Taniguchi et al. review recent advances in stem cell–based models to investigate early human post-implantation development
Abstract
Proper development of the human embryo following its implantation into the uterine wall is critical for the successful continuation of pregnancy. However, the complex cellular and molecular changes that occur during this post-implantation period of human development are not amenable to study in vivo. Recently, several new embryo-like human pluripotent stem cell (hPSC)–based platforms have emerged, which are beginning to illuminate the current black box state of early human post-implantation biology. In this review, we will discuss how these experimental models are carving a way for understanding novel molecular and cellular mechanisms during early human development.
Introduction
Implantation of the human embryo into the uterine wall represents a critical developmental milestone; up to 50% of pregnancies fail during this important peri-implantation period (Macklon et al., 2002). Successful implantation entails both a receptive endometrium and the proper progression of embryonic development, but the exact causes of early implantation failure remain largely unknown because this period is so difficult to study in the human. Thus, the generation of models that allow investigators to mechanistically probe human peri-implantation development is of enormous importance to fertility treatment.
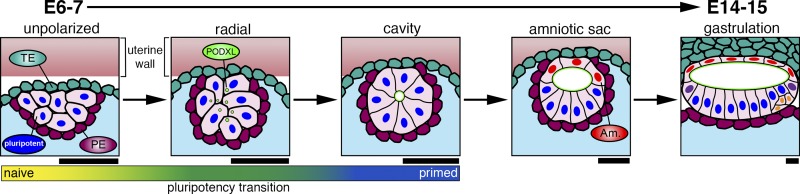
The implanting embryo (referred to at this stage as a blastocyst) contains three morphologically and molecularly distinct cell types: a cluster of pluripotent epiblast cells (precursors to the embryo proper as well as amniotic ectoderm) is surrounded by trophectoderm (TE, which will give rise to placental tissues) and extraembryonic primitive endoderm (ExPE, precursors to the yolk sac; Fig. 1). Excellent reviews on development of this preimplantation blastocyst have been published recently (Frum and Ralston, 2015; Rossant, 2016). As the blastocyst implants, the pluripotent epiblast cells undergo apico-basal polarization to form a cyst with a central lumen, the future amniotic cavity (Fig. 1). Shortly thereafter, the uterine-proximal pole of this initially uniform lumenal cyst of pluripotent cells differentiates into squamous amniotic ectoderm, and a sharp boundary forms between amnion and pluripotent epiblast portions of the cyst. This structure, the amniotic sac (Fig. 1), represents the substrate for the next essential steps of embryonic development, including primitive streak formation and initiation of gastrulation.
Figure 1.
Post-implantation human embryonic development (embryonic day 6–15). As the embryo implants, an initially unpolarized group of pluripotent epiblast cells initiate radial organization and lumen formation, aided by apically charged (PODXL+, green) vesicles, to form a cyst. Cells proximal to the endometrial pole then differentiate to amniotic ectoderm, giving rise to an asymmetric sac. A gradient scale indicates the naive to primed pluripotency transition that accompanies polarization. By embryonic day 15, gastrulation initiates in the posterior epiblast (yellow). Trophectoderm (TE, teal), primitive endoderm (PE, magenta), pluripotent epiblast (blue), amniotic ectoderm (Am., red), blastocoel cavity (aqua), and uterine wall (light pink). Estimated scale bars (25 µm) are shown based on images obtained from http://virtualhumanembryo.lsuhsc.edu.
The complex developmental events that accompany implantation are often referred to as the “black box” of human embryogenesis (Macklon et al., 2002); indeed, it is ethically unacceptable to manipulate this stage in vivo and visualization of the intact embryo is limited by its small size. Though the library of snapshots of human developmental stages provided by the Carnegie collection (Table 1), among others, provides valuable morphological data, dynamics of signaling interactions and fate determinations cannot be gleaned from such images. Recently, several laboratories reported progress in culturing human blastocysts left over from in vitro fertilization procedures (O’Leary et al., 2012, 2013; Deglincerti et al., 2016a; Shahbazi et al., 2016). A small subset of these blastocysts did continue to develop in culture, reaching a stage with an apically polarized epiblast surrounded by cells with a character of TE and ExPE, a testimony to the powers of the early embryo to self-organize. However, no amniotic sac structure was seen, amnion fate determination was not documented, and primitive streak formation was absent. While it is possible that a primitive streak would have formed after 14 d (when the experiments were terminated), exploring this is currently impermissible, given the Warnock 14-d rule (Table 1) that prohibits research on human embryos ex vivo past 14 d (Hurlbut et al., 2017; Pera, 2017). Nevertheless, these improvements to blastocyst culture will enhance our understanding of some aspects of human development up to 14 d.
Table 1. Glossary.
| Term | Definition |
|---|---|
| Endometrium | Innermost lining of the uterus; provides the surface for blastocyst implantation |
| Blastocyst | Pre-implantation embryo; consists of three cell types: trophoblast, primitive endoderm, and inner cell mass |
| Inner cell mass | Unpolarized pluripotent stem cells that are considered to be in the naive pluripotent state |
| Epiblast | Pluripotent stem cells that transition from naive to primed state as cells of the ICM undergo apico-basal polarization |
| Trophectoderm/trophoblast | Extraembryonic cells that give rise to the chorion |
| Extraembryonic primitive endoderm | Extraembryonic cells that give rise to the yolk sac |
| Amniotic ectoderm | Derived from epiblast cells underlying the invading trophectoderm during implantation |
| Amniotic sac | An asymmetric cyst formed by lumenal polarization of epiblast cells, with squamous amnion cells on one side and pluripotent epiblast cells on the other side |
| Amniotic cavity | Lumenal cavity enclosed by the amniotic sac |
| Pro-amniotic/epiblast cavity | Lumenal cavity surrounded by recently polarized epiblast cells, before amnion fate determination |
| Gastrulation | Developmental process by which all three embryonic germ layers are established |
| Primitive streak | Streak-shaped domain that forms in the posterior of the embryonic disc, marking the beginning of gastrulation |
| Carnegie collection | Collection of human embryos held at the Carnegie Institution of Washington |
| Warnock 14-d rule | Rule that limits the research on human embryos to the first 14 d of development, based on the 1984 Report of the Committee of Inquiry into Human Fertilization and Embryology, chaired by Mary Warnock |
| Turing patterning | Reaction-diffusion–based activator/inhibitor model of patterning, first proposed by Alan Turing in 1952 |
Many of our ideas about mechanisms of post-implantation biology have come via extrapolation of studies done in mice. However, at this stage human and mouse embryos have significantly different embryonic organization. For example, the amniotic ectoderm forms at a different location and time, and the mouse lacks an equivalent to the amniotic sac (Rossant and Tam, 2017). Other species may serve as better models. Indeed, the guinea pig embryo mirrors that of the human, forming an obvious amniotic sac (Huber, 1918). Unfortunately, this model is not yet well developed for experimental studies in developmental biology. Recent work in monkeys, however, is providing exciting new data (Sasaki et al., 2016) in a model that is evolutionarily much closer to humans and morphologically very similar during post-implantation development. This model is amenable to exploration with the full toolkit of modern biology, including morphological, immunological, in situ, and single-cell RNA sequencing (RNaseq) methods (Sasaki et al., 2016). Yet, efforts toward mechanistic dissection in the primate model will likely stay limited because it is expensive, time consuming, presently difficult to perturb genetically, and may present its own ethical issues.
In a continuing effort to find platforms for experimental exploration of human post-implantation biology, researchers have begun to exploit human embryonic stem cells (hESC; Thomson et al., 1998) and human-induced pluripotent stem cells (hiPSC; Takahashi et al., 2007). Easily expandable in culture and readily manipulatable with genetic techniques, these models take advantage of the remarkable self-organizing ability of human pluripotent stem cells (hPSC) in both 2D and 3D settings. New work using such models has accelerated our understanding of patterning and morphogenesis during post-implantation development. This review highlights the recent mechanistic discoveries in the field of human post-implantation development that have resulted from studies using hPSC-based in vitro systems, from amniotic cavity formation to gastrulation and axis formation. We concentrate on the cell biological insights and conceptual advances provided by specifically human models and consider both their advantages and limitations for answering fundamental questions of early human embryology. Parallel studies in mice are actively ongoing and except where they impinge on human mechanisms, are not discussed here; a recent excellent review can be found elsewhere (Shahbazi and Zernicka-Goetz, 2018). Progress in illuminating the black box of human post-implantation development afforded by these and other models will hopefully soon lead to strategies to diagnose or someday, even prevent, some causes of infertility.
Polarization and formation of the pro-amniotic cavity
Upon implantation of the blastocyst, the aggregate of epiblast cells undergoes a dramatic reorganization: cells polarize along their apico-basal axis and adopt a rosette-like structure with a central shared lumen (Fig. 1). Lumenogenesis is not driven by cavitation, as once thought (Coucouvanis and Martin, 1995). Rather, as they radially organize, epiblast cells exhibit intracellular collections of podocalyxin (Podxl; Bedzhov and Zernicka-Goetz, 2014), a sialoprotein that is critical for lumenogenesis in other cell types (Bryant et al., 2010; Klinkert et al., 2016; Mrozowska and Fukuda, 2016). Though the details are unclear, a central lumen forms as these PODXL aggregates are delivered to the center of the rosette (Fig. 1). Interestingly, a similar mechanism seems to drive lumen formation in the extraembryonic ectoderm of the mouse. The two cavities (epiblast and extraembryonic ectoderm) then merge to form the murine pro-amniotic cavity (Bedzhov and Zernicka-Goetz, 2014; Shahbazi and Zernicka-Goetz, 2018).
Implanting human embryos exhibit morphologically similar lumen-initiating events in the epiblast. Indeed, when human blastocysts are cultured under implantation-like conditions, they attach to the substrate in an oriented manner, with the epiblast pole nearest the substrate (Deglincerti et al., 2016a; Shahbazi et al., 2016). They then continue to develop a yolk sac cavity and a pro-amniotic cavity, organize the epiblast as a bi-laminar disc and specify diverse TE fates (Deglincerti et al., 2016a; Shahbazi et al., 2016). Thus, for both mouse and human blastocysts, maternal tissue is not required for initial post-implantation events. However, cues from the physical attachment of the blastocyst to a substrate are essential for the continuation of embryonic development beyond the blastocyst stage.
More recently, epiblast polarization and formation of the pro-amniotic cavity have been investigated using mouse and human PSC. When cultured as single cells suspended in Matrigel or Geltrex, these cells readily form lumens, with the ability to undergo lumenogenesis linked to the progressing pluripotency state of the cells (Bedzhov and Zernicka-Goetz, 2014; Taniguchi et al., 2015; Shahbazi et al., 2016). That is, pluripotent cells of the inner cell mass of preimplantation embryos exist in a “naive” state, which is transcriptionally and epigenetically distinguishable from the “primed” state characteristic of post-implantation epiblast cells (Kinoshita and Smith, 2018). In mouse as well as human PSC, lumen formation is suppressed if cells are maintained in a Nanoghigh naive state, though some signs of polarization are observed (radial organization, Golgi positioning, and formation of tight junctions; Shahbazi et al., 2017). However, when PSC transition from the naive to the primed state, they acquire a robust ability to form lumens. Mechanistically, the naive to primed transition in both mouse and human epiblast cells is associated with an increase in the expression of cell polarity proteins, such as Cingulin (Cgn) and Podxl, that aid in cortical vesicle docking (Shahbazi et al., 2017). Furthermore, deletion of Cgn in mouse ESC impairs lumenogenesis and leads to cytoplasmic accumulation of Podxl (Shahbazi et al., 2017). These findings divide the process of amniotic cavity formation into two separate events: a rosette-like organization of cells and the subsequent activation of the vesicular transport machinery to establish the lumenal domain. While the former event occurs in naive epiblast cells, the latter plays out as these cells transition to the primed state (Fig. 1).
The process of vesicular trafficking to form a lumen has been well studied in diverse epithelial cell types, including the well-established MDCK.2 and Caco-2 models. Some of the molecular players are shared between these systems and primed PSC, including Rho-GTPases and integrins (Yu et al., 2005; Bedzhov and Zernicka-Goetz, 2014; Rodriguez-Boulan and Macara, 2014; Taniguchi et al., 2015). In all of these cell types, singly plated cells reproducibly form a lumen upon the first cell division (Bedzhov and Zernicka-Goetz, 2014; Taniguchi et al., 2015). An actin-, Podxl-, and aPKCζ-rich domain is seen at the shared cytokinetic membrane, immediately after the completion of mitosis. This domain has been termed the apical membrane initiation site (Rodriguez-Boulan and Macara, 2014). In the case of PSC, the two-celled lumenal structures quickly grow into multi-cellular lumenal cysts in which all cells retain pluripotency (hPSC-cyst; Taniguchi et al., 2015). This robust lumen-forming tendency of primed PSC may be related to establishment of the pro-amniotic cavity.
Interestingly, the mechanism of apical membrane initiation site formation appears to differ in hPSC compared with MDCK.2 or Caco-2 cells. In MDCK.2 cells, polarization begins at mitosis; apical polarity components first accumulate at two opposite poles of the dividing cell and, as the cell completes division, these components are trafficked along the spindles of each nascent daughter to the forming cytokinetic plane (Schlüter et al., 2009; Klinkert et al., 2016; Mangan et al., 2016). In contrast, isolated PSC begin the polarization process before their entry into mitosis (Bedzhov and Zernicka-Goetz, 2014; Taniguchi et al., 2017). In singly plated PSC, the first sign of apical polarization is the perinuclear accumulation of membrane bound vesicles carrying PODXL and other apical components (Taniguchi et al., 2017). These vesicles fuse to form an intracellular structure called an apicosome several hours before cell division begins. The apicosome has characteristics of an intracellular lumen, complete with microvilli, primary cilium, and high calcium concentration. During mitosis, the apicosome is largely inherited by one daughter cell and is subsequently delivered to the cytokinetic plane, where it establishes a shared lumen between the two daughters. Interestingly, when PSC are plated as clusters, individual cells in the clusters form apicosomes within hours after plating. As cells rearrange to form rosettes, apicosomes are actively trafficked to form a common lumen at the center (Taniguchi et al., 2017). Though these steps in apicosome trafficking have not yet been visualized in vivo, cytoplasmic accumulations of Podxl-charged vesicles have been documented at the future apical poles of the forming rosette in the mouse epiblast; these intracellular collections of apical material precede lumen formation (Fig. 1; Bedzhov and Zernicka-Goetz, 2014). Additionally, the functional role for cortical vesicle docking that characterizes the transition of epiblast cells transition from the naive to primed state (Shahbazi et al., 2017) provides further evidence that distinct polarized vesicular trafficking events are critical for establishment of the pro-amniotic cavity. Thus, it appears that, unlike established de novo lumen-forming mechanisms (cavitation and hollowing), embryonic epiblast cavity formation involves a novel process of apicosome-driven lumenogenesis; it will be important to determine the molecular underpinnings and the functional consequences of this important event.
Amniogenesis and amniotic sac formation: establishment of the amnio/embryonic axis
Images in the Carnegie collection of human embryo development show that upon its initial formation, the pro-amniotic cavity is surrounded by epiblast cells, and the cyst structure is apparently symmetrical. However, as implantation proceeds, epiblast cells that are nearest to cytotrophoblast cells begin to flatten as they take on amnion fate, while cells on the opposite side of the cyst, adjacent to the primitive endoderm, become more columnar, forming the embryonic disc (Fig. 2). This asymmetric amnion-embryonic disc structure, the amniotic sac, has also been documented in developing primate embryos in vivo (Fig. 2; Sasaki et al., 2016), but accessibility to this structure has limited its study. In vitro, it has been shown that a subset of cultured preimplantation human blastocysts will further develop to form a pro-amniotic cavity, though a patterned amniotic sac–like structure does not emerge before 14 d, the designated termination time of such cultures (Deglincerti et al., 2016a; Shahbazi et al., 2016). However, recent work indicates that hPSC, when cultured under specific conditions, can organize to form an asymmetrically patterned cyst with squamous amnion-like cells on one side and columnar pluripotent embryonic disc–like cells on the other, a structure that is termed “post-implantation amniotic sac embryoid” or PASE (Shao et al., 2017b).
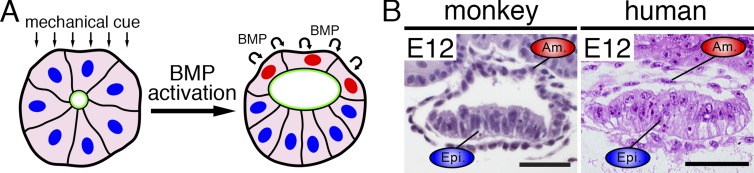
Figure 2.
BMP-dependent amniotic sac formation. (A) During PASE formation, mechanically activated BMP signaling induces amniogenesis, resulting in an amniotic sac–like structure. (B) Amniotic sacs of primate embryos in vivo; left: cynomolgous monkey embryo, right: human embryo. Images adapted with permission: cynomolgous monkey (Sasaki et al., 2016). Human (Carnegie stage 5c human embryo section; http://virtualhumanembryo.lsuhsc.edu). Bars, 50 µm.
Interestingly, PASE development occurs spontaneously when hPSC are cultured in mTeSR (without added growth factors) on top of a thick bed of Geltrex (extracellular matrix) and overlaid with dilute Geltrex (Shao et al., 2017b). The initial cue for PASE development appears to be mechanical (Fig. 2 A), since reducing the thickness of the underlying gel bed (read by cells as a stiffer substrate) or plating cells on a hard (vs. soft) polydimethylsiloxane micro-post substrate results in development of symmetrical, pluripotent cysts, without amnion. Live imaging reveals that during PASE morphogenesis, amniogenesis initiates focally on the side of the cyst that faces the gel bed and then spreads laterally. Downstream of this mechanical input, activation of BMP signaling is required for amniogenesis (Fig. 2 A), since addition of the BMP antagonist, NOGGIN, or the BMP inhibitor, LDN193189 completely abolishes amnion differentiation under permissive mechanical conditions and instead results in formation of symmetrical, fully pluripotent cysts (Shao et al., 2017b). Forming amnion cells secrete both BMP4 and its inhibitor, NOGGIN, and the balance of these factors appears to be critical for establishing the boundary between amnion and epiblast cells. Indeed, in this culture system, the spreading amnion fate encompasses the entire cyst in most cases; 95% of the cysts undergo progressive cellular flattening, lose pluripotency, and acquire morphological and transcriptomic features consistent with amniotic (hPSC-amnion; Shao et al., 2017a). These key fate-identifying transcriptomic features include the expression of ITGB6, VTCN1, GABRP, MUC16, POSTN, TFAP2A, and TFAP2B, as previously reported for first trimester human amnion (Roost et al., 2015; Slieker et al., 2015).
The critical role for endogenous activation of BMP/SMAD signaling during amnion differentiation in the PASE model is consistent with findings in cynomolgous monkey embryos in vivo; squamous amnion cells in developing primate amniotic sacs express BMP ligands and BMP target genes (Sasaki et al., 2016). Moreover, even though the mouse does not develop an amniotic sac structure at all and begins amnion formation after gastrulation begins, BMP signaling is critical for amniogenesis in that model as well. In mice, BMP ligands are expressed at the embryonic/extraembryonic border of the cup-shaped cylinder, and Smad5 mutants show defects in that border, as well as aberrant amnion and chorion development (Pereira et al., 2011; Dobreva et al., 2018). A recent detailed clonal analysis of amniotic ectoderm progenitors in the mouse shows that these cells arise from the epiblast cells at this border and expand both anteriorly and posteriorly (Dobreva et al., 2018). Loss of Smad5 compromises both clonal expansion of these progenitors and squamous morphogenesis of their progeny. A similar phenotype is seen in mice lacking Hand1, a known target of BMP signaling (Firulli et al., 1998).
Apart from the apparent conservation of a requirement for BMP signaling during amniogenesis, our current understanding of the mechanisms underlying human amniotic ectoderm formation and maturation is rudimentary at best. The possibility that a mechanical signal could initiate amniogenesis in mouse (Pereira et al., 2011; Dobreva et al., 2018) and human (Shao et al., 2017a) seems likely, but it is not clear how that signal is generated or how it activates downstream BMP signaling. Moreover, though RNaseq studies suggest that BMP activates de novo expression of several transcription factors (Shao et al., 2017a), the specific roles that these factors play in amniogenesis remain to be elucidated.
Gastrulation
Following establishment of the amniotic sac, the next major event in human development is gastrulation. The onset of gastrulation is marked by formation of the primitive streak, a depression within the disc-shaped epiblast through which cells begin to migrate (Fig. 1). In the mouse, primitive streak formation is induced by a cascade of several developmental signals (e.g., BMP, WNT, TGFβ/NODAL, and FGF) that break the morphological symmetry of the epiblast (Ben-Haim et al., 2006; Tam and Loebel, 2007; Robertson, 2014). Gastrulation is marked by epithelial-mesenchymal transition (EMT) of some of the epiblast cells. As they break away from the epithelium, the mobile cells begin to express mesodermal and endodermal markers, while cells that remain in the epiblast epithelium take on ectodermal fate. The resulting three germ layers form an organized trilaminar structure and, through directed migration and regional signaling, simultaneously establish the body axes of the developing embryo. Although gastrulation is well-studied in multiple model systems, from fly and sea urchin to chick and mouse (Keller, 2005; Leptin, 2005; Stern, 2006; Lyons et al., 2012), this process has remained unexplored in the human. However, the recent finding that hPSC can replicate several of the complex events accompanying gastrulation (Fig. 3 and discussed below) is allowing an unprecedented view into some of the key signaling requirements and molecular fate determinants that may underlie this process in the human embryo.
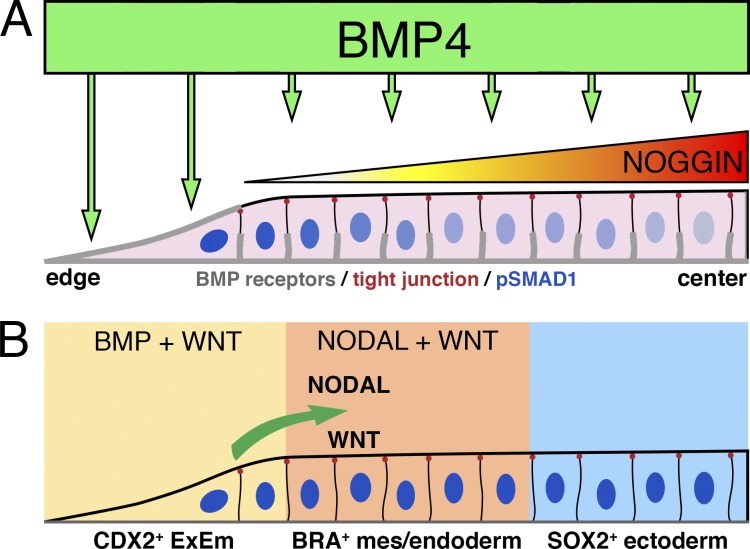
Figure 3.
Formation of primary germ layer–like cells in a micropattern platform orchestrated by BMP signaling. (A) BMP signaling is most prominent in the edge of micropattern colonies (indicated by nuclear pSMAD1, blue) as basolaterally localized BMP receptors are exposed to exogenous BMP4 ligand (“edge effect”). BMP4 signaling induces the expression of its antagonist NOGGIN, resulting in a NOGGIN gradient that is highest at the colony center. BMP receptors (gray), tight junction (red dots), and nuclear pSMAD1 (blue; Warmflash et al., 2014; Etoc et al., 2016). (B) Adjacent to the colony edge, BMP-dependent NODAL and WNT expression establishes a mes-/endoderm cell population; center cells are maintained as SOX2+ neuroectoderm-like state.
In discussing the various cell-based models of gastrulation and patterning, it is important to separate the concepts of marker activation, symmetry breaking, and axis formation, features that are closely intertwined in the intact embryo but can be disconnected in vitro. Indeed, before gastrulation, the intact primate embryo is a flat disc of pluripotent cells; a proximal/distal axis (as defined in the mouse) is missing and none of the epiblast cells express the posterior marker, BRA (Sasaki et al., 2016). The first activation of BRA expression in the epiblast occurs at the forming primitive streak, as cells begin to undergo the EMT movements of gastrulation. This marker activation occurs on the posterior side of the embryo, breaking its previous symmetry with respect to BRA expression (Sasaki et al., 2016). Directed migration of gastrulating cells, in concert with patterning cues, then elongates the anterior/posterior (A/P) axis and subdivides it into a linear arrangement of segments expressing a sequence of Hox genes while simultaneously imparting dorsal/ventral information. In contrast, it should be noted that in many stem cell systems, activation of BRA expression occurs in a symmetrical or rotationally invariant manner (Shao et al., 2017b; Simunovic et al., 2018). Thus, while symmetry is broken by BRA activation, axial patterning per se is absent. Careful attention to these concepts in the various cell-based models will aid in the further exploration of their origin and interdependence.
Micropattern: modeling germ layer formation and patterning in 2D
Several recent studies have made use of cell culture substrates that consist of micropatterned arrays of shapes coated with ECM proteins that allow cell attachment (e.g., Matrigel, fibronectin, and laminin) on a surface that otherwise prevents cell adhesion. Micropatterns provide control over the spatial distribution of cells and have become a fundamental tool to examine mechanical and signaling properties that regulate the differentiation of hPSC and other stem cell types (Guilak et al., 2009; Deglincerti et al., 2016b; Heemskerk et al., 2017). When hPSC are grown on micropatterns (Matrigel or laminin-521) and BMP4 is added to the medium, each colony self-organizes into a radially symmetrical pattern, consisting of (from center to edge) ectoderm, mesoderm, endoderm, and CDX2+ extraembryonic cells (Fig. 3; Warmflash et al., 2014). Emergence of similar patterns using micropatterned mouse Epiblast-like cells was recently documented, though this study used different combinations of growth factors (BMP4 and WNT3A) and observed somewhat different colony fates (Morgani et al., 2018). Nevertheless, radial patterning of cell fates in both species is accompanied by EMT and stratification of cell types within the mesendodermal cell population, indicating a remarkable degree of conservation of self-organizational behavior during these complex processes. Importantly, in the case of the mouse, cell responses observed in vitro can be verified by in vivo analyses, a critical validation step that will not be possible in the human.
Patterning depends on colony size. As colony size is reduced, central fates are progressively lost while the width of outer regions is maintained. The smallest colonies (<250 µm) exhibit edge fates only, demonstrating that the patterning mechanism has a fixed length scale (patterning does not scale with colony size) and that full pattern development requires a minimal colony diameter (Warmflash et al., 2014). Moreover, it shows that the pattern is formed from the edge, suggesting a crucial role for the tissue edge in patterning (Fig. 3). The reason for this appears to lie in the signaling dynamics underlying pattern formation, which begins at the edge and spreads toward the middle. During patterning, SMAD1/5/9 activity concentrates at the colony edge, indicating increased BMP signal transduction in outer regions of the colony, while SMAD2 activity, indicating ACTIVIN/NODAL signaling, is highest in a ring away from the edge, where primitive streak-like fates are observed (Fig. 3 B; Warmflash et al., 2014). Inhibition of ACTIVIN/NODAL signaling abolishes this SMAD2 activity as well as differentiation of endoderm and mesoderm (Warmflash et al., 2014). WNT inhibition similarly prevents mesoderm and endoderm differentiation (Martyn et al., 2018), as well as endogenous NODAL signaling (Chhabra et al., 2018). Thus, work with human stem cell colonies strongly suggests that the hierarchy of BMP to WNT to NODAL signaling that initiates gastrulation in the mouse embryo is preserved in the human embryo and that the same molecular circuits responsible for patterning the embryo in vivo are likely responsible for patterning stem cells in vitro.
The rapid concentration of SMAD1 signaling at the colony edge is due to two factors: a differential localization of BMP receptors in the colony and the formation of a NOGGIN gradient across the colony (Fig. 3 A; Etoc et al., 2016). Away from the colony edge, increasing cell density results in relocalization of BMP receptors to lateral surfaces below the tight junctions of the packed epithelial cells, where they are inaccessible to apically added BMP4. Concurrently, BMP-dependent production of NOGGIN leads to a gradient that is lowest on the edge and prevents response to BMP4 through the minority of remaining apical receptors in the colony center. In contrast, edge cells experience low levels of NOGGIN while their receptors are completely exposed to BMP added to the culture medium. This property of differential receptor localization can therefore establish a density-dependent prepattern in edge cells (Fig. 3). Interestingly, in addition to their differential receptor localization, edge cells also exhibit different tensile properties, with increased focal adhesions and actin cables, greater myosin activity, and enhanced traction forces. It has been suggested that these distinct features might provide a mechano-signal that could encourage rapid differentiation of these edge cells (Rosowski et al., 2015).
Real-time analysis of the response of SMAD proteins during hPSC differentiation has shed further light on patterning and differentiation dynamics within hPSC microcolonies (Heemskerk et al., 2017; Nemashkalo et al., 2017). Using an engineered hESC line in which DNA encoding EGFP is incorporated into SMAD4 locus (creating a SMAD4-EGFP fusion protein) nuclear levels of fluorescent SMAD4 were measured over time in hPSC to visualize BMP4 and NODAL signaling (Heemskerk et al., 2017; Nemashkalo et al., 2017). While SMAD4 nuclear movement occurs during signaling with both of these two ligands, the response to BMP4 is accompanied by nuclear movement of SMAD1, while the response to NODAL can be tracked by SMAD2 (Heemskerk et al., 2017). Live imaging of SMAD4-GFP, combined with SMAD1 staining shows that the BMP4 response in micropatterned colonies is stable and restricted to edge cells by 12 h after stimulation. At ∼25–30 h, a front of nuclear SMAD4-EGFP begins to move toward the center of the colony at a rate of one cell per hour. This wave coincides with nuclear SMAD2 staining (a sign of ACTIVIN/NODAL signaling) and is followed by activation of endodermal and mesodermal markers in the inner cells. The fact that NODAL signaling is highly dynamic and sweeps across the colony as a wave, rather than forming a stable signaling gradient similar to BMP, is at first surprising, but further studies suggest an underlying reason for this (Heemskerk et al., 2017). That is, the dynamics of SMAD4 signaling differ dramatically, depending on whether the ligand is BMP or NODAL. The BMP response directly reflects the presence of the ligand and is characterized by sustained nuclear SMAD4 localization and stable target gene expression. However, the ACTIVIN/NODAL SMAD4 response is transient (peaking and then diminishing), with some target genes showing transient dynamics mirroring SMAD4 and others showing stable transcription. This transient or adaptive response appears to correspond to the ability of cells to sense the rate of increase in ACTIVIN/NODAL signal rather than its concentration per se. This suggests that rapid increases in NODAL signaling, rather than specific concentrations of the ligand may be important in pattern formation and provides a possible explanation for the wave of NODAL signaling seen in self-organized patterning, as this causes individual cells to see such rapid increases. WNT signaling also shows wave-like dynamics, and that propagation of this wave requires continuous BMP activity (Chhabra et al., 2018). In contrast, the NODAL wave requires WNT initially, but becomes independent of upstream signals at later times (Nemashkalo et al., 2017). These rich signaling dynamics add a previously unappreciated layer of complexity to these signaling cascades. It will be of great interest to develop a SMAD4-EGFP reporter mouse model to explore the dynamics of signal spreading in vivo and clarify how the cellular signaling steps involved in self-organization evolve during gastrulation.
Interestingly, the fact that BMP4 not only activates the WNT/NODAL cascade but also promotes expression of its own inhibitor, NOGGIN, is specific to human ESC (Etoc et al., 2016) and does not apply to mouse Epiblast-like cells (Morgani et al., 2018). Adding to this difference from mouse cells, BMP may also activate itself in hESC, giving this activator-inhibitor system all the necessary properties to enable pattern formation through a Turing-like mechanism (Turing, 1952; Etoc et al., 2016; Tewary et al., 2017). In support of this, under specific conditions, a periodic spot pattern, consistent with patterning through the Turing mechanism, forms in colony centers (Etoc et al., 2016; Tewary et al., 2017). Thus, despite the deceptively simple-looking 2D system offered by micropatterns, overall self-organization of these patterns requires a remarkably complex balance of signaling pathways.
While certain hallmarks of gastrulation in vivo are not seen in cultured mouse or human micropatterned colonies—most importantly, trilaminar organization of the primary germ layers and A/P axis formation—this powerful model will continue to lend new insights into the signaling dynamics that initiate germ layer patterning. Indeed, when whole patterned colonies (induced by WNT and ACTIVIN) are transplanted onto the marginal zone of an early chick embryo, they induce host cells to form a secondary axis and contribute directly to that new axis (Martyn et al., 2018). These properties suggest that some cells within the colony are acting as a classically defined Spemann’s organizer, a signaling center that acts to organize the body axis (Beddington, 1994; Crease et al., 1998; Gritsman et al., 2000; Kinder et al., 2001; Spemann and Mangold, 2001; Martyn et al., 2018). This transplantation scenario provides yet another model system to investigate and dissect cascading developmental signals in the implanting embryo.
3D models of gastrulation
Recently, 3D models of human gastrulation have been developed to complement the 2D systems (Shao et al., 2017b; Simunovic et al., 2018). As described above, PASE are generated from dissociated hPSC, which are plated on a thick bed of Geltrex, in mTeSR1 pluripotency medium containing diluted Geltrex (Fig. 2 A; Shao et al., 2017b). No additional signaling factors, such as BMP4, are added, although mTeSR1 does contain FGF2 and TGFβ1, sufficient to maintain pluripotency (Ludwig et al., 2006). Under these conditions, the cells readily form cysts, some of which spontaneously become asymmetric structures that closely resemble the amniotic sac, with squamous amnion cells at one pole and tall, pluripotent cells at the other. On days 4–5, at the pluripotent pole, basal lamina breakdown, and cell dissemination can be observed; these events are hallmarks of gastrulation. Migrating cells express nuclear pSMAD1/5, SNAI1, BRA, and CDX2, but not FOXA2 and SOX17, consistent with differentiation to mesodermal cells of the posterior primitive streak (Shao et al., 2017b). Genetic deletion of SNAI1, a major regulator of EMT during gastrulation, inhibits these gastrulation-like movements. Though only 5–10% of the total cysts grown in such cultures develop as asymmetric PASE (the rest become fully amnion cysts), dozens of asymmetric PASE can be observed within each culture, and their morphological and topological resemblance to the human amniotic sac is particularly striking. Thus, this model provides a valuable tool for the further analysis of amnion fate determination, establishment of the amnion/epiblast border, and initiation of gastrulation. Further refinements of this system may allow emergence of endoderm and/or establishment of axial patterning.
An alternative 3D system that utilizes a hydrogel matrix in mouse embryonic fibroblast–conditioned HUSEM medium supplemented with 20 ng/ml FGF2 has been developed (Simunovic et al., 2018). This system gives rise to fully symmetric pluripotent cysts after 3 d. Subsequent addition of BMP4 to the medium leads to spontaneous symmetry breaking, formation of a primitive streak-like structure, basement membrane disruption, EMT, cell dissemination, and mesoderm/endoderm marker expression. In an arrangement that is similar to 2D micropatterned colonies, cells that are SOX2+ (anterior, early ectoderm) occupy a different domain than those that are BRA+ (posterior primitive streak/mesoderm) or SOX17+ (endoderm). Though domains are rather loosely organized and lack the sharper boundaries between germ layers seen in 2D micropatterns, this “epiblast model” permits analysis of factors that control symmetry breaking. Indeed, genetic deletion of DKK1, a WNT inhibitor, leads to complete conversion of the model epiblast to BRA+ cells, confirming a key role for a WNT-based activator–inhibitor interaction in domain patterning (Simunovic et al., 2018). In contrast to 2D micropatterns, deletion of CER1 and LEFTY1, inhibitors of ACTIVIN/NODAL signaling, did not perturb patterning, perhaps because LEFTY2 was still present. Additionally, in contrast to 2D micropatterns or PASE, this model contains no extraembryonic tissues (at least as assessed by GATA3 staining; Simunovic et al., 2018). Its patterning therefore appears to be dependent on stochastic BMP4-driven induction of a focus of mesoderm in the context of a cyst, and localized spread of that pattern.
In vitro model validation and beyond
The embryoid-based model systems discussed here provide ample evidence of the remarkable ability of pluripotent stem cells (PSC) to organize into complex structures that mirror several characteristics of early post-implantation embryos. The next step is to validate findings from any of these hPSC-derived models in vivo, a challenge that, given ethical considerations, must rely on the overlay of information gleaned from mouse or primate. This effort will be aided by single-cell analysis tools that allow quantitative transcriptomic and epigenomic characterization of heterogeneous cell populations as they transition from one state to another. Single-cell RNaseq data are already enabling cross-species comparisons of mouse (Scialdone et al., 2016; Mohammed et al., 2017) and primate embryos (Sasaki et al., 2016; Nakamura et al., 2017) as well as early preimplantation human embryos (Yan et al., 2013; Blakeley et al., 2015; Li et al., 2018; Zhong et al., 2018; Zhu et al., 2018). For post-implantation human embryos, however, these analyses have been necessarily limited to embryos older than 4 wk (Guo et al., 2015; Li et al., 2017; Fan et al., 2018; Gao et al., 2018; Menon et al., 2018), the earliest point at which abortions are normally performed. Interestingly, an analysis of human and mouse preimplantation blastocysts (containing TE, epiblast, and ExPE cells), uncovered significant differences in gene expression, even though the blastocysts at this stage are morphologically very similar (Blakeley et al., 2015). For example, in mouse TE, Cdx2 and Id2 are activated early, followed by Eomes and Elf5. However, in human TE, CDX2 is activated later, while the other genes are not expressed at appreciable levels. Additional genes, such as KLF17, are expressed in epiblast cells of the mouse, but not the human. Further application of these single-cell sequencing techniques to the various models of hPSC-based embryoids in vitro will provide important verification of the presence of specific cell states/types in these systems. Moreover, recent bioinformatic advances now allow the estimation of developmental pseudotiming, using differential gene expression (Trapnell et al., 2014; Haghverdi et al., 2016; Qiu et al., 2017) or RNA dynamics (La Manno et al., 2018) to order progressive fate stages. Given the controlled, progressive development of embryoid systems in vitro, application of pseudotiming methods to the single-cell output has the potential to reveal the molecular dynamics of cell state/fate determination. This information in turn will guide CRISPR/Cas9-based genome editing experiments that can probe the functional underpinnings of embryoid development.
Stem cell models provide several important technical advantages, relative to in vivo studies: the culture environment can be manipulated with high precision in ways that are unthinkable in vivo and their geometry provides ideal conditions for high resolution microscopy. Systems such as micropatterns are also high throughput, allowing collection and quantification of data from hundreds to thousands of embryoids simultaneously. Such quantitative data significantly increase the number of phenotypes we can resolve, revealing both relative magnitude and hierarchy of response, data that could be productively exploited in drug screens. Similarly, live imaging combined with signaling reporters allows detection of subtle influences modulating pathway activity. Although 3D embryoid models are generally less reproducible, hampering their application in large scale screens, they are nevertheless amenable to genetic manipulation and in particular cases, such as the PASE, can provide a system that is topologically closer to actual embryonic structures seen in vivo.
Interpreting the higher resolution picture of biology afforded by the improved quantitative data from stem cell systems requires higher resolution modeling tools. This typically means moving from logical models (diagrams with arrows, which can also be mathematically formalized; Wang et al., 2012) to systems of differential equations that predict values of measured variables in space and time, i.e., “mathematical modeling.” Such models can distinguish qualitatively different mechanisms based only on quantitatively different phenotypes. They can also guide future experiments by determining what parameters should be measured to distinguish among different mechanisms or by making precise predictions about the outcome of experiments (e.g., to aid in increasing the yield of some cell type during reprogramming). Often however, models are under-constrained. That is, the large number of unknown parameters makes it possible to fit a range of different models to existing data, with diminishing predictive power. It is therefore often most informative to determine which models are ruled out by the data rather than which models are consistent with it. Additionally, progress can be made by constructing simplified phenomenological models that provide predictive power and insight at the expense of abstracting away molecular details (Corson and Siggia, 2017). As an example, modeling suggested that adaptive response through degradation of receptors predicts a refractory period (inability to respond to fresh ligand) that is much longer than the time scale of adaptation, contrary to what is observed (Heemskerk et al., 2017). Ruling out this possibility then suggested a negative feedback or incoherent feedforward mechanism; although these mechanisms are qualitatively different, both provide enough parameter freedom to explain all available data. Future experiments can now be directed at distinguishing between these mechanisms.
The complexity of spatial patterning and the quantitative data we can obtain from stem cell models make it both necessary and possible to gain additional insight using mathematical modeling. For explaining the final pattern of cell fates, there is still insufficient data to rule out many models, but we appear to be on the cusp of understanding the mechanisms underlying the observed signaling patterns in gastruloids. Several qualitatively different models for gastrulation-like patterning in micropatterned stem cells have been suggested in the literature, including patterning that is completely controlled by a static signaling gradient created by a reaction diffusion system with (Tewary et al., 2017) or without (Etoc et al., 2016) Turing instability, as well as patterning by waves of signaling in a more clock and wavefront like manner (Chhabra et al., 2018). While fundamentally different, each model is consistent with the accompanying experimental data and in some sense refines previous models. Downstream of these dynamic signaling patterns, complex cell responses lead to the localized organization of cell fates. Though we expect the patterning mechanisms to be the same in micropatterned colonies as in spherical cysts in 3D culture and actual embryos, it will be important to confirm this.
An important aspect of human development that is not yet well-captured in hPSC models is establishment of the body axes. At least in mice, axis formation begins before the formation of the primitive streak and is dictated by a signaling center located in an extraembryonic tissue, the anterior visceral endoderm (AVE; Beddington and Robertson, 1999). The AVE secretes antagonists of BMP, TGFβ/NODAL, and WNT signaling such as CER1, LEFTY1, and DKK1, to specify anterior identity and inhibit formation of the primitive streak on the anterior side of the embryo (Arnold and Robertson, 2009). A similar asymmetrically located group of LEFTY1-/DKK1-expressing cells has been identified by in situ analysis in the ExPE of developing cynomolgous monkey embryos (Sasaki et al., 2016), but the functional role of those cells in establishing the A/P axis has yet to be confirmed.
Although these findings reinforce the idea that signals from the ExPE are critical for positioning the primitive streak and limiting its extent, an hESC model lacking this tissue nevertheless exhibits localized expression of primitive streak markers (Simunovic et al., 2018). Moreover, mouse gastruloids (small aggregates of mESC, consisting of 300 ± 100 cells in the absence of any extraembryonic tissues) not only show asymmetric positioning of primitive streak markers, but subsequently undergo a remarkable degree of axial patterning, expressing a linear sequence of Hox genes along the A/P axis (van den Brink et al., 2014; Turner et al., 2017; Beccari et al., 2018). On this basis, it has been proposed that the extraembryonic tissues may function to bias or reinforce axial patterning cues that are already contained within the epiblast (Turner et al., 2017).
Further improvements in axis elaboration may require rebuilding the embryo by adding back additional extraembryonic tissues. Indeed, murine embryo-like structures called “ETS-embryos” can be derived by coculturing mESC with trophoblast stem cells (TSC) but in the absence of ExPE, and these structures show signs of A/P patterning and specification of nascent mesoderm (Harrison et al., 2017, 2018). Going further, adding mouse ExVE or XEN cells to mESC and TSC generated “ETX embryos” (Sozen et al., 2018). About 20% of the resulting structures bear a striking morphological similarity to intact embryos, with an AVE-like domain expressing Lefty1 and Cer1, as well as a primitive streak structure, with disseminating mesodermal and endodermal lineages, though the organization of gastrulating cells remains imperfect.
Currently, none of the in vitro human PSC models contain TE or ExPE. Although it will be controversial, it is likely that refined 3D models of human embryos will soon be developed, similar to those seen in the mouse systems. Indeed, human TSC were recently derived for long-term culture directly from culturing human blastocysts (Okae et al., 2018) as well as from the first trimester placenta (Haider et al., 2018). However, lines of human XEN cells have not yet been established in culture (Ralston, 2018).
Apart from rebuilding embryos, which is likely to have ethical limitations, significant improvements in the in vitro model systems can likely be generated by applying localized external signals in proper sequence. This could be achieved by a wide range of promising technologies including customized substrates, microfluidics, and/or optogenetics. Alternatively, an interesting Notch receptor–based activation system has been recently designed that has the ability to enhance or inhibit specific cell-cell signals at precise times and locations (Morsut et al., 2016).
As we continue the investigation of early human embryogenesis using synthetic human embryoids, it is important to anticipate ethical concerns and adopt appropriate guidelines for the ongoing work. Several excellent reviews and commentaries on this topic, as well as proposed guidelines have been published (Daley et al., 2016; Hyun et al., 2016; Hurlbut et al., 2017; Pera, 2017). The Warnock 14-d rule says that research on human embryos ex vivo is prohibited past 14 d (essentially the point at which twinning is no longer possible) or formation of the primitive streak (initiation of gastrulation), whichever comes first (Hurlbut et al., 2017; Pera, 2017). Certainly, gastrulation-like movements, emergence of germ layers, and even some degree of axial patterning and organizer potential have already been achieved in culture by several laboratories using gastruloids, micropatterns, PASE, and model epiblasts. Though one can argue that the development of these structures from hPSC takes place on a different timeline (compared with in vivo) and none of these models are initiated by a fertilization-like event, it does appear, at least from a morphological and cell fate standpoint, that the 14-d rule is already being pushed beyond its intended limit (formation of the primitive streak). But is 14 d really biologically relevant as an end point? And if we extend the end point beyond 14 d, what is the new end point and why? If we now begin to add extraembryonic cell types to improve embryo patterning, or transplant patterned embryoids onto living embryos to induce neural tissue (Martyn et al., 2018), when does the model gain embryonic potential and how do we know that? And finally, from a practical standpoint, even with revised ethical standards in place, if we are successful in developing better embryoids, how will we validate our work, without comparison to real human embryos?
Given the current black box state of early post-implantation biology, it is important to come back to the fact that hPSC-based models, in addition to their value for biological understanding of human development, pose immense clinical potential for diagnosis and, eventually, treatment of couples with chronic infertility. Furthermore, such model platforms will allow high-throughput chemical and genetic screening assays to predict human reproductive success or examine the effect of drugs/toxins on embryonic development. Indeed, a computational quantitative morphometric system was recently applied to a micropattern platform for teratogen screening during mesendoderm differentiation (Xing et al., 2015). In future pursuits, it will be important to continue to leverage the power of these embryoid platforms to reap their huge potential benefits, without compromising our ethical standards.
Acknowledgments
We apologize to fellow investigators whose work was not cited due to space constraints. We thank Ryan Townshend for his help in generating schematic figures.
D.L. Gumucio was funded by National Institutes of Health grant R01-DK089933. I. Heemskerk was funded by Society in Science – Branco Weiss fellowship, administered by ETH Zurich.
The authors declare no competing financial interests.
K. Taniguchi, I. Heemskerk, and D.L. Gumucio wrote and edited the manuscript.
References
- Arnold S.J., and Robertson E.J.. 2009. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10:91–103. 10.1038/nrm2618 [DOI] [PubMed] [Google Scholar]
- Beccari L., Moris N., Girgin M., Turner D.A., Baillie-Johnson P., Cossy A.C., Lutolf M.P., Duboule D., and Arias A.M.. 2018. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature. 562:272–276. 10.1038/s41586-018-0578-0 [DOI] [PubMed] [Google Scholar]
- Beddington R.S. 1994. Induction of a second neural axis by the mouse node. Development. 120:613–620. [DOI] [PubMed] [Google Scholar]
- Beddington R.S., and Robertson E.J.. 1999. Axis development and early asymmetry in mammals. Cell. 96:195–209. 10.1016/S0092-8674(00)80560-7 [DOI] [PubMed] [Google Scholar]
- Bedzhov I., and Zernicka-Goetz M.. 2014. Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell. 156:1032–1044. 10.1016/j.cell.2014.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haim N., Lu C., Guzman-Ayala M., Pescatore L., Mesnard D., Bischofberger M., Naef F., Robertson E.J., and Constam D.B.. 2006. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev. Cell. 11:313–323. 10.1016/j.devcel.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Blakeley P., Fogarty N.M., del Valle I., Wamaitha S.E., Hu T.X., Elder K., Snell P., Christie L., Robson P., and Niakan K.K.. 2015. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. 142:3151–3165. 10.1242/dev.123547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D.M., Datta A., Rodríguez-Fraticelli A.E., Peränen J., Martín-Belmonte F., and Mostov K.E.. 2010. A molecular network for de novo generation of the apical surface and lumen. Nat. Cell Biol. 12:1035–1045. 10.1038/ncb2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra S., Liu L., Goh R., and Warmflash A.. 2018. The timing of signaling events in the BMP, WNT, and Nodal cascade determines self-organized fate patterning in human gastruloids. bioRxiv. doi:10.1101/440164 (Preprint posted October 10, 2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corson F., and Siggia E.D.. 2017. Gene-free methodology for cell fate dynamics during development. eLife. 6:e30743 10.7554/eLife.30743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucouvanis E., and Martin G.R.. 1995. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell. 83:279–287. 10.1016/0092-8674(95)90169-8 [DOI] [PubMed] [Google Scholar]
- Crease D.J., Dyson S., and Gurdon J.B.. 1998. Cooperation between the activin and Wnt pathways in the spatial control of organizer gene expression. Proc. Natl. Acad. Sci. USA. 95:4398–4403. 10.1073/pnas.95.8.4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley G.Q., Hyun I., Apperley J.F., Barker R.A., Benvenisty N., Bredenoord A.L., Breuer C.K., Caulfield T., Cedars M.I., Frey-Vasconcells J., et al. 2016. Setting Global Standards for Stem Cell Research and Clinical Translation: The 2016 ISSCR Guidelines. Stem Cell Reports. 6:787–797. 10.1016/j.stemcr.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deglincerti A., Croft G.F., Pietila L.N., Zernicka-Goetz M., Siggia E.D., and Brivanlou A.H.. 2016a Self-organization of the in vitro attached human embryo. Nature. 533:251–254. 10.1038/nature17948 [DOI] [PubMed] [Google Scholar]
- Deglincerti A., Etoc F., Ozair M.Z., and Brivanlou A.H.. 2016b Self-Organization of Spatial Patterning in Human Embryonic Stem Cells. Curr. Top. Dev. Biol. 116:99–113. 10.1016/bs.ctdb.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobreva M.P., Abon Escalona V., Lawson K.A., Sanchez M.N., Ponomarev L.C., Pereira P.N.G., Stryjewska A., Criem N., Huylebroeck D., Chuva de Sousa Lopes S.M., et al. 2018. Amniotic ectoderm expansion in mouse occurs via distinct modes and requires SMAD5-mediated signalling. Development. 145:dev157222 10.1242/dev.157222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etoc F., Metzger J., Ruzo A., Kirst C., Yoney A., Ozair M.Z., Brivanlou A.H., and Siggia E.D.. 2016. A Balance between Secreted Inhibitors and Edge Sensing Controls Gastruloid Self-Organization. Dev. Cell. 39:302–315. 10.1016/j.devcel.2016.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Dong J., Zhong S., Wei Y., Wu Q., Yan L., Yong J., Sun L., Wang X., Zhao Y., et al. 2018. Spatial transcriptomic survey of human embryonic cerebral cortex by single-cell RNA-seq analysis. Cell Res. 28:730–745. 10.1038/s41422-018-0053-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli A.B., McFadden D.G., Lin Q., Srivastava D., and Olson E.N.. 1998. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat. Genet. 18:266–270. 10.1038/ng0398-266 [DOI] [PubMed] [Google Scholar]
- Frum T., and Ralston A.. 2015. Cell signaling and transcription factors regulating cell fate during formation of the mouse blastocyst. Trends Genet. 31:402–410. 10.1016/j.tig.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Yan L., Wang R., Li J., Yong J., Zhou X., Wei Y., Wu X., Wang X., Fan X., et al. 2018. Tracing the temporal-spatial transcriptome landscapes of the human fetal digestive tract using single-cell RNA-sequencing. Nat. Cell Biol. 20:721–734. 10.1038/s41556-018-0105-4 [DOI] [PubMed] [Google Scholar]
- Gritsman K., Talbot W.S., and Schier A.F.. 2000. Nodal signaling patterns the organizer. Development. 127:921–932. [DOI] [PubMed] [Google Scholar]
- Guilak F., Cohen D.M., Estes B.T., Gimble J.M., Liedtke W., and Chen C.S.. 2009. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 5:17–26. 10.1016/j.stem.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Yan L., Guo H., Li L., Hu B., Zhao Y., Yong J., Hu Y., Wang X., Wei Y., et al. 2015. The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell. 161:1437–1452. 10.1016/j.cell.2015.05.015 [DOI] [PubMed] [Google Scholar]
- Haghverdi L., Büttner M., Wolf F.A., Buettner F., and Theis F.J.. 2016. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods. 13:845–848. 10.1038/nmeth.3971 [DOI] [PubMed] [Google Scholar]
- Haider S., Meinhardt G., Saleh L., Kunihs V., Gamperl M., Kaindl U., Ellinger A., Burkard T.R., Fiala C., Pollheimer J., et al. 2018. Self-Renewing Trophoblast Organoids Recapitulate the Developmental Program of the Early Human Placenta. Stem Cell Reports. 11:537–551. 10.1016/j.stemcr.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.E., Sozen B., Christodoulou N., Kyprianou C., and Zernicka-Goetz M.. 2017. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science. 356:eaal1810 10.1126/science.aal1810 [DOI] [PubMed] [Google Scholar]
- Harrison S.E., Sozen B., and Zernicka-Goetz M.. 2018. In vitro generation of mouse polarized embryo-like structures from embryonic and trophoblast stem cells. Nat. Protoc. 13:1586–1602. 10.1038/s41596-018-0005-x [DOI] [PubMed] [Google Scholar]
- Heemskerk I., Burt K., Miller M., Chhabra S., Guerra M.C., and Warmflash A.. 2017. Morphogen dynamics control patterning in a stem cell model of human embryo. bioRxiv. doi: (Preprint posted October 13, 2017) 10.1101/202366 [DOI] [Google Scholar]
- Huber G.C. 1918. On the anlage and morphogenesis of the chorda dorsalis in mammalia, in particular the guinea pig (Cavia cobaya). Anat. Rec. 14:217–264. 10.1002/ar.1090140402 [DOI] [Google Scholar]
- Hurlbut J.B., Hyun I., Levine A.D., Lovell-Badge R., Lunshof J.E., Matthews K.R.W., Mills P., Murdoch A., Pera M.F., Scott C.T., et al. 2017. Revisiting the Warnock rule. Nat. Biotechnol. 35:1029–1042. 10.1038/nbt.4015 [DOI] [PubMed] [Google Scholar]
- Hyun I., Wilkerson A., and Johnston J.. 2016. Embryology policy: Revisit the 14-day rule. Nature. 533:169–171. 10.1038/533169a [DOI] [PubMed] [Google Scholar]
- Keller R. 2005. Cell migration during gastrulation. Curr. Opin. Cell Biol. 17:533–541. 10.1016/j.ceb.2005.08.006 [DOI] [PubMed] [Google Scholar]
- Kinder S.J., Tsang T.E., Wakamiya M., Sasaki H., Behringer R.R., Nagy A., and Tam P.P.. 2001. The organizer of the mouse gastrula is composed of a dynamic population of progenitor cells for the axial mesoderm. Development. 128:3623–3634. [DOI] [PubMed] [Google Scholar]
- Kinoshita M., and Smith A.. 2018. Pluripotency Deconstructed. Dev. Growth Differ. 60:44–52. 10.1111/dgd.12419 [DOI] [PubMed] [Google Scholar]
- Klinkert K., Rocancourt M., Houdusse A., and Echard A.. 2016. Rab35 GTPase couples cell division with initiation of epithelial apico-basal polarity and lumen opening. Nat. Commun. 7:11166 10.1038/ncomms11166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M.E., Lönnerberg P., Furlan A., et al. 2018. RNA velocity of single cells. Nature. 560:494–498. 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leptin M. 2005. Gastrulation movements: the logic and the nuts and bolts. Dev. Cell. 8:305–320. 10.1016/j.devcel.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Li L., Dong J., Yan L., Yong J., Liu X., Hu Y., Fan X., Wu X., Guo H., Wang X., et al. 2017. Single-Cell RNA-Seq Analysis Maps Development of Human Germline Cells and Gonadal Niche Interactions. Cell Stem Cell. 20:858–873.e4. 10.1016/j.stem.2017.03.007 [DOI] [PubMed] [Google Scholar]
- Li L., Guo F., Gao Y., Ren Y., Yuan P., Yan L., Li R., Lian Y., Li J., Hu B., et al. 2018. Single-cell multi-omics sequencing of human early embryos. Nat. Cell Biol. 20:847–858. 10.1038/s41556-018-0123-2 [DOI] [PubMed] [Google Scholar]
- Ludwig T.E., Bergendahl V., Levenstein M.E., Yu J., Probasco M.D., and Thomson J.A.. 2006. Feeder-independent culture of human embryonic stem cells. Nat. Methods. 3:637–646. 10.1038/nmeth902 [DOI] [PubMed] [Google Scholar]
- Lyons D.C., Kaltenbach S.L., and McClay D.R.. 2012. Morphogenesis in sea urchin embryos: linking cellular events to gene regulatory network states. Wiley Interdiscip. Rev. Dev. Biol. 1:231–252. 10.1002/wdev.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macklon N.S., Geraedts J.P., and Fauser B.C.. 2002. Conception to ongoing pregnancy: the ‘black box’ of early pregnancy loss. Hum. Reprod. Update. 8:333–343. 10.1093/humupd/8.4.333 [DOI] [PubMed] [Google Scholar]
- Mangan A.J., Sietsema D.V., Li D., Moore J.K., Citi S., and Prekeris R.. 2016. Cingulin and actin mediate midbody-dependent apical lumen formation during polarization of epithelial cells. Nat. Commun. 7:12426 10.1038/ncomms12426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyn I., Kanno T.Y., Ruzo A., Siggia E.D., and Brivanlou A.H.. 2018. Self-organization of a human organizer by combined Wnt and Nodal signalling. Nature. 558:132–135. 10.1038/s41586-018-0150-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon R., Otto E.A., Kokoruda A., Zhou J., Zhang Z., Yoon E., Chen Y.C., Troyanskaya O., Spence J.R., Kretzler M., and Cebrián C.. 2018. Single-cell analysis of progenitor cell dynamics and lineage specification in the human fetal kidney. Development. 145:dev164038 10.1242/dev.164038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H., Hernando-Herraez I., Savino A., Scialdone A., Macaulay I., Mulas C., Chandra T., Voet T., Dean W., Nichols J., et al. 2017. Single-Cell Landscape of Transcriptional Heterogeneity and Cell Fate Decisions during Mouse Early Gastrulation. Cell Reports. 20:1215–1228. 10.1016/j.celrep.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani S.M., Metzger J.J., Nichols J., Siggia E.D., and Hadjantonakis A.K.. 2018. Micropattern differentiation of mouse pluripotent stem cells recapitulates embryo regionalized cell fate patterning. eLife. 7:e32839 10.7554/eLife.32839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsut L., Roybal K.T., Xiong X., Gordley R.M., Coyle S.M., Thomson M., and Lim W.A.. 2016. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell. 164:780–791. 10.1016/j.cell.2016.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrozowska P.S., and Fukuda M.. 2016. Regulation of podocalyxin trafficking by Rab small GTPases in 2D and 3D epithelial cell cultures. J. Cell Biol. 213:355–369. 10.1083/jcb.201512024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Yabuta Y., Okamoto I., Sasaki K., Iwatani C., Tsuchiya H., and Saitou M.. 2017. Single-cell transcriptome of early embryos and cultured embryonic stem cells of cynomolgus monkeys. Sci. Data. 4:170067 10.1038/sdata.2017.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemashkalo A., Ruzo A., Heemskerk I., and Warmflash A.. 2017. Morphogen and community effects determine cell fates in response to BMP4 signaling in human embryonic stem cells. Development. 144:3042–3053. 10.1242/dev.153239 [DOI] [PubMed] [Google Scholar]
- O’Leary T., Heindryckx B., Lierman S., van Bruggen D., Goeman J.J., Vandewoestyne M., Deforce D., de Sousa Lopes S.M., and De Sutter P.. 2012. Tracking the progression of the human inner cell mass during embryonic stem cell derivation. Nat. Biotechnol. 30:278–282. 10.1038/nbt.2135 [DOI] [PubMed] [Google Scholar]
- O’Leary T., Heindryckx B., Lierman S., Van der Jeught M., Duggal G., De Sutter P., and Chuva de Sousa Lopes S.M.. 2013. Derivation of human embryonic stem cells using a post-inner cell mass intermediate. Nat. Protoc. 8:254–264. 10.1038/nprot.2012.157 [DOI] [PubMed] [Google Scholar]
- Okae H., Toh H., Sato T., Hiura H., Takahashi S., Shirane K., Kabayama Y., Suyama M., Sasaki H., and Arima T.. 2018. Derivation of Human Trophoblast Stem Cells. Cell Stem Cell. 22:50–63.e6. 10.1016/j.stem.2017.11.004 [DOI] [PubMed] [Google Scholar]
- Pera M.F. 2017. Human embryo research and the 14-day rule. Development. 144:1923–1925. 10.1242/dev.151191 [DOI] [PubMed] [Google Scholar]
- Pereira P.N.G., Dobreva M.P., Graham L., Huylebroeck D., Lawson K.A., and Zwijsen A.N.. 2011. Amnion formation in the mouse embryo: the single amniochorionic fold model. BMC Dev. Biol. 11:48 10.1186/1471-213X-11-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., and Trapnell C.. 2017. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 14:979–982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A. 2018. XEN and the Art of Stem Cell Maintenance: Molecular Mechanisms Maintaining Cell Fate and Self-Renewal in Extraembryonic Endoderm Stem (XEN) Cell Lines. Adv. Anat. Embryol. Cell Biol. 229:69–78. 10.1007/978-3-319-63187-5_6 [DOI] [PubMed] [Google Scholar]
- Robertson E.J. 2014. Dose-dependent Nodal/Smad signals pattern the early mouse embryo. Semin. Cell Dev. Biol. 32:73–79. 10.1016/j.semcdb.2014.03.028 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., and Macara I.G.. 2014. Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 15:225–242. 10.1038/nrm3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roost M.S., van Iperen L., Ariyurek Y., Buermans H.P., Arindrarto W., Devalla H.D., Passier R., Mummery C.L., Carlotti F., de Koning E.J., et al. 2015. KeyGenes, a Tool to Probe Tissue Differentiation Using a Human Fetal Transcriptional Atlas. Stem Cell Reports. 4:1112–1124. 10.1016/j.stemcr.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosowski K.A., Mertz A.F., Norcross S., Dufresne E.R., and Horsley V.. 2015. Edges of human embryonic stem cell colonies display distinct mechanical properties and differentiation potential. Sci. Rep. 5:14218 10.1038/srep14218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J. 2016. Making the Mouse Blastocyst: Past, Present, and Future. Curr. Top. Dev. Biol. 117:275–288. 10.1016/bs.ctdb.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Rossant J., and Tam P.P.L.. 2017. New Insights into Early Human Development: Lessons for Stem Cell Derivation and Differentiation. Cell Stem Cell. 20:18–28. 10.1016/j.stem.2016.12.004 [DOI] [PubMed] [Google Scholar]
- Sasaki K., Nakamura T., Okamoto I., Yabuta Y., Iwatani C., Tsuchiya H., Seita Y., Nakamura S., Shiraki N., Takakuwa T., et al. 2016. The Germ Cell Fate of Cynomolgus Monkeys Is Specified in the Nascent Amnion. Dev. Cell. 39:169–185. 10.1016/j.devcel.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Schlüter M.A., Pfarr C.S., Pieczynski J., Whiteman E.L., Hurd T.W., Fan S., Liu C.J., and Margolis B.. 2009. Trafficking of Crumbs3 during cytokinesis is crucial for lumen formation. Mol. Biol. Cell. 20:4652–4663. 10.1091/mbc.e09-02-0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialdone A., Tanaka Y., Jawaid W., Moignard V., Wilson N.K., Macaulay I.C., Marioni J.C., and Göttgens B.. 2016. Resolving early mesoderm diversification through single-cell expression profiling. Nature. 535:289–293. 10.1038/nature18633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M.N., and Zernicka-Goetz M.. 2018. Deconstructing and reconstructing the mouse and human early embryo. Nat. Cell Biol. 20:878–887. 10.1038/s41556-018-0144-x [DOI] [PubMed] [Google Scholar]
- Shahbazi M.N., Jedrusik A., Vuoristo S., Recher G., Hupalowska A., Bolton V., Fogarty N.N.M., Campbell A., Devito L., Ilic D., et al. 2016. Self-organization of the human embryo in the absence of maternal tissues. Nat. Cell Biol. 18:700–708. 10.1038/ncb3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M.N., Scialdone A., Skorupska N., Weberling A., Recher G., Zhu M., Jedrusik A., Devito L.G., Noli L., Macaulay I.C., et al. 2017. Pluripotent state transitions coordinate morphogenesis in mouse and human embryos. Nature. 552:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Taniguchi K., Gurdziel K., Townshend R.F., Xue X., Yong K.M.A., Sang J., Spence J.R., Gumucio D.L., and Fu J.. 2017a Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nat. Mater. 16:419–425. 10.1038/nmat4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Taniguchi K., Townshend R.F., Miki T., Gumucio D.L., and Fu J.. 2017b A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat. Commun. 8:208 10.1038/s41467-017-00236-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic M., Metzger J.J., Etoc F., Yoney A., Ruzo A., Martyn I., Croft G., Brivanlou A.H., and Siggia E.D.. 2018. Molecular mechanism of symmetry breaking in a 3D model of a human epiblast. bioRxiv. doi:10.1101/330704 (Preprint posted May 29, 2018) [DOI] [PubMed] [Google Scholar]
- Slieker R.C., Roost M.S., van Iperen L., Suchiman H.E., Tobi E.W., Carlotti F., de Koning E.J., Slagboom P.E., Heijmans B.T., and Chuva de Sousa Lopes S.M.. 2015. DNA Methylation Landscapes of Human Fetal Development. PLoS Genet. 11:e1005583 10.1371/journal.pgen.1005583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozen B., Amadei G., Cox A., Wang R., Na E., Czukiewska S., Chappell L., Voet T., Michel G., Jing N., et al. 2018. Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol. 20:979–989. 10.1038/s41556-018-0147-7 [DOI] [PubMed] [Google Scholar]
- Spemann H., and Mangold H.. 2001. Induction of embryonic primordia by implantation of organizers from a different species. 1923. Int. J. Dev. Biol. 45:13–38. [PubMed] [Google Scholar]
- Stern C.D. 2006. Evolution of the mechanisms that establish the embryonic axes. Curr. Opin. Genet. Dev. 16:413–418. 10.1016/j.gde.2006.06.005 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S.. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 131:861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Tam P.P., and Loebel D.A.. 2007. Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 8:368–381. 10.1038/nrg2084 [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Shao Y., Townshend R.F., Tsai Y.H., DeLong C.J., Lopez S.A., Gayen S., Freddo A.M., Chue D.J., Thomas D.J., et al. 2015. Lumen Formation Is an Intrinsic Property of Isolated Human Pluripotent Stem Cells. Stem Cell Reports. 5:954–962. 10.1016/j.stemcr.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K., Shao Y., Townshend R.F., Cortez C.L., Harris C.E., Meshinchi S., Kalantry S., Fu J., O’Shea K.S., and Gumucio D.L.. 2017. An apicosome initiates self-organizing morphogenesis of human pluripotent stem cells. J. Cell Biol. 216:3981–3990. 10.1083/jcb.201704085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewary M., Ostblom J., Prochazka L., Zulueta-Coarasa T., Shakiba N., Fernandez-Gonzalez R., and Zandstra P.W.. 2017. A stepwise model of reaction-diffusion and positional information governs self-organized human peri-gastrulation-like patterning. Development. 144:4298–4312. 10.1242/dev.149658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., and Jones J.M.. 1998. Embryonic stem cell lines derived from human blastocysts. Science. 282:1145–1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., and Rinn J.L.. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32:381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turing A.M. 1952. The Chemical Basis of Morphogenesis. Philos T Roy Soc B. 237:37–72. 10.1098/rstb.1952.0012 [DOI] [Google Scholar]
- Turner D.A., Girgin M., Alonso-Crisostomo L., Trivedi V., Baillie-Johnson P., Glodowski C.R., Hayward P.C., Collignon J., Gustavsen C., Serup P., et al. 2017. Anteroposterior polarity and elongation in the absence of extra-embryonic tissues and of spatially localised signalling in gastruloids: mammalian embryonic organoids. Development. 144:3894–3906. 10.1242/dev.150391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink S.C., Baillie-Johnson P., Balayo T., Hadjantonakis A.K., Nowotschin S., Turner D.A., and Martinez Arias A.. 2014. Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development. 141:4231–4242. 10.1242/dev.113001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.S., Saadatpour A., and Albert R.. 2012. Boolean modeling in systems biology: an overview of methodology and applications. Phys. Biol. 9:055001 10.1088/1478-3975/9/5/055001 [DOI] [PubMed] [Google Scholar]
- Warmflash A., Sorre B., Etoc F., Siggia E.D., and Brivanlou A.H.. 2014. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods. 11:847–854. 10.1038/nmeth.3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J., Toh Y.C., Xu S., and Yu H.. 2015. A method for human teratogen detection by geometrically confined cell differentiation and migration. Sci. Rep. 5:10038 10.1038/srep10038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J., et al. 2013. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20:1131–1139. 10.1038/nsmb.2660 [DOI] [PubMed] [Google Scholar]
- Yu W., Datta A., Leroy P., O’Brien L.E., Mak G., Jou T.S., Matlin K.S., Mostov K.E., and Zegers M.M.. 2005. Beta1-integrin orients epithelial polarity via Rac1 and laminin. Mol. Biol. Cell. 16:433–445. 10.1091/mbc.e04-05-0435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Zhang S., Fan X., Wu Q., Yan L., Dong J., Zhang H., Li L., Sun L., Pan N., et al. 2018. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 555:524–528. 10.1038/nature25980 [DOI] [PubMed] [Google Scholar]
- Zhu P., Guo H., Ren Y., Hou Y., Dong J., Li R., Lian Y., Fan X., Hu B., Gao Y., et al. 2018. Single-cell DNA methylome sequencing of human preimplantation embryos. Nat. Genet. 50:12–19. 10.1038/s41588-017-0007-6 [DOI] [PubMed] [Google Scholar]