Abstract
We report an important but long-overlooked manifestation of low-resolution power of 16S rRNA sequence analysis at the species level, namely, in 16S rRNA-based phylogenetic trees polyphyletic placements of closely-related species are abundant compared to those in genome-based phylogeny. This phenomenon makes the demarcation of genera within many families ambiguous in the 16S rRNA-based taxonomy. In this study, we reconstructed phylogenetic relationship for more than ten thousand prokaryote genomes using the CVTree method, which is based on whole-genome information. And many such genera, which are polyphyletic in 16S rRNA-based trees, are well resolved as monophyletic clusters by CVTree. We believe that with genome sequencing of prokaryotes becoming a commonplace, genome-based phylogeny is doomed to play a definitive role in the construction of a natural and objective taxonomy.
Keywords: Archaea and bacteria taxonomy, Phylogeny, CVTree, Whole-genome sequence, 16S rRNA sequence
Introduction
The use of small subunit (SSU) rRNA as molecular marker by Carl Woese and coworkers in the 1970s [1] has been a great success in prokaryotic taxonomy. Nowadays, the major references to prokaryotic taxonomy such as The Bergey’s Manual, including both the 2nd hardcopy edition [2] and the online electronic edition (BMSAB) [3], the multi-volume treatise The Prokaryotes IV [4], and the List of Prokaryotic Names with Standing in Nomenclature (LPSN) [5], are all based on 16S rRNA sequence analysis. At the same time, it has been recognized that the SSU rRNA sequences lack resolution at the species level and below (see, e.g., [6], [7], [8], [9]). However, to the best of our knowledge, a more severe consequence of the low resolution of 16S rRNA sequence analysis has not been reported in the literature so far, namely, redundant polyphyletic placements of species in 16S rRNA trees prevent correct definition of many genera. In contrast, many such genera are well-defined as monophyletic clusters in whole-genome-based phylogeny. In the present work, we demonstrate this phenomenon with a number of examples.
Methods
We use the All-Species Living Tree [9], [10], [11], abbreviated as LVTree, as reference of phylogenetic information from 16S rRNA sequence analysis. The latest release of LVTree, LTPs128 of February 2017, was based on 475 archaeal and 12,478 bacterial 16S rRNA sequences. We display and manipulate LVTree using the LVTree Viewer [12].
Whole-genome-based phylogenetic trees were constructed by implementing the alignment-free Composition Vector approach [13], [14], [15], [16]. In fact, in order to generate data for this paper, we use a more powerful version of the publicly-available CVTree3 Web Server [16]. It is capable to deal with 10,000–15,000 genomes in a single run within reasonable CPU time. These genomes were picked up from a collection of more than 125,000 prokaryotic genomes downloaded from IMG [17], RefSeq [18], NCBI [19], and occasionally, PATRIC [20] or EzBioCloud [21]. It is a good practice to put any group of species under study in the background of a large number of genomes with a wide taxonomic distribution. A typical CVTree job used in the present study contains 254 archaeal and 8036 bacterial genomes with K = 6.
A guiding principle in evaluating the quality of a taxon is monophyly. Historically, the notion of monophyly originated from zoology and was associated with sexual reproduction. We apply it to prokaryotes in a pragmatic way by restricting the discussion to an input dataset and a reference taxonomy. A tree branch is said to be monophyletic if it contains exclusively species from a given taxon according to the reference taxonomy. For example, if all 144 leaves of a branch come from the same family, say, Acetobacteraceae, and no members of this family appear in other branches, we write the family as Acetobacteraceae{144}, where 144 is the number of 16S rRNA sequences in LVTree or number of genomes in CVTree. A taxon is said to be well-defined if it is monophyletic.
Both CVTree Web Server and LVTree Viewer report automatically whether a taxon is monophyletic or not, at all taxonomic ranks from phylum down to species. Comparison of CVTree and LVTree phylogenies with taxonomy is carried out in a family-by-family manner. LVTree Release 128 contains 358 families. Among them, 68 monospecific families are trivially monophyletic containing only a single species, 180 are monophyletic, and the remaining 110 families are non-monophyletic. The aforementioned typical CVTree job contains 313 families, of which 76 are trivially monophyletic, 139 monophyletic, and 98 non-monophyletic. Some non-monophyletic families may become well-defined by making just a few obvious lineage modifications. Table 1 lists a number of families containing a comparatively large number of subordinate genera and species.
Table 1.
Number of organisms in some families well-defined in both LVTree and CVTree up to probable minor lineage modifications
| Family | LVTree | CVTree | Remark | Ref. |
|---|---|---|---|---|
| Acetobacteraceae | 144 | 233 | Stella transferred to Rhodospirillaceae | [22] |
| Bifidobacteriaceae | 68 | 119 | ||
| Caulobacteraceae | 51 | 85 | ||
| Corynebacteriaceae | 98 | 103 | ||
| Flavobacteriaceae | 671 | 188 | ||
| Leuconostocaceae | 46 | 75 | ||
| Methanobacteriaceae | 46 | 70 | Re-assigning Methanothermus, see text | |
| Pasteurellaceae | 83 | 97 | ||
| Staphylococcaceae | 95 | 88 | ||
| Streptococcaceae | 118 | 222 | ||
| Veillonellaceae | 74 | 151 | Retained as part of Negativicutes, see text |
In order to demonstrate the main conclusion of this paper, namely, there are abundant polyphyletic placements of species across genera in LVTree compared to predominant monophyletic genera in CVTree, we elaborate three groups of examples. These include, (1) straightforward cases without invoking lineage modifications; (2) cases requiring minor lineage modifications; and (3) a case that which at first glance speaks in favor of LVTree but a recent taxonomic proposal has eventually made it a supporter of CVTree.
Results
Straightforward cases
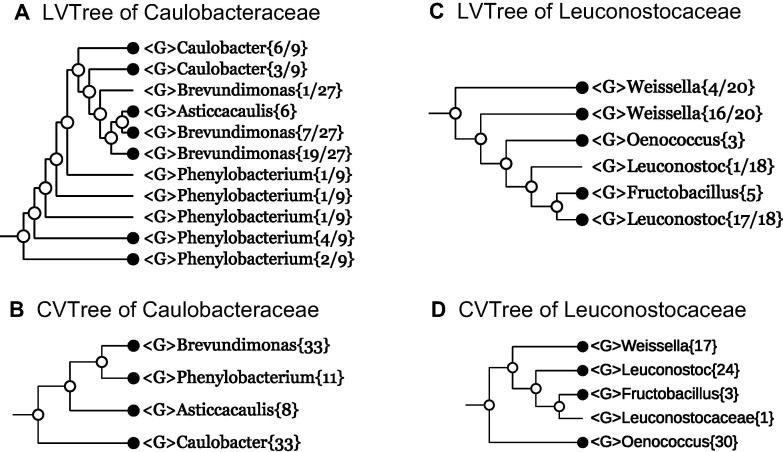
Example 1 Caulobacteraceae
According to BMSAB [3], Caulobacteraceae is the only family in the order Caulobacterales in class Alphaproteobacteria of the phylum Proteobacteria. Organisms of this family have been grouped together owing to their specific way of asymmetric cell division long before molecular means of characterizing bacteria has been developed. Being the first example of this study, we present some more details behind the construction of phylogenetic trees. The family Caulobacteraceae contains four genera, but major taxonomic references list different number of species as shown in Table 2. A few comments on Table 2 are appropriate:
Table 2.
Number of species in the constituent genera of Caulobacteraceae as listed in major taxonomic references
| Genus |
BMSAB[3] 2005/2017 |
The Prokaryotes IV[4] 2015 |
LPSN[5] Dec 2017 |
EzBioCloud[21] Oct 2017 |
|---|---|---|---|---|
| Asticcacaulis | 2 | 4 | 6 | 6 |
| Brevundimonas | 9 | 21 | 28 | 29 |
| Caulobacter | 4 | 6 | 9 | 9 |
| Phenylobacterium | 1 | 7+1* | 11 | 11+1* |
Note: 1* denotes the species Phenylobacterium zucineum, which has not be validly published by BMSAB and LPSN. BMSAB, Bergey’s Manual of Systematics of Archaea and Bacteria; LPSN, List of Prokaryotic Names with Standing in Nomenclature.
First, the electronic edition of BMSAB [3] appeared online in 2015, but most of its texts remained the same as in the volumes of The Bergey’s Manual of Systematic Bacteriology, 2nd edition [2]. Though partial updates of the electronic edition have been released four times a year, it may take many years to have all parts of BMSAB updated. In particular, the files related to Caulobacteraceae in BMSAB were identical to those of Bergey’s Manual of 2005. This explains why the numbers of species in the first column of Table 2 are the lowest ones.
Second, the corresponding volume of The Prokaryotes IV [4], published in 2014, was organized by families and contained more updated information. In particular, the genus Phenylobacterium included a species P. zucineum [23], which is considered to be not validly published by BMSAB and LPSN, despite the fact that its finished genome is available for almost 10 years [24]. This is marked by “+1” in the last row of Table 2.
Third, although both LPSN [5] and EzBioCloud [21] reflect the content of International Journal of Systematic and Evolutionary Microbiology, EzBioCloud adds more information on sequenced prokaryotic genomes, which is useful for the inspection of whole-genome-based CVTree.
While BMSAB and LPSN contain only validly-published names, especially those of type strains, the dataset behind CVTree includes many genomes with unclassified lineages. For example, Caulobacterales_bacterium_RIFOXYB1_FULL_67_16 is classified only to the order and Caulobacteraceae_bacterium_PMMR1 only to the family level. There are many more genomes classified to the species level without validly-published names, e.g., Brevundimonas_sp_Root1423. CVTree is capable of assigning most of them to a proper genus, as summarized in Table 3.
Table 3.
Number of representatives in the constituent genera of Caulobacteraceae in LVTree and CVTree used in the present work
| Genus | No. of 16S rRNA sequences in LVTree | No. of genomes in CVTree |
|---|---|---|
| Asticcacaulis | 6 | 4 genomes from 4 species; 4 genomes from unclassified species |
| Brevundimonas | 27 | 16 genomes from 14 species; 17 genomes from unclassified species |
| Caulobacter | 9 | 12 genomes from 4 species; 21 genomes from unclassified species |
| Phenylobacterium | 9 | 3 genomes from 3 species; 8 genomes from unclassified species |
Figure 1 shows the maximally-collapsed Caulobacteraceae branch in both LVTree (Figure 1A) and CVTree (Figure 1B). Only numbers of organisms are indicated in the figure. The detailed names with strain tags can be found in the fully-expanded figures (Figures S1 and S2). In order to avoid confusion, a remark must be made concerning Streptomyces longisporoflavus, which appeared in 27 species of the genus Brevundimonas. Its 16S rRNA sequence (GenBank accession No. DQ442520, 2006) apparently came from a Brevundimonas strain mislabeled as a Streptomyces. Although the authors of the original 16S rRNA submitted a new sequence (GenBank accession No. NR_115963) in 2015, they did not make a formal emendation to replace the old one. This problem was pointed out in Chapter 7 of The Prokaryotes IV [4] without drawing a conclusion. We have performed BLAST comparison of the two 16S rRNA sequences and confirmed the correctness of NR_115963 for Streptomyces longisporoflavus [12]. However, a piece of validly-published information, though incorrect, may remain there as long as no one makes a formal emendation. Therefore, the wrong Streptomyces longisporoflavus label still exists in the literature, e.g., in Figure 7.1 of The Prokaryotes IV [4]. We mention in passing that, all the four genera in Figure 7.1 of The Prokaryotes IV [4] are monophyletic, contradicting the LVTree (Figure 1A) but agreeing with the CVTree (Figure 1B). To this end, it must be noticed that in almost all phylogenetic trees given in The Prokaryotes IV [4], the input data and method of tree inference were indicated in figure captions except for Figure 7.1. Therefore, one must assume that this figure represented a consensus branching scheme, not what followed from a single phylogenetic tree based on 16S rRNA sequence analysis.
Figure 1.
Collapsed trees of families Caulobacteraceae and Leuconostocaceae
Branches are collapsed at genus level (denoted by G) for both 16S rRNA-based LVTree and whole-genome-based CVTree for every family. A solid circle at the end of the branch denoted that there are more than one genomes in the branch. Numbers in a bracket represent the total number of taxa in a genus (denominator) and those included in the branch (numerator), while only the total number of taxa is shown when a branch is monophyletic.
The contrast of LVTree and CVTree is noticeable in Figure 1A and B. While in 16S rRNA-based LVTree only one genus Asticcacaulis is monophyletic, all four genera are well-defined in whole-genome-based CVTree.
Example 2 Leuconostocaceae
Now we turn to the family Leuconostocaceae which is represented by 46 16S rRNA sequences in LVTree (Figure 1C) and by 75 genomes in CVTree (Figure 1D). As in LPSN, there are five valid genera, named Convivina, Fructobacillus, Leuconostoc, Oenococcus, and Weissella in this family. The genus Convivina was not involved in this analysis as only one genome of the genus was published recently [25]. Among the rest four genera, only Oenococcus and Fructobacillus are monophyletic on the 16S rRNA-based LVTree, while the other two polyphyletic genera are represented in form of Leuconostoc{17+1} and Weissella{16+4}. On the contrary, all four genera are monophyletic in CVTree. It is worth noting that an unclassified species Leuconostocaceae sp. R53105 is placed as a sister branch of the genus Fructobacillus, implying its possible classification as a member of Fructobacillus or a new genus. Expanded versions of these two phylogenetic trees with full names and strain tags are given in Figures S3 and S4.
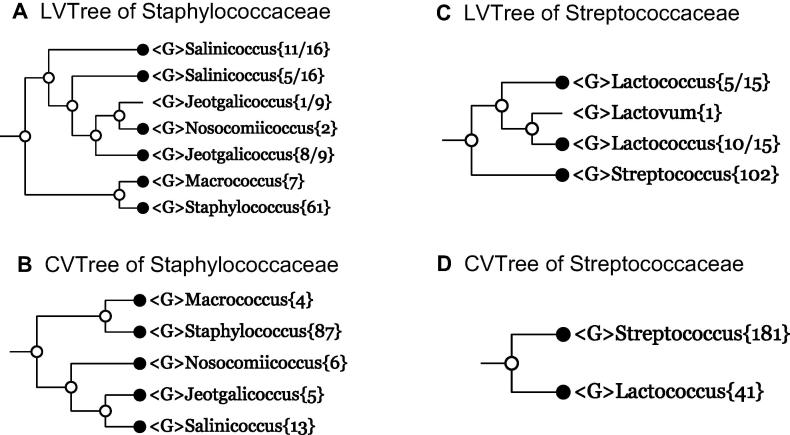
Example 3 Staphylococcaceae
The family Staphylococcaceae contains the notorious species Staphylococcus aureus whose methicillin-resistant strains (MRSA) cause severe cross-infections in hospitals. Owing to its clinical importance, more than 8000 genomes of this species have been sequenced. It is remarkable that all these genomes form a monophyletic cluster in CVTree. However, as epidemiologic studies of pathogens go beyond the scope of this work, we only retain a few tens of S. aureus strains as members of the genus Staphylococcus.
In 16S rRNA-based LVTree, although the family Staphylococcaceae{95} appears as a monophyletic cluster, it does contain two polyphyletically-placed genera, Salinicoccus and Jeotgalicoccus. Contrary to LVTree (Figure 2A), in whole-genome-based CVTree (Figure 2B), all subordinate genera in the family Staphylococcaceae{115} appear monophyletic on their own.
Figure 2.
Collapsed trees of families Staphylococcaceae and Streptococcaceae
Branches are collapsed at genus level for both 16S rRNA-based LVTree and whole-genome-based CVTree for every family.
Example 4 Streptococcaceae
This is a trivial case. In LVTree, the main cluster of family Streptococcaceae{118} consists of three genera: Streptococcus{102}, Lactococcus{5+10}, and Lactovum{1} (Figure 2C). However, in CVTree, the family Streptococcaceae{222} consists of a monophyletic cluster made of two monophyletic genera: Streptococcus{181} and Lactococcus{41} (Figure 2D). The monophyly of Lactococcus being violated by insertion of a monospecific genus Lactovum (as shown in LVTree) was proposed in 2005. There are two possibilities for Lactovum: either it is a disguised Lactococcus, or it actually makes a new genus, thus causing Lactococcus species placement to be polyphyletic. Since no sequenced genome is available so far, one does not have enough information to draw conclusions.
Example 5 Corynebacteriaceae
This is another trivial case as the family essentially contains only a single genus Corynebacterium. There was a monospecific genus Turicella proposed in 1994, which violated monophyly of the genus Corynebacterium in both LVTree and CVTree. As we have pointed out recently [26], Turicella could not make an independent genus and should be considered as a synonym to Corynebacterium. Therefore, the family Corynebacteriaceae contains only a single monophyletic genus Corynebacterium in both LVTree and CVTree, and there is no polyphyly in both trees.
The comparisons in all the five examples above are made under the assumption that the corresponding taxonomy is correct and no lineage modifications are needed. However, as taxonomy has always been a work in progress, revisions happen constantly as a rule. Therefore, we turn to the second group of examples that require minor lineage modifications. In fact, this second group of examples represents commonplace in prokaryotic taxonomy.
Cases requiring minor lineage modifications
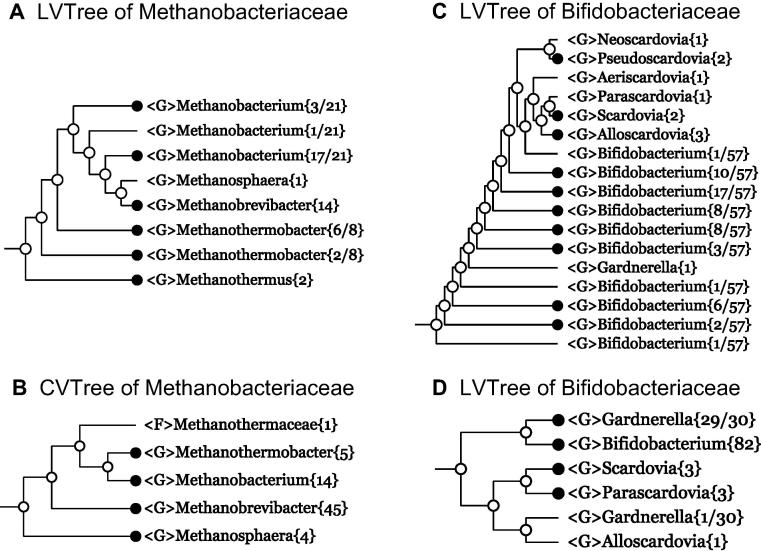
Example 6 Methanobacteriaceae
Our next example comes from Archaea. In LVTree the family Methanobacteriaceae{44} consists of a monophyletic cluster made of four genera: Methanosphaera{1}, Methanobrevibacter{14}, Methanothermobacter{2+6}, and Methanobacterium{1+3+17} (Figure 3A). The last two genera turn out to be polyphyletic. For example, Methanothermobacter{2+6} means that the genus Methanothermobacter comprises two parallel branches represented by 2 and 6 sequences of 16S rRNA, respectively. Please note that next to the monophyletic cluster Methanobacteriaceae{44}, there is a genus Methanothermus{2}, belonging to the family Methanothermaceae, which was proposed in 1981 [27] together with its type genus Methanothermus. Since then, no new genus has been discovered and described in the family.
Figure 3.
Collapsed trees of families Methanobacteriaceae and Flavobacteriaceae
Branches are collapsed at genus level for both 16S rRNA-based LVTree and whole-genome-based CVTree for every family. F and G denote family and genus, respectively.
In whole-genome-based CVTree, the family Methanobacteriaceae is represented by 68 genomes from four genera Methanothermobacter{5}, Methanobacterium{14}, Methanobrevibacter{45}, and Methanosphaera{4} (Figure 3B). However, these four genera do not form a monophyletic cluster, as the family Methanothermaceae with its only type genus Methanothermus gets deeply inside the cluster above. Pursuing monophyly as a guiding principle, this fact suggests a plausible revision: including Methanothermus as a part of the family Methanobacteriaceae and dropping the family name Methanothermaceae from the prokaryotic nomenclature. This lineage modification does not contradict the branching scheme in LVTree, i.e., it is acceptable in both LVTree and CVTree. This explains the numbers 46 and 70 in the Methanobacteriaceae row of Table 1.
Example 7 Bifidobacteriaceae
An inspection of family Bifidobacteriaceae{58} in LVTree reveals clearly polyphyly of the genus Bifidobacterium{1+1+6+1+3+8+8+17+10+1} (Figure 3C). In sharp contrast, genus Bifidobacterium{82} in CVTree is manifestly monophyletic (Figure 3D). A few words on the monospecific genus Gardnerella. Ever since the genus and species was proposed in 1980 [28], Gardnerella remains monospecific. In LVTree, it gets deeply into the genus Bifidobacterium. In CVTree, it stands next to the monophyletic Bifidobacterium cluster and might be absorbed into the latter without causing taxonomic contradiction. Not being related to the main theme of this paper, we leave this problem open. Another part of the family Bifidobacteriaceae is made of several genera from the Scardovia group, mostly polyphyletic in LVTree (Figure 3C) and seemingly monophyletic in CVTree (Figure 3D). A convincing elucidation of the situation requires more data.
Example 8 Acetobacteraceae
Now let us consider the family Acetobacteraceae. In both LVTree (Figure 4A) and CVTree (Figure 4B), species from two genera Gluconacetobacter and Komagataeibacter are heavily intermixed. In fact, the genus Gluconacetobacter was proposed in 1997 [29]. Later on, some species of this genus were taken out to form a new genus Komagataeibacter, as new combinations [30] and transfer from the former to the latter continued, e.g., in 2014 [31]. All these proposals were made by the same leading author Y. Yamada and his collaborators by comparing incomplete 16S rRNA sequences [29–31]. However, it is a sobering fact that in CVTree, species from the two genera Gluconacetobacter and Komagataeibacter, taken together, do make a monophyletic cluster. This fact hints strongly on the rationality of making the two genera a single one by retaining only the name Gluconacetobacter, which has the priority of being introduced first [29]. With this lineage modification done, the Acetobacteraceae branch appears as shown for LVTree (Figure 4A) and CVTree (Figure 4B), respectively.
Figure 4.
Collapsed trees of families Acetobacteraceae and Pasteurellaceae
Branches are collapsed at genus level for both 16S rRNA-based LVTree and whole-genome-based CVTree for every family. G and S denote genus and species, respectively.
Although the genus Gluconacetobacter{45} comes out as a monophyletic group in CVTree, its counterpart appears as six juxtaposed polyphyletic leaves, or, in our notations, as Gluconacetobacter{15+2+1+1+3+3}. It seems that this fact has misled the original authors to introduce a new genus but could not yet resolve the problem. Another non-monophyletic group in both LVTree and CVTree is formed by Roseomonas species interspersed with organisms from other genera. In particular, LVTree contains many genus names that are absent in CVTree, due to the lack of sequenced genomes. One must await new emerging data to complete the evaluation of branching schemes in LVTree and CVTree. Nonetheless, for the time being, CVTree behaves “better” by accommodating only one polyphyletic cluster of Roseomonas.
Example 9 Pasteurellaceae
Now we turn to a more complicated case. As shown in Figure 4C and D, the family Pasteurellaceae{83} in LVTree has different taxa number as {97} in CVTree, which is the most intricate branching figure given explicitly in this paper. Suffice it to look at how species from the three genera Pasteurella, Haemophilus, and Actinobacillus are mixed up in LVTree. Their interrelationship cannot be simply characterized as polyphyletic. However, the branching scheme in CVTree brings about some enlightenment. The genus Actinobacillus{11} is monophyletic, and the genus Haemophilus{9/10} is de facto monophyletic, if taking into account the assignment of Haemophilus ducreyi to a new unclassified genus by EzBioCloud [21]. Only the Pasteurella species come out polyphyletically. There is good hope that based on whole-genome analysis, the taxonomy of Pasteurellaceae will be brought to a better shape. In addition, we note that the newly proposed genus Rodentibacter [22] makes the Pasteurella species fewer in both LVTree and CVTree.
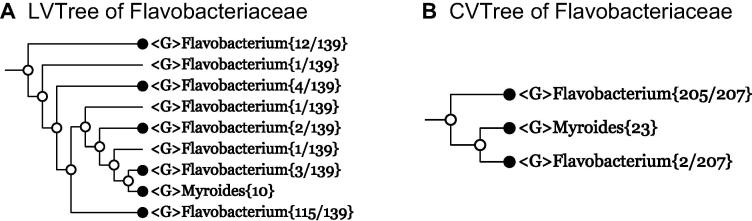
Example 10 Flavobacteriaceae
Now we look at an even more complicated case in Flavobacteriaceae. In LVTree, this family is represented by 671 species from 131 genera after assigning Pibocella to the genus Maribacter according to EzBioCloud [21]. The branching scheme is not shown because even the maximallycollapsed tree contains 189 lines. Although about 1/3 of the genera presented in LVTree do not have a genome sequenced, there are many sequenced genomes that are classified only to the species level without a validly-published name. These organisms are excluded from the LVTree dataset by design. However, as they do not violate monophyly of many genera in CVTree, it is easy to construct a whole-genome-based tree with a total genome number comparable with the number of 16S rRNA sequences present in LVTree (671) (Figure S5). In fact, we have a monophyletic family Flavobacteriaceae{818} in a CVTree (Figure S6). In order to highlight the difference between these two kinds of trees, it is instructive to pay attention to some local part. For example, Figure 5A shows the vicinity of the two genera Flavobacterium and Myroides in LVTree. The insertion of the genus Myroides made the genus Flavobacterium forming eight groups. The Flavobacterium species are clearly polyphyletic compared to the same vicinity in CVTree (Figure 5B). Anyway, CVTree comes out closer to monophyly than LVTree does.
Figure 5.
Collapsed tree of two genera, Flavobacterium and Myroides of family Flavobacteriaceae
Branches are collapsed at genus level (denoted by G) for both 16S rRNA-based LVTree and whole-genome-based CVTree. The collapsed trees of LVTree and CVTree for all genera of the family are shown in Figures S5 and S6, respectively.
The special case of class Negativicutes
Being stained Gram-positive makes an important part of the definition of species in the phylum Firmicutes. However, there is a group of Gram-negative organisms embedded in the generally Gram-positive sea of Firmicutes. The taxonomic placement of this group has undergone long debates and, eventually, a new class Negativicutes in the phylum Firmicutes was proposed in 2010 [32].
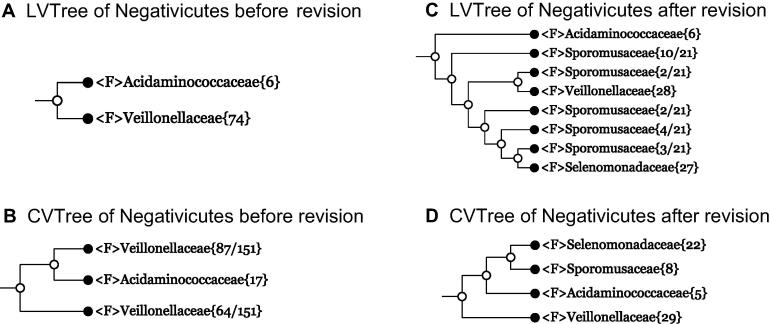
As the last example in this paper, we consider the class Negativicutes. Not long ago, the 16S rRNA-based LVTree (Release 123; September 2015) followed the taxonomy that this class consisted of a single order Selenomonadales, which in turn was made of two monophyletic families Acidaminococcaceae and Veillonellaceae (Figure 6A). In contrast, according to this taxonomy, the whole-genome-based CVTree led to a polyphyletic family Veillonellaceae (Figure 6B). Therefore, LVTree seems to be “better” than CVTree in the sense of monophyly of the family Veillonellaceae. However, this was caused by the fact that the placement of about 20 genera in Veillonellaceae was questionable. These genera should be considered as Selenomonadales Incertae sedis, as indicated in Figure 35.1 on p. 434 of the corresponding volume of The Prokaryotes IV [33], but ignored in the dataset behind LVTree. This was the situation when the class Negativicutes was defined as containing only a single order Selenomonadales.
Figure 6.
Collapsed tree of class Negativicutes before and after taxonomic revision
Branches are collapsed at family level (denoted by F) for 16S rRNA-based LVTree and whole-genome-based CVTree.
About the same time, a detailed taxonomic analysis using genomic data [34] arrived at the conclusion that the class Negativicutes actually contains three orders instead of one, that is, Veillonellales, Acidaminococcales, and Selenomonadales, with the last one consisting of two families Selenomonadaceae and Sporomusaceae. At present, both the LVTree Release 128 (February 2017) and CVTree adopted this validly published classification. This being done, the collapsed trees shown in Figure 6A and B transform into those shown in Figure 6C and D, respectively. In CVTree, all orders and families are now monophyletic. However, with this new classification, the family Sporomusaceae in LVTree becomes polyphyletic. Therefore, taxonomic proposal [34] again makes CVTree superior compared to LVTree. In other words, it supports our statement that whole-genome-based phylogeny agrees better with taxonomy in the sense of accommodating more monophyletic taxa.
Discussion
In this study, phylogenetic relationship for ten families and one class of prokaryotes is reconstructed based on alignment-free analysis upon whole-proteome information using CVTree, to provide detailed and comprehensive information for further comparisons with 16S rRNA-based phylogeny upon ten families and one class. This work is not simply a collection of examples. Using these examples, we intent to call attention on some principles in prokaryotic phylogeny and taxonomy.
We look at some problems at large for prokaryotic phylogeny and taxonomy, as the intention of this study goes far beyond the collection of examples. In 1987, an Ad Hoc Committee wrote in its report [35]: “There was general agreement that the complete deoxyribonucleic acid (DNA) sequence would be the reference standard to determine phylogeny and that phylogeny should determine taxonomy. Furthermore, nomenclature should agree with (and reflect) genomic information.”
Taxonomy came much earlier than phylogeny. Taxonomy is the classification of organisms by assigning them to discrete levels, i.e., from domain to species. A great achievement was made by Carl Woese and his colleagues [36] to propose the division of life into three domains based on small subunit rRNA sequences. The proposal greatly enhanced people’s acknowledgment of “the tree of life”, to which the increasing bacterial genomes from the end of the last century raise strong controversies instead of providing support [37]. As different genes may tell different stories, horizontal gene transfer, gene duplication and loss, incomplete lineage sorting, and other possibilities all together bring challenges to the development of objective taxonomic system guided by whole-genome information.
Compared with taxonomy, phylogeny is more definitive in nature. Given an input dataset, be it a collection of 16S rRNA sequences or a collection of genomes, and a fixed method of inference of phylogenetic information, be it based on sequence-alignment or alignment-free, it produces a phylogenetic tree, i.e., a branching scheme of the input data. There is no way to do fine adjustment of the input data or the final results. Phylogeny cannot produce nomenclature on its own, but provides standard for hierarchical classification of organisms, ruling by their evolutionary histories.
Does phylogeny represent relation among individual organisms or among populations? The notion of type strain was associated with individual organisms, but taxonomy always deals with population. In the long run “type strains” may be replaced by “type genomes”. By defining distance between genomes in the genome space, it is possible to make this approach quantitative. DNA–DNA hybridization gives some “distance” between genomes, but cannot be used incrementally to build an entire distance matrix, while CVTree can. We will elaborate this point in forthcoming publications.
Authors’ contributions
BH designed the study. GZ built and maintained the web server, collected data, and carried out the calculation. GZ and BH performed the analysis. GZ, JQ, and BH wrote the manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Project; Grant No. 2013CB834100) and the National Natural Science Foundation of China (Grant No. 11474068). Authors thanks the support of the State Key Laboratory of Applied Surface Physics and the Department of Physics, Fudan University, China.
Handled by Yu Xue
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2018.06.005.
Supplementary material
The following are the Supplementary data to this article:
A fully expanded branch of Caulobacteraceae cut from a LVTree based on 12,953 prokaryotic 16S rRNA sequences Branches are expanded at species level (denoted by S).
A fully expanded branch of Caulobacteraceae cut from a CVTree based on 8290 prokaryotic genomes Branches are expanded at species and strain level (denoted by S and T, respectively).
A partially expanded branch of Leuconostocaceae cut from a LVTree based on 12,953 prokaryotic 16S rRNA sequences Solid circle at the end of the branch denotes that there are more than one genomes in the branch. Branches are expanded at species and genus level (denoted by S and G, respectively).
A partially expanded branch of Leuconostocaceae cut from a CVTree based on 8290 prokaryotic genomes Solid circle at the end of the branch denotes that there are more than one genomes in the branch.
Collapsed tree of 16S rRNA-based LVTree for family Flavobacteriaceae {671} Solid circle at the end of the branch denotes that there are more than one genomes in the branch. The monophyletic and non- monophyletic branches are indicated in red and blue lines, respectively.
Collapsed tree of CVTree for family Flavobacteriaceae {818} Solid circle at the end of the branch denotes that there are more than one genomes in the branch. The monophyletic and non- monophyletic branches are indicated in red and blue lines, respectively.
References
- 1.Fox G.E., Pechman K.R., Woese C.R. Comparative cataloging of 16S ribosomal ribonucleic acid: molecular approach to procaryotic systematics. Int J Syst Evol Microbiol. 1977;27:44–57. [Google Scholar]
- 2.Brenner D.J., Krieg N.R., Staley J.T. Springer-Verlag; New York: 2005. Bergey’s manual of systematic bacteriology. [Google Scholar]
- 3.Whitman W.B. Wiley Online Library; 2015. Bergey's manual of systematics of Archaea and Bacteria. [Google Scholar]
- 4.Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F. Springer, Berlin Heidelberg; New York: 2014. The Prokaryotes: Alphaproteobacteria and Betaproteobacteria. [Google Scholar]
- 5.Parte A.C. LPSN-list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2014;42:D613–D616. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox G.E., Wisotzkey J.D., Jurtshuk P., Jr. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 7.Sneath P.H.A. Evidence from Aeromonas for genetic crossing-over in ribosomal sequences. Int J Syst Bacteriol. 1993;43:626–629. doi: 10.1099/00207713-43-3-626. [DOI] [PubMed] [Google Scholar]
- 8.Staley J.T. The bacterial species dilemma and the genomic-phylogenetic species concept. Philos Trans R Soc Lond B Biol Sci. 2006;361:1899–1909. doi: 10.1098/rstb.2006.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yarza P., Richter M., Peplies J., Euzeby J., Amann R., Schleifer K.H. The all-species living tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31:241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Munoz R., Yarza P., Ludwig W., Euzéby J., Amann R., Schleifer K.H. Release LTPs104 of the all-species living tree. Syst Appl Microbiol. 2011;34:169–170. doi: 10.1016/j.syapm.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Yarza P., Sproer C., Swiderski J., Mrotzek N., Spring S., Tindall B.J. Sequencing orphan species initiative (SOS): filling the gaps in the 16S rRNA gene sequence database for all species with validly published names. Syst Appl Microbiol. 2013;36:69–73. doi: 10.1016/j.syapm.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Zuo G., Zhi X., Xu Z., Hao B. LVTree viewer: an interactive display for the all-species living tree incorporating automatic comparison with prokaryotic systematics. Genomics Proteomics Bioinformatics. 2016;14:94–102. doi: 10.1016/j.gpb.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi J., Luo H., Hao B. CVTree: a phylogenetic tree reconstruction tool based on whole genomes. Nucleic Acids Res. 2004;32:W45–W47. doi: 10.1093/nar/gkh362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi J., Wang B., Hao B.L. Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach. J Mol Evol. 2004;58:1–11. doi: 10.1007/s00239-003-2493-7. [DOI] [PubMed] [Google Scholar]
- 15.Xu Z., Hao B. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 2009;37:W174–W178. doi: 10.1093/nar/gkp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo G., Hao B. CVTree3 web server for whole-genome-based and alignment-free prokaryotic phylogeny and taxonomy. Genomics Proteomics Bioinformatics. 2015;13:321–331. doi: 10.1016/j.gpb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen I.A., Markowitz V.M., Chu K., Palaniappan K., Szeto E., Pillay M. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017;45:D507–D516. doi: 10.1093/nar/gkw929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatusova T., Ciufo S., Fedorov B., O'Neill K., Tolstoy I. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 2014;42:D553–D559. doi: 10.1093/nar/gkt1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwala R., Barrett T., Beck J., Benson D.A., Bollin C., Bolton E. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wattam A.R., Davis J.J., Assaf R., Boisvert S., Brettin T., Bun C. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017;45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon S.H., Ha S.M., Kwon S., Lim J., Kim Y., Seo H. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adhikary S., Nicklas W., Bisgaard M., Boot R., Kuhnert P., Waberschek T. Rodentibacter gen. nov including Rodentibacter pneumotropicus comb. nov., Rodentibacter heylii sp nov., Rodentibacter myodis sp nov., Rodentibacter ratti sp nov., Rodentibacter heidelbergensis sp nov., Rodentibacter trehalosifermentans sp nov., Rodentibacter rarus sp nov., Rodentibacter mrazii and two genomospecies. Int J Syst Evol Microbiol. 2017;67:1793–1806. doi: 10.1099/ijsem.0.001866. [DOI] [PubMed] [Google Scholar]
- 23.Zhang K., Han W.D., Zhang R., Xu X.L., Pan Q.R., Hu X. Phenylobacterium zucineum sp nov., a facultative intracellular bacterium isolated from a human erythroleukemia cell line K562. Syst Appl Microbiol. 2007;30:207–212. doi: 10.1016/j.syapm.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Luo Y.F., Xu X.L., Ding Z.H., Liu Z., Zhang B., Yan Z.Y. Complete genome of Phenylobacterium zucineum — a novel facultative intracellular bacterium isolated from human erythroleukemia cell line K562. BMC Genomics. 2008;9:386. doi: 10.1186/1471-2164-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Praet J., Meeus I., Cnockaert M., Houf K., Smagghe G., Vandamme P. Novel lactic acid bacteria isolated from the bumble bee gut: Convivina intestini gen. nov., sp nov., Lactobacillus bombicola sp nov., and Weissella bombi sp nov. Antonie Van Leeuwenhoek. 2015;107:1337–1349. doi: 10.1007/s10482-015-0429-z. [DOI] [PubMed] [Google Scholar]
- 26.Zuo G., Hao B. On monospecific genera in prokaryotic taxonomy. Synth Syst Biotechnol. 2017;2:226–235. doi: 10.1016/j.synbio.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stetter K.O., Thomm M., Winter J., Wildgruber G., Huber H., Zillig W. Methanothermus fervidus, sp nov, a novel extremely thermophilic methanogen isolated from an Icelandic hot spring. Zentralbl Bakteriol Mikrobiol Hyg C. 1981;2:166–178. [Google Scholar]
- 28.Greenwood J.R., Pickett M.J. Transfer of Haemophilus vaginalis Gardner and Dukes to a new genus Gardnerella: G. vaginalis (Gardner and Dukes) comb. nov. Int J Syst Bacteriol. 1980;30:170–178. [Google Scholar]
- 29.Yamada Y., Hoshino K., Ishikawa T. Taxonomic studies of acetic acid bacteria and allied organisms.11. The phylogeny of acetic acid bacteria based on the partial sequences of 16S ribosomal RNA: the elevation of the subgenus Gluconoacetobacter to the generic level. Biosci Biotechnol Biochem. 1997;61:1244–1251. doi: 10.1271/bbb.61.1244. [DOI] [PubMed] [Google Scholar]
- 30.Yamada Y., Yukphan P., Huong T.L.V., Muramatsu Y., Ochaikul D., Tanasupawat S. Description of Komagataeibacter gen. nov., with proposals of new combinations (Acetobacteraceae) J Gen Appl Microbiol. 2012;58:397–404. doi: 10.2323/jgam.58.397. [DOI] [PubMed] [Google Scholar]
- 31.Yamada Y. Transfer of Gluconacetobacter kakiaceti, Gluconacetobacter medellinensis and Gluconacetobacter maltaceti to the genus Komagataeibacter as Komagataeibacter kakiaceti comb. nov., Komagataeibacter medellinensis comb. nov and Komagataeibacter maltaceti comb. nov. Int J Syst Evol Microbiol. 2014;64:1670–1672. doi: 10.1099/ijs.0.054494-0. [DOI] [PubMed] [Google Scholar]
- 32.Marchandin H., Teyssier C., Campos J., Jean-Pierre H., Roger F., Gay B. Negativicoccus succinicivorans gen. nov., sp. nov., isolated from human clinical samples, emended description of the family Veillonellaceae and description of Negativicutes classis nov., Selenomonadales ord. nov. and Acidaminococcaceae fam. nov. in the bacterial phylum Firmicutes. Int J Syst Evol Microbiol. 2010;60:1271–1279. doi: 10.1099/ijs.0.013102-0. [DOI] [PubMed] [Google Scholar]
- 33.Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F. Springer-Verlag, Berlin Heidelberg; New York: 2014. The prokaryotes: firmicutes and tenericutes. [Google Scholar]
- 34.Campbell C., Adeolu M., Gupta R.S. Genome-based taxonomic framework for the class Negativicutes: division of the class Negativicutes into the orders Selenomonadales emend., Acidaminococcales ord. nov. and Veillonellales ord. nov. Int J Syst Evol Microbiol. 2015;65:3203–3215. doi: 10.1099/ijs.0.000347. [DOI] [PubMed] [Google Scholar]
- 35.Wayne L., Brenner D., Colwell R., Grimont P., Kandler O., Krichevsky M. Report of the ad hoc committee on reconciliation of Approaches to bacterial systematics. Int J Syst Evol Microbiol. 1987;37:463–464. [Google Scholar]
- 36.Fox G.E., Magrum L.J., Balch W.E., Wolfe R.S., Woese C.R. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc Natl Acad Sci U S A. 1977;74:4537–4541. doi: 10.1073/pnas.74.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doolittle W.F. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A fully expanded branch of Caulobacteraceae cut from a LVTree based on 12,953 prokaryotic 16S rRNA sequences Branches are expanded at species level (denoted by S).
A fully expanded branch of Caulobacteraceae cut from a CVTree based on 8290 prokaryotic genomes Branches are expanded at species and strain level (denoted by S and T, respectively).
A partially expanded branch of Leuconostocaceae cut from a LVTree based on 12,953 prokaryotic 16S rRNA sequences Solid circle at the end of the branch denotes that there are more than one genomes in the branch. Branches are expanded at species and genus level (denoted by S and G, respectively).
A partially expanded branch of Leuconostocaceae cut from a CVTree based on 8290 prokaryotic genomes Solid circle at the end of the branch denotes that there are more than one genomes in the branch.
Collapsed tree of 16S rRNA-based LVTree for family Flavobacteriaceae {671} Solid circle at the end of the branch denotes that there are more than one genomes in the branch. The monophyletic and non- monophyletic branches are indicated in red and blue lines, respectively.
Collapsed tree of CVTree for family Flavobacteriaceae {818} Solid circle at the end of the branch denotes that there are more than one genomes in the branch. The monophyletic and non- monophyletic branches are indicated in red and blue lines, respectively.