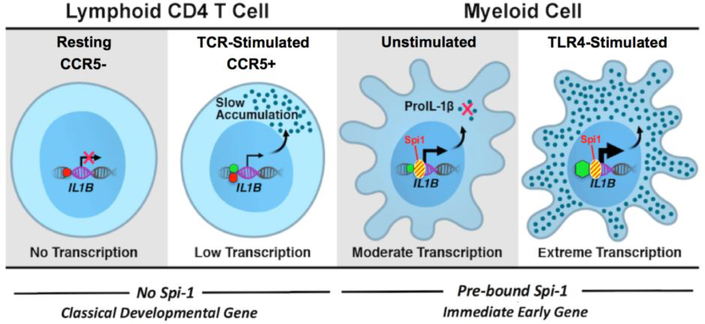
Abstract
Interleukin 1β is a pro-inflammatory cytokine important for both normal immune responses and chronic inflammatory diseases. The regulation of the 31 kDa proIL-1β precursor coded by the IL1B gene has been extensively studied in myeloid cells, but not in lymphoid-derived CD4 T cells. Surprisingly, we found that some CD4 T cell subsets express higher levels of proIL-1β than unstimulated monocytes, despite relatively low IL1B mRNA levels. We observed a significant increase in IL1B transcription and translation in CD4 T cells upon ex vivo CD3/CD28 activation, and a similar elevation in the CCR5+ effector memory population compared to CCR5- T cells in vivo. The rapid and vigorous increase in IL1B gene transcription for stimulated monocytes has previously been associated with the presence of Spi-1/PU.1 (Spi1), a myeloid-lineage transcription factor, pre-bound to the promoter. In the case of CD4 T cells, this increase occurred despite the lack of detectable Spi1 at the IL1B promoter. Additionally, we found altered epigenetic regulation of the IL1B locus in CD3/CD28-activated CD4 T cells. Unlike monocytes, activated CD4 T cells possess bivalent H3K4me3+/H3K27me3+ nucleosome marks at the IL1B promoter, reflecting low transcriptional activity. These results support a model in which the IL1B gene in CD4 T cells is transcribed from a low-activity bivalent promoter independent of Spi1. Accumulated cytoplasmic proIL-1β may ultimately be cleaved to mature 17 kDa bioactive IL-1β, regulating T cell polarization and pathogenic chronic inflammation.
Keywords: Spi1/PU.1, Interleukin 1beta, Bivalent Promoter, T Cell Receptor Activation
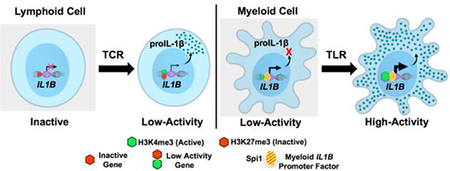
Graphical abstract
1. Introduction
ProIL-1β, the interleukin 1β protein precursor, is encoded by the IL1B gene within an IL1 family gene cluster located on human chromosome 2. ProIL-1β is principally produced by activated monocytes/macrophages. These cells become activated by pathogen-associated molecule patterns (PAMPs) recognized by pattern recognition receptors (PRRs), and/or secondary damage signals (DAMPs, danger-associated molecular patterns)[1, 2]. Following activation, proIL-1β is synthesized as a precursor protein with very low biological activity. It is then processed into highly active mature IL-1β, either intracellularly by the caspase-1 inflammasome or extracellularly by other proteases[3–5]. Most of our current understanding of IL1B transcriptional and translational regulation stems from studies of myeloidlineage cells. IL1B gene expression has not extensively been examined in lymphoid cells.
Spi-1/PU.1 (Spi1) is a transcription factor involved in genome-wide development and maintenance of cells in the macrophage lineage[6]. Spi1 often associates with inducible transcription factors, such as NFκB and C/EBPβ, on lipopolysaccharide (LPS)-responsive promoters and enhancers in human and murine macrophages[7, 8]. In the monocyte/macrophage lineage, Spi1 constitutively binds to the IL1B promoter at two distinct sites located between −50 to −39 and −115 to −97 relative to the transcription start site[9, 10]. In non-myeloid cells, its ectopic expression can result in IL1B transcription in the presence of an activation signal for NF-κB and C/EBPβ[9–11]. Additionally, Spi1 can act as a “pioneer factor”, binding nucleosome-occluded DNA and facilitating chromatin accessibility for LPS-responsive transcription factors in activated monocytes[12, 13]. Further, it directly recruits TATA-binding protein (TBP), which is involved in forming the pre-initiation complex (PIC) that helps to recruit RNA Polymerase II (Pol II) to gene promoters[9, 14].
IL-1β is expressed at extremely high levels in myeloid-derived cells in response to microbial invasion and tissue injury[9]. Although activated monocytes are a major source of IL-1β; NK cells, B cells, dendritic cells, fibroblasts, and epithelial cells also express this protein, but at much lower levels[15]. ProIL-1β has been previously detected in human lymphoid CD4 T cells expressing chemokine receptor 5 (CCR5+)[16]. This proIL-1β can be cleaved and released as highly active mature IL-1β following abortive HIV infection. Recent reports have further shown that stimulation of the T cell antigen-receptor (TCR), when combined with varied co-stimulation can induce the production of proIL-1β in CD4 T cells. Specifically, mouse CD4 T cells that were TCR-activated in vitro by CD3/CD28 crosslinking were found to produce proIL-1β mRNA and protein[17]. Human CD4 T cells also produced high levels of proIL-1β when CD3 stimulation was combined with anti-CD46 complement receptor activation[18]. This study further showed that NLRP3 inflammasome activation in these cells caused cleavage of proIL-1β to highly bioactive mature IL-1β form, supporting polarization of type-1 T-helper cells in an autocrine manner [18]. While these studies highlight the biologic importance of lymphocyte-derived IL-1β, little is known about the regulation of the IL1B gene in CD4 T cells.
As stated above, IL1B transcription depends on the Spi1 transcription factor, which is highly expressed in monocytes. However, it is not known whether proIL-1β expression depends on Spi1 in CD4 T cells. The status of IL1B gene transcription and its epigenetic landscape in lymphoid CD4 T cells is also unknown. Thus, we set out to measure the regulation of IL1B in lymphoid-derived CD4 T cells, including Pol II and Spi1 engagement and specific epigenetic marks on IL1B chromatin, in both ex vivo CD3/CD28-activated CD4 T cells and in vivo differentiated memory CCR5+ CD4 T cells isolated from human lymph nodes.
2. Materials and Methods
2.1. Cell Culture, Reagents and Treatment Conditions
Cell lines were obtained from American Type Culture Collection (ATCC). THP-1 cells were cultured in RPMI media (Cellgro) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone), 1% Penicillin/Streptomycin Solution (Cellgro) and 500 μl of 2-mercaptoethanol (Invitrogen). HEK293 cells were grown in EMEM (Cellgro) containing 10% heat-inactivated fetal bovine serum and 1% Penicillin/Streptomycin Solution. In mRNA and ChIP analysis, THP-1 cells were stimulated with 1ug/ml of E.coli 055:B5 Lipopolysaccharide (LPS) (L2880, Sigma-Aldrich) for 1.5 hours. Human tonsils were obtained from the Cooperative Human Tissue Network and processed as previously described[16, 19]. Briefly, single cell suspensions were prepared and purified by density gradient-centrifugation with FicollPaque Hypaque (GE Healthcare Life Sciences). CD4 T cells were enriched by negative selection using EasySep Human CD4+ T Cell Enrichment Kits (STEMCELL Technologies), per the manufacturer’s protocol. Following isolation, cells were cultured in RPMI (Gibco), plus 10% heat-inactivated FBS (Atlas) and 1X Penicillin/Streptomycin/L-Glutamine (Gibco). For CD3/CD28 stimulation, antiCD3/CD28 magnetic beads (Dynabeads, Thermo Fisher Scientific) were added at a ratio of one bead per cell. Ex vivo anti-CD3/CD28 treatment of CD4 T cells results in antigen-independent activation of the CD3 T cell receptor and the CD28 co-receptor. This is in contrast to normal in vivo activation involving cellular presentation of antigen by major histocompatibility complex II to the T cell receptor complex, in the context of costimulatory and cytokine signals.
2.2. Cell Sorting and Flow Cytometric Analysis
Human tonsils were obtained from the Cooperative Human Tissue Network and processed as previously described[16, 19]. These tissue specimens are de-identified before receipt by the laboratory, and have been classified exempt from human subjects research by the Human Research Protection Program Institutional Review Board of the University of California, San Francisco. Briefly, single cell suspensions were prepared and purified by density gradient-centrifugation with Ficoll-Paque Hypaque (GE Healthcare Life Sciences). For ChIP analyses, CD4 T cells were isolated from HLAC by positive selection using CD4 microbeads (Miltenyi) per manufacturer’s protocol. Isolated CD4 T cells were stained with APCCD4, APC/Cy7-CD3, and PE-CCR5 (clone 2D7/CCR5) (BD Biosciences) on ice for 3 hours. Stained cells were sorted using FACSAriaII (BD Biosciences) to isolate CD3+CD4+CCR5+, and CD3+CD4+CCR5- T cell populations. For ChIP analyses the sorted cells were cross-linked immediately with 1% formaldehyde (as described below) and pellets were stored at −80 °C. For mRNA analyses cells were lysed in TRIzol (Invitrogen). For protein analyses, CD4 T cells were enriched by negative selection using EasySep Human CD4+ T Cell Enrichment Kits (STEMCELL Technologies), per the manufacturer’s protocol. The CD4 T cells were labelled on ice for 30 minutes with APC/Cy7-CD3, PE/Cy7-CD4, and Brilliant Violet 421-CCR5 (BioLegend). The cells were then FACS sorted as above. Sorted cells were lysed with RIPA buffer: 50mM Tris (Sigma), 150 mM NaCl (Sigma), 0.1% SDS (BioRad), 0.5% sodium deoxycholate (Sigma), 1% Triton X100 (Sigma), and Complete protease inhibitor (Roche), and the lysates were frozen at −80˚C.
2.3. Western Blot Analysis
Cell lysates prepared as above were loaded in 4–12% Bis-Tris NuPAGE gels (ThermoFisher) for gel electrophoresis and transferred to PVDF membrane (EMD Millipore). Membranes were blocked for 1 hour at room temperature in blocking buffer: PBS + 0.05% Tween20 (Sigma) + 5% non-fat dry milk (BioRad). Primary antibodies were diluted in blocking buffer and incubated overnight at 4°C. ProIL-1b was detected by a C-terminal-reactive mAb at 1 μg/mL (mAb 201, R&D Systems). Blots were washed and probed with secondary Goat Anti-Mouse IgG, Human ads-HRP (SouthernBiotech), diluted 1:20,000 in PBS + 0.05% Tween20 + 5% bovine serum albumin (Axenia BioLogix) for 2 hours at room temperature. HRP was detected using Western Lightning ECL Pro Enhanced Luminal Reagent (PerkinElmer) and Hyperfilm ECL (GE/Amersham).
2.4. mRNA Expression Analysis
1×106 THP-1 and HEK293 cells were plated into 6-well plates (Thermo Fisher). Following the LPS treatments, cells were pelleted and supernatant was removed. The cell pellet was re-suspended in 500 μl of TRIzol reagent (Invitrogen). For ex vivo differentiation experiments, naïve or anti-CD3/CD28-treated T cells were cultured for 3 days, counted, and 5×106 cells were resuspended in 500 μL of TRIzol reagent. For in vivo analyses, T cells were sorted, as described above, and 5×106 CCR5- cells and 2×106 CCR5+ cells were immediately resuspended in 500 μl of TRIzol reagent. Following the addition of 170 μl of Chloroform (Fisher), the samples were vortexed well and incubated at room temperature for 15 minutes. Then, the samples were centrifuged at 13200 RPM for 15 minutes at 4°C. Aqueous layer was transferred into a fresh tube and combined with 500 μl of isopropanol (Fisher) and 1 μl of Glycogen (Ambion). The samples were incubated at room temperature for 10 minutes and then centrifuged at 13200 RPM for 10 minutes at 4°C. The RNA pellets were washed with 500 μl of 75% Ethanol (Pharmaco-AAPER) and centrifuged for 10 min in room temperature at 14000 RPM. Then, the ethanol was aspirated and the pellets were allowed to air-dry for 10–15 minutes. Air-dried pellets were re-suspended in 25 μl of DEPCtreated water (Ambion). The samples were incubated at 65°C for 10 minutes and subjected to DNAse treatments using Turbo DNA-free reagents (Ambion) according to the manufacturer instructions in order to eliminate genomic DNA contamination. RNA concentration, 260/280 and 260/230 values were measured using NanoDrop 1000 spectrophotometer (Thermo Fisher). mRNA was converted into cDNA using GoScript Reverse Transcription System (A5001, Promega). cDNA was analyzed using quantitative PCR (qPCR) carried out in a StepOnePlus Applied Biosystems Real Time Instrument (Thermo Fisher). Relative expression levels were calculated using ΔCt method. Primer Sequences used for mRNA analysis are indicated in Table 1A.
2.5. Chromatin Immuno-precipitation (ChIP)
ChIP was performed using a modification of the Millipore/Upstate protocol (MCPROTO407). In brief, a total of 1×107 THP-1 monocytes, 2.5×107 naïve CD4 T cells, 2.5×107 CD3/CD28-activated CD4 T cells, 5.9×107 CCR5- CD4 T cells and 1.58×107 CCR5+ CD4 T cells were fixed in 1% formaldehyde (Fisher) for 10 min at room temperature. Cross-linking was inhibited by addition of glycine to a final concentration 0.125 M. Cell pellets were washed twice with ice cold PBS and resuspended in SDS Lysis Buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1) supplemented with Protease inhibitor cocktail (Sigma) and 1mM phenylmethylsulfonyl fluoride (PMSF, Fluka). Samples were sonicated (to generate DNA fragments of 250 base pairs (bp) average length) on ice using a Fisher Scientific Sonic Dismembrator (Model 100), as follows: 15×25 strokes at 100% power followed by 3×25 stokes at 50% power and centrifuged at 12000 RPM for 10 min. Chromatin was diluted 7-fold in ChIP Dilution Buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH8.1, 167 mM NaCl. Total equivalence of 3×106 cells was used for Pol II and Spi1 pull downs for both ex vivo and in vivo CD3/CD28-activated T cells. For histone modifications ChIP analyses, 3×106 cells were used for ex vivo whereas 1.5×106 cells were used for in vivo CD3/CD28-activated CD4 T cells. Supernatants were incubated at 4˚C overnight with Santa Cruz antibodies Pol II (sc-899x), PU.1 (sc-352x), control IgG (sc2027x); Active Motif antibodies H3K9ac (61251), H3K4me3 (39915), H3K27me3 (39155), H3K36me3 (61101) and Cell Signaling Technology antibody Histone 3 (9715S). Aliquots for INPUT and nonspecific IgG control samples were included with each experiment. Samples were precipitated using 25 μl of Magna ChIP Protein A+G Magnetic beads, at 4˚C for 3 hours, and subsequently washed with following solutions: once with Low-Salt Buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20 mM TrisHCl, pH 8.1, 150 mM NaCl), once with High-Salt Buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20 mM Tris-HCl, pH 8.1, 550 mM NaCl), once with LiCl Wash Buffer (0.25 M LiCl, 1% IGEPAL-CA630, 1% deoxycholic acid, 1mM EDTA, 10 mM Tris, pH 8.1), and twice with TE Buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). Immunocomplexes were eluted for 4 hours at 65˚C with 200 μl of ChIP Elution Buffer (1% SDS, 0.1M NaHCO3). To reverse the cross-linking, eluted samples were treated with 10 μl of 5 M NaCl and subsequently incubated at 65˚C for ≥4 hours. DNA was purified using a GeneJET PCR Purification kit (K0702, Thermo Scientific). Primer Sequences used for ChIP analysis are indicated in Table 1B. The size of the PCR products ranges between 80 and 150 bp. Twenty micro liter qPCR reactions containing 2x Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific), 250 nM of primers, and 3 μl of precipitated DNA were set up in Fast 96-Well Reaction Plates (Applied Biosystems). qPCR reactions were carried out in a StepOnePlus Applied Biosystems Real Time Instrument. Fold enrichment was calculated based on Ct as 2(ΔCt), where ΔCt = (Ct Input – Ct IP). Final enrichment values were adjusted by subtraction of the nonspecific IgG antibody binding.
3. Results
3.1. Human lymphoid T cells express proIL-1β after T cell antigen-receptor activation.
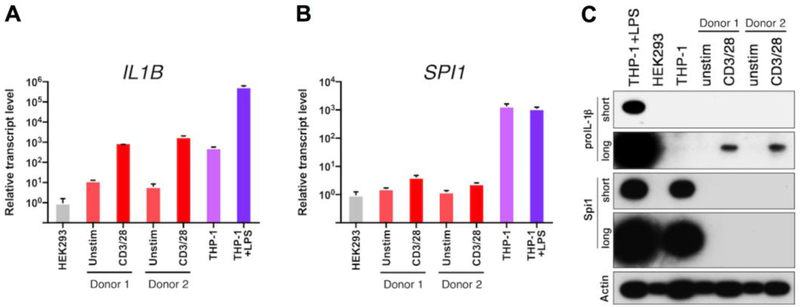
To better understand the process of proIL-1β expression in lymphoid CD4 tonsillar T cells, antiCD3/CD28 beads were used in order to stimulate the T cell antigen receptors. After three days of activation, IL1B mRNA significantly increased in the ex vivo-activated tonsillar CD4 T cells, as compared to unstimulated T cells and the THP-1 human monocytic cell line, a well-studied benchmark population for IL1B gene expression, as well as the negative control human kidney epithelial HEK293 (Fig. 1A). The mRNA kinetic profile for IL1B revealed highest transcription after 3 days following the CD3/CD28 activation (Supplementary Fig. 1A). However, LPS-treated THP-1 cells expressed approximately 500fold more IL1B mRNA than the 3-day CD3/CD28 activated CD4 T cells. Of note, the level of SPI1 mRNA, which encodes Spi1, a myeloid-lineage transcription factor required for vigorous IL1B gene expression in activated monocytes[9], was slightly increased following the activation of CD3/CD28 on CD4 T cells (Fig. 1B, Supplementary Fig. 1B), although Spi1 protein was undetectable (Fig. 1C). By contrast, THP-1 monocytes expressed high levels of constitutive SPI1 mRNA and protein, which did not increase after LPS treatment.
Figure 1. CD4 T cells express proIL-1β following TCR stimulation.
Relative mRNA expression for (A) IL1B and (B) SPI1 in purified CD4 T cells from human lymphoid tissue (two individual patient donors), stimulated for 3 days with anti-CD3/CD28 beads, compared to resting CD4 T cells, HEK293 cells, and THP-1 cells (unstimulated or LPStreated for 1.5 h). Data were normalized to the HEK293 cell sample; standard error represents technical replicates. (C) Western blot analysis of proIL-1β and Spi1 proteins from samples shown in A and B. The proIL-1β and Spi1 blots are shown at short and long exposures to illustrate the relative abundance of these proteins in CD4 T cells. Membranes were stripped and re-probed for β-actin. Note that the β-actin levels vary between cell types and increases with TCR stimulation. Sample inputs for western blots were normalized by cell equivalents: 1.5×105 for all lanes. For conciseness and clarity, only cropped western blot images are displayed. The full-length images of these blots are provided in Supplementary Fig. 3A. Standard error for each donor from lymphocyte populations represents technical replicates, and control cell lines from cultured biologic replicates.
Western blot analysis revealed that CD3/CD28 activated T cells expressed significantly higher levels of proIL-1β compared to their unstimulated counterparts (Fig. 1C). Importantly, unstimulated THP-1 monocytes did not express proIL-1β protein (Fig. 1C). As expected, CD4 T cells expressed much lower levels of proIL-1β protein than LPS-treated THP-1 monocytes. Together, these results demonstrate that antigen receptor stimulation of human CD4 lymphocytes can induce de novo expression of IL1B gene transcription and protein synthesis. Flow cytometry analysis for proIL-1β revealed maximal protein after 3 days of CD3/CD28 activation with significant increase in induction over naïve T cells after 1 day of activation (Supplementary Fig. 2D). This flow cytometry analysis also revealed that increase in proIL-1β production in activated CD4 T cells is likely from majority of the T cells expressing the protein at low levels and not from few high producers (Supplementary Fig. 2E). Interestingly, among the CD3/CD28 activated T cells, proIL-1β is produced by both CCR5+ and CCR5- cells (Supplementary Fig. 2F).
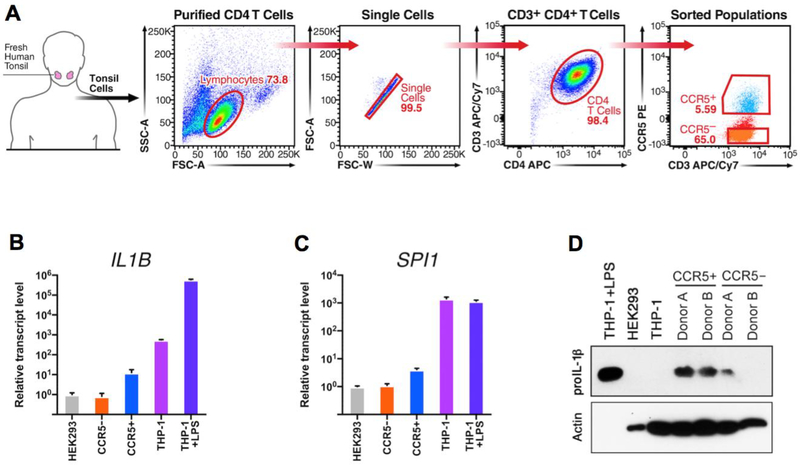
Previous work has shown that CCR5+ cells express proIL-1β protein, which can be cleaved and released during pyroptotic cell death[16]. In vivo CCR5+ CD4 T cells are predominantly effector memory cells, expanded from previous antigen-mediated activation. Lymphoid-derived CCR5+ and CCR5- CD4 T cells were purified from fresh human tonsil tissue by sequential magnetic and flow sorting (Fig. 2A). All preparations of these cells were completely devoid of monocytes (Supplementary Fig. 2A). Cells from six donors were combined and compared with negative control HEK293 cells, as well as both unstimulated and LPS-stimulated THP-1 cells. IL1B mRNA was approximately 15-fold higher in CCR5+ CD4 T cells than their CCR5- counterparts (Fig. 2B). Interestingly, IL1B mRNA was much lower in CCR5+ T cells than unstimulated THP-1 monocytes, which express measurable levels of IL1B mRNA[20]. SPI1 mRNA was also slightly higher in CCR5+ cells than in CCR5- T cells (Fig. 2C). As previously described[9], THP-1 monocytic cells expressed high levels of constitutive SPI1 mRNA, independent of LPS stimulation. Western blot analysis revealed that CCR5+ T cells expressed higher levels of proIL-1β protein than CCR5- T cells, unstimulated THP-1, and HEK293 cells (Fig. 2D). Anticipating that LPStreated THP-1 cells would produce high levels of proIL-1β protein, lysate for this positive control was loaded at one-tenth of that used for all other samples. This revealed an intense band for proIL-1β protein at ~31 kDa for LPS-treated THP-1. Again, proIL-1β protein was completely undetectable in unstimulated THP-1 cells, but was present in CCR5+ CD4 T cells (Fig. 2D). This result contrasts with the 25-fold greater abundance of IL1B mRNA in unstimulated THP-1 compared to CCR5+ T cells (Fig. 2B), reflecting different mechanisms of post-transcriptional regulation of IL1B mRNA between these two cell types.
Figure 2: in vivo differentiated CCR5+ CD4 T cells express proIL-1β.
(A) Shown is a representative flow-sort from an individual CCR5+ CD4 T cell isolation. Cells were sorted from the CCR5+ and CCR5- gates as shown, after performing negative-selection magnetic enrichment of CD4 T cells from fresh human tonsil. Relative mRNA expression levels for (B) IL1B and (C) SPI1 from CCR5+ and CCR5- CD4 T cells, HEK293 cells, and both unstimulated and LPS-stimulated THP-1. For mRNA analysis, multiple donors were pooled and the standard error represents technical replicates. The standard error for THP-1 and HEK293 reference samples were calculated from biological replicates. (D) Western blot for proIL-1β from cells prepared as above described for two individual patient donors. Samples were normalized by cell equivalents: 1.5×104 cell equivalents for THP-1+LPS and 1.5×105 in all other lanes. Membranes were stripped and re-probed for β-actin. The β-actin band for LPS stimulated THP-1 is not visible on the blot because only 1/10th of the cells were added to prevent over-saturation of pro-IL1β band. For conciseness and clarity, only cropped western blot images are displayed. The full-length images of these blots are provided in Supplemental Fig. 3B.
3.2. Spi1 is not a detectable regulator of IL1B gene transcription in activated T cells.
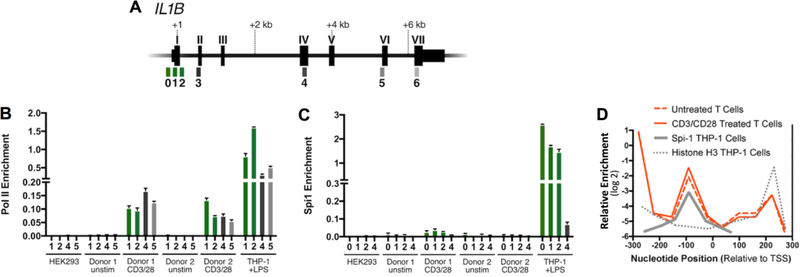
To further understand de novo transcription of IL1B after CD3/CD28 activation of CD4 T cells, the presence of Pol II and Spi1 on the IL1B gene was investigated using chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR). Amplicons for chromatin immunoprecipitation (ChIP) were designed to span the IL1B promoter and structural gene in order to measure the enrichment profile for Pol II and Spi1, as previously described[9] (Fig. 3A). We found that RNA Polymerase II (Pol II) on the IL1B gene in ex vivo CD3/CD28-activated T cells was significantly enriched at the promoter and throughout the gene body, compared with unstimulated CD4 T cells (Fig. 3B). Pol II engagement was not detected on the IL1B gene in negative control HEK293 cells. As expected, LPS-treated THP-1 monocytes had the highest levels of Pol II at both the promoter and throughout the body of the gene.
Figure 3. Occupancy of Pol II and Spi1 on the IL1B gene.
(A) Gene schematics with exons labeled as Roman numerals, and qPCR amplicons indicated by Arabic numbers referenced to positions listed in Supplementary Table 1. (B) Pol II and (C) Spi1 ChIP for indicated CD4 T cell populations. For comparison, IL1B-refractory HEK293 cells were used as a negative control, and THP-1 cells stimulated with LPS for 1.5 hours were used as a positive control. CD4 T cells were purified from human lymphoid tissue and activated for 3 days with anti-CD3/CD28 beads and compared to unstimulated CD4 T cells. Promoter-proximal amplicons are shown as green bars, whereas downstream amplicons are shown as gray scale bars. Standard error for each donor from lymphocyte populations represents technical replicates, and control cell lines from cultured biologic replicates. (D) Histone H3 nucleosome ChIP at IL1B promoter in ex vivo CD3/CD28-activated and unstimulated CD4 T cells as described within, are qualitatively compared to the previously reported Spi1 binding site and nucleosome distribution at the IL1B promoter in THP-1 cells[9]. This reveals that a nucleosome is positioned over the Spi1 recognition site in T cells. It has also been reported that Spi1-binding displaces this histone in THP-1 cells, but not in both Spi1-negative 293 cells and the HUT102 T cell line[9]. The qPCR amplicons used are listed in Supplementary Table 1.
In monocytes, Spi1 constitutively binds at the IL1B promoter and is required for transcription[9]. Because SPI1 mRNA was slightly up-regulated in CD4 T cells after ex vivo CD3/CD28 activation, Spi1 protein engagement at the IL1B promoter was measured. Spi1 protein was not enriched at the promoter region of IL1B in either CD3/CD28-activated or CCR5+ CD4 T cells, when compared to the body of the gene (Fig. 3C, Supplementary Fig. 4C). This is supported by the lack of detectable Spi1 protein in T cells (Fig. 1C). Spi1 was also not significantly enriched at the promoter in IL1B-refractory HEK293 cells. However, Spi1 was more abundant at the promoter of LPS-stimulated THP-1 cells than at the downstream gene body (Fig. 3C). Histone H3 ChIP nucleosome profiles were also generated for the IL1B promoter in activated CD4 T cells and compared to those of THP-1 cells, Hut102 T cells, and HEK293 cells from our previous work[9]. These revealed that both unstimulated and stimulated primary T cells, like HEK293 and Hut102 T cells, possess a nucleosome positioned over the Spi1 binding site, supporting a Spi1-independent model for IL1B gene transcription in CD4 T cells (Fig. 3D).
3.3. IL1B is transcribed from a bivalent H3K4me3/H3K27me3 promoter in CD3/CD28-activated T cells.
The ex vivo CD3/CD28-activated lymphoid CD4 T cells were examined for the presence of chromatin marks that correlate with either permissive/active or repressed/inactive genes at the IL1B locus. Specifically, H3K4me3 and H3K9ac at promoter-proximal regions are generally associated with actively transcribed genes[21], whereas H3K27me3 is generally associated with inactive genes[22]. Additionally, H3K36me3 located at downstream regions of genes is generally associated with active transcript elongation by Pol II[23].
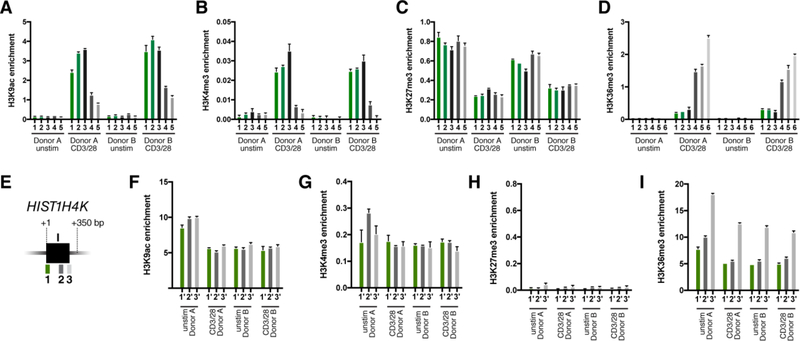
Following ex vivo CD3/CD28 activation, the H3K9ac and H3K4me3 marks significantly increased (Fig. 4A and B), consistent with transcriptional activity. The H3K27me3 repressive mark only slightly decreased (Fig. 4C), while the H3K36me3 mark increased along the length of the gene, indicating ongoing transcription (Fig. 4D). However, comparing these IL1B histone modifications to those of constitutively expressed “housekeeping” HISTH4K gene in the same cells, the activating H3K9ac and H3K4me3 marks were approximately 2- and 4-fold lower, respectively, on IL1B than HISTH4K (Fig. 4F and G). Also, the repressive H3K27me3 mark is 10 to 12-fold higher on IL1B in activated T cells than on HISTH4K in the same cells (Fig. 4H). Together, these findings of intermediate levels for both activating and inhibitory marks support a “bivalent” status for the IL1B promoter in CD4 lymphocytes following activation.
Figure 4. CD3/CD28-stimulation of CD4 T cells induces bivalent H3K4me3 +/H3K27me3+ marks.
Histone modification ChIP analysis of IL1B in CD3/CD28-activated and unstimulated CD4 T cell populations from two donor samples as indicated: (A) H3K9Ac; (B) H3K4me3; (C) H3K27me3, and (D) H3K36me3. The qPCR amplicon designations for panels A-D are as described in Fig. 3A. Histone modification ChIP of the HIST1H4K (H4) gene in ex vivo CD3/CD28-activated and resting CD4 T cell populations as indicated: (F) H3K9Ac; (G) H3K4me3; (H) H3K27me3; and (I) H3K36me3. The qPCR amplicon designations for panels F-I are as shown in (E). Promoter-proximal amplicons are shown as green bars, whereas downstream amplicons are shown as gray scale. H3K9ac levels at the IL1B promoter are approximately half of HIST1H4K in activated CD4 cells. H3K4me3 is much lower on IL1B. Note that the inhibitory mark, H3K27me3, is significantly reduced on the IL1B gene following TCR activation, but not as low as it is on HIST1H4K in the same cells. Standard error for each donor represents technical replicates.
The epigenetic status of the IL1B gene in CCR5+ and CCR5- CD4 T cells was also investigated; revealing a slightly enriched H3K9 acetylation at the IL1B promoter in CCR5+ CD4 T cells (Supplementary Fig. 4F). Both active H3K4me3 and repressive H3K27me3 were also enriched in CCR5+ CD4 T cells (Supplementary Fig. 4G and H), a characteristic of low-activity bivalent promoters[24]. In contrast, CCR5- CD4 T cells exhibited the H3K27me3, but not the active H3K4me3 mark at the IL1B promoter. Histone H3K36me3 was not significantly different on IL1B in either CCR5+ or CCR5- populations (Supplementary Fig. 4I). The control “housekeeping” HIST1H4K gene for both activated CCR5+ and naïve CCR5- cells revealed a promoter signature characteristic of actively expressed genes (H3K4me3+/H3K27me3Low), with CCR5- revealing higher levels of H3K4me3 than CCR5+ (Supplementary Fig. 4G and H). Histone H3K36me3 revealed the classic increase of gene body over promoter at the transcriptionally active HIST1H4K gene (Supplementary Fig. 4I).
4. Discussion
Recent findings of proIL-1β production and functionality in CD4 T cells highlight the importance of lymphocyte-derived IL-1β, a previously unappreciated phenomenon. We set out to investigate the regulation of the IL1B gene expression in CD4 T cells, as previous reports have all focused on these processes in myeloid-lineage cells. In the absence of stimulation, low-level IL1B transcription in resting monocytic cells does not result in IL-1β protein expression because of a specific post-transcriptional translation blockade[20] (Fig. 1). LPS-TLR-stimulation of THP-1 cells causes a massive increase in IL1B transcription, overcoming the post-transcriptional block, resulting in abundant translation of proIL-1β. By contrast, the vast majority of unstimulated lymphoid CD4 T cells have little or no IL1B transcription, while TCR activation causes de novo transcription and translation of IL1B to proIL-1β (Fig. 1A and C, Supplementary Fig. 1A). It is important to note that the low expression of proIL-1β protein in CD4 T cells is not from few high producers but from majority of the activated cells expressing the protein at low levels (Supplementary Fig. 2E). Interestingly, both CCR5+ and CCR5- CD4 T cells are expressing proIL-1β after 3 days of CD3/CD28 activation. Thus, it is likely that proIL-1β expression is induced independently of CCR5, following TCR activation.
Previous studies demonstrated that vigorous IL1B transcription in activated monocytes has an absolute requirement for the myeloid lineage-determining Spi1 transcription factor[9]. Although Spi1 is expressed in early stage, pre-commitment (CD4/8 double-negative) thymocytes, it is lost in CD4/8 double-positive and single-positive naïve T cells[25]. Spi1 can be re-expressed in polarized Th2[26] and Th9[27] cells, suggesting that it may play a role in the regulation of IL1B gene expression following activation of CD4 T cells. Accordingly, we sought to compare the status of Spi1 involvement in IL1B gene transcription for activated CD4 T cells. Although, a slightly higher level of SPI1 mRNA was observed in activated T cells, when compared to resting T cells and HEK293 cells, Spi1 protein was not detected in any of these cells (Fig. 1C). Consistent with the role of Spi1 as a required monocyte-macrophage lineage commitment factor for IL1B transcription[9], its basal expression in both unstimulated and LPS-treated THP-1 monocytes were very high when compared to HEK293 and CD4 T cells. The very low level of proIL-1β protein in activated T cells, as compared to LPS-treated THP-1 monocytes, suggested a potentially distinct mechanism for IL1B gene transcription in T cells. The significant increase of Pol II at the promoter and body of the IL1B gene in activated T cells revealed by ChIP-qPCR provides further support for de novo IL1B transcription following TCR activation (Fig. 2B). Consistent with the mRNA observations, this level of Pol II engagement at the IL1B gene is much lower than that observed on LPStreated THP-1 cells, resulting in significantly lower IL1B gene transcription in T cells than in activated monocytes. Additionally, Spi1 ChIP did not reveal enrichment at the IL1B promoter in activated T cells, arguing that transcription of IL1B in lymphocytes is Spi1-independent. This result is in striking contrast to monocytes, for which IL1B gene expression is absolutely dependent on Spi1 and THP-1 cells exhibit a robust constitutive presence of Spi1 at the canonical DNA recognition site on the IL1B promoter[9] (Fig. 2C).
Additionally, the IL1B promoter in both unstimulated and activated T cells reveals strong nucleosome enrichment positioned directly over the Spi1 DNA binding site (Fig. 3D), consistent with the lack of involvement of Spi1 in CD4 T cell transcription of IL1B. We have previously reported the presence of a stable nucleosome directly over the Spi1 binding site at the IL1B promoter in the Hut102 T cell line that does not express IL1B mRNA[9]. The winged helix-turn-helix (wHTH) DNA binding domain of Spi1, like other wHTH proteins[28], possesses potential “pioneer” activity for opening chromatin by nucleosome displacement. Consequently, the presence of a strong nucleosome signal over the high avidity Spi1 DNA binding sequence at the IL1B promoter is consistent with the absence of Spi1 activity in CD4 T cells.
As previously demonstrated[16], the majority of CD4 T cells producing proIL-1β in vivo are marked by expression of the chemokine receptor CCR5 (Fig. 2D). Interestingly, the relative IL1B mRNA expression in CCR5+ lymphocytes, though 15-fold higher than their CCR5- counterparts, was lower than that of unstimulated THP-1 monocytes. This observation is consistent with previous studies demonstrating that the IL1B gene is transcribed from a Spi1-poised promoter in unstimulated THP-1 monocytes[9] that expresses a low basal level of IL1B mRNA[29]. Despite active transcription of IL1B in unstimulated THP-1 cells, the intracellular proIL-1β protein level in CCR5+ CD4 T cells was higher. This finding is not surprising, since unstimulated monocytes are known to exhibit a constitutive background level of IL1B mRNA transcript in the absence of detectable protein, due to a translational blockade that is reversed by LPS treatment[20]. Therefore, if CCR5+ CD4 T cells lack a post-transcriptional blockade, a low-level translation of proIL-1β may occur as a result of a low level of IL1B transcription. Supporting a model of low IL1B transcription rate in CCR5+ cells, we detected only slight Pol II enrichment at IL1B promoter-proximal regions, as compared to CCR5- CD4 T cells (Supplementary Fig. 4B), but differences in Pol II enrichment between these T cells populations were not detected throughout the length of the IL1B gene body, as observed in CD3/CD28 ex vivo activated T cells. The same chromatin samples were also evaluated for the actively transcribed HIST1H4K housekeeping gene. Both CCR5+ and CCR5- cells showed significant engagement of Pol II on HIST1H4K (Supplementary Fig. 4D). Although Pol II is likely transcribing IL1B at a very low level, over time, accumulation from low-rate transcription/translation could explain the substantial pool of intracellular proIL-1β detected in CCR5+ CD4 T cells (Fig 2D).
As observed following ex vivo CD3/CD28 activation, SPI1 gene mRNA levels were slightly higher in CCR5+ cells than in both CCR5- and HEK293 cells. However, in light of the lack of detectable Spi1 in activated CD4 T cells (Fig. 1C) and negative detection of IL1B in Spi1 ChIP of CCR5+ CD4 T cells (Supplementary Fig. 4C), it is unlikely that Spi1 is responsible for the low level IL1B transcription observed in CCR5+ CD4 T cells in vivo.
It is well established that CCR5+ CD4 T lymphocytes are the primary cellular targets of HIV infection[30–34]. However, in ex vivo cultures from fresh human tonsil or spleen tissues[35], the majority of these cells are non-permissive for HIV replication, with the cytosolic viral DNA intermediates initiating an innate immune response that leads to the activation of caspase-1. This results in abortive HIV infection[36], along with the death of abortively-infected CD4+ T-cells via caspase 1-mediated pyroptosis (an inflammatory form of programmed cell death)[16]. As a result, dying CCR5+ CD4 T cells release mature IL-1β protein into the extracellular space, potentially driving a localized inflammatory response[16]. Such an inflammatory response likely drives a vicious pathogenic cycle, where pyroptotic CD4 T cells release bioactive IL-1β and inflammatory mediators including chemokines that attract more cells into the infected lymph nodes to die, driving even more inflammation[37].
Resident CCR5+ CD4 T cells in lymphoid tissues are primarily memory cells. Unlike naïve CD4 T cells (which do not express CCR5), these CCR5+ CD4 T cells have previously undergone TCR activation before developing into activated/memory T cells. As our results show that CD3/CD28 activation of resting CD4 cells drives proIL-1β expression, it is likely that CCR5+ cells express proIL-1β as a consequence of prior, in vivo TCR activation. Memory CD4 T cells continually recirculates within lymphoid tissues, scanning for presentation of their cognate antigen[38–40]. CCR5 expression has been previously characterized as a marker of Th1 lymphocytes[41]. CCR5+ CD4 T cells could contribute substantially to chronic inflammation through activation of caspase-1 and release of bioactive IL-1β.
Our previous work explored epigenetic regulation of IL1B in myeloid cells[9]. Genes in embryonic and hematopoietic stem cells[42–44], as well as differentiated T cells[44], have been previously reported to be marked by active and repressive histone modifications, with the ratio of these marks modulating gene transcription. The new evidence presented in this study suggests that the IL1B gene in CD4 T cells following TCR-activation is modified to a bivalent H3K4me3+/H3K27me3+ status (Fig. 4) that likely supports of a low level of transcriptional activity[24]. By contrast, the IL1B gene in unstimulated T cells is marked solely by high levels of monovalent H3K27me3 (i.e., no detectable promoter-localized H3K4me3). This supports the observed lack of IL1B transcription in naïve/unstimulated T cells. Additionally, a significantly higher level of H3K9ac is observed at the IL1B promoter in activated CD4 T cells, supporting active transcription. Finally, progressively increasing H3K36me3 through the IL1B gene body is consistent with ongoing, de novo Pol II transcription [9]. These marks in activated CD4 T cells differ from myeloid cells: unstimulated THP-1 cells possess monovalent H3K4me3, the active mark, which increases along with H3K9ac when stimulated with LPS[9].
Validation of our results is found in the epigenetic analysis of the short, intronless HIST1H4K gene from the same chromatin samples. This “housekeeping” gene is actively transcribed, displaying high-level monovalent H3K4me3, with minimal enrichment of H3K27me3 in both resting and activated T cells (Fig. 4). Also, as expected for a constitutively active gene, strong enrichment for H3K9ac was observed at the HIST1H4K promoter, and H3K36me3 increased towards the 3’ end of the gene body. Notably, the histone marks associated with transcriptional activity were all significantly higher on the constitutively expressed HIST1H4K gene than the IL1B gene from the same activated T cell chromatin samples. The repressive H3K27me3 histone mark at the IL1B promoter is much higher than that on the HIST1H4K gene, supporting the model that IL1B in activated T cells is transcribed from a low-activity bivalent H3K4me3+/H3K27me3+ promoter. The Spi1-independent nature of T cell IL1B expression may explain this observation, whereas Spi1 activity in monocytes supports monovalent H3K4me3+ and high transcription.
Furthermore, the epigenetic study of CCR5+ T cells also reveals a bivalent profile at (H3K4me3+/H3K27me3+) of the IL1B promoter. Similar to unstimulated CD4 T cells, the IL1B gene in CCR5- cells contains only high levels of monovalent H3K27me3, the transcriptional repressive mark (Supplementary Fig. 4G and H). Also, the H3K36me3 comparison did not reveal a progressive increase toward the 3’ end of the IL1B, as was observed in ex vivo activated T cells. H3K36me3 did present a classic profile for the HIST1H4K gene in both CCR5+ and CCR5- cell types. It is striking that the relative level of modification for this mark throughout IL1B in both CCR5+ and CCR5- T cells is comparable and appears elevated throughout the gene with respect to that of HIST1H4K. Interestingly, literature suggests that H3K36me3 modification patterns are not exclusively associated with gene activation, and can be related to other processes[45]. Consequently, although the pattern of this modification did not provide information on the transcriptional status of IL1B in either CCR5+ or CCR5- T cells, it is not evidence against active IL1B transcription in CD4 T cells. Generally, the epigenetic studies of CCR5+ vs. CCR5- CD4 T cells were limited by the low amounts of primary T cell chromatin samples with six donor samples being pooled in order to generate sufficient material for analysis.
In conclusion, we have extended the current understanding of IL1B gene transcription in human CD4 T lymphocytes, by investigating the involvement of Spi1 and the nature of chromatin organization for the IL1B gene in these newly recognized cellular sources of IL-1β protein expression. The proposed model argues that the IL1B gene in TCR-activated CD4 T cells is transcribed from a low-activity bivalent (H3K4me3+/H3K27me3+) promoter in a Spi1-independent fashion that results in the accumulation of intracellular proIL-1β (Fig. 5). By contrast, myeloid cells transcribe the IL1B gene constitutively at a low level in a Spi1-dependent manner but have a post-transcriptional block on translation. TLR activation of monocytes drives extreme transcription of Spi1-dependent IL1B, while releasing translation inhibition. These findings are highly relevant to further our understanding of adaptive immunity, since CD4 lymphocytes, as a source of IL-1β, can shape immune response polarization and inflammation in disease.
Figure 5. Monocytes and T cells have distinct mechanisms for IL1B gene regulation.
LPS-treated monocytes have high levels of active monovalent H3K4me3 ( ) but lack inhibitory H3K27me3 (
) but lack inhibitory H3K27me3 ( ) epigenetic histone modifications at the IL1B promoter. These modifications, along with activated NF-κB and C/EBPβ [9], drive a vigorous Pol II engagement with extremely high transcriptional activity (
) epigenetic histone modifications at the IL1B promoter. These modifications, along with activated NF-κB and C/EBPβ [9], drive a vigorous Pol II engagement with extremely high transcriptional activity ( ) and correspondingly high levels of cytoplasmic proIL-1β protein (
) and correspondingly high levels of cytoplasmic proIL-1β protein ( ). Unstimulated monocytes contain lower levels of monovalent H3K4me3 at the IL1B promoter and are deficient in inhibitory marks, resulting in weak Pol II engagement and extremely low transcription levels compared to stimulated monocytes. Accumulation of proIL-1β in these cells is inhibited due to a translational blockade (
). Unstimulated monocytes contain lower levels of monovalent H3K4me3 at the IL1B promoter and are deficient in inhibitory marks, resulting in weak Pol II engagement and extremely low transcription levels compared to stimulated monocytes. Accumulation of proIL-1β in these cells is inhibited due to a translational blockade ( ). The myeloid competence factor, Spi1 (
). The myeloid competence factor, Spi1 ( ), is constitutively bound at a constant level to the IL1B promoter in these cells [9]. In contrast to the immediate-early LPS responsive IL1B gene in monocytes, slowly-activated developmentally-regulated genes possess bivalent promoters with both active H3K4me3 and inhibitory H3K27me3 epigenetic histone modifications. The IL1B promoter in lymphoid CCR5+ CD4 T cells contains such a bivalent H3K4me3+/H3K27me3+ mark, resulting in decreased Pol II engagement and significantly lower (~25‑ times) transcription activity, as compared to resting monocytes. Interestingly, despite such a low level of transcription, intracellular proIL-1β accumulates in CCR5+ CD4 T cells in amounts higher than unstimulated monocytes. CCR5- CD4 T cells exclusively contain the inhibitory H3K27me3, which does not support active IL1B transcription until after the engagement of the T cell receptor (TCR).
), is constitutively bound at a constant level to the IL1B promoter in these cells [9]. In contrast to the immediate-early LPS responsive IL1B gene in monocytes, slowly-activated developmentally-regulated genes possess bivalent promoters with both active H3K4me3 and inhibitory H3K27me3 epigenetic histone modifications. The IL1B promoter in lymphoid CCR5+ CD4 T cells contains such a bivalent H3K4me3+/H3K27me3+ mark, resulting in decreased Pol II engagement and significantly lower (~25‑ times) transcription activity, as compared to resting monocytes. Interestingly, despite such a low level of transcription, intracellular proIL-1β accumulates in CCR5+ CD4 T cells in amounts higher than unstimulated monocytes. CCR5- CD4 T cells exclusively contain the inhibitory H3K27me3, which does not support active IL1B transcription until after the engagement of the T cell receptor (TCR).
Supplementary Material
Highlights.
TCR activation induces IL1B gene transcription in CD4 T cells
Unlike monocytes, IL1B in T cells is independent of critical myeloid Spi1 protein
Spi1 binding site on IL1B promoter is blocked by a nucleosome in CD4 T cells
Distinctly, IL1B gene in CD4 T cells utilizes a bivalent H3K4me3/H3k27me3 promoter
Acknowledgments
Financial Support
This work was supported by National Institutes of Health grants R21 AI102782, 1DP 1036502, R01 DA044605, and U19 AI0961133 (to W.C.G); NIH P30 AI027763 (to T.A.P.); NIH S10 RR028962 (to Gladstone Institutes); and R01 AR057310 (to D.L.G.). We also gratefully acknowledge The James B. Pendleton Charitable Trust (to Gladstone Institutes), The Charles Henry Leach II Fund and Duquesne University, Biological Sciences HAW Award (to P.E.A).
Footnotes
Competing Interests: The authors declare that they have no competing interests.
Additional Information
Supplementary information accompanies this paper.
Data Availability: All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Eder C, Mechanisms of interleukin-1beta release, Immunobiology 214(7) (2009) 543–53. [DOI] [PubMed] [Google Scholar]
- [2].Petrilli V, Dostert C, Muruve DA, Tschopp J, The inflammasome: a danger sensing complex triggering innate immunity, Curr Opin Immunol 19(6) (2007) 615–22. [DOI] [PubMed] [Google Scholar]
- [3].Dubyak GR, P2X7 receptor regulation of non-classical secretion from immune effector cells, Cell Microbiol 14(11) (2012) 1697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Malhotra V, Unconventional protein secretion: an evolving mechanism, EMBO J 32(12) (2013) 1660–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dinarello CA, Interleukin-1 in the pathogenesis and treatment of inflammatory diseases, Blood 117(14) (2011) 3720–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lawrence T, Natoli G, Transcriptional regulation of macrophage polarization: enabling diversity with identity, Nature reviews. Immunology 11(11) (2011) 750–61. [DOI] [PubMed] [Google Scholar]
- [7].Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, Ragoussis J, Natoli G, Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages, Immunity 32(3) (2010) 317–28. [DOI] [PubMed] [Google Scholar]
- [8].Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK, Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities, Mol Cell 38(4) (2010) 576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Adamik J, Wang KZ, Unlu S, Su AJ, Tannahill GM, Galson DL, O’Neill LA, Auron PE, Distinct mechanisms for induction and tolerance regulate the immediate early genes encoding interleukin 1beta and tumor necrosis factor alpha, PloS one 8(8) (2013) e70622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kominato Y, Galson D, Waterman WR, Webb AC, Auron PE, Monocyte expression of the human prointerleukin 1 beta gene (IL1B) is dependent on promoter sequences which bind the hematopoietic transcription factor Spi-1/PU.1, Mol Cell Biol 15(1) (1995) 58–68. [PMC free article] [PubMed] [Google Scholar]
- [11].Shirakawa F, Saito K, Bonagura CA, Galson DL, Fenton MJ, Webb AC, Auron PE, The human prointerleukin 1 beta gene requires DNA sequences both proximal and distal to the transcription start site for tissue-specific induction, Mol Cell Biol 13(3) (1993) 1332–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Marecki S, McCarthy KM, Nikolajczyk BS, PU.1 as a chromatin accessibility factor for immunoglobulin genes, Mol Immunol 40(10) (2004) 723–31. [DOI] [PubMed] [Google Scholar]
- [13].Natoli G, NF-kappaB and chromatin: ten years on the path from basic mechanisms to candidate drugs, Immunol Rev 246(1) (2012) 183–92. [DOI] [PubMed] [Google Scholar]
- [14].Hagemeier C, Bannister AJ, Cook A, Kouzarides T, The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB, Proc Natl Acad Sci U S A 90(4) (1993) 1580–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Libby P, Ordovas JM, Auger KR, Robbins AH, Birinyi LK, Dinarello CA, Endotoxin and tumor necrosis factor induce interleukin-1 gene expression in adult human vascular endothelial cells, Am J Pathol 124(2) (1986) 179–85. [PMC free article] [PubMed] [Google Scholar]
- [16].Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Munoz-Arias I, Greene WC, Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection, Nature 505(7484) (2014) 509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Martin BN, Wang C, Zhang CJ, Kang Z, Gulen MF, Zepp JA, Zhao J, Bian G, Do JS, Min B, Pavicic PG Jr., El-Sanadi C, Fox PL, Akitsu A, Iwakura Y, Sarkar A, Wewers MD, Kaiser WJ, Mocarski ES, Rothenberg ME, Hise AG, Dubyak GR, Ransohoff RM, Li X, T cellintrinsic ASC critically promotes T(H)17-mediated experimental autoimmune encephalomyelitis, Nat Immunol 17(5) (2016) 583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Arbore G, West EE, Spolski R, Robertson AA, Klos A, Rheinheimer C, Dutow P, Woodruff TM, Yu ZX, O’Neill LA, Coll RC, Sher A, Leonard WJ, Kohl J, Monk P, Cooper MA, Arno M, Afzali B, Lachmann HJ, Cope AP, Mayer-Barber KD, Kemper C, T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4(+) T cells, Science (New York, N.Y.) 352(6292) (2016) aad1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Galloway NL, Doitsh G, Monroe KM, Yang Z, Munoz-Arias I, Levy DN, Greene WC, Cellto-Cell Transmission of HIV-1 Is Required to Trigger Pyroptotic Death of Lymphoid-Tissue-Derived CD4 T Cells, Cell reports 12(10) (2015) 1555–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kaspar RL, Gehrke L, Peripheral blood mononuclear cells stimulated with C5a or lipopolysaccharide to synthesize equivalent levels of IL-1 mRNA show unequal IL-1beta protein accumulation but similar polyribosome profiles, J Immunol 153(1) (1994) 277–86. [PubMed] [Google Scholar]
- [21].Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K, Combinatorial patterns of histone acetylations and methylations in the human genome, Nature genetics 40(7) (2008) 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K, A model for transmission of the H3K27me3 epigenetic mark, Nature cell biology 10(11) (2008) 1291–300. [DOI] [PubMed] [Google Scholar]
- [23].Henikoff S, Shilatifard A, Histone modification: cause or cog?, Trends in genetics : TIG 27(10) (2011) 389–96. [DOI] [PubMed] [Google Scholar]
- [24].Voigt P, Tee WW, Reinberg D, A double take on bivalent promoters, Genes & development 27(12) (2013) 1318–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rothenberg EV, Champhekar A, Damle S, Del Real MM, Kueh HY, Li L, Yui MA, Transcriptional establishment of cell-type identity: dynamics and causal mechanisms of T-cell lineage commitment, Cold Spring Harbor symposia on quantitative biology 78 (2013) 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chang H-A, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, Klemsz MJ, Kaplan MH, PU.1 expression delineates heterogeneity in primary Th2 cells, Immunity 22 (2005) 693–703. [DOI] [PubMed] [Google Scholar]
- [27].Chang H-C, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi A-N, Han L, Nguyen ET, Robertsen MJ, Perumal N, Tepper RS, Nutt SI, Kaplan MH, The transcription factor PU.1 is required for the development of IL-9-producing T Cells and allergic inflammation, Nature Immunol 11(6) (2010) 527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS, The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation, Mol Cell 62(1) (2016) 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chanput W, Mes J, Vreeburg RA, Savelkoul HF, Wichers HJ, Transcription profiles of LPSstimulated THP-1 monocytes and macrophages: a tool to study inflammation modulating effects of foodderived compounds, Food & function 1(3) (2010) 254–61. [DOI] [PubMed] [Google Scholar]
- [30].Roy AM, Schweighardt B, Eckstein LA, Goldsmith MA, McCune JM, Enhanced replication of R5 HIV-1 over X4 HIV-1 in CD4(+)CCR5(+)CXCR4(+) T cells, J Acquir Immune Defic Syndr. 40(3) (2005) 267–75. [DOI] [PubMed] [Google Scholar]
- [31].Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR, The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes, Proc Natl Acad Sci U S A. 94(5) (1997) 1925–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Eckstein DA, Penn ML, Korin YD, Scripture-Adams DD, Zack JA, Kreisberg JF, Roederer M, Sherman MP, Chin PS, Goldsmith MA, HIV-1 actively replicates in naive CD4(+) T cells residing within human lymphoid tissues, Immunity 15(4) (2001) 671–82. [DOI] [PubMed] [Google Scholar]
- [33].Jekle A, Keppler OT, De Clercq E, Schols D, Weinstein M, Goldsmith MA, In vivo evolution of human immunodeficiency virus type 1 toward increased pathogenicity through CXCR4-mediated killing of uninfected CD4 T cells, J Virol 77(10) (2003) 5846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schweighardt B, Roy AM, Meiklejohn DA, Grace EJ 2nd, Moretto WJ, Heymann JJ, Nixon DF, R5 human immunodeficiency virus type 1 (HIV-1) replicates more efficiently in primary CD4+ Tcell cultures than X4 HIV-1, J Virol 78(17) (2004) 9164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Glushakova S, Baibakov B, Margolis LB, Zimmerberg J, Infection of human tonsil histocultures: a model for HIV pathogenesis, Nat Med. 1(12) (1995) 1320–2. [DOI] [PubMed] [Google Scholar]
- [36].Doitsh G, Cavrois M, Lassen KG, Zepeda O, Yang Z, Santiago ML, Hebbeler AM, Greene WC, Abortive HIV Infection Mediates CD4 T Cell Depletion and Inflammation in Human Lymphoid Tissue, Cell. 143(5) (2010) 789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Doitsh G, Greene WC, Dissecting How CD4 T Cells Are Lost During HIV Infection, Cell host & microbe 19(3) (2016) 280–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lanzavecchia A, Sallusto F, Dynamics of T lymphocyte responses: intermediates, effectors, and memory cells, Science (New York, N.Y.) 290(5489) (2000) 92–7. [DOI] [PubMed] [Google Scholar]
- [39].Mackay CR, Immunological memory, Advances in immunology 53 (1993) 217–65. [DOI] [PubMed] [Google Scholar]
- [40].Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A, Two subsets of memory T lymphocytes with distinct homing potentials and effector functions, Nature 401(6754) (1999) 708–12. [DOI] [PubMed] [Google Scholar]
- [41].Loetscher P, Uguccioni M, Bordoli L, Baggiolini M, Moser B, Chizzolini C, Dayer JM, CCR5 is characteristic of Th1 lymphocytes, Nature 391(6665) (1998) 344–5. [DOI] [PubMed] [Google Scholar]
- [42].Vaquerizas JM, Torres-Padilla ME, Developmental biology: Panoramic views of the early epigenome, Nature 537(7621) (2016) 494–496. [DOI] [PubMed] [Google Scholar]
- [43].Shen X, Orkin SH, Glimpses of the epigenetic landscape, Cell stem cell 4(1) (2009) 1–2. [DOI] [PubMed] [Google Scholar]
- [44].Roh TY, Cuddapah S, Cui K, Zhao K, The genomic landscape of histone modifications in human T cells, Proc Natl Acad Sci U S A 103(43) (2006) 15782–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wagner EJ, Carpenter PB, Understanding the language of Lys36 methylation at histone H3, Nature reviews. Molecular cell biology 13(2) (2012) 115–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.