The untargeted discovery and replication of a blood protein panel shows promise for predicting preclinical Alzheimer’s disease.
Abstract
A blood-based assessment of preclinical disease would have huge potential in the enrichment of participants for Alzheimer’s disease (AD) therapeutic trials. In this study, cognitively unimpaired individuals from the AIBL and KARVIAH cohorts were defined as Aβ negative or Aβ positive by positron emission tomography. Nontargeted proteomic analysis that incorporated peptide fractionation and high-resolution mass spectrometry quantified relative protein abundances in plasma samples from all participants. A protein classifier model was trained to predict Aβ-positive participants using feature selection and machine learning in AIBL and independently assessed in KARVIAH. A 12-feature model for predicting Aβ-positive participants was established and demonstrated high accuracy (testing area under the receiver operator characteristic curve = 0.891, sensitivity = 0.78, and specificity = 0.77). This extensive plasma proteomic study has unbiasedly highlighted putative and novel candidates for AD pathology that should be further validated with automated methodologies.
INTRODUCTION
The search for a blood-based signature that can predict the onset of Alzheimer’s disease (AD) has gathered considerable momentum in recent years. Much effort has been dedicated to the discovery of single- and multianalyte protein markers to differentiate AD from age-matched cognitively unimpaired individuals [reviewed in (1)]. A major concern for the field has been the lack of reproducibility for these putative candidates, and these inconsistencies might be explained by substantial preanalytical variations among research cohorts. However, it is likely that the inherent heterogeneity of AD is partially accountable for the failure in replicating “case-versus-control” studies (2). Given the long preclinical phase in AD where the accumulation of pathology is thought to begin 15 to 20 years before clinical presentation (3), a considerable proportion of seemingly unimpaired individuals will harbor substantial amyloid β (Aβ) pathology. Consequently, within a clinical diagnosis–dependent study design, individuals in the preclinical disease phase would be classified as “healthy controls,” despite underlying Aβ pathology being present. Increasingly, “endophenotype” studies are using surrogate markers of neocortical Aβ burden [positron emission tomography (PET) imaging or cerebrospinal fluid (CSF) measures] to identify blood-based biomarkers indicative of ongoing disease pathogenesis. Tools to identify and monitor Aβ burden are of critical importance, given the increased focus on anti-Aβ therapeutics. Retrospective imaging from these early trials confirmed that a considerable proportion of recruited participants did not exhibit the target pathology or were too advanced in the disease course (4). This was a major concern and highlighted the importance of a biomarker-driven participant selection, and consequently, these trials are targeting individuals with biomarker evidence of pathology with no or little cognitive deficits. This selection process, based on PET imaging and/or CSF Aβ, is likely to be unfeasible to implement widely, particularly for population-based screening. Therefore, a minimally invasive and accessible blood-based prediction of Aβ burden would be of considerable use in therapeutic stratification and clinical management at the earliest stage.
Ultrasensitive immunoassay and immunoprecipitation mass spectrometry (MS) methods have recently reported plasma Aβ ratios as being able to predict Aβ PET (5, 6). However, there has been limited investigation using untargeted methods in the discovery of novel blood markers that could reflect Aβ burden. Studies using Aβ endophenotype stratification have seldom investigated preclinical AD and have used a range of technologies, including two-dimensional gel electrophoresis (7, 8) and immunoassay panels (9, 10), but these approaches have limitations (e.g., predetermined targets, large sample volumes, and large variability). At the discovery level, MS has the key advantage of unbiased measurement of features present within a sample without prior knowledge of its contents. This enables MS to be compatible with a hypothesis-free investigation, the results of which could be confirmed using targeted proteomics. The foremost criticism of shotgun MS workflows is poor sensitivity owing to the large dynamic range of protein abundances, which results in an inadequate coverage of the plasma proteome. The marked improvement in MS technology over recent years has renewed the interest in untargeted plasma proteins in many disease areas, and combined with immunodepletion of highly abundant proteins and extensive fractionation methods, it is possible to identify more than 1000 proteins (11). With this in mind, the aim of this study was to perform an extensive untargeted proteomic discovery in plasma to predict Aβ burden in preclinical disease. We primarily focused on 238 cognitively unimpaired individuals from the Australian Imaging, Biomarker and Lifestyle Flagship Study of Ageing (AIBL) and the Kerr Anglican Retirement Village Initiative in Ageing Health (KARVIAH) studies who had undergone PET to determine their Aβ status. Moreover, we took a comprehensive approach to replicating our findings in an independent sample set using the same methodological approach.
RESULTS
Subject demographics
The demographic and clinical characteristics of the cognitively unimpaired individuals are presented in Table 1. In AIBL, 100 participants were classified as Aβ negative (Aβ−) and 44 participants as Aβ positive (Aβ+) by 11C-PiB standardized uptake values ratio (SUVR) cutoff of 1.4. In KARVIAH, 59 participants were classified as Aβ− and 35 participants as Aβ+ by 18FBB (Florbetaben) SUVR cutoff of 1.35. There was a significant increase in the number APOE ε4 carriers in both Aβ+ groups, and the Aβ+ groups also had a tendency to be older. There was no significant difference in cognitive performance between Aβ− and Aβ+ groups (Table 1). A secondary analysis included a further 46 AIBL participants with a diagnosis of mild cognitive impairment (MCI; n = 21) or AD (n = 25). The characteristics of the full cohort including these subjects are presented in Table 2.
Table 1. Subject demographics for cognitively unimpaired individuals (AIBL, n = 144; KARVIAH, n = 94).
MMSE, Mini Mental State Examination; ns, not significant.
| AIBL discovery cohort (11C-PiB) | KARVIAH replication cohort (18FBB) | |||||
| Aβ− | Aβ+ | P value | Aβ− | Aβ+ | P value | |
| Number of subjects (n) | 100 | 44 | 59 | 35 | ||
| Aβ PET SUVR [means (SD)] |
1.16 (0.1) | 1.90 (0.3) | 1.11 × 10−54 | 1.16 (0.1) | 1.70 (0.2) | 8.35 × 10−23 |
| Gender; females [n (%)] |
51 (50.0) | 21 (50.0) | ns | 40 (67.8) | 21 (60.0) | ns |
| Age in years [means (SD)] |
70.8 (7.1) | 75.5 (6.9) | 0.001 | 77.5 (5.5) | 79.6 (5.3) | ns |
| APOE ε4 carrier [n (%)] | 27 (27) | 27 (61.4) | 7.70 × 10−4 | 6 (10.2) | 15 (42.9) | 0.001 |
| MMSE [means (SD)] | 28.9 (1.2) | 28.5 (1.3) | ns | 29.4 (1.6) | 29.1 (1.4) | ns |
Table 2. Subject demographics for the mixed diagnosis cohort (AIBL, n = 190; KARVIAH, n = 94).
n/a, not available.
| AIBL discovery cohort (11C-PiB) | KARVIAH replication cohort (18FBB) | |||||
| Aβ− | Aβ+ | P value | Aβ− | Aβ+ | P | |
| Number of subjects (n) | 108 | 82 | 59 | 35 | ||
| Aβ PET SUVR [means (SD)] |
1.16 (0.1) | 2.09 (0.4) | 7.44 × 10−60 | 1.16 (0.1) | 1.70 (0.2) | 8.35 × 10−23 |
| Gender; females [n (%)] |
54 (50.0) | 40 (48.7) | ns | 40 (67.8) | 21 (60.0) | ns |
| Age in years [means (SD)] |
71.17 (7.2) | 74.04 (7.7) | 0.011 | 77.5 (5.5) | 79.6 (5.3) | ns |
| Clinical diagnosis [n (%)] Cognitively unimpaired MCI AD |
100 (92.6) 7 (6.5) 1 (0.9) |
44 (53.7) 14 (17.1) 24 (29.2) |
3.38 × 10−11 | 59 (100.0) n/a n/a |
35 (100.0) n/a n/a |
n/a |
| APOE ε4 carrier [n (%)] | 28 (25.9) | 54 (65.9) | 5.91 × 10−5 | 6 (10.2) | 15 (42.9) | 0.001 |
| MMSE [means (SD)] | 28.7 (1.4) | 26.1 (4.0) | 5.96 × 10−7 | 29.4 (1.6) | 29.1 (1.4) | ns |
Plasma protein metrics
A total of 2356 individual protein groups at 5% false discovery rate (FDR) were measured across all experiments. The lowest observed concentration was 4.3 pg/ml (multiple epidermal growth factor–like domains protein 8), with 29% of protein groups measured in this study had reported concentrations in a reference database (Human Plasma Proteome Project; www.hupo.org/plasma-proteome-project/). A total of 560 protein groups were consistently measured across >75% of the sample set (table S1), and these protein groups were taken forward for statistical evaluation.
Plasma proteins associated with Aβ burden in a preclinical disease
Aβ PET SUVR measures from 238 cognitively unimpaired individuals (Table 1) were categorized into binary “Aβ−” or “Aβ+” classification, and adjusted scores for 560 plasma proteins were analyzed for their association with Aβ pathology. A total of 37 protein groups were found to be nominally associated with Aβ groups at the uncorrected P value of <0.05 (Fig. 1 and table S2A). After Benjamini-Hochberg multiple testing correction (FDR), five protein groups remained associated with Aβ classification (Q = <0.05; Table 3). An increased expression for Aβ A4 precursor protein (APP), neurogenin-2 (NGN2), neurofilament light polypeptide (NfL), and Aβ APP–binding family B member 3 (APBB3) in Aβ+ individuals was observed with a medium-to-large effect size (Fig. 1). A decreased protein expression in the Aβ+ group was found for RE1-silencing transcription factor (REST) with a small effect size (Cohen’s d = 0.46). After adjusting for the influence of APOE genotype, APP, NGN2, and NfL remained statistically increased in the Aβ+ group despite a weaker association (Q = <0.05; Table 3). Synaptosomal-associated protein 25 (SNAP25) was the only protein group to become nominally associated with Aβ burden after APOE adjustment but did not pass FDR (Fig. 1 and table S2A). Only four protein groups were uniquely significantly associated with Aβ+ in cognitively unimpaired individuals: DENN domain-containing protein 3 (DENN3), sentrin-specific protease 5 (SENP5), zinc finger CCCH domain–containing protein 13 (ZCH13), and cilia- and flagella-associated protein 43 (WDR96). In addition, NGN2, helicase-like transcription factor (HLTF), forkhead-associated domain-containing protein 1 (FHAD1), ribosomal protein S6 kinase alpha-3 (RPS6KA3), and signal-induced proliferation-associated 1–like protein 3 (SIPAIL3) had greater effect sizes in the cognitively unimpaired group despite being statistically significant in both analyses (Fig. 1).
Fig. 1. Pyramid plot to display the effect sizes (Cohen’s d) of protein significantly (P = <0.05) associated with Aβ burden (Aβ− versus Aβ+).
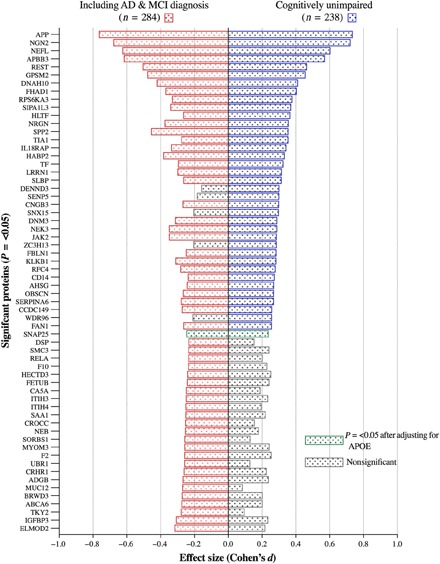
On the right are proteins associated with cognitively unimpaired individuals and the association with the addition of individuals with MCI and AD on the left. Gray bars illustrate a nonsignificant effect size.
Table 3. GLM-adjusted protein groups significantly associated with Aβ SUVR in cognitively unimpaired participants stratified by Aβ+/− classification after multiple testing correction.
Protein groups were also associated with Aβ SUVR with an adjustment for APOE genotype. All protein groups that are nominally associated with Aβ in cognitively unimpaired (P > 0.05) are shown in table S1.
| UniProt ID |
Protein group description |
Aβ+ group | Adjusted for APOE ε4 status | |||||||
| t |
Mean difference |
P value | Q | t |
Mean difference |
P value | Q value | |||
| P05067 | APP | ↑ | −5.651 | −0.290 | 4.57 × 10−08 | 2.08x10−05 | −4.686 | −0.241 | 4.71 × 10−06 | 0.001 |
| Q9H2A3 | NGN2 | ↑ | −5.556 | −0.319 | 7.43 × 10−08 | 2.08x10−05 | −4.787 | −0.276 | 2.99 × 10−06 | 0.001 |
| P07196 | NfL | ↑ | −4.639 | −0.229 | 5.80 × 10−06 | 0.001 | −3.716 | −0.184 | 2.53 × 10−04 | 0.047 |
| O95704 | APBB3 | ↑ | −4.389 | −0.274 | 1.71 × 10−05 | 0.002 | −3.374 | −0.210 | 0.001 | ns |
| Q13127 | REST | ↓ | 3.570 | 0.142 | 4.33 × 10−04 | 0.048 | 3.385 | 0.135 | 0.001 | ns |
Plasma proteins associated with Aβ burden including individuals with MCI and AD
We then combined an additional 46 participants from AIBL with an MCI, and AD diagnosis was included in a secondary analysis (table S3). A total of 56 protein groups were found to be statistically different between Aβ− and Aβ+ groups at the uncorrected P value of <0.05 (Fig. 1). After FDR, eight protein groups remained associated with Aβ classification (Q = <0.05; table S3A). These included APP, NGN2, NfL, APBB3, and REST, with the addition of dynein heavy chain 10 (DNAH10), G protein–signaling modulator 2 (GPSM2), and secreted phosphoprotein 24 (SPP2).
Most of protein groups had larger effect sizes when including Aβ+ individuals with AD/MCI, including APP, NfL, REST, SPP2, and NRGN (neurogranin). There were an additional 24 protein groups that were statistically significant in the AD/MCI group compared with the cognitively unimpaired. However, several of these proteins were similar in effect size, and therefore, the reduced sample size in the cognitively unimpaired meant that they did not reach statistical significance [e.g., prothrombin (F2); Fig. 1]. Protein groups that had substantially greater effect in AD/MCI were nonreceptor tyrosine-protein kinase (TKY2) and mucin-12 (MUC12).
Plasma protein classifier for Aβ positivity
Univariate analysis revealed several single plasma protein groups to be nominally associated with Aβ classification, with a small number of protein groups being strongly associated with Aβ status with medium-to-large effect sizes (Tables 3 and 4). While this univariate analysis studies associations one protein at a time, classifier methods do so with all proteins simultaneously, which enables detecting proteins that are related to disease only when considered in combination with other proteins. Owing to this, classifiers are able to achieve the greater sensitivity and specificity required for clinical implementation, although the proteins they select may not always coincide with those found by univariate methods.
Table 4. GLM-adjusted protein groups significantly associated with Aβ SUVR in all subjects stratified by Aβ+/− classification after Benjamini-Hochberg multiple testing corrections.
Protein groups were also associated with Aβ SUVR with an adjustment for APOE genotype. All protein groups that are nominally associated with Aβ (P > 0.05) are shown in table S2 (A and B).
| UniProt ID |
Protein group description |
Aβ+ group | Adjusted for APOE ε4 status | |||||||
| t | Mean difference | P value | Q | t | Mean difference | P value | Q value | |||
| P05067 | Aβ A4 protein | ↑ | −6.419 | −0.279 | 5.79 × 10−10 | 3.24 × 10−07 | −5.019 | −0.220 | 9.22 × 10−07 | 0.001 |
| Q9H2A3 | NGN2 | ↑ | −5.702 | −0.283 | 2.98 × 10−08 | 8.34 × 10−06 | −4.624 | −0.231 | 5.75 × 10−06 | 0.002 |
| P07196 | NfL | ↑ | −5.258 | −0.219 | 2.88 × 10−07 | 5.38 × 10−05 | −3.928 | −0.164 | 1.08 × 10−04 | 0.015 |
| O95704 | APBB3 | ↑ | −5.189 | −0.272 | 4.05 × 10−07 | 5.67 × 10−05 | −3.710 | −0.194 | 2.49 × 10−04 | 0.028 |
| Q13127 | REST | ↓ | 4.247 | 0.143 | 2.94 × 10−05 | 3.29 × 10−03 | 3.984 | 0.135 | 8.64 × 10−05 | 0.015 |
| P81274 | GPSM2 | ↓ | 4.016 | 0.124 | 7.61 × 10−05 | 0.007 | 3.564 | 0.111 | 4.28 × 10−04 | 0.040 |
| Q13103 | SPP2 | ↓ | 3.833 | 0.108 | 1.56 × 10−04 | 0.013 | 3.310 | 0.094 | 0.001 | 0.084 |
| Q8IVF4 | DNAH10 (axonemal) |
↑ | −3.548 | −0.090 | 4.54 × 10−04 | 0.032 | −3.165 | −0.081 | 0.002 | 0.120 |
The feature selection method “Ridge,” followed by a support vector machine (SVM) analysis, was used to create classifiers predicting Aβ+ in the AIBL cohort. We used either demographics only [gender, age, and APOE ɛ4 count (“demographic data”)] and/or proteins (“full data”). Each scenario produced 50 classifier models, with each classifier including one additional feature into the model. We tested each classifier in the independent KARVIAH dataset and calculated the area under the receiver operator characteristic (ROC) curve (AUC) to detect the best model at predicting Aβ positivity in each scenario. The optimal model to predict Aβ positivity in cognitively unimpaired individuals produced a testing AUC of 0.891 [cutoff = 0.63, specificity = 0.77, sensitivity = 0.78, positive predictive value (PPV) = 0.85, and negative predictive value (NPV) = 0.68; Fig. 2]. This classifier included 10 protein features (Table 5) and two demographic features (APOE ε4 count and age). A classifier that included participants with MCI and AD produced a testing AUC of 0.904 (cutoff = 0.687, specificity = 0.80, sensitivity = 0.81, PPV = 0.87, and NPV = 0.72; Fig. 3). This classifier included one demographic feature (APOE ε4 count) and nine protein features, of which eight proteins were common with the cognitively unimpaired classifier (Table 6). The optimum demographic data model demonstrated a testing AUC of 0.725 for the cognitively unimpaired cohort.
Fig. 2. Protein classifier to predict Aβ positivity in cognitively unimpaired individuals.
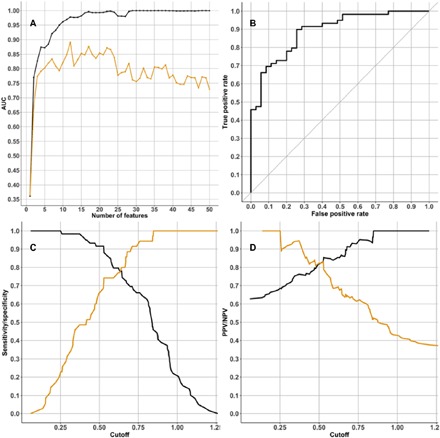
(A) Graph showing the AUC statistic of the 50 classifier models produced using the “cognitively unimpaired cohort” training dataset. The AUC when testing each classifier model in the training dataset is in black, and the AUC when testing the classifier model in the testing dataset (KARVIAH) is in orange. On the x axis is the number of features used in each classifier model. For the classifier with the best AUC in the testing dataset (this was the classifier that used 12 features; Table 5), three graphs access the classifier’s performance: (B) ROC curve, (C) sensitivity and specificity plotted in black and orange, respectively, and (D) PPV and NPV plotted in black and orange, respectively.
Table 5. Feature list for multianalyte classifier predicting elevated Aβ burden in cognitively unimpaired cohort.
The classifier was training in the AIBL cohort (n = 144), achieving a testing AUC of 0.891 in the KARVIAH cohort (n = 94). GPCR, G protein–coupled receptor; sens, sensitivity; spec, specificity.
|
Feature no. |
Cognitively unimpaired classifier for Aβ burden (Testing AUC = 0.891, sens = 0.78, spec = 0.77, NPV = 0.68, PPV = 0.85) |
||
| UniProt ID |
Feature description |
Tryptic amino acid peptide sequences contributing to protein scores | |
| 1 | P00734 | Prothrombin | 87–94, 98–116, 125–133, 178–198, 199–224, 217–224, 225–243, 248–263, 315–327, 328–344, 345–363, 354–363, 441–447, 447–452, 467–468, 518–537, 544–560, and 560–575 |
| 2 | Q8IZF3 | Adhesion GPCR F4 |
100–107, 315–328, 657–689, and 673–698 |
| 3 | P05067 | Aβ A4 protein | 677–687 and 688–699 |
| 4 | Q9H2A3 | NGN2 | 114–120 |
| 5 | n/a | APOE ε4 count | n/a |
| 6 | Q8IVF4 | DNAH10 (axonemal) |
207–223, 528–540, 879–915, 1133–1148, 1524–1528, 1870–1927, 2404–2436, 2906–2941, 2923–2945, and 4100–4139 |
| 7 | Q13127 | REST | 353–369, 727–742, and 882–896 |
| 8 | P07196 | NfL | 137–144, 154–164, 318–331, and 447–462 |
| 9 | P51812 | RPS6KA3 | 80–100, 109–132, 193–216, 226–242, 274–282, 508–525, and 570–591 |
| 10 | P81274 | GPSM2 | 267–278, 318–347, and 422–433 |
| 11 | B1AJZ9 | FHAD1 | 861–868, 876–887, 897–905, 931–942, 1030–1052, 1059–1070, and 1104–1139 |
| 12 | n/a | Age of participant |
n/a |
Fig. 3. Protein classifier to predict Aβ positivity that includes participants with MCI and AD.
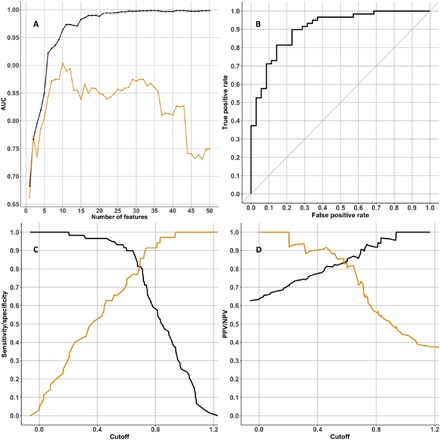
(A) Graph showing the AUC statistic of the 50 classifier models produced using the “mixed diagnosis cohort” training dataset. The AUC when testing each classifier model in the training dataset is in black, and the AUC when testing the classifier model in the testing dataset (KARVIAH) is in orange. On the x axis is the number of features used in each classifier model. For the classifier with the best AUC in the testing dataset (this was the classifier that used 10 features; Table 6), three graphs access the classifier’s performance: (B) ROC curve, (C) sensitivity and specificity plotted in black and orange, respectively, (D) PPV and NPV plotted in black and orange, respectively.
Table 6. Feature list for multianalyte classifier predicting elevated Aβ burden in a mixed diagnosis cohort.
The classifier was training in the AIBL cohort (n = 169), achieving a testing AUC of 0.905 in the KARVIAH cohort (n = 94).
| Feature no. |
Mixed diagnosis classifier for Aβ burden (Testing AUC = 0.904, sens = 0.81, spec = 0.80, NPV = 0.72, PPV = 0.87) |
||
| UniProt ID |
Feature description |
Tryptic amino acid peptide sequences contributing to protein scores | |
| 1 | n/a | APOE ε4 count* | n/a |
| 2 | P05067 | Aβ A4 protein* | 677–687 and 688–699 |
| 3 | P07196 | NfL* | 137–144, 154–164, 318–331, and 447–462 |
| 4 | Q9H2A3 | NGN2* | 114-120 |
| 5 | Q8IVF4 | DNAH10 (axonemal)* |
207–223, 528–540, 879–915, 1133–1148, 1524–1528, 1870–1927, 2404–2436, 2906–2941, 2923–2945, and 4100–4139 |
| 6 | Q13127 | REST* | 353–369, 727–742, and 882–896 |
| 7 | O95704 | APBB3 | 155–175 and 414–430 |
| 8 | P81274 | GPSM2* | 267–278, 318–347, and 422–433 |
| 9 | P00734 | Prothrombin* | 87-94, 98–116, 125–133, 178–198, 199–224, 217–224, 225–243, 248–263, 315–327, 328–344, 345–363, 354–363, 441–447, 447–452, 467–468, 518–537, 544–560, and 560–575 |
| 10 | B1AJZ9 | FHAD1* | 861–868, 876–887, 897–905, 931–942, 1030–1052, 1059–1070, and 1104–1139 |
*Proteins also included in the classifier for predicting Aβ in the cognitively unimpaired individuals.
DISCUSSION
Current imaging and CSF measurements of Aβ are considered gold standards for diagnosis of cerebral AD pathophysiology (12). However, PET imaging is costly, access to the ligand is limited, and this imaging is only available in relatively specialized centers. Therefore, it is unlikely to be part of routine clinical assessment of cognitive complaints before therapies become available. Conversely, CSF collection is more readily accessible, less expensive, but generally considered as invasive with a small risk of mild adverse effects by the public or as time consuming by clinicians. Furthermore, neither of these gold standards is suitable for population-based screening for identifying high-risk individuals for early intervention before symptom onset. Thus, a blood-based measure that accurately reflects AD pathology, ideally at the preclinical phase, would have significant advantages in therapeutic trials and clinical decisions in prescribing Aβ-targeting drugs once available.
In the present study, we used a nontargeted proteomic technique that combined extensive prefractionation and high-resolution MS to demonstrate a substantial number of individual plasma proteins to be nominally associated with Aβ burden in the cognitively unimpaired. A distinct group of analytes were shown to be highly related to Aβ burden, with medium-to-large effect sizes. However, the substantial overlap of the group-wise distributions suggested that these markers alone would not reliably distinguish Aβ burden. Therefore, and more notably, using feature selection and SVM, we detailed a multianalyte classifier to predict Aβ burden that was replicated by the identical proteomic workflow in an independent cohort with high accuracy. Although the statistical workflow is complex, this complexity was only necessary when seeking which was the minimal combination of proteins that produced the most accurate biomarker. Once the identity of these proteins has been established, which was one of the objectives of the paper, the classifier is reduced to a simple weighted sum. Namely, each of these selected proteins is multiplied by a number (so-called weight) and the results added together. If this sum surpasses a threshold value (so-called decision threshold), then the subject is classified as Aβ+. We have demonstrated the possible clinical utility of these plasma biomarker panels in two practical scenarios: one preclinical and one mixed diagnosis cohort (AD and MCI). In both scenarios, the high accuracies demonstrate the potential of a blood-based screen to precede or complement CSF and Aβ PET scans in participant selection for clinical trials. Predicting Aβ burden in the mixed cohort was superior (AUC = 0.904, specificity = 0.80, and sensitivity = 0.81), but when assessing Aβ burden at the preclinical stage, an almost identical classifier showed very similar diagnostic performance (AUC = 0.891, specificity = 0.77, and sensitivity = 0.78).
The proteins included in this predictive panel represent a diverse array of pathways, which most are not directly related to Aβ pathology per se. The serine protease prothrombin (a precursor to thrombin) was the highest ranked feature in the cognitively unimpaired cohort. At the univariate level, prothrombin (or coagulation factor II) was shown to be decreased in Aβ+ individuals but had a modest effect size. Multiple lines of evidence support that cerebrovascular disease may play a role in AD and that Aβ may be involved in thrombosis, fibrinolysis, and inflammation via its interaction with the coagulation cascade (13). The APP isoform Kunitz protease inhibitor domain (protease nexin-2) is involved in the regulation of coagulation and in thrombosis pathophysiology (14, 15). Thrombin has previously been reported to induce the release of APP from platelets and its subsequent processing (16), and we have previously reported fibrinogen gamma chain, a protein involved in the intrinsic coagulation cascade and target of prothrombin, as having a modest prediction of Aβ positivity (17). However, this study did not replicate our previous finding. Encouragingly, our study has highlighted the most investigated current plasma biomarkers for AD and/or neurodegeneration, APP and NfL, in an unbiased fashion. These proteins are included in both Aβ burden prediction models but have a greater effect size in the classifier that included individuals with reported cognitive decline, confirming their connection with the more established disease state. Aβ, derived from APP, is readily measured in plasma, but historically, the correlation with AD and/or surrogate Aβ measures has been absent or weak (18). Plasma Aβ concentrations have been interpreted as influenced by production in platelets and other extracerebral tissues (19). However, recent MS studies suggest that a ratio of APP-derived fragments (APP669–711 and Aβ42/Aβ40) identifies Aβ+ individuals with high sensitivity and specificity (6). In part, our data agree with these studies and with prior work that found plasma APP species to be elevated in AD and implicated serine proteases in these changes (20). We have shown that combined intensities from tryptic peptides residing from similar APP fragment regions, as the studies described above were shown to be statistically related to Aβ PET, although large overlap between groups remained. As with other panel-based studies, we also conclude that APP peptides (including Aβ species) have the utility to contribute to a compound panel (9, 21). NfL is an axonal neuron-specific protein actively involved in the pathogenesis of axonal injury and degeneration. NfL is detectable and quantifiable in blood despite being more than 50-fold lower in concentration than in the CSF (22), and here, we have demonstrated NfL peptide measurements using an untargeted MS approach. In previous studies, NfL levels in blood have been found to be increased in AD (23), frontotemporal dementia (24), and progressive supranuclear palsy (25). CSF and plasma concentrations correlate, supporting the notion that plasma NfL reflects CSF concentration and potentially CNS damage (26). NfL, after APP and NGN2, was the most statistically significant finding in our study. We found a highly significant association between NfL and Aβ burden, in keeping with previous studies, although we do not find an age-related increase of plasma NfL (23). As previously hypothesized by others, it is likely that observed elevation of plasma NfL has been included in our prediction classifier(s) as a reflection of CNS injury and not that of Aβ burden itself, even at the preclinical stage. Nonetheless, as the amyloid cascade hypothesis suggests that Aβ deposition is the main initiator behind events that result in neurodegeneration, elevations of NfL in response to Aβ burden, if present, would be an expected observation.
Novel markers of Aβ burden highlighted by this panel study include NGN2, FHAD1, and DNAH10, although further work will be needed to determine the mechanistic relationship between these proteins with Aβ pathogenesis and AD. Furthermore, two protein groups were specifically associated with Aβ burden in cognitively unimpaired individuals (GPR115 and RPS6KA3). GPR115 was the second most important feature, after prothrombin, offering it as a potential marker of early Aβ deposition. Numerous studies have presented evidence that implicate G protein–coupled receptors in the pathogenesis of AD and in multiple stages of the hydrolytic processing of APP (27). At the univariate level, NGN2 was highly associated with Aβ, with a larger effect size in the cognitively unimpaired group. NGN2 is a bHLH transcription factor that was first identified for its ability to promote neuronal differentiation in brain and spinal cord (28). NGN2 also specifies phenotypic features of neurons and regulates axonal guidance (29). NGN2 has been found to correlate with APP expression by an increase in APP transgenic mice (Tg2576) but significantly down-regulated in neural stem/progenitor cells from APP knockout mice (30). In addition, a close relationship between NGN2 and APP was found in our study, but the origin of our NGN2 expression is unclear as limited information is available about peripheral NGN2.
A biologically complex neurodegenerative disease such as AD is unlikely to be caused by a single pathogenic event (31), and the finding of a panel of plasma biomarkers characterizing AD pathology, rather than a single marker, was to be expected. What was encouraging was the high performance and reproducibility of the trained model in an entirely independent cohort, despite a methodically complex procedure. Previous studies have used shotgun MS as a hypothesis-generating tool to identify plasma protein biomarkers of AD pathology (7, 8, 17, 32). In each of these occasions, an attempt was then made to replicate these markers on an orthogonal platform, typically an immuno-based assay [e.g., enzyme-linked immunosorbent assay (ELISA) or Luminex xMAP]. In these cases, translation between techniques has been relatively disappointing and is likely due to key platform differences. Untargeted proteomics by MS involves the analysis of peptides resulting from denatured proteins, while ELISA measures native protein or, more precisely, the region of the intact protein where the epitope is recognized by the antibody resides. Therefore, using different methods will inevitably lead to different results, and this may contribute to why proteins identified by an untargeted approach fail in replication using a targeted approach. To our knowledge, this is the first time that a multianalyte plasma biomarker panel for an AD-related phenotype has been found and independently replicated by a nontargeted MS approach, and the commonality in platform, and certainly the homology in sample preprocessing, between discovery and replication has contributed to this successful result.
There are limitations to this study. First, although generating promising results, the platform used cannot be used as a routine tool, although we have demonstrated it to be robust in an independent cohort. Interlaboratory standardized operating procedures for this analytical process would prove difficult, given the complexed and variable methodological stages. Efforts will need to be made, guided by the MS spectral data, to transform the panel to an alternative platform that is more appropriate for widespread implementation. Second, the proteins measured in this study have been inferred on the basis of the combination of peptides, where a selective reaction monitoring approach as a validation of specific peptides associated with Aβ burden might be more appropriate. Further, it remains unclear whether the classifiers can track longitudinal changes in Aβ, monitor Aβ reduction in a therapeutic trial setting, or differentiate between other dementias that display Aβ pathology. Last, cross-sectional studies have indicated that CSF Aβ42 changes precede changes in Aβ PET (33). This signifies that individuals with AD may be initially classified as “CSF+/PET−” before converting to “CSF+/PET+” at a later stage (34). Future studies could complement the endophenotype approach using PET measures of Aβ load, with studies using CSF measures of both Aβ42 and tau as the comparison variable.
In summary, using an unbiased MS approach, we have found and replicated with high accuracy, specificity, and sensitivity a plasma protein classifier reflecting Aβ burden in a cognitively unimpaired cohort. These predictive panels highlighted novel and established markers for AD. These panels almost certainly need to be refined, simplified, and undoubtedly validated in independent cohorts. Furthermore, efforts need to be made to successfully translate this panel to a simpler automated platform suitable for wider utility. The prediction of Aβ burden in preclinical AD using a blood-based measure offers great potential in preclinical stratification for clinical trials and future diagnostic management.
MATERIALS AND METHODS
Study cohorts, assessments, and biofluid procedures
The AIBL was used as the discovery cohort in the study. The AIBL study is a longitudinal study of aging, neuroimaging, biomarkers, lifestyle, and clinical and neuropsychological analysis, with a focus on early detection and lifestyle intervention (www.aibl.csiro.au). Specifics regarding participant recruitment, study design, and clinical assessments were previously described (35). Plasma samples from 190 AIBL participants with baseline Aβ PET imaging (11C-PiB) were selected, with a focus on individuals without cognitive deficits at baseline. Further details about the 11C-PiB imaging protocols for the AIBL cohort were previously described (36). Participants were categorized as Aβ− and Aβ+ based on the 11C-PiB SUVR cutoff of 1.4 (37). For plasma preparation, whole blood (80 ml) was collected in the morning (overnight fasting) by venepuncture and centrifuged at 200g (20°C) for 10 min. The platelet-rich plasma was further spun at 800g (20°C) for 15 min (9). The KARVIAH was used as an independent replication cohort in this study. Participants recruited to this cohort were residents of Anglicare (Sydney, NSW, Australia), and all volunteers were required to meet the set screening inclusion and exclusion criteria to be eligible for the KARVIAH cohort (38). Selection of 94 KARVIAH participants was dependent on the availability of 18FBB images for each subject, where an Aβ SUVR cutoff score of 1.35 was used to categorize Aβ− and Aβ+ (39). All KARVIAH participants were cognitively unimpaired based on their Mini Mental State Examination score of >26. Plasma preparation for 94 KARVIAH participants followed the same protocol as AIBL participants.
Immunodepletion, enzymatic digestion, and tandem mass tag peptide labeling
All AIBL and KARVIAH samples were randomized before sample preparation. Albumin and immunoglobulin G immuno-depletion was achieved by a commercially available immunoaffinity column (ProteoPrep, Sigma-Aldrich) with a starting volume of 30 μl. For enzymatic digestion, 100 μg of immunodepleted sample was initially incubated with 100 mM triethylammonium bicarbonate (TEAB) and 0.1% (w/v) SDS. Reduction and alkylation were achieved in 1 mM tris (2-carboxyethyl) phosphine for 1 hour at 55°C, followed by incubation in 7.5 mM iodoacetamide. Protein samples were individually digested overnight in 4 μg of trypsin (sequencing grade, Roche) reconstituted in 100 mM TEAB. TMT10plex reagents (Thermo Fisher Scientific), reconstituted in acetonitrile (ACN), were added to the appropriate samples and incubated for 1 hour at room temperature (RT). Samples were treated with 5% hydroxylamine and incubated at RT for a further 15 min. For the 284 samples included in the study, a total of 32 “TMT10plex groups” were created by combining nine clinical samples (AIBL or KARVIAH) and one study reference. The study reference was an equal contribution of all samples examined in the study. After combining, TMT10plex groups were incubated at RT for a further 15 min.
OFFGEL (OGE):Isoelectric peptide fractionation
Peptide separation was achieved using the 3100 OFFGEL Fractionator (Agilent Technologies) with a 24-well setup. Immobilized pH gradient (IPG) gel strips (24 cm; GE Healthcare), with a 3 to 10 linear pH gradient, were rehydrated for 15 min. Before fractionation, TMT10plex groups were dried down and resuspended in OFFGEL stock solution. Next, 150 μl of resuspended TMT10plex peptide sample was loaded into each compartment of the OFFGEL system. Peptide samples were focused until 50 kVh had accumulated (~48 hours). Liquid fractions were collected into separate Eppendorf tubes (primary recovery). To extract larger peptides retained within the IPG strips, 150 μl of ddH20/ACN/formic acid (49:50:1) was added to each compartment and incubated for a further 30 min with occasional pipette mixing. The supernatant (secondary recovery) was retrieved and added to the primary recovery. Each fraction collection was purified by a SOLA horseradish peroxidase solid-phase extraction cartridge (10 mg/1 ml; Thermo Fisher Scientific) before evaporation by centrifugation under a vacuum.
MS (LC-MS/MS) analysis
Before liquid chromatography–tandem MS (LC-MS/MS) analysis, each OFFGEL peptide fraction was reconstituted in ddH20/ACN/formic acid (50:49.9:0.1) and then shaken at 37°C and vortexed thoroughly. Chromatographic separation and mass spectra acquisition (LTQ Orbitrap Velos Pro, Thermo Fisher Scientific) was performed on each fraction and was previously described (17), with a modified MS1 resolution of 60,000 and MS2 resolution of 30,000.
Preprocessing of MS (LC-MS/MS) data
Raw MS data for all fractions within a single TMT10plex were combined and searched as a “MudPIT” against the human UniProtKB/Swiss-Prot database using Mascot and Sequest (Proteome Discoverer version 1.4). The criteria for Mascot were described elsewhere (17) and Sequest also followed these settings. Peptide spectrum matches (PSMs) were rejected if identified with only low confidence (≥5% FDR) and/or missing quantification channels [e.g., not all peaks for tandem mass tags (TMTs) visible in spectra]. Raw intensity values of TMTs from PSMs passing filters were used for quantification. Preprocessing steps were applied to translate PSM intensities to relative protein abundance. This was a three-step process and was modified from a previously described R script (PRQ, or Pre-processing for Relative Quantification) (17). First, sequences with an intensity <50 were removed from the analysis. Second, median normalization within each TMT10plex was applied. This was achieved by calculating the ratio between all the intensities within a sample and the corresponding reference sample and then dividing each intensity by the calculated ratio, obtaining an intensity ratio. Each intensity ratio is, in turn, divided by the corresponding reference sample, and all results corresponding to the same peptide were summed into a peptide score. Third, the mean peptide scores are calculated to obtain protein abundance measurements. Last, a protein matrix table was then produced, which collated all identified protein groups and their corresponding TMT scores for each individual across the whole study. A protein group is defined as proteins that are identified by the same set or subset of assigned peptides.
Statistical analysis
Principal components analysis demonstrated a significant variation in protein abundance scores between the 32 TMT10plex groups. Many experimental factors are intrinsic with TMT10plex group: off-gel electrophoresis (OGE) fractionation date, TMT labeling date, and date of MS acquisition, which are prone to significant variation. Therefore, an initial generalized linear model (GLM) correcting for only TMT10plex group was performed before further investigating the effect of other covariates on protein group ratios. All GLM protein abundance scores were then log10-transformed to achieve normal distribution. Covariates including age, gender, and center (AIBL or KARVIAH) were investigated. We found that most proteins were significantly affected by these covariates, and therefore, subsequent GLM to include these confounders was appropriate. Missing protein abundance observations were imputed with the R package “mice” (version 2.46.0) using four imputations, four iterations, and 50 other proteins as a predictor. The 50 predictor proteins were chosen using the feature selection method LASSO via the R package “glmnet” (version 2.0-13) (40) with an α penalty cost of 1, the result of which was a list of proteins ranked by their correlation with the protein to impute; the top 50 predictor proteins were selected from this list.
Univariate analysis
Univariate analysis specializes on finding proteins that, by themselves, correlate with a given outcome once the effect of a number of covariates is eliminated. This analysis was performed in SPSS version 24 (IBM). For univariate analysis only, AIBL and KARVIAH samples were combined for the association with Aβ PET. All univariate analyses were performed on the imputed GLM-adjusted data described above. Student’s t test and Pearson’s correlation were performed to assess protein abundances with Aβ as a binary (Aβ+/−) and continuous measure (SUVR). A partial correlation and binary logistic regression were also performed to examine the influence of APOE genotype on the results. Benjamini-Hochberg Q values were calculated as a multiple testing correction. This univariate pipeline was performed for two datasets: (i) those classified as cognitively unimpaired (n = 238; Table 1) and (ii) all individuals included in the study, including those with MCI and AD (n = 284; Table 2).
Multivariate biomarker classifier
In the case of our study, and given a large set of proteins and an outcome, multivariate analysis specializes on finding which is the combination of proteins that best predicts the outcome. Many, but not necessarily all, of these proteins will also correlate with the outcome by themselves. All analyses in this section were performed using R (version 3.3.3). For the purpose of multianalyte classification of Aβ positivity, imputed protein abundances that were only GLM adjusted for TMT10plex group were used, thus allowing demographic variables to be independently chosen in feature selection. GLM was achieved using the R package “stats” (version 3.3.3).
Biomarker classifier development took place in three steps: (i) feature selection, (ii) training, and (iii) testing. Feature selection and training used only the AIBL cohort, while the KARVIAH cohort was kept for independent testing. Two training datasets were created, cognitively unimpaired individuals (n = 144; Table 1) and one including those with MCI and AD (n = 169; Table 2). This format would allow us to observe the influence of disease on the ability of a classifier model to predict Aβ positivity. The testing dataset contained a total of 94 samples from KARVIAH (Table 1). Two feature sets were used, either demographic variables only (gender, age at sampling, and APOE ɛ4 status) or both demographic and protein scores. Analysis was performed four times, each with a different combination of training dataset and feature set. For each analysis, the aforementioned three steps (feature selection, training, and testing) were performed over 50 iterations, where, in each iteration, the algorithm was allowed to use one protein more than in the previous iteration, starting with 1 protein and finishing with 50. Feature selection was performed using a training dataset and the R package glmnet (version 2.0-13) (40) with an α penalty cost of zero (“Ridge regression”), this resulted in a list of features ranked by their correlation with Aβ positivity, of which the top x was selected for subsequent training (where x = iteration number). Training was performed using the selected features, the same training dataset, and SVM from the R package “e1071” (version 1.6-8), this resulted in a model to classify Aβ positivity. Testing involved evaluating the accuracy at which the classifier could predict Aβ positivity in the testing dataset (KARVIAH); this was achieved by calculating the AUC using the R package “ROCR” (version 1.0.7). The training and testing AUC of all 50 classifiers was plotted using the R package “ggplot2” (version 2.2.1). Following these three steps, we selected the classifier with the highest AUC in the testing dataset as the best performing, and to assess this further, we used ROCR to calculate six statistics for a range of cutoff values and plotted these over three images using ggplot2. These statistics included the true positive rate, false positive rate, sensitivity, specificity, PPV, and NPV. The best cutoff value was chosen as the intersection of the sensitivity and specificity.
Supplementary Material
Acknowledgments
Funding: We express our appreciation to all participants in the AIBL and KARVIAH studies. This paper presents independent research funded by Butterfield Trust via Rosetree Trust UK. N.J.A. was supported by Rosetree Trust for this study. P.C. is supported by KaRa Institute of Neurological Diseases (KaRa MINDS) and Macquarie University. K.G. is funded by Anglicare (Sydney, Australia). H.Z. is a Wallenberg Academy Fellow and acknowledges support from the UK Dementia Research Institute. A.I.B. is funded by the National Health and Medical Research Council (GNT1103703, GNT1132604, and GNT1103703) and the Cooperative Research Centre for Mental Health. S. Lovestone is funded by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre and named in inventor on biomarker intellectual property protected by Proteome Science and King’s College London (unrelated to this study). R.M. is funded jointly by Edith Cowan University and Macquarie University. A.H. is funded by the Research Centre for Mental Health and Biomedical Research Unit for dementia. All other authors acknowledge that they received no funding in support of this research. This study represents independent research part funded by the NIHR Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care. Competing interests: S. Lovestone is currently an employee of Johnson and Johnson. All other authors declare that they have no competing interests. Author contributions: N.J.A., S. Lovestone, R.M., and A.H. contributed to the study concept and design. N.J.A. and S. Lynham were responsible for the data acquisition. N.J.A., A.J.N.-H., I.S.B., and A.H. carried out the data analysis and drafted the manuscript. All authors contributed to the sample selection and data interpretation and revised the manuscript. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw MS/MS data and additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/2/eaau7220/DC1
Table S1. Residual scores for all 560 protein groups.
Table S2. The association of plasma protein groups with Aβ SUVR in cognitively unimpaired subjects.
Table S3. The association of plasma protein groups with Aβ SUVR in all subjects.
REFERENCES AND NOTES
- 1.Baird A. L., Westwood S., Lovestone S., Blood-based proteomic biomarkers of Alzheimer’s disease pathology. Front. Neurol. 6, 236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bazenet C., Lovestone S., Plasma biomarkers for Alzheimer’s disease: Much needed but tough to find. Biomark. Med. 6, 441–454 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Jack C. R. Jr., Knopman D. S., Jagust W. J., Petersen R. C., Weiner M. W., Aisen P. S., Shaw L. M., Vemuri P., Wiste H. J., Weigand S. D., Lesnick T. G., Pankratz V. S., Donohue M. C., Trojanowski J. Q., Tracking pathophysiological processes in Alzheimer’s disease: An updated hypothetical model of dynamic biomarkers. Lancet Neurol. 12, 207–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salloway S., Sperling R., Fox N. C., Blennow K., Klunk W., Raskind M., Sabbagh M., Honig L. S., Porsteinsson A. P., Ferris S., Reichert M., Ketter N., Nejadnik B., Guenzler V., Miloslavsky M., Wang D., Lu Y., Lull J., Tudor I. C., Liu E., Grundman M., Yuen E., Black R., Brashear H. R., Bapineuzumab 301 and 302 Clinical Trial Investigators , Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer’s disease. N. Engl. J. Med. 370, 322–333 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janelidze S., Stomrud E., Palmqvist S., Zetterberg H., van Westen D., Jeromin A., Song L., Hanlon D., Tan Hehir C. A., Baker D., Blennow K., Hansson O., Plasma β-amyloid in Alzheimer’s disease and vascular disease. Sci. Rep. 6, 26801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura A., Kaneko N., Villemagne V. L., Kato T., Doecke J., Doré V., Fowler C., Li Q.-X., Martins R., Rowe C., Tomita T., Matsuzaki K., Ishii K., Ishii K., Arahata Y., Iwamoto S., Ito K., Tanaka K., Masters C. L., Yanagisawa K., High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature 554, 249–254 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Thambisetty M., Tripaldi R., Riddoch-Contreras J., Hye A., An Y., Campbell J., Sojkova J., Kinsey A., Lynham S., Zhou Y., Ferrucci L., Wong D. F., Lovestone S., Resnick S. M., Proteome-based plasma markers of brain amyloid-β deposition in non-demented older individuals. J. Alzheimers Dis. 22, 1099–1109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westwood S., Leoni E., Hye A., Lynham S., Khondoker M. R., Ashton N. J., Kiddle S. J., Baird A. L., Sainz-Fuertes R., Leung R., Graf J., Hehir C. T., Baker D., Cereda C., Bazenet C., Ward M., Thambisetty M., Lovestone S., Blood-based biomarker candidates of cerebral amyloid using PiB PET in non-demented elderly. J. Alzheimers Dis. 52, 561–572 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnham S. C., Faux N. G., Wilson W., Laws S. M., Ames D., Bedo J., Bush A. I., Doecke J. D., Ellis K. A., Head R., Jones G., Kiiveri H., Martins R. N., Rembach A., Rowe C. C., Salvado O., Macaulay S. L., Masters C. L., Villemagne V. L., Alzheimer’s Disease Neuroimaging Initiative, Australian Imaging, Biomarkers and Lifestyle Study Research Group , A blood-based predictor for neocortical Aβ burden in Alzheimer’s disease: Results from the AIBL study. Mol. Psychiatry 19, 519–526 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Kiddle S. J., Thambisetty M., Simmons A., Riddoch-Contreras J., Hye A., Westman E., Pike I., Ward M., Johnston C., Lupton M. K., Lunnon K., Soininen H., Kloszewska I., Tsolaki M., Vellas B., Mecocci P., Lovestone S., Newhouse S., Dobson R., Alzheimers Disease Neuroimaging Initiative , Plasma based markers of [11C] PiB-PET brain amyloid burden. PLOS ONE 7, e44260 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geyer P. E., Kulak N. A., Pichler G., Holdt L. M., Teupser D., Mann M., Plasma proteome profiling to assess human health and disease. Cell Syst. 2, 185–195 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Blennow K., Mattsson N., Schöll M., Hansson O., Zetterberg H., Amyloid biomarkers in Alzheimer’s disease. Trends Pharmacol. Sci. 36, 297–309 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Ahn H. J., Chen Z. L., Zamolodchikov D., Norris E. H., Strickland S., Interactions of β-amyloid peptide with fibrinogen and coagulation factor XII may contribute to Alzheimerʼs disease. Curr. Opin. Hematol. 24, 427–431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu F., Davis J., Miao J., Previti M. L., Romanov G., Ziegler K., van Nostrand W. E., Protease nexin-2/amyloid β-protein precursor limits cerebral thrombosis. Proc. Natl. Acad. Sci. U.S.A. 102, 18135–18140 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu W., Li H., Navaneetham D., Reichenbach Z. W., Tuma R. F., Walsh P. N., The Kunitz protease inhibitor domain of protease nexin-2 inhibits factor XIa and murine carotid artery and middle cerebral artery thrombosis. Blood 120, 671–677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bush A. I., Martins R. N., Rumble B., Moir R., Fuller S., Milward E., Currie J., Ames D., Weidemann A., Fischer P., The amyloid precursor protein of Alzheimer’s disease is released by human platelets. J. Biol. Chem. 265, 15977–15983 (1990). [PubMed] [Google Scholar]
- 17.Ashton N. J., Kiddle S. J., Graf J., Ward M., Baird A. L., Hye A., Westwood S., Wong K. V., Dobson R. J., Rabinovici G. D., Miller B. L., Rosen H. J., Torres A., Zhang Z., Thurfjell L., Covin A., Hehir C. T., Baker D., Bazenet C., Lovestone S., AIBL Research Group , Blood protein predictors of brain amyloid for enrichment in clinical trials? Alzheimers Dement. 1, 48–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olsson B., Lautner R., Andreasson U., Öhrfelt A., Portelius E., Bjerke M., Hölttä M., Rosén C., Olsson C., Strobel G., Wu E., Dakin K., Petzold M., Blennow K., Zetterberg H., CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: A systematic review and meta-analysis. Lancet Neurol. 15, 673–684 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Zetterberg H., Plasma amyloid β—Quo vadis? Neurobiol. Aging 36, 2671–2673 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Bush A. I., Whyte S., Thomas L. D., Williamson T. G., van Tiggelen C. J., Currie J., Small D. H., Moir R. D., Li Q.-X., Rumble B., Mönning U., Beyreuther K., Masters C. L., An abnormality of plasma amyloid protein precursor in Alzheimer’s disease. Ann. Neurol. 32, 57–65 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Rembach A., Faux N. G., Watt A. D., Pertile K. K., Rumble R. L., Trounson B. O., Fowler C. J., Roberts B. R., Perez K. A., Li Q. X., Laws S. M., Taddei K., Rainey-Smith S., Robertson J. S., Vandijck M., Vanderstichele H., Barnham K. J., Ellis K. A., Szoeke C., Macaulay L., Rowe C. C., Villemagne V. L., Ames D., Martins R. N., Bush A. I., Masters C. L., AIBL research group , Changes in plasma amyloid beta in a longitudinal study of aging and Alzheimer’s disease. Alzheimers Dement. 10, 53–61 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Kuhle J., Barro C., Andreasson U., Derfuss T., Lindberg R., Sandelius Å., Liman V., Norgren N., Blennow K., Zetterberg H., Comparison of three analytical platforms for quantification of the neurofilament light chain in blood samples: ELISA, electrochemiluminescence immunoassay and Simoa. Clin. Chem. Lab. Med. 54, 1655–1661 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Mattsson N., Andreasson U., Zetterberg H., Blennow K., Alzheimer’s Disease Neuroimaging Initiative , Association of plasma neurofilament light with neurodegeneration in patients with Alzheimer disease. JAMA Neurol. 74, 557–566 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohrer J. D., Woollacott I. O. C., Dick K. M., Brotherhood E., Gordon E., Fellows A., Toombs J., Druyeh R., Cardoso M. J., Ourselin S., Nicholas J. M., Norgren N., Mead S., Andreasson U., Blennow K., Schott J. M., Fox N. C., Warren J. D., Zetterberg H., Serum neurofilament light chain protein is a measure of disease intensity in frontotemporal dementia. Neurology 87, 1329–1336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rojas J. C., Karydas A., Bang J., Tsai R. M., Blennow K., Liman V., Kramer J. H., Rosen H., Miller B. L., Zetterberg H., Boxer A. L., Plasma neurofilament light chain predicts progression in progressive supranuclear palsy. Ann. Clin. Transl. Neurol. 3, 216–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gisslén M., Price R. W., Andreasson U., Norgren N., Nilsson S., Hagberg L., Fuchs D., Spudich S., Blennow K., Zetterberg H., Plasma concentration of the neurofilament light protein (NFL) is a biomarker of CNS injury in HIV infection: A cross-sectional study. EBioMedicine 3, 135–140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thathiah A., De Strooper B., The role of G protein-coupled receptors in the pathology of Alzheimer’s disease. Nat. Rev. Neurosci. 12, 73–87 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Li S., Mattar P., Zinyk D., Singh K., Chaturvedi C. P., Kovach C., Dixit R., Kurrasch D. M., Ma Y. C., Chan J. A., Wallace V., Dilworth F. J., Brand M., Schuurmans C., GSK3 temporally regulates neurogenin 2 proneural activity in the neocortex. J. Neurosci. 32, 7791–7805 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kele J., Simplicio N., Ferri A. L., Mira H., Guillemot F., Arenas E., Ang S.-L., Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development 133, 495–505 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Bolos M., Hu Y., Young K. M., Foa L., Small D. H., Neurogenin 2 mediates amyloid-β precursor protein-stimulated neurogenesis. J. Biol. Chem. 289, 31253–31261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovacs G. G., Milenkovic I., Wöhrer A., Höftberger R., Gelpi E., Haberler C., Hönigschnabl S., Reiner-Concin A., Heinzl H., Jungwirth S., Krampla W., Fischer P., Budka H., Non-Alzheimer neurodegenerative pathologies and their combinations are more frequent than commonly believed in the elderly brain: A community-based autopsy series. Acta Neuropathol. 126, 365–384 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Hye A., Lynham S., Thambisetty M., Causevic M., Campbell J., Byers H. L., Hooper C., Rijsdijk F., Tabrizi S. J., Banner S., Shaw C. E., Foy C., Poppe M., Archer N., Hamilton G., Powell J., Brown R. G., Sham P., Ward M., Lovestone S., Proteome-based plasma biomarkers for Alzheimer’s disease. Brain 129, 3042–3050 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Palmqvist S., Mattsson N., Hansson O., Alzheimer’s Disease Neuroimaging Initiative , Cerebrospinal fluid analysis detects cerebral amyloid-β accumulation earlier than positron emission tomography. Brain 139, 1226–1236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmqvist S., Schöll M., Strandberg O., Mattsson N., Stomrud E., Zetterberg H., Blennow K., Landau S., Jagust W., Hansson O., Earliest accumulation of β-amyloid occurs within the default-mode network and concurrently affects brain connectivity. Nat. Commun. 8, 1214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellis K. A., Bush A. I., Darby D., de Fazio D., Foster J., Hudson P., Lautenschlager N. T., Lenzo N., Martins R. N., Maruff P., Masters C., Milner A., Pike K., Rowe C., Savage G., Szoeke C., Taddei K., Villemagne V., Woodward M., Ames D., AIBL Research Group , The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: Methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease. Int. Psychogeriatr. 21, 672–687 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Rowe C. C., Ellis K. A., Rimajova M., Bourgeat P., Pike K. E., Jones G., Fripp J., Tochon-Danguy H., Morandeau L., O’Keefe G., Price R., Raniga P., Robins P., Acosta O., Lenzo N., Szoeke C., Salvado O., Head R., Martins R., Masters C. L., Ames D., Villemagne V. L., Amyloid imaging results from the Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging. Neurobiol. Aging 31, 1275–1283 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Chetelat G., Villemagne V. L., Villain N., Jones G., Ellis K. A., Ames D., Martins R. N., Masters C. L., Rowe C. C., AIBL Research Group , Accelerated cortical atrophy in cognitively normal elderly with high β-amyloid deposition. Neurology 78, 477–484 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Goozee K., Chatterjee P., James I., Shen K., Sohrabi H. R., Asih P. R., Dave P., ManYan C., Taddei K., Ayton S. J., Garg M. L., Kwok J. B., Bush A. I., Chung R., Magnussen J. S., Martins R. N., Elevated plasma ferritin in elderly individuals with high neocortical amyloid-β load. Mol. Psychiatry 23, 1807–1812 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Goozee K., Chatterjee P., James I., Shen K., Sohrabi H. R., Asih P. R., Dave P., Ball B., ManYan C., Taddei K., Chung R., Garg M. L., Martins R. N., Alterations in erythrocyte fatty acid composition in preclinical Alzheimer’s disease. Sci. Rep. 7, 676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman J., Hastie T., Tibshirani R., Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010). [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/2/eaau7220/DC1
Table S1. Residual scores for all 560 protein groups.
Table S2. The association of plasma protein groups with Aβ SUVR in cognitively unimpaired subjects.
Table S3. The association of plasma protein groups with Aβ SUVR in all subjects.


