Chikungunya virus (CHIKV) is an understudied threat to human health. During the 2015 chikungunya epidemic in Managua, Nicaragua, we estimated the ratio of symptomatic to asymptomatic CHIKV infections, which is important for understanding transmission dynamics and the public health impact of CHIKV. This index cluster study identified and monitored persons at risk of infection, enabling capture of asymptomatic infections. We estimated that 31 (49%) of 63 at-risk participants had asymptomatic CHIKV infections, which is significantly outside the 3% to 28% range reported in literature reviews. However, recent seroprevalence studies, including two large pediatric cohort studies in the same setting, had also found percentages of inapparent infections outside the 3% to 28% range. Bayesian and simulation analyses, informed by a systematic literature search, revealed that the percentage of inapparent infections in epidemic settings varies by CHIKV phylogenetic lineage. Our study quantifies and provides the first epidemiological evidence that chikungunya epidemic characteristics are strongly influenced by CHIKV lineage.
KEYWORDS: Bayesian analysis, chikungunya virus, S:A ratio, epidemics, index cluster study, lineage
ABSTRACT
In late 2013, chikungunya virus (CHIKV) was introduced into the Americas, leading to widespread epidemics. A large epidemic caused by the Asian chikungunya virus (CHIKV) lineage occurred in Managua, Nicaragua, in 2015. Literature reviews commonly state that the proportion of inapparent CHIKV infections ranges from 3 to 28%. This study estimates the ratio of symptomatic to asymptomatic CHIKV infections and identifies risk factors of infection. In October to November 2015, 60 symptomatic CHIKV-infected children were enrolled as index cases and prospectively monitored, alongside 236 household contacts, in an index cluster study. Samples were collected upon enrollment and on day 14 or 35 and tested by real-time reverse transcription-PCR (rRT-PCR), IgM capture enzyme-linked immunosorbent assays (IgM-ELISAs), and inhibition ELISAs to detect pre- and postenrollment CHIKV infections. Of 236 household contacts, 55 (23%) had experienced previous or very recent infections, 41 (17%) had active infections at enrollment, and 21 (9%) experienced incident infections. Vehicle ownership (multivariable-adjusted risk ratio [aRR], 1.58) increased the risk of CHIKV infection, whereas ≥4 municipal trash collections/week (aRR, 0.38) and having externally piped water (aRR, 0.52) protected against CHIKV infection. Among 63 active and incident infections, 31 (49% [95% confidence interval {CI}, 36%, 62%]) were asymptomatic, yielding a ratio of symptomatic to asymptomatic infections of 1:0.97 (95% CI, 1:0.56, 1:1.60). Although our estimate is outside the 3% to 28% range reported previously, Bayesian and simulation analyses, informed by a systematic literature search, suggested that the proportion of inapparent CHIKV infections is lineage dependent and that more inapparent infections are associated with the Asian lineage than the East/Central/South African (ECSA) lineage. Overall, these data substantially improve knowledge regarding chikungunya epidemics.
IMPORTANCE Chikungunya virus (CHIKV) is an understudied threat to human health. During the 2015 chikungunya epidemic in Managua, Nicaragua, we estimated the ratio of symptomatic to asymptomatic CHIKV infections, which is important for understanding transmission dynamics and the public health impact of CHIKV. This index cluster study identified and monitored persons at risk of infection, enabling capture of asymptomatic infections. We estimated that 31 (49%) of 63 at-risk participants had asymptomatic CHIKV infections, which is significantly outside the 3% to 28% range reported in literature reviews. However, recent seroprevalence studies, including two large pediatric cohort studies in the same setting, had also found percentages of inapparent infections outside the 3% to 28% range. Bayesian and simulation analyses, informed by a systematic literature search, revealed that the percentage of inapparent infections in epidemic settings varies by CHIKV phylogenetic lineage. Our study quantifies and provides the first epidemiological evidence that chikungunya epidemic characteristics are strongly influenced by CHIKV lineage.
INTRODUCTION
Chikungunya is an understudied alphaviral disease caused by chikungunya virus (CHIKV), spread to humans primarily by infected Aedes aegypti and Aedes albopictus mosquitoes (1). Acute-phase chikungunya-associated morbidity is substantial, with crippling arthralgia, high fever (typically 102°F to 104°F [39°C to 40°C]), a macropapular skin rash, and severe fatigue (2). Most cases recover within several weeks; however, joint and musculoskeletal pain may last for weeks to years postinfection.
Phylogenetic analyses reveal three distinct CHIKV lineages: the West African, Asian, and East/Central/South African (ECSA) lineages. The ECSA lineage includes the Indian Ocean lineage (IOL) subgroup, now recognized as a strain of ECSA. Recent evidence suggests that the lineages may differentially activate inflammatory responses in mouse models (3, 4) and vary in virulence and cross-protective ability in mice and nonhuman primates (5). Mutations conferring a differential capacity for viral fitness and dissemination in Aedes albopictus (e.g., the IOL strain of the ECSA lineage) are also lineage specific (6).
Historically endemic to parts of West Africa and Asia (7), CHIKV now circulates in over 60 countries (8). In the last 10 to 15 years, the virus has spread into the Pacific islands and, most recently, the Americas, where >2 million suspected cases of autochthonously transmitted chikungunya occurred from 2014 to 2016 (9). The Asian CHIKV lineage predominated in epidemics throughout the Americas and was responsible for the two chikungunya epidemics in Managua, Nicaragua, during 2014 and 2015 (10–14).
Within the chikungunya field, there is some uncertainty regarding the balance of symptomatic and inapparent/asymptomatic infections. The proportion of inapparent infections (i.e., infected persons who do not seek medical care) in epidemic settings is regularly reported in literature reviews to range from 3 to 28% (15–19), based on early seroprevalence surveys in human populations. Nevertheless, recent studies, including four large cohort studies, have characterized chikungunya epidemics with proportions of inapparent infections higher than the reported upper range of 28% (10, 14, 20–27). In particular, a large (n = 4,210) seroprevalence study conducted in Managua during its first chikungunya epidemic showed that 59% of CHIKV infections in children and 65% of CHIKV infections in adults were inapparent (10). Some authors (10, 24, 26) have hypothesized lineage-specific differences in the proportions of inapparent CHIKV infections. At present, no studies have examined the literature systematically to evaluate this preexisting hypothesis and quantify possible lineage-dependent differences.
Assessing the percentage of truly asymptomatic infections during any epidemic has important public health implications, as the balance between symptomatic and asymptomatic infections impacts detection, transmission dynamics, and clinical outcomes (28). However, asymptomatic infections are difficult to identify in epidemic settings. For ethical and logistical reasons, traditional cohort studies infrequently obtain viremic blood samples from participants who do not report signs and symptoms consistent with an infection. When serological evidence of a prior infection is discovered in such participants (generally months after their infection and the end of the epidemic), they are often retroactively classified as having had an inapparent infection, since their history of disease cannot be ascertained with complete certainty (i.e., not reporting signs/symptoms is different from not experiencing signs/symptoms). The outcome of infection in these studies is then characterized by a proxy measure, the ratio of symptomatic to inapparent infections (S:I ratio). Chikungunya typically manifests with severe, acute signs and symptoms that are likely to be reported to health professionals; hence, the proportion of inapparent CHIKV infections serves as an approximate measure for the proportion of asymptomatic CHIKV infections.
We undertook this study primarily to quantify the percentage of truly asymptomatic CHIKV infections during Managua’s second chikungunya epidemic. To capture asymptomatic infections while minimizing bias due to incomplete sign/symptom recall, we used a household index cluster design (29). In this study design, laboratory-confirmed (index) cases are used to identify household members at risk of infection, as they live with an infected individual. Household contacts of the index cases are treated as the primary study sample, prospectively monitored, and repeatedly assessed for evidence of incident infection and the occurrence of signs and symptoms. Laboratory testing enables the identification of preenrollment and postenrollment infections, including asymptomatic infections that occur within the study period. In addition, collection of sign and symptom data from household contacts, ambidirectional determination of infection status, and detailed follow-up permit ascertainment of participants’ infection history with more granularity possible than with traditional serosurveys. Here, we estimate the ratio of symptomatic to asymptomatic CHIKV infections (S:A ratio), elucidate sign and symptom cooccurrence, characterize the probability of infection and disease occurrence across age, identify risk factors associated with infection and disease outcomes, and quantify differences in the proportions of inapparent CHIKV infections between the Asian CHIKV lineage and the ECSA CHIKV lineage.
RESULTS
Study population.
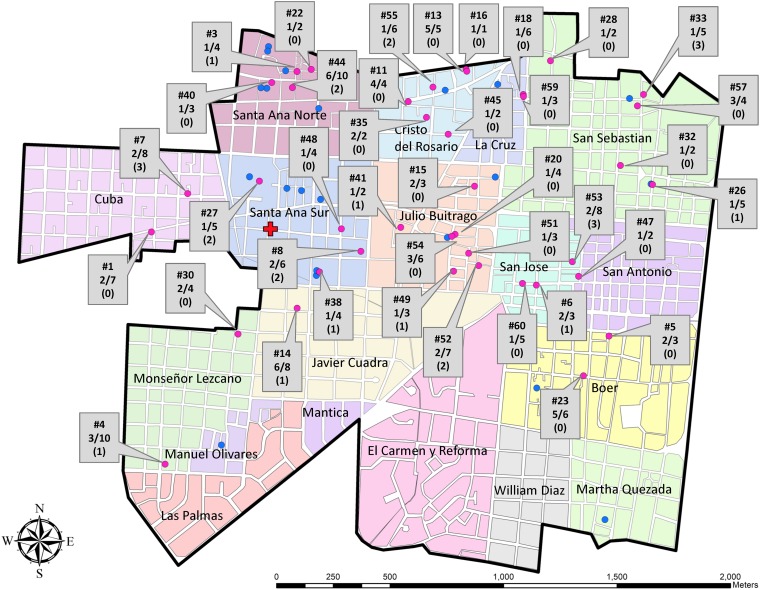
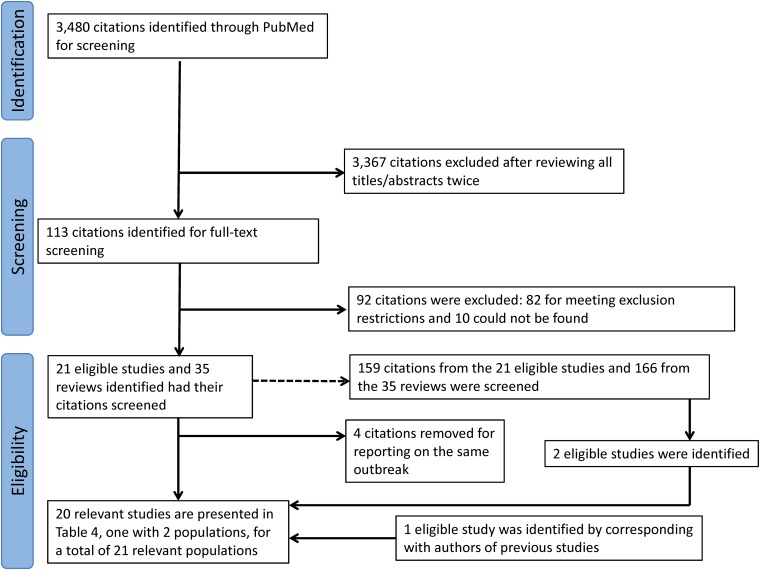
During October and November 2015, we enrolled 60 CHIKV real-time reverse transcription-PCR (rRT-PCR)-positive index cases who presented to Health Center Sócrates Flores Vivas (HCSFV) with signs and symptoms meeting the study case definition. During the initial visit to households of index cases, we enrolled 236 (70.9%) of 333 potential household contacts across the study area (Fig. 1).
FIG 1.
Geographic distribution in District II of Managua, Nicaragua, of laboratory-confirmed CHIKV infections per household in the study. The blocks corresponding to each neighborhood within the study site are colored and labeled. The study health center is indicated by a red cross. Participating homes that did not experience an infection among at-risk individuals are shown in blue, and homes with infections among at-risk individuals are shown in pink. The box label by each home with an infection indicates, in descending order, the house number (e.g., #7); the proportion of recent, active, and incident infections out of the total number of contacts (e.g., 2/8); and the number of previous infections in that household (e.g., 0). The households’ longitude and latitude values have been jittered to protect participants’ confidentiality.
Demographics and serology.
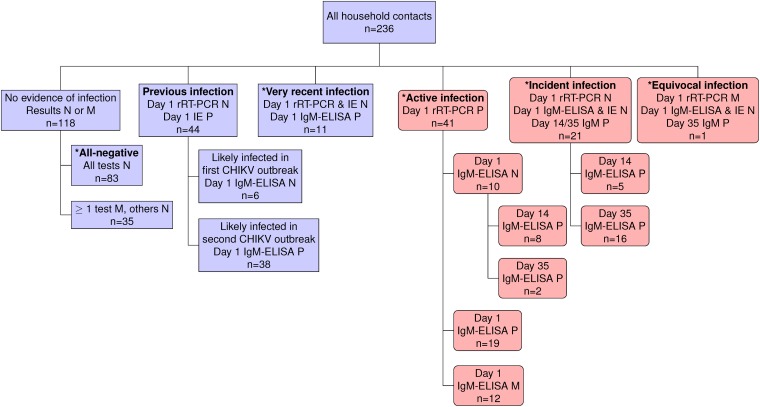
The ratio of females to males for both index cases and household contacts was ∼6:4 (Table 1). There was no evidence of CHIKV infection in 118 (50%) household contacts, 83 of whom had samples that were negative on every test performed (the all-negative reference group) (Fig. 2). The remaining 35 subjects had at least 1 missing sample but were negative on all of the samples tested; these contacts were excluded from further analysis. Forty-four (18.6%) of the 236 contacts’ baseline samples tested positive by an inhibition enzyme-linked immunosorbent assay (IE), indicative of a previous CHIKV infection. Eleven contacts (4.6%) were classified as having had a very recent infection at baseline. Day 1 samples from 41 contacts (17.4%) were positive by rRT-PCR, indicative of an active infection at baseline. Twenty-one contacts (8.9%) seroconverted as determined by an IgM capture enzyme-linked immunosorbent assay (IgM-ELISA) during the study period. Only one household contact was classified as experiencing an equivocal infection due to a missing rRT-PCR result. Contacts with a previous infection (mean age, 30 years; P value of <0.003) or an active infection at baseline (mean age, 33 years; P value of <0.004) had significantly different age distributions than all-negative contacts (mean age, 20 years). There were no other differences between participants’ age and sex. Our statistical findings regarding age and sex differences were unchanged upon using the Wilcoxon rank sum test and Fisher’s exact test, respectively.
TABLE 1.
Demographics and clinical manifestations of the index cases and household contacts
| Demographic or clinical manifestation | Value for group |
|||||||
|---|---|---|---|---|---|---|---|---|
| Index cases (n = 60) | All household contacts (n = 236) | Contacts with all-negative test results (n = 83) | Contacts with previous infections (n = 44) | Contacts with very recent infections (n = 11) | Contacts with active infection (n = 41) | Contacts with incident infections (n = 21) | Contacts with equivocal infections (n = 1) | |
| No. (%) of individuals of age (yr) | ||||||||
| ≤16 | 60 (100) | 82 (34.7) | 39 (47.0) | 8 (18.2) | 7 (63.6) | 8 (19.5) | 8 (38.1) | 1 (100.0) |
| >16 | 0 (0.0) | 154 (65.3) | 44 (53.0) | 36 (81.9) | 4 (36.4) | 33 (80.5) | 13 (61.9) | 0 (0.0) |
| Mean, median age (yr) | 8.6, 9.0 | 28.4, 26.0 | 23.5, 20.0 | 32.3, 15.0 | 14.5, 12.0 | 36.3, 33.0 | 29.6, 23.0 | 2.0, 2.0 |
| No. (%) of individuals of sex | ||||||||
| Female | 36 (60.0) | 150 (63.6) | 54 (65.1) | 33 (75.0) | 7 (63.6) | 25 (61.0) | 12 (57.1) | 1 (100.0) |
| Male | 24 (40.0) | 86 (36.4) | 29 (34.9) | 11 (25.0) | 4 (36.4) | 16 (39.0) | 9 (42.9) | 0 (0.0) |
| No. (%) of individuals with clinical manifestationa | ||||||||
| Classic chikungunya | 60 (100.0) | 10 (12.0) | 25 (61.0) | 4 (19.0) | 0 (0.0) | |||
| Undifferentiated fever | 0 (0.0) | 4 (4.8) | 1 (2.4) | 1 (4.8) | 1 (100.0) | |||
| Asymptomatic | 0 (0.0) | 69 (83.1) | 15 (36.6) | 16 (76.2) | 0 (0.0) | |||
Clinical manifestations are not presented for contacts with either a previous or a very recent infection as these contacts’ sign/symptom duration may not have overlapped the study period, hence leading to incomplete recall of signs/symptoms.
FIG 2.
Classification scheme based on CHIKV infection status of the 236 household contacts. Shown is a classification scheme of the household contacts with mutually exclusive categories. Day 1 refers to the baseline. The designation of particular categories was based only on the listed molecular or serological tests. For example, the classification of active infections was based solely on a positive rRT-PCR result on day 1, irrespective of day 1, 14, or 35 ELISA results. Based on viremia dynamics, individuals with very recent infections were likely infected within a week of the baseline visit. Those with equivocal infections met the definition of either an active or incident infection but were missing the day 1 rRT-PCR sample. Contacts in pink boxes (n = 63), unlike those in purple boxes (n = 173), were used to calculate the S:A ratio because their viremic period (and, hence, symptomatic period) overlapped the study period. Contact categories marked by an asterisk (n = 157) were at risk of a CHIKV infection after or slightly before study initiation. P, positive; N, negative; M, missing.
Disease presentation.
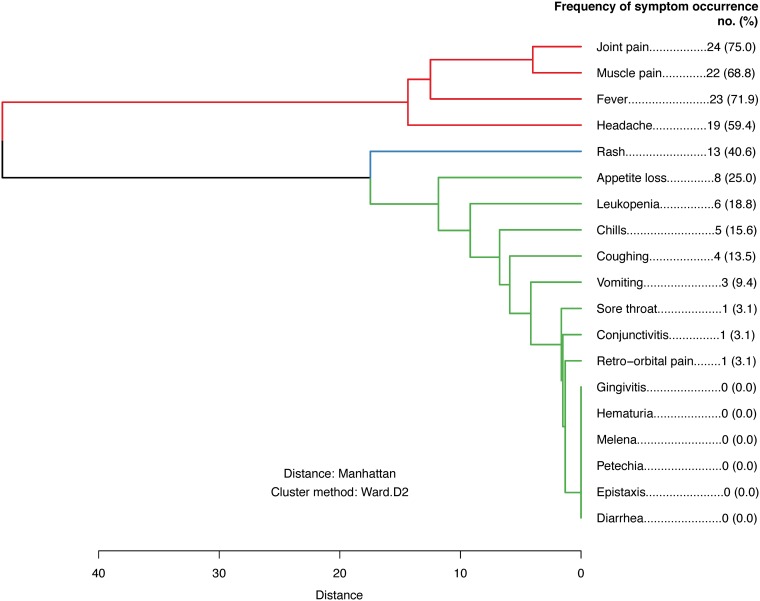
All index cases (60/60; 100%) presented with classic chikungunya (Table 1). Among household contacts, there were 63 active and incident CHIKV infections. Of these, 29 displayed classic chikungunya signs and symptoms, 3 experienced undifferentiated fever, and 31 were asymptomatic; this translates to an overall S:A ratio of 1:0.97 (95% confidence interval [CI], 1:0.56, 1:1.60), or a 49% (95% CI, 36%, 62%) probability of asymptomatic infection within our sample (Table 2). Among the 12 individuals with rRT-PCR-confirmed active infections with missing/incomplete serology data, 6 presented with classical chikungunya, while 6 were asymptomatic. The crude risk difference for CHIKV infection status (exposure) and symptomatic status (outcome) was 34% (95% CI, 19%, 48%), and its age- and sex-adjusted analogue was 31% (95% CI, 15%, 47%). Among the 32 symptomatic CHIKV infections, joint pain (75%), fever (72%), and muscle pain (69%) were the 3 most commonly reported signs/symptoms, followed by headache (60%). These 4 signs/symptoms cooccurred as one distinct symptom cluster (Fig. 3).
TABLE 2.
Point and interval estimates of the S:A ratios for the 63 active and incident infectionsa
| Parameter | Value for group |
||||||
|---|---|---|---|---|---|---|---|
| Overall | Children (≤16 yr old) | Adults (>16 yr old) | Males | Females | Individuals with active infection | Individuals with incident infection | |
| No. of households in the bootstrap procedure | 34 | 15 | 29 | 16 | 28 | 24 | 15 |
| No. of contacts with an active or incident infection | 63 | 17 | 46 | 25 | 38 | 41 | 21 |
| S:A ratiob | 32:31, 1:0.97 | 6:11, 1:1.83 | 26:20, 1:0.77 | 10:15, 1:1.50 | 22:16, 1:0.73 | 26:15, 1:0.53 | 5:16, 1:3.20 |
| 95% BCa CI for S:A ratio | 1:0.56, 1:1.60 | NV | 1:0.40, 1:1.38 | 1:0.69, 1:3.33 | 1:0.36, 1:1.29 | 1:0.26, 1:1.05 | NV |
| % of individuals with asymptomatic presentation given infection (95% BCa CI) | 49.2 (35.8, 61.5) | 64.7 (NV) | 43.5 (27.5, 58.0) | 60.0 (40.7, 76.9) | 42.1 (26.3, 56.4) | 36.6 (20.6, 51.3) | 76 (NV) |
Bias-corrected and accelerated bootstrap confidence intervals (BCa CI) were estimated after nonparametrically bootstrapping at the household level 10,000 times. NV, nonvalid BCa CI.
Number of symptomatic to asymptomatic CHIKV infections in the category, ratio re-expressed as a function of 1 symptomatic CHIKV infection.
FIG 3.
Sign and symptom cooccurrence dendrogram for the 32 symptomatic infections. The hierarchical clustering dendrogram conveys how signs and symptoms cooccurred simultaneously in the 32 individuals with symptomatic active and incident infections. Particular signs and symptoms that appear closer together on the dendrogram are more likely to be either jointly present or jointly absent in symptomatic individuals than signs and symptoms that are further apart on the dendrogram. Use of the modern Ward agglomerative method identified three clusters: (i) joint pain, muscle pain, fever, and headache (in red); (ii) rash (in blue); and (iii) all other signs and symptoms (in green). The occurrence frequency for each individual sign and symptom is additionally listed on the right.
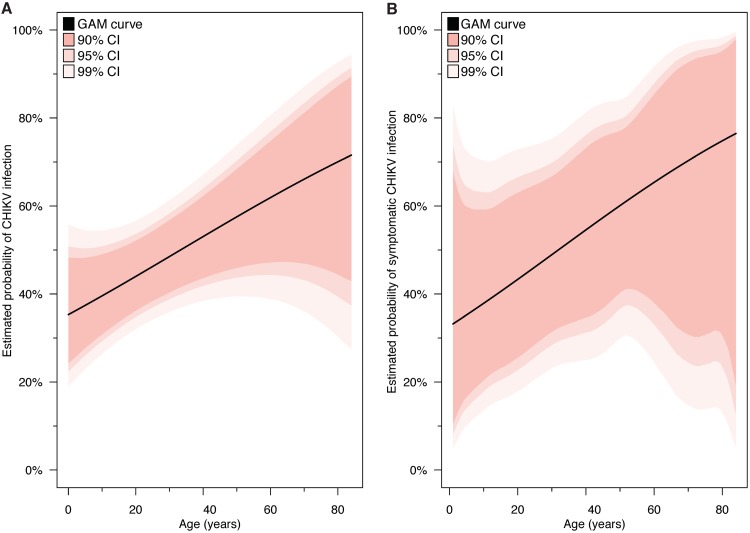
Age trend analysis.
Among those at risk of CHIKV infection after or slightly before study initiation (n = 157), the probability of CHIKV infection increased linearly with age (Fig. 4A). Among the 63 active and incident infections, the probability of having a symptomatic outcome also increased linearly with age (Fig. 4B).
FIG 4.
Age trend analysis of CHIKV infections and symptomatic CHIKV infections. Generalized additive model (GAM) curves illustrate the marginal trend between age and the probability of infection (A) and age and the probability of a symptomatic outcome, given infection (B). The first curve was fit among the 157 household contacts who were at risk of CHIKV infection after or slightly before study initiation. The second curve was fit among the 63 individuals with active and incident CHIKV infections. Confidence intervals (CI) account for the clustered data structure, having been nonparametrically bootstrapped at the household level 10,000 times.
Risk factor analysis.
Risk factor analyses (Table 3) showed that having a high frequency of trash collection (≥4 times/week) (multivariable-adjusted risk ratio [aRR], 0.38; 95% CI, 0.19, 0.76) and having externally piped water (aRR, 0.52; 95% CI, 0.32, 0.84) were significantly protective with respect to very recent, active, and incident CHIKV infections (model 1; n = 157) that occurred after or slightly before study initiation. Vehicle ownership (aRR, 1.58; 95% CI, 1.10, 2.27) was associated with a significantly increased risk of CHIKV infection across study households. No risk factors were significantly associated with incident infection (model 2; n = 104); however, the directionality of association for external piped water and vehicle ownership was preserved. We did not find any factors predictive of sign/symptom occurrence among household contacts with active and incident infections (model 3; n = 63) or strictly among those with incident infections (model 4; n = 21).
TABLE 3.
Results of risk factor analyses for CHIKV infection and symptomatic status in different participant subgroups using modified Poisson regressiona
| Model | Outcome | Study population (no. of individuals with outcome/total no. of individuals) | Variable selected | aRR | 95% CI |
|---|---|---|---|---|---|
| 1 | Infection after or slightly before study initiation | 157 contacts at risk of infection after or slightly before study initiation (74/157) | High frequency of weekly trash collection | 0.38 | 0.19, 0.76 |
| Externally piped water | 0.52 | 0.32, 0.84 | |||
| Ownership of vehicle | 1.58 | 1.10, 2.27 | |||
| Having at least 1 tire in the yard | 1.35 | 0.73, 2.49 | |||
| At or below primary education | 1.02 | 0.71, 1.47 | |||
| High frequency of yearly abatement useb | 0.97 | 0.58, 1.64 | |||
| 2 | Incident infection | 104 contacts at risk of an infection after study initiation (21/104) | Ownership of a vehicle | 2.10 | 0.93, 4.76 |
| Externally piped water | 0.40 | 0.14, 1.10 | |||
| At or below primary education | 0.58 | 0.27, 1.21 | |||
| Having at least 1 tire in the yard | 0.61 | 0.16, 2.38 | |||
| 3 | Symptomatic status | 63 contacts with active and incident infections (32/63) | — | — | — |
| 4 | Symptomatic status | 21 contacts with incident infections (5/21) | — | — | — |
Variables for inclusion in the model were chosen on the basis of a best-subset variable selection procedure. See the text for the candidate set of predictors. Dashes indicate that the variable selection procedure did not select any variables; hence, no predictors were found for models 3 and 4. aRR, multivariable-adjusted risk ratio from a modified Poisson model; CI, confidence interval.
A high frequency of an event was defined as an event that occurred ≥4 times.
Variability of the S:A ratio.
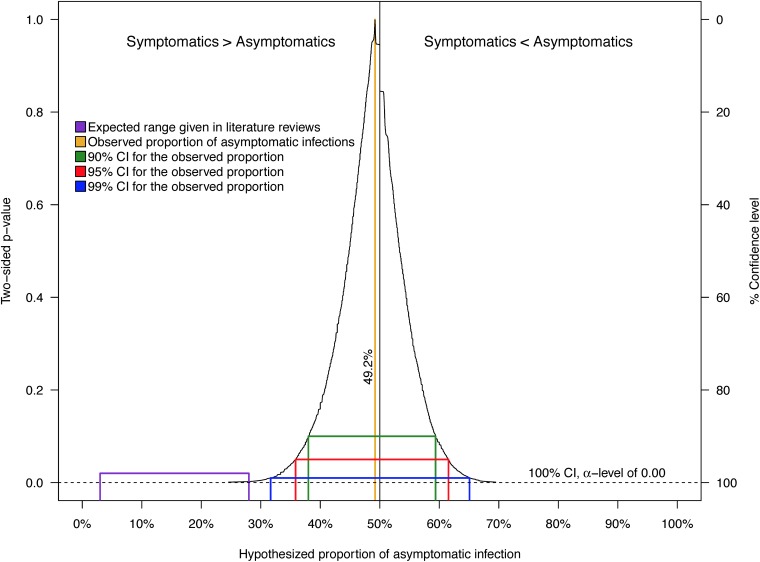
The overall S:A ratio of 1:0.97 represents a weighted average of stratum-specific S:A ratios. The stratified point estimates for the S:A ratio by infection classification and demographic strata ranged from 1:0.5 to 1:3.2 (i.e., stratified proportions of asymptomatic infection ranged from 37% to 76%) (Table 2). Only the sampling distributions for incident infections and children were too non-normal to estimate unbiased bias-corrected and accelerated bootstrap confidence intervals (BCa CIs) (data not shown). As a result of some strata having small sample sizes and, hence, invalid BCa CIs, statistical tests were not performed to compare stratum-specific S:A ratios. CI functions depict CIs at all possible α values around a point estimate and thereby visualize the variability of the data; in doing so, they avoid the arbitrary selection of an α value of 0.05 for the construction of interval estimates. In the CI function for the overall proportion of asymptomatic CHIKV infections (Fig. 5), values closer to the point estimate of 49.2% are more compatible with the data than those further away from it. As the 3 to 28% range is outside even 99% CIs, our data are not compatible with the expected range. Specifically, the one-sided P value comparing our point estimate (49.2%) against the upper limit of the expected range (28%) is <0.002. Thus, despite the small sample size on which it is based (i.e., 63 active and incident infections), our estimate of the proportion of asymptomatic CHIKV infections is significantly different from and highly incompatible with the expected range of 3 to 28%, as previously reported (15–19).
FIG 5.
Confidence interval function for the proportion of infections based on the 63 household contacts with active and incident CHIKV infections. Shown is a simultaneous depiction of 1,000 bias-corrected and accelerated (BCa) confidence intervals (CIs) as well as every two-sided P value associated with null values for hypothetical proportions of infection, given the data, across the full range of α values. For the CI function, values closer to the point estimate of 49.2% are more compatible with the data than those further away. The purple line represents the range for the proportion of inapparent CHIKV infections (3 to 28%) commonly reported in literature reviews.
Bayesian analysis of the proportion of asymptomatic infection.
We conducted a simple Bayesian analysis on the overall proportion of asymptomatic CHIKV-infected household contacts to reduce the variability in our estimate and incorporate the full extent of what is known about inapparent infections into our analysis. A systematic literature search of studies documenting chikungunya epidemics in initially CHIKV-naive populations was conducted, and data were extracted (Table 4) to inform prior distributions. We used the systematic literature search to inform prior distributions (Table 5), rather than estimating a prior based on subjective confidence in the 3 to 28% range, as our estimates from the first chikungunya epidemic in Managua were far outside this range (10). Moreover, because CHIKV is an understudied virus, we were concerned that the use of subjective priors might exceed the extent of established knowledge regarding CHIKV infections. As the effect of clustering in our data was small (intraclass correlation coefficient of 0.10 [data not shown]), the Bayesian analysis was performed assuming that our data were independent.
TABLE 4.
Data extracted from the systematic literature search conducted to identify studies reporting the percentage of inapparent CHIKV infections after a chikungunya epidemic in an initially CHIKV-naive populationa
| Identified study, yr (reference) | Yr of symptom collection | Outbreak location(s) | Virus lineageb | Responsible vector(s)b,c | % of CHIKV-positive participants (no. of CHIKV-positive participants/total no. of participants) | % of participants with inapparent infections (no. of participants with inapparent infections/total no. of CHIKV-positive participants)d | Sampling method |
|---|---|---|---|---|---|---|---|
| Gordon et al., 2018 (2014 children) (14)e | 2014 (during outbreak) | Managua, Nicaragua | Asian (likely) | A. aegypti (more likely) and A. albopictus (less likely) | 6.1 (204/3,361) | 54.5 (108/198) | Age stratified, systematic |
| Gordon et al., 2018 (2015 children)e (14) | 2015 (during outbreak) | Managua, Nicaragua | Asian (likely) | A. aegypti (more likely) and A. albopictus (less likely) | 21.3 (689/3,230) | 39.4 (254/645) | Age stratified, systematic |
| Cunha et al., 2017f (23) | 2016 (18 mo postoutbreak) | District of Chapada, Riachão do Jacuípe, Brazil | Non-IOL-strain ECSA (likely) | A. aegypti (likely) | 20.0 (24/120) | 45.8 (11/24) | Systematic, random |
| Bloch et al., 2016e (20) | 2014 (during outbreak) | Puerto Rico, USA | Asian (likely) | A. aegypti (likely) | 24.1 (56/232) | 37.5 (21/56) | Geographic (150 m around index cases’ homes) |
| Yoon et al., 2015g (24) | 2012–2013 (during outbreak) | Cebu City, Philippines | Asian | A. aegypti and A. albopictus (equally likely) | 12.5 (107/853) | 81.3 (87/107) | Age stratified, systematic |
| Srikiatkhachorn et al., 2016e (25) | 2013–2014 (during outbreak) | Cebu City, Philippines | Asian (likely) | A. aegypti and A. albopictus (equally likely) | 4.7 (21/443) | 71.4 (15/21) | Age stratified, systematic |
| Kuan et al., 2016 (adults)h (10) | 2016 (3 mo postoutbreak) | Managua, Nicaragua | Asian (likely) | A. aegypti (more likely) and A. albopictus (less likely) | 13.1 (111/848) | 64.9 (72/111) | Systematic, random |
| Galatas et al., 2016g (21) | 2012 (during outbreak) | Trapeang Roka, Cambodia | IOL strain ECSA (likely) | A. aegypti (likely) | 44.7 (190/425) | 5.3 (10/190) | Convenience (villagers available on day of investigation) |
| Gay et al., 2016 (26) | 2014 (during outbreak) | Saint Martin, France | Asian (likely) | A. aegypti (likely) | 20.7 (42/203) | 40.5 (17/42) | Convenience (laboratory clients) |
| Nakkhara et al., 2013 (22) | 2010–2011 (2 yr postoutbreak) | Thung Nari subdistrict, Pa Bon district, Thailand | IOL strain ECSA (likely) | A. albopictus (more likely) and A. aegypti (less likely) | 61.9 (314/507) | 47.1 (148/314) | Convenience (adults only) |
| Schwarz et al., 2012 (85) | 2010 (1–2 mo postoutbreak) | Mananjary and Manakaa, Madagascar | IOL strain ECSA (likely) | A. albopictus (more likely) and A. aegypti (less likely) | 32.3 (144/446) | 20.8 (30/144) | Convenience (pregnant women) |
| Ayu et al., 2010 (86) | 2007 (1 yr postoutbreak) | Bagan Panchor, Malaysia | Asian | A. aegypti and A. albopictus (equally likely) | 55.6 (40/72) | 17.5% (7/40) | Convenience (villagers who agreed to participate) |
| Sissoko et al., 2010 (87) | 2006 (9 mo after peak of outbreak) | Mayotte, France | IOL strain ECSA (likely) | A. albopictus (likely) | 38.1 (440/1,154) | 27.7 (122/440) | Multistage, cluster |
| Moro et al., 2010 (88) | 2007 (3–5 mo postoutbreak) | Castiglione di Cervia, Italy | IOL strain ECSA (likely) | A. albopictus (likely) | 10.2 (33/325) | 18.2 (6/33) | Age stratified, random |
| Manimunda et al., 2010 (89) | 2008 (during outbreak) | Dakshina Kannada District, Karnataka State, India | IOL strain ECSA (likely) | A. albopictus (likely) | 62.2 (224/360) | 6.3 (14/224) | Systematic, poststratified by age and sex |
| Leo et al., 2009 (90) | 2008 (during outbreak) | Singapore | IOL strain ECSA | A. aegypti (likely) | 0.5 (13/2,626) | 23.1 (3/13) | Geographic (150 m around index cases’ homes) |
| Gérardin et al., 2008 (91) | 2006 (1–3 mo postoutbreak) | La Réunion, France | IOL strain ECSA (likely) | A. albopictus (likely) | 39.6 (967/2,442) | 12.6 (116/921) | Multistage, random |
| Sergon et al., 2008f (27) | 2004 (2 mo after peak of outbreak) | Lamu Island, Kenya | IOL strain ECSA (likely) | A. aegypti (likely) | 75 (215/288) | 45.1 (97/215) | Multistage, random |
| Queyriaux et al., 2008i (30) | 2006 (during outbreak) | La Réunion, France | IOL strain ECSA (likely) | A. albopictus (likely) | 19.3 (128/662) | 3.1 (4/128) | Convenience (French soldiers) |
| Sergon et al., 2007 (92) | 2005 (during outbreak) | Grande Comore Island, Comoros | IOL strain ECSA (likely) | A. aegypti (likely) | 63.1 (209/331) | 14.4 (30/209) | Multistage, systematic |
| Retuya, 1998 (31) | 1996 (during outbreak) | Barangay Pulo, Indang, Cavite, Philippines | Unknown | A. aegypti and A. albopictus (equally likely) | 52.3 (156/298) | 23.1 (36/156) | Convenience (villagers who agreed to participate) |
A previous version of this table was used in another publication (93), from which the data were adapted with permission. Abbreviations: ECSA, East/Central/South African CHIKV lineage; IOL, Indian Ocean strain of the ECSA lineage.
The viral lineage responsible for the outbreak, when not explicitly confirmed through sequencing in the paper, was derived from other reported studies that genotyped virus isolates from either (i) the outbreak in question or (ii) e-mail correspondence with the study authors. Similarly, the responsible vector, when not explicitly confirmed through entomological investigation in the paper, was derived from either (i) the global compendium of Aedes aegypti and Aedes albopictus occurrence data set (82) or (ii) e-mail correspondence with the study authors. In these instances, the cell with the given information is labeled as “likely.”
When various Aedes species were circulating in the area of the epidemic (based on the text in the paper, correspondence with a study author, or the global Aedes occurrence compendium data set), the more populous subspecies was considered and labeled as “more likely” to be responsible for the epidemic, and vice versa.
Due to peculiarities in individual studies, the numerator of the “proportion of CHIKV-positive participants” column may not equal the denominator of the “proportion of inapparent infections” column.
Data in the study were supplemented by data provided by the first, senior, or corresponding author.
The proportion of inapparent infections was back-calculated from the reported proportion of symptomatic infections.
Data regarding the proportion of CHIKV-positive individuals or the proportion of inapparent infections were supplemented with data from another article reporting the same outbreak. (Data from Galatas et al. [21] were supplemented by data from Ly et al. [81], and data from Yoon et al. [24] were supplemented by data from Srikiatkhachorn et al. [25].)
The proportion of inapparent infections was back-calculated from the point estimate and/or the confidence interval and the assumed statistical distribution for the data, as per the cited study.
The numerator for the proportion of inapparent infections was back-calculated from the reported proportion of symptomatic infections and the sample size, as per the cited study.
TABLE 5.
Data regarding literature-based prior distributions for the probability of inapparent CHIKV infection
| Prior distribution category | Reference(s) summarized by the prior | Prior beta distribution hyperparameters (α, β)a | Approx median (%)h | Precisionb,h | 95% HDCIh (%)c | Mean (%)h | P(x) > 28.0 (%)d,h | P(x) > 49.2 (%)e,h |
|---|---|---|---|---|---|---|---|---|
| Overall | 10, 14, 20–27, 30, 31, 85–92 | 1.49, 3.69 | 25.6 | 30.18 | 0.3, 63.3 | 28.8 | 45.9 | 15.0 |
| Epidemics within the 3–28% range | 21, 30, 31, 85–92 | 2.97, 16.72 | 13.9 | 161.46 | 2.0, 30.5 | 15.1 | 6.9 | 0.1 |
| Epidemics caused by the Asian lineage | 10, 14, 20, 24–26, 86 | 4.95, 5.35 | 47.9 | 45.29 | 19.7, 76.6 | 48.1 | 90.7 | 46.8 |
| Epidemics caused by the ECSA lineage | 21–23, 27, 30, 85, 87–92 | 1.68, 6.39 | 18.2 | 54.98 | 0.3, 46.9 | 20.8 | 26.7 | 3.8 |
| Epidemics caused by the IOL strain of the ECSA lineage | 21, 22, 27, 30, 85, 87–92 | 1.69, 6.50 | 18.0 | 56.15 | 0.3, 46.4 | 20.6 | 26.1 | 3.6 |
| Epidemics caused by non-IOL strains of the ECSA lineagef | 23 | |||||||
| Epidemics with A. aegypti as the primary vector | 10, 14, 20, 21, 23, 26, 90, 92 | 2.17, 3.92 | 33.9 | 30.90 | 3.9, 69.6 | 35.6 | 62.2 | 23.3 |
| Epidemics with A. albopictus as the primary vector | 22, 30, 85, 87–89, 91 | 1.87, 7.43 | 17.8 | 64.09 | 0.6, 44.3 | 20.1 | 24.1 | 2.6 |
| Epidemics in the Americas | 10, 14, 20, 23, 26 | 13.86, 16.94 | 44.9 | 128.47 | 27.9, 62.3 | 45.0 | 97.6 | 31.7 |
| Epidemics outside the Americas | 21, 22, 24, 25, 27, 30, 31, 85–92 | 1.35, 4.29 | 20.4 | 36.49 | 0.0, 55.9 | 23.9 | 35.0 | 8.9 |
| Epidemics in Africa | 27, 30, 85, 87, 91, 92 | 2.51, 10.37 | 17.8 | 88.43 | 1.9, 40.2 | 19.5 | 20.2 | 1.1 |
| Epidemics in Asia | 21, 22, 24, 25, 31, 86, 89, 90 | 1.00, 2.21 | 26.2 | 19.60 | 0.0, 74.3 | 31.2 | 48.4 | 22.4 |
| Epidemic studies with a convenience sample | 21, 22, 26, 30, 31, 85, 86 | 1.21, 3.75 | 20.4 | 32.31 | 0.0, 58.6 | 24.4 | 36.1 | 10.5 |
| Epidemic studies with a nonconvenience sample | 10, 14, 20, 23–25, 27, 87–92 | 1.65, 3.80 | 27.5 | 30.60 | 0.8, 64.3 | 30.2 | 49.3 | 16.3 |
| Updated, overallg | 10, 14, 20–27, 30, 31, 85–92; this study | 1.50, 3.67 | 25.9 | 29.95 | 0.3, 63.6 | 29.0 | 46.5 | 15.3 |
| Updated, epidemics caused by the Asian lineageg | 10, 14, 20, 24–26, 86; this study | 5.20, 5.61 | 48.0 | 47.31 | 20.4, 76.0 | 48.1 | 91.3 | 46.9 |
Hyperparameters are the parameters that specify the particular probability distribution to be used as a prior distribution. Beta hyperparameters were estimated by numerically optimizing the likelihood after weighting each identified study’s contribution by the number of CHIKV-infected individuals.
Precision is the reciprocal of variance. In Bayesian analyses, precision is a preferred way of quantifying the variability in a distribution. Higher values indicate a less variable distribution.
Values for 95% HDCIs (highest-density credible intervals) are usually presented for the medians of posterior distributions and not prior distributions. However, we list them here to quantify the variability around the medians for the probability of inapparent CHIKV infections given the state of the literature.
Probability of observing a proportion of inapparent CHIKV infections, under this prior, that exceeds the upper limit of the expected range, 28.0%.
Probability of observing a proportion of inapparent CHIKV infection, under this prior, that exceeds the observed proportion in this study, 49.2%.
Hyperparameters for a prior distribution could not be estimated via maximum likelihood or method-of-moments estimators from the single non-IOL ECSA study identified in the systematic search, so no parameters could be calculated.
These data were derived by treating the present study as another study contributing to the understanding of the proportion of inapparent CHIKV infections. Results from the present study were incorporated along with those of the identified studies into a prior distribution, as before. Because our index cluster study concerns an epidemic caused by the Asian lineage, only the overall and Asian lineage priors are updated. Data in this column could be used as prior distributions for future Bayesian studies, public health planning, or mathematical modeling studies focusing on epidemics of CHIKV-specific lineages.
Statistics were calculated from the prior beta distribution with the given hyperparameters in the respective row.
All of the identified studies captured inapparent infections instead of asymptomatic infections as a result of having more traditional study designs; however, because chikungunya signs/symptoms are typically severe and hence are likely to be reported to medical professionals, the percentage of inapparent CHIKV infections captured in the identified studies is likely comparable to the percentage of asymptomatic CHIKV infections that authors would have captured with intensive blood sampling of their participants, all else being equal. Because of the expected similarity in the percentages of inapparent and asymptomatic CHIKV infections that could be reported for a given chikungunya outbreak, the Bayesian analyses were informed by prior distributions estimated from the inapparent CHIKV infections in the literature with our study data based on asymptomatic CHIKV infections. Our discussion of asymptomatic CHIKV infections is thus limited to our own study sample, whereas our discussion of inapparent CHIKV infections concerns findings from the systematic literature search.
We did not find any study documenting the proportion of inapparent CHIKV infections caused by the West African CHIKV lineage. All but one of the ESCA lineage studies meeting our criteria corresponded to outbreaks caused by the IOL strain.
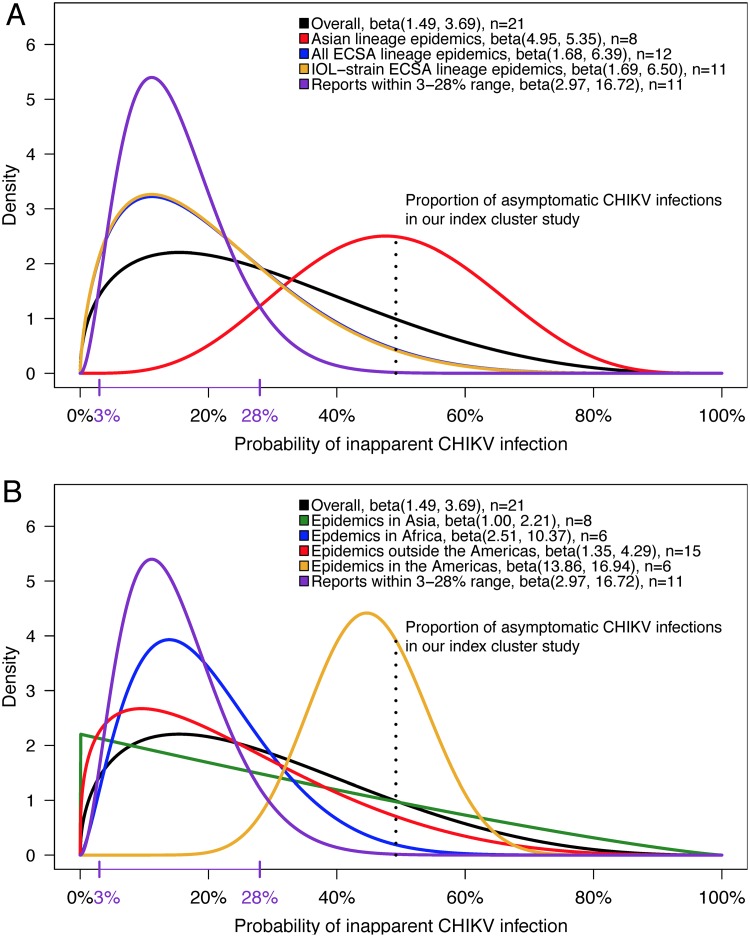
The median of the overall posterior distribution (Table 6), using all 20 studies that we identified to inform the overall prior distribution (Table 4), was 48% (95% highest-density credible interval [HDCI], 36%, 60%), very similar to our overall frequentist estimate of 49% (95% CI, 36%, 62%) (Table 2). We observed an unexpectedly small degree of shrinkage (i.e., movement from the frequentist point estimate [49%] to the median of the overall posterior [48%]) toward the expected 3 to 28% range. This indicated that the overall prior did not, contrary to our expectation, allocate the vast majority of its area under the curve (AUC) to the 3 to 28% range (Fig. 6). Instead, 46% of the AUC for the overall prior distribution curve lies above 28% (Table 5), implying that about half of the evidence in the literature pointed toward proportions of inapparent infection above the cited upper limit (15–19). Moreover, this finding indicated that greater variability exists in this parameter than is currently recognized (Fig. 6; Table 5).
TABLE 6.
Data regarding posterior distributions for the literature-based Bayesian analysis of asymptomatic CHIKV infectionsa
| Type of epidemic reports used to parameterize the priorb | Prior beta distribution hyperparameters (α, β)c | Posterior beta distribution parameters (α, β) | Approx median (%)d | 95% HDCI (%)d | P(x) > 28.0% (%)d |
|---|---|---|---|---|---|
| None | 1.00, 1.00 (uninformative prior) | 32.00, 33.00 | 49.2 | 37.2, 61.3 | 100.0 |
| Overall | 1.49, 3.69 | 32.49, 35.69 | 47.6 | 35.9, 59.4 | 100.0 |
| Epidemics within the 3–28% range | 2.97, 16.72 | 33.97, 48.72 | 41.0 | 30.6, 51.6 | 99.4 |
| Epidemics caused by the Asian lineage | 4.95, 5.35 | 35.95, 37.35 | 49.0 | 37.7, 60.4 | 100.0 |
| Epidemics caused by the ECSA lineage | 1.68, 6.39 | 32.68, 38.39 | 45.9 | 34.5, 57.5 | 99.9 |
| Epidemics caused by the IOL strain of the ECSA lineage | 1.69, 6.50 | 32.69, 38.50 | 45.9 | 34.5, 57.4 | 99.9 |
| Epidemics caused by non-IOL strains of the ECSA lineagee | |||||
| Epidemics with A. aegypti as the primary vector | 2.17, 3.92 | 33.17, 35.92 | 48.0 | 36.4, 59.7 | 100.0 |
| Epidemics with A. albopictus as the primary vector | 1.87, 7.43 | 32.87, 39.43 | 45.4 | 34.1, 56.9 | 99.9 |
| Epidemics in the Americas | 13.86, 16.94 | 44.86, 48.94 | 47.8 | 37.8, 57.9 | 100.0 |
| Epidemics outside the Americas | 1.35, 4.29 | 32.35, 36.29 | 47.1 | 35.5, 58.8 | 100.0 |
| Epidemics in Africa | 2.51, 10.37 | 33.51, 42.37 | 44.1 | 33.1, 55.3 | 99.9 |
| Epidemics in Asia | 1.00, 2.21 | 32.00, 34.21 | 48.3 | 36.4, 60.3 | 100.0 |
| Epidemic studies with a convenience sample | 1.21, 3.75 | 32.21, 35.75 | 47.4 | 35.7, 59.2 | 100.0 |
| Epidemic studies with a nonconvenience sample | 1.65, 3.80 | 32.65, 35.80 | 47.7 | 36.0, 59.4 | 100.0 |
Fourteen different Bayesian analyses were run, an overall analysis and 13 analyses with different priors based on different subsets of studies presented in Table 5.
See Table 5.
Hyperparameters are the parameters that specify the particular probability distribution to be used as a prior distribution. Beta hyperparameters were estimated by numerically optimizing the likelihood after weighting each identified study’s contribution by the number of CHIKV-infected individuals.
Statistics were calculated from the posterior beta distribution with the given hyperparameters in the respective row.
Hyperparameters for a prior distribution could not be estimated via maximum likelihood or method-of-moments estimators from the single non-IOL ECSA study identified in the systematic literature search, so no parameters could be calculated.
FIG 6.
Comparison of prior distributions for the probability of inapparent CHIKV infection. Prior distributions, estimated from the literature, from epidemics caused by different lineages (A) and in different geographical settings (B) are shown. A prior distribution for epidemics caused by the non-IOL-strain ECSA lineage, with respect to the proportion of inapparent infections, could not be estimated from the single non-IOL-strain ECSA lineage study that met our inclusion and exclusion criteria. As a result, the prior estimated for IOL strain ECSA lineage outbreaks (A, in yellow) very closely mirrors the prior estimated for all ECSA lineage outbreaks (in blue) faintly visible below it. n, number of distinct populations contributing to the estimation of the prior distribution’s hyperparameters.
The Bayesian sensitivity analysis revealed that CHIKV lineage-specific heterogeneity in the probability of inapparent infection was the likely cause of the overall prior’s high variance (Fig. 6 and 7; Tables 5 and 6). The sensitivity analysis was based on 13 different priors estimated from subsets of selected studies. The resultant medians of the posterior distributions ranged from 41 to 49% (Table 6), suggesting that all priors resulted in a similar posterior distribution for the percentage of asymptomatic CHIKV infections in our study sample.
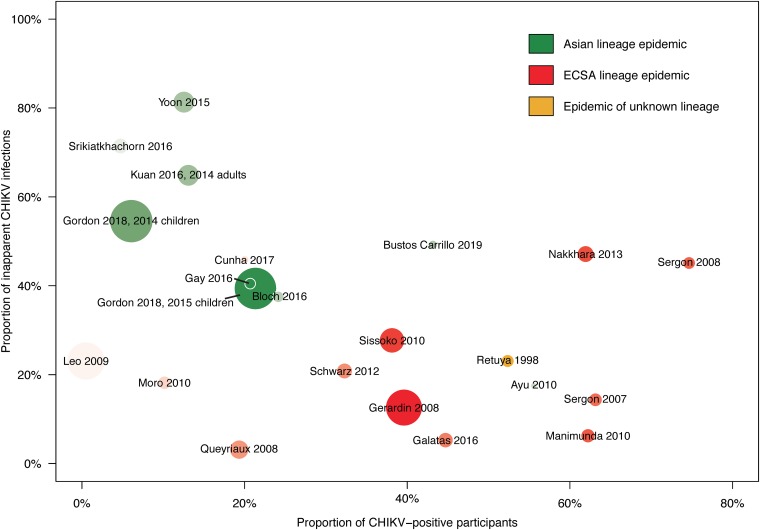
FIG 7.
Risk of CHIKV infection and inapparent clinical presentation in the literature. Scatterplot elements correspond to data in Table 4 (10, 14, 20–27, 30, 31, 85–92). The radius of each study’s data point is scaled to the sample size, and the circle is shaded to the size of the CHIKV-infected population (adapted from reference 93 with permission). Data from the present study are included for visual comparison with previously published studies.
However, we observed unexpected differences among the priors related to CHIKV lineage. In particular, 91% of the AUC for the prior based on Asian lineage CHIKV epidemics (Fig. 6A; Table 5) and 98% of the AUC for the prior estimated for CHIKV epidemics in the Americas (Fig. 6B; Table 5), where the Asian CHIKV lineage predominates, lie above 28%, suggesting that inapparent CHIKV infections caused by the Asian lineage do not conform to the 3 to 28% range. In contrast, prior distributions estimated from all ECSA epidemics, IOL-strain ECSA epidemics, and chikungunya epidemics on the African continent (where the ECSA lineage predominates) showed ∼75% of their AUC below 28% (Fig. 6; Table 5), largely in agreement with the 3 to 28% range. Whereas the prior distribution for the proportion of inapparent infections caused by ECSA lineage symptoms was centered at (i.e., had an approximate median of) 19%, the Asian lineage distribution was centered at 48%, very similar to the observed proportion of asymptomatic infections in our study sample (Table 5). Studies of Asian lineage CHIKV epidemics reported higher (and more-variable) proportions of inapparent infections than epidemics of non-Asian CHIKV epidemics (Fig. 6 and 7; Tables 4 and 5).
As a result of the apparent CHIKV lineage-specific heterogeneity in the probability of inapparent infection, using the Asian lineage CHIKV prior to estimate the median of the posterior distribution, 49% (HDCI, 38%, 60%), is a more appropriate approach than using the overall prior (Table 6). The prior distributions for studies based on convenience and nonconvenience samples (e.g., random samples, systematic samples, and multistage cluster samples, etc.) were quite similar (Table 5), implying that the sampling technique was not driving the lineage-specific differences that we observed.
Differences in CHIKV lineage-specific prior distributions.
The lineage-specific prior distributions were formally compared to characterize and quantify differences in their behaviors. Multiple distance and similarity measures/metrics indicated that the prior distribution for the ECSA lineage’s IOL strain was most compatible with the expected range of 3 to 28%, while the Asian lineage distribution was the most divergent (Table 7). Monte Carlo resampling simulations based on the Asian and ECSA literature-based prior distributions were performed to investigate predicted differences between chikungunya outbreaks caused by the different lineages. If the CHIKV lineage was unrelated to the percentage of inapparent CHIKV infections in epidemic settings, then, on average, Asian lineage epidemics should have a higher percentage of inapparent infections than ECSA lineage CHIKV epidemics 50% of the time. However, Monte Carlo sampling of the Asian and ECSA priors indicates that, on average, the proportion of inapparent infections from Asian lineage CHIKV epidemics exceeds that of ECSA lineage CHIKV epidemics 91% (95% CI, 71%, 99%) of the time. Moreover, the proportion of inapparent infections from Asian lineage CHIKV epidemics exceeds that of ECSA lineage CHIKV epidemics by at least 10, 20, and 30 percentage points approximately 81% (95% CI, 54%, 96%), 66% (95% CI, 36%, 89%), and 46% (95% CI, 17%, 78%) of the time, respectively.
TABLE 7.
Comparison of the prior distributions for inapparent CHIKV infections by way of distance and similarity measures/metricsa
| Prior distribution categoryb | Kullback-Leibler divergence (range, 0, +∞)c | Hellinger distance (range, 0, 1)d | Bhattacharyya coefficient (range, 0, 1)e |
|---|---|---|---|
| Overall | 0.49 | 0.41 | 0.83 |
| Epidemics caused by the Asian lineage | 3.34 | 0.79 | 0.37 |
| Epidemics caused by the ECSA lineage | 0.19 | 0.25 | 0.94 |
| Epidemics caused by the IOL strain of the ECSA lineage | 0.19 | 0.24 | 0.94 |
| Epidemics caused by non-IOL strains of the ECSA lineagef |
The Kullback-Leibler divergence, the Hellinger distance, and the Bhattacharyya coefficient quantify differences in global behaviors between two probability distribution functions.
For these analyses, prior distributions were compared to beta(2.97, 16.72), the prior distribution corresponding to all studies that reported a proportion of inapparent CHIKV infections within the expected 3 to 28% range.
The lower the Kullback-Leibler divergence, the more the probability distribution under consideration behaves in a fashion similar to that of the distribution built from studies conforming to the expected range in expectation, and vice versa.
The lower the Hellinger distance (the probabilistic analogue to the Euclidean distance between two points), the more similar the distribution functions, and vice versa.
Conversely, a low Bhattacharyya coefficient indicates a low degree of overlap between the distribution functions considered, and vice versa.
Hyperparameters for a prior distribution could not be estimated via maximum likelihood or method-of-moments estimators from the single non-IOL-strain ECSA study identified in the systematic literature search, so no parameters could be calculated.
As a result of the identified lineage-specific differences in the proportions of inapparent CHIKV infections, we estimated a second set of prior distributions, incorporating results from this study, for use by future researchers (Table 5). Owing to our study’s small sample size, the updated set of prior distributions closely reflected the prior distributions originally estimated from the literature.
DISCUSSION
We present an index cluster study of chikungunya, which used a household-based design to estimate the ratio of symptomatic to asymptomatic CHIKV infections during a large chikungunya epidemic in Managua, Nicaragua, in 2015. We find laboratory-confirmed evidence for an overall S:A ratio of 1:0.97 among a sample of participants of all ages, equivalent to a 49% probability of asymptomatic CHIKV infection. CHIKV infection and disease had linear associations with age. We did not find risk factors for symptomatic occurrence; however, vehicle ownership (aRR, 1.58), living in a home with a high level of municipal trash collection (aRR, 0.38), and having externally piped water (aRR, 0.52) were associated with CHIKV infection. Finally, we showed that the 3 to 28% range for the proportion of inapparent infections reported in literature reviews (15–18) is misleading, as the range is overly narrow and obscures important lineage-specific differences. We estimated that 49% of CHIKV infections in our sample were asymptomatic using both frequentist and Bayesian methods. Although this finding is discrepant with both epidemics caused by the ECSA lineage and the 3 to 28% range for inapparent CHIKV infections commonly cited in literature reviews, we showed that this finding agrees with other reports of CHIKV epidemics caused by the Asian lineage. These results provide comprehensive epidemiological evidence that chikungunya epidemic characteristics are strongly influenced by CHIKV lineage and provide, to our knowledge, the first quantification of differences between the Asian lineage and the ECSA lineage, particularly its IOL strain, with respect to the proportion of inapparent CHIKV infections at the population level. Importantly, our findings of lineage-specific differences were based on studies with sample populations that had no documented prior exposure to CHIKV, limiting the impact that previous CHIKV exposure could have on the occurrence of disease presentation.
High proportions of inapparent CHIKV infections have been observed in multiple settings with different study designs over the last 15 years (10, 14, 20–27); however, the expected range of 3 to 28% was based (17) only on three studies conducted in the late 1990s and mid-2000s (30–32). This range has repeatedly been cited in subsequent reviews (15, 16, 18) and the Centers for Disease Control and Prevention’s Yellow Book (19), despite recent serosurveys reporting higher levels of inapparent infections. The possibility that the proportion of inapparent infections observed during an epidemic might be lineage dependent was raised first by Yoon et al. (24) and then by others (10, 33). However, while the hypothesis of lineage-specific differences has previously been raised as an explanation for observed discrepancies between chikungunya epidemics, it has not yet been widely accepted enough to be reflected in literature reviews. We argue on the basis of the estimated prior distributions, statistical measures/metrics, and Monte Carlo resampling of the estimated priors that sufficient evidence exists in the literature to warrant inclusion of these epidemiological, lineage-specific differences in the proportion of inapparent infections in future literature reviews.
The prior distribution that we estimated for inapparent infections from Asian lineage CHIKV infections (Fig. 6A) has a median similar to that of the distribution estimated from populations in the Americas (Fig. 6B). We are unable to rule out the possibility that the high proportion of inapparent infections from Asian lineage CHIKV epidemics is due to or is confounded by the genetic background or immunological history of populations in the Americas. However, the immunological and pathophysiological CHIKV lineage-specific differences observed in mouse and nonhuman primate models (3–6), combined with the comprehensive epidemiological evidence identified in the systematic literature search, suggest that CHIKV lineage-specific differences help to explain broadly observed heterogeneity in CHIKV infection outcomes in both human populations and animal models. Although our focus was on the proportion of inapparent infections, lineage-specific differences may also underlie differences in chikungunya severity, as has been postulated by others (34).
Chikungunya virus is not the only arbovirus to display subgroup variation with respect to infection outcomes. Studies of dengue virus (DENV), the most common arboviral human disease, have reported wide ranges for ratios of symptomatic to inapparent infections (S:I ratios) across different years, even in the same population (35, 36). For example, Endy et al. described S:I ratios ranging from approximately 0.4:1 to 0.9:1 in populations of schoolchildren over a 5-year period, and within the same year, S:I ratios ranged from approximately 0.1:1 to 5.0:1 (36). DENV has four serotypes, and serotype-specific differences in clinical manifestations, including severe disease, have been reported in pediatric (37, 38) as well as adult (39, 40) populations. Zika virus (ZIKV), a virus closely related to dengue virus, has shown strain-dependent differences in ex vivo virulence (41), apoptosis induction capacity (42), infectivity (43), as well as disease manifestations in mouse models (44) and potentially humans (41). To the best of our knowledge, S:I ratios for DENV and ZIKV have not yet been examined by serotype and strain, respectively, in human populations. However, given the various subgroup differences previously noted, it would be unsurprising if future studies identified subgroup differences in their S:I ratios.
The large difference between Asian and ECSA lineage chikungunya epidemics has important public health implications, as it implies that the vast majority (82%) of ECSA lineage infections will be symptomatic, whereas only half (52%) of Asian lineage infections will be symptomatic. Our main finding on the dependence of the S:I ratio on CHIKV lineage can help public health officials better plan for chikungunya epidemics of the specific lineage in their geographical vicinity. Our data can also help mathematical modelers accurately capture the transmission dynamics of chikungunya epidemics. Importantly, we recommend that planning for and modeling of chikungunya epidemics be based on lineage-specific data instead of the 3 to 28% range (Table 5). The wide 95% credible intervals that we estimated for these lineage-specific data reflect the paucity of large, published studies. As such, they more accurately reflect the uncertainty of this parameter than is expressed by the 3 to 28% range. Since evidence from the fields of immunology and pathology also suggests lineage-specific differences postinfection (3–6), future work should consider stratifying analyses of postinfection outcomes by CHIKV lineage.
The potential for lineage-specific differences to impact chikungunya disease manifestation also has implications for the development of vaccines to chikungunya. From the studies that we gathered, it is not clear whether the observed lineage-specific differences in epidemic chikungunya behavior are due to lineage-specific virus-induced pathology, immune responses differentially mounted to CHIKVs of different lineages, or some immunopathological combination thereof. Future studies should examine the mechanisms giving rise to the lineage-specific epidemiological differences we identified, as these mechanisms may necessitate the development of multivalent chikungunya vaccines.
Our evidence of lineage-specific differences also has implications for epidemiological comparisons between DENV, ZIKV, and CHIKV. The vast majority of CHIKV infections, as per the state of current literature reviews, are regarded as being symptomatic, in contrast to flaviviruses such as DENV (45) and ZIKV (46), which result in large proportions of inapparent/asymptomatic infections. Our finding that infection with an Asian lineage CHIKV is symptomatic only 52% of the time suggests that the Asian lineage may not be so different from DENV, ZIKV, and other flaviviruses with respect to symptomatic occurrence.
Across households, vehicle ownership was the only variable associated with an increase in the risk of CHIKV infection in our most powered model. However, it is unclear if vehicle ownership functioned more as a proxy for transportation-related increases in the risks of infection or as a proxy for socioeconomic status. A high frequency of trash collection was the most protective variable associated with CHIKV infection. The removal of trash in the peridomestic environment represents an important modifiable risk factor, since Aedes mosquitoes are known to breed in trash and other debris capable of catching rainwater (47). We did not find an association between having tires in the yard and CHIKV infection, even though tires are known to retain rainwater and hence be a risk factor for mosquito-borne infections (48). However, most (89%) at-risk participants had tires in their yard, which may explain our null finding for this known risk factor. Previous work by Nakkhara et al. found that refuse in the peridomestic environment was associated with infection and that age and education were inversely associated with disease manifestation (22). However, our results are not directly comparable with these results. Nakkhara et al. used an inappropriate multinomial regression that assumed that no infection, symptomatic infection, and asymptomatic infection were statistically equitable outcomes. We modeled the infection and disease processes separately, as a symptomatic or asymptomatic outcome is always conditional on infection.
Our study has multiple strengths. The characterization of CHIKV infection histories, the documentation of signs and symptoms in a systematic manner, and the use of statistical methods for clustered data allowed us to derive valid inferences, particularly of the S:A ratio. Importantly, the repeated sample collection and sign/symptom questionnaires also enabled the identification of true, asymptomatic infections, minimizing bias due to incomplete recall of signs and symptoms. Finally, the study design also allowed us to estimate associations for both person- and household-level risk factors of infection.
Our study has several limitations. First, the study population was not selected randomly; thus, the results from our study sample may not generalize to the epidemic experience of Managua’s population. Second, the overall sample size was modest, which prompted us to use a variable selection procedure in the risk factor analysis and a literature-based Bayesian approach to reduce the variability in our frequentist estimate of the proportion of asymptomatic CHIKV infections. The small sample size, combined with the clustered data structure, also prevented us from validly comparing the proportions of asymptomatic infections among household contact subgroups. Third, the study occurred past the period of highest incidence (40 cases per week in the Pediatric Dengue Cohort Study [PDCS]), limiting the amount of incident infections that we could observe. Nevertheless, the study period coincided with a weekly average of 22 new cases being diagnosed within the PDCS, indicating that the study was conducted during a period of high CHIKV transmission in the study area. Fourth, the total number of populations that contributed to the Bayesian analyses (n = 21) was relatively small, limiting the precision of the prior distributions. However, the small number of studies identified through the systematic literature search reflects how understudied CHIKV is relative to other arboviruses such as DENV and ZIKV. Fifth, the studies identified through the literature search each used a slightly different way of classifying a chikungunya case, which could have introduced information bias into the Bayesian analysis. Sixth, we were unable to identify any chikungunya study documenting an outbreak caused by the West African CHIKV lineage that met our inclusion and exclusion criteria, and we could identify only one non-IOL ESCA lineage study that met our criteria. Although most studies of inapparent CHIKV infections have examined ECSA lineage outbreaks (24), our literature search suggests that the West African lineage and non-IOL strains of the ECSA lineage remain understudied, even relative to the Asian CHIKV lineage and the IOL strain of the ECSA CHIKV lineage.
We find epidemiological evidence for lineage-specific differences in the probability of inapparent CHIKV infection and suggest lineage-specific distributions that should replace the overly narrow 3 to 28% range. Future studies should report lineage-specific proportions of asymptomatic or inapparent CHIKV-infected participants and provide statistically appropriate estimates of uncertainty. In addition, future studies should report risk differences for chikungunya disease manifestations; the vast majority of articles that we identified here did not report risk differences. Therefore, we focused on the proportion of asymptomatic/inapparent infections to enable a direct comparison between our findings and those in the literature. Our data provide a baseline for future studies to estimate CHIKV S:A ratios in different settings and should help international organizations and ministries of health better plan for future chikungunya epidemics.
MATERIALS AND METHODS
Ethics statement.
This study was approved by the Institutional Review Boards (IRBs) of the University of California, Berkeley, and the Nicaraguan Ministry of Health. Adult participants and parents or legal guardians of participating children provided written consent. Children aged 6 to 11 years provided oral assent. Children aged 12 to 17 years provided written assent.
Population and study design.
(i) Study site and recruitment. This study was based in 17 of the neighborhoods that constitute the catchment area of the Health Center Sócrates Flores Vivas (HCSFV) in District II of Managua, the low- to middle-income site of the Pediatric Dengue Cohort Study (PDCS) (49). In the HCSFV catchment area, 53% of the population is female and ∼30% of the population is <15 years of age (50). Approximately 95% of homes in the area have potable water and sewage services (10), but because of intermittent water service, many residents store water on-site in barrels or other containers, introducing mosquito breeding sites into the domestic environment. The geographical extent of the study area and household-specific infection information were visualized using ESRI’s ArcGIS software version 10.3.1 (51) (Fig. 1).
The PDCS is an ongoing study of arboviral infections prospectively monitoring ∼3,700 children aged 2 to 14 years. In October and November of 2015, during an explosive chikungunya epidemic (14), residents of the HCSFV catchment area and PDCS participants were eligible to participate in the study. When individuals presented to the HCSFV with chikungunya-like signs/symptoms and were confirmed as being CHIKV positive by real-time reverse transcription-PCR (rRT-PCR) (52), study personnel visited their homes 1 to 2 days later to invite them to participate in the study as an index case. At that time, household members were invited to participate as contacts.
(ii) Study design and sample collection. During the initial home visit (day 1), individual-level data were collected on participants’ age, sex, occupation, and educational level; household-level data were also collected. Data on household size, number of rooms, and number and types of electronic appliances were also collected, along with its Global Positioning System coordinates. On day 1 and on subsequent household visits, a sign-and-symptom questionnaire was administered to index cases and their household contacts, who were also asked to provide blood samples for testing by rRT-PCR, an IgM capture ELISA (IgM-ELSA), and an inhibition ELISA (IE) (Table 8). If the day 1 rRT-PCR test was positive for CHIKV, household contacts were visited around day 14 for a second blood sample to test via IgM-ELISA and IE; otherwise, sera from contacts were collected for testing around day 35. The additional 21 days allowed for initially rRT-PCR-negative contacts to develop an incident infection. Seroconversion from negative to positive using the IgM-ELISA or IE on paired samples indicates an incident infection. Regardless of the day 1 rRT-PCR result, index cases were visited only on day 14 for collection of additional serum.
TABLE 8.
Visit days and symptom data/sample collection for study participants (n = 296) in Managua, Nicaragua, during October to November 2015
| Participant category | Description of data/sample collectionc |
||||
|---|---|---|---|---|---|
| 1–2 days before the day 1 visit | Day 1 visit (day of enrollment) | Day 3 visit | Day 14 visit | Day 35 visit | |
| Index cases (n = 60) | Visited HCSFV and received a diagnosis of symptomatic chikungunya | Reported signs/symptoms in previous 72 h, gave a blood sample | Reported signs/symptoms in previous 72 h | Reported signs/symptoms since the day 3 visit, gave a blood sample | |
| Household contacts (n = 236) | Reported signs/symptoms in previous 72 h, gave a blood sample | Reported signs/symptoms in previous 72 h | Reported signs/symptoms since the day 3 visit, gave a blood samplea | Reported signs/symptoms since the day 3 visit, gave a blood sampleb | |
Among rRT-PCR-positive contacts on the day 1 sample.
Among rRT-PCR-negative contacts on the day 1 sample.
Abbreviations: HCSFV, Health Center Sócrates Flores Vivas; rRT-PCR, real-time reverse transcription-PCR.
Participant classification.
(i) CHIKV infections. To ascertain infection history and for comparative analyses, household contacts were classified on the basis of laboratory test results (Fig. 2). Contacts who were negative by all rRT-PCR tests, IgM ELISAs, and IE tests on all samples were designated as the all-negative reference group. Any contact whose day 1 sample was negative for CHIKV by rRT-PCR but positive by IE was classified as a having had a previous CHIKV infection, including infections that were also positive by an IgM-ELISA. We classified contacts whose day 1 sample was CHIKV positive by an IgM-ELISA but negative by rRT-PCR and IE as having had very recent infections. We classified those contacts whose day 1 sample tested positive by rRT-PCR as having active infections. Based on viremia dynamics, very recently infected individuals were likely infected within a week of those with active infections. We defined those with incident infections as household contacts who were negative by rRT-PCR, IgM-ELISA, and IE at baseline and tested positive by IgM-ELISA on either their day 14 or day 35 sample. We defined those with equivocal infections as contacts who met the definition of either an active infection or incident infection but were missing the day 1 rRT-PCR sample.
Collectively, active, incident, and equivocal infections are referred to as active and incident infections. Analyses examining different outcomes considered different subsets of at-risk participants. When we considered incident infection as the outcome of interest, the at-risk group was restricted to household contacts without any serological or molecular evidence of a past or ongoing CHIKV infection at enrollment (i.e., the all-negative contacts and those with incident infections). When we considered the broader outcome of CHIKV infection after or slightly before study initiation, the at-risk group was restricted to household contacts not classified as having had a previous CHIKV infection at enrollment (i.e., the all-negative contacts and those with recent, active, incident, and equivocal infections).
(ii) Disease presentation. We classified CHIKV-infected participants as experiencing classic chikungunya disease if they reported ≥2 of the following signs/symptoms during the monitoring period: fever, joint pain, headache, muscle aches, retro-orbital pain, rash, leukopenia, conjunctivitis, diarrhea, vomiting, and/or hemorrhagic manifestations (petechia, epistaxis, gingivitis, hematuria, or melena). Those with undifferentiated fever were defined as only experiencing fever. Participants with classical chikungunya or undifferentiated fever were considered to have a symptomatic CHIKV infection (i.e., they met the case definition). Otherwise, CHIKV-infected participants were classified as having an asymptomatic infection. Acute signs and symptoms of chikungunya generally coincide with the viremic period (18, 53); therefore, the S:A ratio was calculated among active and incident infections.
Laboratory methods.
Upon collection, all blood samples were transported to the National Virology Laboratory and processed and/or stored at −80°C. Samples from potential index cases and day 1 samples from household contacts were tested by a pan-DENV-CHIKV rRT-PCR assay (52). Paired day 1 and day 14/35 samples were tested by an in-house IgM-ELISA and an inhibition ELISA (55). The IgM-ELISA has a sensitivity of 95.7% (95% confidence interval [CI], 91.3%, 98.2%) and a specificity of 89.8% (95% CI, 82.0%, 95.0%) compared to the CDC CHIKV rRT-PCR assay (55). The IE has a sensitivity and a specificity of 94.9% (95% CI, 88.6%, 98.3%) (55).
Statistical analyses.
All data analysis was done in R v3.4.2 (56).
(i) Demographics and symptom occurrence. Differences in participants’ age and sex were examined using the nonparametric two-sample Kolmogorov-Smirnov test (57) and the modern N-1 Pearson chi-square test (58), respectively. Differences by age and sex were also examined by the more traditional Wilcoxon rank-sum test and Fisher’s exact test. All-negative household contacts served as the reference group for comparisons of age and sex distributions to other groups of contacts.
To calculate the risk difference for a symptomatic outcome, the all-negative contacts were considered the unexposed group, and the active and incident infections were considered the exposed group. The risk difference was calculated as the difference between the proportion of the exposed group (active and incident infection) meeting the case definition (either classic chikungunya or undifferentiated fever) and the proportion of the unexposed group (all-negative contacts) meeting the case definition. We used a binomial linear model, estimated within a generalized estimating equations (GEE) framework, to adjust for age and sex and account for household-level clustering. The GEE model assumed an exchangeable correlation matrix, used the household identifier (ID) variable as the clustering unit, fixed the scale parameter to 1, and used robust variance estimators (59–62).
Cooccurrence of different signs and symptoms among CHIKV-positive contacts was assessed using a hierarchical clustering dendrogram. The dendrogram was based on the Manhattan distance matrix of sign/symptom occurrence and constructed based on the modern Ward minimum-variance method (63), as previously described (20).
(ii) Trend analysis. We used a logistic generalized additive model (GAM) to characterize the association of age with CHIKV infection and disease occurrence (64). GAMs are flexible semiparametric approaches that, unlike traditional generalized linear models, can capture nonlinear trends in the data. The GAMs were estimated using the mgcv R package (64) with thin-plate regression splines (65). Smoothing parameters were estimated with generalized cross-validation, and the basis dimensions were tested to ensure appropriate smoothing (66, 67). To account for the household-based sampling design, we used the nonparametric bootstrap (68) to estimate a 95% pointwise confidence band instead of the default confidence band provided by the package. Data were bootstrapped at the household level 10,000 times.
(iii) Risk factor analysis. A literature search (described below) and a variable selection procedure were used to select potential predictors of infection and symptomatic outcome. The following variables were included as potential predictors of CHIKV infection after or slightly before study initiation (model 1): age, sex, educational attainment, number of previous infections, externally piped water status, hours without running water, tires in the yard, high frequency of abatement use, high frequency of fumigation, high frequency of municipal trash collection, number of household electronics, ownership of a vehicle, and a crowding measure (household size/total number of rooms in the household). A high frequency of abatement use/fumigation or trash collection was defined a priori as ≥4 times per year or per week, respectively; all variables except age, sex, and educational attainment were at the household level. When strictly examining incident infections as the outcome of interest (model 2), all model 1 variables were considered, except municipal trash collection, as all 104 at-risk persons lived in households with a high level of trash collection services. Model 2 was estimated among the all-negative group and the incident infections. Model 3 considered symptomatic status among all active and incident infections. Its candidate set of predictors included age, sex, the sum of electronics in the household, vehicle ownership, and the crowding measure. Model 4 had the same predictor set but considered only symptomatic status among incident infections.
Because the candidate sets of predictors were large relative to the study sample size, the candidate sets were reduced using a best-subset variable-selection procedure (69) based on Akaike’s information criterion corrected for small sample sizes (70). We estimated multivariate-adjusted risk ratios (aRRs) using modified Poisson regression (59). The modified Poisson model used the same settings as for the binomial linear model described above.
(iv) Variability of the S:A ratio. To account for the clustered data structure, we calculated bias-corrected and accelerated (BCa) CIs for the natural log of the S:A ratio using the nonparametric cluster bootstrap, resampling 10,000 times at the household level (71); estimates were then exponentiated to return them to the natural scale. The S:A ratio was log transformed to attain a bootstrap sampling distribution that was more normal. To ensure that BCa CIs provided reliable inference, we compared the underlying sampling distributions to normal curves, fitting their means and standard deviations to those of the sampling distribution. Due to small sample sizes, no statistical tests comparing the probabilities of asymptomatic infection among different subsets of participants (e.g., males versus females, active versus incident infection, and children versus adults) were performed. To explore the variability of our estimate for the probability of asymptomatic infection, a BCa-based confidence interval function (72–74) was constructed using the boot package (75, 76). To assess the nature of correlation within our household-clustered data, we estimated an intracluster correlation coefficient (ICC) using the ICC R package and its ICCest() function (77) and constructed a CI function for the overall proportion of asymptomatic infections using BCa CIs (taking clustering into account) and exact CIs (assuming independent data).
(v) Literature-based Bayesian analysis. In Bayesian analyses, the estimate of a particular parameter (obtained from the study’s data) is statistically combined with existing knowledge regarding that parameter (the prior distribution) to produce an updated (posterior) distribution for the parameter of interest in the study. Literature-based priors thus summarize, in the form of a statistical distribution, the extent of real knowledge regarding a particular parameter, as reported in the literature. In Bayesian analyses, the median of a posterior distribution is taken as the updated value of the parameter of interest. To make the priors representative of published knowledge, we conducted a systematic search of the literature using PRISMA guidelines (78) to identify previously published studies documenting the proportion of inapparent CHIKV infections (the parameter of interest) in previously CHIKV-naive populations. For the search, we selected only chikungunya outbreak reports on initially CHIKV-naive samples, as the S:A ratio that we calculated was among initially CHIKV-naive samples. This restriction also helped to ensure that the reports that we considered were fairly homogeneous with respect to CHIKV infection history. These studies informed the estimation of Bayesian prior distributions.
As a sensitivity analysis, we estimated a variety of prior distributions using different subsets of identified studies, described in detail below. We characterized the behavior of different priors using measures of centrality, multiple statistical measures/metrics, and Monte Carlo resampling simulations (10,000 random draws per distribution per scenario) (79). To estimate 95% BCa CIs for Monte Carlo comparisons of the Asian and ECSA lineage prior distributions, 10,000 random draws were taken from each distribution, comparisons were made, and the entire procedure was nonparametrically bootstrapped 10,000 times, for a total of 200,000,000 Monte Carlo simulations. The calculated statistical measures/metrics were the Kullback-Leibler divergence (which compares a given distribution to an expected distribution), the Hellinger distance (which measures probabilistic distance between distributions) (80), and the Bhattacharyya coefficient (which measures overlap between distributions). For these measures/metrics, priors were compared to one constructed using all identified studies reporting proportions of inapparent CHIKV infection between 3% and 28%, as this prior represented the strongest literature-based statistical expression for the expected distribution of inapparent infections reported in literature reviews and other authoritative sources (15–19).
(vi) Framework of the systematic literature search. Because we sought to estimate distributions instead of summarizing the substantive content of the identified reports or an overall effect measure, we adapted the PRISMA guidelines (78) that are meant for systematic reviews and meta-analyses. On 29 June 2017, we downloaded the PubMed database returns using the singular keyword “chikungunya.” No restriction (by time, language, or otherwise) was added to the search. We attempted to be as general as possible, as an initial search had revealed several relevant articles that did not include terms such as “serosurvey,” “serology,” or “asymptomatic.”
In all, 3,480 articles were returned by the PubMed search (Fig. 8). We first screened the returns by their title and abstract, being overly generous so as not to exclude potentially relevant studies. We excluded 3,367 articles on this basis, and 113 articles were selected for a full text review. The title/abstract screen was performed twice to increase the probability of finding relevant articles. After reviewing the text of the above-mentioned articles, 21 articles were deemed relevant to our search, including one (14) that included two populations of interest. Whenever more than one study reported on the same outbreak, we selected the most thorough one to include in our analysis; in these cases, all reports were reviewed for information that might supplement data in the most thorough report. (For example, the report by Galatas et al. [21] is a more thorough report of the same Cambodian outbreak than the one by Ly et al. [81]; however, Ly et al. include more information on the proportion of inapparent infections, so those data are presented as if they were reported by Galatas et al.) Four of the 21 articles were removed because they reported on the same chikungunya epidemic. Thirty-five review articles and 159 citations of the 21 reports were reviewed to identify pertinent articles, and 2 additional articles were selected in this manner. The literature was also searched by contacting the corresponding authors of selected articles and asking them if they knew of similar articles; one article was selected as a result of this additional search. For a third round of assessing the literature, we further examined the citations of the above-mentioned 159 articles and 35 review articles but did not identify any further relevant articles.
FIG 8.
Flow diagram of the systematic literature search.
(vii) Inclusion and exclusion criteria. We used exclusion and inclusion criteria to select the final articles. The overarching goals of the exclusion and inclusion criteria were to (i) select studies that documented the laboratory-confirmed percentage of CHIKV infections and the percentage of inapparent CHIKV infections within infected populations; (ii) ensure that the data reported concerned a recent epidemic in a population/sample that had been, mostly or exclusively, CHIKV naive in the preepidemic period; and (iii) include sufficient detail regarding relevant aspects of the study as to be informative with respect to the goals of our study. We focused on studies that described the epidemiology of recent outbreaks in populations that had been CHIKV naive before the epidemic because the S:A ratio that we calculated was estimated among persons who had not been previously infected by CHIKV. Thus, our focus was to ensure the comparability of our study data with literature-based data that would help to estimate prior distributions.
We excluded articles that were case reports, summarized infections in travelers (as their infection experience may not reflect that of people, living in locations of interest for an extended period of time, who lived through a chikungunya outbreak), hospital-based reports, studies that exclusively reported serology results for symptomatic participants, articles that did not examine the seroprevalence of CHIKV after a recent chikungunya outbreak, studies in areas where CHIKV is endemic that relied on IgG-ELISAs as the only measure of postoutbreak infection, case-control studies (which distort the probabilities of interest by virtue of the sampling method), and animal studies. We included articles for consideration of a full text review if they were reviews of chikungunya, seroprevalence studies, outbreak reports, national surveillance reports, articles discussing the burden of chikungunya disease in any population or cohort, or discussions of the epidemiology of chikungunya. To be included in the final set of considered studies, articles needed to confirm CHIKV infection in their participants by serological or molecular methods and reliably report the proportion of CHIKV-infected participants who experienced signs and symptoms consistent with a CHIKV infection.
(viii) Data extracted from identified studies. We extracted the following information for each identified study: author(s) and year of publication, when the symptom information was collected relative to the outbreak, the location of the outbreak, the virus lineage/strain responsible for the outbreak, the mosquito vector responsible for the outbreak, the proportion of CHIKV-positive participants, the proportion of inapparent CHIKV infections, and the sampling method. Whenever some of these data were not available or when a question arose regarding particular aspects of the study, efforts were made to contact the first author, senior author, and/or corresponding author to obtain the necessary information via e-mail correspondence. Virus lineage and the vector, when not explicitly confirmed through sequencing or entomological investigation in the paper, were derived from other published studies that genotyped virus isolates from the outbreak in question, the global compendium of Aedes aegypti and Aedes albopictus occurrence data set (82), or correspondence with the study authors.
(ix) Estimation of the prior and posterior distributions. The probability of inapparent CHIKV infection was assumed to follow a beta distribution, and its hyperparameters were estimated using the ExtDist R package’s (83) numerical optimization of the likelihood method (or the method of moments if the numerical optimization approach failed to converge) after weighting each identified study’s contribution by the number of CHIKV-infected individuals. Because the beta distribution is the conjugate prior for binomial data, the estimated posteriors were also beta distributions.
For the sensitivity analysis, different Bayesian priors were constructed using different subsets of studies to evaluate how, for example, relying on studies in the Americas versus studies outside the Americas would impact the final results. When calculating a prior based on the different vectors, we attributed a study’s data to the most likely vector. Those studies for which the authors or we (based on a review of the global Aedes occurrence compendium data set) deemed both Aedes species to be equally likely vectors for the epidemic were ignored in the calculation of the vector-specific Bayesian priors.
To provide future studies with prior distributions that incorporated data from the literature and this study, we estimated a second set of prior distributions that included findings from this study in the same data set as the findings from the literature. We estimated highest-density credible intervals (HDCIs) using the HDI R package (84).
Data availability.
Data can be shared with outside investigators following approval from the University of California, Berkeley, IRB and Committee for the Protection of Human Subjects. Please contact E.H. to arrange for data access.
ACKNOWLEDGMENTS
We give special thanks to the participating study families, who took time out of their daily lives to contribute to our work. We thank Jairo Carey, William Avilés Monterrey, Sonia Argüello, and Douglas Elizondo for assistance in managing and processing much of the underlying data. We thank Josefina Coloma, Jennifer Ahern, Ellie Matthay, Holly Elser, Brooke Read, Emily Yette, and Art Reingold for feedback and substantial input on prior drafts of the manuscript. We give particular thanks to Patrick Bradshaw, Dana Goin, and Caleb Miles for providing expert consultation on several analyses presented here. Finally, we give special thanks to the phenomenal study team based at the Centro de Salud Sócrates Flores Vivas and the National Virology Laboratory in the Centro Naciónal de Diagnóstico y Referencia of the Nicaraguan Ministry of Health and the Sustainable Sciences Institute in Nicaragua for their years of dedicated hard work.
No author has a commercial or other association that might pose a conflict of interest.
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health via grants P01 AI106695 (to E.H.) and R01 AI099631 (to A.B.).
REFERENCES
- 1.Centers for Disease Control and Prevention. 2015. Chikungunya virus: clinical evaluation and disease. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 2.Nkoghe D, Kassa RF, Caron M, Grard G, Mombo I, Bikie B, Paupy C, Becquart P, Bisvigou U, Leroy EM. 2012. Clinical forms of Chikungunya in Gabon, 2010. PLoS Negl Trop Dis 6:e1517. doi: 10.1371/journal.pntd.0001517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teo T-H, Her Z, Tan JJL, Lum F-M, Lee WWL, Chan Y-H, Ong R-Y, Kam Y-W, Leparc-Goffart I, Gallian P, Rénia L, de Lamballerie X, Ng LFP. 2015. Caribbean and La Réunion chikungunya virus isolates differ in their capacity to induce proinflammatory Th1 and NK cell responses and acute joint pathology. J Virol 89:7955–7969. doi: 10.1128/JVI.00909-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner J, Anraku I, Le TT, Larcher T, Major L, Roques P, Schroder WA, Higgs S, Suhrbier A. 2010. Chikungunya virus arthritis in adult wild-type mice. J Virol 84:8021–8032. doi: 10.1128/JVI.02603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langsjoen RM, Haller SL, Roy CJ, Vinet-Oliphant H, Bergren NA, Erasmus JH, Livengood JA, Powell TD, Weaver SC, Rossi SL. 2018. Chikungunya virus strains show lineage-specific variations in virulence and cross-protective ability in murine and nonhuman primate models. mBio 9:e02449-17. doi: 10.1128/mBio.02449-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsetsarkin KA, Chen R, Weaver SC. 2016. Interspecies transmission and chikungunya virus emergence. Curr Opin Virol 16:143–150. doi: 10.1016/j.coviro.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiferaw B, Lam P, Tuthill S, Choudhry H, Syed S, Ahmed S, Yasmin T. 2015. The chikungunya epidemic: a look at five cases. IDCases 2:89–91. doi: 10.1016/j.idcr.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization. 2016. Chikungunya fact sheet. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 9.Pan American Health Organization. 2017. Number of reported cases of chikungunya fever in the Americas, by country or territory, 2016 cumulative cases. Pan American Health Organization, Washington, DC. [Google Scholar]
- 10.Kuan G, Ramirez S, Gresh L, Ojeda S, Melendez M, Sanchez N, Collado D, Garcia N, Mercado JC, Gordon A, Balmaseda A, Harris E. 2016. Seroprevalence of anti-chikungunya virus antibodies in children and adults in Managua, Nicaragua, after the first chikungunya epidemic, 2014-2015. PLoS Negl Trop Dis 10:e0004773. doi: 10.1371/journal.pntd.0004773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balmaseda A, Gordon A, Gresh L, Ojeda S, Saborio S, Tellez Y, Sanchez N, Kuan G, Harris E. 2016. Clinical attack rate of chikungunya in a cohort of Nicaraguan children. Am J Trop Med Hyg 94:397–399. doi: 10.4269/ajtmh.15-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C, Saborio S, Gresh L, Eswarappa M, Wu D, Fire A, Parameswaran P, Balmaseda A, Harris E. 2016. Chikungunya virus sequences across the first epidemic in Nicaragua, 2014-2015. Am J Trop Med Hyg 94:400–403. doi: 10.4269/ajtmh.15-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa-da-Silva AL, Ioshino RS, Petersen V, Lima AF, Cunha MDP, Wiley MR, Ladner JT, Prieto K, Palacios G, Costa DD, Suesdek L, Zanotto PMDA, Capurro ML. 2017. First report of naturally infected Aedes aegypti with chikungunya virus genotype ECSA in the Americas. PLoS Negl Trop Dis 11:e0005630. doi: 10.1371/journal.pntd.0005630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon A, Gresh L, Ojeda S, Chowell-Puente G, Gonzalez K, Sanchez N, Saborio S, Mercado JC, Kuan G, Balmaseda A, Harris E. 2018. Differences in transmission and disease severity between two successive waves of chikungunya. Clin Infect Dis 67:1760–1767. doi: 10.1093/cid/ciy356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yactayo S, Staples JE, Millot V, Cibrelus L, Ramon-Pardo P. 2016. Epidemiology of Chikungunya in the Americas. J Infect Dis 214:S441–S445. doi: 10.1093/infdis/jiw390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiberville S-D, Moyen N, Dupuis-Maguiraga L, Nougairede A, Gould EA, Roques P, de Lamballerie X. 2013. Chikungunya fever: epidemiology, clinical syndrome, pathogenesis and therapy. Antiviral Res 99:345–370. doi: 10.1016/j.antiviral.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staples JE, Breiman RF, Powers AM. 2009. Chikungunya fever: an epidemiological review of a re‐emerging infectious disease. Clin Infect Dis 49:942–948. doi: 10.1086/605496. [DOI] [PubMed] [Google Scholar]
- 18.Silva LA, Dermody TS. 2017. Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. J Clin Invest 127:737–749. doi: 10.1172/JCI84417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staples JE, Hills SL, Powers AM. 2017. Chapter 3: Chikungunya In Brunette GW (ed), CDC yellow book 2018: health information for international travel. https://wwwnc.cdc.gov/travel/yellowbook/2018/infectious-diseases-related-to-travel/chikungunya. [Google Scholar]
- 20.Bloch D, Roth NM, Caraballo EV, Muñoz-Jordan J, Hunsperger E, Rivera A, Pérez-Padilla J, Rivera Garcia B, Sharp TM. 2016. Use of household cluster investigations to identify factors associated with chikungunya virus infection and frequency of case reporting in Puerto Rico. PLoS Negl Trop Dis 10:e0005075. doi: 10.1371/journal.pntd.0005075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galatas B, Ly S, Duong V, Baisley K, Nguon K, Chan S, Huy R, Ly S, Sorn S, Som L, Buchy P, Tarantola A. 2016. Long-lasting immune protection and other epidemiological findings after chikungunya emergence in a Cambodian rural community, April 2012. PLoS Negl Trop Dis 10:e0004281. doi: 10.1371/journal.pntd.0004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakkhara P, Chongsuvivatwong V, Thammapalo S. 2013. Risk factors for symptomatic and asymptomatic chikungunya infection. Trans R Soc Trop Med Hyg 107:789–796. doi: 10.1093/trstmh/trt083. [DOI] [PubMed] [Google Scholar]
- 23.Cunha RV, Trinta KS, Montalbano CA, Sucupira MVF, de Lima MM, Marques E, Romanholi IH, Croda J. 2017. Seroprevalence of chikungunya virus in a rural community in Brazil. PLoS Negl Trop Dis 11:e0005319. doi: 10.1371/journal.pntd.0005319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon I-K, Alera MT, Lago CB, Tac-An IA, Villa D, Fernandez S, Thaisomboonsuk B, Klungthong C, Levy JW, Velasco JM, Roque VG, Salje H, Macareo LR, Hermann LL, Nisalak A, Srikiatkhachorn A. 2015. High rate of subclinical chikungunya virus infection and association of neutralizing antibody with protection in a prospective cohort in the Philippines. PLoS Negl Trop Dis 9:e0003764. doi: 10.1371/journal.pntd.0003764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srikiatkhachorn A, Alera MT, Lago CB, Tac-An IA, Villa D, Fernandez S, Thaisomboonsuk B, Klungthong C, Levy JW, Velasco JM, Roque VG, Nisalak A, Macareo LR, Yoon I-K. 2016. Resolution of a chikungunya outbreak in a prospective cohort, Cebu, Philippines, 2012–2014. Emerg Infect Dis 22:1852–1854. doi: 10.3201/eid2210.160729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gay N, Rousset D, Huc P, Matheus S, Rosine J, Ledrans M, Cassadou S, Noël H. 2016. Seroprevalence of Asian lineage chikungunya virus infection on Saint Martin Island, 7 months after the 2013 emergence. Am J Trop Med Hyg 94:393–396. doi: 10.4269/ajtmh.15-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sergon K, Njuguna C, Kalani R, Ofula V, Onyango C, Konongoi LS, Bedno S, Burke H, Dumilla AM, Konde J, Njenga MK, Sang R, Breiman RF. 2008. Seroprevalence of Chikungunya virus (CHIKV) infection on Lamu Island, Kenya, October 2004. Am J Trop Med Hyg 78:333–337. doi: 10.4269/ajtmh.2008.78.333. [DOI] [PubMed] [Google Scholar]
- 28.Manica M, Guzzetta G, Poletti P, Filipponi F, Solimini A, Caputo B, Della Torre A, Rosà R, Merler S. 2017. Transmission dynamics of the ongoing chikungunya outbreak in central Italy: from coastal areas to the metropolitan city of Rome, summer 2017. Euro Surveill 22(44):pii=17-00685. doi: 10.2807/1560-7917.ES.2017.22.44.17-00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beckett CG, Kosasih H, Faisal I, Nurhayati, Tan R, Widjaja S, Listiyaningsih E, Ma’roef C, Wuryadi S, Bangs MJ, Samsi TK, Yuwono D, Hayes CG, Porter KR. 2005. Early detection of dengue infections using cluster sampling around index cases. Am J Trop Med Hyg 72:777–782. doi: 10.4269/ajtmh.2005.72.777. [DOI] [PubMed] [Google Scholar]
- 30.Queyriaux B, Simon F, Grandadam M, Michel R, Tolou H, Boutin J-P. 2008. Clinical burden of chikungunya virus infection. Lancet Infect Dis 8:2–3. doi: 10.1016/S1473-3099(07)70294-3. [DOI] [PubMed] [Google Scholar]
- 31.Retuya J. 1998. Chikungunya fever outbreak in an agricultural village in Indang, Cavite, Philippines. Philipp J Microbiol Infect Dis 27:93–96. [Google Scholar]
- 32.Sissoko D, Moendandze A, Malvy D, Giry C, Ezzedine K, Solet JL, Pierre V. 2008. Seroprevalence and risk factors of chikungunya virus infection in Mayotte, Indian Ocean, 2005-2006: a population-based survey. PLoS One 3:e3066. doi: 10.1371/journal.pone.0003066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simmons G, Brès V, Lu K, Liss NM, Brambilla DJ, Ryff KR, Bruhn R, Velez E, Ocampo D, Linnen JM, Latoni G, Petersen LR, Williamson PC, Busch MP. 2016. High incidence of chikungunya virus and frequency of viremic blood donations during epidemic, Puerto Rico, USA, 2014. Emerg Infect Dis 22:1221–1228. doi: 10.3201/eid2207.160116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langsjoen RM, Rubinstein RJ, Kautz TF, Auguste AJ, Erasmus JH, Kiaty-Figueroa L, Gerhardt R, Lin D, Hari KL, Jain R, Ruiz N, Muruato AE, Silfa J, Bido F, Dacso M, Weaver SC. 2016. Molecular virologic and clinical characteristics of a chikungunya fever outbreak in La Romana, Dominican Republic, 2014. PLoS Negl Trop Dis 10:e0005189. doi: 10.1371/journal.pntd.0005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balmaseda A, Standish K, Mercado JC, Matute JC, Tellez Y, Saborío S, Hammond SN, Nuñez A, Avilés W, Henn MR, Holmes EC, Gordon A, Coloma J, Kuan G, Harris E. 2010. Trends in patterns of dengue transmission over 4 years in a pediatric cohort study in Nicaragua. J Infect Dis 201:5–14. doi: 10.1086/648592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Endy TP, Anderson KB, Nisalak A, Yoon I-K, Green S, Rothman AL, Thomas SJ, Jarman RG, Libraty DH, Gibbons RV. 2011. Determinants of inapparent and symptomatic dengue infection in a prospective study of primary school children in Kamphaeng Phet, Thailand. PLoS Negl Trop Dis 5:e975. doi: 10.1371/journal.pntd.0000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balmaseda A, Hammond SN, Pérez L, Tellez Y, Saborío SI, Mercado JC, Cuadra R, Rocha J, Pérez MA, Silva S, Rocha C, Harris E. 2006. Serotype-specific differences in clinical manifestations of dengue. Am J Trop Med Hyg 74:449–456. doi: 10.4269/ajtmh.2006.74.449. [DOI] [PubMed] [Google Scholar]
- 38.Fried JR, Gibbons RV, Kalayanarooj S, Thomas SJ, Srikiatkhachorn A, Yoon I-K, Jarman RG, Green S, Rothman AL, Cummings DAT. 2010. Serotype-specific differences in the risk of dengue hemorrhagic fever: an analysis of data collected in Bangkok, Thailand from 1994 to 2006. PLoS Negl Trop Dis 4:e617. doi: 10.1371/journal.pntd.0000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yung C-F, Lee K-S, Thein T-L, Tan L-K, Gan VC, Wong JGX, Lye DC, Ng L-C, Leo Y-S. 2015. Dengue serotype-specific differences in clinical manifestation, laboratory parameters and risk of severe disease in adults, Singapore. Am J Trop Med Hyg 92:999–1005. doi: 10.4269/ajtmh.14-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nisalak A, Endy TP, Nimmannitya S, Kalayanarooj S, Thisayakorn U, Scott RM, Burke DS, Hoke CH, Innis BL, Vaughn DW. 2003. Serotype-specific dengue virus circulation and dengue disease in Bangkok, Thailand from 1973 to 1999. Am J Trop Med Hyg 68:191–202. doi: 10.4269/ajtmh.2003.68.191. [DOI] [PubMed] [Google Scholar]
- 41.Simonin Y, van Riel D, Van de Perre P, Rockx B, Salinas S. 2017. Differential virulence between Asian and African lineages of Zika virus. PLoS Negl Trop Dis 11:e0005821. doi: 10.1371/journal.pntd.0005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anfasa F, Siegers JY, van der Kroeg M, Mumtaz N, Stalin Raj V, de Vrij FMS, Widagdo W, Gabriel G, Salinas S, Simonin Y, Reusken C, Kushner SA, Koopmans MPG, Haagmans B, Martina BEE, van Riel D. 2017. Phenotypic differences between Asian and African lineage Zika viruses in human neural progenitor cells. mSphere 2:e00292-17. doi: 10.1128/mSphere.00292-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simonin Y, Loustalot F, Desmetz C, Foulongne V, Constant O, Fournier-Wirth C, Leon F, Molès J-P, Goubaud A, Lemaitre J-M, Maquart M, Leparc-Goffart I, Briant L, Nagot N, Van de Perre P, Salinas S. 2016. Zika virus strains potentially display different infectious profiles in human neural cells. EBioMedicine 12:161–169. doi: 10.1016/j.ebiom.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang F, Wang H-J, Wang Q, Liu Z-Y, Yuan L, Huang X-Y, Li G, Ye Q, Yang H, Shi L, Deng Y-Q, Qin C-F, Xu Z. 2017. American strain of Zika virus causes more severe microcephaly than an old Asian strain in neonatal mice. EBioMedicine 25:95–105. doi: 10.1016/j.ebiom.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clapham HE, Cummings DAT, Johansson MA. 2017. Immune status alters the probability of apparent illness due to dengue virus infection: evidence from a pooled analysis across multiple cohort and cluster studies. PLoS Negl Trop Dis 11:e0005926. doi: 10.1371/journal.pntd.0005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li R, Ding J, Ding G, Fan X, He Y, Wang X, Zhang H, Ji J, Li H. 2017. Zika virus infections, a review. Radiol Infect Dis 4:88–93. doi: 10.1016/j.jrid.2017.01.002. [DOI] [Google Scholar]
- 47.Adebote AD, Oniye JS, Ndams SI, Nache KM. 2006. The breeding of mosquitoes (Diptera: Culicidae) in peridomestic containers and implication in yellow fever transmission in villages around Zaria, Northern Nigeria. J Entomol 3:180–188. doi: 10.3923/je.2006.180.188. [DOI] [Google Scholar]
- 48.Enserink M. 2008. Entomology. A mosquito goes global. Science 320:864–866. doi: 10.1126/science.320.5878.864. [DOI] [PubMed] [Google Scholar]
- 49.Kuan G, Gordon A, Aviles W, Ortega O, Hammond SN, Elizondo D, Nunez A, Coloma J, Balmaseda A, Harris E. 2009. The Nicaraguan Pediatric Dengue Cohort Study: study design, methods, use of information technology, and extension to other infectious diseases. Am J Epidemiol 170:120–129. doi: 10.1093/aje/kwp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reyes M, Mercado JC, Standish K, Matute JC, Ortega O, Moraga B, Avilés W, Henn MR, Balmaseda A, Kuan G, Harris E. 2010. Index cluster study of dengue virus infection in Nicaragua. Am J Trop Med Hyg 83:683–689. doi: 10.4269/ajtmh.2010.10-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.ESRI. 2015. ArcGIS for desktop, 10.3.1. Environmental Systems Research Institute, Redlands, CA. [Google Scholar]
- 52.Waggoner JJ, Ballesteros G, Gresh L, Mohamed-Hadley A, Tellez Y, Sahoo MK, Abeynayake J, Balmaseda A, Harris E, Pinsky BA. 2016. Clinical evaluation of a single-reaction real-time RT-PCR for pan-dengue and chikungunya virus detection. J Clin Virol 78:57–61. doi: 10.1016/j.jcv.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kam Y-W, Ong EKS, Rénia L, Tong J-C, Ng LFP. 2009. Immuno-biology of Chikungunya and implications for disease intervention. Microbes Infect 11:1186–1196. doi: 10.1016/j.micinf.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 54.Reference deleted. [Google Scholar]
- 55.Saborío Galo S, González K, Téllez Y, García N, Pérez L, Gresh L, Harris E, Balmaseda Á. 2017. Development of in-house serological methods for diagnosis and surveillance of chikungunya. Rev Panam Salud Publica 41:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R Core Team. 2018. R: a language and environment for statistical computing, 3.4.2. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 57.Conover WJ. 1999. Practical nonparametric statistics, 3rd ed John Wiley & Sons Inc, New York, NY. [Google Scholar]
- 58.Campbell I. 2007. Chi-squared and Fisher-Irwin tests of two-by-two tables with small sample recommendations. Stat Med 26:3661–3675. doi: 10.1002/sim.2832. [DOI] [PubMed] [Google Scholar]
- 59.Zou G, Donner A. 2013. Extension of the modified Poisson regression model to prospective studies with correlated binary data. Stat Methods Med Res 22:661–670. doi: 10.1177/0962280211427759. [DOI] [PubMed] [Google Scholar]
- 60.Liang K-Y, Zeger SL. 1986. Longitudinal data analysis using generalized linear models. Biometrika 73:13–22. doi: 10.1093/biomet/73.1.13. [DOI] [Google Scholar]
- 61.White H. 1980. A heteroskedasticity-consistent covariance matrix estimator and a direct test of heteroskedasticity. Econometrica 48:817–830. doi: 10.2307/1912934. [DOI] [Google Scholar]
- 62.Huber PJ. 1967. The behavior of maximum likelihood estimates under nonstandard conditions, p 221–233. In Lecam L, Neyman J (ed), Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability. University of California Press, Berkeley, CA. [Google Scholar]
- 63.Murtagh F, Legendre P. 2014. Ward’s hierarchical agglomerative clustering method: which algorithms implement Ward’s criterion? J Classif 31:274–295. doi: 10.1007/s00357-014-9161-z. [DOI] [Google Scholar]
- 64.Wood SN. 2011. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J R Stat Soc Series B Stat Methodol 73:3–36. doi: 10.1111/j.1467-9868.2010.00749.x. [DOI] [Google Scholar]
- 65.Wood S. 2003. Thin plate regression splines. J R Stat Soc Series B Stat Methodol 65:95–114. doi: 10.1111/1467-9868.00374. [DOI] [Google Scholar]
- 66.Wood SN. 2017. Generalized additive models: an introduction with R, 2nd ed CRC Press, Boca Raton, FL. [Google Scholar]
- 67.Wood SN. 2004. Stable and efficient multiple smoothing parameter estimation for generalized additive models. J Am Stat Assoc 99:673–686. doi: 10.1198/016214504000000980. [DOI] [Google Scholar]
- 68.Efron B, Tibshirani RJ. 1994. An introduction to the bootstrap. Chapman & Hall, London, United Kingdom. [Google Scholar]
- 69.Calcagno V. 2013. glmulti: model selection and multimodel inference made easy. R package version 1.0.7. https://CRAN.R-project.org/package=glmulti. [Google Scholar]
- 70.Sugiura N. 1978. Further analysis of the data by Akaike’s information criterion and the finite corrections. Commun Stat Theory Methods 7:13–26. doi: 10.1080/03610927808827599. [DOI] [Google Scholar]
- 71.Efron B. 1987. Better bootstrap confidence intervals. J Am Stat Assoc 82:171–185. doi: 10.1080/01621459.1987.10478410. [DOI] [Google Scholar]
- 72.Poole C. 1987. Beyond the confidence interval. Am J Public Health 77:195–199. doi: 10.2105/AJPH.77.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sullivan KM, Foster DA. 1990. Use of the confidence interval function. Epidemiology 1:39–42. doi: 10.1097/00001648-199001000-00009. [DOI] [PubMed] [Google Scholar]
- 74.Rothman KJ, Greenland S, Lash TL. 2008. Modern epidemiology, 3rd ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 75.Canty A, Ripley B. 2017. boot: Bootstrap R (S-Plus) functions. R package version 1.3-20. [Google Scholar]
- 76.Davidson AC, Hinkley DV. 1997. Bootstrap methods and their applications. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 77.Wolak ME, Fairbairn DJ, Paulsen YR. 2012. Guidelines for estimating repeatability. Methods Ecol Evol 3:129–137. doi: 10.1111/j.2041-210X.2011.00125.x. [DOI] [Google Scholar]
- 78.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group . 2009. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bhattacharyya AK. 1943. On a measure of divergence between two statistical populations defined by their probability distributions. Bull Calcutta Math Soc 35:99–109. [Google Scholar]
- 80.Ernst H. 1909. Neue Begründung der Theorie quadratischer Formen von unendlichvielen Veränderlichen. J Reine Angew Math 136:210–271. [Google Scholar]
- 81.Ly S, Sorn S, Tarantola A, Canier L, Buchy P, Duong V, Chuang I, Newell S, Ly S, Sok T, Somease V, Char MC, Nganaria C, Som L, Roces MC. 2012. Chikungunya outbreak—Cambodia, February-March 2012. MMWR Morb Mortal Wkly Rep 61:737–740. [PubMed] [Google Scholar]
- 82.Kraemer MUG, Sinka ME, Duda KA, Mylne AQN, Shearer FM, Barker CM, Moore CG, Carvalho RG, Coelho GE, Van Bortel W, Hendrickx G, Schaffner F, Elyazar IRF, Teng H-J, Brady OJ, Messina JP, Pigott DM, Scott TW, Smith DL, Wint GRW, Golding N, Hay SI. 2015. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife 4:e08347. doi: 10.7554/eLife.08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu H, Godfrey AJR, Govindaraju K, Pirikahu S. 2015. ExtDist: extending the range of functions for probability distributions. R package version 0.6-3. [Google Scholar]
- 84.Meredith M, Kruschke J. 2016. HDInterval: highest (posterior) density intervals. R package version 0.1.3. [Google Scholar]
- 85.Schwarz NG, Girmann M, Randriamampionona N, Bialonski A, Maus D, Krefis AC, Njarasoa C, Rajanalison JF, Ramandrisoa HD, Randriarison ML, May J, Schmidt-Chanasit J, Rakotozandrindrainy R. 2012. Seroprevalence of antibodies against Chikungunya, Dengue, and Rift Valley fever viruses after febrile illness outbreak, Madagascar. Emerg Infect Dis 18:1780–1786. doi: 10.3201/eid1811.111036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ayu SM, Lai LR, Sam I-C, Chan YF, Hairi NN, Ayob A, Hatim A. 2010. Seroprevalence survey of chikungunya virus in Bagan Panchor, Malaysia. Am J Trop Med Hyg 83:1245–1248. doi: 10.4269/ajtmh.2010.10-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sissoko D, Ezzedine K, Moendandzé A, Giry C, Renault P, Malvy D. 2010. Field evaluation of clinical features during chikungunya outbreak in Mayotte, 2005–2006. Trop Med Int Health 15:600–607. doi: 10.1111/j.1365-3156.2010.02485.x. [DOI] [PubMed] [Google Scholar]
- 88.Moro ML, Gagliotti C, Silvi G, Angelini R, Sambri V, Rezza G, Massimiliani E, Mattivi A, Grilli E, Finarelli AC, Spataro N, Pierro AM, Seyler T, Macini P, Chikungunya Study Group . 2010. Chikungunya virus in north-eastern Italy: a seroprevalence survey. Am J Trop Med Hyg 82:508–511. doi: 10.4269/ajtmh.2010.09-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Manimunda SP, Muruganandam N, Sugunan AP, Shriram AN, Chaitanya IK, Rai SK, Sharma S, Vijayachari P, Sudeep AB, Guruprasad DR. 2010. Outbreak of Chikungunya fever, Dakshina Kannada District, South India, 2008. Am J Trop Med Hyg 83:751–754. doi: 10.4269/ajtmh.2010.09-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leo YS, Chow ALP, Tan LK, Lye DC, Lin L, Ng LC. 2009. Chikungunya outbreak, Singapore, 2008. Emerg Infect Dis 15:836–837. doi: 10.3201/eid1505.081390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gérardin P, Guernier V, Perrau J, Fianu A, Le Roux K, Grivard P, Michault A, de Lamballerie X, Flahault A, Favier F. 2008. Estimating Chikungunya prevalence in La Réunion Island outbreak by serosurveys: two methods for two critical times of the epidemic. BMC Infect Dis 8:99. doi: 10.1186/1471-2334-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sergon K, Yahaya AA, Brown J, Bedja SA, Mlindasse M, Agata N, Allaranger Y, Ball MD, Powers AM, Ofula V, Onyango C, Konongoi LS, Sang R, Njenga MK, Breiman RF. 2007. Seroprevalence of Chikungunya virus infection on Grande Comore Island, union of the Comoros, 2005. Am J Trop Med Hyg 76:1189–1193. doi: 10.4269/ajtmh.2007.76.1189. [DOI] [PubMed] [Google Scholar]
- 93.Bustos Carrillo F, Gordon A, Harris E. 2019. Reply to Gérardin et al Clin Inf Dis 68:172–174. doi: 10.1093/cid/ciy535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data can be shared with outside investigators following approval from the University of California, Berkeley, IRB and Committee for the Protection of Human Subjects. Please contact E.H. to arrange for data access.