Dengue, caused by dengue virus, is a rapidly spreading disease, and currently there are no treatments available. Host factors involved in the viral replication of dengue virus are potential antiviral therapeutic targets. Although RHA has been shown to promote the multiplication of several viruses, such as HIV and adenovirus, its role in the flavivirus family, including dengue virus, Japanese encephalitis virus, and emerging Zika virus, remains elusive. The current study revealed that RHA relocalized into the cytoplasm upon DENV infection and associated with viral RNA and nonstructural proteins, implying that RHA was actively engaged in the viral life cycle. We further provide evidence that RHA promoted the viral yields of DENV2 independent of its helicase activity. These findings demonstrated that RHA is a new host factor required for DENV replication and might serve as a target for antiviral drugs.
KEYWORDS: dengue virus, flavivirus, host factor, viral replication
ABSTRACT
Dengue virus (DENV) utilizes host factors throughout its life cycle. In this study, we identified RNA helicase A (RHA), a member of the DEAD/H helicase family, as an important host factor of DENV. In response to DENV2 infection, nuclear RHA protein was partially redistributed into the cytoplasm. The short interfering RNA-mediated knockdown of RHA significantly reduced the amounts of infectious viral particles in various cells. The RHA knockdown reduced the multistep viral growth of DENV2 and Japanese encephalitis virus but not Zika virus. Further study showed that the absence of RHA resulted in a reduction of both viral RNA and protein levels, and the data obtained from the reporter replicon assay indicated that RHA does not directly promote viral protein synthesis. RHA bound to the DENV RNA and associated with three nonstructural proteins, including NS1, NS2B3, and NS4B. Further study showed that different domains of RHA mediated its interaction with these viral proteins. The expression of RHA or RHA-K417R mutant protein lacking ATPase/helicase activity in RHA-knockdown cells successfully restored DENV2 replication levels, suggesting that the helicase activity of RHA is dispensable for its proviral effect. Overall, our work reveals that RHA is an important factor of DENV and might serve as a target for antiviral agents.
IMPORTANCE Dengue, caused by dengue virus, is a rapidly spreading disease, and currently there are no treatments available. Host factors involved in the viral replication of dengue virus are potential antiviral therapeutic targets. Although RHA has been shown to promote the multiplication of several viruses, such as HIV and adenovirus, its role in the flavivirus family, including dengue virus, Japanese encephalitis virus, and emerging Zika virus, remains elusive. The current study revealed that RHA relocalized into the cytoplasm upon DENV infection and associated with viral RNA and nonstructural proteins, implying that RHA was actively engaged in the viral life cycle. We further provide evidence that RHA promoted the viral yields of DENV2 independent of its helicase activity. These findings demonstrated that RHA is a new host factor required for DENV replication and might serve as a target for antiviral drugs.
INTRODUCTION
Dengue virus (DENV) is a mosquito-borne flavivirus with four serotypes of viruses (DENV1 to DENV4). The infection with either DENV serotype leads to dengue fever (DF), dengue hemorrhagic fever (DHF), and dengue shock syndrome (DSS) (1). DENV has been found in more than 100 countries that are located in tropical and subtropical areas and infects more than 60 million people annually (2–4). However, there have been no effective therapeutic drugs available for dengue diseases (2, 5).
The DENV genome is a positive-strand RNA that is 10.4 kb in length (1). Once the DENV virion enters the host cell through attachment and endocytosis, the viral genome RNA that encodes a single polyprotein is released into the cytoplasm. The polyprotein is cleaved into three structural proteins (C, capsid; pr/M, membrane; E, envelope) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) by cellular and viral proteases. The NS proteins induce formation of a replication complex in the perinuclear region of the endoplasmic reticulum (ER) where viral RNA synthesis and protein translation occur (6, 7). The nascent viral RNA then is packaged into structural proteins and transported through the Golgi to the cell surface and released (1, 8).
As its genome is extremely limited, DENV must exploit host proteins to efficiently replicate in the host cells (8, 9). Recently, a variety of novel host factors involved in DENV invasion or replication have been identified (10–13). Two groups have reported that the oligosaccharyltransferase (OST) complex plays a key role in viral RNA replication, and the subunits of OST specifically associate with viral NS2B and NS3 proteins using the CRISPR/Cas9 technique (11, 12). Zhang et al. reported that the signal peptidase complex subunits are a group of crucial host factors that function by promoting the posttranslational processing of viral structural proteins (13). Through RNA affinity chromatography or quantitative thiouridine cross-linking spectrometry technologies, many RNA binding proteins, including DEAD box RNA helicase (DDX6), NF90, and ER13, have been identified to participate in the life cycle of DENV (14–18). Nonetheless, additional host proteins remain to be identified, and their roles in DENV replication have yet to be elucidated.
RNA helicase A (RHA), a member of the DEAD/H family of RNA helicases, also named the DExD/H-box 9 (DHX9) or nuclear DNA helicase II (NDH II), has been shown to play a proviral role in the life cycle of many viruses (19). The RHA protein is composed of two N-terminal double-stranded RNA (dsRNA)-binding domains (dsRBD), a helicase core domain, and a C-terminal arginine-glycine-glycine (RGG) repeat domain. RHA mainly resides in the nuclei and shuttles between the cytoplasm and nucleus. RHA unwinds both DNA and RNA (20) and participates in many biological processes involved in the cellular DNA and RNA metabolic processes, such as RNA processing and transport. Not surprisingly, the functions of RHA are exploited by a number of RNA and DNA viruses, including HIV, hepatitis C virus (HCV), cytomegalovirus, adenovirus, hepatitis E virus, influenza A virus, myxoma virus, classical swine fever virus, and foot-and-mouth-disease viruses (19). The proviral effect of RHA can be exerted through the binding of viral proteins, such as HIV GAG (21), influenza NS1 (22), or myxoma M029 protein (23), or through the binding of viral RNA, such as HIV RNA (20). RHA facilitates viral replication at various steps of virus life cycles, such as RNA synthesis (24), RNA transcription (25), protein translation (26, 27), and viral packaging (21).
Although RHA has been previously reported to be present in the protein complex that binds dengue virus RNA (15), whether RHA plays a role in DENV replication was still unknown. Our study utilized RNA interference (RNAi) to knock down RHA in different cells and then compared the viral replication levels in these cells. Our data demonstrated that RHA enhanced the DENV replication level at the viral RNA synthesis step. In addition, we found that RHA associated with viral RNA and three NS proteins, and its proviral effect was independent of its ATPase/helicase activity.
RESULTS
Nuclear RHA was partially redistributed into the cytoplasm upon DENV2 infection and facilitated viral replication.
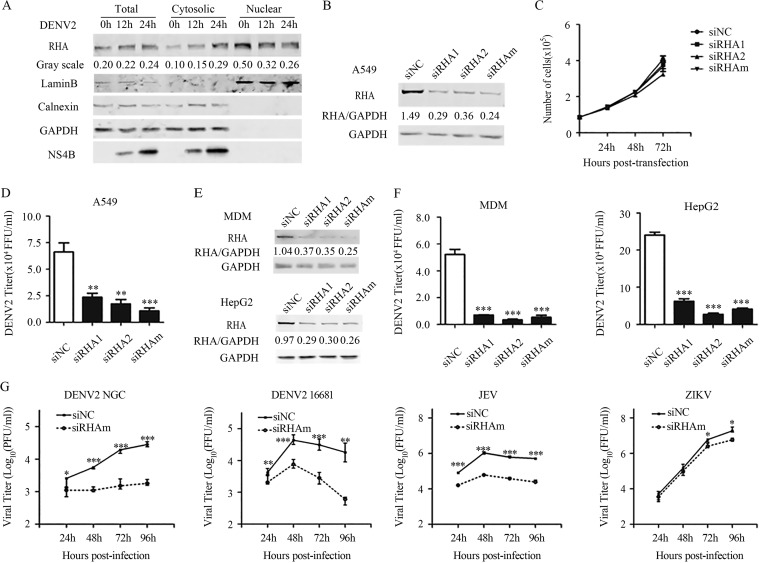
As RHA predominantly resides in the nucleus while DENV replicates in the cytoplasm, we first examined whether RHA translocation occurred upon DENV infection. A549 cells were infected with DENV2 and harvested at 0, 12, and 24 h postinfection (p.i.). The proteins from the cytoplasmic and nuclear fractions were subjected to Western blot analysis. As shown in Fig. 1A, increased levels of RHA protein were detected in the cytoplasmic fractions of the DENV2-infected cells at 12 and 24 h p.i., while a reduction of RHA levels was clearly observed in the nuclear fractions. In the whole-cell extracts and cytoplasmic fractions, the levels of viral NS4B protein increased as time elapsed, validating that these cells were infected.
FIG 1.
RHA was relocalized into cytoplasm upon DENV2 infection and was involved in the viral production. (A) A549 cells were mock infected or infected with DENV2 (MOI of 3). Cells were harvested at 0, 12, and 24 h p.i. for protein extraction. Proteins derived from cytoplasmic fraction and nuclear fraction of the cells were subjected to Western blotting. GAPDH and Lamin B served as the loading control for the cytoplasmic fraction and nuclear fraction. Calnexin served as an indicator of ER. Data are representative of at least three independent experiments. (B) Confirmation of siRNA efficiency. A549 cells were transfected with 16 nmol siNC, siRHA1, siRHA2, or siRHAm (mixture of siRHA1 and siRHA2). At 48 h posttransfection, cells were harvested for Western blotting. GAPDH was probed as an internal control. (C) Cell growth curve of RHA-KD cells. At 1, 2, and 3 days posttransfection with siNC or siRHAm, cell numbers were counted by trypan blue staining. (D) Single-step virus growth assay. A549 cells were transfected with siNC or siRHAm for 48 h, followed by DENV2 infection (MOI of 3). At 24 h p.i., cell supernatants were collected for focus-forming assay. (E) Western blot to examine the siRNA efficacy in monocyte-derived macrophage (MDM) and HepG2 cells. Cells were transfected with 16 nmol siNC, siRHA1, siRHA2, or siRHAm. At 48 h posttransfection, cells were harvested for Western blotting. (F) Single-step virus growth assay. MDM and HepG2 cells were transfected with siNC, siRHA1, siRHA2, or siRHAm for 48 h, followed by DENV2 infection (MOI of 3). At 24 h p.i., cell supernatants were collected for focus-forming assay. (G) Multistep virus growth assay. A549 cells were transfected with siRNA for 48 h, followed by infection of DENV2 NGC, DENV2 16681, JEV, and ZIKV (MOI of 0.01). Samples were collected at 24, 48, 72, and 96 h p.i. The viral titers were measured by standard plaque or focus-forming assay. Data were shown as means ± SD from at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (each by unpaired, two-tailed Student's t test).
To test whether RHA plays a role in DENV replication, we employed an RNA interference (RNAi) strategy to knock down the RHA protein in three permissive cell lines originating from different tissues, including A549, monocyte-derived macrophage (MDM), and HepG2 cells. The silencing efficacy of two different RHA-targeted short interfering RNAs (siRNAs; designated siRHA1 and siRHA2) and their mixture (siRHAm) in A549 cells was confirmed by Western blot analysis. As shown in Fig. 1B, the cells transfected with siRHA1, siRHA2, or siRHAm expressed significantly less RHA protein than those cells transfected with negative-control siRNA (siNC). Transfection of siRHAs did not lead to significant alterations of cell viability and growth (Fig. 1C).
We next compared single-step virus growth in RHA-sufficient and RHA-deficient cells. Cells were transfected with siRNAs, followed by DENV2 infection at 2 days posttransfection. The supernatants were collected at 1 days p.i. and titers determined by a plaque assay or focus-forming assay. The viral yields in the A549 cells transfected with siRHA1, siRHA2, and siRHAm were 2.8-, 3.8-, and 6.2-fold lower, respectively, than those with the siNC-transfected control cells (P values of <0.01, 0.01, and 0.001) (Fig. 1D). In addition, the DENV2 titers in the siRHA1-, siRHA2-, or siRHAm-transfected MDMs were reduced by 7.5-, 15.1-, and 9.7-fold, respectively; similarly, the transfection of siRHA1, siRHA2, and siRHAm in HepG2 cells led to significant reductions of viral yield by 3.8-, 8.8-, and 5.8-fold, respectively (P < 0.001) (Fig. 1E and F).
We then performed a multistep virus growth assay to explore whether RHA affects the replication and transmission of several flavivirus members, including DENV2 NGC and 16681 strains, Japanese encephalitis virus (JEV), and Zika virus (ZIKV). A549 cells were transfected with siRNAs, followed by virus infection at a multiplicity of infection (MOI) of 0.01. The viral supernatants were collected at 1, 2, 3, and 4 days p.i., and then titers were determined. In the absence of RHA, the viral yields of both DENV2 NGC and 16681 at all of the tested time points were significantly reduced (Fig. 1G). Similarly, the viral yields of JEV in RHA-knockdown (KD) cells were downregulated by 5.1-, 18.1-, 16.4-, and 20.3-fold at 1, 2, 3, and 4 days p.i., respectively (P < 0.001) (Fig. 1G). In contrast, the amounts of ZIKV in the RHA-KD cells were comparable to those of control cells at 1 and 2 days p.i. (P > 0.05) (Fig. 1G) and were slightly reduced at 3 and 4 days p.i. (P < 0.05) (Fig. 1G).
Silencing of RHA downregulated viral RNA and protein synthesis.
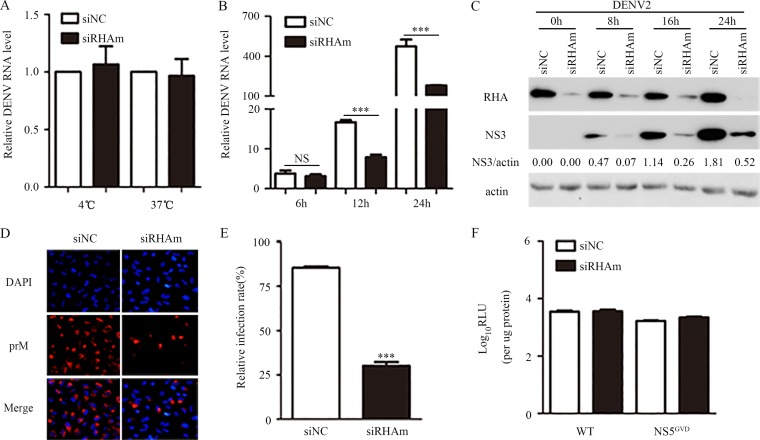
To identify the virus life cycle step that requires RHA, we tested the impact of RHA on viral attachment by inoculating the DENV2 particles onto RHA-sufficient and RHA-deficient cells at 4°C for 1 h. Total RNA was extracted for quantitative real-time PCR (qRT-PCR) to detect viral RNA levels, indicating the amounts of virions that bound to the cell membrane. The PCR data showed that the viral RNA levels were comparable in the siNC- and siRHAm-transfected cells (Fig. 2A), suggesting that RHA does not act at the attachment stage. We then incubated the DENV2 virus at 37°C for 1 h to allow for virions to attach and internalize. The whole cells were collected and total RNA was extracted. Real-time PCRs were performed to detect viral RNA levels to indicate the amounts of internalized virions. The viral RNA levels in the RHA-KD cells were similar to those of the control cells (Fig. 2A), implying that RHA was not involved in viral endocytosis.
FIG 2.
RHA knockdown reduced the viral RNA and protein levels. A549 cells were transfected with siNC or siRHAm for 48 h and applied for the following detections. (A) Entry assay. Cells were inoculated with DENV2 (MOI of 3), followed by incubation at 4°C or 37°C for 1 h. Cells were washed with PBS and harvested for RNA extraction and real-time PCR to measure viral RNA levels. (B) Cells were mock infected or infected with DENV2 and harvested at 6, 12, and 24 h p.i. The viral RNA level was measured by real-time PCR. (C and D) Cells were infected with DENV2 and harvested at 8, 16, and 24 h p.i. for Western blotting to detect NS3 (C) or at 24 h p.i. for IFA to detect prM (D). Actin served as an internal control. (E) At least 200 cells from each sample in three independent experiments were counted. (F) Effect of RHA knockdown on translation of the DENV2 reporter replicon. Cells were transfected with pDENrep-FH (16681) replicon (WT) and the NS5 GVD mutant (NS5GVD) RNAs. The cells were harvested at 6 h posttransfection for luciferase assay. Data are shown as means ± SD from at least three independent experiments. NS, no statistical significance; ***, P < 0.001 by unpaired, two-tailed Student's t test.
We next compared the viral RNA levels at 6, 12, and 24 h p.i. in the control and RHA-KD cells. The RHA knockdown slightly reduced the viral RNA level at 6 h p.i. (P > 0.05) (Fig. 2B) but led to significant reductions at 12 and 24 h p.i. (P < 0.001) (Fig. 2B). The viral protein levels were measured by Western blotting and immunofluorescence microscopy assay (IFA). The Western blot analysis revealed that at 8, 16, and 24 h p.i., the viral NS3 levels in RHA-KD cells were dramatically decreased compared to those of RHA-sufficient cells (Fig. 2C). As a control, the β-actin levels in all samples were comparable. Consistent with this, the IFA images revealed that in RHA-KD cells, the percentage of prM-positive cells was lower than that of control cells (30.1% versus 85.4%; P < 0.001) (Fig. 2D and E).
Furthermore, we utilized subgenomic DENV2 replicons that encode the seven NS proteins with a firefly (FF) reporter to distinguish between the RHA proviral effect on protein translation and RNA replication (Fig. 2F) (28). The in vitro-transcribed replicon RNA of wild-type (WT) DENV2 (16681) or the NS5GVD mutant replicon was introduced into the control or RHA-KD cells. Ideally, two peaks of luciferase activity could be detected with wild-type DENV2 replicon: an early peak between 4 and 8 h posttransfection, indicating the protein translation of the replicon, and a second peak between 48 and 96 h posttransfection that primarily depends on RNA synthesis of the replicon. In contrast, only one early peak of luciferase activity was generated by the NS5GVD mutant DENV replicon because the RNA-dependent RNA polymerase (RdRp) activity of NS5 was abolished, which also served as an indicator of protein translation. Unfortunately, we observed that most of the cells transfected with these replicon RNAs died at 24 and 48 h posttransfection, and the luciferase activities were extremely low (data not shown). Therefore, the presented data only showed the relative luciferase activity measured with the cell lysate harvested at 6 h posttransfection to indicate the protein translation of replicons (Fig. 2F). The levels of relative luciferase activity of WT DENV2 and NS5GVD mutant replicons were comparable in the control and RHA-KD cells (P > 0.05) (Fig. 2F), suggesting that RHA does not promote viral protein translation.
RHA bound to DENV2 and ZIKV RNA.
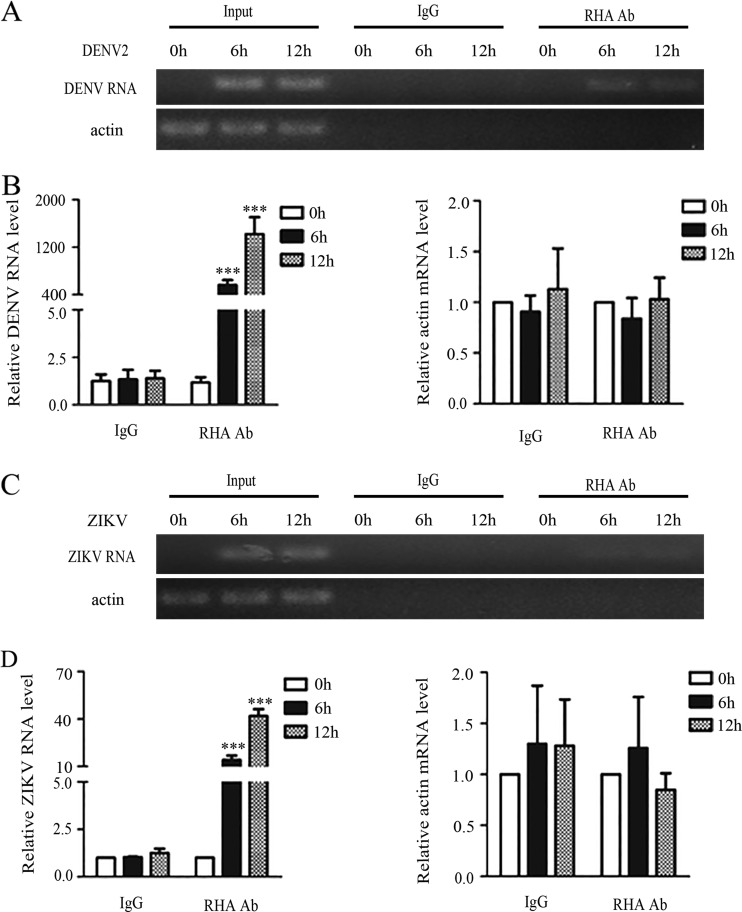
Because RHA promotes viral replication of some viruses by binding to viral RNA (20), we then tested whether RHA binds to DENV2 RNA by RNA immunoprecipitation (RIP) assay. A549 cells were infected with DENV2 or ZIKV and harvested at 0, 6, and 12 h p.i. The immunoprecipitation (IP) assay was performed using control IgG or anti-RHA antibody, followed by total RNA extraction and conventional or quantitative PCR. The PCR data showed that DENV2-specific fragments were detected in the anti-RHA-associated precipitate but not in the control IgG (Fig. 3A). The real-time PCR data confirmed that the viral RNA levels associated with anti-RHA were 479.1 (6 h p.i.)- and 1,220.4-fold (12 h p.i.) higher than those associated with control IgG (P < 0.001 and P < 0.001) (Fig. 3B). As a control, no association between RHA and actin RNA was detected (Fig. 3A and B). In the ZIKV-infected cells, weak bands were detected in the anti-RHA-precipitated samples but not in the control IgG-precipitated samples (Fig. 3C). The ZIKV RNA levels associated with anti-RHA were 14.1-fold (6 h p.i.) and 41.8-fold (12 h p.i.) higher than that of the control (P < 0.001) (Fig. 3D).
FIG 3.
RHA was associated with viral RNA. A549 cells were infected with DENV2 (A and B) or ZIKV (C and D) at an MOI of 3 and harvested at 6 and 12 h p.i. for RIP assay. Whole-cell lysates were prepared for immunoprecipitation using anti-RHA antibody (Ab) or control IgG. Total RNAs in the precipitates were isolated, followed by reverse transcription. Conventional PCR (A and C) or real-time PCR (B and D) using primers specific for the DENV2 or ZIKV genome were performed to measure the relative levels of RHA-associated viral RNA. Data are shown as means ± SD from three independent experiments. ***, P < 0.001 by unpaired, two-tailed Student's t test.
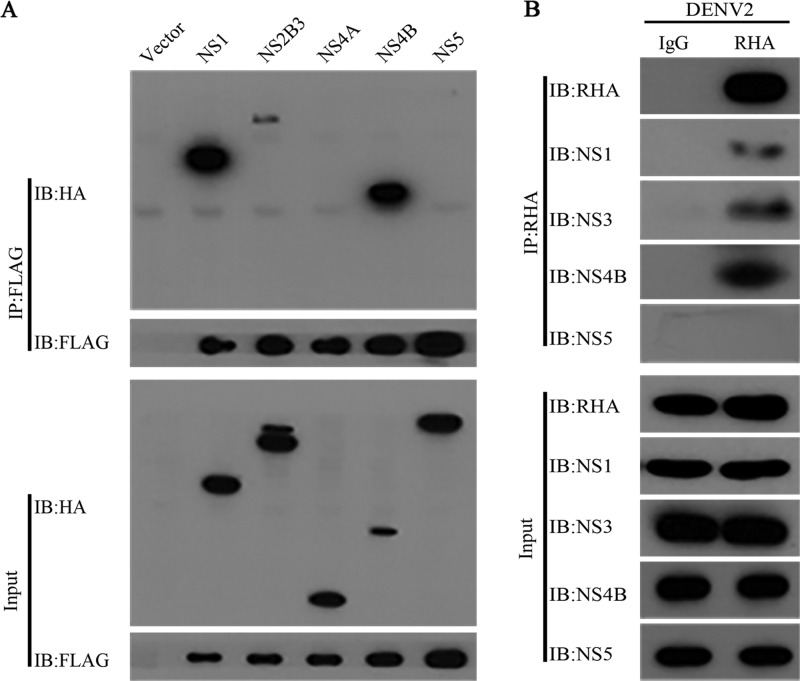
RHA associated with viral NS1, NS2B3, and NS4B but not NS5.
During the DENV2 replication process, viral nonstructural proteins such as NS1, NS3, and NS5 are gathered in the replication complex to facilitate viral RNA replication (1, 8, 9). The data described above revealed that RHA might play a role in RNA synthesis and interacts with viral RNA; therefore, we hypothesized that RHA is recruited into the replication complex and associates with DENV NS proteins. To verify this hypothesis, we examined the association between RHA and viral NS proteins by a co-IP assay. The 293T cells were transfected with the plasmids expressing RHA-FLAG and NS-HA proteins, including NS1, NS2B3, NS4A, NS4B, or NS5. The whole-cell extracts were prepared at 48 h posttransfection and then applied for the co-IP assay using anti-FLAG beads. The Western blot analysis revealed that NS1, NS2B3, and NS4B, but not NS4A and NS5, precipitated with RHA (Fig. 4A). Furthermore, we performed a co-IP assay using the RHA antibody to test whether RHA associated with these viral proteins in DENV2-infected cells. Consistent with our findings, RHA, NS1, NS2B3, and NS4B were found to be present in the immunoprecipitated complex, while NS5 was absent (Fig. 4B).
FIG 4.
RHA interacted with viral NS1, NS3, and NS4B. (A) 293T cells were transfected with plasmids expressing RHA-FLAG and DENV2 NS proteins for 48 h. Whole-cell extracts were prepared for co-IP assay using anti-FLAG antibody. IB, immunoblot. (B) A549 cells were infected with DENV2 (MOI of 3). Whole-cell extracts were prepared at 24 h p.i. for co-IP assay using anti-RHA antibody. Protein complexes were separated by SDS-PAGE and detected by Western blotting with antibodies against the indicated proteins. Data were representative of three independent experiments.
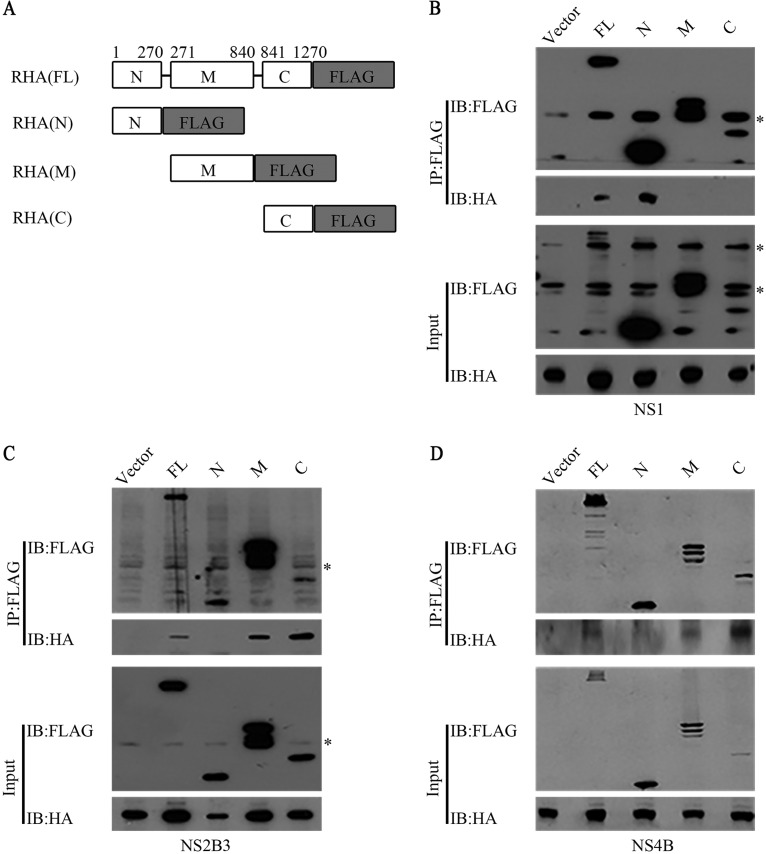
Different domains of RHA mediated its interaction with different NS proteins.
To map the domains of RHA that are involved in its interaction with viral proteins, we constructed three plasmids expressing truncated RHA fragments tagged with FLAG, namely, N-terminal dsRBDs (1 to 270 amino acids [aa]) (RHAN-FLAG), helicase motif (RHAM-FLAG) (271 to 840 aa), and C-terminal domain (RHAC-FLAG) (841 to 1270 aa) (Fig. 5A). The 293T cells were cotransfected with the constructs expressing NS-HA and RHA tagged with FLAG and harvested for a co-IP assay using the FLAG antibody. The Western blot analysis revealed that NS1 precipitated with RHA-FLAG and RHAN-FLAG but not RHAM-FLAG and RHAC-FLAG (Fig. 5B), while NS2B3 and NS4B precipitated with RHA-FLAG, RHAM-FLAG, and RHAC-FLAG but not RHAN-FLAG (Fig. 5C and D). The data revealed that different domains of RHA mediate its interaction with viral proteins.
FIG 5.
Mapping of RHA domains that interacted with viral NS proteins. (A) Schematic representation of full-length RHA-FLAG and three truncated forms of RHA. 293T cells were transfected with plasmids encoding different regions of RHA and DENV2 NS1-HA (B), NS2B3-HA (C), or NS4B-HA (D). The whole-cell extracts were prepared at 48 h posttransfection and immunoprecipitated with FLAG antibody. Western blot was probed with FLAG or HA antibody. RHA-FLAG, full-length RHA tagged with FLAG at C terminus; RHAN-FLAG, N-terminal fragment of RHA tagged with FLAG at C terminus; RHAM-FLAG, intermediate fragment of RHA tagged with FLAG at C terminus; RHAC-FLAG, C-terminal fragment of RHA tagged with FLAG at C terminus. Data are representative of three independent experiments. Asterisks indicate nonspecific bands.
Expression of wild-type or mutant RHA rescued DENV2 replication in RHA-KD cells.
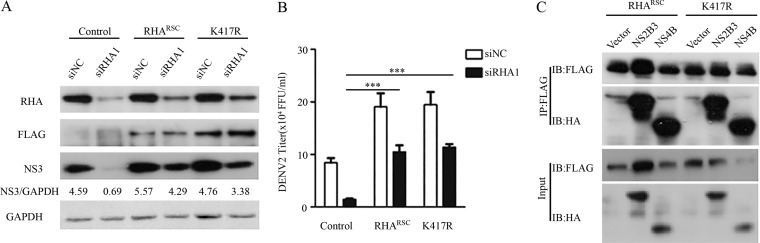
To examine whether the helicase activity of RHA is required for its proviral function, we induced a K417R mutation in RHA, a key site for its ATP-binding and helicase activities (21, 29). In the lentiviral vector CSII-EF-MCS-IRES2-Venus-RHARSC-FLAG or CSII-EF-MCS-IRES2-Venus-RHA-K417R-FLAG, the siRHA1-targeted sequence in the RHA open reading frame (ORF) was mutated into synonymous sites to resist the knockdown effect by siRHA1. The A549 cell pools stably expressing RHARSC and K417R mutants were sorted by flow cytometry and verified by Western blotting. The control cells and RHARSC- and RHA-K417R-expressing cells were transfected with siNC or siRHA1, followed by DENV2 infection. Cells and supernatants were harvested at 24 h p.i. Western blot analysis revealed that the RHARSC-FLAG and K417R-FLAG proteins were comparable in siNC- and siRHA1-transfected cells, indicating they were resistant to the siRHA1 knockdown effect (Fig. 6A). The NS3 protein level was reduced in the siRHA1-transfected control cells, as expected, while in both RHARSC-FLAG- and K417R-FLAG-expressing cells the viral NS3 levels were largely restored (Fig. 6A). Importantly, the expression of RHARSC or the K417R mutant in RHA-KD cells led to loss of inhibition of viral yields mediated by RHA knockdown (P < 0.001) (Fig. 6B). To further verify the role of the K417R mutant in viral replication, we tested whether it interacts with viral proteins by a co-IP assay. The Western blot analysis revealed that the K417R mutant protein, similar to RHARSC, was associated with viral NS2B3 and NS4B (Fig. 6C). These data suggested that the ATPase/helicase activity of RHA was dispensable for RHA to promote DENV2 replication.
FIG 6.
Expression of RHARSC or RHA-K417R rescued viral replication levels. (A and B) Control cells or RHARSC- or RHA-K417R (K417R)-expressing cells were transfected with siNC or siRHA1. At 48 h posttransfection, cells were infected by DENV2 at an MOI of 3. Cells and supernatants were collected at 24 h p.i. for Western blotting (A) and plaque assay (B). Western blot was probed with antibodies against RHA, FLAG, NS3, and GAPDH. (C) Co-IP assay. The RHARSC- or RHA-K417R-expressing cells were infected by DENV2 at an MOI of 3. At 24 h p.i., cells were harvested for co-IP assay using anti-FLAG antibody. Protein complexes were separated by SDS-PAGE and detected by Western blotting with antibodies against the indicated proteins. Data are representative of three independent experiments. ***, P < 0.001 by unpaired, two-tailed Student's t test.
The RHA proviral effect was independent of its role in suppressing IFNs.
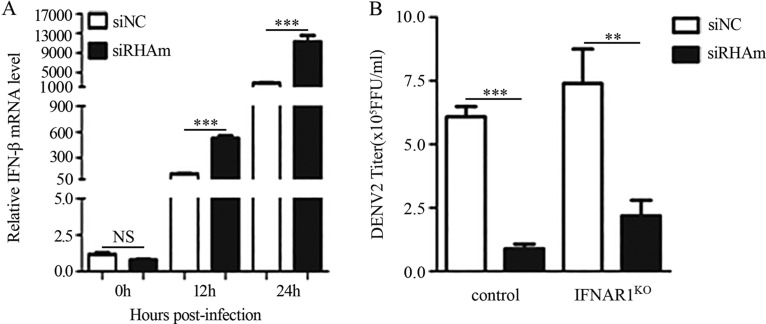
A previous study demonstrated that RHA was involved in the immune response by interacting with IPS-1 (30). Since DENV is sensitive to interferon (IFN) treatment, we hypothesized that RHA promoted viral replication via IFNs. To test this, we first measured the IFN production levels in the control and RHA-KD cells at 0, 12, and 24 h p.i. The real-time PCR data showed that the IFN-β levels in the RHA-KD cells were 4.66- and 5.86-fold higher than that of the control cells (P < 0.001) (Fig. 7A), indicating that RHA negatively regulated the IFN level induced by DENV infection. To assess whether the increased IFNs in RHA-KD cells led to the lower viral replication level in RHA-KD cells, we utilized IFN receptor knockout (IFNAR1KO) cells in which the IFN signaling pathway was blocked. The control cells and IFNAR1KO cells were transfected with siNC or siRHAm, followed by DENV2 infection. At 24 h p.i., the supernatants were collected for viral titration. The RHA knockdown led to comparable reduction of viral yields in control cells (4.66-fold) and IFNAR1KO cells (5.47-fold) (Fig. 7B), suggesting that the proviral effect of RHA was independent of its suppression effect on IFNs.
FIG 7.
RHA proviral function was independent of its regulation of IFN production. A549 cells were transfected with siNC or siRHAm. At 48 h posttransfection, cells were mock infected or infected with DENV2 (MOI of 3). Cells were harvested at 12 and 24 h p.i. for real-time PCR to detect the IFN-β level. (A) Actin was measured as a control. (B) Control and IFNAR1KO A549 cells were transfected with siNC or siRHAm for 48 h, followed by DENV2 infection (MOI of 3). Supernatants were collected at 24 h. The viral titers were measured by focus-forming assay. Data are shown as means ± SD from three independent experiments. NS, no statistical significance. **, P < 0.01; ***, P < 0.001 (both by unpaired, two-tailed Student's t test).
DISCUSSION
DENVs are cytoplasmic RNA viruses that must hijack host cellular proteins for their replication. Previously, several DExD/H RNA helicases have been reported to upregulate or downregulate DENV replication (31), including DDX3 (32), DDX6 (17), DDX3X (33), and DDX21 (34). In this study, we uncovered a new DEAD box member, RHA, as an additional helicase required for efficient replication of DENV.
First, our data obtained with the subcellular fractionation assay clearly showed that RHA levels in the cytoplasmic fractions were increased in response to DENV2 infection, indicating that the RHA protein was redistributed into the cytoplasm. This finding was inconsistent with data reported by Gomila et al. showing that RHA localized in the nucleus in DENV-infected HeLa cells (15). A possible explanation is that compared with the highly concentrated RHA in the nucleus, a small fraction of RHA translocated into the cytoplasm might not be readily detected by IFA.
To demonstrate that RHA was involved in the virus life cycle, we utilized RNAi to knock down the endogenous RHA protein in three types of cells, including lung carcinoma epithelial cells, MDM, and liver carcinoma cells. In all tested cells, RHA silencing resulted in significantly lower viral yields in the single-step growth assay. Furthermore, the multistep viral growth of DENV2 (NGC and 16681 strains) and JEV was significantly reduced in RHA-KD cells, while the ZIKV titers were barely affected in RHA-silenced cells at 24 and 48 h p.i. and were slightly decreased at 72 and 96 h p.i. In combination with the fact that RHA promoted the replication of two other flaviviruses, bovine viral diarrhea virus (35) and hepatitis C virus (36), we deduced that the proviral role of RHA is virus specific, which potentially contributes to different tropisms and pathogenesis of different flaviviruses. Of note, we attempted to establish RHA knockout cells by CRISPR/Cas9 to validate the observations gathered from the RNAi assay. Unfortunately, we were not able to obtain a homozygous RHA-knockout cell clone (unpublished data). Since the RHA knockout mouse embryos are lethal (37), we deduced that RHA is a vital protein such that the RHA knockout cells were unable to survive. In contrast, the residual RHA protein in the RHA-KD cells helps to maintain cell function and viability.
It was not surprising to find that RHA, as an RNA-binding protein, has a strong affinity to DENV2 RNA. This observation was consistent with data reported by Gomila et al., demonstrating that RHA was present in the protein complex that binds to the 3′-terminal stem-loop RNA of DENV2 through RNA affinity chromatography (15). RHA was also found to be one of the 79 DENV RNA-binding proteins by thiouracil cross-linking mass spectrometry (14), and it binds to both the 5ʹ- and 3ʹ-untranslated regions of the bovine viral diarrhea virus genomic RNA (35). All of these observations demonstrated that RHA binds to DENV RNA. Interestingly, RHA was also associated with ZIKV RNA but had a much lower affinity than DENV2, suggesting that RHA is involved in ZIKV replication, while there might be alternative factor(s) for ZIKV such that RHA was dispensable. These findings reinforce our observations that RHA is an essential factor for replication of DENV but not ZIKV.
Interestingly, RHA binds not only to DENV RNA but also to several nonstructural proteins. Moreover, the associations between RHA and viral NS1, NS3, and NS4B were mediated through different regions of RHA. As all of these NS proteins are located in the replication complex during RNA synthesis (9), the association of RHA with NS proteins presents another piece of evidence that RHA was involved in viral replication. Although RHA was associated with the viral protease NS2B3, the RHA protein in the DENV2-infected samples remained intact and the amount of RHA was not altered, suggesting that RHA was not a target of the NS3 protease. In addition, the RHA K417R mutant lacking the ATPase/helicase activity also binds to these proteins, and the expression of RHA K417R in the RHA-KD cells largely rescued the viral protein levels and viral yields. Since NS1, NS3, and NS4B are key elements in the viral replication cycle (1, 38), all of which closely interact with each other to form a complex (7, 39, 40), it was rational to deduce that RHA, a big molecule with RNA and protein binding activities, functions as a scaffold, independent of its helicase activity, thereby connecting the viral RNA and proteins for efficient viral replication.
Furthermore, our work illustrated the step of viral replication where RHA acts. First, we ruled out its involvement in viral entry. We then observed that the RHA knockdown led to significant decreases of both viral protein and RNA levels at 12 and 24 h p.i. In the DENV2 reporter replicon assay, the RHA knockdown did not alter the protein translation of both wild-type and NS5 mutant replicons at 6 h posttransfection, implying that RHA does not affect the protein translation of DENV2. Unfortunately, we were not able to measure the luciferase activities at 24 to 96 h posttransfection due to excessive cell death that occurred in A549 cells upon viral RNA transfection. Based on these observations, we conclude that RHA does not directly promote viral protein synthesis and might be involved in viral RNA synthesis while indirectly affecting viral protein translation.
Finally, we examined the impact of RHA on the innate immune response, since RHA has been shown to regulate the innate immune activation pathway induced by dsRNA and virus infection (30). Unexpectedly, our data showed that RHA negatively regulated IFN induction in response to DENV2 infection. The reasons behind these conflicting observations are currently unknown but may arise from differences in the cells and viruses used in these studies. Of note, the RHA knockdown in the IFNAR1 knockout cells led to a significant reduction of viral yields, similar to that of the wild-type cells, suggesting that the IFNs upregulated by RHA knockdown are not the major mediators that lead to fewer viral particles.
In conclusion, we have demonstrated that in response to DENV infection, nuclear RHA was partially translocated into the cytoplasm. The knockdown of RHA led to lower viral replication levels, including those of viral RNA, protein, and infectious particles. The data obtained from the reporter replicon assay supports a role of RHA in RNA synthesis while indirectly affecting protein translation. Furthermore, the role of RHA in viral RNA replication was further supported by observations that RHA associates with the viral RNA and three NS proteins that were localized in the replication complex. Therefore, our work identified a new host factor for DENV replication and illustrated its function. Further investigations to analyze how RHA promotes viral RNA synthesis will be of value in the understanding of viral replication and in the design of anti-DENV drugs.
MATERIALS AND METHODS
Cell culture.
Human lung carcinoma epithelial cells (A549; ATCC CCL-185), human liver carcinoma cells (HepG2; ATCC HB-8065), human embryonic kidney cells (293T; ATCC CRL-3216), baby hamster kidney cells (BHK21; ATCC CCL-10), and African green monkey kidney cells (Vero; ATCC CCL-81) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% or 10% fetal bovine serum (FBS) (Gibco) at 37°C with 5% CO2. Aedes albopictus cells (C6/36; ATCC CRL-1660) were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS at 28°C. The media were added with 100 U/ml of streptomycin and penicillin (Invitrogen). Peripheral blood mononuclear cells (PBMCs) of healthy donors were isolated by Ficoll (TBD Sciences) density gradient centrifugation. The monocytes were purified from peripheral blood mononuclear cells by anti-human CD14 magnetic particles (BD Biosciences) according to the manufacturer’s protocol. CD14+ monocytes were differentiated into macrophages by incubation in RPMI 1640 medium (Invitrogen) with 10% FBS (Gibco) and 50 ng/ml human GM-CSF (granulocyte-macrophage colony-stimulating factor) (Proteintech), and media were replaced every 3 to 4 days. After incubation for 7 to 10 days, MDM were obtained and used for subsequent experiments as described previously (41).
Antibodies.
Primary antibodies included anti-NDH II (RHA), anti-E protein, anti-Lamin B (Santa Cruz Inc.), anti-prM protein (Abcam), anti-NS2B, anti-NS3, anti-NS4A, anti-NS4B, anti-NS5, anti-HA, anti-β-actin (Sigma), anti-Calnexin, anti-FLAG (CST), and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Proteintech). Anti-NS1 was a gift from Trai-Ming Yeh and Yung-Chun Chuang at National Cheng Kung University (42). Secondary antibodies included IRDye 800 CW-conjugated anti-rabbit IgG, IRDye 680 CW-conjugated anti-mouse IgG (LI-COR), horseradish peroxidase (HRP)-conjugated anti-mouse IgG (CST), and anti-rabbit IgG (Bio-Rad).
Viruses, virus infection, and virus titration.
DENV2 New Guinea C (NGC) was provided by Guangzhou Centers for Disease Control. The DENV2 16681 strains, JEV (14-14-2 vaccine strain), and ZIKV (H/PF/2013 strain) were provided by Michael Diamond at Washington University School of Medicine. Viruses were propagated in C6/36 or Vero cells. The supernatants were collected when cytopathic effect appeared, and the cellular debris was removed by centrifugation. Titers of virus stocks were determined, and stocks were stored at −80°C.
A549, MDM, and HepG2 cells were seeded into 12-well plates. Cells were infected with DENV2 at an MOI of 3. The cells were harvested at the indicated time points for Western blotting or real-time PCR detection. In the single-step virus growth assay, the supernatants were harvested at 24 h p.i. for virus titration. In the multistep virus growth assay, cells were infected with DENV2 at an MOI of 0.01. The supernatants were harvested at 24, 48, 72, and 96 h p.i.
Virus titers of DENV2, JEV, and ZIKV were determined by standard plaque or focus-forming assay (FFA) on BHK21 or Vero cells. In the plaque assay, serial 10-fold dilutions of each sample were prepared, and 100 μl/well of diluted virus was added into the 12-well plates. The incubation media were removed and cultured in a mixture of 2× DMEM (Invitrogen) and 2% agarose (1:1) (Lonza). Visible plaques were counted at 5 to 6 days (DENV2) and 4 to 5 days (JEV) p.i. In the FFA assay, DENV2 (16681) or ZIKV samples were diluted in DMEM supplemented with 2% FBS. Vero cells were inoculated with serial 10-fold dilutions of each virus sample. Methylcellulose overlay containing 1% methylcellulose (Sigma) and 2× DMEM (Invitrogen) was then added and incubated at 37°C. Cells were washed with phosphate-buffered saline (PBS) to remove methylcellulose and fixed with 1% paraformaldehyde (PFA) in PBS at 3 days p.i. Cells were incubated with anti-WNV E18 monoclonal antibody (MAb) or E16 MAb (43), followed by incubation with goat anti-mouse HRP-conjugated secondary antibody (CST). Cells were incubated with TrueBlue peroxidase substrate (KPL). The number of spots was determined by a CTL-BioSpot analyzer.
Subcellular fractionation.
The cells were washed gently with PBS buffer and resuspended in cytoplasmic extract buffer (10 mM HEPES, 60 mM KCl, 1 mM EDTA, 0.075% [vol/vol] NP-40, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). The cytoplasmic extracts were acquired by spinning. The residues were washed with cytoplasmic extract buffer and lysed in nuclear extract buffer (pH 8.0) (20 mM HEPES, 420 mM NaCl, 10 mM KCl, 1 mM EDTA, 1 mM PMSF, and 20% [vol/vol] glycerol). The mixture was spun at 14,000 rpm for 5 min to separate the nuclear extract.
Western blotting.
Cells were grown in 6-well plates and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (pH 7.4) (50 mM Tris-HCl, 0.5% [vol/vol] NP-40, 1% Triton-100, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1% protease inhibitor cocktails, 1 mM Na3VO4, and 1 mM NaF). Proteins were separated on SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked in PBST with 5% bovine serum albumin (BSA) (New England Biolabs) and incubated with the indicated primary antibodies at 4°C overnight. Detection was performed with IRDye 800 CW-conjugated anti-rabbit IgG and IRDye 680 CW-conjugated anti-mouse IgG secondary antibody (LI-COR) according to the manufacturer’s protocols or with horseradish peroxidase-conjugated secondary antibodies (Bio-Rad). Immunoreactive bands were visualized using an Odyssey infrared imaging system (Bio-Rad) as described previously (44).
RNA interference.
The sequences of the two siRNAs targeting human RHA RNA were CAAAUCAUCUGUUAAUUGU and GCUUGUAGAUGACUGGAUU (Invitrogen). A control siRNA with scrambled sequence was used as a negative control (siNC). Transfection was carried out with 16 nmol siRNAs using Lipofectamine 2000 reagent (Invitrogen) by following the manufacturer’s instructions. At 48 h posttransfection, cells were harvested or used for further analysis.
qRT-PCR.
Total cellular RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Total RNA was reverse transcribed using M-MLV reverse transcriptase (Promega). The qRT-PCR analyses were performed with SYBR select master mix for CFX (Applied Biosystems) in an Bio-Rad CFX96 machine (see Table 2 for a list of qRT-PCR primer sequences). Data analyses for differences in gene expression by qRT-PCR were done by using ΔCT (change in threshold cycle) values as described previously (41).
TABLE 2.
Sequences of primers used in qRT-PCR
| Gene name | Sequence (5′–3′) |
|---|---|
| RHA | 5F, TCCTTCTGTGCCAGTTGCACTC |
| 3R, TTGTGGAGGTGACCAAGGAACC | |
| DENV2 C gene | 5F, TCCTAACAATCCCACCAACAGCA |
| 3R, AGTTCTGCGTCTCCTGTTCAAGA | |
| DENV2 5′ UTR | 5F, AGTAGTTAGTCTACGTGGAC |
| 3R, GTCGACACGCGGTTTCTCTC | |
| ZIKV | 5F, GTCAGAGCAGCAAAGACAA |
| 3R, CAGCCTCCTTTCCCTTAACA | |
| β-Actin | 5F, GCTCCTCCTGAGCGCAAG |
| 3R, CATCTGCTGGAAGGTGGACA | |
| IFN-β | 5F, AAACTCATGAGCAGTCTGCA |
| 3R, AGGAGATCTTCAGTTTCGGAGG |
IFA.
A549 cells (8 × 104) were seeded onto coverslips in 24-well plates and infected with DENV2 NGC at an MOI of 3. Cells were washed in PBS and fixed in 4% (vol/vol) paraformaldehyde at 24 h p.i. Cells were permeabilized in 0.02% Triton X-100 for 15 min and incubated with blocking buffer (5% BSA in PBS) for 1 h, followed by incubation with primary antibodies at 4°C overnight. Cells were incubated for 1 h with Cy3-conjugated goat anti-rabbit-IgG (Millipore) and Alexa Fluor 488-conjugated anti-mouse-IgG (R&D). Cells were then stained with 4’,6-diamidino-2-phenylindole (DAPI) (Invitrogen, CA) and visualized using a Zeiss fluorescence microscope as described previously (45).
Plasmid construction and transfection.
To amplify full-length or truncated fragments of the RHA gene (GenBank accession number NM_001357.4), cDNAs prepared from A549 cells were used as a template. The PCR primers included a FLAG-encoding sequence. pSG5-RHARSC-FLAG expression plasmids were mutated at synonymous sites in siRHA1 (CAAATCATCTGTTAATTGT) target sequence, present in the human RHA open reading frame, in order to resist knockdown by siRNA. The amplified fragments were purified and inserted into pSG5 for eukaryotic expression. The Lys at aa 417 of RHA was mutated to Arg by site-directed mutagenesis using pSG5-RHARSC-FLAG as the template and verified by sequencing. RHARSC or RHA-K417R fragment then was amplified by PCR and inserted into the lentiviral vector CSII-EF-MCS-IRES2-Venus.
To obtain IFNAR1 knockout cells, the IFNAR1 single guide RNA (sgRNA) was inserted into the lentiCRISPR v2 vector (52961; Addgene). All primer and sgRNA sequences are listed in Table 1. All plasmids were validated by sequencing and transfected into cells using Lipofectamine 2000 reagent (Invitrogen) by following the manufacturer’s instructions.
TABLE 1.
Sequences of primers used in PCR
| Gene name | Sequence (5′–3′) |
|---|---|
| RHA-FLAG | 5F, ATGGGTGACGTTAAAAATTTTCTGTATGC |
| 3R, CGCGGATCCTTACTTATCGTCGTCATCCTTGTAATCACCACCACCATAGCCGCCACCTCCTCTTCCCTG | |
| RHAN-FLAG | 5F, CCGGAATTCTATGGGTGACGTTAAAAATTTTCTGTATGC |
| 3R, CGCGGATCCCTACTTATCGTCGTCATCCTTGTAATCACCACCACCTGTCTCTCCTTCCTTCTTCTTTGTAAGTCC | |
| RHAM-FLAG | 5F, CCGGAATTCATGGTGGAGCCTTACAAAGTAAACC |
| 3R, CGCGGATCCCTACTTATCGTCGTCATCCTTGTAATCACCACCACCATCAAGCTCTCTAAGAGTGTGTT | |
| RHAC-FLAG | 5F, CCGGAATTCATGGCATTAGATGCCAATGATGAGTTGACTCC |
| 3R, CGCGGATCCTTACTTATCGTCGTCATCCTTGTAATCACCACCACCATAGCCGCCACCTCCTCTTCCCTG | |
| RHA(RSC) mutation | 5F, TGCACTTATCCATAAGTCGTCTGTCAATTGCCCTTTTAGT |
| 3R, ACTAAAAGGGCAATTGACAGACGACTTATGGATAAGTGCA | |
| Lentivirus-RHA | 5F, ATTTGCGGCCGCATGGGTGACGTTAAAAATTTTCTGT |
| (RSC)-FLAG | 3R, CGCGGATCCTTACTTATCGTCGTCATCCTTGTAATCACCACCACCATAGCCGCCACCTCCTCTTCCCTG |
| RHA(K417R) mutation | 5F, CTGGATGTGGGAGAACCACACAGG |
| 3R, CTGGATGTGGGAAAACCACACAGG | |
| Lentivirus-RHA | 5F, ATTTGCGGCCGCATGGGTGACGTTAAAAATTTTCTGT |
| (K417R)-FLAG | 3R, CGCGGATCCTTACTTATCGTCGTCATCCTTGTAATCACCACCACCATAGCCGCCACCTCCTCTTCCCTG |
| RHA siRNA resistance mutation | 5F, CTGATGGGCAGATTGTATTAGTCGACGATTGGATCAAACTGCAAATATCTCAT |
| 3R, ATGAGATATTTGCAGTTTGATCCAATCGTCGACTAATACAATCTGCCCATCAG | |
| NS1-HA | 5F, CGCGGATCCATGGATAGTGGTTGCGTTGT |
| 3R, CCGCTCGAGCTACTTATCGTCGTCATCCTTGTAATCACCACCACCAGCTGTGACCAAGGAGTT | |
| NS2A-HA | 5F, CGCGGATCCATGGGACATGGGCAGATTGACAACTT |
| 3R, CCCAAGCTTCTAAGCGTAATCTGGAACATCGTATGGGTAACCACCACCCCTTTTCTTGTTGGTTCTTGAAAG | |
| NS2B3-HA | 5F, CGCGGATCCATGAATAACCAACGAAAAAAGGCG |
| 3R, CCCAAGCTTCTAAGCGTAATCTGGAACATCGTATGGGTAACCACCACCCGCCATCACTGTTGGAATCAG | |
| NS4A-HA | 5F, CCCGGATCCATGTCCCTGACCCTGAACCTAATCACAGAA |
| 3R, AAGCTTTAGAGCGTAATCTGGAACATCGTATGGGTAACCACCACCTCTTTTCTGAGCTTCTCTGGT | |
| NS4B-HA | 5F, CGCGGATCCATGAACGAGATGGGTTTCCTGGAA |
| 3R, CCCAAGCTTCTAAGCGTAATCTGGAACATCGTATGGGTAACACCACCCCTTCTCGTGTTGGTTGTGTTCTT | |
| NS5-HA | 5F, CCCAAGCTTATGGGAACTGGCAACATAGGAG |
| 3R, GCTCTAGACTAAGCGTAATCTGGAACATCGTATGGGTAACCACCACCCCACAAAACTCCTGCCTCT | |
| IFNAR1 sgRNA | 5F, CACCGGATCTAATGTTAAAGACTGG |
| 3R, AAACCCAGTCTTTAACATTAGATCC |
Generation of RHARSC- and RHA-K417R-expressing cells and rescue assay.
293T cells were transfected with CSII-EF-MCS-IRES2-Venus–RHARSC or CSII-EF-MCS-IRES2-Venus–RHARSC-K417R, psPAX2, and pMD2.G using FuGENE HD reagent (Promega). After 2 days, culture supernatants were passed through a 0.45-μm filter and used for gene transduction. The RHARSC or RHA-K417R gene was transduced into A549 cells, and the Venus-positive cells were sorted by flow cytometry (CytoFLEX). The purity was estimated to be more than 80%.
The RHARSC- and RHA-K417R-transduced A549 cells were transfected with siNC or siRHA1. Cells were infected with DENV2 at an MOI of 3. Cells and the supernatant were harvested at 24 h p.i. for Western blotting or titration.
Replicon assay.
pDENrep-FH (16681) replicon WT and NS5 mutant (GDD→GVD) plasmids were provided by Eng Eong Ooi at Duke-NUS Medical School (28). The DENV2 reporter replicon plasmid was linearized, phenol-chloroform extracted, and precipitated with sodium acetate. The linearized DNA template was transcribed using an mMESSAGE mMACHINE T7 kit (Ambion) according to the manufacturer's protocol. The product of transcription was purified by LiCl precipitation. The reporter replicon RNAs (0.25 μg) were transfected into each well of A549 cells in a 24-well plate using Lipofectamine 2000 reagent (Invitrogen). The cells were harvested at 6 h and lysed for luminometry using Promega Luciferase assay reagent (Promega). Luciferase signals were measured using a GloMax 96 microplate luminometer (Promega).
Co-IP assay.
A549 cells were cotransfected with plasmids expressing NS1-HA, NS2B3-HA, and NS4B-HA, together with RHA-FLAG, RHAN-FLAG, RHAM-FLAG, or RHAC-FLAG fusion proteins by Lipofectamine 2000 reagent (Invitrogen). At 24 h posttransfection, cells were harvested for co-IP assay. Cells were resuspended in 1 ml ice-cold RIPA lysis buffer containing protease inhibitors (Sigma) and phosphatase inhibitor (NaF and Na3VO4). Cells were lysed at 4°C, and lysates were spun for 30 min at 4°C. Twenty μl of anti-HA/anti-FLAG beads (Sigma) was added to the cell lysate and incubated overnight. Beads were washed six times, and precipitated proteins were eluted by boiling in loading buffer.
To perform co-IP assay detecting the interaction between endogenous RHA and viral NS proteins, A549 cells were mock infected or infected with DENV2 at an MOI of 3. At 24 h or the indicated hours postinfection, whole-cell extracts were collected in RIPA lysis buffer and used for co-IP assay using protein A/G agarose (Calbiochem) as described previously (44).
RIP assay.
The interaction of RHA with DENV RNA was detected by RIP assay. The mock- or DENV2- infected cells were lysed in RIP lysis buffer (100 mM KCl, 5 mM MgCl2, 0.5% [vol/vol] NP-40, 10 mM HEPES, 1 mM PMSF, 1% protease inhibitor cocktails, and RNase inhibitor), and the lysates were pretreated with protein A/G agarose (Calbiochem). Immunoprecipitation was carried out using anti-RHA or control rabbit IgG. Immune complexes were precipitated with protein A/G agarose as described above. Viral RNA immunoprecipitation was performed using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Viral RNA levels were analyzed by qRT-PCR (primer sequences are listed in Table 2).
Construction of IFNAR1 knockout cells by the CRISPR/Cas9 system.
293T cells were transfected with LentiCRISPR v2-IFNAR1 sgRNA, psPAX2, and pMD2.G using FuGENE HD transfection reagent (Promega). Supernatants were collected at 2 days posttransfection and passed through a 0.45-μm filter. The LentiCRISPR v2-IFNAR1 sgRNAs were transduced into A549 cells, and IFNAR1-knockout cells were sorted and confirmed by Sanger DNA sequencing. Genomic DNA was extracted using a cell genomic DNA extraction kit (Bioteke). Regions surrounding sgRNA target sequences were amplified by PCR. PCR products were cloned into pMD-18T (TaKaRa) for sequencing.
Statistical analysis.
All statistical analyses of viral RNA levels or viral titers were performed with an unpaired, two-tailed Student's t test. Data were presented as means ± standard deviations (SD) from at least three independent experiments. The differences were considered statistically significant at a P value of <0.05, as described previously (44).
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81371794), the Guangdong Science and Technology Program (2017A050501012 and 2016A020219003), the Natural Science Foundation of Guangdong Province (2017A030313504 and 2014A030311007), and Fundamental Research Funds for the Central Universities (17ykjc01).
We thank Michael S. Diamond at Washington University School of Medicine for his helpful comments and for providing the viruses (JEV, DENV2 16681, and ZIKV) and WNV antibody. We thank Marc R. Montminy at Salk Institute, Ana Fernandez-Sesma at Mount Sinai School of Medicine, and Eng Eong Ooi at Duke-NUS Medical School for generously providing pBK-CMV-RHA, NS2B3-expressing plasmids, and the luciferase DENV2 (16681) replicon. We thank Trai-Ming Yeh and Yung-Chun Chuang at National Cheng Kung University for generously providing the anti-NS1 monoclonal antibody.
REFERENCES
- 1.Simmons CP, Farrar JJ, Nguyen V, Wills B. 2012. Dengue. N Engl J Med 366:1423–1432. doi: 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- 2.Messina JP, Brady OJ, Scott TW, Zou C, Pigott DM, Duda KA, Bhatt S, Katzelnick L, Howes RE, Battle KE, Simmons CP, Hay SI. 2014. Global spread of dengue virus types: mapping the 70 year history. Trends Microbiol 22:138–146. doi: 10.1016/j.tim.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanaway JD, Shepard DS, Undurraga EA, Halasa YA, Coffeng LE, Brady OJ, Hay SI, Bedi N, Bensenor IM, Castaneda-Orjuela CA, Chuang TW, Gibney KB, Memish ZA, Rafay A, Ukwaja KN, Yonemoto N, Murray CJL. 2016. The global burden of dengue: an analysis from the Global Burden of Disease Study 2013. Lancet Infect Dis 16:712–723. doi: 10.1016/S1473-3099(16)00026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fredericks AC, Fernandez-Sesma A. 2014. The burden of dengue and chikungunya worldwide: implications for the southern United States and California. Ann Glob Health 80:466–475. doi: 10.1016/j.aogh.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diamond MS, Pierson TC. 2015. Molecular insight into dengue virus pathogenesis and its implications for disease control. Cell 162:488–492. doi: 10.1016/j.cell.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Blanco MA, Vasudevan SG, Bradrick SS, Nicchitta C. 2016. Flavivirus RNA transactions from viral entry to genome replication. Antiviral Res 134:244–249. doi: 10.1016/j.antiviral.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Brand C, Bisaillon M, Geiss BJ. 2017. Organization of the flavivirus RNA replicase complex. Wiley Interdiscip Rev RNA 8:wrna.1437. doi: 10.1002/wrna.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acosta EG, Kumar A, Bartenschlager R. 2014. Revisiting dengue virus-host cell interaction: new insights into molecular and cellular virology. Adv Virus Res 88:1–109. doi: 10.1016/B978-0-12-800098-4.00001-5. [DOI] [PubMed] [Google Scholar]
- 9.Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. 2018. Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol 16:125–142. doi: 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campos RK, Wong B, Xie X, Lu YF, Shi PY, Pompon J, Garcia-Blanco MA, Bradrick SS. 2017. RPLP1 and RPLP2 are essential flavivirus host factors that promote early viral protein accumulation. J Virol 91:e01706-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin DL, Cherepanova NA, Bozzacco L, MacDonald MR, Gilmore R, Tai AW. 2017. Dengue virus hijacks a noncanonical oxidoreductase function of a cellular oligosaccharyltransferase complex. mBio 8:e00939-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, Carette JE. 2016. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R, Miner JJ, Gorman MJ, Rausch K, Ramage H, White JP, Zuiani A, Zhang P, Fernandez E, Zhang Q, Dowd KA, Pierson TC, Cherry S, Diamond MS. 2016. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535:164–168. doi: 10.1038/nature18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viktorovskaya OV, Greco TM, Cristea IM, Thompson SR. 2016. Identification of RNA binding proteins associated with dengue virus RNA in infected cells reveals temporally distinct host factor requirements. PLoS Negl Trop Dis 10:e0004921. doi: 10.1371/journal.pntd.0004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomila RC, Martin GW, Gehrke L. 2011. NF90 binds the dengue virus RNA 3' terminus and is a positive regulator of dengue virus replication. PLoS One 6:e16687. doi: 10.1371/journal.pone.0016687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips SL, Soderblom EJ, Bradrick SS, Garcia-Blanco MA. 2016. Identification of proteins bound to dengue viral RNA in vivo reveals new host proteins important for virus replication. mBio 7:e01865-15. doi: 10.1128/mBio.01865-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA. 2011. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3' UTR structures. RNA Biol 8:1173–1186. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward AM, Calvert ME, Read LR, Kang S, Levitt BE, Dimopoulos G, Bradrick SS, Gunaratne J, Garcia-Blanco MA. 2016. The Golgi associated ERI3 is a Flavivirus host factor. Sci Rep 6:34379. doi: 10.1038/srep34379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee T, Pelletier J. 2016. The biology of DHX9 and its potential as a therapeutic target. Oncotarget 7:42716–42739. doi: 10.18632/oncotarget.8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing L, Niu M, Kleiman L. 2012. In vitro and in vivo analysis of the interaction between RNA helicase A and HIV-1 RNA. J Virol 86:13272–13280. doi: 10.1128/JVI.01993-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy BB, Hu J, Guo X, Russell RS, Guo F, Kleiman L, Liang C. 2006. Association of RNA helicase a with human immunodeficiency virus type 1 particles. J Biol Chem 281:12625–12635. doi: 10.1074/jbc.M510596200. [DOI] [PubMed] [Google Scholar]
- 22.Lin L, Li Y, Pyo HM, Lu X, Raman SN, Liu Q, Brown EG, Zhou Y. 2012. Identification of RNA helicase A as a cellular factor that interacts with influenza A virus NS1 protein and its role in the virus life cycle. J Virol 86:1942–1954. doi: 10.1128/JVI.06362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman MM, Liu J, Chan WM, Rothenburg S, McFadden G. 2013. Myxoma virus protein M029 is a dual function immunomodulator that inhibits PKR and also conscripts RHA/DHX9 to promote expanded host tropism and viral replication. PLoS Pathog 9:e1003465. doi: 10.1371/journal.ppat.1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L, Tian J, Nan H, Tian M, Li Y, Xu X, Huang B, Zhou E, Hiscox JA, Chen H. 2016. Porcine reproductive and respiratory syndrome virus nucleocapsid protein interacts with Nsp9 and cellular DHX9 to regulate viral RNA synthesis. J Virol 90:5384–5398. doi: 10.1128/JVI.03216-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujii R, Okamoto M, Aratani S, Oishi T, Ohshima T, Taira K, Baba M, Fukamizu A, Nakajima T. 2001. A role of RNA helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J Biol Chem 276:5445–5451. doi: 10.1074/jbc.M006892200. [DOI] [PubMed] [Google Scholar]
- 26.Bolinger C, Sharma A, Singh D, Yu L, Boris-Lawrie K. 2010. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res 38:1686–1696. doi: 10.1093/nar/gkp1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. 2006. RNA helicase A is necessary for translation of selected messenger RNAs. Nat Struct Mol Biol 13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 28.Holden KL, Stein DA, Pierson TC, Ahmed AA, Clyde K, Iversen PL, Harris E. 2006. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3' stem-loop structure. Virology 344:439–452. doi: 10.1016/j.virol.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, Montminy M. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90:1107–1112. doi: 10.1016/S0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z, Yuan B, Lu N, Facchinetti V, Liu YJ. 2011. DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J Immunol 187:4501–4508. doi: 10.4049/jimmunol.1101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranji A, Boris-Lawrie K. 2010. RNA helicases: emerging roles in viral replication and the host innate response. RNA Biol 7:775–787. doi: 10.4161/rna.7.6.14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brai A, Fazi R, Tintori C, Zamperini C, Bugli F, Sanguinetti M, Stigliano E, Este J, Badia R, Franco S, Martinez MA, Martinez JP, Meyerhans A, Saladini F, Zazzi M, Garbelli A, Maga G, Botta M. 2016. Human DDX3 protein is a valuable target to develop broad spectrum antiviral agents. Proc Natl Acad Sci U S A 113:5388–5393. doi: 10.1073/pnas.1522987113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li G, Feng T, Pan W, Shi X, Dai J. 2015. DEAD-box RNA helicase DDX3X inhibits DENV replication via regulating type one interferon pathway. Biochem Biophys Res Commun 456:327–332. doi: 10.1016/j.bbrc.2014.11.080. [DOI] [PubMed] [Google Scholar]
- 34.Dong Y, Ye W, Yang J, Han P, Wang Y, Ye C, Weng D, Zhang F, Xu Z, Lei Y. 2016. DDX21 translocates from nucleus to cytoplasm and stimulates the innate immune response due to dengue virus infection. Biochem Biophys Res Commun 473:648–653. doi: 10.1016/j.bbrc.2016.03.120. [DOI] [PubMed] [Google Scholar]
- 35.Isken O, Grassmann CW, Sarisky RT, Kann M, Zhang S, Grosse F, Kao PN, Behrens SE. 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J 22:5655–5665. doi: 10.1093/emboj/cdg562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He QS, Tang H, Zhang J, Truong K, Wong-Staal F, Zhou D. 2008. Comparisons of RNAi approaches for validation of human RNA helicase A as an essential factor in hepatitis C virus replication. J Virol Methods 154:216–219. doi: 10.1016/j.jviromet.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Lee CG, da Costa Soares V, Newberger C, Manova K, Lacy E, Hurwitz J. 1998. RNA helicase A is essential for normal gastrulation. Proc Natl Acad Sci U S A 95:13709–13713. doi: 10.1073/pnas.95.23.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lescar J, Soh S, Lee LT, Vasudevan SG, Kang C, Lim SP. 2018. The dengue virus replication complex: from RNA replication to protein-protein interactions to evasion of innate immunity. Adv Exp Med Biol 1062:115–129. doi: 10.1007/978-981-10-8727-1_9. [DOI] [PubMed] [Google Scholar]
- 39.Zou J, Lee Le T, Wang QY, Xie X, Lu S, Yau YH, Yuan Z, Geifman Shochat S, Kang C, Lescar J, Shi PY. 2015. Mapping the interactions between the NS4B and NS3 proteins of dengue virus. J Virol 89:3471–3483. doi: 10.1128/JVI.03454-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou J, Xie X, Wang QY, Dong H, Lee MY, Kang C, Yuan Z, Shi PY. 2015. Characterization of dengue virus NS4A and NS4B protein interaction. J Virol 89:3455–3470. doi: 10.1128/JVI.03453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Wu S, Pu J, Huang X, Zhang P. 2015. Dengue virus up-regulates expression of notch ligands Dll1 and Dll4 through interferon-beta signalling pathway. Immunology 144:127–138. doi: 10.1111/imm.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chuang YC, Lin J, Lin YS, Wang S, Yeh TM. 2016. Dengue virus nonstructural protein 1-induced antibodies cross-react with human plasminogen and enhance its activation. J Immunol 196:1218–1226. doi: 10.4049/jimmunol.1500057. [DOI] [PubMed] [Google Scholar]
- 43.Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. 2005. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature 437:764–769. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang P, Li Y, Xia J, He J, Pu J, Xie J, Wu S, Feng L, Huang X, Zhang P. 2014. IPS-1 plays an essential role in dsRNA-induced stress granule formation by interacting with PKR and promoting its activation. J Cell Sci 127:2471–2482. doi: 10.1242/jcs.139626. [DOI] [PubMed] [Google Scholar]
- 45.Xia J, Chen X, Xu F, Wang Y, Shi Y, Li Y, He J, Zhang P. 2015. Dengue virus infection induces formation of G3BP1 granules in human lung epithelial cells. Arch Virol 160:2991–2999. doi: 10.1007/s00705-015-2578-9. [DOI] [PubMed] [Google Scholar]