Abstract
Featuring a circular, knotted structure and diverse bioactivities, cyclotides are a fascinating family of peptides that have inspired applications in drug design. Most likely evolved to protect plants against pests and herbivores, cyclotides also exhibit anti-cancer, anti-HIV, and hemolytic activities. In all of these activities, cell membranes appear to play an important role. However, the question of whether the activity of cyclotides depends on the recognition of chiral receptors or is primarily modulated by the lipid-bilayer environment has remained unknown. To determine the importance of lipid membranes on the activity of the prototypic cyclotide, kalata B1, we synthesized its all-D enantiomer and assessed its bioactivities. After the all-D enantiomer had been confirmed by 1H NMR to be the structural mirror image of the native kalata B1, it was tested for anti-HIV activity, cytotoxicity, and hemolytic properties. The all-D peptide is active in these assays, albeit with less efficiency; this reveals that kalata B1 does not require chiral recognition to be active. The lower activity than the native peptide correlates with a lower affinity for phospholipid bilayers in model membranes. These results exclude a chiral receptor mechanism and support the idea that interaction with phospholipid membranes plays a role in the activity of kalata B1. In addition, studies with mixtures of L and D enantiomers of kalata B1 suggested that biological activity depends on peptide oligomerization at the membrane surface, which determines affinity for membranes by modulating the association–dissociation equilibrium.
Keywords: anti-HIV, chirality, cyclic peptides, membranes, proteins
Introduction
Cyclotides are a remarkable class of plant peptides characterized by their head-to-tail cyclized backbone and knotted arrangement of three conserved disulfide-bonds[1] (Figure 1). This topology gives cyclotides exceptional stability, including resistance to enzymatic, chemical, and thermal degradation.[2–4] More than 200 cyclotides with a range of bioactivities have now been characterized. Although their natural function appears to be plant defense, based on their potent insecticidal activity and high expression levels in plant leaves,[5,6] cyclotides are also pharmaceutically relevant, possessing anti-cancer[7] and anti-HIV activities.[8,9] These activities and the cyclotides’ remarkable stability[2] and tolerance for substitution[10,11] suggest that these molecules are useful molecular frameworks for protein engineering, with applications in medicine and agriculture.[12] One such application was recently demonstrated by the grafting of an anti-angiogenic peptide onto a cyclotide framework to stabilize it for therapeutic use.[13]
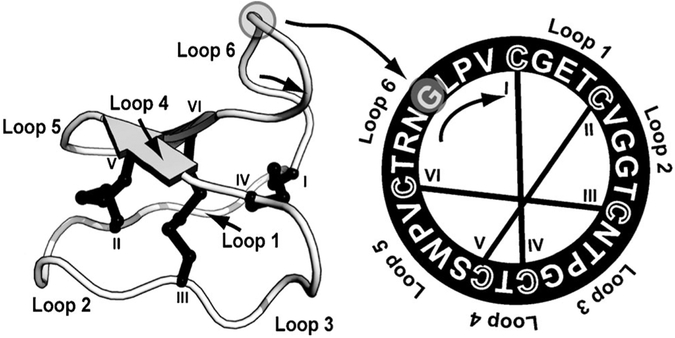
Figure 1.
3D structure (left) and amino acid sequence (right) of native kalata B1 (PDB ID: 1nb1) showing the knotted, cyclic nature of cyclotides. All cyclotides contain six conserved cysteine residues, and the backbone segments in between are termed “loops”. The disulfide connectivity is shown by lines connecting the cysteine residues, the cysteine residues are labeled I–VI, and the six loops are identified. The peptide is processed from a larger linear precursor[6] with ligation at Gly1 to Asn29 (indicated by circles).
The use of cyclotides as protein engineering frameworks was also recently demonstrated by the expression of bioengineered cyclotide combinatorial libraries inside[14] or displayed on the surface of[15] Escherichia coli cells; together with high-throughput screening, this could accelerate drug discovery.[15,16]
Although scores of cyclotides have now been identified, their mechanisms of action remain largely unknown. Understanding these mechanisms is important for the broader application of cyclotides as molecular frameworks. The prototypic cyclotide, kalata B1, is well characterized, but it is still unclear whether a receptor-dependent mechanism is involved in its bioactivities.
The activities ascribed to kalata B1, which include hemolytic,[17] insecticidal,[5,6] and anti-HIV activities,[18] show that different cell types can be targeted by kalata B1 and hint at a receptor-independent mechanism. This hypothesis is supported by electron microscopy studies that revealed the ability of kalata B1 to disrupt the membrane of midgut cells in lepidopteran larvae after cyclotide ingestion;[5] and by electrophysiological measurements on model membranes, which showed membrane leakage through a pore-formation mechanism.[19] An Ala-mutagenesis scan of kalata B1 revealed that its activity can be greatly reduced by single-residue mutations.[20] Despite the finding that no Ala substitution promoted structural perturbations, the substitution of a specific set of individual residues resulted in the loss of the hemolytic and insecticidal activity of kalata B1 mutants. The residues critical for activity are located on one face of the molecule;[20] this implies a recognition process that is highly specific. However, these data did not discriminate between chiral (protein-based) or achiral (membrane-based) specific recognition.
In general, when peptide recognition and membrane binding occur through a receptor-dependent mechanism, a specific chirality is required. Therefore, all-D enantiomers of proteins are expected to be inactive. In the alternative scenario, when peptide–membrane binding is mediated by the lipid bilayer alone, the binding should be essentially independent of peptide chirality and confer similar activity for L and D isomers.[21] To provide insights into the mechanism of action of cyclotides, and particularly the action of kalata B1, we synthesized and characterized an all-D enantiomer (i.e., mirror-image form) of kalata B1.
Results
Synthetic D-kalata B1 is a mirror image of native kalata B1
The all-D isomer of kalata B1 was assembled by solid-phase peptide synthesis (SPPS) and cyclized by using intramolecular native chemical ligation[22,23] (Figure 2). As expected, D-kalata B1 was late-eluting and could not be distinguished from native kalata B1 by RP-HPLC. Cyclotides differ from many proteins in that they become more (rather than less) hydrophobic during oxidative folding.[17] This presumably occurs because hydrophobic residues are excluded from the molecular core by the cystine knot structure and hence form a surface-exposed hydrophobic patch, which causes late elution in HPLC.
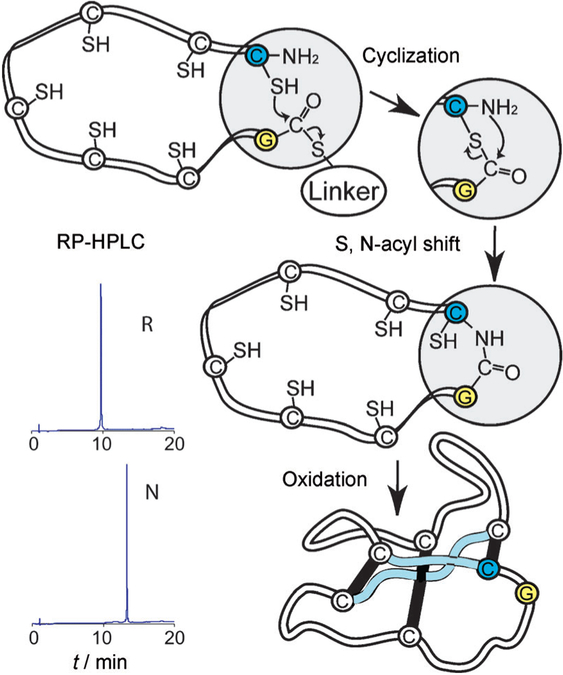
Figure 2.
Scheme for the cyclization and folding of synthetic kalata B1. After the assembly of D amino acids by using manual SPPS with Boc chemistry (see the sequence in Figure 1), D-kalata B1 was cyclized and folded. In a proposed mechanism,[23] the C-terminal thioester, attached through a linker, reacts successively with the cysteine side chains, which act as intermediate nucleophiles and “zip” the activated C terminus towards the N-terminal cysteine, where a final S,N-acyl migration creates a native peptide bond. For clarity, only the last nucleophilic attack is shown. RP-HPLC traces show the increase in hydrophobicity from linear reduced (R) to natively folded (N) protein.
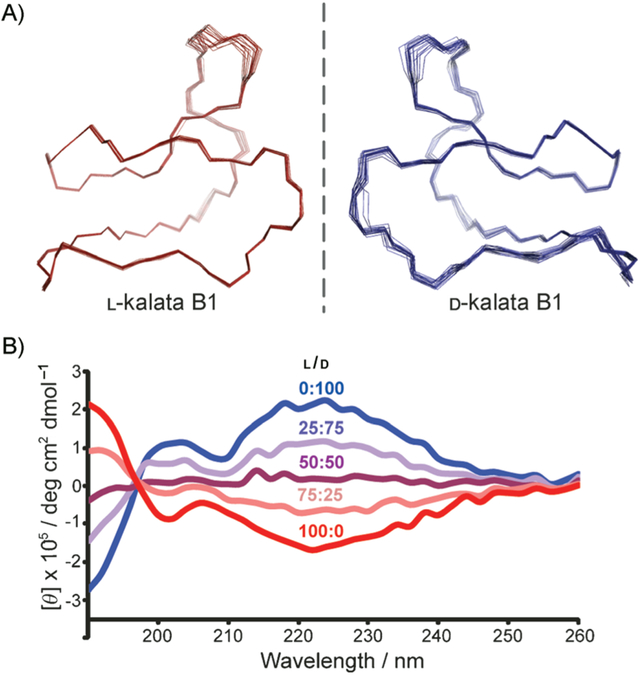
The structure of D-kalata B1 was determined by 1H NMR (spectra provided in Figure S1 and chemical shifts in Table S1 in the Supporting Information). The secondary structure of D-kalata B1 was identical to that of native kalata B1 based on chemical shift index,[24] slow exchange, and coupling patterns (Figure S2). Of the 100 calculated 3D structures, the 20 lowest-energy structures were chosen to represent the D-kalata B1 solution structure (Figure 3). A summary of the structural statistics is given in Table S2. The solution structure of D-kalata B1 is precise, as reflected in the backbone superposition (Figure 3A), and is a mirror image of that of native kalata B1. This mirror image symmetry is reflected in the circular dichroism spectrum by the equal but opposite optical rotation of the two enantiomeric peptides (Figure 3B). When the two isomers were mixed in equal proportions, each CD signal was cancelled by the other.
Figure 3.
Structural comparison of native and D-kalata B1. A) Backbone superposition of the 20 lowest-energy structures of L- and D-kalata B1 as determined by 1H NMR. The orientation is as in Figure 1. B) CD spectra of the two enantiomers in pure solutions (25 μM in H2O) and mixed together in three different ratios.
The anti-HIV activity of kalata B1 does not involve strong chiral recognition
Kalata B1[18] and other cyclotides[8,25,26] have anti-HIV activity, but the mechanism by which they exert this activity has hitherto been unknown, apart from the finding that a cyclic backbone is required.[18] In the current study, an in vitro assay that detects inhibition of the cytopathic effects of HIV-1RF infection on human lymphoblastoid cells[27] revealed increases in both antiviral cytoprotective concentration (EC50), from 0.9 to2.5 μM, and cytotoxicity index concentration (IC50), from 6.3 to10.5 μM, for the all-D isomer compared with native kalata B1.
D-Kalata B1 has slightly weaker hemolytic activity than the native peptide
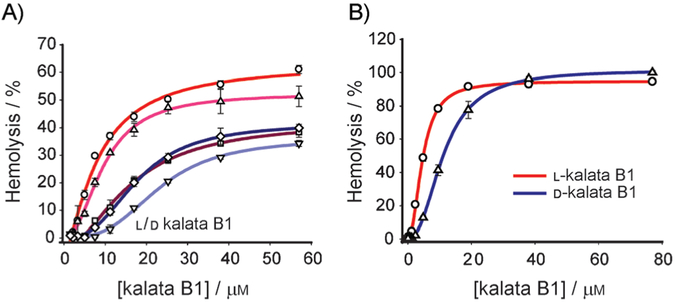
Red blood cells (RBCs) are a valuable model for evaluating the ability of peptides to interact with mammalian membranes,[28] and a test for hemolytic activity was one of the first bioassays used in the discovery of cyclotides.[29] Native kalata B1 is mildly hemolytic;[17,20,30] this suggests a mechanism that involves cell-membrane disruption. Synthetic D-kalata B1 was less active than the native peptide (Figure 4). From the dose–response curves obtained after 1 hour of incubation of the peptides with RBCs (Figure 4A), it was evident that the d enantiomer caused less hemolysis than native kalata B1 at all concentrations tested—IC50=26.0 μM for native kalata B1 and 77.0 μM for the D form. When the incubation time was increased to 14 h (Figure 4B), the two peptides produced a higher hemolysis response (IC50=5.0 μM for the native form and 11.0 μM for D-kalata B1), with comparable hemolysis levels (100%) at the higher concentrations. The viability of 14-hour-old RBCs was confirmed by control experiments in which kalata B1 was incubated for 1 h with 14-hour-old RBCs. Hemolytic levels similar to those with fresh RBCs were obtained for concentrations up to 12.5 μM.
Figure 4.
Hemolytic dose–response curves for native and D-kalata B1. The hemolytic activities of the two enantiomers in pure solutions and mixed together in three different ratios were assayed (○: 0, △: 25, □: 50, ▽: 75 and ◇: 100% D). Peptide solutions were tested in triplicate, with test concentrations ranging from 1.5–85 μM. The solutions were incubated in a 0.25% (v/v) suspension of human RBCs at 37°C A) for 1 h or B) overnight (14 h).
This time dependence suggests that the lysis mechanism depends on a threshold peptide concentration at the membrane surface. We explored the possibility that self-association of kalata B1 is important for its activity by assaying mixed solutions of L- and D-kalata in the following l/d ratios: 75:25, 50:50, and 25%:75%. In these experiments, a 1 h incubation was used, as the differences between pure L and pure D at this incubation time were more prominent. As illustrated in Figure 4A, the 75:25 mixture produced an intermediate curve, with an IC50 of 37.1 μM, the racemic mixture produced a dose–response curve similar to that for the pure all-D solution, IC50=77.8 μM, and the 25:75 mixture gave lower hemolysis levels than the pure all-D solution, IC50=83.2 μM.
D-Kalata B1 has lower affinity for membranes than native L-kalata B1
Using surface plasmon resonance (SPR) with pure phospholipid bilayers, we investigated whether the natural chirality of membrane lipids could explain the differences in bioactivity of the kalata B1 enantiomers. In SPR, peptides are passed over model membranes deposited on the surface of a sensor chip, and the association and dissociation of the peptide and lipid bilayers are monitored in real time.[31] Association (binding) is detected by changes in the refractive index. Measured response units (RU) are proportional to the adsorbed mass on the sensor surface and can be converted to amount of peptide bound to the lipid membrane (1 RU~1 pg mm−2).[32]
We have recently shown that native kalata B1 has selectivity for membranes that include phospholipids containing phosphoethanolamine (PE) headgroups in their composition.[33] Thus, we compared the interaction of kalata B1 with pure palmitoyl oleoyl phosphatidylcholine (POPC) bilayers and a POPC/palmitoyl oleoyl phosphatidylethanolamine (POPE) mixture; both systems have fluid-phase properties.[34,35] Figure 5A shows that kalata B1 does not bind avidly to POPC bilayers but has high affinity for membranes containing POPE; this is in agreement with our previous results. The content of PE phospholipids in eukaryotic cells is around 20%, hence we used a POPC/POPE mixture (4:1 molar ratio) to evaluate differences in membrane affinity for native and D-kalata B1.
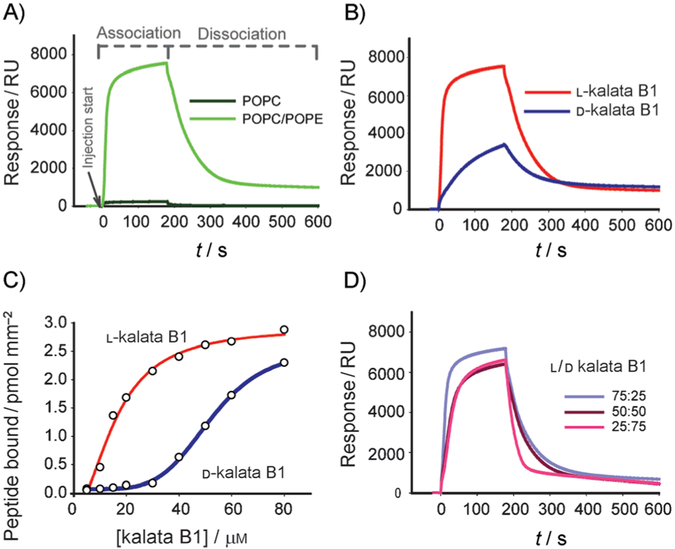
Figure 5.
Membrane binding of native and D-kalata B1 studied by surface plasmon resonance. Kalata B1 and its d isomer were injected for 180 s (association phase) over lipid surfaces deposited on an L1 chip, and dissociation was monitored after the injection had stopped (dissociation phase). A) Sensorgrams obtained with 50 μM native kalata B1 over POPC and POPC/POPE (4:1) lipid surfaces. B) Comparison of 50 μM L- and D-kalata B1 injected over POPC/POPE (4:1). C) Amount of L- and D-kalata B1 bound to the POPC/POPE (4:1) lipid surface at the end of the association phase as a function of different peptide concentrations. D) Sensorgrams obtained with mixtures of L and D isomers of kalata B1 in ratios as indicated on the figure).
Varying concentrations of native and D-kalata B1 injected over POPC/POPE bilayers revealed that the mirror-image isomers differed in their affinity for the membrane (Figure 5B, C and Figure S3A, B). The sensorgrams obtained at a fixed concentration (50 μM, Figure 5B) showed faster association of native kalata B1 than its d-enantiomer (see initial binding rates, Table 1). At the end of the association phase, the amount of bound peptide was twice as high for native kalata B1 as for the d-peptide. Nevertheless, the dissociation rate for the two peptides was comparable (0.0166 vs 0.0184 s−1, Table 1), thus indicating that association rate is the limiting step that accounts for the differences in the overall membrane affinity of the two mirror-image isomers. Once bound to the membrane, the L and D isomers had a comparable tendency to remain bound.
Table 1.
Kinetic parameters for the interaction of L-/D-kalata mixtures with POPC/POPE membrane.
| L-/D-kB1[a] | Initial rate [ngmm−2S−1][b] | Peptide/lipid [mol/mol][c] | kd [× 10−2S−1][d] |
|---|---|---|---|
| 100:0 | 0.419 | 0.23 | 1.66 ± 0.01 |
| 75:25 | 0.330 | 0.20 | 1.62 ± 0.01 |
| 50:50 | 0.160 | 0.18 | 1.79 ± 0.01 |
| 25:75 | 0.120 | 0.18 | 4.42 ± 0.03 |
| 0:100 | 0.068 | 0.08 | 1.84 ± 0.01 |
50 μM of total peptide was injected.
The initial peptide binding rate was calculated from the initial slope of the sensorgram (10 s after injection start); response units (RU) were converted into ngmm−2 (1 RU ~1 pgmm−2).
Peptide/lipid ratio at the end of association phase. The amount of lipid was calculated by converting the RU obtained at the end of lipid deposition into ngmm−2, while the amount of peptide bound was calculated at the end of injection (see Figure S3 for further details on the calculations).
The dissociation constant (kd) was calculated by separately fitting the dissociation phase with the BIAeval software (version 4.1), the standard error is given.
The membrane affinities of the mirror forms of kalata B1 over the concentration range studied (0–80 μM) were assessed by the amount of peptide bound to the membrane at the end of injection (Figure 5C). Whereas the difference between the l and d isomers was large at the lower peptide concentrations, the difference was smaller at higher concentrations, and the amount of bound peptide reached saturation for native kalata B1. These results correlate with the hemolytic data (Figure 4).
Mixtures of L and D isomers suggest that membrane affinity depends on self-assembly of kalata B1
To provide insight into the binding mechanism of the L and D isoforms of kalata B1 and help interpret their differing hemolytic effects, we compared the affinity for POPC/POPE membranes in mixtures of the two enantiomers (L/D 75:25, 50:50 and 25%:75%) at 50 μM total concentration (Figure 5D). At this concentration, binding of the D isoform was detectable, and differences between the two isomers were still evident (Figure 5B, C). The initial binding rate decreases with the increase in D-kalata percentage (Table 1). Interestingly, the dissociation rate was comparable for the pure L and D isomers and the mixtures, except for the 25:75 mixture, which had a faster dissociation rate than any of the other solutions tested, thus suggesting that this mixture was easily removed from the membrane. A lower propensity of the 25:75 mixture to stay bound to the membrane is consistent with the lower hemolysis produced by this mixture (see Figure 4A).
Discussion
In this study we set out to determine if the bioactivity of the prototypic cyclotide kalata B1 involves a chiral receptor-dependent mechanism. We showed that a synthetic D-kalata B1 peptide is the structural mirror image of its native twin and that this inverted peptide has anti-HIV-1 and hemolytic properties, albeit at lower levels than those of the native peptide. These results demonstrate that the bioactivity of kalata B1 does not rely on specific chiral recognition and therefore is not mediated by a receptor-dependent mechanism. Although the hemolytic dose–response curves for the two isomers indicate that the D isomer is less active than native kalata B1, the response curves show that both L- and D-kalata B1 disrupt the membrane of RBCs and suggest an identical mechanism.
The weaker activity of D-kalata B1 correlated with a lower affinity for phospholipids in model membranes. In interacting with phospholipid bilayers comprising POPC/POPE, the D isomer had a slower membrane binding rate (see Figure 5B, C and Table 1), which governs the local peptide concentration on the membrane surface. As the D isomer binds more slowly to the bilayers than native kalata B1, it needs to be present in a higher dose to reach the critical local peptide concentration required for disrupting the membrane. Although a POPC/POPE mixture may be considered a simplistic model, the close correlation of hemolytic effects with membrane affinities for L- and D-kalata B1 (see Figures 4 and 5B, C) shows that activity and membrane affinity are intimately related.
The differences in membrane binding affinity between native and D-kalata B1, which differ structurally only in their chirality, show that the chiral environment created by phospholipid bilayers modulates the bioactivity of kalata B1. The chirality of membrane phospholipids is normally considered weak, but the results here show that it can still modulate activity. Natural phospholipids create a chiral environment,[36,37] and chiral discrimination has been demonstrated for some molecules.[37–39] All phospholipids have a chiral center in their glycerol skeleton immediately adjacent to the hydrophobic acyl chains; this suggests that kalata B1 inserts near to, or in, the hydrophobic region of lipid bilayers. Hence, we suggest that the activity of kalata B1 does not require enantiomeric specificity for a protein receptor but depends on the affinity for lipid membranes, and is governed by specific interactions with PE headgroups and by hydrophobic interactions that are privileged for the native peptide compared with the D isomer.
Overall, our results suggest that L and D isomers of kalata B1 exert their activity by a membrane-leakage mechanism that requires a critical peptide/lipid ratio at the membrane surface. This is supported by 1) the differences in the concentration of kalata B1 required to achieve bioactivity with different target cells (whereas EC50 for anti-HIV activity is 0.9 μM, this concentration produces no significant hemolysis); 2) the importance of lipid composition in modulating the affinity of kalata B1 for (see Figure 5A), and disruption of, model membranes;[19] and 3) the correlation between membrane affinity and activity for L- and D-kalata B1 (see Figures 4 and 5B, C).
The membrane leakage caused by kalata B1 appears to depend on peptide self-association.[19] The association of peptides to form a high concentration at the membrane surface, before or after membrane binding, is a requirement for pore formation and membrane leakage by many peptides.[40–43] The hemolytic effects of the kalata B1 isomers, influenced by incubation time and concentration (see Figure 4), corroborate a mechanism that involves a concentration threshold and suggest a self-association phenomenon. Oligomerization of kalata B1 is further supported by the hemolytic response curves obtained with mixtures of the L and D peptides (Figure 4A), which cannot be explained simply by regarding the peptide as a monomeric active unit. In that case, the mixtures of D and L peptides should have produced intermediate dose–response curves comparable to the intermediate spectral curves obtained in the CD experiment (Figure 3B). The hemolytic response curves from this study, combined with data from membrane-leakage studies showing that kalata B1 forms pores,[19] suggest that kalata B1 self-association is important for bioactivity.
Self-association of kalata B1 is more likely to occur in the membrane than in solution. At millimolar concentrations in aqueous solution, kalata B1 exists mainly as a monomer.[44] Therefore, it seems likely that kalata B1 binds to membranes as a monomer and self-associates after binding. Peptide hydro-phobicity has been correlated with hemolytic properties,[28,40] as peptides with higher hydrophobicity have a greater tendency to aggregate and penetrate deeper into the hydrophobic core of RBCs, inducing hemolysis.[40] The differences in hemolytic activity and membrane affinity between native kalata B1 and its D isomer cannot be explained by differences in their propensity to self-associate in aqueous solution because the mirror-image isomers have identical hydrophobicity and therefore identical self-association properties. Furthermore, the observed comparable membrane-dissociation rates for pure-L and pure-D isomers suggest that both isomers insert at comparable depth into the membrane core. As, in general, deep insertion of peptides into membranes depends on peptide oligomerization,[40] our results suggest that once kalata B1 and the d isomer bind to the membrane surface they have a tendency to self-aggregate and insert into the lipid bilayer.
The hemolytic efficiency of the L/D mixtures, in particular the 25:75 mixture, was lower than that expected from a linear interpolation of data for the two isomers. This finding suggests that oligomerization of monomers with identical chirality is necessary for insertion into the phospholipid bilayer, and consequently for hemolysis. This notion is supported by the faster dissociation of the 25:75 mixture from the membrane (see Figure 5D and Table 1), showing that this mixture does not insert deeply into the hydrophobic core. Hence, for the L/D mixtures, not only is the binding to the membrane relevant for the final activity, but also their propensity to stay bound to the membrane. In the membrane-affinity studies, major differences were only detected for the 25:75 mixture. For the other mixtures, the concentrations of the L isomer, which binds faster to the membrane, were probably high enough for the peptide to oligomerize in the membrane and insert enough into the bilayer to produce a comparable dissociation rate to that of the pure L isomer.
The theory that self-association of kalata B1 at the membrane surface leads to membrane disruption agrees with recent electrophysiological data.[19] Electrophysiological recordings on model membranes showed that, after exposure to kalata B1, an initial period with no variation in current was followed by a step-wise increase in current until the membrane ruptured. This induced membrane disruption suggests that the peptide must associate on the membrane.[19]
Conclusions
In summary, this study shows that D-kalata B1 is active, thereby revealing that recognition by a chiral protein is not a requirement for kalata B1 activity. Furthermore, our results support a mechanism dependent on a peptide–lipid interaction and self-assembly of cyclotides on the lipid membrane. Based on these findings, together with previous reports, we would like to propose a pore-formation mechanism in four steps: 1) binding of kalata B1 to the membrane surface, governed by hydrophobic interactions and facilitated by PE phospholipids; 2) oligomerization of the peptide, once bound to the membrane; 3) insertion into the hydrophobic core, facilitated by self-association rather than bulk peptide concentration at the membrane surface; 4) pore formation as the embedded peptides span the two layers of the membrane. Overall, this process is limited by kalata B1 concentration, incubation time, and the lipid composition of the target membrane, which together modulate the concentration of the peptide on the membrane surface and the self-association process.
These results not only answer an important question about the mechanism of action of the prototypic cyclotide, kalata B1, they also shed light on why the activity of cyclotides varies across the family. We propose that the main differences in activity are modulated by membrane binding affinity, and not by the interaction with a receptor.
Experimental Section
Kalata B1 isolation:
Native kalata B1 was extracted from aerial parts of Oldenlandia affinis, as described previously.[45] RP-HPLC and LC-MS were used to identify and isolate kalata B1.[45] Purity was evaluated by analytical RP-HPLC, and native kalata B1 with a purity ≥95% was used in all studies. The concentration of peptide was determined by absorbance at 280 nm (ε280=5875M−1cm−1).
Peptide synthesis:
D-kalata B1 was assembled from D-amino acids by using SPPS with Boc chemistry, as reported for L-kalata B1,[17] with the following modifications: 1) an S-trityl-β-mercaptopropionic acid linker was attached to the resin, 2) the peptide was cleaved from the resin by using HF/cresol/thiocresol (50:4:1, v/v/v). The yield of peptide with the correct coupling was 31%. Cyclization and disulfide formation were facilitated in one reaction by dissolving the reduced linear peptide (1 mg mL−1) in ammonium carbonate (0.1M, pH 8.2), 50% isopropanol, and reduced glutathione (1 mM), and the mixture was stirred at room temperature with aeration for 16 h. The yield of the cyclization step was 38%. Correctly folded peptide was purified by preparative RP-HPLC, as previously described.[17] Purity was evaluated by analytical RP-HPLC (purity ≥95%). Retention times for the D-kalata B1 and the native kalata B1 were identical.
NMR spectroscopy:
Samples for NMR were prepared by dissolving D-kalata B1 in either H2O/D2O (95:5) or D2O (1.6 mM). Spectra were recorded on Bruker Avance 500 and 600 spectrometers at temperatures of 290–298 K. For resonance assignment and structure determination, two-dimensional spectra were recorded, as previously described.[46]
Structure calculations:
Interproton distance restraints for D-kalata B1 were derived from crosspeaks in NOESY spectra recorded with a mixing time of 200 ms. Spectra were analyzed by using the program Sparky. After initial structure calculations with DYANA,[47] sets of 100 structures were calculated by using a simulated annealing protocol within CNS,[48] as previously described.[46] As the peptide exclusively consisted of all-D amino acid, all angles and stereo-specific assignments of β-protons were reversed in the structure calculations. The program files were modified to convert each amino acid from all-L to all-D. Structures were analyzed by using PROMOTIF[49] and PROCHECK-NMR.[50] To allow correct analysis of the all-D peptide by these programs, all the PDB file coordinates in the final family of 20 structures were reversed to simulate the corresponding all-L peptide. A summary of the statistics is given in Table S2.
Circular dichroism spectroscopy:
Native and D-kalata B1 were dissolved in H2O (25 μM). Mixtures of the two solutions were prepared in 75:25, 50:50 and 25%:75% L/D ratios. CD spectra were recorded on a Jasco J-810 spectropolarimeter. For each sample, five scans from 260–190 nm were performed in a 1 mm path-length cell at 20–22°C.
Anti-HIV assay:
Native and D-kalata B1 were tested for anti-HIV activity by using an in vitro assay that detects inhibition of the cytopathic effects of HIV-1RF infection on human lymphoblastoid cells, as described previously.[27] The EC50 (concentration at which 50% of the uninfected target cells were protected from the lytic effects of HIV infection) and IC50 (concentration at which 50% of the uninfected target cells were killed due to direct cytotoxic effects of the test sample) values were determined.
Hemolytic activity assay:
Peptides were serially diluted in phosphate-buffered saline (PBS) in a 96-well microtiter plate. Fresh RBCs were washed with PBS and centrifuged at 1500 g for 30 s in a microcentrifuge repeatedly until the supernatant was clear. Fresh, 0.25% (v/v), washed RBCs in PBS (100 μL) were added to peptide solution (20 μL, peptide final concentration in the range 1.5–85 μM). The plate was incubated for 1 h or overnight (14 h) at 37°C and centrifuged at 150g for 5 min. A control in which the RBCs were aged for 14 h and incubated for 1 h with kalata B1 was also performed. Each supernatant was transferred to a 96-well, flat-bottomed microtiter plate. The absorbance was measured at 405 nm with an automatic Multiskan Ascent plate reader (Labsystems), and the hemolysis was calculated as the percentage of maximum lysis (Triton-X 100 1% (v/v) as positive control) after adjusting for minimum lysis (PBS as negative control).
Affinity for phospholipid membrane followed by SPR:
The affinity of kalata B1 and D-kalata B1 for lipid membranes was studied by using SPR measurements performed in a Biacore 3000 (Biacore–GE Healthcare). POPC and POPC/POPE (4:1) lipid vesicles were deposited over a L1 sensor chip. Vesicle preparation and general SPR experimental details, including lipid deposition and sensor chip surface regeneration, followed methods previously described.[51] All solutions were freshly prepared and filtered (0.22 μm) before use. The lipid was immobilized reaching a steady-state plateau under all conditions, thus confirming coverage of the chip surface. To calculate the peptide/lipid ratio at the membrane surface, the RU obtained after lipid deposition was converted into ngmm−2 (1 RU ~1 pgnm−2).
Kalata B1 and its D isomer (0–80 μM) were injected over the lipid surfaces (5 μL min−1, 180 s), and dissociation was followed for 600 s per injection cycle. Mixtures of L and D isomers (75:25, 50:50 and 25%:75% L/D ratios) were compared at 50 μM total peptide concentration. The relative affinity for each isomer or mixture was compared based on the amount of kalata B1 bound to the lipid surface by conversion of RU to pgmm−2 (assuming 1000 RU= 1 ng mm−2) at the end of association phase (see Figure S3). The kinetics for the different isomer mixtures was compared by fitting the dissociation phases separately with the BIAeval software (version 4.1).
Supplementary Material
Acknowledgements
Studies in our laboratory on cyclotides are supported by grants from the Australian Research Council (ARC-DP0880105) and the National Health and Medical Research Council (NHMRC). D.J.C. is an NHMRC Professorial Fellow. S.T.H. is a Marie Curie Postdoctoral Fellow awarded by European Commission (EU) (PIOF-GA-2008–220318). N.L.D. is a Queensland Smart State Fellow.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201100450: 1H NMR spectra and secondary structure of D-kalata B1, membrane binding of native kalata B1 and D-kalata B1 (SPR).
References
- [1].Craik DJ, Daly NL, Bond T, Waine C, J. Mol. Biol 1999, 294, 1327–1336. [DOI] [PubMed] [Google Scholar]
- [2].Colgrave ML, Craik DJ, Biochemistry 2004, 43, 5965–5975. [DOI] [PubMed] [Google Scholar]
- [3].Gran L, Lloydia 1973, 36, 207–208.4744557 [Google Scholar]
- [4].Sletten K, Gran L, Medd. Nor. Farm. Selsk 1973, 7–8, 69–82. [Google Scholar]
- [5].Barbeta BL, Marshall AT, Gillon AD, Craik DJ, Anderson MA, Proc. Natl. Acad. Sci. USA 2008, 105, 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jennings C, West J, Waine C, Craik D, Anderson M, Proc. Natl. Acad. Sci. USA 2001, 98, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lindholm P, Gçransson U, Johansson S, Claeson P, Gulbo J, Larsson R, Bohlin L, Backlund A, Mol. Cancer Ther 2002, 1, 365–369. [PubMed] [Google Scholar]
- [8].Gustafson KR, McKee TC, Bokesch HR, Curr. Protein Pept. Sci 2004, 5, 331–340. [DOI] [PubMed] [Google Scholar]
- [9].Gustafson KR, Sowder RCI, Henderson LE, Parsons IC, Kashman Y, Cardellina JHI, McMahon JB, Buckheit RWJ, Pannell LK, Boyd MR, J. Am. Chem. Soc 1994, 116, 9337–9338. [Google Scholar]
- [10].Clark RJ, Daly NL, Craik DJ, Biochem. J 2006, 394, 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Craik DJ, Cemazar M, Wang CK, Daly NL, Biopolymers 2006, 84, 250–266. [DOI] [PubMed] [Google Scholar]
- [12].Craik DJ, Cemazar M, Daly NL, Curr. Opin. Drug Discovery Dev 2006, 9, 251–260. [PubMed] [Google Scholar]
- [13].Gunasekera S, Foley FM, Clark RJ, Sando L, Fabri LJ, Craik DJ,Daly NL, J. Med. Chem 2008, 51, 7697–7704. [DOI] [PubMed] [Google Scholar]
- [14].Austin J, Wang W, Puttamadappa S, Shekhtman A, Camarero JA, ChemBioChem 2009, 10, 2663–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Getz JA, Rice JJ, Daugherty PS, ACS Chem. Biol 2011, 6, 837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jagadish K, Camarero JA, Biopolymers 2010, 94, 611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Daly NL, Love S, Alewood PF, Craik DJ, Biochemistry 1999, 38, 10606–10614. [DOI] [PubMed] [Google Scholar]
- [18].Daly NL, Gustafson KR, Craik DJ, FEBS Lett. 2004, 574, 69–72. [DOI] [PubMed] [Google Scholar]
- [19].Huang YH, Colgrave ML, Daly NL, Keleshian A, Martinac B, Craik DJ, J. Biol. Chem 2009, 284, 20699–20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Simonsen SM, Sando L, Rosengren KJ, Wang CK, Colgrave ML,Daly NL, Craik DJ, J. Biol. Chem 2008, 283, 9805–9813. [DOI] [PubMed] [Google Scholar]
- [21].Wade D, Boman A, Wahlin B, Drain CM, Andreu D, Boman HG, Merrifield RB, Proc. Natl. Acad. Sci. USA 1990, 87, 4761–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dawson PE, Muir TW, Clark-Lewis I, Kent SB, Science 1994, 266, 776–779. [DOI] [PubMed] [Google Scholar]
- [23].Tam JP, Lu YA, Protein Sci. 1998, 7, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wishart DS, Sykes BD, Richards FM, Biochemistry 1992, 31, 1647–1651. [DOI] [PubMed] [Google Scholar]
- [25].Ireland DC, Wang CK, Wilson JA, Gustafson KR, Craik DJ, Biopolymers 2008, 90, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang CK, Colgrave ML, Gustafson KR, Ireland DC, Göransson U,Craik DJ, J. Nat. Prod 2008, 71, 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Weislow OS, Kiser R, Fine DL, Bader J, Shoemaker RH, Boyd MR,Natl J. Cancer Inst 1989, 81, 577–586. [DOI] [PubMed] [Google Scholar]
- [28].Chen Y, Mant CT, Farmer SW, Hancock RE, Vasil ML, Hodges RS,J. Biol. Chem 2005, 280, 12316–12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Schöpke T, Hasan Agha MI, Kraft R, Otto A, Hiller K, Sci. Pharm 1993, 61, 145–153. [Google Scholar]
- [30].Barry DG, Daly NL, Clark RJ, Sando L, Craik DJ, Biochemistry 2003, 42, 6688–6695. [DOI] [PubMed] [Google Scholar]
- [31].Mozsolits H, Aguilar MI, Biopolymers 2002, 66, 3–18. [DOI] [PubMed] [Google Scholar]
- [32].Cooper MA, Anal. Bioanal. Chem 2003, 377, 834–842. [DOI] [PubMed] [Google Scholar]
- [33].Henriques ST, Huang YH, Rosengren KJ, Franquelim HG, Carvalho FA, Johnson A, Sonza S, Tachedjian G, Castanho MA, Daly NL,Craik DJ, J. Biol. Chem 2011, 286, 24231–24241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cannon B, Lewis A, Metze J, Thiagarajan V, Vaughn MW, Somerharju P, Virtanen J, Huang J, Cheng KH, J. Phys. Chem. B 2006, 110, 6339–6350. [DOI] [PubMed] [Google Scholar]
- [35].Domenech O, Redondo L, Picas L, Morros A, Montero MT, Hernandez-Borrell J, J. Mol. Recognit 2007, 20, 546–553. [DOI] [PubMed] [Google Scholar]
- [36].Nandi N, Vollhardt D, Acc. Chem. Res 2007, 40, 351–360. [DOI] [PubMed] [Google Scholar]
- [37].Nakagawa H, Onoda M, Masuoka Y, Yamada K, Chirality 2006, 18, 212–216. [DOI] [PubMed] [Google Scholar]
- [38].Bernardini C, D’Arrigo P, Elemento G, Mancini G, Servi S, Sorrenti A, Chirality 2009, 21, 87–91. [DOI] [PubMed] [Google Scholar]
- [39].Tsuchiya H, Chem. Biol. Interact 2001, 134, 41–54. [DOI] [PubMed] [Google Scholar]
- [40].Glukhov E, Burrows LL, Deber CM, Biopolymers 2008, 89, 360–371. [DOI] [PubMed] [Google Scholar]
- [41].Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS, Antimicrob. Agents Chemother 2007, 51, 1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sengupta D, Leontiadou H, Mark AE, Marrink SJ, Biochim. Biophys. Acta Biomembr 2008, 1778, 2308–2317. [DOI] [PubMed] [Google Scholar]
- [43].Shai Y, Biopolymers 2002, 66, 236–248. [DOI] [PubMed] [Google Scholar]
- [44].Nourse A, Trabi M, Daly NL, Craik DJ, J. Biol. Chem 2004, 279, 562–570. [DOI] [PubMed] [Google Scholar]
- [45].Plan MRR, Göransson U, Clark RJ, Daly NL, Colgrave ML, Craik DJ, ChemBioChem 2007, 8, 1001–1011. [DOI] [PubMed] [Google Scholar]
- [46].Nicke A, Loughnan ML, Millard EL, Alewood PF, Adams DJ, Daly NL, Craik DJ, Lewis RJ, J. Biol. Chem 2003, 278, 3137–3144. [DOI] [PubMed] [Google Scholar]
- [47].G ntert P, Mumenthaler C, W thrich K, J. Mol. Biol 1997, 273, 283–298. [DOI] [PubMed] [Google Scholar]
- [48].Br nger AT, Adams PD, Rice LM, Structure 1997, 5, 325–336. [DOI] [PubMed] [Google Scholar]
- [49].Hutchinson EG, Thornton JM, Protein Sci 1996, 5, 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM, Biomol J. NMR 1996, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- [51].Henriques ST, Pattenden LK, Aguilar MI, Castanho MA, Biophys. J 2008, 95, 1877–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.