Abstract
Mutations are the root source of genetic variation and underlie the process of evolution. Although the rates at which mutations occur vary considerably between species, little is known about differences within species, or the genetic and molecular basis of these differences. Here, we leveraged the power of the yeast Saccharomyces cerevisiae as a model system to uncover natural genetic variants that underlie variation in mutation rate. We developed a high-throughput fluctuation assay and used it to quantify mutation rates in seven natural yeast isolates and in 1040 segregant progeny from a cross between BY, a laboratory strain, and RM, a wine strain. We observed that mutation rate varies among yeast strains and is heritable (H2 = 0.49). We performed linkage mapping in the segregants and identified four quantitative trait loci underlying mutation rate variation in the cross. We fine-mapped two quantitative trait loci to the underlying causal genes, RAD5 and MKT1, that contribute to mutation rate variation. These genes also underlie sensitivity to the DNA-damaging agents 4NQO and MMS, suggesting a connection between spontaneous mutation rate and mutagen sensitivity.
Keywords: Spontaneous mutation rate, QTL mapping, genetic basis, natural variants, yeast, complex traits
MUTATIONS are permanent changes to the genome of an organism that can result from DNA damage that is improperly repaired, from errors in DNA replication (Lieber 2010), or from the movement of mobile genetic elements. Mutations give rise to genetic variants in populations and are the wellspring of evolution (Long et al. 2003). Mutations also play a major role in both inherited diseases and acquired diseases such as cancer (Tomlinson et al. 1996).
The mutation rate can be defined as the number of mutational events per cell division, generation, or unit of time (Baer et al. 2007). Mutation rates tend to be ∼10−9 – 10−10 mutations per base pair, per cell division, for most microbial species (Drake et al. 1998), making them difficult to measure and compare across individuals. As a consequence, the effects of genetic background differences on mutation rates have only been investigated on a small scale (Demerec 1937). Two types of experimental approaches have been used to measure mutation rates in yeast. The first is the fluctuation assay (Luria and Delbrück 1943). This method requires a gene with a selectable phenotype such that loss-of-function mutations in the gene enable the mutants to grow in the corresponding selective conditions. Spontaneous mutation rate is then estimated from the distribution of mutant numbers in parallel cultures. Lang and Murray applied the fluctuation assay to Saccharomyces cerevisiae and estimated the per-base-pair mutation rate in yeast (Lang and Murray 2008). A second method tracks mutation accumulation during experimental evolution and uses whole-genome sequencing to estimate mutation rates (Zhu et al. 2014). This approach also provides information on the number, locations, and types of spontaneous mutations. However, this assay requires growing the mutation accumulation lines over hundreds of generations, as well as sequencing many genomes. Although the fluctuation assay is faster and cheaper, the need for many parallel cultures makes it laborious to extend it to many different strains.
Here, we developed a modified version of the fluctuation assay to enable higher-throughput measurements of spontaneous mutation rates. We used the new assay to quantify mutation rates across genetically distinct yeast strains and observed considerable variation. To find the genes underlying the observed variation, we applied the modified fluctuation assay to a large panel of 1040 segregants from a cross between the laboratory strain BY4724 (hereafter referred to as BY) and the vineyard strain RM11-1a (hereafter referred to as RM). We identified four loci associated with mutation rate variation and narrowed the two loci that contributed the most to mutation rate variation to missense variants in the genes RAD5 and MKT1. We also found interactions between alleles of RAD5 and MKT1.
Materials and Methods
Yeast strains and media
Seven natural S. cerevisiae strains (Supplemental Material, Table S1) were used in this study. The 1040 segregants derived from BY4724 (MATa) and RM11-1a (MATa, MKT1-BY, ho∆::HphMX, flo8∆::NatMX) were generated, genotyped, and described previously (Bloom et al. 2013). The RM::MKT1-BY strain was made previously by our lab. The BY::RAD5-RM strain and the RAD5 variants substitution strains (Table 1) were from Demogines et al. (2008). For fluctuation assay, yeast was grown in synthetic complete liquid medium without arginine (SC-Arg) before plating onto selective plates. For DNA-damaging agent sensitivity assays, yeast were grown in rich YPD medium (1% yeast extract, 2% peptone, and 2% glucose) before plating onto YPD agar plates with DNA-damaging agents. SC-Arg and YPD liquid media and agar plates were made according to Amberg et al. (2005).
Table 1. The allele replacement strains and variant substitution strains.
| Strain | Background | Relevant genotype | Source |
|---|---|---|---|
| YLK802 | RM | MATa, MKT1-BY, ho∆::HphMX, flo8∆::NatMX | Smith and Kruglyak (2008) |
| EAY1463 | BY | MATα, lys2Δ, RAD5-RM::NatMX | Demogines et al. (2008) |
| EAY1471 | BY | MATα, lys2Δ, RAD5-I791S::KanMX | Demogines et al. (2008) |
| EAY2169 | BY | MATα, lys2Δ, RAD5-E783D::KanMX | Demogines et al. (2008) |
Selection agar plate construction
Selective canavanine plates were made from arginine minus synthetic complete agar medium with 60 mg/liter L-canavanine (C1625; Sigma, St. Louis, MO). The canavanine plates were dried by incubating at 30° overnight. Selective plates for the DNA-damaging agent sensitivity assay were made with YPD agar medium containing the respective agents at the concentrations indicated in Table 2. Then, 50 ml of the agar medium was poured into each Nunc OmniTray plates (264728; Thermo Scientific) and placed on a flat surface to solidify. Each experiment was performed with the same batch of selection plates. The concentrations for 4-nitroquinoline 1-oxide (4NQO) (N8141; Sigma), methyl methane sulfonate (MMS) (64382; Sigma), and hydrogen peroxide (H2O2) (216763; Sigma) were 0.1 μg/ml, 0.01% and 4 mM. These concentrations capture the sensitivity difference between the segregants, while maintaining enough colony growth for quantitative trait locus (QTL) mapping.
Table 2. DNA-damaging agents used for the sensitivity assay.
| Agent | Agent characteristic |
|---|---|
| Hydrogen peroxide (H2O2) | Altering DNA structure |
| Methyl methane sulfonate (MMS) | Altering (alkylating) DNA bases |
| 4-Nitroquinoline 1-oxide (4NQO) | Ultraviolet mimetic |
Fluctuation assays
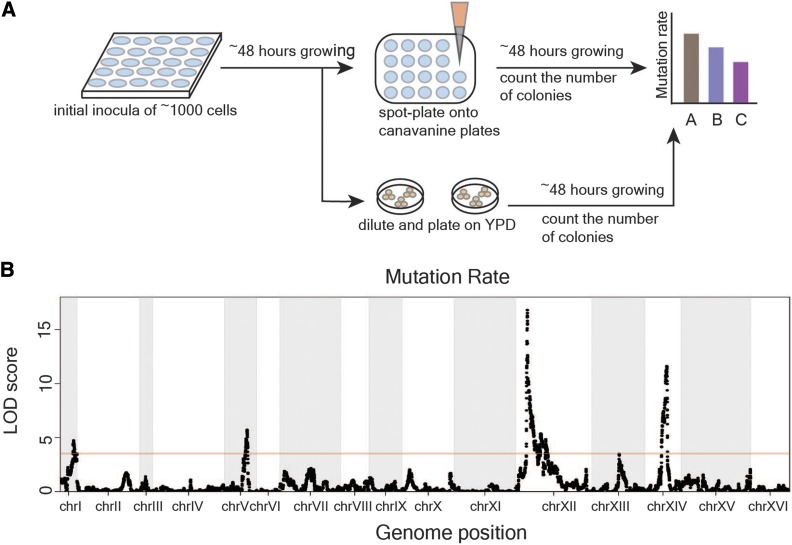
To begin the fluctuation assay, yeast were grown in SC-Arg in 96-well plates (3370; Costar, Cambridge, MA) for ∼48 hr to saturation. Saturated cultures were diluted and pinned into a new 96-well plate with liquid SC-Arg medium. This step ensured a small number of ∼1000 yeast cells in the initial inoculum. Plates were sealed with a Breathe-Easy sealing membrane (Z380059; Sigma) to prevent evaporation, and incubated at 30° with shaking for ∼48 hr. Then, 100 μl saturated cultures were spot-plated onto canavanine plates in a four by six configuration using a Biomek FXP automated workstation. Plates with spot-plated yeast culture were dried in the laminar flow hood (Nuair) for 30 mins or until dry, and incubated at 30° for ∼48 hr. We imaged the plates using an imaging robot (BM3-SC; S&P Robotics), and the number of colonies in each spot was manually counted from the images. An example of the imaged plate is shown in Figure S1.
Mutation rate was estimated using the Ma–Sandri–Sarkar maximum likelihood method, where the numbers of observed colonies on canavanine plates was fitted into the Luria–Delbrück distribution and a single parameter m was calculated (Sarkar et al. 1992). The parameter m represents the expected number of mutation events per culture. For the natural isolates and engineered strains, the mutation rate was calculated from the equation , where N is the average number of cells per culture (as a proxy for the number of cell divisions, given the starting inoculum is much smaller than N). In the segregant panel, we defined a mutation rate score that was calculated as the residual phenotype after regressing out the effect of average number of cells per strain (N) from the estimate of m per strain across all of the segregants.
For each of the seven natural isolate strains, we performed 96 replicates of the fluctuation assay, which means we had 96 estimations of mutation rate. In each replicate three cultures were plated onto canavanine plates, and the number of resistant colonies in these three plates were fitted into the Luria–Delbrück distribution to estimate the mutation events per culture (m). One culture was diluted and plated onto YPD to determine the number of cells per culture (N) in each replicate. Given the number of replicates used for estimate m and N were limited, the mutation rate estimation for the seven natural isolate strains had large variance. For the BY × RM segregants panel, 12 independent replicate cultures were plated onto canavanine plates for every segregant. The number of canavanine resistant colonies in these 12 plates was fitted into the Luria–Delbrück distribution to calculate the number of mutations per culture (m), and one culture was diluted and plated on the YPD plates to determine the number of cells (N). Given only one culture was used to estimate the number of cells (N) for each segregant, the mutation rate estimation from the equation would be largely affected by that one measure of N. The number of cells per culture (N) was measured by counting the number of cells growing on a YPD nonselective plate after a 105 dilution. This dilution was chosen based on pilot experiments in which different dilutions were tested. This 105 dilution was applied to all segregants before plating on YPD to measure N. However, this degree of dilution was not suitable for all individual segregants. Any segregants that had too many colonies (>70) or no colonies growing on the YPD plates could not give us an accurate measurement of N. Thus we removed these 197 segregants from the downstream QTL analysis; only 843 segregants with confident measurements of N were used. To minimize noise driven by the measurements of N, we defined a mutation rate score that regressed out the effect of N. We built a linear model that included the number of cells per culture (N) and a plate effect as additive covariates for the number of mutations per culture (m). The residuals from this linear model were called the “mutation rate score” and used in downstream analyses. For each allele replacement strain (Table 1), 96 replicates of fluctuation analysis were performed, providing 96 estimations of mutation rate. In each replicate, 12 cultures were plated onto canavanine plate to estimate the number of mutations per culture (m), and three cultures were pooled, diluted, and plated on YPD plates to determine the number of cells per culture (N).
Yeast growth measurement for DNA-damaging agent sensitivity assay
The segregant panel were originally stored in 96-well plates (3370; Costar). During the DNA-damaging agent sensitivity assay, individual segregants were inoculated in two plate configurations in 384-well plates (264574; Thermo Scientific) with YPD and grown for ∼48 hr in a 30° incubator without shaking. Saturated cultures were mixed for 1 min at 2000 rpm using a MixMate (Eppendorf), before pinning. The colony handling robot (BM3-SC; S&P Robotics) was used to pin segregants onto selective agar plates with 384 long pins. The plates were incubated at 30° for ∼48 hr and imaged by the colony handling robot (BM3-SC; S&P Robotics). Custom R code (Bloom et al. 2013) was used to determine the size of each colony and the size was used as a proxy for growth in the presence of the DNA-damaging agents.
QTL mapping and detecting QTL–QTL interactions
To control for intrinsic growth rate differences and plate position effects, we normalized the traits for growth by fitting a regression for growth of the yeast that were in the same layout configuration on control plate (YPD agar plates for mutagen sensitivity assay). Residuals were used for QTL mapping. We tested for linkage by calculating logarithm likelihood ratio (LOD scores) for each genotypic marker and trait as , where r is the Pearson correlation coefficient between the segregant genotypes and the segregant mutation rate or DNA-damaging agents sensitivity. The threshold declaring the significant QTL effect was calculated from the empirical null distribution of the maximum LOD score determined from 1000 permutations (Churchill and Doerge 1994). The estimated 5% family-wise error rate significance thresholds were 3.52, 3.62, 3.61, and 3.64 for mutation rate, mutagen sensitivity for 4NQO, MMS, and H2O2 respectively. The 95% confidence intervals were determined using a 1.5 LOD score drop. The code and the data for QTL mapping is available at https://github.com/gouliangke/Mutation-rate/tree/master/qtl_mapping.
We tested for interactions between each QTL pair by comparing a model that includes an interaction term for the two QTL,, with a model that does not, , using the add1 function in R and calculating an F-statistic. Here, is the residuals vector for the mutation rate score, is the genotype vector at the first QTL, is the genotype vector at the second QTL, and and are estimated parameters specific to each locus pair.
Calculating heritability
Broad-sense heritability was calculated using the natural isolate data and a random effect ANOVA. The variance structure of the phenotype is , where is an incidence matrix mapping phenotypes to strain identity and is the identity matrix. The broad-sense heritability was estimated as , where is the genetic variance and is the error variance. Standard errors of variance component estimates were calculated as the square root of the diagonal of the Fisher information matrix from the iteration at convergence of the AI-REML algorithm (Bloom et al. 2015).
Amplicon sequencing of the CAN1 region in segregants
A total of 1040 segregants were assigned into four groups according to their alleles at gene RAD5 and MKT1 (Table S2). We collected the canavanine-resistant colonies from the canavanine plates that we used to measure the mutation rate of segregants in the previous fluctuation analysis. A single canavanine-resistant colony (if any) was picked from each segregant and the picked colonies from the same group were pooled together for DNA extraction. DNA was extracted from the pool using the Qiagen DNeasy Blood & Tissue Kit. The CAN1 region was amplified from the DNA of four groups using the Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific) and eight pairs of designed primers (File S3). The amplicon sequencing library was prepared using the Illumina Nextera DNA Library Prep Kit with the adjusted protocols to skip the Nextera treatment. The library was then sequenced on the MiSeq platform using the MiSeq Reagent V2 Nano Kit. As shown in Table S3, the original aligned read counts for each library varied widely. To eliminate bias caused by read count variation, we adjusted the number of read counts for each sample by randomly downsampling the reads in the fastq files to be the same for all samples, using custom Python code. Then, we processed reads with the following pipeline: read pairing [paired-end reAd mergeR (PEAR)], read trimming (Trimmomatic-0.36), and read alignment [Burrows-Wheeler Aligner (BWA)] to the reference-targeting region using the downsampled fastq files. The adjusted read counts for each sample are shown in Table S3. Custom R code was used to determine the mutation rate spectrum of each group. The code for the mutation rate spectrum analysis is available at https://github.com/gouliangke/Mutation-rate/tree/master/mutation_spectrum.
Data availability
Strains and plasmids are available upon request. File S1 contains the “mutation rate score” for 843 segregants. File S2 contains the mutation rate for allele replacement and variant-engineered strains. File S3 contains the primer sequences used for the amplicon sequencing. Data and code for QTL mapping and mutation spectrum analysis are available at https://github.com/gouliangke/Mutation-rate. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7343036.
Results
High-throughput fluctuation assay for measuring mutation rates
The fluctuation assay for measuring mutation rate involves growing many parallel cultures, each starting from a small number of cells, under nonselective conditions, followed by plating to selective medium to identify mutants. The number of mutations that occurs in each culture should follow the Poisson distribution, as mutations arise spontaneously. However, the number of mutant cells that survive on the selective plates can vary greatly because early mutations are inherited by all offspring of the mutant. This leads to the “jackpot” effect, in which some cultures contain a large number of mutant individuals. The number of observed mutant cells per culture follows the Luria–Delbrück distribution (Luria and Delbrück 1943), and the Ma–Sandri–Sarkar maximum likelihood method can be used to estimate the expected number of mutations per culture from the observed numbers of mutants (Sarkar et al. 1992). The underlying mutation rate is then calculated by dividing the number of mutations per culture by the average number of cells per culture (Luria and Delbrück 1943). Here, we measured rare spontaneous loss-of-function mutations in the gene CAN1, which encodes an arginine permease. Yeast cells carrying loss-of-function mutations in CAN1 can grow on canavanine, an otherwise toxic arginine analog. Typically, fluctuation assays are labor-intensive and have limited throughput because a large number of parallel cultures is required for estimating the mutation rate in each assay, and several replicate assays are needed for a robust measurement of the mutation rate in each strain (Lang and Murray 2008). We modified the fluctuation assay into a high-throughput method for measuring mutation rates in many strains in parallel. We grew cultures in 96-well plates, automated the spotting of cultures, and used a plate-imaging robot to capture images of the mutant colonies on plates (Materials and Methods, Figure 1A). The automated spotting process for 96 strains took only ∼20 min, and the imaging process required even less time. These improvements enabled us to measure the spontaneous mutation rates in the hundreds of strains necessary for genetic mapping.
Figure 1.
Linkage analysis identified four loci underlying mutation rate variation. (A) The fluctuation assay was performed as shown in the workflow. The assay started with a small number of cells growing in 96-well plates in liquid SC-Arg medium for ∼48 hr, followed by plating onto selective agar plates with canavanine. A proportion of the cultures were diluted to measure the number of cells per culture (Materials and Methods). Plates were imaged 2 days after spot-plating, and the number of colonies on canavanine plate was counted. (B) LOD score for mutation rate variation is plotted against the genetic map. The four significant QTL explain 20.7% of the phenotypic variance. The red line indicates a 5% FWER significance threshold (LOD = 3.52).
Spontaneous mutation rate varies among yeast isolates
To investigate mutation rate variation among S. cerevisiae strains, we measured the spontaneous mutation rate of seven yeast isolates using the high-throughput fluctuation assay (Table S1). The seven strains span a large range of yeast genetic diversity (Schacherer et al. 2009). We found that the mutation rates of these strains range from 1.1 × 10−7 to 5.8 × 10−7 mutations per gene per generation, with a median of 1.7 × 10−7 (Figure S2 and Table S1). The median mutation rate was very similar to the previously reported mutation rate at CAN1 (Lang and Murray 2008). In particular, the mutation rate we observed for the BY strain (1.7 × 10−7) is very similar to the previously reported rate, which was measured in strain W303 (1.5 × 10−7) (Lang and Murray 2008), consistent with the fact that W303 shares a large fraction of its genome with BY (Schacherer et al. 2007; Matheson et al. 2017). An ANOVA showed that strain identity explained a significant fraction of the observed variance in mutation rates (F = 69.9, d.f. = 6, P < 2 × 10−16) (Figure S2). The fraction of total variance in mutation rates explained by the repeatability of measurements for each strain, 49% (SE = 0.29), serves as an upper bound for the estimate of the total contribution of genetic differences between strains to trait variation (broad-sense heritability or H2). We observed that RM, a vineyard strain, had a mutation rate higher than all other strains (Figure S2).
Four QTL explain the majority of observed mutation rate variation
To find the genetic factors underlying the difference in mutation rate between BY and RM, we performed QTL mapping in 1040 genotyped haploid segregants from a cross between these strains (Bloom et al. 2013). We measured the mutation rate of each segregant using the high-throughput fluctuation assay (Materials and Methods). We estimated the fraction of phenotypic variance explained by the additive effects of all segregating markers (narrow-sense heritability) to be 30% (Materials and Methods) (Lynch and Walsh 1998). This sets an upper bound for the expectation of the total amount of additive genetic variance that could be explained with a QTL-based model. QTL mapping in the segregant panel identified significant linkage at four distinct loci (Figure 1B). At two of the QTL, on chromosomes XII and I, the RM allele conferred a higher mutation rate, consistent with the higher mutation rate of this strain. At the other two QTL, on chromosomes XIV and V, the BY allele conferred a higher mutation rate (Figure S3), showing that a strain with lower trait value can nevertheless harbor trait-increasing alleles. The four detected QTL explained 20.7% of the phenotypic variance, thus accounting for 69% of the estimated additive heritability. The loci on chromosomes XII, XIV, I, and V explained 8.8, 6.1, 3.1, and 2.6% of the variance, respectively. We tested the four identified QTL for pairwise interactions and found a significant interaction between the QTL on chromosome XII and the QTL on chromosome XIV that explained 1% of the phenotypic variance (F = 8.41, d.f. = 1, Bonferroni-corrected P = 0.023).
Polymorphisms in genes RAD5 and MKT1 underlie the major QTL on chromosomes XII and XIV
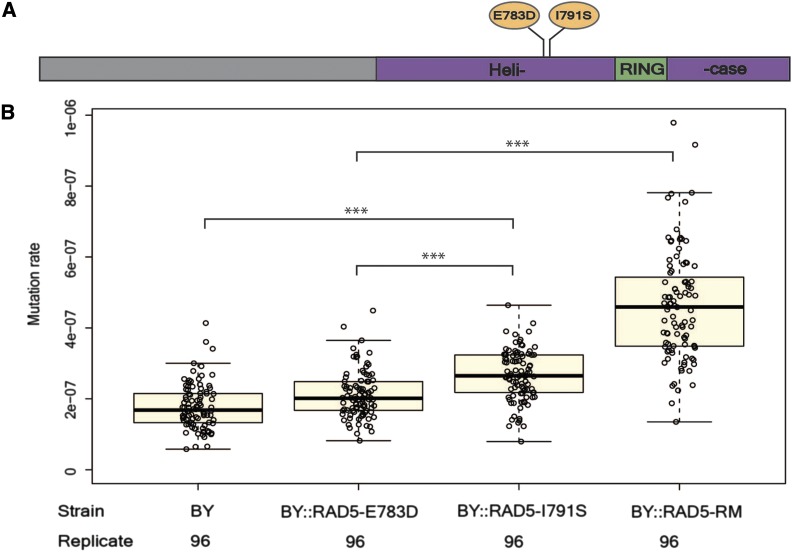
Ten genes fell within the confidence interval of the QTL on chromosome XII. A strong candidate was RAD5, based on previous studies showing that natural variants in RAD5 contribute to sensitivity to the mutagen 4NQO (Demogines et al. 2008). RAD5 encodes a DNA repair protein involved in the error-free DNA damage tolerance (DDT) pathway (Torres-Ramos et al. 2002; Blastyák et al. 2007). The DDT pathway promotes the bypass of single-stranded DNA lesions encountered by DNA polymerases during DNA replication, thus preventing the stalling of DNA replication (Bi 2015). RAD5 plays a crucial role in one branch of the DDT pathway called template switching, in which the stalled nascent strand switches from the damaged template to the undamaged newly synthesized sister strand for extension past the lesion (Bi 2015). Two nonsynonymous substitutions exist between BY and RM strains in RAD5 (Figure 2A), at amino acid positions 783 (glutamic acid in BY and aspartic acid in RM) and 791 (isoleucine in BY and serine in RM). According to Pfam alignments (Sonnhammer et al. 1997), RAD5 contains a HIRAN domain, an SNF2-related N-terminal domain, a RING-type zinc finger domain, and a helicase C-terminal domain (Figure 2A). Both nonsynonymous polymorphisms mapped to the helicase domain of RAD5 (Figure 2A), and no other sequenced strains of S. cerevisiae in the 1002 Yeast Genomes Project contain the aspartic acid 783 and serine 791 variants that are exclusive to the RM strain (Peter et al. 2018). We used protein variation effect analyzer (PROVEAN) (Choi and Chan 2015) to predict whether the two nonsynonymous substitutions have an effect on the biological function of the protein. PROVEAN showed the I791S substitution (score −5.4) might have a strong deleterious effect, while the E783D variant (score −1.8) was not predicted to have a strong effect.
Figure 2.
Polymorphisms in RAD5 underlie mutation rate variation. (A) RAD5 polymorphisms between BY and RM are located in the helicase region. The first letter for each polymorphism indicates the BY polymorphisms (E783, I791) and the second letter indicates the RM polymorphisms (D783, S791). (B) The effect of single RAD5 polymorphism and RAD5 whole-gene replacement was tested in the BY strain background for mutation rate. For each strain, the mutation rates of 96 replicates were measured. Bold lines show the mean. Boxes show the interquartile range. Statistical significance was tested using a permutation t-test. Permutation P value <0.001 is shown as ***.
Nineteen genes fell within the confidence interval of the QTL on chromosome XIV. A strong candidate was MKT1, which was also reported to affect 4NQO sensitivity (Demogines et al. 2008). MKT1 encodes an RNA-binding protein that affects multiple traits and underlies an expression QTL hotspot in yeast (Albert and Kruglyak 2015). The RM allele of MKT1 increases sporulation rate (Deutschbauer and Davis 2005) and improves survival at high temperature (Steinmetz et al. 2002), in low glucose (Parreiras et al. 2011), after exposure to DNA-damaging agents (Demogines et al. 2008), and in high ethanol levels (Swinnen et al. 2012). The coding region of the BY and RM alleles of MKT1 differs by one synonymous polymorphism and two nonsynonymous substitutions. MKT1 has an Xeroderma Pigmentosum Complementation Group G (XPG) domain, which is relevant to DNA repair, and an MKT1 domain, which is related to the maintenance of K2 killer toxin (Wickner 1980). One nonsynonymous variant is in the XPG domain at amino acid position 30 (aspartic acid in BY and glycine in RM), while the other nonsynonymous variant is in the MKT1 domain at position 453 (lysine in BY and arginine in RM). PROVEAN predicted a large effect of the D30G variant (score 6.7) on the function of MKT1, and this variant was previously found to influence sporulation rate (Deutschbauer and Davis 2005), mitochondrial genome stability (Dimitrov et al. 2009), and survival at high temperature (Parreiras et al. 2011). The other variant (K453R) was not predicted to have a strong effect (score 0.6).
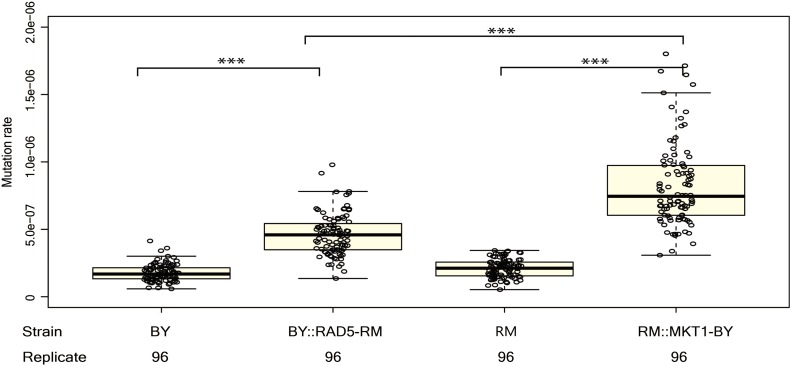
We tested whether RAD5 and MKT1 alleles caused differences in mutation rate by using the fluctuation test on allele replacement strains (Demogines et al. 2008; Smith and Kruglyak 2008) (Table 1). The BY strain carrying the RM allele of RAD5 (BY::RAD5-RM) had a higher mutation rate than the BY strain (permutation t-test, mean difference = 2.9 × 10−7, P < 1 × 10−4), demonstrating that the RM RAD5 allele increases mutation rate (Figure 3). This result is consistent with the observed difference between segregants grouped by parental allele at RAD5 (mean difference = 2.3 × 10−7). The RM strain carrying the BY allele of MKT1 (RM::MKT1-BY) had a higher mutation rate than the RM strain (permutation t-test, mean difference = 6.1 × 10−7, P < 1 × 10−4), showing that the BY MKT1 allele increases mutation rate (Figure 3), consistent with the direction of effect observed in the segregants.
Figure 3.
The RM allele of RAD5 and BY allele of MKT1 increase mutation rate. The mutation rate of two allele replacement strains, the BY strain, and the RM strain are measured and compared. For each strain, 96 replicate measurements for mutation rate were performed. Bold lines show the mean. Boxes show the interquartile range. Statistical significance was tested using permutation t-test. Permutation P value <0.001 is shown as ***.
To gain a finer-level understanding of the two missense variants between BY and RM in the gene RAD5, we tested strains (Demogines et al. 2008) in which these sites in BY were individually replaced with the RM alleles (Table 1) by site-directed mutagenesis. Strains with either variant had a higher mutation rate than BY (permutation t-test, mean difference = 0.9 × 10−7, P < 1 × 10−4 for BY::RAD5-I791S; mean difference = 0.3 × 10−7, P = 6 × 10−4 for BY::RAD5-E783D) (Figure 2B), suggesting that both variants contribute to the higher mutation rate. The BY strain with the I791S substitution had a higher mutation rate than the BY strain with the E783D substitution (permutation t-test, mean difference = 0.6 × 10−7, P < 1 × 10−4) (Figure 2B), consistent with the PROVEAN prediction of a stronger effect for the I791S variant. However, neither variant alone nor the additive effect of the two variants fully recapitulated the increase in mutation rate that we observed when replacing the entire coding region of RAD5 in BY with the RM allele (F = 67.6, d.f. = 1, P = 3.3 × 10−15), suggesting an interaction between the two variants.
Mutation rate shares two large effect QTL with growth on DNA-damaging agents 4NQO and MMS
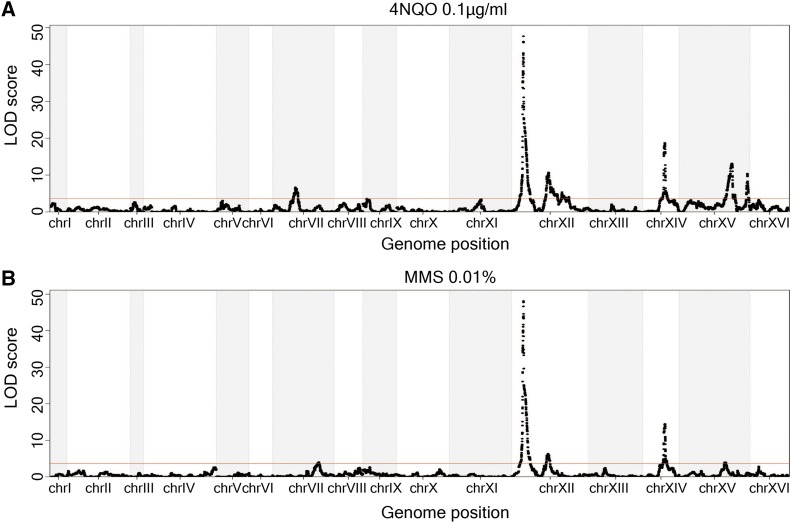
Deficiencies in DNA repair can increase mutation rate (Supek and Lehner 2015; Sabarinathan et al. 2016) and increase sensitivity to DNA-damaging agents such as alkylating compounds and ultraviolet light (Frankfurt 1991; Sun and Moses 1991). We hypothesized that genetic variants that cause deficiencies in DNA repair may underlie QTL for both mutation rate variation and sensitivity to DNA-damaging agents. Previously, Demogines et al. identified a large-effect QTL on chromosome XII for MMS and 4NQO sensitivity in a panel of 123 segregants from a cross between BY and RM (Demogines et al. 2008). Additionally, they identified a QTL on chromosome XIV for 4NQO sensitivity by using backcrossing and bulk segregant analysis. These QTL overlapped with the major QTL that we identified for mutation rate variation, and the underlying causal genes for 4NQO sensitivity were also RAD5 and MKT1.
To follow up on these results, we measured sensitivity to three different DNA damaging agents in our panel of 1040 segregants (Table 2). The compounds assayed included MMS, an alkylating agent that induces DNA double-strand breaks and stalls replication forks (Hampsey 1997); 4NQO, an ultraviolet light mimetic agent (Hampsey 1997); and H2O2, a compound that induces DNA single- and double-strand breaks (Hampsey 1997). We observed that segregants with higher mutation rate, and presumably less-efficient DNA repair systems, were more sensitive to MMS, 4NQO, and H2O2 (Figure S4), consistent with our hypothesis that deficiencies in DNA repair increase the rate of spontaneous mutations and increase sensitivity to DNA-damaging agents. We identified two large-effect QTL for 4NQO and MMS sensitivity that overlapped with the major QTL for mutation rate (Figure 4, A and B). The QTL on chromosome XII and XIV were still observed in the linkage mapping for H2O2, but they had small effects (Figure S5). The large effect QTL detected for H2O2 sensitivity on other chromosomes likely reflects trait-specific effects of variants acting on sensitivity to H2O2 (Figure S5).
Figure 4.
Loci underlying mutation rate variation, 4NQO sensitivity, and MMS sensitivity are overlapping. (A and B) The LOD scores for 4NQO (0.1 µg/ml) sensitivity and MMS (0.01%) sensitivity are plotted against the genetic map. The red line indicates a 5% FWER significance threshold (LOD = 3.62 for 4NQO and LOD = 3.61 for MMS).
Similar mutation spectra in segregants with different RAD5 and MKT1 genotypes
To gain a better understanding of how genetic variation in RAD5 and MKT1 might influence the DNA damage repair process, we characterized the spontaneous mutation spectrum at CAN1 in the segregants. We divided 1040 segregants into four groups based on their genotypes at RAD5 and MKT1 and sequenced pools of CAN1-resistant mutants from each group (Tables S2 and S3). The mutation spectra of the four groups are shown in Figure S6 and Table S4. C:G > T:A transitions were the most frequently observed mutations, A:T > G:C was the rarest transition, and A:T > T:A was the rarest transversion. The spectra for single base-pair substitutions observed here (Figure S7) are similar to previous observations based on whole-genome sequencing of mutation accumulation strains (Zhu et al. 2014). While there were some differences in the relative frequencies of specific mutation types (for instance, more C:G > G:C transversions in segregants with the RM MKT1 allele and more A:T > C:G transversions in segregants with the BY RAD5 allele), these mutation differences were not statistically significant after correction for multiple testing.
Discussion
We developed and implemented a high-throughput fluctuation assay to directly measure mutation rates in yeast. We used this assay to map four QTL that influence differences in the spontaneous mutation rate, and narrowed the two QTL with the largest effects to causal genes and variants. We attempted to gain insight into how these variants might affect the mutation rate by comparing mutational spectra of segregants grouped by genotype, but the differences we observed did not reach statistical significance.
We identified RAD5 as the gene underlying the QTL with the largest effect on mutation rate. RAD5 encodes a DNA helicase and ubiquitin ligase involved in the error-free DDT pathway (Hishida et al. 2009; Unk et al. 2010). We showed that two nonsynonymous variants between BY and RM in the helicase domain affect mutation rate. The Rad5 DNA helicase involves in the replication fork regression process, which was hypothesized to promote DDT and repair during replication (Neelsen and Lopes 2015). The RM allele of RAD5 increases the sensitivity of yeast to 4NQO and MMS (Demogines et al. 2008), probably due to a defect in replication fork regression. Thus the RM allele of RAD5 causes both decreased growth in mutagenic conditions and a higher mutation rate in nonstressful normal conditions.
We furthermore showed that polymorphisms in MKT1 contribute to mutation rate variation. MKT1 is a highly pleiotropic gene that has been shown to affect a number of phenotypes (Steinmetz et al. 2002; Deutschbauer and Davis 2005; Demogines et al. 2008; Smith and Kruglyak 2008; Parreiras et al. 2011; Swinnen et al. 2012; Wang and Kruglyak 2014; Albert and Kruglyak 2015). The BY and RM alleles of MKT1 differ by two nonsynonymous substitutions. One variant (K453R) is located in the MKT1 domain, which is required for activity of the Mkt1 protein in maintaining K2 killer toxin (Vermut et al. 1994). Another variant (D30G) localizes to the XPG-N (the N-terminus of XPG) domain. Four other yeast proteins contain this domain: Exo1, Din7, Rad27 and Rad2. All of these proteins have functions related to DNA repair and cellular response to DNA damage, including DNA double-strand break repair (Exo1) (Tkach et al. 2012), DNA mismatch repair (Exo1, Din7) (Koprowski et al. 2003; Tran et al. 2007), nucleotide excision repair (Rad2) (Northam et al. 2010), ribonucleotide excision repair (Rad27) (Sparks et al. 2012), and large loop repair (Rad27) (Sommer et al. 2008). The internal XPG (XPG-I) domain, together with XPG-N, forms the catalytic domain of the XPG protein in humans. The XPG protein has well-established catalytic and structural roles in nucleotide excision repair, a DNA repair process, and acts as a cofactor for a DNA glycosylase that removes oxidized pyrimidines from DNA (Clarkson 2003). Mutations in the XPG protein commonly cause xeroderma pigmentosum, which often leads to skin cancer (O’Donovan et al. 1994). We hypothesize that Mkt1 has a previously unknown function in DNA damage repair, mediated through its XPG domain.
We found that variants in RAD5 and MKT1 contribute to both mutation rate variation and mutagen sensitivity. These results suggest that spontaneously occurring mutations may have a similar mutation spectrum to those created by 4NQO and MMS, and are potentially repaired by the same mechanisms. Deficient DNA repair can lead to increased sensitivity to agents such as alkylating compounds and ultraviolet light (Frankfurt 1991; Sun and Moses 1991; O’Driscoll et al. 1999), and to higher mutation rates at sites that are less accessible to the DNA repair system (Sabarinathan et al. 2016). Because mutation rates can be difficult to measure, sensitivity to mutagens may serve as a useful proxy.
Recently, Jerison et al. reported heritable differences in adaptability in 230 yeast segregants from the same cross we studied here (Jerison et al. 2017). They measured adaptability as the difference in fitness between a given segregant (“founder”) and a descendant of that founder after 500 generations of experimental evolution. Interestingly, RAD5 fell within one of the QTL found to influence adaptability. Together with our observation that RAD5 influences mutation rate, this finding suggests that differences in mutation rate can affect the adaptability of organisms.
Acknowledgments
We are grateful to members of the Kruglyak laboratory for insightful comments on this manuscript and suggestions for experiments and data analyses. We thank Meru Sadhu for helpful discussion. We would like to especially thank the Alani laboratory in Cornell University for the RAD5 allele replacement and variant substitution strains.
Author contributions: Conceived and designed the experiments: L.G., J.S.B., and L.K. Performed the experiments: L.G. Analyzed the data: L.G. and J.S.B. Wrote the paper: L.G., J.S.B., and L.K.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7343036.
Communicating editor: C. Queitsch
Literature Cited
- Albert F. W., Kruglyak L., 2015. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16: 197–212. 10.1038/nrg3891 [DOI] [PubMed] [Google Scholar]
- Amberg D. C., Burke D. J., Strathern J. N., 2005. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Baer C. F., Miyamoto M. M., Denver D. R., 2007. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat. Rev. Genet. 8: 619–631. 10.1038/nrg2158 [DOI] [PubMed] [Google Scholar]
- Bi X., 2015. Mechanism of DNA damage tolerance. World J. Biol. Chem. 6: 48 10.4331/wjbc.v6.i3.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyák A., Pintér L., Unk I., Prakash L., Prakash S., et al. , 2007. Yeast rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell 28: 167–175. 10.1016/j.molcel.2007.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. S., Ehrenreich I. M., Loo W. T., Lite T.-L. V., Kruglyak L., 2013. Finding the sources of missing heritability in a yeast cross. Nature 494: 234–237. 10.1038/nature11867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J. S., Kotenko I., Sadhu M. J., Treusch S., Albert F. W., et al. , 2015. Genetic interactions contribute less than additive effects to quantitative trait variation in yeast. Nat. Commun. 6: 8712 10.1038/ncomms9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Chan A. P., 2015. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31: 2745–2747. 10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. 10.1534/genetics.107.080101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson S. G., 2003. The XPG story. Biochimie 85: 1113–1121. 10.1016/j.biochi.2003.10.014 [DOI] [PubMed] [Google Scholar]
- Demerec M., 1937. Frequency of spontaneous mutations in certain stocks of drosophila melanogaster. Genetics 22: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demogines A., Smith E., Kruglyak L., Alani E., 2008. Identification and dissection of a complex DNA repair sensitivity phenotype in baker’s yeast. PLoS Genet. 4: e1000123 10.1371/journal.pgen.1000123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer A. M., Davis R. W., 2005. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 37: 1333–1340. 10.1038/ng1674 [DOI] [PubMed] [Google Scholar]
- Dimitrov L. N., Brem R. B., Kruglyak L., Gottschling D. E., 2009. Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics 183: 365–383. 10.1534/genetics.109.104497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., Charlesworth B., Charlesworth D., Crow J. F., 1998. Rates of spontaneous mutation. Genetics 148: 1667–1686. https://doi.org/citeulike-article-id:610966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankfurt O. S., 1991. Inhibition of DNA repair and the enhancement of cytotoxicity of alkylating agents. Int. J. Cancer 48: 916–923. 10.1002/ijc.2910480620 [DOI] [PubMed] [Google Scholar]
- Hampsey M., 1997. A review of phenotypes in Saccharomyces cerevisiae. Yeast 13: 1099–1133. [DOI] [PubMed] [Google Scholar]
- Hishida T., Kubota Y., Carr A. M., Iwasaki H., 2009. RAD6–RAD18–RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light. Nature 457: 612–615. 10.1038/nature07580 [DOI] [PubMed] [Google Scholar]
- Jerison E. R., Kryazhimskiy S., Mitchell J. K., Bloom J. S., Kruglyak L., et al. , 2017. Genetic variation in adaptability and pleiotropy in budding yeast. Elife 6: e27167 10.1101/121749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprowski P., Fikus M. U., Dzierzbicki P., Mieczkowski P., Lazowska J., et al. , 2003. Enhanced expression of the DNA damage-inducible gene DIN7 results in increased mutagenesis of mitochondrial DNA in Saccharomyces cerevisiae. Mol. Genet. Genomics 269: 632–639. 10.1007/s00438-003-0873-8 [DOI] [PubMed] [Google Scholar]
- Lang G. I., Murray A. W., 2008. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178: 67–82. 10.1534/genetics.107.071506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M. R., 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79: 181–211. 10.1146/annurev.biochem.052308.093131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M., Betrán E., Thornton K., Wang W., 2003. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 4: 865–875. 10.1038/nrg1204 [DOI] [PubMed] [Google Scholar]
- Luria S., Delbrück M., 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511. 10.1038/nature10260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits, pp. 980 Sinauer, Sunderland, MA [Google Scholar]
- Matheson K., Parsons L., Gammie A., 2017. Whole-genome sequence and variant analysis of W303, a widely-used strain of Saccharomyces cerevisiae. G3 (Bethesda) 7: 2219–2226. 10.1534/g3.117.040022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen K. J., Lopes M., 2015. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 16: 207–220. 10.1038/nrm3935 [DOI] [PubMed] [Google Scholar]
- Northam M. R., Robinson H. A., Kochenova O. V., Shcherbakova P. V., 2010. Participation of DNA polymerase?? in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics 184: 27–42. 10.1534/genetics.109.107482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan A., Scherly D., Clarkson S. G., Wood R. D., 1994. Isolation of active recombinant XPG protein, a human DNA repair endonuclease. J. Biol. Chem. 269: 15965–15968. [PubMed] [Google Scholar]
- O’Driscoll M., Macpherson P., Xu Y. Z., Karran P., 1999. The cytotoxicity of DNA carboxymethylation and methylation by the model carboxymethylating agent azaserine in human cells. Carcinogenesis 20: 1855–1862. 10.1093/carcin/20.9.1855 [DOI] [PubMed] [Google Scholar]
- Parreiras L. S., Kohn L. M., Anderson J. B., 2011. Cellular effects and epistasis among three determinants of adaptation in experimental populations of Saccharomyces cerevisiae. Eukaryot. Cell 10: 1348–1356. 10.1128/EC.05083-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J., De Chiara M., Friedrich A., Yue J.-X., Pflieger D., et al. , 2018. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556: 339–344. 10.1038/s41586-018-0030-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabarinathan R., Mularoni L., Deu-Pons J., Gonzalez-Perez A., Lopez-Bigas N., 2016. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature 532: 264–267. 10.1038/nature17661 [DOI] [PubMed] [Google Scholar]
- Sarkar S., Ma W. T., Sandri G. H., 1992. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica 85: 173–179. 10.1007/BF00120324 [DOI] [PubMed] [Google Scholar]
- Schacherer J., Ruderfer D. M., Gresham D., Dolinski K., Botstein D., et al. , 2007. Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS One 2: e322 10.1371/journal.pone.0000322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J., Shapiro J. A., Ruderfer D. M., Kruglyak L., 2009. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458: 342–345. 10.1038/nature07670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. N., Kruglyak L., 2008. Gene-environment interaction in yeast gene expression. PLoS Biol. 6: e83 10.1371/journal.pbio.0060083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer D., Stith C. M., Burgers P. M. J., Lahue R. S., 2008. Partial reconstitution of DNA large loop repair with purified proteins from Saccharomyces cerevisiae. Nucleic Acids Res. 36: 4699–4707. 10.1093/nar/gkn446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E. L., Eddy S. R., Durbin R., 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28: 405–420. [DOI] [PubMed] [Google Scholar]
- Sparks J. L., Chon H., Cerritelli S. M., Kunkel T. A., Johansson E., et al. , 2012. RNase H2-Initiated ribonucleotide excision repair. Mol. Cell 47: 980–986. 10.1016/j.molcel.2012.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., et al. , 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330. 10.1038/416326a [DOI] [PubMed] [Google Scholar]
- Sun Y., Moses R. E., 1991. Reactivation of psoralen-reacted plasmid DNA in Fanconi anemia, xeroderma pigmentosum, and normal human fibroblast cells. Somat. Cell Mol. Genet. 17: 229–238. 10.1007/BF01232819 [DOI] [PubMed] [Google Scholar]
- Supek F., Lehner B., 2015. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature 521: 81–84. 10.1038/nature14173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen S., Schaerlaekens K., Pais T., Claesen J., Hubmann G., et al. , 2012. Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res. 22: 975–984. 10.1101/gr.131698.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach J. M., Yimit A., Lee A. Y., Riffle M., Costanzo M., et al. , 2012. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 14: 966–976. 10.1038/ncb2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson I. P., Novelli M. R., Bodmer W. F., 1996. The mutation rate and cancer. Proc. Natl. Acad. Sci. USA 93: 14800–14803. 10.1073/pnas.93.25.14800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ramos C. A., Prakash S., Prakash L., 2002. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 2419–2426. 10.1128/MCB.22.7.2419-2426.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. T., Fey J. P., Erdeniz N., Gellon L., Boiteux S., et al. , 2007. A mutation in EXO1 defines separable roles in DNA mismatch repair and post-replication repair. DNA Repair (Amst.) 6: 1572–1583. 10.1016/j.dnarep.2007.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk I., Hajdú I., Blastyák A., Haracska L., 2010. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst.) 9: 257–267. 10.1016/j.dnarep.2009.12.013 [DOI] [PubMed] [Google Scholar]
- Vermut M., Widner W. R., Dinman J. D., Wickner R. B., 1994. XIV. Yeast sequencing reports. Sequence of MKT1, needed for propagation of M2 satellite dsRNA of the L‐A virus of Saccharomyces cerevisiae. Yeast 10: 1477–1479. 10.1002/yea.320101111 [DOI] [PubMed] [Google Scholar]
- Wang X., Kruglyak L., 2014. Genetic basis of haloperidol resistance in Saccharomyces cerevisiae is complex and dose dependent. PLoS Genet. 10: e1004894 10.1371/journal.pgen.1004894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., 1980. Plasmids controlling exclusion of the K2 killer double-stranded RNA plasmid of yeast. Cell 21: 217–226. 10.1016/0092-8674(80)90129-4 [DOI] [PubMed] [Google Scholar]
- Zhu Y. O., Siegal M. L., Hall D. W., Petrov D. A., 2014. Precise estimates of mutation rate and spectrum in yeast. Proc. Natl. Acad. Sci. USA 111: E2310–E2318. 10.1073/pnas.1323011111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. File S1 contains the “mutation rate score” for 843 segregants. File S2 contains the mutation rate for allele replacement and variant-engineered strains. File S3 contains the primer sequences used for the amplicon sequencing. Data and code for QTL mapping and mutation spectrum analysis are available at https://github.com/gouliangke/Mutation-rate. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7343036.