Abstract
In the filamentous fungus Neurospora crassa, constitutive heterochromatin is marked by tri-methylation of histone H3 lysine 9 (H3K9me3) and DNA methylation. We identified mutations in the Neurospora defective in methylation-1 (dim-1) gene that cause defects in cytosine methylation and implicate a putative AAA-ATPase chromatin remodeler. Although it was well-established that chromatin remodelers can affect transcription by influencing DNA accessibility with nucleosomes, little was known about the role of remodelers on chromatin that is normally not transcribed, including regions of constitutive heterochromatin. We found that dim-1 mutants display both reduced DNA methylation in heterochromatic regions as well as increased DNA methylation and H3K9me3 in some intergenic regions associated with highly expressed genes. Deletion of dim-1 leads to atypically spaced nucleosomes throughout the genome and numerous changes in gene expression. DIM-1 localizes to both heterochromatin and intergenic regions that become hyper-methylated in dim-1 strains. Our findings indicate that DIM-1 normally positions nucleosomes in both heterochromatin and euchromatin and that the standard arrangement and density of nucleosomes is required for the proper function of heterochromatin machinery.
Keywords: heterochromatin, DIM-1, CATP, nucleosome, DNA methylation, Neurospora crassa
NUCLEOSOMES do more than compact DNA. The arrangement of nucleosomes and the post-translational modifications of their constituent histones serve regulatory mechanisms, e.g., to regulate cellular responses to environmental and developmental stimuli through transcriptional control (Vermaak and Wolffe 1998; Clapier and Cairns 2009; Henikoff 2016; Lai and Pugh 2017; Lämke and Bäurle 2017). Conserved chromatin remodelers have been identified that place, remove, or shift nucleosomes on DNA, and that can elicit a cascade of defects if compromised (Soppe et al. 2002; Tsukiyama 2002; Henikoff 2016; Clapier et al. 2017; Hammond et al. 2017; Lai and Pugh 2017). Despite advances identifying and characterizing the role of chromatin remodelers in gene expression, much remains to be learned about how they work (Tsukiyama 2002). Moreover, a major gap in our knowledge concerns possible effects of nucleosome positioning on heterochromatin, which is the largely silent fraction of the genome (Grewal and Jia 2007; Rountree and Selker 2010; Allshire and Madhani 2017). Constitutive heterochromatin is mostly in centromeric regions, and is gene-poor, rich in repeated sequences, generally marked by methylation of lysine 9 of histone H3 (H3K9me) and cytosine methylation, and contains characteristic proteins such as Heterochromatin Protein-1 (HP1) (Henikoff 2000; Freitag et al. 2004; Grewal and Jia 2007; Lewis et al. 2008; Bühler and Gasser 2009; Freitag 2017). This form of heterochromatin is responsible for silencing transcription and recombination of underlying selfish DNA, and plays roles in chromosome segregation and genome organization (Henikoff 2000; Lewis et al. 2008). Facultative heterochromatin is also transcriptionally quiescent but is associated with functional genes and its distribution can change during development and/or as a result of environmental changes. In most eukaryotes, facultative heterochromatin is marked by methylation of lysine 27 of histone H3 (H3K27me) (Trojer and Reinberg 2007; Wutz 2011; Connolly et al. 2013; Jamieson et al. 2013; Freitag 2017; Wiles and Selker 2017). Changes in heterochromatin have been associated with developmental defects, cancer, and embryonic lethality (Ronemus et al. 1996; Okano et al. 1999; Tachibana et al. 2002; Heard 2005; Wutz 2011; Connolly et al. 2013; Elgin and Reuter 2013; Lewis and Allis 2013; Lewis et al. 2013; Timp and Feinberg 2013; Grossniklaus and Paro 2014; Morgan and Shilatifard 2015; Studt et al. 2016).
Information on the genes necessary for the formation and regulation of heterochromatin is limited, prompting investigations in favorable model organisms with genomic features similar to higher metazoans, such as Drosophila melanogaster and the filamentous fungus Neurospora crassa (Rountree and Selker 2010; Elgin and Reuter 2013). Heterochromatin is not essential in Neurospora, which allows for the identification of genes involved in heterochromatin formation and function (Foss et al. 1993; Lewis et al. 2010a,b), including the Defective In Methylation-5 (DIM-5) H3K9 methyltransferase (MTase), and other members of DCDC (DIM-5/-7/-9, CUL4, DDB1dim-8 Complex). All components of DCDC are required for H3K9me3 and localization of HP1, which specifically binds to the H3K9me3 mark (Nielsen et al. 2002; Freitag et al. 2004; Lewis et al. 2010a,b; Honda et al. 2012). HP1 directly recruits the DNA MTase DIM-2 and an HDAC complex, HCHC (HP1/CDP-2/HDA-1/CHAP), to constitutive heterochromatin and is also involved in the DMM complex, which limits spreading of heterochromatin (Honda and Selker 2008; Honda et al. 2010, 2012).
We report the identification and characterization of the Neurospora dim-1 gene, which was originally found because mutations of this gene conferred resistance to the toxicity of 5-azacytidine (5-azaC) in the mutagen-sensitive mus-20 genetic background; the mutant was also found to be partially defective in DNA methylation. We identified the gene, showed that it encodes an AAA-ATPase nucleosome remodeler conserved from yeast to humans, and found that a strain with a deletion of the gene (Δdim-1) has altered nucleosome positions throughout the genome. Loss of DIM-1 causes constitutive heterochromatin to lose much of its cytosine methylation although the underlying H3K9me3 is retained. In addition, intergenic regions exhibit DNA hyper-methylation in dim-1 strains. DIM-1 and the heterochromatin machinery preferentially localize to regions of aberrant hyper-methylation, suggesting that they are targets for DIM-1 activity. We note that these regions have variable and often increased nucleosome density that mirror the nucleosome profile in heterochromatic regions and suggest that nucleosome disorder may lead to the hyper-methylation.
Materials and Methods
N. crassa strains were grown, maintained, and crossed following standard protocols (Davis 2000); strains are listed in Supplemental Material, Table S1. Gene replacement constructs and his-3 targeting constructs were generated as described (Honda and Selker 2009) using oligonucleotides listed in Table S2. Deletion strains were obtained from the Neurospora knockout collection (Colot et al. 2006) stored at the Fungal Genetics Stock Center or remade (Δdim-1::nat1) with the oligonucleotides listed in Table S2 using a similar protocol.
Whole-genome sequencing of dim-1 strains [N654 (heterokaryotic strain) and N1072 (microconidiated, homokaryotic strain)] and a parental strain (N200) was performed with the Nextera kit (Illumina) following the manufacturer’s protocol. Single nucleotide polymorphisms (SNPs) in parental and dim-1 strains, relative to the Neurospora version 10 genome, were identified using Freebayes (https://github.com/ekg/freebayes), SNPs common between output vcf files were removed using vcf-isec in VCFtools (Danecek et al. 2011), genic mutations were identified using SnpEFF (Cingolani et al. 2012), and SNPs residing on Linkage Group III were filtered using SnpSift (Cingolani et al. 2012).
Bisulfite-sequencing was performed as described (Klocko et al. 2015; Honda et al. 2016) and chromatin immunoprecipitation (ChIP)-sequencing was performed as described in Jamieson et al. (2013); one replicate of WT H3K9me3 and the WT H3K27me2me3 data set were previously reported (NCBI GEO GSE68897) (Jamieson et al. 2016). All reads were mapped to the corrected (Galazka et al. 2016) Neurospora genome, version 12 (NC12), and final TDF files were generated across 25 bp windows. To generate ChIP-sequencing and bisulfite-sequencing average enrichment profiles, .bam files were uploaded to the Galaxy web platform public server at usegalaxy.org (Afgan et al. 2018), and the DeepTools program was used (Ramírez et al. 2016). Peaks of ChIP-sequencing enrichment were called with MACS2 (Zhang et al. 2008) or SICER (Xu et al. 2014) using the default settings of each program; SICER detected 119 new peaks while MACS detected 394 new peaks, and while many peaks were in agreement between the two peak call output files, differences may reflect differences in how enrichment over background is calculated or changes in the gaps between called peaks. Peak call output files for each strain and the differences between strains are provided in File S2 in a format so these data can be easily made into bed files for genome browser display. PolyA messenger RNA (mRNA)-sequencing was performed as described (Klocko et al. 2016), using the wild-type (WT) polyA mRNA data set as a control [GSE82222; previously published in Klocko et al. (2016)]; for mRNA-sequencing analysis, a log2 ≥ 2.0 and log2 ≤ −2.0 cut-off with adjusted P-values of 0.05 were used. ChIP-sequencing enriched peaks, bisulfite-sequencing methylated regions, genes with significant expression changes in Δdim-1, and genes with cytosine methylation peaks that change expression are provided in File S2. All individual locus pictures were visualized and generated on Integrative Genomics Viewer (Robinson et al. 2011). Southern blotting was performed as in Miao et al. (2000) with the denoted probe DNA (oligos listed in Table S2). Histone isolation was performed as in Honda and Selker (2009). Western blotting was performed as in Adhvaryu et al. (2011). Mass spectrometry was performed as in Honda et al. (2012). Micrococcal nuclease (MNase) digestion followed by sequencing (MNase-seq) was performed as follows. Isolated nuclei were used for input DNA; nuclei were isolated following the protocol from Klocko et al. (2015). Frozen nuclei in storage buffer were thawed and resuspended in MNase digestion buffer [10 mM Hepes-NaOH, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 50 µg/ml filter-sterilized bovine serum albumin, β-mercaptoethanol 5 mM final)] and pepstatin (10 µg/ml final), leupeptin (10 µg/ml final), and PMSF (1 mM final) were added before use. DNA concentration was determined by removing a 10 µl fraction, adding SDS to 1% to solubilize nuclear membranes and DNA quantified with a Qubit HS in triplicate. Nuclei containing 16 µg of DNA were adjusted to 400 µl with MNase digestion buffer, CaCl2 was added to a final concentration of 2 mM, MNase (New England Biolabs, Beverly, MA) was added to a final concentration of 0.5 unit/µl, and the reaction was carried out at 37° for 20 min. At 2-min intervals, 30 µl aliquots (corresponding to 1.2 µg of DNA) were removed and mixed with 8 µl of 100 mM EGTA to stop the reaction and stored on ice. RNase A (5 µg) was then added to all samples and they were incubated an additional 10 min at 37° before addition of SDS to a final concentration of 1% to solubilize the nuclear membranes. DNA was purified from samples with a Qiagen MinElute Kit, and either used for Southern blotting or sequencing. For MNase-seq, the mono- and di-nucleosome populations, with the associated linker DNA, of two independent biological replicates of the 8- and 10-min samples were pooled for sequencing, reasoning that at earlier time points, the slower-digested euchromatic regions and the rapidly digested heterochromatic regions would be present in our data. While mono- and di-nucleosome populations were analyzed (independently and combined), for simplicity, only data from mono-nucleosomes are shown in the figures. MNase-sequencing libraries were constructed using the Illumina TruSeq kit, following the manufacturer’s protocols with the following modifications. After end repair, DNA was purified using a Qiagen MinElute Kit, as recommended by Lombardi et al. (2014). Libraries containing mono- and di-nucleosomes (and the fragments between these prominent bands) were size-selected with 2% TAE gel electrophoresis following an eight-cycle PCR amplification step; the average size of mono-nucleosomes was ∼270 bp, while the average size of di-nucleosomes was ∼490 bp. Libraries were sequenced on a NextSeq 500 (University of Oregon Genomics Core Facility) as 37 nt paired-end (PE) reads. Reads denoting mono-nucleosomes were mapped to the corrected Neurospora genome version 12 (Galazka et al. 2016) using Bowtie2 (Langmead and Salzberg 2012), with only PE reads with a total distance between reads of 120 and 180 bp to denote mono-nucleosomes used in subsequent analyses; nucleosome positions were determined and nucleosome peaks were smoothed with the program NucWave (Quintales et al. 2014). Nucleosome signals relative to genomic position were determined with the program SitewriterCFD.pl (Kent et al. 2010; Gal et al. 2015) using bed files of the transcriptional start sites (TSSs) all, up-, and downregulated genes, or genes that are near (<500 bp) or distant (∼7500 bp) from cytosine methylation peaks; all region bed files were modified as four-column input site files for SitewriterCFD.pl (provided in File S2). For average enrichment profiles, average nucleosome signal was graphed in Excel, while for heatmaps, individual loci signal matrices were normalized to the average signal (in Excel), clustered (Cluster 3.0) (de Hoon et al. 2004) or reordered relative to other clusters (custom python script; File_S3-Arrange_clusters_Arguments), and plotted as heatmaps (Java TreeView) (Saldanha 2004). SitewriterCFD.pl was also used to calculate cytosine methylation signal relative to genomic position using single base pair resolution input files generated in UNIX by replacing cytosine methylation signal data into a single base zero-signal position file.
Data availability
All ChIP-sequencing, MNase-sequencing, bisulfite-sequencing, and RNA-sequencing data sets, as well as genome position files, have been deposited to the NCBI Gene Expression Omnibus (accession number GSE98911). Supplemental Figure files (Figures S1–S20 and File S1), data sets (File S2), and program (File S3) have been deposited to Figshare. Reagents, including strains, are available upon request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7210190.
Results
Identification of the dim-1 gene
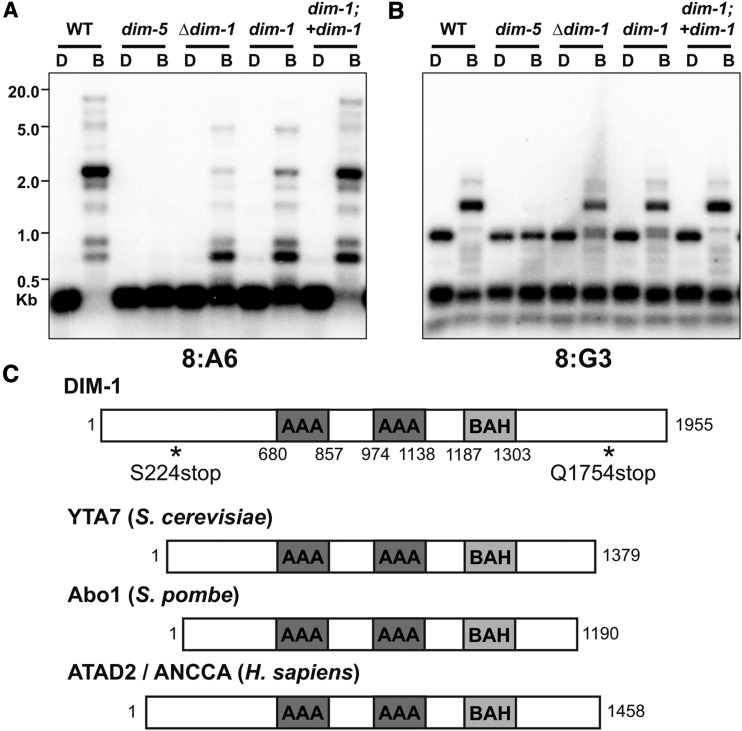
The Neurospora dim-1 strain has a ∼50% reduction in DNA methylation at constitutive heterochromatin (see Figure 1, A and B) (Foss et al. 1993), and was initially mapped to a large region of the right arm of Linkage Group III (Foss et al. 1998). To identify the defect responsible for this methylation defect, we sequenced the genomes of two dim-1 strains (N654 and N1072) derived from one of the three independent isolates from the original selection (Foss et al. 1993). A nonsense mutation (S224stop) was identified in gene NCU06484 in the dim-1 strains and we confirmed that this mutation was absent from the parental mus-20 strain (N200). Sanger sequencing of NCU06484 from an independent dim-1 mutant strain (N1634; Figure S1) revealed a nonsense mutation at Q1754, supporting the idea that NCU06484 is dim-1. The predicted protein encoded by NCU06484 is 1955 amino acids long and has orthologs that share significant sequence similarity in organisms from yeast to humans (File S1). The DIM-1 primary structure includes two AAA-ATPase domains, a weakly predicted BAH domain, and a positively charged N terminus (Figure 1C); the latter two structural elements suggest DIM-1 may interact with histones (Lombardi et al. 2011). This gene product was previously identified as “clock ATPase” (CATP) because of an apparent role in altered circadian rhythm (see Discussion) (Cha et al. 2013). Further, we analyzed DNA methylation in a strain harboring a complete deletion of NCU06484 (N5496) and found a loss of methylation at constitutive heterochromatin comparable to that of the strains harboring the dim-1 nonsense mutations (Figure 1, A and B). Finally, we found that the hypomethylation phenotype of a dim-1 strain was complemented by expression of an ectopic, FLAG-tagged dim-1 gene (Figure 1, A and B and Figure S2), confirming that the mutations within NCU06484 are responsible for the Dim phenotype.
Figure 1.
DNA methylation is abnormal in dim-1 strains at select loci, and DIM-1 has features of ATP-dependent chromatin remodeling factors. (A and B) Southern blot of genomic DNA from the indicated strains digested with either DpnII (D) or its 5mC-sensitive isoschizomer BfuCI (B), and probed for the heterochromatic regions 8:A6 (A) or 8:G3 (B). To test for complementation of the dim-1 mutation, we crossed in a WT allele inserted at the his-3 locus (“+dim-1”) (C). Primary structure schematics of DIM-1 and its homologs from yeasts and humans with known domains are indicated, amino acid coordinates are shown below, and positions of nonsense mutations in dim-1 strains are shown by asterisks.
Hyper-methylation at intergenic regions in dim-1 strains
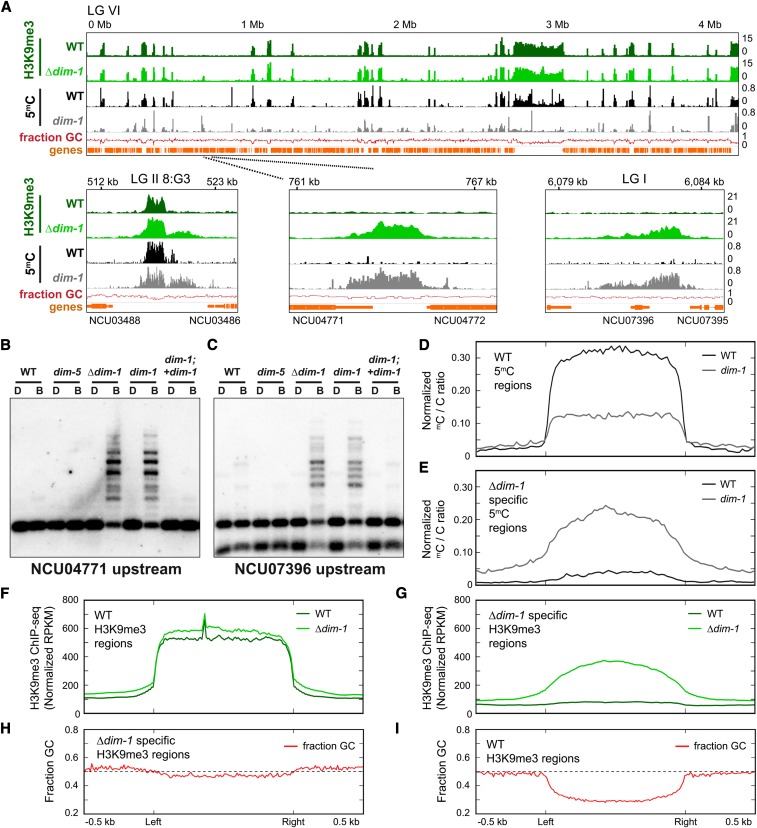
Early assessments of DNA methylation in dim-1 strains by Southern hybridization suggested that the defect does not uniformly reduce DNA methylation (Foss et al. 1998). Unlike effects of reducing methylation with 5-azaC treatment or methionine starvation (Roberts and Selker 1995), differential changes were observed at the sites tested. To characterize the effect of dim-1 on DNA methylation globally, we performed whole genome bisulfite-sequencing on WT and dim-1 strains bearing either the S224stop allele (N1634) or a full deletion of the gene (Δdim-1; N5496). Bisulfite-sequencing confirmed the substantial hypomethylation in dim-1 strains detected by Southern hybridizations at regions of constitutive heterochromatic (Figure 2A and Figures S3, A and B and S4). Surprisingly, however, both dim-1 strains also showed numerous new peaks of cytosine methylation, as illustrated for Linkage Group VI (Figure 2A and Figure S3, A and B; other chromosomes are shown in Figure S4). The hyper-methylation was found predominantly at intergenic regions but also in some gene bodies. Representative examples of this aberrant methylation were confirmed by Southern hybridizations (Figure 2, B and C and Figure S3C); control Southern blots, e.g., with a fragment of the unmethylated am gene body (Figure S3D), verified that the digests were complete. Average enrichment profiles, which display the average sequencing signal across the examined regions, revealed that while overall methylation was reduced ∼50% in constitutive heterochromatin (Figure 2D and Figure S3E top), when all methylated regions are considered (constitutive heterochromatin plus the “neo-heterochromatin” in dim-1 strains), the hyper-methylation and hypomethylation largely cancel each other such that dim-1 and WT strains show similar levels of cytosine methylation (5mC) overall (Figure S3E, middle). Some spreading of 5mC in dim-1 beyond the normal heterochromatin boundaries was also observed, e.g., at 8:G3 (Figure 2A and Figure S3A) and 8:A6 (Figure S3B) and as shown in the average enrichment profiles of aggregated data sets (Figure S3E, middle). Interestingly, close examination of only the regions that became hyper-methylated in dim-1 strains revealed traces of DNA methylation in the WT strain (Figure 2E and Figure S3, A, B, and E bottom). Methylation patterns of strains with deletion or nonsense alleles were very similar but effects of the deletion appeared more pronounced in some assays (Figures S3, A, B, and E and S4). We confirmed that the DNA methyltransferase DIM-2 is required for the aberrant cytosine methylation in Δdim-1 (Figure S5) and found that the hypomethylation of constitutive heterochromatin regions was not rescued by overexpression of dim-2 when a second copy of the gene was inserted at an ectopic locus (Figure S6).
Figure 2.
DNA methylation and H3K9me3 are abnormal in dim-1 strains genome-wide. (A) ChIP-sequencing tracks (merged replicates) for H3K9me3 (green) or bisulfite-sequencing for cytosine methylation (black/gray) for WT or dim-1 strains, and base composition (“fraction GC,” red); all tracks are displayed in 25 bp windows. LG VI is shown, but comparable results were obtained for the other six chromosomes (Figure S4). Selected regions are shown expanded below; different chromosomes [Linkage Groups (LGs)] are identified. Tracks here and in other figures are displayed with the Integrative Genomics Viewer (Robinson et al. 2011) with coordinates relative to the left telomere, and NCU numbers of genes, shown above and below, respectively. (B and C) Southern blots of genomic DNA from the indicated strains digested with either DpnII (D) or its 5mC-sensitive isoschizomer BfuCI (B) and probed for the labeled regions, as in Figure 1A; “upstream” denotes the intergenic region upstream of that gene. (D and E) Average enrichment profiles of bisulfite-sequencing data showing the ratio of methylated-to-unmethylated cytosines across H3K9me3-marked regions in WT strains (D; n = 210) or specifically found in Δdim-1 strains (E; n = 239). Plots show 500 bp upstream and downstream of the left and right borders of constitutive heterochromatic regions, which were scaled to 1 kb for presentation. y-axis denotes the normalized ratio of methylated cytosines/total cytosines per amount of sequencing genome coverage. (F and G) Average enrichment profiles, as in D and E, but of H3K9me3 ChIP-sequencing enrichment in WT and Δdim-1 strains merged from two experiments; y-axis denotes the normalized amount of ChIP-sequencing reads per kilobase per million total reads (RPKM). (H and I) Average enrichment profiles, as in B and C, but of fraction of GC base pairs; horizontal line marks the position where base composition is 50% G+C.
We considered the possibility that the novel methylation was directed by small interfering RNAs, such as Dicer-independent small interfering RNAs (disiRNAs) that have been implicated in low-level methylation associated with antisense or convergent transcription (Lee et al. 2010; Dang et al. 2016). To test this, we deleted the eri-1gene, which encodes the RNase necessary for disiRNA and disiRNA locus DNA methylation (Dang et al. 2016). Loss of ERI-1 did not abolish the peaks of cytosine methylation that appear in dim-1 mutants (Figure S7), ruling out this possibility.
Altered enrichment of post-translational histone marks in dim-1 strains
Considering that cytosine methylation in N. crassa depends on H3K9me3, we compared the distribution of this mark in Δdim-1 and WT strains by ChIP-sequencing. The patterns of H3K9me3 in the strains were similar but the Δdim-1 strain showed additional peaks of H3K9me3 associated with the new peaks of intergenic DNA methylation (Figure 2A and Figures S3B and S4). The new H3K9me3 peaks in dim-1 were often smaller (average of ∼2000 bp) and less intense than those in constitutive heterochromatic regions of WT strains. This general difference was observed with two different peak-call pipelines, MACS2 and SICER (Zhang et al. 2008; Xu et al. 2014), although details varied, with SICER detecting fewer new peaks than MACS2 (File S2). The gain of H3K9me3 peaks did not appreciably increase global H3K9me3 levels (Figure S3F). An average enrichment profile analysis of H3K9me3 ChIP-sequencing from Δdim-1 strains showed nearly equivalent levels of H3K9me3 in constitutive heterochromatic regions as defined in a WT strain (Figure 2F and Figure S8A) plus minor spreading of H3K9me3 beyond the heterochromatic region borders. When the neo-heterochromatin of the Δdim-1 was also considered, however, the mutant showed increased H3K9me3 levels (Figure S8B). Considering just the intergenic regions associated with new DNA methylation revealed substantially increased H3K9me3 (Figure 2G and Figure S8C). As expected, all H3K9me3 associated with neo-heterochromatin was found to require DIM-5 (Figure S9). Unlike the highly AT-rich constitutive heterochromatin sequences that are normally methylated, the base composition was found to be only slightly enriched for A:T base pairs in the regions hyper-methylated in the Δdim-1 strain (Figure 2, H and I). Silencing associated with constitutive heterochromatic regions was not noticeably affected by dim-1 (Figure S10).
We considered whether the mark of facultative heterochromatin, H3K27me2/3, might be also affected by deletion of dim-1. ChIP-sequencing of H3K27me2/3 showed some, but relatively few, differences in the level and/or distribution of this mark in WT and Δdim-1 strains (Figure S11); a peak analysis showed that 11 minor peaks were lost and 45 peaks were gained, (File S2), and some peaks present in both WT and Δdim-1 strains became more and less prominent in the Δdim-1 strain (Figure S11, top and bottom, respectively). In addition, one region of constitutive heterochromatin had a reduction in H3K9me3 but gained H3K27me2/3 (Figure S11, top).
Transcriptional effects of deletion of dim-1
Unlike Neurospora strains bearing a deletion of the DNA MTase gene, dim-2, which grow well (Honda et al. 2012), Δdim-1 strains grow slowly (Figure S12). This suggested that the DNA hyper-methylation observed in euchromatic regions of dim-1 strains affects gene expression. We explored this possibility by carrying out RNA-sequencing on poly-A mRNA from Δdim-1 and WT strains. In the ∆dim-1 strain, 601 genes were significantly upregulated, and 958 genes were significantly downregulated relative to the WT strain (File S2). Given that many of the newly methylated regions are at promoter regions, we wondered if establishment of heterochromatin at promoters repressed gene expression. Of the 958 downregulated genes, only 155 (16.2%) showed cytosine methylation of at least 200 bp within 500 bp of the predicted TSSs (File S2). Consistent with the weak correlation between new cytosine methylation and reduced gene expression, average enrichment profiles of 5mC across up- and downregulated genes in Δdim-1 showed minor changes in methylation and these were not correlated with changes in gene expression (Figure S13, A and B). It remains possible that methylation changes indirectly affect gene expression. Interestingly, the highest expressed quartile of genes in WT showed the greatest new cytosine methylation in the Δdim-1 strain, which is mostly in promoter regions and somewhat in 3′UTR regions (Figure S13C). Silent genes showed little cytosine methylation associated with their 5′ and 3′ regions (Figure S13D).
Loss of DIM-1 alters nucleosome positions genome-wide
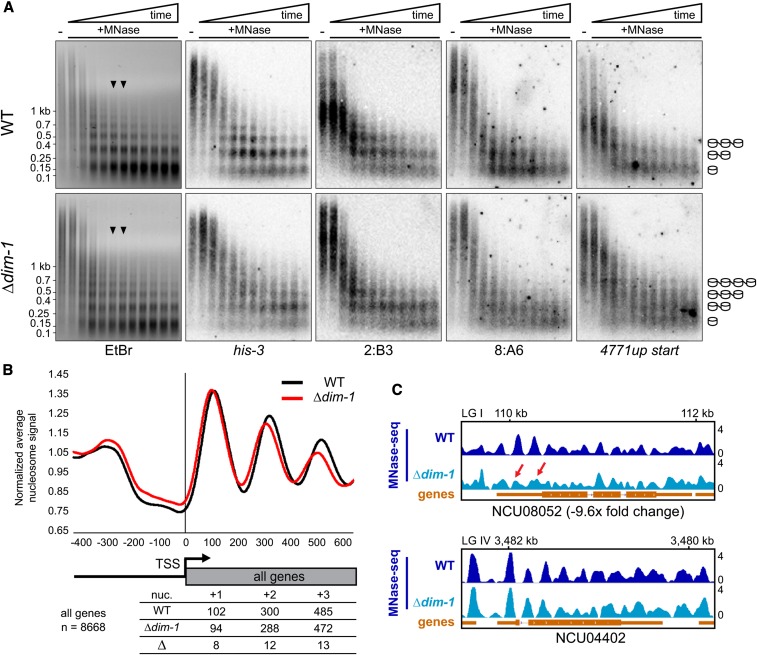
The substantial changes in gene expression detected in dim-1 strains are consistent with the possibility that DIM-1 serves as an ATP-dependent chromatin remodeling factor (“chromatin remodeler”). DIM-1 includes two ATPase domains and two regions that may facilitate histone interactions, a weakly conserved BAH domain and a positively charged N terminus (Figure 1C), and is homologous to chromatin remodelers in other species, including Yta7 in Saccharomyces cerevisiae, Abo1 in Schizosaccharomyces pombe, and ANCCA/ATAD2 in humans (Figure 1C and File S1) (Zou et al. 2007; Revenko et al. 2010; Lombardi et al. 2011; Gal et al. 2015). Indeed, DIM-1 was reported as a CATP involved in the Neurospora circadian clock and has been shown to affect nucleosome occupancy at the C-box enhancer element of the frequency locus (Cha et al. 2013). To further investigate the possibility that DIM-1 is a chromatin remodeler, we tested MNase sensitivity of chromatin from WT and Δdim-1 strains. We first performed MNase time-course experiments, gel-fractionated the products, and probed for various genomic regions. The terminal time points showed predominantly mono- and di-nucleosome populations (Figure 3A, EtBr). Curiously, in both strains, heterochromatic regions (2:B3 and 8:A6) appeared to be digested to mono- and di-populations somewhat faster than the euchromatic region (his-3), perhaps due to differences of base composition or accessibility. In addition, heterochromatic nucleosomes appeared more disordered than their euchromatic counterparts. Nucleosome bands for both the euchromatic (his-3) and heterochromatic (2:B3 and 8:A6) regions appeared “fuzzier” in chromatin from the Δdim-1 strain than those from WT, suggestive of greater nucleosome disorder in the mutant (Figure 3A). Results for an intergenic region that has aberrant hyper-methylation in dim-1 (NCU04771 upstream) were also suggestive of nucleosome alterations in a dim-1 strain, with multiple poorly defined nucleosomes rather than predominately a single nucleosome at this region of WT (Figure 3A, right). The Southern probe covers the nucleosome-free region (NFR) of NCU04771, raising the possibility that this NFR in a Δdim-1 strain has increased, albeit variable, nucleosome occupancy, as has been reported for Yta7 in S. cerevisiae (Lombardi et al. 2011).
Figure 3.
Nucleosome positioning is altered at the TSSs of genes in Δdim-1 strain. (A) Southern blots of DNA from 20-min time course micrococcal nuclease (MNase) digest with WT and Δdim-1 nuclei, probed for the indicated regions; the “NCU04771up start” probe covers the intergenic promoter region of gene NCU04771, including the nucleosome-free region. Arrowheads indicate the time points (8 and 10 min) from which mono- and di-nucleosomes and intervening DNA was purified for paired-end high-throughput sequencing (below). Cartoons at right show interpretation of nucleosome patterns leading to smallest fragments. (B) Average nucleosome enrichment profiles of MNase-sequencing data from WT and Δdim-1 strains normalized to the average signal across the 5′ end of all Neurospora genes, spanning 400 bp upstream to 600 bp downstream of the TSS; numbers below indicate the peak apices, in base pairs from the transcriptional start site (TSS) in WT and Δdim-1 strains, and the difference in the apex position, for the +1, +2, and +3 nucleosomes. (C) Representative examples of nucleosome positions in individual genes with changed (NCU08052), or unchanged (NCU04402; dim-5) expression in a Δdim-1 background. Red arrows highlight nucleosome disorder in a Δdim-1 strain of two nucleosomes that are well-positioned in a WT strain.
To characterize nucleosome positioning genome-wide, we performed paired-end sequencing of mono-nucleosomes isolated from moderately digested chromatin of Δdim-1 and WT strains (time points indicated by arrow heads in Figure 3A). Results from replicates covered the entire genome and were highly reproducible (Figure S14), giving us confidence in the significance of observed differences and allowing us to merge the data for subsequent analyses. Alignment of all genes with the presumptive TSSs (n = 8668) revealed that nucleosome peaks in the Δdim-1 strain were progressively shifted slightly toward the 5′ ends of genes; the nucleosome length was shortened by roughly 6 bp (Figure 3B), which became obvious by the third (+3) nucleosome (Figure 3B). An equivalent effect had been reported for the budding yeast homolog of DIM-1 homolog, Yta7 (Lombardi et al. 2014). Given that Yta7 has been reported to be a regulator of histone H3 (hH3) expression (Gradolatto et al. 2008; Fillingham et al. 2009), we checked whether expression of hH3 (NCU01635) in the Δdim-1 strain was similar to that of a WT strain and found that it was (File S2, RNA-sequencing). We also found that addition of an ectopic hH3 gene did not rescue the Δdim-1 phenotype (Figure S15), suggesting that changes in hH3 levels were not responsible for the differences in nucleosome positioning.
Considering that a global analysis of nucleosome positioning of all genes might hide some DIM-1-specific nucleosome changes, we examined nucleosome positioning at the TSS of subsets of genes, including those whose expression or DNA methylation changed in Δdim-1. Genes with decreased expression showed the most substantial changes in nucleosome positions in average enrichment profiles (Figure S16A). Genes whose expression increased, or did not change, in a Δdim-1 strain seemed to exhibit less change (Figure S16, B and C). Examination of individual genes supported the suggestion that the dim-1 defect leads to differences in nucleosome positioning, as illustrated for downregulated NCU08052 (Figure 3C, top). Genes that did not have substantial changes in expression and those that were upregulated appeared to maintain nucleosome positioning better, as illustrated by NCU04402 (Figure 3C, bottom) and NCU03598 (Figure S16D). We also tested if changes in nucleosome positioning correlated with changes in DNA methylation at nearby genes. An average enrichment profile of 541 genes whose TSS was within 500 bp of a new peak of cytosine methylation in a Δdim-1 strain showed modest changes in nucleosome arrangement (Figure S16E) but similar changes were observed with control regions farther (∼7500 bp) from the new cytosine methylation (Figure S16F).
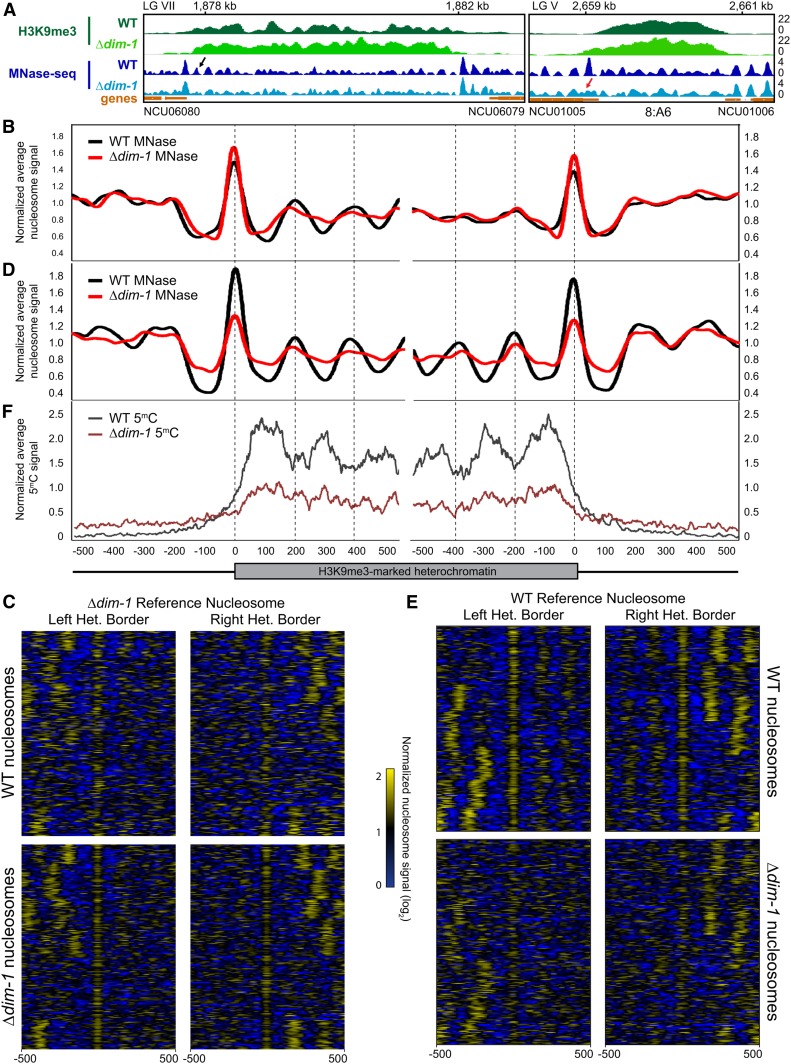
We next investigated nucleosome positions in constitutive heterochromatin of WT and dim-1 strains. Our survey, performed on 210 regions of H3K9me3 in WT, resulted in two conclusions: (1) Relative to euchromatin, nucleosomes in the regions of constitutive heterochromatin appear irregular/disordered; and (2) this disorder is exacerbated by loss of DIM-1. Examination of representative regions of the constitutive heterochromatin reveal that the nucleosome peaks were shorter and broader than in the adjacent chromatin (e.g., see Figure 4A), both in the WT and Δdim-1 strains, presumably reflecting irregularly spaced nucleosomes. Differences between the strains were also evident though somewhat subtle. Especially in the WT strain, the heterochromatin was typically flanked by strong border nucleosomes. Although more disordered, internal nucleosomes sometimes showed some regularity, especially in the WT strain (see Figure 4A, left, black arrow). The relative disorder of nucleosomes in the heterochromatin of the Δdim-1 may partly relate to fact that H3K9me3 exhibits spreading in the mutant beyond the normal heterochromatin boundary (e.g., see Figure 4A, right, red arrow). To further explore the difference in nucleosome arrangement between heterochromatin and euchromatin in Δdim-1 and WT strains, we generated average enrichment profiles and heatmaps to display the nucleosome signals ±500 bp of H3K9me3 regions. Alignments were to the first nucleosome within the H3K9me3 of the Δdim-1 (Figure 4, B and C) or WT (Figure 4, D and E) strains. The relative disorder of nucleosomes greatly limited the periodicity, especially in the Δdim-1 strain. Although even the WT strain showed limited nucleosome order (Figure 4B), the disorder of the dim-1 strain was strikingly greater at both borders, independent of the reference nucleosome (Figure 4, B–E). A control analysis based on random genomic positions did not show patterning (Figure S17). Together, these analyses suggest that heterochromatic regions in WT Neurospora have inherent nucleosome disorder, and that nucleosomes in a Δdim-1 strain are even more disordered.
Figure 4.
Constitutive heterochromatic regions exhibit nucleosome disorder, especially in Δdim-1 strains. (A) Representative examples of nucleosome positioning within constitutive heterochromatin. H3K9me3 ChIP-sequencing (green) and MNase-sequencing (blue) tracks of WT and Δdim-1 strains are shown for a region on LG VII between NCU06080 and NCU06079 and the 8:A6 region on LG V (Selker et al. 2003). The black arrow denotes an internal nucleosome that has periodicity in the WT strain while the red arrow denotes a border nucleosome that becomes disordered in the ?dim-1 strain. (B) Average mono-nucleosome enrichment profiles of MNase-sequencing data (MNase) in WT and Δdim-1 strains at the left and right borders (relative to the left telomere) of constitutive heterochromatic regions longer than at least 1000 base pairs aligned from the first nucleosome peak completely covered by H3K9me3 in Δdim-1 strains. (C) Nucleosome peak signal heatmaps from WT and Δdim-1 strains normalized to the average enrichment signal. Data are shown for ±500 bp for the left and right heterochromatic region borders (n = 210) relative to the first heterochromatic nucleosome peak in Δdim-1 strains. Heterochromatic regions are identically ordered in each heatmap group. (D) Average mono-nucleosome enrichment profiles of MNase-sequencing data (MNase) in WT and Δdim-1 strains at the left and right borders, as in B. Vertical dashed lines mark the apices of WT nucleosome signals. (E) Nucleosome peak signal heatmaps from WT and Δdim-1 strains normalized to the average enrichment signal, as in C, relative to the first heterochromatic nucleosome peak in WT strains. (F) Average enrichment profiles of bisulfite-sequencing showing enrichment of cytosine methylation in WT and Δdim-1 strains at the left and right borders of constitutive heterochromatic regions.
Interestingly, when we aligned bisulfite-sequencing data to the WT constitutive heterochromatin border, cytosine methylation showed an oppositely phased pattern relative to the nucleosome peaks in WT; i.e., it occurs principally in linker DNA, consistent with the possibility that the DNA methyltransferase DIM-2 has greater access to cytosines in linker DNA (Figure 4F, dark gray line). This pattern is not obvious in DNA of the Δdim-1 strain and cytosine methylation is consistently lower across the constitutive heterochromatin region (Figure 4F, maroon line).
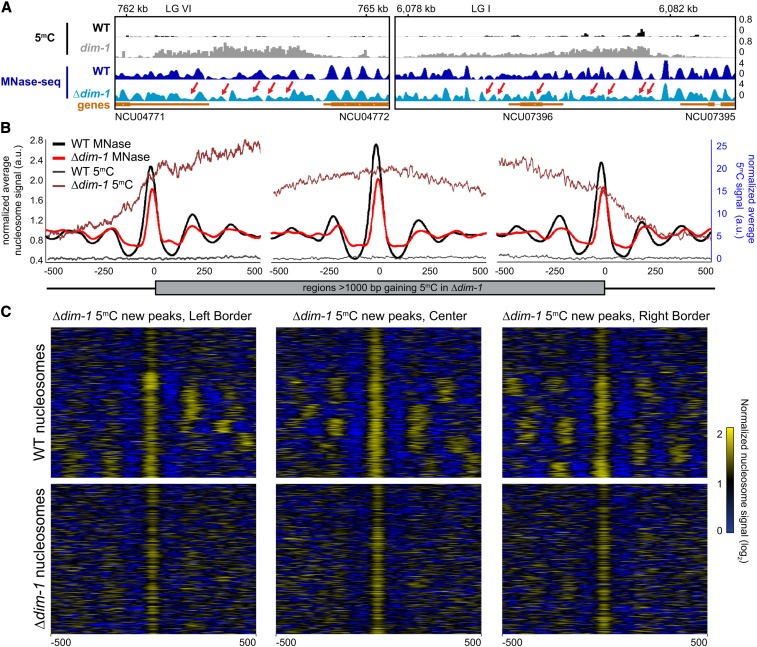
Although we did not observe marked changes in nucleosome positions around the TSSs of genes near neo-heterochromatin, we did find significant changes in the position of nucleosomes in intergenic regions that gain cytosine methylation in dim-1 strains. This was evident both by inspection of individual regions (Figure 5A) and in average enrichment profiles and heatmaps (Figure 5, B and C). Nucleosome peaks in the Δdim-1 strain were more numerous, lower, and broader than in the WT strain (Figure 5A). This is consistent with the increased nucleosome laddering from reduced MNase accessibility observed by Southern hybridizations (Figure 3A) and suggests that nucleosome positions are destabilized by loss of DIM-1. Average enrichment profiles of nucleosome and bisulfite-sequencing signals at the borders of these regions and at a randomly chosen, more central nucleosome showed reduced signals indicating reduced nucleosome positioning, especially of those nucleosomes internal to the neo-heterochromatin borders (Figure 5B), a conclusion reinforced by the heatmaps showing the position of nucleosomes at each region analyzed (Figure 5C). The cytosine methylation that appeared in the Δdim-1 strain showed no clear periodicity comparable to that normally found in heterochromatin. Together, these data reinforce the notion that regions gaining cytosine methylation in a Δdim-1 strain have relatively disordered nucleosomes.
Figure 5.
Intergenic regions that gain cytosine methylation in Δdim-1 exhibit aberrant nucleosome positioning. (A) Representative examples of nucleosome positions within intergenic regions that gain cytosine methylation. Tracks of bisulfite-sequencing for WT (black) and dim-1 (light gray), as well as MNase-sequencing for WT (dark blue) and Δdim-1 (light blue) strains are shown. Red arrows highlight changes in nucleosome positioning in Δdim-1 strain. (B) Average mono-nucleosome enrichment profiles (MNase) and cytosine methylation (5mC) in WT and Δdim-1 strains at the first nucleosome apex in left and right borders of regions that gain methylation in a dim-1 strain, as well as a central reference nucleosome peak in a WT strain (n = 322); the WT data used reference nucleosomes in a WT strain while the dim-1 data used reference nucleosomes from a Δdim-1 strain. (C) Heatmaps of nucleosome signals from WT and Δdim-1 strains normalized to average nucleosome signal examining ±500 bp of the intergenic regions gaining cytosine methylation in Δdim-1 strain shown in B; regions are shown in each heatmap pair in the same order.
We decided to take advantage of the fact that DIM-1 is highly expressed in WT cells (Figure S18A), to seek information on interacting proteins by mass spectroscopy of coimmunopurified proteins. DIM-1-3xFLAG pulled-down histones H1, H2A, H2B, H3, H4, the H2A.Z histone variant, and two uncharacterized proteins encoded by NCU00856 and NCU03126 (Figure S18B). Mass spectroscopy of proteins associated with NCU00856-3xFLAG revealed only interactions with DIM-1 and histones (Figure S19A) and deletion of NCU00856 did not cause a phenotype comparable to that of dim-1 mutants, i.e., aberrant methylation (Figure S19B). The association with histones and the above defects in nucleosome positioning are consistent with the possibility that DIM-1 is chromatin remodeler.
DIM-1 localizes to regions gaining aberrant cytosine methylation
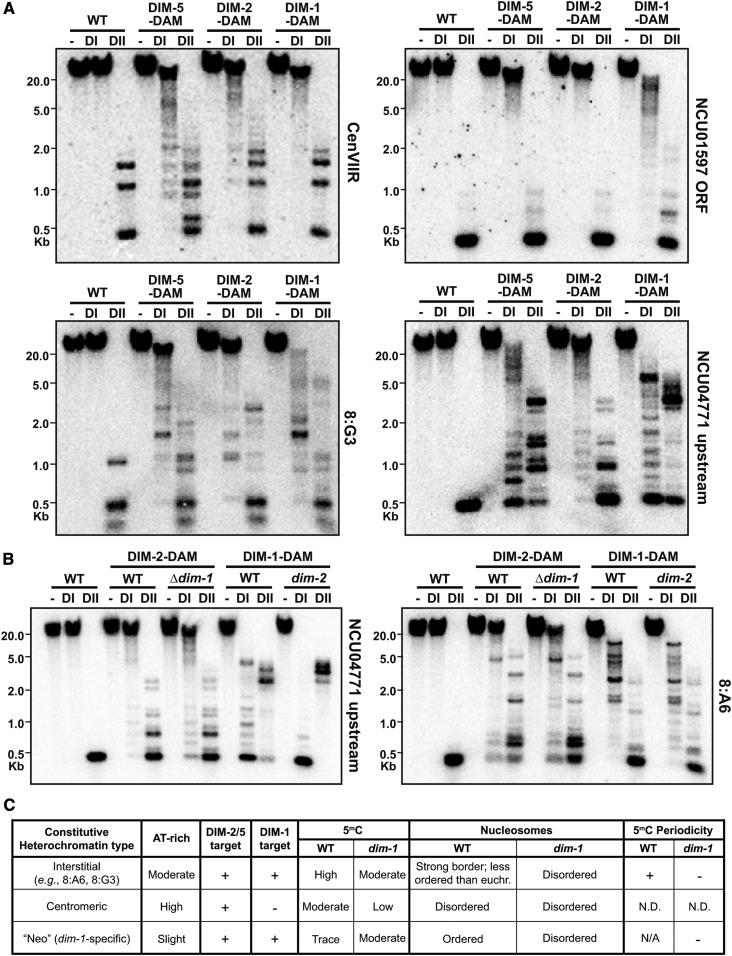
To explore the possibility that the regions of hyper-methylation in dim-1 strains are direct targets of DIM-1 action, we examined DIM-1 localization. Attempts to map DIM-1 by ChIP were unsuccessful, leading us to turn to DamID (Vogel et al. 2007; Lewis et al. 2010b), in which the DNA adenine MTase gene from Escherichia coli was fused to DIM-1 so that its location could be inferred by the pattern of adenine methylation at GATC sites. Genomic DNA was digested with DpnII, which is inhibited by adenine methylation in GATC sites, or by the isoschizomer DpnI, which specifically cleaves when the adenine is methylated. Although little adenine methylation was observed at a centromere or at a euchromatic region not subject to new cytosine methylation (NCU01597), substantial adenine methylation was detected at an interspersed heterochromatic region (8:G3; Figure 6A and Figure S20A) and at euchromatic regions gaining aberrant hyper-methylation in Δdim-1, such as the NCU04771 and NCU07396 upstream regions and the am locus (Figure 6A and Figure S20, B and C). Control DamID constructs for the DNA MTase DIM-2 and the H3K9 MTase DIM-5 gave adenine methylation at centromeres but not NCU01597 (Figure 6A and Figure S20A). Surprisingly, DIM-2-DAM and DIM-5-DAM also localized to the regions that are subject to hyper-methylation, even in dim+ strains (Figure 6A and Figure S20C).
Figure 6.
DIM-1 and heterochromatin machinery localize to heterochromatic regions and intergenic regions. (A) DamID Southern blots of genomic DNA of the indicated dim+ strains digested with DpnI (cuts GAmTC; DI) or its isoschizomer DpnII (inhibited by adenine methylation; DII) or left undigested (−) and probed for the indicated regions. (B) DamID Southern blots, as in A, for different mutant backgrounds expressing the indicated DAM fusion constructs. (C) Table summarizing the phenotypes of a dim-1 strain. N.D., not determined.
To investigate which domains of DIM-1 are required for its function and localization, we individually deleted the positively charged N terminus or the BAH domain of DIM-1. These deletion constructs had reduced expression compared to the WT DIM-1 protein when protein levels were assayed through 3xFLAG immunoblotting. Neither mutant complemented the ∆dim-1 hyper-methylation phenotype, suggesting these domains are important for DIM-1 function or the expression levels were not sufficient (Figure S20D). We used DAM fusion proteins to test localization. The ΔBAH domain DIM-1-DAM construct still exhibited significant localization to promoter regions that are hyper-methylated, indicating the BAH domain is not critical for localization (Figure S20D, middle). The ΔN terminus DIM-1-DAM construct showed no DpnI-specific cleavage, possibly due to low levels of expression.
To explore the relationship between DIM-1 localization and hyper-methylation, we examined DIM-1-DAM localization, as inferred from adenine methylation, in the presence or absence of DIM-2, and similarly examined localization of DIM-2 in the presence or absence of DIM-1. Loss of DIM-1 caused a modest increase in localization of DIM-2-DAM to an upstream region that becomes hyper-methylated, as evidenced by the increased cleavage of DpnI (Figure 6B, left). Conversely, loss of DIM-2 increased DIM-1-DAM targeting, as shown by the near-complete DpnI cleavage (Figure 6B, left). No comparable changes were observed at a constitutive heterochromatic region (Figure 6B, right).
Discussion
We determined that Neurospora dim-1 encodes a putative AAA ATPase chromatin remodeler and results in both hypo- and hyper-methylation; Figure 6C summarizes the phenotypes of a dim-1 strain. The original dim-1 mutants were obtained in a selection scheme based on the observation that DNA repair–defective strains (e.g., a mus-20 mutant) were hyper-sensitive to 5-azaC, potentially due to defective repair of adducts between 5-azaC in DNA and a DNA MTase. However, no MTase mutants were identified and deletion of the gene encoding the only cytosine MTase in the genome, dim-2, did not alleviate the 5-azaC sensitivity of mus-20 strains (Foss et al. 1998). We found that DIM-1 is not only required for WT levels of DNA methylation at constitutive heterochromatin, the first observed phenotype (Foss et al. 1998), but also prevents aberrant hyper-methylation in intergenic regions. These regions of hyper-methylation showed aberrantly positioned nucleosomes as well as greater, but variable, nucleosome density, indicating that DIM-1 is essential for proper nucleosome positioning and occupancy, similar to Yta7 in S. cerevisiae (Lombardi et al. 2011, 2014). While speculative, one possibility for the intergenic methylation is that nucleosome disorder creates a nucleosomal pattern that resembles constitutive heterochromatin, where the underlying AT-rich DNA may allow for nucleosome “slippage” or otherwise signals the heterochromatic machinery. The gain in intergenic methylation was, at best, weakly correlated with high levels of overall gene expression, despite hundreds of genes being downregulated in Δdim-1 strains. This discrepancy may reflect the sensitivity of these promoters to changes in the arrangement of nucleosomes around the TSS and not increased levels of cytosine or histone methylation.
Curiously, even in a WT background, the H3K9 MTase, DIM-5, the DNA MTase, DIM-2, and DIM-1 all strongly localize to the intergenic regions that become aberrantly methylated in dim-1 strains. This gives the impression that the heterochromatin machinery is “poised” at these regions and DIM-1 may regulate nucleosome occupancy at these loci by responding to H3K9me3 and 5mC levels. Interestingly, deletion of dim-2 allows DIM-1 to localize to a greater extent at these intergenic regions. Regions of constitutive heterochromatin in Δdim-1 strains contain near WT levels of H3K9me3 that may remain high due to the presence of ample histone tails for DIM-5 action. These same regions contain roughly half the level of cytosine methylation observed in a WT strain. We noticed that the distribution of DNA methylation in WT Neurospora is higher in linker regions than in the nucleosomal DNA at constitutive heterochromatic regions, as reported for other species (Takeshima et al. 2008; Felle et al. 2011; Huff and Zilberman 2014), raising the possibility that the reduced periodicity and tighter packing of nucleosomes in a Δdim-1 strain reduces DNA methylation at constitutive heterochromatin by limiting accessible linker DNA. Consistent with this, elimination of the linker histone H1 slightly increases DNA methylation in Neurospora constitutive heterochromatin (Seymour et al. 2016). Cytosine methylation and H3K9me3 do not appear to be due to limiting DIM-2 or DIM-5, considering that additional copies of dim-2 and dim-5 did not rescue the methylation defects. Of course, we cannot exclude the possibility that unidentified factor(s) working in conjunction with DIM-1 affect DNA methylation, negatively or positively.
Interestingly, we found indications that heterochromatin is more sensitive than euchromatin to MNase digestion in Neurospora, which contrasts observations in Drosophila (Sun et al. 2001); of possible relevance, Drosophila does not contain a homolog of dim-1 (Zou et al. 2007). The peaks of nucleosomes in Neurospora constitutive heterochromatin tend to be lower and broader than in euchromatin, implying irregular positioning (Lai and Pugh 2017). Because of repeat-induced point mutation (RIP), Neurospora constitutive heterochromatin is characterized by relatively simple, AT-rich DNA, which may contribute to movement of nucleosomes, resulting in the observed disorder (Lai and Pugh 2017); the known preference of MNase for A:T-rich DNA, or conceivably a preference for methylated DNA, could also contribute to the observed pattern of nucleosomes. It is intriguing that heterochromatin borders often have well-positioned nucleosomes, which may play a role in localizing protein complexes, such as the Neurospora DMM complex, that limit heterochromatin spreading (Honda et al. 2010).
The dim-1 gene was independently identified as catp (clock ATPase) because a mutation of this gene resulted in aberrant circadian periodicity (Cha et al. 2013). The authors suggested that increased nucleosome occupancy could affect clock gene expression and reported increased histone H3 occupancy at the critical C-box enhancer element at the frequency gene (Cha et al. 2013). We showed that nucleosome positions are altered in gene bodies, heterochromatic regions, and intergenic regions genome-wide in a Δdim-1 strain, in accordance with observations in other organisms defective in homologs of DIM-1. In S. pombe, deletion of the comparable gene abo1 leads to nucleosome disorder, and lower nucleosome signals are observed at gene bodies and transposable elements underlying heterochromatin (Gal et al. 2015). Moreover, in S. cerevisiae, loss of the DIM-1 homolog Yta7 causes a progressive shift of nucleosomes toward the 5′ end of genes (Lombardi et al. 2014). In a Neurospora Δdim-1 strain, nucleosome positions also shift toward the 5′ end of gene bodies, although the incomplete mapping of Neurospora TSSs undoubtedly contributes some “noise” to our analyses. While DIM-1, like Yta7, appears to maintain NFRs, the mechanism is unknown. NFRs in multiple species seem to depend on short, often directional AT tracts (Iyer and Struhl 1995; Segal and Widom 2009; Krietenstein et al. 2016; Lai and Pugh 2017) but we did not identify any strong NFR motifs in our analyses. Presumably, proper NFR positioning plays a role in the function of transcriptional machinery, accounting for observed changes in dim-1 strains. In S. pombe, ABO1, interacts with the RNA Polymerase II–associated FACT complex to direct nucleosome assembly (Gal et al. 2015). Future studies may reveal specifics of changes in factor binding and gene expression resulting from changes in nucleosomal positioning.
DIM-1 is conserved among eukaryotes, and it is noteworthy that the human homolog ATAD2 (also known as ANCCA) (Zou et al. 2007, 2009; Boussouar et al. 2013) has a high level of sequence identity with DIM-1, Yta7, and ABO1, suggesting that ATAD2 is also an AAA ATPase chromatin remodeler. ATAD2 is a marker for multiple cancerous cell types: changes in expression of this protein is correlated with poor prognosis in multiple cancers (Zou et al. 2007, 2009; Ciró et al. 2009; Caron et al. 2010; Kalashnikova et al. 2010; Liu et al. 2012). Altered levels of ATAD2 cause misregulation of some oncogenes and influence epigenetic marks, such as increased levels of histone acetylation (Zou et al. 2007) and H3K4me3 (Revenko et al. 2010). In addition, ATAD2 overexpression is correlated with higher levels of the PRC2 member EZH2 in tumors (Kalashnikova et al. 2010), suggesting H3K27me levels could be affected. The ATAD2 bromodomain interacts with acetylated N-terminal tails of histone H3 and H4 (Ciró et al. 2009; Caron et al. 2010; Revenko et al. 2010). Although it is known that gene activation is diminished upon knockdown of ATAD2 (Zou et al. 2007, 2009; Revenko et al. 2010), and as such, ATAD2 acts as a transcriptional coactivator (Boussouar et al. 2013), little is known about the in vivo function of the protein. To the best of our knowledge, the level of cytosine methylation/H3K9me3 or the position of nucleosomes have not been examined in ATAD2 deficient cancer cells. While downstream epigenetic changes may be different between oncogenic human cells and Neurospora dim-1 strains, our discovery of aberrant cytosine methylation associated with altered nucleosome positioning in Neurospora Δdim-1 strains suggests that further studies in humans are warranted to learn if aberrant methylation, which is common in cancer, occurs in human cells with altered ATAD2 activity.
Acknowledgments
The authors would like to thank Josh Lowry and Will Storck (University of Oregon) for SNP identification and filtering help, Jeff McKnight (University of Oregon) for his help generating the MNase heatmap and for the use of his custom python script (File S3) for reordering nucleosome signal arrays, Neena Leggett for the initial bioinformatic analysis, all members of the Selker laboratory for helpful comments and discussions, Mark Johnston for title suggestions, and the three anonymous reviewers whose comments/suggestions helped improve the manuscript. Funding for this work was provided by grants from the National Institutes of Health (NIH) to E.U.S. (GM035690 and GM093061) and the Naito Foundation, the Nakajima Foundation, and the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) (24870012), Japan, to S.H. A.D.K. was partly supported by an NIH postdoctoral fellowship (GM097821), and E.T.W. was partly supported by a fellowship from the American Heart Association (14POST20450071).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7210190.
Communicating editor: O. Rando
Literature Cited
- Adhvaryu K. K., Berge E., Tamaru H., Freitag M., Selker E. U., 2011. Substitutions in the amino-terminal tail of Neurospora histone H3 have varied effects on DNA methylation. PLoS Genet. 7: e1002423 10.1371/journal.pgen.1002423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., et al. , 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46: W537–W544. 10.1093/nar/gky379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R. C., Madhani H. D., 2017. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19: 229–244. 10.1038/nrm.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussouar F., Jamshidikia M., Morozumi Y., Rousseaux S., Khochbin S., 2013. Malignant genome reprogramming by ATAD2. Biochim. Biophys. Acta 1829: 1010–1014. 10.1016/j.bbagrm.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Bühler M., Gasser S. M., 2009. Silent chromatin at the middle and ends: lessons from yeasts. EMBO J. 28: 2149–2161. 10.1038/emboj.2009.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron C., Lestrat C., Marsal S., Escoffier E., Curtet S., et al. , 2010. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene 29: 5171–5181. 10.1038/onc.2010.259 [DOI] [PubMed] [Google Scholar]
- Cha J., Zhou M., Liu Y., 2013. CATP is a critical component of the Neurospora circadian clock by regulating the nucleosome occupancy rhythm at the frequency locus. EMBO Rep. 14: 923–930. 10.1038/embor.2013.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciró M., Prosperini E., Quarto M., Grazini U., Walfridsson J., et al. , 2009. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 69: 8491–8498. 10.1158/0008-5472.CAN-09-2131 [DOI] [PubMed] [Google Scholar]
- Clapier C. R., Cairns B. R., 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- Clapier C. R., Iwasa J., Cairns B. R., Peterson C. L., 2017. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18: 407–422. 10.1038/nrm.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357 (erratum: Proc. Natl. Acad. Sci. USA 103: 16614). 10.1073/pnas.0601456103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly L. R., Smith K. M., Freitag M., 2013. The Fusarium graminearum histone H3 K27 methyltransferase KMT6 regulates development and expression of secondary metabolite gene clusters. PLoS Genet. 9: e1003916 10.1371/journal.pgen.1003916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Cheng J., Sun X., Zhou Z., Liu Y., 2016. Antisense transcription licenses nascent transcripts to mediate transcriptional gene silencing. Genes Dev. 30: 2417–2432. 10.1101/gad.285791.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. H., 2000. Neurospora: Contributions of a Model Organism. Oxford University Press, Oxford. [Google Scholar]
- de Hoon M. J. L., Imoto S., Nolan J., Miyano S., 2004. Open source clustering software. Bioinformatics 20: 1453–1454. 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- Elgin S. C. R., Reuter G., 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 5: a017780 10.1101/cshperspect.a017780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle M., Hoffmeister H., Rothammer J., Fuchs A., Exler J. H., et al. , 2011. Nucleosomes protect DNA from DNA methylation in vivo and in vitro. Nucleic Acids Res. 39: 6956–6969. 10.1093/nar/gkr263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J., Kainth P., Lambert J.-P., van Bakel H., Tsui K., et al. , 2009. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell 35: 340–351. 10.1016/j.molcel.2009.06.023 [DOI] [PubMed] [Google Scholar]
- Foss H. M., Roberts C. J., Claeys K. M., Selker E. U., 1993. Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science 262: 1737–1741. 10.1126/science.7505062 [DOI] [PubMed] [Google Scholar]
- Foss H. M., Roberts C. J., Selker E. U., 1998. Mutations in the dim-1 gene of Neurospora crassa reduce the level of DNA methylation. Mol. Gen. Genet. 259: 60–71. 10.1007/s004380050789 [DOI] [PubMed] [Google Scholar]
- Freitag M., 2017. Histone methylation by SET domain proteins in fungi. Annu. Rev. Microbiol. 71: 413–439. 10.1146/annurev-micro-102215-095757 [DOI] [PubMed] [Google Scholar]
- Freitag M., Hickey P. C., Khlafallah T. K., Read N. D., Selker E. U., 2004. HP1 is essential for DNA methylation in Neurospora. Mol. Cell 13: 427–434. 10.1016/S1097-2765(04)00024-3 [DOI] [PubMed] [Google Scholar]
- Gal C., Murton H. E., Subramanian L., Whale A. J., Moore K. M., et al. , 2015. Abo1, a conserved bromodomain AAA-ATPase, maintains global nucleosome occupancy and organisation. EMBO Rep. 17: 79–93. 10.15252/embr.201540476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galazka J. M., Klocko A. D., Uesaka M., Honda S., Selker E. U., et al. , 2016. Neurospora chromosomes are organized by blocks of importin alpha-dependent heterochromatin that are largely independent of H3K9me3. Genome Res. 26: 1069–1080. 10.1101/gr.203182.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradolatto A., Rogers R. S., Lavender H., Taverna S. D., Allis C. D., et al. , 2008. Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics 179: 291–304. 10.1534/genetics.107.086520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S. I. S., Jia S., 2007. Heterochromatin revisited. Nat. Rev. Genet. 8: 35–46. 10.1038/nrg2008 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U., Paro R., 2014. Transcriptional silencing by polycomb-group proteins. Cold Spring Harb. Perspect. Biol. 6: a019331 10.1101/cshperspect.a019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C. M., Strømme C. B., Huang H., Patel D. J., Groth A., 2017. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18: 141–158. 10.1038/nrm.2016.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E., 2005. Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Curr. Opin. Genet. Dev. 15: 482–489. 10.1016/j.gde.2005.08.009 [DOI] [PubMed] [Google Scholar]
- Henikoff S., 2000. Heterochromatin function in complex genomes. Biochim. Biophys. Acta 1470: O1–O8. [DOI] [PubMed] [Google Scholar]
- Henikoff S., 2016. Mechanisms of nucleosome dynamics in vivo. Cold Spring Harb. Perspect. Med. 6: a026666 10.1101/cshperspect.a026666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S., Selker E. U., 2008. Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora crassa. Mol. Cell. Biol. 28: 6044–6055. 10.1128/MCB.00823-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S., Selker E. U., 2009. Tools for fungal proteomics: multifunctional Neurospora vectors for gene replacement, protein expression and protein purification. Genetics 182: 11–23. 10.1534/genetics.108.098707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S., Lewis Z. A., Huarte M., Cho L. Y., David L. L., et al. , 2010. The DMM complex prevents spreading of DNA methylation from transposons to nearby genes in Neurospora crassa. Genes Dev. 24: 443–454. 10.1101/gad.1893210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S., Lewis Z. A., Shimada K., Fischle W., Sack R., et al. , 2012. Heterochromatin protein 1 forms distinct complexes to direct histone deacetylation and DNA methylation. Nat. Struct. Mol. Biol. 19: 471–477. 10.1038/nsmb.2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S., Bicocca V. T., Gessaman J. D., Rountree M. R., Yu E. Y., et al. , 2016. Dual chromatin recognition by the histone deacetylase complex HCHC is required for proper DNA methylation in Neurospora crassa. Proc. Natl. Acad. Sci. USA 113: E6135–E6144 [corrigenda: Proc. Natl. Acad. Sci. USA 114: E1037 (2017)]. 10.1073/pnas.1614279113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff J. T., Zilberman D., 2014. Dnmt1-independent CG methylation contributes to nucleosome positioning in diverse eukaryotes. Cell 156: 1286–1297. 10.1016/j.cell.2014.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V., Struhl K., 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 14: 2570–2579. 10.1002/j.1460-2075.1995.tb07255.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson K., Rountree M. R., Lewis Z. A., Stajich J. E., Selker E. U., 2013. Regional control of histone H3 lysine 27 methylation in Neurospora. Proc. Natl. Acad. Sci. USA 110: 6027–6032. 10.1073/pnas.1303750110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson K., Wiles E. T., McNaught K. J., Sidoli S., Leggett N., et al. , 2016. Loss of HP1 causes depletion of H3K27me3 from facultative heterochromatin and gain of H3K27me2 at constitutive heterochromatin. Genome Res. 26: 97–107. 10.1101/gr.194555.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalashnikova E. V., Revenko A. S., Gemo A. T., Andrews N. P., Tepper C. G., et al. , 2010. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. 70: 9402–9412. 10.1158/0008-5472.CAN-10-1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent N. A., Adams S., Moorhouse A., Paszkiewicz K., 2010. Chromatin particle spectrum analysis: a method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic Acids Res. 39: e26 10.1093/nar/gkq1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klocko A. D., Rountree M. R., Grisafi P. L., Hays S. M., Adhvaryu K. K., et al. , 2015. Neurospora importin α is required for normal heterochromatic formation and DNA methylation. PLoS Genet. 11: e1005083 10.1371/journal.pgen.1005083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klocko A. D., Ormsby T., Galazka J. M., Leggett N. A., Uesaka M., et al. , 2016. Normal chromosome conformation depends on subtelomeric facultative heterochromatin in Neurospora crassa. Proc. Natl. Acad. Sci. USA 113: 15048–15053. 10.1073/pnas.1615546113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietenstein N., Wal M., Watanabe S., Park B., Peterson C. L., et al. , 2016. Genomic nucleosome organization reconstituted with pure proteins. Cell 167: 709–721.e12. 10.1016/j.cell.2016.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W. K. M., Pugh B. F., 2017. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 18: 548–562. 10.1038/nrm.2017.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämke J., Bäurle I., 2017. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 18: 124 10.1186/s13059-017-1263-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-C., Li L., Gu W., Xue Z., Crosthwaite S. K., et al. , 2010. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol. Cell 38: 803–814. 10.1016/j.molcel.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P. W., Allis C. D., 2013. Poisoning the “histone code” in pediatric gliomagenesis. Cell Cycle 12: 3241–3242. 10.4161/cc.26356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P. W., Müller M. M., Koletsky M. S., Cordero F., Lin S., et al. , 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861. 10.1126/science.1232245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis Z. A., Honda S., Khlafallah T. K., Jeffress J. K., Freitag M., et al. , 2008. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 19: 427–437. 10.1101/gr.086231.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis Z. A., Adhvaryu K. K., Honda S., Shiver A. L., Knip M., et al. , 2010a DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet. 6: e1001196 10.1371/journal.pgen.1001196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis Z. A., Adhvaryu K. K., Honda S., Shiver A. L., Selker E. U., 2010b Identification of DIM-7, a protein required to target the DIM-5 H3 methyltransferase to chromatin. Proc. Natl. Acad. Sci. USA 107: 8310–8315. 10.1073/pnas.1000328107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lee W., Jiang Z., Chen Z., Jhunjhunwala S., et al. , 2012. Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res. 22: 2315–2327. 10.1101/gr.140988.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi L. M., Ellahi A., Rine J., 2011. Direct regulation of nucleosome density by the conserved AAA-ATPase Yta7. Proc. Natl. Acad. Sci. USA 108: E1302–E1311. 10.1073/pnas.1116819108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi L. M., Davis M. D., Rine J., 2014. Maintenance of nucleosomal balance in cis by conserved AAA-ATPase Yta7. Genetics 199: 105–116. 10.1534/genetics.114.168039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao V. P., Freitag M., Selker E. U., 2000. Short TpA-rich segments of the ζ-η region induce DNA methylation in Neurospora crassa. J. Mol. Biol. 300: 249–273. 10.1006/jmbi.2000.3864 [DOI] [PubMed] [Google Scholar]
- Morgan M. A., Shilatifard A., 2015. Chromatin signatures of cancer. Genes Dev. 29: 238–249. 10.1101/gad.255182.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. R., Nietlispach D., Mott H. R., Callaghan J., Bannister A., et al. , 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416: 103–107. 10.1038/nature722 [DOI] [PubMed] [Google Scholar]
- Okano M., Bell D. W., Haber D. A., Li E., 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- Quintales L., Vázquez E., Antequera F., 2014. Comparative analysis of methods for genome-wide nucleosome cartography. Brief. Bioinform. 16: 576–587. 10.1093/bib/bbu037 [DOI] [PubMed] [Google Scholar]
- Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., et al. , 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenko A. S., Kalashnikova E. V., Gemo A. T., Zou J. X., Chen H.-W., 2010. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol. Cell. Biol. 30: 5260–5272. 10.1128/MCB.00484-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C. J., Selker E. U., 1995. Mutations affecting the biosynthesis of S-adenosylmethionine cause reduction of DNA methylation in Neurospora crassa. Nucleic Acids Res. 23: 4818–4826. 10.1093/nar/23.23.4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus M. J., Galbiati M., Ticknor C., Chen J., Dellaporta S. L., 1996. Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654–657. 10.1126/science.273.5275.654 [DOI] [PubMed] [Google Scholar]
- Rountree M. R., Selker E. U., 2010. DNA methylation and the formation of heterochromatin in Neurospora crassa. Heredity 105: 38–44. 10.1038/hdy.2010.44 [DOI] [PubMed] [Google Scholar]
- Saldanha A. J., 2004. Java Treeview–extensible visualization of microarray data. Bioinformatics 20: 3246–3248. 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- Segal E., Widom J., 2009. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 19: 65–71. 10.1016/j.sbi.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Tountas N. A., Cross S. H., Margolin B. S., Murphy J. G., et al. , 2003. The methylated component of the Neurospora crassa genome. Nature 422: 893–897. 10.1038/nature01564 [DOI] [PubMed] [Google Scholar]
- Seymour M., Ji L., Santos A. M., Kamei M., Sasaki T., et al. , 2016. Histone H1 limits DNA methylation in Neurospora crassa. G3 (Bethesda) 6: 1879–1889. 10.1534/g3.116.028324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe W. J. J., Jasencakova Z., Houben A., Kakutani T., Meister A., et al. , 2002. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21: 6549–6559. 10.1093/emboj/cdf657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studt L., Rösler S. M., Burkhardt I., Arndt B., Freitag M., et al. , 2016. Knock-down of the methyltransferase Kmt6 relieves H3K27me3 and results in induction of cryptic and otherwise silent secondary metabolite gene clusters in Fusarium fujikuroi. Environ. Microbiol. 18: 4037–4054. 10.1111/1462-2920.13427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F. L., Cuaycong M. H., Elgin S. C., 2001. Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin. Mol. Cell. Biol. 21: 2867–2879. 10.1128/MCB.21.8.2867-2879.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., et al. , 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16: 1779–1791. 10.1101/gad.989402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H., Suetake I., Tajima S., 2008. Mouse Dnmt3a preferentially methylates linker DNA and is inhibited by histone H1. J. Mol. Biol. 383: 810–821. 10.1016/j.jmb.2008.03.001 [DOI] [PubMed] [Google Scholar]
- Timp W., Feinberg A. P., 2013. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer 13: 497–510. 10.1038/nrc3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P., Reinberg D., 2007. Facultative heterochromatin: is there a distinctive molecular signature? Mol. Cell 28: 1–13. 10.1016/j.molcel.2007.09.011 [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., 2002. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat. Rev. Mol. Cell Biol. 3: 422–429. 10.1038/nrm828 [DOI] [PubMed] [Google Scholar]
- Vermaak D., Wolffe A. P., 1998. Chromatin and chromosomal controls in development. Dev. Genet. 22: 1–6. [DOI] [PubMed] [Google Scholar]
- Vogel M. J., Peric-Hupkes D., van Steensel B., 2007. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2: 1467–1478. 10.1038/nprot.2007.148 [DOI] [PubMed] [Google Scholar]
- Wiles E. T., Selker E. U., 2017. H3K27 methylation: a promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 43: 31–37. 10.1016/j.gde.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A., 2011. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 12: 542–553. 10.1038/nrg3035 [DOI] [PubMed] [Google Scholar]
- Xu S., Grullon S., Ge K., Peng W., 2014. Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol. Biol. 1150: 97–111. 10.1007/978-1-4939-0512-6_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J. X., Revenko A. S., Li L. B., Gemo A. T., Chen H.-W., 2007. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc. Natl. Acad. Sci. USA 104: 18067–18072. 10.1073/pnas.0705814104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J. X., Guo L., Revenko A. S., Tepper C. G., Gemo A. T., et al. , 2009. Androgen-induced coactivator ANCCA mediates specific androgen receptor signaling in prostate cancer. Cancer Res. 69: 3339–3346. 10.1158/0008-5472.CAN-08-3440 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All ChIP-sequencing, MNase-sequencing, bisulfite-sequencing, and RNA-sequencing data sets, as well as genome position files, have been deposited to the NCBI Gene Expression Omnibus (accession number GSE98911). Supplemental Figure files (Figures S1–S20 and File S1), data sets (File S2), and program (File S3) have been deposited to Figshare. Reagents, including strains, are available upon request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7210190.