Abstract
Polycomb group (PcG) proteins repress master regulators of development and differentiation through organization of chromatin structure. Mutation and dysregulation of PcG genes cause developmental defects and cancer. PcG proteins form condensates in the cell nucleus, and these condensates are the physical sites of PcG-targeted gene silencing via formation of facultative heterochromatin. However, the physiochemical principles underlying the formation of PcG condensates remain unknown, and their determination could shed light on how these condensates compact chromatin. Using fluorescence live-cell imaging, we observed that the Polycomb repressive complex 1 (PRC1) protein chromobox 2 (CBX2), a member of the CBX protein family, undergoes phase separation to form condensates and that the CBX2 condensates exhibit liquid-like properties. Using site-directed mutagenesis, we demonstrated that the conserved residues of CBX2 within the intrinsically disordered region (IDR), which is the region for compaction of chromatin in vitro, promote the condensate formation both in vitro and in vivo. We showed that the CBX2 condensates concentrate DNA and nucleosomes. Using genetic engineering, we report that trimethylation of Lys-27 at histone H3 (H3K27me3), a marker of heterochromatin formation produced by PRC2, had minimal effects on the CBX2 condensate formation. We further demonstrated that the CBX2 condensate formation does not require CBX2–PRC1 subunits; however, the condensate formation of CBX2–PRC1 subunits depends on CBX2, suggesting a mechanism underlying the assembly of CBX2–PRC1 condensates. In summary, our results reveal that PcG condensates assemble through liquid–liquid phase separation (LLPS) and suggest that phase-separated condensates can organize PcG-bound chromatin.
Keywords: Polycomb, epigenetics, heterochromatin, chromatin, gene regulation, histone methylation, histone, chromatin modification, chromatin structure, chromatin regulation, CBX2, liquid-liquid phase separation, PcG, phase separation, PRC1
Introduction
The genome in eukaryotic cells can be broadly classified as euchromatin (active transcription) and heterochromatin (repression and silencing) (1, 2). Heterochromatin can be further described broadly as constitutive and facultative heterochromatin (3–5). Constitutive heterochromatin is observed at and near centromeres and telomeres (5). Facultative heterochromatin is found at a specific subset of genes encoding regulators of development and differentiation (3, 4). Another example of facultative heterochromatin is X chromosome inactivation in female mammals (6). Facultative heterochromatin represses gene expression in part through compacting chromatin to reduce the accessibility of DNA (3–5). Facultative heterochromatin is decorated by the trimethylation of Lys-27 at histone H3 (H3K27me3),3 which is the catalytic product of Polycomb repressive complex (PRC) 2, one complex of Polycomb group (PcG) proteins (3, 4). H3K27me3 is the binding site for PRC1 (another complex of PcG proteins) that can compact chromatin, forming particular chromatin compartments (3, 4). The repressed, compacted chromatin domains are preserved during cell division (3, 4). These functions and behaviors of facultative heterochromatin have raised several fundamental questions. For example, what are the properties of facultative heterochromatin? How is chromatin compaction achieved? How do PcG proteins contribute to establishing facultative heterochromatin compartments? How do PcG proteins maintain facultative heterochromatin compartments? These questions are essential for understanding how facultative heterochromatin functions and preserves cell identity.
Biochemical and genetic studies of PcG proteins have begun addressing some of the questions raised above. For example, in terms of understanding the properties of facultative heterochromatin, it has been shown that PcG proteins directly regulate chromatin structure and chemically modify the histones of facultative heterochromatin (7). PRC2 methylates histone H3 on lysine 27 by the catalytic subunit enhancer of zeste homolog 2 (8–12). Embryonic ectoderm development (EED), one core subunit of PRC2, binds H3K27me3, which allosterically stimulates PRC2 activity, and this stimulation can increase the local H3K27me3 level (13, 14). H3K27me3 provides a binding site for chromobox 7 (CBX7) and CBX8 of the CBX family proteins (CBX2/4/6/7/8) (15), which recruit canonical CBX7–PRC1 and CBX8–PRC1 to chromatin. PRC1 complexes can ubiquitinate histone H2A at lysine 119 by the catalytic subunit ring finger protein 2 (RING1B) (16, 17), which can stimulate PRC2 activity (18, 19). Thus, a feedback loop between PRC1 and PRC2 is created to reinforce the epigenetic modifications of facultative heterochromatin. In terms of understanding compaction, it has been demonstrated that the compaction function of PRC1 in mammals is facilitated by CBX2 of the CBX protein family (20, 21). In Drosophila, the protein posterior sex combs exerts chromatin compaction (22, 23). Mutation of the CBX2 residues that are required for the compaction leads to homeotic transformations that are similar to those observed with PcG loss-of-function mutations (21). In terms of understanding facultative heterochromatin compartments, studies have demonstrated that, in the cell nucleus, PcG proteins form microscopically visible condensates (24–28). PcG condensates function as specific nuclear compartments for target gene silencing (24–28). Overall, these biochemical and genetic studies suggest that PRC1 and PRC2 coordinate to establish and maintain facultative heterochromatin. Despite these exciting advances, the fundamental physicochemical principles that underpin how PcG proteins establish, maintain, and regulate facultative heterochromatin remain incompletely understood. Understanding these questions is critical for appreciating how PcG proteins control development and differentiation.
Spatial and temporal compartmentalization of intracellular components into organelles in eukaryotic cells is a generic theme for organizing biochemical reactions (29–34). These organelles can be membrane-bound or membraneless. A large number of membraneless compartments, including the nucleolus, stress granules, Cajal bodies, promyelocytic leukemia nuclear bodies, and others, are condensates formed by condensation of cellular components through liquid–liquid phase separation (LLPS) (29–33). The forces that drive LLPS are multivalent interactions among proteins and other macromolecular polymers such as RNA and DNA (29–31). Phase-separated condensates have been shown to be involved in multiple cellular processes and functions (29–31). Over the past year, phase-separated membraneless condensates have been suggested to be implicated in transcriptional activation and repression (35–41). A phase-separated model has been emerging to explain transcriptional activation: transcription factors and coactivators phase separate to form condensates that interact with condensates of RNA polymerase II to efficiently activate transcription (35–39). Phase-separated condensates also function in transcriptional repression. Heterochromatin protein 1α phase separates to form condensates that compartmentalize constitutive heterochromatin (40, 41). Facultative heterochromatin represents one major class of chromatin structures. Whether the PcG proteins that are responsible for the formation of facultative heterochromatin phase separate to form condensates remains unknown.
Here, we provide the first experimental evidence that the PRC1 protein CBX2 phase separates to form condensates that can concentrate DNA and nucleosomes. The conserved residues within the CBX2 intrinsically disordered region (IDR) are required for the CBX2 condensate formation both in vitro and in vivo. We show that H3K27me3 contributes little to the CBX2 condensate formation. We demonstrate that CBX2 determines the condensate formation of CBX2–PRC1; however, the CBX2–PRC1 subunits are not required for the formation of CBX2 condensates. Thus, we provide a general experimental framework that can explain how PcG condensates assemble and organize PcG-bound chromatin. Our results provide a starting point for further exploring how phase separation facilitates efficient and specific control of transcription.
Results
CBX2 forms condensates in cells
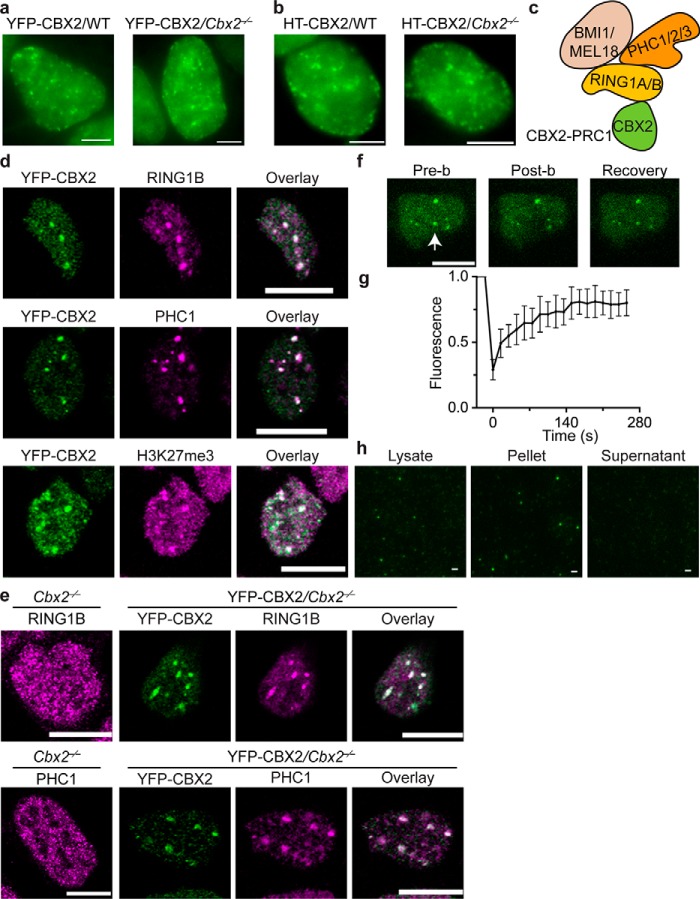
We first investigated whether CBX2 forms condensates in living cells. We integrated YFP-Cbx2 and HaloTag (HT)-Cbx2 into the genome of PGK12.1 mouse embryonic stem (wildtype (WT) mES) cells, respectively. The expression of these fusion genes is under the control of an inducible, tetracycline response element–tight promoter. To observe the cellular distribution of HT-CBX2, we labeled the fusion protein by HaloTag® TMR ligand in living cells. Both YFP-CBX2 and HT-CBX2 formed condensates in living WT mES cells (Fig. 1, a and b), which is consistent with the previous observations reporting that both exogenous and endogenous CBX2 forms condensates (42–44). About half of cells contained microscopically visible CBX2 condensates. The average area of condensates was 0.19 μm2 (∼250 nm in radius), and their fluorescence intensity was ∼1.5-fold higher than the average intensity. It is possible that the formation of CBX2 fusion condensates in mES cells is due to their overexpression. To resolve this possibility, we integrated YFP-CBX2 and HT-CBX2 into the genome of Cbx2−/− mES cells. Under the basal expression, the protein level of YFP-CBX2 was similar to that of the endogenous protein CBX2 (45). Without doxycycline induction, the distribution of YFP-CBX2 and HT-CBX2 in Cbx2−/− mES cells was similar to that in WT mES cells (Fig. 1, a and b). These data indicate that CBX2 forms microscopically visible condensates in living cells.
Figure 1.
CBX2 phase separates to form condensates in cells. a, live-cell epifluorescence images of YFP-CBX2 in WT and Cbx2−/− mES cells. Scale bars, 5.0 μm. b, live-cell epifluorescence images of HT-CBX2 in WT and Cbx2−/− mES cells. HT-CBX2 was labeled with HaloTag TMR ligand. Scale bars, 5.0 μm. c, schematic representation of the core subunits of the CBX2–PRC1 complex. d, confocal fluorescence images of immunostained YFP-CBX2 and H3K27me3 in WT mES cells as well as endogenous RING1B and PHC1. We stained the cells using antibodies against YFP (green), RING1B (magenta), PHC1 (magenta), and H3K27me3 (magenta). Overlay images are shown. Scale bars, 10 μm. e, confocal fluorescence images of immunostained RING1B and PHC1 in Cbx2−/− mES cells as well as in YFP-CBX2/Cbx2−/− mES cells. We stained the cells using antibodies against YFP (green), RING1B (magenta), and PHC1 (magenta). Overlay images are shown. Scale bars, 10 μm. f, representative FRAP images of YFP-CBX2 stably expressed in WT mES cells. The images were taken before (Pre-b) and after (Post-b) photobleaching. The condensate that was bleached is indicated by a white arrowhead. Scale bar, 10 μm. g, FRAP curve of YFP-CBX2 in WT mES cells that stably express YFP-CBX2. The FRAP curve was obtained from averaging data from 10 cells. Error bars represent S.D. h, epifluorescence imaging of YFP-CBX2 condensates isolated from cells. We cross-linked cells stably expressing YFP-CBX2 with formaldehyde. Lysate was prepared. Both lysate and resuspended pellet contained YFP-CBX2 condensates; however, the supernatant did not have condensates. Scale bars, 2.0 μm.
CBX2 forms a stable PRC1 complex (CBX2–PRC1), including polyhomeotic homolog 1 (PHC1) and RING1B (Fig. 1c) (7, 46), so we investigated whether YFP-CBX2 condensates colocalize with CBX2–PRC1 subunits. We stained endogenous RING1B and PHC1 as well as YFP-CBX2. RING1B and PHC1 formed condensates in cells (Fig. 1d), consistent with the previous reports (25, 43, 47, 48). YFP-CBX2 condensates colocalized with condensates of RING1B and PHC1 (Fig. 1d). Because H3K27me3 marks PcG-targeted genes, we investigated whether CBX2 condensates colocalize with H3K27me3. Immunofluorescence of H3K27me3 and YFP-CBX2 showed that CBX2 condensates colocalize with chromatin with the dense H3K27me3 mark (Fig. 1d), suggesting that PcG-targeted genes are recruited to CBX2 condensates or vice versa. Thus, our results show that CBX2 condensates colocalize with CBX2–PRC1 subunits and H3K27me3-marked chromatin regions.
To determine whether the condensate formation of RING1B and PHC1 depends on CBX2, we first stained RING1B and PHC1 in Cbx2−/− mES cells. In contrast to WT mES cells, RING1B and PHC1 exhibited a granular distribution in Cbx2−/− mES cells, and their large condensates disappeared (Fig. 1e). We then stained RING1B and PHC1 in YFP-Cbx2/Cbx2−/− mES cells. RING1B and PHC1 formed condensates that colocalize with YFP-CBX2 condensates (Fig. 1e). Our results indicate that the formation of RING1B and PHC1 condensates depends on CBX2.
Next, we interrogated whether CBX2 condensates exhibit liquid-like features that are characterized with rapid exchange kinetics, which can be studied by measuring the recovery rate using fluorescence recovery after photobleaching (FRAP). We performed FRAP experiments on condensates of YFP-CBX2 stably expressed in mES cells. FRAP showed that 80% of YFP-CBX2 within condensates is recovered within 3 min (Fig. 1, f and g), consistent with our previous reports (42, 49). These results indicate that CBX2 within condensates dynamically exchanges with surrounding environments and has liquid-like properties in cells.
If CBX2 condensates were liquid-like, reducing the concentration of YFP-CBX2 would dissolve these condensates. We lysed cells stably expressing YFP-CBX2 in lysis buffer to cause a local decrease of YFP-CBX2 through diffusion. We did not detect YFP-CBX2 condensates in the lysate. We expected that formaldehyde cross-linking would preserve CBX2 condensates. After sonication and centrifugation, YFP-CBX2 condensates would be within the pellets. To test this speculation, prior to lysis, we cross-linked cells with formaldehyde and prepared lysates from the cross-linked cells. These lysates were subjected to sonication. Using fluorescence microscopy, we observed CBX2 condensates within the sonicated lysate (Fig. 1h). After centrifugation, we did not observe CBX2 condensates in the supernatant but instead observed them in the resuspended pellets (Fig. 1h). These data further support that CBX2 condensates form in the cells and possess liquid-like properties.
CBX2 phase separates to form condensates in vitro
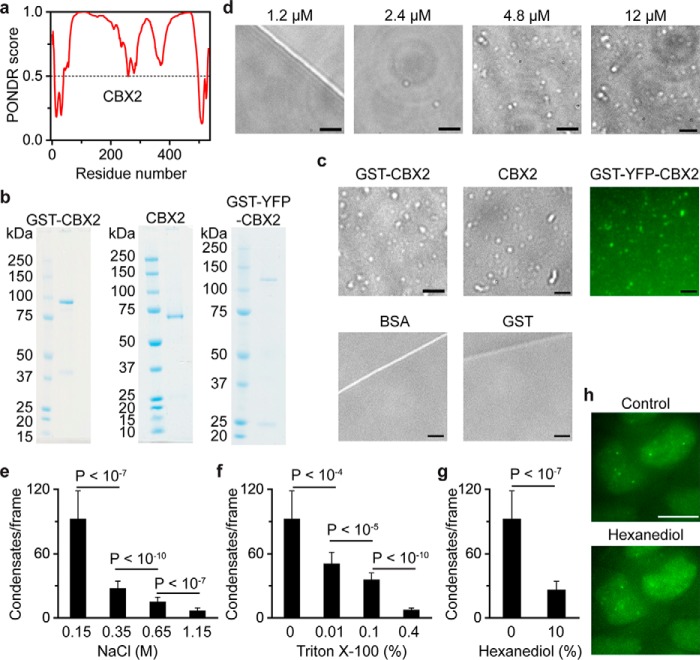
Proteins can undergo LLPS through IDRs, leading to formation of condensate in aqueous solution (29–31). We analyzed the properties of the primary sequence of CBX2 and found that 59% of the CBX2 sequence is intrinsically disordered as predicted by MobiDB 3 (Fig. 2a) (50). CBX2 is the protein that condenses chromatin through the highly basic residues within the IDR (20, 21). Thus, we reasoned that CBX2 could form condensates in vitro through LLPS. To test this hypothesis, we expressed and purified recombinant GST-CBX2-FLAG (GST-CBX2) from Escherichia coli at high salt concentration or in the presence of glutathione (GSH) (Fig. 2b). We found that both high salt and GSH prevent aggregation of CBX2. We placed the tags at the respective N-terminal and C-terminal ends of CBX2 to remove truncated CBX2 during the purification. We dialyzed the high salt of GST-CBX2 fusion to 140 mm NaCl at 4 °C overnight and transferred 10 μl of sample to a coverslip. After condensates settled down on the surface of the coverslip, we performed differential interference contrast (DIC) imaging and observed CBX2 condensates with a size of a few hundred nanometers (Fig. 2c). Next, we generated CBX2-FLAG (CBX2) without GST fusion (Fig. 2b). CBX2 also underwent LLPS to form condensates (Fig. 2c), suggesting that the condensate formation is not driven by GST. To determine the identity of these condensates, we produced GST-YFP-CBX2-FLAG (GST-YFP-CBX2) (Fig. 2b). Fluorescence imaging showed that GST-YFP-CBX2 assembles into condensates (Fig. 2c). Under the same conditions, GST and BSA did not form condensates (Fig. 2c). LLPS typically depends on the concentration of components in the system, so we performed the condensate formation assay with varying concentrations of CBX2, ranging from 1.2 to 12 μm. We found that the phase separation of CBX2 is concentration-dependent (Fig. 2d). Thus, our results demonstrate that CBX2 can undergo LLPS to form condensates in vitro.
Figure 2.
CBX2 phase separates to form condensates in vitro. a, CBX2 is an intrinsically disordered protein predicted by MobiDB 3 (50). A PONDR score greater than 0.5 indicates intrinsically disordered regions. 59% of CBX2 sequence is intrinsically disordered. b, SDS-PAGE analysis of recombinant CBX2 proteins. Left, recombinant GST-CBX2-FLAG (GST-CBX2). Middle, recombinant CBX2-FLAG (CBX2). Right, recombinant GST-YFP-CBX2-FLAG (GST-YFP-CBX2). The molecular mass ladder is shown at the left of the gel image. c, representative DIC images of GST-CBX2 (4.8 μm) and CBX2 (4.8 μm) condensates as well as the control BSA (10 μm) and GST (10 μm) on the surface of a coverslip. A representative epifluorescence image of GST-YFP-CBX2 (4.8 μm) condensates is shown. Scale bars, 2.0 μm. d, dependence of the formation of CBX2 condensates on its concentrations shown above the images. Representative DIC images of condensates on the surface of coverslip are shown. Scale bars, 2.0 μm. e–g, increasing concentrations of NaCl, Triton X-100, and hexanediol dissolve CBX2 condensates. We incubated CBX2 (4.8 μm) condensates with the indicated concentrations of NaCl, Triton X-100, and hexanediol for 30 min on ice. The mixture was loaded to a coverslip for imaging. We counted condensates using ImageJ. The data were from at least 10 frames for each sample. Error bars represent S.D. p values were calculated based on Student's t test. h, representative epifluorescence images of WT mES cells stably expressing YFP-CBX2 before and after treatment with 10% hexanediol for 5.0 min. Scale bar, 10 μm.
Classical polymer theory predicts that polymers undergo LLPS through multivalency-driven interactions such as cation–π, electrostatic, dipolar, and hydrophobic interactions (29–31). Thus, we investigated whether NaCl and Triton X-100 dissolve CBX2 condensates. We found that treatment of CBX2 condensates with increasing concentrations of NaCl and Triton X-100 causes a reduction in the number of CBX2 condensates (Fig. 2, e and f). Hexanediol is known to dissolve liquid-like condensates (35, 37), possibly through disruption of hydrophobic interactions. We found that treatment of CBX2 condensates with hexanediol results in a reduction in the number of condensates in vitro (Fig. 2g). Treatment of mES cells expressing YFP-CBX2 with hexanediol caused mild effects on the formation of CBX2 condensates (Fig. 2h), suggesting that other noncovalent interactions are also involved in the phase separation of CBX2. These results indicate that multivalent interactions contribute to the LLPS of CBX2.
CBX2 condensates concentrate DNA and nucleosomes
One of the characteristic properties of cellular compartmentalization is that compartments can increase the local concentration of resident biochemical molecules (29–34). PcG condensates are the physical sites of PcG-mediated silencing, involving organization of the PcG-bound chromatin (24–26). Given that CBX2 can directly bind DNA (51), we investigated whether CBX2 condensates can concentrate DNA. We labeled 24-bp dsDNA with fluorescent dye and mixed them with CBX2. Dye-labeled DNA did not form condensates; however, in the presence of CBX2, DNA was concentrated, and CBX2 condensates colocalized with the concentrated DNA (Fig. 3a). Previous studies have shown that CBX2 can compact chromatin on its own (20), so we tested whether CBX2 can concentrate core nucleosome particles. We prepared dye-labeled nucleosomes and mixed them with CBX2. After dialysis, we observed that CBX2 condensates colocalize with the concentrated dye-labeled nucleosomes (Fig. 3a). Under the same conditions, CBX2 condensates could not enrich the dye (Fig. 3a). Thus, these results suggest that CBX2 condensates can concentrate DNA and nucleosomes in vitro.
Figure 3.
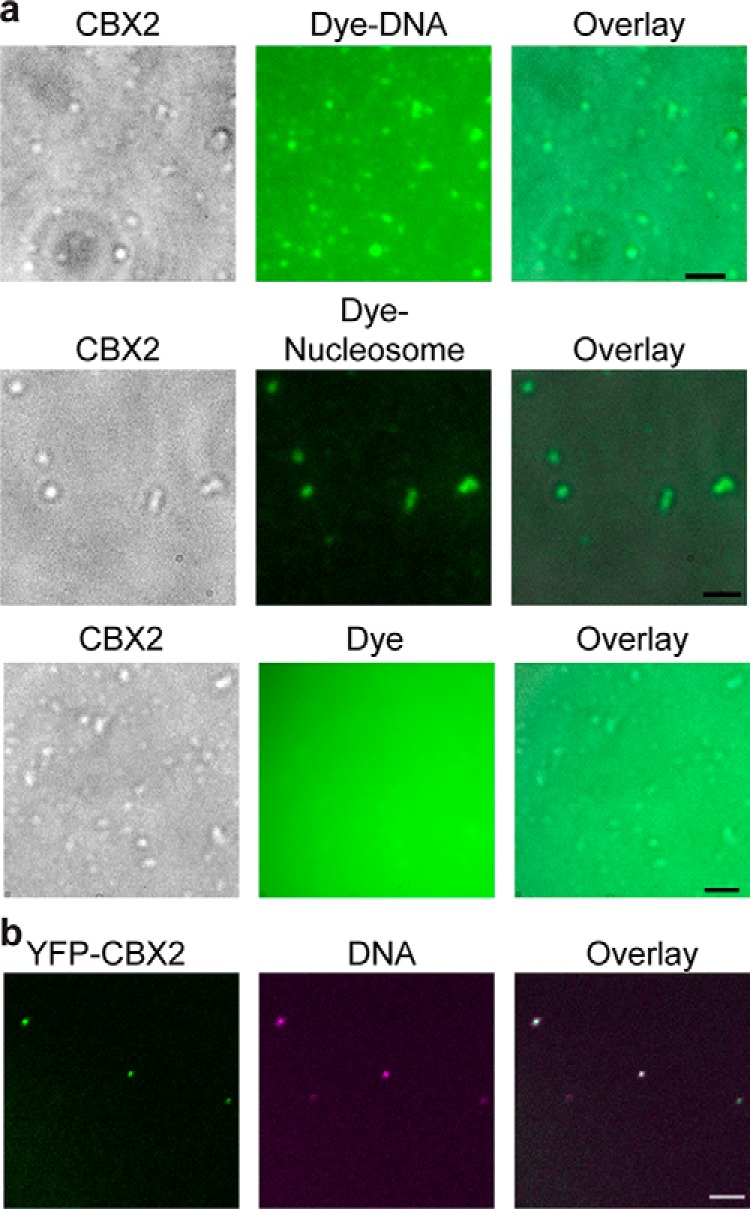
CBX2 condensates concentrate DNA and nucleosomes. a, micrographs of phase-separated CBX2 (4.8 μm) condensates with Alexa Fluor 488 (1.0 μm) or Alexa Fluor 488–labeled DNA (0.5 μm) as well as CBX2 (2.4 μm) condensates with Cy5-labeled nucleosomes (40 nm). DIC images of CBX2 condensates on the surface of coverslips are shown. Alexa Fluor 488, Alexa Fluor 488–labeled DNA, and Cy5-labeled nucleosomes are shown in fluorescence images. Overlay images are also shown. Scale bars, 2 μm. b, epifluorescence imaging of DNA and YFP-CBX2 condensates isolated from cells. We cross-linked mES cells stably expressing YFP-CBX2 with formaldehyde. Lysate was prepared. Resuspended pellets were stained with Hoechst. An overlay image is shown. Scale bar, 5.0 μm.
Given that PcG condensates are the repressive compartments for PcG-targeted genes, we should be able to detect DNA within CBX2 condensates isolated from cells. We cross-linked cells stably expressing YFP-CBX2 with formaldehyde. After sonication and centrifugation, we resuspended the pellet and stained DNA with Hoechst. Fluorescence images showed that YFP-CBX2 condensates contain concentrated DNA labeled by Hoechst (Fig. 3b). Our data indicate that CBX2 condensates can enrich chromatin/DNA within cells.
Conserved residues within the IDR of CBX2 contribute to LLPS in vitro and in living cells
The driving forces of phase separation of proteins containing IDRs are noncovalent interactions, particularly cation–π and electrostatic interactions (29–34). The cation–π interactions occur between aromatic residues and Lys or Arg residues (52–57). We found that CBX2 contains a high content of Lys and Arg; however, the frequency of aromatic residues is lower than their respective average frequency in vertebrate proteins. Because proteins whose phase separations are promoted by the cation–π interactions contain a high content of aromatic residues (52), we expect that cation–π interactions may not be the major driving forces for the phase separation of CBX2. Instead, we found that, in CBX2, many positively and negatively charged residues are grouped into a series of clusters across the sequence (Fig. 4b). Alternative clusters of positive and negative charges can form an electrostatic interaction, which is one of the major driving forces that promote the phase separation of IDR-containing proteins (41, 56, 58–62). It is interesting to note that the three conserved regions, AT-hook (ATH), ATH-like 1 (ATHL1), and ATHL2 (63), are positively charged clusters (Fig. 4, a and b). We substituted residues PRG (amino acids 77–79) for AAA to generate CBX2ATH, PRG (amino acids 134–136) for AAA to generate CBX2ATHL1, and RKKRGRK (amino acids 161–167) for AAAAGAA to generate CBX2ATHL2 (Fig. 4b). The net positive charge per residue of the mutated regions of CBX2ATH and CBX2ATHL1 was slightly reduced compared with CBX2, whereas the net positive charge per residue of the mutated region of CBX2ATHL2 was completely eliminated (Fig. 4b). We generated these mutant proteins and compared their ability to form condensates with CBX2 in vitro. Our analysis indicated that the phase-separation ability of the three mutants is greatly reduced compared with CBX2 (Fig. 4, c and d). CBX2ATH and CBX2ATHL1 had a better capacity to phase separate than CBX2ATHL2 (Fig. 4, c and d), consistent with the complete loss of positive charge in the ATHL2 of CBX2ATHL2. Within the IDR of CBX2, there is a conserved serine-rich region (SRR) consisting of a stretch of consecutive 19 residues of serine and threonine (63). We substituted SKSKSSSSSSSSTSSSSSS (amino acids 102–120) for SKSKASASASASTASASAA to generate CBX2SRR (Fig. 4a). CBX2SRR greatly reduced its ability to phase separate compared with CBX2 (Fig. 4, c and d). Thus, these data indicate that these conserved residues within the IDR of CBX2 promote LLPS in vitro.
Figure 4.
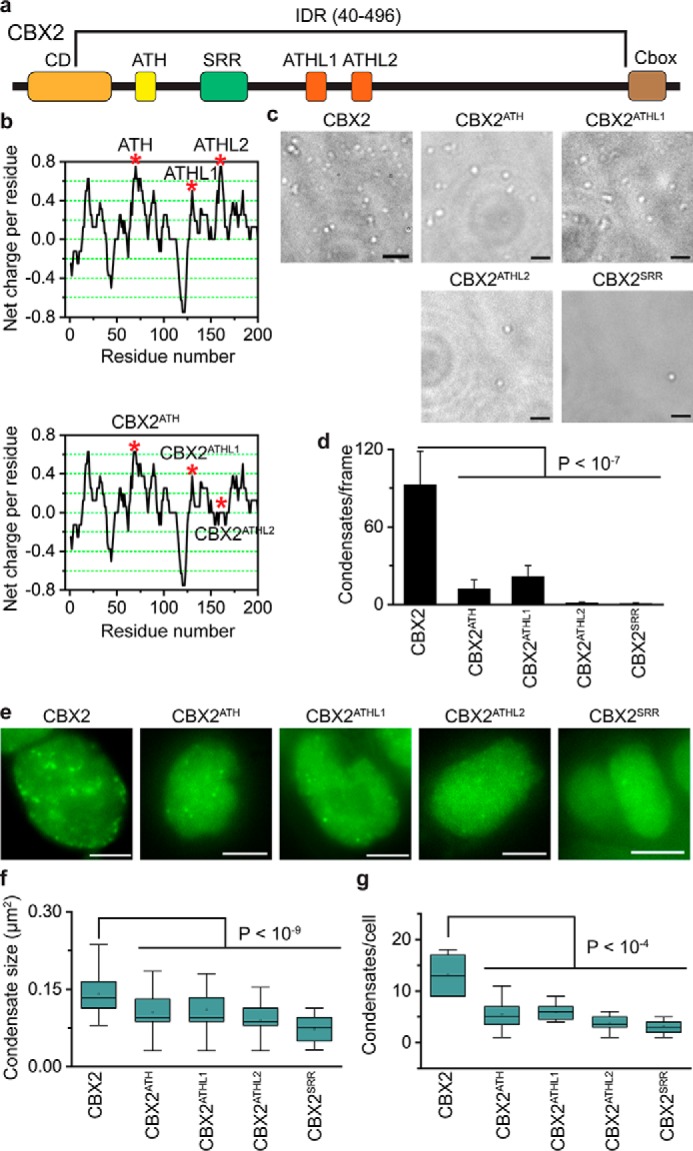
Conserved residues promote the LLPS of CBX2. a, schematic representation of CBX2. The IDR was predicted by MobiDB 3 (50). Conserved regions include CD, ATH, SRR, ATHL1, ATHL2, and chromobox (Cbox) (63). b, charge distribution of CBX2 and its variants calculated by EMBOSS charge. The net charge per residue was averaged over a sliding window of eight residues. The three covered regions, ATH, ATHL1, and ATHL2, are positively charged (top panel). The three conserved regions were mutated to generate CBX2ATH, CBX2ATHL1, and CBX2ATHL2 (bottom panel). The red asterisks indicate the three conserved regions that are mutated. c, representative DIC images of condensates of CBX2 and its variants (CBX2ATH, CBX2ATHL1, CBX2ATHL2, and CBX2SRR) on the surface of coverslips. The formation of condensates was carried out at a concentration of 4.8 μm for both CBX2 and its variants. d, quantification of condensates per frame from c. The data were from at least 10 frames for each sample. Error bars represent S.D. The p value was calculated based on Student's t test. e, representative epifluorescence images of WT mES cells stably expressing HT-CBX2 replicated from Fig. 1b and its variants (HT-CBX2ATH, HT-CBX2ATHL1, HT-CBX2ATHL2, and HT-CBX2SRR), respectively. We labeled HT-CBX2 and its variants by HaloTag TMR ligand and then performed live-cell epifluorescence imaging of cells with similar fluorescence intensity. Scale bars, 5.0 μm. f, box plot of the condensate sizes for HT-CBX2 and its variants (HT-CBX2ATH, HT-CBX2ATHL1, HT-CBX2ATHL2, and HT-CBX2SRR) from e. Data were obtained from at least 10 cells. Error bars represent upper and lower adjacent values. The p value was calculated based on Student's t test. g, box plot of the number of condensates for HT-CBX2 and its variants (HT-CBX2ATH, HT-CBX2ATHL1, HT-CBX2ATHL2, and HT-CBX2SRR) from e. Data were obtained from at least 10 cells. Error bars represent upper and lower adjacent values. The p value was calculated based on Student's t test.
To investigate whether these conserved residues of CBX2 contribute to LLPS in vivo, we established WT mES cells stably expressing HT-CBX2ATH, HT-CBX2ATL1, HT-CBX2ATL2, or HT-CBX2SRR. These CBX2 mutants were labeled with HaloTag TMR ligand. We performed live-cell imaging of these mutants (Fig. 4e). To compare their distribution, we imaged cells with similar fluorescence intensity. Quantitative analysis showed that the size and the number of condensates of these CBX2 mutants are significantly reduced compared with WT CBX2 (Fig. 4, f and g). We also noted that the size and the number of condensates of CBX2ATHL2 and CBX2SRR were slightly smaller than for CBX2ATH and CBX2ATHL1 (Fig. 4, f and g), consistent with in vitro analysis. Thus, our data demonstrate that the conserved residues within the IDR that are critical for the LLPS of CBX2 in vitro are also critical for the formation of CBX2 condensates in vivo.
H3K27me3 has minimal effects on the formation of CBX2 condensates
The PRC2-catalyzed product H3K27me3 is the marker of PcG-targeted chromatin (7). H3K27me3 has been hypothesized to be the mark for recruiting CBX–PRC1 to chromatin (Fig. 5a) (64). Thus, we asked whether H3K27me3 affects the formation of CBX2 condensates in vivo. To this end, we integrated HT-CBX2 into the genome of Eed−/− mES cells. EED is the core component of PRC2, and Eed knockout results in a complete loss of H3K27me3 (15). Live-cell imaging of HT-CBX2 labeled with HaloTag TMR ligand showed that HT-CBX2 forms condensates in Eed−/− mES cells (Fig. 5c). Quantitative analysis indicated that the size and the number of CBX2 condensates in Eed−/− mES cells are not significantly different from those in WT mES cells (Fig. 5, d and e). Previous studies have shown that the aromatic cage, consisting of three aromatic residues, of the chromodomain (CD) of CBX2 is critical for the H3K27me3 binding in vitro (65). We mutated the cage residue Phe-12 of CBX2 to Ala (CBX2F12A) (Fig. 5b). We stably expressed HT-CBX2F12A in WT mES cells. Live-cell imaging showed that HT-CBX2F12A forms condensates (Fig. 5c). The size and the number of HT-CBX2F12A were similar to HT-CBX2 (Fig. 5, d and e). These data suggest that H3K27me3 contributes little to the formation of CBX2 condensates in living cells.
Figure 5.
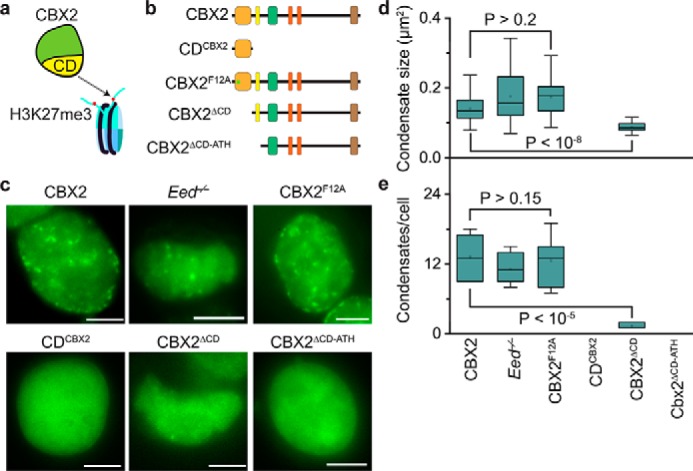
H3K27me3 has minor effects on the CBX2 condensate formation. a, a hypothetic model for targeting CBX2 to chromatin. It has been proposed that CBX2 is recruited to chromatin through the interactions between CD and H3K27me3. b, schematic representation of CBX2 and its variants used in the study. CD is the binding domain for H3K27me3 in vitro. The residue Phe-12 is the key residue involved in the H3K27me3 binding in vitro. c, representative epifluorescence images for HT-CBX2 in WT mES cells replicated from Fig. 1b; for HT-CBX2 in Eed−/− mES cells; and for HT-CBX2F12A, HT-CDCBX2, HT-CBX2ΔCD, and HT-CBX2ΔCD-ATH in WT mES cells. We labeled HT-CBX2 fusions by HaloTag TMR ligand and performed live-cell epifluorescence imaging of cells with similar fluorescence intensity. Scale bars, 5.0 μm. d, box plot of the condensate sizes for HT-CBX2 in WT mES cells replicated from Fig. 4f; for HT-CBX2 in Eed−/− mES cells; and for HT-CBX2F12A, HT-CDCBX2, HT-CBX2ΔCD, and HT-CBX2ΔCD-ATH in WT mES cells. Data were obtained from at least 10 cells. Error bars represent upper and lower adjacent values. The p value was calculated based on Student's t test. e, box plot of the number of condensates for HT-CBX2 in WT mES cells replicated from Fig. 4g; for HT-CBX2 in Eed−/− mES cells; and for HT-CBX2F12A, HT-CDCBX2, HT-CBX2ΔCD, and HT-CBX2ΔCD-ATH in WT mES cells. Data were obtained from at least 10 cells. Error bars represent upper and lower adjacent values. The p value was calculated based on Student's t test.
Because the CD of CBX2 is the binding domain for H3K27me3 in vitro (65, 66) (Fig. 5a), we investigated the effects of the CD on the formation of CBX2 condensates in living cells. We fused CD with HT, generating HT-CDCBX2 (Fig. 5b). We also deleted CD to generate HT-CBX2ΔCD (Fig. 5b). We established mES cells that stably express HT-CDCBX2 and HT-CBX2ΔCD, respectively. HT-CDCBX2 did not form condensates in living cells (Fig. 5c). HT-CBX2ΔCD phase separated to form condensates (Fig. 5c); however, their size and number were significantly reduced compared with HT-CBX2 (Fig. 5, d and e). Given that ATH is adjacent to CD and binds DNA (51) (Fig. 5b), we deleted both CD and ATH to generate HT-CBX2ΔCD-ATH (Fig. 5b), which was then stably integrated into the genome of mES cells. HT-CBX2ΔCD-ATH did not phase separate to form condensates within living cells (Fig. 5c). Thus, these results indicate that the interactions of H3K27me3 and CBX2 are not required for the formation of CBX2 condensates; however, the amino acid residues within CD are required for the condensate formation.
Depletion of CBX2–PRC1 subunits does not prevent the formation of CBX2 condensates
CBX2 phase separates on its own in vitro, so we speculated that removal of CBX2–PRC1 subunits would not prevent the formation of CBX2 condensates in vivo. To address this, we integrated HT-CBX2 into the genome of Ring1a−/−/RIing1bfl/fl; Rosa26::CreERT2 and Bmi1−/−/Mel18−/− mES cells, respectively. Ring1b in Ring1a−/−/Ring1bfl/fl; Rosa26::CreERT2 mES cells was depleted by administrating hydroxytamoxifen as described previously (15, 49). Live-cell imaging showed that depletion of Ring1a and Ring1b or Mel18 and Bmil1 does not disperse CBX2 condensates (Fig. 6a). Instead, we found that CBX2 condensates in these double-knockout mES cells are typically more numerous and larger compared with WT mES cells (Fig. 6a). Some CBX2 condensates in the double-knockout mES cells were irregular instead of the usual droplet-like shape, suggesting that these CBX2–PRC1 subunits may influence the material properties of CBX2 condensates. Quantitative analysis demonstrated that both the size and the number of CBX2 condensates in the double-knockout mES cells are significantly larger than those in WT mES cells (Fig. 6, b and c). These data indicate that CBX2–PRC1 subunits contribute less to the formation of CBX2 condensates in vivo but may regulate their material properties.
Figure 6.
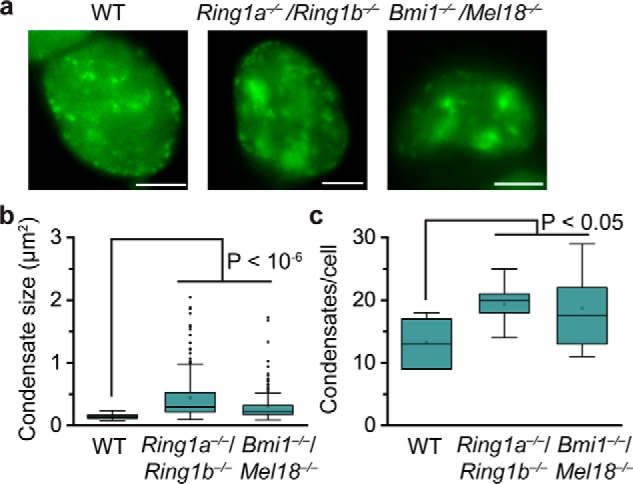
Depletion of CBX2–PRC1 subunits does not prevent the formation of CBX2 condensates. a, representative epifluorescence images for HT-CBX2 in WT mES cells replicated from Fig. 1b, for HT-CBX2 in Ring1a−/−/Ring1b−/− mES cells, and for HT-CBX2 in Bmi1−/−/Mel18−/− mES cells. We labeled HT-CBX2 by HaloTag TMR ligand and performed live-cell epifluorescence imaging of cells with similar fluorescence intensity. Scale bars, 5.0 μm. b, box plot of the condensate sizes for HT-CBX2 in WT mES cells replicated from Fig. 4f, for HT-CBX2 in Ring1a−/−/Ring1b−/− mES cells, and for HT-CBX2 in Bmi1−/−/Mel18−/− mES cells. Data were obtained from at least 10 cells. Error bars represent upper and lower adjacent values. The p value was calculated based on Student's t test. c, box plot of the number of condensates for HT-CBX2 in WT mES cells replicated from Fig. 4g, for HT-CBX2 in Ring1a−/−/Ring1b−/− mES cells, and for HT-CBX2 in Bmi1−/−/Mel18−/− mES cells. Data were obtained from at least 10 cells. Error bars represent upper and lower adjacent values. The p value was calculated based on Student's t test.
Discussion
Numerous studies have demonstrated that PcG proteins form microscopically visible condensates in primary and transformed cells, both in flies and mammals (24–28). These condensates have been shown to be the physical sites for repressing PcG-targeted genes (24–28). Consistent with these previous observations, we demonstrate that PcG protein CBX2 forms condensates in mES cells that colocalize with H3K27me3-dense chromatin regions and CBX2–PRC1 subunits. We further show that CBX2 can undergo LLPS in vitro in the absence of other proteins. It is striking that CBX2 mutants that lack the capacity of LLPS in vitro have a similar deficient ability to form condensates in vivo. The strong correlation between in vitro and in vivo data indicates that CBX2 condensates in living cells form through LLPS. This is further supported by our following observations: CBX2 condensates in living cells exhibit rapid exchange dynamics, a hallmark of liquid-like condensates, and our data show that CBX2 condensates concentrate DNA and nucleosomes. Previous studies have shown that CBX2 can compact chromatin (20, 21). Similar to heterochromatin protein 1α condensates compaction of heterochromatin (40, 41), we propose that CBX2 compacts chromatin by forming phase-separated condensates.
The phase behavior of proteins containing IDRs can be described by the theory of associative polymers (52, 67). Associative polymers phase separate through interactions between associative motifs called stickers that are separated from one another by spacers (52, 67). Spacers can impart the material properties of polymers and modulate the phase-separation ability of polymers (52, 67). The stickers can be residues that involve cation–π, electrostatic, hydrophobic, or dipolar interactions (29, 30, 52). In the case of CBX2, the stickers appear to be appositively charged clusters. Perturbation of these charged clusters reduces the phase separation of CBX2 both in vitro and in vivo, which is consistent with the notion that the phase separation of IDR-containing proteins can be promoted by interactions between blocks of appositively charged residues (41, 56, 58–62). The SRR also appears to be a sticker because substitution of the Ser residues of SRR with Ala prevents the phase separation of CBX2 both in vitro and in vivo. Because the content of aromatic residues is low and there is no apparent pattern for aromatic residues across the CBX2 sequence, we hypothesize that these aromatic residues are unlikely to be stickers. However, further experiments are required to test this hypothesis.
Our data suggest a scaffold–client model for the assembly of CBX2–PRC1 condensates (30, 68). Our results indicate that CBX2 is the scaffold, and the other subunits of CBX2–PRC1 are clients. Three lines of evidence support this model. First, the depletion of Ring1a and Ring1b or Bmi1 and Mel18 does not prevent the formation of CBX2 condensates in cells. Second, the depletion of Cbx2 dissolves condensates of PHC1 and RING1B in cells. Finally, CBX2 can phase separate to form condensates in the absence of CBX2–PRC1 subunits in vitro. Previous studies have mapped the physical interactions between the subunits within the CBX–PRC1 complexes. It has been suggested that one of RING1A/RING1B, MEL18/BMI1, and PHC1/PHC2/PHC3 combines to form a stable heterotrimeric protein complex (69, 70). We expect that the trimeric protein complex could be recruited into CBX2 condensates through the C-terminal domain of RING1B interactions with the chromobox (Cbox) motif of CBX2 (71, 72). The absence of CBX2–PRC1 subunits, leading to irregular shapes of CBX2 condensates, suggests that the trimeric client has critical roles in regulating the material properties of CBX2–PRC1 condensates as well as the assembly of PcG condensates.
How do CBX2–PRC1 condensates compact and organize PcG-bound chromatin domains? There should be at least two kinds of organization of PcG-bound chromatin domains: condensing PcG-bound chromatin and establishing long-range interactions of distal PcG-bound regions (7). CBX2 is the PRC1 protein that is responsible for compaction of chromatin in vertebrates (20, 21). The highly charged positive residues within the IDR of CBX2 that are required for the compaction (20) are also required for the phase separation, suggesting that the phase separation is coupled to the chromatin compaction. The chromatin compaction by CBX2 depends on direct binding to nucleosomes but is independent of H3K27me3 (20). CBX4/6/7/8 proteins also have a high content of IDRs; however, unlike CBX2, they do not contain a high content of highly charged basic residues and cannot compact chromatin (20). We suggest that CBX4/6/7/8 may have less capacity to phase separate than CBX2. We hypothesize that CBX2–PRC1 condensates exert compaction of chromatin by binding DNA elements underlying PcG-targeted domains. Our data and previous observations support this hypothesis. Our data demonstrate that CBX2 condensates can concentrate DNA and core-unmodified nucleosomes and that H3K27me3 has minimal effects on the formation of CBX2–PRC1 condensates. Our previous live-cell single-molecule tracking showed that H3K27me3 has negligible effects on the chromatin-bound level of CBX2 (15). Previous studies demonstrated that CBX2 directly binds DNA with a Kd value hundreds-fold smaller than its binding to H3K27me3 in vitro (51, 65, 66, 73) and that the compaction function is independent of histone tails (20, 21, 23). Thus, these data suggest a model in which CBX2–PRC1 condensates compact chromatin through CBX2 directly binding chromatin.
What are the molecular factors that bring distal PcG-bound regions into CBX2–PRC1 condensates for compaction? We propose a bridge model in which CBX7–PRC1 or CBX8–PRC1 acts as the molecular bridge that recruits H3K27me3-marked domains into CBX2–PRC1 condensates. CBX7–PRC1 and CBX8–PRC1 bind to H3K27me3-chromatin via CBX7 and CBX8 interactions with H3K27me3 and then recruit H3K27me3-chromatin into CBX2–PRC1 condensates through the polymerization ability of PHC proteins. The model is supported by our previous live-cell single-molecule tracking results that demonstrate that the removal of H3K27me3 greatly reduces the bound level of CBX7 and CBX8 (15) and by the fact that the sterile α motif domain of PHC proteins can form long filaments via head-to-tail intermolecular interactions (74, 75). Thus, our model suggests that the phase separation of CBX2–PRC1, CBX7–PRC1, CBX8–PRC1, and H3K27me3 coordinates to organize chromatin structure, thereby regulating gene activity. The model can explain why the presence of PHC protein, RING1B, and H3K27me3 is required for compaction of chromatin (25, 27, 28, 76, 77) and be tested in the future.
In summary, our results demonstrate, for the first time, that PcG condensates form through LLPS, and these condensates can concentrate nucleosomes and DNA. Our data show that the charged basic residues of CBX2 that are responsible for compaction of chromatin promote LLPS of CBX2. We further show that H3K27me3 has minimal effects on the formation of CBX2–PRC1 condensates. Our data suggest a scaffold–client model that underpins how CBX2–PRC1 condensates assemble and compact chromatin. Together, these results provide a starting point for conceptualizing the roles of PcG proteins in the assembly, structure, and functions of facultative heterochromatin.
Materials and methods
Cell culture
PGK12.1 mES cells (78) were provided by Dr. Neil Brockdorff (University of Oxford, UK). Cbx2−/− mES cells (79), Eed−/− mES cells (80), Ring1a−/−/RIing1bfl/fl; Rosa26::CreERT2 mES cells (80), and Bmi11−/−/Mel18−/− mES cells (80) were provided by Dr. Haruhiko Koseki (RIKEN Center for Integrative Medical Sciences, Japan). To deplete Ring1b, Ring1a−/−/Ring1bfl/fl; Rosa26::CreERT2 mES cells were treated with 4-hydroxytamoxifen (H7904, Sigma-Aldrich) for 2.0 days under a concentration of 1.0 μm as described previously (15, 49). Hereafter, we refer to Ring1a−/−/Ring1bfl/fl; Rosa26::CreERT2 mES cells as Ring1a−/−/Ring1b−/− mES cells. Dr. Tom Kerppola (University of Michigan, Ann Arbor, MI) provided HEK293T cells. mES cells were grown in the mES cell medium (DMEM (D5796, Sigma-Aldrich) supplemented with 15% FBS (97068-085, VWR), 0.1 mm nonessential amino acids (11140050, Life Technologies), 100 units/ml penicillin-streptomycin (15140-122, Life Technologies), 55 μm β-mercaptoethanol (21985-023, Life Technologies), 2 mm glutamine (G7513, Life Technologies), and 103 units/ml leukemia inhibitor factor at 37 °C in 5% CO2. HEK293T cells were maintained in the HEK293T cell medium (DMEM (D5796; Sigma) supplemented with 10% FBS, 100 units/ml penicillin-streptomycin, 2 mm glutamine, and 55 μm β-mercaptoethanol at 37 °C in 5% CO2.
Plasmids
The pTRIPZ (M1)-HT-Cbx2 plasmid harbors a puromycin resistance gene (15). To generate Cbx2 variants fused with HT, we replaced the Cbx2 sequence in the plasmid pTRIPZ (M1)-HT-Cbx2 with the Cbx2 variant sequence. We generated the following CBX2 variants: 1) CBX2ATH, substitution of PRG with AAA (amino acids 77–79); 2) CBX2ATHL1, substitution of PRG with AAA (amino acids 134–136); 3) CBX2ATHL2, substitution of RKKRGRK with AAAAGAA (amino acids 161–167); 4) CBX2SRR, substitution of SKSKSSSSSSSSTSSSSSS with SKSKASASASASTASASAA (amino acids 102–120); 5), CBX2F12A, substitution of Phe-12 with Ala; 6) CDCBX2, amino acids 1–65 of CBX2; 7) CBX2ΔCD, deletion of the CD (amino acids 1–65); and 8) CBX2ΔCD-ATL, deletion of both the CD and the ATL motif (amino acids 1–88).
To generate recombinant CBX2 in E. coli, we amplified the Cbx2 sequence by PCR and inserted it into the downstream GST sequence within the pGEX-6P-1-GST vector (GE Healthcare) to generate pGEX-6P-1-GST-CBX2. To facilitate double-affinity purification, we added a FLAG tag downstream of the Cbx2 sequence to generate pGEX-6P-1-GST-CBX2-FLAG. We amplified the YFP sequence to insert it upstream of the Cbx2 sequence to generate pGEX-6P-1-GST-YFP-CBX2-FLAG. To generate plasmids for expressing CBX2 variants in E. coli, we amplified the sequence encoding the Cbx2 variants by PCR and used them to replace the Cbx2 sequence in the plasmid pGEX-6P-1-GST-CBX2-FLAG. We generated the following CBX2 variants: 1) CBX2ATH, substitution of PRG with AAA (amino acids 77–79); 2) CBX2ATHL1, substitution of PRG with AAA (amino acids 134–136); 3) CBX2ATHL2, substitution of RKKRGRK with AAAAGAA (amino acids 161–167); and 4) CBX2SRR, substitution of SKSKSSSSSSSSTSSSSSS with SKSKASASASASTASASAA (amino acids 102–120).
Establishing cell lines
24 h before transfection, HEK293T cells were seeded into a 100-mm dish to reach 85–90% confluence at the time of transfection. Cells were cotransfected with 21 μg of pTRIPZ (M) containing the fusion gene, 21 μg of psPAX2, and 10.5 μg of pMD2.G using calcium phosphate precipitation. 12 h after transfection, the medium was replaced with fresh medium. 50 h after the medium change, the medium was harvested and centrifuged at 1000 × g to remove cell debris. Cells were mixed with the harvested medium in the presence of 8.0 μg/ml Polybrene (H9268, Sigma-Aldrich). 48–72 h after transduction, 1.0–2.0 μg/ml puromycin (P8833, Sigma-Aldrich) was added to cells. Cells were selected in the presence of puromycin for at least 1 week.
Generating recombinant protein of CBX2 and its variants
Recombinant CBX2 and its variants were generated and purified according to previous reports with modifications (81). The pGEX-6P-1-GST-FLAG vector containing the Cbx2 fusion gene was transformed into RosettaTM 2 (pLysS) host strains (71403, Novagen). A single colony was used to inoculate 5.0 ml of LB medium. Following overnight culture at 37 °C, 1.0 ml of the overnight culture was transferred into 1.0 liter of LB medium. After 6-h culture at 37 °C, the protein expression was induced overnight at 18 °C in the presence of 1.0 mm isopropyl 1-thio-β-d-galactopyranoside (IB02105, IBI Scientific). After centrifugation, cell pellets were resuspended in 25 ml of lysis buffer (50 mm HEPES, pH 7.5, 1.6 m KCl, 0.5 mm MgCl2, 0.5 mm EDTA, 0.06% Nonidet P-40, 1.0 mm DTT, 1.0 mg/ml lysozyme, 20 μg/ml RNase A, protease inhibitor (S8830, Sigma-Aldrich), and 0.2 mm phenylmethylsulfonyl fluoride). After three freeze-thaw cycles using liquid nitrogen, cells were disrupted using a sonicator (VCX130, Vibra-CellTM) for 3.0 min at 45% amplitude using 15-s on and 45-s off cycles. Cell debris was removed by centrifugation at 10,000 × g for 20 min at 4 °C. To precipitate nucleic acids, 10% polyethylenimine in 20 mm HEPES, pH 7.5, was added to the lysate to achieve a final concentration of 0.15%. The mixture was incubated for 30 min at 4 °C. After centrifugation at 20,000 × g for 20 min, the supernatant was incubated with 0.5 ml of prewashed GSH-Sepharose 4B beads (17-0756-01, GE Healthcare) for 1.0 h at 4 °C. After washing four times with washing buffer (20 mm HEPES, pH 7.5, 500 mm KCl, 0.2 mm EDTA, 1.0 mm DTT, and 0.2 mm phenylmethylsulfonyl fluoride), recombinant protein was eluted with 1.0 ml of 40 mm reduced l-GSH (G4251, Sigma-Aldrich). Alternatively, recombinant protein was eluted by incubating with 80 units of PreScission protease (27-0843-01, GE Healthcare), which cleaves the GST tag, at 4 °C overnight. Eluted protein was incubated with 100 μl of anti-FLAG-M2 affinity resin (A2220, Sigma) for 2.0 h at 4 °C. After washing four times with washing buffer supplemented with 1.0 m KCl, recombinant protein was eluted with 0.4 mg/ml FLAG peptide (F3290, Sigma-Aldrich) dissolved in washing buffer supplemented with 1.0 m KCl. Recombinant protein was resolved by SDS-PAGE to determine its purity and identity.
In vitro condensate formation
After purification, KCl was added to the purified protein to reach a final concentration of 2.0 m, and then the mixture was concentrated to 30–50 μl by using an Amicon centrifugal tube (UFC500324, Millipore). Protein concentration was quantified by the Coomassie (Bradford) protein assay (1856209, Thermo Scientific). 30 μl of protein samples were dialyzed with Spectra/Pro 1 dialysis tubing (132645, Spectrum Labs) in 1.0 liter of dialysis buffer (10 mm phosphate buffer, pH 7.4, containing 2.7 mm KCl and 137 m NaCl) or dialysis buffer supplemented with 1.0 mm DTT at 4 °C. After changing buffer once, dialysis was performed overnight. 10 μl of the dialyzed sample were added to a coverglass dish made as described previously (82). After all condensates had settled down to the surface of the coverslip, DIC or fluorescence images of condensates were acquired using an Axio Observer D1 microscope (Zeiss, Germany) equipped with a 100×/1.40 numerical aperture oil immersion objective with additional 2.5× magnification and an Evolve electron-multiplying charge-coupled density camera (512 × 512; Photometrics, Tucson, AZ). For the excitation and emission of YFP, a Brightline® single-band laser filter set (Semrock; excitation filter, FF02-482/18-25; emission filter, FF01-525/25-25; dichroic mirror, Di02-R488-25) was used. The number of condensates per frame was counted by using ImageJ.
To determine the critical/saturation concentration of phase separation of CBX2, a series of concentrations of CBX2 (1.2, 2.4, 4.8, and 12 μm) was dialyzed under the same conditions, and the number of condensates per frame was counted as described above. To investigate driving forces that contribute to the formation of CBX2 condensates, to 10 μl of the dialyzed sample, NaCl, Triton X-100, and 1,6-hexanediol (240117, Sigma-Aldrich) were added, respectively. The mixture was incubated at 4 °C for 30 min. Condensates were imaged and analyzed as described above.
To prepare CBX2 condensates that concentrate DNA, 4.8 μm CBX2 was mixed with Alexa Fluor 488–labeled DNA (0.5 μm) or Alexa Fluor 488 (1.0 μm), respectively. To prepare CBX2 condensates that concentrate nucleosomes, 2.4 μm CBX2 was mixed with Cy5-labeled mononucleosomes (40 nm). The mixture was dialyzed as described above. DIC and fluorescence images were taken using an Axio Observer D1 microscope as described above. For the excitation and emission of Alexa Fluor 488, a Brightline single-band laser filter set (Semrock; excitation filter, FF02-482/18-25; emission filter, FF01-525/25-25; dichroic mirror, Di02-R488-25) was used. For the excitation and emission of Cy5, a Brightline long-pass laser filter set (Semrock; excitation filter, FF01-640/14-25; emission filter, BLP01-635R-25; dichroic mirror, Di02-R635-25) was used. Images were processed and presented using Photoshop.
Live-cell imaging of condensates
Transgenic mES cells harboring HT-Cbx2 and its variants were seeded to a gelatin-coated coverglass-bottom dish. After 24-h culture at 37 °C in 5% CO2, cells were incubated with 10–20 nm HaloTag TMR ligand (G8251, Promega) for 15 min at 37 °C in 5% CO2. After 15-min incubation, cells were washed with cell culture medium and incubated in the cell culture medium for 1.0 h at 37 °C in 5% CO2. The medium was replaced with the live-cell imaging medium (FluoroBrite DMEM, A1896701, Life Technologies) supplemented with 10% FBS. Cells were maintained at 37 °C using a heater controller (TC-324, Warner Instruments) during imaging. Images of cells were acquired using an Axio Observer D1 microscope as described above. For the excitation and emission of TMR, a Brightline single-band laser filter set (Semrock; excitation filter, FF01-561/14; emission filter, FF01-609/54; dichroic mirror, Di02-R561-25) was used. Visible condensates were counted using ImageJ. Images were presented using Photoshop.
Live-cell imaging of YFP-CBX2 treated with 1,6-hexanediol
We seeded mES cells stably expressing YFP-CBX2 to a gelatin-coated coverglass-bottom dish 24 h before the imaging. Cell culture medium was replaced with the live-cell imaging medium and maintained at 37 °C using a heater controller. Hexanediol was added to the medium to reach a final concentration of 10%. An image stack was taken at every 2-min interval for 20 min using an Axio Observer D1 microscope as described above. For the excitation and emission of YFP, a YFP-2427B filter set (Semrock; excitation filter, FF01-500/24; emission filter, FF01-542/25; dichroic mirror, FF520-Di02) was used.
Immunofluorescence
Cells stably expressing YFP-CBX2 were seeded to coverslips and cultured for 24 h. Cells were fixed with 1.0% paraformaldehyde for 10 min at room temperature. After treatment with 0.2% Triton X-100 for 10 min, cells were washed with basic buffer (10 mm PBS, pH 7.2, 0.05% Tween 20, and 0.1% Triton X-100) and incubated with basic blocking buffer (basic buffer supplemented with 3% goat serum and 3% BSA) overnight. Primary antibodies anti-PHC1 (39723, Active Motif; 1:200 dilution), anti RING1B (D-319, MBL; 1:200 dilution), anti-H3K27me3 (9733, Cell Signaling Technology; 1:200 dilutions), and anti-GFP (A11122, Life Technology; 1:500 dilution) were added to cells and incubated for 2.0 h at room temperature. After washing with basic blocking buffer, cells were incubated with secondary antibodies Alexa Fluor 488–labeled anti-rabbit (A-11008, Life Technologies; 1:1000 dilutions) and/or Alexa Fluor 647–labeled anti-mouse (A32728, Invitrogen; 1:1000 dilution) for 2.0 h at room temperature. Cells were washed and mounted with ProLong Antifade reagents (P7481, Life Technologies) and imaged using a Zeiss LSM 700 Observer Z1 equipped with a 100× oil objective (numerical aperture, 1.4) and an electron-multiplying charge-coupled density camera. For Alexa Fluor 488, 514-nm excitation and 527-nm emission filters were used. For Alexa Fluor 647, 639-nm excitation and 665-nm emission filters were used.
FRAP
Cells stably expressing YFP-CBX2 were seeded to a 35-mm gelatin-coated coverglass-bottom dish. Cells were maintained as described for live-cell imaging. FRAP imaging was performed using a Zeiss LSM 700 Observer with the following parameters: pinhole, full open; scan speed, 177.32 μs/pixel. Before photobleaching, two images were taken. Immediately after photobleaching, 20 images were taken with 15-s intervals. The images were analyzed using ImageJ. We used TurboReg to correct images for movement in the xy plane. After correcting fluctuations in background and total signal, the fluorescence intensities were normalized to the signal before photobleaching to obtain the fluorescence recovery as described previously (42, 49).
Author contributions
R. T., S. K., K. B., H. N. D., T. N. H., H. W., and X. R. formal analysis; R. T., S. K., K. B., T. Y., H. N. D., T. N. H., C. Y. Z., B. M., and X. R. investigation; R. T., S. K., K. B., T. Y., H. N. D., T. N. H., C. Y. Z., B. M., and X. R. methodology; T. Y., H. W., and X. R. resources; H. W. and X. R. supervision; X. R. conceptualization; X. R. data curation; X. R. funding acquisition; X. R. validation; X. R. visualization; X. R. writing-original draft; X. R. project administration; X. R. writing-review and editing.
Acknowledgments
We thank Dr. Haruhiko Koseki for providing Cbx2−/− mES cells, Eed−/− mES cells, Ring1a−/−/Ring1bfl/fl; Rosa26::CreERT2 mES cells, and Bmi1−/−/Mel18−/− mES cells and Dr. Neil Brockdorff for providing PGK12.1 mES cells.
This work was supported by NCI, National Institutes of Health Grant R03CA191443 (to X. R.), National Science Foundation Grant CHE-1500285 (to H. W.), NIGMS, National Institutes of Health Grant R01GM098401 (to T. Y.), and the Office of Research Services (ORS) at the University of Colorado Denver. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- H3K27me3
- trimethylation of Lys-27 at histone H3
- CBX
- chromobox
- EED
- embryonic ectoderm development
- IDR
- intrinsically disordered region
- LLPS
- liquid–liquid phase separation
- PcG
- Polycomb group
- PHC
- polyhomeotic homolog
- PRC
- Polycomb repressive complex
- RING1B
- ring finger protein 2
- mES
- mouse embryonic stem
- HT
- HaloTag
- YFP
- yellow fluorescent protein
- FRAP
- fluorescence recovery after photobleaching
- GST
- glutathione S-transferase
- DIC
- differential interference contrast
- ATH
- AT-hook
- ATHL
- ATH-like
- SRR
- serine-rich region
- CD
- chromodomain
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- TMR
- tetramethylrhodamine.
References
- 1. Brown S. W. (1966) Heterochromatin. Science 151, 417–425 [DOI] [PubMed] [Google Scholar]
- 2. Larson A. G., and Narlikar G. J. (2018) The role of phase separation in heterochromatin formation, function, and regulation. Biochemistry 57, 2540–2548 10.1021/acs.biochem.8b00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trojer P., and Reinberg D. (2007) Facultative heterochromatin: is there a distinctive molecular signature? Mol. Cell 28, 1–13 10.1016/j.molcel.2007.09.011 [DOI] [PubMed] [Google Scholar]
- 4. Reinberg D., and Vales L. D. (2018) Chromatin domains rich in inheritance. Science 361, 33–34 10.1126/science.aat7871 [DOI] [PubMed] [Google Scholar]
- 5. Allshire R. C., and Madhani H. D. (2018) Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19, 229–244 10.1038/nrm.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heard E. (2005) Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Curr. Opin. Genet. Dev. 15, 482–489 10.1016/j.gde.2005.08.009 [DOI] [PubMed] [Google Scholar]
- 7. Schuettengruber B., Bourbon H. M., Di Croce L., and Cavalli G. (2017) Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell 171, 34–57 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 8. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., and Zhang Y. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- 9. Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., and Pirrotta V. (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196 10.1016/S0092-8674(02)00975-3 [DOI] [PubMed] [Google Scholar]
- 10. Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., and Reinberg D. (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16, 2893–2905 10.1101/gad.1035902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Müller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A., Wild B., Miller E. L., O'Connor M. B., Kingston R. E., and Simon J. A. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208 10.1016/S0092-8674(02)00976-5 [DOI] [PubMed] [Google Scholar]
- 12. Shen X., Liu Y., Hsu Y. J., Fujiwara Y., Kim J., Mao X., Yuan G. C., and Orkin S. H. (2008) EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502 10.1016/j.molcel.2008.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Margueron R., Justin N., Ohno K., Sharpe M. L., Son J., Drury WJ 3rd, Voigt P., Martin S. R., Taylor W. R., De Marco V., Pirrotta V., Reinberg D., and Gamblin S. J. (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 10.1038/nature08398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu C., Bian C., Yang W., Galka M., Ouyang H., Chen C., Qiu W., Liu H., Jones A. E., MacKenzie F., Pan P., Li S. S., Wang H., and Min J. (2010) Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2). Proc. Natl. Acad. Sci. U.S.A. 107, 19266–19271 10.1073/pnas.1008937107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhen C. Y., Tatavosian R., Huynh T. N., Duc H. N., Das R., Kokotovic M., Grimm J. B., Lavis L. D., Lee J., Mejia F. J., Li Y., Yao T., and Ren X. (2016) Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin. Elife 5, e17667 10.7554/eLife.17667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., and Zhang Y. (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 10.1038/nature02985 [DOI] [PubMed] [Google Scholar]
- 17. de Napoles M., Mermoud J. E., Wakao R., Tang Y. A., Endoh M., Appanah R., Nesterova T. B., Silva J., Otte A. P., Vidal M., Koseki H., and Brockdorff N. (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7, 663–676 10.1016/j.devcel.2004.10.005 [DOI] [PubMed] [Google Scholar]
- 18. Kalb R., Latwiel S., Baymaz H. I., Jansen P. W., Müller C. W., Vermeulen M., and Müller J. (2014) Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 21, 569–571 10.1038/nsmb.2833 [DOI] [PubMed] [Google Scholar]
- 19. Blackledge N. P., Farcas A. M., Kondo T., King H. W., McGouran J. F., Hanssen L. L., Ito S., Cooper S., Kondo K., Koseki Y., Ishikura T., Long H. K., Sheahan T. W., Brockdorff N., Kessler B. M., et al. (2014) Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445–1459 10.1016/j.cell.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grau D. J., Chapman B. A., Garlick J. D., Borowsky M., Francis N. J., and Kingston R. E. (2011) Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 25, 2210–2221 10.1101/gad.17288211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lau M. S., Schwartz M. G., Kundu S., Savol A. J., Wang P. I., Marr S. K., Grau D. J., Schorderet P., Sadreyev R. I., Tabin C. J., and Kingston R. E. (2017) Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science 355, 1081–1084 10.1126/science.aah5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. King I. F., Emmons R. B., Francis N. J., Wild B., Müller J., Kingston R. E., and Wu C. T. (2005) Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol. Cell. Biol. 25, 6578–6591 10.1128/MCB.25.15.6578-6591.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Francis N. J., Kingston R. E., and Woodcock C. L. (2004) Chromatin compaction by a polycomb group protein complex. Science 306, 1574–1577 10.1126/science.1100576 [DOI] [PubMed] [Google Scholar]
- 24. Pirrotta V., and Li H. B. (2012) A view of nuclear Polycomb bodies. Curr. Opin. Genet. Dev. 22, 101–109 10.1016/j.gde.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Isono K., Endo T. A., Ku M., Yamada D., Suzuki R., Sharif J., Ishikura T., Toyoda T., Bernstein B. E., and Koseki H. (2013) SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell 26, 565–577 10.1016/j.devcel.2013.08.016 [DOI] [PubMed] [Google Scholar]
- 26. Kondo T., Isono K., Kondo K., Endo T. A., Itohara S., Vidal M., and Koseki H. (2014) Polycomb potentiates meis2 activation in midbrain by mediating interaction of the promoter with a tissue-specific enhancer. Dev. Cell 28, 94–101 10.1016/j.devcel.2013.11.021 [DOI] [PubMed] [Google Scholar]
- 27. Kundu S., Ji F., Sunwoo H., Jain G., Lee J. T., Sadreyev R. I., Dekker J., and Kingston R. E. (2017) Polycomb repressive complex 1 generates discrete compacted domains that change during differentiation. Mol. Cell 65, 432–446.e5 10.1016/j.molcel.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wani A. H., Boettiger A. N., Schorderet P., Ergun A., Münger C., Sadreyev R. I., Zhuang X., Kingston R. E., and Francis N. J. (2016) Chromatin topology is coupled to Polycomb group protein subnuclear organization. Nat. Commun. 7, 10291 10.1038/ncomms10291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shin Y., and Brangwynne C. P. (2017) Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- 30. Banani S. F., Lee H. O., Hyman A. A., and Rosen M. K. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hyman A. A., Weber C. A., and Jülicher F. (2014) Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- 32. Boeynaems S., Alberti S., Fawzi N. L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., Tompa P., and Fuxreiter M. (2018) Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Woodruff J. B., Hyman A. A., and Boke E. (2018) Organization and function of non-dynamic biomolecular condensates. Trends Biochem. Sci. 43, 81–94 10.1016/j.tibs.2017.11.005 [DOI] [PubMed] [Google Scholar]
- 34. Meldi L., and Brickner J. H. (2011) Compartmentalization of the nucleus. Trends Cell Biol. 21, 701–708 10.1016/j.tcb.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cho W. K., Spille J. H., Hecht M., Lee C., Li C., Grube V., and Cisse I. I. (2018) Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G. M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., and Tjian R. (2018) Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 10.1126/science.aar2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sabari B. R., Dall'Agnese A., Boija A., Klein I. A., Coffey E. L., Shrinivas K., Abraham B. J., Hannett N. M., Zamudio A. V., Manteiga J. C., Li C. H., Guo Y. E., Day D. S., Schuijers J., Vasile E., et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boehning M., Dugast-Darzacq C., Rankovic M., Hansen A. S., Yu T., Marie-Nelly H., McSwiggen D. T., Kokic G., Dailey G. M., Cramer P., Darzacq X., and Zweckstetter M. (2018) RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 25, 833–840 10.1038/s41594-018-0112-y [DOI] [PubMed] [Google Scholar]
- 39. Lu H., Yu D., Hansen A. S., Ganguly S., Liu R., Heckert A., Darzacq X., and Zhou Q. (2018) Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323 10.1038/s41586-018-0174-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Strom A. R., Emelyanov A. V., Mir M., Fyodorov D. V., Darzacq X., and Karpen G. H. (2017) Phase separation drives heterochromatin domain formation. Nature 547, 241–245 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Larson A. G., Elnatan D., Keenen M. M., Trnka M. J., Johnston J. B., Burlingame A. L., Agard D. A., Redding S., and Narlikar G. J. (2017) Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ren X., Vincenz C., and Kerppola T. K. (2008) Changes in the distributions and dynamics of polycomb repressive complexes during embryonic stem cell differentiation. Mol. Cell. Biol. 28, 2884–2895 10.1128/MCB.00949-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alkema M. J., Bronk M., Verhoeven E., Otte A., van 't Veer L. T., Berns A., and van Lohuizen M. (1997) Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian Polycomb complex. Gene Dev 11, 226–240 10.1101/gad.11.2.226 [DOI] [PubMed] [Google Scholar]
- 44. Schoorlemmer J., Marcos-Gutiérrez C., Were F., Martínez R., García E., Satijn D. P., Otte A. P., and Vidal M. (1997) Ring1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J. 16, 5930–5942 10.1093/emboj/16.19.5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tatavosian R., Zhen C. Y., Duc H. N., Balas M. M., Johnson A. M., and Ren X. (2015) Distinct cellular assembly stoichiometry of Polycomb complexes on chromatin revealed by single-molecule chromatin immunoprecipitation imaging. J. Biol. Chem. 290, 28038–28054 10.1074/jbc.M115.671115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Levine S. S., Weiss A., Erdjument-Bromage H., Shao Z., Tempst P., and Kingston R. E. (2002) The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22, 6070–6078 10.1128/MCB.22.17.6070-6078.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Satijn D. P., Gunster M. J., van der Vlag J., Hamer K. M., Schul W., Alkema M. J., Saurin A. J., Freemont P. S., van Driel R., and Otte A. P. (1997) RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol. Cell. Biol. 17, 4105–4113 10.1128/MCB.17.7.4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gunster M. J., Satijn D. P., Hamer K. M., den Blaauwen J. L., de Bruijn D., Alkema M. J., van Lohuizen M., van Driel R., and Otte A. P. (1997) Identification and characterization of interactions between the vertebrate polycomb-group protein BMI1 and human homologs of polyhomeotic. Mol. Cell. Biol. 17, 2326–2335 10.1128/MCB.17.4.2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhen C. Y., Duc H. N., Kokotovic M., Phiel C. J., and Ren X. (2014) Cbx2 stably associates with mitotic chromosomes via a PRC2- or PRC1-independent mechanism and is needed for recruiting PRC1 complex to mitotic chromosomes. Mol. Biol. Cell 25, 3726–3739 10.1091/mbc.e14-06-1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Piovesan D., Tabaro F., Paladin L., Necci M., Micetic I., Camilloni C., Davey N., Dosztányi Z., Mészáros B., Monzon A. M., Parisi G., Schad E., Sormanni P., Tompa P., Vendruscolo M., et al. (2018) MobiDB 3.0: more annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res. 46, D471–D476 10.1093/nar/gkx1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kawaguchi T., Machida S., Kurumizaka H., Tagami H., and Nakayama J. I. (2017) Phosphorylation of CBX2 controls its nucleosome-binding specificity. J. Biochem. 162, 343–355 10.1093/jb/mvx040 [DOI] [PubMed] [Google Scholar]
- 52. Wang J., Choi J. M., Holehouse A. S., Lee H. O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D., Poser I., Pappu R. V., Alberti S., and Hyman A. A. (2018) A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 10.1016/j.cell.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Boeynaems S., Bogaert E., Kovacs D., Konijnenberg A., Timmerman E., Volkov A., Guharoy M., De Decker M., Jaspers T., Ryan V. H., Janke A. M., Baatsen P., Vercruysse T., Kolaitis R. M., Daelemans D., et al. (2017) Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e5 10.1016/j.molcel.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee K. H., Zhang P., Kim H. J., Mitrea D. M., Sarkar M., Freibaum B. D., Cika J., Coughlin M., Messing J., Molliex A., Maxwell B. A., Kim N. C., Temirov J., Moore J., Kolaitis R. M., et al. (2016) C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788.e17 10.1016/j.cell.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jiang H., Wang S., Huang Y., He X., Cui H., Zhu X., and Zheng Y. (2015) Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell 163, 108–122 10.1016/j.cell.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nott T. J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T. D., Bazett-Jones D. P., Pawson T., Forman-Kay J. D., and Baldwin A. J. (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pak C. W., Kosno M., Holehouse A. S., Padrick S. B., Mittal A., Ali R., Yunus A. A., Liu D. R., Pappu R. V., and Rosen M. K. (2016) Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell 63, 72–85 10.1016/j.molcel.2016.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mitrea D. M., Cika J. A., Stanley C. B., Nourse A., Onuchic P. L., Banerjee P. R., Phillips A. H., Park C. G., Deniz A. A., and Kriwacki R. W. (2018) Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat. Commun. 9, 842 10.1038/s41467-018-03255-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Feric M., Vaidya N., Harmon T. S., Mitrea D. M., Zhu L., Richardson T. M., Kriwacki R. W., Pappu R. V., and Brangwynne C. P. (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C. C., Eckmann C. R., Myong S., and Brangwynne C. P. (2015) The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 10.1073/pnas.1504822112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Patel A., Lee H. O., Jawerth L., Maharana S., Jahnel M., Hein M. Y., Stoynov S., Mahamid J., Saha S., Franzmann T. M., Pozniakovski A., Poser I., Maghelli N., Royer L. A., Weigert M., et al. (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- 62. Altmeyer M., Neelsen K. J., Teloni F., Pozdnyakova I., Pellegrino S., Grøfte M., Rask M. B., Streicher W., Jungmichel S., Nielsen M. L., and Lukas J. (2015) Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 8088 10.1038/ncomms9088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Senthilkumar R., and Mishra R. K. (2009) Novel motifs distinguish multiple homologues of Polycomb in vertebrates: expansion and diversification of the epigenetic toolkit. BMC Genomics 10, 549 10.1186/1471-2164-10-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Blackledge N. P., Rose N. R., and Klose R. J. (2015) Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat. Rev. Mol. Cell Biol. 16, 643–649 10.1038/nrm4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kaustov L., Ouyang H., Amaya M., Lemak A., Nady N., Duan S., Wasney G. A., Li Z., Vedadi M., Schapira M., Min J., and Arrowsmith C. H. (2011) Recognition and specificity determinants of the human Cbx chromodomains. J. Biol. Chem. 286, 521–529 10.1074/jbc.M110.191411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bernstein E., Duncan E. M., Masui O., Gil J., Heard E., and Allis C. D. (2006) Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 26, 2560–2569 10.1128/MCB.26.7.2560-2569.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Semenov A. N., and Rubinstein M. (1998) Thermoreversible gelation in solutions of associative polymers. 1. Statics. Macromolecules 31, 1373–1385 10.1021/ma970616h [DOI] [Google Scholar]
- 68. Banani S. F., Rice A. M., Peeples W. B., Lin Y., Jain S., Parker R., and Rosen M. K. (2016) Compositional control of phase-separated cellular bodies. Cell 166, 651–663 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hemenway C. S., Halligan B. W., and Levy L. S. (1998) The Bmi-1 oncoprotein interacts with dinG and MPh2: the role of RING finger domains. Oncogene 16, 2541–2547 10.1038/sj.onc.1202042 [DOI] [PubMed] [Google Scholar]
- 70. Satijn D. P., and Otte A. P. (1999) RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol. Cell. Biol. 19, 57–68 10.1128/MCB.19.1.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang R., Ilangovan U., Robinson A. K., Schirf V., Schwarz P. M., Lafer E. M., Demeler B., Hinck A. P., and Kim C. A. (2008) Structural transitions of the RING1B C-terminal region upon binding the polycomb cbox domain. Biochemistry 47, 8007–8015 10.1021/bi800857f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang R., Taylor A. B., Leal B. Z., Chadwell L. V., Ilangovan U., Robinson A. K., Schirf V., Hart P. J., Lafer E. M., Demeler B., Hinck A. P., McEwen D. G., and Kim C. A. (2010) Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 18, 966–975 10.1016/j.str.2010.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tardat M., Albert M., Kunzmann R., Liu Z., Kaustov L., Thierry R., Duan S., Brykczynska U., Arrowsmith C. H., and Peters A. H. (2015) Cbx2 targets PRC1 to constitutive heterochromatin in mouse zygotes in a parent-of-origin-dependent manner. Mol. Cell 58, 157–171 10.1016/j.molcel.2015.02.013 [DOI] [PubMed] [Google Scholar]
- 74. Kim C. A., Gingery M., Pilpa R. M., and Bowie J. U. (2002) The SAM domain of polyhomeotic forms a helical polymer. Nat. Struct. Biol. 9, 453–457 10.1038/nsb802 [DOI] [PubMed] [Google Scholar]
- 75. Kim C. A., and Bowie J. U. (2003) SAM domains: uniform structure, diversity of function. Trends Biochem. Sci. 28, 625–628 10.1016/j.tibs.2003.11.001 [DOI] [PubMed] [Google Scholar]
- 76. Eskeland R., Leeb M., Grimes G. R., Kress C., Boyle S., Sproul D., Gilbert N., Fan Y., Skoultchi A. I., Wutz A., and Bickmore W. A. (2010) Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 38, 452–464 10.1016/j.molcel.2010.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schoenfelder S., Sugar R., Dimond A., Javierre B. M., Armstrong H., Mifsud B., Dimitrova E., Matheson L., Tavares-Cadete F., Furlan-Magaril M., Segonds-Pichon A., Jurkowski W., Wingett S. W., Tabbada K., Andrews S., et al. (2015) Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat. Genet. 47, 1179–1186 10.1038/ng.3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Penny G. D., Kay G. F., Sheardown S. A., Rastan S., and Brockdorff N. (1996) Requirement for Xist in X chromosome inactivation. Nature 379, 131–137 10.1038/379131a0 [DOI] [PubMed] [Google Scholar]
- 79. Katoh-Fukui Y., Tsuchiya R., Shiroishi T., Nakahara Y., Hashimoto N., Noguchi K., and Higashinakagawa T. (1998) Male-to-female sex reversal in M33 mutant mice. Nature 393, 688–692 10.1038/31482 [DOI] [PubMed] [Google Scholar]
- 80. Endoh M., Endo T. A., Endoh T., Fujimura Y., Ohara O., Toyoda T., Otte A. P., Okano M., Brockdorff N., Vidal M., and Koseki H. (2008) Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135, 1513–1524 10.1242/dev.014340 [DOI] [PubMed] [Google Scholar]
- 81. Huynh T. N., and Ren X. (2017) Producing GST-Cbx7 fusion proteins from Escherichia coli. Bio Protoc. 7, e2333 10.21769/BioProtoc.2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Duc H. N., and Ren X. (2017) Labelling HaloTag fusion proteins with HaloTag ligand in living cells. Bio Protoc. 7, e2526 10.21769/BioProtoc.2526 [DOI] [PMC free article] [PubMed] [Google Scholar]