Abstract
During protein synthesis genetic instructions are passed from DNA via mRNA to the ribosome to assemble a protein chain. Occasionally, stop codons in the mRNA are bypassed and translation continues into the untranslated region (3′-UTR). This process, called translational readthrough (TR), yields a protein chain that becomes longer than would be predicted from the DNA sequence alone. Protein sequences vary in propensity for translational errors, which may yield evolutionary constraints by limiting evolutionary paths. Here we investigated TR in Saccharomyces cerevisiae by analysing ribosome profiling data. We clustered proteins as either prone or non-prone to TR, and conducted comparative analyses. We find that a relatively high frequency (5%) of genes undergo TR, including ribosomal subunit proteins. Our main finding is that proteins undergoing TR are highly expressed and have a higher proportion of intrinsically disordered C-termini. We suggest that highly expressed proteins may compensate for the deleterious effects of TR by having intrinsically disordered C-termini, which may provide conformational flexibility but without distorting native function. Moreover, we discuss whether minimizing deleterious effects of TR is also enabling exploration of the phenotypic landscape of protein isoforms.
INTRODUCTION
Mutations occurring in the DNA are the main source of evolutionary novelty. The average genotypic base-substitutional rate for prokaryotes is estimated to be 0.5 (SE = 0.2) × 10−9 per site per DNA replication (1,2) and even higher in unicellular eukaryotes (1.6 × 10−9), due to a greater effect of drift on smaller populations (1,2). While the genotypic error rate is low, the error rate during protein synthesis is estimated to be 10−3 to 10−4 misreadings per codon in Escherichia coli (3). Phenotypic mutation is here defined as mutations occurring during protein synthesis (translational errors), which cause an alteration to a protein. In the following, we consider such a change as a ‘molecular phenotype’ which may or may not affect the organismal phenotype. There is an upper limit for the load of phenotypic mutation that a cell can handle and a lower limit, where the error rate is minuscule at the cost of synthesis efficiency (4). Within the limits of the upper and lower threshold for phenotypic mutations that a single-cell system can handle, a mathematical model by Bürger et al. (4) finds that there is still evolutionarily leeway to reduce the error rate. However, there is seemingly no selective pressure to reduce the phenotypic mutation rate (4), which is surprising given that mutations may be deleterious and decrease the fitness of the organism. According to the drift-barrier hypothesis, the ability of selection to increase the fidelity of replication, transcription, and translation is limited and should scale positively with the effective population size of the organism in question (1,2). However, regardless what processes underpin the relatively high phenotypic mutation rate, it remains an open question what impact phenotypic mutations have on protein evolution. If errors are unavoidable, an alternative may be to evolve towards increased tolerance of errors. Proteins that are tolerant towards errors, a property that is known as robustness, will fold and function in the presence of many phenotypic mutations. Selection pressure for protein robustness has been been predicted (5,6) and confirmed (7) to increase protein thermostability (8–11).
Increasing tolerance towards phenotypic mutations would not only neutralize the cost of errors, but potentially also facilitate the evolutionary emergence of novel traits. Protein isoforms generated by erroneous translation have been proposed to be used as a mechanism for exploration of the phenotypic landscape (12,13), where phenotypic mutations act as intermediate stepping stones for traits not yet encoded in the DNA (12). Taken together, tolerance for errors would facilitate rapid adaptation, for which there is accumulating support (13,14). A recent study in fungi has shown how phenotypic mutations may yield protein isoforms that are functional and increase fitness in stressful conditions (13). However, gaps remain regarding the context of translational errors with respect to structural features and what selection may act upon. Knowing what properties are responsible for the robustness of a protein facing phenotypic mutations elucidates the evolutionary paths available for novel features to evolve.
Here, our aim is to understand the biophysical and evolutionary context of phenotypic mutations on protein evolution, specifically so called ‘translational readthrough’. During protein synthesis the genetic instructions are passed from DNA via mRNA to the ribosome to assemble a protein chain. The ribosome terminates protein synthesis upon encountering a stop codon in the mRNA. Recent research finds that stop codons are occasionally ignored by the ribosome and that translation continues into the untranslated region (3′-UTR). This process, called translational readthrough (TR), yields a protein chain that becomes longer than one would predict from the DNA sequence alone. Recent studies indicate functionality in proteins that undergo TR (13,15,16). Here, we take advantage of the availability of public ribosome profiling data to investigate TR and what features co-occur with TR in Saccharomyces cerevisiae by analyzing ribosome profiling data from Nedialkova and Leidel (17). We analyse the physical features of genes experiencing TR, and try to elucidate selective pressures and evolutionary constraint.
MATERIALS AND METHODS
Mapping of reads
The ribosome profiling data and respective RNAseq data was retrieved by Nedialkova and Leidel (17) from Gene Expression Omnibus (18). The reads were trimmed and mapped accordingly to (17). Adaptors described in the original publications were trimmed from filtered reads. Reads below 26 nucleotides were not considered. Trimmed reads were filtered by mapping them to reference RNA (rRNA) using Bowtie, version 1.0.0 (19). The remaining unaligned reads were aligned with reference genome with TopHat (20) version 2.0.12. As there is always a risk of detecting spurious reads in the 3′-UTR that is not necessarily a yield from translational readthrough, extra measures were taken to align reads to the 3′-UTR. For mapping reads to the 3′-UTR, we used bowtie allowing only one mismatch and no multimapping (bowtie -S -m 1 –best –seed 21 -n 0 -e 1 -p 22). Genome for S. cerevisiae S288C was downloaded from Ensembl with annotations (21).
Detecting translational readthrough
Only expressed genes that have annotated 3′-UTR were included in further analyses. Annotations by Yassour et al. (22) were used for mapping reads to the 3′-UTR. HTSeq was used (23) to retrieve the count number of reads mapped with genes and respective 3′-UTR, using strict-mode that excludes overlapping reads.
Genes that consistently were showing translational readthrough (TR) in all replicates were grouped as ‘leaky genes’. Genes displaying TR in some but not all replicates were grouped as ‘semi-leaky genes’. Genes with annotated 3′-UTR without any count hits, consistently between replicates, were grouped as ‘non-leaky genes’.
Mapped reads to 3′-UTR can indicate continued translation of the mRNA beyond the first stop codon, but these reads can also be mere noise. Several measures were made to ensure reads mapped to the 3′-UTR were justifiably counted as TR. Firstly, annotated 3′-UTRs that are overlapping with a gene on the same strand were excluded. 3′-UTRs with a sequence length shorter than 30 nucleotides were excluded as they infer high stochasticity when calculating coverage.
Before TR rate was estimated, an initial threshold was set for at least 5 reads to be registered as mapped to the 3′-UTR for each replicate. This is a common lower threshold when considering gene expression (24). TR rate was calculated as the following: The sequence hit count (obtained by HTSeq) was normalised by dividing read length with the sequence length, as done by (25). The normalized hit count for the 3′-UTR was divided with the normalized hit count value of the protein coding sequence (CDS), yielding relative expression of 3′-UTR. Genes displaying spurious translation by relative expression of one or above were excluded. Relative expression over or near one, effectively implies that the 3′-UTR is being expressed as high as the CDS.
Assuring that the reads were accurately indicating TR, we controlled for background noise and that the TR followed the appropriate open reading frame (ORF). We estimated background noise by quantifying the coverage of riboreads that aligned to tRNA, that were aligned by the same stringent criteria as 3′-UTR. tRNA is not translated by the ribosome. We therefore interpret riboreads aligned to tRNA as noise—either caused by ribosomes that spuriously bind to RNA or imperfect alignment. By dividing the read count with sequence length we retrieved the normalised coverage for tRNAs. The highest value—between the replicates, not the mean of the replicates—was used as a threshold for noise: all genes that had a read coverage in the 3′-UTR equal or lower to the tRNA coverage (our threshold) were excluded from our analyses. After this step the leaky set contained 408 genes. Lastly, we control for that our indicated TR follow the appropriate ORF. Ribosome profiling data enables calculation of the ribosomal reading frame of the mRNA (26). However, the data we made use of had a relatively modest digestion step (17), which is known to make the reading frame imprecise. We have no ambition of establishing the reading frame, but rather establish real translation from spurious binding of the ribosome. We controlled, by an in house script, that the reads aligned with the open reading frame up until next stop codon in frame in the 3′-UTR. If the coverage was higher or equal beyond the first stop codon encountered in the 3′-UTR, they were dismissed from further analyses as ambiguous. After these steps the leaky set contains 323 genes and the semi-leaky set contains 44 genes.
Essential genes
Essential gene list (Essential ORFs) was retrieved from the Saccharomyces Genome Deletion Project (27). An essential gene indicate that by a knock-out, the cell will die as the gene’s designated function is vital. The term ‘essential gene’ is one way to assess the relative biological importance of a protein’s functionality. However, the term is discussed further in (28). We scanned our gene sets against the essential gene list by an in house script and conducted enrichment tests by Fischer exact test (using the statistical python module scipy.stats - http://www.scipy.org/).
Gene ontology and pathway enrichment analyses
Gene ontology analyses were performed for leaky genes against all expressed genes as background, using TopGO (29) in the R environment version 3.3.0 (30). To make the word cloud we used tagcloud (https://CRAN.R-project.org/package=tagcloud). The pathway enrichment analysis was performed on the online platform DAVID (31,32).
Gene expression, RNA stability, disorder
For each replicate of both footprints and RNA-seq, gene expression was calculated as Transcript Per Million (TPM). Translational efficiency (TE) was calculated as described by Ingolia et al. (26), dividing TPM of the ribosome profiling reads by the TPM of the RNA-seq reads. Sequence length was measured in nucleotides of the CDS (not including UTR). The replicates were investigated for significant distribution differences by a Kolmogorov-Smirnov test and found to be non-significant. Thereafter, we used the mean of the replicates, for TPM and TE, in further analyses.
To analyse the mRNA of yeast we relied on the annotations by Yassour et al. (22) to retrieve the 5′-UTR and 3′-UTR. By an in house script the UTRs were added to the CDS to remake the full mRNA. To retrieve data on mRNA structural stability we made use of RNA minimum free energy (mfe). We used RNAfold (33) to retrieve mfe of the mRNA (we excluded those where UTRs were not available). The significance of a mfe-value is commonly estimated by comparing it to random sequences of the same length and base composition. We generated random sequences (re-shuffled native sequences) by using an algorithm (34) as implemented in the MEME suite (35). The ratio of mfe of the native mRNA was calculated by using the ratio Z = (x − μ)/σ. x represents the mfe of the mRNA, μ the standard deviation and σ the arithmetic mean obtained of the mfe of the shuffled controls. Moreover, we investigated for enriched motifs in the start of the 3′-UTR (first 30 nucleotides) using DREME, which is part of the MEME suite (35). DREME scans the region for RNA secondary structures and compares if the given cluster, which was the leaky set, are enriched for motifs, relative to the non-leaky set. Previous studies have reported on stop codon context with respect to TR (36). We investigated stop codons as well as the nucleotide after the stop codon by an in house script. We compared and clustered the genes by presence of TR.
We used the IUPred short algorithm to predict intrinsic disorder in the protein sequences based on the frequency of disorder-promoting amino acids (37), which uses 0.5 as the threshold for a sequence to be disordered. For practices on disorder, see (38–41). As a measurement of folding potential, we employed Seg-HCA, which analyses clusters of hydrophobic amino acids (42,43).
3′-UTR extension
We retrieved the translated 3′-UTR as estimated by translational readthrough (TR). The presence of stop codons throughout the 3′-UTR sequence makes it questionable to analyse as a translated protein. Therefore, only the sequence from the initial stop codon until the next stop codon in the first reading frame (continuation of the CDS) were kept of the 3′-UTR. This was used as an imitation of what the 3′-UTR peptide may look like, assuming no frameshift. Both the extended peptide - translated 3′-UTR fused with the parent peptide - and the extension itself were analysed for disorder.
Codon usage
We analyzed codon usage for the proteins within all three sets. We made use of Codon Adaptation index (CAI) (44). We used codonW version 1.4.2 (http://codonw.sourceforge.net/) to conduct the analyses. We analysed all CDS in all three sets. Moreover, we analysed the last 30 nucleotides of each CDS in all sets as an estimation of the C-termini.
Search for protein domains
To investigate if TR would yield a functional protein domain, we investigated protein domains in the 3′-UTR. Studies have shown that a protein product yielded by frameshift, yield a very similar isoform to the encoded one (13), especially in disordered regions (45). We therefore generated multiple strand specific open reading frames (ORFs) by using gffread from cufflinks (46). The generated ORFs were all scanned by pfam (47). This was done for all protein sequences in all sets.
RESULTS AND DISCUSSION
Detecting translational readthrough
Ribosome profiling data for wildtype yeast were retrieved from NCBI GEO database, published by Nedialkova and Leidel (17) (Materials and Methods). Continued translation beyond the stop codon may indicate translational readthrough (TR). To detect TR we looked for reads mapped to the 3′-UTR of expressed genes and then we clustered them by occurrence of TR (Supplementary Figures S1 and S10). Genes that consistently were showing TR in all replicates were grouped as ‘leaky genes’. Genes displaying TR in some but not all replicates were grouped as ‘semi-leaky genes’. Genes with an annotated 3′-UTR without count hits in any replicate were grouped as ‘non-leaky genes’. The set of “leaky genes“ contained 323 genes, which is 5% of the annotated genes. In the leaky set there are 22 proteins (or 0,3% of all annotated proteins) that have a TR rate equal or >3%.
Translational readthrough occurs regardless of protein function
We wondered if leaky genes might have a functional bias, i.e. if TR occurs in proteins with specific functions. We find 70 essential genes in the leaky set, 207 in the non-leaky set and 18 in the semi-leaky set. A gene ontology (GO) and a pathway enrichment analysis (Materials and Methods) displayed that proteins of the leaky set are enriched for the GO term translation, amongst other terms (Supplementary Figure S3). We found that the biggest cluster are all ribosomal subunit proteins (Supplementary Table S9). These results are corroborated by a previous study investigating proteins prone to TR (48), that also found proteins prone to TR to be involved with translation and the ribosome apparatus. A separate GO-analysis for proteins with a TR rate of 3% or higher from the leaky set (22 proteins) displayed a variety of biological functions including metabolic and stress regulation (Supplementary Figure S4). The subset with TR rate greater than 3% did not have any significantly enriched pathways. Many of these genes are classified as non-essential or even as hypothetical proteins. However, five of these highly leaky proteins are in fact included in the essential gene list (Supplementary Table S8).
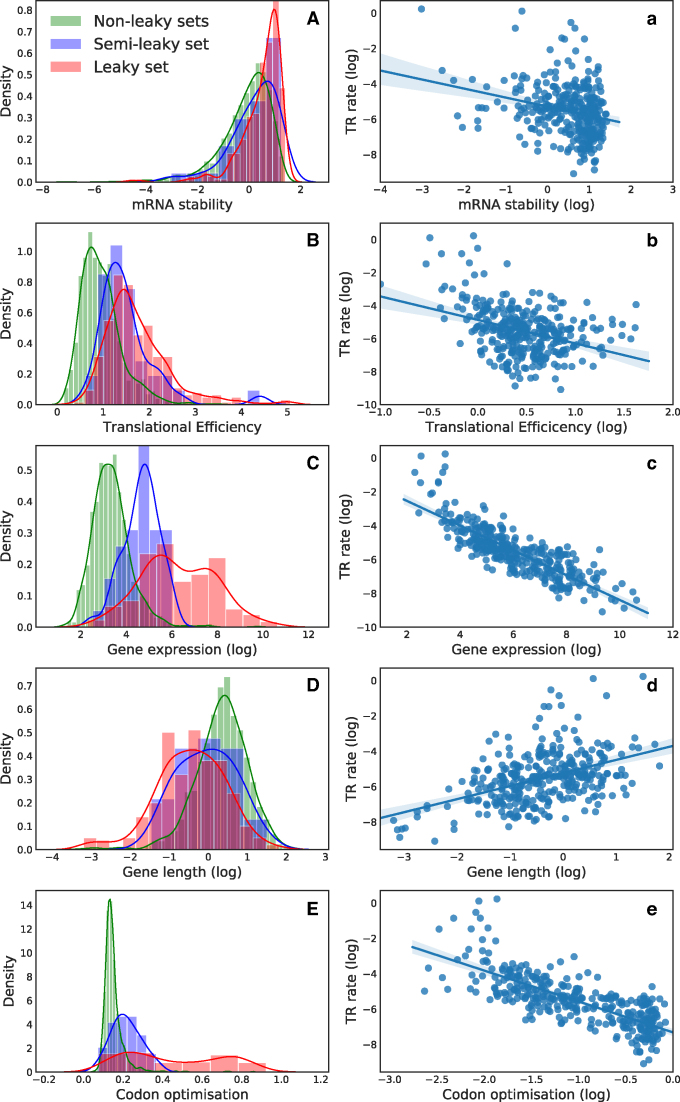
We also investigated gene expression and found that leaky genes have an overall higher gene expression (Figure 1 C). Previous research on highly expressed genes reports that ‘protein synthesis’ is among the most enriched categories in the yeast transcriptome (49). If TR is a result of gene expression—and not protein functionality—one would expect a functional bias toward translation.
Figure 1.
Density distributions of characteristics in sets (A–E). (A) mRNA stability (as minimum free energy) for each of the sets. The mRNA is more unstable the closer to 0. (B) Translational efficiency of the proteins in each set. (C) Gene expression (Transcript per million) for proteins in each set (log transformed). (D) Gene length (log transformed) for CDS in each set. (E) Frequency of optimal codon amongst sets. Ratio goes from 0 to 1, where 0 is non-optimized and 1 is fully optimized. Correlation plots of TR rate against different variables (A–E). TR rate is always depicted on the Y-axis and the other variable on the X-axis. a: TR rate versus mRNA stability. b: TR rate versus Translational Efficiency. c: TR rate versus gene expression. d: TR rate versus gene (CDS) length. e: TR rate versus codon usage (calculated according to Cai, see Materials and Methods).
In conclusion, most proteins undergoing the highest levels of TR are non-essential. As expected, we find most essential genes in the non-leaky set. However, we find the leaky set to contain essential genes, e.g. ribosomal and chaperon subunit proteins, in addition to be enriched for the GO biological process term ‘translation’. As the leaky set has an overall high gene expression, we believe that our findings—of essential proteins and the enriched involvement of translation—are a reflection of high gene expression, rather than of protein functionality.
Protein characteristics
Given how our results of functional bias towards protein synthesis may be explained by gene expression, we asked if leaky genes might have some structural bias related to gene expression and translation. Translation has been found to be affected by mRNA stability, whereas GC-content and sequence length affect mRNA stability. We analysed several features in all three sets (leaky, semi-leaky and non-leaky): GC-content of both CDS and 3′-UTR, translational efficiency, gene expression (Transcripts Per Million), sequence length of CDS, and mRNA structural stability by minimum free energy (see Materials and Methods), henceforth referred to as mRNA stability.
Translational readthrough rate is most strongly reflected in gene expression
The leaky set has relatively higher GC-content, shorter sequence length, higher gene expression, and lower mRNA stability than the other two sets. At the other end of the scale, we find the non-leaky set to have longer genes, lower gene expression, GC-content and higher mRNA stability (Figure 1 and Supplementary Figure S5). For the measured parameters, the values of proteins belonging to the semi-leaky set are in between as of a gradual transition between the leaky and non-leaky sets.
When analysing all the sets, TR correlates weakly and negatively with all factors except gene length. As the non-leaky set also contains the highest quantity of genes, the statistical analyses become zero-inflated when correlating with TR, for which the rate is 0 (Table 1). When only analysing the error prone proteins (semi-leaky and leaky sets), we find that gene length correlates positively with TR rate, whereas translational efficiency and gene expression correlate negatively and more strongly with TR rate. In other words, the rate of TR increases with sequence length but decreases with gene expression. The fact that TR rate increases with sequence length makes sense intuitively, assuming that a longer protein would see a relatively small effect of an extension by TR, compared to a shorter protein.
Table 1.
Spearman rank correlations between TR rate and various variables
| Variables | Rho | P-value | Rho* | P-value* |
|---|---|---|---|---|
| GC CDS | 0.297 | 0.0 | −0.317 | 0.0 |
| Gene length | −0.356 | 0.0 | 0.429 | 0.0 |
| mRNA stability | 0.308 | 0.0 | −0.304 | 0.0 |
| Gene expression | 0.619 | 0.0 | −0.848 | 0.0 |
| CAI CDS | 0.563 | 0.0 | −0.79 | 0.0 |
| CAI end | 0.43 | 0.0 | −0.652 | 0.0 |
| Translational efficiency | 0.496 | 0.0 | −0.35 | 0.0 |
Data for columns with asterisk (*) indicate that the non-leaky set is excluded (contains only proteins from leaky and semi-leaky sets). mRNA stability is measured as minimum free energy. Gene expression is calculated as Transcript Per Million (TPM). CAI stands for Codon Adaptation Index—a parameter for estimating codon usage. CAI CDS is for the CDS, whereas CAI end includes only the last 30 nucleotides (see Materials and Methods).
RNA structural stability has previously been found to be under translational selection, and we therefore next asked if there is also a link between RNA stability and translational fidelity e.g. TR. Using default parameters of RNAfold (see Materials and Methods) we estimated structural stability of the mRNA. We find mRNA stability to correlate moderately with gene expression (Supplementary Figure S8) and TR rate (Table 1) whereas the strongest correlation with mRNA stability is sequence length (Supplementary Figure S8). The weak correlation between TR rate and mRNA stability can be explained by the fact that error prone proteins have relatively short sequences compared to those in the non-leaky set. Additionally, to investigate the region that is directly affected by TR—the C-terminus—we also conducted an mRNA stability analysis for the last 30 nucleotides of the CDS, as well as the first 30 nucleotides the 3′-UTR. We find no meaningful difference between the sets (Supplementary Figure S7), which suggests that the mRNA stability in the vicinity of the stop codon does not affect the occurrence of TR. We do not deem there to be a significant relationship between TR rate and GC-content given the weak correlation (Table 1 and Supplementary Figure S5).
Previous studies have reported on stop codon context with respect to translational readthrough (36). We did not find any enriched nucleotide context, neither with respect to stop codons (Supplementary Table S2), nucleotide after the stop codons (Supplementary Table S3) or with respect to TR rate (Supplementary Table S4). Investigating enriched motifs in the first 30 nucleotides of the 3′-UTR by the MEME suite (35) did not yield any enriched motifs.
The majority of genes undergoing TR, does so at a very low TR rate. Our data suggest therefore that deep coverage is needed to detect most of TR. In other words, the connection between gene expression, translational efficiency and TR rate may be a data sampling artefact derived by high gene expression. However, it has been predicted that highly expressed genes are—by a higher expression level—prone to undergo erroneous translation simply by higher translation exposure (8,9). Our results are in accordance with this assumption that the majority of erroneous translational events are found among highly expressed genes. Whether the connection between high expression and TR is a sampling artefact or a real biological connection remains unresolved until more data sets of deep coverage are available.
In conclusion, the variables measured—gene expression, translational efficiency, sequence length, GC-content and mRNA stability—are not complete predictors of TR. Partial correlations did not increase the strength in describing TR for any the variables (Supplementary Table S7). Nonetheless, gene expression is the strongest indicator of TR: high gene expression increases the probability of TR, whereas the TR rate decreases with increased gene expression (Figure 1).
Error prone genes are codon optimized
As we found a significant correlation between TR and gene expression, we next analysed the sets for optimal codon usage. Codon usage is known to be under selection and strongly affect gene expression (50). We used the Codon Adaptation Index (CAI) to calculate optimal codon usage. Optimal codon usage corresponds to cognate tRNA species that are more abundant and that are associated with efficient translation (50). The ribosome slows down over non-optimal codons and, conversely, translates faster over optimal codons. Studies on this topic concluded that the distributions of non-optimal and optimal codons in mRNAs are non-random with respect to proper protein folding, and that the codon distribution maximizes translation fidelity and efficiency (51–53). For example, non-optimal codons are commonly found between regions coding for secondary structures of encoded proteins (54,55). It has been suggested that the structuring of non-optimal and optimal codons promotes co-translational folding (56–58) because usage of optimised codons is foremost associated with fast translation and thus, highly expressed genes (59,60). This is in accordance with our findings: we find gene expression to correlate positively with codon optimisation (Supplementary Figure S8) and again we find a significant difference between sets (Table Supplementary S6) with genes in the leaky set being codon optimised (Figure 1E). When considering only error prone proteins (leaky and semi-leaky sets), codon optimisation correlates negatively with TR rate (Table 1). We find the same trend, but somewhat weaker, for the last 30 nucleotides (Supplementary Figure S5 and Table 1). Like the CAI value based on the whole CDS, the CAI value for the C-termini correlates with TR rate, but less strongly.
Next to fast elongation, it has been suggested that optimal codon usage reduces the frequency of nonsense errors (3,61,62) but it is debated whether it also reduces missense errors, where an amino acid—different from than what is encoded—gets incorporated into the peptide chain by a non-cognate tRNA (50,63). The negative correlation indicates that optimised codon usage may offer a selective advantage by lowering the error rate in highly expressed genes. Due to their high abundance, highly expressed genes will contribute more in absolute numbers to the phenotypic mutational load, than lowly expressed genes (10). Harmful phenotypic mutations are predicted to impose selection for compensatory mechanisms: Either, the mutational load leads to selection for error avoidance, e.g. increased proofreading mechanisms, or the mutational load leads to selection for elevated tolerance towards errors, also known as increased robustness (10).
In conclusion, we have shown that TR takes place in short and highly expressed genes with optimised codon usage. Moreover, we show how optimised codon usage of the CDS correlates negatively with TR rate. We hypothesise that highly expressed genes cannot avoid TR altogether but that high optimised codon usage may decrease the TR rate (51–53).
Error prone proteins have highly disordered C-termini
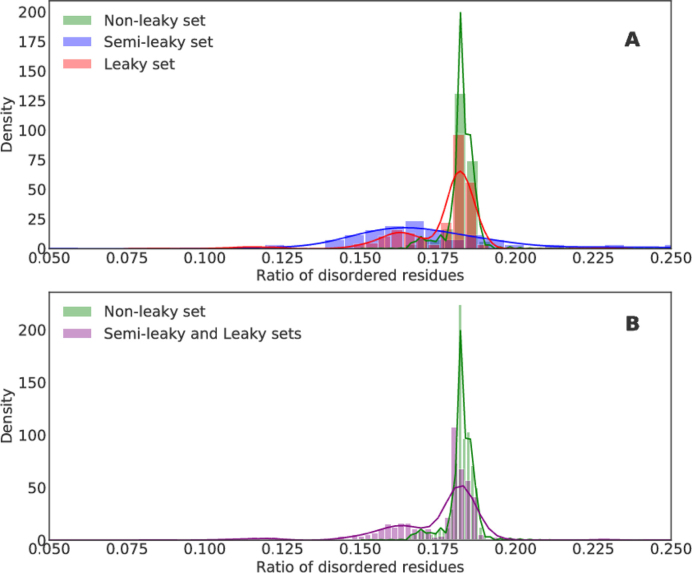
To investigate if there are any patterns of physical properties that are common to error prone proteins, we analysed protein sequence features: ratio of disordered residues (IUPred (37)), disordered binding sites (Anchor (64)), and hydrophobic clusters (see Materials and Methods). The analyses were conducted for each sequence separately in all sets. No meaningful correlation was found between TR rate and protein structural features such as ratio of disordered residues, hydrophobic clusters, disordered binding sites. We did not find the distributions of disordered residues to differ between the sets (see Figure 2).
Figure 2.
Ratio of disordered residues of full protein sequences. The -axis displays density of sequences and the X-axes display ratio of disordered residues. The colours display what set the proteins belong too. (A) The leaky set (red) is mostly overlapping with the semi-leaky set (blue), but also overlapping with the non-leaky set (green). (B) The leaky and semi-leaky sets are clustered as one (purple), whereas non-leaky is maintained unaltered (green). The leaky and semi-leaky sets have a significantly higher proportion of disordered residues (Mann Whitney test, p-value 0.005, U-value 966).
Moreover, we analysed the last 30 amino acids of the peptide chains separately. All sets have a high frequency of disordered C-termini (see Figure 3 and Supplemental Figure S6.), but the leaky set has a significantly higher proportion than the non-leaky set (Mann Whitney one-sided rank test, pvalue 0.03). We found five of our proteins to be curated in the DisProt database (65) (see Table S1). In other words, we find that the leaky and semi-leaky sets have significantly more disordered C-termini than proteins belonging to the non-leaky set.
Figure 3.
Ratio of disordered residues of last 30 amino acids of protein sequences in sets. The Y-axis displays density of sequences and the X-axis displays ratio of disordered residues. Leaky and semi-leaky sets are clustered as one (purple), whereas non-leaky is maintained unaltered (green). Many proteins of both error prone and non-leaky set have intrinsically disordered C-termini, but the C-termini of error-prone proteins are more disordered.
Due to the lack of structural constraints of intrinsically disordered regions, a missense error, i.e. shifting the reading frame, would not significantly disrupt the structure and errors are therefore believed to have a near-neutral effect (39,40,66). However, by mutating native protein sequences into random sequences, Schaefer et al. (67) found that forming secondary structures is an intrinsic feature of peptides, whereas maintaining long disordered regions appears hard to maintain by evolution (67). On the other hand, the study by Schaefer et al. (67) found this to apply foremost to long disordered regions. Short disordered regions seem to be functionally robust with introduction of single mutations, and thereby more easily maintained (67). In other words, the impact of mutations and translational errors on proteins with intrinsically disordered regions vary with respect to region length and context. To infer about the impact of TR, we investigated the predicted extensions from TR for intrinsic disorder and sequence length (Materials and Methods). Next to being very short, we found the predicted extensions to be ordered, see Supplementary Figures S6 and S9. This is in accordance with the results of Schaefer et al. (67), assuming that the translated 3′-UTR yields random peptides. A more recent study found that next to be able to form secondary structures, random sequences are well tolerated in vivo (68). We can only cautiously speculate what the full impact are of the extensions. As the extensions are ordered and short, we have no support to assume the extensions would be interactive. We suspect that the extensions have a low impact on native protein functionality. Moreover, when a protein is yielded by the ribosome, it starts at the N-terminus, which is little influenced by the remaining peptide chain. The C-terminus, on the other hand, is suggested to be under influence of the already folded part of the protein and does not influence the protein fold as the N-terminus (69,70). As the termini have been found to be located on the surface in most proteins, especially C-terminus (71), we speculate that alterations of the C-terminus are effectively near-neutral with respect to the protein fold. Our study highlights that the majority of proteins that undergo TR have an intrinsically disordered C-terminus. The full impact and effect of TR undoubtedly deserves further research.
Furthermore, we find the C-termini of the leaky set to be more codon optimised than the other sets (Supplementary Figure S5). This is in variation to previous research that found disordered regions to be encoded by non-optimal codons in yeast (57,66). As already described, optimal codon usage has previously been found to induce fast translation. Intrinsically disordered proteins have been found to be unstable with the risk of aggregation and are tightly regulated (72). Given that many of the leaky proteins have relatively high disorder, it may be advantageous to release the C-terminus quickly from the ribosome to allow the peptide to quickly complete its folding structure.
In conclusion, we hypothesize that the effect of TR may be near-neutral as the erroneous elongation by TR occurs at an already highly disordered region. This would explain why it is also present in essential proteins and the relatively high frequency of TR rate.
Investigation of homology and divergence
No conservation found of the 3′-UTRs
Given our results of protein feature analysis, we looked for conserved regions of the 3′-UTR by a homology analysis. It is conceivable that the 3′-UTRs were once part of a functional protein and that TR is due to a lack of fine tuning to an evolutionarily recent inserted stop codon. Protein domains may get lost in protein evolution (73) and investigation of the ortholog’s 3′-UTR may display the remnants of a former domain. Highly leaky genes (TR rate above 5%) were investigated for homology. Orthologs to the leaky genes were searched for by a blastp against 50 fungi species. Each ortholog’s transcript was blasted against the 3′-UTR nucleotide-sequence with a cut off value at 1e-5. No significant homology to the focal 3′-UTR was found in the ortholog transcripts to support the notion of a recently inserted stop codon. As a previous study investigating TR in yeast (48) but with different orthologs, we did not find the 3′-UTR to be conserved. In other words, no support was found to indicate evolutionary conservation or that leaky genes have evolved from longer genes.
Protein domains and motifs
We next asked if the elongation by TR would yield a functional protein. The presence of protein domains in the 3′-UTR would imply that TR leads to a functionally folded elongation. Moreover, protein domains in the 3′-UTR would imply selection for a functional extension. Alternatively, the 3′-UTRs were once part of a longer functional protein and TR is due to a lack of fine tuning to an inserted stop codon. Domain losses has previously been suggested to be frequent at the C-terminus and to be explained by an introduced stop (or start) codon (73). Implementing a more targeted approach than homology search, by using Hidden Markov Models in pfam, we searched for protein domains in multiple ORFs of the CDS (Materials and Methods), as TR has been found to occasionally be caused by frame-shift (e.g. 13). This was done for all three sets (leaky, semi-leaky and non-leaky). We did not find protein domains, suggesting that continued translation of the parent-gene beyond the stop codon into the 3′-UTR does not include a functionally annotated protein domain. In conclusion, we do not find support for selection of functional extensions for proteins undergoing TR.
CONCLUSION
We have shown that TR rate is seemingly related to gene expression and peptide structure, specifically intrinsic disorder. The evolutionary rate of proteins has repeatedly been found to correlate with peptide structure and gene expression (74,75). We did not find an overall trend for difference of evolutionary rate when comparing proteins that are error prone and not error prone by our branch-site test. However, there is a significant difference of codon usage between proteins, reflected by propensity for TR. In other words, our findings indicate there may be selection on traits influencing translation and TR. In addition to TR being associated with high gene expression, we found that TR is foremost associated with proteins having high intrinsic disorder—most profoundly at the C-termini. This finding raises multiple questions for further exploration.
There is no gradual increase of disorder that correlates with gene expression to support a symmetric relationship between gene expression and intrinsic disorder. However, a highly expressed gene is by exposure at high risk of mistranslation (8,9). Accordingly, highly expressed proteins are expected to be under selection for translational robustness (8,9). One study, confirming this expectation, on antibiotic resistance in E. coli found that once an error occurs—given short evolutionarily time-scale and in large populations—the system first reduces the consequences of translational errors rather than reducing the errors themselves (14). In other words, that proteins evolve towards error tolerance rather than error mitigation (14,76). Our results corroborate these assumptions on selective pressure and evolutionary constraints. We hypothesize that intrinsically disordered C-termini make error prone proteins functionally robust in the occurrences of TR. Intrinsically disordered regions can act as dynamic switches in response to environmental changes, e.g. shift in pH, metabolite concentrations or post-translational modifications, and deliver an alternative protein conformation, by being phosphorylised by enzymes (41,77–80).
There is a strong evolutionary link between disorder propensity and secondary structure (81,82). However, assuming that intrinsically disordered C-termini can elevate protein robustness and be advantageous, it is peculiar that we do not find intrinsically disordered C-termini to be present in all proteins. As already stated, high gene expression imposes an elevated risk of phenotypic mutations. We suggest intrinsically disordered C-termini are present mostly in highly expressed genes because of their relatively higher exposure risk of errors.
Moreover, highly expressed genes are found to evolve slowly independently of protein function (8,9). Selection for translational robustness has been suggested to explain the constrained sequence evolution for highly expressed genes (8,9). Addressing what possibilities exist for proteins to evolve, phenotypic mutations may provide an opportunity for genes under constrained selection to explore alternative isoforms. According to the look ahead-effect (12), phenotypic mutations may act as intermediate stepping stones for traits not yet encoded in the DNA, enabling rapid adaptation by exploration of the phenotypic landscape. The look-ahead-effect has been partially supported by recent experimental research in fungi (13), but demands further experimental validation. Alternative conformations, e.g. provided by intrinsic disorder, provide the most accessible solution when new protein functions are needed (83). Previous research has found that intrinsically disordered regions may diverge and evolve more rapidly than structured regions (84,85), facilitate innovation (86) and protein expansion (43,87), and to be indispensable to non-adaptive evolutionary processes (81). We hypothesize that intrinsically disordered C-termini may not only act to increase protein robustness, but potentially also facilitate the exploration of protein isoforms in evolutionarily constrained genes without significantly distorting the protein structure.
In conclusion, we have shown that error prone proteins are codon optimised, are highly expressed and have intrinsically disordered C-terminus. We suggest that intrinsic disorder may play an instrumental role in protein robustness when facing phenotypic mutations as TR. To investigate the effect of intrinsically disordered regions experimentally, it should be possible to e.g. remove or add an intrinsically disordered C-terminus and measure how it relates to gene expression, TR rate, and fitness. The full nature of the relationship between intrinsic disorder and phenotypic mutations, with regard to protein robustness and evolution, invites further research.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Sebastian Leidel for generous advise regarding ribosome profiling. We also thank Brennen Heames and Daniel Dowling for valuable input that lifted the paper’s quality. The work has been conducted within the framework of the Münster Graduate School of Evolution.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Studienstiftung des Deutschen Volkes sponsor the research scholarship for April Snofrid Kleppe.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lynch M. The cellular, developmental and population-genetic determinants of mutation-rate evolution. Genetics. 2008; 180:933–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lynch M. The origins of genome architecture 2007. Science. 2007; 302:1401–1404. [DOI] [PubMed] [Google Scholar]
- 3. Kramer E.B., Farabaugh P.J.. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007; 13:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burger R., Willensdorfer M., Nowak M.A.. Why are phenotypic mutation rates much higher than genotypic mutation rates. Genetics. 2006; 172:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bornberg-Bauer E., Chan H.S.. Modeling evolutionary landscapes: mutational stability, topology, and superfunnels in sequence space. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:10689–10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Nimwegen E., Crutchfield J.P., Huynen M.. Neutral evolution of mutational robustness. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:9716–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goldsmith M., Tawfik D.S.. Potential role of phenotypic mutations in the evolution of protein expression and stability. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:6197–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Drummond D.A., Wilke C.O.. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008; 134:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Drummond D.A., Bloom J.D., Adami C., Wilke C.O., Arnold F.H.. Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilke C.O., Drummond D.A.. Population genetics of translational robustness. Genetics. 2006; 173:473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Willensdorfer M., Burger R., Nowak M.A.. Phenotypic mutation rates and the abundance of abnormal proteins in yeast. PLoS Comput. Biol. 2007; 3:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Whitehead D.J., Wilke C.O., Vernazobres D., Bornberg-Bauer E.. The look-ahead effect of phenotypic mutations. Biol. Direct. 2008; 3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yanagida H., Gispan A., Kadouri N., Rozen S., Sharon M., Barkai N., Tawfik D.S.. The evolutionary potential of phenotypic mutations. PLoS Genet. 2015; 11:e1005445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bratulic S., Gerber F., Wagner A.. Mistranslation drives the evolution of robustness in TEM-1 -lactamase. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:12758–12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Freitag J., Ast J., Bolker M.. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature. 2012; 485:522–525. [DOI] [PubMed] [Google Scholar]
- 16. Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S.. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013; 2:e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nedialkova D.D., Leidel S.A.. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015; 161:1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim D., Salzberg S.L.. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011; 12:R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J., Billis K., Carvalho-Silva D., Cummins C., Clapham P. et al.. Ensembl 2017. Nucleic Acids Res. 2017; 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yassour M., Pfiffner J., Levin J.Z., Adiconis X., Gnirke A., Nusbaum C., Thompson D.A., Friedman N., Regev A.. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol. 2010; 11:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anders S., McCarthy D.J., Chen Y., Okoniewski M., Smyth G.K., Huber W., Robinson M.D.. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013; 8:1765–1786. [DOI] [PubMed] [Google Scholar]
- 25. Namy O., Hatin I., Rousset J.P.. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001; 2:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H. et al.. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999; 285:901–906. [DOI] [PubMed] [Google Scholar]
- 28. Zhang Z., Ren Q.. Why are essential genes essential?—The essentiality of Saccharomyces genes. Microb. Cell. 2015; 2:280–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alexa A., Rahnenfuhrer J., Lengauer T.. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006; 22:1600–1607. [DOI] [PubMed] [Google Scholar]
- 30. R Core Team R: A Language and Environment for Statistical Computing. 2016; Vienna: R Foundation for Statistical Computing. [Google Scholar]
- 31. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 32. Huang da W., Sherman B.T., Lempicki R.A.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Altschul S.F., Erickson B.W.. Significance of nucleotide sequence alignments: a method for random sequence permutation that preserves dinucleotide and codon usage. Mol. Biol. Evol. 1985; 2:526–538. [DOI] [PubMed] [Google Scholar]
- 35. Bailey T.L., Johnson J., Grant C.E., Noble W.S.. The MEME Suite. Nucleic Acids Res. 2015; 43:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beznoskova P., Wagner S., Jansen M.E., von der Haar T., Valášek L.S.. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 2015; 43:5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dosztanyi Z., Csizmok V., Tompa P., Simon I.. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005; 21:3433–3434. [DOI] [PubMed] [Google Scholar]
- 38. Ferron F., Longhi S., Canard B., Karlin D.. A practical overview of protein disorder prediction methods. Proteins. 2006; 65:1–14. [DOI] [PubMed] [Google Scholar]
- 39. Habchi J., Tompa P., Longhi S., Uversky V.N.. Introducing protein intrinsic disorder. Chem. Rev. 2014; 114:6561–6588. [DOI] [PubMed] [Google Scholar]
- 40. Lieutaud P., Ferron F., Uversky A.V., Kurgan L., Uversky V.N., Longhi S.. How disordered is my protein and what is its disorder for? A guide through the “dark side” of the protein universe. Intrinsically Disord. Proteins. 2016; 4:e1259708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K., Fuxreiter M., Gough J., Gsponer J., Jones D.T. et al.. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014; 114:6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Faure G., Callebaut I.. Comprehensive repertoire of foldable regions within whole genomes. PLoS Comput. Biol. 2013; 9:e1003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bitard-Feildel T., Heberlein M., Bornberg-Bauer E., Callebaut I.. Detection of orphan domains in Drosophila using “hydrophobic cluster analysis”. Biochimie. 2015; 119:244–253. [DOI] [PubMed] [Google Scholar]
- 44. Sharp P.M., Li W.H.. The codon Adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987; 15:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kovacs E., Tompa P., Liliom K., Kalmar L.. Dual coding in alternative reading frames correlates with intrinsic protein disorder. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:5429–5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al.. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pancsa R., Macossay-Castillo M., Kosol S., Tompa P.. Computational analysis of translational readthrough proteins in Drosophila and yeast reveals parallels to alternative splicing. Sci. Rep. 2016; 6:32142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jansen R., Gerstein M.. Analysis of the yeast transcriptome with structural and functional categories: characterizing highly expressed proteins. Nucleic Acids Res. 2000; 28:1481–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hanson G., Coller J.. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018; 19:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Komar A.A., Lesnik T., Reiss C.. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999; 462:387–391. [DOI] [PubMed] [Google Scholar]
- 52. Zhang G., Hubalewska M., Ignatova Z.. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 2009; 16:274–280. [DOI] [PubMed] [Google Scholar]
- 53. Thanaraj T.A., Argos P.. Protein secondary structural types are differentially coded on messenger RNA. Protein Sci. 1996; 5:1973–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang G., Ignatova Z.. Generic algorithm to predict the speed of translational elongation: implications for protein biogenesis. PLoS ONE. 2009; 4:e5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chaney J.L., Steele A., Carmichael R., Rodriguez A., Specht A.T., Ngo K., Li J., Emrich S., Clark P.L.. Widespread position-specific conservation of synonymous rare codons within coding sequences. PLoS Comput. Biol. 2017; 13:e1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weinberg D.E., Shah P., Eichhorn S.W., Hussmann J.A., Plotkin J.B., Bartel D.P.. Improved Ribosome-Footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 2016; 14:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhou M., Wang T., Fu J., Xiao G., Liu Y.. Nonoptimal codon usage influences protein structure in intrinsically disordered regions. Mol. Microbiol. 2015; 97:974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pechmann S., Frydman J.. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 2013; 20:237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994; 136:927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Powell J.R., Moriyama E.N.. Evolution of codon usage bias in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:7784–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kramer E.B., Vallabhaneni H., Mayer L.M., Farabaugh P.J.. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA. 2010; 16:1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang Y., Koonin E.V., Lipman D.J., Przytycka T.M.. Selection for minimization of translational frameshifting errors as a factor in the evolution of codon usage. Nucleic Acids Res. 2009; 37:6799–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dix D.B., Thompson R.C.. Codon choice and gene expression: synonymous codons differ in translational accuracy. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:6888–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dosztanyi Z., Meszaros B., Simon I.. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009; 25:2745–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Piovesan D., Tabaro F., Mičetić I., Necci M., Quaglia F., Oldfield C.J., Aspromonte M.C., Davey N.E., Davidović R., Dosztanyi Z. et al.. DisProt 7.0: a major update of the database of disordered proteins. Nucleic Acids Res. 2017; 45:D1123–D1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Homma K., Noguchi T., Fukuchi S.. Codon usage is less optimized in eukaryotic gene segments encoding intrinsically disordered regions than in those encoding structural domains. Nucleic Acids Res. 2016; 44:10051–10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schaefer C., Schlessinger A., Rost B.. Protein secondary structure appears to be robust under in silico evolution while protein disorder appears not to be. Bioinformatics. 2010; 26:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tretyachenko V., Vymětal J., Bednarova L., Kopecky V., Hofbauerova K., Jindrova H., Hubalek M., Souček R., Konvalinka J., Vondrašek J. et al.. Random protein sequences can form defined secondary structures and are well-tolerated in vivo. Sci. Rep. 2017; 7:15449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ellis J.J., Huard F.P., Deane C.M., Srivastava S., Wood G.R.. Directionality in protein fold prediction. BMC Bioinformatics. 2010; 11:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Saunders R., Deane C.M.. Protein structure prediction begins well but ends badly. Proteins. 2010; 78:1282–1290. [DOI] [PubMed] [Google Scholar]
- 71. Jacob E., Unger R.. A tale of two tails: why are terminal residues of proteins exposed. Bioinformatics. 2007; 23:e225–e230. [DOI] [PubMed] [Google Scholar]
- 72. Babu M.M., van der Lee R., de Groot N.S., Gsponer J.. Intrinsically disordered proteins: regulation and disease. Curr. Opin. Struct. Biol. 2011; 21:432–440. [DOI] [PubMed] [Google Scholar]
- 73. Weiner J., Beaussart F., Bornberg-Bauer E.. Domain deletions and substitutions in the modular protein evolution. FEBS J. 2006; 273:2037–2047. [DOI] [PubMed] [Google Scholar]
- 74. Wolf M.Y., Wolf Y.I., Koonin E.V.. Comparable contributions of structural-functional constraints and expression level to the rate of protein sequence evolution. Biol. Direct. 2008; 3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Drummond D.A., Wilke C.O.. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009; 10:715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wilke C.O. Evolutionary paths of least resistance. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:12553–12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tsai C.J., Ma B., Sham Y.Y., Kumar S., Nussinov R.. Structured disorder and conformational selection. Proteins. 2001; 44:418–427. [DOI] [PubMed] [Google Scholar]
- 78. Tompa P. The principle of conformational signaling. Chem. Soc. Rev. 2016; 45:4252–4284. [DOI] [PubMed] [Google Scholar]
- 79. McDowell G.S., Hindley C.J., Lippens G., Landrieu I., Philpott A.. Phosphorylation in intrinsically disordered regions regulates the activity of Neurogenin2. BMC Biochem. 2014; 15:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kasahara K., Shiina M., Higo J., Ogata K., Nakamura H.. Phosphorylation of an intrinsically disordered region of Ets1 shifts a multi-modal interaction ensemble to an auto-inhibitory state. Nucleic Acids Res. 2018; 46:2243–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ahrens J.B., Nunez-Castilla J., Siltberg-Liberles J.. Evolution of intrinsic disorder in eukaryotic proteins. Cell. Mol. Life Sci. 2017; 74:3163–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ahrens J., Dos Santos H.G., Siltberg-Liberles J.. The nuanced interplay of intrinsic disorder and other structural properties driving protein evolution. Mol. Biol. Evol. 2016; 33:2248–2256. [DOI] [PubMed] [Google Scholar]
- 83. Tawfik D.S. Messy biology and the origins of evolutionary innovations. Nat. Chem. Biol. 2010; 6:692–696. [DOI] [PubMed] [Google Scholar]
- 84. Brown C.J., Takayama S., Campen A.M., Vise P., Marshall T.W., Oldfield C.J., Williams C.J., Dunker A.K.. Evolutionary rate heterogeneity in proteins with long disordered regions. J. Mol. Evol. 2002; 55:104–110. [DOI] [PubMed] [Google Scholar]
- 85. Brown C.J., Johnson A.K., Daughdrill G.W.. Comparing models of evolution for ordered and disordered proteins. Mol. Biol. Evol. 2010; 27:609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Montanari F., Shields D.C., Khaldi N.. Differences in the number of intrinsically disordered regions between yeast duplicated proteins, and their relationship with functional divergence. PLoS ONE. 2011; 6:e24989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Light S., Sagit R., Sachenkova O., Ekman D., Elofsson A.. Protein expansion is primarily due to indels in intrinsically disordered regions. Mol. Biol. Evol. 2013; 30:2645–2653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.