Abstract
Identifying the neurotransmitters used by specific neurons is a critical step in understanding the function of neural circuits. However, methods for the consistent and efficient detection of neurotransmitter markers remain limited. Fluorescence in situ hybridization (FISH) enables direct labeling of type-specific mRNA in neurons. Recent advances in FISH allow this technique to be carried out in intact tissue samples such as whole-mount Drosophila melanogaster brains. Here, we present a FISH platform for high-throughput detection of eight common neurotransmitter phenotypes in Drosophila brains. We greatly increase FISH throughput by processing samples mounted on coverslips and optimizing fluorophore choice for each probe to facilitate multiplexing. As application examples, we demonstrate cases of neurotransmitter coexpression, reveal neurotransmitter phenotypes of specific cell types, and explore the onset of neurotransmitter expression in the developing optic lobe. Beyond neurotransmitter markers, our protocols can in principle be used for large-scale FISH detection of any mRNA in whole-mount fly brains.
Keywords: neurotransmitter, Drosophila, fluorescence in situ hybridization, gene expression, mRNA
A critical step in understanding the function of neural circuits is to identify the neurotransmitters used by specific neurons. Typical indicators of transmitter phenotypes are genes with specific roles in transmitter synthesis, vesicular transport, or recycling. While such markers are known for common transmitters, their reliable detection in specific cell types remains challenging, in particular if a large number of specimens need to be examined.
Common methods for detecting neurotransmitter markers are sequencing transcriptomics, immunolabeling, and fluorescent in situ hybridization (FISH). RNA sequencing (RNAseq)-based methods can provide comprehensive catalogs of gene expression including of neurotransmitter markers (Henry et al. 2012; Konstantinides et al. 2015; Croset et al. 2018; Davie et al. 2018; Davis et al. 2018). However, these approaches either examine dissociated single cells, which can be difficult to map to specific cell types, or depend on genetic markers, which are not always available, to isolate neuronal populations. In addition, both single-cell and population transcriptomics result in the loss of spatial information (Buxbaum et al. 2015; Lein et al. 2017) and incur high costs if examining a small number of mRNAs across many conditions. Immunohistochemistry preserves spatial information and can be applied on a large scale. However, high-quality antibodies are often not readily available and antibody generation can be time-consuming (Fritschy 2008). In addition, proteins that primarily localize to fine neurites may be difficult to assign to specific cells by immunolabeling.
FISH is a powerful method for detecting endogenous mRNA sequences in intact tissues (Zhao et al. 2003; Lécuyer et al. 2007; Raj et al. 2008; Moffitt et al. 2016; Shah et al. 2016; Yang et al. 2017). Recent improvements enable the localization of mRNA, including of indicators of neurotransmitter phenotypes, in whole-mount Drosophila tissues (Long et al. 2017). However, validated FISH probes are only available for marking neurons expressing acetylcholine, glutamate, and γ-aminobutyric acid (GABA), and current FISH protocols are not efficient for processing large numbers of specimens. Here, we extend detection to dopaminergic, serotoninergic, tyraminergic, octopaminergic, and histaminergic neurons with validated FISH probes, and describe a high-throughput, optimized FISH procedure for detecting these neurotransmitter cell types. The approach is particularly suitable for rapidly identifying neurotransmitter markers expressed by neurons labeled by libraries of genetic markers (e.g., GAL4 or LexA driver collections). To demonstrate the utility of our FISH platform, we map neurotransmitter markers to specific cell types using split-GAL4 lines and examine the onset of neurotransmitter expression in the developing fly visual system.
Materials and Methods
Fly stocks
TH-GAL4 was from Friggi-Grelin et al. (2003). Tdc2-GAL4 was from Cole et al. (2005). SerT-GFP was SerTMI02578 from Nagarkar-Jaiswal et al. (2015). HdcJk910 was from Burg et al. (1993). UAS-7xHaloTag::CAAX in VK00005 for Figure 2 and Figure 3, and Supplemental Material, Figure S6 were from Sutcliffe et al. (2017). UAS-myr-HaloTag for Figure S3 was from Kohl et al. (2014). R58E02-GAL4 was from Liu et al. (2012). SS02425 was from Davie et al. (2018). Wild-type flies were Canton-S. Split-GAL4 stock SS02565 consists of R55C09-p65ADZp in VK00027 and VT040566-ZpGDBD in attP2 (Luan et al. 2006; Pfeiffer et al. 2010; Dionne et al. 2018; Tirian and Dickson 2018). Split-GAL4 stock SS45407 consists of VT012639-p65ADZp in attP40 and VT000608-ZpGdbd in attP2. Split-GAL4 stock SS51118 consists of VT050405-p65ADZp in attP40 and VT007068-ZpGDBD in attP2.
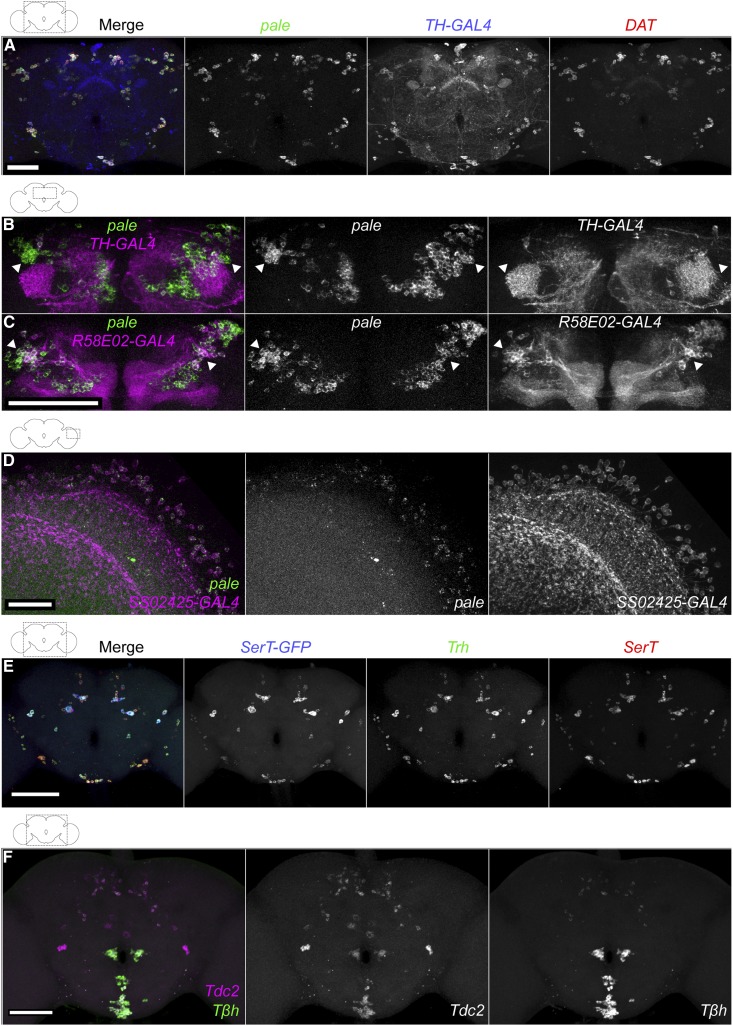
Figure 2.
Validation of FISH probes for identification of dopaminergic, serotoninergic, tyraminergic, and octopaminergic neurons. (A) Simultaneous detection of pale and DAT mRNAs with TH-GAL4. A TH-GAL4; UAS-HaloTag brain was labeled with ATTO 647N HaloTag ligand (blue), and FISH probes for pale (Quasar 570; green) and DAT (CAL Fluor 610 Red; magenta) mRNAs, shown together (left) and as individual grayscale images. Samples were processed using microcentrifuge protocol. Bar, 50 μm. (B and C) pale and GAL4 marker expression in the dorsal anterior brain, focusing on neurons of the protocerebral anterior medial cluster. In TH-GAL4 (B) and R58E02-GAL4 (C), UAS-HaloTag brains were labeled with AF488 HaloTag ligand (magenta) and pale FISH probe (Cy5; green), shown together (left) and as individual grayscale images. Images are maximum intensity z-projections through the anterior central brain. Arrowheads indicate neurons without overlap in (B) and neurons with overlap in (C). Bar, 50 μm. Movies of (B and C) are in Files S8 and S9, respectively. (D) pale expression in Mi15 medulla neurons. An SS02425-GAL4, UAS-HaloTag brain was labeled with ATTO 647N HaloTag ligand (magenta) and pale FISH probe (Quasar 570; green) together (left) or as individual grayscale images (Davis et al. 2018). Bar, 20 μm. (E) A SerT-GFP brain was labeled with FISH probes for GFP (AF488; blue), Trh (Cy3; green), and SerT (Cy5; red) mRNAs. Each channel is shown in gray on the right. Bar, 100 μm. (F) Simultaneous detection of Tdc2 and Tβh mRNAs. A wild-type brain was labeled with FISH probes for Tdc2 (Cy5; magenta) and Tβh (Cy3; green). The channels are shown to the right in gray. Bar, 50 μm. Movie is in File S11. The overlap of Tdc2 and Tβh FISH probes with Tdc2-GAL4 is shown in Figure S3C.
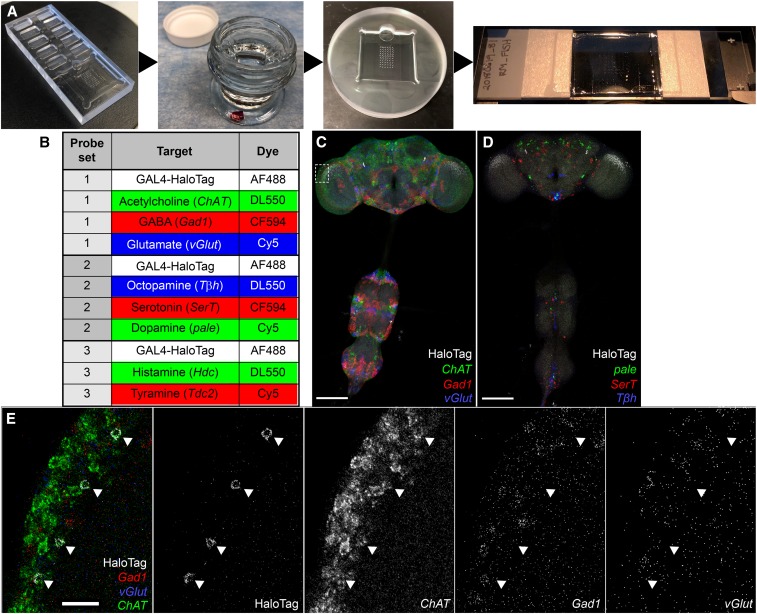
Figure 3.
High-throughput FISH platform for identifying neurotransmitter phenotypes in Drosophila CNS. (A) Key steps and equipment of the high-throughput FISH platform. Samples are mounted on a coverslip using a plexiglass mounting T-dish (see Materials and Methods), using the printed grid beneath the coverslip as a guide. Most processing steps occur by moving coverslips between jars of solution. Hybridization is carried out with the coverslip resting on spacers to either side of a custom hybridization chamber, trapping ∼150 μl of hybridization solution with the samples between the coverslip and the bottom of the chamber. For imaging, the coverslip is mounted in distyrene, plasticizer, and xylene on a slide with a split coverslip for spacers. A schematic of the hybridization chamber is shown in Figure S5A. (B) Neurotransmitter marker detection with optimized FISH probe sets and fluorophore selection. Each set permits detection of two or three FISH probes together with a HaloTag reporter. Movies of optimized probe sets without HaloTag reporter are in Files S1–S6. (C–E) Neurotransmitter detection using the FISH platform. Identifying the neurotransmitter phenotypes of a population of medulla neurons. SS02565, UAS-HaloTag brains were labeled with AF488 HaloTag ligand (white) and (C) FISH probes for Gad1 (CF594; red), vGlut (Cy5; blue), and ChAT (DL550; green) mRNAs or (D) SerT (CF594; red), pale (Cy5; blue), and Tβh (DL550; green) mRNAs. Bar, 100 μm. (E) Boxed region from (C) was imaged with a 63× objective. Individual channels are shown to the right in gray. Arrowheads indicate the location of HaloTag-labeled cell bodies. Movie is in File S7. Bar, 10 μm.
CNS preparation
To label cells with HaloTag ligand, specific GAL4 driver lines were crossed to UAS-HaloTag. Flies were reared on standard corn meal molasses food at 22–25°. Approximately 3–5-day-old adult flies were used for the studies, except for the optic lobe developmental samples, for which tissues were collected at the specified developmental stages. Dissection was carried out in phosphate-buffered saline (PBS) or cold S2 medium (Schneider’s Insect Medium, S01416; Sigma [Sigma Chemical], St. Louis, MO). After dissection, brain tissues were transferred to 2% paraformaldehyde in S2 medium. Samples underwent fixation followed by one to four 15-min washes in PBS + 0.5% Triton X-100 (PBT), then were labeled with 2 μM HaloTag ligand in PBT for 15 min. Samples were washed twice in PBT then dehydrated in a 30, 50, 75, and 100% ethanol series. Samples can be stored in 100% EtOH at 4° for up to 2 weeks. HaloTag ligands were fused to either JF646 (Grimm et al. 2015), AF488 (G1001; Promega, Madison, WI), or ATTO 647N (Meissner et al. 2018).
FISH protocol
FISH labeling followed the protocol of Long et al. (2017) with the following modifications, unless otherwise specified. After initial dehydration (and associated tissue shrinkage), samples were mounted in 75% ethanol on poly-l-lysine-coated coverslips. They were returned to 4° 100% ethanol for storage until beginning the main FISH protocol. One to four coverslips were moved between jars (W900180-6; Wheaton, Millville, NJ) containing 10 ml of solution for most processing steps. Multiple jars were processed in parallel when needed. Hybridization was performed in custom plexiglass chambers modified from https://hhmi.flintbox.com/public/project/26606/ and related designs [Figure 3A, Figure S5A, and Wu et al. (2016)]. The 22 × 22-mm coverslip is held above the bottom of the chamber by 0.5 × 5.5 × 22-mm spacers on each side, leaving an ∼0.5 × 11 × 22-mm space for the samples and 150–180 μl of hybridization solution. A hole at the top allows for overflow, and access for coverslip addition and removal. The 20-hr hybridization reaction was carried out with the chambers inside a humidified polypropylene container (2249-6; Ted Pella). FISH probes were labeled with one of the following fluorophores: Cy3 (GE PA13101), Cy5 (GE PA15101), Quasar 570 (LGC Biosearch Technologies), CF594 (92132; Biotium), Alexa Fluor 594 (AF594) (A20004; Thermo Fisher Scientific), DyLight550 (DL550) (62262; Thermo Fisher Scientific), or CAL Fluor 610 Red (LGC Biosearch Technologies). Please see Supplemental Material for step-by-step coverslip FISH protocol. For serotonin immunostaining with FISH, the tissues were first exposed to 1:50 mouse anti-serotonin (MS-1431-S0; Thermo Fisher Scientific) overnight at 4°. After washing the primary antibody, brain tissues were prepared for FISH. Next, 1:400 AF568 goat anti-mouse secondary (A-11031; Thermo Fisher Scientific) was added in the second step of hybridization and incubated along with FISH probes. After the series of wash steps described in the FISH protocol, the tissues were fixed and mounted with distyrene, plasticizer, and xylene (DPX).
Confocal imaging
Samples were imaged on Zeiss ([Carl Zeiss], Thornwood, NY) LSM 710, 780, or 880 confocal microscopes and Zeiss ZEN software. Excitation and detection bands for each dye were generally as follows: AF488 with a 488-nm laser and 498–543-nm detection, DL550 and Cy3 with a 561-nm laser and 569–595-nm detection, CF594 with a 594-nm laser and 600–638-nm detection, and ATTO 647N and Cy5 with a 633-nm laser and 638–735-nm detection. Due to microscope limitations, most four-color images were captured as two separate image stacks, the first with 488- and 594-nm channels, and the second with 488-, 561-, and 633-nm channels. The two stacks were merged after imaging for further analysis. Confocal parameters for each image were individually optimized for signal quality unless stated otherwise. All confocal images are full maximum-intensity projections unless stated otherwise. Images were processed with Fiji software (Schindelin et al. 2012).
FISH probe sequences
Probe sequences are listed in Table S1.
Data availability
All fly strains are available in the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu) or upon request. FISH probe sequences are listed in Table S1. All data necessary for confirming the conclusions of the article are present within the article and figures. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7455137.
Results and Discussion
Detection of dopaminergic, serotoninergic, tyraminergic, octopaminergic, and histaminergic neurons in the intact Drosophila CNS
We previously reported the identification of cholinergic, glutamatergic, and GABAergic neurons in whole-mount Drosophila brains by FISH with probes for Gad1, vGlut and ChAT mRNAs (Long et al. 2017). This method was based on hybridizing mRNA to multiple short, singly labeled oligonucleotides, combined with tissue-specific treatments for improved FISH in the Drosophila brain. To expand this approach to additional neurotransmitters, we developed and validated FISH probes to detect dopaminergic, serotoninergic, octopaminergic, tyraminergic, and histaminergic neurons by probing mRNAs that are specific to their synthesis or transport (Figure 1).
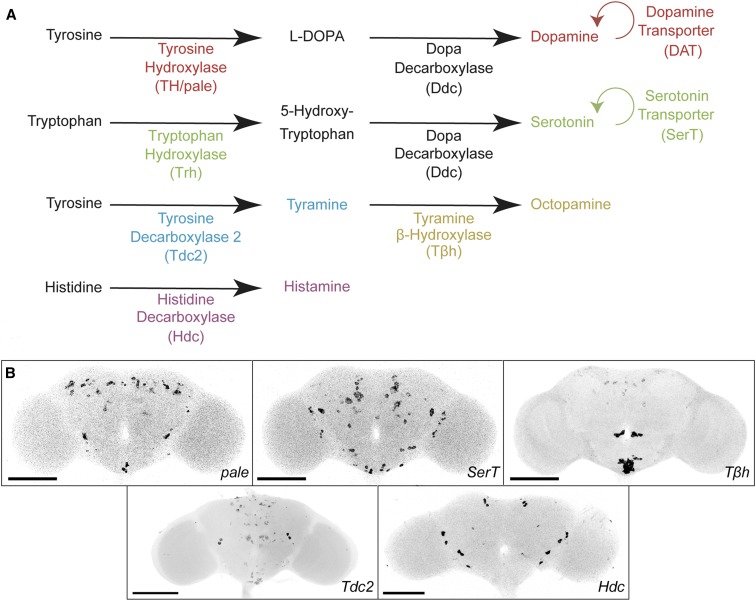
Figure 1.
Detection of dopaminergic, serotoninergic, tyraminergic, octopaminergic, and histaminergic neurons in whole-mount Drosophila brains using FISH. (A) Biosynthetic pathway of neurotransmitters. To detect neurotransmitter-specific cell types, FISH probes were targeted to mRNAs for specific pathways. Colors indicate pairs of targeted neurotransmitters and mRNAs. (B) Confocal images of TH/pale, SerT, Tβh, Tdc2, and Hdc expression patterns in the brain. CNS images of these samples are in Figure S1. Samples were imaged with a 20× objective and intensity values were inverted. Bar, 100 μm.
To identify dopaminergic neurons, we probed pale mRNA, encoding tyrosine hydroxylase (TH), a key enzyme for dopamine biosynthesis (Neckameyer and White 1993) (Figure 1A). We also probed DAT mRNA, encoding a dopamine transporter that mediates reuptake of dopamine from the synaptic cleft (Penmatsa et al. 2013; Wang et al. 2015). To validate our approach, we compared pale and DAT expression patterns with a GAL4 reporter line for tyrosine hydroxylase (TH-GAL4) expressing the HaloTag protein (Friggi-Grelin et al. 2003; Kohl et al. 2014; Sutcliffe et al. 2017). We observed widespread overlap (Figure 2A), with only a few exceptions, such as a small group of cells in the superior medial brain (Figure S2A).
TH-GAL4 was not observed in the dopaminergic protocerebral anterior medial (PAM) cluster or the medulla region of the optic lobe, but the pale probe was present, consistent with previous reports (Budnik and White 1988; Liu et al. 2012; Davie et al. 2018) (Figure 2B and File S8). On the other hand, lines R58E02-GAL4 and SS02425 have been reported to label the PAM cluster and dopaminergic Mi15 neurons in the optic lobe, respectively (Liu et al. 2012; Davis et al. 2018). We observed extensive colocalization of our pale FISH probes with the R58E02 and SS02425 driver lines (Figure 2, C and D, and File S9). We identified an average of 723 ± 92 pale-expressing neurons in the whole brain, including 257 + 11 across both central brain hemispheres and 218 ± 26 in a single optic lobe (n = 4 central brains and 7 optic lobes; count ± SE), consistent with previous reports (Mao and Davis 2009). Most pale-positive optic lobe neurons are small and had relatively weak FISH signals, hindering precise counts.
To identify serotoninergic neurons, we probed SerT mRNA, encoding a serotonin transporter that returns serotonin from the synaptic cleft to presynaptic neurons (Figure 1A) (Giang et al. 2011). To validate the probe, we compared it to three other reporters for serotonin expression: (1) SerT-GFP, in which the endogenous SerT locus was modified to express a SerT-GFP fusion protein (Venken et al. 2011; Nagarkar-Jaiswal et al. 2015); (2) a serotonin antibody, which has been reported to show identical cell-type specificity as the SerT protein (Giang et al. 2011); and (3) a Trh FISH probe. Trh mRNA encodes tryptophan hydroxylase, a key enzyme involved in serotonin synthesis (Figure 1A) (Neckameyer and White 1992; Coleman and Neckameyer 2005; Neckameyer et al. 2007). We observed consistent colocalization across several combinations of the reporters (Figure 2E and Figure S2, B and C), although the wide range and moderate background of antibody labeling makes its full coexpression less certain. In addition, we identified an average of 98 ± 12 cells in the brain with SerT FISH signal (n = 6; count ± SE), in agreement with previous reports (Vallés and White 1988). Together with previous work on SerT as a marker for serotoninergic cells (Giang et al. 2011), these results indicate that our SerT FISH probes specifically identify serotoninergic neurons.
Coexpression of serotonin and tyrosine hydroxylase proteins in PPL1 neurons was recently reported (Niens et al. 2017). To determine whether we can also detect these coexpressing neurons, we simultaneously probed pale and SerT mRNAs. We observed two pairs of neurons with consistent overlapping signals within the PPL1 cluster region (Figure S3A). We also observed SerT mRNA overlap with TH-GAL4/UAS-HaloTag (Figure S3B).
To identify tyraminergic and octopaminergic neurons, we probed Tdc2 (Tyrosine decarboxylase 2) and Tβh (Tyramine β hydroxylase) mRNAs. We identified an average of ∼116 ± 7 Tdc2 and 90 ± 5 Tβh neurons in the brain (n = 4–6; count ± SE). We observed Tdc2 expression in 19.0 ± 1.2 ventral and 13.3 ± 0.7 anterior large cell bodies, along with weakly expressing neurons in the dorsal and posterior brain. We identified Tβh expression in 23.4 ± 2.4 ventral and 10.5 ± 0.5 anterior large cell bodies, along with weakly expressing neurons in the lateral and posterior central brain.
The biosynthetic pathway of octopamine in neurons is controlled by both Tdc2 and Tβh: first Tdc2 converts tyrosine to tyramine, then Tβh converts tyramine to octopamine (Figure 1A). Thus, Tdc2 is predicted to label both tyraminergic and octopaminergic neurons, whereas Tβh should be specific to octopaminergic neurons. We examined the colocalization of Tdc2 and Tβh FISH probes with each other and Tdc2-GAL4/UAS-HaloTag, which labels many tyraminergic/octopaminergic neurons (Cole et al. 2005). We observed extensive overlap in the ventral and anterior brain, and more limited overlap in the dorsal and posterior brain (Figure 2F, Figure S3C, and File S11). Most of the nonoverlapping cells showed Tdc2 but not Tβh signals, and are therefore presumably tyraminergic, but a few weakly labeled cells appeared to only express Tβh. Cole et al. (2005) and Busch et al. (2009) also observed inconsistencies between Tdc2-GAL4 and octopamine immunoreactivity in the dorsal and posterior brain, including octopaminergic neurons not labeled by Tdc2-GAL4. Although the biosynthetic pathway would predict that all Tβh neurons also express Tdc2, this may not be universally true, although it could also be explained by a difference in timing of expression.
For the identification of histaminergic neurons, we probed Hdc mRNA, encoding histidine decarboxylase, which catalyzes the decarboxylation of histidine to form histamine (Figure 1A). The cell bodies of the most common histaminergic neurons in flies, photoreceptor cells, are located outside the brain and were removed during dissection. However, histamine is also present in some central brain neurons. In wild-type flies we identified an average of 20.8 ± 1.4 cells with a Hdc FISH signal (n = 9; count ± SE), with a distribution similar to previous reports (Figure 1B and Figure S4B) (Nässel 1999). Furthermore, we observed a decrease in Hdc FISH signal in HdcJK910 mutant flies (Figure S4, C and D) (Melzig et al. 1996) (Burg et al. 1993). Thus, our Hdc probe appears to specifically label histaminergic neurons.
Together, these results establish a FISH probe set for major neurotransmitter markers in Drosophila. Because all the tested probes showed the expected specificity, this also suggests that mRNA detection with our approach is a reliable tool for cell-type identification based on marker expression in general.
High-throughput FISH efficiently detects neurotransmitter markers
While our earlier detection of cholinergic, glutamatergic, and GABAergic neurons in a single brain showed the multiplex capability of the described FISH method, the approach is not optimized for processing large numbers of specimens simultaneously (Long et al. 2017). To increase throughput, we adapted an approach previously used for large-scale immunostaining, in which fly brains of different genotypes were mounted on a coverslip for parallel labeling (Wu et al. 2016). Multiple coverslips, each with up to 60 identified CNSs, can easily be moved between jars for most steps of the FISH process. We created a hybridization chamber with a minimal reaction volume, based on an existing plexiglass mounting dish design (Li et al. 2014; Wu et al. 2016). The chamber holds the samples in a space of ∼150 μl underneath the coverslip, which rests on spacers above the base of the chamber (Figure 3A). The coverslip processing approach maintains the original FISH procedure but is less laborious than the separate processing of individual brains (Figure S5).
We optimized the fluorophore combination for the neurotransmitter markers used in combination. Figure 3B show the fluorophores for each FISH probe set. The rationale behind this improved scheme is as follows. (1) We reserve the 488-nm laser channel for a cell identity marker, which gives the flexibility to use GFP and derivatives for cell labeling if needed. (2) CF594 works well in DPX-mounted Drosophila brain tissues for standard confocal microscopy. It gives a brighter signal and less cross talk than AF594. (3) DL550 performs similarly to Cy3. This gives flexibility to label probes with an alternative fluorophore if the other fluorophore is not available. (4) Fluorophores and FISH probes were paired to balance overall signal levels of each combination. We tested our fluorophore combination using the previously reported FISH probes for Gad1, vGlut and ChAT mRNAs. The similar expression patterns validated the optimized fluorophore combinations (Figure 3C and Files S1 and S2).
With the above optimization, we can simultaneously detect four molecular markers using standard confocal microscopes. To identify the neurotransmitter phenotype for a given cell type, we can rapidly screen through eight different neurotransmitter probes with three parallel FISH experiments. An example is shown in Figure 3C and D. We used a split-GAL4 driver (SS02565) to express a HaloTag reporter (UAS-HaloTag::CAAX) (Sutcliffe et al. 2017) in a specific cell population in the optic lobe. HaloTag was labeled with AF488 HaloTag ligand, then samples were colabeled with Gad1, vGlut and ChAT, or pale, Tβh, and SerT FISH probes in two separate experiments with the optimized fluorophore-labeling combination described above. The overlap between AF488 HaloTag ligand and DL550 ChAT FISH signals suggests that the neurons of interest are cholinergic, and thus likely to activate their immediate downstream targets (Figure 3C and E). Examples with two additional GAL4 lines are shown in Figure S6.
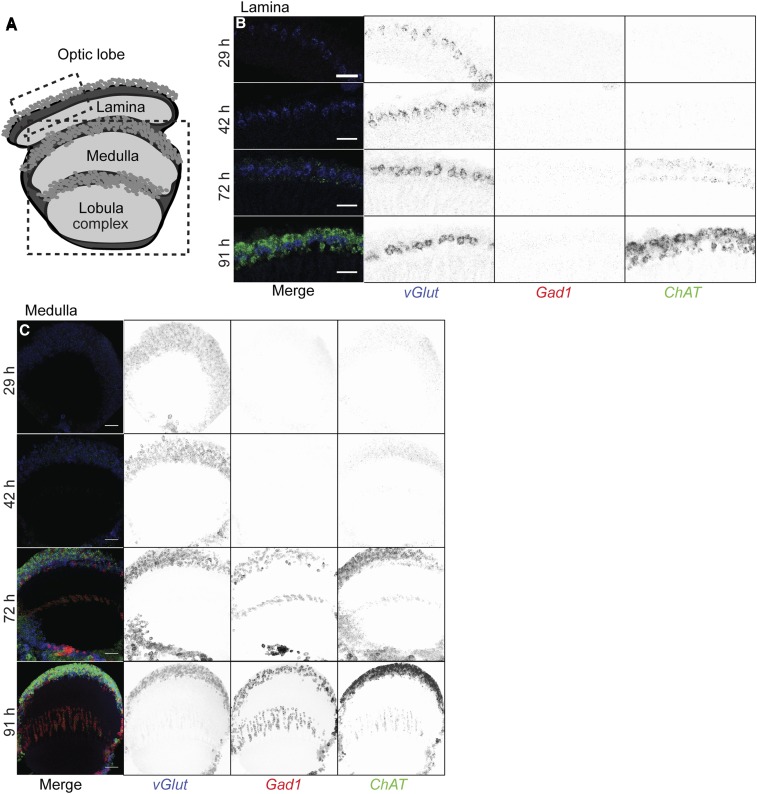
While we focused on the adult CNS, this FISH approach is also suitable to investigate neurotransmitter identity in larval and pupal brains. To illustrate the use of FISH for developing neurons, we examined the expression pattern of Gad1, vGlut and ChAT in the visual system at different pupal stages (Figure 4). All three mRNAs were already detectable during the early-to-midpupal stage, in agreement with RNAseq results showing pupal stage vGlut and ChAT expression in some optic lobe cell types (Tan et al. 2015). Interestingly, vGlut expression appeared earlier than the other two markers.
Figure 4.
Developmental time course of vGlut, Gad1 and ChAT expression in the optic lobe. (A) Schematics of optic lobe subregions. Boxes indicate approximate region of the lamina and medulla imaged. (B and C) Distribution of vGlut (Cy5; blue), Gad1 (CF594; red), and ChAT (DL550; green) transcripts in the lamina (B) or medulla (C) at different developmental stages (hours after puparium formation at 25°). Lamina images (B) show single 63× confocal sections. Bar, 20 µm. Medulla images (C) are maximum intensity projections through 25 z-planes for a total depth of 20 µm. Merged images are shown to the left, with split channels inverted in gray to the right. Bar, 10 µm.
Conclusions
In this study, we introduce a high-throughput, multiplexable FISH platform for the identification of eight different neurotransmitter cell types in the intact Drosophila CNS. The ability to identify neurotransmitter types reliably provides pivotal information for the understanding of neural circuit function. That being said, expression of the markers used here does not always indicate the transmitter phenotype of a cell [for example, a neuron might lack a biosynthetic enzyme but acquire a transmitter via uptake from the extracellular space, as reported for some GABAergic neurons in C. elegans (Gendrel et al. 2016)]. Similarly, other potential transmitters that are less-well characterized in flies [e.g., glycine (Frenkel et al. 2017)] are not covered by our probe set. However, these limitations are not specific to our FISH approach and could potentially be addressed in the future by examining additional mRNAs with the same methods.
With the coverslip-processing approach and optimized fluorophore labeling combinations, we can map neurotransmitter markers to specific cell types labeled with Drosophila genetic driver lines using standard confocal microscopy. While increasing numbers of neural cell types are being described anatomically, many of their transmitter phenotypes remain unknown. High-resolution images from this work can be used to suggest neurotransmitters within a region of interest, which can then be mapped to specific GAL4-labeled cells by colocalization with FISH. We expect that our FISH protocol for mapping neurotransmitters will find widespread use in Drosophila neuroscience studies.
The same FISH approach can be used to study other gene expression patterns in Drosophila. This may be of particular utility in the developing brain, where dynamic expression patterns require analyses of many different time points, as illustrated here by capturing early neurotransmitter marker expression in the visual system during development. Finally, our protocols could serve as a basis for further development by research groups interested in the analysis of gene expression in thick tissue sections (> 200 μm), which are comparable in thickness to whole-mount fly brains.
Acknowledgments
We thank Luke Lavis and Kaiyu Wang for comments on the manuscript; Kari Close and Alyson Petruncio for assistance performing the FISH procedure; other members of Janelia Project Technical Resources and FlyLight for assistance with CNS dissection; Jennifer Jeter, other members of FlyLight, and Damien Alcor for assistance with imaging; Michael Reiser for his contribution to the octopamine FISH probes; Kaiyu Wang and Barry Dickson for providing SS45407 and SS51118 GAL4 lines; the FlyLight Steering Committee (Gerry Rubin, Barry Dickson, Reed George, Wyatt Korff, and Jim Truman) for valuable suggestions; and Timothée Lionnet for the initial discussion of the project. R.H.S. received salary support from National Institutes of Health grant NS-083086. Funding for this work was provided by the Howard Hughes Medical Institute.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7455137.
Communicating editor: L. Luo
Literature Cited
- Budnik V., White K., 1988. Catecholamine-containing neurons in Drosophila melanogaster: distribution and development. J. Comp. Neurol. 268: 400–413. 10.1002/cne.902680309 [DOI] [PubMed] [Google Scholar]
- Burg M. G., Sarthy P. V., Koliantz G., Pak W. L., 1993. Genetic and molecular-identification of a Drosophila histidine-decarboxylase gene required in photoreceptor transmitter synthesis. EMBO J. 12: 911–919. 10.1002/j.1460-2075.1993.tb05732.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch S., Selcho M., Ito K., Tanimoto H., 2009. A map of octopaminergic neurons in the Drosophila brain. J. Comp. Neurol. 513: 643–667. 10.1002/cne.21966 [DOI] [PubMed] [Google Scholar]
- Buxbaum A. R., Haimovich G., Singer R. H., 2015. In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 16: 95–109 (erratum: Nat. Rev. Mol. Cell Biol. 16: 513). 10.1038/nrm3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. H., Carney G. E., McClung C. A., Willard S. S., Taylor B. J., et al. , 2005. Two functional but noncomplementing Drosophila tyrosine decarboxylase genes: distinct roles for neural tyramine and octopamine in female fertility. J. Biol. Chem. 280: 14948–14955. 10.1074/jbc.M414197200 [DOI] [PubMed] [Google Scholar]
- Coleman C. M., Neckameyer W. S., 2005. Serotonin synthesis by two distinct enzymes in Drosophila melanogaster. Arch. Insect Biochem. Physiol. 59: 12–31. 10.1002/arch.20050 [DOI] [PubMed] [Google Scholar]
- Croset V., Treiber C. D., Waddell S., 2018. Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. Elife 7: e34550. 10.7554/eLife.34550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie K., Janssens J., Koldere D., De Waegeneer M., Pech U., et al. , 2018. A single-cell transcriptome atlas of the aging Drosophila brain. Cell 174: 982–998.e20. 10.1016/j.cell.2018.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis F. P., Nern A., Picard S., Reiser M. B., Rubin G. M., et al. , 2018. A genetic, genomic, and computational resource for exploring neural circuit function. bioRxiv. Available at https//.org/10.1101/385476. 10.1101/385476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne H., Hibbard K. L., Cavallaro A., Kao J. C., Rubin G. M., 2018. Genetic reagents for making split-GAL4 lines in Drosophila. Genetics 209: 31–35. 10.1534/genetics.118.300682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel L., Muraro N. I., Gonzalez A. N. B., Marcora M. S., Bernabo G., et al. , 2017. Organization of circadian behavior relies on glycinergic transmission. Cell Rep. 19: 72–85. 10.1016/j.celrep.2017.03.034 [DOI] [PubMed] [Google Scholar]
- Friggi-Grelin F., Coulom H., Meller M., Gomez D., Hirsh J., et al. , 2003. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J. Neurobiol. 54: 618–627. 10.1002/neu.10185 [DOI] [PubMed] [Google Scholar]
- Fritschy J. M., 2008. Is my antibody-staining specific? How to deal with pitfalls of immunohistochemistry. Eur. J. Neurosci. 28: 2365–2370. 10.1111/j.1460-9568.2008.06552.x [DOI] [PubMed] [Google Scholar]
- Gendrel M., Atlas E. G., Hobert O., 2016. A cellular and regulatory map of the GABAergic nervous system of C. elegans. Elife 5: e17686. 10.7554/eLife.17686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giang T., Ritze Y., Rauchfuss S., Ogueta M., Scholz H., 2011. The serotonin transporter expression in Drosophila melanogaster. J. Neurogenet. 25: 17–26. 10.3109/01677063.2011.553002 [DOI] [PubMed] [Google Scholar]
- Grimm J. B., English B. P., Chen J. J., Slaughter J. P., Zhang Z. J., et al. , 2015. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12: 244–250. 10.1038/nmeth.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry G. L., Davis F. P., Picard S., Eddy S. R., 2012. Cell type-specific genomics of Drosophila neurons. Nucleic Acids Res. 40: 9691–9704. 10.1093/nar/gks671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl J., Ng J., Cachero S., Ciabatti E., Dolan M. J., et al. , 2014. Ultrafast tissue staining with chemical tags. Proc. Natl. Acad. Sci. USA 111: E3805–E3814. 10.1073/pnas.1411087111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinides N., Rossi A. M., Desplan C., 2015. Common temporal identity factors regulate neuronal diversity in fly ventral nerve cord and mouse retina. Neuron 85: 447–449. 10.1016/j.neuron.2015.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., et al. , 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187. 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Lein E., Borm L. E., Linnarsson S., 2017. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358: 64–69. 10.1126/science.aan6827 [DOI] [PubMed] [Google Scholar]
- Li H. H., Kroll J. R., Lennox S. M., Ogundeyi O., Jeter J., et al. , 2014. A GAL4 driver resource for developmental and behavioral studies on the larval CNS of Drosophila. Cell Rep. 8: 897–908. 10.1016/j.celrep.2014.06.065 [DOI] [PubMed] [Google Scholar]
- Liu C., Placais P. Y., Yamagata N., Pfeiffer B. D., Aso Y., et al. , 2012. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature 488: 512–516. 10.1038/nature11304 [DOI] [PubMed] [Google Scholar]
- Long X., Colonell J., Wong A. M., Singer R. H., Lionnet T., 2017. Quantitative mRNA imaging throughout the entire Drosophila brain. Nat. Methods 14: 703–706. 10.1038/nmeth.4309 [DOI] [PubMed] [Google Scholar]
- Luan H., Peabody N. C., Vinson C. R., White B. H., 2006. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52: 425–436. 10.1016/j.neuron.2006.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z., Davis R. L., 2009. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front. Neural Circuits 3: 5 10.3389/neuro.04.005.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner G. W., Grimm J. B., Johnston R. M., Sutcliffe B., Ng J., et al. , 2018. Optimization of fluorophores for chemical tagging and immunohistochemistry of Drosophila neurons. PLoS One 13: e0200759 10.1371/journal.pone.0200759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzig J., Buchner S., Wiebel F., Wolf R., Burg M., et al. , 1996. Genetic depletion of histamine from the nervous system of Drosophila eliminates specific visual and mechanosensory behavior. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 179: 763–773. 10.1007/BF00207355 [DOI] [PubMed] [Google Scholar]
- Moffitt J. R., Hao J., Bambah-Mukku D., Lu T., Dulac C., et al. , 2016. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc. Natl. Acad. Sci. USA 113: 14456–14461. 10.1073/pnas.1617699113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkar-Jaiswal S., Lee P. T., Campbell M. E., Chen K., Anguiano-Zarate S., et al. , 2015. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife 4: e05338. 10.7554/eLife.05338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel D. R., 1999. Histamine in the brain of insects: a review. Microsc. Res. Tech. 44: 121–136. [DOI] [PubMed] [Google Scholar]
- Neckameyer W. S., White K., 1992. A single locus encodes both phenylalanine hydroxylase and tryptophan hydroxylase activities in Drosophila. J. Biol. Chem. 267: 4199–4206. [PubMed] [Google Scholar]
- Neckameyer W. S., White K., 1993. Drosophila tyrosine hydroxylase is encoded by the pale locus. J. Neurogenet. 8: 189–199. 10.3109/01677069309083448 [DOI] [PubMed] [Google Scholar]
- Neckameyer W. S., Coleman C. M., Eadie S., Goodwin S. F., 2007. Compartmentalization of neuronal and peripheral serotonin synthesis in Drosophila melanogaster. Genes Brain Behav. 6: 756–769. 10.1111/j.1601-183X.2007.00307.x [DOI] [PubMed] [Google Scholar]
- Niens J., Reh F., Coban B., Cichewicz K., Eckardt J., et al. , 2017. Dopamine modulates serotonin innervation in the Drosophila brain. Front. Syst. Neurosci. 11: 76 10.3389/fnsys.2017.00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmatsa A., Wang K. H., Gouaux E., 2013. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature 503: 85–90. 10.1038/nature12533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Ngo T. T., Hibbard K. L., Murphy C., Jenett A., et al. , 2010. Refinement of tools for targeted gene expression in Drosophila. Genetics 186: 735–755. 10.1534/genetics.110.119917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., van den Bogaard P., Rifkin S. A., van Oudenaarden A., Tyagi S., 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5: 877–879. 10.1038/nmeth.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S., Lubeck E., Zhou W., Cai L., 2016. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 92: 342–357. 10.1016/j.neuron.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe B., Ng J., Auer T. O., Pasche M., Benton R., et al. , 2017. Second-generation Drosophila chemical tags: sensitivity, versatility, and speed. Genetics 205: 1399–1408. 10.1534/genetics.116.199281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Zhang K. X., Pecot M. Y., Nagarkar-Jaiswal S., Lee P. T., et al. , 2015. Ig superfamily ligand and receptor pairs expressed in synaptic partners in Drosophila. Cell 163: 1756–1769. 10.1016/j.cell.2015.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirian L., Dickson B., 2018. The VT GAL4, LexA, and split-GAL4 driver line collections for targeted expression in the Drosophila nervous system. bioRxiv. Available at https//.org/10.1101/198648. 10.1101/198648 [DOI] [Google Scholar]
- Vallés A. M., White K., 1988. Serotonin-containing neurons in Drosophila melanogaster: development and distribution. J. Comp. Neurol. 268: 414–428. 10.1002/cne.902680310 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Schulze K. L., Haelterman N. A., Pan H., He Y., et al. , 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8: 737–743. 10.1038/nmeth.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. H., Penmatsa A., Gouaux E., 2015. Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature 521: 322–327. 10.1038/nature14431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Nern A., Williamson W. R., Morimoto M. M., Reiser M. B., et al. , 2016. Visual projection neurons in the Drosophila lobula link feature detection to distinct behavioral programs. Elife 5: e21022. 10.7554/eLife.21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Titlow J., Ennis D., Smith C., Mitchell J., et al. , 2017. Single molecule fluorescence in situ hybridisation for quantitating post-transcriptional regulation in Drosophila brains. Methods 126: 166–176. 10.1016/j.ymeth.2017.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Kilman V. L., Keegan K. P., Peng Y., Emery P., et al. , 2003. Drosophila clock can generate ectopic circadian clocks. Cell 113: 755–766. 10.1016/S0092-8674(03)00400-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All fly strains are available in the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu) or upon request. FISH probe sequences are listed in Table S1. All data necessary for confirming the conclusions of the article are present within the article and figures. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7455137.