Abstract
The sodium potassium pump (Na/K-ATPase) ensures the electrochemical gradient of a cell through an energy-dependent process that consumes approximately one third of regenerated ATP. We report that the G protein-coupled receptor GPR35 interacted with the α chain of Na/K-ATPase and promotes its ion transport and Src signaling activity in a ligand-independent manner. Deletion of Gpr35 increased baseline Ca2+ to maximal levels and reduced Src activation and overall metabolic activity in macrophages and intestinal epithelial cells (IECs). In contrast, a common T108M polymorphism in GPR35 was hypermorphic and had the opposite effects to Gpr35 deletion on Src activation and metabolic activity. The T108M polymorphism is associated with ulcerative colitis and primary sclerosing cholangitis, inflammatory diseases with a high cancer risk. GPR35 promoted homeostatic IEC turnover, whereas Gpr35 deletion or inhibition by a selective pepducin prevented inflammation-associated and spontaneous intestinal tumorigenesis in mice. Thus, GPR35 acts as a central signaling and metabolic pacesetter, which reveals an unexpected role of Na/K-ATPase in macrophage and IEC biology.
Introduction
Genetic studies of complex diseases have been highly successful in identifying disease-associated single nucleotide polymorphisms (SNPs), but discerning the biology responsible for these associations has proven to be a formidable challenge. Indeed, for only a handful of disease-associated SNPs have the underlying mechanisms been elucidated. These examples, however, have shed new light on disease pathogenesis and highlighted the importance of transitioning our knowledge from SNPs to the underlying biology responsible for altering disease risk.
We were intrigued by a protein coding variant (rs3749171, leading to a T108M substitution) in the G protein-coupled receptor (GPCR) gene GPR35, which is associated with risk for primary sclerosing cholangitis (PSC) and ulcerative colitis (UC) across multiple ethnicities (1–4). One advantage of investigating coding variants is that they are much more likely to be causal than non-coding associations.
GPCRs are a large family of receptors, with over 300 non-olfactory members, and yet in general they remain relatively poorly understood. For instance, the endogenous ligands for over 100 GPCRs remain unknown or disputed, and the basic biological function of the majority is unknown. Despite this, GPCRs have been the class of targets for which small molecule therapeutics have been most successfully developed, even though this extends only to a minority of GPCRs (5, 6). Several candidate ligands of GPR35 have been suggested, foremost the tryptophan metabolite kynurenic acid (KYNA) (7) and the chemokine CXCL17 (8). However, KYNA exhibits astounding species-dependent differences in potency, which is particularly low (EC50 >10–3 M) for human GPR35 (9, 10), and CXCL17 does not act as ligand in some experimental systems (11) and is a member of a family (chemokine receptors) that are not predicted to cluster with GPR35’s predicted core ligand binding domain (12). As such, both putative ligands remain disputed (5). Consistent with being associated with inflammatory diseases with high cancer risk, GPR35 is expressed in a tissue-specific pattern, with high expression in myeloid and intestinal epithelial cells (IECs) (7) – cell-types implicated in both UC and PSC – and is also highly expressed in several cancers (13, 14).
PSC and UC are chronic inflammatory conditions of the bile ducts and the large intestine, respectively. (15, 16) Both diseases have an extreme cumulative risk for the development of cancer (17, 18) Although PSC and UC are both considered autoimmune diseases, a classification that is supported by strong genetic association signals within the MHC region (19–21), the mechanistic basis of these disorders remains poorly understood, and the basis for the excessive cancer association is particularly enigmatic.
Here we discovered that GPR35 interacted with and promoted the activity of Na/K-ATPase, the central ion pump that maintains a cell’s electrochemical gradient. We reveal the mechanism by which the T108M mutation not only altered risk of UC and PSC, but also contributed directly to the cancer risk associated with these diseases. We showed that this Na/K-ATPase modulation was ligand-independent. Furthermore, we demonstrated that the disease-associated variant was hypermorphic and elicited an increase in Na/K-ATPase activity with profound consequences for cellular membrane potential, Ca2+ homeostasis, and metabolism. This increase in Na/K-ATPase activity directly activated the kinase Src, thereby altering signaling through unrelated receptors and increasing cell proliferation and tumor formation. Finally, we described a pepducin therapeutic that targeted GPR35 and protected mice from tumor formation, in a manner similar to genetic deletion of Gpr35, which could represent a targeted therapeutic in carriers of this mutation.
Results
GPR35 deficiency increases cytoplasmic Ca2+ levels
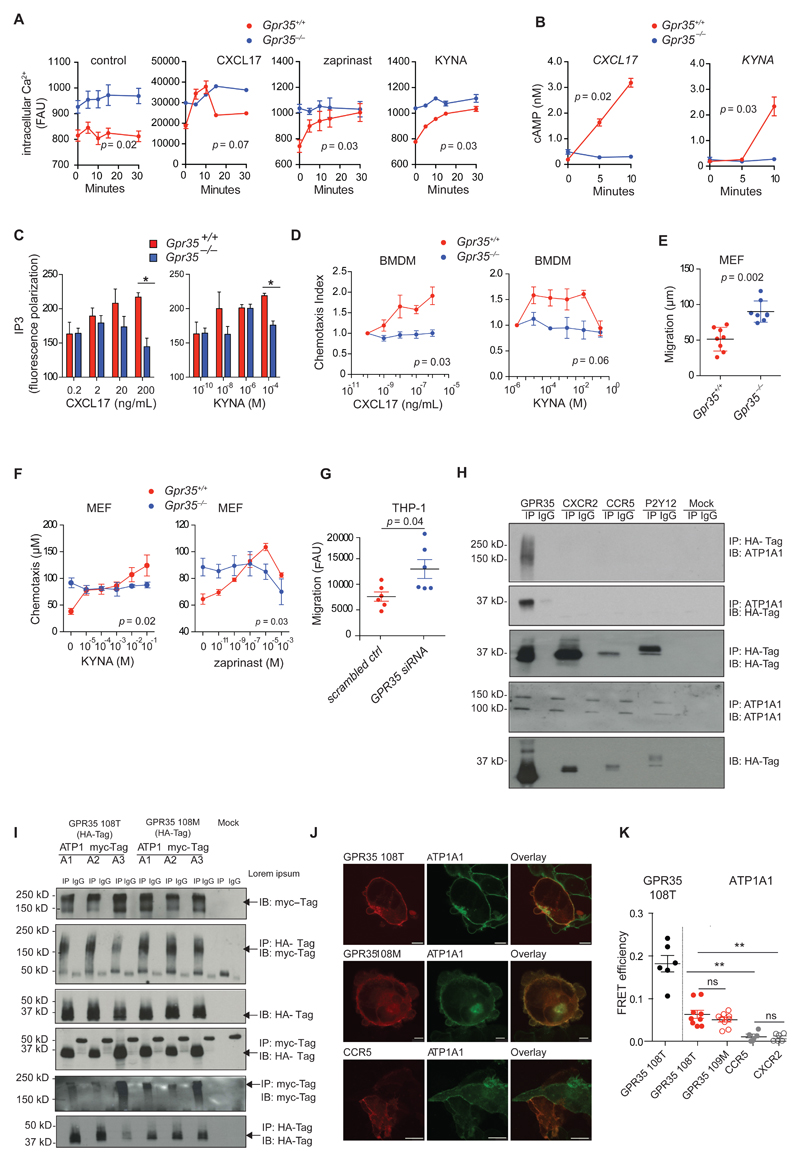
Kynurenic acid (KYNA) and CXCL17 have been reported to trigger GPR35 activation in cells transfected with this receptor (9, 22). KYNA, CXCL17 and zaprinast (a phosphodiesterase inhibitor that is a GPR35 agonist (23)) elicited intracellular Ca2+ release, cAMP production, and inositol-trisphosphate (IP3) generation in murine bone marrow-derived macrophages (BMDMs), which was absent in Gpr35–/– BMDMs (Fig. 1, A to C). Further consistent with receptor activation, the small GTPase Rho was gradually activated by both KYNA and CXCL17 in wild-type BMDMs, but not in Gpr35–/– BMDMs (fig. S1a). KYNA also elicited transient activation of the small GTPase Rac-1 in wild-type, but not Gpr35–/– BMDMs (fig. S1a). Consistent with the important role of Rac-1 on the cytoskeleton, fluorescent phalloidin imaging demonstrated actin filament reorganization upon KYNA stimulation in wild-type, but not Gpr35–/– BMDMs (fig. S1b). CXCL17 and KYNA also elicited a concentration-dependent chemotactic response in wild-type, but not Gpr35–/– BMDMs (Fig. 1D). Together, these results confirm in a genetic model that KYNA and CXCL17 trigger GPR35 signaling.
Figure 1. GPR35 deficiency re-sets resting cytoplasmic Ca2+ levels and GPR35 interacts with the α-chain of Na/K-ATPase.
A, Intracellular Ca2+ release in BMDMs at basal level (control) and after stimulation with CXCL17 (200 ng/mL), zaprinast (10 μM) or KYNA (300 μM); n=3 mice/group. B, Cyclic AMP (cAMP) production in BMDMs after stimulation with CXCL17 (200 ng/mL) or KYNA (300 μM); n=3 mice/group. C, Dose dependent inositol trisphosphate (IP3) generation in BMDMs after stimulation with CXCL17 (0.2 - 200 ng/mL) or KYNA (0.1 nM - 100 μM) for 20 min; n=3 mice/group. D, Directed migration (chemotaxis) of BMDMs towards different concentrations of CXCL17 and KYNA expressed as the chemotaxis index of stimulated compared to unstimulated cells; n=3 mice/group. E, Undirected migration of MEFs expressed as distance migrated into membrane; n=7 independent experiments for Gpr35+/+ and n=8 independent experiments for Gpr35-/-/group (we have used 3 Gpr35+/+ and 3 Gpr35-/- clones). F, Migratory distance of MEFs towards KYNA and Zaprinast; n=3 clones. G, Undirected migration of THP1 cells transfected with either GPR35 siRNA or scrambled siRNA (scrambled ctrl); n=6 independent experiments/group. Data are shown as mean ± s.e.m. Statistical significance calculated by Mann Whitney U test after Kruskal Wallis one-way ANOVA. H, Immunoprecipitates of HA tagged GPCRs probed for ATP1A1 (upper panel). Immunoprecipitates of ATP1A1 probed for HA (middle panel). Immunopreciptation using normal IgG antibody (IgG) used as a control. Western blot of HA-tagged GPCRs in whole cell lysates shown below. n=3 independent experiments I, Immunoprecipitates of HA-tagged GPR35108T or GPR35108M variant probed for myc-tagged ATP1A isoforms. Western Blot of myc-tagged ATP1A isoforms in whole cell lysates shown above (upper two panels). Immunoprecipitates of myc-tagged ATP1A isoforms (ATPA1-3) probed for HA. Western Blot of HA-tagged GPR35108T or GPR35108M variant in whole cell lysates shown above (lower two panels). Immunopreciptation using normal IgG antibody (IgG) used as a control. Arrows indicate the relevant band. n=3 independent experiments J, Fluorescent microscopy of ATP1A1-Venus (green) and Cerulean tagged GPR35108T or GPR35108M variant (red) co-expressed in HEK293 cells. Cerulean tagged CCR5 (red) used as a control. Scale bar, 5 μm. K, Fluorescence resonance energy transfer (FRET) efficiency measuring proximity of fluorescent-labelled GPR35108T or GPR35108M variant and ATP1A1 co-expressed in HEK293 cells. Fluorescent-labelled CCR5 and CXCR2 used as a control. GPR35 homo dimers used as a positive control. Data points represents individual acceptor photo bleaching events. n=6 events from 3 independent experiments for GPR35 homodimers and CCR5 and CXCR2 control, n=9 events from 3 independent experiments for GPR38108M and GPR35 108T. All data represented as mean ± s.e.m. Statistical significance was calculated by two-way ANOVA followed by post-hoc comparison of means, ** (p<0.01).
However, we observed that cytoplasmic Ca2+ levels were greatly increased in Gpr35–/– compared to wild-type BMDMs (Fig. 1A). These resting levels in Gpr35–/– BMDMs were as high as the peak intracellular Ca2+ achieved in wild-type BMDMs upon stimulation with KYNA, CXCL17, and zaprinast. In contrast, baseline cAMP and IP3 concentrations were indistinguishable between Gpr35–/– and wild-type BMDMs (Fig. 1, B and C). We also observed increased baseline (meaning, ligand-independent) migration in Gpr35–/– compared to wild-type mouse embryonic fibroblasts (MEFs), although only wild-type MEFs demonstrated chemotaxis to KYNA and zaprinast (Fig. 1, E and F). Increased baseline migration was also observed in GPR35-silenced THP1 (a human macrophage cell line) compared to control-silenced cells (Fig 1G). Collectively, these data demonstrate that even without GPR35 ligands, the genetic deletion of Gpr35 induces a phenotype analogous to that observed following ligand stimulation, suggesting that GPR35 has a constitutive, ligand-independent function.
GPR35 interacts with the α chain of Na/K-ATPase
To better understand the constitutive function of GPR35, we sought to examine the interactome of GPR35, because GPCR function often depends on protein-protein interactions (24). To perform an unbiased proteomics survey for interaction partners, we used human embryonic kidney HEK293T cells transfected with human GPR35 cDNAs as baits that were engineered to express Strep or hemagglutinin (HA) tags at the C terminus. In two independent experiments, the most abundant peptide hit for tagged GPR35 mapped to Na/K-ATPase subunit α1 (Uniprot P05023 isoform 4; encoded by ATP1A1), and was not present in HEK293T cells transfected with tag only, or in mock-transfected cells (table S1).
We confirmed this interaction in HEK293T cells transfected with GPR35-HA by probing HA immunoprecipitates for ATP1A1 to detect endogenous protein (Fig. 1H). Moreover, a band consistent with GPR35-HA was detected by immunoblotting for HA in ATP1A1 immunoprecipitates (Fig. 1H). In contrast to GPR35-HA, HA-tagged GPCRs CXCR2, CCR5 or P2Y12 did not co-immunoprecipitate with ATP1A1 (Fig. 1H), indicating that the interaction between Na/K-ATPase and GPR35 was specific for this particular GPCR.
Na/K-ATPase consists of a catalytic α subunit (encoded by 4 genes, ATP1A1 to ATP1A4) and a regulatory β subunit (encoded by 4 genes, ATP1B1 to ATP1B4), which is important for the recruitment of the α subunit to the plasma membrane (25, 26). ATP1A1 is ubiquitously expressed and the main α chain of heterodimeric Na/K-ATPase, whereas other α chains exhibit a more restricted expression pattern (27, 28). We therefore asked whether GPR35-Na/K-ATPase interaction was restricted to the ATP1A1 isoform only. To address this question, we co-transfected HEK293T cells with Myc-tagged α1, α2, or α3 chains (α4 is only expressed in testis, where GPR35 is not present (26)) together with either HA-tagged GPR35108T (a non-risk variant) or GPR35108M (a risk variant). HA immunoprecipitates for both GPR35108T-HA and GPR35108M-HA pulled down all three Na/K-ATPase α chains as detected by immunoblotting for Myc (Fig. 1I). Conversely, Myc immunoprecipitates for α chain constructs pulled down GPR35-HA variants (Fig. 1I). GPR35108T and GPR35108M co-immunoprecipitated to a similar extent with the individual Na/K-ATPase α chains (Fig. 1I). These data suggested that GPR35 biochemically interacts with ATP1A1, ATP1A2, and ATP1A3 chains, and this interaction is not affected by the GPR35 risk variant.
To confirm the intimate relationship of GPR35 and ATP1A1, we used photo-bleaching confocal fluorescent resonance energy transfer (FRET) microscopy of HEK293T cells transfected with a full-length ATP1A1 tagged C-terminally in-frame with Venus (ATP1A1-Venus) as the fluorescent donor, and a construct of full-length GPR35 tagged C-terminally in-frame with Cerulean (GPR35-Cerulean) as the fluorescent acceptor. A FRET signal typically occurs when fluorescent donors and acceptors are closer than 100 Å. The fluorescence signals of GPR35, ATP1A1 and the control CCR5 were located to the plasma membrane (Fig. 1J), with FRET efficiency between GPR35 and ATP1A1 of 7% (Fig. 1K) consistent with a distance equivalent to approximately 75 Å. For comparison, FRET efficiency between GPR35-Venus and GPR35-Cerulean was 18% (65 Å). In contrast, no FRET signal above background was obtained with HEK293T cells transfected with the chemokine receptor CCR5-Cerulean (1%) or CXCR2-Cerulean (0.7%) together with ATP1A1-Venus (Fig. 1K). The FRET signal obtained with GPR35108T-Cerulean was similar from GPR35108M-Cerulean upon co-transfection with ATP1A1-Venus (Fig. 1K). Together these data establish an intimate spatial relationship between the α1 subunit of Na/K-ATPase and GPR35, whether it is wild-type or the GPR35T108M variant. Furthermore, a close spatial relationship did not exist between GPR35 and either CXCR2 or CCR5, suggesting that the interaction between Na/K-ATPase and GPR35 is specific.
GPR35 promotes Na/K-ATPase function
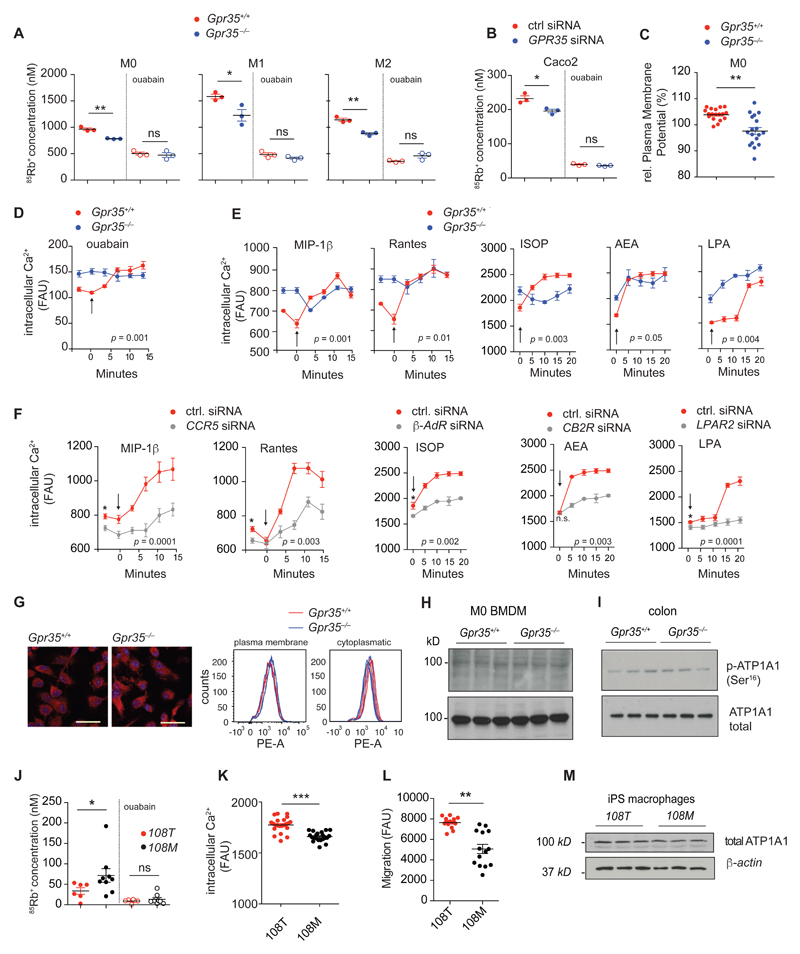
Considering the spatial relationship between GPR35 and ATP1A1, we hypothesized that GPR35 affects Na/K-ATPase function in macrophages and intestinal epithelial cells. Na/K-ATPase (EC 3.6.3.9) is the ubiquitously present cation pump (‘sodium-potassium pump’), which is essential for maintenance of the electrochemical gradient across the plasma membrane (26, 28). We directly assessed Na/K-ATPase pump function through inductively coupled plasma mass spectrometry (ICP-MS) using Rubidium (85Rb+) uptake in BMDMs and the colonic epithelial cell line Caco2. 85Rb+ is a tracer ion for K+ at physiological concentrations of extracellular K+, which does not natively occur in biological systems (29). Macrophages exhibit different functional states, which can be approximated in vitro by differentiation under M0 (baseline), M1 (‘inflammatory’, IFNγ+LPS) and M2 (‘regenerative’, IL-4) conditions (30). Gpr35–/– BMDMs differentiated under M0, M1 and M2 conditions transported less 85Rb+ across the plasma membrane compared to their corresponding wild-type cells. Moreover, selective inhibition of Na/K-ATPase with ouabain (31) decreased 85Rb+ uptake further, reaching similarly low levels in wild-type and Gpr35–/– M0, M1 and M2 macrophages (Fig. 2A). We next asked whether GPR35-dependent regulation of Na/K-ATPase extended to other cells that highly express the receptor, such as intestinal epithelial cells (IECs). Indeed a 75% knock-down (fig S2a) of GPR35 mRNA expression by siRNA in Caco2 cells resulted in a 15% reduction in 85Rb+ uptake compared to cells transfected with control siRNA (Fig. 2B). Similar to BMDMs, ouabain reduced 85Rb+ uptake to similarly low levels in GPR35-silenced and control Caco2 cells (Fig. 2B). Consistent with the effect of GPR35 on Na/K-ATPase function, the plasma membrane potential (Δψp) was reduced in Gpr35–/– compared to wild-type BMDMs (Fig. 2C). These data demonstrate that in macrophages and IECs, GPR35 promotes Na/K-ATPase ion pump activity and thereby determines the electrochemical gradient across the plasma membrane.
Figure 2. GPR35 sets resting intracellular Ca2+ by promoting Na/K-ATPase function.
A, Intracellular 85Rb+ concentration (nM) measured by ICP-MS in M0, M1 and M2 BMDMs (n=3 mice/group) and B, Caco2 cells transfected with GPR35 siRNA or control siRNA (ctrl) (n=3 independent experiments/group), after 2 hours of incubation in Rb85+ containing media in the absence or presence of 1 mM ouabain. C, Plasma membrane potential (Δψp) in M0 BMDMs. Δψp is expressed as relative change (%) of fluorescent units (RFU) at 516 nm. n=18 cells from 3 mice/group. d-e, Intracellular Ca2+ release in BMDMs under baseline conditions and after stimulation with ouabain (1 mM), MIP-1β (10 nM), RANTES (10 nM), isoproterenol (ISOP) (1 μM), andanamide (AEA) (10 μg/mL) or lysophosphatidic acid (LPA) (1 μM); n=3 mice/group. Arrow represents time point at which stimuli were injected. f, Intracellular Ca2+ release at basal level and after stimulation with MIP-1 β (10 nM), RANTES (10 nM), isoproterenol (ISOP) (1 μM), andanamide (AEA) (10 μg/mL) or lysophosphatidic acid (LPA) (1 μM) in BMDMs of wild type mice transfected with siRNA targeting the corresponding cognate receptor (grey) or control siRNA (ctrl) (red); n=3 mice/group. Arrow represents time point at which stimuli were injected. g, Immunofluorescence detection of ATP1A1 (red) showing equal expression and localization on the cell membrane of BMDMs of Gpr35+/+ and Gpr35–/– mice. Nuclei stained with DAPI (blue). Scale bar, 25μm. Representative images (left panels); n=3 mice per group. Quantification of membrane-bound (non-permeabilized) and cytoplasmic (permeabilized) ATP1A1 of BMDMs by FACS (right panels); n=3 mice/group. h-i, Western blot detection of ATP1A1 and p-ATP1A1 (Ser16) in BMDMs and colon; n=3 mice j, Intracellular 85Rb+ concentration (nM) measured via ICP-MS after 2 hours of incubation in 85Rb+ containing media in the absence or presence of 1 mM ouabain in human iPSC-derived M0 macrophages comparing GPR35108T (red), (n=6 sets of cells) and GPR35108M variant (black), (n=9 sets of cells); k, Basal intracellular calcium level in human iPSC derived M0 macrophages. n=7 individual experiments per group (3 technical replicates/individual experiment). l, Undirected migration of human iPSC derived M0 macrophages, n=7 individual experiments/group (2 technical replicates/individual experiment). m, Western blot of ATP1A1 in iPSC derived M0 macrophages. n=3 independent experiments. All data represented as mean ± s.e.m. Statistical significance was calculated by two-tailed Student’s t test using individual data points for 85Rb+ uptake and membrane potential assays and Mann Whitney U after Kruskal Wallis for intracellular Ca2+ measurements, * (p<0.05), ** (p<0.01).
GPR35 sets resting intracellular Ca2+ by promoting Na/K-ATPase function
Intracellular Ca2+ is critical for many signaling pathways and resting levels are controlled indirectly by the Na/K-ATPase (through the Na/Ca exchanger) (32). We therefore investigated whether GPR35 was involved in regulating this process. Cardiac glycosides such as ouabain or digoxin increase cardiac contractility by increasing intracellular Ca2+ through inhibition of Na/K-ATPase (33). Consistent with this effect, ouabain increased cytoplasmic Ca2+ in wild-type BMDMs to levels similar to that observed in Gpr35–/– BMDMs at baseline (Fig. 2D). In contrast, direct inhibition of Na/K-ATPase with ouabain did not further increase cytoplasmic Ca2+ in Gpr35–/– BMDMs (Fig. 2D). These data show that GPR35 maintains low intracellular Ca2+ levels by stimulating Na/K-ATPase.
We therefore hypothesized that absence of GPR35 may affect signaling through unrelated GPCRs that rely on triggering a Ca2+ response. The CCR5 ligands MIP-1β and RANTES, the β-adrenoreceptor agonist isoproterenol, the endocannabinoid anandamide and lysophosphatidic acid (LPA) triggered hardly any Ca2+ response in Gpr35–/– BMDMs, in contrast to the effect in wild-type cells (Fig. 2E). The increase in resting Ca2+ to maximum levels appeared to be specific for Gpr35 deletion, because siRNA-mediated knock-down (fig S2b) of mRNAs encoding other, unrelated GPCRs such as Ccr5, Adrb2 (which encodes the β2-adrenoreceptor), cannabinoid receptor 2 (Cnr2), and LPA receptor 2 (Lpar2) in BMDMs did not affect, or minimally lowered resting intracellular Ca2+ (Fig. 2F). Together, these data indicate that Ca2+ signaling is altered in the absence of GPR35 due to decreased Na/K-ATPase function, which blunts signaling through unrelated GPCRs.
The GPR35T108M polymorphism augments Na/K-ATPase activity
The activity of Na/K-ATPase is controlled both transcriptionally and post-translationally (25, 34, 35). We therefore investigated whether the effects of GPR35 deficiency might occur by regulating these processes. However, plasma membrane and intracellular pools of ATP1A1 (Fig. 2G) and total cellular ATP1A1 protein levels were similar between Gpr35–/– and wild-type BMDMs (Fig. 2H) and in colon epithelial scrapings (Fig. 2I). ATP1A1 phosphorylation, which has previously been associated with the recruitment of new pumps to the cell membrane (36, 37), was also not affected by Gpr35 genotype (Fig. 2, H and I). This finding suggested that the GPR35-dependent regulation of Na/K-ATPase was not mediated by transcription or post-translational control and thus that a protein-protein interaction might be involved.
We reasoned that if regulation of Na/K-ATPase activity by GPR35 is the physiologically critical function of this GPCR, GPR35T108M would affect ion transport. The human induced pluripotent stem cell (iPSC) line HPSI0114i-kolf_2 (‘KOLF2’) has been derived from a male individual of European descent and is homozygous for the protective C allele (encoding threonine at amino acid position 108). Using CRISPR/Cas9 gene editing, we introduced a single-nucleotide mutation in multiple derivative KOLF2 iPSC lines (‘KOLF2-108M’) to make them homozygous for the risk allele (ATG instead of ACG). We differentiated the parent and derivative KOLF2 lines toward macrophages (38) and subjected them to 85Rb+ uptake assays. KOLF2-108M macrophages exhibited significantly increased 85Rb+ uptake compared to parental KOLF2 macrophages (Fig. 2J), indicative of increased ion transport activity. Inhibition of Na/K-ATPase with ouabain decreased 85Rb+ uptake to similarly low levels in both cell lines (Fig. 2J). Consequently, basal intracellular Ca2+ levels were lower in KOLF-108M compared to KOLF-108T derived macrophages (Fig. 2K). Random migration of KOLF-108M macrophages was lower than of KOLF-108T-derived cells (Fig. 2L). As expected, this phenotype was not associated with altered levels of ATP1A1 protein in KOLF-108M compared to KOLF-108T macrophages (Fig 2M). Hence, changes in 85Rb+ transport, baseline Ca2+ levels and random migration in T108M carrying cells were reminiscent of GPR35 deficiency, demonstrating the hypermorphic nature of the GPR35T108M polymorphism.
GPR35 controls macrophage and intestinal epithelial cell metabolism
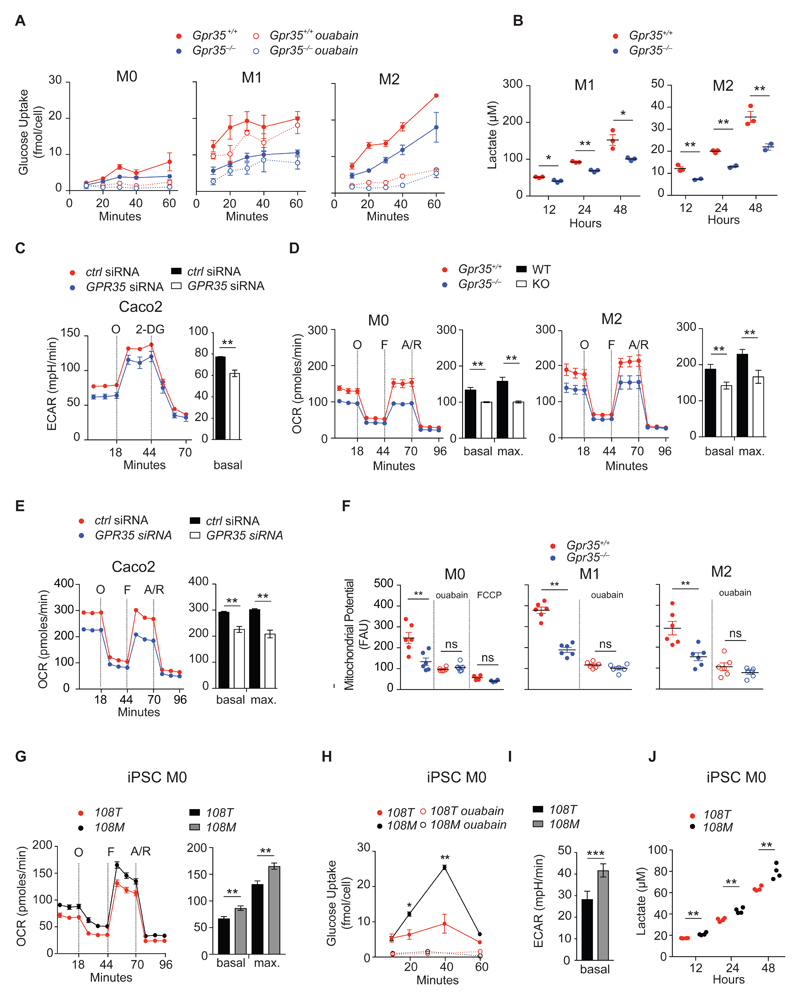
Na/K-ATPase activity accounts for ~30% of a cell’s overall energy consumption, which in some cell types is preferentially fueled by ATP regenerated by aerobic glycolysis (25, 39–45). We therefore hypothesized that reduced Na/K-ATPase activity in the absence of GPR35 might lead to decrease demand for glucose. Glucose uptake was indeed profoundly reduced in Gpr35–/– BMDMs compared to wild-type cells, irrespective of polarizing condition (Fig. 3A). Inhibition of Na/K-ATPase with ouabain almost completely abrogated glucose uptake in both wild-type and Gpr35–/– BMDMs differentiated under M0 or M2 conditions, whereas ouabain proportionally decreased glucose uptake in Gpr35+/+ and Gpr35–/– M1 BMDMs (Fig. 3A). Correspondingly, the production of lactic acid, the end product of aerobic glycolysis, was lower in Gpr35–/– compared to Gpr35+/+ BMDMs (Fig. 3B). This finding demonstrated that GPR35 promoted glucose uptake and oxidation in a Na/K-ATPase-dependent manner. In IECs, GPR35-silenced Caco2 cells exhibited lower extracellular acidification rates (ECAR) (Fig. 3C), which is a measure of aerobic glycolysis in metabolic flux studies, thus indicating that GPR35 also promoted glycolysis in this cell type.
Figure 3. GPR35 controls macrophage and intestinal epithelial cell metabolism.
A, Glucose uptake in M0, M1 and M2 BMDMs in the absence or presence of ouabain (1 mM). Glucose uptake expressed as intracellular 2-deoxy-D-glucose-6-phosphate (2DG6P) concentration detected per cell; n=3 mice/group. B, Lactate production measured in M1 and M2 BMDMs; n=3 mice/group. C, ECAR measured during sequential injection of oligomycin (O) and 2-deoxyglucose (2-DG) in Caco2 cells transfected with control (ctrl) siRNA (red) or GPR35 siRNA (blue). Bar chart shows basal ECAR; n=3 mice/group. D, Cellular respiration measured as basal and maximal oxygen consumption rate (OCR) during sequential treatment with oligomycin (O), FCCP (F) and antimycin/rotenone (A/R) in M0 and M2 BMDMs. Bar chart indicates basal and maximal OCR after FCCP injection; n=3 mice/group. E, Basal and maximal OCR measured during sequential treatment with oligomycin (O), FCCP (F) and antimycin/rotenone (A/R) in Caco2 cells transfected with either control (ctrl) siRNA (red) or GPR35 siRNA (blue). Bar chart indicates basal and maximal OCR after FCCP stimulation; n=3 independent experiments/group. F, Mitochondrial membrane potential (ΔΨm) in M0, M1 and M2 BMDMs in the absence or presence of 1 mM ouabain. Depolarisation with 1 μM FCCP used as a control. ΔΨm expressed as relative fluorescent units (RFU) at 590 nm; n=6 mice/group. G, Glucose uptake in human iPSC derived M0 macrophages in the absence or presence of ouabain (1 mM); n=3 independent experiments. H, Basal ECAR measured in human iPSC derived M0 macrophages; n=8 sets of cells/group. I, Lactate production in human iPSC derived M0 macrophages; n=3-4 sets of cells/group. J, Basal and maximal OCR during sequential treatment with oligomycin (O), FCCP (F) and antimycin/rotenone (A/R) in iPSC-derived human macrophages homozygous for the GPR35108T (red) or GPR35T108M (black) alleles. Bar chart indicates basal and maximal OCR after FCCP stimulation; n=8 sets of cells/group. All data represented as mean ± s.e.m. Statistical significance was calculated by two-tailed Student’s t test using individual data points, * (p<0.05), ** (p<0.01).
Oxygen consumption rate (OCR), a measure of mitochondrial oxidative phosphorylation (OXPHOS) (46) was also reduced in Gpr35–/– M0 and M2 BMDMs compared to wild-type BMDMs (Fig. 3d). Specifically, both basal and maximum OCR were lower in Gpr35–/– M0 and M2 BMDMs, whereas OCR was low in M1 BMDMs of either genotype (fig. S3) as expected (47). GPR35 also augmented OXPHOS in Caco2 cells as revealed through Gpr35 silencing (Fig. 3E). Consistent with impaired OXPHOS, mitochondrial membrane potential (ΔΨm) was reduced in Gpr35–/– M0 and M2 BMDMs compared to their Gpr35+/+ controls (Fig. 3F). This effect depended on Na/K-ATPase pump function, because ouabain reduced ΔΨm to similarly low levels as achieved by complete dissipation of the mitochondrial electrochemical proton gradient with FCCP (Fig. 3F). Together, these results showed that augmentation of Na/K-ATPase activity by GPR35 involved both aerobic glycolysis and also oxidative phosphorylation to meet energetic demands, highlighting the effect of this receptor on energy metabolism.
We next sought to understand the effect of the disease-associated SNP on this effect. Glucose uptake in KOLF-108M-derived macrophages was increased compared to KOLF-108T cells (Fig 3G). Consistent with a higher energetic demand of KOLF-108M- compared to KOLF-108T-derived cells, macrophages exhibited a higher basal ECAR (Fig. 3H) and increased lactate production (Fig. 3I). KOLF-108M-derived macrophages also exhibited increased baseline and maximum OCR compared to KOLF2-108T-derived cells (Fig. 3J). These data are consistent with the hypermorphic nature of the GPR35T108M variant and demonstrate that this variant increases energy demand because of increased Na/K-ATPase activity.
GPR35 modulates Na/K-ATPase-dependent signaling
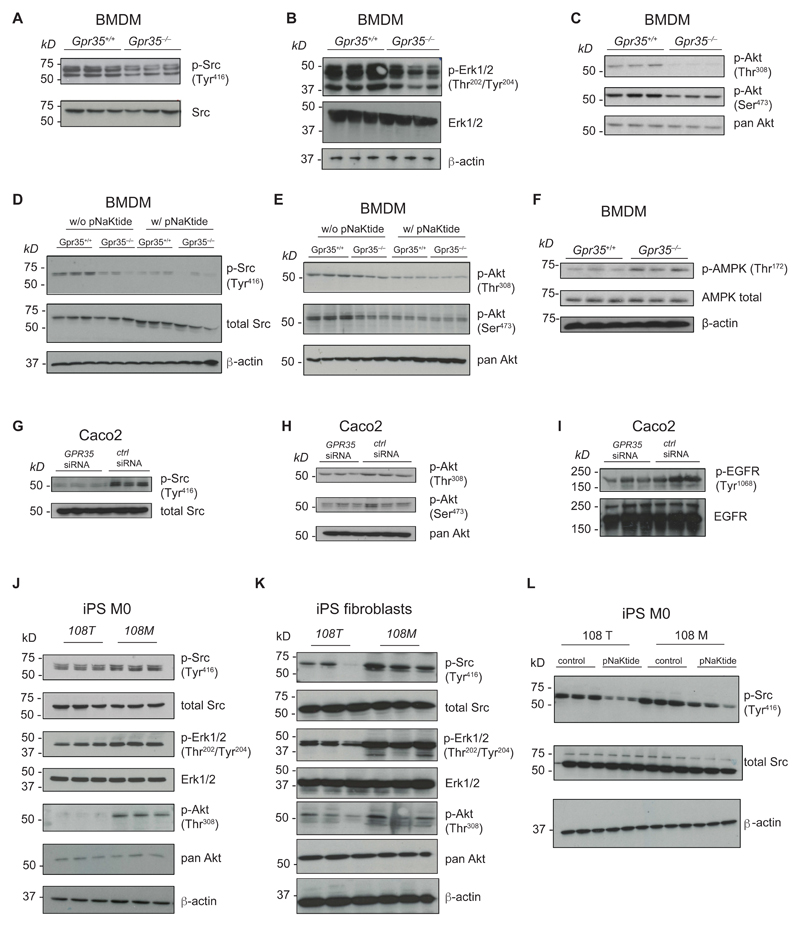
Na/K-ATPase is also an important signaling scaffold because ATP1A1 binds a portion of cellular Src (25, 28, 48). Src is a member of the Src family non-receptor protein tyrosine kinases and plays a critical role in signal transduction of many extracellular stimuli such as cytokines, growth factors, and stress responses (49). Binding of cardiotonic steroids such as ouabain to Na/K-ATPase activates Src (25, 50–54). We therefore assessed whether GPR35 modulated basal Src activity in a Na/K-ATPase dependent manner. Indeed, Gpr35–/– BMDMs exhibited decreased phosphorylation of Src Tyr416 compared to Gpr35+/+ BMDMs (Fig. 4A). Phosphorylation at Tyr416 in the activation loop of the kinase domain activates Src and thereby induces down-stream signaling. (55).
Figure 4. GPR35 modulates Na/K ATPase dependent signaling.
A-C, Western Blot detection of total Src, p-Src (Tyr416), p-Erk1/2 (Thr202/Tyr204), pan-Akt, p-Akt (Thr308) and p-Akt (Ser473) in BMDMs. n=3 mice/genotype. D-E, Western Blot detection of total Src, p-Src (Tyr416), pan-Akt, p-Akt (Thr308) and p-Akt (Ser473) in BMDMs with or without treatment with pNaKtide (1 μM). n=3 mice. f, Detection of total AMPK and p-AMPK Thr172 in BMDMs. n=3 mice/genotype. G-I, Western Blot detection of total Src, p-Src (Tyr416), pan-Akt, p-Akt (Thr308), p-Akt (Ser473), EGFR and p-EGFR (Tyr1068) in Caco2 cells transfected with either control (ctrl) siRNA or GPR35 siRNA. n=3 independent experiments/group. J-K, Western blot detection of total Src, p-Src (Tyr416), p-Erk1/2 (Thr202/Tyr204), total Erk1/2, p-Akt (Thr308) and pan-Akt in human iPSC derived M0 macrophages (J) and human iPSC derived fibroblasts (K). n=3 independent experiments/group. L, Western blot detection of p-Src (Tyr416) and total Src in human iPSC derived M0 macrophages in the absence (control) or presence of treatment with pNaktide (1 μM). n=3 independent experiments/group.
Na/K-ATPase-dependent activation of Src can transactivate the epidermal growth factor receptor (EGFR) and mitogen-activated protein kinases (MAPK), phospholipase C γ (PLC-γ) and the IP3 receptor, and phosphoinositol-3-kinase (PI3K) and Akt (25, 56–60). In Gpr35–/– BMDMs, phosphorylation of Thr202/Tyr204 of the MAPK extracellular signal-regulated kinases (ERK)1/2 was reduced compared to wild-type BMDMs (Fig. 4B). Phosphorylation of ERK at these residues fully activates the kinase (61). Phosphorylation of Akt Thr308 and Ser473 were absent and substantially reduced, respectively, in Gpr35–/– compared to wild-type BMDMs (Fig. 4C), suggesting lower glycolysis and less proliferative signaling in cells lacking GPR35. Baseline phosphorylation of Src and consequently Akt in wild-type BMDMs depended on the Na/K-ATPase – Src complex, because it was almost completely inhibited by pNaKtide (Fig. 4, D and E), a cell-permeable selective inhibitor of Na/K-ATPase-dependent activation of Src that does not affect cytoplasmic Src (62). Furthermore, pNaKtide did not further reduce the low levels of residual Src and Akt phosphorylation observed in Gpr35–/– BMDMs (Fig. 4DE). Consistent with reduced Akt Thr308 and Ser473 in Gpr35–/– BMDMs, the phosphorylation of Thr172 in 5'-adenosine monophosphate-activated protein kinase (AMPK) was increased in Gpr35–/– compared to wild-type BMDMs (Fig. 4F), indicating perturbed energy homeostasis in the absence of GPR35. Phosphorylation of AMPK at Thr172 which is located in the activation chain of its α subunit is required for its activation and is frequently induced by the kinases LKB1 or IκBK (63–65).
The strong baseline phosphorylation of Tyr416 in Src in control-silenced Caco2 IECs was almost absent in GPR35-silenced cells (Fig. 4G). Correspondingly, Akt Thr308 and Ser473 phosphorylation was reduced in GPR35-silenced Caco2 compared to control-silenced cells (Fig. 4H). Furthermore, activating EGFR Tyr1068 phosphorylation was reduced in Caco2 cells with knock-down of GPR35 compared to control cells (Fig. 4I). These results indicate that regulation of these phosphorylation events by GPR35 extended beyond macrophages to IECs. Furthermore, these data suggest that GPR35 maintains a baseline tone of Src activation by the Na/K-ATPase-associated plasma membrane pool of Src. GPR35 thereby determines the degree of baseline activation of cardinal signaling nodes such as Akt, ERK1/2, and EGFR.
Human macrophages and fibroblasts differentiated from KOLF2-108M iPSCs displayed increased Src Tyr416 phosphorylation compared to their corresponding KOLF2-108T-derived cells (Fig 4, J and K). pNaKtide inhibited Src Tyr416 phosphorylation in both KOLF2-108M and KOLF2-108T iPSCs-derived macrophages (Fig. 4L), which demonstrated the involvement of Na/K-ATPase in Src activation. Corresponding to increased Src Tyr416 phosphorylation, ERK1/2 Thr202/Tyr204 and Akt Thr308 phosphorylation was increased in KOLF-108M- compared to KOLF-108T-derived macrophages and fibroblasts (Fig. 4, J and K). Collectively, these findings demonstrate that the GPR35 T108M risk variant that is associated with risk for UC and PSC augments Src, ERK1/2 and Akt signaling in a Na/K-ATPase-dependent manner. Hence, GPR35 modulation of Na/K-ATPase-dependent signaling extends to many cell types expressing this receptor, is conserved between species, and is independent of GPR35’s cognate ligand(s).
GPR35 maintains IEC turn-over under homeostatic conditions
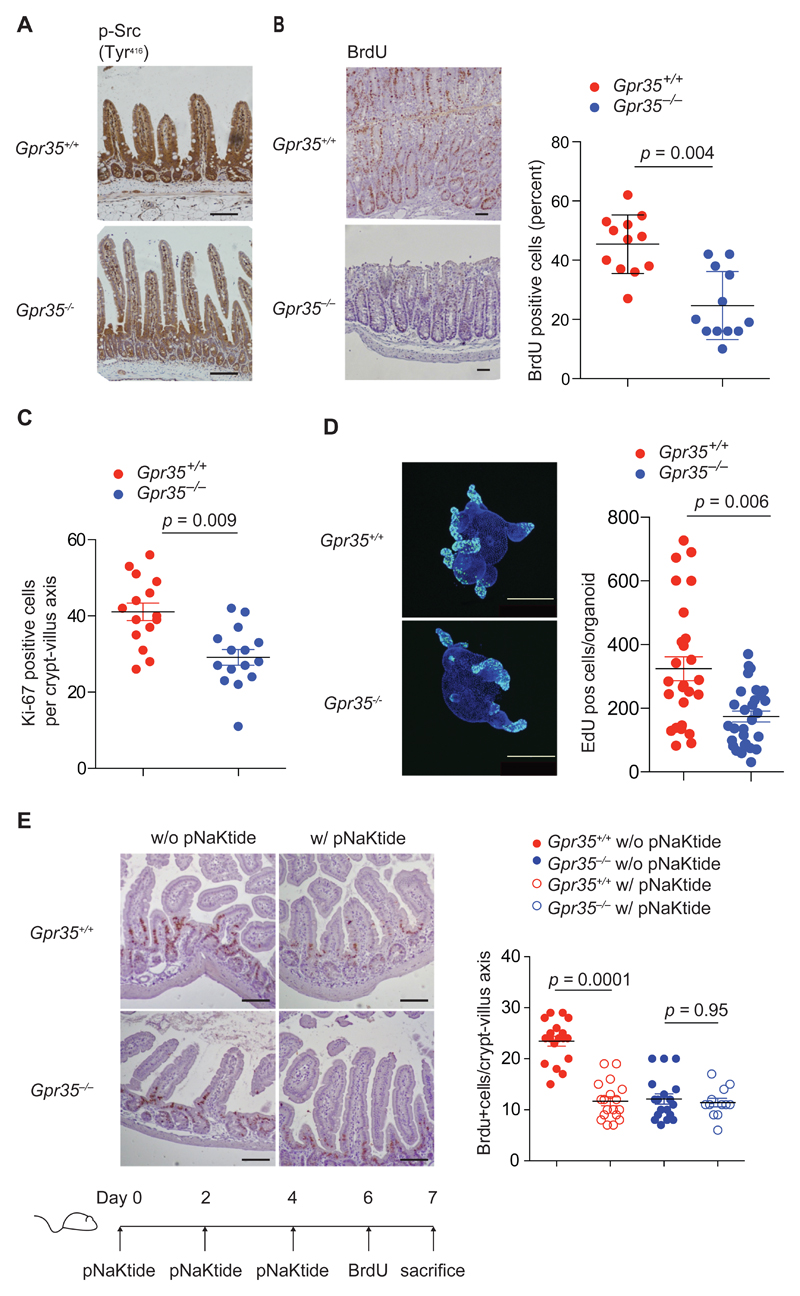
EGFR/Src−Ras−ERK and PI3K1A−PDK−Akt signaling are critically involved in proliferation of many cell types (57–60, 66). The intensity of immunostaining for phospho-Tyr416 Src was reduced in the intestinal epithelium of Gpr35–/– compared to that of wild-type mice (Fig. 5A), consistent with our observations in Caco2 cells (Fig. 4G). Hence we investigated the role of GPR35 in the baseline turn-over of IECs. The number of IECs that had migrated along the crypt-villous axis after a 24h pulse of BrdU was reduced by more than 50% in Gpr35–/– compared to wild-type mice (Fig. 5B). This effect corresponded to a comparable decrease in the number of IECs staining for the proliferation marker Ki-67 in Gpr35–/– compared to wild-type mice (Fig. 5C). Colonic intestinal organoids derived from Gpr35–/– mice exhibited similarly decreased proliferation as determined by fewer cells incorporating EdU compared to cultures derived from wild-type mice (Fig. 5D). These data demonstrated a ~40% reduction in the turn-over of the intestinal epithelium in the absence of GPR35. Moreover, administration of pNaKtide (67) resulted in a reduction in IEC turn-over in wild-type mice to levels observed in vehicle-treated Gpr35–/– mice, whereas pNaKtide did not cause any further reduction in GPR35-deficient mice (Fig. 5E). Together, these data identify a Na/K-ATPase–Src-dependent mechanism through which GPR35 promotes the homeostatic renewal of the intestinal epithelium.
Figure 5. GPR35 maintains intestinal epithelial cell turnover under homeostatic conditions.
a, Immunohistochemistry of p-Src (Tyr416) in intestinal sections from Gpr35+/+ and Gpr35–/–mice. Representative images of 6 mice per group. Scale bars represent 100 μm. b, Immunohistochemistry of BrdU incorporation after a 24 hour pulse in intestinal sections from Gpr35+/+ and Gpr35–/– mice. Representative images (left panel). Quantification of cells staining positive for BrdU (right panel). Each data point represents one crypt villus axis from 3 individual mice per group. c, Quantification of cells staining positive for Ki-67 using immunohistochemistry in intestinal sections from Gpr35+/+ and Gpr35–/– mice. Each data point represents one crypt villus axis; n=3 mice per group. d, Representative immunofluorescent images of colonic organoid cultures stained with EdU (green) for proliferative cells (left panel). Quantification of EdU positive cells in Gpr35+/+ and Gpr35–/– organoids (right panel). Each data point represents one organoid. Scale bars, 250 μm; n=3 mice per group. e, Representative images showing immunohistochemistry of BrdU incorporation in intestinal sections of Gpr35+/+ and Gpr35–/– mice after treatment with pNaKtide. Scale bars 100 μm (left panel). Timeline of pNaKtide and BrdU injections shown below. Quantification of cells staining positive for BrdU using immunohistochemistry (right panel). Each data point represents one crypt villus axis; n=3 mice per group. All data represented as mean ± s.e.m. Statistical significance was calculated by Mann Whitney U after Kruskal Wallis.
Loss of GPR35 protects from intestinal tumorigenesis
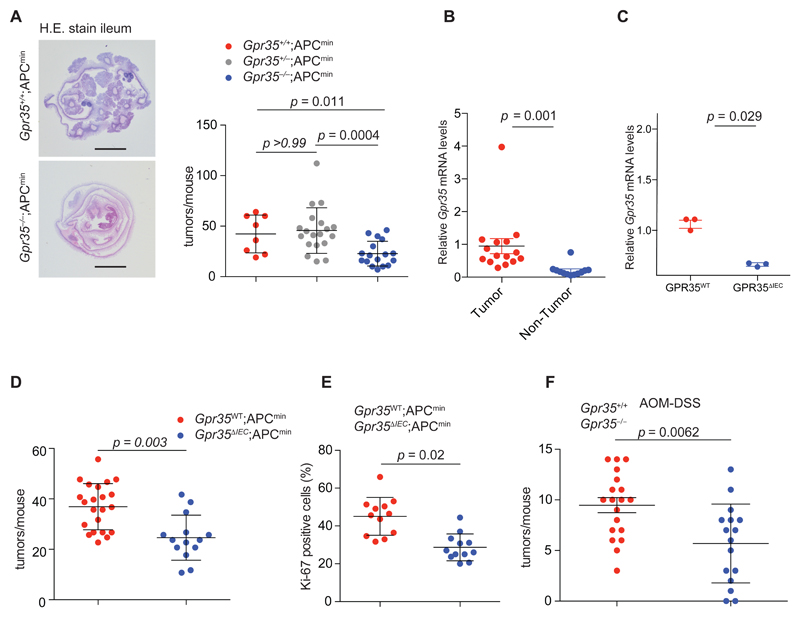
Based on the effects of GPR35 on IEC turnover, we hypothesized that GPR35 might also affect intestinal tumor development. We crossed Gpr35–/– mice with Apcmin mice, which carry a hypomorphic variant of Apc, whose human orthologue APC causes adenomatous polyposis coli and is a major early driver of spontaneous colorectal cancer (CRC) (68, 69). Gpr35–/–;Apcmin mice developed 40% fewer intestinal tumors (namely non-invasive adenomas), compared to Apcmin mice with intact GPR35 (Fig. 6A), indicating that GPR35 plays an important role in intestinal tumor promotion. GPR35 is highly abundant in IECs. Furthermore Gpr35 mRNA expression was significantly higher in tumors compared to healthy intestinal tissue (Fig. 6B). We hence predicted an IEC-intrinsic role of GPR35. We therefore generated mice with a floxed Gpr35 allele (‘Gpr35flox’), and crossed them with IEC-specific Cre recombinase transgenic mice (‘Vil-Cre’) (70) and the Apcmin line to obtain Vil-Cre;Gpr35flox/flox;Apcmin mice (‘Gpr35ΔIEC;Apcmin’) and Vil-Cre-negative GPR35-wild-type littermates (‘Gpr35WT;Apcmin’). Gpr35 mRNA expression was significantly reduced or not detectable in Gpr35ΔIEC mice (Fig 6C). At week 15, Gpr35ΔIEC;Apcmin mice had developed 30% fewer tumors compared to Gpr35WT;Apcmin mice (Fig. 6D). Correspondingly, the percentage of proliferative cells staining positive for Ki-67 were significantly lower in Gpr35ΔIEC;Apcmin compared to Gpr35WT;Apcmin mice (Fig. 6E).
Figure 6. Loss of GPR35 protects from intestinal tumorigenesis.
A, Haemotoxylin and eosin (H&E) stain of ileal swiss rolls from Gpr35+/+;Apcmin and Gpr35–/–;Apcmin mice (left). Total number of intestinal tumors in 15-week-old Gpr35+/+;Apcmin (red, n=8), Gpr35+/–;Apcmin (grey, n=19) and Gpr35–/–;Apcmin (blue, n=17) littermates (right). Scale bar, 5 mm. B, Gpr35 mRNA expression of tumor tissue compared to healthy intestinal tissue. n=15 tissue samples/group. C, mRNA expression in Gpr35+/+ mice compared to Gpr35 ΔIEC mice, n=3 mice/group. D, Total number of intestinal tumors in 15-week-old Gpr35WT;Apcmin (red, n=22) and Gpr35ΔIEC;Apcmin (blue, n=14) littermates. E, Quantification of cells staining positive for Ki-67 using immunohistochemistry in intestinal sections of 15-week-old Gpr35WT;Apcmin (n=12) and Gpr35 ΔIEC;Apcmin (n=12) littermates. F, Number of tumors in the large intestine of Gpr35+/+ (red, n=19) and Gpr35–/– (blue, n=16) mice in the AOM-DSS model. All data represented as mean ± s.e.m. Statistical significance was calculated using the Mann Whitney U test after Kruskal Wallis one-way ANOVA.
In contrast to sporadic CRC, APC mutations are less common in colitis-associated cancer (CAC), which is instead characterized by field cancerization and a distinct mutational spectrum (71–73). CAC can be modelled in mice by administration of the carcinogen azoxymethane (AOM) followed by 3 cycles of the barrier-breaching chemical dextran sodium sulfate (DSS) (74, 75). Gpr35–/– mice subjected to AOM-DSS developed 35% fewer colorectal tumors compared to littermate wild-type and Gpr35+/+ mice (Fig. 6F). Together, these data demonstrate that GPR35 promotes intestinal tumorigenesis in both, spontaneous and inflammation-driven tumor models.
Pharmacological targeting of GPR35 selectively modulates receptor activity
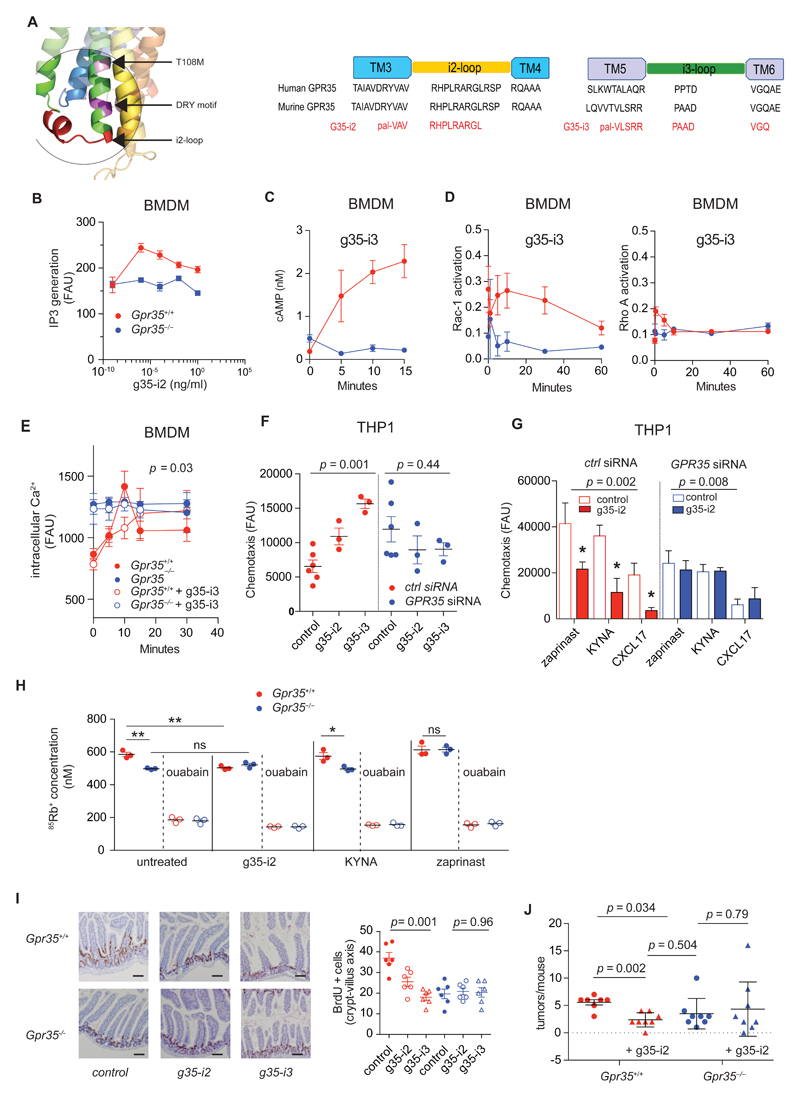
Short lipid-coupled peptides (‘pepducins’) with sequence identity to intracellular loops can selectively activate or inhibit GPCRs as has been demonstrated for several classes of G protein coupled receptors (76). The GPR35T108M polymorphism and adjacent DRY motif are close to the i2 loop of GPR35 (Fig. 7A). We therefore designed a series of palmitoylated peptides that targeted the i1, i2, or i3 loop and initially screened them in wild-type compared to Gpr35–/– BMDMs for their effect on Ca2+ and chemotactic responses (fig S4, A and B). g35-i2 and g35-i3, which target the i2 and i3 loop, respectively, were characterized further (Fig. 7A). g35-i2 triggered a transient increase in IP3 in wild-type BMDMs, which was absent in Gpr35–/– BMDMs (Fig. 7B). Similarly, g35-i3 triggered cAMP production (Fig. 7C), and transient Rac-1 and RhoA activation (Fig. 7D) in wild-type, but not in Gpr35–/– BMDMs. Both pepducins increased intracellular Ca2+ over time in wild-type BMDMs, which reached the higher baseline Ca2+ levels in Gpr35–/– cells, without affecting intracellular Ca2+ in Gpr35–/– cells (Fig. 7E). g35-i2 and g35-i3 were active across species because both induced a chemotactic response in human THP1 cells transfected with control siRNA, but not in those transfected with GPR35 siRNA, hence demonstrating specificity (Fig. 7F). To assess whether g35-i2 could modulate the response to known agonists, we pre-incubated THP1 cells with pepducin before adding KYNA, CXCL17 and zaprinast. g35-i2 blocked migration to these agonists in control-silenced, but not in GPR35-silenced THP1 cells (Fig. 7G). We were interested to explore whether pepducins would affect Na/K-ATPase-dependent ion transport. Indeed, g35-i2 reduced ouabain-inhibitable 85Rb+ transport in wild-type BMDMs to levels similar to those in untreated Gpr35–/– BMDMs, whereas no further reduction in 85Rb+ transport was detected in Gpr35–/– BMDMs when incubated with g35-i2 (Fig. 7H). In contrast, KYNA or Zaprinast did not affect 85Rb+ uptake in wild-type BMDMs, and the difference in ion transport between wild-type and Gpr35–/– cells was maintained in KYNA-treated cells (Fig. 7H). Zaprinast increased 85Rb+ uptake in Gpr35–/– BMDMs (Fig. 7H), an effect that may be related to its promiscuity and clinical use as a phosphodiesterase inhibitor (23). Together, these data show that g35-i2 inhibits Na/K-ATPase ion transport function, which distinguishes this agonist from previously reported GPR35 agonists.
Figure 7. Pharmacological targeting of GPR35 selectively modulates receptor activity and decreases tumor burden in colitis-associated cancer.
A, Structural model of GPR35 focused on the second intracellular loop (left panel). The position of the T108M variant is marked in pink and the DRY motif is marked in purple. The g35-i2 pepducin targeted region is marked in red (i2-loop). The amino acid sequence of g35-i2 (middle panel) and g35-i3 (right panel) pepducins mapped to human and murine GPR35 i2 and i3 loops. B, Dose dependent IP3 generation in BMDMs after stimulation with g35-i2 pepducin; n=3 mice/group. C, cAMP production in BMDMs after stimulation with g35-i3 pepducin; n=3 mice/group. D, Activated Rac-1 and Rho A levels in BMDMs after stimulation g35-i3 pepducin; n=3 mice/group. E, Intracellular Ca2+ release in BMDMs basally and after stimulation with g35-i2 (closed circles) or g35-i3 pepducin (open circles); n=3 mice/group. F, Chemotaxis of THP-1 cells transfected with either GPR35 siRNA Rac-1 siRNA (ctrl) stimulated with g35-i2 or g35-i3 pepducin. Untreated cells are used as a control. G, Directed migration of THP-1 cells transfected with either GPR35 siRNA (blue) or control siRNA (ctrl) (red) towards zaprinast (10 μM), KYNA (300 μM) or CXCL17 (200 ng/mL) in the presence (closed bars) or absence (open bars) of g35-i2 pepducin. n=3 independent experiments/group. H, Intracellular 85Rb+ concentration (nM) measured by ICP-MS in the absence or presence of 1 mM ouabain in BMDMs after 1 hour of incubation in 85Rb+ containing media in the presence of g35-i2 pepducin, KYNA (300 μM) or zaprinast (10 μM); n=3 mice/group. I, Representative images showing immunohistochemistry of BrdU incorporation in intestinal sections of Gpr35+/+ and Gpr35–/– mice after treatment with g35-i2 (5 mg/kg) or g35-i3 (5mg/kg) pepducins (left panel). Quantification of cells staining positive for BrdU using immunohistochemistry (right panel). Each data point represents one crypt villus axis; n=3 mice per group. Scale bar represents 100μm. J, Number of colonic tumors in vehicle treated (n=7) or g35-i2 pepducin treated (n=8) Gpr35+/+ (red) mice compared to vehicle treated (n=8) or g35-i2 pepducin treated (n=8) Gpr35–/– (blue) mice in the AOM-DSS model. All data represented as mean ± s.e.m. Statistical significance was calculated using Mann Whitney U after Kruskal Wallis. * (p<0.05), ** (p<0.01).
g35-i2 decreases tumor burden in colitis-associated cancer
We next investigated whether g35-i2 and g35-i3 would affect homeostatic turn-over of IECs in vivo. Intraperitoneal administration of either pepducin reduced BrdU+ cells along the crypt-villus axis after a 24h pulse of BrdU in wild-type mice to levels observed in vehicle-treated Gpr35–/– mice (Fig. 7I). In contrast, g35-i2 and g35-i3 did not affect IEC turn-over in Gpr35–/– mice (Fig. 7I), thereby also demonstrating specificity of these pepducins in vivo. Finally, we chose the g35-i2 pepducin to explore GPR35 as a pharmacological target in CAC. Wild-type and Gpr35–/– mice undergoing the AOM/DSS model were administered g35-i2 intraperitoneally, starting with the first cycle of DSS. g35-i2 reduced tumor numbers by 57.4% in wild-type mice, which is a reduction similar to that observed in vehicle-treated Gpr35–/– mice (Fig. 7J). In contrast, no additional decrease was observed in g35-i2-treated Gpr35–/– mice (Fig. 7J). Thus, GPR35-selective pepducins inhibit IEC turn-over and reduce tumor burden in experimental CAC.
Discussion
Here we identified GPR35 as a key promoter of Na/K-ATPase ion pumping function and Src signaling activity. This property did not apply to other GPCRs we tested and did not require cognate ligand engagement. It appeared to be distinct from canonical signaling that is triggered by the proposed agonists of GPR35. These ligand-independent GPR35 activities on Na/K-ATPase function were enhanced with the disease risk-associated GPR35T108M variant. GPR35-dependent alterations in Na/K-ATPase ion pump function re-set baseline intracellular Ca2+ levels; in the absence of GPR35, baseline Ca2+ levels were increased to levels otherwise only observed upon receptor triggering, whereas baseline Ca2+ levels were lowered with the GPR35T108M variant. Through Na/K-ATPase, GPR35 also set baseline Src kinase activity, which was lowered in the absence of GPR35, and increased by the GPR35T108M variant. Consequently, signaling induced by various ligands unrelated to GPR35 that trigger Ca2+ and Src activation were affected by GPR35.
We showed that through these mechanisms, GPR35 potently promoted intestinal epithelial turnover and intestinal tumourigenesis. Elegant studies in Drosophila and mice with conditional epithelial deletion has revealed that Src drives intestinal stem cell proliferation through upregulation of EGFR and activation of Ras/MAPK and Stat3 signaling (66). Although mammalian SFKs Src, Fyn and Yes are redundant for intestinal homeostasis, Src has a non-redundant role in damage-induced intestinal regeneration (66, 77). Compared to normal epithelium, Src expression is increased in ~80% of human CRCs and associated with poor prognosis (78, 79), and putative activating Src mutations are common (80). In addition to its long-appreciated role in advanced tumor stages and metastasis (81), Src is also required for early intestinal tumorigenesis caused by APC mutation (66), which may be of particular importance in the tumor-prone epithelium in UC and PSC. Nonetheless, the protective effect of Gpr35 deletion in the CRC Apcmin and the AOM/DSS CAC model was likely not only a consequence of reduced Na/K-ATPase-dependent Src signaling, but may also be determined by an altered Ca2+ signaling threshold and reduced metabolic activity present in GPR35-deficient intestinal epithelium. In this context, GPR35-related activation of Akt, which plays an evolutionarily conserved role in cellular metabolism, proliferation, and cell survival (82), may similarly contribute to the tumor-promoting ability of GPR35.
Direct protein-protein interaction is important in regulating Na/K-ATPase activity although the precise mechanisms remain mostly unresolved and diverse domains within ATP1A1 have been implicated (25) (83) (84). It will be important to elucidate the structural basis of Na/K-ATPase modulation by GPR35, including its mode of tonic activation of Src. Finally, it remains conceivable that endogenous GPR35 ligands, different from those that induce canonical signaling, may exist that modulate GPR35’s Na/K-ATPase-promoting activity.
Targeting GPR35 therapeutically might therefore be attractive, especially in patients at high risk for cancer such as in UC and PSC, in which GPR35T108M is the strongest genetic risk factor outside the MHC. Here we characterized GPR35-specific pepducins that blocked both agonist-triggered GPR35 activation and also GPR35-mediated promotion of Na/K-ATPase ion transport and signaling activity. In contrast to agonist or antagonist binding to the orthosteric binding pocket of GPCRs, pepducins translocate across the cell membrane and target the intracellular surface of the receptor (76, 85, 86). Pepducins can allosterically modulate receptor signaling and can also function in a biased manner to selectively promote the interaction with specific G proteins over others, G protein receptor kinases, and β-arrestins (76, 87, 88), although the precise mechanism of action remains unclear. The effect of g35-i2 on Na/K-ATPase suggests that pepducins may also block the interaction of GPCRs with transmembrane proteins. A first-in-human study of a pepducin (PZ-128) has shown promising results for proteinase activated receptor 1 (PAR1) blockade in acute coronary syndrome (89). In conclusion, studying in-depth a single ‘risk gene’ of two complex inflammatory diseases that are tumor-prone, we discovered a GPCR that directly promotes a fundamental process of a cell, established hypermorphic function of the variant associated with disease risk, characterised its implications for tumor biology, and developed a candidate therapeutic that is effective in vivo.
Materials and Methods
Mice
Ear-biopsy genomic DNA was used for routine genotyping of all mice. All mice were bred and maintained in specific pathogen-free conditions at the Central Biomedical Services (CBS) facility, University of Cambridge. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted and all were conducted with approval of the UK home office. Gpr35-/- mice were obtained from the KOMP repository (clone ID 646854). Conditional knock-out mice were generated by outbreeding the FlpO recombinase mice and then breeding in VillinCre transgene positive mice. Mice littered at Mendelian ratios and developed normally without any spontaneous disease emerging under specific pathogen free conditions. Six- to twelve-week-old mice that were age and sex matched as described in the relevant methods sections were used for all experiments unless otherwise stated.
ApcMin/+ model of sporadic colon cancer
ApcMin/+ mice were obtained from the Jackson Laboratory and intercrossed with mice lacking Gpr35 globally or conditionally in their intestinal epithelial cells. At the age of 15 weeks mice were sacrificed and after flushing with PBS the intestine was longitudinally cut, formalin fixed and tumor count was assessed microscopically. Samples were embedded as swiss rolls in either paraffin or OCT for further analysis.
AOM/DSS model of colitis associated cancer
6-8-wk-old mice were injected intraperitoneally with 12.5 mg/kg AOM (Sigma-Aldrich). Colitis was induced by two cycles of 2.5% DSS (MP Biomedicals) in drinking water for 5 days, followed by a 16 day tap water period (75). The final DSS cycle (2%) was administered for 4 days, followed by a 10 day tap water period. Tumor count and tumor area were determined at day 61 microscopically in longitudinally cut and formalin fixed specimen.
Murine colonic crypt derived organoids
The colon was flushed with cold PBS, cut longitudinally and into 5 mm pieces. To dissociate the crypts, pieces were placed in 25mM EDTA/PBS for 1h under constant gentle shaking at 4°C. Samples were washed multiple times with cold PBS followed by centrifugation at 1000 rpm. Isolated crypts were washed, counted and pelleted. A total of 200 crypts were mixed with 40 μl of Matrigel (Corning 354230) and plated in 24-well plates. After polymerization of Matrigel, 500 μl of IntestiCult Organoid Growth Medium (Stem Cell #06005) containing 100 ng/ml Wnt-3a (Peprotech 315-20) was added. Medium was changed every 3 days. For passaging, organoids were removed from Matrigel and mechanically dissociated into single-crypt domains, then transferred to fresh Matrigel. Passaging was performed once per week with a 1:2 split ratio.
EdU staining
For EdU incorporation, 40 μl of matrigel containing organoids was seeded in 8-well chamber-slides. 10 μM EdU was added to the wells 6 h prior to fixation. The proliferation marked by Edu incorporation was detected with Click-iT Edu Alexa Fluor 488 Imaging Kit (Invitrogen, C10337) according to the supplier’s instructions.
BrdU incorporation
2.5 mg BrdU was injected i.p 24 h before euthanizing mice. Paraffin embedded samples were then de-paraffinised in xylene and de-hydrated with ethanol. Cells positive for BrdU were detected using a BrdU In-Situ Detection Kit (BD Pharmingen, 550803). Proliferation was revealed as BrdU+ cells along the crypt–villus.
Crispr/Cas9 editing of human iPSC line
The rs3749171 mutation in the human GPR35 gene was generated by a single base substitution (C>T) using CRISPR/Cas9-induced homology directed repair in the KOLF2 human iPSC line. This was achieved by nucleofection of 106 cells with Cas9-crRNA-tracrRNA ribonucleoprotein (RNP) complexes. Synthetic RNA oligonucleotides (target site: 5’- CCTGGTCACGGCCATCGCCG -3’ or 5’ CACATAGCGGTCCACGGCGA 3’, 225 pmol crRNA/tracrRNA) were annealed by heating to 95°C for 2 min in duplex buffer (IDT) and cooling slowly, followed by addition of 122 pmol recombinant eSpCas9_1.1 protein (in 10 mM Tris-HCl, pH 7.4, 300 mM NaCl, 0.1 mM EDTA, 1 mM DTT), incubation at room temperature for 20 minutes, and addition of 500 pmol of a 100 nt ssDNA oligonucleotide (IDT Ultramer) as a homology-directed repair template to introduce the desired base change. After recovery, plating at single cell density and colony picking into 96 well plates, 480 clones were screened for heterozygous and homozygous mutations by high throughput sequencing of amplicons spanning the target site using an Illumina MiSeq instrument. Final cell lines were further validated by Sanger sequencing. Two independently targeted clones homozygous for 108T or 108M were used in downstream assays.
Bone marrow derived macrophages (BMDMs)
Mouse femurs and tibias were flushed with macrophage culture medium (RPMI-1640 containing 100 U/mL of penicillin-streptomycin, 1 mM HEPES pH 7.4 and 10% FBS) and the bone marrow was filtered through a 70 μm cell strainer. Cells were thereafter incubated in macrophage culture medium supplemented with M-CSF (100 ng/mL) for 6 days. BMDMs were the reseeded and polarized overnight toward M1 or M2 with IFN-γ (50 ng/mL) plus LPS (20 ng/mL) or with IL-4 (20 ng/mL), respectively.
iPSC culture and macrophage differentiation
The KOLF-2 human induced pluripotent stem cell line was maintained in mTeSR-E8 medium (Stemcell Technologies cat# 05990) on Vitronectin (rhVTN-N) coated plates (Gibco #A14700). For macrophage differentiation, pluripotent colonies were lifted using ReLeSR (Stemcell Technologies cat#05872) and plated onto irradiated MEFs on gelatin coated 100 mm tissue culture plates in Advanced DMEM-F12 (ThermoFisher Cat#12634028), 20% knock-out serum replacement (Gibco #10828028), 1% L-glutamine, 1% Penicillin/streptomycin and 7 μl/L 2-mercaptoethanol supplemented with 4 ng/mL rhFGF basic (R&D systems Cat#233-FB-025). Once colonies were large but not touching each other, they were lifted using 1mg/ml collagenase type IV (Gibco #17104-019) and 1 unit/mL dispase II (GIbco #17105-041) diluted in a 1:1 ratio in Advanced DMEM-F12. Detached colonies were gently added to a 15 mL tube using a 10 mL pipette to avoid breaking them up and were centrifuged at 300G for 3 minutes. After removing the supernatant, the colonies were gently re-suspended in fresh Advanced DMEM-F12 to wash and recentrifuged. This wash step was carried out a total of three times. Colonies were then transferred to non-adherent 100mm plates in 13ml of Advanced DMEM-F12 (ThermoFisher Cat#12634028), 20% knock-out serum replacement (Gibco #10828028), 1% L-glutamine, 1% Penicillin/streptomycin and 7μl/L 2-mercaptoethanol without FGF. After four days embryoid bodies had formed and these were collected in suspension, centrifuged and then plated in gelatin coated plates in X-Vivo-15 media (Lonza #BE02-060Q), 1% L-glutamine, 1% Penicillin/Streptomycin and 7μl/L 2-mercaptoethanol supplemented with 50 ng/mL human M-CSF and 0.5 mg/mL IL-3. Media was changed twice weekly. After 20-30 days macrophage precursors began to appear in the media and were collected in suspension, strained using a 40 μm cell strainer and plated on standard tissue culture plates in RPMI 1640, 10% FBS and 1% L-glutamine supplemented with 100 ng/mL M-CSF. After 7 days, mature macrophages were used for assays.
iPSC derived intestinal organoids
The KOLF-2 human induced pluripotent cell line was differentiated into intestinal organoids as previously described (90). Organoids were maintained in Matrigel as spheroids and passaged every 5-7 days for a minimum of 10 passages to allow maturation before being used in assays. For protein extraction, organoids were removed from matrigel by mechanical dissociation in cold cell recovery solution (Corning #354253). The suspension was transferred to a 15 mL tube and incubated on ice for 30 minutes. The suspension was then centrifuged and the pellet resuspended in 1 mL cold cell recovery solution. After a further centrifugation, the pellet was resuspended in cold PBS and transferred to a 1.5 mL microcentrifuge tube before being centrifuged at 20,000 G for 3 mins. The pellet was then resuspended in 80 μL 10mM Tris, 1% SDS, pH 7.4 supplemented with protease and phosphatase inhibitors. This resuspension was then incubated at 95 C for 30 mins before being sonicated for 5 seconds. Samples were then centrifuged and the supernatant used for western blotting.
To culture the organoids in a monolayer, tissue culture plates were first coated with 0.5% (v/v) Matrigel in Advanced DMEM F12 medium for 1 hour at 37°C. Organoids were then released from Matrigel and broken into fragments by mechanical dissociation in ice cold PBS. The suspension was then centrifuged at 300 G for 3 mins before being resuspended and incubated in TrypLE Express (Gibco #12605036) for 10 mins at 37 C with intermittent gentle agitation to avoid clumping. The single cell suspension was then centrifuged and resuspended in organoid growth media before being counted and seeded.
Intracellular Ca2+ assaysIntracellular Ca2+ was measured in BMDMs using Fluo-8 Calcium Flux Assay Kit (ABCAM, ab112129). In brief macrophages were incubated with inactive calcium dye which gets activated after penetrating the cell membrane. When active the dye binds to calcium released from the endoplasmatic reticulum and fluorescence was measured in a fluorescent plate reader with an excitation wavelength of 482nm and emission was measured at 530 nm.
IP3 assay
IP3 accumulation in macrophages was measured using the HitHunter® IP3 Fluorescent Polarization Assay (GE Healthcare, 90-0037-02). M0 macrophages were cultured in RPMI1640 containing 10% FBS and 100 ng/mL M-CSF, serum starved overnight and a series of different concentrations potential endogenous agonists were added to designated wells in a 96 well plate. The reaction was quenched after 1 minute and the tracer was added followed by an IP3 binding protein. Fluorescence was excited with a wavelength of 483 nm and emission was measured at 530 nm.
cAMP assay
Production of cAMP was measured using the cAMP-Glo Assay (Promega, V1501). In brief, 4×104 macrophages were seeded per well in a 96 well plate. Cells were treated with test compounds for the indicated time prior to lysis and detection of luminescence in a plate format luminometer. Values are given as relative luminescence units (RLU).
Rac1- and RhoA G-LISA activation assays
Overnight serum-starved BMDMs were exposed to test compounds for 5 sec, 30 sec, 5 min, 30 min and 60 min. Thereafter cells were immediately lysed in ice-cold lysis buffer containing protease inhibitors, sodium fluoride, sodium pyrophosphate, p-nitrophenyl phosphate and microcystin LR and G-LISA assays were performed according to the manufacturer’s instructions. Plates were read at 490 nm and values are given as extinction coefficient.
Migration and chemotaxis in vitro assays
Migration was measured in 48 well micro Boyden chambers equipped with a 5 μm–pore-size cellulose nitrate filter which separated the upper and the lower chambers (Neuroprobe, Gaithersburg, MD). BMDMs were resuspended in RPMI 1640, 0.5% BSA (1x106 cells/mL). After a migration period of 4 h, the nitrocellulose filters were dehydrated, fixed, and stained with hematoxylin. The migration depth of the cells into the filters was quantified by microscopy, measuring the distance (in micrometers) from the surface of the filter to the leading front of three cells. Data are expressed as a chemotaxis index, which is the ratio between the distance of directed migration and random migration of monocytes into the nitrocellulose filters.
Alternatively, THP1 cell migration was assessed using the Neuroprobe ChemoTx system (Neuroprobe, Gaithersburg, MD). THP1 cells were loaded with calcein-AM and then migrated towards pepducins or chemokines in the lower wells of the chamber. In inhibition experiments, THP1 cells were pre-incubated with the pepducins and then migrated towards chemokines or GPR35 agonists.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde containing either 0.2% or 1% Triton X-100 for 15 min at 20 °C or 100% methanol at −20 °C for 15 min. Fixed cells or deparaffinised slides were washed with PBS and non-specific binding was blocked with either 5% normal serum or 0.5% milk before incubation with primary antibodies for 1 h at 20 °C. Unbound antibody was removed by washing with PBS and secondary antibody was bound for 30 min at 20 °C or overnight at 4 °C. Coverslips were then mounted with Prolong mounting media containing DAPI. Fluorescence was visualized with a Leica SP5 confocal microscope.
Fluorescence resonance energy transfer (FRET)
For construction method of vectors encoding C-terminal Cerulean (Addgene #27795) or Venus (Addgene #27793) fluorescent protein tagged versions of GPR35-T108T, GPR35-T108M, ATP1A1, CCR5 and CXCR2 see Section Cloning and Constructs. 24 hours prior to transfection, HEK293T cells were seeded onto Poly-L-Lysine coated glass coverslips in 6-well plates at a density of 0.5 x 106 per well. 2500 ng plasmid DNA was transfected using Lipofectamine 2000 (ThermoFisher #11668027) in Opti-MEM Reduced Serum Media (ThermoFisher 31985062) according to the manufacturer’s protocol. Pairs of different constructs were mixed 1:1 prior to transfection. 24 hours post-transfection, cells were washed with PBS and fixed with 4% paraformaldehyde in 0.12 M sucrose for 15 minutes, followed by three further PBS washes. Coverslips were then lifted and mounted onto microscope slides. FRET measurements were performed using a Leica TCS SP5 confocal microscope (Leica Biosystems) and Leica Application Suite Advanced Fluorescence software. FRET measurements were taken using the acceptor photobleaching method. Briefly, cells expressing similar fluorescence levels of both transfected constructs were identified. Baseline cerulean “donor” emission was measured during excitation at 405nm wavelength at 10% laser power. The venus “acceptor” protein was then photobleached using 20 pulses of 514nm wavelength at 98% laser power so that it was no longer able to accept energy from the cerulean “donor”. The cerulean emission during excitation at 405nm wavelength at 10% laser power was then re-measured. FRET efficiency was then calculated from pre- and post-photobleaching emission values. Measurements from non-bleached cells within the same field were used to normalise.
Lactate Assay
Murine bone marrow derived macrophages and iPSC derived macrophages were plated in 96 well plates at a density of 50,000 cells per well. After 24 hours media was removed and replaced with 100 μL per well of RPMI, 1% L-glutamine, 1% Penicillin/Streptomycin and 10% dialysed FBS (Gibco #26400044). At set timepoints, 5 μL of media was then sampled and diluted in 95 μL PBS. Lactate concentration was measured using the Lactate-Glo Assay (Promega, J5021) according to the manufacturer’s instructions.
Immunohistochemistry
Paraffin sections were pre-treated with xylene and dehydrated in ethanol. Antigen retrieval was performed using citrate or EDTA buffer for 15 min at sub-boiling temperature in a microwave, followed by blocking of endogenous peroxidases activity. Primary antibody was incubated overnight at 4°C and secondary biotinylated antibody was detected with streptavidin-HRP (Vector Laboratories). Sections were developed using a DAB Peroxidase Substrate kit (Vector Laboratories).
FACS
BMDMs were seeded in V-well plates, centrifuged at 600 g 6 min and supernatant removed. Cells were then incubated with Fc-Block in FACS buffer for 15 min followed by incubation with the primary antibody (ATP1A1) in 50 μL FACS buffer for 1 hour at 4°C. Cells were then fixed in 2% paraformaldehyde for 10 min at RT. Cells were then permeabilised and incubated for 15 min at RT. At this point primary antibody was added for cytoplasmic staining for 1 hour 4°C. Secondary antibody was added in 50 μL FACS buffer for 1 hour 4°C. Cells were then resuspended in FACS buffer and measured using BD Biosciences FACSCantoII.
RNA extraction and RT-qPCR
RNA was isolated using the RNeasy Mini kit (QIAGEN). RNA was then reverse transcribed with M-MLV RT (Invitrogen), and SYBR-Green (Eurogentec) qPCR was performed using MX-3000 (Agilent Technologies). Target gene expression is expressed as ratio to housekeeping gene expression. For oligonucleotide sequences, see table S2.
Western blotting
Cells were lysed in RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS). Protein content was then tested using the BCA assay (Pierce) and equal amounts of lysates were boiled in Laemmli buffer for 10 min at 95°C. Samples were then subjected to SDS-PAGE. After blotting onto Hybond-P polyvinylidene fluoride membranes (GE Healthcare) blots were blocked with 5% milk in TBS-T, and primary antibody, in 5% BSA in TBS-T was added at 4ºC overnight. The protein was then detected by using a HRP-conjugated secondary antibody and visualized with LumiGLO (Cell Signaling Technology).
Cloning and Constructs
The Human GPR35a CDS was obtained from RefSeq entry NM_005301. Primers with EcoRI and BamHI (1 and 2 in table S2, respectively) linker sequences were used to amplify the GPR35 coding sequence from human cDNA. After double digestion of the resulting PCR fragment with EcoRI and BamHI, the DNA was cloned into the pEXPR-IBA103 human expression vector (IBA Life Sciences, Goettingen, Germany). This resulted in a C-terminal TwinStrep-tagged GPR35 expression system.
We generated an identical construct for the GPR35aT108M variant through site-directed mutagenesis. First, two overlapping PCR fragments were generated using the mutagenic primers separately (3 and 4 in table S2), combined with the previously mentioned EcoRI/BamHI linker primers (1 and 2 in table S2) respectively. In a second step, full length GPR35a coding sequence was generated with the linker primers (1 and 2 in table S2) only. The resulting fragment was cloned into pEXPR-IBA103.
Constructs for FRET experiments were generated using vector backbones mVenusN1 and mCeruleanN1 that were a gift from Steven Vogel (Addgene plasmids #27793 and 27795). We generated GPR35a and GPR35aT108M PCR fragments with EcoRI/BamHI linker sequences using the primer pair 5 and 6 (table S2) for C-terminal fusion. Resulting PCR fragments and vector backbones were digested with EcoRI/BamHI. ATP1A1 cDNA (RefSeq entry NM_000701.7) was amplified using the primer pair 7 and 8 (table S2) for C-terminal fusion. PCR fragments of ATP1A1 were cloned after XhoI/XmaI digestion. CXCR2 cDNA was amplified using primers 9 and 10 (table S2) and CCR5 using primers 11 and 12 (table S2). Both cDNAs were cloned after XhoI/HindIII digestion. We used N-terminal HA-tagged expression vectors for GPR35 (#GPR035TN00), CXCR2 (#CXCR20TN00), CCR5 (#CCR050TN00) and P2Y12R (#P2Y120TN00) obtained from cDNA Resource Center (Bloomsburg University, USA). Myc-DDK-tagged expression vectors for ATP1A1 (RC201009), ATP1A3 (RC203198) and untagged expression vector for ATP1A2 (SC119715) were obtained as lyophilised DNA (OriGene, Rockville, MD). ATP1A2 cDNA was amplified using primers 13 and 14 (table S2), digested with Sgf1/Mlu1 and ligated into pre-digested pCMV6-Entry. All expression vectors were cloned and stored in DH5alpha. All constructs were verified by Sanger sequencing.
Heterologous expression and pull-down assays
HEK293T cells were grown at 37 ºC, 5% CO2 in DMEM Medium supplied with 10% FBS and 1% PenStrep (all ThermoFisher Scientific, USA). Upon reaching 70% confluency HA- and Strep-tagged constructs were transiently transfected using Lipofectamine 3000 (ThermoFisher Scientific, USA) according to the supplier’s recommendation. Cells were lysed after 48 hours with lysis buffer (150 mM KCl, 0.5% CHAPS, 50 mM HEPES (pH 7.4)) containing Halt Protease Inhibitor Cocktail (ThermoFisher Scientific, USA). Lysates were centrifuged at 3000 g for 10 minutes at 4°C. Supernatants were collected and incubated with 1 U/mL of Avidin to block endogenous biotin for 15 min at 4 °C. Afterwards samples were centrifuged again at 3000 g for 10 minutes at 4 °C. Protein concentration was measured by analysing absorbance at 280 nm and set to 5 mg/mL. Strep-tagged protein was purified from 10 mg total protein using 1 mL StrepTactin columns (IBA Life Sciences, Goettingen, Germany). The purification procedure was carried out as recommended by the supplier and purified protein eluted with 5 mM Desthiobiotin. The eluate was analysed by immunoblot and silver-stained SDS PAGE.
SDS gels were submitted to the Cambridge Centre for Proteomics, University of Cambridge. The protein bands were excised, containing proteins reduced, alkylated and subsequently digested enzymatically. Samples were analysed by reverse-phase HPLC tandem MS. Resulting spectra were analysed using the MASCOT database and interpreted using the Uniprot database.
Immunoprecipitation
HEK293T cells were transiently transfected with HA-tagged constructs. After 24 hours cells were lysed in CHAPS buffer (150 mM KCl, 0.5% CHAPS, 50 mM HEPES (pH 7.4)) containing Halt Protease Inhibitor Cocktail and Phosphatase Inhibitor (ThermoFisher Scientific, USA). The lysates were centrifuged for 10 min at 3000 g at 4 °C. The supernatants were collected and protein concentration measured using absorbance at 280 nm. Protein concentration was normalized across samples to 1 μg/μL and 200 μg used per immunoprecipitation experiment. Protein was precipitated with HA Antibody (CST Technologies #3724) or ATP1A1 Antibody (ThermoFisher Scientific, MA3-928). Precipitation was carried out over night at 4 °C under constant shaking. IgG Isotype control antibody was used as a control. Protein-Antibody complexes were then conjugated to 70 μL Protein A sepharose beads each for 1 hour at 4 °C under constant shaking. Samples were then washed three times with 500 μL of TBST buffer containing 0.05% Tween 20. Protein complexes were finally eluted with 50 μL 0.2 M Glycine (pH 2). Eluates were immediately neutralized by addition of 5 μL 1 M Tris-HCl (pH 7.5). 4X Laemmli buffer containing 10 % 2-Mercaptoethanol was added to eluates and lysates prior SDS-PAGE and Immunoblot analysis.
Gene silencing
siRNA was transfected using Lipofectamine 2000 (ThermoFisher Scientific) according to the manufacturer’s protocol for experiments involving Caco2 cells. THP1 and primary BMDMs were silenced using Amaxa Nucleofector Kit (Lonza VPA-1109 and VPA-1103) as per the manufacturer’s protocol.
Rubidium Uptake Assay
Rubidium uptake was determined as described in Figueroa et al. (91). Specifically, cells were seeded in 6 well plates and allowed to attach overnight in normal culture medium. Immediately prior to the assay, cells were washed three times with 1 ml of rubidium assay buffer (130 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 0.5 mM NaH2PO4, 10 mM d-glucose, 12 mM NaHCO3, 10 mM HEPES; pH 7.4). Cells were then incubated for 1 or 2 hours at 37 °C in rubidium assay buffer containing 270 μM RbCl. Incubation was carried out with and without 1 mM Ouabain. Subsequently, cells were washed three times in rubidium buffer and scraped into 1 mL of ultrapure water. 10 μL of 100 ppB Cerium was added as an internal control. Afterwards, 1.4 g of Nitric Acid 70 % (SigmaAldrich #225711) was added and samples boiled at 100 °C for 30 min. Then 250 μL H2O2 (SigmaAldrich #95321) was added and samples boiled again at 100 °C for 30 min. As a last step, samples were diluted 1:10 with nitric acid 1 % prior to analysis by ICP-MS. 85Rb+ concentrations measured were normalized to sulfur (S) concentration and the internal Cerium control.
Seahorse extracellular flux analysis
Cells were seeded into 96 well XFe Seahorse plates (Agilent, USA) in normal culture medium. 1 hour prior to the start of extracellular flux measurements, the medium was changed to Agilent Seahorse XF Base Medium (103334-100) supplemented with 1 mM L-glutamine (ThermoFisher Scientific, USA) and 10 mM D-glucose (SigmaAldrich, UK). The pH of the medium was set to 7.4 after warming to 37 ºC. OCR and ECAR were measured using the XFe96 Seahorse extracellular flux analyser (Agilent, USA) over the course of 2 hours. Oligomycin (1 μM), FCCP (1.5 μM) and Rotenone/Antimycin A (both 1 μM) (all Sigma Aldrich, UK) were injected sequentially to allow assessment of respiratory chain functionality. Glycolytic function was measured in a separate protocol. Cells were incubated in seahorse assay medium without glucose. Glycolysis was assessed through measurement of ECAR after sequential injections with D-glucose (10 mM), Oligomycin (1 μM) and 2-deoxyglucose (100 mM) (SigmaAldrich, UK). Spare respiratory capacity (SRC) was computed as the difference between basal OCR and maximal OCR after FCCP injection. Maximal glycolytic capacity was computed as the difference between ECAR after injection with D-glucose and ECAR after injection with Oligomycin.
Glucose uptake assay
The Glucose Uptake-Glo Assay (Promega, USA) was used according to the supplier’s recommendation. Briefly, BMDMs were seeded into in white 96 well plates (Nunc, Denmark) at a cell density of 5 x104 cells per well and polarised as described above. Cells were washed once with PBS and then incubated with PBS containing 1 mM 2-deoxyglucose (2-DG) for the given time points (10 – 60 min). The reaction was stopped with stop buffer and immediately after this pH was neutralized with neutralization buffer. The detection reagent for 2-deoxy glucose-6-phosphate (2DG6P) was added to the wells. After incubation for 1 hour, luminescence (Centro LB 690 Luminometer, Berthold) was measured using 0.3 second integration every 2 minutes for 15 minutes. A standard curve of 2DG6P was used to extrapolate the concentration of 2DG6P in the sample.
Membrane potential
Cell membrane potential was measured by incubating cells with 1.9 μM DiBAC4(3) (molecular probes, B438) for 20 minutes. A fluorescent plate reader was then used to excite the dye at 493nm and measure emission at 516nm. Mitochondrial membrane potential was measured by incubating cells with 10 μg/mL JC-1 (molecular probes T3168) for 10 minutes at 37°C before measuring emission at 530nm and 535nm upon excitation at 485nm and 535nm respectively.
Antibodies, plasmids and reagents
Antibodies directed against HA (3724), ATP1A1 phospho-Ser16 (4020S), β-actin (4970S), polyclonal normal IgG (2729S), AMPK (2532), AMPK phospho-Thr172 (2535), pan Akt (4685S), Akt phospho-Thr308 (5056S), Akt phospho-Ser473 (4058), Src (2109S), Src phospho-Tyr416 (2101), EGFR (4267), EGFR phospho-Tyr1068 (8543S), p44/42 MAPK (Erk1/2) (4695), p44/42 MAPK (Erk1/2) phospho-Thr202/Tyr204 (4094S) and anti-Ki-67 (11882) were purchased from Cell Signaling Technology. Antibodies directed against c-myc-tag (MA1980) and ATP1A1 (MA3-928) were purchased from Thermo Fisher Scientific. The antibody directed against mouse normal IgG (sc-2025) was purchased from Santa Cruz Biotechnology. Pepducins and pNaktide were custom synthesised by Peptide 2.0 Inc.
The pEXPR-IBA 103 Strep-tag plasmid (2-3503-000) was purchased from IBA Life Sciences. HA tagged plasmids (GPR035TN00, CXCR20TN00 and CCR050TN00) were purchased from the cDNA Resource Centre. Plasmids for ATP1A1 (RC201009), ATP1A2 (SC119715) and ATP1A3 (RC203198) were purchased from Origene. mVenusN1 (27793) and mCeruleanN1 (27795) were purchased from Addgene. The HPSI0114i-kolf_2 (KOLF-2) (ECACC 77650100) iPSC cell line has previously established by the Wellcome Trust Sanger Institute. Other cell lines were purchased from ATCC.
Statistical analysis
All statistical analysis was performed using Prism 7 (GraphPad) software. The statistical tests used in each experiment are described in the respective figure legends.
Supplementary Material
Table S2. Oligonucleotide sequences.
One-sentence summary.
A GPCR−Na/K-ATPase interaction alters metabolism and proliferation to enhance intestinal tumor formation.
Editor’s summary.
Altering membrane potential for cancer
Disease-associated polymorphisms can give insight into protein function, particularly for receptors whose ligands are unknown or controversial. Polymorphisms in the G protein–coupled receptor GPR35 are associated with increased risk for certain inflammatory diseases of the bile duct and large intestine that have increased cancer risk. Schneditz et al. found that GPR35 promoted the activity of Na/K-ATPase, a ubiquitous and essential transmembrane pump that sets the membrane potential in cells, in a manner that did not rely on its proposed ligands. A disease-associated variant of GPR35 induced a more pronounced increase in Na/K-ATPase activity. This stimulation of Na/K-ATPase activity enhanced glycolysis and proliferation in intestinal epithelial cells. Deficiency in Na/K-ATPase or treatment with a pepducin targeting Gpr35 decreased tumor burden in mouse models of intestinal cancer. Thus, GPR35 coordinates regulation of membrane potential by Na/K-ATPase with metabolism and proliferation.
Acknowledgments
We thank Jason Day and the Department of Earth Sciences, University of Cambridge for help with ICP-MS, the BRC Cell Phenotyping Hub for support with FACS and confocal microscopy, and Catherine Turberfield for initial proteomics studies. We thank Alexey Shiryaev and Sarah Kempster for pilot experiments, and Joya Bhattacharyya for assistance with phenotyping. We are very thankful to James Lee for critically reading and input into the manuscript.
Funding: This paper was supported by the Wellcome Trust (Career Re-entry Fellowship to N.C.K. 103077/Z/13/Z, Senior Investigator Award 106260/Z/14/Z to A.K., PhD training fellowship for clinicians UNS59491 to J.E.E.), the European Research Council under the European Community's Seventh Framework Programme Consolidator Grant n° 648889 to A.K., and Scientia Fellowship (FP7-PEOPLE-2013-COFUND) grant agreement n° 609020 to G.S., the Addenbrooke’s Charitable Trust (ACT 25/16A) to J.E.E., and an UniNA and Compagnia Di San Paolo ‘STAR program for young researchers’ fellowship to E.P.
Footnotes
Author contributions: N.C.K., G.S., J.E.E. and E.P., together with S.S., M.Z.C., and D.A.M, performed the experiments; K.L., A.B. and S.M. designed, generated and confirmed the genotype of CRISPR-Cas9–generated cell lines; S.M. and G.D. helped with the generation of macrophages from iPSC. M.A. and T.D.L. generated mice with a floxed Gpr35 allele. N.C.K. and A.K., together with G.S., J.E.E., and E.P. designed the experiments. N.C.K., T.H.K. and A.K. devised and coordinated the project. N.C.K. and A.K., with input from all authors, interpreted the data and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The mass spectrometry proteomics data have been deposited to the ProteomeXchanger Consortium through the PRIDE partner repository with the dataset identifier PXD011269. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References
- 1.Ellinghaus D, et al. Genome-wide association analysis in primary sclerosing cholangitis and ulcerative colitis identifies risk loci at GPR35 and TCF4. Hepatology. 2013;58:1074–1083. doi: 10.1002/hep.25977. [DOI] [PubMed] [Google Scholar]
- 2.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu JZ, et al. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nature genetics. 2013;45:670–675. doi: 10.1038/ng.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji SG, et al. Genome-wide association study of primary sclerosing cholangitis identifies new risk loci and quantifies the genetic relationship with inflammatory bowel disease. Nat Genet. 2017;49:269–273. doi: 10.1038/ng.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milligan G. G protein-coupled receptors not currently in the spotlight: free fatty acid receptor 2 and GPR35. Br J Pharmacol. 2017 doi: 10.1111/bph.14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Audet M, Bouvier M. Restructuring G-protein- coupled receptor activation. Cell. 2012;151:14–23. doi: 10.1016/j.cell.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, et al. Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J Biol Chem. 2006;281:22021–22028. doi: 10.1074/jbc.M603503200. [DOI] [PubMed] [Google Scholar]
- 8.Maravillas-Montero JL, et al. Cutting edge: GPR35/CXCR8 is the receptor of the mucosal chemokine CXCL17. J Immunol. 2015;194:29–33. doi: 10.4049/jimmunol.1401704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins L, et al. Agonist activation of the G protein-coupled receptor GPR35 involves transmembrane domain III and is transduced via Galpha(1)(3) and beta-arrestin-2. Br J Pharmacol. 2011;162:733–748. doi: 10.1111/j.1476-5381.2010.01082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milligan G. Orthologue selectivity and ligand bias: translating the pharmacology of GPR35. Trends in pharmacological sciences. 2011;32:317–325. doi: 10.1016/j.tips.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Park SJ, Lee SJ, Nam SY, Im DS. GPR35 mediates lodoxamide-induced migration inhibitory response but not CXCL17-induced migration stimulatory response in THP-1 cells; is GPR35 a receptor for CXCL17? Br J Pharmacol. 2018;175:154–161. doi: 10.1111/bph.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ngo T, et al. Orphan receptor ligand discovery by pickpocketing pharmacological neighbors. Nature chemical biology. 2017;13:235–242. doi: 10.1038/nchembio.2266. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Okumura S, et al. Cloning of a G-protein-coupled receptor that shows an activity to transform NIH3T3 cells and is expressed in gastric cancer cells. Cancer science. 2004;95:131–135. doi: 10.1111/j.1349-7006.2004.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo YJ, Zhou YJ, Yang XL, Shao ZM, Ou ZL. The role and clinical significance of the CXCL17-CXCR8 (GPR35) axis in breast cancer. Biochem Biophys Res Commun. 2017;493:1159–1167. doi: 10.1016/j.bbrc.2017.09.113. [DOI] [PubMed] [Google Scholar]
- 15.Karlsen TH, Folseraas T, Thorburn D, Vesterhus M. Primary sclerosing cholangitis - a comprehensive review. J Hepatol. 2017;67:1298–1323. doi: 10.1016/j.jhep.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 16.Danese S, Fiocchi C. Ulcerative colitis. The New England journal of medicine. 2011;365:1713–1725. doi: 10.1056/NEJMra1102942. [DOI] [PubMed] [Google Scholar]
- 17.Weismuller TJ, et al. Patient Age, Sex, and Inflammatory Bowel Disease Phenotype Associate With Course of Primary Sclerosing Cholangitis. Gastroenterology. 2017;152:1975–1984 e1978. doi: 10.1053/j.gastro.2017.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boonstra K, et al. Population-based epidemiology, malignancy risk, and outcome of primary sclerosing cholangitis. Hepatology. 2013;58:2045–2055. doi: 10.1002/hep.26565. [DOI] [PubMed] [Google Scholar]
- 19.Schrumpf E, et al. HLA antigens and immunoregulatory T cells in ulcerative colitis associated with hepatobiliary disease. Scandinavian journal of gastroenterology. 1982;17:187–191. doi: 10.3109/00365528209182038. [DOI] [PubMed] [Google Scholar]
- 20.Melum E, et al. Genome-wide association analysis in primary sclerosing cholangitis identifies two non-HLA susceptibility loci. Nat Genet. 2011;43:17–19. doi: 10.1038/ng.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins L, et al. Identification of novel species-selective agonists of the G-protein-coupled receptor GPR35 that promote recruitment of beta-arrestin-2 and activate Galpha13. Biochem J. 2010;432:451–459. doi: 10.1042/BJ20101287. [DOI] [PubMed] [Google Scholar]
- 23.Taniguchi Y, Tonai-Kachi H, Shinjo K. Zaprinast, a well-known cyclic guanosine monophosphate-specific phosphodiesterase inhibitor, is an agonist for GPR35. FEBS Lett. 2006;580:5003–5008. doi: 10.1016/j.febslet.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 24.Hilger D, Masureel M, Kobilka BK. Structure and dynamics of GPCR signaling complexes. Nat Struct Mol Biol. 2018;25:4–12. doi: 10.1038/s41594-017-0011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinhard L, Tidow H, Clausen MJ, Nissen P. Na(+),K (+)-ATPase as a docking station: protein-protein complexes of the Na(+),K (+)-ATPase. Cell Mol Life Sci. 2013;70:205–222. doi: 10.1007/s00018-012-1039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clausen MV, Hilbers F, Poulsen H. The Structure and Function of the Na,K-ATPase Isoforms in Health and Disease. Frontiers in physiology. 2017;8:371. doi: 10.3389/fphys.2017.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanco G. Na,K-ATPase subunit heterogeneity as a mechanism for tissue-specific ion regulation. Semin Nephrol. 2005;25:292–303. doi: 10.1016/j.semnephrol.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Lingrel JB. The physiological significance of the cardiotonic steroid/ouabain-binding site of the Na,K-ATPase. Annu Rev Physiol. 2010;72:395–412. doi: 10.1146/annurev-physiol-021909-135725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKay NG, Kirby RW, Lawson K. Rubidium efflux as a tool for the pharmacological characterisation of compounds with BK channel opening properties. Methods Mol Biol. 2008;491:267–277. doi: 10.1007/978-1-59745-526-8_21. [DOI] [PubMed] [Google Scholar]
- 30.Murray PJ, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aperia A, Akkuratov EE, Fontana JM, Brismar H. Na+-K+-ATPase, a new class of plasma membrane receptors. Am J Physiol Cell Physiol. 2016;310:C491–495. doi: 10.1152/ajpcell.00359.2015. [DOI] [PubMed] [Google Scholar]
- 32.Kato A, Romero MF. Regulation of electroneutral NaCl absorption by the small intestine. Annu Rev Physiol. 2011;73:261–281. doi: 10.1146/annurev-physiol-012110-142244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blaustein MP, et al. Pivotal role of alpha2 Na(+) pumps and their high affinity ouabain binding site in cardiovascular health and disease. J Physiol. 2016;594:6079–6103. doi: 10.1113/JP272419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G, Kawakami K, Gick G. Regulation of Na,K-ATPase alpha1 subunit gene transcription in response to low K(+): role of CRE/ATF- and GC box-binding proteins. J Cell Physiol. 2007;213:167–176. doi: 10.1002/jcp.21107. [DOI] [PubMed] [Google Scholar]
- 35.Li Z, Langhans SA. Transcriptional regulators of Na,K-ATPase subunits. Front Cell Dev Biol. 2015;3:66. doi: 10.3389/fcell.2015.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Therien AG, Blostein R. Mechanisms of sodium pump regulation. Am J Physiol Cell Physiol. 2000;279:C541–566. doi: 10.1152/ajpcell.2000.279.3.C541. [DOI] [PubMed] [Google Scholar]
- 37.Yingst DR, Massey KJ, Rossi NF, Mohanty MJ, Mattingly RR. Angiotensin II directly stimulates activity and alters the phosphorylation of Na-K-ATPase in rat proximal tubule with a rapid time course. Am J Physiol Renal Physiol. 2004;287:F713–721. doi: 10.1152/ajprenal.00065.2004. [DOI] [PubMed] [Google Scholar]
- 38.Hale C, et al. Induced pluripotent stem cell derived macrophages as a cellular system to study salmonella and other pathogens. PloS one. 2015;10:e0124307. doi: 10.1371/journal.pone.0124307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul RJ, Bauer M, Pease W. Vascular smooth muscle: aerobic glycolysis linked to sodium and potassium transport processes. Science. 1979;206:1414–1416. doi: 10.1126/science.505014. [DOI] [PubMed] [Google Scholar]
- 40.Lynch RM, Balaban RS. Coupling of aerobic glycolysis and Na+-K+-ATPase in renal cell line MDCK. Am J Physiol. 1987;253:C269–276. doi: 10.1152/ajpcell.1987.253.2.C269. [DOI] [PubMed] [Google Scholar]
- 41.James JH, et al. Linkage of aerobic glycolysis to sodium-potassium transport in rat skeletal muscle. Implications for increased muscle lactate production in sepsis. J Clin Invest. 1996;98:2388–2397. doi: 10.1172/JCI119052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss J, Hiltbrand B. Functional compartmentation of glycolytic versus oxidative metabolism in isolated rabbit heart. J Clin Invest. 1985;75:436–447. doi: 10.1172/JCI111718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung J, Yoon T, Choi EC, Lee K. Interaction of cofilin with triose-phosphate isomerase contributes glycolytic fuel for Na,K-ATPase via Rho-mediated signaling pathway. J Biol Chem. 2002;277:48931–48937. doi: 10.1074/jbc.M208806200. [DOI] [PubMed] [Google Scholar]
- 44.Campbell JD, Paul RJ. The nature of fuel provision for the Na+,K(+)-ATPase in porcine vascular smooth muscle. The Journal of physiology. 1992;447:67–82. doi: 10.1113/jphysiol.1992.sp018991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhar-Chowdhury P, Malester B, Rajacic P, Coetzee WA. The regulation of ion channels and transporters by glycolytically derived ATP. Cell Mol Life Sci. 2007;64:3069–3083. doi: 10.1007/s00018-007-7332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu M, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 47.O'Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui X, Xie Z. Protein Interaction and Na/K-ATPase-Mediated Signal Transduction. Molecules. 2017;22 doi: 10.3390/molecules22060990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roskoski R., Jr Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol Res. 2015;94:9–25. doi: 10.1016/j.phrs.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 50.Tian J, et al. Binding of Src to Na+/K+-ATPase forms a functional signaling complex. Mol Biol Cell. 2006;17:317–326. doi: 10.1091/mbc.E05-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L, et al. Comparative properties of caveolar and noncaveolar preparations of kidney Na+/K+-ATPase. Biochemistry. 2011;50:8664–8673. doi: 10.1021/bi2009008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Irtegun S, Wood RJ, Ormsby AR, Mulhern TD, Hatters DM. Tyrosine 416 is phosphorylated in the closed, repressed conformation of c-Src. PLoS One. 2013;8:e71035. doi: 10.1371/journal.pone.0071035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J, et al. Ouabain interaction with cardiac Na+/K+-ATPase initiates signal cascades independent of changes in intracellular Na+ and Ca2+ concentrations. J Biol Chem. 2000;275:27838–27844. doi: 10.1074/jbc.M002950200. [DOI] [PubMed] [Google Scholar]
- 54.Morth JP, et al. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 55.Roskoski R., Jr Src protein-tyrosine kinase structure and regulation. Biochem Biophys Res Commun. 2004;324:1155–1164. doi: 10.1016/j.bbrc.2004.09.171. [DOI] [PubMed] [Google Scholar]
- 56.Comellas AP, et al. Insulin regulates alveolar epithelial function by inducing Na+/K+-ATPase translocation to the plasma membrane in a process mediated by the action of Akt. J Cell Sci. 2010;123:1343–1351. doi: 10.1242/jcs.066464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prassas I, Diamandis EP. Novel therapeutic applications of cardiac glycosides. Nat Rev Drug Discov. 2008;7:926–935. doi: 10.1038/nrd2682. [DOI] [PubMed] [Google Scholar]
- 58.Kometiani P, Liu L, Askari A. Digitalis-induced signaling by Na+/K+-ATPase in human breast cancer cells. Mol Pharmacol. 2005;67:929–936. doi: 10.1124/mol.104.007302. [DOI] [PubMed] [Google Scholar]
- 59.Liu L, Zhao X, Pierre SV, Askari A. Association of PI3K-Akt signaling pathway with digitalis-induced hypertrophy of cardiac myocytes. Am J Physiol Cell Physiol. 2007;293:C1489–1497. doi: 10.1152/ajpcell.00158.2007. [DOI] [PubMed] [Google Scholar]
- 60.Wu J, et al. Cell signaling associated with Na(+)/K(+)-ATPase: activation of phosphatidylinositide 3-kinase IA/Akt by ouabain is independent of Src. Biochemistry. 2013;52:9059–9067. doi: 10.1021/bi4011804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iwamoto N, et al. Context-specific flow through the MEK/ERK module produces cell- and ligand-specific patterns of ERK single and double phosphorylation. Sci Signal. 2016;9 doi: 10.1126/scisignal.aab1967. ra13. [DOI] [PubMed] [Google Scholar]
- 62.Li Z, et al. NaKtide, a Na/K-ATPase-derived peptide Src inhibitor, antagonizes ouabain-activated signal transduction in cultured cells. J Biol Chem. 2009;284:21066–21076. doi: 10.1074/jbc.M109.013821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hawley SA, et al. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J Biol Chem. 1996;271:27879–27887. doi: 10.1074/jbc.271.44.27879. [DOI] [PubMed] [Google Scholar]
- 64.Lizcano JM, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Antonia RJ, Baldwin AS. IKK promotes cytokine-induced and cancer-associated AMPK activity and attenuates phenformin-induced cell death in LKB1-deficient cells. Sci Signal. 2018;11 doi: 10.1126/scisignal.aan5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cordero JB, et al. c-Src drives intestinal regeneration and transformation. EMBO J. 2014;33:1474–1491. doi: 10.1002/embj.201387454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu J, et al. Attenuation of Na/K-ATPase Mediated Oxidant Amplification with pNaKtide Ameliorates Experimental Uremic Cardiomyopathy. Sci Rep. 2016;6 doi: 10.1038/srep34592. 34592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 69.Vogelstein B, Kinzler KW. The Path to Cancer --Three Strikes and You're Out. N Engl J Med. 2015;373:1895–1898. doi: 10.1056/NEJMp1508811. [DOI] [PubMed] [Google Scholar]
- 70.Madison BB, et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 71.Robles AI, et al. Whole-Exome Sequencing Analyses of Inflammatory Bowel Disease-Associated Colorectal Cancers. Gastroenterology. 2016;150:931–943. doi: 10.1053/j.gastro.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yaeger R, et al. Genomic Alterations Observed in Colitis-Associated Cancers Are Distinct From Those Found in Sporadic Colorectal Cancers and Vary by Type of Inflammatory Bowel Disease. Gastroenterology. 2016;151:278–287 e276. doi: 10.1053/j.gastro.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140:1807–1816. doi: 10.1053/j.gastro.2011.01.057. [DOI] [PubMed] [Google Scholar]
- 74.Wirtz S, et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc. 2017;12:1295–1309. doi: 10.1038/nprot.2017.044. [DOI] [PubMed] [Google Scholar]
- 75.Greten FR, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 76.O'Callaghan K, Kuliopulos A, Covic L. Turning receptors on and off with intracellular pepducins: new insights into G-protein-coupled receptor drug development. J Biol Chem. 2012;287:12787–12796. doi: 10.1074/jbc.R112.355461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Imada S, et al. Role of Src family kinases in regulation of intestinal epithelial homeostasis. Mol Cell Biol. 2016 doi: 10.1128/MCB.00311-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sirvent A, Benistant C, Roche S. Oncogenic signaling by tyrosine kinases of the SRC family in advanced colorectal cancer. American journal of cancer research. 2012;2:357–371. [PMC free article] [PubMed] [Google Scholar]
- 79.Sousa-Victor P, Jasper H. Epithelial regeneration and cancer: news from the Src front. EMBO J. 2014;33:1423–1424. doi: 10.15252/embj.201488815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Irby RB, et al. Activating SRC mutation in a subset of advanced human colon cancers. Nat Genet. 1999;21:187–190. doi: 10.1038/5971. [DOI] [PubMed] [Google Scholar]
- 81.Yeatman TJ. A renaissance for SRC. Nat Rev Cancer. 2004;4:470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 82.Manning BD, Toker A. AKT/PKB Signaling: Navigating the Network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Geering K. FXYD proteins: new regulators of Na-K-ATPase. Am J Physiol Renal Physiol. 2006;290:F241–250. doi: 10.1152/ajprenal.00126.2005. [DOI] [PubMed] [Google Scholar]
- 84.Jung J, et al. Translationally controlled tumor protein interacts with the third cytoplasmic domain of Na,K-ATPase alpha subunit and inhibits the pump activity in HeLa cells. J Biol Chem. 2004;279:49868–49875. doi: 10.1074/jbc.M400895200. [DOI] [PubMed] [Google Scholar]
- 85.Covic L, Gresser AL, Talavera J, Swift S, Kuliopulos A. Activation and inhibition of G protein-coupled receptors by cell-penetrating membrane-tethered peptides. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:643–648. doi: 10.1073/pnas.022460899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaneider NC, Agarwal A, Leger AJ, Kuliopulos A. Reversing systemic inflammatory response syndrome with chemokine receptor pepducins. Nat Med. 2005;11:661–665. doi: 10.1038/nm1245. [DOI] [PubMed] [Google Scholar]
- 87.Carr R, 3rd, et al. Development and characterization of pepducins as Gs-biased allosteric agonists. J Biol Chem. 2014;289:35668–35684. doi: 10.1074/jbc.M114.618819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quoyer J, et al. Pepducin targeting the C-X-C chemokine receptor type 4 acts as a biased agonist favoring activation of the inhibitory G protein. Proc Natl Acad Sci U S A. 2013;110:E5088–5097. doi: 10.1073/pnas.1312515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gurbel PA, et al. Cell-Penetrating Pepducin Therapy Targeting PAR1 in Subjects With Coronary Artery Disease. Arterioscler Thromb Vasc Biol. 2016;36:189–197. doi: 10.1161/ATVBAHA.115.306777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Forbester JL, Hannan N, Vallier L, Dougan G. Derivation of Intestinal Organoids from Human Induced Pluripotent Stem Cells for Use as an Infection System. Methods Mol Biol. 2016 doi: 10.1007/7651_2016_7. [DOI] [PubMed] [Google Scholar]
- 91.Figueroa JA, Stiner CA, Radzyukevich TL, Heiny JA. Metal ion transport quantified by ICP-MS in intact cells. Sci Rep. 2016;6 doi: 10.1038/srep20551. 20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.