Abstract
Mitochondrial genome variation and its effects on phenotypes have been widely analyzed in higher eukaryotes but less so in the model eukaryote Saccharomyces cerevisiae. Here, we describe mitochondrial genome variation in 96 diverse S. cerevisiae strains and assess associations between mitochondrial genotype and phenotypes as well as nuclear-mitochondrial epistasis. We associate sensitivity to the ATP synthase inhibitor oligomycin with SNPs in the mitochondrially encoded ATP6 gene. We describe the use of iso-nuclear F1 pairs, the mitochondrial genome equivalent of reciprocal hemizygosity analysis, to identify and analyze mitochondrial genotype-dependent phenotypes. Using iso-nuclear F1 pairs, we analyze the oligomycin phenotype-ATP6 association and find extensive nuclear-mitochondrial epistasis. Similarly, in iso-nuclear F1 pairs, we identify many additional mitochondrial genotype-dependent respiration phenotypes, for which there was no association in the 96 strains, and again find extensive nuclear-mitochondrial epistasis that likely contributes to the lack of association in the 96 strains. Finally, in iso-nuclear F1 pairs, we identify novel mitochondrial genotype-dependent nonrespiration phenotypes: resistance to cycloheximide, ketoconazole, and copper. We discuss potential mechanisms and the implications of mitochondrial genotype and of nuclear-mitochondrial epistasis effects on respiratory and nonrespiratory quantitative traits.
Keywords: Saccharomyces cerevisiae, mitochondrial genome variation, phenotypic variation, transgression, nuclear-mitochondrial epistasis, Introgression
MITOCHONDRIAL genome polymorphisms and nuclear-mitochondrial epistasis have been extensively studied in multicellular model organisms (Ballard and Melvin 2010; Joseph et al. 2013) and in humans. In humans, polymorphisms in both the maternally inherited mitochondrial genome and the nuclear genome are major sources of drug-induced and inherited mitochondrial diseases (Carelli et al. 2003; Thorburn 2004; Dimauro and Davidzon 2005; Taylor and Turnbull 2005; Graziewicz et al. 2006; Mancuso et al. 2007; Barnhill et al. 2012; Schapira 2012; Singh et al. 2014; Lodi et al. 2015). Relative to its size (16,569 bp; 37 genes), human mitochondrial genome polymorphisms are responsible for a large proportion of these mitochondrial diseases. The large proportion of mitochondrial genotype-dependent human mitochondrial diseases may reflect nuclear-mitochondrial epistasis that arises because all mitochondrially encoded RNAs and proteins form complexes with at least one, and often multiple, nuclearly encoded proteins. Both the human and Saccharomyces cerevisiae mitochondrial genomes contain a small subset of the genes required for mitochondrial translation and for respiration and in both species most of the 500–1500 member mitochondrial proteome is nuclearly encoded (Steinmetz et al. 2002a; Sickmann et al. 2003; Perocchi et al. 2008; Rhee et al. 2013). Thus, S. cerevisiae is an excellent model for studies of human mitochondrial and nuclear-mitochondrial gene product functions (Steinmetz et al. 2002a; Schwimmer et al. 2006; Perocchi et al. 2008; Baile and Claypool 2013; Rutter and Hughes 2015), as well as for nuclear-mitochondrial pathogenic polymorphisms (Stumpf and Copeland 2011; Montanari et al. 2013; Kabala et al. 2014; Lodi et al. 2015).
In addition to being a model for mitochondrial and nuclear-mitochondrial gene product functions, S. cerevisiae is a well-established model for population and quantitative genetics studies (Liti and Louis 2012; Fay 2013; Strope et al. 2015; Peter et al. 2018). However, S. cerevisiae population and quantitative genetics studies have almost exclusively focused on nuclear genome sequences and phenotypic contributions with relatively few studies considering the phenotypic contributions of mitochondrial genome variation (Codón et al. 1995; Dimitrov et al. 2009; Edwards et al. 2014; Paliwal et al. 2014; Wolters et al. 2018). We previously described the nuclear genome sequences, phenotypes, and nuclear genotype-phenotype associations in the 100-genomes collection of S. cerevisiae strains (Strope et al. 2015). In this work, we annotate the mitochondrial genomes of 93 S. cerevisiae strains sequenced by Strope et al. (2015). In addition, we test for associations of previously determined phenotypes (Strope et al. 2015) and novel respiration phenotypes with S. cerevisiae mitochondrial genome variation. Finally, we identify mitochondrial genotype-dependent contributions to multiple respiration and nonrespiration phenotypes, as well as extensive nuclear-mitochondrial epistasis.
Materials and Methods
Strains
The S. cerevisiae strains listed in Supplemental Material, Table S1 have been deposited in and should be requested from the Fungal Genetics Stock Center (http://www.fgsc.net). For additional descriptions of the sequenced S. cerevisiae strains or genetic backgrounds in Table S1, see Strope et al. (2015).
Mitochondrial genome assemblies, annotation, and phylogenies
We isolated and sequenced genomic DNA from 93 S. cerevisiae strains as described previously (Strope et al. 2015). We assembled the mitochondrial genomes from the 93 S. cerevisiae strains using the de novo assembler ABySS (v.1.3.4), with parameters “k” (the k-mer length), “n” (the minimum number of pairs needed to join contigs), and “c” (the minimum mean k-mer coverage of a unitig) optimized for each strain. Assembly of mitochondrial genomes was carried out using a range of parameters, using ABySS, and also velvet, with the goal of obtaining complete genomes. In most cases, multiple assemblies were combined. In some cases, it was necessary to use an exhaustive assembly algorithm (pondslime) that uses quality scores, to overcome the problems of assembly of highly AT-rich sequences with numerous sequence errors. For the mitochondrial genomes, N50 scores in the 5–15 kb range were generally good. Higher N50 scores often involved chimeric assemblies due to inappropriate joining of AT-rich runs by the nonquality score–based assemblers. Once the mitochondrial genome assemblies were complete, they were checked using Pilon (Broad Institute) as well as checked for paired-end pairing errors, “gene errors” (whether the eight conserved protein-encoding genes and the ribosomal rDNA/transfer RNA (tRNA) encoding genes were all present and intact), and completeness. Completeness was determined using BWA to generate a .sam file for each set of sequences aligned against the genome assembled from it. Sequence reads not aligning were assembled (velvet) and used to determine if pieces of the mitochondrial genomes had been inadvertently left out.
We identified mitochondrial sequences by high read depths relative to single copy nuclear genes and by homology to previously sequenced Saccharomyces mitochondrial genomes, genes, and introns. We also used BLAST, ssearch36, LAGAN, EMBOSS tools, and Perl scripts to identify mitochondrial genes and sequence polymorphisms relative to the reference S288c mitochondrial genome. Table files were created with the mitochondrial gene coordinates for each strain. We used the NCBI tool tbl2asn (http://www.ncbi.nlm.nih.gov/genbank/tbl2asn2/) to annotate the mitochondrial genome of each strain.
We used BWA and samtools to estimate the mitochondrial genome copy number of these 93 S. cerevisiae strains, which were all grown under the same conditions, by read depths of the mitochondrial 21S recombinant DNA gene relative to MDN1, a single-copy nuclear gene. 21S was used for estimating mitochondrial depth of coverage because it is by far the largest mitochondrial gene (4438 bp with and 3731 bp without the SCE1 intron), is found in all mitochondrial genomes, and is conserved. MDN1 was chosen as the nuclear marker for estimating depth of coverage because it is the largest protein coding gene in the nuclear genome and does not contain repetitive sequences.
The mitochondrial genome sequences of 128 strains in 8 Saccharomyces species (Foury et al. 1998; Fritsch et al. 2014; Strope et al. 2015; Leducq et al. 2017; Sulo et al. 2017) were screened for ATP6 and ENS2 gene sequences. Evolutionary histories of these genes were inferred by using the maximum likelihood (ML) method, using MEGA6 (Tamura et al. 2013). For each gene family, we calculated Bayesian information criterion (BIC) scores for 24 different nucleotide substitution models using their sequence alignments in MEGA6 (Tamura et al. 2013). The nucleotide substitution model with the lowest BIC scores are considered to describe the substitution pattern the best, which was used in the ML phylogenetic inference. Specifically, the Hasegawa–Kishino–Yano model (Hasegawa et al. 1985) was used for ATP6 and the Tamura 3-parameter model (Tamura 1992) was used for ENS2. Nonuniformity of evolutionary rates among sites were modeled by using a discrete γ-distribution (+G) with five rate categories, and by assuming that a certain fraction of sites were evolutionarily invariable (+I).
Iso-nuclear F1 strain construction
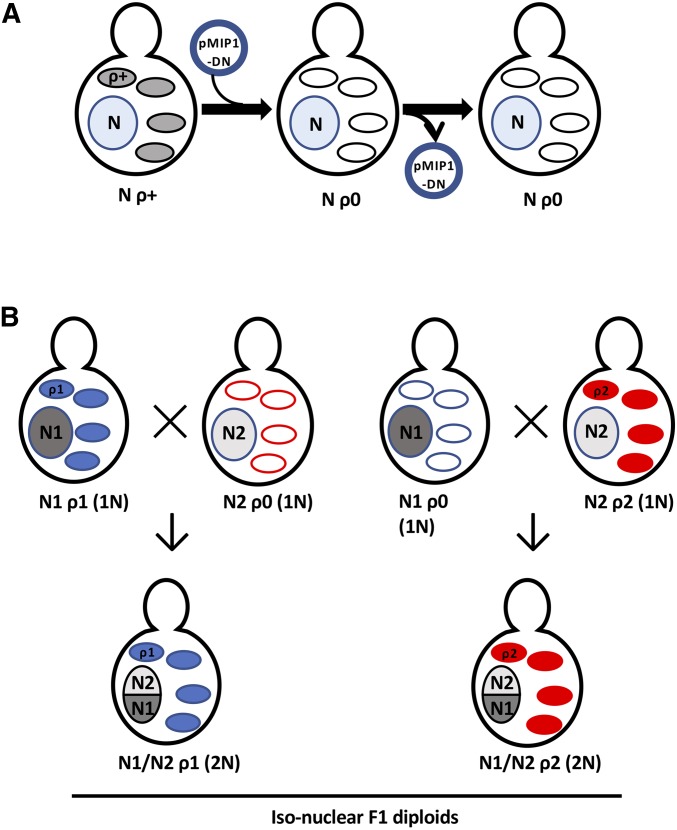
We constructed iso-nuclear (i.e., isogenic nuclear genomes) F1 pairs, the mitochondrial genome equivalent of reciprocal hemizygosity analysis (Steinmetz et al. 2002b), to identify mitochondrial genotype-dependent phenotypes and to assess nuclear-mitochondrial epistasis. To construct iso-nuclear F1 pairs, we first used the MIP1DN-containing plasmid pLND46 (Dimitrov et al. 2009) to eliminate the mitochondrial genome from haploid ρ+ strains (Table S1); for each haploid ρ+ strain, we independently generated two ρ0 derivatives. The complete absence of mitochondrial DNA in ρ0 strains was confirmed by the absence of mitochondrial nucleoids (MacAlpine et al. 2000). We crossed each of two independently generated ρ0 haploid strains, from which pLND46 had been lost, with haploid ρ+ strains from different genetic backgrounds to create two independent pairs of iso-nuclear F1 diploids: N1 ρ1 × N2 ρ0 → N1/N2 ρ1 and N1 ρ0 × N2 ρ2 → N1/N2 ρ2. We compared the phenotypes of iso-nuclear F1 diploids to identify mitochondrial genotype-dependent contributions to phenotypes (Figure 1).
Figure 1.
Generation and phenotypic comparison of iso-nuclear F1 diploids. (A) The MIP1DN- and centromere-containing plasmid pLND46 (Dimitrov et al. 2009) (pMIP1-DN) was introduced into haploid ρ+ strains. MIP1DN induction led to the complete loss of the mitochondrial genome, generating ρ0 petites that were then screened for plasmid loss. Nucleus, N-containing blue circle; ovals, parental ρ+ (shaded) and ρ0 (open) mitochondria. (B) Pairwise crosses between haploid ρ+ strains and ρ0 strains of opposite mating types differing in nuclear (N1 vs. N2) and mitochondrial (ρ1 vs. ρ2) DNA backgrounds generated pairs of iso-nuclear F1 diploids (N1/N2) that carried the parental mitochondrial genomes (ρ1 or ρ2), thus allowing for systematic phenotypic comparison of distinct mitochondrial genomes. Mitochondria: solid blue ovals, ρ1; solid red ovals, ρ2; open ovals, ρ0.
Media and phenotypic analysis
We performed high-throughput phenotypic analysis of diploid S. cerevisiae strains as described previously (Strope et al. 2015). Briefly, we grew strains in 80 µl yeast peptone dextrose (YPD) (1% yeast extract, 2% bacto peptone, 2% dextrose) in 384-well plates at 30° for 48 hr. We arrayed strains by robotic pinning (BM5 robot; S&P Robotics) onto rectangular agar plates (catalog number 78116; Greiner Bio-One), at a density of 1536 spots or colonies per plate. To minimize position and neighbor effects, we arrayed each strain in a 6 × 4 block of colonies, with the eight internal colonies from each 6 × 4 block being used to determine phenotypes. We incubated plates at 30° for 1–4 days, unless otherwise specified, that we digitally imaged at 24-hr intervals using the BM5 robot digital camera. We quantified digital images of colony areas using ImageJ 1.47v (http://imagej.nih.gov/ij/index.html) with a Patch Detector Plus plug-in (University of Graz Microscopy Facility; http://microscopy.uni-graz.at/index.php?item=new1). We used the eight internal colonies from each 6 × 4 block to calculate median colony areas for each strain on experimental and control plates; phenotypes were subsequently quantified using the ratio of colony size under experimental vs. control conditions. Iso-nuclear F1 strain pairs were analyzed similarly, except for each being arrayed in 3 × 4 blocks. We also phenotypically analyzed some sets of strains by 10-fold spot dilutions (initial cell density: 107 cells/ml) onto control and experimental media to assess phenotypes.
Phenotypes were determined on plate media containing 2% agar. Ethanol, the utilization of which requires respiratory competence, and all inhibitors were added to media after autoclaving. Sensitivity to inhibitors of respiration/mitochondrial functions were determined on yeast peptone ethanol (YPE) (1% yeast extract, 2% bacto peptone, 2% ethanol) and/or SE synthetic ethanol (SE) (0.67% yeast nitrogen base without amino acids, 2% ethanol) plates . Growth at low (15°; up to 7 days incubation) and high (39°; up to 7 days incubation) temperatures (30° Control) was tested on YPD, YPEG (YPE + 2% glycerol), synthetic dextrose (SD) (0.67% yeast nitrogen base without amino acids, 2% dextrose), and SEG (SE + 2% glycerol) plates. Sensitivity to nonrespiration inhibitors (cycloheximide, ketoconazole, copper) was determined on YPD and SD plates. Inhibitors, inhibitor concentrations, media used, inhibitor targets, and mitochondrial genes in which resistance mutations previously have been identified are listed in Table S2.
Genotype-phenotype associations
We tested for associations between nuclear genotypes and respiration/mitochondrial inhibitor phenotypes (Table S3), as described previously (Strope et al. 2015). We tested for associations between genotypes at 180 mitochondrial genome sites (Table S4) in 96 S. cerevisiae strains and previously determined phenotypes (Strope et al. 2015), as well as respiration/mitochondrial inhibitor phenotypes (Table S3). Mitochondrial genotypes in the 96 S. cerevisiae mitochondrial genome sequences examined included only variation at biallelic sites with a minor allele frequency of ≥5%.
The program GEMMA version 0.94beta (Zhou and Stephens 2012) was used to conduct association tests. This program takes a linear mixed-model approach to controlling population structure using a relatedness matrix (normally constructed from genotype data using GEMMA). To ensure that this matrix accurately reflected relatedness between strains, we constructed the matrix using 171,345 nuclear and mitochondrial biallelic SNPs with minor allele frequency ≥5% (Strope et al. 2015). To establish significance of genotype-phenotype associations, we used a threshold of P < 4 × 10−7, which was used by Strope et al. (2015) and corresponds to an approximate Bonferroni correction for association mapping using whole-genome genotypes.
Strain and plasmid availability
All strains listed in Table S1 of this work have been deposited into and are available from the Fungal Genetics Stock Center. All plasmids generated as part of this work have been deposited into and are available from Addgene (http://www.addgene.org/John_McCusker/). The MIP1DN-containing plasmid pLND46 (Dimitrov et al. 2009) was obtained from, and should be requested from, D. Gottschling (CalicoLabs).
Data availability
All S. cerevisiae nuclear and mitochondrial genome sequence data has been previously published and deposited (https://doi.org/10.1101/gr.185538.114); see Table S19 of Strope et al. (2015) for GenBank accession and Sequence Read Archive numbers. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7361240.
Results
S. cerevisiae mitochondrial genome assemblies, sizes, copy numbers, and gene/intron annotations
We assembled the mitochondrial genomes from 93 S. cerevisiae strains (Strope et al. 2015). With the likely exception of YJM1242, where read depths suggested that the region encompassing the mitochondrial genes tM(CAU)Q1-COX2-RF1-tF(GAA)Q-tT(UAG)Q2-tV(UAC)Q-COX3-tM(CAU)Q2 may be duplicated, S. cerevisiae mitochondrial genome sizes ranged from 73,450 to 92,176 bp (median = 82,308 bp) (Table S5). With the possible exception of the aforementioned YJM1242, the S. cerevisiae mitochondrial genomes had the same gene order as the S288c mitochondrial genome (Foury et al. 1998). Mitochondrial genome copy number in the 93 S. cerevisiae strains ranged from 11 to 77 copies (median = 24) (Table S5). There was a negative association between mitochondrial genome size and copy number (Pearson’s correlation = −0.31; P = 0.00264). We annotated mitochondrial genes (21S, 15S, RPM1, VAR1, COX1, COX2, COX3, COB, ATP6, ATP8, OLI1, RF1, ENS2, and 24 tRNAs) in the 93 S. cerevisiae (Strope et al. 2015) and the three previously published S. cerevisiae mitochondrial genome sequences (Foury et al. 1998; Fritsch et al. 2014) for SNPs and indels (Table S6). We also annotated introns, the presence of which varied widely between strains and in their contributions to mitochondrial genome sizes (Table S7).
ENS2 and ATP6 genotype-phenotype associations and phylogenies
None of the previously determined 49 phenotypes of the 100-genomes strains (Strope et al. 2015), including those requiring respiratory competence [i.e., growth on ethanol/glycerol (15, 30, and 39°; rich and defined media) and sporulation (multiple media; 25 and 30°)], showed mitochondrial genotype associations. Thus, for closer focus on the mitochondrial genome, we determined 14 additional phenotypes on media with ethanol or ethanol plus glycerol as the sole carbon source(s), the utilization of which requires respiratory competence (Table S2 and Table S3). With respect to nuclear genotypes, we found multiple phenotype associations (Table S8), including oligomycin sensitivity with loss-of-function polymorphisms in the general stress response regulator-encoding gene WHI2, the analysis of which is described in the Supplemental Material.
For these additional 14 respiration phenotypes, we found two mitochondrial genotype associations, both for sensitivity to the ATP synthase inhibitor oligomycin. First, the presence of ENS2 sequences associated with oligomycin sensitivity (Table S8). When functional, the mobile ENS2 gene encodes a site-specific endonuclease that has a 26 bp recognition sequence in the ATP6 ORF (Nakagawa et al. 1992). We annotated ENS2 (Nakagawa et al. 1992) in the 96 S. cerevisiae strains for presence/absence and, when present, for full-length, potentially functional ENS2 ORFs, SNPs, and indels (Table S6). In contrast to the presence/absence association, there was no oligomycin association with full-length, potentially functional ENS2 ORFs (Table S9).
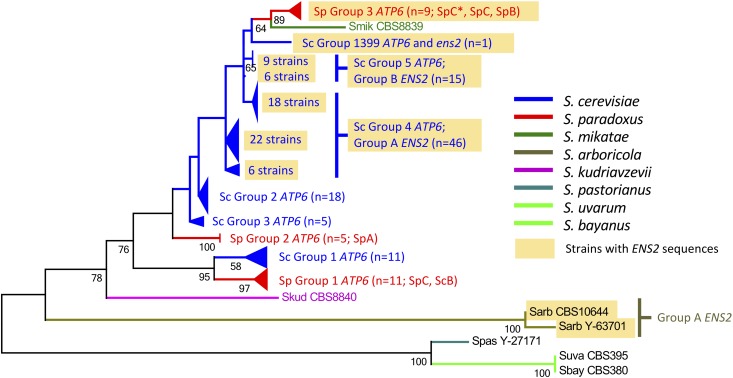
We searched the mitochondrial genome sequences of seven other Saccharomyces species (Leducq et al. 2017; Sulo et al. 2017) and identified ens2 sequences in Saccharomyces arboricola and Saccharomyces paradoxus (Figure 2). Similar to S. cerevisiae (Figure 3 and Table S6), ENS2 was located immediately downstream of ATP6 in S. arboricola (two of two strains) as well as in some S. paradoxus (9 of 25) strains; there was evidence of ENS2 introgression (Figure 1, Figure S1, and Figure S2). We determined the ENS2 phylogeny and identified ENS2 group A, group B, group 1399, and group S. paradoxus; full-length, potentially functional ENS2 ORFs were present only in S. cerevisiae (Figure 2, Figure S1, and Table S6). There was evidence of ENS2 introgression (Figure 1 and Figure S1).
Figure 2.
Compressed ATP6 molecular phylogeny in Saccharomyces sensu stricto species. ATP6 genes were identified from the mitochondrial genome sequences of 128 strains in 8 S. sensu stricto species examined, sequences of which were obtained from Strope et al. (2015), Leducq et al. (2017), Sulo et al. (2017). The rate variation model allowed for some sites to be evolutionarily invariable (+I; 84.3517% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All codon positions were included for analyses. All positions containing gaps and missing data were eliminated. Light brown shading denotes ENS2-containing groups. SpA, SpB, SpC, and SpC* correspond to the S. paradoxus population nomenclature of Leducq et al. (2017). The three S. paradoxus ATP6 groups (S. paradoxus groups 1 ATP6 ens20, 2 ATP6 ens20, and 3 ATP6-ens2) and six S. cerevisiae ATP6 groups (S. cerevisiae group 1 ATP6 ens20, group 2 ATP6 ens20, group 3 ATP6 ens20, group 4 ATP6-ENS2, group 5 ATP6-ENS2, and group 1399 ATP6-ens2) are labeled. For the strains that comprise the three S. paradoxus ATP6 groups and the six S. cerevisiae ATP6 groups, see Figure S1, Figure S2, and Table S6.
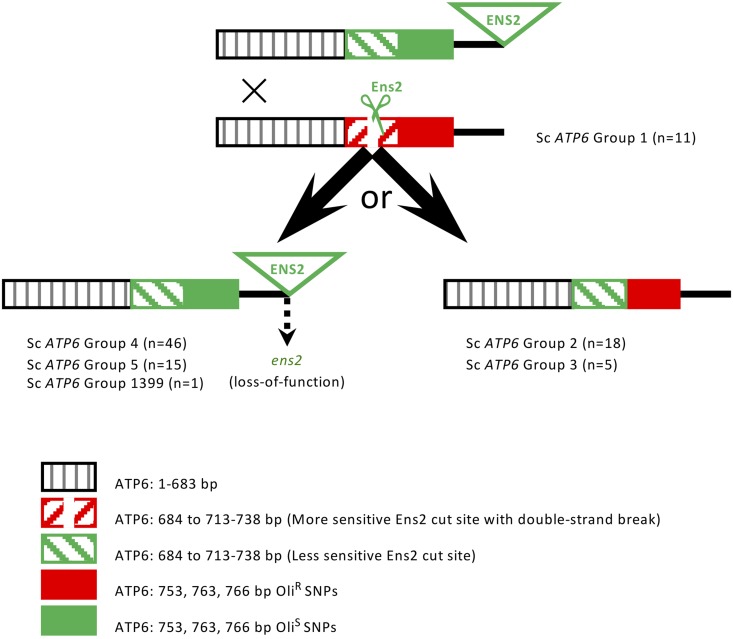
Figure 3.
ATP6-ENS2 vs. ATP6-ens20 structures and deduced ⍴+ ATP6-ENS2 ✕ ⍴+ ATP6-ens20 recombination products. Structures of ATP6-ENS2 [ATP6: less-sensitive Ens2 recognition sequence (713–738 bp) and OliS SNPs (753, 763, 766 bp); ENS2 with full-length ORF] and S. cerevisiae group 1 ATP6 ens20 [ATP6: more-sensitive Ens2 recognition sequence (713–738 bp) and OliR SNPs (753, 763, 766 bp); no ENS2 sequences] are shown. The Ens2 recognition sequences, and their relative sensitivities to Ens2, are as previously identified (Nakagawa et al. 1992). The distance between the most distal of the Ens2 recognition sequence SNPs (738 bp) and the most proximal of the oligomycin phenotype-associated SNPs (753 bp) is 15 bp. The distance between the most distal of the oligomycin phenotype-associated SNPs (766 bp) and the 5′ end of ENS2 ORF that, with the exception of YJM1399 (64 bp), is 75–76 bp downstream of the TAA stop codon of the 780 bp ATP6 ORF, is 89–90 bp. Recombination breakpoints proximal to the Ens2 recognition sequences are proposed to be proximal to the 684 and 693 bp SNPs based on linkage disequilibrium (Table S6). ATP6 and ENS2 genotypes and deduced recombination products can be seen in tabular form in Table S6. ENS2 loss-of-function is proposed to occur postmating. For further details, see Discussion.
Oligomycin phenotype also associated with SNPs in the ORF of the ATP synthase subunit-encoding gene ATP6 (Table S8). The 753, 763, and 766 bp SNPs in the ATP6 ORF most highly associated with oligomycin phenotype were in linkage disequilibrium. We annotated the ATP synthase subunit-encoding gene ATP6 in the 96 S. cerevisiae mitochondrial genome sequences and identified six groups (S. cerevisiae groups 1–5 and 1399) (Figure 1, Figure S1, Figure S2, and Table S6).
We examined the ATP6 ORFs of the 96 S. cerevisiae strains for the more sensitive Ens2 recognition sequence previously identified in an ens20 (i.e., no ens2 sequences) strain (TCATTCAGGGATATGTGTGGGCTATT) and the less-sensitive Ens2 recognition sequence previously identified in an ENS2-containing strain (TTATCCAATCTTATGTTTGACTTATC) (Table S6) (Nakagawa et al. 1992). The three S. cerevisiae ENS2 groups (A, B, and 1399), ens2 sequence presence/absence, the two classes of Ens2 site-specific endonuclease recognition sequences, and the three most highly oligomycin phenotype-associated SNPs coincided with the six S. cerevisiae ATP6 groups (Figure 2, Figure S1, Figure S2, and Table S6). Finally, we searched the recently published mitochondrial genome sequences of seven other Saccharomyces species (Leducq et al. 2017; Sulo et al. 2017) for ATP6 and constructed phylogenies. We identified three S. paradoxus ATP6 groups (S. paradoxus groups 1, 2, and 3). In S. cerevisiae and S. paradoxus, there was evidence of ATP6 introgression (Figure 2, Figure S1, and Figure S2).
Mitochondrial genotype-dependent oligomycin and other respiration inhibitor phenotypes in iso-nuclear F1 pairs
We first performed a small-scale (12 genetic backgrounds, 8 iso-nuclear F1 pairs) experiment (Figure 1) to test the effects of ATP6 SNP genotypes on oligomycin phenotypes. In six of these eight iso-nuclear F1 diploid pairs, ATP6 SNP genotype had the predicted effect on oligomycin phenotype, either mitochondrial genotype-dependent oligomycin resistance/sensitivity phenotypes (n = 4; ATP6R vs. ATP6S) or equivalent oligomycin phenotypes (n = 2; ATP6S vs. ATP6S) (Figure 4, Figure S3, Figure S4, and Table S10). However, the remaining two iso-nuclear F1 diploid pairs, both ATP6S vs. ATP6S, nonetheless had mitochondrial genotype-dependent oligomycin resistance/sensitivity phenotypes, consistent with epistasis, with one (YJM1083/YJM627) also being transgressive, i.e., opposite to that predicted by the parental phenotypes (Table S10). We next tested the ability of these eight iso-nuclear F1 diploid pairs to identify mitochondrial genotype-dependent phenotypes for which there were no mitochondrial genotype-phenotype associations. Again, we observed multiple mitochondrial genotype-dependent phenotypes, with some being transgressive (Figure 4, Figures S3–S6, and Table S10).
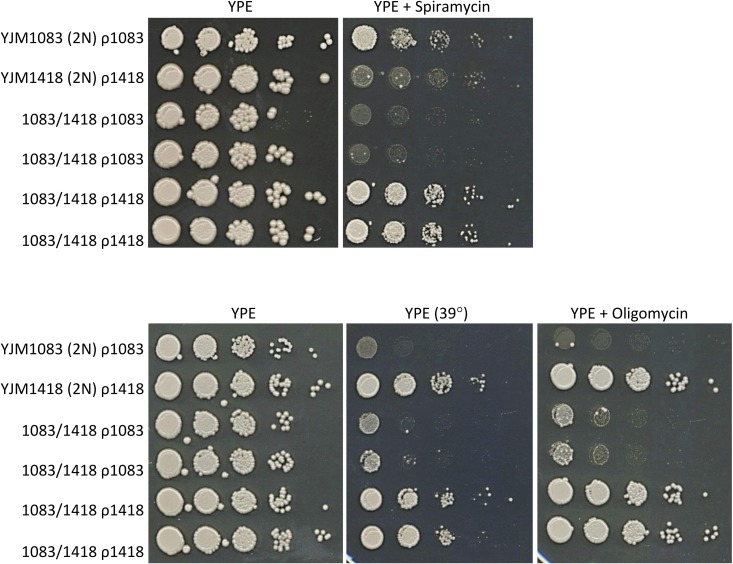
Figure 4.
Mitochondrial, genotype-dependent spiramycin, 39°, and oligomycin respiration phenotypes. Spot dilution phenotypes on YPE of parental diploids [YJM1083 ρ1083 (ATP6S SNPs) and YJM1418 ρ1418 (ATP6R SNPs)] and two independently made iso-nuclear F1 diploid pairs (1083/1418 ρ1083 and 1083/1418 ρ1418). Mitochondrial genotypes are denoted as ρ1083 and ρ1418.
Because the small-scale experiment was conducted with only 12 genetic backgrounds and 8 iso-nuclear F1 diploid pairs, we expanded our analysis by selecting 14 genetic backgrounds from the 100-genomes strains, haploid derivatives of which we crossed to create a 14 × 14 matrix of diploids: the 14 recreated diploid parent strains plus 91 pairs of iso-nuclear F1 diploids. Similar to the small-scale experiment, we identified iso-nuclear F1 diploid pairs with mitochondrial genotype-dependent oligomycin phenotypes; in some cases, these were consistent with their ATP6 SNP genotypes (i.e., ATP6R vs. ATP6S) while others were consistent with epistasis (i.e., ATP6S vs. ATP6S and ATP6R vs. ATP6R). Also consistent with epistasis, some ATP6R vs. ATP6S iso-nuclear F1 diploid pairs had equivalent oligomycin phenotypes. Finally, we identified mitochondrial genotype-dependent myxothiazol phenotypes, with many iso-nuclear F1 pairs exhibiting transgression (Table S11).
Mitochondrial genotype-dependent nonrespiration inhibitor phenotypes in iso-nuclear F1 pairs
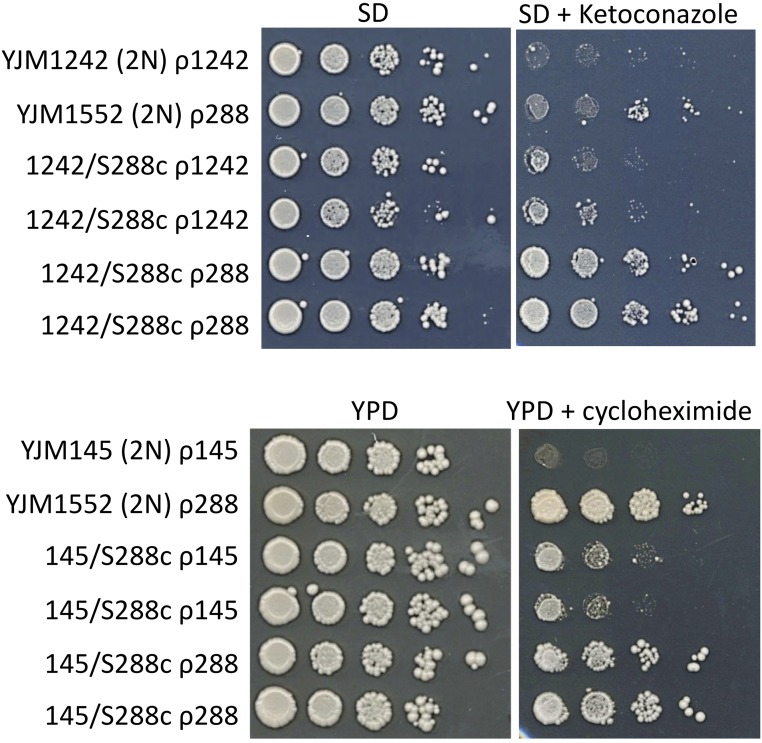
To test if mitochondrial genotype influenced nonrespiration phenotypes, we determined the spot dilution phenotypes of the aforementioned eight iso-nuclear F1 diploid pairs, and the parent diploids, on SD and/or YPD media, which contained fermentable dextrose as the sole carbon source, for low (15°) and high (39°) temperature growth, as well as for cycloheximide, ketoconazole, and copper resistance. In some of these eight iso-nuclear F1 pairs, we identified mitochondrial genotype-dependent growth temperature, ketoconazole, cycloheximide, and copper resistance phenotypes (Figure 5, Figure 6, and Table S10). The mitochondrial genotype-dependent copper phenotypes in the YJM1083/YJM1418 iso-nuclear F1 pairs were transgressive. Similar to the small-scale experiment, in the 14 × 14 matrix of diploids we identified mitochondrial genotype-dependent phenotypic differences for ketoconazole and cycloheximide resistance; one iso-nuclear F1 pair had transgressive cycloheximide phenotypes (Table S11). Nuclear-mitochondrial genotype epistasis, including transgression, that affects respiratory and nonrespiratory phenotypes is discussed below.
Figure 5.
Mitochondrial genotype-dependent, nonrespiration ketoconazole and cycloheximide resistance phenotypes. Spot dilution phenotypes on SD (±ketoconazole) and YPD (±cycloheximide) of parental diploids (YJM1242 ρ1242 and YJM1552 ρ288, the diploid YJM1552 is isogenic with S288c; YJM145 ρ145 and YJM1552 ρ288, the diploid YJM145 is isogenic with YJM789) and two independently made iso-nuclear F1 diploid pairs (1242/S288c ρ1242 and 1242/S288c ρ288; 145/S288c ρ145 and 145/S288c ρ288). Mitochondrial genotypes are denoted as ρ1242, ρ288, ρ145, and ρ1418.
Figure 6.
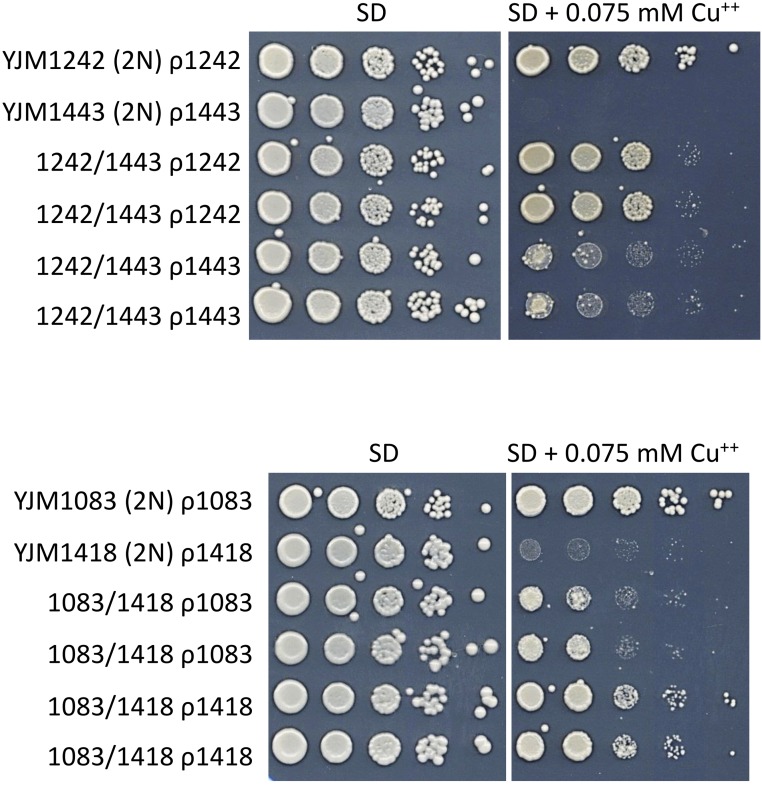
Mitochondrial genotype-dependent, nonrespiration copper resistance phenotypes. Spot dilution phenotypes on SD (±0.075 mM CuSO4) of parental diploids (YJM1242 ρ1242 and YJM1443 ρ1443; YJM1083 ρ1083 and YJM1418 ρ1418) and two independently made, iso-nuclear F1 diploid pairs (1242/1443 ρ1242 and 1242/1443 ρ1443; 1083/1418 ρ1083 and 1083/1418 ρ1418). Mitochondrial genotypes are denoted as ρ1242, ρ1443, ρ1083, and ρ1418. In the 1083/1418 iso-nuclear F1 pairs, mitochondrial genotype-dependent copper resistance is transgressive.
Discussion
The vast diversity in the S. cerevisiae mitochondrial genomes in our study population included size, intron content, and copy number variation, as well as SNPs/indels in the 36 core genes [RNA-encoding (n = 27): 21S, 15S, RPM1, 24 tRNAs; protein-encoding (n = 9): VAR1, COX1, COX2, COX3, COB, ATP6, ATP8, OLI1, RF1]. Some aspects of these S. cerevisiae mitochondrial genomes have been previously described (Wolters et al. 2015; Peris et al. 2017; Repar and Warnecke 2017). Therefore, we focus this discussion on the novel ENS2 and ATP6 genotype-oligomycin phenotype associations; ENS2 and ATP6 phylogenies and introgression; Ens2-mediated recombination; the identification and characterization of mitochondrial genotype-dependent phenotypes in iso-nuclear F1 pairs; and nuclear-mitochondrial epistasis, as well as its implications for the analysis of quantitative traits.
ENS2 loss-of-function polymorphisms and oligomycin phenotype
With the exception of some group A and group B ENS2 S. cerevisiae strains, most ens2 ORFs had loss-of-function polymorphisms (Figure S1 and Table S6). What might be the basis for the high frequency of ens2 loss-of-function polymorphisms? When functional, ENS2 encodes a site-specific endonuclease and is mobile (Nakagawa et al. 1992; Morishima et al. 1993). Similarly, some mitochondrial homing introns encode site-specific endonucleases that promote intron mobility. When a site-specific endonuclease-encoding homing intron is fixed, selection for mobility and endonuclease function is lost and, as a result, the homing endonuclease-encoding intron will be lost or accumulate loss-of-function polymorphisms (Burt and Koufopanou 2004). Thus, one hypothesis for the high frequency of ens2 loss-of-function polymorphisms is lack of selection for Ens2 site-specific endonuclease activity and ENS2 mobility. A comparison between the mobile SCE1 intron, which encodes the site-specific endonuclease I-SceI, and the mobile ENS2 gene in S. cerevisiae is informative with respect to the lack of selection hypothesis. Like the homing SCE1 intron (Table S7), the mobile ENS2 gene is not fixed in S. cerevisiae (Table S6) (ENS2 is also not fixed in S. paradoxus (Figure 1, Figure S1, and Figure S2). However, in contrast to the homing SCE1 intron that integrates at (and disrupts) the I-SceI recognition sequence in SCE1-free 21S and has no polymorphisms in S. cerevisiae (Table S7), ENS2 does not integrate at (and disrupt) the Ens2 recognition sequence in ATP6 (i.e., ENS2 is not homing) and most ens2 ORFs have inactivating polymorphisms (Table S6). In addition, the less sensitive Ens2 recognition sequence in ATP6 is not fixed in S. cerevisiae (Figure 3 and Table S6). These results argue against the hypothesis that lack of selection for Ens2 site-specific endonuclease activity and ENS2 mobility is responsible for the high frequency of ens2 loss-of-function polymorphisms.
A second hypothesis for the high frequency of ens2 loss-of-function polymorphisms is that there is selection against Ens2 function. Selection against Ens2 function may be a consequence of Ens2-Ssc1 heterodimer formation that might reduce the availability of Ssc1, a nuclearly encoded, mitochondrially localized, essential gene product. Alternatively, selection against functional Ens2 may be a consequence of its site-specific endonuclease activity, which cuts a 26 bp sequence in ATP6, and/or the endonuclease activity of functional Ens2-Ssc1 heterodimers, which cut >30 sites in the mitochondrial genome (Morishima et al. 1990; Kawasaki et al. 1991; Nakagawa et al. 1992; Shibata et al. 1995; Mizumura et al. 1999). That is, functional Ens2 and Ens2-Ssc1 heterodimers presumably introduce deleterious double-stranded DNA breaks in ENS2-containing mitochondrial genomes. The less-sensitive Ens2 recognition sequence in functional ENS2-containing strains should confer relative resistance to Ens2-mediated double-stranded breaks in ATP6. However, the less-sensitive Ens2 recognition sequence would have no effect on Ens2-Ssc1 heterodimer-mediated double-stranded breaks elsewhere in the mitochondrial genome or on reduced availability of Ssc1. Only, ens2 loss-of-function would protect against reduced availability of Ssc1 as well as Ens2- and Ens2-Ssc1–mediated double-stranded DNA breaks in the mitochondrial genome.
Although the basis for the high frequency remains to be determined, the ens2 loss-of-function polymorphisms were informative with respect to the ens2 sequence-oligomycin sensitivity association (Table S8). Specifically, consistent with Ens2 function(s) not contributing to oligomycin phenotype, full-length, presumably functional ENS2 ORFs showed no oligomycin association (Table S9). The basis for the ENS2 sequence presence-oligomycin sensitivity association, including introgression, is discussed below.
ATP6 phylogeny, ENS2-independent and -dependent introgression of ATP6, Ens2-mediated recombination, and oligomycin phenotype in S. cerevisiae
The complex ATP6 phylogeny showed evidence of both ENS2-independent and -dependent introgression of ATP6. With respect to ENS2-independent ATP6 introgression, we identified ATP6ens20 groups in S. cerevisiae (S. cerevisiae group 1, 2, and 3 ATP6) and in S. paradoxus (S. paradoxus group 1 and 2 ATP6) (Figure 2 and Figure S2). While directionality cannot be assessed, introgression of ATP6ens20 appears to have occurred more than once between S. cerevisiae and S. paradoxus. Because of the absence of ENS2, these ATP6ens20 introgressions are presumed to be ENS2-independent.
With respect to the hypothesis that cointrogression of ATP6-ENS2 may be ENS2-dependent, we identified ATP6-ENS2 groups in S. cerevisiae (S. cerevisiae group 4 ATP6/group A ENS2; S. cerevisiae group 5 ATP6/group B ENS2; and S. cerevisiae group 1399 ATP6/group 1399 ens2), S. arboricola (S. arboricola ATP6/group A ENS2), and in S. paradoxus (S. paradoxus group 3 ATP6/S. paradoxus ens2) (Figure 1 and Figure S1). From the perspective of ATP6-ENS2 cointrogression, intraspecific ATP6-ENS2 × ATP6ens20 results in Ens2-dependent, highly biased inheritance of ENS2 and of a mutation in the ENS2-linked ATP6 (Nakagawa et al. 1992; Morishima et al. 1993; Shibata et al. 1995). The presence of full-length, potentially functional ENS2 ORFs only in S. cerevisiae suggests ENS2-mediated introgression from S. cerevisiae to S. arboricola (ENS2 only) and to S. paradoxus (ATP6-ENS2). Consistent with the ENS2-mediated introgression hypothesis, with the sole exception of S. arboricola, there is strong correspondence between the ENS2 and ATP6 phylogenies (Figure 1, Figure S1, and Table S6). One hypothesis for the exception of S. arboricola is that S. cerevisiae ATP6 may be incompatible with S. arboricola mitochondrially and/or nuclearly encoded components of ATP synthase.
To assess the hypothesized Ens2-mediated mobility and recombination in S. cerevisiae, we examined the Ens2 recognition sequences in ATP6 [713–738 bp; 11 SNPs that, with the exception of the 721 bp (n = 5; S. cerevisiae group 3 ATP6) and 723 bp (n = 1; S. cerevisiae group 1 ATP6] SNPs, were in linkage disequilibrium); two proximal SNPs that were in linkage disequilibrium (684 and 693 bp); the three most highly oligomycin phenotype-associated ATP6 SNPs that were in linkage disequilibrium (753, 763, and 766 bp); and the presence/absence of ENS2. Similar to experimental ATP6-ENS2 × ATP6ens20 crosses (Nakagawa et al. 1992), the data suggests that in ATP6-ENS2 × ATP6ens20 heteroplasmic zygotes, Ens2-mediated double-stranded DNA breaks at the more sensitive Ens2 recognition sequence of ATP6ens20 (i.e., S. cerevisiae group 1 ATP6) were repaired and replaced, in most cases, with ATP6 sequences encompassing the 684 and 693 bp proximal SNPs, the less-sensitive Ens2 recognition sequence, the distal OliS-associated SNPs, and ENS2 (Figure 3). That is, in most cases, ENS2 has driven its movement and, minimally, a 684–766 bp region of ATP6 into the S. cerevisiae population, as exemplified by the ENS2-containing S. cerevisiae group 4 (n = 46), 5 (n = 15), and YJM1399 (n = 1) ATP6 strains (note that the T513G oligomycin resistance mutation in ATP6 that exhibits biased inheritance in experimental ATP6-ENS2 × ATP6ens20 crosses (Nakagawa et al. 1992) is proximal to this minimal 684–766 bp region). However, in the ens20 S. cerevisiae group 2 ATP6 (n = 18) and group 3 ATP6 (n = 5) strains, repair and replacement excluded the OliS SNPs and ENS2 (Figure 3 and Table S6). The oligomycin-resistant phenotypes of these S. cerevisiae group 2 and group 3 ATP6 strains further strengthens the hypothesis that the nonsynonymous 763 and/or 766 bp ATP6 SNPs contribute to oligomycin phenotype.
The mechanistic basis for the oligomycin phenotype-ATP6 genotype association remains to be determined. However, oligomycin sensitivity is affected by the level of F0F1 ATP synthase activity (Pagliarani et al. 2013). That is, reduced ATP synthase activity results in increased oligomycin sensitivity. Thus, one hypothesis is that the oligomycin sensitivity-ATP6 genotype association is due to nonsynonymous ATP6 SNPs that reduce Atp6 stability, the assembly of Atp6 into F0, and/or the assembly of F0 with F1.
Iso-nuclear F1 pairs bypass the limitations of association and illustrate the complexity of mitochondrial genotype-dependent phenotypes
In addition to sample size (n = 96), our identification of mitochondrial genotype-phenotype associations may have been limited by epistasis. To bypass these limitations, we performed more focused experiments to identify and analyze mitochondrial genotype-dependent phenotypes. Previous studies have used kar1-mediated mitochondrial genome transfer into novel haploid or homozygous diploid nuclear genetic backgrounds to identify mitochondrial genotype-dependent phenotypes (Codón et al. 1995; Dimitrov et al. 2009; Edwards et al. 2014; Paliwal et al. 2014; Spirek et al. 2014). However, we did not use kar1-mediated mitochondrial genome transfer (Conde and Fink 1976) due to concerns about the potentially confounding cotransfer of 2µ plasmid, amyloid and nonamyloid prions, RNA viruses and satellites, and whole chromosomes (Dutcher 1981; Sigurdson et al. 1981; Tartakoff et al. 2018). Rather than kar1-mediated mitochondrial genome transfer, we used iso-nuclear F1 pairs, in which the parental mitochondrial genotypes are fixed, to identify mitochondrial genotype-dependent phenotypes. Similar to reciprocal hemizygosity analysis in the nuclear genomes of multiply heterozygous F1 diploids (Steinmetz et al. 2002b), any difference in the phenotypes of the iso-nuclear F1 diploids must be mitochondrial genotype-dependent (Figure 1). Also, in contrast to kar1-mediated mitochondrial genome transfer into novel haploid or homozygous diploid nuclear genetic backgrounds, the nuclear genomes of iso-nuclear F1 diploids closely mimic the nuclear genomes of multiply heterozygous outbred species, such as humans; the multiply heterozygous nuclear genomes of diploid S. cerevisiae isolates that occur in nature (McCusker et al. 1994; Muller and McCusker 2009; Esberg et al. 2011; Magwene et al. 2011; Granek et al. 2013; Peter et al. 2018); and the nuclear genomes of multiply heterozygous F1 in quantitative genetics studies of S. cerevisiae and other species.
Respiration phenotypes
We first analyzed iso-nuclear F1 pairs to experimentally assess the association between oligomycin phenotype and, using the three most highly associated ATP6 SNPs that are in linkage disequilibrium, ATP6 genotype (Figure 4, Figure S3, Figure S4, Table S10, and Table S11). Relative to the straightforward oligomycin phenotype-ATP6 genotype association in our 96 strains, analysis of oligomycin phenotype and ATP6 genotype in iso-nuclear F1 pairs identified substantial complexity. We feel confident in excluding the other two mitochondrially encoded components of ATP synthase, OLI1 and ATP8, as contributing to the complexity of the oligomycin phenotype in either the 96 parental strains or in iso-nuclear F1 because they each have only one common synonymous SNP and no nonsynonymous SNPs (Table S6). Our iso-nuclear F1 results are consistent with oligomycin phenotype complexity being due to epistatic nuclear-mitochondrial genotype interactions.
We also analyzed iso-nuclear F1 pairs to determine whether other respiration phenotypes, for which there were no mitochondrial genotype associations in our 96 strains, were mitochondrial genotype-dependent. Indeed, we found many iso-nuclear F1 pairs with mitochondrial genotype-dependent respiration inhibitor phenotype(s) (Figure 4, Figure S3–S5, Table S10, and Table S11), some of which exhibited transgression. Broadly, transgression can be viewed as occurring when a quantitative trait locus (in this case, the mitochondrial genome) or gene from a less fit parent increases fitness and, conversely, the locus or gene from the more fit parent decreases fitness. Although typically observed in segregating progeny (Rieseberg et al. 1999, 2003; Goulet et al. 2017), transgressive alleles have been identified in reciprocally hemizygous F1; for example, alleles of END3 (Steinmetz et al. 2002b; Sinha et al. 2006). We hypothesize that extensive nuclear-mitochondrial epistasis, including transgression, is a likely contributor to the lack of other mitochondrial genotype-phenotype associations in the 96 strains.
Nonrespiration phenotypes
Our analysis of iso-nuclear F1 pairs also identified mitochondrial genotype-dependent effects on the nonrespiration growth temperature, cycloheximide, ketoconazole, and copper resistance phenotypes (Figure 5, Figure 6, Table S10, and Table S11). With the exception of high-temperature growth (Paliwal et al. 2014; Spirek et al. 2014), mitochondrial genotype-dependent effects on these nonrespiration phenotypes are novel. What might be the basis for mitochondrial genotype-dependent effects on nonrespiration phenotypes?
While distinct from the naturally occurring ρ+ mitochondrial genotype variation analyzed in this work, mitochondrial genotype-dependent effects on nonrespiration phenotypes have been extensively studied in ρ+ vs. ρ0 S. cerevisiae strains. In ρ0 strains, mitochondrial dysfunction activates a retrograde signaling pathway that affects nuclear gene expression and nonrespiration phenotypes (Butow and Avadhani 2004; Jazwinski 2013; da Cunha et al. 2015), including increased cycloheximide resistance in ρ0 strains (Hallstrom and Moye-Rowley 2000; Zhang and Moye-Rowley 2001; Moye-Rowley 2003, 2005; Liu and Butow 2006) (in contrast to cycloheximide, to the best of our knowledge there are no descriptions of retrograde signaling effects on S. cerevisiae azole resistance). Thus, one hypothesis is that epistatic nuclear-mitochondrial genotype interactions in some ρ+ iso-nuclear F1 pairs results in mitochondrial dysfunction sufficient to activate a retrograde signaling pathway in one of the iso-nuclear F1 resulting in differential, mitochondrial genotype-dependent, nonrespiration cycloheximide resistance phenotypes.
Alternatively, there is a nonretrograde signaling hypothesis for mitochondrial genotype-dependent cycloheximide resistance. The cytosolic translation inhibitor cycloheximide has been shown to rescue some nuclear mutations that cause mitochondrial dysfunction, possibly by reducing the toxic cytosolic accumulation of nuclearly encoded, mitochondrially targeted proteins (Wang et al. 2008; Wang and Chen 2015; Wrobel et al. 2015; de Taffin de Tilques et al. 2018; Guaragnella et al. 2018). Thus, epistatic nuclear-mitochondrial genotype interactions in some ρ+ iso-nuclear F1 pairs may result in cycloheximide-remediable mitochondrial dysfunction in one of the iso-nuclear F1, and consequently, mitochondrial genotype-dependent cycloheximide resistance. Indeed, for one of the iso-nuclear F1 pairs, YJM1450/YJM1479, the mitochondrial genotype-dependent cycloheximide resistance phenotypes were transgressive, consistent with nuclear-mitochondrial epistasis.
We also observed mitochondrial genotype-dependent copper resistance phenotypes in iso-nuclear F1 pairs (Figure 6 and Table S10). To the best of our knowledge, there are no descriptions of retrograde signaling effects on S. cerevisiae copper resistance. However, laboratory strains, in which most and possibly all of the work on S. cerevisiae ρ+ vs. ρ0 and retrograde signaling has been performed, have multiple copies of the copper resistance-conferring, copper metallothionein-encoding gene CUP1; for example, the very commonly used S288c and W303 laboratory strains have 14 tandem copies of CUP1 (Zhao et al. 2014). In the 100-genomes strains, the sole and very strong association with copper resistance was CUP1 copy number, which ranged from one copy to 18 tandem copies (Strope et al. 2015). Thus, one hypothesis is that the high CUP1 copy number in commonly used laboratory strains may be responsible, at least in part, for the lack of previously described ρ+ vs. ρ0 or retrograde signaling effects on S. cerevisiae copper resistance. Conversely, low CUP1 copy number may be necessary to detect mitochondrial genotype-dependent copper resistance phenotypes in iso-nuclear F1 pairs. Consistent with the CUP1 copy number hypothesis, the parent strains of the iso-nuclear F1 pairs that have mitochondrial genotype-dependent copper resistance phenotypes have low CUP1 copy numbers: YJM1083 (CUP1: n = 3)/YJM1418 (CUP1: n = 1) and YJM1242 (CUP1: n = 1)/YJM1443 (CUP1: n = 1) (Strope et al. 2015).
Although the low CUP1 copy number hypothesis suggests a reasonable prerequisite for observing mitochondrial genotype-dependent copper resistance, it is not a mechanistic hypothesis. A mechanistic hypothesis for mitochondrial genotype-dependent copper resistance may involve mitochondrially encoded, copper-containing gene product(s) in a mitochondrially localized, copper-dependent enzyme. Of the known copper-containing and copper-dependent enzymes (Festa and Thiele 2011), only the mitochondrially localized cytochrome c oxidase contains mitochondrially encoded gene products (Cox1, Cox2, Cox3). Because cytochrome c oxidase also contains nuclearly encoded gene products, and requires many additional nuclearly encoded gene products for its synthesis, assembly, and activity, its hypothesized effect on copper resistance may be particularly susceptible to nuclear-mitochondrial epistasis. Indeed, for the YJM1083/YJM1418 iso-nuclear F1 pairs (Figure 6), the mitochondrial genotype-dependent copper resistance phenotypes were transgressive, consistent with nuclear-mitochondrial epistasis. Thus, one hypothesis is that epistatic nuclear-mitochondrial genotype interactions in some iso-nuclear F1 pairs, possibly involving the synthesis, assembly, and/or activity of cytochrome c oxidase, may activate a signaling pathway in one of the iso-nuclear F1, resulting in mitochondrial genotype-dependent copper resistance.
In conclusion, naturally occurring mitochondrial genome variation has major phenotypic effects in model systems (Ballard and Melvin 2010; Joseph et al. 2013) and in humans (Taylor and Turnbull 2005; Wallace 2010; Schon et al. 2012; Dowling 2014). However, with the exception of a relatively small number of studies (Codón et al. 1995; Dimitrov et al. 2009; Edwards et al. 2014; Paliwal et al. 2014; Wolters et al. 2018), the phenotypic contributions of naturally occurring S. cerevisiae mitochondrial genome variation have only infrequently been considered. As we show with iso-nuclear F1 pairs, multiple respiration and nonrespiration phenotypes are strongly influenced by naturally occurring mitochondrial genome variation and by nuclear-mitochondrial epistasis. Even for the presumably straightforward oligomycin phenotype-ATP6 genotype association, our iso-nuclear F1 analysis showed a high level of complexity. Because of its effect on complex respiration and nonrespiration phenotypes, mitochondrial genotype is likely a major contributor to S. cerevisiae quantitative traits. That is, the S. cerevisiae mitochondrial genome, which contains 36 core genes (21S, 15S, RPM1, VAR1, COX1, COX2, COX3, COB, ATP6, ATP8, OLI1, RF1, 24 tRNAs) and up to 16 mobile and variably present genes (ENS2 and introns in the 21S, COB, and COX1 genes), is a 73,450–92,176 bp quantitative trait locus.
In higher eukaryotes and many lower eukaryotes, mitochondrial genome inheritance is uniparental; that is, the mitochondrial genome (ρ) is a fixed, nonrecombinant locus. In contrast, S. cerevisiae mitochondrial genome inheritance is biparental (Berger and Yaffe 2000). That is, N1 ρ1 × N2 ρ2 results in N1/N2 heteroplasmic ρ1 + ρ2 zygotes, which tend to produce parental ρ genomes from terminal buds and recombinant ρ genomes from medial buds (Berger and Yaffe 2000). Heteroplasmic N1/N2 ρ1 + ρ2 zygotes produce homoplasmic ρ+ cells via high levels of ρ1×ρ2 recombination (Fritsch et al. 2014) and rapid ρ genome segregation (Birky et al. 1978; Zinn et al. 1987). Recovery of ρ genotypes from heteroplasmic ρ1 + ρ2 zygotes may be biased by prezygotic differences in ρ1 vs. ρ2 copy numbers, the frequency of terminal vs. medial budding in zygotes, the presence in one of the two parental ρ genomes of ENS2 and any of the numerous mobile introns, as well as both nuclear-mitochondrial and mitochondrial-mitochondrial epistasis. Thus, N1 ρ1 × N2 ρ2, which has been the standard approach in S. cerevisiae quantitative trait studies, yields N1/N2 that are homoplasmic for multiple different mitochondrial genotypes, which negatively affects reproducibility. Fortunately, S. cerevisiae mitochondrial genotype can be easily controlled, or fixed, as we have done with iso-nuclear F1 pairs. In addition to allowing the identification and analysis of mitochondrial genotype-dependent phenotypes, controlling S. cerevisiae mitochondrial genotype will also enable the identification of mitochondrial genotype-independent vs. mitochondrial genotype-dependent quantitative trait genes in the nuclear genome.
Acknowledgments
The authors thank D. Gottschling (CalicoLabs) for the MIP1DN-containing plasmid pLND46, and thank T. Petes, S. Jinks-Robertson, and anonymous reviewers for their comments on and suggestions for this work. This work was supported by National Institutes of Health grant R01 GM098287 awarded to JHM. P.K.S. was supported as a fellow of the Tri-Institutional Molecular Mycology and Pathogenesis Training Program, supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases (T32 award AI052080-11). National Institutes of Health grant F32 GM110997 supported D.A.S.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7361240.
Communicating editor: L. Steinmetz
Literature Cited
- Baile M. G., Claypool S. M., 2013. The power of yeast to model diseases of the powerhouse of the cell. Front. Biosci. (Landmark Ed) 18: 241–278. 10.2741/4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard J. W., Melvin R. G., 2010. Linking the mitochondrial genotype to the organismal phenotype. Mol. Ecol. 19: 1523–1539. 10.1111/j.1365-294X.2010.04594.x [DOI] [PubMed] [Google Scholar]
- Barnhill A. E., Brewer M. T., Carlson S. A., 2012. Adverse effects of antimicrobials via predictable or idiosyncratic inhibition of host mitochondrial components. Antimicrob. Agents Chemother. 56: 4046–4051. 10.1128/AAC.00678-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger K. H., Yaffe M. P., 2000. Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol. 8: 508–513. 10.1016/S0966-842X(00)01862-X [DOI] [PubMed] [Google Scholar]
- Birky C. W., Strausberg R. L., Perlman P. S., 1978. Vegetative segregation of mitochondria in yeast: estimating parameters using a random model. Mol. Gen. Genet. 158: 251–261. 10.1007/BF00267196 [DOI] [Google Scholar]
- Burt A., Koufopanou V., 2004. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr. Opin. Genet. Dev. 14: 609–615. 10.1016/j.gde.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Butow R. A., Avadhani N. G., 2004. Mitochondrial signaling: the retrograde response. Mol. Cell 14: 1–15. 10.1016/S1097-2765(04)00179-0 [DOI] [PubMed] [Google Scholar]
- Carelli V., Giordano C., d’Amati G., 2003. Pathogenic expression of homoplasmic mtDNA mutations needs a complex nuclear-mitochondrial interaction. Trends Genet. 19: 257–262. 10.1016/S0168-9525(03)00072-6 [DOI] [PubMed] [Google Scholar]
- Codón A. C., Gasent-Ramírez J. M., Benítez T., 1995. Factors which affect the frequency of sporulation and tetrad formation in Saccharomyces cerevisiae baker’s yeasts. Appl. Environ. Microbiol. 61: 630–638 (erratum: Appl. Environ. Microbiol. 61: 1677). http://www.ncbi.nlm.nih.gov/pubmed/7574601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde J., Fink G. R., 1976. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. U.S.A. 73: 3651–3655. 10.1073/pnas.73.10.3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha F. M., Torelli N. Q., Kowaltowski A. J., 2015. Mitochondrial retrograde signaling: triggers, pathways, and outcomes. Oxid. Med. Cell. Longev. 2015: 482582 10.1155/2015/482582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taffin de Tilques M., Lasserre J. P., Godard F., Sardin E., Bouhier M., et al. , 2018. Decreasing cytosolic translation is beneficial to yeast and human Tafazzin-deficient cells. Microb. Cell 5: 220–232. 10.15698/mic2018.05.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimauro S., Davidzon G., 2005. Mitochondrial DNA and disease. Ann. Med. 37: 222–232. 10.1080/07853890510007368 [DOI] [PubMed] [Google Scholar]
- Dimitrov L. N., Brem R. B., Kruglyak L., Gottschling D. E., 2009. Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics 183: 365–383. 10.1534/genetics.109.104497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling D. K., 2014. Evolutionary perspectives on the links between mitochondrial genotype and disease phenotype. Biochim. Biophys. Acta 1840: 1393–1403. 10.1016/j.bbagen.2013.11.013 [DOI] [PubMed] [Google Scholar]
- Dutcher S. K., 1981. Internuclear transfer of genetic information in kar1–1/KAR1 heterokaryons in Saccharomyces cerevisiae. Mol. Cell. Biol. 1: 245–253. 10.1128/MCB.1.3.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M. D., Symbor-Nagrabska A., Dollard L., Gifford D. K., Fink G. R., 2014. Interactions between chromosomal and nonchromosomal elements reveal missing heritability. Proc. Natl. Acad. Sci. USA 111: 7719–7722. 10.1073/pnas.1407126111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg A., Muller L. A., McCusker J. H., 2011. Genomic structure of and genome-wide recombination in the Saccharomyces cerevisiae S288C progenitor isolate EM93. PLoS One 6: e25211 10.1371/journal.pone.0025211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., 2013. The molecular basis of phenotypic variation in yeast. Curr. Opin. Genet. Dev. 23: 672–677. 10.1016/j.gde.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa R. A., Thiele D. J., 2011. Copper: an essential metal in biology. Curr. Biol. 21: R877–R883. 10.1016/j.cub.2011.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F., Roganti T., Lecrenier N., Purnelle B., 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440: 325–331. 10.1016/S0014-5793(98)01467-7 [DOI] [PubMed] [Google Scholar]
- Fritsch E. S., Chabbert C. D., Klaus B., Steinmetz L. M., 2014. A genome-wide map of mitochondrial DNA recombination in yeast. Genetics 198: 755–771. 10.1534/genetics.114.166637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet B. E., Roda F., Hopkins R., 2017. Hybridization in plants: old ideas, new techniques. Plant Physiol. 173: 65–78. 10.1104/pp.16.01340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granek J. A., Murray D., Kayrkçi Ö., Magwene P. M., 2013. The genetic architecture of biofilm formation in a clinical isolate of Saccharomyces cerevisiae. Genetics 193: 587–600. 10.1534/genetics.112.142067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziewicz M. A., Longley M. J., Copeland W. C., 2006. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem. Rev. 106: 383–405. 10.1021/cr040463d [DOI] [PubMed] [Google Scholar]
- Guaragnella N., Coyne L. P., Chen X. J., Giannattasio S., 2018. Mitochondria-cytosol-nucleus crosstalk: learning from Saccharomyces cerevisiae. FEMS Yeast Res. 18: foy088. 10.1093/femsyr/foy088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallstrom T. C., Moye-Rowley W. S., 2000. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275: 37347–37356. 10.1074/jbc.M007338200 [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T., 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160–174. 10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- Jazwinski S. M., 2013. The retrograde response: when mitochondrial quality control is not enough. Biochim. Biophys. Acta 1833: 400–409. 10.1016/j.bbamcr.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B., Corwin J. A., Li B., Atwell S., Kliebenstein D. J., 2013. Cytoplasmic genetic variation and extensive cytonuclear interactions influence natural variation in the metabolome. eLife 2: e00776 10.7554/eLife.00776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabala A. M., Lasserre J. P., Ackerman S. H., di Rago J. P., Kucharczyk R., 2014. Defining the impact on yeast ATP synthase of two pathogenic human mitochondrial DNA mutations, T9185C and T9191C. Biochimie 100: 200–206. 10.1016/j.biochi.2013.11.024 [DOI] [PubMed] [Google Scholar]
- Kawasaki K., Takahashi M., Natori M., Shibata T., 1991. DNA sequence recognition by a eukaryotic sequence-specific endonuclease, Endo.SceI, from Saccharomyces cerevisiae. J. Biol. Chem. 266: 5342–5347. http://www.ncbi.nlm.nih.gov/pubmed/2002067 [PubMed] [Google Scholar]
- Leducq J. B., Henault M., Charron G., Nielly-Thibault L., Terrat Y., et al. , 2017. Mitochondrial recombination and introgression during speciation by hybridization. Mol. Biol. Evol. 34: 1947–1959. 10.1093/molbev/msx139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Louis E. J., 2012. Advances in quantitative trait analysis in yeast. PLoS Genet. 8: e1002912 10.1371/journal.pgen.1002912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Butow R. A., 2006. Mitochondrial retrograde signaling. Annu. Rev. Genet. 40: 159–185. 10.1146/annurev.genet.40.110405.090613 [DOI] [PubMed] [Google Scholar]
- Lodi T., Dallabona C., Nolli C., Goffrini P., Donnini C., et al. , 2015. DNA polymerase γ and disease: what we have learned from yeast. Front. Genet. 6: 106 10.3389/fgene.2015.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine D. M., Perlman P. S., Butow R. A., 2000. The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway. EMBO J. 19: 767–775. 10.1093/emboj/19.4.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwene P., Kayikci O., Granek J. A., Reininga J. M., Scholl Z., et al. , 2011. Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 108: 1987–1992. 10.1073/pnas.1012544108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso M., Filosto M., Choub A., Tentorio M., Broglio L., et al. , 2007. Mitochondrial DNA-related disorders. Biosci. Rep. 27: 31–37. 10.1007/s10540-007-9035-2 [DOI] [PubMed] [Google Scholar]
- McCusker J. H., Clemons K. V., Stevens D. A., Davis R. W., 1994. Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics 136: 1261–1269. http://www.genetics.org/content/136/4/1261.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumura H., Shibata T., Morishima N., 1999. Stable association of 70-kDa heat shock protein induces latent multisite specificity of a unisite-specific endonuclease in yeast mitochondria. J. Biol. Chem. 274: 25682–25690. 10.1074/jbc.274.36.25682 [DOI] [PubMed] [Google Scholar]
- Montanari A., Zhou Y. F., D’Orsi M. F., Bolotin-Fukuhara M., Frontali L., et al. , 2013. Analyzing the suppression of respiratory defects in the yeast model of human mitochondrial tRNA diseases. Gene 527: 1–9. 10.1016/j.gene.2013.05.042 [DOI] [PubMed] [Google Scholar]
- Morishima N., Nakagawa K., Yamamoto E., Shibata T., 1990. A subunit of yeast site-specific endonuclease SceI is a mitochondrial version of the 70-kDa heat shock protein. J. Biol. Chem. 265: 15189–15197. http://www.ncbi.nlm.nih.gov/pubmed/2203771 [PubMed] [Google Scholar]
- Morishima N., Nakagawa K., Shibata T., 1993. A sequence-specific endonuclease, Endo.SceI, can efficiently induce gene conversion in yeast mitochondria lacking a major exonuclease. Curr. Genet. 23: 537–541. 10.1007/BF00312648 [DOI] [PubMed] [Google Scholar]
- Moye-Rowley W. S., 2003. Transcriptional control of multidrug resistance in the yeast Saccharomyces. Prog. Nucleic Acid Res. Mol. Biol. 73: 251–279. 10.1016/S0079-6603(03)01008-0 [DOI] [PubMed] [Google Scholar]
- Moye-Rowley W. S., 2005. Retrograde regulation of multidrug resistance in Saccharomyces cerevisiae. Gene 354: 15–21. 10.1016/j.gene.2005.03.019 [DOI] [PubMed] [Google Scholar]
- Muller L. A., McCusker J. H., 2009. Microsatellite analysis of genetic diversity among clinical and nonclinical Saccharomyces cerevisiae isolates suggests heterozygote advantage in clinical environments. Mol. Ecol. 18: 2779–2786. 10.1111/j.1365-294X.2009.04234.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K., Morishima N., Shibata T., 1992. An endonuclease with multiple cutting sites, Endo.SceI, initiates genetic recombination at its cutting site in yeast mitochondria. EMBO J. 11: 2707–2715. 10.1002/j.1460-2075.1992.tb05336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarani A., Nesci S., Ventrella V., 2013. Modifiers of the oligomycin sensitivity of the mitochondrial F1F0-ATPase. Mitochondrion 13: 312–319. 10.1016/j.mito.2013.04.005 [DOI] [PubMed] [Google Scholar]
- Paliwal S., Fiumera A. C., Fiumera H. L., 2014. Mitochondrial-nuclear epistasis contributes to phenotypic variation and coadaptation in natural isolates of Saccharomyces cerevisiae. Genetics 198: 1251–1265. 10.1534/genetics.114.168575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris D., Arias A., Orlić S., Belloch C., Péréz-Través L., et al. , 2017. Mitochondrial introgression suggests extensive ancestral hybridization events among Saccharomyces species. Mol. Phylogenet. Evol. 108: 49–60. 10.1016/j.ympev.2017.02.008 [DOI] [PubMed] [Google Scholar]
- Perocchi F., Mancera E., Steinmetz L. M., 2008. Systematic screens for human disease genes, from yeast to human and back. Mol. Biosyst. 4: 18–29. 10.1039/B709494A [DOI] [PubMed] [Google Scholar]
- Peter J., De Chiara M., Friedrich A., Yue J. X., Pflieger D., et al. , 2018. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556: 339–344. 10.1038/s41586-018-0030-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repar J., Warnecke T., 2017. Mobile introns shape the genetic diversity of their host genes. Genetics 205: 1641–1648. 10.1534/genetics.116.199059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H. W., Zou P., Udeshi N. D., Martell J. D., Mootha V. K., et al. , 2013. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339: 1328–1331. 10.1126/science.1230593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Archer M. A., Wayne R. K., 1999. Transgressive segregation, adaptation and speciation. Heredity 83: 363–372. 10.1038/sj.hdy.6886170 [DOI] [PubMed] [Google Scholar]
- Rieseberg L. H., Widmer A., Arntz A. M., Burke J. M., 2003. The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1141–1147. 10.1098/rstb.2003.1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J., Hughes A. L., 2015. Power(2): the power of yeast genetics applied to the powerhouse of the cell. Trends Endocrinol. Metab. 26: 59–68. 10.1016/j.tem.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira A. H., 2012. Mitochondrial diseases. Lancet 379: 1825–1834. 10.1016/S0140-6736(11)61305-6 [DOI] [PubMed] [Google Scholar]
- Schon E. A., DiMauro S., Hirano M., 2012. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat. Rev. Genet. 13: 878–890. 10.1038/nrg3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer C., Rak M., Lefebvre-Legendre L., Duvezin-Caubet S., Plane G., et al. , 2006. Yeast models of human mitochondrial diseases: from molecular mechanisms to drug screening. Biotechnol. J. 1: 270–281. 10.1002/biot.200500053 [DOI] [PubMed] [Google Scholar]
- Shibata T., Nakagawa K., Morishima N., 1995. Multi-site-specific endonucleases and the initiation of homologous genetic recombination in yeast. Adv. Biophys. 31: 77–91. 10.1016/0065-227X(95)99384-2 [DOI] [PubMed] [Google Scholar]
- Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., et al. , 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 100: 13207–13212. 10.1073/pnas.2135385100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson D. C., Gaarder M. E., Livingston D. M., 1981. Characterization of the transmission during cytoductant formation of the 2 micrometers DNA plasmid from Saccharomyces. Mol. Gen. Genet. 183: 59–65. 10.1007/BF00270139 [DOI] [PubMed] [Google Scholar]
- Singh R., Sripada L., Singh R., 2014. Side effects of antibiotics during bacterial infection: mitochondria, the main target in host cell. Mitochondrion 16: 50–54. 10.1016/j.mito.2013.10.005 [DOI] [PubMed] [Google Scholar]
- Sinha H., Nicholson B. P., Steinmetz L. M., McCusker J. H., 2006. Complex genetic interactions in a quantitative trait locus. PLoS Genet. 2: e13 10.1371/journal.pgen.0020013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirek M., Polakova S., Jatzova K., Sulo P., 2014. Post-zygotic sterility and cytonuclear compatibility limits in S. cerevisiae xenomitochondrial cybrids. Front. Genet. 5: 454 10.3389/fgene.2014.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz L. M., Scharfe C., Deutschbauer A. M., Mokranjac D., Herman Z. S., et al. , 2002a. Systematic screen for human disease genes in yeast. Nat. Genet. 31: 400–404. http://www.ncbi.nlm.nih.gov/pubmed/12134146 [DOI] [PubMed] [Google Scholar]
- Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., et al. , 2002b. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330. 10.1038/416326a [DOI] [PubMed] [Google Scholar]
- Strope P. K., Skelly D. A., Kozmin S. G., Mahadevan G., Stone E. A., et al. , 2015. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 25: 762–774. 10.1101/gr.185538.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf J. D., Copeland W. C., 2011. Mitochondrial DNA replication and disease: insights from DNA polymerase gamma mutations. Cell. Mol. Life Sci. 68: 219–233. 10.1007/s00018-010-0530-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulo P., Szaboova D., Bielik P., Polakova S., Soltys K., et al. , 2017. The evolutionary history of Saccharomyces species inferred from completed mitochondrial genomes and revision in the ‘yeast mitochondrial genetic code’. DNA Res. 24: 571–583. 10.1093/dnares/dsx026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., 1992. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 9: 678–687. 10.1093/oxfordjournals.molbev.a040752 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartakoff A. M., Dulce D., Landis E., 2018. Delayed encounter of parental genomes can lead to aneuploidy in Saccharomyces cerevisiae. Genetics 208: 139–151. 10.1534/genetics.117.300289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. W., Turnbull D. M., 2005. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 6: 389–402. 10.1038/nrg1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn D. R., 2004. Mitochondrial disorders: prevalence, myths and advances. J. Inherit. Metab. Dis. 27: 349–362. 10.1023/B:BOLI.0000031098.41409.55 [DOI] [PubMed] [Google Scholar]
- Wallace D. C., 2010. Mitochondrial DNA mutations in disease and aging. Environ. Mol. Mutagen. 51: 440–450. 10.1002/em.20586 [DOI] [PubMed] [Google Scholar]
- Wang X., Chen X. J., 2015. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 524: 481–484. 10.1038/nature14859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zuo X., Kucejova B., Chen X. J., 2008. Reduced cytosolic protein synthesis suppresses mitochondrial degeneration. Nat. Cell Biol. 10: 1090–1097. 10.1038/ncb1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters J. F., Chiu K., Fiumera H. L., 2015. Population structure of mitochondrial genomes in Saccharomyces cerevisiae. BMC Genomics 16: 451 10.1186/s12864-015-1664-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters J. F., Charron G., Gaspary A., Landry C. R., Fiumera A. C., et al. , 2018. Mitochondrial recombination reveals mito-mito epistasis in yeast. Genetics 209: 307–319. 10.1534/genetics.117.300660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L., Topf U., Bragoszewski P., Wiese S., Sztolsztener M. E., et al. , 2015. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524: 485–488. 10.1038/nature14951 [DOI] [PubMed] [Google Scholar]
- Zhang X., Moye-Rowley W. S., 2001. Saccharomyces cerevisiae multidrug resistance gene expression inversely correlates with the status of the F(0) component of the mitochondrial ATPase. J. Biol. Chem. 276: 47844–47852. 10.1074/jbc.M106285200 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Strope P. K., Kozmin S. G., McCusker J. H., Dietrich F. S., et al. , 2014. Structures of naturally evolved CUP1 tandem arrays in yeast indicate that these arrays are generated by unequal nonhomologous recombination. G3 (Bethesda) 4: 2259–2269. 10.1534/g3.114.012922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M., 2012. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44: 821–824. 10.1038/ng.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn A. R., Pohlman J. K., Perlman P. S., Butow R. A., 1987. Kinetic and segregational analysis of mitochondrial DNA recombination in yeast. Plasmid 17: 248–256. 10.1016/0147-619X(87)90033-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All S. cerevisiae nuclear and mitochondrial genome sequence data has been previously published and deposited (https://doi.org/10.1101/gr.185538.114); see Table S19 of Strope et al. (2015) for GenBank accession and Sequence Read Archive numbers. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7361240.