Abstract
BRAC1 has multiple important interactions with triple-negative breast cancer, the specific molecular characteristics of this interaction, however, have not yet been completely elucidated. By examining cell signaling pathways, important information for comprehending the potential mechanisms of this cancer may become known. The aim of the present study was to identify the effects of BRAC1 and to find the signaling pathway(s) involved in the pathogenic mechanism of triple-negative breast cancer. In this study, GSE27447 microarray data were obtained from the Gene Expression Omnibus (GEO) database of the National Center for Biotechnology Information, and differentially expressed genes (DEGs) from GSE27447 were distinguished by Significant Analysis of Microarray. Gene ontology (GO) analysis was carried out on 132 upregulated and 198 downregulated genes with DAVID. The signaling was forecast by the Kyoto Encyclopedia of Genes and Genomes (KEGG). Transcription factors were recognized by TFatS. The BRAC1 relevant protein-protein interaction networks (PPI) were fixed by STRING and visualized by CytoScape. Overall, the upregulated DEGs, which included CR2, IGHM, PRKCB, CARD11, PLCG2, CD79A, IGKC and CD27, were primarily enriched in the terms associated with immune responses, and the downregulated DEGs, which included STARD3, ALDH8A1, SRD5A3, CACNA1H, UGT2B4, SDR16C5 and MED1, were primarily enriched in the hormone metabolic process. In addition, 13 pathways, such as the B-cell receptor-signaling pathway, the hormone synthesis signaling pathway and the oxytocin-signaling pathway, were chosen. MYC, SP1 and CTNNB1 were determined to be enriched in triple-negative breast cancer. A total of 8 genes were identified to be downregulated in the BRAC1-related PPI network. The results of the present study show a fresh angle on the molecular mechanism of triple-negative breast cancer and indicate a possible target for its treatment.
Keywords: triple-negative breast cancer, differentially expressed genes, gene ontology analysis, signaling pathway, protein-protein interaction network
Introduction
Breast cancer is a malignant tumor with high incidence and heterogeneity in females worldwide (1). In 2000, Perou et al (2) divided breast cancer into luminal-like, basal-like, human epidermal growth factor receptor 2, and normal breast-like, according to the clinical detection of molecular markers. In addition, breast cancer can be classified into triple-negative breast cancer (TNBC), based on immunohistochemical techniques and hormone receptor status, which is negative for the estrogen receptor, progesterone receptor and human epidermal growth factor receptor 2 (HER2), and non-negative breast cancer (non-TNBC) (3). TNBC is a specific type of breast cancer that accounts for 15–20% of total breast cancer cases, having unique biological characteristics and clinical pathological features, such as not expressing Estrogen receptor (ER), progesterone receptor (PR) and human epiderma lgrowth factor receptor 2 (her-2) (4). TNBC is characterized by high recurrence, early metastasis and a poor prognosis (5,6). Considering that there is no corresponding hormone receptor or HER2 expression, traditional therapies, such as endocrine therapy, anti-HER2 targeted therapy and chemotherapy, are not effective for the treatment of TNBC (7). Therefore, there is a need to examine the etiopathogenesis and molecular mechanism of TNBC.
Over the past 20 years, our research has focused exclusively on surgery and chemotherapy to understand the molecular subtypes that cause major changes in clinical practice (8). Genome-wide analysis has been extensively used to identify important cancer genes in humans. For example, gene expression profiles have been constructed in order to explore a variety of changes in different types of cancer, including colorectal, gastric and breast cancer (9–11). Furthermore, according to reports, genetically modified core pathways and regulatory systems become obvious in cases in which the coding area of the genome is analyzed, as the cause of tumorigenesis can be explained by the dysregulation of core pathways and processes (12,13). Therefore, different signaling pathways serve important roles in the pathogenesis of cancer. It was observed that EGFR was closely associated with the proliferation, invasion and vascular formation of cancer, and it was markedly overexpressed in basal-like TNBC, which was significantly negatively correlated with clinical prognosis (14,15). MAPK signaling pathways regulate tumor proliferation and survival studies have demonstrated that MAPK was abnormally activated in the occurrence and development of TNBC, which may provide TNBC cells with the ability to proliferate and resist apoptosis (16–18). The PI3K/AKT/mTOR signaling pathway controls the proliferation, metabolism, survival and movement of cancer and has frequent communication with the MAPK signaling pathway (19–22). Other signaling pathways, such as DNA repair, are also reportedly dysregulated in TNBC (23).
Breast cancer susceptibility gene 1 (BRCA1) is a well-established tumor suppressor gene, which is linked to hereditary breast cancer (24). A person with a BRCA1 mutation may have a 60–80% chance of developing breast cancer, which is called BRCA1-related breast cancer (25). According to immunohistochemical results, the majority of BRCA1-associated breast cancer cases are TNBC (26). It has been suggested that these two types of tumors may have the same or similar carcinogenic pathways (27), thus providing an important basis to identify potential therapeutic targets. Transcription factors (TFs) regulate DNA expression by binding to target gene promoter regions (28). Previous studies have recognized that there are numerous cancer-associated TFs, such as transmembrane serine protease 2 (TMPRSS2) and ETS in prostate cancer, and KLF4 and KLF5 in esophageal cancer; these TFs influence the proliferation, apoptosis and invasion of cancer cells (29,30). The transcriptome section of human TNBC has described previously (31). However, the precise molecular mechanisms have not been completely elucidated. It is possible to examine the BRCA1-associated network, which may represent an essential mechanism in facilitating TNBC metabolism, using existing data. In the present study, differentially expressed genes (DEGs) between TNBC and non-TNBC were ascertained, and functional specifications including, gene ontology and signalling pathways analysis, forecast of transcription factors and protein-protein interaction (PPI) networks were subsequently conducted. The TFs were later determined, and a BRCA1-connected protein-protein interaction (PPI) network was constructed. Using this bioinformatics knowledge, the influences of both BRCA1 and associated signaling pathways were analyzed with respect to the etiopathogenesis of TNBC.
Materials and methods
Affymetrix microarray data
The Affymetrix microarray data were obtained from the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) using the series accession number, GSE27447 (32). In total, 14 pre-treated non-triple-negative breast tumors and 5 triple-negative breast tumors were collected based on the GPL6244 (HuGene-1_0-st) Affymetrix Human Gene 1.0 ST Array.
Gene ontology and signaling pathways impact the analysis of DEGs
The DEGs were ascertained by means of Significant Analysis of Microarray (SAM) (33). GO analysis determined that there were 132 upregulated and 198 downregulated DEGs (https://www.ncbi.nlm.nih.gov/geo/), applying the Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://david.ncifcrf.gov/) (34). Using the KEGG pathway map tool, the signaling pathways that the DEGs would likely impact were predicted.
Forecast of transcription factors
The transcription factor target genes associated with DEGs were evaluated by using a bioinformatics tool named TFatS (www.tfacts.org) (35). The 132 upregulated and 198 downregulated genes were imported to TfactS to determine the TFs.
Protein-protein interaction (PPI) network for BRCA1
BRCA1 and DEGs were mapped to the CHARACTER STRING database to discover the possible interacting proteins. CHARACTER STRING (https://string-db.org/) is a repository of forecasted protein interrelationships (36). The PPI network was subsequently visualized by Cytoscape v3. 5.1 software.
Patient samples
Patients with breast cancer were chosen from the pathological diagnosis of Changzheng hospital, including three TNBC patient samples and three non-TNBC patient samples from ACKERMAN PATHOLOGY & DIAGNOSTICS center, and total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. The present study was approved by the Ethics Committee of Changzheng Hospital, Second Military Medical University (Shanghai, China). Patients provided written informed consent.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
According to the instructions included in the PrimeScript™ RT reagent Kit (Perfect Real Time) (TaKaRa Bio, Inc., Otsu, Japan) kit, the RT reaction was performed for the mRNA (37°C for 15 min or 85°C for 5 sec) and miRNA (42°C for 60 min or 70°C for 10 min). Then, they were subjected to an RT reaction using the ABI 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific, Inc.). The miRNA primers were designed by Guangzhou RiboBio Co., Ltd (Guangzhou, China). The fold-changes for miRNA and mRNA expression were calculated by the 2−ΔΔCq method. The amplification primers of CR2, PRKCB, CARD11, PLCG2, CD79A, CD27, STARD3, SRD5A3, CACNA1H, UGT2B4, SDR16C5 and MED1 were then designed (Table I).
Table I.
Primers used in RT-qPCR.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| CR2 | GGCTACCTTATGGCTGGAGAG | AGAGTCACAGTAGTCCCAAACC |
| PRKCB | GCCTACCCCAAGGTCCATGT | CTTGGTCATGAGCCCTTTG |
| CARD11 | CAGGGTGCCTGCCTCATAG | TATAGGGAGAAGCAAGGCAGGG |
| PLCG2 | CTGGCAACCGACTCAAAGGA | GCTGATGCTGTTTCTTCGGG |
| CD79A | CTACGGCTTCTCCAGCTGAAT | CAGCTGAATGTCTTCCTCACA |
| CD27 | ATGGAAAGGGAAGCACGTC | TTGGCCAACTCCTCTCCTAA |
| STARD3 | GACCTGGTTCCTTGACTTCAA | CGGCAAGACGTTTATCCTGAA |
| SRD5A3 | TTTAATCAGGCCCTGTCTGC | GGGGTATAGAAATGGAATGGAGA |
| CACNA1H | GGATCCCAAGCTTGGTACCG | CTTCATGGCCCTCTAGAGGATCC |
| UGT2B4 | TGGACTCATCACCTGACTCATGTAA | GTCAAAGAGACTGCAGGAACATGA |
| SDR16C5 | TGGAAACTCTTAAAGTTTTGCTCCTTACAATC | GAAAGATATTCATGGTGAATTCGAATC |
| MED1 | GAGACTCCGCCCACTTACCTG | GGACACACTTCAAACTGGAGG |
| GAPDH | ACGGCAAGTTCAACGGCACAG | CCACGACATACTCAGCACCAGC |
Western blot analysis
Patient sample was homogenized in a lysis buffer (P0013; Beyotime Institute of Biotechnology, Haimen, China) supplemented with 1% protease inhibitor cocktail (Pierce; Thermo Fisher Scientific, Inc.). Antibodies against the following proteins were used for Western blot analysis: MYC (1:1,000 dilution; no. #2272; Cell Signalling Technology, Inc., Danvers, MA, USA), SP1 (1:1,000 dilution; no. #5931; Cell Signalling Technology, Inc.,), β-actin (1:1,000 dilution; no. #4967; Cell Signalling Technology, Inc.,) and CTNNB1 (1:1,000 dilution; Phospho-β-Catenin (Ser33/37) Antibody no. #2009; Cell Signalling Technology, Inc.). Following centrifugation at 12,000 × g for 30 min at 4°C, the supernatant was collected, and the protein concentration was determined using an enhanced bicinchoninic acid protein assay kit according to the manufacturer's protocol (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The membrane proteins were transferred to a polyvinylidene difluoride (PVDF) membrane following electrophoresis with 10% SDS-PAGE. The membranes were blocked with 5% non-fat dry milk in Tris-buffered saline prior to overnight incubation at 4°C, incubated with the primary antibodies and were then incubated with the secondary antibodies (Abmart, Shanghai China) for 60 min at 37°C. Following washing with TBST 3 times for 10 min each, the membranes were developed with an enhanced chemiluminescent ECL assay kit (Santa Cruz Biotechnology, Inc., Dallas, TX, USA).
Statistical analysis
Data are presented as the mean ± standard error of the mean. The primary data were formatted into expression measurements and normalized by the robust multi-array average (RMA) algorithm (23). The criteria for screening DEGs were |logFC|>1.5 and the false discovery rate, (FDR) <0.05. An unpaired student's t-test was used to analyze data between two groups. P<0.05 was considered to indicate a statistically significant difference. All analyses were performed using SPSS v.22.0 (IBM Corp., Armonk, NY, USA).
Results
Discernment and GO analysis of DEGs
On the basis of the SAM analysis, a total of 132 upregulated and 198 downregulated DEGs were identified. The GO analysis was subsequently conducted (Table II). The results demonstrated that the upregulated DEGs, which included CR2, IGHM, PRKCB, CARD11, PLCG2, CD79A, IGKC and CD27, were relative to the immune response, such as lymphocyte activation (P=1.49×10−11), leukocyte activation (P=4.68×10−11) and B-cell activation (P=6.02× 10−8) (Table IIA). The downregulated DEGs, which included STARD3, ALDH8A1, SRD5A3, CACNA1H, UGT2B4, SDR16C5 and MED1, were primarily enriched in the hormone metabolic process (P=5.45×10−5) (Table IIB).
Table II.
GO terms of DEGs.
| A, The top 10 GO terms of the 132 upregulated DEGs | ||||
|---|---|---|---|---|
| Category | Term | Count | P-value | Genes |
| GO:0046649 | Lymphocyte activation | 24 | 1.49×10−11 | GAPT, TCF7, CR2, IGHM |
| GO:0045321 | Leukocyte activation | 25 | 4.68×10−11 | GAPT, TCF7, CR2, IGHM |
| GO:0002684 | Positive regulation of immune system process | 27 | 2.66×10−10 | BLK, CR2, IGHM, PRKCB |
| GO:0050778 | Positive regulation of immune response | 23 | 3.37×10−10 | CR2, IGHM, PRKCB, CARD11 |
| GO:0006955 | Immune response | 33 | 1.46×10−9 | IL16, BLK, PAX5, GPRC5B |
| GO:0002376 | Immune system process | 42 | 2.11×10−9 | IL16, BLK, PAX5, GPRC5B |
| GO:0001775 | Cell activation | 25 | 2.52×10−9 | GAPT, TCF7, CR2, IGHM |
| GO:0002682 | Regulation of immune system process | 30 | 6.97×10−9 | BLK, CR2, IGHM, PRKCB |
| GO:0050776 | Regulation of immune response | 24 | 1.34×10−8 | CR2, IGHM, PRKCB, CARD11 |
| GO:0042113 | B cell activation | 13 | 6.02×10−8 | GAPT, CR2, IGHM, PRKCB |
| B, Top 10 GO terms of the 198 downregulated DEGs | ||||
| Category | Term | Count | P-value | Genes |
| GO:0042445 | Hormone metabolic process | 10 | 5.45×10−5 | STARD3, TG, ALDH8A1, FOXA1 |
| GO:0044699 | Single-organism process | 143 | 4.38×10−4 | PDP1, ALDH8A1, ADCY1, TSPAN1 |
| GO:0034754 | Cellular hormone metabolic process | 7 | 5.22×10−4 | STARD3, ALDH8A1, SRD5A3, CACNA1H |
| GO:0010817 | Regulation of hormone levels | 14 | 5.62×10−4 | ALDH8A1, TG, FOXA1, IYD |
| GO:0009952 | Anterior/posterior pattern specification | 9 | 6.17×10−4 | HOXC10, MSX2, PCGF2, HOXC11 |
KEGG pathways analysis
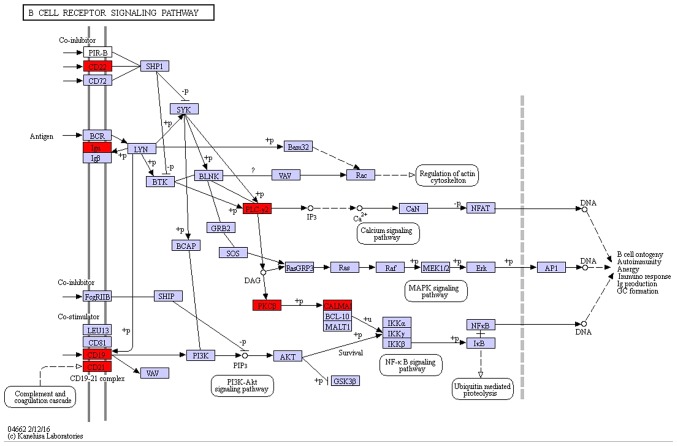
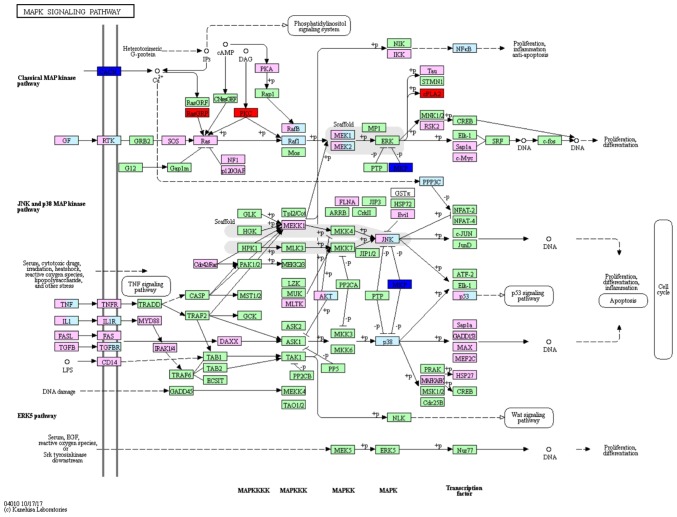
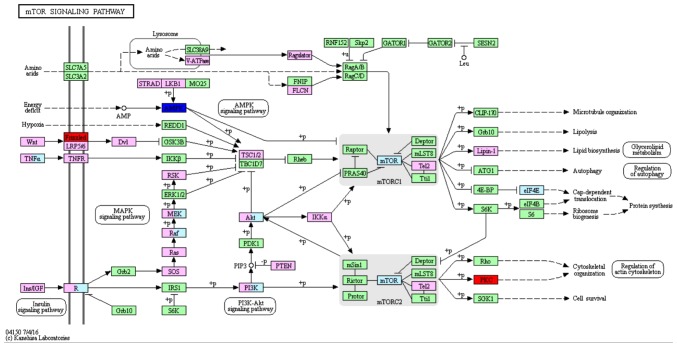
On the basis of the SPIA analysis, a total of 13 KEGG signaling pathways were examined to determine if they were dysregulated in TNBC (Table III). The B cell receptor signaling pathway (P=3.99×10−3) (Fig. 1), hormone synthesis signaling pathway (P=2.21×10−2) and oxytocin signaling pathway (P=3.41×10−2) were each identified to exhibit significant differences. MAPK/RAS and mTOR signaling pathways that correlated with cancer were selected for subsequent investigation. Certain protein kinase genes, such as GRP, PKC and CPLA2, were upregulated, and MKP and CACN were downregulated, in the MAPK signaling pathway (Fig. 2). In the mTOR signaling pathway, PKC was upregulated and AMPK was downregulated (Fig. 3).
Table III.
Fourteen pathways identified based on KEGG.
| Pathway | Count | P-value | Genes |
|---|---|---|---|
| B cell receptor signaling pathway | 6 | 0.003988 | CARD11, CD19, CR2, PLCG2, CD22, CD79A |
| Hormone synthesis | 5 | 0.022133 | TG, ADCY1, CREB3L4, PRKCB, IYD |
| Oxytocin signaling pathway | 7 | 0.034094 | ADCY1, PLA2G4A, RGS2, RYR3, CACNG4, PRKAA2, PRKCB |
| Hematopoietic cell lineage | 5 | 0.041129 | CD19, CR2, CD3E, MS4A1, CD22 |
| Platelet activation | 6 | 0.049048 | ADCY1, PLA2G4A, FGA, RASGRP1, PLCG2, RASGRP2 |
| Non-small cell lung cancer | 4 | 0.054172 | ERBB2, PLCG2, RARB, PRKCB |
| Calcium signaling pathway | 7 | 0.056584 | ADCY1, CHRM3, ERBB2, RYR3, PLCG2, CACNA1H, PRKCB |
| Wnt signaling pathway | 6 | 0.060444 | TCF7, DKK1, SFRP1, WIF1, FZD7, PRKCB |
| Inflammatory mediator regulation of TRP channels | 5 | 0.063253 | ADCY1, PLA2G4A, P2RY2, PLCG2, PRKCB |
| Melanogenesis | 5 | 0.067119 | ADCY1, TCF7, CREB3L4, FZD7, PRKCB |
| Circadian rhythm | 3 | 0.081308 | RORC, PRKAA2, BHLHE41 |
| Primary immunodeficiency | 3 | 0.095274 | CD19, CD3E, CD79A |
| MAPK signaling pathway | 8 | 0.096136 | DUSP4, PLA2G4A, RASGRP1, RASGRP2, DUSP10, CACNG4, CACNA1H, PRKCB |
Figure 1.
B-cell receptor pathway, which may be dysregulated in TNBC. Red boxes indicate upregulated genes, purple boxes indicate no differentially expressed genes. TNBC, triple negative breast cancer.
Figure 2.
MAPK signaling pathway, which may be dysregulated in TNBC. Red boxes denote upregulated genes, and blue boxes denote downregulated genes, green and pink boxes represent indicate no differentially expressed genes. TNBC, triple negative breast cancer.
Figure 3.
mTOR signaling pathway, which may be dysregulated in TNBC. Red boxes indicate upregulated genes and blue boxes indicate downregulated genes and green and pink boxes represent indicate no differentially expressed genes.
Transcription factor-regulated DEGs
TFactS analysis was performed to ascertain transformation in the degree of transcription factor activity in upregulated and downregulated genes in TNBC (Table IV). The results indicated that MYC, SP1 and CTNNB1 were stimulated in TNBC. In total, 14 target genes of CTNNB1 were identified, including 5 upregulated and 9 downregulated genes, while for SP1, 3 target genes were upregulated and 6 genes were downregulated. Additionally, 4 target genes of MYC were upregulated and 2 genes were downregulated.
Table IV.
Results of the TfactS analysis.
| Gene name | TF | Regulation |
|---|---|---|
| WIF1 | CTNNB1 | Up |
| FZD7 | CTNNB1 | Up |
| QPCT | CTNNB1 | Up |
| TCF7 | CTNNB1 | Up |
| SOX10 | CTNNB1 | Up |
| INHBB | CTNNB1 | Down |
| SEMA3C | CTNNB1 | Down |
| DKK1 | CTNNB1 | Down |
| MSX2 | CTNNB1 | Down |
| AR | CTNNB1 | Down |
| CEACAM6 | CTNNB1 | Down |
| MUC6 | CTNNB1 | Down |
| SCGB2A2 | CTNNB1 | Down |
| CEACAM5 | CTNNB1 | Down |
| CD19 | SP1 | Up |
| UGT8 | SP1 | Up |
| PLA2G4A | SP1 | Up |
| PAPSS2 | SP1 | Down |
| KRT19 | SP1 | Down |
| AR | SP1 | Down |
| CEACAM5 | SP1 | Down |
| FOXA1 | SP1 | Down |
| SLC9A2 | SP1 | Down |
| RARB | MYC | Up |
| SPIB | MYC | Up |
| PLA2G4A | MYC | Up |
| RGS2 | MYC | Up |
| RPL23 | MYC | Down |
| CEACAM5 | MYC | Down |
Verifying the expression of DEGs and transcription factors
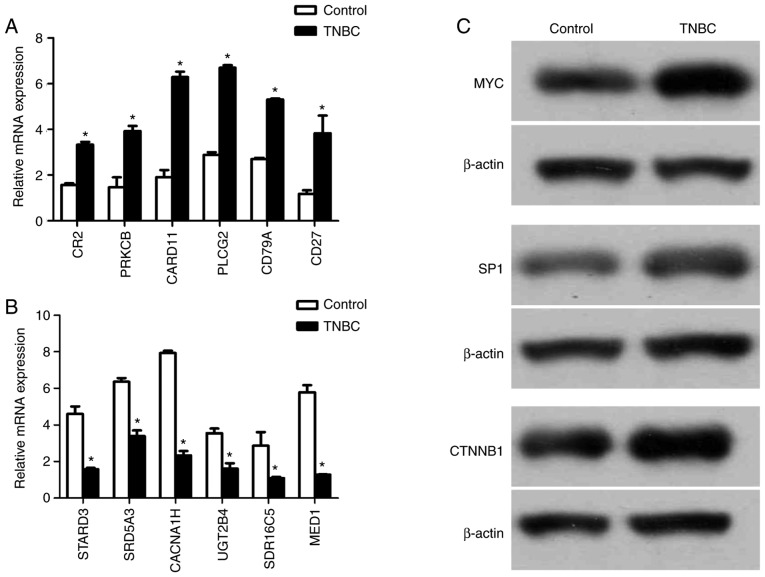
The RT-qPCR results demonstrated that the expression of CR2, PRKCB, CARD11, PLCG2, CD79A and CD27 was increased, and the expression of STARD3, SRD5A3, CACNA1H, UGT2B4, SDR16C5 and MED1 was decreased in TNBC-patient samples (Fig. 4A and B). The western blotting results revealed that the expression of MYC, SP1 and CTNNB1 was increased in TNBC-patient samples (Fig. 4C).
Figure 4.
The expression of DEGs and transcription factors. (A) RT-qPCR confirmed the result of upregulated genes' mRNA expression (n=3). *P<0.05. (B) RT-qPCR confirmed the result of downregulated genes' mRNA expression (n=3). *P<0.05. (C) Western blot confirmed the result of TFs. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; DEG's differentially expressed genes; TF, transcription factor.
BRCA1 associated with PPI network
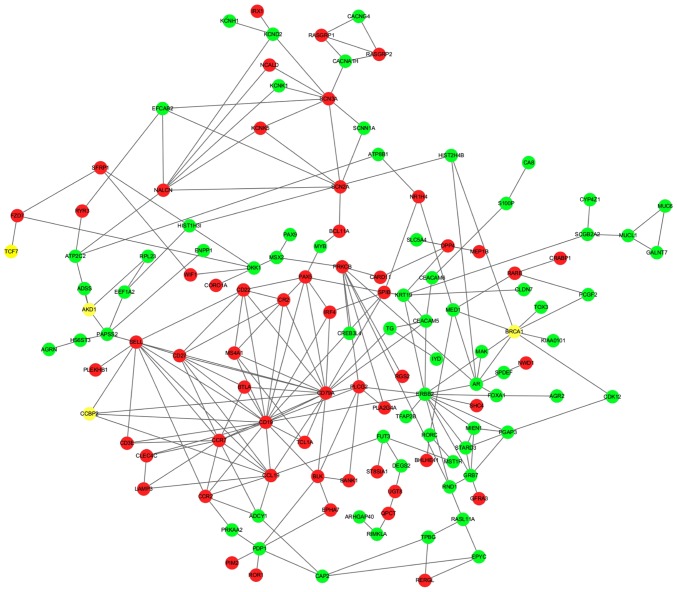
Metastasis is the main cause of cancer-related death (27). Mutations in BRCA1, one of nucleoprotein species, have been discovered to improve tumorigenesis (23,24). To examine the role of BRCA1 in TNBC, a BRCA1-related PPI network was constructed (Fig. 5). The results demonstrated that 8 proteins, AR, CDK12, KIAA0101, PCGF2, TOX3, HIST2H4B, MED1 and ERBB2, which were encoded by downregulated genes, interacted with BRCA1, while no proteins encoded by upregulated genes interacted with BRCA1. Additionally, an important interaction with BRCA1 in the expression levels of the androgen receptor (AR) was observed.
Figure 5.
BRCA1 related protein-protein interaction network. The interaction network was predicated by STRING and visualized by Cytoscape software. Red indicates upregulated genes and green indicates downregulated genes and yellow indicates no differentially expressed genes.
Discussion
Breast cancer is the most common type of cancer in females worldwide, and is characterized by a high mortality rate (37,38). TNBC is the most dangerous type of breast cancer and is characterised by high recurrence, early metastasis and a poor prognosis (5,6). Although it has been the focus of numerous studies, the potential mechanisms of TNBC have not been completely elucidated. In the present study, the DEGs of TNBC and non-TNBC samples were identified, and the functions were predicted through GO analysis and signaling pathways. A PPI network associated with BRCA1 was also identified. Taken together the BRAC1-associated interactions offer novel insights into the pathogenesis of TNBC.
GO analysis revealed that the upregulated genes were primarily concentrated in the immune response, including lymphocyte activation, cell cycle progression, leukocyte activation and B cell activation. Complement receptor type 2 (CR2; CD21), which binds fragments of C3, may ligate CD19 or CD23 to present antigens to B cells (39). This outcome can facilitate the interaction of B cells with other cells essential for cellular activation (39). CR2 also serves an important role in enhancing humoral immunity to T-dependent and T-independent foreign antigens, and in regulating T-cell immunity to self and non-self-antigens. CR2 is an important receptor that amplifies B lymphocyte activation by bridging the innate and adaptive immune systems (40). Variations or deletions of the CR2 gene in humans are associated with a variety of autoimmune and inflammatory conditions, including but not limited to rheumatoid arthritis, systemic lupus erythmatosus, psoriatic arthritis, reactive arthritis, Crohn's disease, ulcerative colitis and scleroderma (40). In aggressive adult T-cell leukemia/lymphoma, a multivariate analysis incorporating clinical factors and genetic alterations demonstrated that being at high-risk, including patient age, the presence of PRKCB mutations and PD-L1 amplifications, were all independent poor prognosticators listed in the Japan Clinical Oncology Group prognostic index (41). Through pathway analysis, these upregulated genes closely associated with immune related pathways, such as the B cell receptor-signalling pathway. Conversely, a possible target for further clinical verification is conducive to TNBC immunotherapy.
The results of the present study revealed that downregulated genes, including ALDH8A1 and SRD5A3, in TNBC were significantly concentrated in the hormone metabolic biological process. Retinoic acid (RA) is required for cellular differentiation and is known to arrest tumor development (42). RA is synthesized from retinaldehyde by retinaldehyde dehydrogenases, specifically ALDH1A1, ALDH1A2, ALDH1A3 and ALDH8A1 (42). ALDH8A1 is considered to be a candidate marker to identify and isolate normal and cancer stem cells (42). In addition, SRD5A3 is associated with the production of 5 α-dihydrotestosterone and the activation of the androgen-androgen receptor-pathway is a promising molecular target for prostate cancer therapy (43).
Signaling pathways serve an important role in the investigation of cancer pathogenesis (44). According to the latest research, CD19, CD22 and CD79A were concentrated in the B cell receptor-signaling pathway (44–46). These genes encode a cell surface molecule that assembles with the antigen receptor of B-lymphocytes in order to decrease the threshold for antigen receptor-dependent stimulation (44–46). In there present study, these DEGs were closly associated with potential therapeutic targets for TNBC. Previous study (47) has demonstrated that the most prominent signaling pathway that is dysregulated in TNBC is the PI3K/AKT/mTOR signaling pathway. PKC (protein kinase C) is an intracellular effector of the C kinase pathway. PKC is associated with the mTOR signaling pathway, which is involved in the regulation of metabolism and gene expression, including serine/threonine residue phosphorylation, cell proliferation and nuclear type changes (48). It has been previously demonstrated that the attenuation of the mTOR signaling pathway presents an obvious curative possible for the diagnosis of cancer. The present study provides detailed insights into the novel cytotoxic mechanism of an anti-tumor compound originated from the herbal plant, which may be useful in promoting autophagy mediated- cell death in cancer cell that is resistant to apoptosis. Mechanistically, tetrandrine induces autophagy that depends on mTOR inactivation (49). The results of the present study indicated that the B-cell receptor signaling pathway by enriched genes was differentially significant, when compared with the PI3K/AKT/mTOR signaling pathway in TNBC.
Transcription factors are targets for certain anticancer drugs; however, a limited list of transcription factors are overactive in most human cancer cells, which makes them targets for the development of anticancer drugs. That they are the most direct and hopeful targets for treating cancer is proposed, and this is supported by the fact that there are many more human oncogenes in signalling pathways than there are oncogenic transcription factors (50). NF-κB, is associated with control of cell cycle, apoptosis, migration and cell differentiation, and is one of the most appropriate transcription factors as a target for the development of cancer immunotherapeutics (51,52). HIF-1α and STAT have also been reported to be involved in the development of cancer therapeutics (53,54). Specific transcription factors including, MYC, SP1 and CTNNB1, have been demonstrated to function as regulators of certain DEGs (55). In the Wnt/APC/β-catenin signaling pathway in colorectal cancer, the activation and overexpression of MYC, marked as a downstream step, is crucial for tumour metastasis (55,56). Sp1, a stress-responsive factor, inhibits the sensitivity of cancer cells to chemotherapy by regulating stemness gene expression in glioblastoma (57). CTNNB1 is the key regulatory factor of the Wnt/β-catenin signaling pathway, and the overexpression pattern promotes the growth of hepatocellular carcinoma cells by targeting CD44 and c-Myc protein (58). These findings may help to develop drugs for tumor therapy.
The present study has helped to elucidate the molecular mechanisms of tumor development and metastasis. The BRCA1 mutation is associated with neoplastic transformation (59), and the results of the present study suggest that BRCA1 may interact with DEGs to participate in the occurrence of TNBC. Among the mass of genes associated with BRCA1, the interaction with the androgen receptor (AR) is significant. Several lines of evidence suggest that androgen may have direct effects on breast cancer cells, by binding to their selective androgen receptors (ARs) (60). Therefore, in the case of oestrogen receptor negative, evidence has revealed that TNBC is associated with the hormone pathway. The network of BRCA1 was predicated in order to help elucidate the mechanism of tumor cell metastasis, and provide therapeutic targets for the development of cancer therapeutics. However, there limitations to the present research, as all differentially expressed genes and transcription factors did not undergo RT-qPCR and western blotting for further verification.
The present study demonstrated that the B cell receptor-signaling pathway and hormone synthesis-signaling pathway are of vital importance in the development of TNBC. In addition, numerous genes that may function as potential targets for TNBC have been identified. Nevertheless, to reveal TNBC's potential molecular mechanisms, further research should be performed to investigate other signaling pathways and key cancer-associated proteins.
Acknowledgements
The authors would like to thank Ackerman Pathology and Diagnostics Center (Shanghai China) for providing specimen tissues.
Funding
The present study was funded by Chinese National Science Foundation (grant no. 81502260, 2016).
Availability of data and materials
The Affymetrix microarray data were obtained from the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) using the series accession number, GSE27447. Patient samples and three non-TNBC patient samples from ACKERMAN PATHOLOGY & DIAGNOSTICS Centre.
Authors' contributions
FQ and WXQ performed the majority of experiments; FQ, WXQ and YSZ provided vital reagents and analytical tools and were also involved in editing the manuscript; FQ and WXQ designed the study and wrote the manuscript. YSZ analyzed and interpreted the data, and agreed to be accountable for all aspects of the work in ensuring that question related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Changzheng Hospital, Second Military Medical University (Shanghai, China). Patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chu J, Bae H, Seo Y, Cho SY, Kim SH, Cho EY. The prognostic impact of synchronous ipsilateral multiple breast cancer: Survival Outcomes according to the Eighth American Joint Committee on cancer staging and molecular subtype. J Pathol Transl Med. 2018;52:396–403. doi: 10.4132/jptm.2018.10.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Zubeda S, Kaipa PR, Shaik NA, Mohiuddin MK, Vaidya S, Pavani B, Srinivasulu M, Latha MM, Hasan Q. Her-2/neu status: A neglected marker of prognostication and management of patients with breast cancer in India. Asian Pac J Cancer Prev. 2013;14:2231–2235. doi: 10.7314/APJCP.2013.14.4.2231. [DOI] [PubMed] [Google Scholar]
- 4.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: A population-based study from the California cancer Registry. Cancer. 2010;109:1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 5.Haque R, Ahmed SA, Inzhakova G, Shi J, Avila C, Polikoff J, Bernstein L, Enger SM, Press MF. Impact of breast cancer subtypes and treatment on survival: An analysis spanning two decades. Cancer Epidemiol Biomarkers Prev. 2012;21:1848–1855. doi: 10.1158/1055-9965.EPI-12-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 7.Bosch A, Eroles P, Zaragoza R, Viña JR, Lluch A. Triple-negative breast cancer: Molecular features, pathogenesis, treatment and current lines of research. Cancer Treat Rev. 2010;36:206–215. doi: 10.1016/j.ctrv.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanza G, Ferracin M, Gafà R, Veronese A, Spizzo R, Pichiorri F, Liu CG, Calin GA, Croce CM, Negrini M. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer. 2007;6:54. doi: 10.1186/1476-4598-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol. 2003;129:199–221. doi: 10.1007/s00432-003-0431-0. [DOI] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Network: Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Sommer RJ. Antagonism of LIN-17/Frizzled and LIN-18/Ryk in nematode vulva induction reveals evolutionary alterations in core developmental pathways. PLoS Biol. 2011;9:e1001110. doi: 10.1371/journal.pbio.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams CB, Soloff AC, Ethier S, Yeh ES. Perspectives on epidermal growth factor receptor regulation in triple-negative breast cancer: Ligand-mediated mechanisms of receptor regulation and potential for clinical targetin. Adv Cancer Res. 2015;127:253–281. doi: 10.1016/bs.acr.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Tao JJ, Castel P, Radosevic-Robin N, Elkabets M, Auricchio N, Aceto N, Weitsman G, Barber P, Vojnovic B, Ellis H, et al. Antagonism of EGFR and HER3 enhances the response to inhibitors of the PI3K-Akt pathway in triple-negative breast cancer. Sci Signal. 2014;7:ra29. doi: 10.1126/scisignal.2005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, Galloway R, Grandjean G, Jacob J, Humphries J, Bartholomeusz C, Goodstal S, Lim B, Bartholomeusz G, Ueno NT, Rao A. Comprehensive two- and three-dimensional RNAi screening identifies PI3K inhibition as a complement to MEK inhibitor AS703026 for combination treatment of triple-negative breast cancer. J Cancer. 2015;6:1306–1319. doi: 10.7150/jca.13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Lee J, Jeon M, Lee JE, Nam SJ. MEK-dependent IL-8 induction regulates the invasiveness of triple-negative breast cancer cells. Tumour Biol. 2016;37:4991–4999. doi: 10.1007/s13277-015-4345-7. [DOI] [PubMed] [Google Scholar]
- 19.Aksamitiene E, Kiyatkin A, Kholodenko BN. Cross-talk between mitogenicRas/MAPK and survival PI3K/Akt pathways: A fine balance. Biochem Soc Trans. 2012;40:139–146. doi: 10.1042/BST20110609. [DOI] [PubMed] [Google Scholar]
- 20.Fruman DA, Rommel C. PI3K and cancer: Lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13:140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganesan P, Moulder S, Lee JJ, Janku F, Valero V, Zinner RG, Naing A, Fu S, Tsimberidou AM, Hong D, et al. Triple-negative patients with breast cancer treated at MD Anderson cancer center in phase I trials: Improved outcomes with combination chemotherapy and targeted agents. Mol Cancer Ther. 2014;13:3175–3184. doi: 10.1158/1535-7163.MCT-14-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juvekar A, Burga LN, Hu H, Lunsford EP, Ibrahim YH, Balmañà J, Rajendran A, Papa A, Spencer K, Lyssiotis CA, et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2012;2:1048–1063. doi: 10.1158/2159-8290.CD-11-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murai J, Huang SN, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul A, Paul S. The breast cancer susceptibility genes (BRCA) in breast and ovarian cancers. Front Biosci (Landmark Ed) 2014;19:605–618. doi: 10.2741/4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tischkowitz MD, Foulkes WD. The basal phenotype of BRCA1-related breast cancer: Past, present and future. Cell Cycle. 2006;5:963–967. doi: 10.4161/cc.5.9.2713. [DOI] [PubMed] [Google Scholar]
- 26.He BS, Pan YQ, Lin K, Ying HQ, Wang F, Deng QW, Sun HL, Gao TY, Wang SK. Evaluation the susceptibility of five polymorphisms in microRNA-binding sites to female breast cancer risk in Chinese population. Gene. 2015;573:160–165. doi: 10.1016/j.gene.2015.07.052. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Fang C, Xu X, Li A, Cai Q, Long X. Androgen receptor, EGFR, and BRCA1 as biomarkers in triple-negative breast cancer: A meta-analysis. Biomed Res Int. 2015;2015:357485. doi: 10.1155/2015/357485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sankpal UT, Goodison S, Abdelrahim M, Basha R. Targeting Sp1 transcription factors in prostate cancer therapy. Med Chem. 2011;7:518–525. doi: 10.2174/157340611796799203. [DOI] [PubMed] [Google Scholar]
- 29.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 30.Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, Apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4:1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- 31.Yang L, Wu X, Wang Y, Zhang K, Wu J, Yuan YC, Deng X, Chen L, Kim CC, Lau S, et al. FZD7 has a critical role in cell proliferation in triple negative breast cancer. Oncogene. 2011;30:4437–4446. doi: 10.1038/onc.2011.145. [DOI] [PubMed] [Google Scholar]
- 32.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Essaghir A, Toffalini F, Knoops L, Kallin A, van Helden J, Demoulin JB. Transcription factor regulation can be accurately predicted from the presence of target gene signatures in microarray gene expression data. Nucleic Acids Res. 2010;38:e120. doi: 10.1093/nar/gkq149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 37.Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM, Goss PE. Breast cancer in China. Lancet Oncol. 2014;15:e279–e289. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 38.Fearon DT, Carter RH. The CD19/CR2/TAPA-1 complex of B lymphocytes: Linking natural to acquired immunity. Annu Rev Immunol. 1995;13:127–149. doi: 10.1146/annurev.iy.13.040195.001015. [DOI] [PubMed] [Google Scholar]
- 39.Donius LR, Weis JH. Quantification of complement receptor 2 calcium signaling enhancement using flow cytometry. Methods Mol Biol. 2014;1100:311–317. doi: 10.1007/978-1-62703-724-2_25. [DOI] [PubMed] [Google Scholar]
- 40.Kataoka K, Iwanaga M, Yasunaga JI, Nagata Y, Kitanaka A, Kameda T, Yoshimitsu M, Shiraishi Y, Sato-Otsubo A, Sanada M, et al. Prognostic relevance of integrated genetic profiling in adult T-cell leukemia/lymphoma. Blood. 2018;131:215–225. doi: 10.1182/blood-2017-01-761874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh S, Arcaroli J, Thompson DC, Messersmith W, Vasiliou V. Acetaldehyde and retinaldehyde-metabolizing enzymes in colon and pancreatic cancers. Adv Exp Med Biol. 2015;815:281–294. doi: 10.1007/978-3-319-09614-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uemura M, Tamura K, Chung S, Honma S, Okuyama A, Nakamura Y, Nakagawa H. Novel 5 alpha-steroid reductase (SRD5A3, type-3) is overexpressed in hormone-refractoryprostate cancer. Cancer Sci. 2008;99:81–86. doi: 10.1111/j.1349-7006.2007.00656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 44.Chung HJ, Chi HS, Cho YU, Lee EH, Jang S, Park CJ, Seo EJ. Prognostic effect of cytoplasmic CD79a expression in acute myeloid leukemia with t(8;21) Korean J Lab Med. 2007;27:388–393. doi: 10.3343/kjlm.2007.27.6.388. (In Korean) [DOI] [PubMed] [Google Scholar]
- 45.Berahovich R, Xu S, Zhou H, Harto H, Xu Q, Garcia A, Liu F, Golubovskaya VM, Wu L. FLAG-tagged CD19-specific CAR-T cells eliminate CD19-bearing solid tumor cells in vitro and in vivo. Front Biosci (Landmark Ed) 2017;22:1644–1654. doi: 10.2741/4563. [DOI] [PubMed] [Google Scholar]
- 46.Bednar KJ, Shanina E, Ballet R, Connors EP, Duan S, Juan J, Arlian BM, Kulis MD, Butcher EC, Fung-Leung WP, et al. Human CD22 inhibits murine B cell receptor activation in a human CD22 transgenic mouse model. J Immunol. 2017;199:3116–3128. doi: 10.4049/jimmunol.1700898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Gao M, Lin Z, Chen L, Jin Y, Zhu G, Wang Y, Jin T. DEK promoted EMT and angiogenesis through regulating PI3K/AKT/mTOR pathway in triple-negative breast cancer. Oncotarget. 2017;8:98708–98722. doi: 10.18632/oncotarget.21864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szewczyk B, Pochwat B, Rafało A, Palucha-Poniewiera A, Domin H, Nowak G. Activation of mTOR dependent signaling pathway is a necessary mechanism of antidepressant-like activity of zinc. Neuropharmacology. 2015;99:517–526. doi: 10.1016/j.neuropharm.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 49.Wong VKW, Zeng W, Chen J, Yao XJ, Leung ELH, Wang QQ, Chiu P, Ko BCB, Law BYK. Tetrandrine, an activator of autophagy, induces autophagic cell death via PKC-α inhibition and mTOR-dependent mechanisms. Front Pharmacol. 2017;8:351. doi: 10.3389/fphar.2017.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 51.O'neill LA, Kaltschmidt C. NF-κB: A crucial transcription factor for glial and neuronal cell function. Trends Neurosci. 1997;20:252–258. doi: 10.1016/S0166-2236(96)01035-1. [DOI] [PubMed] [Google Scholar]
- 52.Barnes PJ. Nuclear factor-kappa B. Int J Biochem Cell Biol. 1997;29:867–870. doi: 10.1016/S1357-2725(96)00159-8. [DOI] [PubMed] [Google Scholar]
- 53.Zhu X, Mao Z, Na Y, Guo Y, Wang X, Xin D. Significance of pituitary tumor transforming gene 1 (PTTG1) in prostate cancer. Anticancer Res. 2006;26:1253–1259. [PubMed] [Google Scholar]
- 54.Gorynia S, Bandeiras TM, Pinho FG, McVey CE, Vonrhein C, Round A, Svergun DI, Donner P, Matias PM, Carrondo MA. Structural and functional insights into a dodecameric molecular machine-the RuvBL1/RuvBL2 complex. J Struct Biol. 2011;176:279–291. doi: 10.1016/j.jsb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 55.Bièche I, Laurendeau I, Tozlu S, Olivi M, Vidaud D, Lidereau R, Vidaud M. Quantitation of MYC gene expression in sporadic breast tumors with a real-time reverse transcription-PCR assay. Cancer Res. 1999;59:2759–2765. [PubMed] [Google Scholar]
- 56.Le Floch N, Rivat C, De Wever O, Bruyneel E, Mareel M, Dale T, Gespach C. The proinvasive activity of Wnt-2 is mediated through a noncanonical Wnt pathway coupled to GSK-3beta and c-Jun/AP-1 signaling. FASEB J. 2005;19:144–146. doi: 10.1096/fj.04-2373fje. [DOI] [PubMed] [Google Scholar]
- 57.Chang KY, Huang CT, Hsu TI, Hsu CC, Liu JJ, Chuang CK, Hung JJ, Chang WC, Tsai KK, Chuang JY. Stress stimuli induce cancer-stemness gene expression via Sp1 activation leading to therapeutic resistance in glioblastoma. Biochem Biophys Res Commun. 2017;493:14–19. doi: 10.1016/j.bbrc.2017.09.095. [DOI] [PubMed] [Google Scholar]
- 58.Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol. 2014;11:340–349. doi: 10.1038/nrgastro.2014.6. [DOI] [PubMed] [Google Scholar]
- 59.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 60.Azariadis K, Kiagiadaki F, Pelekanou V, Bempi V, Alexakis K, Kampa M, Tsapis A, Castanas E, Notas G. Androgen triggers the pro-migratory CXCL12/CXCR4 axis in AR-positive breast cancer cell lines: Underlying mechanism and possible implications for the use of aromatase inhibitors in breast cancer. Cell Physiol Biochem. 2017;44:66–84. doi: 10.1159/000484584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Affymetrix microarray data were obtained from the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) using the series accession number, GSE27447. Patient samples and three non-TNBC patient samples from ACKERMAN PATHOLOGY & DIAGNOSTICS Centre.