Abstract
Tugarinovia (Family Asteraceae) is a monotypic genus. It’s sole species, Tugarinovia mongolica Iljin, is distributed in the northern part of Inner Mongolia, with one additional variety, Tugarinovia mongolica var ovatifolia, which is distributed in the southern part of Inner Mongolia. The species has a limited geographical range and declining populations. To understand the phylogeographic structure of T. mongolica, we sequenced two chloroplast DNA regions (psbA-trnH and psbK-psbI) from 219 individuals of 16 populations, and investigated the genetic variation and phylogeographic patterns of T. mongolica. The results identified a total of 17 (H1-H17) chloroplast haplotypes. There were no haplotypes shared between the northern (T. mongolica) and southern groups (T. mongolica var. ovatifolia), and they formed two distinct lineages. The regional split was also supported by AMOVA and BEAST analyses. AMOVA showed the main variation that occurred between the two geographic groups. The time of divergence of the two groups can be dated to the early Pleistocene epoch, when climate fluctuations most likely resulted in the allopatric divergence of T. mongolica. The formation of the desert blocked genetic flow and enhanced the divergence of the northern and southern groups. Our results indicate that the genetic differences between T. mongolica and T. mongolica var. ovatifolia are consistent with previously proposed morphological differences. We speculate that the dry, cold climate and the expansion of the desert during the Quaternary resulted in the currently observed distribution of extant populations of T. mongolica. In the northern group, the populations Chuanjinsumu, Wuliji and Yingen displayed the highest genetic diversity and should be given priority protection. The southern group showed a higher genetic drift (FST = 1, GST = 1), and the inbreeding load (HS = 0) required protection for each population. Our results propose that the protection of T. mongolica should be implemented through in situ and ex situ conservation practices to increase the effective population size and genetic diversity.
Introduction
In recent years, phylogeographic studies of the arid region of Northwest China have increased and mainly focus on the impact of the Quaternary climate fluctuations on species’ phylogeographic patterns [1–3]. An increasing number of studies have shown that the deserts have an impact on the genetic structure and phylogeographic pattern of species, causing the speciation and population differentiation of many desert species [1, 4–6]. Evidence from pollen records indicates that ice sheets did not appear in arid Northwest China during the Quaternary [7]. However, glacial and interglacial cycles affected the evolutionary processes of species in this region [1, 8–10], through allopatric divergence [2, 11], range fragmentation, and regional range expansion [12, 13]. Additionally, the uplift of the Tibetan Plateau and global Pleistocene cooling promoted the formation and subsequent evolution of the desert [14, 15]. Several previous studies have shown that the increased aridification and desert expansion led to the speciation, habitat fragmentation, and diversification of desert plant species, as well as the distribution of montane plants on both sides of the desert [1, 2, 4, 16]. In addition, the desert zone acted as a geographical barrier that hindered gene flow among populations, which led to high genetic diversity among the populations and low genetic diversity within populations in arid Northwest China [4, 5, 16, 17]. However, few researchers have investigated the effects of desert formation on the evolutionary process of regional species in this arid region.
The arid regions of western Inner Mongolia contain many endemic species, several of which are considered endangered [18–21]. Specifically, the Alxa-Helan Mountain Range is considered to be one of eight high-diversity areas in China [22, 23]. Tugarinovia is a monotypic genus (Tugarinovia mongolica Iljin) with one additional variety (T. mongolica var. ovatifolia). T. mongolica, which is a member of the China Species Red List [24], is endemic to the gravel slopes of Inner Mongolia [25]. It is a perennial low herb with a dioecious reproductive system that flowers and fruits between May and June [26]. T. mongolica var. ovatifolia shows differences in morphology and habitat [26–28]. The major morphological differences between the two varieties are leaf and inflorescence size [29]. Based on field observations and herbaria specimen records, we believe this genus possesses disjunct distributions on the two sides of the desert. We find that T. mongolica is mostly distributed in the northern regions of the Alxa Desert, whereas its variety T. mongolica var. ovatifolia only occurs in narrow swaths southeast of the Alxa Desert. Currently, the combination of narrow distribution and overgrazing has resulted in a rapid decline of the species. Previous studies of T. mongolica have concentrated on embryology, taxonomy, origin, migratory route, and distribution patterns [29–32] but, to our knowledge, there have been no discussions of intraspecific taxonomy, phylogeography, or any aspect of conservation genetics.
In this study, we sequenced two chloroplast DNA sequences (psbA-trnH and psbK-psbI) to investigate the phylogeographic pattern of 16 populations of the genus Tugarinovia throughout its distributional range. Our study had the following aims. First, determine whether intraspecific phenotype variations of the genus are consistent with genetic differentiation. Second, identify whether Quaternary climate fluctuations (such as aridification and desert formation) affect the differentiation of Tugarinovia. Third, based on the genetic diversity and genetic structure of T. mongolica and T. mongolica var. ovatifolia populations, propose effective protection measures for the species.
Materials and methods
Sample collection
The study area was not a nature reserve and no specific permissions were required by the authoritative organization. Only one leaf was used as experimental material, so there was no serious damage to the target plant during field sampling. We collected leaf samples of 16 natural populations, which covered nearly the entire region occupied by T. mongolica from the northern part of the Alxa Desert to east of the southern part of the Alxa Desert, Inner Mongolia (Table 1, Fig 1). Ten populations belonging to T. mongolica (1–10) were sampled from northern Alxa, and six populations belonging to T. mongolica var. ovatifolia (11–16) from southeastern Alxa. We collected between 11 and 22 individuals from each population. Fresh leaves were sampled and dried immediately using silica gel in the field. Then, one sample of each population was deposited as a voucher specimen at the Herbarium of Xinjiang Institute of Ecology and Geography, Chinese Academy of Science (XJBI). We selected Brachylaena huillensis and Atractylodes lancea as outgroups in the phylogenetic analysis [31].
Table 1. Sample and genetic variation information for 16 populations of Tugarinovia mongolica in northwest China.
| Species name | Population | Sample Location | Latitude/Longitude(N/E) | Altitude(m) | N | Haplotypes | Hd(±SD) | π(±SD) |
|---|---|---|---|---|---|---|---|---|
| Overall | 219 | 17 | 0.9086±0.0070 | 0.0092±0.0047 | ||||
| NG | North Group | 139 | 11 | 0.8250±0.0148 | 0.0065±0.0034 | |||
| 1 HLT | Hailiutu,NM | 41.60°/108.51° | 1346 | 11 | H1 | 0 | 0 | |
| 2 DLS | Delingshan,NM | 41.29°/108.67° | 1120 | 10 | H1 | 0 | 0 | |
| 3 CJSM | Chuanjinsumu,NM | 41.89°/108.22° | 1336 | 12 | H1,H2,H3 | 0.6818±0.0910 | 0.0055±0.0032 | |
| 4 BYH | Bayinhua,NM | 42.13°/110.05° | 1267 | 22 | H4 | 0 | 0 | |
| T. mongolica | 5 BYHT | Bayinhatai,NM | 41.54°/108.66° | 1341 | 16 | H2 | 0 | 0 |
| 6 SQ | Shuiquan,NM | 41.31°/108.43° | 1047 | 12 | H1,H2 | 0.1667±0.1343 | 0.0015±0.0011 | |
| 7 SZ | Saizhen,NM | 41.54°/106.95° | 1582 | 15 | H5 | 0 | 0 | |
| 8 BYT | Baoyintu,NM | 41.71°/106.99° | 1396 | 16 | H1,H5,H6 | 0.5750±0.0799 | 0.0012±0.0009 | |
| 9 WLJ | Wuliji,NM | 40.82°/104.47° | 1448 | 11 | H5,H7,H8 | 0.6909±0.0861 | 0.0041±0.0026 | |
| 10 YG | Yingen,NM | 40.80°/104.79° | 1338 | 14 | H1,H5,H7,H8,H9,H10,H11 | 0.8462±0.0742 | 0.0131±0.0071 | |
|
T. mongolica var. ovatifolia |
SG | South Group | 80 | 6 | 0.8408±0.0085 | 0.0045±0.0025 | ||
| 11 LSM | Lashenmiao,NM | 39.29°/106.83° | 1134 | 11 | H12 | 0 | 0 | |
| 12 DZT | Dizhentai,NM | 39.68°/106.85° | 1172 | 13 | H13 | 0 | 0 | |
| 13 BRBL | Barunbieli,NM | 38.39°/105.72° | 1576 | 17 | H14 | 0 | 0 | |
| 14 QPJ | Qipanjing,NM | 39.47°/107.08° | 1426 | 13 | H15 | 0 | 0 | |
| 15 QLG | Qianligou,NM | 39.80°/107.01° | 1518 | 13 | H16 | 0 | 0 | |
| 16 HBW | Haibowan,NM | 39.65°/106.85° | 1178 | 13 | H17 | 0 | 0 |
Hd: haplotype diversity, π: nucleotide diversity. Bold letters indicate that the population has high genetic diversity.
Fig 1. Study regions of Tugarinovia mongolica.
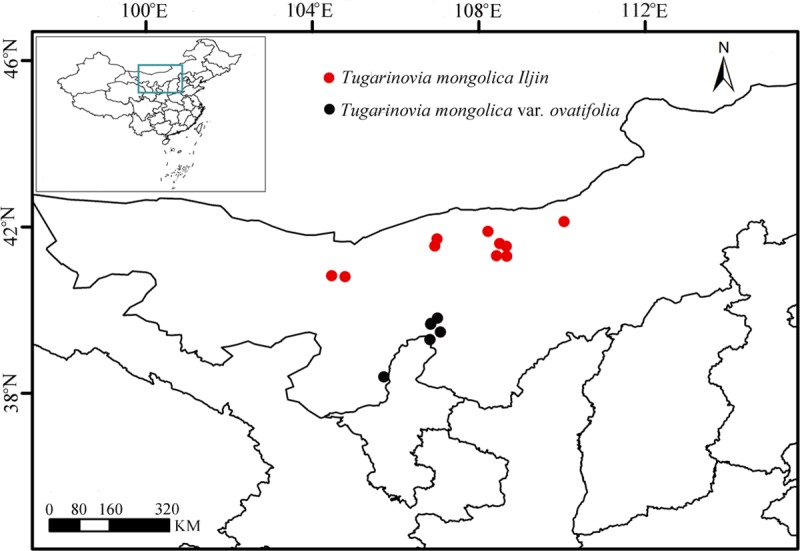
Red dots represent T. mongolica, and black dots represent T. mongolica var. ovatifolia.
DNA extraction, PCR amplification, and sequencing
The total genomic DNA was extracted using a modified cetyltrimethy ammonium bromide (CTAB) protocol [33]. Initially, we screened ten pairs of chloroplast DNA regions (trnS-trnG, psbA-trnH, psbK-psbI, rps16-trnK, rpl32-trnL, ycf6-psbM, trnC-rpoB, trnV, trnD-trnT and trnL-trnF); however, only two plastid intergenic spacers (psbA-trnH and psbK-psbI) were found to be highly variable among individuals in the populations. Polymerase chain reaction (PCR) amplifications were performed in a total volume of 25 UL reactions with the amplification of two cpDNA regions, which were conducted by the following procedure: 95°C for 4 min, 36 cycles at 94°C for 30 s, 52°C for 30 s, 72°C for 1 min and, finally, 72°C for 10 min. PCR products were detected on 1.0% agarose gel and were purified using the QIAquick Gel Extraction Kit (Qiagen). These were then sequenced using an ABI Prism 3730 automated sequencer from Sangon Biotech Co., Ltd., Shanghai, China. Sequencing alignments were carried out in CLUSTAL W [34] and were both refined and adjusted manually. Finally, sequences representing all haplotypes were submitted to GenBank with accession numbers MK299501-MK299518.
Genetic diversity and population structure
To understand the levels of genetic diversity of the species, haplotype diversity (Hd) and nucleotide diversity (π) for each population, the two geographic groups, and across all populations were calculated in ARLEQUIN 3.5 [35]. The total genetic diversity (HT), within-population genetic diversity (HS), and genetic differentiation (NST, GST) were estimated using the program Permut 1.0 with 1,000 permutation tests [36]. We used the parameters NST and GST to check whether a phylogeographic structure existed for all populations and the two geographic groups. Analysis of molecular variance (AMOVA) [37] was also performed to estimate the genetic structure by Arlequin 3.5, with significance tests based on 1,000 permutations [35]. The phylogenetic relationship among the haplotypes was constructed using Network v. 5.0 [38], followed by the median-joining (MJ) algorithm. Gaps were treated as a single mutation event. The spatial analysis of molecular variance (SAMOVA) was used to test the spatial genetic structure of cpDNA genetic variation using SAMOVA v. 1.0, where these analyses were performed for the range of 2 ≤ K ≤ 8 [39]. Finally, the best grouping was determined when the number of groups retained was maximized, FCT. However, this configuration was excluded when a single population appeared in the geographic group [40, 41].
Population demographic analyses
To test whether the species experienced demographic expansions, the parameters of Tajima’s D and Fu’s FS were estimated [42, 43]. A significant Tajima’s D value or a significant and large negative Fu’s FS value indicated that the population had experienced demographic expansion. The sum of squared deviations (SSD) and raggedness index of Harpending (HRag) were also calculated [44]. At the same time, we calculated a mismatched distribution of pairwise differences [45]. The SSD value was measured using the P-value, for which a nonsignificant P-value and unimodal distribution of pairwise differences indicated that the population experienced recent expansion, whereas a significant P-value and multimodal mismatched distribution of pairwise differences indicated that the population did not experience recent expansion. Significant tests of the above analyses were estimated using Arlequin 3.5, with 1,000 permutation tests [35].
Divergence time estimation
We estimated divergence times among the cpDNA haplotypes using BEAST v. 1.6.1 [46]. Since there were no fossil records, we used the reported substitution rate (1 and 3×10−9 s/s/y) based on the cpDNA substitution rates of most angiosperm species [47]. Based on the uncertainty of the rate values, we used a mean of 2×10−9 and an SD of 6.080×10−10 of the distribution to estimate the divergence times in this study [4, 48, 49]. We used the GTR substitution model and a coalescent tree prior. The Markov chain Monte Carlo (MCMC) permutations were run for 10,000,000 generations, sampling every 1,000 generations. The effective sample sizes (ESS) for the relevant estimated parameters were well above 200 by TRACER v. 1.5. We applied FigTree v. 1.3.1 to edit trees.
Results
Sequence analysis and haplotype distribution
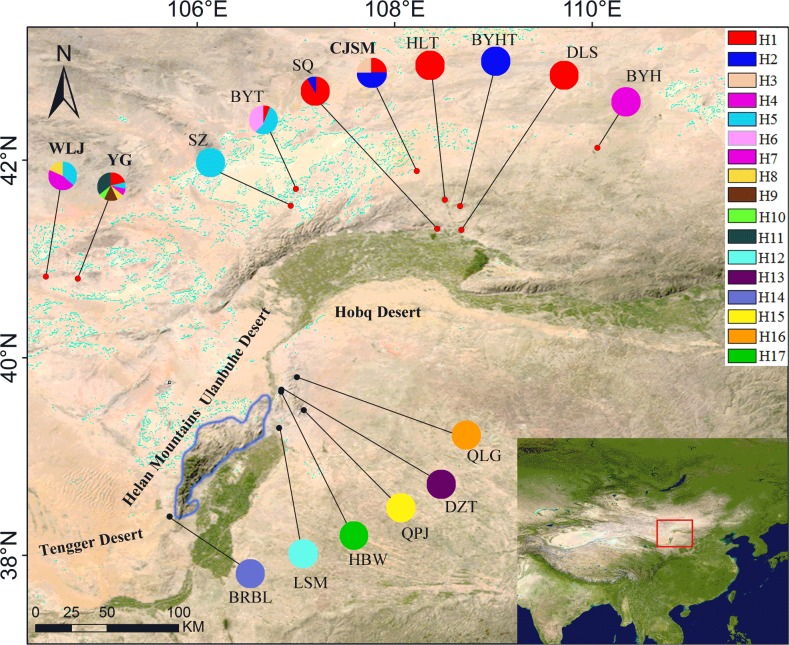
The sequences of psbA-trnH and psbK-psbI were both successfully amplified and sequenced in the 219 individuals from the 16 natural populations. The total length of the combined alignments was 897 bp. We were able to detect 21 variable sites, which included 16 substitutions and 5 indels (S1 Table). A total of 17 haplotypes were identified from all variable sites (Table 1). The results from the network analysis indicated that two clades, a northern group (NG) and southern group (SG) existed, with the northern group including 11 haplotypes (H1-H11) and the southern group including 6 haplotypes (H12-H17) (Figs 2 and 3). Importantly, no haplotype was shared between the two regions (Fig 3). In the northern group, haplotypes H1 and H5 were widespread, while haplotypes H3, H4, H6 and H9, H10, and H11 were fixed in the populations of CSJM, BYH, BYT and YG, and in the southern group, each of the 6 populations corresponded to a specific haplotype (Fig 2).
Fig 2. Geographic distribution of 17 cpDNA haplotypes detected in 16 populations of Tugarinovia mongolica from Inner Mongolia.
Pie charts show the frequency of haplotype in each population. Red dots represent the population of the northern group (NG), and black dots represent the population of the southern group (SG). The blue line outlines represent the location of Helan Mountain. The nomenclature of NG and SG populations (See Table 1) is: HLT, Hailiutu; DLS, Delingshan; CJSM, Chuanjinsumu; BYH, Bayinhua; BYHT, Bayinhatai; SQ, Shuiquan; SZ, Saizhen; BYT, Bayintu; WLJ, Wuliji; YG, Yingen; LSM, Lashenmiao; DZT, Dizhentai; BRBL, Barunbieli; QPJ, Qipanjing; QLG, Qianligou; HBW, Haibowan.
Fig 3. Statistical parsimony network of 17 haplotypes of Tugarinovia mongolica based on two cpDNA regions.
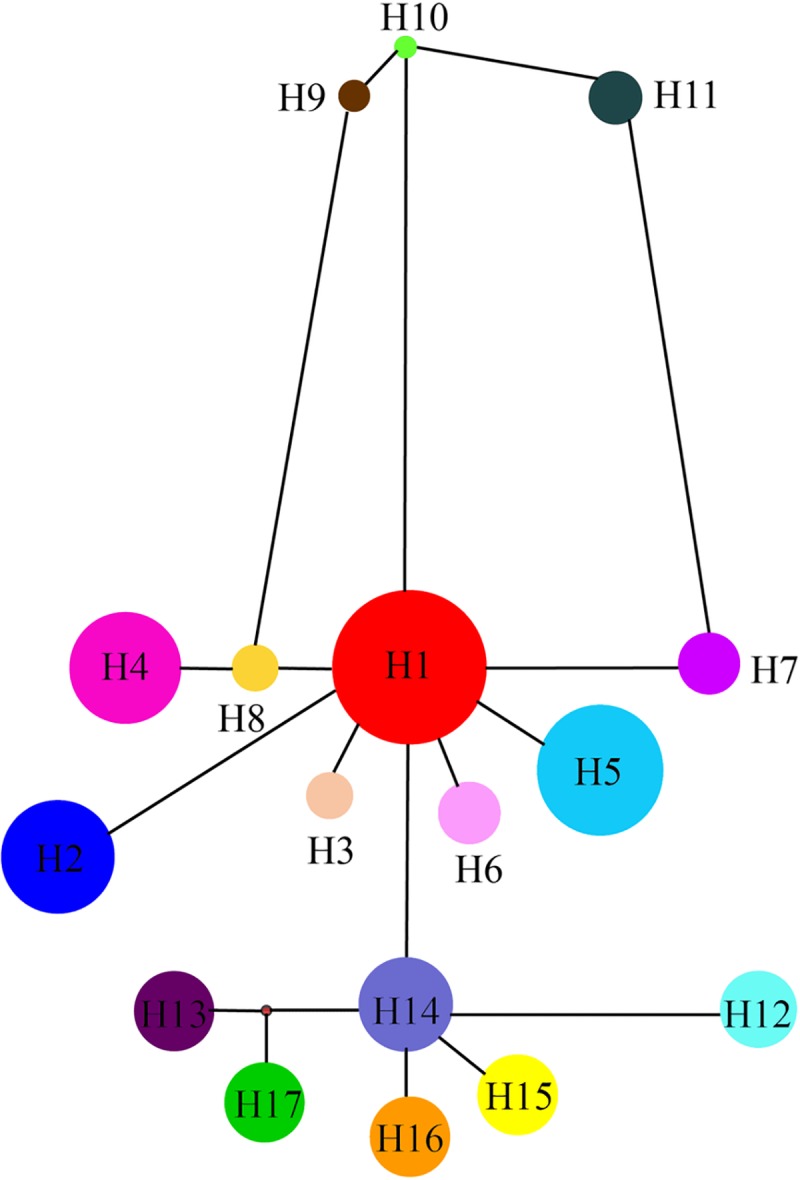
The size of each circle is proportional to the frequency of the haplotype. The haplotypes H1-H11 are found exclusively in the northern group (NG), while haplotypes H12-H17 are found exclusively in the southern group (SG).
Genetic diversity and population structure
The results of the SAMOVA also showed that as the number of groups increased to three, a single population emerged. We found that the optimal population grouping pattern of K = 2 was optimal: (1) populations 1–10 belonged to the northern group (T. mongolica), and (2) populations 11–16 belonged to the southern group (T. mongolica var. ovatifolia).
The total haplotype diversity (Hd = 0.9086±0.0070) was high, and the haplotype diversity of the southern group (Hd = 0.8408±0.0085) was slightly higher than that of the northern group (Hd = 0.8250±0.0148) (Table 1). The total nucleotide diversity (π) was 0.0092±0.0047, with the northern group (π = 0.0065±0.0034) exhibiting higher diversity than that of the southern group (π = 0.0045±0.0025) (Table 1). Throughout all populations of T. mongolica, the total genetic diversity, HT, was 0.947 and the average within-population diversity, HS, was 0.185. Although NST (0.841) was greater than GST (0.805), there was no significant difference between these two values (P>0.05). At the regional level, HT and HS were 0.858 and 0.296, and NST (0.623) was less than GST (0.655) for the northern group; for the southern group, the genetic diversity, HT, was 1; the average within-population diversity, HS, was 0; and NST (1) was equal to GST (1) (Table 2). We found no significant phylogeographical structures, either regionally or overall, in T. mongolica.
Table 2. Estimation of gene diversity (HS, HT) and gene differentiation (GST, NST) values of the total populations, the northern group (NG) and southern group (SG).
| Region | N | HS | HT | GST | NST |
|---|---|---|---|---|---|
| All region | 219 | 0.185(0.0782) | 0.947(0.0290) | 0.805(0.0840) | 0.841(0.0747) |
| NG | 139 | 0.296(0.1126) | 0.858(0.0638) | 0.655(0.1267) | 0.623(0.1022) |
| SG | 80 | 0 | 1 | 1 | 1 |
For the whole population, the results of AMOVA revealed that most of the total variance that occurred among the groups, among populations within the groups, and within the populations were small. A strong population genetic structure was detected in the species (FST = 0.88853, P<0.0001). In the northern group, variations among populations and within populations were 66.15% and 33.85%, respectively, whereas in the southern group, all variations were mainly presented among populations, with no variation within populations (FST = 1, P<0.0001) (Table 3).
Table 3. Results of analysis of molecular variance of cpDNA sequence data of Tugarinovia mongolica.
| Source of variation | d.f. | SS | VC | PV(%) | Fixation index |
|---|---|---|---|---|---|
| Among groups | 1 | 349.346 | 3.13894 | 52.86 | FCT = 0.52857** |
| Among populations within groups | 14 | 416.909 | 2.13763 | 36 | FSC = 0.76356** |
| Within populaitons | 203 | 134.375 | 0.66194 | 11.15 | FST = 0.88853** |
| Total | 218 | 900.630 | 5.93851 | ||
| NG | |||||
| Among populations | 9 | 262.359 | 2.03571 | 66.15 | |
| Within populaitons | 129 | 134.375 | 1.04166 | 33.85 | FST = 0.66151** |
| Total | 138 | 396.734 | 3.07737 | ||
| SG | |||||
| Among populations | 5 | 154.550 | 2.32668 | 100 | |
| Within populaitons | 74 | 0.000 | 0.00000 | 0 | FST = 1.00000** |
| Total | 79 | 154.550 | 2.32668 |
Degrees of freedom (d.f.), sum of squares (SS), variance components (VC), percentage of variation (PV).
**, p<0.001, 1000 permutations.
NG: northern group, SG: southern group.
Population demographic analyses
Demographic analysis of all populations and the two groups showed that the values of Fu’s FS and Tajiam’s D were positive and not significant (S2 Table), which indicated that neither all populations nor the two groups experienced range expansion. We found further support from the mismatched distribution for all populations and for the two geographical groups, which were both multimodal (S1 Fig). Although the SSD value and raggedness index (P>0.05) showed a sudden expansion model, the results of Fu’s FS, Tajiam’s D and the mismatch analysis indicated that recent range expansion did not occur in T. mongolica (S2 Table, S1 Fig).
Divergence time estimation
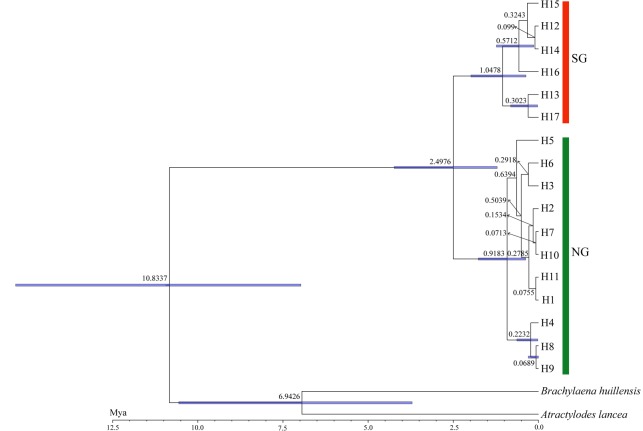
We found that the divergence time between the northern group (T. mongolica) and southern group (T. mongolica var. ovatifolia), which was determined from the BEAST analysis, occurred at 2.4976 (95%HPD: 1.2094–4.2318) Mya (Fig 4), during the early Pleistocene epoch.
Fig 4. The divergence time (Mya) of 17 cpDNA haplotypes of the northern group (NG) and southern group (SG) of Tugarinovia mongolica based on BEAST analysis.
Discussion
Allopatric divergence of Tugarinovia mongolica in Inner Mongolia
Based on the haplotype network and BEAST analysis of T. mongolica, there were two distinct clades that were clearly distributed in the northern and southern regions of Inner Mongolia (Figs 3 and 4). Furthermore, the AMOVA results showed that most of the genetic variation occurred between these two groups, along with a significant phylogeographic break that occurred between them. The SAMOVA showed the same results, divided into two groups, one northern group and one southern group. These results clearly indicate an allopatric divergence in T. mongolica. Several causes of this divergence could be: habitat fragmentation resulting from periodic oscillations of the Quaternary climate [50–52], lack of long distance dispersal [53], and geographic isolation [1, 4, 54, 55].
Based on morphological characteristics of T. mongolica, there are clear differences between T. mongolica and its variety T. mongolica var. ovatifolia. The leaves of T. mongolica are pinnately divided, long oval or rectangular, whereas the leaves of T. mongolica var. ovatifolia are ovate or oval, with a nondivided leaf margin and larger capitulum. We found evidence that T. mongolica var. ovatifolia in the southern group should be recognized as an independent species based on our molecular results, which is consistent with the morphological classification proposed by Zhao [29].
High genetic differentiation due to vicariance
All populations of T. mongolica showed high levels of haplotype diversity (Hd = 0.9086) and low nucleotide diversity (π = 0.0092). Low nucleotide diversity is usually associated with a low seed amount and a small effective population size in some endangered species [18]. However, the low nucleotide diversity that was observed for T. mongolica may be attributed to the small effective population sizes that are associated with the reproductive barrier (male sterility). In general, species that have narrow distributions and small effective population sizes show high genetic differentiation among populations [17, 18], which we detected in this species (Table 2). T. mongolica showed strong genetic differentiation (GST = 0.805) and low genetic diversity within populations (HS = 0.185). The above results indicated a high level of genetic differentiation among populations in T. mongolica that was due to restricted gene flow.
The divergence between the northern and southern groups can be traced back to the early Pleistocene epoch (Fig 4) during the development of arid conditions that led to the formation of the deserts that are located in Northwest China [15]. We speculate that the creation of the extreme climate may have resulted in the diversification of T. mongolica. In addition, the divergence time of two geographic groups (Fig 4) is consistent with the formation of the Hobq Desert [56, 57], which, as a geographical barrier, may have blocked the genetic flow between the northern and southern groups. Previous studies have shown that the desert, as a geographical barrier, promotes the allopatric divergence of species [4]. The pollination and fertilization requirements of T. mongolica make long-distance dispersal between the southern and northern groups impossible, because they were separated by deserts. Thus, we speculate that the desert may have acted as a geographic barrier that blocked gene flow between the two geographic groups, thereby resulting in allopatric divergence. Consequently, populations became isolated and independent in the northern and southern regions of the Alxa Desert.
In this study, each population of the southern group contained one specific haplotype, and most populations in the northern group shared one haplotype (Fig 2). The differentiation of haplotypes within the two groups occurred in the middle and late Pleistocene (Fig 4), and this period coincides with the formation period of the desert [14, 15]. The expansion of the desert may have led to habitat fragmentation [1]. Here, we use desert expansion to explain the fragmented distribution of T. mongolica. In addition, the biological characteristics (dioecious with male sterility and pistil abortion) of T. mongolica could have resulted in the distribution of extant populations. The fragmentation distribution of T. mongolica var. ovatifolia may also be related to desert expansion, but evidence of male sterility and pistil abortion in this group requires further research.
Implications for Tugarinovia mongolica conservation
The results of genetic diversity and population structure are important to consider for the implementation of effective conservation strategies, particularly for endemic and endangered species [58, 59]. The risk of extinction is higher for species with narrow distributions and small population sizes, especially if the gene flow among populations is lower than those with large and stable populations. In addition, small population sizes are sensitive to reduced genetic diversity through genetic drift and inbreeding [60, 61].
According to the analysis of cpDNA data, the high genetic drift load (FST = 0.88853, GST = 0.805) and inbreeding load (HS = 0.185) showed a significant extinction risk in the genus Tugarinovia (Table 2). This extinction risk is particularly noticeable in the populations of T. mongolica var. ovatifolia, which showed fragmented distributions, small population sizes, high genetic drift load (FST = 1, GST = 1), and high inbreeding load (HS = 0) (Tables 2 and 3). All of the above indices can increase sensitivity to environmental changes and the risk of extinction. In addition, populations LSM, DZT, BRBL, QPJ, QLG and HBW of the southern group exhibited unique haplotypes, which offer some protection from extinction for the population of T. mongolica var. ovatifolia. In the northern group, the CJSM, WLJ and YG populations of T. mongolica exhibited higher genetic diversity than other populations. Since T. mongolica is a critically endangered, protected species with a second-class national priority [24], it is recommended that the hotspots of populations that contain the highest genetic diversity be protected [62, 63].
To mitigate genetic drift and the inbreeding load and increase the effective population size of T. mongolica, we propose establishing the following conservation measures. First, enact additional in situ conservation measures for the species, such as the creation of additional nature reserves in the northern and southeast Alxa Desert, especially the CJSM, WLJ, and YG populations of the northern areas. (The Wuhai location has established nature reserves for some endangered and rare species [19])’. In particular, nature reserves for T. mongolica var. ovatifolia, as an independent species with an unique haplotype, should be established. Second, a protocol should be developed for ex situ conservation actions, such as seed collection from natural populations and reproduction in botanical gardens or other places, which can ensure maximum conservation of the genetic diversity of species in those particular areas.
Supporting information
(DOC)
(DOCX)
(DOCX)
(TIF)
Acknowledgments
We thank Hong-Xiang Zhang and Zhi-Bin Wen (Xinjiang Institute of Ecology and Geography, CAS) for their kind help with molecular data analysis. We also thank the professor Yi-Zhi Zhao (College of life science, Inner Mongolia University) for his kind help with sampling collection (2015–2016) in Inner Mongolia.
Data Availability
All files are available from the GenBank database (accession numbers MK299501-MK299518).
Funding Statement
This study is financially supported by Biodiversity Conservation Strategy Program of Chinese Academy of Sciences (ZSSD-012), and China National Key Basic Research Program (2014CB954201).
References
- 1.Meng HH, Gao XY, Huang JF, Zhang ML. Plant phylogeography in arid Northwest China: Retrospectives and perspectives. Journal of Systematics and Evolution. 2015;53(1):33–46. 10.1111/jse.12088 [DOI] [Google Scholar]
- 2.Wang Q, Abbott RJ, Yu QS, Lin K, Liu JQ. Pleistocene climate change and the origin of two desert plant species, Pugionium cornutum and Pugionium dolabratum (Brassicaceae), in northwest China. New Phytologist. 2013;199(1):277–87. 10.1111/nph.12241 . [DOI] [PubMed] [Google Scholar]
- 3.Zhang YH, Yu QS, Zhang Q, Hu XK, Hu J, Fan BL. Regional-scale differentiation and phylogeography of a desert plant Allium mongolicum (Liliaceae) inferred from chloroplast DNA sequence variation. Plant Systematics and Evolution. 2017;303(4):451–66. 10.1007/s00606-016-1383-6 [DOI] [Google Scholar]
- 4.Wang P, Zhang XZ, Tang N, Liu JJ, Xu LR, Wang K. Phylogeography of Libanotis buchtormensis (Umbelliferae) in Disjunct Populations along the Deserts in Northwest China. PloS one. 2016;11(7):e0159790 10.1371/journal.pone.0159790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo YP, Zhang R, Chen CY, Zhou DW, Liu JQ. Allopatric divergence and regional range expansion of Juniperus sabina in China. Journal of Systematics and Evolution. 2010;48(3):153–60. [Google Scholar]
- 6.Xu Z, Zhang ML. Phylogeography of the arid shrub Atraphaxis frutescens (Polygonaceae) in northwestern China: evidence from cpDNA sequences. Journal of Heredity. 2015;106(2):184–95. 10.1093/jhered/esu078 . [DOI] [PubMed] [Google Scholar]
- 7.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405(6789):907–13. 10.1038/35016000 [DOI] [PubMed] [Google Scholar]
- 8.Meng HH, Zhang ML. Phylogeography of Lagochilus ilicifolius (Lamiaceae) in relation to Quaternary climatic oscillation and aridification in northern China. Biochemical Systematics and Ecology. 2011;39(4–6):787–96. 10.1016/j.bse.2011.07.015 [DOI] [Google Scholar]
- 9.Liu JQ, Sun YS, Ge XJ, Gao LM, Qiu YX. Phylogeographic studies of plants in China: Advances in the past and directions in the future. Journal of Systematics and Evolution. 2012;50(4):267–75. 10.1111/j.1759-6831.2012.00214.x [DOI] [Google Scholar]
- 10.Shi XJ, Zhang ML. Phylogeographical structure inferred from cpDNA sequence variation of Zygophyllum xanthoxylon across north-west China. Journal of plant research. 2015;128(2):269–82. 10.1007/s10265-014-0699-y . [DOI] [PubMed] [Google Scholar]
- 11.Zhang HX, Zhang ML. Identifying a contact zone between two phylogeographic lineages of Clematis sibirica (Ranunculeae) in the Tianshan and Altai Mountains. Journal of Systematics and Evolution. 2012;50(4):295–304. 10.1111/j.1759-6831.2012.00198.x [DOI] [Google Scholar]
- 12.Jia DR, Liu TL, Wang LY, Zhou DW, Liu JQ. Evolutionary history of an alpine shrub Hippophae tibetana (Elaeagnaceae): allopatric divergence and regional expansion. Biological Journal of the Linnean Society. 2011;102(1):37–50. 10.1111/j.1095-8312.2010.01553.x [DOI] [Google Scholar]
- 13.Li ZH, Chen J, Zhao GF, Guo YP, Kou YX, Ma YZ, et al. Response of a desert shrub to past geological and climatic change: A phylogeographic study of Reaumuria soongarica (Tamaricaceae) in western China. Journal of Systematics and Evolution. 2012;50(4):351–61. 10.1111/j.1759-6831.2012.00201.x [DOI] [Google Scholar]
- 14.Wang Y, Li S, Wang JH, Yan MC. The uplift of the Qinghai-Xizang(Tibetan) plateau and its effect on the formation and evolution of China desert. Arid Zone Research. 1996;13(2):20–4. [Google Scholar]
- 15.Zhang LX, Song YQ. The effect of the Qinghai-Tibet plateau uplift on the space-time distributing pattern of Chinese desert and desertification. China Population, Resources and Environment. 2001;11(4):98–101. [Google Scholar]
- 16.Meng HH, Zhang ML. Diversification of plant species in arid Northwest China: species-level phylogeographical history of Lagochilus Bunge ex Bentham (Lamiaceae). Molecular Phylogenetics Evolution. 2013;68(3):398–409. 10.1016/j.ympev.2013.04.012 . [DOI] [PubMed] [Google Scholar]
- 17.Jia J, Zeng LQ, Gong X. High Genetic Diversity and Population Differentiation in the Critically Endangered Plant Species Trailliaedoxa gracilis (Rubiaceae). Plant Molecular Biology Reporter. 2016;34(1):327–38. 10.1007/s11105-015-0924-4 [DOI] [Google Scholar]
- 18.Ge XJ, Hwang CC, Liu ZH, Huang CC, Huang WH, Hung KH, et al. Conservation genetics and phylogeography of endangered and endemic shrub Tetraena mongolica (Zygophyllaceae) in Inner Mongolia China. BMC Genetics. 2011;12:1 10.1186/1471-2156-12-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su ZH, Zhang ML. Chloroplast phylogeography of Helianthemum songaricum (Cistaceae) from northwestern China: implications for preservation of genetic diversity. Conservation Genetics. 2011;12(6):1525–37. 10.1007/s10592-011-0250-9 [DOI] [Google Scholar]
- 20.Ma SM, Zhang ML. Phylogeography and conservation genetics of the relic Gymnocarpos przewalskii (Caryophyllaceae) restricted to northwestern China. Conservation Genetics. 2012;13(6):1531–41. 10.1007/s10592-012-0397-z [DOI] [Google Scholar]
- 21.Su ZH, Pan BR, Zhang ML, Shi W. Conservation genetics and geographic patterns of genetic variation of endangered shrub Ammopiptanthus (Fabaceae) in northwestern China. Conservation Genetics. 2015;17(2):485–96. 10.1007/s10592-015-0798-x [DOI] [Google Scholar]
- 22.Wang HS, Zhang YL. The bio-diversity and characters of spermatophytic genera endemic to China. Acta Botanica Yunnanica 1994;16(3):209–20. [Google Scholar]
- 23.Zhu ZY, Ma YQ, Liu ZL, Zhao YZ. Endemic plants and floristic characteristics in Alashan-Ordos biodiversity center. Journal of Arid Land Resources and Environment. 1999;13(2):1–16. [Google Scholar]
- 24.Fu LG. The Red Book of Chinese Plants-Rare and Endangered Plants Science Press, Beijing. 1992: 236–7.
- 25.Zhao YZ. Endemic genera and their basic characteristics of the Mongolian planteau plants. Acta Scientiarum Naturalium Universitatis NeiMongol. 1997;28(4):547–52. [Google Scholar]
- 26.Shi Z. Flora of China Beijing: Science Press and St Louis: Missouri Botanical Garden Press; 2011:41–2. [Google Scholar]
- 27.Shi Z. Flora Reipublicae Popularis Sinicae Beijing: Science Press; 1979;75:246–8. [Google Scholar]
- 28.Fu XQ. Flora Intramongolica Hohhot: Inner Mongolia Peoples Publishing House; 1992;Ⅱ:704–7. [Google Scholar]
- 29.Zhao YZ. The classification and its geographycal distribution of Tugarinovia. Acta Botanica Boreali-Occidentalia Sinica. 2000;20(5):873–5. [Google Scholar]
- 30.Ma H, Wang YC, Cao R, Guo XL. The embryological study of Tugarinovia mongolicaⅠ.Megasporogenesis、microsporogenesis and development of gametophytes. Acta Botanica Boreali-Occidentalia Sinica. 2000;20(3):461–6. [Google Scholar]
- 31.Zhu XM. Study on origin and migration of three desert plants: Gymnocarpos przewalskii(Caryophyllaceae), Tetraena mongolica(Zygophyllaceae), Tugarinovia mongolica(Compositae) Inner Mongolia University; 2008. [Google Scholar]
- 32.Ma SM. Distribution patterns of endemic plants in central Asian desert Xinjiang Institute of Ecology and Geography Chinese Academy of Sciences; 2011. [Google Scholar]
- 33.Doyle JJ, Doyle JL. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 1987;19:11–5. [Google Scholar]
- 34.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular ecology resources. 2010;10(3):564–7. 10.1111/j.1755-0998.2010.02847.x . [DOI] [PubMed] [Google Scholar]
- 36.Pons O, Petit RJ. Measwring and testing genetic differentiation with ordered versus unordered alleles. Genetics. 1996;144:1237–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial-DNA restriction data. Genetics. 1992;131:479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16(1):37–48. 10.1093/oxfordjournals.molbev.a026036 [DOI] [PubMed] [Google Scholar]
- 39.Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Molecular Ecology. 2002;11(12):2571–81. [DOI] [PubMed] [Google Scholar]
- 40.Beatty GE, Provan J, Comes HP. Post-glacial dispersal, rather than in situ glacial survival, best explains the disjunct distribution of the Lusitanian plant species Daboecia cantabrica(Ericaceae). Journal of Biogeography. 2013;40(2):335–44. 10.1111/j.1365-2699.2012.02789.x [DOI] [Google Scholar]
- 41.Santiso X, Lopez L, Retuerto R, Barreiro R. Population Structure of a Widespread Species under Balancing Selection: The Case of Arbutus unedo L. Front Plant Sci. 2015;6:1264 10.3389/fpls.2015.01264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147(2):915–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harpending HC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Human Biology. 1994;66:591–600. [PubMed] [Google Scholar]
- 45.Schneider S, Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics. 1999;152(3):1079–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Academy of Sciences. 1987;84(24):9054–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia DR, Abbott RJ, Liu TL, Mao KS, Bartish IV, Liu JQ. Out of the Qinghai-Tibet Plateau: evidence for the origin and dispersal of Eurasian temperate plants from a phylogeographic study of Hippophae rhamnoides (Elaeagnaceae). New Phytologist. 2012;194(4):1123–33. 10.1111/j.1469-8137.2012.04115.x . [DOI] [PubMed] [Google Scholar]
- 49.Zhang HX, Zhang ML, Sanderson SC. Retreating or standing: responses of forest species and steppe species to climate change in arid Eastern Central Asia. PloS one. 2013;8(4):e61954 10.1371/journal.pone.0061954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Comes HP, Kadereit JW. The effect of Quaternary climatic changes on plant distribution and evolution. Trends in plant science. 1998;3(11):432–8. [Google Scholar]
- 51.Qiu YX, Fu CX, Comes HP. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world's most diverse temperate flora. Molecular Phylogenetics Evolution. 2011;59(1):225–44. 10.1016/j.ympev.2011.01.012 . [DOI] [PubMed] [Google Scholar]
- 52.Bai WN, Wang WT, Zhang DY. Phylogeographic breaks within Asian butternuts indicate the existence of a phytogeographic divide in East Asia. The New phytologist. 2016;209(4):1757–72. 10.1111/nph.13711 . [DOI] [PubMed] [Google Scholar]
- 53.Li WJ, Sui XL, Kuss P, Liu YY, Li AR, Guan KY. Long-Distance Dispersal after the Last Glacial Maximum (LGM) Led to the Disjunctive Distribution of Pedicularis kansuensis (Orobanchaceae) between the Qinghai-Tibetan Plateau and Tianshan Region. PloS one. 2016;11(11):e0165700 10.1371/journal.pone.0165700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen LY, Muchuku JK, Yan X, Hu GW, Wang QF. Phylogeography of Haplocarpha rueppelii (Asteraceae) suggests a potential geographic barrier for plant dispersal and gene flow in East Africa. Science Bulletin. 2015;60(13):1184–92. 10.1007/s11434-015-0832-x [DOI] [Google Scholar]
- 55.Su ZH, Lu W, Zhang ML. Phylogeographical patterns of two closely related desert shrubs, Nitraria roborowskii and N. sphaerocarpa (Nitrariaceae), from arid north-western China. Botanical Journal of the Linnean Society. 2016;180:334–47. [Google Scholar]
- 56.Dong GR, Li BS, Gao SY, Wu Z, Shao YJ. The Quaternary ancient eolian sands in the Ordos Platau. Acta Geographica Sinica. 1983;38(4):341–7. [Google Scholar]
- 57.Li BF, Sun DH, Xu WH, Wang F, Liang BQ, Ma ZW, et al. Paleomagnetic chronology and paleoenvironmental records from drill cores from the Hetao Basin and their implications for the formation of the Hobq Desert and the Yellow River. Quaternary Science Reviews. 2017;156:69–89. 10.1016/j.quascirev.2016.11.023 [DOI] [Google Scholar]
- 58.Frankel OH, Brown AHD, Burdon JJ. The conservation of plant biodiversity. Cambridge University Press, Cambridge: 1995. [Google Scholar]
- 59.Bevill RL, Louda SM. Comparisons of related rare and common species in the study of plant rarity. Conservation Biology. 1999;13(3):493–8. [Google Scholar]
- 60.Anthropogenic Lande R., ecological and genetic factors in extinction and conservation. Researches on population ecology. 1998;40(3):259–69. [Google Scholar]
- 61.Frankham R, Briscoe DA, Ballou JD. Introduction to conservation genetics Cambridge university press; 2002. [Google Scholar]
- 62.Aoki K, Suzuki T, Hsu TW, Murakami N. Phylogeography of the component species of broad-leaved evergreen forests in Japan, based on chloroplast DNA variation. Journal of plant research. 2004;117(1):77–94. 10.1007/s10265-003-0132-4 . [DOI] [PubMed] [Google Scholar]
- 63.Huang JC, Wang WK, Peng CI, Chiang TY. Phylogeography and conservation genetics of Hygrophila pogonocalyx (Acanthaceae) based on atpB-rbcL noncoding spacer cpDNA. Journal of plant research. 2005;118(1):1–11. 10.1007/s10265-004-0185-z . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)
(DOCX)
(TIF)
Data Availability Statement
All files are available from the GenBank database (accession numbers MK299501-MK299518).