The symptoms of the disease caused by the ebolaviruses Ebola, Bundibugyo, and Sudan are similar, and their areas of endemicity overlap. However, because of the limited antigenic relatedness of the ebolavirus glycoprotein (GP) used in all candidate vaccines against these viruses, they protect only against homologous and not against heterologous ebolaviruses. Therefore, a broadly specific pan-ebolavirus vaccine is required, and this might be achieved by administration of a cocktail of vaccines. The effects of cocktail administration of ebolavirus vaccines on the antibody repertoire remain unknown. Here, an in-depth analysis of the antibody responses to administration of a cocktail of human parainfluenza virus type 3-vectored vaccines against individual ebolaviruses was performed, which included analysis of binding to GP, neutralization of individual ebolaviruses, epitope specificity, Fc-mediated functions, and protection against the three ebolaviruses. The results demonstrated potent and balanced responses against individual ebolaviruses and no significant reduction of the responses compared to that induced by individual vaccines.
KEYWORDS: antibody repertoire, Ebola virus, immune response, immunization, neutralizing antibodies
ABSTRACT
Ebolaviruses Zaire (EBOV), Bundibugyo (BDBV), and Sudan (SUDV) cause human disease with high case fatality rates. Experimental monovalent vaccines, which all utilize the sole envelope glycoprotein (GP), do not protect against heterologous ebolaviruses. Human parainfluenza virus type 3-vectored vaccines offer benefits, including needle-free administration and induction of mucosal responses in the respiratory tract. Multiple approaches were taken to induce broad protection against the three ebolaviruses. While GP consensus-based antigens failed to elicit neutralizing antibodies, polyvalent vaccine immunization induced neutralizing responses to all three ebolaviruses and protected animals from death and disease caused by EBOV, SUDV, and BDBV. As immunization with a cocktail of antigenically related antigens can skew the responses and change the epitope hierarchy, we performed comparative analysis of antibody repertoire and Fc-mediated protective mechanisms in animals immunized with monovalent versus polyvalent vaccines. Compared to sera from guinea pigs receiving the monovalent vaccines, sera from guinea pigs receiving the trivalent vaccine bound and neutralized EBOV and SUDV at equivalent levels and BDBV at only a slightly reduced level. Peptide microarrays revealed a preponderance of binding to amino acids 389 to 403, 397 to 415, and 477 to 493, representing three linear epitopes in the mucin-like domain known to induce a protective antibody response. Competition binding assays with monoclonal antibodies isolated from human ebolavirus infection survivors demonstrated that the immune sera block the binding of antibodies specific for the GP glycan cap, the GP1-GP2 interface, the mucin-like domain, and the membrane-proximal external region. Thus, administration of a cocktail of three ebolavirus vaccines induces a desirable broad antibody response, without skewing of the response toward preferential recognition of a single virus.
IMPORTANCE The symptoms of the disease caused by the ebolaviruses Ebola, Bundibugyo, and Sudan are similar, and their areas of endemicity overlap. However, because of the limited antigenic relatedness of the ebolavirus glycoprotein (GP) used in all candidate vaccines against these viruses, they protect only against homologous and not against heterologous ebolaviruses. Therefore, a broadly specific pan-ebolavirus vaccine is required, and this might be achieved by administration of a cocktail of vaccines. The effects of cocktail administration of ebolavirus vaccines on the antibody repertoire remain unknown. Here, an in-depth analysis of the antibody responses to administration of a cocktail of human parainfluenza virus type 3-vectored vaccines against individual ebolaviruses was performed, which included analysis of binding to GP, neutralization of individual ebolaviruses, epitope specificity, Fc-mediated functions, and protection against the three ebolaviruses. The results demonstrated potent and balanced responses against individual ebolaviruses and no significant reduction of the responses compared to that induced by individual vaccines.
INTRODUCTION
Ebola virus (EBOV), of the genus Ebolavirus, family Filoviridae, causes a severe disease in humans with case fatality rates approaching 90% and in nonhuman primates with fatality rates approaching 100% (1–3). Besides EBOV, there are four additional ebolaviruses: Bundibugyo virus (BDBV), Sudan virus (SUDV), Tai Forest virus (TAFV), and Reston virus (RESTV). Out of the five, three (EBOV, SUDV, and BDBV) have caused large human outbreaks. The viruses are highly contagious, and transmission primarily occurs via direct contact with infected persons or animals, their bodily fluids, or cadavers. Transmission by droplets of respiratory secretions has been observed in nonhuman primates (NHPs) in laboratory settings, and guinea pigs and NHPs directly exposed to aerosolized EBOV have developed lethal infections; however, the role of this route of transmission in natural outbreaks in humans is unclear (4–7).
Currently, there are no approved vaccines for any of the ebolaviruses. Several vaccine strategies have been tested over the years and have had various degrees of success in animal models, but they have primarily been directed only at EBOV. The most promising vaccine candidates are based on viral vectors, including various adenoviruses, vesicular stomatitis virus (VSV), and human parainfluenza virus type 3 (HPIV3), which have proven effective at protecting NHPs from a lethal EBOV challenge (8–15) and have gone on to clinical trials. The recent widespread outbreak of EBOV in West Africa has directed a lot of attention and effort toward developing a practical, effective vaccine against the virus. However, SUDV and BDBV pose a substantial threat for future outbreaks and necessitate similar vaccine efforts. Moreover, the three ebolaviruses produce similar clinical symptoms, and their areas of endemicity overlap (16). Therefore, a single effective vaccine that protects against the three ebolavirus species is needed to protect vulnerable populations and health care workers. This need is especially present at the earliest time points of an outbreak, when a rapid response is required but a reliable identification of the viral species may not be available.
Multivalent vaccine approaches have been successful against several viral infections, such as the quadrivalent MMRV vaccine against the measles, mumps, rubella, and varicella viruses (17) and the nine-valent vaccine against multiple types of human papillomaviruses (18). However, with certain viruses, most notably, polio virus and dengue virus, multivalent approaches result in an imbalanced response to vaccine components, where the response to one or more components is reduced by the heterologous response to another immunodominant component of the mixture (19–22). Multivalent approaches have been tested with filoviruses. A bivalent, virus-like particle vaccine against EBOV and a Marburg virus (MARV), which belongs to a separate filovirus genus, protected guinea pigs from lethal challenges with either virus, without a perceived imbalanced response to either component (23). In contrast, a trivalent VSV-vectored vaccine with EBOV, SUDV, and MARV components protected macaques from death caused by each of the three viruses, but animals exposed to SUDV showed signs of disease before recovering (12). This finding likely was due to a more dominant response to the EBOV and MARV components and was remedied by vaccination with VSV-SUDV first, followed by a bivalent dose of the EBOV and MARV vaccine components 2 weeks later.
Neutralizing antibodies are critical for the efficacy of a filovirus vaccine to control infection. However, there are examples of nonneutralizing monoclonal antibodies (MAbs) that protect from lethal filovirus challenge (24). This can be explained by the fact that, in addition to mediating neutralization through the antibody Fab domain, antibodies can contribute to viral control through recruitment of innate immune effector functions meditated by Fc domains (25, 26). Specifically, EBOV infection can be affected by antibody-dependent cellular cytotoxicity (ADCC) by natural killer (NK) cells or antibody-dependent cellular phagocytosis (ADCP) of viral particles or infected cells by Fc receptor-bearing monocytes, macrophages, and neutrophils. In fact, the role of NK cells in controlling EBOV infections in mice via some of these mechanisms was demonstrated (27, 28). Moreover, the production and secretion of cytokines and chemokines, such as gamma interferon (IFN-γ) and macrophage inflammatory protein 1β (MIP-1β), by NK and other Fc receptor-bearing cells serve to recruit additional innate immune cells to the site of infection through chemoattractant activity and may also activate infiltrating cells to better prime the adaptive immune response. In addition, IgG and IgM can activate the soluble complement system via the classical and lectin pathways, respectively, contributing to neutralization of virions, as well as induction of innate and adaptive immunity (29).
No previous filovirus vaccine study utilizing a multivalent vaccine regimen examined in great depth the contribution of individual components within the cocktail toward the immune response balance. The filovirus glycoprotein (GP) is the major antibody protective antigen possessing both variable and conserved epitopes across ebolavirus species (30–32), which could influence the antibody response pattern when used in combination with each other. Subsequently, in addition to assessing trivalent and monovalent vaccine efficacy, a major goal of this study was to investigate if the response to the trivalent ebolavirus vaccine is qualitatively or quantitatively different from that to monovalent filovirus vaccines. We previously demonstrated that the HPIV3-vectored EBOV vaccine HPIV3/EboGP (referred to here as HPIV3-EBOV) effectively protects guinea pigs and NHPs against a lethal challenge (13, 14). Delivery of the vaccine to the lungs of NHPs in the form of an aerosol induced a strong cell-mediated and antibody response in serum and the respiratory tract and protected against a lethal dose of EBOV delivered by the intramuscular (i.m.) route (33). In this study, we used the same HPIV3 vector to design, generate, and test multiple single-component vaccines and a multivalent vaccine. The latter strategy resulted in robust immune responses and protection from death and disease caused by EBOV, SUDV, and BDBV in vaccinated guinea pigs and ferrets. Furthermore, we evaluated the multivalent approach for signs of an imbalanced response using peptide arrays specific for each component and assessed the ability of the vaccine to generate Fc-mediated cellular responses and activate complement.
RESULTS
Consensus-based antigens fail to elicit a neutralizing response to three ebolaviruses.
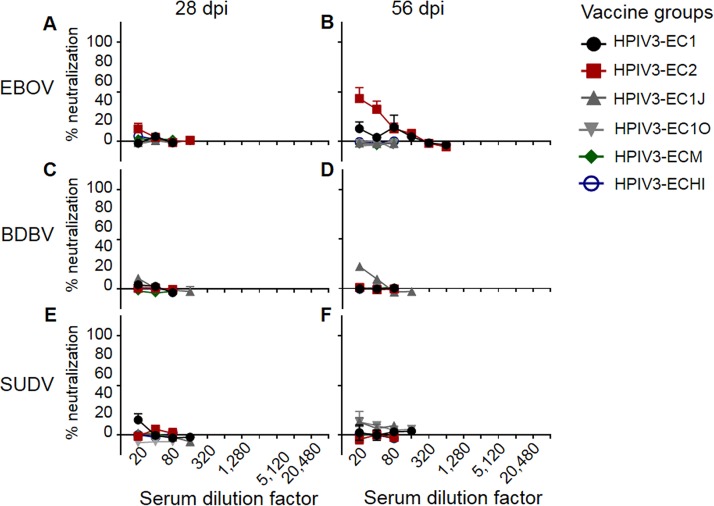
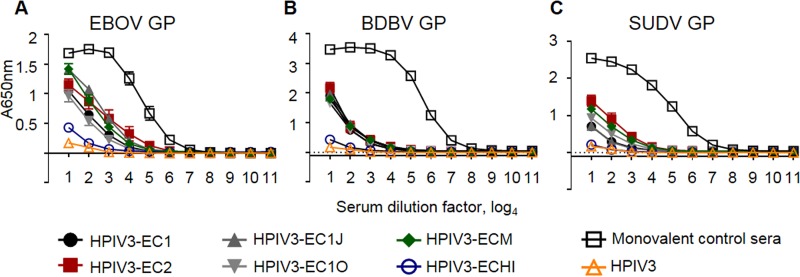
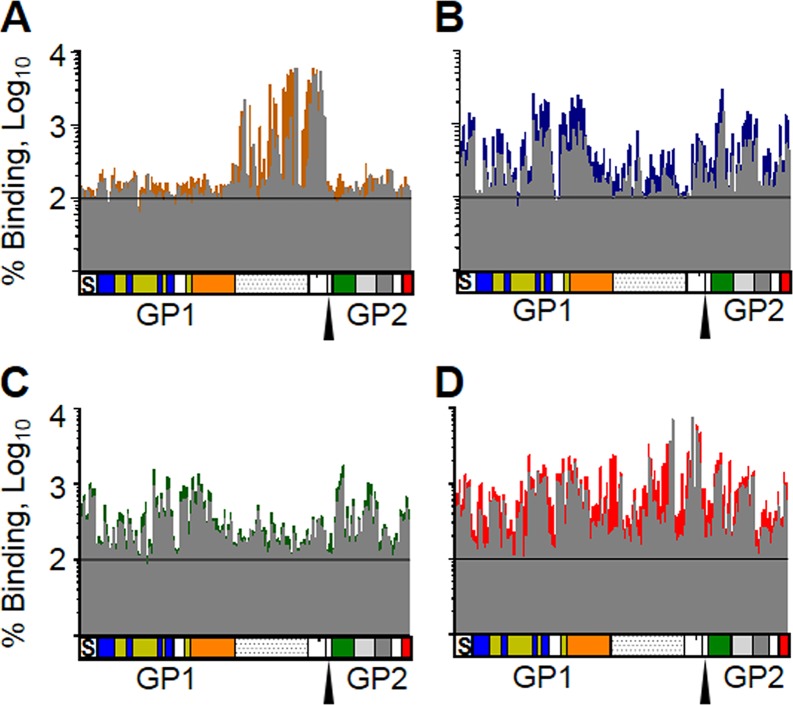
Initially, we attempted to generate a single-component, broadly neutralizing ebolavirus vaccine. To achieve this, we generated six consensus-based antigens based on multilevel consensus generation in an effort to reduce sampling bias, similar to approaches taken against highly pathogenic avian influenza viruses (34). Briefly, 52 partial and whole ebolavirus GP sequences were used to generate a phylogenetic tree (see Fig. S1 in the supplemental material). Based on the tree, the sequences were divided into groups that aligned along species and outbreaks. EBOV sequences were grouped into either lineage A (groups 1 and 2) and lineage B (groups 3 and 4) (35) or 4 groups that divided each lineage into 2 groups that corresponded to distinct outbreaks. Consensus sequences were generated for each group by the use of BLOSUM62 (36). An intermediate consensus sequence, EC1, was generated by creating a second-level EBOV consensus from the lineage A consensus and the lineage B consensus. The final EC1 sequence was generated by creating a consensus from this second-level EBOV consensus, a BDBV/TAFV consensus, and an SUDV consensus (Fig. S1B). EC2 was created similarly, but the EBOV consensus was generated by using consensus sequences from each of the four groups to generate a second-level EBOV consensus (Fig. S1B). EC1J and EC1O are based on the EC1 sequence, but the mucin-like domain, amino acids 309 to 495 (EC1J) or 315 to 506 (EC1O), was removed. ECHI was generated by replacing amino acids in EC1 with residues that have the highest immunogenicity in the predicted epitope regions of the EBOV, BDBV, and SUDV GPs. ECM was created by replacing sequences from EC1J with sequences corresponding to predicted EBOV, BDBV, and SUDV epitopes, creating a mosaic protein. Recombinant HPIV3-vectored vaccine constructs were generated similarly to the HPIV3-EBOV construct (13), which included the generation of transcriptional cassettes for expression of the designed antigens by flanking their open reading frames with HPIV3-specific gene-start and gene-end transcriptional signals, their incorporation into the P-M intergenic sequence of the antigenomic cDNA, and recovery of the recombinant vaccine viruses. Groups of guinea pigs were intranasally inoculated with 4 × 105 PFU of each HPIV3 consensus-based antigen at days 0 and 28. Analysis of sera collected on days 0, 28, and 56 by plaque reduction assays showed that none of the six consensus antigens elicited a fully neutralizing response against BDBV, EBOV, or SUDV by day 28 (Fig. 1A, C, and E). Most of the serum samples collected on day 56 failed to achieve effective neutralization of all three viruses as well; however, EC1J neutralized 22% of BDBV and EC2 neutralized 43% of EBOV at a 1:20 dilution (Fig. 1B, D, and F). Analysis of binding of the immune sera to individual ebolavirus GPs by enzyme-linked immunosorbent assay (ELISA) demonstrated that the response to all six antigens resulted in antibody binding to individual ebolavirus GPs which was ∼100- to 1,000-fold lower than that of monovalent sera (Fig. 2). These data demonstrate that such a consensus-based approach cannot be used for generation of a pan-ebolavirus vaccine, most likely due to the high level of diversity of ebolavirus GP proteins or due to the insufficient level of their expression in cells.
FIG 1.
Consensus-based antigens fail to elicit neutralizing antibody titers. Groups of guinea pigs were vaccinated with the indicated vaccines. Sera collected at days 28 and 56 were serially diluted (2-fold dilution starting at 1:20) and tested for the ability to neutralize EBOV (A and B), BDBV (C and D), or SUDV (E and F). Mean values ± SEM are based on the results for 5 animals per group. dpi, days postinfection.
FIG 2.
Immunization with consensus sequence-based antigens elicits immune sera with lower binding to GP than immunization with monovalent homologous vaccines. Groups of five guinea pigs were vaccinated with the indicated vaccine constructs, and sera collected at day 56 were analyzed by ELISA for the ability to bind EBOV GP (A), BDBV GP (B), or SUDV GP (C). The monovalent control sera are from guinea pigs immunized with HPIV3-vectored constructs expressing GP of EBOV (A), BDBV (B), or SUDV (C) from the experiment whose results are shown in Fig. 4. Mean values ± SEM are based on the results for 5 animals per group.
The trivalent vaccine induces robust balanced neutralization of multiple ebolaviruses without a significant skew.
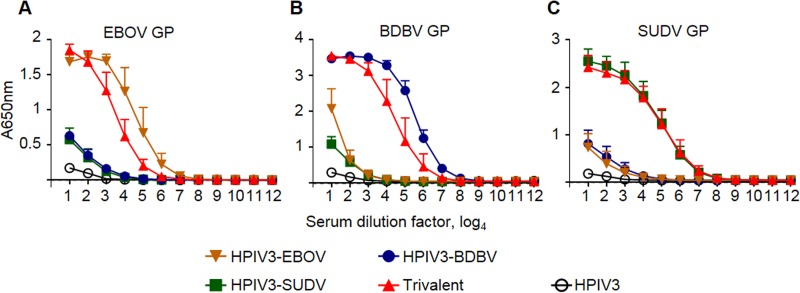
As the consensus-based approach failed, we next focused on generation of a multivalent vaccine using the cocktail approach. HPIV3-vectored BDBV and SUDV components were generated similarly to the HPIV3-EBOV construct (13), as described above for the consensus-based antigens. Protein expression of the GP antigens by the individual vaccine constructs and the trivalent cocktail was confirmed by flow cytometry (Fig. 3A to C) and Western blot analyses (Fig. 3D) using a rabbit polyclonal anti-EBOV GP antibody for EBOV GP, the human MAb BDBV52 (30) for BDBV GP, and the murine MAb 2H5 (IBT Bioservices, Rockville, MD, USA) for SUDV GP. In both assays, the BDBV GP and SUDV GP proteins were expressed at comparable levels in infected cells with the monovalent or cocktail vaccines, while EBOV GP expression was higher in infected cells with the monovalent EBOV construct than the trivalent cocktail. Guinea pigs were inoculated by the intranasal route with 4 × 105 PFU of each of the monovalent vaccine constructs or a mixture of the three constructs at 4 × 105 PFU of each construct, resulting in a total dose of 1.2 × 106 PFU; 28 days later, second vaccine doses were administered. Sera collected on day 28 from animals individually immunized with each of the three vaccine constructs fully neutralized the homologous virus but not the two heterologous viruses. In contrast, sera from animals vaccinated with the trivalent vaccine fully neutralized EBOV (1:160) and SUDV (1:80) and partially neutralized BDBV (84% neutralization at 1:20) (Fig. 4A, C, and E). Sera collected after the second vaccination, at day 56, showed increased neutralization of homologous viruses following immunization with HPIV3-BDBV and HPIV3-EBOV, which was observed up to a dilution of 1:640, while a second dose of HPIV3-SUDV showed no increase of the response compared to that at day 28. Sera collected at day 56 from animals immunized with the trivalent vaccine were able to fully neutralize BDBV (1:160), EBOV (1:320), and SUDV (1:160) (Fig. 4B, D, and F).
FIG 3.
Vaccine constructs express ebolavirus GP. GP expression by the vaccine constructs was assessed by flow cytometry (A to C) and by Western blotting (D). LLC-MK2 cells were infected with the monovalent vaccines, the trivalent vaccine, or the empty HPIV3 vector. (A) Representative primary flow cytometry data. SSC, side scatter; FSC forward scatter. (B) Flow cytometry histogram analysis evaluating the expression of GP by each vaccine construct. (C) Expression levels of GP by each single HPIV3-vectored vaccine construct, EBOV (brown), BDBV (blue), or SUDV (green), compared to that by the trivalent vaccine (red). Mean values ± SEM are based on the results for triplicate samples. (D) Western blot analysis with purified GP proteins as positive controls and actin or GAPDH as loading controls.
FIG 4.
The trivalent vaccine induces neutralization similar to that induced by the individual vaccine components. Groups of five guinea pigs were vaccinated with the indicated vaccines. Sera collected at days 28 and 56 were analyzed for the ability to neutralize EBOV (A and B), BDBV (C and D), or SUDV (E and F). Mean values ± SEM are based on the results for 5 animals per group.
Sera collected at day 56 were also tested for GP binding by ELISA. The trivalent sera bound to EBOV GP and BDBV GP at slightly reduced (4-fold) levels compared to the respective monovalent sera and to HPIV3-SUDV at a level similar to that for the respective monovalent sera. The cross-bindings of the three monovalent sera were minimal (Fig. 5). Thus, the binding patterns matched the homologous neutralization pattern (Fig. 4). These data indicate that each of the individual components induces a strongly neutralizing response against a homologous virus, accompanied by strong binding. Importantly, these data also demonstrate that the trivalent formulation induces a balanced neutralizing antibody response similar to that induced by the monovalent vaccines, with only a slight reduction of the BDBV-specific response.
FIG 5.
The trivalent vaccine has binding characteristics similar to those of the individual single-component vaccines by ELISA. Groups of guinea pigs were immunized with the indicated vaccines, and sera collected at day 56 were analyzed by ELISA for the ability to bind EBOV GP (A), BDBV GP (B), or SUDV GP (C). Mean values ± SEM are based on the results for 5 animals per group.
The lack of cross-neutralization of monovalent sera is consistent with their predominant binding to linear epitopes in the MLD.
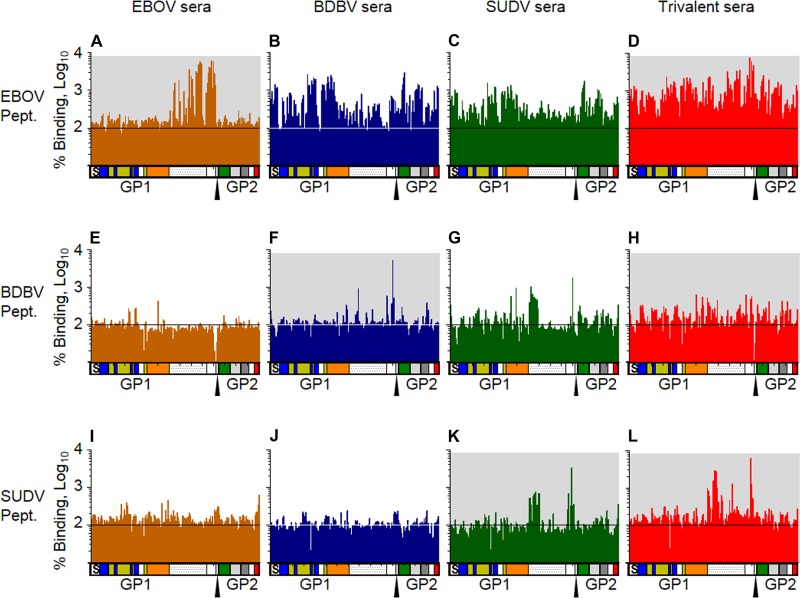
To characterize the binding of the immune sera to linear epitopes, serum samples were analyzed by peptide microarrays. Sets of 167 15-mer peptides offset by 4 amino acids spanning the GP sequences of EBOV, BDBV, and SUDV were tested. Comparison of guinea pig sera collected on day 28 or 56 (Fig. 6) showed only a modest increase of binding at the later time point: 1.4-, 1.8-, 1.3-, or 1.9-fold for EBOV, BDBV, SUDV or the trivalent vaccine, respectively. Comparison of the binding of day 56 sera to peptides of all three ebolaviruses (Fig. 7) revealed that, in general, the majority of EBOV homologous binding (HPIV3-EBOV serum binding to EBOV peptides) occurred in the mucin-like domain (MLD), with minimal binding occurring across the other parts of the protein. The HPIV3-EBOV sera showed the highest levels of binding above the background across the entire MLD. The HPIV3-BDBV sera showed the smallest amount of binding over the background signal, with only two peptides (peptide 88 [residues 349 to 364] and peptide 122 [residues 484 to 499]), both of which are located in the MLD, showing an approximately 10-fold or higher increase in binding. The HPIV3-SUDV sera showed three peaks in the N-terminal third of the MLD (residues 313 to 367) and another peak at the extreme C-terminal end of the MLD adjacent to the cleavage site (residues 469 to 495).
FIG 6.
Trivalent vaccine sera bind linear EBOV GP peptides similarly to single-component vaccine sera. Sera from animals vaccinated with either HPIV3-EBOV (A), HPIV3-BDBV (B), HPIV3-SUDV (C), or the trivalent vaccine (D) were analyzed for binding to peptides matching the protein sequence of EBOV GP. Binding of sera collected on day 28 (gray) is overlaid on binding of sera collected on day 56 (color) to highlight regions that showed increased binding after the second vaccine dose. Values represent the average binding of sera from 3 vaccinated animals as a percentage of the binding of sera drawn prior to vaccination. Each column represents the results for a 15-residue peptide with a sequence matching the sequence of EBOV GP. The diagrams below the graphs show the domains of GP in a linear fashion that correspond to the binding data. S, signal peptide; blue, base; yellow, head; orange, glycan cap; dotted region, mucin domain; green, fusion loop; light gray, HR1; dark gray, HR2; red, transmembrane and cytoplasmic tail. The furin cleavage site is marked by the triangle.
FIG 7.
Trivalent vaccine sera bind GP peptides similarly to single-component vaccine sera. Sera collected at day 56 after immunization with the EBOV vaccine (orange), BDBV vaccine (blue), SUDV vaccine (green), or trivalent vaccine (red) were analyzed for binding to peptides matching the protein sequence of EBOV GP (A to D), BDBV GP (E to H), or SUDV GP (I to L). Values represent the average binding of sera from 3 vaccinated animals as a percentage of the binding of preimmune sera. Each column represents the results for a 15-residue peptide with a sequence matching the sequence of GP. The diagrams below each graph show the domains of GP in a linear fashion that correspond to the binding data. S, signal peptide; blue, base; yellow, head; orange, glycan cap; dotted region, mucin domain; green, fusion loop; light gray, HR1; dark gray, HR2; red, transmembrane and cytoplasmic tail. The furin cleavage site is marked by the triangle. Graphs with a gray background indicate that the vaccine or its component is homologous with the peptide array sequence.
Heterologous binding profiles showed differing levels of binding. Sera from EBOV- and BDBV-immunized guinea pigs showed very limited binding to SUDV peptides. The HPIV3-EBOV-vaccinated group also showed very limited binding to BDBV. Unexpectedly, HPIV3-SUDV sera bound BDBV peptides in a manner similar to that in which they bound SUDV peptides. Both HPIV3-SUDV and HPIV3-BDBV sera had increased binding across the entire EBOV GP, with the exception of the MLD, essentially the inverse binding profile of HPIV3-EBOV sera to EBOV peptides. These data are consistent with the lack of similarity between the MLDs of the three filoviruses, resulting in a greater detection of non-MLD-bound peptides.
Trivalent sera showed a binding pattern similar to that of a combination of the three single-component vaccine groups, for the most part. For example, HPIV3-EBOV sera bound EBOV peptides across the MLD at a 10- to 60-fold increase over the background but did not bind in other regions. HPIV3-BDBV and HPIV3-SUDV sera bound EBOV peptides at about a 10-fold increase to all areas except the MLD. The trivalent sera bound EBOV MLD with a similar intensity and pattern as the EBOV sera, but they also bound the rest of the peptides with a similar intensity as the BDBV and SUDV sera. Thus, the binding profile of the trivalent sera with EBOV peptides looked similar to that of a composite of the profiles of the sera from animals receiving each of the three individual vaccines (Fig. 8). However, one exception was observed: the trivalent sera demonstrated a 10-fold increase in binding to the peptides corresponding to the N-terminal end of the MLD (residues 313 to 335) over what any of the monovalent vaccines elicited (Fig. 8D). Thus, at least based on binding to linear epitopes, in particular for EBOV, the lack of cross-neutralization by monovalent vaccine sera is related to the immunodominance of the MLD, which has almost no similarity between the individual ebolaviruses. These data also demonstrate that the vaccines induce antibody responses that cover a diverse set of epitopes across the whole GP protein and that the trivalent vaccine induces antibodies with a binding profile similar to that of the monovalent vaccines and does not induce a skewed response.
FIG 8.
Immune sera from animals immunized with the trivalent vaccine bind peptides in a similar pattern as a composite of EBOV, BDBV, and SUDV vaccine sera. The results for all three single-component-vaccinated animal sera (HPIV3-EBOV, HPIV3-BDBV, and HPIV3-SUDV; gray) were merged and overlaid with those for a trivalent-vaccinated animal serum (red). (A and B) Day 28 sera; (C and D) day 56 sera. The responses in the trivalent vaccine-immunized animals are shown in front of the responses in the 3 individual vaccine groups in panels A and C, so any visualized gray indicates areas where the trivalent vaccine induced less binding than the composite of the three individual vaccines. In contrast, the responses in the individual vaccine groups are displayed in front of the responses in the trivalent vaccine group in panels B and D, so any visualized red indicates areas where the trivalent vaccine induced more binding than the composite of the individual vaccines.
Both monovalent and trivalent vaccines induce broad antibody responses that recognize a diverse set of known epitopes across the whole GP.
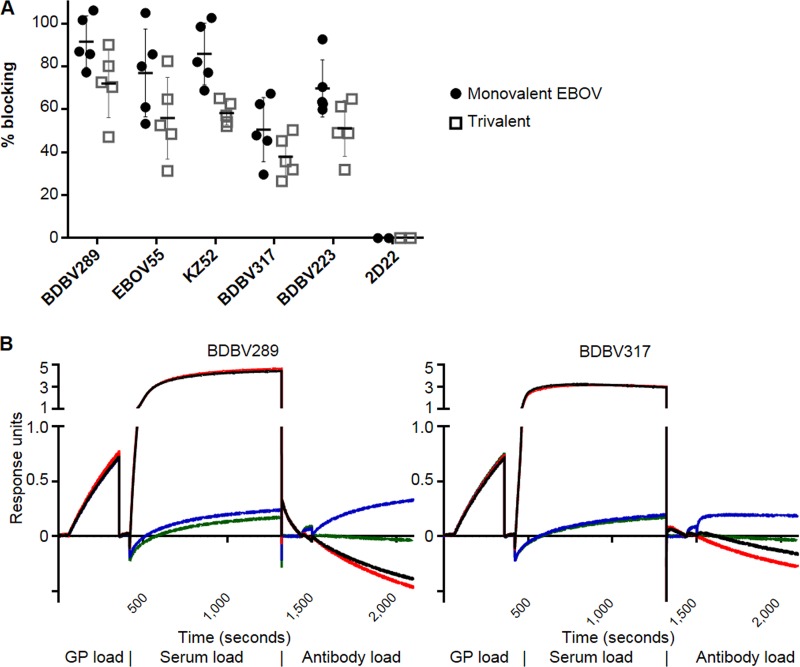
Peptide microarrays are a useful tool for identifying linear epitopes but fail to probe for conformational epitopes. Thus, we conducted competition binding assays between MAbs with known epitopes and our monovalent or trivalent sera, using biolayer interferometry. Knowledge of ebolavirus epitopes is skewed heavily toward EBOV; therefore, the study was targeted at known EBOV epitopes and tested the monovalent EBOV and trivalent sera. The following MAbs were selected for this analysis (Fig. 9A and B): (i) BDBV289, isolated from a human survivor of BDBV infection, which recognizes amino acids 273 to 310, located in the glycan cap (30); (ii) EBOV55, isolated from a survivor of EBOV infection using the previously published approach (30, 37), which binds amino acids 397 to 402 in MLD; (iii) KZ52, which binds amino acids 42, 43, 505 to 514, and 549 to 556 in the GP1-GP2 interface (38); (iv) BDBV317, the binding of which is prevented by a mutation of amino acid 633 in the membrane-proximal external region (MPER) (37); and (v) BDBV223, whose binding is prevented by a mutation at amino acid 634 in MPER. Both EBOV monovalent and trivalent vaccines induced antibodies which blocked the binding of all five tested MAbs to some extent (between 38% and 92%; Fig. 9A). The trivalent vaccine sera showed 21% to 32% less blocking than the monovalent vaccine sera (P < 0.05 for all pairs except BDBV317).
FIG 9.
Immune sera from animals vaccinated with HPIV3-EBOV or the trivalent vaccine block the binding of monoclonal antibodies specific for diverse GP epitopes. EBOV GP was bound to biosensors and then incubated with the indicated immune sera. The sensors were then incubated with antibodies known to bind GP. (A) The amounts of binding were measured by biolayer interferometry and are shown as percentages of the binding of a negative control (empty HPIV3 vector control sera). 2D22 is a human antibody specific for dengue virus. Mean values ± SEM are based on the results for 5 animals per group. Differences between blocking by mono- and trivalent sera were significant (P < 0.05) for all monoclonal antibodies except BDBV317. (B) Representative data curves for two samples. The negative-control value was determined by subtracting the value on the green line (no serum, no antibody) from the value on the blue line (no serum with antibody). The sample value was determined by subtracting the value for the red line (with serum, no antibody) from the value for the black line (with serum and antibody). The percent blocking was calculated by converting the sample value to a percentage of the negative-control value and subtracting from 100%.
Taken together, the data suggest that despite the immunodominance of MLD, the HPIV3-EBOV vaccine induces a response that covers a diverse set of epitopes across the whole GP protein. The trivalent vaccine sera blocked the binding of various MAbs at a moderately reduced rate compared to that for the monovalent HPIV3-EBOV vaccine sera, demonstrating a mitigated response against individual components of the vaccine.
Vaccine-induced antibodies are potent mediators of Fc-dependent innate immune effector functions.
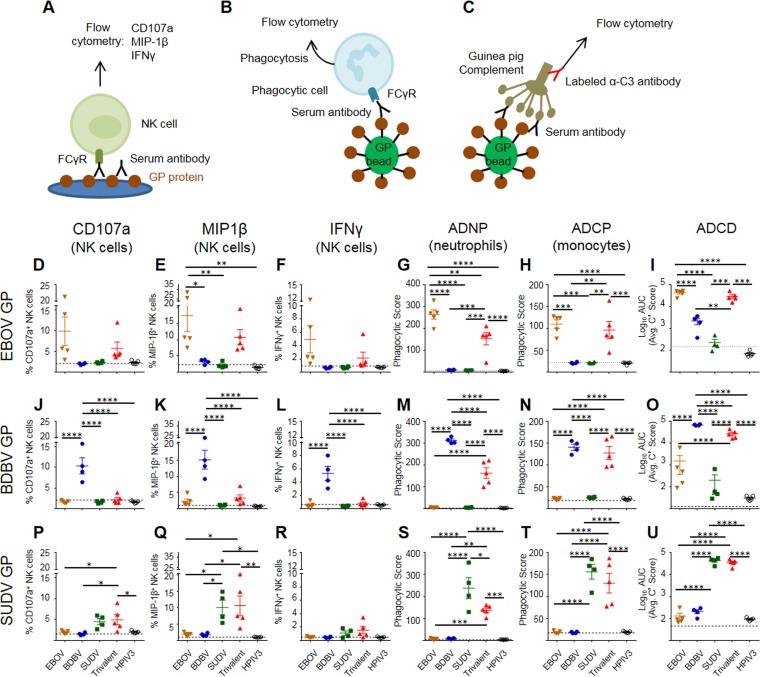
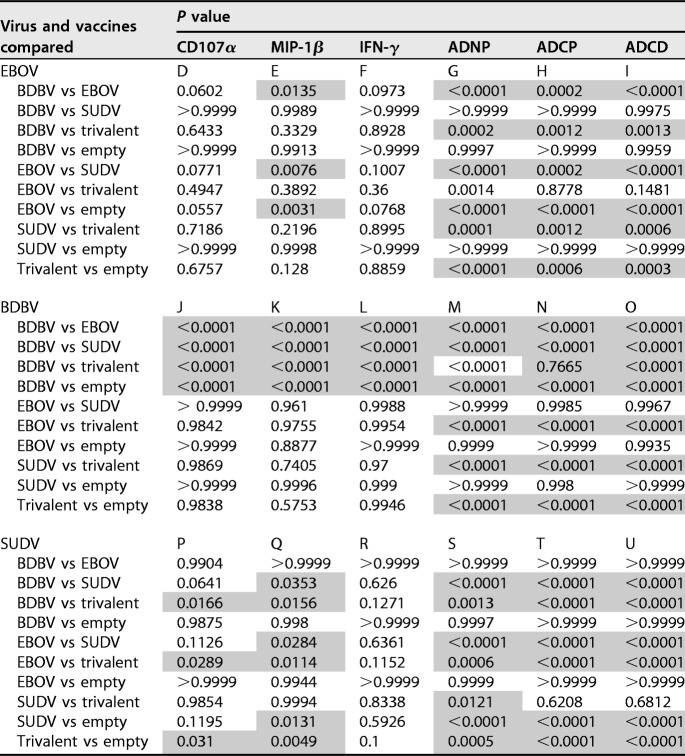
To determine whether vaccination induced antibodies capable of activating innate immune effector functions in several different types of innate immune cells, we assayed the sera collected on day 56 from vaccinated guinea pigs for activation of NK cells, induction of phagocytosis by monocytes and neutrophils, and complement deposition (Fig. 10A to C; the gating strategies are shown in Fig. S2). The degranulation of cytotoxic granules from NK cells is marked by surface expression of CD107a (39). Analysis of NK cells demonstrated that sera from guinea pigs vaccinated with a monovalent vaccine were able to induce expression of CD107a (Fig. 10D, J, and P), albeit at a reduced level compared to that for the EBOV-specific MAb c13C6, which is glycosylated with a glycan known to enhance ADCC (26, 40), when plated on the homologous GP protein (Fig. S2B). Trivalent sera were capable of inducing degranulation above background levels when NK cells were plated on SUDV GP- and EBOV GP-coated plates (with no significant difference in the latter case due to the high sample-to-sample variability) but not when plated on BDBV GP-coated plates (Table 1). In addition, when stimulated with homologous GPs, the EBOV and SUDV monovalent vaccines elicited CD107a responses which were similar to those induced by the trivalent vaccine, whereas the BDBV monovalent vaccine elicited significantly higher responses than the trivalent vaccine. For all three GP proteins, secretion of MIP-1β by NK cells (Fig. 10E, K, and Q) was detectable in both the monovalent and trivalent vaccine groups, yet a trend similar to that for CD107a was observed, with the levels of MIP-1β secretion in the animals receiving the EBOV and SUDV monovalent vaccines being similar to those in the group receiving the trivalent vaccine. Again, the BDBV monovalent vaccine elicited significantly higher levels of MIP-1β than the trivalent vaccine. The secretion of IFN-γ by NK cells (Fig. 10F, L, and R) was detected for the EBOV and SUDV monovalent vaccines (although for EBOV, the levels did not reach statistical significance due to high variation between samples) but not for the BDBV vaccine. Monovalent sera showed no effect on NK cell activation with heterologous GP proteins. Overall, the trivalent vaccine induced activation of EBOV-specific and SUDV-specific NK cells at a level comparable to that for the respective monovalent vaccines, while the induction of BDBV-specific NK cells was lower than that by the monovalent vaccine. This difference between the BDBV vaccine on the one hand and the EBOV and SUDV vaccines on the other hand is consistent with the reduced induction of neutralizing antibody responses observed for the monovalent BDBV vaccine but not for the monovalent EBOV or SUDV vaccine compared to that observed for the trivalent vaccine (Fig. 4).
FIG 10.
Monovalent and trivalent vaccines can induce highly functional antibodies capable of recruiting different Fc-dependent innate immune effector functions from several types of immune cells and activating the complement system. (A) Sera from guinea pigs immunized with the indicated HPIV3-vectored vaccines were diluted 1:100 and incubated on plates coated with EBOV GP, BDBV GP, or SUDV GP. Freshly isolated human NK cells were added to the plates and assessed for the expression of three activation markers: CD107a (D, J, P), MIP-1β (E, K, Q), and IFN-γ (F, L, R). The same sera were also incubated with GP-coated fluorescent beads (B), which were incubated with either human neutrophils (G, M, S) or THP-1 monocytes (H, N, T). Cells were then analyzed to quantify the phagocytic intake of the fluorescent bead/antibody complexes. (C) GP-bound beads were incubated with guinea pig sera and exposed to guinea pig complement. C3 deposition was measured by flow cytometry using a fluorescent anti-guinea pig C3 antibody (I, O, U). Dashed lines indicate the levels of no-antibody controls. Data are the mean ± SEM. P values were determined by 1-way ANOVA followed by the Tukey posttest. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. The results of a complete pairwise statistical analysis are shown in Table 1. ADNP, antibody-dependent neutrophil phagocytosis; ADCP, antibody-dependent cellular phagocytosis; ADCD, antibody-dependent complement deposition.
TABLE 1.
P values were determined by one-way ANOVA with Tukey’s multiple-comparison post hoc test on antibody-mediated cellular response data presented in the panels in Fig. 10 indicated in the subheads. Significant differences at P values of <0.05 are shaded. ADNP, antibody-dependent neutrophil phagocytosis; ADCP, antibody-dependent cellular phagocytosis; ADCD, antibody-dependent complement deposition; trivalent; trivalent vaccine; empty, empty vector.
As phagocytosis of virus and/or infected cells may be a mechanism by which antibodies contribute to viral control, we next tested the ability of the immune sera to induce the phagocytosis of GP-coated beads by neutrophils and monocytes. Briefly, fluorescein isothiocyanate (FITC)-positive (FITC+) streptavidin beads were coated with biotinylated EBOV GP, SUDV GP, or BDBV GP to generate antigen-coated beads and incubated with diluted serum samples prior to addition of phagocytic cells (neutrophils or monocytes) for either 1 h (neutrophils) or 18 h (monocytes) (Fig. 10B). The uptake of the beads by cells was determined by flow cytometry, and the phagocytic score was calculated.
Neutrophils are the most abundant innate immune cells in the blood, are highly phagocytic, and are capable of killing infected cells through several different mechanisms, including production of reactive oxygen species and neutrophil extracellular traps (41, 42). Vaccine-induced antibodies from all three monovalent vaccine groups were robust inducers of neutrophil-mediated phagocytosis of homologous GP-coated beads and were capable of inducing high levels of phagocytosis by nearly all neutrophils assayed (Fig. 10G, M, and S). For all three types of GP beads, the phagocytic score of homologous beads was significantly higher in the monovalent vaccine groups than in the trivalent vaccine groups. In addition, strong monocyte-mediated phagocytosis of homologous GP-coated beads was induced by all three monovalent vaccine groups and the trivalent vaccine group (Fig. 10H, N, and T). Interestingly, there was no effect of the monovalent vaccines on the neutrophil- or monocyte-mediated phagocytosis of heterologous GP-coated beads.
Finally, antibody-dependent complement activation was tested by analyzing C3 deposition onto GP-coated beads (Fig. 10C). High levels of C3 deposition onto homologous beads were observed in all three monovalent vaccine groups and the trivalent vaccine group (Fig. 10I, O, and U), with slightly higher C3 deposition in the BDBV monovalent vaccine group than in the trivalent vaccine group. There was no significant deposition on heterologous GP beads in the BDBV and SUDV monovalent vaccine groups compared to the empty vector vaccine groups, but there was some deposition on BDBV GP beads with the EBOV sera. Together, these data indicate that vaccination with either monovalent or trivalent vaccines induces highly functional antibodies capable of recruiting multiple different Fc-dependent innate immune effector functions from several types of immune cells and activating the complement system, all of which likely contribute to the protective efficacy afforded by the vaccines.
The trivalent vaccine is fully protective against EBOV, SUDV, and BDBV lethal challenges.
We chose to test the protective efficacy of our vaccines delivered as one dose rather than two doses, based on the immunogenicity results described in the previous section. Thus, as the trivalent vaccine induced comparable levels of neutralizing antibodies against each of the three ebolaviruses (Fig. 4), the protective efficacy of the vaccine against EBOV and SUDV was tested in guinea pigs (Fig. 11) with the respective guinea pig-adapted filoviruses (43, 44), while the protective efficacy of the vaccine against wild-type BDBV was tested in ferrets (44, 45) (Fig. 12). Animals were vaccinated once intranasally with a vaccine dose of 4 × 105 PFU per guinea pig and 1 × 107 PFU per ferret; the control groups received an equivalent dose of the empty HPIV3 vector. At 28 days postvaccination, guinea pigs were challenged intraperitoneally (i.p.) with a target dose of 103 PFU of guinea pig-adapted EBOV or guinea pig-adapted SUDV and ferrets were challenged i.m. with a target dose of 103 PFU of wild-type BDBV. One PFU of guinea pig-adapted EBOV contains multiple 50% lethal doses for guinea pigs (46; Anthony Sanchez, personal communication); the 50% lethal doses of guinea pig-adapted SUDV for guinea pigs and of wild-type BDBV for ferrets are unknown.
FIG 11.
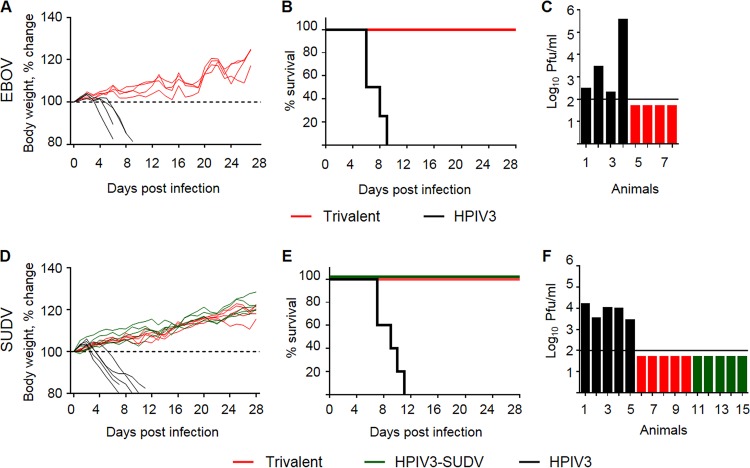
The trivalent vaccine completely protects guinea pigs from death and disease caused by EBOV and SUDV. Groups of guinea pigs were vaccinated with the empty HPIV3 vector (black), the trivalent vaccine (red), or HPIV3-SUDV (green). At 4 weeks postvaccination, guinea pigs were challenged with guinea pig-adapted EBOV (A to C) or guinea pig-adapted SUDV (D to F). (A, D) Percent change in body weight in vaccinated and control animals. (B, E) Percent survival of vaccinated and control animals. P was 0.0062 for the trivalent vaccine versus the empty HPIV3 vector (B); P was 0.0017 by the log-rank (Mantel-Cox) test for HPIV3-SUDV versus the empty HPIV3 vector and the trivalent vaccine versus the empty HPIV3 vector (E). (C, F) Viremia of individual animals at 6 days postchallenge. The limit of detection is shown by a horizontal line.
FIG 12.
The monovalent BDBV and trivalent vaccines elicit binding and neutralizing antibodies and completely protect ferrets from death and disease caused by BDBV. Groups of ferrets were vaccinated with HPIV3-BDBV, the trivalent vaccine, or the empty HPIV3 vector. Sera collected at day 28 were analyzed for the ability to bind to EBOV GP (A), BDBV GP (B), and SUDV GP (C) as well as to neutralize EBOV (D), BDBV (E), or SUDV (F). Mean values ± SEM are displayed. At 4 weeks postvaccination, ferrets were challenged with BDBV. (G) Percent change in body weight in vaccinated and control animals. (H) Percent survival of vaccinated and control animals. For both HPIV3-BDBV versus the empty HPIV3 vector and the trivalent vaccine versus the empty HPIV3 vector, P was 0.0016 by the log-rank (Mantel-Cox) test. (I) Viremia of individual animals at 6 days postchallenge. The limit of detection is shown by a horizontal line.
For the EBOV challenge, guinea pigs in the control group began losing weight at day 4 postchallenge (Fig. 11A). Other signs of disease, including scruffy coat, lethargy, and failure to respond to stimulus, began at day 6. Two control animals reached a moribund state and were euthanized at day 6, one at day 8 and one at day 9 (Fig. 11B). All animals had lost 15% to 20% of their body weight at the time of euthanasia. Viremia was detected in the control animals at day 6. The two animals that were euthanized at day 6 had the highest titers (2.8 × 103 and 3.6 × 105 PFU/ml of serum), while the two that were euthanized later were less viremic (2 × 102 and 3 × 102 PFU/ml). The animals in the trivalent vaccine group had no detectable viremia (Fig. 11C), showed no signs of disease, and maintained or steadily gained body weight during the challenge period (Fig. 11A).
For the SUDV challenge, guinea pigs in the control group began losing weight at 3 days postchallenge (Fig. 11D). Other signs of disease began at day 5; two control animals were euthanized at day 7, and one guinea pig each was euthanized on days 9, 10, and 11 (Fig. 11E). Viremia was detected only in the control animals, with the maximum level at day 6 ranging from 2.7 × 103 to 1.6 × 104 PFU/ml (Fig. 11F). Animals vaccinated with either the SUDV monovalent vaccines or the trivalent vaccine were not positive for viremia, showed no signs of disease, and increased in weight during the challenge period (Fig. 11D).
Ferrets vaccinated with the trivalent vaccine elicited antibody responses with homologous binding and neutralization levels equivalent to those induced by the HPIV3-BDBV monovalent vaccine (Fig. 12B and E). The trivalent vaccine also elicited binding (Fig. 12A and C) and neutralizing (Fig. 12D and F) antibodies specific for EBOV and SUDV. Following challenge with BDBV, ferrets in the control group began losing weight at day 3 (Fig. 12G). Other signs of disease began at day 7 and progressed extremely rapidly; one control animal died during the day and three control animals were euthanized by the end of day 7, while the last control ferret was euthanized on day 8 (Fig. 12H). Viremia was detected only in the control animals, all of which were positive by day 6, with the maximum viremia measured at day 7 ranging from 1.1 × 105 to 8.4 × 105 PFU/ml (Fig. 12I). Ferrets vaccinated with either the BDBV monovalent vaccines or the trivalent cocktail vaccine did not test positive for viremia, showed no signs of disease (data not shown), and maintained their body weight or steadily gained body weight during the challenge phase (Fig. 12G).
Combined, these data demonstrate that the HPIV3-vectored trivalent ebolavirus vaccine was capable of fully protecting guinea pigs against challenge with EBOV and SUDV and ferrets against challenge with BDBV, while the monovalent HPIV3-based vaccines were also protective against their targeted viruses.
DISCUSSION
HPIV3-vectored vaccines against emerging viral infections can be useful for containment of outbreaks and vaccination of health care personnel, as they have certain advantages. First, delivery of the vaccine to the respiratory tract would not require trained medical personnel and can be used for immunization of large numbers of people in case of an outbreak. Second, upon delivery to the respiratory tract, this type of vaccine induces strong local cell-mediated and antibody responses in the respiratory tract, in addition to the systemic immune response (33). As aerosols of EBOV (4, 5) and possibly other emerging viruses are infectious, the strong local immune response in the respiratory tract is likely to provide an additional layer of protection against aerosolized infection. The initial goal of the current study was to develop a consensus-based single-component vaccine that elicits broad-spectrum protection against several ebolaviruses; however, none of the generated antigens induced antibody responses with adequate levels of binding or neutralizing antibodies. The approach used was based on a methodology similar to that reported by Giles and Ross, who developed computationally optimized broadly reactive antigens (COBRA) based on consensus sequences of H5 influenza A virus hemagglutinin (HA) antigens (34). That HA antigen induced antibodies that broadly neutralized many clades of H5N1 viruses. The difference between this work and ours is likely related to sequence similarity since the sequence similarity across all clades of H5 HA exceeds 90%, whereas the sequences of ebolavirus species are far less conserved. Comparing the GP sequences of the specific strains used in this study demonstrated that SUDV shares only 55% identity with BDBV and EBOV, while BDBV and EBOV are 66% identical. Furthermore, the amino acid sequence identity of the linear epitopes revealed by the peptide array in wild-type GPs to that in the consensus-based vaccines was found to be low (Table 2). The consensus sequence generated by these divergent sequences was too dissimilar from the sequences of individual GPs to generate antibodies capable of neutralizing individual ebolavirus species.
TABLE 2.
Amino acid sequence identity analysis of consensus GP vaccines
| Consensus vaccine | % amino acid sequence identity to GP reference sequence at the indicated MLD residues |
||||||
|---|---|---|---|---|---|---|---|
| EBOV |
BDBV |
SUDV |
|||||
| Residues 389–403 | Residues 401–415 | Residues 477–491 | Residues 349–364 | Residues 484–499 | Residues 313–367 | Residues 469–495 | |
| EC1 | 40.00 | 26.67 | 33.33 | 56.25 | 68.75 | 50.91 | 51.85 |
| EC2 | 33.33 | 26.67 | 33.33 | 37.50 | 68.75 | 50.91 | 51.85 |
| ECHI | 40.00 | 26.67 | 33.33 | 56.25 | 68.75 | 50.91 | 51.85 |
| ECM | 40.00 | 26.67 | 33.33 | 56.25 | 68.75 | 50.91 | 51.85 |
| EC1J | NAa | NA | NA | NA | 37.50 | NA | 3.70 |
| EC1O | NA | NA | NA | NA | NA | NA | NA |
NA, not applicable since EC1J and EC1O lack the mucin-like domain (MLD).
The lack of a response to consensus antigens led us to evaluate wild-type antigens in mono- or trivalent formulations. All monovalent HPIV3-ebolavirus GP vaccine constructs elicited strong immune responses to the respective homologous ebolaviruses and minimal cross-reactive responses to heterologous ebolaviruses. These diverse responses included strong neutralizing antibodies, ADCC and ADCP activities, and activation of complement. The trivalent vaccine induced, with only a few exceptions, a response notably similar to that of the monovalent components. All three monovalent vaccine components generated similar neutralizing responses 28 days after a single dose of the vaccine (Fig. 4A, C, and E). However, after an identical booster vaccine at day 28, SUDV sera showed no increase in neutralization, while BDBV and EBOV sera had an 8-fold increase (Fig. 4B, D, and F). This failure of the SUDV vaccine boost to increase neutralization was also seen in the trivalent vaccine group. Because the failure of boosting occurred only with the SUDV immunogen, we rule out vector-induced preexisting immunity as a cause, but it is possible that the response at the saturation level was reached after a single dose of HPIV3-SUDV, as previously observed with some other filovirus vaccines (47). Additionally, in guinea pigs but not in ferrets, the trivalent vaccine group showed a noticeable reduction in neutralization of BDBV compared to the monovalent vaccine group (Fig. 4C to E). In guinea pigs, the lowest (1:20) dilution of serum from the trivalent vaccine group failed to fully neutralize BDBV after a single dose, and the approximately 6-fold reduction in neutralization was seen after the second dose as well. Concomitantly, trivalent sera were able to bind EBOV and SUDV peptides similarly to the monovalent components but had reduced binding of BDBV peptides compared to the monovalent sera (Fig. 7). Several multivalent vaccines against various pathogens, including dengue virus, polio virus, and the cocktail of VSV-vectored vaccines against EBOV, SUDV, and MARV, have produced skewed responses to individual components (12, 19–22). In this study, the HPIV3-vectored, multivalent vaccine appeared to only slightly skew the response away from the BDBV component in an animal species-dependent manner.
Wilson et al. identified several murine MAbs that bound to linear epitopes and protected mice from a lethal EBOV challenge (24). These linear epitopes (amino acids 401 to 417, 389 to 405, and 477 to 493) have been studied extensively, and antibodies binding them have been categorized as competition groups I, II, and III, respectively. Antibodies to these epitopes have been used successfully in a cocktail format to treat EBOV infections, most notably, in the MB-003 treatment (48). In our study, these epitopes are most closely represented by peptides 98 (amino acids 389 to 403, group II), 101 (amino acids 401 to 415, group I), and 120 (amino acids 477 to 491, group III) on the EBOV peptide microarray (Fig. 7A). These peptides have three of the five highest binding scores at 35.6-fold, 19.5-fold, and 60-fold increases over the values for negative serum, respectively, and the epitopes likely contribute to the protective effects of the EBOV component. Interestingly, peptides 98 and 101 of the BDBV and SUDV arrays are completely dissimilar to their EBOV counterparts, and binding to these peptides was not observed. However, peptide 120 of BDBV and SUDV shares some similarity with peptide 120 of EBOV (44% and 55%, respectively) and also shows high levels of binding (53-fold and 34-fold increases, respectively). The BDBV and SUDV antibodies that bind this epitope likely function in a similar fashion and have a similar protective effect. The competition group III epitope has been well characterized as a site of vulnerability in the EBOV GP useful as a therapeutic target (48). This study provides the first evidence of an epitope in BDBV and SUDV at the same location.
Similar to peptide 120, peptide 88 (amino acids 349 to 363) has high levels of binding for all three ebolavirus peptide arrays. EBOV sera bound EBOV peptide 88 at a level 18.9-fold over that for the background, while BDBV bound BDBV peptide 88 at a 9-fold higher level and SUDV bound SUDV peptide 88 at a 7-fold higher level (Fig. 7A, F, and K). This peptide corresponds to a region of the GP protein (amino acids 301 to 359) that a previous study identified as being highly reactive with sera from symptomatically and asymptomatically infected people during the 1996 and 2001 EBOV outbreaks in Gabon (49). Additionally, trivalent sera bound the N-terminal portion of the MLD in this EBOV immunogenic region (amino acids 315 to 335) at an approximately 10-fold greater level over monovalent EBOV sera, indicating a contribution to protection against all ebolaviruses and a potential advantage of the trivalent approach.
The peptide array (Fig. 7) demonstrated unexpected features of the antibody response. First, we observed an asymmetric binding of immune sera induced by the BDBV and SUDV vaccines to the MLD of homologous and heterologous viruses; SUDV-immune sera bound both the SUDV and BDBV MLD, but BDBV-immune sera bound only the BDBV MLD and not the SUDV MLD. Second, the EBOV-immune sera showed very limited binding to BDBV, while the SUDV-immune sera bound BDBV and SUDV equally. The cross-binding pattern does not correspond to the phylogenetic tree (see Fig. S1 in the supplemental material), which shows that BDBV is closer to EBOV than to SUDV. These discrepancies can be related to slight differences in the corresponding epitopes of EBOV, BDBV, and SUDV GPs, which may result in different levels of cross-reactivity of the induced antibodies.
We previously isolated multiple MAbs from survivors of BDBV and EBOV infections (30), some of which, along with the MAb KZ52 previously generated from bone marrow RNA of a survivor of EBOV infection (50), were used in this study in biolayer interferometry competition binding assays. The data showed that the trivalent sera had coverage of epitopes targeted by all these MAbs. Three of these MAbs, BDBV289, BDBV223, and KZ52, were demonstrated to be protective in rodent models of EBOV (30, 51), suggesting the contribution of antibodies targeting these epitopes to the protection conferred by the trivalent vaccine.
Importantly, the study used two complementary methods to determine the precise antigenic specificity of the immune sera: peptide array analysis and competition with MAbs with known epitopes analyzed by biolayer interferometry. The first method demonstrated a high-level binding to the homologous MLD, while binding to other regions of GP was more obvious with trivalent sera or with heterologous peptides. This finding can be explained by a greater affinity or a greater abundance of MLD MAbs absorbing a disproportionally high fraction of secondary antibodies that leads to a reduced detection of non-MLD antibodies. The second method demonstrated the presence of antibodies specific to all regions of GP without comparing their relative abundance. Using a combination of methods for these studies is important, as any single method may produce skewed results. In addition, based on these two methods, vaccine-elicited antibodies targeted GP epitopes from the three tiers recently defined by Saphire et al. (52): MLD (tier 1), the glycan cap (tier 1), the GP1-GP2 interface (tier 2), and MPER (tier 3). Tier 1 epitopes were more recognized by antibodies elicited by VSV-EBOV (53), and we obtained a similar preponderant recognition of tier 1 epitopes in our study.
While a neutralizing antibody response is vitally important for the efficacy of most vaccines, induction of an antibody-mediated cellular response via cytolysis, phagocytosis, as well as activation of complement can also play a critical role in vaccine-induced protection. The EBOV and SUDV monovalent sera caused a partial degranulation of human NK cells that was not statistically significantly different from the degranulation elicited by the trivalent sera, as measured by CD107a expression. In contrast, the degranulation caused by the BDBV monovalent sera was not detected with the trivalent sera. MIP-1β is a chemoattractant for NK cells, monocytes, and other immune cells. The MIP-1β expression pattern by NK cells was similar to the one observed for CD107a, with the BDBV monovalent vaccine inducing significantly higher MIP-1β expression than the trivalent vaccine, while both the EBOV and SUDV monovalent vaccines elicited MIP-1β expression similar to that elicited by the trivalent vaccine. Additionally, neither vaccine induced the strong expression of IFN-γ, a hallmark of NK cell activity; however, previous reports have shown that IFN-γ expression is not necessary for a protective NK cell response (28). A recent study of the Fc-mediated protective effects of a large panel of MAbs against EBOV demonstrated that while activation of NK cells facilitates antibody-mediated protection, it is not absolutely required (54).
Interestingly, the induction of Fc-mediated phagocytosis appeared to be different with neutrophils and monocytes. All three monovalent vaccines induced higher levels of neutrophil phagocytosis of coated beads with the homologous GP than the trivalent vaccine. In contrast, there was no difference in monocyte phagocytosis of coated beads with the homologous GP between the monovalent and trivalent vaccines. Together, these results indicate that the protective efficacy of the HPIV3-vectored ebolavirus vaccines likely results from both the direct neutralization by Fab domains and the Fc-mediated protective effects.
In conclusion, this is the first in-depth investigation of the effects of combining monovalent ebolavirus vaccines on antibody responses, including a detailed analysis of the antibody repertoire, Fc-mediated protective mechanisms, and a potential imbalance between the responses to individual components. This study demonstrated for the first time that a single dose of a monovalent SUDV or BDBV HPIV3-vectored vaccine completely protects against the corresponding virus and that a single dose of the trivalent vaccine completely protects against all three filoviruses. The results presented here and in previous studies on HPIV3-vectored EBOV vaccines point toward a single-dose, needle-free vaccine that is fully protective against the three ebolaviruses and that elicits strong systemic humoral and cellular responses, as well as a mucosal immune response in the respiratory tract.
MATERIALS AND METHODS
Biosafety.
All work with filoviruses was performed within the Galveston National Laboratory biosafety level 4 (BSL-4) laboratories. All staff had the appropriate training and U.S. government permissions and registrations for work with filoviruses.
Ethics statement.
This study was carried out in strict accordance with the recommendations described in the Guide for the Care and Use of Laboratory Animals of the National Research Council (55) and the United States Department of Agriculture. All animal work was approved by the University of Texas Medical Branch (UTMB) Institutional Animal Care and Use Committee. The facilities at UTMB are accredited by the American Association for Accreditation of Laboratory Animal Care (accreditation number 000870). All efforts were made to minimize animal suffering, and all procedures involving potential pain were performed with the appropriate anesthetic or analgesic. The number of animals used in this study was scientifically justified based on statistical analyses of virological and immunological outcomes.
Generation of trivalent vaccine constructs.
The HPIV3-vectored vaccine constructs expressing BDBV and SUDV were cloned as previously described (13, 56). Briefly, the transcription initiation and transcription termination sequences were cloned into the genome of the JS strain of HPIV3 between the P and M genes via a modified AflII site. Filovirus GP sequences were generated by PCR using cDNA produced from the following virus stocks: for EBOV, Ebola virus/H.sapiens-tc/COD/1976/Yambuku-Mayinga (GenBank accession number NC_002549.1); for BDBV, Bundibugyo virus/H.sapiens-tc/UGA/200706291/Butalya-811249 (GenBank accession number KU182911.1); and for SUDV, Sudan virus/H.sapiens-tc/UGA/2000/Gulu-200011676 (GenBank accession number MH121163.1). The GP cDNAs were cloned between the added start/stop sequences. The chimeric viruses were recovered by transfection into BSR-T7 cells and propagated in LLC-MK2 cells at 48 h (43). To reach viral stock titers sufficiently high for intranasal immunization of ferrets, the supernatants of infected LLC-MK2 cells were concentrated with Centricon Plus 70 devices (EMD Millipore, Burlington, MA) according to the manufacturer’s instructions.
Generation of consensus-based antigens.
Six artificial antigens were developed to elicit broad-spectrum protection. Consensus-based antigens were designed using multilevel consensus generations similar to Giles and Ross’s COBRA design (34). All known (as of June 2013) full-length ebolavirus GP sequences were used to develop a phylogenetic tree (by use of the DNAStar MegAlign program). As expected, the sequences separated into species-specific and outbreak-specific clusters (see Fig. S1 in the supplemental material). The EBOV sequences were grouped into either 2 (EC1) or 4 (EC2) groups. A first-level consensus sequence for each group was generated. These group consensus sequences were then used to create an EBOV second-level consensus sequence. For EC1, the single TAFV sequence was grouped into the BDBV sequences to generate a single second-level consensus BDBV/TAFV sequence. For EC2, a BDBV-only first-level consensus sequence was generated and used to create a second-level consensus sequence with TAFV. All SUDV sequences were used to generate a second-level consensus sequence for both EC1 and EC2 (Fig. S1). Two MLD deletion constructs were generated by removing MLD amino acids from the EC1 sequence. EC1J had residues 309 to 495 removed, similar to the construct of Jeffers et al. (57), while EC1O had residues 315 to 506 removed, similar to the construct of Ou et al. (58). A mosaic antigen, ECM, was generated by replacing residues in EC1 with predicted epitopes specific to EBOV, BDBV, and SUDV. Finally, ECHI was generated in a fashion similar to that used to generate EC1; however, mismatches were solved by selecting the amino acid with the highest predicted immunogenicity. Consensus antigens were cloned, and viruses were recovered and propagated similarly to the wild-type GPs. Analysis of the amino acid sequence identity between different consensus sequences and the reference GP sequences was performed with Geneious software (version 11.1.5; Biomatters Ltd., Auckland, New Zealand).
GP expression by infected cells.
For the Western blot detection of GP expression by infected cells, monolayers of LLC-MK2 cells in six-well plates were infected with either wild-type HPIV3 (control), each monovalent vaccine construct (HPIV3-EBOV GP, HPIV3-SUDV GP, or HPIV3-BDBV GP), or a trivalent cocktail containing all three monovalent vaccine constructs at a multiplicity of infection (MOI) of 2 for 24 h at 32°C. Then, cells were lysed in radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Waltham, MA) and run on a 4 to 12% SDS-PAGE gel in the presence of purified GP proteins as the respective controls: EBOV GP (catalog number 0501-015; IBT Bioservices, Rockville, MD, USA), BDBV GP (catalog number 0505-015; IBT Bioservices), and SUDV GP (catalog number 0502-015; IBT Bioservices). After transfer to nitrocellulose, the Western blots were stained with the following antibody combinations: for EBOV GP, rabbit anti-EBOV GP at 1 μg/ml (catalog number 0301-015; IBT Bioservices) followed by goat anti-rabbit IgG 800 CW at 1:15,000 (catalog number 926-32311; LI-COR, Lincoln, NE); for BDBV GP, human MAb BDBV52 at 1 μg/ml followed by goat anti-human IgG 800 CW at 1:15,000 (catalog number 925-32232; LI-COR); and for SUDV GP, mouse MAb 2H5 specific for SUDV GP at 1 μg/ml (catalog number 0202-029; IBT Bioservices) followed by goat anti-mouse IgG 800 CW at 1:15,000 (catalog number 925-32210; LI-COR). As loading controls, actin was used for the EBOV and BDBV GP Western blots and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used for SUDV GP. Mouse anti-actin was used at 1:2,500 (catalog number MA5-11869; Invitrogen), and donkey anti-mouse IgG 680 RD was used at 1:15,000 (catalog number 925-68072; LI-COR). Rabbit anti-GAPDH was used at 1:1,000 (catalog number G9545-200UL; Sigma), and donkey anti-rabbit IgG 680 RD was used at 1:15,000 (catalog number 925-68073; LI-COR). Western blots were visualized with a LI-COR Odyssey Fc imaging system (LI-COR, Lincoln, NE).
For flow cytometry detection of GP in infected cells, monolayers of LLC-MK2 cells in 12-well plates were infected with either wild-type HPIV3 (control), each monovalent vaccine construct (HPIV3-EBOV GP, HPIV3-SUDV GP, or HPIV3-BDBV GP), or the trivalent cocktail containing all three monovalent vaccine constructs at an MOI of 1 for 48 h at 32°C. Then, the cells were harvested, washed twice with phosphate-buffered saline (PBS) and once with PBS supplemented with 2% fetal bovine serum (FBS; Thermo Fisher Scientific), and stained for 20 min at room temperature with one of the primary antibodies, rabbit polyclonal antibody specific for EBOV GP (catalog number 0301-015; IBT Bioservices), MAb BDBV52, or a mouse MAb specific for SUDV GP (catalog number 0202-029; IBT Bioservices), at a dilution of 1:200. Next, the cells were washed three times with PBS supplemented with 2% FBS and stained for 20 min at room temperature with one of the secondary antibodies, donkey anti-rabbit IgG-Alexa Fluor 405 (catalog number 175651; Abcam, Cambridge, UK), goat anti-human IgG-Alexa Fluor 488 (catalog number A-11013; Thermo Fisher Scientific), or chicken anti-mouse IgG-Alexa Fluor 647 (catalog number A-21463; Thermo Fisher Scientific), at a dilution of 1:200. Cells were washed twice with PBS with 2% FBS and once with PBS. Lastly, GP expression on the cells was analyzed by flow cytometry analysis using a BD LSRFortessa (BD Biosciences, San Jose, CA) flow cytometer; 3 × 104 events per samples were acquired.
Vaccination and filovirus challenge.
Three-week-old HPIV3-naive female Dunkin-Hartley guinea pigs were acquired from Charles Rivers Laboratories (Kingston, NY) and housed at UTMB for 1 week prior to vaccination. For blood collections and vaccine inoculations, the animals were anesthetized with 5% isoflurane. Groups of 4 to 5 animals were inoculated with 4 × 105 PFU of each of the monovalent vaccine constructs or a mixture of the three constructs at 4 × 105 of each construct, resulting in a total dose of 1.2 × 106 total PFU, in 200 μl phosphate-buffered saline via the intranasal route (100 μl in each nostril) on days 0 and 28. Retro-orbital blood collections were performed on days −1 (1 day prior to the first vaccine inoculation), 28, and 56. For the challenge studies, at 4 weeks postvaccination, vaccinated and control animals were exposed to the targeted dose of 103 PFU of guinea pig-adapted EBOV strain Mayinga or guinea pig-adapted SUDV strain Boniface, delivered by i.p. injection. Animals were monitored up to three times daily for weight loss and signs of disease. Animals that had reached the moribund state (which was defined as reaching one of the following conditions: failure to move upon stimulation, an inability to reach food or water, greater than 20% body weight loss, or paralysis) were euthanized. Retro-orbital blood collections from surviving animals were performed at days 3, 6, 9, 12 or 13, and 28 postchallenge. All remaining animals were euthanized at 28 days postinfection by CO2 asphyxiation, and the chest was opened under veterinarian direction.
Female ferrets (weight, 0.6 kg to 1.0 kg) were acquired from Marshall BioResources (North Rose, NY) and housed at UTMB 1 week prior to vaccination. For blood collections and vaccine inoculations, animals were anesthetized with 5% isoflurane. Groups of 5 ferrets were inoculated with 1 × 107 PFU of each of the monovalent vaccine constructs or a mixture of the three constructs at 1 × 107 of each construct, resulting in a total dose of 3 × 107 total PFU, in 1.0 ml phosphate-buffered saline via the intranasal route (500 μl in each nostril) on day 0. One ferret immunized with HPIV3-BDBV was removed from the experiment due to a condition not related to the study. Blood collections were performed on day −1 (1 day prior to vaccine inoculation) and on day 28. At 4 weeks postvaccination, vaccinated and control animals were exposed to the targeted dose of 103 PFU BDBV strain Uganda by i.m. injection. Animals were monitored up to three times daily for weight loss and signs of disease. Animals that had reached a moribund state (which was defined as one of the following conditions: failure to move upon stimulation, visible ecchymosis, respiratory rate of >80 beats per minute, open-mouth breathing, or severe dehydration, evidenced by sunken eyes and prominent vertebrae, dorsal pelvis, and/or ribs) were euthanized. Collection of blood from surviving animals was performed at days 3, 6, 9, 13, and 28 postchallenge. All remaining animals were euthanized at 28 days after BDBV challenge.
Plaque reduction assay.
Sera collected from animals were tested for virus-neutralizing capabilities against EBOV strain Mayinga, BDBV strain Uganda, and SUDV strain Gulu. Briefly, 10-fold-diluted sera were further diluted in a 2-fold serial fashion. Fifty microliters of each serum dilution was mixed with 200 PFU of virus in 50 μl. The serum/virus mixtures were incubated for 1 h at 37°C. Fifty microliters (100 PFU) of the mixtures was then added to Vero E6 cell monolayers in flat-bottom 96-well plates and the cells were incubated for 1 h at 37°C. Serum/virus mixtures were removed from the cells, which were then covered with 1% methylcellulose in minimal essential medium with 2% FBS (Gibco, Grand Island, NY). The cells were fixed in 10% formalin for 2 days and removed from the BSL-4 laboratory, and plaques were visualized by immunostaining and counted as previously described (33). Briefly, EBOV plaques were stained with goat anti-EBOV serum at a dilution of 1:2,000 (a gift of T. G. Ksiazek, UTMB) and horseradish peroxidase (HRP)-conjugated bovine anti-goat IgG at a dilution of 1:2,000 (catalog number 805-035-180; Jackson ImmunoResearch, West Grove, PA). BDBV and SUDV plaques were stained with 1 μg/ml human MAb BDBV52 or BDBV43 (30), respectively, and HRP-conjugated goat anti-human IgG at a dilution of 1:2,000 (catalog number 474-1002; KPL, Gaithersburg, MD).
ELISA.
Sera collected from animals were tested for their ability to bind the GPs of the three ebolaviruses by enzyme-linked immunosorbent assay (ELISA). For the guinea pig sera, the ELISA plates were coated overnight at room temperature with 8 ng of recombinant EBOV GP, BDBV GP, or SUDV GP (His-tagged glycoproteins minus the transmembrane domain produced in Sf9 insect cells; IBT Bioservices) diluted in PBS. The plates were washed five times in PBS with 0.1% Tween 20 and blocked with 3% dry milk in PBS for 1 h at 37°C. After blocking, 4-fold dilutions of guinea pig sera were prepared in blocking buffer and incubated on the GP-coated plates for 1 h at 37°C. The plates were washed as described above and incubated with a 1:5,000 dilution of horseradish peroxidase-conjugated goat anti-guinea pig immunoglobulin antibody (Jackson ImmunoResearch, West Grove, PA) for 1 h at 37°C. The plates were washed as described above and developed using a SureBlue ELISA substrate system (KPL, Gaithersburg, MD), and the color intensity was measured on a BioTek Synergy HT microplate reader (Winooski, VT) at 630 nm.
For the ferret sera, ELISA plates were coated overnight at room temperature with 6.25 ng of recombinant EBOV GP, BDBV GP, or SUDV GP (IBT Bioservices) diluted in PBS. The plates were washed five times in PBS with 0.1% Tween 20 and blocked for 1 h at 37°C in PBS with 5% dry milk and 2.5% bovine serum albumin (BSA). After blocking, 4-fold dilutions of ferret serum were prepared in blocking buffer and incubated on the GP-coated plates for 1 h at 37°C. The plates were washed as described above and incubated with a 1:10,000 dilution of horseradish peroxidase-conjugated goat anti-ferret IgG antibody (Abcam, Cambridge, MA) for 1 h at 37°C. The plates were washed as described above and developed using the SureBlue ELISA substrate system (KPL), and the color intensity was measured on the BioTek Synergy HT microplate reader (Winooski, VT) at 630 nm.
Peptide microarrays.
A total of 167 overlapping, 15-residue peptides were designed to cover the sequence of the GP of EBOV (Ebola virus/H.sapiens-tc/COD/1976/Yambuku-Mayinga). An additional 167 peptides were designed similarly to match the BDBV (Bundibugyo virus/H.sapiens-tc/UGA/200706291/Butalya-811249) or SUDV (Sudan virus/H.sapiens-tc/UGA/2000/Gulu-200011676) sequence. The first peptide corresponded to residues 1 to 15, and each successive peptide began 4 residues downstream (producing peptides from residues 5 to 19, 9 to 23, etc.). The peptides were produced and immobilized on glass slides by JPT Peptide Technologies (Berlin, Germany). Each of the peptides was spotted via its N terminus in triplicate to form 1 block of spots, and 21 blocks were spotted onto a slide. Dilutions of 1:200 of day 0, 28, and 56 serum samples were incubated for 1 h at 30°C, followed by 4 washes in JPT washing buffer (1× Tris-buffered saline [TBS] buffer [20 mM Tris, 136 mM NaCl, pH 7.4] plus 0.1% Tween 20 [TBS-T]). One block incubated in washing buffer was used as a negative control. All blocks were incubated in 0.1 μg/ml anti-guinea pig-Cy5-conjugated antibodies (Jackson ImmunoResearch, West Grove, PA), followed by four washes. After an additional wash in deionized water, the slide was dried by centrifugation. Fluorescent readings were recorded for each spot on a GenePix 4000b microarray scanner (Molecular Devices, Sunnyvale, CA) and analyzed by GenePix Pro 6 software (Molecular Devices, Sunnyvale, CA). Sera from 3 animals per group were tested, and the results were normalized as a percentage of the value for the negative control.
Biolayer interferometry competition assay.
We used an Octet RED96 (FortéBio, Fresno, CA) instrument for biolayer interferometry antibody competition assays. All samples were diluted in 1× kinetics buffer (FortéBio) to a final volume of 200 μl. The assay was performed with agitation at 1,000 rpm and 28°C in black 96-well plates (Greiner Bio-One, Monroe, NC). GP was biotinylated using an EZ-Link Micro-PEG4-Biotinylation kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions and was immobilized on streptavidin sensors (FortéBio) at a capture level of 0.5 nm. The probes were dipped into serum samples diluted 1:5 for 900 s to saturate the immobilized GP. The probes were then washed twice for 60 s each time, and the reactivity of competing human MAbs (25 nM 2D22, which is specific for dengue virus serotype II [59], used as a negative control; BDBV289; EBOV55; KZ52; and BDBV223 and 300 nM BDBV317) for GP was assessed for 600 s. The difference in the level of competitor binding to GP was calculated by subtracting the level of competitor MAb binding observed after preincubation with serum from vaccinated animals from the level of MAb binding observed after incubation with serum from empty vector-vaccinated animals. Data analysis was carried out using Octet software, version 7.0.
Antibody-dependent NK cell degranulation.
Recombinant EBOV GP, BDBV GP, or SUDV GP lacking the transmembrane domain (IBT Bioservices) was used for all experiments to assess the induction of innate immune effector functions. Recombinant antigen was coated onto MaxiSorp 96-well plates (Nunc) at 300 ng/well at 4°C for 18 h. The wells were washed three times with PBS and blocked with 5% bovine serum albumin in PBS. Sera from immunized guinea pigs diluted 1:50 in PBS were added, and the plates were incubated for 2 h at 37°C. Unbound antibodies were removed by washing three times with PBS, and human NK cells freshly isolated from the peripheral blood of human donors by negative selection (Stem Cell Technologies, Vancouver, Canada) were added at 5 × 104 cells/well in the presence of 4 μg/ml brefeldin A (Sigma-Aldrich, St. Louis, MO), 5 μg/ml GolgiStop protein transport inhibitor (Life Technologies, Carlsbad, CA), and anti-CD107a antibody (1:40 phycoerythrin [PE]-Cy5; clone H4A3; BD Biosciences). The plates were incubated for 5 h. Cells were stained with anti-CD3 (1:100 Alexa Fluor 700; clone UCHT1; BD Biosciences), anti-CD16 (1:100 allophycocyanin [APC]-Cy7; clone 3G8; BD Biosciences), and anti-CD56 (1:100 PE-Cy7; clone B159; BD Biosciences), followed by fixation and permeabilization with the Fix & Perm reagent (Life Technologies) according to the manufacturer’s instructions to stain for intracellular IFN-γ (1:50 APC; clone B27; BD Biosciences) and MIP-1β (1:50 PE; clone D21-1351; BD Biosciences). Cells were analyzed on a BD LSRII flow cytometer, and a minimum of 10,000 events were recorded and analyzed. The gating strategy is presented in Fig. S2A. The EBOV GP-specific humanized murine IgG1 c13C6 (IBT Bioservices) was used at 5 μg/ml as a positive control, and naive guinea pig sera were used as a negative control (dilution, 1:50).
Antibody-mediated neutrophil phagocytosis.
Recombinant EBOV GP, BDBV GP, or SUDV GP was biotinylated and coupled to 1-μm FITC+ NeutrAvidin beads (Life Technologies). Briefly, the proteins were biotinylated using Sulfo-NHS-LC-LC biotin (catalog number 21338; Thermo Fisher Scientific), and excess biotin was removed using a Zeba desalting column (catalog number 89883; Thermo Fisher Scientific). Biotinylated protein was incubated with NeutrAvidin-coated fluorescent beads (catalog number F8776; Thermo Fisher Scientific) at 4°C overnight (1 μg biotinylated protein per 1 μl of beads). The beads were washed twice with 0.1% BSA in PBS and resuspended in 100 μl of 0.1% BSA in PBS per 1 μl of coupled beads. Sera from vaccinated guinea pigs or MAbs were diluted 1:100 (guinea pig serum) or to 5 μg/ml (MAbs) in cell culture medium and incubated with GP-coated beads for 2 h at 37°C. Freshly isolated neutrophils from donor peripheral blood were added at a concentration of 5.0 × 104 cells/well, and the plates were incubated for 1 h at 37°C. The cells were stained at 1:100 with CD66b (Pacific Blue; clone G10F5; BioLegend, San Diego, CA), CD3 (Alexa Fluor 700; clone UCHT1; BD Biosciences), and CD14 (APC-Cy7; clone MφP9; BD Biosciences), fixed with 4% paraformaldehyde, and analyzed by flow cytometry on a BD LSRII flow cytometer, and a minimum of 30,000 events were recorded and analyzed. The gating strategy is presented in Fig. S2B. Neutrophils were defined as positive for a high side scatter area (SSC-Ahigh), CD66b+, CD3−, and CD14−. The phagocytic score was determined using the following formula: [(percentage of FITC+ cells) × (median fluorescent intensity [MFI] of the FITC+ cells)]/10,000.
Antibody-mediated cellular phagocytosis by human monocytes.
Recombinant EBOV GP, BDBV GP, or SUDV GP was biotinylated and coupled to 1-μm FITC+ NeutrAvidin beads (Life Technologies). Serum samples from vaccinated guinea pigs or MAbs were diluted 1:500 or 5 μg/ml, respectively, in culture medium and incubated with GP-coated beads for 2 h at 37°C, followed by addition of cells for 18 h, as previously described (60). Cells were fixed with 4% paraformaldehyde and analyzed on a BD LSRII flow cytometer, and a minimum of 10,000 events were recorded and analyzed. The gating strategy is presented in Fig. S2C. The phagocytic score was determined as described above.
Antibody-mediated complement deposition.
Recombinant EBOV GP, BDBV GP, or SUDV GP was biotinylated and coupled to 1-μm red fluorescent NeutrAvidin beads (Life Technologies). Serum from vaccinated guinea pigs was diluted 1:10, 1:100, and 1:200 in culture medium and incubated with GP-coated beads for 2 h at 37°C, followed by the addition of reconstituted guinea pig complement (Cedarlane Labs, Burlington, NC) diluted in gelatin Veronal buffer containing magnesium and calcium (Boston Bioproducts, Boston, MA) and incubation for 20 min at 37°C. The beads were washed twice with phosphate-buffered saline containing 15 mM EDTA and stained with an anti-guinea pig C3 antibody conjugated to FITC (1:100; MP Biomedicals, Santa Ana, CA). C3 deposition onto the beads was analyzed on a BD LSRII flow cytometer, and a minimum of 10,000 events were recorded and analyzed. The gating strategy is presented in Fig. S2D. The complement score was calculated according to the following formula: [(geometric mean fluorescent intensity of FITC of all beads) × (percentage of FITC+ beads)]/10,000.
Statistics.
All statistical calculations were performed using GraphPad Prism software (version 6.07) for Windows. Descriptive statistics (mean ± standard error of the mean [SEM]) are provided for continuous variables, as noted. Significance was calculated by analysis of variance (ANOVA) with Tukey’s multiple-comparison test. Survival curves were compared by the log-rank (Mantel-Cox) test. A P value of <0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jessica Graber and the Animal Resources Center of the Galveston National Laboratory for assisting with all animal work. We thank Britney Ross for assisting with the generation of the vaccine constructs. We also thank James Zhu for assisting with the analysis of the peptide microarray.
This work was supported by National Institute of Allergy and Infectious Diseases (NIAID), NIH, grant 1R01AI102887-01A1; NIAID, NIH, grant U19 AI109711; and Defense Threat Reduction Agency grant HDTRA1-13-1-0034 to A.B.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01845-18.
REFERENCES
- 1.CDC. 2016. 2014 Ebola outbreak in West Africa—case counts. http://www.cdc.gov/vhf/ebola/outbreaks/2014-west-africa/case-counts.html.
- 2.Kuhn JH. 2008. Filoviruses, 1st ed Springer, Vienna, Austria. [Google Scholar]
- 3.Feldmann H, Sanchez A, Geisbert TW. 2013. Filoviridae: Marburg and Ebola viruses, p. 923–956. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed.), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Jaax N, Jahrling P, Geisbert T, Geisbert J, Steele K, McKee K, Nagley D, Johnson E, Jaax G, Peters C. 1995. Transmission of Ebola virus (Zaire strain) to uninfected control monkeys in a biocontainment laboratory. Lancet 346:1669–1671. doi: 10.1016/S0140-6736(95)92841-3. [DOI] [PubMed] [Google Scholar]
- 5.Johnson E, Jaax N, White J, Jahrling P. 1995. Lethal experimental infections of rhesus monkeys by aerosolized Ebola virus. Int J Exp Pathol 76:227–236. [PMC free article] [PubMed] [Google Scholar]
- 6.Reed DS, Lackemeyer MG, Garza NL, Sullivan LJ, Nichols DK. 2011. Aerosol exposure to Zaire ebolavirus in three nonhuman primate species: differences in disease course and clinical pathology. Microbes Infect 13:930–936. doi: 10.1016/j.micinf.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Wong G, Qiu X, Richardson JS, Cutts T, Collignon B, Gren J, Aviles J, Embury-Hyatt C, Kobinger GP. 2015. Ebola virus transmission in Guinea pigs. J Virol 89:1314–1323. doi: 10.1128/JVI.02836-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nazir SA, Metcalf JP. 2005. Innate immune response to adenovirus. J Investig Med 53:292–304. doi: 10.2310/6650.2005.53605. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan NJ, Geisbert TW, Geisbert JB, Shedlock DJ, Xu L, Lamoreaux L, Custers JH, Popernack PM, Yang ZY, Pau MG, Roederer M, Koup RA, Goudsmit J, Jahrling PB, Nabel GJ. 2006. Immune protection of nonhuman primates against Ebola virus with single low-dose adenovirus vectors encoding modified GPs. PLoS Med 3:e177. doi: 10.1371/journal.pmed.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan NJ, Sanchez A, Rollin PE, Yang ZY, Nabel GJ. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 408:605–609. doi: 10.1038/35046108. [DOI] [PubMed] [Google Scholar]
- 11.Jones SM, Feldmann H, Ströher U, Geisbert JB, Fernando L, Grolla A, Klenk HD, Sullivan NJ, Volchkov VE, Fritz EA, Daddario KM, Hensley LE, Jahrling PB, Geisbert TW. 2005. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat Med 11:786–790. doi: 10.1038/nm1258. [DOI] [PubMed] [Google Scholar]
- 12.Geisbert TW, Geisbert JB, Leung A, Daddario-DiCaprio KM, Hensley LE, Grolla A, Feldmann H. 2009. Single-injection vaccine protects nonhuman primates against infection with Marburg virus and three species of Ebola virus. J Virol 83:7296–7304. doi: 10.1128/JVI.00561-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bukreyev A, Yang L, Zaki SR, Shieh WJ, Rollin PE, Murphy BR, Collins PL, Sanchez A. 2006. A single intranasal inoculation with a paramyxovirus-vectored vaccine protects guinea pigs against a lethal-dose Ebola virus challenge. J Virol 80:2267–2279. doi: 10.1128/JVI.80.5.2267-2279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bukreyev A, Rollin PE, Tate MK, Yang L, Zaki SR, Shieh WJ, Murphy BR, Collins PL, Sanchez A. 2007. Successful topical respiratory tract immunization of primates against Ebola virus. J Virol 81:6379–6388. doi: 10.1128/JVI.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanley DA, Honko AN, Asiedu C, Trefry JC, Lau-Kilby AW, Johnson JC, Hensley L, Ammendola V, Abbate A, Grazioli F, Foulds KE, Cheng C, Wang L, Donaldson MM, Colloca S, Folgori A, Roederer M, Nabel GJ, Mascola J, Nicosia A, Cortese R, Koup RA, Sullivan NJ. 2014. Chimpanzee adenovirus vaccine generates acute and durable protective immunity against ebolavirus challenge. Nat Med 20:1126–1129. doi: 10.1038/nm.3702. [DOI] [PubMed] [Google Scholar]
- 16.CDC. 2016. About Ebola virus disease. CDC, Atlanta, GA. [Google Scholar]
- 17.Knuf M, Habermehl P, Zepp F, Mannhardt W, Kuttnig M, Muttonen P, Prieler A, Maurer H, Bisanz H, Tornieporth N, Descamps D, Willems P. 2006. Immunogenicity and safety of two doses of tetravalent measles-mumps-rubella-varicella vaccine in healthy children. Pediatr Infect Dis J 25:12–18. [DOI] [PubMed] [Google Scholar]
- 18.Tunis MC, Deeks SL, National Advisory Committee on Immunization. 2016. Summary of the National Advisory Committee on Immunization's updated recommendations on human papillomavirus (HPV) vaccines: nine-valent human papillomavirus (HPV) of minimum intervals between doses in the HPV immunization schedule. Can Commun Dis Rep 42:149–151. doi: 10.14745/ccdr.v42i07a03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plotkin SA, Koprowski H, Stokes J Jr.. 1959. Clinical trials in infants of orally administered attenuated poliomyelitis viruses. Pediatrics 23:1041–1062. [PubMed] [Google Scholar]
- 20.Perkins FT, Yetts R, Gaisford W. 1963. Response of 3-months-old infants to 3 doses of trivalent oral poliomyelitis vaccine. Br Med J 1:1573–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guy B, Barban V, Mantel N, Aguirre M, Gulia S, Pontvianne J, Jourdier TM, Ramirez L, Gregoire V, Charnay C, Burdin N, Dumas R, Lang J. 2009. Evaluation of interferences between dengue vaccine serotypes in a monkey model. Am J Trop Med Hyg 80:302–311. [PubMed] [Google Scholar]
- 22.Anderson KB, Gibbons RV, Edelman R, Eckels KH, Putnak RJ, Innis BL, Sun W. 2011. Interference and facilitation between dengue serotypes in a tetravalent live dengue virus vaccine candidate. J Infect Dis 204:442–450. doi: 10.1093/infdis/jir279. [DOI] [PubMed] [Google Scholar]
- 23.Swenson DL, Warfield KL, Negley DL, Schmaljohn A, Aman MJ, Bavari S. 2005. Virus-like particles exhibit potential as a pan-filovirus vaccine for both Ebola and Marburg viral infections. Vaccine 23:3033–3042. doi: 10.1016/j.vaccine.2004.11.070. [DOI] [PubMed] [Google Scholar]
- 24.Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, Hart MK. 2000. Epitopes involved in antibody-mediated protection from Ebola virus. Science 287:1664–1666. [DOI] [PubMed] [Google Scholar]
- 25.Gunn BM, Alter G. 2016. Modulating antibody functionality in infectious disease and vaccination. Trends Mol Med 22:969–982. doi: 10.1016/j.molmed.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeitlin L, Pettitt J, Scully C, Bohorova N, Kim D, Pauly M, Hiatt A, Ngo L, Steinkellner H, Whaley KJ, Olinger GG. 2011. Enhanced potency of a fucose-free monoclonal antibody being developed as an Ebola virus immunoprotectant. Proc Natl Acad Sci U S A 108:20690–20694. doi: 10.1073/pnas.1108360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams KJ, Qiu X, Fernando L, Jones SM, Alimonti JB. 2015. VSVDeltaG/EBOV GP-induced innate protection enhances natural killer cell activity to increase survival in a lethal mouse adapted Ebola virus infection. Viral Immunol 28:51–61. doi: 10.1089/vim.2014.0069. [DOI] [PubMed] [Google Scholar]
- 28.Warfield KL, Perkins JG, Swenson DL, Deal EM, Bosio CM, Aman MJ, Yokoyama WM, Young HA, Bavari S. 2004. Role of natural killer cells in innate protection against lethal Ebola virus infection. J Exp Med 200:169–179. doi: 10.1084/jem.20032141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricklin D, Hajishengallis G, Yang K, Lambris JD. 2010. Complement: a key system for immune surveillance and homeostasis. Nat Immunol 11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flyak AI, Shen X, Murin CD, Turner HL, David JA, Fusco ML, Lampley R, Kose N, Ilinykh PA, Kuzmina N, Branchizio A, King H, Brown L, Bryan C, Davidson E, Doranz BJ, Slaughter JC, Sapparapu G, Klages C, Ksiazek TG, Saphire EO, Ward AB, Bukreyev A, Crowe JE Jr. 2016. Cross-reactive and potent neutralizing antibody responses in human survivors of natural ebolavirus infection. Cell 164:392–405. doi: 10.1016/j.cell.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holtsberg FW, Shulenin S, Vu H, Howell KA, Patel SJ, Gunn B, Karim M, Lai JR, Frei JC, Nyakatura EK, Zeitlin L, Douglas R, Fusco ML, Froude JW, Saphire EO, Herbert AS, Wirchnianski AS, Lear-Rooney CM, Alter G, Dye JM, Glass PJ, Warfield KL, Aman MJ. 2016. Pan-ebolavirus and pan-filovirus mouse monoclonal antibodies: protection against Ebola and Sudan viruses. J Virol 90:266–278. doi: 10.1128/JVI.02171-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furuyama W, Marzi A, Nanbo A, Haddock E, Maruyama J, Miyamoto H, Igarashi M, Yoshida R, Noyori O, Feldmann H, Takada A. 2016. Discovery of an antibody for pan-ebolavirus therapy. Sci Rep 6:20514. doi: 10.1038/srep20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer M, Garron T, Lubaki NM, Mire CE, Fenton KA, Klages C, Olinger GG, Geisbert TW, Collins PL, Bukreyev A. 2015. Aerosolized Ebola vaccine protects primates and elicits lung-resident T cell responses. J Clin Invest 125:3241–3255. doi: 10.1172/JCI81532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giles BM, Ross TM. 2011. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29:3043–3054. doi: 10.1016/j.vaccine.2011.01.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittmann TJ, Biek R, Hassanin A, Rouquet P, Reed P, Yaba P, Pourrut X, Real LA, Gonzalez JP, Leroy EM. 2007. Isolates of Zaire ebolavirus from wild apes reveal genetic lineage and recombinants. Proc Natl Acad Sci U S A 104:17123–17127. doi: 10.1073/pnas.0704076104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henikoff S, Henikoff JG. 1993. Performance evaluation of amino acid substitution matrices. Proteins 17:49–61. doi: 10.1002/prot.340170108. [DOI] [PubMed] [Google Scholar]
- 37.Flyak AI, Kuzmina N, Murin CD, Bryan C, Davidson E, Gilchuk P, Gulka CP, Ilinykh PA, Shen X, Huang K, Ramanathan P, Turner H, Fusco ML, Lampley R, Kose N, King H, Sapparapu G, Doranz BJ, Ksiazek TG, Wright DW, Saphire EO, Ward AB, Bukreyev A, Crowe JE Jr. 2018. Broadly neutralizing antibodies from human survivors target a conserved site in the Ebola virus glycoprotein HR2-MPER region. Nat Microbiol 3:670–677. doi: 10.1038/s41564-018-0157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alter G, Malenfant JM, Altfeld M. 2004. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods 294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 40.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP. 2014. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borregaard N. 2010. Neutrophils, from marrow to microbes. Immunity 33:657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 42.Nauseef WM. 2007. How human neutrophils kill and degrade microbes: an integrated view. Immunol Rev 219:88–102. doi: 10.1111/j.1600-065X.2007.00550.x. [DOI] [PubMed] [Google Scholar]
- 43.Connolly BM, Steele KE, Davis KJ, Geisbert TW, Kell WM, Jaax NK, Jahrling PB. 1999. Pathogenesis of experimental Ebola virus infection in guinea pigs. J Infect Dis 179(Suppl 1):S203–S217. doi: 10.1086/514305. [DOI] [PubMed] [Google Scholar]
- 44.Wong G, He S, Wei H, Kroeker A, Audet J, Leung A, Cutts T, Graham J, Kobasa D, Embury-Hyatt C, Kobinger GP, Qiu X. 2016. Development and characterization of a guinea pig-adapted Sudan virus. J Virol 90:392–399. doi: 10.1128/JVI.02331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cross RW, Mire CE, Borisevich V, Geisbert JB, Fenton KA, Geisbert TW. 2016. The domestic ferret (Mustela putorius furo) as a lethal infection model for 3 species of ebolavirus. J Infect Dis 214:565–569. doi: 10.1093/infdis/jiw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volchkov VE, Chepurnov AA, Volchkova VA, Ternovoj VA, Klenk HD. 2000. Molecular characterization of guinea pig-adapted variants of Ebola virus. Virology 277:147–155. doi: 10.1006/viro.2000.0572. [DOI] [PubMed] [Google Scholar]
- 47.Pratt WD, Wang D, Nichols DK, Luo M, Woraratanadharm J, Dye JM, Holman DH, Dong JY. 2010. Protection of nonhuman primates against two species of Ebola virus infection with a single complex adenovirus vector. Clin Vaccine Immunol 17:572–581. doi: 10.1128/CVI.00467-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pettitt J, Zeitlin L, Kim DH, Working C, Johnson JC, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, Pauly MH, Whaley KJ, Ingram MF, Zovanyi A, Heinrich M, Piper A, Zelko J, Olinger GG. 2013. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Sci Transl Med 5:199ra113. doi: 10.1126/scitranslmed.3006608. [DOI] [PubMed] [Google Scholar]
- 49.Becquart P, Mahlakoiv T, Nkoghe D, Leroy EM. 2014. Identification of continuous human B-cell epitopes in the VP35, VP40, nucleoprotein and glycoprotein of Ebola virus. PLoS One 9:e96360. doi: 10.1371/journal.pone.0096360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maruyama T, Parren PW, Sanchez A, Rensink I, Rodriguez LL, Khan AS, Peters CJ, Burton DR. 1999. Recombinant human monoclonal antibodies to Ebola virus. J Infect Dis 179(Suppl 1):S235–S239. doi: 10.1086/514280. [DOI] [PubMed] [Google Scholar]
- 51.Parren PW, Geisbert TW, Maruyama T, Jahrling PB, Burton DR. 2002. Pre- and postexposure prophylaxis of Ebola virus infection in an animal model by passive transfer of a neutralizing human antibody. J Virol 76:6408–6412. doi: 10.1128/JVI.76.12.6408-6412.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saphire EO, Schendel SL, Fusco ML, Gangavarapu K, Gunn BM, Wec AZ, Halfmann PJ, Brannan JM, Herbert AS, Qiu X, Wagh K, He S, Giorgi EE, Theiler J, Pommert KBJ, Krause TB, Turner HL, Murin CD, Pallesen J, Davidson E, Ahmed R, Aman MJ, Bukreyev A, Burton DR, Crowe JE Jr, Davis CW, Georgiou G, Krammer F, Kyratsous CA, Lai JR, Nykiforuk C, Pauly MH, Rijal P, Takada A, Townsend AR, Volchkov V, Walker LM, Wang CI, Zeitlin L, Doranz BJ, Ward AB, Korber B, Kobinger GP, Andersen KG, Kawaoka Y, Alter G, Chandran K, Dye JM, Viral Hemorrhagic Fever Immunotherapeutic Consortium. 2018. Systematic analysis of monoclonal antibodies against Ebola virus GP defines features that contribute to protection. Cell 174:938–952.e13. doi: 10.1016/j.cell.2018.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khurana S, Fuentes S, Coyle EM, Ravichandran S, Davey RT Jr, Beigel JH. 2016. Human antibody repertoire after VSV-Ebola vaccination identifies novel targets and virus-neutralizing IgM antibodies. Nat Med 22:1439–1447. doi: 10.1038/nm.4201. [DOI] [PubMed] [Google Scholar]
- 54.Gunn BM, Yu WH, Karim MM, Brannan JM, Herbert AS, Wec AZ, Halfmann PJ, Fusco ML, Schendel SL, Gangavarapu K, Krause T, Qiu X, He S, Das J, Suscovich TJ, Lai J, Chandran K, Zeitlin L, Crowe JE Jr, Lauffenburger D, Kawaoka Y, Kobinger GP, Andersen KG, Dye JM, Saphire EO, Alter G. 2018. A role for Fc function in therapeutic monoclonal antibody-mediated protection against Ebola virus. Cell Host Microbe 24:221–233.e5. doi: 10.1016/j.chom.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 56.Skiadopoulos MH, Surman SR, Riggs JM, Orvell C, Collins PL, Murphy BR. 2002. Evaluation of the replication and immunogenicity of recombinant human parainfluenza virus type 3 vectors expressing up to three foreign glycoproteins. Virology 297:136–152. [DOI] [PubMed] [Google Scholar]
- 57.Jeffers SA, Sanders DA, Sanchez A. 2002. Covalent modifications of the Ebola virus glycoprotein. J Virol 76:12463–12472. doi: 10.1128/JVI.76.24.12463-12472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ou W, Delisle J, Jacques J, Shih J, Price G, Kuhn JH, Wang V, Verthelyi D, Kaplan G, Wilson CA. 2012. Induction of ebolavirus cross-species immunity using retrovirus-like particles bearing the Ebola virus glycoprotein lacking the mucin-like domain. Virol J 9:32. doi: 10.1186/1743-422X-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fibriansah G, Ibarra KD, Ng TS, Smith SA, Tan JL, Lim XN, Ooi JS, Kostyuchenko VA, Wang J, de Silva AM, Harris E, Crowe JE Jr, Lok SM. 2015. Dengue virus. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 349:88–91. doi: 10.1126/science.aaa8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ackerman ME, Moldt B, Wyatt RT, Dugast AS, McAndrew E, Tsoukas S, Jost S, Berger CT, Sciaranghella G, Liu Q, Irvine DJ, Burton DR, Alter G. 2011. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J Immunol Methods 366:8–19. doi: 10.1016/j.jim.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.