NT functions as a compatibility factor. Our data suggest that extracellular bioactive nucleotides and their perception play an important role in fungus–root interactions and that fungal‐derived enzymes can modify apoplastic metabolites to promote fungal accommodation.
Keywords: DORN1, extracellular nucleotides, lectin receptor kinase LecRK‐I.9, Piriformospora indica, purinergic signaling
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Plant Biology
Introduction
ATP is a coenzyme that serves as a universal energy currency and as a building block of nucleic acids and secondary metabolites. Additionally, ATP and adenosine are important extracellular regulators in plants and animals 1, 2, 3, 4, 5. In plants, it has been shown that cells release ATP during growth, physical injury, and in response to various abiotic and biotic stresses and microbe‐derived elicitors 4, 5, 6, 7, 8, 9, 10, 11, 12, 13. Accumulation of extracellular ATP (eATP) triggers the production of reactive oxygen species (ROS), nitric oxide, callose deposition, cytoplasmic calcium increase, transient phosphorylation of MPK3 and MPK6, and expression of genes involved in plant stress response and immunity 8, 14. To be physiologically relevant, eATP must be sensed by specific receptors at the cell surface. Recently, the Arabidopsis thaliana lectin receptor kinase DORN1 (DOes not Respond to Nucleotides 1, At5g60300), also known as the LecRK‐I.9 (lectin receptor kinase I.9), was identified as the first plant purinoceptor essential for the plant response to eATP, unveiling an eATP signaling pathway in plants 8. The RXLR‐dEER effector protein IPI‐O secreted by the oomycete pathogen Phytophthora infestans targets LecRK‐I.9/DORN1 15, 16, 17, 18, suggesting that some microbes have the tools to manipulate host eATP perception/signaling. LecRK‐I.9/DORN1 mutant plants show enhanced susceptibility to pathogen infections such as the oomycete P. brassicae and the bacterium Pseudomonas syringae (Pst). Consistently, overexpression of LecRK‐I.9/DORN1 increases plant resistance to Phytophthora spp. and Pst 19. Taken together, these results suggest a role for an eATP‐receptor protein in plant immunity.
There is increasing evidence that fungi, both pathogenic and mutualistic, have large repertoires of secreted effectors. Some were functionally shown to be able to manipulate the host cell physiology, suppress plant defense, and ultimately promote fungal colonization 20. Among fungi, compatibility factors are mainly reported from biotrophic and hemibiotrophic leaf pathogens 21. In contrast, only few factors have been functionally characterized from root mutualistic fungi 22, 23, 24, 25. Therefore, the basis and mode of action of mutualistic fungal compatibility factors remain poorly understood.
The filamentous root endophyte Serendipita indica (syn. Piriformospora indica) belongs to the order Sebacinales (Basidiomycota). Serendipita indica colonizes the root epidermal and cortex cells without penetrating the central cylinder and displays a biphasic colonization strategy 26, 27, 28, 29. During the initial phase of biotrophic colonization, the fungus invades the root cells inter‐ and intracellularly. Subsequently, S. indica switches to a host cell death‐associated phase, although a defined switch to necrotrophy with massive cell death does not occur 26, 27, 29. Serendipita indica colonization exhibits various effects on host plants including enhanced growth, improved assimilation of nitrate and phosphate, increased tolerance to abiotic stresses, and resistance against pathogens 30, 31, 32, 33. Since S. indica establishes symbiotic interactions with a wide range of experimental hosts, including the dicot model plant A. thaliana and the monocot cereal crop Hordeum vulgare (barley), it represents an excellent model system to study the role of extracellular bioactive nucleotides and eATP‐mediated plant responses in the roots of unrelated hosts.
In order to identify S. indica secreted effectors, proteins present in the apoplastic fluid (APF) of colonized barley roots were analyzed at three different symbiotic stages by liquid chromatography–tandem mass spectrometry (LC‐MS/MS). One secreted fungal protein consistently found in the apoplast at all time points is a predicted 5′‐nucleotidase. The gene encoding this enzyme is induced during colonization of both barley and A. thaliana but not in axenic culture. Animal ecto‐5′‐nucleotidases (E5′NTs) have been considered to play a key role in the conversion of AMP to adenosine, counteracting eATP release from stimulated cells and further purinergic signaling together with ecto‐nucleotide pyrophosphatase/phosphodiesterase (E‐NPP), ecto‐nucleoside triphosphate diphosphohydrolase (E‐NTPDase), and alkaline phosphatases (AP) 34, 35. The importance of bioactive nucleotide‐triggered signaling and fungal extracellular E5′NT activity during plant–fungus interactions is unknown. We show here that S. indica E5′NT is capable of hydrolyzing ATP, ADP, and AMP to adenosine and phosphate, altering eATP levels and plant responses to fungal colonization. Considering the important role E5′NT plays in S. indica accommodation at early symbiotic stages, we propose that modulation of extracellular nucleotide levels plays a role in compatibility during plant–fungus interactions.
Results
Identification of fungal proteins in the apoplast of barley roots
In order to identify soluble secreted candidate effector proteins during S. indica root colonization, the proteins present in the APF of barley roots at three different symbiotic stages, 5, 10, and 14 days postinoculation (representing the biotrophic, early, and late cell death‐associated phases) were analyzed together with the proteins found in the culture filtrate (CF) obtained from S. indica axenically grown in liquid complex medium (CM). To assess possible cytoplasmic contaminations, a strain constitutively expressing an S. indica codon‐optimized GFP under the SiGPD promoter 36 was used. The absence of fungal cytoplasmic GFP protein contamination due to cell lysis was corroborated by Western blotting of the apoplastic protein samples using anti‐GFP antibody (Appendix Fig S1).
LC‐MS/MS analysis after tryptic digestion led to the identification of 102 S. indica proteins putatively targeted to the apoplast of which 33 were present at all three time points in at least one of the biological replicates (Table EV1, Dataset EV1 and Fig 1A and B). Predictions using ApoplastP (http://apoplastp.csiro.au/) and SecretomeP (http://www.cbs.dtu.dk/services/SecretomeP-1.0/) indicated that 48 of the 102 S. indica proteins are putatively targeted to the plant apoplast (47%) with nine proteins predicted to be secreted via a non‐canonical secretion pathway (Table EV2). No peptides for GFP were found in any of the apoplastic fluid samples, confirming the GFP Western blotting analysis. Twenty proteins were uniquely identified at 5 dpi, 4 at 10 dpi, and 21 at 14 dpi, suggesting differential secretion of S. indica proteins at different stages of colonization (Table EV1).
Figure 1. Identification of S. indica apoplastic proteins.
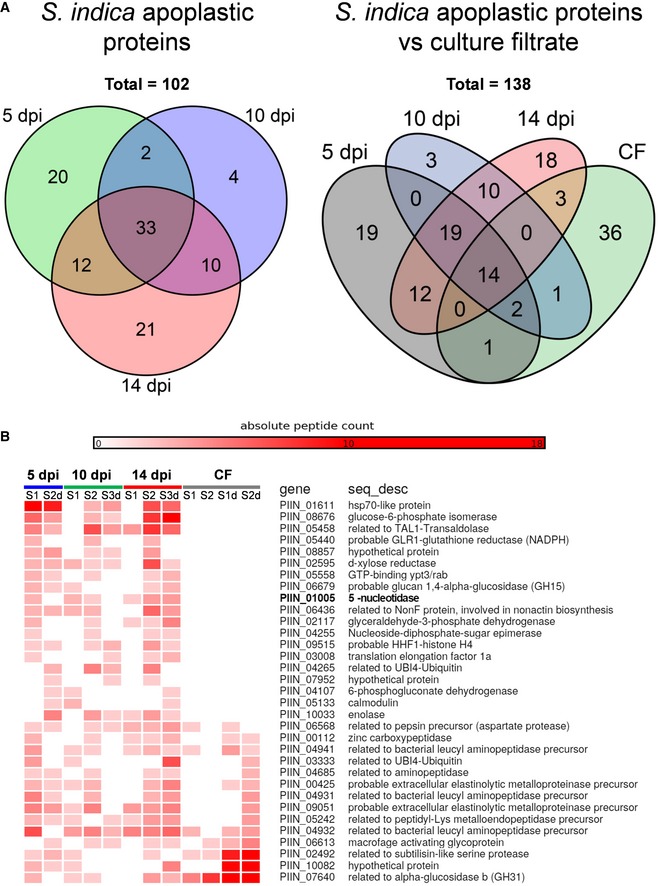
- Distribution of S. indica apoplastic proteins identified by LC‐MS/MS analysis from different symbiotic stages (5, 10, and 14 dpi) and in relation to proteins identified in CF of S. indica axenically grown in CM. In total, 102 S. indica proteins were identified in the APF of colonized barley roots. Of these, 33 proteins were present at all three time points. Twenty proteins were unique at 5 days postinfection (dpi), 4 at 10 dpi, and 21 at 14 dpi.
- Heat map showing absolute counts of unique peptides for S. indica apoplastic proteins present at all three time points and in culture filtrate. S1/S2/S3 = biologically independent samples 1, 2, and 3; d = deglycosylated.
Twenty‐one of the 102 identified apoplastic proteins were also found in the culture filtrate of S. indica grown in CM, which included a large proportion of enzymes acting on peptide bonds (Fig 1A and B). The majority of these proteases might conceivably play a role in nutrition rather than in suppression of host defense because of the large overlap with the culture filtrate proteome and because amino acid uptake seems to be an active process during root colonization based on transcriptomic data 27, 28, 37. Four peptidases (PIIN_00628, PIIN_02239, PIIN_02952, and PIIN_07002), however, were specifically present in the apoplastic fluid, which makes them good candidate effectors possibly involved in host defense suppression (Table EV1).
Remarkably, 19 of the 33 common proteins found in the APF at all three time points were not present in the CF samples, implying an induced secretion for these proteins during interaction with the plant roots (Table EV1, and Fig 1B). We did not identify any peptides derived from the S. indica bacterial endosymbiont Rhizobium radiobacter (syn. Agrobacterium tumefaciens, syn. Agrobacterium fabrum) 38, 39 in the APF or CF samples, suggesting that R. radiobacter’s contribution to successful symbiosis does not involve accumulation of bacterial proteins in the apoplast of barley.
In a first step for functional interpretation of the resultant fungal protein list from the APF and CF, we assigned Gene Ontology terms 40, 41, 42. GO term enrichment analysis for biological processes showed an over‐representation of GO terms associated with proteolysis in the culture filtrate obtained from axenically grown S. indica, whereas the apoplastic fungal proteins were related to purine metabolism and specifically to ATP metabolic processes (Table EV3, Appendix Figs S2 and S3).
Among the fungal proteins specifically present in the apoplastic fluid of colonized roots, several ATP‐scavenging enzymes, including a 5′‐nucleotidase (PIIN_01005) and a nucleoside diphosphate kinase (PIIN_00784), were identified (Table EV1 and Appendix Figs S2 and S4). Twenty‐one orthologues of the S. indica apoplastic proteins were also found in the APF of rice leaves infected by the rice blast pathogen Magnaporthe oryzae, including an orthologue of the secreted S. indica nucleoside diphosphate kinase (Table EV4 and Appendix Fig S2 asterisks) 43. The presence of fungal‐derived ATP‐scavenging enzymes in the apoplast of colonized plants indicates that extracellular nucleotides may be important during plant–fungus interactions.
eATP plays a role during fungal colonization in barley and in Arabidopsis
The enrichment for GO terms related to ATP metabolic processes among the fungal‐derived apoplastic proteins led us to test whether root colonization by S. indica triggers changes in eATP levels at different symbiotic stages and in different plant species. We used a specific luminescent ATP detection assay to assess eATP levels of Arabidopsis seedlings grown in liquid medium or barley root apoplastic fluid samples both from S. indica‐colonized and from mock‐treated plants at 3, 5, 7, and 10 dpi. It was not possible to extract apoplastic fluid from the roots of Arabidopsis seedlings for these experiments because this young tissue is very delicate and fragile. All our efforts to collect apoplastic fluids ended up with cytoplasmic contaminations (measured by the LC‐MS/MS proteomic approach) in this host. The results show that eATP levels are significantly higher in S. indica‐colonized roots compared to mock‐treated plants, especially at early symbiotic stages where the biotrophic interaction is still predominant (Fig 2A and B). Thus, there is release/accumulation of eATP during S. indica biotrophic colonization of both Arabidopsis and barley.
Figure 2. Serendipita indica colonization leads to increased eATP levels in barley and Arabidopsis roots.
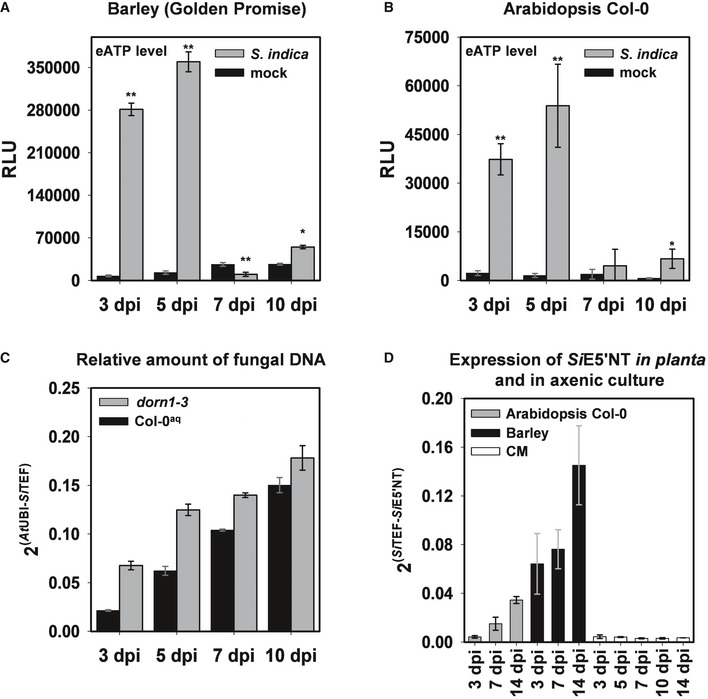
- eATP levels in APF from S. indica‐inoculated and mock‐treated barley root samples collected at the predominantly biotrophic phase (3 and 5 dpi) as well as the predominantly cell death‐associated phase (7 and 10 dpi). Error bars show the standard error of the mean obtained from three independent biological replicates at P < 0.05 (*), 0.01 (**) analyzed by Student's t‐test. RLU: relative light units.
- eATP levels measured from culture medium collected from mock‐treated and S. indica‐colonized Arabidopsis seedlings at 3, 5, 7, and 10 dpi. Error bars represent ±SE of the mean from three independent biological replicates. RLU: relative light units. Asterisks indicate significance at P < 0.05 (*), 0.01 (**) analyzed by Student's t‐test.
- S. indica colonization of Arabidopsis dorn1‐3 mutant and the parental Col‐0aq lines quantified by qPCR at 3, 5, 7, and 10 dpi. The ratio of fungal (SiTEF) to plant (AtUBI) amplicons representing fungal colonization levels in plant root tissue was calculated using gDNA as template and the 2−ΔCT method. Error bars represent standard error of the mean of three technical replicates. The experiment was repeated three times for 3, 5, and 7 dpi with similar outcomes.
- Transcript levels of S. indica E5′NT during colonization of barley and Arabidopsis at different symbiotic stages and in axenic culture. Error bars represent standard error of the mean of three independent biological replicates. CM = complex medium.
To investigate whether eATP detection plays a role during root colonization, we used an eATP‐insensitive A. thaliana mutant, dorn1‐3 defective in the receptor LecRK‐I.9 8. After confirming the inability of the dorn1‐3 mutant to respond to eATP and eADP with cytoplasmic calcium influx (Appendix Fig S5), colonization of this line along with the appropriate aequorin‐expressing Col‐0 control (Col‐0aq) was assessed by qPCR at different time points (3, 5, 7, and 10 dpi). We observed that the dorn1‐3 mutant supports significantly more fungal colonization in comparison with Col‐0aq control, especially at early symbiotic stages (Fig 2C). Importantly, these observations strongly indicate that detection of eATP by the host affects root colonization.
Serendipita indica PIIN_01005 is an ecto‐5′‐nucleotidase capable of hydrolyzing ATP, ADP, and AMP
Since our data show that detection of eATP plays a role during root–S. indica interaction, we hypothesized that this fungus has evolved an intrinsic mechanism to manipulate extracellular nucleotides levels to counteract eATP‐mediated host immune responses. The secreted S. indica PIIN_01005, consistently found in the apoplastic fluid but not in the culture filtrate samples, is a suitable candidate for such a mechanism because of its putative hydrolytic activity on nucleotides. Microarray analysis of fungal transcripts 27, 28, 37 and gene expression experiments on colonized H. vulgare and A. thaliana root tissue showed that PIIN_01005 is induced during root colonization in both experimental hosts but not in CM (Table EV1 and Fig 2D), confirming our results of the LC‐MS/MS analysis. Moreover, the expression of this gene can be induced by the addition of the nucleotides AMP and ATP to the medium (Appendix Fig S6).
Ecto‐5′‐nucleotidases have been found in bacteria, animals, and fungi and display significant differences in the range of substrates hydrolyzed and localization. Studies of mammalian Ecto‐5′‐nucleotidases have shown that a membrane‐bound form and soluble forms exist. A C‐terminal glycosylphosphatidylinositol (GPI) anchor for PIIN_01005 was predicted by the program FragAnchor 44. The presence of this fungal protein in the apoplastic fluid of barley indicates that a soluble form might originate during interaction in planta from cleavage of the anchor by phosphatidylinositol‐specific phospholipase or by proteolytic cleavage as suggested for some animal 5′‐nucleotidases 45, 46, 47. Most fungi have at least one copy of a cytosolic 5′NT without predicted signal peptide and GPI anchor, whereas the secreted ecto‐enzyme members are discontinuously distributed within the fungal kingdom (Fig 3A). E5′NT members are not represented in the available genomes for the Ustilaginomycotina, Pucciniomycotina, and Saccharomycotina and only rarely found in the Eurotiomycetes. However, they are widely distributed in two fungal Ascomycota classes, the Sordariomycetes with 107 from the 121 taxa examined and the Dothideomycetes with 24 from 37 taxa. In the Basidiomycota, E5′NT members are only present in the Agaricomycotina (Fig 3A). These fungal classes are among the largest groups of fungi with a high level of ecological diversity, including many plant and insect pathogens infecting a broad range of hosts.
Figure 3. Serendipita indica PIIN_01005 encodes a secreted ecto‐5′‐nucleotidase (SiE5′NT).
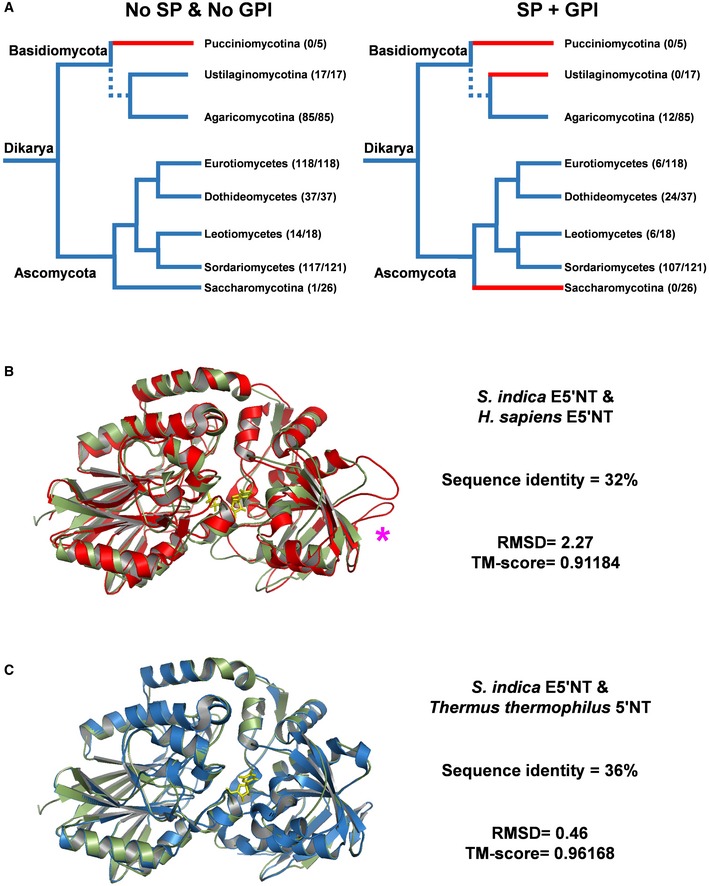
- Distribution of SiE5′NT orthologues across higher fungi. End nodes are color‐coded based on the presence (blue) or absence (red) of 5′NT genes in a particular fungal taxon. Numbers in parentheses besides the nodes specify the number of species that have 5′NT genes with respect to the total number of genomes analyzed. Left tree: distribution of 5′NT without signal peptide (SP) and GPI anchor. Right tree: E5′NT with SP and GPI anchor. The distribution shows that E5′NT genes are mostly present in Ascomycota such as Sordariomycetes (107/121), followed by Dothideomycetes (24/37) and Leotiomycetes (6/18). Few species of Eurotiomycetes (6/118) possess an E5′NT orthologue. In Basidiomycota, E5′NT members are only found in the class of Agaromycotina (12/85).
- Comparison of the SiE5′NT structural homology model (green) with the crystal structure of human E5′NT (PDB id 4H2I, red) with 32% sequence identity. *: position of the loop involved in dimerization of human E5′NT.
- Comparison of the SiE5′NT structural homology model (green) with the crystal structure of Thermus thermophiles 5′NT (2Z1A, blue) with 36% sequence identity.
Sequence alignment with homologous E5ʹNT protein sequences from animals and bacteria showed that the loop structure formed by the amino acid residues Cys476‐Pro428 in human E5ʹNT is absent from bacterial and S. indica E5ʹNT proteins (Fig 3B, Appendix Fig S7). It has previously been shown that this loop structure is required for dimerization of human E5ʹNT and that this dimeric state is important for the enzyme's narrow substrate specificity 48. In contrast, bacterial E5ʹNT enzymes function as monomers and have broad substrate specificity being able to hydrolyze ATP, ADP, AMP, and other 5′‐ribo and 5′‐deoxyribonucleotides 49. Our structural predictions (Fig 3B and C) suggest that the fungal E5ʹNT is monomeric and likely capable of hydrolyzing different nucleotides.
In order to evaluate potential substrates for SiE5′NT, full‐length S. indica E5′NT was expressed in A. thaliana and in the phytopathogenic fungus Ustilago maydis, neither of which has a gene encoding a secreted E5′NT predicted in their genome. Ecto‐5′‐nucleotidase activity was determined by the rate of inorganic phosphate release after incubation of 50 or 100 μM of either ATP, ADP, or AMP with washed culture suspensions of U. maydis and with preparations of plasma membrane proteins from A. thaliana transgenic lines, respectively. Intact cells of the U. maydis control strains expressing mCherry, E5′NT without the GPI anchor, or E5′NT without the secretory signal peptide (SP) and the GPI anchor under the synthetic otef promoter were able to hydrolyze ATP and ADP to a certain extent, whereas hydrolysis of AMP occurred at a very low rate. The U. maydis strain expressing the full‐length S. indica ecto‐5′‐nucleotidase showed significantly higher hydrolysis rates for all three nucleotides tested compared to the control strains. This indicates that as for the monomeric bacterial variant, ATP, ADP, and AMP are potential substrates for SiE5′NT (Appendix Fig S8) and that SiE5′NT has extracellular activity. This was confirmed by the activity measurements of ectopically expressed SiE5′NT in A. thaliana. Three A. thaliana transgenic lines in Col‐0 background expressing either full‐length E5′NT (303 1L3), SPE5′NT:mCherry:E5′NT fusion (304 6L8), or cytoplasmic mCherry (305 2L1) under the control of the 35S promoter were selected based on their expression levels in the T3 generation (Appendix Fig S9A and B). Ecto‐5′‐nucleotidase activity in the membrane fractions demonstrated that expression of the native form of SiE5′NT in A. thaliana leads to the production of a membrane‐localized enzyme capable of hydrolyzing ATP, ADP, and AMP (Fig 4A). In contrast, the membrane fraction of the A. thaliana line expressing SPE5′NT:mCherry:E5′NT fusion did not show increased activity compared to the mCherry control line, suggesting that modification at the N‐terminus of the protein impacts protein activity or function. Interestingly, the absence of hydrolytic activity on AMP for A. thaliana lines expressing mCherry or SPE5′NT:mCherry:E5′NT fusion implies that no E5′NT‐like activity is naturally present at the membrane under this growth condition in A. thaliana. The lines expressing SiE5′NT were otherwise morphologically similar to the control lines in having the same shoot fresh weight (Appendix Fig S9C and D) but produced significantly fewer seeds during propagation (Appendix Fig S9E).
Figure 4. The ecto‐5′‐nucleotidase activity of SiE5′NT in the apoplast of Arabidopsis leads to enhanced S. indica colonization similar to that of the dorn1‐3 (ATP receptor) mutant line.
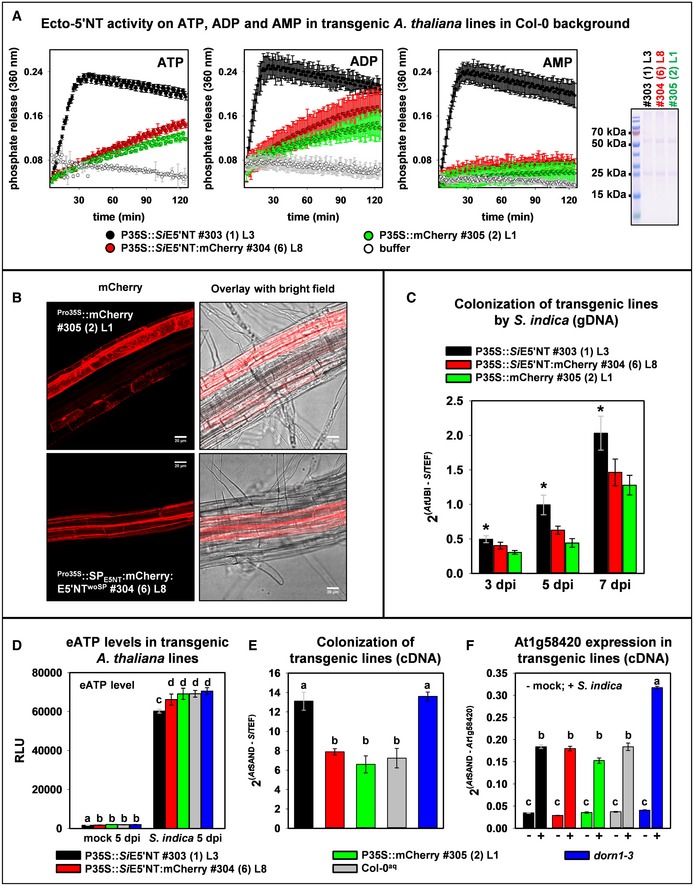
- Ecto‐5′‐nucleotidase activity measured in membrane protein preparations of Arabidopsis plants expressing Pro35S::E5′NT (#303), Pro35S::SPE5′NT:mCherry:E5′NTwoSP (#304), or Pro35S::mCherry (#305). E5′NT activity was measured after incubation with 100 μM of either ATP, ADP, or AMP. In the membrane protein preparations from Pro35S::E5′NT (#303) lines, phosphate release was specifically increased upon incubation with purines. Error bars represent the standard error of the mean from three technical repetitions. The Coomassie‐stained SDS–PAGE shows the protein pattern of the membrane fractions for the individual transgenic lines. Equal volumes were loaded. The experiment was repeated two times with similar results.
- Confocal microscopy images of Arabidopsis roots expressing either cytosolic mCherry (#305) or Pro35S::SPE5′NT:mCherry:E5′NTwoSP (#304) showing secretion of the E5′NT fusion protein. mCherry images show z‐stacks of 14 image planes of 1 μm each. Scale bar = 20 μm.
- The transgenic Arabidopsis line Pro35S::E5′NT (#303) expressing untagged full‐length SiE5′NT was better colonized by S. indica. Error bars of the qPCR data represent ± SE of the mean from three independent biological replicates. Asterisks indicate significance (Student's t‐test, *P < 0.05).
- S. indica induced eATP release in different Arabidopsis transgenic lines. Culture medium was collected from mock‐treated or S. indica‐inoculated seedlings growing in liquid medium at 5 dpi, and released eATP was measured. RLU: relative light units. Error bars represent ± SE of the mean from three biological replicates. Letters indicate significance to all other samples within the same treatment group (ANOVA, P < 0.05).
- S. indica colonization of transgenic lines at 5 dpi. Error bars represent ± SE of the mean from three biological replicates. Letters indicate significance to all other samples within the same treatment group (ANOVA, P < 0.05).
- Expression analysis of the eATP responsive gene At1g58420 measured by qRT–PCR. Error bars represent ± SE of the mean from three independent biological replicates (independent from those shown in Fig. 2C). Letters indicate significant groups (ANOVA, P < 0.01, for the line 305 P < 0.05).
To confirm secretion of the fungal enzyme in A. thaliana transgenic lines, confocal microscopy was carried out with the SPE5′NT:mCherry:E5′NT fusion line (304 6L8). A fluorescence signal in the red channel was visible at the periphery of the plant cells but not in the cytoplasm (Fig 4B, Appendix Fig S10A). Arabidopsis roots were treated with a 0.5 M sorbitol solution to trigger effusion of water from the cytoplasm leading to a volumetric extension of the apoplastic space and plasmolysis. After plasmolysis, the fluorescence signal in the red channel was visible at the membranes and cell walls (Appendix Fig S10B), indicating that the full‐length mCherry‐tagged E5′NT fusion protein is correctly secreted and that the GPI anchor is functional. However, we cannot exclude that the E5′NT part of the fusion protein is partially misfolded leading to the observed impaired hydrolytic activity in this line. No auto‐fluorescence signals were detected in the UV channel and in the control Col‐0 WT line (Appendix Fig S10A and B).
SiE5′NT affects eATP levels and fungal colonization
To investigate the role of SiE5′NT in fungal accommodation in plant roots, the A. thaliana line 303 1L3 constitutively expressing the enzymatically active SiE5′NT (Fig 4A) was used along with the SPE5′NT:mCherry:E5′NT fusion line 304 6L8 and mCherry line 305 2L1 for colonization assays at 3, 5, and 7 dpi (Fig 4C and Appendix Fig S11A–C). We observed a significantly enhanced fungal colonization at all time points in the SiE5′NT 303 1L3 plants with respect to A. thaliana lines with no enhanced ecto‐5′NT enzyme activities (Fig 4C). This phenotype was confirmed in independent transgenic lines (Appendix Fig S11A–C). Correspondingly, the SiE5′NT plants accumulated significantly lower levels of ATP in the growth medium than the control lines at 5 dpi, the time point at which we observed the highest levels of eATP accumulation upon fungal colonization (Fig 4D). Interestingly, mutation of the DORN1 receptor did not affect eATP concentrations (Fig 4D), suggesting that perception of eATP is not involved in the regulation of its extracellular abundance.
To define the transcriptional response of the A. thaliana transgenic SiE5′NT line to fungal colonization at 5 dpi, we determined the expression of AT1g58420, a marker gene reported to be induced by ATP and wounding 8. In our experiments, AT1g58420 was highly responsive to S. indica colonization (Fig 4E and F). Analysis of the expression profiles of this gene upon different treatments showed that AT1g58420 is also responsive to other danger‐ and microbe‐associated molecular patterns (DAMPs and MAMPs), including chitin and flagellin (Appendix Fig S12, and public microarrays, GENEVESTIGATOR, https://genevestigator.com/gv/). Mutation of the DORN1 receptor was shown to abolish ATP‐induced gene expression and to reduce wounding‐induced gene expression 8. Upon colonization with S. indica, the dorn1‐3 mutant plants displayed an increased expression of the marker gene At1g58420 compared to the colonized control line Col‐0aq , which correlates to the increased fungal biomass (Fig 4E and F). This indicates that induction of this marker gene by S. indica is not necessarily related to increased eATP levels and its detection by DORN1. Interestingly, despite a similar level of fungal colonization, the SiE5′NT plants had a significantly lower induction of this marker gene compared to the dorn1‐3 mutant plants (Fig 4E and F). Expression of two additional marker genes responsive to eATP, CPK28, and RBOHD 8 was also analyzed. These genes showed a similar pattern as observed for the AT1g58420 marker gene with lower induction for the SiE5′NT plants compared to the dorn1‐3 mutant plants, but their induction in response to fungal colonization was not as high as for the marker gene AT1g58420 (Appendix Fig S13).
Finally, to test whether SiE5′NT would also increase susceptibility to fungal disease, we additionally assessed colonization of Arabidopsis roots by the pathogen Colletotrichum incanum which colonizes members of the Brassicaceae, Fabaceae, and Solanaceae and which severely inhibits Arabidopsis growth 50. The SiE5′NT plants had a significantly higher colonization by this pathogen (Appendix Fig S14).
Phosphate released from the hydrolysis of eATP could serve as a nutrient
Extracellular SiE5′NT activity leads to significantly decreased amounts of free eATP in the apoplast of colonized root tissue and, consequently, an increased availability of phosphate (Pi). It has previously been shown that S. indica transfers phosphate to the host and improves plant growth more effectively at low phosphate compared to high phosphate conditions at later colonization stages 51, 52. To evaluate the effect of an additional apoplastic Pi‐source on the nutrient exchange between plant and fungus, we employed a computational cell biology approach and simulated the dynamics of a network of proton pumps and proton‐coupled transporters during the early biotrophic interaction phase. We first used a model where the plant provides sugar to the fungus while the fungus provides phosphate to the plant as described for classical mycorrhizal associations 53. When apoplastic ATP levels and subsequent Pi levels increase, the computational simulations predict that the sugar flux from the plant to the fungus is not affected but the phosphate fluxes change (Fig 5). An external Pi‐source provokes an increased Pi‐uptake by the plant and a reduced Pi‐release or even an uptake of Pi by the fungus. Thus, the hydrolysis of eATP to adenosine and phosphate reduces the amount of Pi that the fungus has to provide to obtain sugar from the plant and could also serve for the Pi‐nutrition of the fungus in the early colonization phase without affecting sugar transfer. These new insights might guide future experiments to clarify whether the activity of SiE5′NT in the apoplast could serve both nutritional needs and modulation of host immunity.
Figure 5. Simulation of the effect of eATP release and its cleavage in the apoplast on the nutrient exchange between plant and fungus.
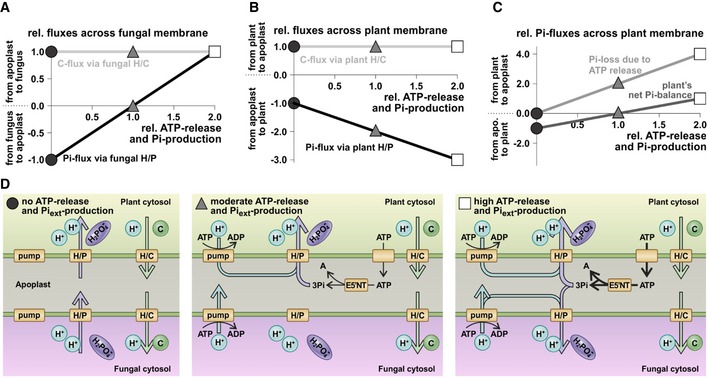
- Relative fluxes of phosphates (Pi) and sugars (C) across the fungal plasma membrane. Without additional Pi‐source in the apoplast (value 0 at the x‐axis, black dots), there is a constant flux of phosphate via the proton‐coupled phosphate (H/P) transporter from the fungus to the apoplast and a constant flux of sugar from the apoplast to the fungus. For better comparison, these fluxes were normalized to 1 and −1, respectively, and all other fluxes were calculated relative to these control values. With increasing ATP‐release and Pi‐production, the Pi‐efflux gets smaller and is zero at a relative Pi‐production rate of 1 (gray triangles). At higher ATP‐release and Pi‐production rates, the fungus imports Pi, i.e., the transport direction of phosphate has been inverted in comparison with the control condition (white square). The C‐flux via the proton‐coupled sugar (H/C) transporter is not affected by the additional Pi‐source (horizontal gray line).
- Relative fluxes of Pi and C across the plant plasma membrane. Without additional Pi‐source in the apoplast (value 0 at the x‐axis, black dots), there is a constant flux of sugar via the H/C transporter from the plant to the apoplast (and thereafter to the fungus) and a constant flux of phosphate (coming from the fungus) via the H/P transporter from the apoplast to the plant. With increasing ATP‐release and Pi‐production, the Pi‐influx via the H/P transporter increases while the C‐flux via the H/C transporter is unaffected by the additional Pi‐source (horizontal gray line).
- Phosphate fluxes across the plant plasma membrane. Besides the Pi‐uptake via the H/P transporter (B, black line, for clarity not shown in C), the plant loses Pi due to the ATP‐release (light gray line). The difference between Pi‐uptake and Pi‐loss is the net Pi‐balance of the plant (dark gray line). Without ATP‐release (value 0 at the x‐axis, black dots), the plant gains Pi. At a relative ATP‐release and Pi‐production of 1 (gray triangles), the plant release as much phosphate as it takes up, while at higher ATP‐release values, the plant loses Pi.
- Schematic representation of the fluxes for three scenarios: (i) no ATP‐release and Pi‐production (left, black dots in A‐C); (ii) moderate ATP‐release, adenosine (A), and Pi‐production (middle, gray triangles in A–C); and (iii) high ATP‐release, A, and Pi‐production (right, white squares in A–C). If there is no ATP‐release (left), there is a constant flux of Pi from the fungus via the apoplast to the plant and a constant flux of sugars in the inverse direction. The energy for these fluxes is provided by the phosphate gradient between fungus and plant and the sugar gradient between plant and fungus. At a moderate ATP‐release and Pi‐production rate (middle, value 1 on the x‐axis, gray triangles in A–C), there is no Pi‐flux from the fungus to the apoplast anymore. The unchanged uptake of sugars is now energized by the proton pump of the fungus. In this condition, the plant retrieves the cleaved Pi originating from the ATP‐release via the H/P cotransporter, a transport which is energized by both the sugar gradient and the proton pump. At a high ATP‐release and Pi‐production rate (right, value 2 on the x‐axis, white squares in A–C), there is a larger Pi‐uptake by the plant and a Pi‐uptake by the fungus. Both processes are energized by larger activities of the proton pumps.
Discussion
To establish an integrated and holistic view of the role of the metabolic interplay between plants and their microbes, it is of paramount importance to investigate the mechanisms by which plant‐associated microbes manipulate plant‐derived metabolites and how plant metabolism is linked to immunity. Apoplastic communication and its metabolic fluxes have crucial functions in mediating microbial accommodation. During root colonization, plants and microbes secrete a number of proteins to the apoplast that are important determinants for the outcome of the interaction. Plant cells release small amounts of ATP into their extracellular matrix as they grow 7. The eATP level can modulate the rate of cell growth in diverse tissues. Besides the physiological role in growth modulation, eATP is released to the extracellular environment in response to biotic stresses modulating defense responses, e.g., upon wounding and also when their plasma membranes are stretched during delivery of secretory vesicles 4. This implicates a regulatory role for enzymes that can hydrolyze extracellular nucleotides and thus limit their accumulation in the extracellular environment. In plants, the level of extracellular nucleotides is controlled by various phosphatases, prominent among which are apyrases EC 3.6.1.5 (nucleoside triphosphate diphosphohydrolases, NTPDases) 4. In animals, ecto‐nucleotidases play a pivotal role in purinergic signal transmission where the major extracellular purine and pyrimidine compounds known to elicit cell surface receptor‐mediated signals are ATP, ADP, UTP, UDP, UDP‐glucose, some additional nucleotide sugars, some dinucleoside polyphosphates, and the nucleoside adenosine 2, 54. The four major groups of ecto‐nucleotidases involved in extracellular purinergic signaling include the ecto‐nucleoside triphosphate diphosphohydrolases (E‐NTPDases), ecto‐5′‐nucleotidase (eN), ecto‐nucleotide pyrophosphatase/phosphodiesterases (E‐NPPs), and alkaline phosphatases (APs) 55. Depending on subtype, ecto‐nucleotidases typically hydrolyze nucleoside tri‐, di‐, and monophosphates and dinucleoside polyphosphates producing nucleoside diphosphates, nucleoside monophosphates, nucleosides, phosphate, and inorganic pyrophosphate (PPi) 55. The extracellular membrane‐bound ecto‐5′‐nucleotidase CD73 is anchored to the plasma membrane via a glycosyl phosphatidylinositol (GPI) anchor 55, 56. This enzyme hydrolyzes AMP to adenosine where ADP and ATP are competitive inhibitors, with inhibition constants in the low micromolar range 48. The CD73 together with the ecto‐apyrase CD39 that catalyzes the phosphohydrolysis of ATP and ADP to AMP is part of a cascade to terminate the action of nucleotides as extracellular signaling molecules by decreasing the levels of eATP and eADP and by generating adenosine. In animals, this latter bioactive nucleoside can activate the purinergic ligand‐gated ion channel P2X and the G protein‐coupled P2Y receptors, counteracting ATP‐induced signaling 55. A purinergic receptor for extracellular adenosine or other bioactive nucleosides is not known in plants, and its presence at the host cell surface needs to be proven. Yet, it is tempting to speculate that a similar mechanism is also in place in plants where metabolites derived from the hydrolysis of extracellular nucleotides modulate eATP‐triggered immunity once the signaling is no longer required.
Taken together, our work shows that eATP perception plays an important role in plant–S. indica interaction in the roots and that S. indica has an intrinsic mechanism to modulate bioactive extracellular nucleotide levels in the apoplast (Fig 6). It remains to be proven if the effects mediated by S. indica ecto‐5′‐nucleotidase in planta are due to the depletion of the signaling molecule eATP or to accumulation of hydrolysis products, such as adenosine, a potent immune suppressor in animal systems, and/or phosphate, a regulator of compatibility in mycorrhizal associations. The contribution of a potential immunomodulatory agent derived from the action of the SiE5′NT is supported by the reduced response of the marker gene At1g58420 in the SiE5′NT plants compared to the dorn1‐3 mutant plants. Additionally, in support of the hypothesis that hydrolysis products may mediate colonization, Daumann et al 57 have shown in a recent study that accumulation of adenosine in the apoplast of Arabidopsis can promote pathogenic fungal infection in plants. Metabolomics data obtained from APF of colonized roots by sebacinoid fungi showed an accumulation of adenosine at 6 dpi compared to non‐colonized roots (Gerd Balcke, Hanna Rövenich, and Alga Zuccaro, personal communication). Thus, it would be important to analyze the role of purine nucleosides in plant root–fungus interactions. Although our data indicate that the activity of the SiE5′NT is important for its function in the apoplast, we cannot fully exclude the possibility that an additional function independent of the hydrolytic activity is responsible for the effects observed in this study. Enzymatic inactivation by targeted point mutations in the catalytic domain could help address this possibility.
Figure 6. Schematic model showing the potential interference of SiE5′NT with apoplastic eATP signaling and nucleotide salvage pathway in Arabidopsis .
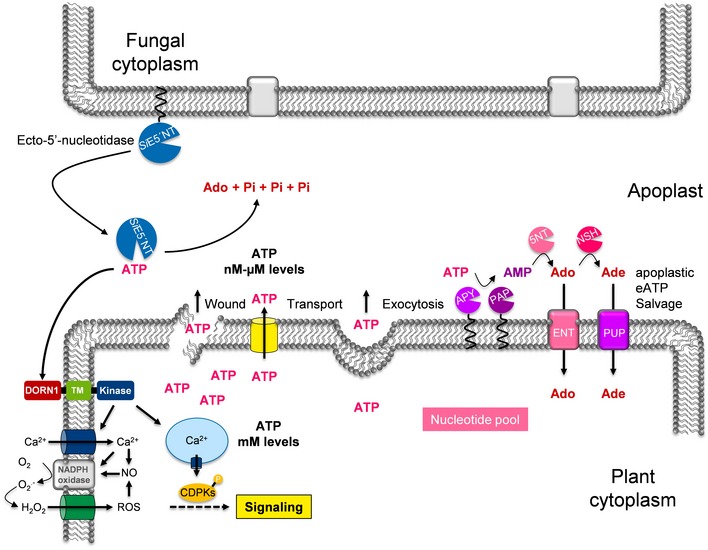
The schema was modified from Ref. 5. ENT = equilibrative nucleoside transporters, PAP = purple acid phosphatases, Apy = ectoapyrases or nucleoside triphosphate diphosphohydrolases (NTPDases), NSH = nucleoside hydrolases, 5NT = hypothetical 5′‐nucleotidases, PUP = hypothetical purine permeases.
Modeling of phosphorus/carbon exchange between the plant and the fungus in the presence of an apoplastic Pi‐source derived from eATP hydrolysis suggests that in the early phase of interaction, the fungus could reduce phosphate transfer to the plant without affecting the carbohydrate flux from the plant and eventually use this phosphate as a nutrient to support its own growth. Secreted E5′NT homologues are present also in fungal pathogens as C. incanum and other endophytes of Arabidopsis, such as C. tofieldiae, where their expression is induced during colonization 50. Fungal genes encoding enzymes involved in ATP‐scavenging were also found in the apoplastic fluid of rice leaves infected with the rice pathogens M. oryzae and C. miyabeanus 43, 58, suggesting that extracellular purine‐based biomolecules play a role also in other plant–fungus interactions. Hydrolysis of plant‐derived primary metabolites with immune modulating functions in the apoplast could thus represent a general mechanism of plant‐associated fungi in modulating plant nucleotides signaling as well as serving nutritional needs.
Materials and Methods
Fungal strains and culturing techniques
Serendipita indica (syn. Piriformospora indica, DSM11827, Leibniz Institute DSMZ—German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) and Serendipita indica GoGFP strain expressing cytosolic GFP 36 were grown in liquid CM supplemented with 2% glucose and incubated at 28°C with constant agitation at 120 rpm, or on solid medium supplemented with 1.5% agar 59. Ustilago maydis SG200 60 cultures were propagated on potato dextrose agar (Difco) or YEPSLight liquid medium at 28°C 61. Colletotrichum incanum strain MAFF238704 was kindly provided by Prof. Paul Schulze‐Lefert, MPIPZ, Cologne, and propagated on solid CM supplemented with 1.5% agar in darkness at 25°C 50.
Plant growth conditions and fungal inoculation
Barley seeds (Hordeum vulgare cv. Golden Promise) were surface sterilized as described previously 36 and germinated for 3 days in the dark on wet filter paper. Germlings were transferred into jars with 1/10 PNM agar 59 and inoculated with 3 ml (500,000 spores/ml) S. indica chlamydospores in 0.002% Tween‐20 aqueous solution and cultivated in a growth chamber with a day : night cycle of 16 h : 8 h (light intensity 108 μmol/m2/s and temperature of 22°C: 18°C). Tween water‐treated germlings were used as control. Three independent biological replicates were used. For each biological replicate, four to five barley seedlings in one jar were pooled at each time point.
Arabidopsis seeds were surface sterilized with 70% ethanol for 10 min followed by 100% ethanol for 7 min and then air‐dried. The sterilized seeds were transferred to half‐strength Murashige and Skoog (1/2MS) medium containing 1% sucrose and 0.4% Gelrite (Duchefa), incubated for 3 days in the dark at 4°C, and subsequently grown for 7 days with a day : night cycle of 8 h: 16 h (light intensity, 47 μmol/m2/s) at 24°C. Three independent biological replicates were used for all Arabidopsis experiments unless otherwise stated in the legend. For each biological replicate, three plates containing 20 plants each were pooled. For colonization studies, 500,000 spores/ml of S. indica chlamydospores in 0.002% Tween‐20 aqueous solution was applied to the roots of each plant of 7‐day‐old seedlings. Tween water‐treated seedlings were used as control.
For colonization analysis using C. incanum, Arabidopsis seedlings were grown for 10 days under a day: night cycle of 8‐h: 16‐h regime (light intensity 111 μmol/m2/s and temperature of 22°C:18°C). Roots were drop‐inoculated with 200,000 spores/ml of C. incanum suspended in sterile distilled water. Water‐treated seedlings served as control. Three independent biological replicates were used each representing a pool of roots from 20 plants.
Identification of S. indica proteins in apoplastic fluid and culture filtrate
Barley seeds (Hordeum vulgare L. cv Golden Promise) were surface sterilized for 1 min under light shaking in 70% ethanol followed by 1 h in 12% sodium hypochlorite. Subsequently, seeds were washed repeatedly for 1 h with sterile distilled water. After germination for 3 days at 22°C in complete darkness on wet filter paper, germinated seeds were transferred into jars with 1/10 PNM 59 under sterile conditions and inoculated with 3 ml of chlamydospores (concentration 500,000 spores/ml) of S. indica GoGFP strain 36 and cultivated in a growth chamber with a day : night cycle of 16 h: 8 h (light intensity, 108 μmol/m2/s) and temperature of 22°C:18°C. For APF extraction, three time points reflecting different interaction stages have been chosen: biotrophic phase (5 dpi) as well as cell death‐associated phase (10 and 14 dpi). After respective growth time, roots from 300 seedlings for each treatment and time point were thoroughly washed and cut into pieces of 2 cm length (2nd to 4th cm of the root). Samples were vacuum‐infiltrated (Vacuubrand, CVC 3000, VWR) with deionized water 5 × 15 min at 250 mbar with 1‐min atmospheric pressure break. Bundled infiltrated roots were centrifuged in 5‐ml syringe barrels at 2,000 rpm in a swing bucket rotor (800 g) for 15 min at 4°C as described in Ref. 25. Pooled APF samples were stored at −20°C. In order to exclude cytoplasmic contamination, apoplastic fluid samples were tested in immunoblots with GFP antibody. For Western blotting, proteins from unstained gels were transferred onto nitrocellulose membranes using a semidry blotting system from Bio‐Rad (Hercules, USA). Anti‐GFP antibody was obtained from Cell Signaling Technology (Danver, USA) and used at a 1:2,000 dilution. An anti‐mouse antibody purchased from Sigma (Munich, Germany) was used as secondary antibody at a dilution of 1:2,000. For higher peptide mass accuracy, part of the APF samples was deglycosylated with Protein Deglycosylation Mix (P6039S, New England Biolabs, Ipswich, USA) under denaturing conditions. In agreement with Ref. 62, at the centrifugation force used the samples were free from obvious symplastic contamination and were thus regarded as being of apoplastic origin.
Secreted proteins from culture filtrate were prepared as follows: S. indica cultures grown for 7 days in liquid CM were passed through a miracloth filter (Merck Millipore, Darmstadt), a folding filter type 600P (Carl Roth, Karlsruhe, Germany), and finally a 0.45‐μm syringe filter (Millex‐GP; Merck Millipore, Darmstadt, Germany) to remove small mycelium fragments and spores. Proteins were precipitated by addition of 10% (v/v, final concentration) trichloroacetic acid and incubation at −20°C overnight and subsequent centrifugation at 40,000 g. For SDS–PAGE, pelleted secreted proteins resolved in 50 μl of 1× SDS sample buffer and 15 μl were used. For mass spectrometric analysis, pelleted secreted proteins were solved in 50 μl TBS (50 mM Tris pH 7.5, 150 mM NaCl). Half of the samples were deglycosylated with the Protein Deglycosylation Mix (New England Biolabs, Ipswich, USA) under denaturing conditions according to manufacturer's protocol. Proteins were then analyzed via liquid chromatography–electron spray ionization–tandem mass spectrometry (LC‐ESI‐MS/MS; LTQ Orbitrap Discovery; Thermo Fisher Scientific, Waltham, USA) after tryptic digest (FASP™ Protein Digestion Kit; Expedeon, Swavesey, UK) in the facilities of CEACAD/CMMC Proteomics Facility in Cologne, Germany. Peptide masses were compared to S. indica in silico trypsin‐digested proteome (Genbank, NCBI) with an inclusion of GFP, with a barley dataset downloaded from the IPK Gatersleben homepage (http://webblast.ipk-gatersleben.de/barley_ibsc/downloads/) and with Rhizobium radiobacter F4 (syn. Agrobacterium tumefaciens, syn. Agrobacterium fabrum) dataset downloaded from Genbank, NCBI (WGS project JZLL01000000, https://www.uniprot.org/proteomes/UP000033487). The exact instrument settings can be found in the Supplementary Appendix containing the processed data. Mass spectrometric raw data were processed with Proteome Discoverer (v1.4, Thermo Scientific) and SEQUEST. Briefly, MS2 spectra were searched against the respective databases, including a list of common contaminants. The minimal peptide length was set to seven amino acids, and carbamidomethylation at cysteine residues was considered as a fixed modification. Oxidation (M) and Acetyl (Protein N‐term) were included as variable modifications. The match‐between‐runs option was enabled. Fungal‐derived proteins matched to peptides were analyzed for the presence of signal peptides and exclusiveness for the three specific time points of APF extraction.
Construction of plasmids
Full‐length E5′NT (PIIN_01005) was amplified from cDNA of barley roots inoculated with S. indica at 5 dpi by respective oligonucleotides (Table EV5) and cloned into the TOPO® cloning plasmid (Thermo Fisher Scientific, Waltham, USA) resulting in TOPO‐E5′NT. For the expression of various E5′NT constructs under the control of the synthetic otef promoter which exhibits strong constitutive expression 63, full‐length E5′NT, E5′NTwoGPI, and E5′NTwoSPwoGPI were amplified from the plasmid TOPO‐E5′NT using Phusion DNA polymerase (NEB) by respective oligonucleotides (Table EV5) and cloned into the BamHI‐NotI digested plasmid Potef::Yup1(RFP)2 64 using Gibson assembly 65, generating the plasmids Potef::E5′NT, Potef::E5′NTwoGPI, and Potef::E5′NTwoSPwoGPI, respectively.
For the construction of SPE5′NT:mCherry:E5′NTwoSP, SiE5′NT without signal peptide (E5′NT woSP) was amplified from the plasmid TOPO‐E5′NT using Phusion DNA polymerase (NEB) by respective oligonucleotides (Table EV5) and cloned into the BamHI digested plasmid ProUmPit2::SPDld1:mCherry:Dld1woSP (Nostadt et al, unpublished data), generating the plasmid ProUmPit2::SPDld1:mCherry:E5′NTwoSP. Subsequently, ProUmPit2::SPDld1:mCherry:E5′NTwoSP was digested with SacII and NcoI and ligated with a DNA fragment encoding the E5′NT signal peptide (SPE5′NT) digested with the same restriction enzymes, replacing the Dld1 signal peptide (SPDld1) and generating the construct ProUmPit2::SPE5′NT:mCherry:E5′NTwoSP. For expression in A. thaliana under the control of CaMV 35S promoter (Pro35S), full‐length E5′NT, SPE5′NT:mCherry:E5′NTwoSP, and mCherry were amplified from the plasmids TOPO‐E5′NT and ProUmPit2::SPE5′NT:mCherry:E5′NTwoSP, respectively, using Phusion DNA polymerase (NEB) by respective oligonucleotides (Table EV5) and cloned into the BamHI digested plasmid pCXSN 66, using Gibson assembly, generating the plasmids Pro35S::E5′NT, Pro35S::SPE5′NT:mCherry:E5′NTwoSP, and Pro35S::mCherry, respectively.
Ustilago maydis transformation
The U. maydis strains were transformed by integration of the p123‐derived plasmids into the ip locus in the haploid solopathogenic strain SG200 as previously described 67.
Ustilago maydis cell surface 5′‐nucleotidase activity assay
For the cell surface 5′‐nucleotidase activity assay, Potef::E5′NT, Potef::E5′NTwoGPI Potef::E5′NTwoSPwoGPI along with the control Potef::mCherry U. maydis strains were grown overnight in YEPSLight. Then, the cultures were diluted in PD broth and grown until OD600 reached 0.8. The cells were harvested through centrifugation and washed two times with 0.9% NaCl and finally re‐suspended in equal volume of sterile double deionized water. Then 100 μl of cell suspension from each U. maydis strains was incubated with 50 μM of ATP, ADP, or AMP for 30 min, and the hydrolysis of these nucleotides was measured by quantifying the amount of phosphate released using EnzChek® Phosphate Assay Kit (Thermo Fisher Scientific).
Arabidopsis thaliana transformation
The plasmids Pro35S::E5′NT (#303), Pro35S::SPE5′NT:mCherry:E5′NTwoSP (#304), and Pro35S::mCherry (#305) were transformed into the Agrobacterium strain GV3101 by electroporation using Gene Pulser Xcell Electroporation System (Bio‐Rad Laboratories, Hercules, CA, USA) following the manufacturer indications. The cells were plated on LB medium containing 25 μg/ml of rifampicin and 50 μg/ml of kanamycin. Agrobacterium‐mediated transformation of A. thaliana was carried out by means of the floral dip method 68, 69. Stratified seeds from T0 plants were grown in 1/2 MS containing 30 mg/l hygromycin under short day condition. Putative transgenic lines were selected by their hygromycin resistance. Putative transgenic lines were transferred to soil and 2 weeks later tested by PCR using a gene‐specific primer (Table EV5). The T3 transgenic lines from 35Spro::E5′NT (#303) and 35Spro::SPE5′NT:mCherry:E5′NTwoSP (#304) were confirmed again by both hygromycin resistance and PCR using a gene‐specific primer. In addition, the elevated expression level of E5′NT in 35Spro::E5′NT (#303) and in 35Spro::SPE5′NT:mCherry:E5′NTwoSP (#304) as well as the mCherry in 35Spro::mCherry (#305) lines was analyzed by quantitative real‐time PCR. The transgenic lines which had the highest transcript levels were selected for further studies.
ATP assay
To test whether colonization of S. indica in Arabidopsis and barley triggers the release of eATP, 7‐day‐old Arabidopsis seedlings and 3‐day‐old barley seedlings were treated either with a S. indica spore suspension in 0.01% tween water (5 × 105 spores/ml) or with 0.01% tween water for 2.5 h. The Arabidopsis treated seedlings were transferred to 60‐mm petri plates (3 plates per treatment). Each plate contained 3 ml of liquid 1/2 MS (ammonium‐free) medium and 15 treated seedlings. Subsequently, seedlings were grown for 3, 5, 7, and 10 days postinoculation (dpi) with constant light at 22°C. Three individual samples of 50‐μl media were collected from each plate, flash frozen in liquid nitrogen, and stored in −80°C until eATP measurements were carried out. Apoplastic fluid from colonized and non‐colonized barley roots was collected as mentioned above, flash frozen in liquid nitrogen, and stored in −80°C until eATP measurements were carried out. The measurement of eATP in the growth media and in the APF was performed using the ENLITEN ATP Assay System from Promega. Reactions were carried out in 96‐well plates, and luminescence was detected using a microplate reader (TECAN Safire).
Membrane protein purification
Membrane‐bound proteins were extracted from 1 g shoot material from the respective A. thaliana lines (25 days old) grown in soil. The material was ground in liquid nitrogen and re‐suspended in 5 ml 0.05 M Tris–HCl buffer containing 1 mM MgCl2 (pH 7.5). After centrifugation for 20 min at 12,000 g, the pellet was re‐suspended in 5 ml Tris buffer containing 1% IGEPAL CA‐630 (Sigma), gently stirred at 4°C for 30 min, and subsequently centrifuged at 5,000 g for 10 min. The supernatant was cleared by ultracentrifugation at 100,000 g for 45 min. The resulting supernatant was used for subsequent enzyme activity assays.
Measurement of E5′NT and apyrase activity assay in A. thaliana lines
The activity of purified, membrane‐bound apyrases and the overexpressed SiE5′NT was monitored using the EnzChek® Phosphate Assay Kit (Thermo Fisher Scientific) according to manufacturer's instruction. 15 μl of purified membrane proteins was used for the enzyme kinetics assay. 100 μM ATP, ADP, or AMP (in 10 mM Tris/MES buffer containing 2 mM MgCl2 and 30 mM KCl; pH 6.5) were used as substrates. The background absorbance at 360 nm was monitored for 5 min before the addition of the substrates. Subsequently, kinetics were measured for 2 h.
Real‐time PCR analysis
DNA isolation was performed from ~200 mg of ground material. The powder was incubated for 5 min at RT under slight rotation with ~500 μl CTAB buffer (2% hexadecyl trimethyl‐ammonium bromide, 100 mM Tris–HCl, 20 mM EDTA, 1.4 M NaCl, and 1% polyvinyl pyrrolidone vinylpyrrolidine homopolymer Mw 40,000, pH 5.0). Subsequently, 250 μl chloroform : isoamyl‐alcohol (24:1) was added followed by an additional incubation for 5 min at RT and centrifugation. The water phase was collected, and polysaccharides were precipitated using ethanol before final DNA precipitation using 1 volume of isopropanol. Total RNA was isolated from 200 mg of ground fungal material or colonized and non‐colonized plant root material using the TRIzol® reagent (Invitrogen, USA) as described previously 36. Contaminating genomic DNA was removed by DNase I treatments (Thermo Fisher Scientific, USA) according to manufacturer's protocol. First‐strand cDNA was synthesized with 1 μg of total RNA primed with Oligo‐dT and random hexamer using First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA). The resulting cDNA preparations were diluted to a final concentration of 2.5 ng/μl, and 4 μl of each cDNA sample was used for qRT–PCR. Serendipita indica translation elongation factor 1 (SiTEF) and A. thaliana SAND or UBI gene primers were used for qPCR amplifications. The reactions were performed in 96‐well reaction plate (Bio‐Rad, USA) on a Real‐Time PCR System (Bio‐Rad, USA). Each well contained 7.5 μl of 2× SYBR® Green QPCR Master Mix (Promega, USA), 4 μl of cDNA or 10 ng of DNA, and 0.7 μl of each primer from a 10 μM stock, in a final volume of 15 μl. Transcripts of each gene were amplified using the primers described in Table EV5. The threshold cycles (C T) of each gene were averaged from three replicate reactions, and the relative expression or plant to fungus ratio from gDNA was calculated using the 2−ΔCT method 70.
E5′NT expression levels in axenic culture in response to nucleotides
In order to test whether SiE5′NT expression is induced in the presence of nucleotides, WT S. indica spores (500,000 spores/ml) were used to inoculate liquid CM medium and grown for 5 days at 28°C with 110 rpm shaking. Subsequently, the culture was filtrated through miracloth, and the mycelium was crushed and regenerated in 100 ml fresh CM medium for 2 days. After regeneration, 5 ml of fungal suspension was diluted with 45 ml of CM medium supplemented with 100 μM of the respective nucleotides. At the indicated time points, the mycelium was harvested by filtration, washed with water, and flash frozen in liquid nitrogen before processing according to the Real‐Time PCR Analysis Protocol listed above. Three to six independent biological replicates for each time point/nucleotide combination were performed.
Calcium influx quantification
All calcium influx experiments were conducted using plants that ectopically express aequorin as Ca2+ reporter (Choi et al 2014). Seven‐day‐old seedlings grown on 1/2 MS plates containing 1% sucrose were transferred to 96‐well plates containing 50 μl 10 μM coelenterazine (Promega) in reconstitution buffer (2 mM MES/KOH containing 10 mM CaCl2; pH 5.7). After 16 h, the luminescence background was measured for 5 min followed by the addition of different adenylates (final concentration 100 μM ATP, ADP, AMP, or adenosine). Luminescence was monitored for 30 min.
Confocal microscopy
Confocal images were recorded using a TCS‐SP8 confocal microscope (Leica, Bensheim, Germany). mCherry fluorescence was imaged with an excitation of 561 nm and a detection bandwidth set to 580–630 nm. DAPI and auto‐fluorescence of the samples were assessed by laser light excitation at 405 nm with a detection bandwidth set to 415–465 nm.
Homology modeling of SiE5′NT
In order to carry out the homology modeling of SiE5′NT, best template was selected through PSI BLAST against the PDB database (http://www.rcsb.org/pdb/home/home.do). The best hit with high sequence identity and atomic resolution < 1.8 Angstroms was selected as template. The three‐dimensional structure of SiE5′NT was generated using a restrained‐based approach in MODELLER 9v11 71. Initial refinement of the 3D model generated was carried out with the help of loop refinement protocol of MODELLER. The assessment of the final structural model was carried out with PROCHECK 72, ProSA 73, and QMEAN 74 analyses. The structure was visualized using PyMOL (http://www.pymol.org/).
Computational cell biology
The nutrient exchange of phosphates (Pi) and sugars (C) between plant and fungus was essentially performed as described previously in mycorrhizal interactions 53, in a minimal model combining proton‐coupled phosphate (H/P) and sugar (H/C) transporters and proton pump‐driven background conductances. The minimal model was enlarged by adding the release of ATP from the plant into the apoplast and its subsequent cleavage into phosphates and adenosine (A) by E5′NT enzymes. The Pi and C exchange between the plant and the fungus was simulated for different values of ATP‐release and Pi‐production. The ATP‐release‐ and Pi‐production rate were normalized that at the relative value of 1, the fungus does not export Pi. The mathematical description of these processes results in redundant parameters, which were combined. Therefore, ATP‐release and cleavage were considered as one process in this study, which was modeled as a constant Pi‐production rate: d[Pi]apo = const×dt. Following the mathematical description of all transporters, an in silico cellular system was programmed and computational cell biology (dry‐lab) experiments were performed using the VCell Modeling and Analysis platform developed by the National Resource for Cell Analysis and Modeling, University of Connecticut Health Center 75.
Author contributions
AZ and SN conceived the project, designed the experiments, and wrote the paper with contributions from SW and GL. SN, SW, XQ, RN, FG, FS, and GL performed experiments and analyzed the data. ID produced the P/C model.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Dataset EV1
Review Process File
Acknowledgements
We are indebted to Prof. Gary Stacey for providing the A. thaliana dorn1‐3 mutant and Col‐0aq lines. Heidi Widmer is acknowledged for her excellent support in the laboratory. Ganga Jeena is acknowledged for her support with bioinformatics analysis. We are also grateful to Dr. Hanna Rövenich for her critical reading of the manuscript. ID was supported by the Fondecyt grant No. 1150054 of the Comisión Nacional de Investigación Científica y Tecnológica (CONICYT) of Chile. AZ acknowledge support from the Cluster of Excellence on Plant Science (CEPLAS, EXC 1028), DFG ZU 263/2‐1, DFG ZU 263/3‐1, and institutional funds of the Max Planck Society.
EMBO Reports (2019) 20: e47430
References
- 1. Burnstock G (1972) Purinergic nerves. Pharmacol Rev 24: 509–581 [PubMed] [Google Scholar]
- 2. Burnstock G (2012) Purinergic signalling: its unpopular beginning, its acceptance and its exciting future. BioEssays 34: 218–225 [DOI] [PubMed] [Google Scholar]
- 3. Chivasa S, Slabas AR (2012) Plant extracellular ATP signalling: new insight from proteomics. Mol BioSyst 8: 445–452 [DOI] [PubMed] [Google Scholar]
- 4. Clark GB, Morgan RO, Fernandez MP, Salmi ML, Roux SJ (2014) Breakthroughs spotlighting roles for extracellular nucleotides and apyrases in stress responses and growth and development. Plant Sci 225: 107–116 [DOI] [PubMed] [Google Scholar]
- 5. Tanaka K, Choi J, Cao Y, Stacey G (2014) Extracellular ATP acts as a damage‐associated molecular pattern (DAMP) signal in plants. Front Plant Sci 5: 446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Song CJ, Steinebrunner I, Wang X, Stout SC, Roux SJ (2006) Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in Arabidopsis . Plant Physiol 140: 1222–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roux SJ, Steinebrunner I (2007) Extracellular ATP: an unexpected role as a signaler in plants. Trends Plant Sci 12: 522–527 [DOI] [PubMed] [Google Scholar]
- 8. Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY, Stacey G (2014) Identification of a plant receptor for extracellular ATP. Science 343: 290–294 [DOI] [PubMed] [Google Scholar]
- 9. Tanaka K, Gilroy S, Jones AM, Stacey G (2010) Extracellular ATP signaling in plants. Trends Cell Biol 20: 601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao YR, Halane MK, Gassmann W, Stacey G (2017) The role of plant innate immunity in the legume‐rhizobium symbiosis. Annu Rev Plant Biol 68: 535–561 [DOI] [PubMed] [Google Scholar]
- 11. Kim SY, Sivaguru M, Stacey G (2006) Extracellular ATP in plants. Visualization, localization, and analysis of physiological significance in growth and signaling. Plant Physiol 142: 984–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu SJ, Wu JY (2008) Extracellular ATP‐induced NO production and its dependence on membrane Ca(2+) flux in Salvia miltiorrhiza hairy roots. J Exp Bot 59: 4007–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dark A, Demidchik V, Richards SL, Shabala S, Davies JM (2011) Release of extracellular purines from plant roots and effect on ion fluxes. Plant Signal Behav 6: 1855–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lim MH, Wu J, Yao JC, Gallardo IF, Dugger JW, Webb LJ, Huang J, Salmi ML, Song JW, Clark G et al (2014) Apyrase suppression raises extracellular ATP levels and induces gene expression and cell wall changes characteristic of stress responses1[C][W][OPEN]. Plant Physiol 164: 2054–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bouwmeester K, de Sain M, Weide R, Gouget A, Klamer S, Canut H, Govers F (2011) The lectin receptor kinase LecRK‐I.9 Is a novel phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog 7: e1001327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y, Nsibo DL, Juhar HM, Govers F, Bouwmeester K (2016) Ectopic expression of Arabidopsis L‐type lectin receptor kinase genes LecRK‐I.9 and LecRK‐IX.1 in Nicotiana benthamiana confers phytophthora resistance. Plant Cell Rep 35: 845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gouget A, Senchou V, Govers F, Sanson A, Barre A, Rouge P, Pont‐Lezica RP, Canut H (2006) Lectin receptor kinases participate in protein‐protein interactions to mediate plasma membrane‐cell wall adhesions in Arabidopsis . Plant Physiol 140: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Senchou V, Weide R, Carrasco A, Bouyssou H, Pont‐Lezica R, Govers F, Canut H (2004) High affinity recognition of a phytophthora protein by Arabidopsis via an RGD motif. Cell Mol Life Sci 61: 502–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Balague C, Gouget A, Bouchez O, Souriac C, Haget N, Boutet‐Mercey S, Govers F, Roby D, Canut H (2017) The Arabidopsis thaliana lectin receptor kinase LecRK‐I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol Plant Pathol 18: 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rovenich H, Boshoven JC, Thomma BP (2014) Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr Opin Plant Biol 20: 96–103 [DOI] [PubMed] [Google Scholar]
- 21. Lo Presti L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R, Merchant S (2015) Fungal effectors and plant susceptibility. Annu Rev Plant Biol 66: 513–545 [DOI] [PubMed] [Google Scholar]
- 22. Kloppholz S, Kuhn H, Requena N (2011) A secreted fungal effector of glomus intraradices promotes symbiotic biotrophy. Curr Biol 21: 1204–1209 [DOI] [PubMed] [Google Scholar]
- 23. Plett JM, Daguerre Y, Wittulsky S, Vayssieres A, Deveau A, Melton SJ, Kohler A, Morrell‐Falvey JL, Brun A, Veneault‐Fourrey C et al (2014) Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc Natl Acad Sci USA 111: 8299–8304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Plett JM, Kemppainen M, Kale SD, Kohler A, Legue V, Brun A, Tyler BM, Pardo AG, Martin F (2011) A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol 21: 1197–1203 [DOI] [PubMed] [Google Scholar]
- 25. Wawra S, Fesel P, Widmer H, Timm M, Seibel J, Leson L, Kesseler L, Nostadt R, Hilbert M, Langen G et al (2016) The fungal‐specific beta‐glucan‐binding lectin FGB1 alters cell‐wall composition and suppresses glucan‐triggered immunity in plants. Nat Commun 7: 13188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deshmukh S, Huckelhoven R, Schafer P, Imani J, Sharma M, Weiss M, Waller F, Kogel KH (2006) The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc Natl Acad Sci USA 103: 18450–18457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lahrmann U, Ding Y, Banhara A, Rath M, Hajirezaei MR, Doehlemann S, von Wiren N, Parniske M, Zuccaro A (2013) Host‐related metabolic cues affect colonization strategies of a root endophyte. Proc Natl Acad Sci USA 110: 13965–13970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zuccaro A, Lahrmann U, Gueldener U, Langen G, Pfiffi S, Biedenkopf D, Wong P, Samans B, Grimm C, Basiewicz M et al (2011) Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica . PLoS Pathog 7: e1002290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qiang X, Zechmann B, Reitz MU, Kogel KH, Schafer P (2012) The mutualistic fungus Piriformospora indica colonizes Arabidopsis roots by inducing an endoplasmic reticulum stress‐triggered caspase‐dependent cell death. Plant Cell 24: 794–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lahrmann U, Zuccaro A (2012) Opprimo ergo sum‐evasion and suppression in the root endophytic fungus Piriformospora indica . Mol Plant Microbe Interact 25: 727–737 [DOI] [PubMed] [Google Scholar]
- 31. Oberwinkler F, Riess K, Bauer R, Selosse M‐A, Weiß M, Garnica S, Zuccaro A (2013) Enigmatic sebacinales. Mycol Prog 12: 1–27 [Google Scholar]
- 32. Qiang X, Weiss M, Kogel KH, Schafer P (2012) Piriformospora indica‐a mutualistic basidiomycete with an exceptionally large plant host range. Mol Plant Pathol 13: 508–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vahabi K, Sherameti I, Bakshi M, Mrozinska A, Ludwig A, Reichelt M, Oelmuller R (2015) The interaction of Arabidopsis with Piriformospora indica shifts from initial transient stress induced by fungus‐released chemical mediators to a mutualistic interaction after physical contact of the two symbionts. BMC Plant Biol 15: 58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Antonioli L, Pacher P, Vizi ES, Hasko G (2013) CD39 and CD73 in immunity and inflammation. Trends Mol Med 19: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matsuoka I, Ohkubo S (2004) ATP‐ and adenosine‐mediated signaling in the central nervous system: adenosine receptor activation by ATP through rapid and localized generation of adenosine by ecto‐nucleotidases. J Pharmacol Sci 94: 95–99 [DOI] [PubMed] [Google Scholar]
- 36. Hilbert M, Voll LM, Ding Y, Hofmann J, Sharma M, Zuccaro A (2012) Indole derivative production by the root endophyte Piriformospora indica is not required for growth promotion but for biotrophic colonization of barley roots. New Phytol 196: 520–534 [DOI] [PubMed] [Google Scholar]
- 37. Lahrmann U, Strehmel N, Langen G, Frerigmann H, Leson L, Ding Y, Scheel D, Herklotz S, Hilbert M, Zuccaro A (2015) Mutualistic root endophytism is not associated with the reduction of saprotrophic traits and requires a noncompromised plant innate immunity. New Phytol 207: 841–857 [DOI] [PubMed] [Google Scholar]
- 38. Guo HJ, Glaeser SP, Alabid I, Imani J, Haghighi H, Kampfer P, Kogel KH (2017) The abundance of endofungal bacterium rhizobium radiobacter (syn. Agrobacterium tumefaciens) increases in its fungal host Piriformospora indica during the tripartite sebacinalean symbiosis with higher plants. Front Microbiol 8: 629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sharma M, Schmid M, Rothballer M, Hause G, Zuccaro A, Imani J, Kaempfer P, Domann E, Schaefer P, Hartmann A et al (2008) Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cell Microbiol 10: 2235–2246 [DOI] [PubMed] [Google Scholar]
- 40. Conesa A, Gotz S, Garcia‐Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- 41. Zheng Q, Wang XJ (2008) GOEAST: a web‐based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res 36: W358–W363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Supek F, Bosnjak M, Skunca N, Smuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6: e21800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim SG, Wang Y, Lee KH, Park ZY, Park J, Wu J, Kwon SJ, Lee YH, Agrawal GK, Rakwal R et al (2013) In‐depth insight into in vivo apoplastic secretome of rice‐Magnaporthe oryzae interaction. J Proteomics 78: 58–71 [DOI] [PubMed] [Google Scholar]
- 44. Poisson G, Chauve C, Chen X, Bergeron A (2007) FragAnchor: a large‐scale predictor of glycosylphosphatidylinositol anchors in eukaryote protein sequences by qualitative scoring. Genomics Proteomics Bioinformatics 5: 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vogel M, Kowalewski H, Zimmermann H, Hooper NM, Turner AJ (1992) Soluble low‐Km 5′‐nucleotidase from electric‐ray (Torpedo‐Marmorata) electric organ and bovine cerebral‐cortex is derived from the glycosyl‐phosphatidylinositol‐anchored ectoenzyme by phospholipase‐C cleavage. Biochem J 284: 621–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klemens MR, Sherman WR, Holmberg NJ, Ruedi JM, Low MG, Thompson LF (1990) Characterization of soluble vs membrane‐bound human placental 5′‐nucleotidase. Biochem Biophys Res Commun 172: 1371–1377 [DOI] [PubMed] [Google Scholar]
- 47. Fini C, Talamo F, Cherri S, Coli M, Floridi A, Ferrara L, Scaloni A (2003) Biochemical and mass spectrometric characterization of soluble ecto‐5 ‘‐nucleotidase from bull seminal plasma. Biochem J 372: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Knapp K, Zebisch M, Pippel J, El‐Tayeb A, Muller CE, Strater N (2012) Crystal structure of the human ecto‐5′‐nucleotidase (CD73): insights into the regulation of purinergic signaling. Structure 20: 2161–2173 [DOI] [PubMed] [Google Scholar]
- 49. Thammavongsa V, Missiakas DM, Schneewind O (2013) Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science 342: 863–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hacquard S, Kracher B, Hiruma K, Munch PC, Garrido‐Oter R, Thon MR, Weimann A, Damm U, Dallery JF, Hainaut M et al (2016) Survival trade‐offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat Commun 7: 11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kumar M, Yadav V, Kumar H, Sharma R, Singh A, Tuteja N, Johri AK (2011) Piriformospora indica enhances plant growth by transferring phosphate. Plant Signal Behav 6: 723–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yadav V, Kumar M, Deep DK, Kumar H, Sharma R, Tripathi T, Tuteja N, Saxena AK, Johri AK (2010) A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J Biol Chem 285: 26532–26544 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53. Schott S, Valdebenito B, Bustos D, Gomez‐Porras JL, Sharma T, Dreyer I (2016) Cooperation through competition ‐ dynamics and microeconomics of a minimal nutrient trade system in arbuscular mycorrhizal symbiosis. Front Plant Sci 7: 912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Burnstock G (2007) Physiology and pathophysiology of purinergic neurotransmission. Physiol Rev 87: 659–797 [DOI] [PubMed] [Google Scholar]
- 55. Zimmermann H, Zebisch M, Strater N (2012) Cellular function and molecular structure of ecto‐nucleotidases. Purinergic Signal 8: 437–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Strater N (2006) Ecto‐5′‐nucleotidase: structure function relationships. Purinergic Signal 2: 343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Daumann M, Fischer M, Niopek‐Witz S, Girke C, Mohlmann T (2015) Apoplastic nucleoside accumulation in Arabidopsis leads to reduced photosynthetic performance and increased susceptibility against Botrytis cinerea . Front Plant Sci 6: 1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim JY, Wu JN, Kwon SJ, Oh H, Lee SE, Kim SG, Wang YM, Agrawal GK, Rakwal R, Kang KY et al (2014) Proteomics of rice and Cochliobolus miyabeanus fungal interaction: insight into proteins at intracellular and extracellular spaces. Proteomics 14: 2307–2318 [DOI] [PubMed] [Google Scholar]
- 59. Basiewicz M, Weiss M, Kogel K‐H, Langen G, Zorn H, Zuccaro A (2012) Molecular and phenotypic characterization of Sebacina uermifera strains associated with orchids, and the description of Piriformospora williamsii sp nov. Fungal Biol 116: 204–213 [DOI] [PubMed] [Google Scholar]
- 60. Kamper J, Kahmann R, Bolker M, Ma LJ, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Muller O et al (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis . Nature 444: 97–101 [DOI] [PubMed] [Google Scholar]
- 61. Molina L, Kahmann R (2007) An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 19: 2293–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yu Q, Tang C, Chen Z, Kuo J (1999) Extraction of apoplastic sap from plant roots by centrifugation. New Phytol 143: 299–304 [Google Scholar]
- 63. Wang L, Berndt P, Xia X, Kahnt J, Kahmann R (2011) A seven‐WD40 protein related to human RACK1 regulates mating and virulence in Ustilago maydis . Mol Microbiol 81: 1484–1498 [DOI] [PubMed] [Google Scholar]
- 64. Lenz JH, Schuchardt I, Straube A, Steinberg G (2006) A dynein loading zone for retrograde endosome motility at microtubule plus‐ends. EMBO J 25: 2275–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gibson DG, Smith HO, Hutchison CA, Venter JC, Merryman C (2010) Chemical synthesis of the mouse mitochondrial genome. Nat Methods 7: 901–U905 [DOI] [PubMed] [Google Scholar]
- 66. Chen S, Songkumarn P, Liu J, Wang GL (2009) A versatile zero background T‐vector system for gene cloning and functional genomics. Plant Physiol 150: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Loubradou G, Brachmann A, Feldbrugge M, Kahmann R (2001) A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis . Mol Microbiol 40: 719–730 [DOI] [PubMed] [Google Scholar]
- 68. Zhang XR, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium‐mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
- 69. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- 70. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(‐Delta Delta C) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 71. Webb B, Sali A (2017) Protein structure modeling with MODELLER. Methods Mol Biol 1654: 39–54 [DOI] [PubMed] [Google Scholar]
- 72. Laskowski RA, Macarthur MW, Moss DS, Thornton JM (1993) Procheck ‐ a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- 73. Wiederstein M, Sippl MJ (2007) ProSA‐web: interactive web service for the recognition of errors in three‐dimensional structures of proteins. Nucleic Acids Res 35: W407–W410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Benkert P, Kunzli M, Schwede T (2009) QMEAN server for protein model quality estimation. Nucleic Acids Res 37: W510–W514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Loew LM, Schaff JC (2001) The virtual cell: a software environment for computational cell biology. Trends Biotechnol 19: 401–406 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Dataset EV1
Review Process File


