The roles of environmental reservoirs, including wild birds, in the molecular epidemiology of Campylobacter jejuni have not been assessed in depth. Our results showed that game birds may pose a risk for acquiring campylobacteriosis, because they had C. jejuni genomotypes highly similar to human isolates detected previously. Therefore, hygienic measures during slaughter and meat handling warrant special attention. On the contrary, a unique phylogeny was revealed for the western jackdaw isolates, and certain genomic characteristics identified among these isolates are hypothesized to affect their host specificity and virulence. Comparative genomics within sequence types (STs), using whole-genome multilocus sequence typing (wgMLST), and phylogenomics are efficient methods to analyze the genomic relationships of C. jejuni isolates.
KEYWORDS: Campylobacter jejuni, antimicrobial resistance, comparative genomics, cytolethal distending toxin, mallard duck, pheasant, public health, western jackdaw, whole-genome sequencing
ABSTRACT
Poultry are considered a major reservoir and source of human campylobacteriosis, but the roles of environmental reservoirs, including wild birds, have not been assessed in depth. In this study, we isolated and characterized Campylobacter jejuni from western jackdaws (n = 91, 43%), mallard ducks (n = 82, 76%), and pheasants (n = 9, 9%). Most of the western jackdaw and mallard duck C. jejuni isolates represented multilocus sequence typing (MLST) sequence types (STs) that diverged from those previously isolated from human patients and various animal species, whereas all pheasant isolates represented ST-19, a common ST among human patients and other hosts worldwide. Whole-genome MLST revealed that mallard duck ST-2314 and pheasant ST-19 isolates represented bacterial clones that were genetically highly similar to human isolates detected previously. Further analyses revealed that in addition to a divergent ClonalFrame genealogy, certain genomic characteristics of the western jackdaw C. jejuni isolates, e.g., a novel cdtABC gene cluster and the type VI secretion system (T6SS), may affect their host specificity and virulence. Game birds may thus pose a risk for acquiring campylobacteriosis; therefore, hygienic measures during slaughter and meat handling warrant special attention.
IMPORTANCE The roles of environmental reservoirs, including wild birds, in the molecular epidemiology of Campylobacter jejuni have not been assessed in depth. Our results showed that game birds may pose a risk for acquiring campylobacteriosis, because they had C. jejuni genomotypes highly similar to human isolates detected previously. Therefore, hygienic measures during slaughter and meat handling warrant special attention. On the contrary, a unique phylogeny was revealed for the western jackdaw isolates, and certain genomic characteristics identified among these isolates are hypothesized to affect their host specificity and virulence. Comparative genomics within sequence types (STs), using whole-genome multilocus sequence typing (wgMLST), and phylogenomics are efficient methods to analyze the genomic relationships of C. jejuni isolates.
INTRODUCTION
With more than 200,000 annually reported cases, Campylobacter jejuni continues to be the most common cause of human bacterial gastroenteritis in the European Union (EU), including in Finland (1). Poultry have been recognized to be a major reservoir and source of human campylobacteriosis (1). However, in Finland, the prevalence of Campylobacter-positive broiler flocks has remained low since 2004, when systematic reporting started (2), while simultaneously, the incidence in the human population has shown an increasing trend (incidence 69/100,000 in 2004 and 85/100,000 in 2016) (3). Besides poultry, wild birds have often been shown to carry C. jejuni, and hence are considered to act as wildlife reservoirs, causing fecal contamination of the environments of food production farms and possibly transmitting Campylobacter to domestic animals as well as to humans (4). C. jejuni has been found in various wild bird species (4–7); however, relatively little is known about its occurrence in western jackdaws and game birds.
Western jackdaw (Corvus monedula) belongs to the family of crows and is one of the most common bird species in some urban and agricultural surroundings in Finland, and thus also being a concern from the public health perspective as a potential reservoir of zoonotic pathogens, including Campylobacter jejuni. The Finnish jackdaw population has increased rapidly during the last thirty years (8). Jackdaws form large flocks producing noise and considerable fecal loads that contaminate the environment, including the surroundings of food production farms and city parks. The western jackdaw is an omnivore, feeding on plants and invertebrates as well as food waste. Jackdaw flocks also feed in farmland and may cause marked agricultural damage (8). Jackdaws are partial migratory birds; adults overwinter near their nests, whereas juveniles migrate to the southern parts of the Baltic Sea.
Besides natural wild game bird populations that are hunted during hunting season, several bird species, including waterfowl, pheasants, and pigeons, are bred and raised on specific farms and released for hunting purposes. Mallard duck (Anas platyrhynchos) is the most common game bird in Finland, with 210,000 to 300,000 birds hunted annually (www.riista.fi/en/). Mallard ducks live close to the water systems and feed on plants and invertebrates. Most of the Finnish natural mallard duck population migrates in late autumn to Central Europe, but some flocks stay in Finland year round. The hunting season lasts approximately 4 months (August to December), and meat is mostly consumed by individual hunters (Ministry of Agriculture and Forestry [www.mmm.fi]).
Pheasants (Phasianus colchicus) belong to the same family as partridges and are a common domesticated game bird in Europe and in the United States (9). In the United Kingdom and Ireland alone, up to 30 million pheasants are hunted annually (10). These birds live typically in open spaces and fields and feed on plants, grain, seeds, insects, and invertebrates. In Finland, 15,000 to 75,000 pheasants are hunted annually (www.riista.fi/en/), and the meat is mostly consumed by individual hunters but is also sold to restaurants (Ministry of Agriculture and Forestry [www.mmm.fi]).
Multilocus sequence typing (MLST) has been an invaluable molecular typing method, providing essential knowledge about C. jejuni types occurring in various hosts and sources worldwide. MLST is, however, limited to the characterization and discrimination of isolates according to sequence type (ST) (11–13); thus, more accurate methods, such as whole-genome MLST (wgMLST), are increasingly being used (11, 14) to compare genetically related isolates in more detail and to be able to identify clones potentially originating from the same source. In previous studies, most of the STs found among wild birds, including mallard ducks (5), barnacle geese (7), starlings (15), and several other bird species (16), have been considered to represent mainly host-associated STs, differing from those STs reported in human patients or domestic animals. Thus, wild birds are commonly considered to have a minor role in human campylobacteriosis. However, certain STs and generalist lineages that are common in human patients (e.g., ST-45 CC) have been detected in several wild bird species as well (5–7, 16, 17), indicating that wild birds are potential reservoirs for certain common STs also infecting humans. For example, ST-45, ST-677, and ST-267, which have been common STs in human infections in Finland (18, 19), have been found among both blackbird and chicken isolates from Sweden (6). However, to our knowledge, there have been no comprehensive wgMLST-level studies, and only one study including comparative genomic analyses on wild bird isolates (American crows) has been published to date (17).
The aim of this study was to assess the occurrence, population genetics, and diversity of Campylobacter spp. in western jackdaws and game birds (i.e., mallard duck and pheasant) in Finland using whole-genome sequencing. We further explored the presence of genomic features associated with virulence and antimicrobial resistance among the isolates to evaluate their importance from a public health perspective.
RESULTS
Occurrence and multilocus sequence typing.
Campylobacter isolates were obtained from 91 western jackdaw (43%), 79 farmed game mallard duck (79%), 9 pheasant (9%), and 3 wild mallard duck (38%) samples. All isolates were identified as C. jejuni.
Whole-genome sequencing (WGS) was successful for 87 (of 91) western jackdaw C. jejuni isolates. MLST showed great diversity, as 62 different sequence types (STs) were found, 46 of which were novel to the PubMLST database (https://pubMLST.org/campylobacter) (see Table S1 in the supplemental material). ST-1282 and ST-6460 were the most frequently found STs and also the most widely distributed both temporally and geographically. Among mallard ducks, 36 of 38 selected representative isolates (35 farmed, chosen based on pulsed-field gel electrophoresis [PFGE] screening, and 3 wild mallard duck isolates) were successfully sequenced. Altogether, 16 STs were identified, 4 of which were novel to the PubMLST database. Taking into account the combined results of PFGE and MLST, the most common STs in mallard ducks were ST-2314 (n = 26), ST-1299 (n = 16), ST-991 (n = 7), ST-2839 (n = 7), and ST-995 (n = 5). All of the nine pheasant isolates represented ST-19 (ST-21 CC).
Phylogenomics.
A phylogenomic tree based on the core genome alignment of altogether 1,261 C. jejuni strains (see Data Set S1 in the supplemental material), including the ones sequenced in this study and those collected from public databases, is shown in Fig. S1. The western jackdaw isolates clustered together in a diffuse branch also including American crow, a few chicken isolates, and a very limited number of genomes representing sandhill crane, mallard duck, and other mammals. All the other isolates, even those inside the branch, were clearly distinct from the western jackdaw isolates, which also were highly diverse. The mallard duck isolates also showed great diversity; however, the majority clustered together with C. jejuni strains isolated from a few chickens and a barnacle goose. The pheasant isolates formed a more homogenous cluster and shared the same branch with a higher proportion of agricultural C. jejuni isolates from chicken, some bovines, and other mammals.
ClonalFrame genealogy.
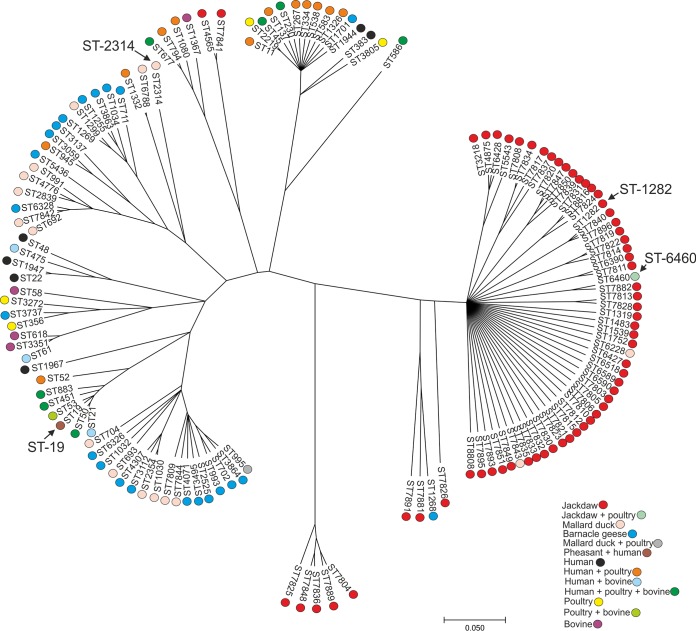
The ClonalFrame genealogy representing the phylogeny of the STs is shown in Fig. 1. ClonalFrame revealed that most of the STs occurring in western jackdaws were in their own diffuse cluster and differed from those that were detected from other hosts in Finland, including other wild bird species. Exceptions were ST-4565 and ST-7841, which were clearly distinct from other western jackdaw STs and located in the same branch with the STs detected from humans, poultry, and bovines (ST-677 CC and related STs), and also ST-6460, which was a common ST among the western jackdaw isolates but was also found previously in poultry in Finland.
FIG 1.
ClonalFrame genealogy of MLST allele sequences in different hosts detected in this study (western jackdaws and mallard ducks) and previously in Finland (2, 7, 18, 19, 28, 29). Different hosts and combinations of hosts are indicated with different colors, and those STs that were further analyzed in wgMLST are indicated with arrows.
STs detected from mallard ducks occurred mostly in the same branches as STs detected previously from barnacle geese. Exceptions were ST-6228 and ST-7843, which were in the same cluster with western jackdaw STs, ST-995, which had also been isolated from poultry, and ST-6788, which was closely related to ST-1332, previously isolated from human patients and poultry. The most commonly found ST among farmed mallard ducks, ST-2314, was not closely related to any other ST previously reported in Finland.
ST-19 (ST-21 CC), represented by the pheasant isolates, had previously been isolated from two human patients in Finland, and it has been common in several other countries in various sources, including humans, poultry, and bovines (www.pubMLST.org/campylobacter).
Whole-genome MLST.
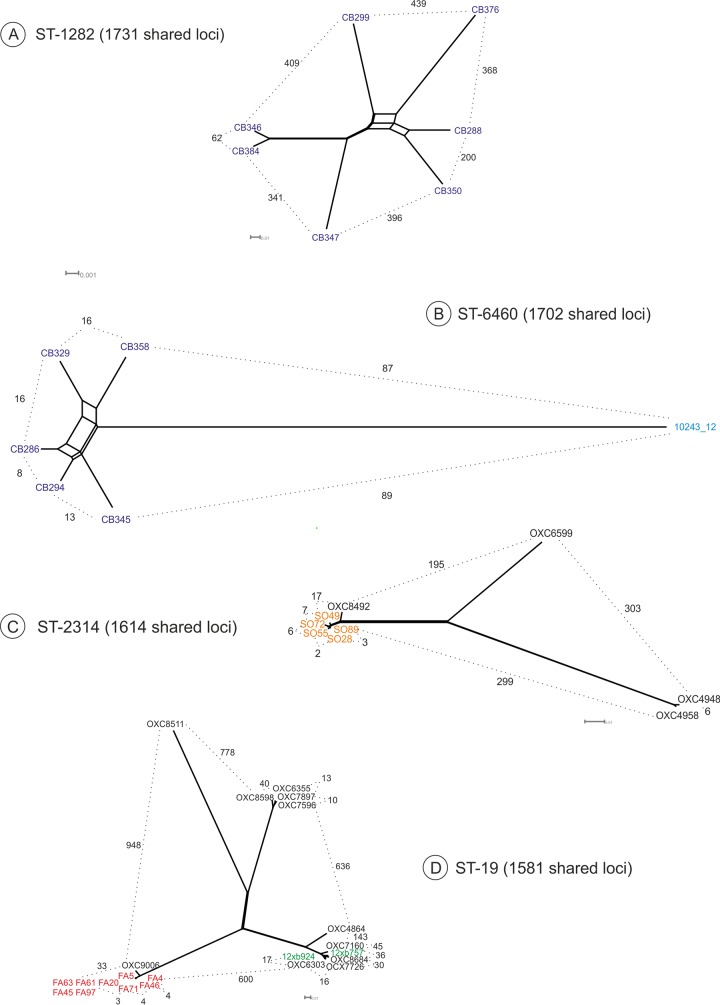
The wgMLST analysis of ST-1282, the most common ST among western jackdaw isolates and obtained from two different locations on five separate occasions, revealed high numbers of allelic differences (62 to 439) between the isolates (Fig. 2A). The two isolates with the fewest allelic differences (n = 62), CB346 and CB384, were isolated from the same town and taken five months apart, while two isolates (e.g., CB299 and CB288) collected from the same sampling site on successive sampling days had 313 allelic differences. The wgMLST of ST-6460, representing the second most common ST, revealed 87 to 89 allelic differences between the western jackdaw isolates and a chicken isolate obtained in 2012 from the Finnish Campylobacter monitoring program (Fig. 2B). Western jackdaw ST-6460 isolates were more closely related to each other, as only 8 to 16 allelic differences occurred among the isolates, which had been collected from two different locations on three separate occasions (Fig. 2B). Although originating from different towns, C. jejuni pairs CB294/CB286 and CB329/CB358 in Fig. 2B were isolated on the same days from the same sampling points.
FIG 2.
wgMLST results for the allelic differences among shared loci of ST-1282 (A), ST-6460 (B), ST-2314 (C), and ST-19 (D) isolates using GeP (14). Numbers between strains indicate allelic differences. Jackdaw isolates are indicated in dark blue, chicken isolate in light blue, mallard duck isolates in orange, pheasant isolates in red, and clinical isolates from the United Kingdom (www.pubMLST.org/campylobacter) in black and from Finland in green.
The wgMLST analysis of ST-2314 isolates from Finnish mallard ducks and human patients from the United Kingdom (isolated in 2010, 2011, and 2013) is shown in Fig. 2C. The isolates formed three clusters, differing by 195 to 303 alleles. Between the mallard duck ST-2314 isolates, only 2 to 7 allelic differences were detected, and 17 to 21 allelic differences (approximately 1% of the 1,614 shared loci) were found between the five mallard duck isolates and a clinical isolate from the United Kingdom (OXC8492) in 2013 (Fig. 2C).
The wgMLST of ST-19, describing the allelic differences between the C. jejuni isolates from pheasants and human patients, collected in the United Kingdom in 2010 to 2014 (www.pubMLST.org/campylobacter) and in Finland in 2012 (19), is shown in Fig. 2D. The isolates formed four distinct (from 600 to 945 allelic differences) clusters. One of the clusters that consisted of all of the genetically highly related pheasant isolates (differing by 3 to 4 alleles) and one clinical isolate obtained from the United Kingdom in 2014 (OXC9006), which diverged from each other by 33 to 37 allelic differences, accounted for 2% of the 1,581 shared loci (Fig. 2D).
Description of genetic features.
(i) Integrated elements. C. jejuni integrated elements 1, 2, 3, and 4 (CJIE1, -2, -3, and -4, respectively) were present in 20% (17/87), 86% (77/87), 85% (74/87), and 14% (12/87) of the western jackdaw isolates, respectively. However, there were large deletions and other major differences in the genomic structures of CJIE2 in the majority of the isolates compared with that of reference strain RM1221 (20). Among the mallard duck isolates, integrated elements CJIE1, -2, -3, and -4 were present in 3% (1/36), 39% (14/36), 81% (29/36), and 3% (1/36) of the isolates, respectively. The only integrated element present in pheasant isolates was CJIE4, which was present in 100% of the isolates.
(ii) Type VI secretion system.
A total of 43 of 87 (49%) of the western jackdaw isolates carried a type VI secretion system (T6SS), similar in gene synteny and sequence (96% query coverage and 96% identity in BLASTN) to the T6SS of strain 108 (21). Interestingly, the majority (72%) of the mallard duck isolates also carried the T6SS, whereas none of the pheasant isolates carried either the CJIE3 or T6SS.
(iii) Cytolethal distending toxin.
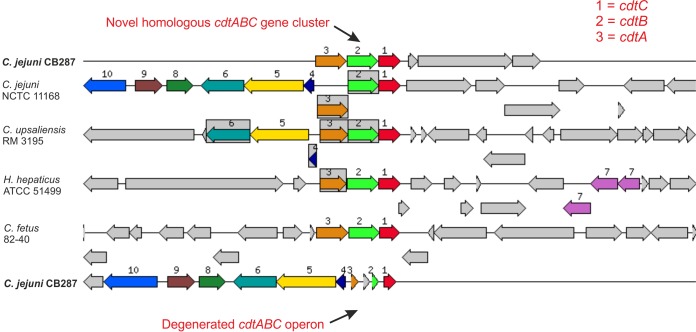
Figure 3 shows an alignment of the cytolethal distending toxin (cdtABC) gene clusters of western jackdaw isolate CB287 against those of the reference strain NCTC 11168. The cdtABC operon, located in the same genomic site as in NCTC 11168, was complete in only two of the western jackdaw isolates, missing completely in three isolates, and degenerated and thus likely dysfunctional in most (82 of 87) of the western jackdaw isolates (e.g., CB287) (Fig. 3). However, another homologous cdtABC gene cluster (69% identity, 84% query coverage against other C. jejuni sequences in nonredundant nucleotide/protein sequences database [nr] using BLASTN) was present in another location of the genome in 59% (51/87) of the western jackdaw isolates (e.g., CB287) (Fig. 3). For further analysis, a phylogenetic neighbor-joining tree was constructed based on the conserved amino acid sequences of the cdtB gene. The novel jackdaw CdtB sequence was relatively distinct from those of other C. jejuni strains and even from other Campylobacter spp., but highly similar to those of crow C. jejuni isolates from California, USA (Fig. 4). The two western jackdaw isolates that had intact cdtABC operons, located in a similar position as in strain NCTC 11168, represented ST-4565 and ST-7841, which were clearly distinct from the majority of the western jackdaw STs that were also in the ClonalFrame genealogy and located in the generalist clade (Fig. 1). Similarly, the CdtB phylogenetic tree (Fig. 4) showed that the sequences of these two isolates were more closely related to those of other C. jejuni strains, including those from the pheasant isolates from our study and the generalist C. jejuni isolates from crows in a previous study (17).
FIG 3.
Seed Viewer homology-based sequence alignment of the two cdtABC gene clusters (top and bottom rows) of western jackdaw strain CB287. The chromosomal region of the focus gene (top) is compared with those from four similar organisms (including Campylobacter fetus, Campylobacter upsaliensis, and Helicobacter hepaticus). The graphic is centered on the focus gene (cdtC), which is red and numbered 1. Sets of genes with similar sequences are grouped with matching numbers and colors. Genes that are not conserved remain unnumbered and are shown in gray.
FIG 4.
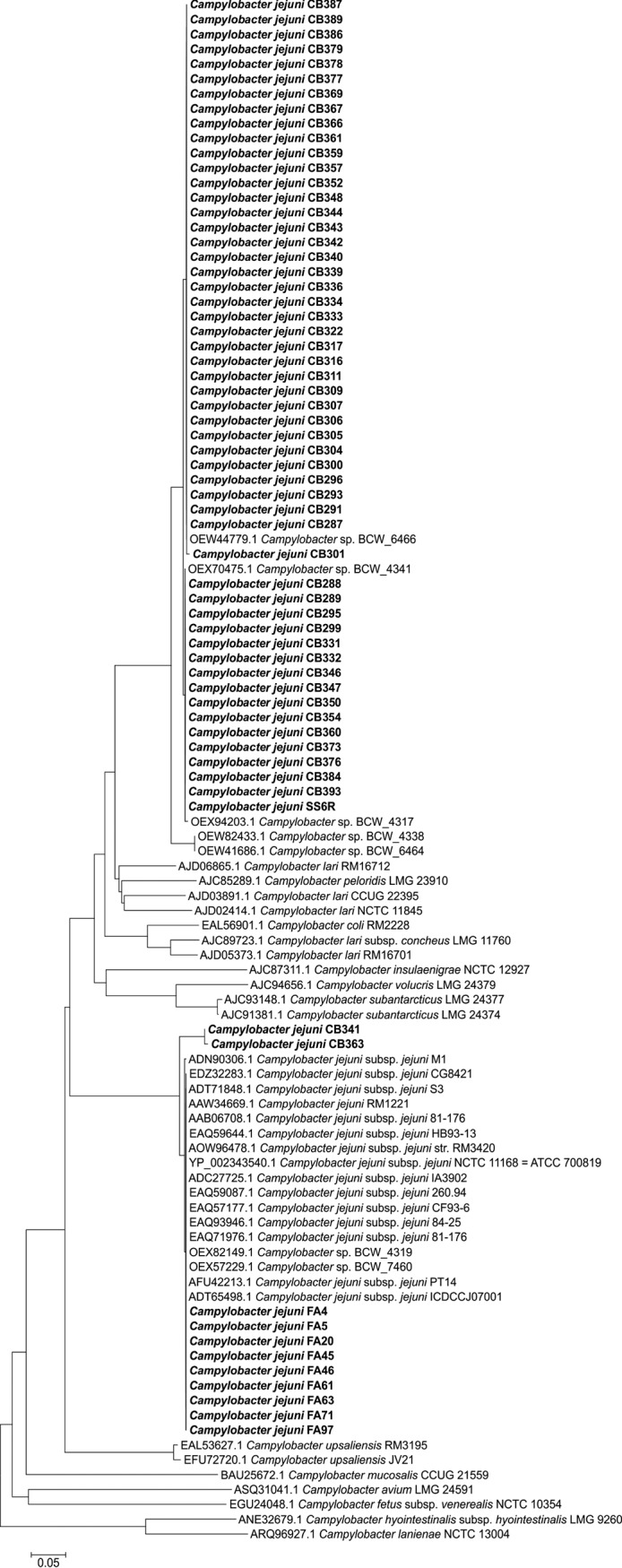
Neighbor-joining tree representing the alignment of CdtB amino acid sequences. Accession numbers, species, and strain numbers are indicated for each sequence. Strains from this study are indicated in boldface and with CB for western jackdaw, SO or SS for mallard duck, and FA for pheasant in the strain name.
The cdtABC operon was also degenerated in all mallard duck isolates, except one isolate that carried the novel cdtABC gene cluster, homologous to most of the western jackdaw isolates. This mallard duck isolate represented ST-6228, which was located in the same clade as the western jackdaw STs in the ClonalFrame genealogy (Fig. 1) and had an identical or closely related CdtB amino acid sequence to those of the western jackdaw and crow isolates (Fig. 4). In contrast to those in western jackdaw and mallard duck isolates, the cdtABC operon (similar to strain NCTC 11168) was intact and most likely functional among all pheasant isolates (ST-19).
(iv) Antimicrobial resistance-associated genetic markers.
The tetO gene encoding tetracycline resistance ribosomal protection protein (TetO) was found to be present in 14% (12/87) of the jackdaw isolates but only in 1 of the 36 mallard duck isolates (3%) and none of the pheasant isolates. The 23S rRNA gene in positions 2074 and 2075 was wild type, suggesting a susceptibility to macrolides. Gene AadE, conferring streptomycin resistance, was absent among all bird isolates. No mutations associated with ciprofloxacin resistance in the gyrA gene were detected among mallard ducks, but one T86I substitution in gyrA was present in a western jackdaw isolate. Similarly, the T86I mutation in gyrA was found among all pheasant isolates.
The neighbor-joining tree representing the alignment of family class D beta-lactamase (blaOXA) gene sequences is shown in Fig. S2. All pheasant isolates carried the blaOXA-61 family class D beta-lactamase gene with sequences identical to that in C. jejuni type strain NCTC 11168. In contrast, blaOXA-184 family class D beta-lactamase was found to be present in 69% (60/87) of the western jackdaw isolates and in all mallard duck isolates. Furthermore, based on the beta-lactamase gene sequences among the western jackdaws and mallard ducks, they formed separate branches in the neighbor-joining tree (Fig. S2), with the exception of the sequences in two mallard duck isolates, SS6R (ST-6228) and SO-68 (ST-7843), located in the same cluster as the majority of the western jackdaw STs in the ClonalFrame analysis (Fig. 1).
DISCUSSION
Birds are well-recognized natural reservoirs of C. jejuni, and different wild bird species have been found to carry C. jejuni, among other Campylobacter spp., with variable frequencies. Thus, wild birds may pose a risk for public health either indirectly by transmitting C. jejuni to other animals, food production farms, and environmental waters, and through these, also to humans, or directly, when bird meat is consumed as game (4). This study is the first to characterize C. jejuni isolates from western jackdaws and game birds using MLST and comparative genomics in order to elucidate their potential public health significance. Western jackdaws have not been investigated extensively. In this study, we found that 43% (n = 212) of our western jackdaw isolates carried C. jejuni. In addition to western jackdaws, 76% (n = 108) of the mallard ducks carried C. jejuni. An earlier study from New Zealand reported that 23% (n = 702) of wild mallard ducks were positive for C. jejuni (5), and in a study from the United Kingdom, high frequencies (93.3 to 100%) of farmed mallard ducks carried Campylobacter, emphasizing their role as a risk for human health (22). The relatively high frequency and diversity of C. jejuni among mallard ducks in this study may at least partly be a consequence of mixing with natural C. jejuni populations from other waterfowl, since the farmed mallard ducks were raised in natural ponds, which were cohabited by wild birds. In contrast to that in western jackdaws and mallard ducks, a low prevalence of C. jejuni was found among farmed pheasants (9%). In a previous study from the Czech Republic, 70% of pheasants that were farmed with intensive production were positive for Campylobacter sp., 41% being C. jejuni (23). However, in the same study, a lower frequency (25.7%) was detected in wild pheasants; only 16% were C. jejuni positive (23). The colonization dynamics of C. jejuni in wild birds is complicated and may depend on, for instance, host ecology, the age of the bird, social behavior, and season (24).
Only limited epidemiological data exist on cases where western jackdaws, mallard ducks, or pheasants have been suspected as a source of human infections. However, from the 1980s to 1990s in the United Kingdom, several milkborne outbreaks were associated with western jackdaws and magpies that were pecking milk bottles located outside the front doors of houses (25, 26). Also, in a Finnish epidemiological study in the 1990s, we found two patients suspected of having acquired campylobacteriosis after contact with pheasants (27).
The high occurrence of novel or rarely detected STs, which have only infrequently been associated with human disease, suggests that while western jackdaws are not a major source, they are a potential source of campylobacteriosis in humans. The most commonly detected STs were ST-1282 and ST-6460, the former was previously isolated from a wild bird in Sweden and environmental water in Luxembourg (www.pubMLST.org). ST-6460, in turn, has been detected infrequently, in a chicken slaughter batch in Finland in 2012 (2) and in a human patient in Sweden (www.pubMLST.org). Other STs found among western jackdaws and have been isolated in human stools include ST-1539 (United Kingdom and Netherlands), ST-6589 (Sweden), and ST-6590 (Sweden) (www.pubMLST.org/campylobacter). However, overall STs previously reported in human disease accounted for only 9 of 87 isolates (10%), and those reported previously in chickens (ST-6460 and ST-5543) accounted for 7 of 87 isolates (8%), supporting the result that jackdaws are not a major source of C. jejuni in human infections. Chicken farming is located in the same area in western Finland where the sampling of western jackdaws took place, suggesting that western jackdaw flocks living close to chicken farms may pose a risk of contamination.
Despite the high frequency (76%) of C. jejuni among Finnish mallard ducks, all STs differed from those previously found among Finnish human patients with infections acquired from domestic sources (18, 19, 28, 29). However, the five most common STs (ST-2314, ST-1299, ST-2839, ST-991, and ST-995) were previously reported in the pubMLST database, obtained from different geographical areas from human stools (except ST-2839, unknown source) and from other wild birds (except ST-2314 and ST-2839), suggesting that these STs may have a wider host range and the capability of causing human infections. Furthermore, the ST-2314 mallard duck isolates, representing one-third of all of the mallard duck isolates based on PFGE types (data not shown), differed from each other in the wgMLST analysis by only 2 to 7 alleles, indicating that these isolates represent the same clone, which was also found to be genetically highly similar to a human isolate previously isolated in the United Kingdom. In addition, both the farmed and wild mallard ducks in this study carried ST-995 (UA), which has also been detected among mallard ducks in Sweden (16) and New Zealand (5) (see Fig. S3 in the supplemental material), suggesting a strong host association of this ST with mallard ducks.
All C. jejuni isolates from pheasants represented ST-19, which has frequently been isolated from human patients with gastroenteritis as well as from a wide variety of different hosts worldwide (22, 30–32). Since all the pheasants carried the same clone of C. jejuni (3 to 4 allelic differences in the wgMLST analysis) but were hunted on three separate occasions and from two different locations, the birds had most likely been colonized already in the breeding farm at a very early stage, before they were moved to the separate hunting farms.
The phylogenomic tree based on the core genomes clearly indicated that although the western jackdaw isolates form a very distinct set of strains from the rest of the C. jejuni population, they do not form a cohesive single population. ClonalFrame genealogy further confirmed that the majority of the C. jejuni STs from western jackdaws clustered separately from the other STs previously detected in human patients, poultry, bovines, and other wild birds in Finland (2, 7, 18, 19, 28, 29), confirming the suggestion that these STs have evolved in western jackdaws and are not frequently transferred between different animal host species. Only two STs (ST-7841 and ST-4565) were located in the same branch, which also comprised STs isolated from human patients or poultry. ClonalFrame also showed that most of the mallard duck STs formed two branches that also contained STs detected previously from Finnish barnacle geese (7), which may indicate that these STs have adapted more widely to waterfowl. This was supported by the phylogenomic analysis. In conclusion, the ClonalFrame genealogy in our study showed concordance with those of earlier studies (6, 7, 16), revealing that wild birds are mostly colonized by host-adapted STs but also have a minor set of STs shared with other hosts, including domestic animals and human patients.
Comparative genomics using wgMLST was performed to explore genomic diversity within the most frequently detected STs. In conclusion, wgMLST using Genome profiler (GeP) (14) to detect closely related C. jejuni isolates revealed that birds living in flocks in close contact are colonized by both genetically highly related (e.g., ST-6460, ST-2314, and ST-19) and diverse (e.g., ST-1282) STs. Each strain and ST will most likely have its own capability to evolve, as shown earlier for ST-45 CC (12) and for ST-2314 and ST-19 in this study; however, more spatial and temporal data from each genetic lineage are needed to make robust conclusions. Our results add to the evidence that comparative genomics within STs, using wgMLST, is a suitable method to analyze the genomic relationships of C. jejuni isolates. These results further confirm the results of our previous studies, where we found that isolates originating from the same outbreak usually have only a few allelic differences, whereas isolates from unassociated sources more likely have tens to several hundreds of allelic differences (33–35).
Even though several putative virulence factors of C. jejuni have been suggested to be associated with the pathogenesis of human gastroenteritis (20, 21, 36, 37), their functions are not well known and it remains unclear whether all strains have the capability to cause human infections (38). Most of the STs detected among western jackdaw and mallard duck isolates were novel; thus, they have not been associated previously with human infections or detected in other animals. Therefore, we studied the genetic characteristics of these isolates to screen for virulence-associated genes in their genomes. One of the most recently described putative virulence-associated systems in C. jejuni (39) and in Campylobacter coli (40) is T6SS. In our study, a complete T6SS gene cluster was found to be common among western jackdaw (46%) and mallard duck (72%) isolates. The high frequency of T6SS in our study is an interesting result, because T6SS has been recognized to contribute to bacterial pathogenesis via toxic effects on host cells or competing bacterial species (39). The C. jejuni T6SS has been found to have pleiotropic effects, ranging from adaptation to host cell adherence, in vivo colonization, invasion, and cytotoxicity toward human erythrocytes (39, 41, 42). In addition, patients harboring a bacterial strain with T6SS often had bacteremia (39), suggesting more severe disease. The T6SS gene cluster has been found with variable frequencies in different studies, most probably depending on the origin of the strain. Commonly, approximately 10% of studied strains harbor T6SS, containing 13 open reading frames (ORFs) similar to those described in C. jejuni strain 108 (39). T6SS was not detected among common STs and clonal lineages occurring in human patients and animal sources in the United Kingdom (e.g., ST-45 CC and ST-21 CC), but it was more often detected among human patient isolates from Vietnam and Pakistan, representing rare STs not commonly detected in Europe (42). This finding corroborates our results, because most of the western jackdaw and mallard duck isolates had novel STs, which were not detected previously in human patients or other animal hosts. More studies on patients infected with uncommon STs containing T6SS are warranted to strengthen the evidence of an association between bacteremia and certain genetic lineages of C. jejuni.
Another virulence-associated system known to be common in C. jejuni is the cytolethal distending toxin (CDT), encoded by a three component operon cdtABC (37). CdtB is a DNase that causes DNA double-strand breaks in the nucleus, resulting in cell cycle arrest at the G2/M stage and apoptosis (43). Previous PCR-based studies directed at the CDT operon or a single gene of the operon have shown that more than 95% of C. jejuni strains are positive (36). In the present study, all except two of our western jackdaw isolates carried a cdtABC operon, but surprisingly, it was degenerated in most (94%) of the isolates. The two isolates with an intact copy of the cdtABC clustered more closely with the generalist clade in the ClonalFrame genealogy and also in the CdtB phylogeny. In addition, the gene cluster was totally missing from three of the jackdaw isolates. Interestingly, almost all isolates with a degenerated cdtABC locus also had another intact homologous cdtABC gene cluster. The newly identified cdtABC gene cluster was consistently present only in the western jackdaw clade in the ClonalFrame genealogy. This is the first time, to our knowledge, that another complete cdtABC gene cluster has been reported to be present in C. jejuni. Further analysis of the CdtB sequences of the newly identified gene cluster revealed that the sequences of C. jejuni isolates from western jackdaws were identical or closely related to those of crow isolates collected in California, USA (17). A comparison of the CdtB sequences also revealed that the jackdaw and crow sequences were more closely related to those of Campylobacter lari than to those of other C. jejuni isolates, including the pheasant isolates from this study. This is an interesting finding and warrants further studies. All mallard duck isolates carried only the degenerated cdtABC locus. Whether the degeneration of the cdtABC locus is connected only to wild bird-associated genetic lineages remains unanswered and awaits future research.
The tetO gene, predicting resistance against tetracycline (TET), was found to be present among 14% of the western jackdaw isolates. Among mallard ducks, the tetO gene was present in only one isolate, and in none of the pheasant isolates, suggesting low resistance against TET. The T86I mutation in gyrA, previously linked to high fluoroquinolone-resistance in Campylobacter strains (44, 45), was found in only one jackdaw isolate. Among mallard ducks, gyrA was found to be wild type; however, among pheasants, the T86I substitution in gyrA was found in all isolates, predicting resistance against ciprofloxacin. The MIC value of ciprofloxacin was confirmed to be ≥16 μg/ml and 64 μg/ml for nalidixic acid (NAL) using VetMIC Camp EU (SVA, Sweden). In a previous study, no resistance against ciprofloxacin (CIP) was found in wild bird isolates in Finland (46), indicating that the results of the present study are mostly in line with the low occurrence of fluoroquinolone resistance among wild birds, since the pheasant isolates most likely represented a single clone. It is intriguing why this single clone was so successful and no other types were found among the pheasant isolates. The farm raising the pheasants for hunting did not use any antibiotics, and this may be excluded as a reason for the clonal expansion of this resistant type. Contact with wildlife is likely in this production system from very early stages on, however, only after the birds are released to the surroundings at 3 to 4 weeks old. Interestingly, a small proportion of the hatched pheasant eggs are imported from France on a yearly basis; this may be a possible source of resistance, since the birds carried the same clone on separate farms even though they had been separated as 1-day-old chicks. Furthermore, ST-19 is a common generalist type known to colonize a variety of hosts, including wild birds in Canada and ducks in the United Kingdom (https://pubmlst.org/campylobacter/), and carries the CIP-NAL resistance phenotype (47, 48). According to EFSA and ECDC, resistance against ciprofloxacin is generally high among human, chicken, and turkey C. jejuni isolates in the EU and shows a rising trend (49). Further studies will reveal if fluoroquinolone resistance is also on the rise among C. jejuni isolates in wild bird populations. A previous study (50) reported that certain strains with this mutation in gyrA showed no biological cost and that fluoroquinolone-resistant strains outcompeted susceptible ones in vivo in chickens in the absence of selective pressure, which might also explain the clonal expansion of successful clones. Genomic markers, conferring streptomycin (AadE) and macrolide (23S rRNA gene mutation in positions 2074 and 2075) resistance, were absent among all bird isolates. Overall, antimicrobial resistance has remained low in Finland among the C. jejuni isolates of domestic origin relative to that in several other countries (46) and seems to also be low in birds migrating to Finland.
β-Lactams (including penicillin, amoxicillin, and ampicillin) have been widely used in veterinary and clinical medicine; however, emerging resistance has compromised their use. High-level β-lactam resistance and beta-lactamase activity have been shown to be linked to an upregulated blaOXA-61 gene in C. jejuni (51). Interestingly, among our C. jejuni collection, the blaOXA-184 family (instead of blaOXA-61) class D beta-lactamase was present in 69% of the western jackdaw isolates, and it was also present in all mallard duck isolates. In contrast, all pheasant isolates carried the OXA-61 family class D beta-lactamase. Further phylogenetic analysis revealed that the blaOXA-184 sequences differed markedly between mallard duck and western jackdaw isolates, suggesting an evolutionary link with the host. Previously, the OXA-184 family beta-lactamases were also found to be associated with crow isolates rather than OXA-61, which has been associated with strains isolated from either domestic animals or human patients (17). These results reveal that OXA-184 family class D beta-lactamase is, for an unknown reason, associated with wild bird isolates, but future studies will extend our knowledge of their evolution and functionality.
In conclusion, phylogenetic studies performed on MLST data and CdtB and beta-lactamase gene sequences support a unique evolutionary history of C. jejuni in western jackdaws. Due to a high frequency of novel STs and STs only rarely detected among human patients and broiler batches, western jackdaws are considered to be an infrequent sources of campylobacteriosis in humans and of contamination of poultry production. Although the STs among western jackdaws and mallard ducks appear host associated, they may have the capability to transfer to domestic animals as well as to occasionally cause human infections. Game birds especially may pose a higher risk for acquiring campylobacteriosis, and both the carcasses and meat should be handled accordingly. More evidence is anticipated to accumulate when MLST, particularly wgMLST, is applied more extensively on different sources worldwide.
MATERIALS AND METHODS
Bacterial isolates.
Campylobacter sp. was isolated from western jackdaws from two small towns, located in Southern Finland (Lahti) and Western Finland (Seinäjoki), during a five-month period from September 2014 to February 2015. Fecal droppings (n = 212) were collected by placing a plastic carpet under the known roosting trees of jackdaws. When jackdaws had settled in their roosting trees, the flock was scared away to cause automatic defecation by the birds. Individual droppings were immediately swabbed into transport tubes (TS0001; Oxoid, Thermo Fisher Scientific, Vantaa, Finland) and delivered to the laboratory chilled (4°C). Contamination of the collected fecal material is unlikely, as we checked that no other bird species were present in the same tree before scaring the jackdaw flocks away. Of note, hooded crows especially tend to roost with jackdaws.
Samples from game birds that were farmed for hunting were collected during the hunting season in Southern Finland. The birds were shot by licensed hunters, and no animals were killed for the purpose of this study. Mallard ducks (n = 100) were hunted from September to November in 2014 and pheasants (n = 100) from November to December in 2013. The birds were plucked (mallard ducks) or skinned (pheasants) and eviscerated immediately after hunting. Intestinal samples were collected during evisceration, and transported to the laboratory chilled (4°C). The mallard ducks were hunted on the surroundings of a single farm. The birds were hatched on another breeding farm, and 3- to 4-week-old birds were transferred and released to the natural ponds of the hunting farm, where they were raised by regular feeding. Pheasants were hatched on the same farm that the mallard ducks were raised on. Six batches stayed on the same farm for hunting, and three batches of 1-day-old chicks were transferred to yet another hunting farm, where they were raised. In both hunting farms, the 4-week-old birds were released into the surroundings, where their feeding was continued.
In addition to farmed game mallard ducks, fresh fecal samples (n = 8) from the wild mallard duck population, living in a lake of an urban park in Southern Finland, were collected from individual droppings into transport tubes (TS0001; Oxoid, Thermo Fisher Scientific, Vantaa, Finland) in November 2014.
Fecal samples were cultivated on mCCDA plates (Oxoid Ltd., Hampshire, UK). Typical colonies were Gram stained, and genomic DNA was extracted using a PureLink genomic DNA minikit (Invitrogen, Waltham, MA, USA). The Campylobacter species was identified by species-specific PCR (52). DNA of all C. jejuni isolates from western jackdaws, pheasants, and wild mallard ducks was subjected to WGS.
For C. jejuni-positive game mallard duck isolates, pulsed-field gel electrophoresis (PFGE), using KpnI restriction enzyme, was first used to screen the diversity of PFGE types among the isolates. PFGE was performed as described previously (53). Thirty-five isolates, representing all of the different PFGE profiles (some in duplicates or triplicates), were selected and their DNA was subjected to WGS.
Whole-genome sequencing.
Whole-genome sequencing (WGS) was performed using Illumina HiSeq technology at the Institute for Molecular Medicine Finland (FIMM). Raw data were assembled into contigs using the INNUca pipeline (https://github.com/B-UMMI/INNUca), and genome sequences (n = 6) that did not pass the quality threshold were excluded from further analyses.
Phylogenetic analyses.
A phylogenomic tree was constructed based on the core genomes of 1,261 C. jejuni strains selected from the INNUENDO Campylobacter jejuni data set (54) and the genomes sequenced in this study (see Data set S1 in the supplemental material). The genomes were annotated using Prokka 1.12 (55). A pan genome analysis was performed using Roary 3.7.0 (56) with 95% protein identity as the cutoff for defining gene clusters and the core genes. FastTree 2.1 (57) was used with the Jukes-Cantor model of nucleotide evolution for building the approximation of a maximum likelihood (ML) phylogenetic tree based on the core genome alignment (99% shared loci in Roary analysis, including 942 core genes). When necessary, the jobs were run in parallel using GNU Parallel (58). iTOL v4.2.3 (59) was used for visualization. The tree was rooted at midpoint for better visualization.
ClonalFrame genealogy was used to describe the phylogeny of the C. jejuni MLST allele sequences collected in this study and also MLST sequence data of C. jejuni isolates from our previous studies originating from several sources (poultry, bovines, barnacle geese, and humans) in Finland (2, 7, 18, 19, 28, 29). ClonalFrame was run using default parameters (60); the consensus tree was displayed in MEGA6 and labeled using CorelDRAW X8.
A GoeBURST full minimum spanning tree (MST) was constructed using PHYLOViZ (61). Besides our wild bird isolates, MLST data for mallard ducks from Sweden (16) and New Zealand (5) were obtained from the PubMLST database (www.pubMLST.org/campylobacter).
Comparative genomics.
Genome profiler (GeP) (14) was used in wgMLST to further compare the draft genomes. In addition to our isolates, WGS data from clinical isolates from the United Kingdom were obtained from the PubMLST database (www.pubMLST.org/campylobacter) for reference. The clinical ST-19 isolates from the United Kingdom were chosen to represent all available subtypes as defined by using coregenome (cg) MLST for 700 loci (https://github.com/mickaelsilva/chewBBACA_deprecated/wiki) and different years. Allelic differences between strains of each ST, obtained using GeP, were visualized using SplitsTree4 (62) and edited in CorelDRAW X8.
The RAST (Rapid Annotation using Subsystem Technology) server (63) was used for preliminary annotation and genomic characterization of the isolates. Sequence-based comparisons, genome browsing, and alignments for visualization of synteny of homologous genomic regions were performed using the SEED viewer (64, 65) with default parameters. Novel open reading frames, genes associated with cytolethal distending toxin (cdtABC) operon, integrated elements (CJIE1, -2, -3, and -4), type VI secretion system (T6SS), family class D beta-lactamases, and antimicrobial resistance were further analyzed using the InterProScan (66) and by BLASTN and/or BLASTP searches against the nonredundant nucleotide/protein sequences database (nr) in GenBank. Phylogenetic analyses of genes of interest were performed by aligning the amino acid sequences of the cdtB gene and nucleotide sequences of family class D beta-lactamases using MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/), and MEGA6 (67) was used to construct a phylogenetic neighbor-joining tree based on the alignments. The resulting phylogenetic trees were text edited in CorelDRAW 2017.
Accession number(s).
Whole-genome sequences of the bird isolates that represented novel STs were deposited in the pubMLST database (www.pubMLST.org/campylobacter) to assign new sequence types and allelic profiles.
The whole-genome sequences of all of the isolates sequenced in this study have been deposited in GenBank under BioProject accession no. PRJNA430314. The relevant BioSample (SAMN08398758 to SAMN08398888) and sequence accession numbers are listed in Table S1. NCBI Prokaryotic Genomes Annotation Pipeline (PGAP) was used to annotate the genomes.
Supplementary Material
ACKNOWLEDGMENTS
Financial support for this work was from the Jenny and Antti Wihuri Foundation, the Finnish Foundation of Veterinary Research, and the Academy of Finland grant on behalf of CoE-MiFoSa (no. 11411405). All jackdaw samples were collected in the Jackdaw damage management project funded by the Ministry of the Environment, Finland. Campylobacter strains were isolated from the jackdaw samples and draft genome-sequenced in the Bacteria hazardous to humans and production animals in feces of jackdaw project (1832/312/2014) funded by The Development Fund for Agriculture and Forestry, Ministry of Agriculture and Forestry, Finland.
We thank laboratory technician Urszula Hirvi for assistance in the laboratory. We also thank CSC-Tieteen tietotekniikan keskus Oy for providing access to cloud computing resources.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02365-18.
REFERENCES
- 1.European Food Safety Authority (EFSA), European Centre for Disease Prevention and Control (ECDC). 2016. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J 14:4634. doi: 10.2903/j.efsa.2016.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llarena AK, Huneau A, Hakkinen M, Hänninen M-L. 2015. Predominant Campylobacter jejuni sequence types persist in Finnish chicken production. PLoS One 10:e0116585. doi: 10.1371/journal.pone.0116585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaakola S, Lyytikäinen O, Rimhanen-Finne R, Salmenlinna S, Savolainen-Kopra C, Liitsola K, Jalava J, Toropainen M, Nohynek H, Virtanen M, Löflund J-E, Kuusi M, Salminen M. 2017. Tartuntataudit Suomessa 2016. THL – Raportti 5/2017. Department of Health and Welfare, Helsinki, Finland: http://urn.fi/URN:ISBN:978-952-302-890-6. [Google Scholar]
- 4.Waldenström J, Griekspoor P. 2014. Ecology and host associations of Campylobacter in wild birds, p 265–284. In Sheppard S. (ed), Campylobacter ecology and evolution, 1st ed Caister Academic Press, Norfolk, UK. [Google Scholar]
- 5.Mohan V, Stevenson M, Marshall J, Fearnhead P, Holland BR, Hotter G, French NP. 2013. Campylobacter jejuni colonization and population structure in urban populations of ducks and starlings in New Zealand. Microbiologyopen 2:659–673. doi: 10.1002/mbo3.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griekspoor P, Hansbro PM, Waldenström J, Olsen B. 2015. Campylobacter jejuni sequence types show remarkable spatial and temporal stability in blackbirds. Infect Ecol Epidemiol 5:28383. doi: 10.3402/iee.v5.28383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llarena AK, Skarp-de Haan CP, Rossi M, Hänninen M-L. 2015. Characterization of the Campylobacter jejuni population in the barnacle geese reservoir. Zoonoses Public Health 62:209–221. doi: 10.1111/zph.12141. [DOI] [PubMed] [Google Scholar]
- 8.Pohja-Mykrä M, Koskinen S, Mykrä S, Nieminen T, Sillanpää H (ed). 2016. Naakka ja ihminen. Naakkojen aiheuttamien haittojen hallinta. Suomen Ympäristö, Ministry of the Environment, Helsinki, Finland. [Google Scholar]
- 9.Sokos CK, Birtsas PK, Tsachalidis EP. 2008. The aims of galliforms release and choice of techniques. Wildlife Biol 14:412–422. doi: 10.2981/0909-6396-14.4.412. [DOI] [Google Scholar]
- 10.Matheson SM, Donbavand J, Sandilands V, Pennycott T, Turner SP. 2015. An ethological approach to determining housing requirements of gamebirds in raised laying units. Appl Anim Behav Sci 165:17–24. doi: 10.1016/j.applanim.2015.02.001. [DOI] [Google Scholar]
- 11.Cody AJ, McCarthy ND, Jansen van Rensburg M, Isinkaye T, Bentley SD, Parkhill J, Dingle KE, Bowler IC, Jolley KA, Maiden MC. 2013. Real-time genomic epidemiological evaluation of human Campylobacter isolates by use of whole-genome multilocus sequence typing. J Clin Microbiol 51:2526–2534. doi: 10.1128/JCM.00066-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llarena AK, Zhang J, Vehkala M, Välimäki N, Hakkinen M, Hänninen M-L, Roasto M, Mäesaar M, Taboada E, Barker D, Garofolo G, Cammà C, Di Giannatale E, Corander J, Rossi M. 2016. Monomorphic genotypes within a generalist lineage of Campylobacter jejuni show signs of global dispersion. Microb Genom 2:e000088. doi: 10.1099/mgen.0.000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cody AJ, Bray JE, Jolley KA, McCarthy ND, Maiden MCJ. 2017. Core genome multilocus sequence typing scheme for stable, comparative analyses of Campylobacter jejuni and C. coli human disease isolates. J Clin Microbiol 55:2086–2097. doi: 10.1128/JCM.00080-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Halkilahti J, Hänninen M-L, Rossi M. 2015. Refinement of whole-genome multilocus sequence typing analysis by addressing gene paralogy. J Clin Microbiol 53:1765–1767. doi: 10.1128/JCM.00051-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colles FM, McCarthy ND, Howe JC, Devereux CL, Gosler AG, Maiden MC. 2009. Dynamics of Campylobacter colonization of a natural host, Sturnus vulgaris (European starling). Environ Microbiol 11:258–267. doi: 10.1111/j.1462-2920.2008.01773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griekspoor P, Colles FM, McCarthy ND, Hansbro PM, Ashhurst-Smith C, Olsen B, Hasselquist D, Maiden MCJ, Waldenström J. 2013. Marked host specificity and lack of phylogeographic population structure of Campylobacter jejuni in wild birds. Mol Ecol 22:1463–1472. doi: 10.1111/mec.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weis AM, Storey DB, Taff CC, Townsend AK, Huang BC, Kong NT, Clothier KA, Spinner A, Byrne BA, Weimer BC. 2016. Genomic comparison of Campylobacter spp. and their potential for zoonotic transmission between birds, primates, and livestock. Appl Environ Microbiol 82:7165–7175. doi: 10.1128/AEM.01746-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Haan CPA, Kivisto R, Hakkinen M, Rautelin H, Hanninen ML. 2010. Decreasing trend of overlapping multilocus sequence types between human and chicken Campylobacter jejuni isolates over a decade in Finland. Appl Environ Microbiol 76:5228–5236. doi: 10.1128/AEM.00581-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovanen SM, Kivistö RI, Rossi M, Schott T, Kärkkäinen UM, Tuuminen T, Uksila J, Rautelin H, Hänninen M-L. 2014. Multilocus sequence typing (MLST) and whole-genome MLST of Campylobacter jejuni isolates from human infections in three districts during a seasonal peak in Finland. J Clin Microbiol 52:4147–4154. doi: 10.1128/JCM.01959-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, DeBoy RT, Parker CT, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Sullivan SA, Shetty JU, Ayodeji MA, Shvartsbeyn A, Schatz MC, Badger JH, Fraser CM, Nelson KE. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol 3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ugarte-Ruiz M, Stabler RA, Dominguez L, Porrero MC, Wren BW, Dorrell N, Gundogdu O. 2015. Prevalence of type VI secretion system in Spanish Campylobacter jejuni isolates. Zoonoses Public Health 62:497–500. doi: 10.1111/zph.12176. [DOI] [PubMed] [Google Scholar]
- 22.Colles FM, Ali JS, Sheppard SK, McCarthy ND, Maiden MC. 2011. Campylobacter populations in wild and domesticated mallard ducks (Anas platyrhynchos). Environ Microbiol Rep 3:574–580. doi: 10.1111/j.1758-2229.2011.00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nebola M, Borilova G, Steinhauserova I. 2008. Prevalence of Campylobacter subtypes in pheasants (Phasianus colchicus spp. torquatus) in the Czech Republic. Vet Med (Praha) 52:496–501. doi: 10.17221/2063-VETMED. [DOI] [Google Scholar]
- 24.Taff CC, Weis AM, Wheeler S, Hinton MG, Weimer BC, Barker CM, Jones M, Logsdon R, Smith WA, Boyce WM, Townsend AK. 2016. Influence of host ecology and behavior on Campylobacter jejuni prevalence and environmental contamination risk in a synanthropic wild bird species. Appl Environ Microbiol 82:4811–4820. doi: 10.1128/AEM.01456-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Southern JP, Smith RM, Palmer SR. 1990. Bird attack on milk bottles: possible mode of transmission of Campylobacter jejuni to man. Lancet 336:1425–1427. [DOI] [PubMed] [Google Scholar]
- 26.Hudson SJ, Lightfoot NF, Coulson JC, Russell K, Sisson PR, Sobo AO. 1991. Jackdaws and magpies as vectors of milkborne human Campylobacter infection. Epidemiol Infect 107:363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hänninen M-L, Pajarre S, Klossner ML, Rautelin H. 1998. Typing of human Campylobacter jejuni isolates in Finland by pulsed-field gel electrophoresis. J Clin Microbiol 36:1787–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kärenlampi R, Rautelin H, Schönberg-Norio D, Paulin L, Hänninen M-L. 2007. Longitudinal study of Finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl Environ Microbiol 73:148–155. doi: 10.1128/AEM.01488-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Haan CP, Kivistö RI, Hakkinen M, Corander J, Hänninen M-L. 2010. Multilocus sequence types of Finnish bovine Campylobacter jejuni isolates and their attribution to human infections. BMC Microbiol 10:200. doi: 10.1186/1471-2180-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rotariu O, Dallas JF, Ogden ID, Macrae M, Sheppard SK, Maiden M, Gormley FJ, Forbes KJ, Strachan NJ. 2009. Spatiotemporal homogeneity of Campylobacter subtypes from cattle and sheep across NE and SW Scotland. Appl Environ Microbiol 75:6275–6281. doi: 10.1128/AEM.00499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cody AJ, McCarthy NM, Wimalarathna HL, Colles FM, Clark L, Bowler IC, Maiden MC, Dingle KE. 2012. A longitudinal 6-year study of the molecular epidemiology of clinical Campylobacter isolates in Oxfordshire, United Kingdom. J Clin Microbiol 50:3193–3201. doi: 10.1128/JCM.01086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mossong J, Mughini-Gras L, Penny C, Devaux A, Olinger C, Losch S, Cauchie HM, van Pelt W, Ragimbeau C. 2016. Human campylobacteriosis in Luxembourg, 2010–2013: a case-control study combined with multilocus sequence typing for source attribution and risk factor analysis. Sci Rep 6:20939. doi: 10.1038/srep20939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Revez J, Llarena AK, Schott T, Kuusi M, Hakkinen M, Kivistö R, Hänninen M-L, Rossi M. 2014. Genome analysis of Campylobacter jejuni strains isolated from a waterborne outbreak. BMC Genomics 15:768. doi: 10.1186/1471-2164-15-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Revez J, Zhang J, Schott T, Kivistö R, Rossi M, Hänninen M-L. 2014. Genomic variation between Campylobacter jejuni isolates associated with milk-borne-disease outbreaks. J Clin Microbiol 52:2782–2786. doi: 10.1128/JCM.00931-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovanen S, Kivistö R, Llarena AK, Zhang J, Kärkkäinen UM, Tuuminen T, Uksila J, Hakkinen M, Rossi M, Hänninen M-L. 2016. Tracing isolates from domestic human Campylobacter jejuni infections to chicken slaughter batches and swimming water using whole-genome multilocus sequence typing. Int J Food Microbiol 226:53–60. doi: 10.1016/j.ijfoodmicro.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Mortensen NP, Schiellerup P, Boisen N, Klein BM, Locht H, Abuoun M, Newell D, Krogfelt KA. 2011. The role of Campylobacter jejuni cytolethal distending toxin in gastroenteritis: toxin detection, antibody production, and clinical outcome. APMIS 119:626–634. doi: 10.1111/j.1600-0463.2011.02781.x. [DOI] [PubMed] [Google Scholar]
- 37.Lai CK, Chen YA, Lin CJ, Lin HJ, Kao MC, Huang MZ, Lin YH, Chiang-Ni C, Chen CJ, Lo UG, Lin LC, Lin H, Hsieh JT, Lai CH. 2016. Molecular mechanisms and potential clinical applications of Campylobacter jejuni cytolethal distending toxin. Front Cell Infect Microbiol 6:9. doi: 10.3389/fcimb.2016.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolton DJ. 2015. Campylobacter virulence and survival factors. Food Microbiol 48:99–108. doi: 10.1016/j.fm.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 39.Bleumink-Pluym NMC, van Alphen LB, Bouwman LI, Wösten MMSM, van Putten JPM. 2013. Identification of a functional type VI secretion system in Campylobacter jejuni conferring capsule polysaccharide sensitive cytotoxicity. PLoS Pathog 9:e1003393. doi: 10.1371/journal.ppat.1003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghatak S, He Y, Reed S, Strobaugh T Jr, Irwin P. 2017. Whole genome sequencing and analysis of Campylobacter coli YH502 from retail chicken reveals a plasmid-borne type VI secretion system. Genom Data 11:128–131. doi: 10.1016/j.gdata.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lertpiriyapong K, Gamazon ER, Feng Y, Park DS, Pang J, Botka G, Graffam ME, Ge Z, Fox JG. 2012. Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS One 7:e42842. doi: 10.1371/journal.pone.0042842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrison JW, Dung TT, Siddiqui F, Korbrisate S, Bukhari H, Tra MP, Hoang NV, Carrique-Mas J, Bryant J, Campbell JI, Studholme DJ, Wren BW, Baker S, Titball RW, Champion OL. 2014. Identification of possible virulence marker from Campylobacter jejuni isolates. Emerg Infect Dis 20:1026–1029. doi: 10.3201/eid2006.130635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickett CL, Pesci EC, Cottle DL, Russell G, Erdem AN, Zeytin H. 1996. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB gene. Infect Immun 64:2070–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Huang WM, Taylor DE. 1993. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob Agents Chemother 37:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han J, Wang Y, Sahin O, Shen Z, Guo B, Shen J, Zhang Q. 2012. A fluoroquinolone resistance associated mutation in gyrA affects DNA supercoiling in Campylobacter jejuni. Front Cell Infect Microbiol 2:21. doi: 10.3389/fcimb.2012.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olkkola S, Nykasenoja S, Raulo S, Llarena AK, Kovanen S, Kivistö R, Myllyniemi AL, Hänninen M-L. 2016. Antimicrobial resistance and multilocus sequence types of Finnish Campylobacter jejuni isolates from multiple sources. Zoonoses Public Health 63:10–19. doi: 10.1111/zph.12198. [DOI] [PubMed] [Google Scholar]
- 47.Kovač J, Čadež N, Stessl B, Stingl K, Gruntar I, Ocepek M, Trkov M, Wagner M, Smole Možina S. 2015. High genetic similarity of ciprofloxacin-resistant Campylobacter jejuni in central Europe. Front Microbiol 6:1169. doi: 10.3389/fmicb.2015.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Habib I, Miller WG, Uyttendaele M, Houf K, De Zutter L. 2009. Clonal population structure and antimicrobial resistance of Campylobacter jejuni in chicken meat from Belgium. Appl Environ Microbiol 75:4264–4272. doi: 10.1128/AEM.00168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.European Food Safety Authority (EFSA), European Centre for Disease Prevention and Control (ECDC). 2018. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J 16:5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, Zhang Q. 2005. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci U S A 102:541–546. doi: 10.1073/pnas.0408966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng X, Brown S, Gillespie B, Lin J. 2014. A single nucleotide in the promoter region modulates the expression of the beta-lactamase OXA-61 in Campylobacter jejuni. J Antimicrob Chemother 69:1215–1223. doi: 10.1093/jac/dkt515. [DOI] [PubMed] [Google Scholar]
- 52.Denis M, Soumet C, Rivoal K, Ermel G, Blivet D, Salvat G, Colin P. 1999. Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett Appl Microbiol 29:406–410. doi: 10.1046/j.1472-765X.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- 53.Kärenlampi R, Rautelin H, Hakkinen M, Hänninen M-L. 2003. Temporal and geographical distribution and overlap of Penner heat-stable serotypes and pulsed-field gel electrophoresis genotypes of Campylobacter jejuni isolates collected from humans and chickens in Finland during a seasonal peak. J Clin Microbiol 41:4870–4872. doi: 10.1128/JCM.41.10.4870-4872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossi M, Silva MSD, Ribeiro-Gonçalves BF, Silva DN, Machado MP, Oleastro M, Borges V, Isidro J, Viera L, Barker DO, Llarena A, Halkilahti J, Jaakkonen A, Kivistö R, Kovanen S, Nieminen T, Hänninen M-L, Salmenlinna S, Hakkinen M, Garaizar J, Bikandi J, Hilbert F, Taboada EN, Carriço JA. 2018. INNUENDO whole genome and core genome MLST schemas and datasets for Campylobacter jejuni. Zenodo doi: 10.5281/zenodo.1322564. [DOI] [Google Scholar]
- 55.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 56.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tange O. 2011. GNU Parallel: the command-line power tool. ;login: 36:42–47. [Google Scholar]
- 59.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Francisco AP, Bugalho M, Ramirez M, Carrico JA. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 63.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata RD, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.