The mitochondrial PPR protein EMP12 is required for the splicing of cis- and trans-introns of nad2, complex I biogenesis, and seed development in maize.
Keywords: EMP12, maize, mitochondrion, PPR, RNA splicing, seed development
Abstract
Plant mitochondrial genes contain cis- and trans-group II introns that must be spliced before translation. The mechanism by which these introns are spliced is not well understood. Several families of proteins have been implicated in the intron splicing, of which the pentatricopeptide repeat (PPR) proteins are proposed to confer the substrate binding specificity. However, very few PPRs are characterized. Here, we report the function of a P-type PPR protein, EMP12, and its role in seed development. EMP12 is targeted to mitochondria. Loss-of-function mutation in Emp12 severely arrests embryo and endosperm development, causing embryo lethality. The trans-splicing of mitochondrial nad2 intron 2 and cis-splicing of nad2 intron 4 are abolished, whereas the cis-splicing of nad2 intron 1 is reduced in emp12 mutants. As a result, complex I assembly is disrupted, and its activity is strongly reduced in the mutants. The expression of the alternative oxidase and several components of other mitochondrial complexes is increased, possibly in response to the defective complex I. These results suggest that Emp12 is required for the trans-splicing of nad2 intron 2 and cis-splicing of nad2 introns 1 and 4, and is important to complex I biogenesis, and embryogenesis and endosperm development in maize.
Introduction
Plant mitochondrial genes have prokaryotic characteristics resulting from their origin from endosymbiosis of α-proteobacteria. Subsequently, they evolved novel features of RNA metabolism to adapt to the eukaryotic host cell environments. Mitochondria have lost most of the bacterial genes or transferred genes to the nucleus of the host cell during evolution (Gray et al., 1999; Hammani and Giegé, 2014). Therefore, the mitochondrial genome retains only a small percentage of genes, encoding proteins, tRNAs, and rRNAs that are essential for the oxidative phosphorylation system (OXPHOS) and the translation machinery (Richardson et al., 2013). The maize mitochondrial genome contains 58 genes, 18 of which encode subunits of complex I, III, IV, and V (atp1 double copies) in the OXPHOS system, 4 are involved in the cytochrome c maturation process, 9 genes encode ribosomal proteins, and 21 genes encode tRNAs required for 14 amino acids. In addition, there are three rRNA genes (rrn5, rrn18, and rrn26), a maturase (mat-r) gene residing within the fourth intron of the nad1 transcript (Clifton et al., 2004), and a transporter gene (mttB) in the genome (Clifton et al., 2004). Some genes are transcribed as long polycistronic RNA precursors. To form mature transcripts, these precursor RNAs undergo extensive post-transcriptional processing including RNA editing, which converts the cytidines to uridines (C-to-U) (Takenaka et al., 2013a); intron splicing that removes the cis- and trans-introns and joins the exons (Brown et al., 2014); RNA maturation that trims the 5' or 3' end of precursor mRNAs, and translation regulation that is facilitated by specific RNA-binding proteins (Colas des Francs-Small, et al., 2014; Haïli et al., 2016).
Some mitochondrial genes are interrupted by introns. Based on the structure and splicing mechanism, these introns are classified as group I or group II introns, the latter of which are prevalent in plant mitochondria (Bonen, 2011). Group II introns are large ribonucleoproteins consisting of a catalytic RNA (ribozyme) and an intron-encoded maturase protein with reverse transcriptase activity (Novikova and Belfort, 2017). Structurally, group II introns have six domains (DI–DVI), in which DI, DV, and DVI are essential for splicing (Novikova and Belfort, 2017). There are 19 group II introns in the genes of nad1, nad2, nad4, nad5, and nad7 (encoding components of complex I), and 3 introns in cox2 (cytochrome c oxidase 2 of complex IV), ccmFc (component of cytochrome c maturation), and rps3 (protein translation) in the maize mitochondrial genome (Clifton et al., 2004; Brown et al., 2014). Most introns are in the cis configuration, but some are in trans which require trans-splicing. Trans-introns are believed to result from the DNA rearrangement, causing a break in DIV of the intron and splitting the transcript into two, such that the two exons with the flanking half-intron are transcribed independently in the genome (Malek and Knoop, 1998; Bonen, 2011).
In contrast to bacteria, plant mitochondrial group II introns have lost the activity of self-splicing because of degeneracy and loss of the cognate maturase, leaving only an immobile maturase gene (matR) encoded in nad1 intron 4 (de Longevialle et al., 2010; Sultan et al., 2016). To facilitate the splicing, nucleus-encoded RNA-binding cofactors are recruited, which are from different protein families. For instance, the plant organellar RNA recognition (PORR) protein, WTF9, is required for the splicing of rpl2 and ccmFc introns (Colas des Francs-Small et al., 2012). Similarly, the REGULATOR OF CHROMOSOME CONDENSATION-like protein, RUG3, is associated with the splicing of nad2 intron 2 and intron 3 (Kühn et al., 2011). A member of the mitochondrial transcription termination factor (mTERF) protein family, mTERF15, is involved in the splicing of the nad2 cis-intron 3 (Hsu et al., 2014). Moreover, a chloroplast RNA splicing and ribosome maturation (CRM) protein, mCSF1 (Zmudjak et al., 2013), a putative DEAD-box RNA helicase PMH2 (Köhler et al., 2010), an RAD-52-like protein ODB1 (Samach et al., 2011), and two nucleus-encoded maturases (Keren et al., 2009, 2012) are required for the splicing of mitochondrial introns. In addition to these splicing factors, the prevalent RNA-binding proteins are from the large family of pentatricopeptide repeat (PPR) proteins (Barkan et al., 2014).
PPRs belong to the α-solenoid superfamily of helical repeat proteins, with a large number in nearly all eukaryotic lineages (Fujii and Small, 2011; Barkan and Small, 2014). The structure of PPRs is defined as tandem repeats of a degenerate 35 amino acid repeat motif and a right-handed superhelix that facilitates RNA binding (Ke et al., 2013; Yin et al., 2013; Barkan et al., 2014). PPR proteins are divided into the P- and PLS-subfamily based on the diversity of the C-terminal motifs. The P-subfamily contains the canonical P-motif, whereas the PLS-subfamily additionally harbors longer (L) or shorter (S) variant PPR motifs and additional C-terminal domains (E, E+, and DYW) (Lurin et al., 2004). In land plants, >450 PPR proteins have been found (Fujii and Small, 2011) and they are mainly localized to plastids and mitochondria (Colcombet et al., 2013). Some mitochondrial PPR proteins have been functionally characterized in Arabidopsis, Physcomitrella, rice, and maize (Barkan and Small, 2014; Colas des Francs-Small and Small, 2014). The P-subfamily PPRs usually facilitate RNA intron splicing (Brown et al., 2014; Colas des Francs-Small, et al., 2014; Hsu et al., 2014; Hsieh et al., 2015; Xiu et al., 2016; Cai et al., 2017; Chen et al., 2017; Qi et al., 2017a; Ren et al., 2017; Dai et al., 2018; Sun et al., 2018), RNA stability (Colas des Francs-Small et al., 2014; Lee et al., 2017; Wang et al., 2017; Zhang et al., 2017) or RNA cleavage and translation (Colas des Francs-Small et al., 2014; Haïli et al., 2016), whereas the PLS-subfamily is predominantly involved in RNA editing (Takenaka et al., 2013a; Barkan and Small, 2014; Sun et al., 2015; Qi et al., 2017b; Wang et al., 2017; Yang et al., 2017; Li et al., 2018) and occasionally in RNA splicing (Chateigner-Boutin et al., 2011; Ichinose et al., 2012). However, very few PPRs involved in intron splicing have been functionally characterized in Arabidopsis, as disruption of their functions often causes embryo lethality (Colas des Francs-Small and Small, 2014).
The Nad2 protein is similar to the MRP family of Na+/H+ antiporters and is a likely site for proton transfer in complex I (Hirst, 2013). Previous genetic evidence has pointed to the involvement of RNA helix proteins from distinctive families for splicing of nad2 introns, including RUG3 (Kühn et al., 2011) and mTERF15 (Hsu et al., 2014) in Arabidopsis, loss of function of which results in reduced complex I activity and retarded growth. The maize kernel mutants are ideal materials to study embryo-lethal genes because of the large size and availability of homozygous endosperms and embryos. Currently, three PPR-mediated nad2 intron splicing events in maize have been described (Xiu et al., 2016; Cai et al., 2017; Dai et al., 2018). Maize EMP16, a P-subfamily of PPR proteins, harboring 11 PPR motifs, is involved in the cis-splicing of nad2 intron 4, the lack of which precludes its normal intron splicing and complex I assembly, and, in turn, probably the defect in embryogenesis and endosperm development (Xiu et al., 2016). Mutation of another P-subfamily PPR protein, EMP10, causes loss of nad2 intron 1 splicing, which severely affects complex I activity, and the embryos in emp10 are blocked in the proembryo stage, producing non-viable maize kernels (Cai et al., 2017). Loss of DEK37 expression in the dek37 mutants leads to reduced nad2 intron 1 splicing, such that the embryogenesis and endosperm development are relatively alleviated, displaying a small kernel phenotype (Dai et al., 2018).
In this study, we characterized a mitochondrial PPR protein designated EMP12 affecting the splicing of nad2 introns in maize. Disruption of Emp12 is lethal, giving rise to aborted embryogenesis and endosperm development. The splicing efficiency of nad2 cis-intron 1 and intron 4, and trans-intron 2 is reduced in emp12 mutants, leading to the disassembly of complex I and a reduced complex I activity. Our results imply that EMP12 plays an essential role in nad2 intron splicing, mitochondria functions, and embryo and endosperm development in maize.
Materials and methods
Plant materials
The maize kernel mutants, emp12-673 (UFMu-02085) and emp12-20 (UFMu-07644), in a W22 background were obtained from the Maize Genetics Cooperation Stock Center. The mutants were isolated from the UniformMu transposon tagging population and sequenced by high-throughput Mu-TAIL (thermal asymmetric interlaced) (Settles et al., 2004; McCarty et al., 2005). The Mu insertion was verified by genomic PCR amplification using EMP12-R: AAGCACACCATCTAATGTGTTATCACTATC and specific TIR8 primers (Tan et al., 2011). Subsequently the PCR products were recovered and subjected to sequencing to confirm the Mu insertion position. The Mu active line was introgressed into the W22 inbred background. Primers EMP12-F, CACCATGCTCTTCCTCGTCCGGCG; and EMP12-R2, GGAGCAGGTTGTGGGTCTTCGTGC were used for detection of Emp12 expression in different tissues. EMP12-F; EMP12-F2, AAGACCCACAACCTGCTCCTCCGTG; EMP12-F3, CACTGCGATCCATGCTGTTGGGATG; and EMP12-R were used for detection of Emp12 expression in emp12 mutants. Ubiquitin was used as an internal control that was amplified by primers Ubi-RTF, GCTGGAGGTCGAGAGTAGCGACAC; and Ubi-RTR, TTGACCTCAGCTCGTTGCTGTGG. Primers of Ubiquitin for qRT-PCR analysis were essentially according to Chen et al. (2017). Sequence data for maize Emp12 was from the GenBank database under accession number GRMZM2G023071 and for alternative oxidases (AOXs) under accession numbers AY059646, AY059647, and AY059648.
Subcellular localization of EMP12
The full-length coding sequence of Emp12 was amplified from the maize cDNA of the W22 inbred line using primers EMP12-F and EMP12-R. The cDNA was cloned into pENTR/D-TOPO vector (ThermoFisher Scientific, http://www.thermofisher.com) and the binary pGWB5 vector. The fused Emp12-GFP was infiltrated into tobacco (Nicotiana tabacum) epidermal cells as described in Sun et al. (2015). The fluorescence signals of EMP12–green fluorescent protein (GFP) were detected at 28 h under the Olympus FluoView FV1000 confocal microscope (Olympus, http://www.olympus-global.com). The leaf slices expressing EMP12–GFP signals were dipped in 30 nM MitoTracker solution (ThermoFisher Scientific) at room temperature for 30 min before confocal microscope detection. The excitation wavelengths of GFP and MitoTracker (containing chlorophyll) were 488 nm and 559 nm, respectively.
Light microscopy of cytological sections
The emp12-673 kernels were harvested from self-pollinated heterozygous maize plants at 12 days after pollination (DAP) and 16 DAP. Sectioned kernels were fixed, dehydrated, and stained with Johansen’s Safranin O, and observed under a microscope as described previously (Liu et al., 2013).
Mitochondrial RNA transcript analysis
Total RNAs of embryo and endosperm of kernels at 12 DAP were extracted by using the TRIzol reagent (ThermoFisher Scientific), and subsequently digested with DNase (NEB, USA) and purified using the Ambion PureLink Plant RNA Kit (ThermoFisher Scientific). The cDNA was transcribed using random hexamer primers. Analyses of mitochondrial gene expression and intron splicing were performed in emp12-673 and emp12-20 mutants by reverse transcription–PCR (RT–PCR) and quantitative real-time PCR (qRT-PCR) using the primers listed previously (Xiu et al., 2016; Yang et al., 2017). qRT-PCR analyses were performed using SYBR Green Master Mix (Roche) using a LightCycler (Roche). The flanking exon–exon primers were used for detection of spliced RNA, and the exon–intron flanking primers were used for detection of unspliced RNA. The splicing efficiency is shown as a ratio of spliced to unspliced forms of each transcript in emp12 mutants normalized to wild-type (WT) maize kernels (Colas des Francs-Small et al., 2014).
Mitochondrial protein and complexes analysis
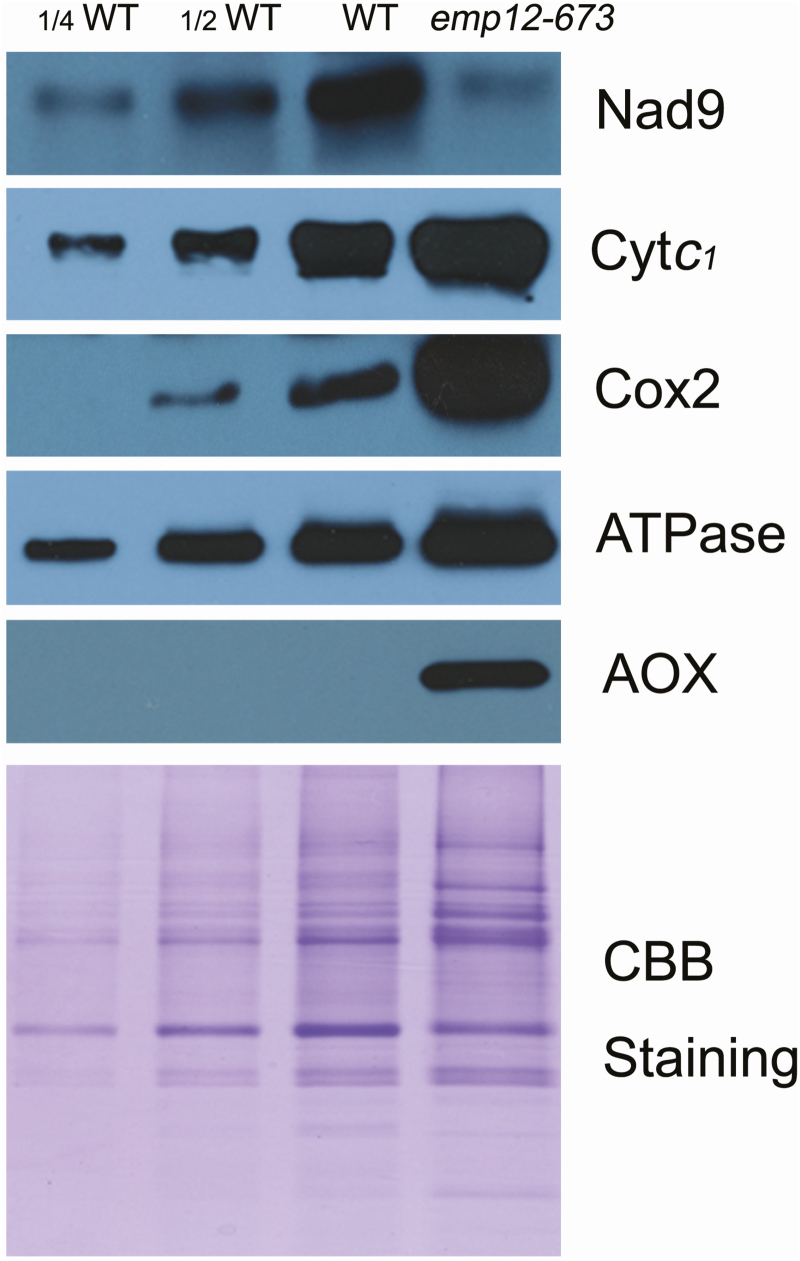
Fresh embryo and endosperm of maize kernels between 12 and 14 DAP were ground in extraction buffer [0.3 M sucrose, 10 mM KH2PO4, pH 7.5, 5 mM tetrasodium pyrophosphate, 2 mM EDTA, 1% (w/v) BSA, 1% (w/v) polyvinylpyrrolidone 40, and 20 mM ascorbic acid] by using a porcelain mortar at 4 °C. The homogenate was filtered through two layers of Miracloth (Calbiochem Co., La Jolla, CA, USA) and centrifuged for 5 min at 3000 g. Crude mitochondria were obtained by centrifugation of the clear supernatant at 20 000 g for 15 min. The mitochondrial membrane proteins were measured by the Bradford assay (Bio-Rad) and a total of 8 µg of denatured proteins were subjected to SDS–PAGE for western blotting analysis (Sun et al., 2015). A 100 µg aliquot of mitochondrial crude membrane proteins was solubilized in 1% N-dodecylmaltoside and separated by 3–12% blue native gel electrophoresis (BN-PAGE) (ThermoFisher Scientific) as described in Sun et al. (2015). The gel strips were stained by Coomassie Brilliant Blue R-250 (CBB) and in-gel nitroblue tetrazolium (NBT)-NADH as described in Meyer et al. (2009). The gel strips were incubated in 50 mM Tris–HCl, pH 6.8, 8 M urea, 1% (w/v) SDS, and 0.5% (w/v) β-mercaptoethanol for 30 min to denature the complexes and subjected to PVDF (polyvinylidene difluoride) membrane transfer and western blotting by incubating the antiserum against Nad9 (complex I/NADH dehydrogenase subunit 9) (Lamattina et al., 1993), maize cytochorme c1 (Cytc1), Arabidopsis Cox2 (Agrisera), ATPase α-subunit (ATP-A), and AOX for detection of complex I, III, VI, and V, and total AOXs, respectively (Xiu et al., 2016).
Results and Discussion
Embryo and endosperm development are arrested in emp12
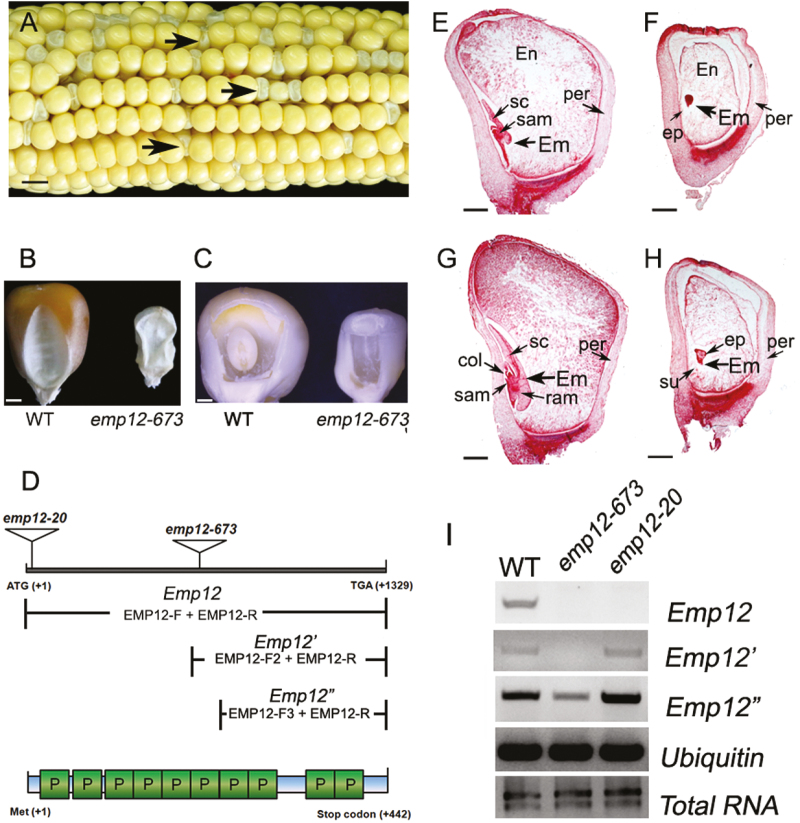
During the screen of seed mutants from the UniformMu population (McCarty et al., 2005), we identified the emp12 mutant. The emp12 mutant displays a severe empty pericarp phenotype (Fig. 1A–C) and cannot survive, suggesting that it is embryo-lethal. This mutant was put in the massive extraction of flanking sequences by high-throughput Mu-TAIL (Settles et al., 2004). Sequence analysis indicates that the Mu element is inserted in the coding region 673 bp downstream of the ATG codon of a putative PPR gene (GRMZM2G023071) (Fig. 1D). Hence we named it emp12-673. The selfed progeny of heterozygous emp12-673/+ plants displayed a 1:3 ratio [emp:(WT + heterozygotes), 140:426, χ2=0.02] in the WT and empty pericarp kernels, indicating that the mutation is monogenic, recessive, and nuclear. Co-segregation analysis was performed to test the linkage of emp12-673 using Emp12-specifc and Mu TIR8 primers (Tan et al., 2011). No recombination was detected from a segregating population from a self-progeny of an emp12-673/Emp12 plant, suggesting that the Mu insertion is tightly linked to the Emp12 mutation (see Supplementary Fig. S1 at JXB online). At 12 DAP, the emp12 kernels were much smaller than those of the WT, displaying obscure embryo structures and vitreous endosperm (Fig. 1C). Sectioned homozygous emp12-673 and WT kernels under microscopy indicated that the emp12-673 mutant kernel displays a remarkable developmental retardation of the embryo and endosperm at 12 DAP (Fig. 1F). In the WT, the embryo had already formed a scutellum and shoot apical meristem, and there were clearly visible endosperm cells (Fig. 1E). At 16 DAP, the WT kernels exhibited significant growth and the pericarp clung tightly to the endosperm (Fig. 1G), while the emp12-673 kernels grew more slowly, displaying a more crumpled empty pericarp. The emp12-673 endosperm accumulated less starch and the embryo development stagnated at the transitional stage, remaining as an undifferentiated embryo and suspensor (Fig. 1H). Taken together, the Emp12 mutation arrests embryo development at the transition stage and severely delays embryo and endosperm development, suggesting an essential role for Emp12 in embryogenesis and endosperm development.
Fig. 1.
The maize Emp12 gene is involved in embryogenesis and endosperm development. (A) A self-pollinated ear segregating for emp12-673 mutant kernels at 15 days after pollination (DAP). Arrows show the emp maize kernels. Scale bar=0.5 cm. (B) The dried kernels of emp12-673 mutants and the wild type (WT). Scale bar=2 mm. (C) The embryo and endosperm of emp12-673 mutant and WT kernels at 12 DAP. Arrows indicate the embryo (Em). Scale bar=2 mm. (D) Schematic diagram of the Emp12 gene and its protein structure, showing the Mu insertion sites of emp12-673 and emp12-20. The expression of full-length and partial Emp12 (Emp12' and Emp12'') downstream of the insertion sites was detected by RT–PCR analysis, with the combinations of primers EMP12-F, EMP12-F2, EMP12-F3, and EMP12-R. PPR motifs (P) of EMP12 are predicted by TPRpred (https://toolkit.tuebingen.mpg.de/#/tools/tprpred). (E–H) Light microscopy of cytological sections of WT (E, G) and emp12-673 mutant kernels (F, H) are longitudinally sectioned early at 12 DAP (E, F) and late at 16 DAP (G, H). En, endosperm; Em, embryo; per, pericarp; sc, scutellum; su, suspensor; col, coleoptile; ep, embryo proper; sam, shoot apical meristem; ram, root apical meristem. Scale bar=1 mm. (I) RT–PCR analysis of full-length Emp12 and truncated Emp12' and Emp12'' expression indicated in (D) was performed in the emp12-673 and emp12-20 mutants and WT siblings at 12 DAP, with normalization by Ubiquitin primers.
To determine whether the mutation in GRMZM2G023071 accounts for the emp12 phenotype, another independent mutant of Emp12 from the UniformMu population was analyzed. This Mu element inserted at +20 bp downstream of the ATG in Emp12 (emp12-20) as indicated by the linkage and genomic PCR analysis (Fig. 1D). The selfed progeny of emp12-20 heterozygotes separated emp kernels as emp12-673. Furthermore, an allelism test by reciprocal crosses between emp12-673/+ and emp12-20/+ heterozygotes produced emp mutant kernels, confirming that each allele could not complement each other, hence GRMZM2G023071 is the causative gene for the emp12 phenotype. RT–PCR amplification of the Emp12 transcripts in these two alleles failed to detect WT Emp12 transcripts (Fig. 1I). However, we did detect the transcripts downstream of the Mu insertion, suggesting the expression of the Mu downstream region and/or the transcript containing the Mu insertion. In any case, the result indicates that the WT EMP12 cannot be produced in the two emp12 alleles. In summary, the disruption of Emp12 results in arrested embryogenesis and endosperm development.
EMP12 is a P-subfamily PPR protein that localizes in mitochondria
Emp12 (GRMZM2G023071) is an intronless gene encoding a P-subfamily PPR protein. This PPR harbors 442 amino acids and is predicted to have 10 putative PPR motifs (Lurin et al., 2004; Cheng et al., 2016). EMP12 is closely related to Sb06g030430 from Sorghum bicolor (93%) and LOC_Os04g55090 from Oryza sativa (84%). However, no close homolog of EMP12 in was found Arabidopsis thaliana (Supplementary Fig. S2). The expression of Emp12 is ubiquitous in a range of vegetative and reproductive tissues, showing relatively higher levels in leaves, stems, roots, silk, and developing kernels (Supplementary Fig. S3).
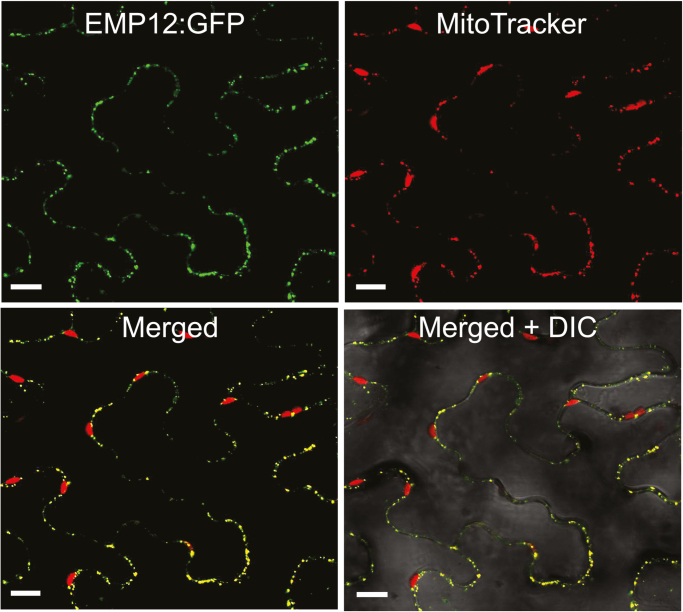
The EMP12 protein contains an N-terminal signal peptide that is targeted to mitochondria (http://www.cbs.dtu.dk/services/TargetP/; Small et al., 2004). To investigate the subcellular targeting of EMP12, the full-length Emp12 sequence was fused to GFP and transiently expressed in N. tabacum epidermal cells. Confocal laser scanning microscopy indicated that the fluorescence signals of green EMP12:GFP co-localize with the specific mitochondrial marker, MitoTracker, but not with chloroplasts (Fig. 2), suggesting that EMP12 locates exclusively to the mitochondrion.
Fig. 2.
EMP12 localizes in the mitochondrion. Full-length Emp12 was fused to green fluorescent protein (GFP) and introduced into Nicotiana tabacum epidermal cells, and fluorescence signals were detected under a confocal microscope. EMP12:GFP co-localizes with the specific mitochondrial marker, MitoTracker, but not with the chlorophyll fluorescence. DIC, differential interference contrast. Scale bar=10 µm.
Splicing of nad2 introns 1, 2, and 4 is defective in emp12
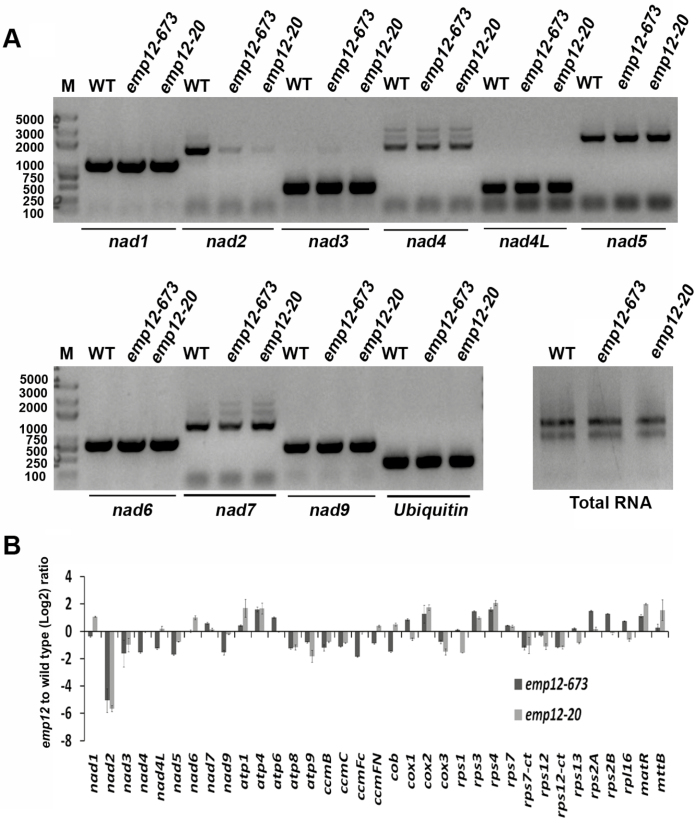
EMP12 belongs to the P-subfamily PPR proteins that are involved in intron splicing, RNA stability, and RNA maturation (Barkan and Small, 2014). To gain insight into EMP12 function, the expression of mitochondrial transcripts was measured by RT–PCR and qRT-PCR analysis between the WT and emp12 mutant kernels (Fig. 3). The results indicated that the expression of the nad2 transcript which encodes the complex I/NADH dehydrogenase subunit2 (Nad2) was significantly reduced in the two emp12 mutant alleles. However, no distinguishable differences were found in the expression of other mitochondrial transcripts (Fig. 3), suggesting a defective RNA processing of the nad2 transcript in emp12 mutants.
Fig. 3.
The emp 12 mutants only affect expression of nad2 in the mitochondria. Total RNA was extracted from fresh maize kernels at 12 DAP and reverse transcribed using hexamer primers. RT–PCR analysis was performed by using three biological replicates and was normalized to Ubiquitin. (A) Transcript analysis of nad genes in emp12 mutant alleles. (B) Expression levels of mitochondrial transcripts were quantified by qRT-PCR analysis. The transcript abundance was plotted as emp12/wild-type log2 ratios using Ubiquitin for normalization.
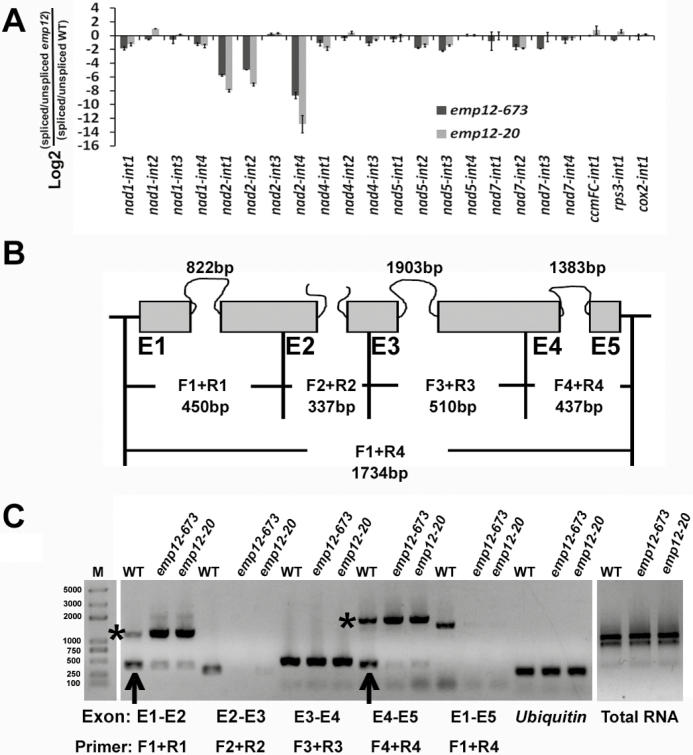
The nad2 transcript in maize contains four introns; intron 2 is trans-spliced while the rest are cis-spliced (Fig. 4B). Since nad2 is greatly reduced in emp12 mutants, possible splicing defects in nad2 introns were monitored by using both exon–exon and exon–intron flanking primers (Xiu et al., 2016; Yang et al., 2017). The results show that in emp12-673 and emp12-20 alleles, the trans-splicing of intron 2 and cis-splicing of intron 4 of the nad2 transcript were lost, whereas the cis-splicing of intron 1 of nad2 was reduced (Fig. 4A), pointing to the requirement for Emp12 in nad2 intron splicing. Moreover, no differences were found in the introns of the other five mitochondrial transcripts (Fig. 4A). Prediction of the binding sites by the recognition code at positions 6 and 1' indicated that these three introns might share similar binding sequences that are not present in other introns (Barkan et al., 2012; Takenaka et al., 2013b) (Supplementary Fig. S4). Therefore, the reduced expression of nad2 in emp12 mutants is associated with the splicing defects of three nad2 introns.
Fig. 4.
Emp12 is required for intron 1, intron 2, and intron 4 splicing of mitochondrial nad2. (A) qRT-PCR analysis of all group II introns in maize mitochondrial genes. Total RNA was isolated from the emp12-673 and emp12-20 mutant kernels at 12 DAP. Values represent the log2 ratio of spliced to unspliced forms for each transcript in the mutants compared with WT maize kernels. Each value is the mean of at least three biological replicates. (B) Structure of the maize nad2 gene. Exons are shown as filled gray boxes. The closed and open lines stand for cis- and trans-introns. Primers (F1+R1, F2+R2, F3+R3, F4+R4, and F1+R4) indicate the PCR products by using flanking exon–exon primers as described previously (Xiu et al., 2016). E1–E5, exon1–exon5. (C) RT–PCR analysis of the intron splicing of nad2 introns using exon–exon primers as indicated in (B). Arrows and asterisks indicate the spliced and unspliced PCR products, respectively.
Complex I biogenesis is reduced in emp12
To gain insight into whether the splicing defect of nad2 introns affects the function of the respiration chain in the emp12 mutants, representative mitochondrion-encoded protein components of each of the mitochondrial complexes in the maize kernels were first monitored by western blot analysis. The results showed that the protein abundance of Nad9 (complex I/NADH dehydrogenase subunit 9) (Lamattina et al., 1993), which is a peripheral membrane subunit of complex I, was severely reduced in emp12-673. This scenario is also seen in other nad2 intron splicing mutants such as emp16 (Xiu et al., 2016), suggesting that the lack of nad2 splicing results in the lost stability of peripheral proteins of complex I. In contrast, a core membrane subunit from complex III, Cytc1, was strongly increased. In addition, Cox2 (cytochrome oxidase subunit2) from complex IV, and mitochondrial ATP synthase α-subunit from complex V, were also increased in emp12-673 mutants (Fig. 5). A possible explanation is that the mitochondrial respiratory chain contains 92 subunits, comprising both mitochondrion- and nucleus-encoded components (Jacoby et al., 2012; Subrahmanian et al., 2016). In emp12 mutants, complex I is affected, so expression of proteins from other complexes or branched electron transport chains would be enhanced to adapt to the altered electron transfer state and NADH accumulation.
Fig. 5.
Protein abundance of the mitochondrial respiration chain is affected in the emp12 mutants. Freshly prepared mitochondrial membrane proteins in emp12 and WT maize kernels were subjected to SDS–PAGE, and the proteins were then transferred to a polyvinylidene difluoride (PVDF) membrane and probed with antibodies against Nad9, Cytc1 (cytochrome c1), Cox2, Cytc (cytochrome c), ATPase, and AOX (alternative oxidase). Coomassie Brilliant Blue- (CBB) stained gels are shown to demonstrate that equal amounts of proteins were loaded.
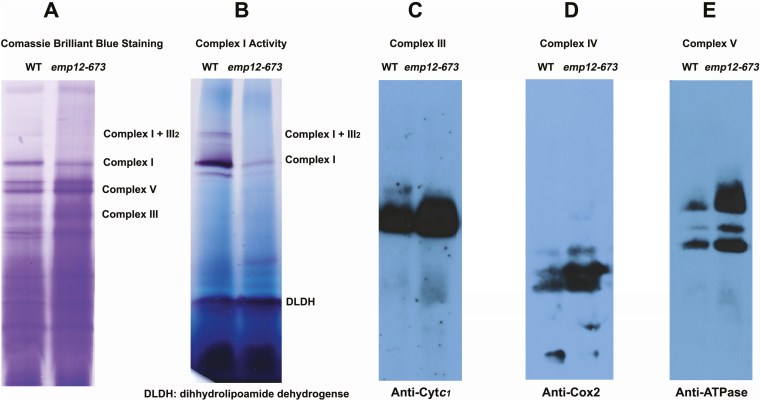
The assembly and amount of respiratory complexes of emp12 were further determined by BN-PAGE using dissolved crude mitochondrial membrane proteins from embryo and endosperm. Complex I in emp12 was strongly reduced, leaving little assembled complex I as indicated by the CBB and in-gel NBT-NADH activity staining. In addition, the supercomplex (I+III2) in emp12-673 was strongly reduced, suggesting that the assembly of complex I in emp12 mitochondria is severely impeded (Fig. 6A, B). In contrast, as shown by CBB staining and western blotting, complex III, IV, and V accumulated to levels greater than found in the WT (Fig. 6C–E). A similar shift in the relative amounts of these complexes was also noted with other complex I mutants such as nmat1, 2, 4 mutants in Arabidopsis (Keren et al., 2009, 2012; Cohen et al., 2014) and emp16 and emp8 mutants in maize (Xiu et al., 2016; Sun et al., 2018).
Fig. 6.
The Emp12 mutation impairs mitochondrial biogenesis in emp12 mutants. The crude mitochondrial membrane was freshly extracted from the emp12 mutant kernels. The complexes were solubilized by 1% dodecyl maltoside and subjected to blue native gel electrophoresis (BN-PAGE) as described in Sun et al. (2015). (A) The mitochondrial membrane complexes were separated by the BN gels and stained with Coomassie Brilliant Blue. (B) Detection of NADH dehydrogenase activity of complex I. Dihydrolipoamide dehydrogenase (DLDH) was used as a loading control. (C–E) Accumulation of respiratory chain complex III, IV, and V in emp12 mutant kernels. The BN gels were denatured, transferred onto the PVDF membranes, and probed with antibodies against Cytc1 for complex III (C), Cox2 for complex IV (D), and ATPase (α-subunit) for complex V (E).
Alternative oxidase is activated in emp12
AOX drains the electrons from the ubiquinone pool, by-passing the cytochrome c pathway for ATP synthesis (Moore and Siedow, 1991; Kühn et al., 2015). Both RT–PCR and qRT-PCR analyses indicated that among the three AOX genes, the AOX2 transcript in emp12 mutant alleles was strongly increased in comparison with the WT, whereas AOX1 and AOX3 showed an indistinguishable expression (Supplementary Fig. S5). Western blotting confirmed that the maize AOX proteins in the emp12 mutant were strongly increased (Fig. 6), which is consistent with the scenarios occurring in other complex I mutants (Li et al., 2014; Xiu et al., 2016; Chen et al., 2017; Cai et al., 2017; Qi et al., 2017a, b, Ren et al., 2017; Zhang et al., 2017; Dai et al., 2018; Sun et al., 2018). These results indicate that the alternative pathway is activated to reduce levels of reactive oxygen species (ROS) when electron flow is improperly maintained through the cytochrome c pathway in emp12 mutants (Wagner and Moore, 1997).
Splicing of one intron of nad2 involves the co-ordination of more than one splicing factor
EMP12 was found to act on nad2 intron 4 splicing, whereas EMP16 also specifically participates in the splicing of this intron, implying that more than one splicing factor is involved in the splicing of a specific intron (Supplementary Fig. S4B). EMP12 is also involved in the cis-splicing of nad2 intron 1, which has been described to require other PPR proteins, such as EMP10 (Cai et al., 2017), DEK37 (Dai et al., 2018), and EMP8 (Sun et al., 2018) in maize. In addition to maize, it has also been found that in Arabidopsis, splicing factors from various families, namely the PPR protein MTSF1 (Haïli et al., 2013), together with the CRM protein mCSF1 (Zmudjak et al., 2013), the DEAD-box protein PMH2 (Köhler et al., 2010), a maturase nMAT1 (Keren et al., 2012), and a RAD-52-like protein ODB1 (Samach et al., 2011), are involved in the nad2 intron 1 splicing. Other nad2 introns, such as cis-intron 3, require the Arabidopsis PPR protein ABO5 (Liu et al., 2010), mCSF1 (Zmudjak et al., 2013), the TERF family protein mTERF15 (Hsu et al., 2014), and the RUG protein RUG3 (Kühn et al., 2011) for the splicing. These results suggest that splicing of a specific intron involves the co-ordination of specialized and general RNA-binding proteins. In addition, these splicing factors, particularly PPRs, showed distinct disparity (or at least lack of distinct sequence conservation) on one intron in monocots (i.e. maize) and dicots (i.e. Arabidopsis). It may reflect the evolutionary divergence and the complexity of splicing. The requirement for a splicing factor in organelles is probably a co-evolutionary result between the mutation in the intron and the corresponding recruitment of a nuclear-encoded protein such as the PPR proteins. In addition, it is also probable that the splicing of a specific intron requires multiple PPR proteins. If the mutation in the intron occurs after the divergence of monocots and eudicots, the splicing PPRs could be different in monocots and eudicots although they function on the same intron. If the mutation is prior to the divergence, the PPR proteins could be similar.
PPRs probably recognize the intron-binding sequences independently or, alternatively, they most probably form a highly dynamic complex similar to the nuclear spliceosome. Yeast two-hybrid analyses between EMP12 and EMP16 revealed no direct interaction (Supplementary Fig. S6). However, PPRs have been described to form a complex with other unknown proteins. PNM1, a dual-targeted PPR protein, has been implicated in a 120 kDa complex (Hammani et al., 2011; Senkler et al., 2017). GRP23 (Ding et al., 2006) was recently identified in a 160 kDa complex (Senkler et al., 2017) and is in a complex including PMH2 and nMAT2 in mitochondria (Zmudjak et al., 2017). Two PPR proteins, DYW2 and NUWA (He et al., 2017), constitute the main components of the editosome in mitochondria. They interact with the mitochondrial PPR protein SLO2 and chloroplast PPR protein CLB19 for RNA editing. NUWA is thought to act as a general bridge for the editing of SLO2 and CLB19 at specific sites (Andres-Colas et al., 2017; Guillaumot et al., 2017). It is most likely that EMP12 constitutes a complex similar to the highly dynamic nuclear spliceosome for each intron, either transiently or stably, as it specifically acts on three introns of the nad2 transcript, to maintain the configuration in a ribozyme active state.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The Mu insertion in Emp12 linked with the empty pericarp phenotype in the emp12-673 allele.
Fig. S2. The amino acid alignment of EMP12 homologs.
Fig. S3. qRT-PCR analysis of Emp12 expression in different tissues and kernels at different developing stages.
Fig. S4. Predicted binding sites of EMP12 and EMP16 in nad2 introns.
Fig. S5. AOX2 expression is increased in emp12 mutants.
Fig. S6. EMP12 did not interact with EMP16 as demonstrated by yeast two-hybrid assay.
Acknowledgements
We thank Professor Larkin Curtis Hannah (University of Florida) for editing the manuscript, and Dr Tsuyoshi Nakagawa (Shimane University, Japan) for the pGWB vectors. This work was supported by the National Natural Science Foundation of China (project nos 31630053, 91735301, 91435201, 31500199, and 31700210) and the Natural Science Foundation of Shandong Province (ZR2017BC084).
Author contributions
FS and BT conceived and designed the experiments: FS, ZX, RJ, YL, XZ, and YY performed the experiments; FS, ZX, WY, and BT analyzed the data; XL contributed reagents/materials/analysis tools; and FS and BT wrote the manuscript.
References
- Andres-Colas N, Zhu Q, Takenaka M, De Rybel B, Weijers D, and Van Der Straeten D. 2017. Multiple PPR protein interactions are involved in the RNA editing system in Arabidopsis mitochondria and plastids. Proceedings of the National Academy of Sciences, USA 114, 8883–8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. 2012. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genetics 8, e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I. 2014. Pentatricopeptide repeat proteins in plants. Annual Review of Plant Biology 65, 415–442. [DOI] [PubMed] [Google Scholar]
- Bonen L. 2011. RNA splicing in plant mitochondria. In: Kempken F, ed. Plant mitochondria. New York: Springer, 131–155. [Google Scholar]
- Brown GG, Colas des Francs-Small C, Ostersetzer-Biran O. 2014. Group II intron splicing factors in plant mitochondria. Frontiers in Plant Science 5, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Li S, Sun F, Sun Q, Zhao H, Ren X, Zhao Y, Tan BC, Zhang Z, Qiu F. 2017. Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. The Plant Journal 91, 132–144. [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, des Francs-Small CC, Delannoy E, Kahlau S, Tanz SK, de Longevialle AF, Fujii S, Small I. 2011. OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. The Plant Journal 65, 532–542. [DOI] [PubMed] [Google Scholar]
- Chen X, Feng F, Qi W, Xu L, Yao D, Wang Q, Song R. 2017. Dek35 encodes a PPR protein that affects cis-splicing of mitochondrial nad4 intron 1 and seed development in maize. Molecular Plant 10, 427–441. [DOI] [PubMed] [Google Scholar]
- Cheng S, Gutmann B, Zhong X, et al. 2016. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. The Plant journal 85, 532–547. [DOI] [PubMed] [Google Scholar]
- Clifton SW, Minx P, Fauron CM, et al. 2004. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiology 136, 3486–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Zmudjak M, Colas des Francs-Small C, et al. 2014. nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. The Plant Journal 78, 253–268. [DOI] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Falcon de Longevialle A, Li Y, Lowe E, Tanz SK, Smith C, Bevan MW, Small I. 2014. The pentatricopeptide repeat proteins TANG2 and ORGANELLE TRANSCRIPT PROCESSING439 are involved in the splicing of the multipartite nad5 transcript encoding a subunit of mitochondrial complex I. Plant Physiology 165, 1409–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Small I. 2014. Surrogate mutants for studying mitochondrially encoded functions. Biochimie 100, 234–242. [DOI] [PubMed] [Google Scholar]
- Colcombet J, Lopez-Obando M, Heurtevin L, Bernard C, Martin K, Berthomé R, Lurin C. 2013. Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biology 10, 1557–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Luan S, Chen X, Wang Q, Feng Y, Zhu C, Qi W, Song R. 2018. Maize Dek37 encodes a P-type PPR protein that affects cis-splicing of mitochondrial nad2 intron 1 and seed development. Genetics 208, 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle AF, Small ID, Lurin C. 2010. Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Molecular Plant 3, 691–705. [DOI] [PubMed] [Google Scholar]
- Ding YH, Liu NY, Tang ZS, Liu J, Yang WC. 2006. Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. The Plant Cell 18, 815–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francs-Small CC, Kroeger T, Zmudjak M, Ostersetzer-Biran O, Rahimi N, Small I, Barkan A. 2012. A PORR domain protein required for rpl2 and ccmF(C) intron splicing and for the biogenesis of c-type cytochromes in Arabidopsis mitochondria. The Plant Journal 69, 996–1005. [DOI] [PubMed] [Google Scholar]
- Fujii S, Small I. 2011. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytologist 191, 37–47. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. 1999. Mitochondrial evolution. Science 283, 1476–1481. [DOI] [PubMed] [Google Scholar]
- Guillaumot D, Lopez-Obando M, Baudry K, et al. 2017. Two interacting PPR proteins are major Arabidopsis editing factors in plastid and mitochondria. Proceedings of the National Academy of Sciences, USA 114, 8877–8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haïli N, Arnal N, Quadrado M, Amiar S, Tcherkez G, Dahan J, Briozzo P, Colas des Francs-Small C, Vrielynck N, Mireau H. 2013. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Research 41, 6650–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haïli N, Planchard N, Arnal N, Quadrado M, Vrielynck N, Dahan J, des Francs-Small CC, Mireau H. 2016. The MTL1 pentatricopeptide repeat protein is required for both translation and splicing of the mitochondrial NADH DEHYDROGENASE SUBUNIT7 mRNA in Arabidopsis. Plant Physiology 170, 354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Giegé P. 2014. RNA metabolism in plant mitochondria. Trends in Plant Science 19, 380–389. [DOI] [PubMed] [Google Scholar]
- Hammani K, Gobert A, Hleibieh K, Choulier L, Small I, Giegé P. 2011. An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. The Plant Cell 23, 730–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Sun Y, Yang Q, et al. 2017. A novel imprinted gene NUWA controls mitochondrial function in early seed development in Arabidopsis. PLoS Genetics 13, e1006553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J. 2013. Mitochondrial complex I. Annual Review of Biochemistry 82, 551–575. [DOI] [PubMed] [Google Scholar]
- Hsieh WY, Liao JC, Chang CY, Harrison T, Boucher C, Hsieh MH. 2015. The SLOW GROWTH3 pentatricopeptide repeat protein is required for the splicing of mitochondrial NADH dehydrogenase subunit7 intron 2 in Arabidopsis. Plant Physiology 168, 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YW, Wang HJ, Hsieh MH, Hsieh HL, Jauh GY. 2014. Arabidopsis mTERF15 is required for mitochondrial nad2 intron 3 splicing and functional complex I activity. PLoS One 9, e112360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose M, Tasaki E, Sugita C, Sugita M. 2012. A PPR-DYW protein is required for splicing of a group II intron of cox1 pre-mRNA in Physcomitrella patens. The Plant Journal 70, 271–278. [DOI] [PubMed] [Google Scholar]
- Jacoby RP, Li L, Huang S, Pong Lee C, Millar AH, Taylor NL. 2012. Mitochondrial composition, function and stress response in plants. Journal of Integrative Plant Biology 54, 887–906. [DOI] [PubMed] [Google Scholar]
- Ke J, Chen RZ, Ban T, et al. 2013. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nature Structural & Molecular Biology 20, 1377–1382. [DOI] [PubMed] [Google Scholar]
- Keren I, Bezawork-Geleta A, Kolton M, Maayan I, Belausov E, Levy M, Mett A, Gidoni D, Shaya F, Ostersetzer-Biran O. 2009. AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15, 2299–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Tal L, des Francs-Small CC, Araújo WL, Shevtsov S, Shaya F, Fernie AR, Small I, Ostersetzer-Biran O. 2012. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. The Plant Journal 71, 413–426. [DOI] [PubMed] [Google Scholar]
- Köhler D, Schmidt-Gattung S, Binder S. 2010. The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana. Plant Molecular Biology 72, 459–467. [DOI] [PubMed] [Google Scholar]
- Kühn K, Carrie C, Giraud E, Wang Y, Meyer EH, Narsai R, des Francs-Small CC, Zhang B, Murcha MW, Whelan J. 2011. The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. The Plant Journal 67, 1067–1080. [DOI] [PubMed] [Google Scholar]
- Kühn K, Yin G, Duncan O, et al. 2015. Decreasing electron flux through the cytochrome and/or alternative respiratory pathways triggers common and distinct cellular responses dependent on growth conditions. Plant Physiology 167, 228–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamattina L, Gonzalez D, Gualberto J, Grienenberger JM. 1993. Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. European Journal of Biochemistry 217, 831–838. [DOI] [PubMed] [Google Scholar]
- Lee K, Han JH, Park YI, Colas des Francs-Small C, Small I, Kang H. 2017. The mitochondrial pentatricopeptide repeat protein PPR19 is involved in the stabilization of NADH dehydrogenase 1 transcripts and is crucial for mitochondrial function and Arabidopsis thaliana development. New Phytologist 215, 202–216. [DOI] [PubMed] [Google Scholar]
- Li XJ, Zhang YF, Hou M, et al. 2014. Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). The Plant Journal 79, 797–809. [DOI] [PubMed] [Google Scholar]
- Li XL, Huang WL, Jiang RC, Sun F, Wang HC, Zhao J, Xu CH, Tan BC. 2018. EMP18 functions in mitochondrial atp6 and cox2 transcript editing and is essential to seed development in maize. New Phytologist doi: 10.1111/nph.15425. [DOI] [PubMed] [Google Scholar]
- Liu Y, He J, Chen Z, Ren X, Hong X, Gong Z. 2010. ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. The Plant Journal 63, 749–765. [DOI] [PubMed] [Google Scholar]
- Liu YJ, Xiu ZH, Meeley R, Tan BC. 2013. Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. The Plant Cell 25, 868–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, et al. 2004. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. The Plant Cell 16, 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek O, Knoop V. 1998. Trans-splicing group II introns in plant mitochondria: the complete set of cis-arranged homologs in ferns, fern allies, and a hornwort. RNA 4, 1599–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Settles AM, Suzuki M, et al. 2005. Steady-state transposon mutagenesis in inbred maize. The Plant Journal 44, 52–61. [DOI] [PubMed] [Google Scholar]
- Meyer EH, Tomaz T, Carroll AJ, Estavillo G, Delannoy E, Tanz SK, Small ID, Pogson BJ, Millar AH. 2009. Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiology 151, 603–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AL, Siedow JN. 1991. The regulation and nature of the cyanide-resistant alternative oxidase of plant mitochondria. Biochimica et Biophysica Acta 1059, 121–140. [DOI] [PubMed] [Google Scholar]
- Novikova O, Belfort M. 2017. Mobile group II introns as ancestral eukaryotic elements. Trends in Genetics 33, 773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Tian Z, Lu L, Chen X, Chen X, Zhang W, Song R. 2017a. Editing of mitochondrial transcripts nad3 and cox2 by Dek10 is essential for mitochondrial function and maize plant development. Genetics 205, 1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Yang Y, Feng X, Zhang M, Song R. 2017b. Mitochondrial function and maize kernel development requires Dek2, a pentatricopeptide repeat protein involved in nad1 mRNA splicing. Genetics 205, 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Pan Z, Zhao H, Zhao J, Cai M, Li J, Zhang Z, Qiu F. 2017. EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize. Journal of Experimental Botany 68, 4571–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. 2013. The ‘fossilized’ mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biology 11, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Melamed-Bessudo C, Avivi-Ragolski N, Pietrokovski S, Levy AA. 2011. Identification of plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. The Plant Cell 23, 4266–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkler J, Senkler M, Eubel H, Hildebrandt T, Lengwenus C, Schertl P, Schwarzländer M, Wagner S, Wittig I, Braun HP. 2017. The mitochondrial complexome of Arabidopsis thaliana. The Plant Journal 89, 1079–1092. [DOI] [PubMed] [Google Scholar]
- Settles AM, Latshaw S, McCarty DR. 2004. Molecular analysis of high-copy insertion sites in maize. Nucleic Acids Research 32, e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. 2004. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4, 1581–1590. [DOI] [PubMed] [Google Scholar]
- Subrahmanian N, Remacle C, Hamel PP. 2016. Plant mitochondrial complex I composition and assembly: a review. Biochimica et Biophysica Acta 1857, 1001–1014. [DOI] [PubMed] [Google Scholar]
- Sultan LD, Mileshina D, Grewe F, et al. 2016. The reverse transcriptase/RNA maturase protein MatR is required for the splicing of various group II introns in Brassicaceae mitochondria. The Plant Cell 28, 2805–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Wang X, Bonnard G, Shen Y, Xiu Z, Li X, Gao D, Zhang Z, Tan BC. 2015. Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required for ccmFN editing, mitochondrial function and seed development in maize. The Plant Journal 84, 283–295. [DOI] [PubMed] [Google Scholar]
- Sun F, Zhang XY, Shen Y, Wang HC, Liu R, Wang XM, Gao DH, Yang YZ, Liu YW, Tan BC. 2018. EMPTY PERICARP8 is required for the splicing of three mitochondrial introns and seed development in maize. The Plant Journal 95, 919–932. [DOI] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Brennicke A, Graichen K. 2013a. Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One 8, e65343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Verbitskiy D, Härtel B, Brennicke A. 2013b. RNA editing in plants and its evolution. Annual Review of Genetics 47, 335–352. [DOI] [PubMed] [Google Scholar]
- Tan BC, Chen Z, Shen Y, Zhang Y, Lai J, Sun SS. 2011. Identification of an active new mutator transposable element in maize. G3 1, 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AM, Moore AL. 1997. Structure and function of the plant alternative oxidase: its putative role in the oxygen defence mechanism. Bioscience Reports 17, 319–333. [DOI] [PubMed] [Google Scholar]
- Wang C, Aubé F, Planchard N, Quadrado M, Dargel-Graffin C, Nogué F, Mireau H. 2017. The pentatricopeptide repeat protein MTSF2 stabilizes a nad1 precursor transcript and defines the 3' end of its 5'-half intron. Nucleic Acids Research 45, 6119–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhong M, Shuai B, Song J, Zhang J, Han L, Ling H, Tang Y, Wang G, Song R. 2017. E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytologist 214, 1563–1578. [DOI] [PubMed] [Google Scholar]
- Xiu Z, Sun F, Shen Y, Zhang X, Jiang R, Bonnard G, Zhang J, Tan BC. 2016. EMPTY PERICARP16 is required for mitochondrial nad2 intron 4 cis-splicing, complex I assembly and seed development in maize. The Plant Journal 85, 507–519. [DOI] [PubMed] [Google Scholar]
- Yang YZ, Ding S, Wang HC, Sun F, Huang WL, Song S, Xu C, Tan BC. 2017. The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytologist 214, 782–795. [DOI] [PubMed] [Google Scholar]
- Yin P, Li Q, Yan C, et al. 2013. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171. [DOI] [PubMed] [Google Scholar]
- Zhang YF, Suzuki M, Sun F, Tan BC. 2017. The mitochondrion-targeted PENTATRICOPEPTIDE REPEAT78 protein is required for nad5 mature mRNA stability and seed development in maize. Molecular Plant 10, 1321–1333. [DOI] [PubMed] [Google Scholar]
- Zmudjak M, Colas des Francs-Small C, Keren I, Shaya F, Belausov E, Small I, Ostersetzer-Biran O. 2013. mCSF1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in Arabidopsis. New Phytologist 199, 379–394. [DOI] [PubMed] [Google Scholar]
- Zmudjak M, Shevtsov S, Sultan LD, Keren I, Ostersetzer-Biran O. 2017. Analysis of the roles of the Arabidopsis nMAT2 and PMH2 proteins provided with new insights into the regulation of group II intron splicing in land-plant mitochondria. International Journal of Molecular Sciences 18, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.