Abstract
Rotavirus A (RVA) exhibits a wide genotype diversity globally. Little is known about the genetic composition of genotype P[6] from Africa. This study investigated possible evolutionary mechanisms leading to genetic diversity of genotype P[6] VP4 sequences.
Phylogenetic analyses on 167 P[6] VP4 full-length sequences were conducted, which included six porcine-origin sequences. Of the 167 sequences, 57 were newly acquired through whole genome sequencing as part of this study. The other 110 sequences were all publicly-available global P[6] VP4 full-length sequences downloaded from GenBank. The strength of association between the phenotypic features and the phylogeny was also determined.
A number of reassortment and mixed infections of RVA genotype P[6] strains were observed in this study. Phylogenetic analyses demostrated the extensive genetic diversity that exists among human P[6] strains, porcine-like strains, their concomitant clades/subclades and estimated that P[6] VP4 gene has a higher substitution rate with the mean of 1.05E-3 substitutions/site/year. Further, the phylogenetic analyses indicated that genotype P[6] strains were endemic in Africa, characterised by an extensive genetic diversity and long-time local evolution of the viruses. This was also supported by phylogeographic clustering and G-genotype clustering of the P[6] strains when Bayesian Tip-association Significance testing (BaTS) was applied, clearly supporting that the viruses evolved locally in Africa instead of spatial mixing among different regions.
Overall, the results demonstrated that multiple mechanisms such as reassortment events, various mutations and possibly interspecies transmission account for the enormous diversity of genotype P[6] strains in Africa. These findings highlight the need for continued global surveillance of rotavirus diversity.
Keywords: Rotaviruses, Diversity, Africa, Bayesian, Phylogenetic, NGS
1. Introduction
Rotavirus A (RVA) is a major cause of severe dehydrating diarrhea in infants and young ones of several mammals and avian species worldwide (Estes and Greenberg, 2013). Annually, RVA cause approximately 215,000 deaths in children below the age of 5 years, primarily in African and Asian countries. The largest mortality due to rotavirus-associated diarrhea occurs in sub-Saharan Africa, where 121,000 children die annually (Tate et al., 2016a). Rotavirus vaccines are recommended by the World Health Organization (WHO) for all countries and provide the best protection against rotavirus-associated mortality. Vaccines also play a vital role in improving the economy as cost-effective measures due to improved childhood health in the countries where they are administered (Path, 2016). Although efficacy trials have shown that rotavirus vaccines could be less efficacious in developing countries compared to developed countries (Armah et al., 2010; Madhi et al., 2010), recent vaccine efficacy data have reassured that these vaccines are still highly effective in Africa as well as other continents (Armah et al., 2016; Groome et al., 2014; Parashar et al., 2016; Platts-Mills et al., 2017; O’Ryan et al., 2015; Santos et al., 2016; Tate et al., 2016b; Velázquez et al., 2017a,b). However, factors such as genetic diversity, nutrition, gut microbiome and maternal antibodies from breastfeeding can affect vaccine performance, leading to lower efficacy as evidenced in Africa and Asia. Despite the lower efficacy in Africa and Asia, the vaccine impact is higher in these low-income areas, because of the higher burden of disease compared to the developed world (Rota Council, 2016).
The rotavirus genome consists of 11 segments of double-stranded RNA encoding six structural and six non-structural viral proteins (VP1-VP4, VP6, VP7 and NSP1-NSP6) (Estes and Greenberg, 2013). Accumulation of point mutations and reassortment events between RVA segments are the major mechanisms driving the evolution of RVAs. The outer capsid proteins (VP4 and VP7) form the basis of RVA binary classification into G (Glycoprotein) and P (Protease sensitive) types. At least 27 G-genotypes and 37 P-genotypes have been ratified by the Rotavirus Classification Working Group (RCWG) (Matthijnssens et al., 2008; Trojnar et al., 2013). Another 7G- and 13 P-types have been proposed by the RCWG, as of June 2017 (https://rega.kuleuven.be/cev/viralmetagenomics/virus-dassification). Globally, RVAs present with a wide variety of different genotypes that are rather unique. The most prevalent circulating VP7 genotypes are Gl, G2, G3, G4, G9 and G12 in combination with VP4 genotypes P[4] and P[8] (Bànyai et al., 2012; Dóró et al., 2014). The most common VP4 genotypes are P[8], P [4] and P[6], respectively. Genotype P [6] strains are independent of G-type combinations and little is known about their genetic composition. However, in sub-Saharan Africa, genotype P[6] is highly prevalent and has been detected in combination with Gl, G2, G3, G4, G5, G6, G8, G9 and G12 suggesting that genotype P[6] should be considered common in Africa (Steele and Ivanoff, 2003; Armah et al., 2001; Seheri et al., 2014). Unlike the genotype P[4] strains, which are mostly associated with DS-l-like constellation and genotype P[8] strains with Wa-like constellation, the P[6] strains are associated with both the DS-l-like and Wa-like genetic backbones (Matthijnssens and Van Ranst, 2012).
For most human rotavirus gene segments, genotypes 1 (Wa-like) or 2 (DS-l-like) are often present. All genotype P[6] strains have zoonotic potential as demonstrated in human rotavirus surveillance studies. Genotype P [6] strains are believed to be of porcine origin (Hwang et al., 2012; Wang et al., 2010). However, they have been documented to be circulating for many years in human populations (Martella et al., 2006). The P genotype is believed to influence different histo-blood group antigen binding sites enhancing susceptibility of rotavirus infections (Hu et al., 2012). Both P[4] and P[8] strains recognize the more common antigen, Leb as the H-type 1 antigens, while the P[6] rotaviruses exclusively recognize the H-type 1 antigen. Subsequently, individuals who are non-secretors and have an inactive α−1, 2 fucosyl-transferase do not synthesize the H-type 1 antigen and are believed to be non-susceptible to infection with P[6] rotaviruses. Therefore, it is believed that the P[6] strains preferentially infect Lewis-negative children (Nordgren et al., 2014). The fact that the non-secretor phenotype is found in approximately 20% of European and North American populations, while the number of non-secretors is hardly observed in Africa, might be a possible reason why the genotype P[6] strains are more prevalent in Africa compared to the developed world (Heylen et al., 2016).
Little is known about the genetic composition of African P[6] strains. There is inadequate complete genome sequences of P[6] strains available in GenBank and this study provides almost double the number of available P [6] VP4 full-length sequences in the public databases. The comprehensive sequence record of genotype P[6] strains is instrumental in understanding their significance in Africa and other parts of the world. The few studies that investigated genotype P[6] strains in Africa have demonstrated that they are constantly evolving through reassortment events (Dennis et al., 2014a; Ndze et al., 2014; Nyaga et al., 2013, 2014; Nakagomi et al., 2017) and are independent from their G type combinations (Heylen et al., 2016). This study analysed the African P[6] strains by exploring their phylogenetic relationship with different G-type combinations and also investigated the evolutionary mechanisms leading to genetic diversity.
2. Methods
2.1. Study design
Archived stool samples initially screened as RVA-positive using commercially available rotavirus kit (ProSpecT Rotavirus Microplate Kit, England) and characterised further by polyacrylamide gel electro-phoresis (PAGE) as long and short patterns, were selected from the stool bank of the South African Medical Research Council Diarrheal Pathogens Research Unit, a World Health Organization Rotavirus Regional Reference Laboratory (WHO RRL) in South Africa. From these, the VP4 encoded gene of 57 samples that were previously genotyped through convention methods as genotype P[6] strains, were subjected to whole genome sequencing and analyses pipeline to determine the genotype constellations and evolutionary mechanisms of their V4 gene by Bayesian analysis (Magagula et al., 2015; Nyaga et al., 2013, 2014, 2015, 2016; Stucker et al., 2015). The 57 P[6] strains were collected from 13 countries: Botswana (n = 1), Cameroon (n = 1), Ethiopia (n = 5), Gambia (n = 3), Ghana (n = 1), Guinea Bissau (n = 2), Uganda (n = 2), Senegal (n = 3), South Africa (n = 25), Swaziland (n = 1), Togo (n = 5), Zambia (n = 7) and Zimbabwe (n = 1).
2.2. Whole genome sequencing of rotaviruses
The dsRNA genome was extracted according to previously described methods (Nyaga et al., 2013, 2014). In brief, extracted RNA was used in each of 11 one-step RT-PCR reactions (QIAGEN OneStepRT-PCR Kit, QIAGEN, Hilden, Germany) to amplify full-length RNA for all 11 genome segments. Primers used to amplify all gene segments were described previously (Magagula et al., 2015; Nyaga et al., 2015). PCR products were cleaned by treatment with exonudease I (New England BioLabs, Ipswich, MA, USA). The products were quantitated using a SYBR green dsDNA detection assay (SYBR Green I Nudeic Acid Gel Stain, Thermo Fisher Scientific, Waltham, MA, USA). All 11 RT-PCR products for each genome were pooled in equimolar amounts in preparation for sequencing using Illumina and/or Ion Torrent platforms.
Illumina libraries were prepared using the Nextera DNA Sample Preparation Kit (Illumina, Inc., San Diego, CA, USA). Briefly, gDNA was normalized to 0.2 ng/μl and tagmented at 55 °C for 5 min. Tagmented DNA was ligated to Illumina sequencing adapters and barcoded through a PCR amplification step, which incorporated tagmented DNA combined with Nextera PCR MasterMix, Nextera PCR primer cocktail and an indexing primer set (Integrated DNA Technologies, Coralville, IA, USA) to a total volume of 50 μl per reaction. After PCR amplification, each library was cleaned two times with Ampure XP Reagent (Beckman Coulter, Inc., Brea, CA, USA) to remove all leftover primers and to size selected an appropriate fragment size by removing the small and large DNA fragments. The quality of the library size was validated using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The cleaned library was sequenced on the Illumina MiSeq instrument (Illumina, Inc., San Diego, CA, USA) with 300 bp paired-end reads using the MiSeq Reagent Kit v3 for 600 cycles.
Ion Torrent libraries were prepared by shearing pooled RVA amplicons and ligating barcoded adaptors to the sheared DNA using the Ion Xpress Plus Fragment Library Kit (Thermo Fisher Scientific, Waltham, MA, USA), to create a 200-bp fragment library size. Equal volumes of pooled libraries were deaned using Ampure XP beads (Beckman Coulter, Inc., Brae, CA, USA). Real-time PCR was performed on the pooled, barcoded libraries to evaluate the library and adaptor quality and to determine the template dilution factor for emulsion PCR. The pool was normalized and amplified on Ion Sphere Particles (ISPs) using the Ion One Touch instrument (Thermo Fisher Scientific, Waltham, MA, USA). The pool was cleaned with Ampure XP Reagent (Beckman Coulter, Inc., Brea, CA, USA) and enriched for template-positive ISPs on the Ion One Touch ES instrument (Thermo Fisher Scientific, Waltham, MA, USA). Sequencing was performed on the Ion Torrent PGM platform using an Ion 316 chip (Thermo Fisher Scientific, Waltham, MA, USA).
The reads generated by the Ion Torrent PGM and Illumina MiSeq platforms were de novo assembled using the clc_novo_assemble program within the CLCBio software suite (QIAGEN, Hilden, Germany) to form contigs. The resulting contigs were mapped against their closest reference sequences for reference-based assembly using the clc_.ref_assemble_long algorithm. Any locus with an observed variant that was detected in the majority of Ion Torrent and Illumina sequencing reads was reflected in the assembled consensus sequence of the segment. Consensus sequences were submitted to NCBI GenBank and assigned accession numbers (Fig. 2; Supplementary data 1).
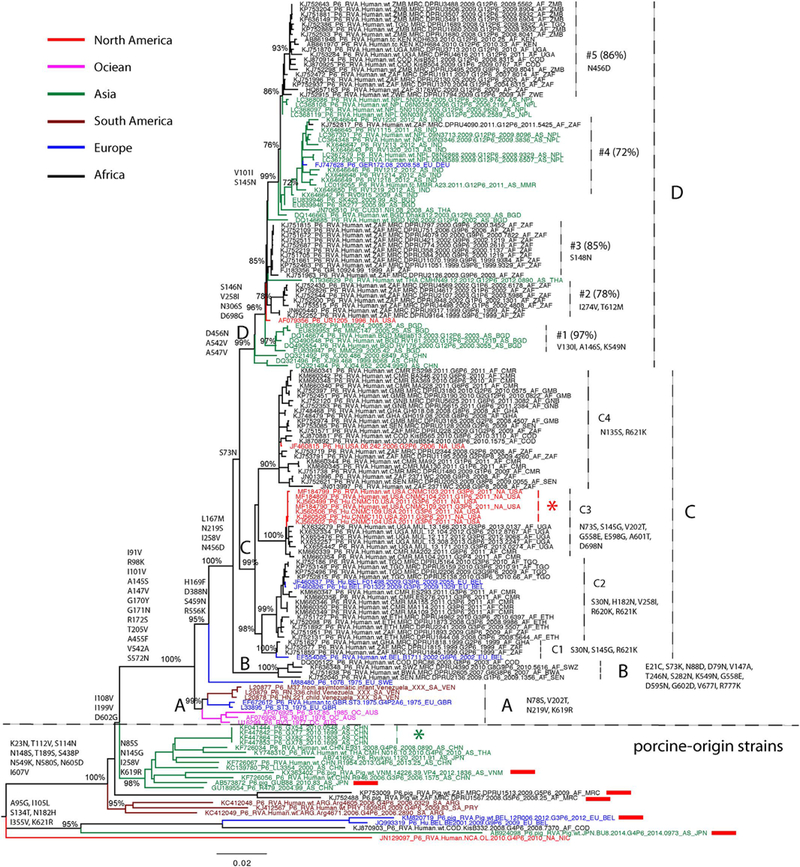
Fig. 2.
Maximum likelihood tree of RVA VP4 segment P[6] genotype. Major nodes with bootstrap value over 70% are indicated in the tree. Sequences from different continents are colored and described in the legend key. Clades, subclades, and monophyletic groups are marked in the tree. Sequences isolated from pigs are also marked by red rectangles following the taxa names. Amino acid substitutions that contribute to clade diversity are mapped to the branches of the tree. The scale bar represents genetic distance.
2.3. Identification of genotype constellations
RotaC, V 2.0 (Maes et al., 2009), an online classification genotyping tool for RVA strains was used to assign genotypes to each of the genome segments with the results used to generate the genome constellations.
2.4. Phylogenetic analyses
To understand the evolutionary dynamics of genotype P[6] strains in Africa, phylogenetic analyses on 167 P[6] VP4 full-length sequences were conducted, which included some porcine-origin strains. Out of the 167 sequences, 57 were newly acquired through whole genome sequencing as part of this study. The other 110 sequences were publicly-available P[6] VP4 full-length sequences downloaded from GenBank (http://www.ncbi.nlm.nih.gov/genbank/), with the associated sampling date and location information available as of Mar 15, 2018. Sequences were aligned using the MUSCLE algorithm in MEGA 6.0 with manual adjustment (Tamura et al., 2013). The Hasegawa-Kishino-Yano (HKY) nucleotide substitution model with a gamma distribution of among-site rate variation (HKY + Γ) was selected as the best-fit model by Modeltest in MEGA 6.0 and used in Maximum Likelihood phylogeny construction with 1000 bootstrap replicates (in PhyML3.0) and the Bayesian Markov chain Monte Carlo (BMCMC) tree inference (in BEAST vl.8.2 package (http://tree.bio.ed.ac.uk/software/BEAST) (Drummond and Rambaut, 2007). The known porcine-origin and phylogenetically distinct sequences in maximum likelihood phylogeny were excluded from analysis in the BMCMC phylogeny inference. A total of 128 sequences were included in the BEAST analysis. A relaxed molecular dock (uncorrelated lognormal, UCLN) and Gaussian Markov Random Fields (GMRF) Bayesian sky ride coalescent tree was employed in the BEAST run for 1 × 108 steps. Trees were sampled every 1 × 104 steps, with the first 1000 trees discarded as bum-in. The results were evaluated using the Tracer program vl.6.0 (http://tree.bio.ed.ac.uk/software/tracer) to ensure that stationarity was achieved. The posterior distribution of BMCMC trees was summarized as the maximum dade credibility (MCC) tree and generated by using TreeAnnotator vl.8.2 (available within BEAST vl.8.2 package), with the first 10% of trees removed as bum-in.
2.5. Association between virus phylogeny, geography and G genotype of P [6] strains
To determine whether there was phylogenetic clustering by geographic region and G genotypes, viruses were grouped into nine regions of their origin (Asia, North America, Europe, South America, Oceania, Eastern Africa, Western Africa, Central Africa and Southern Africa) and grouped by G genotype separately. The strength of association between the phenotypic features described above and the phylogeny was determined using two phylogeny-trait association statistics, the parsimony score (PS) and the association index (AI) tests, both of which were implemented in the Bayesian Tip-association Significance testing (BaTS) program (Parker et al., 2008). A significance levd of p < 0.05 was used in both statistics. A null distribution of these statistics was determined using the posterior distribution of P[6] phylogenetic trees obtained from the BEAST package described above.
3. Results
3.1. Genome constellation analysis, zoonotic and reassortment events in P [6] strains in Africa
All 57 newly-sequenced P[6] VP4 genomes in this study were subjected to genotyping analysis and the complete genotype constellations are shown in Fig. 1. The new P[6] strains were associated with seven G-genotypes, G1 (n = 7), G2 (n = 7), G3 (n = 7), G6 (n = 2), G8 (n = 1), G9 (n = 12) and G12 (n = 14) and seven mixed-infections. All G12 and G1 strains exhibited genotype 1 constellation, while G2, G6, G8and G3 strains demonstrated genotype 2 constellation. The G9 strains were mostly under genotype 2 genome constellation, with two exceptions: 1) two of the G9 strains (RVA/Human-wt/ZAF/MRC-DPRU9164/1999/ G9P[6] and RVA/Human-wt/ZAF/MRC-DPRU9317/1999/G9P[6]) possessed a Wa-like reassortant, within the NSP2, N1 genotype, in a DS-1-like genetic backbone; and 2) RVA/Human-wt/ZMB/MRC-DPRU3495/2009/G9P[6] was under a pure Wa-like constellation. The genetic diversity of strain RVA/Human-wt/ZAF/MRC-DPRU9317/ 1999/G9P[6] was described previously (Nyaga et al., 2013). Notably, all G9P[6] within the DS-1 constellation were collected between 1999 and 2003, whereas strain RVA/Human-wt/ZMB/MRC-DPRU3495/ 2009/G9P[6], the only G9 under a pure Wa-like constellation, was collected almost a decade later, in 2009. This could be the approximate amount of time it took for segment reassortment events to take place for a complete shift of the circulating genotype constellations of the G9P [6] to shift from genotype 2 to genotype 1. This shift could have resulted in the more prevalent G9P[8] strain that circulates more rapidly across human populations under the Wa-like genome constellation in the recent past (Matthijnssens and Van Ranst, 2012).
Fig 1.
The complete genome constellations of 57 P(6) study strains. The Wa- or DS-l-like genogroups were assigned to the study strains if at least seven genome segments belonged to the respective Wa- or DS-l-like genotype. Colours were added to visualize and differentiate genotypes within the constellation: genogroup 1 is Wa-like and genogroup 2 is DS-l-like. VP, viral structural protein; NSP, viral non-structural protein.
There were seven mixed infections observed in this study (Fig. 1). For mixed infections, two or more sequences of different genotypes were assembled for the same genome segment (e.g. G1 and G3 for VP7, II and 12 for VP6, R1 and R2 for VP1, Ml and M2 for VP3, N1 and N2 for the NSP2 gene segments of strain RVA/Human-wt/ETH/MRC-DPRU1893/2009/GlG3P[6]). Mixed infections are often prone to various forms of mutations including gene reassortment, thereby contributing to rotavirus genetic diversity. Two mixed infection strains RVA/Human-wt/BWA/MRC-DPRU2605/2007/G6G8P[6] and RVA/ Human-wt/TGO/MRC-DPRU2206/2009/G3G9P[6] carried a pure DS-1-like constellation, while other mixed infection strains suggested reassortment events since G1 and G12 exhibited Wa-like constellations.
The G12 strain was the most prevalent in combination with genotype P[6], detected in six African countries (Gambia, South Africa, Togo, Uganda, Zambia and Zimbabwe) between 2004 and 2011. The second most prevalent G genotype, the G9, was also typed from strains from many countries: South Africa, Swaziland, Togo and Zambia. There were nine Gl, (Wa-like), ten G2 (DS-l-like) and nine G3 (DS-l-like) strains investigated. The Gl strains were from Cameroon, Senegal and South Africa while the G2 strains were from Ghana, Gambia, Senegal and South Africa. The G3 strains were only identified from two countries; Togo and Ethiopia. We also detected three genotype G6 strains from Guinea Bissau and Botswana and three genotype G8 strains each from Senegal, Swaziland and Botswana. The detection of genotypes previously associated with animal rotaviruses, especially the G6P[6] and G8P[6] in humans, most probably through interspecies transmission, could be a direct or indirect result of the high number of mixed infections among the circulating strains as reported in the annual surveillance studies from Africa (Fig. 1). The combination of G6 with P[6] in human population is unique because both are traditionally animal strains, which is an indication that interspecies transmission is probably common in Africa.
3.2. Phylogenetic analysis of the VP4 segments of P[6] strains
To gain better insight of the evolutionary relationships between our collection of genotype P[6] VP4 sequences in Africa and those that had previously been generated worldwide, a maximum likelihood phylogenetic tree was constructed with all available sequences isolated from humans. Six porcine-origin P[6] strains were also included in the tree (Fig. 2). The long internal and external branches of the phylogenetic tree confirmed the extensive genetic diversity that exists among P[6] strains. Some P[6] strains isolated from humans clustered with the strains isolated from pigs, located at the bottom part of the tree, indicating that they were porcine-origin or porcine-like human P[6] strains (Fig. 2). These porcine-like strains have sporadically been isolated from different parts of the world since 2000, including South America (Argentina and Uruguay, marked in cayenne), Central America (Nicaragua, marked in red), Africa (Democratic Republic of the Congo, marked in black), Europe (Belgium, marked in blue) and Asia (Japan and China, marked in green). A group of P[6] strains were isolated in southern China in 2010 (marked by green star), providing evidence of rapid local transmission of the porcine-origin strain in humans (Fig. 2).
In human P[6] strains, there are four major clades (A, B, C and D) co-circulating worldwide. The prototype human P[6] sequences in clade A were isolated from Venezuela (marked in cayenne), the United Kingdom (marked in blue) and Australia (marked in magenta) in the 1970s and may have died out in the 1980’s since viruses belonging to clade A could not be detected after 1990. clade B includes four sequences from Africa, while the majority of sequences are in clade C and D. We observed a significant genetic diversity in both clades: four subclades (C1-C4) in clade C and multiple monophyletic groups (#1-#5) in clade D (Fig. 2, nodes supported by bootstrap value over 70% in the maximum likdihood phylogeny). Amino acid substitutions that have become fixed into the evolutionary history of genotype P[6] strains were mapped to the branches of maximum likelihood phylogeny (Fig. 2). Differences between porcine-like strains and human strains included 12 amino acid substitutions: I91V, R98K, I101V, A145S, A147V, G170Y, G171N, R172S, T205V, A455F, V542A, and S572N. Additional amino acid substitutions for different clades, subclades and monophyletic groups were also mapped to the maximum likelihood tree (Fig. 2).
In Africa, human P[6] strains were first reported in 1999 and three clades (B, C and D) are co-circulating and phylogenetically distinct from P[6] prototype strains (clade A) reported in the 1970s in three subclades from Venezuela, Britain and Australia, as well as the porcine-origin strains collected worldwide after 2000 (Fig. 2). Clade B contains four strains from Africa and clade C contains the majority of strains from Africa together with some non-African strains. The non-African sequences present in clade C lend additional support of viral movements between Africa and Europe (Belgium, marked in blue) and North America (USA, marked in red). A monophyletic group of sequences in subclade C3 collected in the USA (marked by red star) shows the evidence of local transmission of P [6] in Washington DC, USA and they are genetically similar to the strains that circulated in Cameroon during the same time period (2010 – 2011) and in Uganda during 2012–2013. clade D, which is characterised by a ladder shape and shorter branch length, has less genetic diversity than other clades. Other than the monophyletic groups of African sequences (#2, #3and #5 in Fig. 2), monophyletic groups of non-African strains, #1 and #4, in Asia (China, Nepal, India, Myanmar, Bangladesh and Thailand), North America (USA) and Europe (Belgium) presented relatively higher genetic diversity. The German and South Africa strains in monophyletic group #4 provide evidence for viral movements between Asia and Africa or Europe. However, because of the limited sampling in these non-African regions, the viral migration patterns between Africa and other continents remain indecisive (Fig. 2).
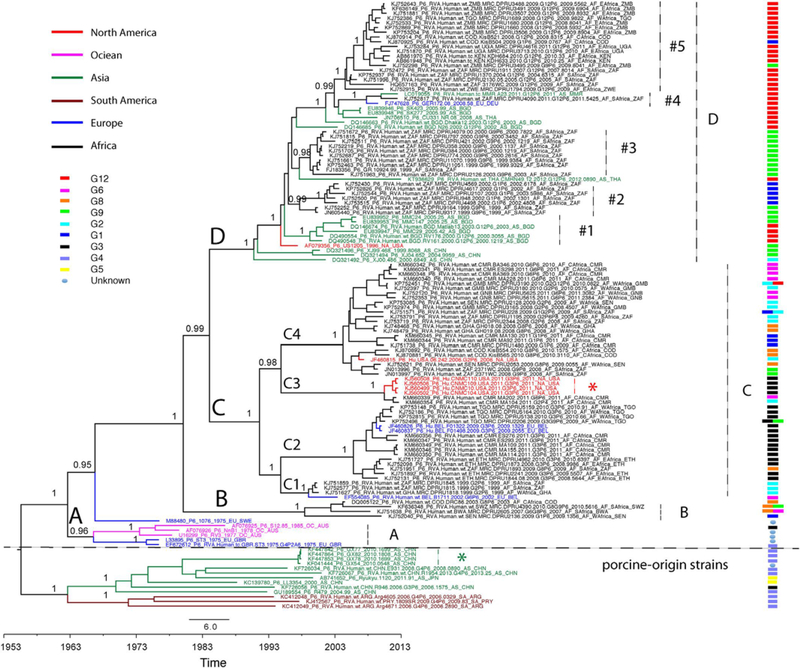
In addition, a time-scaled maximum clade credibility (MCC) phylogenetic tree was generated to estimate the evolutionary history of genotype P[6] strains, especially in Africa. This Bayesian Markov Chain Monte Carlo (BMCMC) phylogeny showed a highly consistent topology to the non-temporal maximum likelihood phylogeny (Fig. 3). The long branch lengths of porcine-like strains and human strains in clades A-D represented a relatively high degree of genetic divergence, and indicates that the P[6] strains have been circulating for many years prior to being detected or sampled. The Bayesian approach allows for estimation of the divergence time between porcine-like strains and human strains as well as the evolutionary history of the P[6] strains in Africa. The time to the most recent common ancestor (tMRCA) of porcine-like P[6] stains and human P[6] strains was estimated to be August 1965 with a 95% highest posterior density (HPD) between November 1959 and September 1970. The tMRCA of African P[6] strains was estimated as January 1979 with a 95% HPD between May 1972 and January 1985. Accordingly, the mean evolutionary rate of all genotype P[6] strains was estimated to be 1.05E-3 substitutions/site/year with a 95% HPD between 8.68E-4 and 1.25E-3 substitutions/site/year.
Fig. 3.
Time-scaled maximum clade credibility (MCC) tree of RVA VP4 segment P[6] genotype. Major nodes with posterior probabilities over 0.95 are indicated in the tree. Sequences from different continents are colored and described in the legend key. clades, subclades, and monophyletic groups are marked in the tree. G genotype of the sequences are also indicated at the right side of the tree and described in the legend key. The scale bar represents times in years.
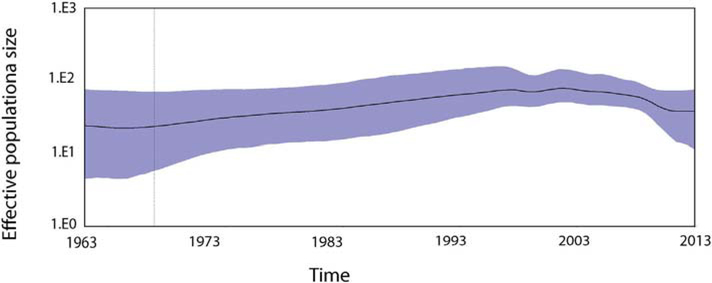
The demographic analysis by sky ride plot suggests the constant dynamic model of the P[6] strains in the population over the years (Fig. 4). The effective population size of P[6] strains remains the same level in > 50 years and it does not change even after more P[6] strains were isolated in Africa since 1999.
Fig. 4.
Past population dynamics estimated by Bayesian colascent skyline plot. The y-axis is estimated effective population size on a log-scale, and the x-axis is time in years. The black line is the median estimates of the estimated effective population size and the blue area shows the upper and the lower estimates of 95% confidence interval.
3.3. Phylogeographic clustering and G-genotype clustering of P[6] strains
To determine the phylogeographic structure of P[6] strain, particularly in Africa, a Bayesian Tip-association Significance testing (BaTS) for phylogeny-trait association tests on BMCMC phylogenies of P[6] strains were performed (Table 1). The results confirmed that there was a significant phylogenetic clustering by geographical location (p-values for AI and PS < 0.001), indicating stronger spatial clustering of the virus by sampling country than expected by chance alone. Additionally, BaTS analysis also revealed that the topology of phylogenetic trees of P [6] strains was influenced by G genotype (p-values for AI and PS < 0.001).
Table 1.
Results of the phylogeny-trait association tests for G genotype and geography of P[6] strains.
| Comparison | Statistica | Observed valueb | Null valueb | P |
|---|---|---|---|---|
| G genotype | AI | 3.24 (2.76–3.75) | 13.01 (12.04–13.92) | < 0.0001 |
| PS | 34.80 (34.0–35.0) | 86.18 (81.98–89.93) | < 0.0001 | |
| Region | AI | 2.27 (1.91–2.67) | 12.55 (11.44–13.52) | < 0.0001 |
| PS | 34.08 (33.0–35.0) | 82.35 (77.94–86.87) | < 0.0001 |
AI, association index; PS, parsimony score.
Value with 95% confidence interval.
4. Discussion
This study reports on 57 newly acquired complete genome sequences of P[6] strains from Africa. The new data significantly increased the number of publicly-available P[6] VP4 full-length sequences and for the first time allowed a comprehensive study of the evolution of P[6] strains in Africa. Genotype P[6] strains in combination with a variety of G-genotypes are detected at high frequencies in Africa (Steele and Ivanoff, 2003; Bànyai et al., 2012; Seheri et al., 2014). The wide range of G-types in in this study in combination with genotype P[6] indicates the high degree of genetic diversity of P[6] strains in Africa. From the total number of samples analysed, most were associated with DS-l-like genogroup, with the whole genome constellation bearing a DS-l-like genetic backbone (n = 35/57), while the minority had the Wa-like backbone (n = 22/57). Interestingly, reassortant gene segments were found from both genome constellations. The majority of the G genotypes circulating with the P[6] were G12 and G9 which are regarded as emerging and re-emerging strains that circulate most rapidly across human populations (Matthijnssens and Van Ranst, 2012).
The detection of uncommon strains like RVA/Human-wt/GNB/ MRC-DPRU5625/2011/G6P [6] and RVA/Human-wt/GNB/MRC-DPRU5615/2011/G6P[6] (both from Guinea Bissau) with constellations of G6-P[6]-I2-R2-C2-M2-A2-N2-T2-E2-H2 and mixed infections of strain RVA/Human-wt/BWA/MRC-DPRU2605/2007/G6G8P[6] from Botswana with a constellation of G6,G8-P[6]-I2-R2-C2-M2-A2-N2-T2-E2-H2, is noteworthy. The G6P[6] strains had only previously been reported from a child who migrated to Belgium from Mali in 2003 (Rahman et al., 2003). A decade later, G6P[6] strains were pre-dominantly seen in studies from Burkina Faso and Cameroon (Nordgren et al., 2012; Ndze et al., 2014). Our findings from the two strains from Guinea Bissau and one from Botswana and also G6P[6] strains reported in other studies, may suggest that the G6P[6] strain with a DS-l-like background is an important emerging pathogen in paediatric RVA diarrhea cases, especially in central, southern and west Africa. A whole genome analysis study of G6P[6] strains from Ghana reported that such viruses evolved from a single ancestral VP7 sequence, originating from a human G6P[9] strain that was estimated to have been collected in 1998 (Agbemabiese et al., 2015).
Genotype G3 is mostly and normally in combination with P[8] under the Wa-like backbone (Matthijnssens and Van Ranst, 2012). Here, genotype G3 in combination with P[6] were under the DS-l-like genogroup constellation and only identified from two countries (Ethiopia and Togo). It is speculated that genotype G3, just like the G4 could be vanishing from circulation in many African countries. The study genotypes G1 and G2 in combination with P[6] were under pure Wa-like and DS-l-like genogroup constellations, respectively.
The comparative occurrence of human P[6] strains outside of Africa is relatively low, albeit outbreaks of P[6] strains have been described in some developed countries (clark et al., 2011; Martella et al., 2008). The P[6] strains are regarded as a major P-type in porcine rotaviruses (Ciarlet et al., 1995; Lorenzetti et al., 2011). The fact that genotype P [6] is more frequently detected in developing countries is also believed to be due to the lifestyles attributed to rural populations in developing countries, where human and animals often live in close proximity. The prevailing rotavirus strains change annually and geographically (Kirkwood, 2010; Seheri et al., 2014). This diversity of circulating rotaviruses is generated by several mechanisms which were evident within the P[6] strains studied. They included reassortment events that lead to viruses with novel genetic and antigenic characteristics like was the case for strains MRC-DPRU9164 and MRC-DPRU9317, as well as interspecies transmission from animals to a human host as was the case for strains MRC-DPRU5615, MRC-DPRU5625 and MRC-DPRU2605, most probably due to living in dose proximity with the animals. Further, nudeotide and amino add mutations that may eventually lead to changes in receptors that facilitate interspecies transmission could easily occur in cases where multiple strains infed a single host.
A large number of mixed infections (n = 7) were reported in combination with genotype P[6] in this study. Mixed infections have been observed in high frequencies in several RVA studies across Africa (Todd et al., 2010; Dóró et al., 2014; Seheri et al., 2014). We propose that mixed infections and also the reassortant strains observed are part of the evolutionary mechanisms that make genotype P[6] more genetically prevalent in Africa, as compared to other regions around the globe. The diverse genotype combinations may represent potential reassortants between human and animal RVA strains because of persistent coinfections, as evidenced by the detection of > 11 genetically-distinct segments from a subset of samples.
Besides the reassortment events, which are facilitated by a high frequency of mixed infections, the evolution of rotavirus is also shaped by a high substitution rate due to random mutations introduced by an error-prone polymerase. Previous studies measured the evolutionary rate of the human VP4 gene at 5.8E-4 substitutions/site/year (Jenkins et al., 2002), which is lower than that of human RVA VP7 gene, 1.45– 1.87E-3 substitutions/site/year for genotypes G2, G9 and G12 strains (Dennis et al., 2014b; Matthijnssens et al., 2010). However, our Bayesian analysis estimated that the P[6] VP4 gene has a substitution rate with a mean of 1.05E-3 substitutions/site/year, which is higher than the rate that had previously been estimated for the VP4 gene employing a different maximum likelihood method (Jenkins et al., 2002). Nevertheless, the observed substitution rate (1.05E-3 substitutions/site/year) appears well within the normal range of the evolutionary rates of rotavirus genes including the VP4 genes when compared to other published evolutionary rates calculated by the similar, more contemporary Bayesian methods (Duffy et al., 2008; Barr and Fearns, 2010; Zeller et al., 2015).
The different substitution rates for the VP4 gene could be due to a higher number of sequences in the current study and is much closer to the observed rate for the VP7 gene. Although the evolutionary rate of P [6] VP4 gene is lower than the substitution rate of HA gene of human influenza A virus (5E-3 substitutions/site/year) (Rambaut et al., 2008) and V3 gene of HIV-1 (6.7E-3 substitutions/site/year) (Leitner and Albert, 1999), it is comparable to the rates of Dengue virus (IE-3 substitutions/site/year) (Chu et al., 2013) and Zika virus (IE-3 substitutions/site/year) (Faria et al., 2016). It is also higher than many other RNA viruses, such as human respiratory syncytial virus A (RSVA) (5E-4 substitutions/site/year) (Schobel et al., 2016), Chikungunya virus (4E-4 substitutions/site/year) (Chen et al., 2016), West Nile virus (5E-4 substitutions/site/year) (Añez et al., 2013) and Ebola virus (8E-4 substitutions/site/year) (Gire et al., 2014).
Apart from the global spread of P[8] and G[2] (Dennis et al., 2014b; Mijatovic-Rustempasic et al., 2014) and the recent emergence of G12 strains (Matthijnssens et al., 2010), our phylogenetic analyses confirm the endemic infection of human RVA P[6] strains, which is supported by the extensive genetic diversity and long-time local evolution of these viruses. In the study, we defined clades (A, B, C and D), subclades in clade C and monophyletic groups in clade D. The long internal branches of clades and subclades in phylogenetic trees demonstrated extensive genetic diversity and confirmed that P[6] strains have been circulating for many years prior to being detected or sampled. The phylogenetic analyses also confirmed the human-origin porcine-like strains, suggesting the relatively recent and direct transmission of porcine strains to humans worldwide (Asia, Europe, Africa, North America and South America). The most recent common ancestor of porcine-origin (or porcine-like) strains and human strains were predicted to have existed since the 1950s by Bayesian analysis, indicating approximately 20 years of un-sampled circulation. Despite their widespread circulation in Europe, Oceaniaand South America; certain clade A P[6] strains appeared to have died out over time. The P[6] strains collected in Africa after the late 1990s belonged to clades B, C and D, with the exception of porcine-like strains, which were phylogenetically distinct from clade A. The origin of clades B, C and D was dated to the 1970s, also indicating around 20 years of un-sampled circulation. Despite the genotype P6 strains being isolated from outside of Africa, they most likely originated in Africa before spreading to other continents, clades B and C may have an African origin. However, non-African strains towards the bottom of clade D in the phylogenetic trees present the possibility that clade D strains were introduced into Africa from other regions (likely Asia), although the possibility we of an African origin could not completely be ruled out due to the limited sampling in non-African regions, especially before the late 1990s. Endemic infection of P[6] strains, especially in Africa, is also supported by phylogeographic clustering and G-genotype clustering of P[6] strains, lending further support to the idea that these viruses evolved locally rather than having undergone spatial mixing in the various regions. Furthermore, the constant demographic dynamic model of P[6] in the past 60 years, as shown by the Bayesian sky ride plot, provided evidence that genotype P[6] is endemic in Africa. There was no evidence for large outbreaks or massive rapid spread of P[6] strains in Africa.
5. Conclusion
The diversity of the rotavirus genome, predominantly the wide multiplicity of the G and P genotype combinations, could be among the factors that possibly constrain the absolute control of rotavirus disease by the currently licenced rotavirus vaccines, especially in sub-Saharan Africa. Such genetic diversity is recognised to be generally greater in developing countries of Africa and Asia where rotavirus vaccine efficacy is still lower than in industrialized countries (Armah et al., 2016; Cunliffe et al., 2012; Groome et al., 2014; Parashar et al., 2016; Santos et al., 2016; Tate et al., 2016b).
The collection of comprehensive VP4 sequence data is instrumental in revealing the significance of genotype P[6] in Africa and other parts of the world. The full-genome and the Bayesian analyses of the VP4 gene indicated that genotype P[6] is endemic in Africa and not due to outbreaks. Multiple mechanisms such as reassortment events, interspecies transmission and random mutations (e.g. amino acid substitutions) are likely the primary contributing factors to rapid spread of the P(6) strains and rotavirus diversity in Africa. These mechanisms are particularly common in viruses with segmented genomes and could be occurring between different P and G genotypes circulating in Africa. This could potentially have a negative impact on the efficacy of the current RVA vaccines. The results underscore the importance of complete genome characterization of RVA strains and the need for continued surveillance of RVA globally.
Supplementary Material
Acknowledgments
The financial assistance of the South African Medical Research Council (SAMRC) is hereby acknowledged. Research reported in this publication was supported in part by the SAMRC under a Self-Initiated Research Grant. The project was also funded in part by US federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under Award Number U19AI110819. The views and opinions expressed in this article are not those of the funding and affiliated institutions but are of the authors of this publication.
We thank representatives from Ministries of Health from all participating countries who sent samples during the African Rotavirus Surveillance Network training workshops at the MRC-DPRU laboratory, which is a WHO-RRL in South Africa and also the dedicated team at MRC-DPRU, especially Ina Peenze, Kebareng Rakau and Nkululeko Magagula.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2018.05.013.
References
- Agbemabiese CA, Nakagomi TV, Suzuki Y, Armah G, Nakagomi O, 2015. Aug. Evolution of a G6P[6] rotavirus strain isolated from a child with acute gastroenteritis in Ghana, 2012. J. Gen. Virol 96 (8), 2219–2231. [DOI] [PubMed] [Google Scholar]
- Añez G, Grinev A, Chancey C, Ball C, Akolkar N, Land KJ, Winkelman V, Strainer SL, Kramer LD, Rios M, 2013. May 30 Evolutionary dynamics of West Nile virus in the United States, 1999–2011: phylogeny, selection pressure and evolutionary time-scale analysis. PLoS Negl Trop. Dis 7 (5), e2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armah GE, Pager CT, Asmah RH, et al. , 2001. Prevalence of unusual human rotavirus strains in Ghanaian children. J. Med. Virol 63, 67–71. [PubMed] [Google Scholar]
- Armah GE, Sow SO, Breiman RF, Dallas MJ, Tapia MD, Feikin DR, Binka FN, Steele AD, Laserson KF, Ansah NA, Levine MM, Lewis K, Coia ML, Attah-Poku M, Ojwando J, Rivers SB, Victor JC, Nyambane G, Hodgson A, Schödel F, Ciarlet M, Neuzil KM, 2010. August 21 Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet 376 (9741), 606–614. [DOI] [PubMed] [Google Scholar]
- Armah G, Pringle K, Enweronu-Laryea CC, Ansong D, Mwenda JM, Diamenu SK, Narh C, Lartey B, Binka F, Grytdal S, Patel M, Parashar U, Lopman B, 2016. May 1 Impact and effectiveness of monovalent rotavirus vaccine against severe rotavirus diarrhea in Ghana. Clin. Infect. Dis 62 (Suppl. 2), S200–S207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bànyai K, Làzlò B, Duque J, Steele AD, Nelson EA, Gentsch JR, Parashar UD, 2012. Systemic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine 30 (1), A122–A130. [DOI] [PubMed] [Google Scholar]
- Barr JN, Feams R, 2010. Jun. How RNA viruses maintain their genome integrity. J. Gen. Virol 91 (Pt 6), 1373–1387. [DOI] [PubMed] [Google Scholar]
- Chen R, Puri V, Fedorova N, Lin D, Hari KL, Jain R, Rodas JD, Das SR, Shabman RS, Weaver SC, 2016 Nov 14. Comprehensive genome scale phylogenetic study provides new insights on the global expansion of chikungunya virus. J. Virol 90 (23), 10600–10611 (Print 2016. December 1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu PY, Ke GM, Chen PC, Liu LT, Tsai YC, Tsai JJ, 2013. September 9 Spatiotemporal dynamics and epistatic interaction sites in dengue virus type 1: a comprehensive sequence-based analysis. PLoS One 8 (9), e74165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarlet M, Ludert JE, Liprandi F, 1995. Comparative amino add sequence analysis of the major outer capsid protein (VP7) of porcine rotaviruses with G3 and G5 serotype spedfidties isolated in Venezuela and Argentina. Arch. Virol 140 (3), 437–451. [DOI] [PubMed] [Google Scholar]
- Clark HF, Lawley D, DiStefano D, Maliga M, Kilby B, Kulnis G, Mallette L, DiNubile MJ, 2011. Jun. An unusual outbreak of rotavirus genotype G2P[6] during the 2005–2006 epidemic season in Philadelphia. Diagn. Microbiol. Infect. Dis 70 (2), 218–222. [DOI] [PubMed] [Google Scholar]
- Cunliffe NA, Witte D, Ngwira BM, Todd S, Bostock NJ, Turner AM, Chimpeni P, Vidor JC, Steele AD, Bouckenooghe A, Neuzil KM, 2012. April 27 Efficacy of human rotavirus vaccine against severe gastroenteritis in Malawian children in the first two years of life: a randomized, double-blind, placebo controlled trial. Vaccine. 30 (Suppl l:A36–43). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis FE, Fujii Y, Haga K, et al. , 2014a. Identification of novel Ghanaian G8P[6] humanbovine reassortant rotavirus strain by next generation sequencing. PLoS One 9 el00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis AF, McDonald SM, Payne DC, Mijatovic-Rustempasic S, Esona MD, Edwards KM, Chappell JD, Patton JT, 2014. Aprb. Molecular epidemiology of contemporary G2P[4] human rotaviruses codrculating in a single U.S. community: footprints of a globally transitioning genotype. J. Virol 88 (7), 3789–3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dóró R, László 2, Martella V, Leshem E, Gentsch J, Parashar U, Bányai K, 2014. Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: is there evidence of strain selection from vaccine pressure? Infect. Genet. EvoL 28, 446–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, 2007. November 8 BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol 7, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S, Shackelton LA, Holmes EC, 2008. Apr. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet 9 (4), 267–276. [DOI] [PubMed] [Google Scholar]
- Estes MK, Greenberg HB, Knipe DM, Howley PM, 2013. Rotaviruses In: Fields Virology, sixth ed. vol. 2 Lippincott, Williams and Wilkins, Philadelphia, PA: (pp.). [Google Scholar]
- Faria NR, Azevedo RDSDS, Kraemer MUG, Souza R, Cunha MS, Hill SC, Thézé J, Bons all MB, Bowden TA, Rissanen I, Rocco IM, Nogueira JS, Maeda AY, Vasami FGDS, Macedo FLL, Suzuki A, Rodrigues SG, Cruz ACR, Nunes BT, Medeiros DBA, Rodrigues DSG, Queiroz ALN, da Silva EVP, Henriques DF, da Rosa EST, de Oliveira CS, Martins LC, Vasconcelos HB, Casseb LMN, Simith DB, Messina JP, Abade L, Lourengo J, Alcantara LCJ, de Lima MM, Giovanetti M, Hay S, de Oliveira RS, Lemos PDS, de Oliveira LF, de Lima CPS, da Silva SP, de Vasconcelos JM, Franco L, Cardoso JF, Vianez-Júnior JLDSG, Mir D, Bello G, Delatorre E, Khan K, Creatore M, Coelho GE, de Oliveira WK, Tesh R, Pybus OG, Nunes MRT, Vasconcelos PFC, 2016. April 15 Zika virus in the Americas: early epidemiological and genetic findings. Science 352 (6283), 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, Wohl S, Moses LM, Yozwiak NL, Winnicki S, Matranga CB, Malboeuf CM, Qu J, Gladden AD, Schaffner SF, Yang X, Jiang PP, Nekoui M, Colubri A, Coomber MR, Fonnie M, Moigboi A, Gbakie M, Kamara FK, Tucker V, Konuwa E, Saffa S, Sellu J, Jalloh AA, Kovoma A, Koninga J, Mustapha I, Kargbo K, Foday M, Yillah M, Kanneh F, Robert W, Massally JL, Chapman SB, Bochicchio J, Murphy C, Nusbaum C, Young S, Birren BW, Grant DS, Scheiffelin JS, Lander ES, Happi C, Gevao SM, Gnirke A, Rambaut A, Garry RF, Khan SH, Sabeti PC, 2014. September 12 Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345 (6202), 1369–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groome MJ, Page N, Córtese MM, Moyes J, Zar HJ, Kapongo CN, Mulligan C, Diedericks R, Cohen C, Fleming JA, Seheri M, Mphahlele J, Walaza S, Kahn K, Chhagan M, Steele AD, Parashar UD, Zell ER, Madhi SA, 2014. Nov. Effectiveness of monovalent human rotavirus vaccine against admission to hospital for acute rotavirus diarrhoea in South African children: a case-control study. Lancet Infect. Dis 14 (11), 1096–1104. [DOI] [PubMed] [Google Scholar]
- Heylen E, Zeller M, Ciarlet M, Lawrence J, Steele D, Van Ranst M, Matthijnssens J, 2016. Human P[6] rotaviruses from Sub-Saharan Africa and Southeast Asia are closely related to those of human P[4] and P[8] rotaviruses circulating worldwide. J. Infect. Dis 214 (7), 1039–1049. [DOI] [PubMed] [Google Scholar]
- Hu L, Crawford SE, Czako R, et al. , 2012. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 485, 256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang KP, Wu FT, Bányai K, Wu HS, Yang DC, Huang YC, Lin JS, Hsiung CA, Huang JC, Jiang B, Gentsch JR, 2012. Jul. Identification of porcine rotavirus-like genotype P[6] strains in Taiwanese children. J. Med. Microbiol 61 (Pt 7), 990–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC, 2002. Feb. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol 54 (2), 156–165. [DOI] [PubMed] [Google Scholar]
- Kirkwood CD, 2010. September 1 Genetic and antigenic diversity of human rotaviruses: potential impact on vaccination programs. J. Infect. Dis 202 (Suppl), S43–S48. [DOI] [PubMed] [Google Scholar]
- Leitner T, Albert J, 1999. September 14 The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc. Natl. Acad. Sd. U. S. A 96 (19), 10752–10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzetti E, da Silva Medeiros TN, Alfieri AF, Alfieri AA, 2011. December 29 Genetic heterogeneity of wild-type G4P[6] porcine rotavirus strains detected in a diarrhea outbreak in a regularly vaccinated pig herd. Vet. Microbiol 154 (1–2), 191–196. [DOI] [PubMed] [Google Scholar]
- Madhi SA, Cunliffe NA, Steele D, Witte D, Kirsten M, Louw C, Ngwira B, Victor JC, Gillard PH, Cheuvart BB, Han HH, Neuzil KM, 2010. January 28 Effect of human rotavirus vaccine on severe diarrhea in African infants. N. Engl. J. Med 362 (4), 289–298. [DOI] [PubMed] [Google Scholar]
- Maes P, Matthijnssens J, Rahman M, Van Ranst M, 2009. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 9, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magagula NB, Esona MD, Nyaga MM, Stucker KM, Halpin RA, Stockwell TB, Seheri ML, Steele AD, Wentworth DE, Mphahlele MJ, 2015. Jan. Whole genome analyses of G1P[8] rotavirus strains from vaccinated and non-vaccinated South African children presenting with diarrhea. J. Med. Virol 87 (1), 79–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V, Banyai K, Ciarlet M, et al. , 2006. Relationships among porcine and human P[6] rotaviruses: evidence that the different human P[6] lineages have originated from multiple interspecies transmission events. Virology 344, 509–519. [DOI] [PubMed] [Google Scholar]
- Martella V, Colombrita D, Lorusso E, Draghin E, Fiorentini S, De Grazia S, Bányai K, Ciarlet M, Caruso A, Buonavoglia C, 2008. Oct. Detection of a porcine-like rotavirus in a child with enteritis in Italy. J. Clin. Microbiol 46 (10), 3501–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Van Ranst M, 2012. Genotype constellation and evolution of group a rotaviruses infecting humans. Curr. Opin. Virol 2, 426–433. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gomara M, Maes P, Patton JT, Rahman M, Van Ranst M, 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-l-like and bovine rotavirus strains. J. Virol 82, 3204–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Heylen E, Zeller M, Rahman M, Lemey P, Van Ranst M, 2010. Oct. Phylodynamic analyses of rotavirus genotypes G9 and G12 underscore their potential for swift global spread. Mol. Biol. Evol 27 (10), 2431–2436. [DOI] [PubMed] [Google Scholar]
- Mijatovic-Rustempasic S, Teel EN, Kerin TK, Hull JJ, Roy S, Weinberg GA, Payne DC, Parashar UD, Gentsch JR, Bowen MD, 2014. Jan. Genetic analysis of G12P[8] rotaviruses detected in the largest U.S. G12 genotype outbreak on record. Infect. Genet. Evol 21, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagomi T, Do LP, Agbemabiese CA, Kaneko M, Gauchan P, Doan YH, Jere KC, Steele AD, Iturriza-Gomara M, Nakagomi O, Cunliffe NA, 2017. Jan. Whole-genome characterisation of G12P[6] rotavirus strains possessing two distinct genotype constellations co-circulating in Blantyre, Malawi, 2008. Arch. Virol 162 (1), 213–226. [DOI] [PubMed] [Google Scholar]
- Ndze VN, Esona MD, Achidi EA, et al. , 2014. Full genome characterization of human rotavirus a strains isolated in Cameroon, 2010–2011: diverse combinations of the G and P genes and lack of reassortment of the backbone genes. Infect. Genet. Evol 28, 537–560. [DOI] [PubMed] [Google Scholar]
- Nordgren J, Bonkoungou IJ, Nitiema LW, Sharma S, Ouermi D, Simpore J, Barro N, Svensson L, 2012. Dec. Rotavirus in diarrheal children in rural Burkina Faso: high prevalence of genotype G6P[6]. Infect. Genet. EvoL 12 (8), 1892–1898. [DOI] [PubMed] [Google Scholar]
- Nordgren J, Sharma S, Bucardo F, et al. , 2014. Both Lewis and secretor status mediate susceptibility to rotavirus infections in a rotavirus genotype-dependent manner. Clin. Infect. Dis 59, 1567–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyaga MM, Jere KC, Peenze I, Mlera L, van Dijk AA, Seheri ML, Mphahlele MJ, 2013. Sequence analysis of the whole genomes of five African human G9 rotavirus strains. Infect. Genet. Evol 16, 62–77. [DOI] [PubMed] [Google Scholar]
- Nyaga MM, Stucker KM, Esona MD, Jere KC, Mwinyi B, Shonhai A, Tsolenyanu E, Mulindwa A, Chibumbya JN, Adolfine H, Halpin RA, Roy S, Stockwell TB, Berejena C, Seheri ML, Mwenda JM, Steele AD, Wentworth DE, Mphahlele MJ, 2014. Whole-genome analyses of DS-l-like human G2P[4] and G8P [4] rotavirus strains from Eastern, Western and Southern Africa. Virus Genes 49 (2), 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyaga MM, Jere KC, Esona MD, Seheri ML, Stucker KM, Halpin RA, Akopov A, Stockwell TB, Peenze I, Diop A, Ndiaye K, Boula A, Maphalala G, Berejena C, Mwenda JM, Steele AD, Wentworth DE, Mphahlele MJ, 2015. Whole genome detection of rotavirus mixed infections in human, porcine and bovine samples co-infected with various rotavirus strains collected from sub-Saharan Africa. Infect. Genet. EvoL 31, 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyaga MM, Peenze I, Potgieter CA, Seheri LM, Page NA, Yinda CK, Steele AD, Matthijnssens J, Mphahlele MJ, 2016. Complete genome analyses of the first porcine rotavirus group H identified from a South African pig does not provide evidence for recent interspecies transmission events. Infect. Genet. EvoL 38, 1–7. [DOI] [PubMed] [Google Scholar]
- O’Ryan M, Giaquinto C, Benninghoff B, 2015. Human rotavirus vaccine (Rotarix): focus on effectiveness and impact 6 years after first introduction in Africa. Expert Rev. Vaccines. 14 (8), 1099–1112. [DOI] [PubMed] [Google Scholar]
- Parashar UD, Johnson H, Steele AD, Tate JE, 2016. May 1 Health impact of rotavirus vaccination in developing countries: progress and way forward. Clin. Infect. Dis 62 (Suppl. 2), S91–S95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, Rambaut A, Pybus OG, 2008. May. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol 8 (3), 239–246. [DOI] [PubMed] [Google Scholar]
- Path, 2016. http://www.path.org/vaccineresources/details.php?i=2235.
- Platts-Mills JA, Amour C, Gratz J, Nshama R, Walongo T, Mujaga B, Maro A, McMurry TL, Liu J, Mduma E, Houpt ER, 2017. May 29 Impact of rotavirus vaccine introduction and post-introduction etiology of diarrhea requiring hospital admission in Haydom, Tanzania, a rural African setting. Clin. Infect. Dis 10.1093/cid/cix494. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, De Leener K, Goegebuer T, Wollants E, Van der Donck I, Van Hoovels L, Van Ranst M, 2003. May. Genetic characterization of a novel, naturally occurring recombinant human G6P[6] rotavirus. J. Clin. Microbiol 41 (5), 2088–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC, 2008. May 29 The genomic and epidemiological dynamics of human influenza A virus. Nature 453 (7195), 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota Council, 2016. http://rotacouncil.org/news/inside-the-12th-annual-intemational-rotavirus-symposium/.
- Santos VS, Marques DP, Martins-Filho PR, Cuevas LE, Gurgel RQ, 2016. August 12 Effectiveness of rotavirus vaccines against rotavirus infection and hospitalization in Latin America: systematic review and meta-analysis. Infect Dis Poverty. 5 (1), 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobel SA, Stucker KM, Moore ML, Anderson LJ, Larkin EK, Shankar J, Bera J, Puri V, Shilts MH, Rosas-Salazar C, Halpin RA, Fedorova N, Shrivastava S, Stockwell TB, Peebles RS, Hartert TV, Das SR, 2016. May 23 Respiratory syncytial virus whole-genome sequencing identifies convergent evolution of sequence duplication in the C-terminus of the G gene. Sci. Rep 6, 26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seheri M, Nemarude L, Peenze I, Netshifhefhe L, Nyaga MM, Ngobeni HG, Maphalala G, Maake LL, Steele AD, Mwenda JM, Mphahlele JM, 2014. Update of rotavirus strains circulating in Africa from 2007 through 2011. Pediatr. Infect. Dis. J 33 (Suppl. 1), S76–S84. [DOI] [PubMed] [Google Scholar]
- Steele AD, Ivanoff B, 2003. Rotavirus strains circulating in Africa during 1996–1999: emergence of G9 strains and P[6] strains. Vaccine 21 (5–6), 361–367. [DOI] [PubMed] [Google Scholar]
- Stucker KM, Stockwell TB, Nyaga MM, Halpin RA, Fedorova N, Akopov A, Ngoveni H, Peenze I, Seheri ML, Mphahlele MJ, Wentworth DE, 2015. Complete genomic sequence for an avian group G rotavirus from South Africa. Genome Announc. 3 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, 2013. Dec. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol 30 (12), 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JE, Burton AH, Boschi-Pinto C, Parashar UD, 2016a. World Health Organization-Coordinated Global Rotavirus Surveillance N. Global, Regional, and National Estimates of Rotavirus Mortality in Children. [DOI] [PubMed] [Google Scholar]
- Tate JE, Ngabo F, Donnen P, Gatera M, Uwimana J, Rugambwa C, Mwenda JM, Parashar UD, 2016. May lb. Effectiveness of pentavalent rotavirus vaccine under conditions of routine use in Rwanda. Clin. Infect. Dis 62 (Suppl. 2), S208–S212. [DOI] [PubMed] [Google Scholar]
- Todd S, Page NA, Duncan Steele A, Peenze I, Cunliffe NA, 2010. September 1 Rotavirus strain types circulating in Africa: Review of studies published during 1997–2006. J. Infect. Dis 202 (Suppl:S34–42). [DOI] [PubMed] [Google Scholar]
- Trojnar E, Sachsenroder J, Twardziok S, Reetz J, Otto PH, Johne R, 2013. Identification of an avian group A rotavirus containing a novel VP4 gene with a close relationship to those of mammalian rotaviruses. J. Gen. Virol 94 (Pt 1), 136–142. [DOI] [PubMed] [Google Scholar]
- Velázquez Raúl F., Linhares Alexandre C., Muñoz Sergio, Seron Pamela, Lorca Pedro, DeAntonio Rodrigo, Ortega-Barria Eduardo, 2017a. Efficacy, safety and effectiveness of licensed rotavirus vaccines: a systematic review and meta-analysis for Latin America and the Caribbean. BMC Pediatr. 17 (14) (Published online 2017 Jan 13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez RF, Linhares AC, Muñoz S, Seron P, Lorca P, DeAntonio R, Ortega-Barria E, 2017. January 13b. Efficacy, safety and effectiveness of licensed rotavirus vaccines: a systematic review and meta-analysis for Latin America and the Caribbean. BMC Pediatr. 17 (1), 14 10.1186/sl2887-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Kobayashi N, Nagashima S, Zhou X, Ghosh S, Peng JS, Hu Q, Zhou DJ, Yang ZQ, 2010. May. Full genomic analysis of a porcine-bovine reassort ant G4P[6] rotavirus strain R479 isolated from an infant in China. J. Med. Virol 82 (6), 1094–1102. [DOI] [PubMed] [Google Scholar]
- Zeller M, Donato C, Trovão NS, Cowley D, Heylen E, Donker NC, McAllen JK, Akopov A, Kirkness EF, Lemey P, Van Ranst M, Matthijnssens J, Kirkwood CD, 2015. August 8 Genome-wide evolutionary analyses of G1P[8] strains isolated before and after rotavirus vaccine introduction. Genome Biol. Evol 7 (9), 2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.