G protein-coupled receptors (GPCRs) are crucial sensors of extracellular signals in eukaryotes, and direct measurement of GPCR-mediated signaling is useful for high-throughput mutational studies. However, this is particularly difficult for the light-activated GPCR rhodopsin...
Keywords: Visual degenerative disease, retinitis pigmentosa, G protein-coupled receptor, disease model, rhodopsin
Abstract
G protein-coupled receptors (GPCRs) are crucial sensors of extracellular signals in eukaryotes, with multiple GPCR mutations linked to human diseases. With the growing number of sequenced human genomes, determining the pathogenicity of a mutation is challenging, but can be aided by a direct measurement of GPCR-mediated signaling. This is particularly difficult for the visual pigment rhodopsin—a GPCR activated by light—for which hundreds of mutations have been linked to inherited degenerative retinal diseases such as retinitis pigmentosa. In this study, we successfully engineered, for the first time, activation by human rhodopsin of the yeast mating pathway, resulting in signaling via a fluorescent reporter. We combine this novel assay for rhodopsin light-dependent activation with studies of subcellular localization, and the upregulation of the unfolded protein response in response to misfolded rhodopsin protein. We use these assays to characterize a panel of rhodopsin mutations with known molecular phenotypes, finding that rhodopsin maintains a similar molecular phenotype in yeast, with some interesting differences. Furthermore, we compare our assays in yeast with clinical phenotypes from patients with novel disease-linked mutations. We demonstrate that our engineered yeast strain can be useful in rhodopsin mutant classification, and in helping to determine the molecular mechanisms underlying their pathogenicity. This approach may also be applied to better understand the clinical relevance of other human GPCR mutations, furthering the use of yeast as a tool for investigating molecular mechanisms relevant to human disease.
THE diversity of biologically relevant signals that G protein-coupled receptors (GPCRs) detect make them critical to how cells sense and respond to their environment. Missense mutations within this large family of cell surface receptors can therefore have serious physiological effects, and many human diseases have been linked with missense mutations in GPCRs (Heng et al. 2013). Over 63,000 missense mutations have been identified in human GPCRs via large-scale human genome studies (Pándy-Szekeres et al. 2018), but the majority have not been studied in detail, so the significance to human health remains unclear. Missense mutations can disrupt GPCR function in many ways, ranging from ligand binding, to protein stability, to changes in downstream signaling and interactions with negative regulators (Stoy and Gurevich 2015). Consequently, disease can arise through many different molecular phenotypes, and not all mutations are disease causing.
To better understand how human missense mutations contribute to disease, the yeast Saccharomyces cerevisiae has emerged as a powerful tool for characterizing human protein function due to conserved molecular pathways, rapid growth, and ease of genetic manipulation (Laurent et al. 2016). These benefits have facilitated yeast models of human disease (Outeiro and Lindquist 2003; Perocchi et al. 2008) and studies of pathogenic human mutations (Sun et al. 2016; Yang et al. 2017). Synthetic biology approaches, where human protein function and interactions are quantified by yeast-based gene circuits, have enabled high-throughput experiments at an impressive scale (Starita et al. 2015; Sokolina et al. 2017; Weile et al. 2017; Woodsmith et al. 2017). With genetic engineering, human GPCRs can be functionally linked to the yeast mating pathway, and mating-responsive reporter genes have allowed for detailed studies of GPCR activation (Liu et al. 2016). As human GPCRs retain their natural preferences for ligands and G proteins in yeast (Brown et al. 2000), this application of synthetic biology combines the high-throughput capabilities of yeast-based studies with the ability to rapidly characterize GPCR function in a cellular context. This has facilitated screens of chemical libraries for novel GPCR ligands (Campbell et al. 1999; Horswill et al. 2007), and screens of mutated GPCRs to characterize specific protein domains or to engineer novel function (Erlenbach et al. 2001; Armbruster et al. 2007; Liu et al. 2015).
Despite the power of yeast-based studies of human GPCRs, only a small proportion of GPCRs have been functionally linked to the yeast mating pathway, and all have been ligand-activated (Liu et al. 2016). Unlike these GPCRs, rhodopsin exists as a covalent complex between its light-sensitive chromophore 11-cis retinal and the seven-helix transmembrane opsin apoprotein (Smith 2010). Light exposure isomerizes the chromophore, which induces a conformational change in rhodopsin’s transmembrane helices, activating the associated heterotrimeric G protein, transducin, and triggering the visual transduction cascade that eventually results in a signal to the brain that light has been perceived (Smith 2010). Not surprisingly, missense mutations in rhodopsin are often associated with retinal diseases in humans (Athanasiou et al. 2018). Retinitis pigmentosa (RP) is a highly heterogeneous, degenerative retinal disorder that results in vision impairment and in some cases eventually blindness, affecting ∼1 in 4000 people worldwide (Fahim et al. 2000). The heterogeneous nature of this degenerative disease has contributed to the difficulties in developing effective prognoses and treatment. Missense mutations in rhodopsin have been associated with 20–30% of autosomal dominant RP and 1% of autosomal recessive RP cases, making rhodopsin one of the most important RP genes (Fahim et al. 2000). Over 150 rhodopsin missense mutations have been associated with disease (Stenson et al. 2014), and an additional >200 uncharacterized mutations in rhodopsin exons are listed in the Genome Aggregation Database (Lek et al. 2016).
Without a tool to rapidly assess rhodopsin function, this increasing availability of genetic information has yet to lead to a better understanding of pathogenicity of mutations associated with RP and other inherited visual diseases. Determining the impact of missense mutations on the ability of rhodopsin to respond to light currently relies on in vitro biochemical assays that are labor intensive, requiring mammalian cell culture to produce one mutant protein at a time, followed by immunofluorescence or immunoaffinity purification (Sung et al. 1991; Reeves et al. 1996). These technical challenges are compounded by the diverse molecular phenotypes of rhodopsin mutations, ranging from constitutively active, to improper subcellular localization, to disrupted post-translational modifications (Athanasiou et al. 2018). To efficiently characterize the wide variety of patient-derived mutations, a rapid method that reliably recapitulates light-dependent signaling of rhodopsin is needed.
Here, we use synthetic biology approaches to engineer rhodopsin coupling to the yeast mating pathway, demonstrating for the first time successful rhodopsin light-activated signal transduction that can be rapidly quantified using a fluorescent reporter gene of mating pathway activation. We compared our novel yeast-based assay to more established mammalian cell-based methods, using a panel of previously studied rhodopsin mutations. We found that measurements of rhodopsin activation in yeast resemble in vitro results using rhodopsin purified from mammalian cells, with some exceptions. We also found that a yeast-based reporter of the unfolded protein response (UPR) produced results consistent with previous studies in mammalian cells quantifying the effects of rhodopsin pathogenic mutants on cellular stress. Finally, we used our combined approaches in yeast to investigate recently identified rhodopsin mutations in patients with retinal disease, and were able to propose pathogenic classifications that are supported by mammalian cell and clinical data.
Materials and Methods
Yeast strain engineering
The parent yeast strain for all strain engineering was CB008, genotype W303 MATa, far1Δ, his3, trp1, leu2, ura3 (Supplemental Material, Table S1 contains all strain genotypes). All gene knock-outs and knock-ins were conducted using homologous recombination of selectable markers. pFUS1-mCherry was integrated at the MFA2 locus using plasmid pJW609 containing the KanR marker. pFUS1 was defined as the 1636 bp immediately upstream of the Fus1 start codon, the mCherry sequence used is from Keppler-Ross et al. (2008), and ∼1 kb homology regions were used. STE2 and SST2 were targeted for deletion using TRP1 and HygB selectable markers respectively, each with 180 bp of flanking homology regions identical to the sequences flanking the ORF. The five C-terminal amino acids of Gpa1 (KIGII) was replaced with a Gpa1-Gαt (transducin) chimera, containing the C-terminal amino acids from mammalian Gαt (DCGLF), using plasmid pBS600 designed for this study (Figure S1), containing selectable marker LEU2, and a sequence homologous to the 800 bp 3′ to the natural Gpa1 gene. The C. albicans Adh terminator was used downstream of both the pFUS1-mCherry and Gpa1-Gαt gene cassettes. Strains were confirmed by PCR and flow cytometry.
Rhodopsin mutation selection and patient phenotyping
Rhodopsin mutations were selected from across phenotypic classes, as reported in a recent comprehensive review (Athanasiou et al. 2018), and with at least one of the following assays previously published: transducin activation, localization in mammalian cells, or spectroscopy indicating chromophore binding. The patient cases were selected from an internal database and the phenotype information was collected retrospectively. Other than basic demographic and genetic information, we collected information about visual acuity (VA), color vision, Goldmann visual fields (GVF), electroretinography (ERG), and imaging. Imaging included fundus photography (VisucamNM/FA; Carl Zeiss Meditec, Dublin, CA and Optos) and optical coherence tomography (OCT, Cirrus from Carl Zeiss Meditec). Genetic testing was done using gene panels based sequencing by CLIA-approved laboratories. This study was approved by the Human Research Ethics Board of the hospital for Sick Children and met the Tenets of the declaration of Helsinki.
Cloning and mutagenesis
The human rhodopsin sequence (RefSeq NP_000530.1) was amplified using Pfu polymerase (Thermo) from plasmid pJET HuRh (Morrow et al. 2017), using primers to insert flanking AarI restriction sites. Following AarI digestion, the rhodopsin sequence was ligated to the yeast centromere plasmids pRS316 and pRS313, which each contained the TDH3 promoter (pTDH3; alternatively called pGPD). For mammalian expression, rhodopsin mutations were introduced into the wild-type bovine rhodopsin sequence in the p1D4 vector for immunoaffinity purification or the pGFP vector for SK-N-SH immunofluorescence microscopy (Figure S2). Mutagenesis was conducted via PCR following the QuikChange site-directed mutagenesis protocol (Agilent) and using PfuUltra II Fusion HS DNA Polymerase (Agilent). Mutagenesis primers were designed with 20 or 21 nucleotides identical to human or bovine rhodopsin flanking the mutant nucleotide(s).
Yeast plasmid transformation
Yeast strain BS017 or yJW1200 were transformed with individual plasmids by a standard lithium-acetate method and plated on selective media (SD-URA or SD-HIS, respectively).
Rhodopsin purification
Culturing and transfection of HEK293T cells was performed as previously described (Bhattacharyya et al. 2017). Briefly, p1D4 vector containing a rhodopsin gene was transfected with Lipofectamine 2000 (Invitrogen) and cells were harvested after 48 hours. Rhodopsin was regenerated with 11-cis retinal for 2 hours before solubilization in 1% dodecyl maltoside (Anatrace) and immunoaffinity purified with 1D4 monoclonal antibody (Molday and MacKenzie 1983). The ultraviolet-visible absorption spectra of the rhodopsin proteins were recorded using a Cary 4000 double beam spectrophotometer (Agilent). Pigments were light bleached with a Fiber-Lite MI-150 high intensity illuminator (Dolan-Jenner) for 60 sec at 20°. Dark-light difference spectra were calculated by subtracting the light-bleached absorbance spectra from the dark spectra.
Yeast light activation assay
Yeast strain BS017 transformed with a human rhodopsin mutant gene in the pRS316 pTDH3 vector was incubated overnight in SD-URA media in a 30° shaker. The same strain transformed with a plasmid not containing the rhodopsin sequence (Vector) was used as a negative control. Cells were diluted to OD600 0.05 in fresh media containing 5 μM 9-cis retinal (Sigma-Aldrich) and incubated for 2 hours, protected from light, in a 30° shaker; 9-cis retinal is a common alternative to the natural chromophore 11-cis retinal, and gives comparable in vitro results (Opefi et al. 2013). LightSafe 50-ml centrifuge tubes (Sigma-Aldrich) were used for 5 ml cultures, and 96-well deep well blocks (VWR) wrapped in aluminum foil for 600 μl cultures. Indicated cultures were then exposed to light using a Fiber-Lite MI-150 high intensity illuminator (Dolan-Jenner) set to full intensity for 15 min at room temperature. After 100 μl samples were taken for analysis, an additional 5 μM 9-cis retinal was added to indicated cultures—to both light exposed and cultures kept in the dark—and placed back in the 30° shaker. Light exposure followed by retinal addition was conducted every hour for a total of 6 hours following the first light exposure. Cells were then treated with the protein synthesis inhibitor cycloheximide, to a final concentration of 10 μg/ml. The mCherry fluorescence of at least 6000 cells was measured for each sample with a Miltenyi Biotec MACSQuant VYB. The mean mCherry fluorescence was determined using FlowJo. After subtracting the mCherry fluorescence signal of the Vector control, fluorescence values were normalized to the wild-type rhodopsin control used in the same experiment, to allow comparisons between experiments performed on different days.
Immunofluorescence microscopy
SK-N-SH neuroblastoma (ATCC HTB-11) cells were grown and cultured in full media [DMEM (Life Technologies), 10% FBS (Invitrogen), and 1% Penicillin-Streptomycin (Invitrogen)] at 37° in 5% CO2 and seeded into 24-well plates with coverslips (Sarstedt) while under five passages. Once cells reached ∼75% confluence, they were transfected with 645 ng of pGFP plasmid containing the appropriate bovine rhodopsin gene, using Lipofectamine 2000 (Invitrogen) protocols. After 24 hours, half the wells were incubated with Wheat germ Agglutinin (Invitrogen) in HBSS for 10 min at 37° to label the plasma membrane. All cells were then rinsed with PBS and fixed with 2% paraformaldehyde in PBS. To label cells with the endoplasmic reticulum marker antibody anti-calreticulin (1:400; Abcam), cells were washed and permeabilized in PBS containing 1% bovine serum albumin (Sigma) and 0.1% saponin (PBS-BS). Anti-calreticulin was diluted in PBS-BS and incubated for 1 hours at room temperature. After washing with PBS-BS, secondary antibody (Cy3-conjugated goat anti-rabbit IgG, 1:200; Jackson Immunoresearch) was diluted in PBS-BS and added to the wells for 1 hours. Nuclei were stained with Hoechst (1:1000 in PBS, Hoechst type 33258 Invitrogen) for 10 min. Cells were mounted with ProLong Gold Antifade (Thermo), a coverslip was applied, and allowed to cure for 24 hours in the dark prior to imaging on Leica TCS SP8 confocal microscope. ImageJ was used to construct Z-stacks, maximum projection images, and scale bars.
Yeast microscopy
Yeast strain BS017 expressing human rhodopsin C-terminally tagged with GFP were grown to log phase in selective media, then plated on glass-bottomed dishes (Greiner Bio-One) treated with 1 mg/ml concanavalin A (Sigma-Aldrich). The centromere plasmid pRS316 pTDH3 was used, and the GFP sequence used is from Moser et al. (2013). A short amino acid linker (GGERGS) was introduced between the final rhodopsin residue and first GFP residue. Images of the cells adhered to the dishes were acquired using a Leica TCS SP8 confocal microscope with a 100× 1.4NA objective and a hybrid detector (Leica). GFP fluorescence was analyzed using a custom Fiji-MATLAB pipeline (File S1 and File S2), which was similar to analyses performed to quantify fluorescence peaks in mammalian cells (Cheng et al. 1999). On Fiji using batch mode (Schindelin et al. 2012), yeast cell maps were generated by first removing high-frequency noise using ROF Denoise (theta = 25) and preparing for thresholding using Enhance Contrast (0.3 saturation with Normalization). Manual thresholding, filling holes, and finally selecting of cells using the watershed plugin generated final cell maps. Cell membrane maps were defined in MATLAB as the seven outer pixels (∼0.7 µm) of each cell map. Fluorescent patches at the cell membrane were defined in MATLAB as pixels with fluorescence intensity at least 2.0 SD above the edge mean. Cells with patchy membrane expression of rhodopsin were defined in MATLAB as cells with at least 10% of their cell membrane containing membrane patches. Bootstrapped SE were generated in MATLAB by using the Statistics and Machine Learning Toolbox functions (Mathworks).
UPR activation assay
Yeast strain yJW1200 was generously provided by the Weissman laboratory, University of California, San Francisco. This strain contains a 4× repeat of the unfolded protein response element upstream of GFP, and the constitutive TEF2 promoter upstream of RFP (Jonikas et al. 2009). yJW1200 transformed with a human rhodopsin mutant gene in the pRS313 pTDH3 vector was incubated overnight in 3 ml SD-HIS media in a 30° shaker. The same strain transformed with a plasmid not containing the rhodopsin sequence (Vector) was used as a negative control. Cells were diluted to OD600 0.2 in 600 μl fresh media and incubated for 4 hours in a 30° shaker. Cells were then treated with the protein synthesis inhibitor cycloheximide, to a final concentration of 10 μg/ml. The GFP and RFP signal of at least 10,000 cells was measured for each sample with a Miltenyi Biotec MACSQuant VYB. The mean GFP and RFP fluorescence was determined using FlowJo. Fluorescence values were normalized to the wild-type rhodopsin control used in the same experiment, to allow comparisons between experiments performed on different days.
Statistical analyses and graphs
Statistical analyses were performed using Prism (GraphPad), using one-way ANOVA for UPR comparisons to wild-type rhodopsin, and for yeast light and dark activation comparisons to wild-type rhodopsin. Student’s t-test was used to compare the results of one mutant or condition to one other mutant or condition. Graphs were generated using Prism or Excel (Microsoft).
Data availability statement
Strains and plasmids are available upon request. All supplemental figures, tables, and files have been uploaded to figshare. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7414109.
Results
Human rhodopsin functionally couples to the yeast mating pathway and signal transduction is dependent on light
We have engineered, for the first time, a vertebrate rhodopsin that can successfully couple to the yeast mating pathway. This was accomplished by first knocking out the genes encoding Far1, to prevent cell cycle arrest, and the GTPase-activating protein Sst2, a negative regulator of the mating pathway (Brown et al. 2000). The endogenous mating pathway GPCR Ste2 was also knocked out, to prevent unnecessary interactions with downstream mating pathway proteins. Next, a chimeric G alpha protein and the fluorescent protein mCherry under the regulation of the mating-responsive FUS1 promoter (pFUS1) were inserted into the yeast genome (Figure 1 and Table S1). As rhodopsin is known to interact only with G alpha proteins containing the same five amino acid C-terminal sequence [transducin in rod photoreceptors and Gαi1 in engineered systems (Maeda et al. 2014; Sun et al. 2015)], only one Gpa1 chimera (Gpa1-Gαt) was required for our study. To ensure functional coupling between Gpa1-Gαt and human rhodopsin, we first expressed a known constitutively active rhodopsin mutant, E113Q M257Y (Han et al. 1998). Consistent and high levels of mCherry fluorescence were observed, regardless of the presence of retinal chromophore (Figure S3), indicative of productive rhodopsin expression and the ability to activate the yeast mating pathway.
Figure 1.
Representation of the engineered mating pathway. Rhodopsin activation was functionally coupled to the expression of a fluorescent reporter protein, mCherry, utilizing the mating-responsive promoter pFUS1. Modifications to the mating pathway included the knockout of negative regulator Sst2, the gene encoding Far1, which halts cell growth in the wild-type mating pathway, and the endogenous mating pathway GPCR Ste2. The chimeric G alpha protein (Gpa1-Gαt) contains the five C-terminal amino acids of the G alpha subunit of human transducin (Gαt).
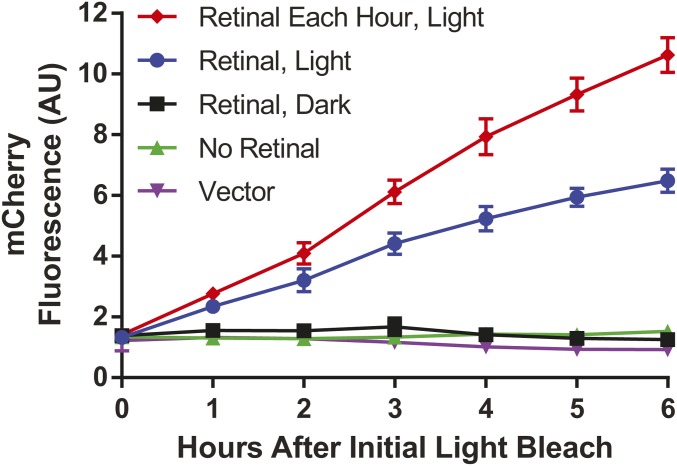
Wild-type human rhodopsin was then expressed using the same strain, and, when incubated with retinal, induced the expression of mCherry only in response to light (Figure 2); 5 μM 9-cis retinal in culture media was sufficient to elicit this light-dependent response—a concentration also used for the heterologous expression of rhodopsin using mammalian cells (Opefi et al. 2013). The 5- to 10-fold increase in mCherry fluorescence in response to GPCR activation was comparable to previous reported activation of the natural mating pathway GPCR Ste2 (Ishii et al. 2008; Kompella et al. 2017). Increasing the retinal concentration did not increase activation of the mating pathway, indicating rhodopsin molecules were saturated with chromophore (Figure S4). However, mating pathway response was enhanced when retinal was added after each hourly light exposure, to account for the lack of retinal recycling enzymes in yeast.
Figure 2.
Light-dependent activation of the mating pathway. Human rhodopsin was found to activate the mating pathway only in response to light, requiring the presence of retinal chromophore. Adding retinal after each hourly light exposure improved the overall response ∼1.6-fold. Incubating with the same concentration of retinal but keeping the culture in the dark did not result in mating pathway activation. Vector: Yeast transformed with a plasmid not containing the rhodopsin gene. Data points represent results of four individual colonies, each in a 5 ml culture. Error bars represent SD.
Magnitude of light-activated signal transduction in yeast comparable to assays of rhodopsin expressed in mammalian cells
After establishing light-dependent activation of rhodopsin in yeast, we next sought to compare these new yeast-based methods to traditional in vitro methods utilizing protein purified from mammalian cells. Previously characterized rhodopsin mutations P23H, M39R, and G51A were specifically chosen to establish a gradient of phenotypic severity. P23H is the most common RP-associated rhodopsin mutation in North America (Dryja et al. 1990; Mendes et al. 2005), and has been characterized in a number of cell and animal models. P23H rhodopsin consistently displays poor stability (Krebs et al. 2010; Chen et al. 2014), aggregation in the ER (Chiang et al. 2012b), and disrupted transducin activation (Opefi et al. 2013; Chen et al. 2014), leading to severe retinal degeneration (Cideciyan et al. 1998; Athanasiou et al. 2017; LaVail et al. 2018). The less severe M39R mutation, which is also associated with RP, has been studied using both bovine and human rhodopsin genes displaying a more severe cytosolic aggregation phenotype in the human gene background (Davies et al. 2012; Ramon et al. 2014). As M39R rhodopsin is expressed more productively by mammalian cells than P23H rhodopsin, and a proportion remains able to form a light-responsive complex with retinal, it was selected as an intermediate RP-associated rhodopsin mutation (Davies et al. 2012; Ramon et al. 2014). G51A is the most common nonsynonymous rhodopsin mutation in humans (Lek et al. 2016), displays a less severe phenotype in vitro and in patients (Cideciyan et al. 1998; Bosch et al. 2003), and may be an asymptomatic variant (Athanasiou et al. 2018). These three rhodopsin mutants were each expressed using the mCherry reporter yeast strain. Following exposure to light, the relative mCherry fluorescence was observed as follows: P23H < M39R < G51A = WT (Figure 3A).
Figure 3.
Characterization of rhodopsin light-dependent function. (A) Response to light from yeast-expressed rhodopsin mutants, indicating a similar magnitude of response as the mammalian cell-expressed protein. WT: Wild-type human rhodopsin. Yeast data points represent results of nine individual colonies, each in a 600 μl culture, minus the mCherry fluorescence of the same strain transformed with empty plasmid control (Vector), and normalized to wild-type. * P < 0.05 vs. WT or between indicated mutants. (B) Difference spectra of mammalian cell-expressed rhodopsin mutants in response to light. The peak at 500 nm indicates a light-dependent response.
Next, we compared rhodopsin activation in yeast to traditional in vitro methods for determining rhodopsin function in response to light. The same three mutants were purified following heterologous expression in mammalian cells, then regenerated with retinal. By recording the absorption spectra before and after exposure to light, difference spectra showing the response of rhodopsin to light could be measured. This method has been used extensively to characterize missense mutations suspected to cause inherited retinal disease, as a measure of the ability of rhodopsin to properly fold and respond to light (Sung et al. 1991; Opefi et al. 2013). The relative response to light displayed a similar range of function as the yeast-expressed mutants (Figure 3B). This suggested not only that the function of yeast-expressed rhodopsin was similarly impacted by pathogenic mutations, but also that the relative activation of the mating pathway in yeast is comparable to the severity of the mutant as measured in vitro using rhodopsin purified from mammalian cells.
Pathogenic mutations known to disrupt rhodopsin stability or G protein coupling prevent light-activated signal transduction in yeast
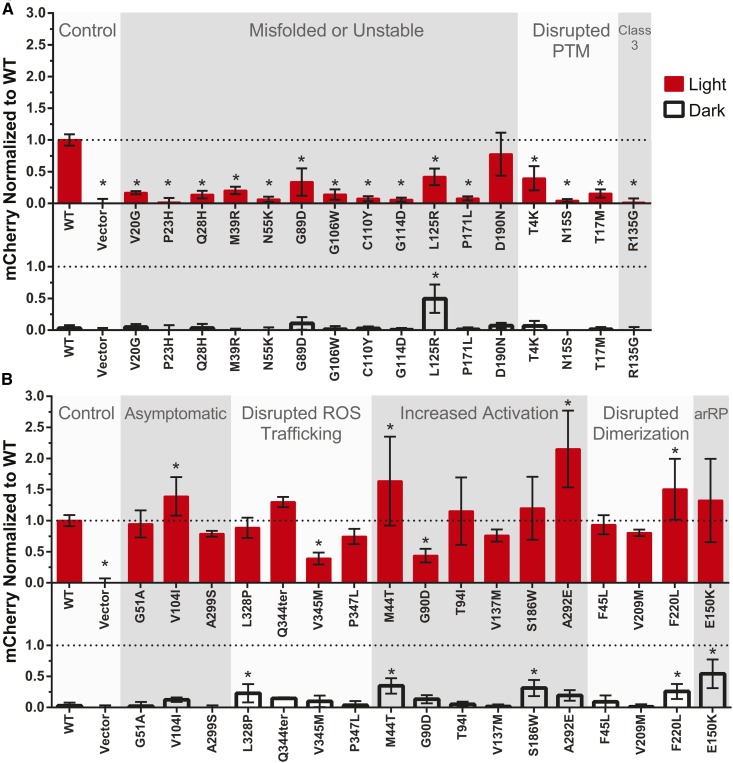
After establishing yeast as a platform for quantifying light-dependent rhodopsin activation, we investigated a larger panel of rhodopsin mutations to understand how the wide range of known functional phenotypes translates to a response in yeast. Efforts have been made to classify mutations based on these phenotypes, which range from completely inactive to constitutively active (Figure S5 and Table S2), as discussed in detail in a recent review (Athanasiou et al. 2018). However, as this new yeast assay examines rhodopsin signaling in an engineered cellular system, it was unknown how results would compare to traditional in vitro methods using purified mammalian cell-expressed rhodopsin. We first focused on pathogenic mutations known to disrupt rhodopsin folding and stability, as we hypothesized these loss-of-function mutations would be easier to distinguish from wild type. These mutations were placed into three groups based on previous characterization: mutations intrinsically disrupting rhodopsin stability; mutations indirectly affecting stability by disrupting a post-translational modification (glycosylation) motif; or mutations disrupting G protein coupling, leading to constitutive endocytosis (Class 3).
When expressed in yeast and exposed to light, rhodopsin function was significantly disrupted by each of the mutations known to result in misfolding or instability, with the exception of D190N (Figure 4A). D190N had previously been shown to be a less severe RP-linked mutation (Fishman et al. 1992; Tsui et al. 2008; Liu et al. 2013), and ERG data from patients matches our observation that this missense mutation does not completely disrupt rhodopsin function (Sancho-Pelluz et al. 2012). The G89D and L125R mutations also had a reduced but measurable response to light when expressed in yeast, which fits with previous trends observed (Kaushal and Khorana 1994; Bosch et al. 2003). Interestingly, L125R lead to signaling in the dark as well, equivalent to the mutant’s light-dependent activation, which has not previously been reported.
Figure 4.
The light-activated signal transduction of pathogenic rhodopsin mutants in yeast. (A) Missense mutations that intrinsically disrupt rhodopsin stability, or that indirectly affect stability by disrupting a post-translational modification (PTM) motif. Class 3 denotes a unique category of mutations at site R135, which disrupt G protein coupling and lead to constitutive endocytosis in mammalian cells. (B) Pathogenic and asymptomatic variants that increase or do not disrupt rhodopsin activation by light. WT: Wild-type human rhodopsin; Vector: yeast transformed with a plasmid not containing the rhodopsin gene; arRP: autosomal recessive RP. Data points represent results of nine individual colonies, each in a 600 μl culture, minus the mCherry fluorescence of Vector, and normalized to wild-type. Error bars represent the 95% CI, * P < 0.05 vs. WT.
Mutations in the N-terminal cap of rhodopsin (V20G, P23H, and Q28H) have been functionally characterized in detail, and are known to poorly activate transducin in response to light (Opefi et al. 2013). Similarly, mutations M39R, N55K, G106W, C110Y, G114D, and P171L have each been shown to disrupt or prevent productive formation of a opsin-retinal complex (Davies et al. 2012; Ramon et al. 2014; Sung et al. 1991, 1993; Hwa et al. 1999; Andrés et al. 2003), which matched our observation that light-activated signal transduction was significantly impaired in yeast.
Mutations T4K, N15S, and T17M prevent glycosylation at residues N2 and N15, resulting in a severe reduction in rhodopsin stability (Kaushal et al. 1994; Opefi et al. 2013). The NXS/T glycosylation consensus sequence is recognized across eukaryotes (Lam et al. 2013), and a previous study indicated that yeast-expressed bovine rhodopsin was glycosylated (Mollaaghababa et al. 1996). The reduced stability of unglycosylated rhodopsin is known to prevent productive transducin activation in vitro (Opefi et al. 2013), and comparable reductions in signaling were observed in our yeast assay. A general trend of T4K (40% wild-type activity) being less severe than N15S (4%) and T17M (15%) was observed, similar to previous studies, which indicated glycosylation is more important on N15 than it is on N2 (Kaushal et al. 1994; Tam and Moritz 2009; Opefi et al. 2013).
Of the rhodopsin mutations we studied with impaired signaling, R135G is unique as it does not cause misfolding and it does not prevent the formation of a stable complex with retinal, when using rhodopsin purified from mammalian cells (Sung et al. 1993). R135G mutates the highly conserved E/DRY motif, where R135 is the arginine residue in this motif and is crucial for G protein coupling (Acharya and Karnik 1996; Rovati et al. 2007). In addition, when heterologously expressed in mammalian cells, R135 mutations cause rhodopsin to be hyper-phosphorylated, leading to aggregation with visual arrestin and constitutively undergoing endocytosis (Chuang et al. 2004). With two unique molecular mechanisms contributing to pathogenicity, R135 mutants have been placed in their own “Class 3” category (Chuang et al. 2004; Athanasiou et al. 2018). The observed absence of signaling in yeast was in line with the reported in vitro transducin activation defect using mammalian cell-derived R135G rhodopsin (Min et al. 1993; Acharya and Karnik 1996). However, as our light-activated signal transduction assay could not distinguish between disrupted G protein coupling vs. aberrant endocytosis, this assay alone could not determine the molecular mechanism behind the lack of R135G signaling in yeast.
Pathogenic mutations that enhance or do not disrupt transducin activation respond similarly in yeast
Based on the wild-type-like activity of G51A we observed in yeast, and the reduced or inactive response of misfolded and unstable mutants, we hypothesized that rhodopsin mutations that maintain or increase light-dependent activation in vitro may behave similarly in yeast. Constitutively active rhodopsin mutants are also associated with disease, causing congenital stationary night blindness (CSNB) and RP (Park 2014). We specifically selected a panel of rhodopsin mutations to characterize in yeast where activity was known to vary greatly, from asymptomatic, to constitutively active, to increased signaling in the dark.
Across this diversity of function, yeast-expressed rhodopsin again behaved comparably to rhodopsin purified from mammalian cells, with both wild-type-like signals and increased signaling observed depending on the mutation (Figure 4B). We grouped mutations known to increase downstream transducin activation in vitro, although this increased activity can occur in the light, dark, or in both states (Park 2014). The M44T mutation showed a significantly higher response than wild-type, at over 1.6-fold wild type, which matches in vitro transducin activation data for M44T (Andrés et al. 2003). T94I trended higher than wild type, and is also believed to cause CSNB due to constitutive activation (119% WT signaling in vitro) (Gross et al. 2003), but the increase we observed vs. wild type (115% WT signaling in yeast) was not statistically significant. V137M has been reported to activate transducin 1.25-fold greater than wild type (Andrés et al. 2003), but we did not observe an increase in rhodopsin activity. The V137M mutation is known to have highly variable clinical phenotypes (Ayuso et al. 1996), and it has been suggested to be an asymptomatic variant (Rakoczy et al. 2011).
Of the rhodopsin mutations with known increased activity that we studied, S186W is unique as it is believed to cause autosomal dominant RP due to increased signaling in the dark (Liu et al. 2013). This is a result of reduced thermal stability of the inactive dark state, where spontaneous thermal isomerization of the chromophore leads to signaling in the dark (Liu et al. 2013), a phenomenon called “dark noise” (Luo et al. 2011). The increased signaling that we observed in the dark for S186W, equivalent to 30% of activated wild-type rhodopsin, fit with this proposed mechanism of RP pathogenesis. E150K was also found to have significantly elevated signaling in the dark, when expressed in yeast. This data is in line with a mouse model of E150K, which was found to have elevated photoreceptor signaling in the dark, also believed to be due to reduced thermal stability of the dark state (Zhang et al. 2013). The E150K mutation is associated with autosomal recessive RP (arRP) and was previously shown to have a 1.3-fold increased activation of transducin after light exposure (Zhang et al. 2013). An identical value was observed using our yeast-based assay but was not statistically significant.
To determine if dark state signaling in yeast was retinal-dependent, we investigated signaling without the addition of retinal for selected mutants (Figure S6). Elevated dark state signaling appeared to be retinal-dependent for only M44T, L125R, E150K, and S186W. This fits with the proposed mechanism of thermal isomerization of retinal contributing to dark noise for the E150K and S186 mutants (Liu et al. 2013; Zhang et al. 2013), while providing new insight on the M44T and L125R mutants.
Some rhodopsin mutations may cause disease by preventing the formation of rhodopsin homodimers (Ploier et al. 2016), but their pathogenicity is debated due to their relatively high frequency in sequenced human genomes (>1:80,000) (Athanasiou et al. 2018). The F45L and V209M mutations were found to activate the mating pathway at wild-type levels when exposed to light, matching published in vitro transducin activation assays (Ploier et al. 2016). F220L was found to have a 1.5-fold greater response, which was unexpectedly higher than the wild-type-like value reported for the F220C mutation (Ploier et al. 2016). Similarly, although V104I is considered asymptomatic, light activation of the mating pathway was ∼1.3-fold greater than wild-type. This mutation does not segregate with RP in genetic studies (Macke et al. 1993), but transducin activation assays have not previously been performed, so it is unclear how our results in yeast relate to a potential human phenotype.
Mutations in the C-terminus of rhodopsin disrupt trafficking to the rod outer segment (ROS), but do not affect trafficking in other mammalian cell types and do not affect transducin activation in vitro (Sung et al. 1994), therefore we did not expect their function to differ from wild-type rhodopsin in yeast. Interestingly, V345M significantly affected light-activated signaling in yeast, despite the mutation occurring in the C-terminus, which is not believed to be required for G protein activation. A study of transducin activation using V345M rhodopsin purified from mammalian cells has not been performed to compare to our yeast results.
There were two examples where light-activated signaling in yeast differed from reported in vitro results using rhodopsin purified from mammalian cells. The A292E mutation is associated with CSNB, and has constitutive activation in vitro in the absence of retinal, but reduced light-dependent transducin activation when retinal is supplied (Gross et al. 2003). In our yeast signaling assay, a 2.1-fold increase in light-activated signaling vs. wild type was observed, greater than any other mutation we studied. A292E signaling in the dark with retinal added was not significantly increased, and was equivalent to not adding retinal. G90D, another constitutively active mutant associated with CSNB (Rao et al. 1994), activated the mating pathway at only 44% of wild-type response. This result was similar to one transducin activation study using G90D (59% of wild type) (Zvyaga et al. 1996), while others have shown wild-type-like responses to light but constitutive activation in the absence of retinal, similar to A292E (Rao et al. 1994; Gross et al. 2003).
Light-activated signal transduction in yeast correlates with published assays of rhodopsin function
Of the 33 mutations and controls studied, 23 had previously reported measurements of light-dependent activation of transducin in vitro, or measurements of photoreceptor activity by ERG (Table S2). When plotted together, our yeast-based measurements closely matched this available data on rhodopsin signaling, approaching a 1:1 ratio (Figure S7). This held true for a diverse array of phenotypes, ranging from pathogenic due to inactivation, or pathogenic due to constitutive activation, to asymptomatic. A292E was found to be an outlier from this trend. The rate of light-dependent transducin activation has been reported at ∼80% of wild type, although this mutant has constitutive activation in the absence of retinal in vitro (Gross et al. 2003). As signaling in yeast occurs in a cellular context, vs. traditional in vitro assays that use purified protein, the increase in light-activated signaling we observed for A292E may have been a combination of signaling both with and without retinal bound, or may represent a unique signaling state in yeast. Overall, however, the general trend strongly supports the use of yeast to quantify rhodopsin-mediated G protein signaling in response to light, as the yeast-based assay was comparable to more laborious assays for characterizing patient derived mutations.
Subcellular localization of rhodopsin is comparable between yeast and mammalian cells
As the majority of disease-linked rhodopsin mutations cause the receptor to misfold (Athanasiou et al. 2018), comparing the subcellular localization of rhodopsin using mammalian cells is a common technique to study protein trafficking and ER retention, and is predictive of pathogenicity (Sung et al. 1991; Behnen et al. 2018). To establish phenotypes in mammalian cells to compare to, we again used the rhodopsin mutations P23H, M39R, and G51A to create a gradient of phenotypic severity. Rhodopsin mutants were expressed in SK-N-SH neuroblastoma cells with a C-terminal GFP tag that has previously been shown to not affect rhodopsin stability in vitro or in vivo (Moritz et al. 2001)
The P23H mutation has been characterized in a number of cell models, consistently showing poor plasma membrane expression regardless of cell type (Sung et al. 1991; Chiang et al. 2012b, 2015). When expressed in SK-N-SH neuroblastoma cells, immunocytochemistry revealed that P23H rhodopsin did not localize to the plasma membrane, forming aggregates in the cytosol and colocalized with an ER-specific marker (Figure 5A and Figure S8). M39R rhodopsin displayed a nearly wild-type phenotype, colocalizing with a plasma membrane marker but with some evidence of mutant rhodopsin retained within the cell (Figure S8), similar to a previous study (Davies et al. 2012). G51A displayed robust wild-type-like localization on the plasma membrane, consistent with a recent report (Behnen et al. 2018). This comparative range of subcellular localization matched what we had previously observed in assays of rhodopsin function for these three mutations.
Figure 5.
Representative subcellular localization of rhodopsin mutants (A) Comparative subcellular localization of rhodopsin mutants in SK-N-SH and yeast cells. PM merge: Fluorescence of a plasma membrane marker, merged with GFP-tagged rhodopsin. Images represent maximal projections, with scale bars representing 30 μm and 5 μm respectively. (B) Quantified localization of GFP-tagged rhodopsin mutants on the yeast plasma membrane. Yeast cells with patchy membrane expression of rhodopsin had at least 10% of their cell membrane containing separated patches of rhodopsin. Error bars represent the bootstrapped SE.
We next sought to determine if the same rhodopsin mutants expressed in yeast trafficked to the plasma membrane in a similar manner. The same yeast strain used to functionally couple human rhodopsin to the mating pathway was used to express select rhodopsin mutants with GFP fused to the C-terminus. Similar techniques have been used to investigate productive expression of other human GPCRs in yeast, by observing expression on the plasma membrane or the presence of aggregates (O’Malley et al. 2009). Mirroring our mammalian cell-based observations, P23H and M39R were poorly distributed across the yeast plasma membrane, with highly localized “patchy” expression, while G51A localized consistently to the membrane (Figure 5A). The peri-nuclear ring observed is similar to the localization of other human GPCRs expressed in yeast, indicating the ER membrane (O’Malley et al. 2009; Hashi et al. 2018). Although P23H was observed to form aggregates in the ER of our mammalian cells, this was not observed in yeast; however, M39R did appear to form aggregates in the cytosol of yeast.
We expanded this microscopy-based analysis to additional rhodopsin mutants expressed in yeast (Figure S9). Image analysis software was used to quantify rhodopsin distribution on the plasma membrane of each yeast cell, identifying localized regions or “patches” where rhodopsin appeared to aggregate (Figure 5B). Wild-type rhodopsin was observed to have a low number of cells displaying a “patchy” phenotype, suggesting even distribution across the plasma membrane. Some mutants associated with misfolding or reduced stability (i.e., N15S, M39R, L125R) exhibited incomplete distribution on the plasma membrane, characterized by a “patchy” phenotype. Mutations in the C-terminus of rhodopsin (Q344ter, V345M, P347L) disrupt the VXPX motif, which is crucial for trafficking rhodopsin in photoreceptors (Wang and Deretic 2014). However, mutations within this motif do not affect rhodopsin localization in mammalian cells that are not photoreceptors (Sung et al. 1991, 1993), and were not found to affect rhodopsin localization in yeast. In general, these findings indicated that rhodopsin maintains its subcellular localization when expressed in yeast, which is likely dependent on rhodopsin folding and stability, just as it is with heterologous expression in mammalian cells.
The yeast unfolded protein response is upregulated by misfolded rhodopsin mutants
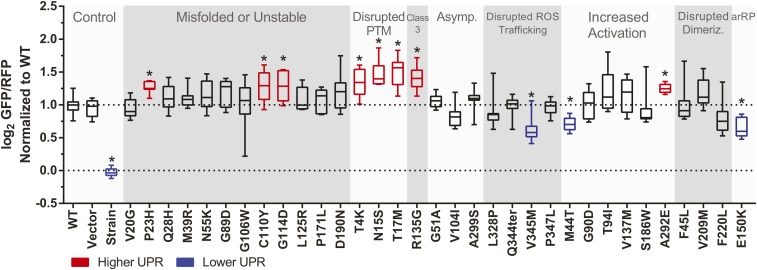
After discovering that the subcellular localization of rhodopsin and changes in responses to light were preserved in yeast, we investigated additional molecular pathways known to be affected by pathogenic rhodopsin mutations. Rhodopsin mutations P23H and T17M have been shown to activate the UPR in mammalian systems, indicative of severe misfolding (Lin et al. 2007; Kunte et al. 2012). P23H has been shown to preferentially activate the IRE1 UPR pathway in mammalian cells (Chiang et al. 2015)—a pathway also present in yeast. Similar to mammalian IRE1, yeast IRE1 serves as a sensor of misfolded protein in the ER, which activates the transcription factor HAC1 (Kimata and Kohno 2011). Yeast strains and plasmids have been devised utilizing a HAC1-responsive promoter to express reporter genes, which have been used to predict productive expression of other human GPCRs in yeast, where greater UPR activation was associated with GPCR misfolding and aggregation (O’Malley et al. 2009). Due to the conserved pathway, and knowing results established with other GPCRs, we hypothesized that the severity of misfolded rhodopsin could be quantified using a yeast-based sensor of UPR upregulation. The strain designed by Jonikas et al. (2009) has an additional gene cassette constitutively expressing RFP, which can be used to correct for changes in global protein expression (Jonikas et al. 2009). We used this strain to study the 33 selected rhodopsin mutations, plus controls, to quantify their effect on UPR upregulation.
Expressed in the UPR-reporter strain, P23H and T17M recapitulated the expected elevated UPR activation in yeast, in addition to several other mutations known to disrupt rhodopsin stability (Figure 6). T4K and N15S, which like T17M disrupt glycosylation, also upregulated the UPR, suggesting a crucial stabilizing nature of these posttranslational modifications. C110Y prevents the formation of a critical disulfide bond in rhodopsin, severely impacting stability and function (Hwa et al. 1999), and the observed increased UPR in yeast matches in silico predictions that this mutant is highly unstable (Rakoczy et al. 2011). Elsewhere in transmembrane helix three, the G114D and R135G mutations similarly increased the UPR. In mammalian cells, R135G is hyper-phosporylated and aggregates in endosomes (Chuang et al. 2004), but accumulation in the ER or activation of the mammalian UPR has not been reported. That R135G activated the yeast UPR suggests that this mutant may be misfolded in yeast.
Figure 6.
UPR activation in yeast relative to wild-type rhodopsin. GFP expression is a reporter of UPR upregulation, while RFP is expressed constitutively to help correct for changes in global protein expression. WT: Wild-type human rhodopsin; Vector: yeast transformed with a plasmid not containing the rhodopsin gene; Strain: the yJW1200 strain not transformed with plasmid and grown in rich media; arRP: autosomal recessive retinitis pigmentosa. Data points represent results of nine individual colonies, each in a 600 μl culture, normalized to wild-type. Boxes extend from the 25th to 75th percentile, the line across the box represents the median value. Bars represent the min and max recorded values. * P < 0.05 vs. WT.
The constitutively active mutant A292E showed an upregulation in the UPR that has not previously been reported, which may be due to the replacement of a small uncharged residue with a large negatively charged residue proximal to the retinal binding pocket. A reduced UPR for mutations M44T, E150K, and V345M compared to wild type was observed, but the physiological relevance is unclear. Interestingly, culturing in selective media alone was sufficient to upregulate the UPR, revealed by the difference between the vector control and the untransformed strain grown in rich medium. A log2 GFP/RFP ratio of ∼1.0 was previously shown to indicate moderate UPR upregulation (Jonikas et al. 2009), which was observed for the Vector control prior to normalizing the data. Expressing wild-type rhodopsin gave a similar value, which suggested baseline UPR upregulation was due to growth in selective media and not to the overexpression of rhodopsin.
Yeast assays of rhodopsin mutations V81Δ, A164E, and A164V found in retinal disease patients
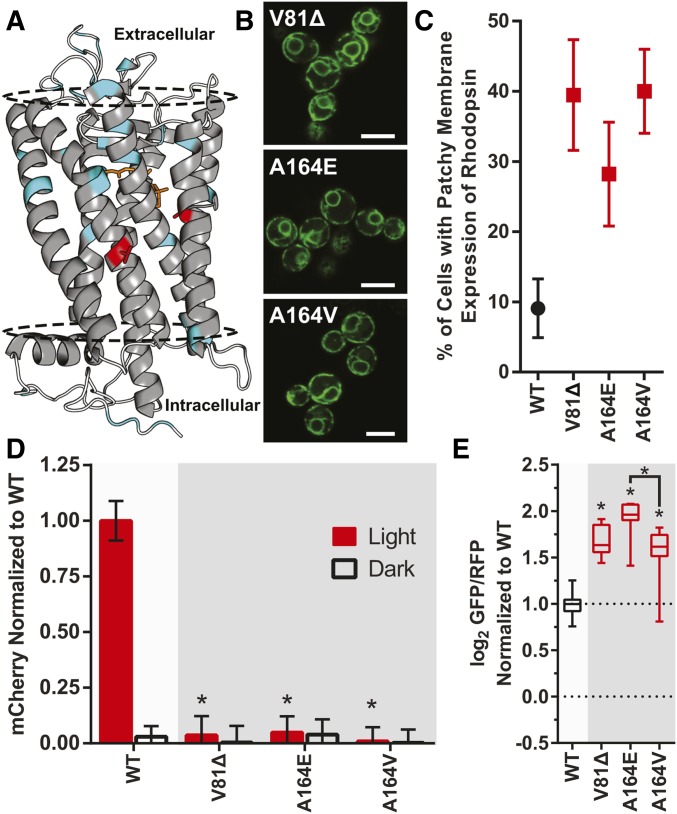
Having established a suite of new yeast-based techniques to investigate rhodopsin functional and molecular phenotypes, we applied these approaches to study three rhodopsin mutations found in degenerative retinal disease patients diagnosed with RP. One of the rhodopsin mutations, V81Δ, is a new mutation that has not been previously reported (Case 1), and two of the other mutations are previously reported but have had little (A164V, Case 2), or no (A164E, Case 3) experimental characterization with respect to those rhodopsin mutations. Missense mutations A164E and A164V have been previously linked to autosomal dominant RP (Fuchs et al. 1994; Hwa et al. 1997), but the molecular mechanism underlying the disruption of rhodopsin function at this site remains to be elucidated. Studies of A164V suggest that helical packing may be an issue (Hwa et al. 1997; Stojanovic et al. 2003), but the effects of introducing a charged residue at this site have not yet been investigated. Mutations at the same residue in rhodopsin can have highly heterogenous phenotypes (Bosch et al. 2003), so we sought to compare Al64E to A164V in greater detail. We identified a new V81Δ mutation in a patient (Case 1) with early-onset autosomal dominant RP (Table S3). This mutation completely removes the V81 codon from the rhodopsin DNA sequence, resulting in a deleted amino acid in the central part of the second transmembrane domain (Figure 7A). Such an amino acid deletion could disrupt alpha helix formation and stability in the membrane, and is likely to lead to a severe molecular phenotype for V81Δ rhodopsin.
Figure 7.
Characterization of novel rhodopsin mutants using yeast. (A) Crystal structure of rhodopsin, highlighting residues V81 and A164 in red. The other rhodopsin mutants characterized in this study are highlighted in cyan, and the approximate location of the membrane is indicated by the dotted line (1U19.pdb). Quantified phenotype and functional assays of V81Δ, A164E, and A164V in yeast (B) Representative subcellular localization, Bar, 5 μm, (C) quantified consistency of rhodopsin localization on the yeast plasma membrane, (D) light-activated signal transduction, and (E) UPR activation. WT: Wild-type human rhodopsin. The number of biological replicates and error bars are identical to previous figures. * P < 0.05 vs. WT in all panels unless otherwise indicated.
We characterized V81Δ, A164E, and A164V using the yeast-based methods for investigating rhodopsin molecular phenotype and function, with experiments conducted in the same manner as previously described (Figure 7, B–E). Plasma membrane localization of all three mutants were poor, with 28–40% of yeast cells displaying patchy expression of rhodopsin. Light-activated signal transduction in yeast was completely abolished for each, similar to the other mutants we studied known to be unstable or misfold. UPR activation was elevated for all three mutants, with A164E higher than any other rhodopsin mutant we studied. Together, our results suggest that all three of these mutants have a severe phenotype in yeast, based on decreased function, subcellular localization, and UPR activation, where we may expect a difference in severity between the A164 mutants based on UPR activation.
Yeast assays comparable to mammalian cell data for rhodopsin mutations V81Δ, A164E, and A164V
We compared our yeast-based assays of V81Δ, A164E, and A164V to expression in mammalian cells. A164V colocalized with the plasma membrane marker, suggesting a less severe phenotype in mammalian cells, which contrasted with the yeast data (Figure 8A). However, A164E and V81Δ were completely retained inside mammalian cells, colocalizing with the ER marker, indicating they were severely misfolded. Following immunoaffinity purification from mammalian cells, the three RP-associated mutations also showed a range in their response to light (Figure 8B). Heterologous expression of V81Δ produced no functional protein, demonstrating a very severe phenotype. A164E, while not as severe a phenotype as V81Δ, expressed poorly and produced limited functional protein. A164V expression did produce a high amount of functional protein, consistent with another in vitro study of this mutation (Stojanovic et al. 2003). The sum of this cellular and functional data suggested the same functional trend we first predicted using yeast-based methods, although there was more evidence of functional A164V protein in mammalian cells.
Figure 8.
Characterization of novel rhodopsin mutants using mammalian cell expression. (A) Comparative subcellular localization of rhodopsin mutants in SK-N-SH cells. PM: Fluorescence of a plasma membrane marker; ER: fluorescence of a ER-specific marker. Bar, 30 μm. (B) Difference spectra of mammalian cell-expressed rhodopsin mutants in response to light. The peak at 500 nm indicates a light-dependent response.
Patient clinical data supports functional trend predicted by yeast and mammalian cells
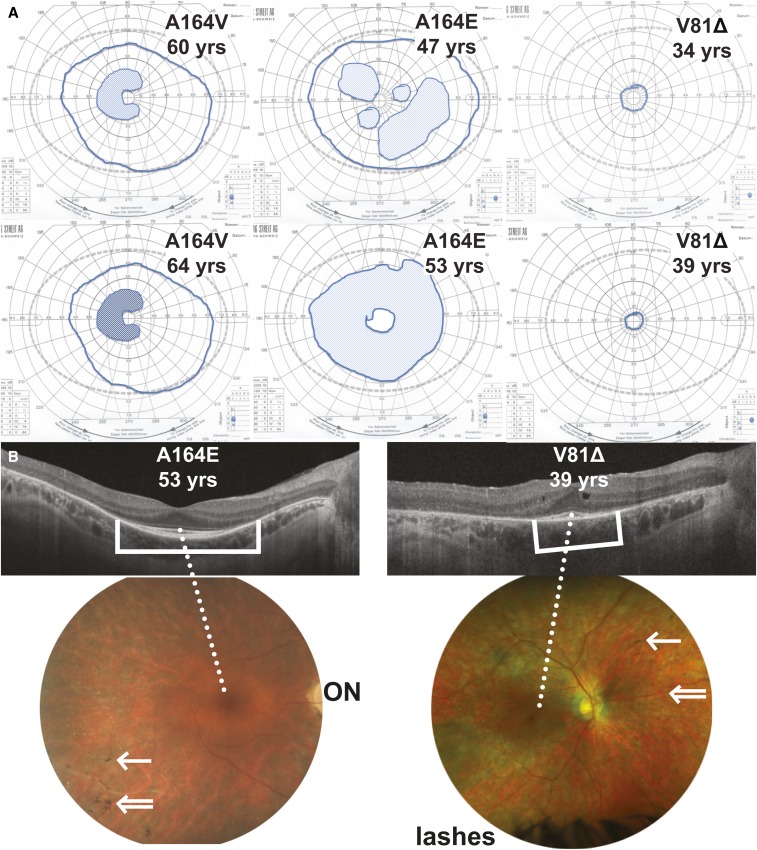
Next, we looked at patient clinical data. The comparative severity in vitro was found to follow the same trend as available patient phenotype information (Table S3). The most severe phenotype was exhibited by the patient with the V81Δ mutation (Case 1). This patient first had symptoms ∼10 years of age, with difficulty adapting to a dim lit environment (nyctalopia). This slowly progressed, and at 39 years she has moderate visual acuity loss (20/50), mildly abnormal color vision, constriction of the visual field to the central 5°. At age 26 years, electroretinography already documented severe reduction of rod and cone function. The phenotype of the patient with the A164V mutation (Case 2) is milder than that of the patient with the A164E mutation. Although Case 2 had symptoms of nyctalopia since childhood, the progression of his disease was extremely slow. At age 64 years his electroretinogram was recordable and only mildly abnormal. His central visual acuity at 67 years was 20/40 and, despite a paracentral scotoma (area of decreased vision), he maintained a peripheral field. In contrast, the patient with the A164E mutation (Case 3) has a good central visual acuity and normal color vision at age 53 years. However, her paracentral scotoma were more severe and progressed to form an annular scotoma at the age of 53 years. Unlike Case 1 (V81Δ), she also preserved some good peripheral field of vision at the age of 45 years, and her ERG was only moderately abnormal.
The V81Δ patient is the youngest of the three with a highly reduced retinal function, while the A164V patient is the eldest of the three with the best retinal function (Figure 9). A164E again appeared intermediate to both. These results show that the overall trend of clinical severity was accurately predicted by combining both sets of yeast-derived and mammalian cell-derived data. The yeast methods provided additional information on UPR upregulation, which also supported the difference in severity between A164E and A164V mutations, while being less labor intensive than mammalian microscopy and expression methods.
Figure 9.
Clinical assessment of patients with rhodopsin mutations V81Δ, A164E, and A164V. (A) Goldmann visual fields of the right eye at two time points. Normal fields would reach the gray dotted line. The solid blue line outlines the actual field. The hatched areas are scotoma, i.e., areas of loss in sensitivity. Darker areas refer to denser scotoma. (B) Structural retinal phenotype of the right eye from cases carrying the A164E and V81Δ mutations. Optical coherence tomography (OCT) above showing the different retinal layers. Brackets show area of preserved outer retina; A164E > V81Δ. Unlike for A164E, the OCT of V81Δ shows disturbed lamination of the retina with degenerative cysts, reflecting more advanced disease. The retinal photograph below centered on the posterior pole. Photograph on the right is taken with a wider field camera. ON: Optic nerve. The dotted white line indicated the foveal area at the center of the macula. Double white arrow indicates vessel attenuation, while single arrow shows typical pigmentary deposits (few in these cases). The width of the central visual field corresponds to the area of preserved outer retina on the OCT.
Discussion
Yeast provide new methods to investigate rhodopsin structure and function
In this study, we have engineered the yeast S. cerevisiae to characterize both known and novel pathogenic mutations of the visual pigment rhodopsin. In comparing our new assays to traditional mammalian cell-based approaches, we demonstrate that the molecular phenotypes of this light-activated human GPCR are similar in yeast, and that these phenotypes reflect patient clinical data. There are a number of advantages and differences when compared to traditional techniques, as these yeast-based rhodopsin assays are performed in a cellular context, which provides a new perspective on signal transduction pathways, subcellular localization, and UPR upregulation.
A direct measurement of downstream pathway activation in response to light was achieved by functionally coupling rhodopsin to the yeast mating pathway. This required productive in vivo activation of a G protein, mimicking the initial step that occurs in human photoreceptors, even if downstream signaling differs. By using yeast, many rhodopsin mutations could be studied in the same experiment, with multiple replicates, without the laborious purification steps that are traditionally required for in vitro transducin activation assays (Reeves et al. 1996). Not only did yeast-expressed rhodopsin maintain its ability to respond to light, but the magnitude of signal pathway activation was comparable to many previous studies of mutations expressed in mammalian systems. We were also able to observe rhodopsin mutations that resulted in the activation of dark state rhodopsin, which has previously required sensitive spectroscopic assays using immunoaffinity purified rhodopsin (Liu et al. 2013), or gene knock-in animal models (Zhang et al. 2013). This included providing new data on L125R signaling in the dark, a mutant poorly expressed in mammalian cells, which has made previous characterization of function challenging (Stojanovic et al. 2003). Mutations at site L125 have been shown to reduce the thermal stability of rhodopsin (Andrés et al. 2001), which could lead to dark noise through thermal isomerization of the bound chromophore (Luo et al. 2011). This mechanism was supported by our finding that L125R dark noise was retinal-dependent in yeast, which could be a result of thermal isomerization. Thus, yeast may serve as a platform for studying difficult-to-express rhodopsin proteins, allowing the rapid quantification of downstream G protein activation under various conditions.
Mutations known to disrupt rhodopsin folding or stability tended to have incomplete localization on the yeast plasma membrane, similar to mammalian-cell-expressed rhodopsin. Although there was significant heterogeneity between the investigated mutants, this observation suggests that the biochemical properties of rhodopsin were conserved in yeast, despite known differences in glycosylation patterns (Mollaaghababa et al. 1996). We quantified membrane localization using a novel automated image analysis procedure, which may be useful for studies of other GPCRs and associated pathogenic mutations when expressed in yeast. However, not all human GPCRs are expressed productively in yeast (O’Malley et al. 2009), so these methods should first be validated with the wild-type receptor.
The majority of disease-linked rhodopsin mutations that have been identified cause the protein to misfold, which can activate the UPR in the ER, and eventually lead to photoreceptor cell death (Athanasiou et al. 2018). Modulating the UPR has been investigated as a potential treatment for RP (Tam et al. 2010; Chiang et al. 2012a; Parfitt et al. 2014), but the effect on UPR upregulation had only been determined for three rhodopsin mutations (Lin et al. 2007; Kunte et al. 2012; Marsili et al. 2015). Determining UPR upregulation by mutant rhodopsin has previously required microscopy, immunoblot, and qPCR methods (Kunte et al. 2012; Marsili et al. 2015). We took advantage of an engineered yeast strain that possesses a reporter linked to the IRE1 UPR pathway, which is the only one of three UPR pathways that is conserved between mammals and yeast (Kimata and Kohno 2011). This yeast-based reporter of UPR activity enables the use of flow cytometry, a more simple and high-throughput method, and offers the advantage of measuring UPR upregulation directly in live cells. We found that upregulation of this pathway was associated with certain rhodopsin mutants known to misfold or with reduced stability, which may be predictive of the molecular mechanism contributing to retinal degeneration.
However, yeast do not contain the PERK and ATF6 UPR pathways found in mammalian cells (Kimata and Kohno 2011). Thus, although the yeast-based methods provide insight into UPR upregulation, these studies would need to be combined with mammalian cells to better understand how all UPR pathways may be affected by rhodopsin and other GPCR mutations. This may also explain why nine of the 15 mutants that are believed to misfold did not cause an increase UPR activation in yeast, suggesting significant heterogeneity between rhodopsin mutations. Determining the contribution a missense mutation makes to UPR upregulation is highly relevant to pharmacogenomics, as an inability of rhodopsin to respond to light may not necessarily indicate that the pathogenic mutation can be rescued by UPR modulation.
Overall, many known rhodopsin phenotypes were recapitulated in yeast, a requirement for any assay of human gene function seeking to determine the clinical relevance of patient derived missense mutations (Amendola et al. 2016). There are also important differences to consider when comparing our yeast-based assays to mammalian cell-based assays. Importantly, when a lack of signaling is observed in yeast, it is difficult to separate rhodopsin mutants that misfold from mutants that fold properly but do not productively activate the downstream pathway. Although our quantified microscopy data supported the notion that rhodopsin mutants with inconsistent plasma membrane expression in yeast are also mutants known to be unstable or misfolded, unique aspects of yeast cellular machinery or post-translational processing may influence these mutants. Results from the R135G mutation highlight these differences, where this mutant activated the UPR in yeast but does not aggregate in the ER of mammalian cells (Chuang et al. 2004).
The signaling phenotypes of the G90D and A292E mutants in yeast also differed from the reported constitutive activity in vitro when using purified protein (Gross et al. 2003). That A292E signaled higher and G90D lower than expected suggests that constitutive activity in yeast is dependent on cellular conditions that would not be revealed in an in vitro assay using purified protein, such as a renewing supply of both rhodopsin and G protein. It is interesting to note, however, that A292E has the highest constitutive activity reported of a CSNB-associated mutation in vitro (Gross et al. 2003), which fits the trend observed in yeast. That light-dependent signaling in yeast was affected by the V104I, F220L, and V345M mutations was unexpected, which provides interesting new data for these previously uncharacterized mutants that should be followed up in mammalian cell-based assays. Thus, these yeast-based methods provide complimentary but independent data to traditional mammalian-cell and in vitro biochemical techniques, offering a unique perspective on rhodopsin structure and function in a cellular context.
Characterizing novel rhodopsin mutations with yeast
Determining mutant function rapidly and accurately has become increasingly important with the rise of whole genome sequencing, and the ever-expanding rise in gene mutations with an unknown impact on human health. Rhodopsin mutations linked to inherited retinal disease have been used as examples of how many gene mutations discovered in patients are rarely characterized, and that the molecular basis for pathology is poorly understood (Davies 2014; Chiang and Gorin 2016). Animal models and traditional in vitro assays provide detailed information, but they have not kept pace with the hundreds (>350) of rhodopsin mutations identified to date. This is true of many other genetic diseases, but new methods of functional characterization are helping to address this (Starita et al. 2017), including using yeast-based assays (Sun et al. 2016; Yang et al. 2017).
We investigated the use of yeast to characterize novel and understudied pathogenic mutations, and compared to clinical data for patients with varying severity of RP. Our yeast-based approaches predicted severe phenotypes for the V81Δ and A164 mutations, which included determinations of their light-activation, subcellular localization, and UPR upregulation. By combining yeast and mammalian cell-based assays, the relative severity of these mutations was revealed, as compared with clinical phenotypes measuring decline in visual function in patients. The V81Δ rhodopsin mutation showed the most severe phenotype in our combined assays and the most extensive visual deterioration clinically, in contrast to A164V, which had the mildest phenotype both clinically and experimentally, and A164E, which was found to be intermediate.
The difference in severity for the two missense mutations found at site 164 highlights the heterogenic nature of RP, and the importance of characterizing individual disease phenotypes. These results indicate that yeast-based approaches could be useful not only for investigating the molecular basis of retinal disease, but also for better prediction of mutation pathogenicity, to help improve the accuracy of prognoses for patients associated with specific mutations in rhodopsin. Integrating the results of both UPR and light-activation assays (Figure S10) may also help determine the molecular mechanism of disease, to differentiate between mutations that cause severe misfolding, vs. disruptions in retinal binding or stability that prevent activation but do not cause cell stress.
Recent successes in gene therapy for inherited retinal disease means that mutation classification is of utmost importance, to determine if such therapy is required (U.S. Food and Drug Administration 2017). This issue is particularly important to address for degenerative diseases, such as inherited retinal disease, where early intervention is crucial. A method to determine the functional consequences of mutations throughout rhodopsin, rapidly and accurately, would therefore be highly beneficial.
The methods presented here could also extend to functionally characterizing mutations of many other GPCRs. Indeed, although over 30 human GPCRs have been functionally linked to the yeast mating pathway, no previous yeast-based study has focused on direct functional characterization of human GPCR mutations. As discussed in a recent review of GPCR pharmacogenomics, characterizing GPCR mutations could lead to a better understanding of disease and drug responses in patients (Hauser et al. 2018). Missense mutations that modulate interactions with downstream signaling and regulatory proteins are known to play a role in this, so assays that accurately reflect GPCR function and interactions in a cellular context, such as the yeast assays presented here, will be key to understanding the impact of GPCR mutations on human health.
Acknowledgments
This study was supported by NSERC Discovery Grants to B.S.W.C. (RGPIN-2015-06279), and S.P (RGPIN-2015-05114), and a Canada Foundation for Innovation Grant to S.P. (#34473).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7414109.
Communicating editor: M. Schuldiner
Literature Cited
- Acharya S., Karnik S. S., 1996. Modulation of GDP release from transducin by the conserved Glu134-Arg135 sequence in rhodopsin. J. Biol. Chem. 271: 25406–25411. 10.1074/jbc.271.41.25406 [DOI] [PubMed] [Google Scholar]
- Amendola L. M., Jarvik G. P., Leo M. C., McLaughlin H. M., Akkari Y., et al. , 2016. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am. J. Hum. Genet. 99: 247 10.1016/j.ajhg.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés A., Kosoy A., Garriga P., Manyosa J., 2001. Mutations at position 125 in transmembrane helix III of rhodopsin affect the structure and signalling of the receptor. Eur. J. Biochem. 268: 5696–5704. 10.1046/j.0014-2956.2001.02509.x [DOI] [PubMed] [Google Scholar]
- Andrés A., Garriga P., Manyosa J., 2003. Altered functionality in rhodopsin point mutants associated with retinitis pigmentosa. Biochem. Biophys. Res. Commun. 303: 294–301. 10.1016/S0006-291X(03)00328-0 [DOI] [PubMed] [Google Scholar]
- Armbruster B. N., Li X., Pausch M. H., Herlitze S., Roth B. L., 2007. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. USA 104: 5163–5168. 10.1073/pnas.0700293104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D., Aguila M., Opefi C. A., South K., Bellingham J., et al. , 2017. Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration. Hum. Mol. Genet. 26: 305–319. 10.1093/hmg/ddw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D., Aguila M., Bellingham J., Li W., McCulley C., et al. , 2018. The molecular and cellular basis of rhodopsin retinitis pigmentosa reveals potential strategies for therapy. Prog. Retin. Eye Res. 62: 1–23. 10.1016/j.preteyeres.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayuso C., Trujillo M. J., Robledo M., Ramos C., Benitez J., et al. , 1996. Novel rhodopsin mutation in an autosomal dominant retinitis pigmentosa family: phenotypic variation in both heterozygote and homozygote Val137Met mutant patients. Hum. Genet. 98: 51–54. 10.1007/s004390050158 [DOI] [PubMed] [Google Scholar]
- Behnen P., Felline A., Comitato A., Di Salvo M. T., Raimondi F., et al. , 2018. A small chaperone improves folding and routing of rhodopsin mutants linked to inherited blindness. iScience 4: 1–19. 10.1016/j.isci.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya N., Darren B., Schott R. K., Tropepe V., Chang B. S. W., 2017. Cone-like rhodopsin expressed in the all-cone retina of the colubrid pine snake as a potential adaptation to diurnality. J. Exp. Biol. 220: 2418–2425. 10.1242/jeb.156430 [DOI] [PubMed] [Google Scholar]
- Bosch L., Ramon E., Del Valle L. J., Garriga P., 2003. Structural and functional role of helices I and II in rhodopsin. A novel interplay evidenced by mutations at Gly-51 and Gly-89 in the transmembrane domain. J. Biol. Chem. 278: 20203–20209. 10.1074/jbc.M301319200 [DOI] [PubMed] [Google Scholar]
- Brown A. J., Dyos S. L., Whiteway M. S., White J. H., Watson M. A., et al. , 2000. Functional coupling of mammalian receptors to the yeast mating pathway using novel yeast/mammalian G protein alpha-subunit chimeras. Yeast 16: 11–22. [DOI] [PubMed] [Google Scholar]
- Campbell R. M., Cartwright C., Chen W., Chen Y., Duzic E., et al. , 1999. Selective A1-adenosine receptor antagonists identified using yeast Saccharomyces cerevisiae functional assays. Bioorg. Med. Chem. Lett. 9: 2413–2418. 10.1016/S0960-894X(99)00398-4 [DOI] [PubMed] [Google Scholar]
- Chen Y., Jastrzebska B., Cao P., Zhang J., Wang B., et al. , 2014. Inherent instability of the retinitis pigmentosa P23H mutant opsin. J. Biol. Chem. 289: 9288–9303. 10.1074/jbc.M114.551713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Song L. S., Shirokova N., Gonzalez A., Lakatta E. G., et al. , 1999. Amplitude distribution of calcium sparks in confocal images: theory and studies with an automatic detection method. Biophys. J. 76: 606–617. 10.1016/S0006-3495(99)77229-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang J., Gorin M. B., 2016. Challenges confronting precision medicine in the context of inherited retinal disorders. Expert Rev. Precis. Med. Drug Dev. 1: 195–205. 10.1080/23808993.2016.1152159 [DOI] [Google Scholar]
- Chiang W. C., Hiramatsu N., Messah C., Kroeger H., Lin J. H., 2012a. Selective activation of ATF6 and PERK endoplasmic reticulum stress signaling pathways prevent mutant rhodopsin accumulation. Invest. Ophthalmol. Vis. Sci. 53: 7159–7166. 10.1167/iovs.12-10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang W. C., Messah C., Lin J. H., 2012b. IRE1 directs proteasomal and lysosomal degradation of misfolded rhodopsin. Mol. Biol. Cell 23: 758–770. 10.1091/mbc.e11-08-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang W. C., Kroeger H., Sakami S., Messah C., Yasumura D., et al. , 2015. Robust endoplasmic reticulum-associated degradation of rhodopsin precedes retinal degeneration. Mol. Neurobiol. 52: 679–695. 10.1007/s12035-014-8881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J. Z., Vega C., Jun W., Sung C. H., 2004. Structural and functional impairment of endocytic pathways by retinitis pigmentosa mutant rhodopsin-arrestin complexes. J. Clin. Invest. 114: 131–140. 10.1172/JCI200421136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cideciyan A. V., Hood D. C., Huang Y., Banin E., Li Z. Y., et al. , 1998. Disease sequence from mutant rhodopsin allele to rod and cone photoreceptor degeneration in man. Proc. Natl. Acad. Sci. USA 95: 7103–7108. 10.1073/pnas.95.12.7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies W. I., 2014. Challenges using diagnostic next-generation sequencing in the clinical environment for inherited retinal disorders. Per. Med. 11: 99–111. 10.2217/pme.13.95 [DOI] [PubMed] [Google Scholar]
- Davies W. I., Downes S. M., Fu J. K., Shanks M. E., Copley R. R., et al. , 2012. Next-generation sequencing in health-care delivery: lessons from the functional analysis of rhodopsin. Genet. Med. 14: 891–899. 10.1038/gim.2012.73 [DOI] [PubMed] [Google Scholar]
- Dryja T. P., McGee T. L., Hahn L. B., Cowley G. S., Olsson J. E., et al. , 1990. Mutations within the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. N. Engl. J. Med. 323: 1302–1307. 10.1056/NEJM199011083231903 [DOI] [PubMed] [Google Scholar]
- Erlenbach I., Kostenis E., Schmidt C., Serradeil-Le Gal C., Raufaste D., et al. , 2001. Single amino acid substitutions and deletions that alter the G protein coupling properties of the V2 vasopressin receptor identified in yeast by receptor random mutagenesis. J. Biol. Chem. 276: 29382–29392. 10.1074/jbc.M103203200 [DOI] [PubMed] [Google Scholar]
- Fahim, A. T., S. P. Daiger, and R. G. Weleber, 2000 Nonsyndromic Retinitis Pigmentosa Overview. Available at: https://www.ncbi.nlm.nih.gov/books/NBK1417/. Accessed: March 20, 2017.
- Fishman G. A., Vandenburgh K., Stone E. M., Gilbert L. D., Alexander K. R., et al. , 1992. Ocular findings associated with rhodopsin gene codon 267 and codon 190 mutations in dominant retinitis pigmentosa. Arch. Ophthalmol. 110: 1582–1588. 10.1001/archopht.1992.01080230082026 [DOI] [PubMed] [Google Scholar]
- Fuchs S., Kranich H., Denton M. J., Zrenner E., Bhattacharya S. S., et al. , 1994. Three novel rhodopsin mutations (C110F, L131P, A164V) in patients with autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 3: 1203 10.1093/hmg/3.7.1203 [DOI] [PubMed] [Google Scholar]
- Gross A. K., Rao V. R., Oprian D. D., 2003. Characterization of rhodopsin congenital night blindness mutant T94I. Biochemistry 42: 2009–2015. 10.1021/bi020613j [DOI] [PubMed] [Google Scholar]
- Han M., Smith S. O., Sakmar T. P., 1998. Constitutive activation of opsin by mutation of methionine 257 on transmembrane helix 6. Biochemistry 37: 8253–8261. 10.1021/bi980147r [DOI] [PubMed] [Google Scholar]
- Hashi H., Nakamura Y., Ishii J., Kondo A., 2018. Modifying expression modes of human neurotensin receptor type 1 alters sensing capabilities for agonists in yeast signaling biosensor. Biotechnol. J. 13: e1700522 10.1002/biot.201700522 [DOI] [PubMed] [Google Scholar]
- Hauser A. S., Chavali S., Masuho I., Jahn L. J., Martemyanov K. A., et al. , 2018. Pharmacogenomics of GPCR drug targets. Cell 172: 41–54.e19. 10.1016/j.cell.2017.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng B. C., Aubel D., Fussenegger M., 2013. An overview of the diverse roles of G-protein coupled receptors (GPCRs) in the pathophysiology of various human diseases. Biotechnol. Adv. 31: 1676–1694. 10.1016/j.biotechadv.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Horswill J. G., Bali U., Shaaban S., Keily J. F., Jeevaratnam P., et al. , 2007. PSNCBAM-1, a novel allosteric antagonist at cannabinoid CB1 receptors with hypophagic effects in rats. Br. J. Pharmacol. 152: 805–814. 10.1038/sj.bjp.0707347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwa J., Garriga P., Liu X., Khorana H. G., 1997. Structure and function in rhodopsin: packing of the helices in the transmembrane domain and folding to a tertiary structure in the intradiscal domain are coupled. Proc. Natl. Acad. Sci. USA 94: 10571–10576. 10.1073/pnas.94.20.10571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwa J., Reeves P. J., Klein-Seetharaman J., Davidson F., Khorana H. G., 1999. Structure and function in rhodopsin: further elucidation of the role of the intradiscal cysteines, Cys-110, -185, and -187, in rhodopsin folding and function. Proc. Natl. Acad. Sci. USA 96: 1932–1935. 10.1073/pnas.96.5.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii J., Tanaka T., Matsumura S., Tatematsu K., Kuroda S., et al. , 2008. Yeast-based fluorescence reporter assay of G protein-coupled receptor signalling for flow cytometric screening: FAR1-disruption recovers loss of episomal plasmid caused by signalling in yeast. J. Biochem. 143: 667–674. 10.1093/jb/mvn018 [DOI] [PubMed] [Google Scholar]
- Jonikas M. C., Collins S. R., Denic V., Oh E., Quan E. M., et al. , 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323: 1693–1697. 10.1126/science.1167983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal S., Khorana H. G., 1994. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry 33: 6121–6128. 10.1021/bi00186a011 [DOI] [PubMed] [Google Scholar]
- Kaushal S., Ridge K. D., Khorana H. G., 1994. Structure and function in rhodopsin: the role of asparagine-linked glycosylation. Proc. Natl. Acad. Sci. USA 91: 4024–4028. 10.1073/pnas.91.9.4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler-Ross S., Noffz C., Dean N., 2008. A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genetics 179: 705–710. 10.1534/genetics.108.087080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y., Kohno K., 2011. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr. Opin. Cell Biol. 23: 135–142. 10.1016/j.ceb.2010.10.008 [DOI] [PubMed] [Google Scholar]
- Kompella P. S., Moses A. M., Peisajovich S. G., 2017. Introduction of premature stop codons as an evolutionary strategy to rescue signaling network function. ACS Synth. Biol. 6: 446–454. 10.1021/acssynbio.6b00142 [DOI] [PubMed] [Google Scholar]
- Krebs M. P., Holden D. C., Joshi P., Clark C. L., III, Lee A. H., et al. , 2010. Molecular mechanisms of rhodopsin retinitis pigmentosa and the efficacy of pharmacological rescue. J. Mol. Biol. 395: 1063–1078. 10.1016/j.jmb.2009.11.015 [DOI] [PubMed] [Google Scholar]
- Kunte M. M., Choudhury S., Manheim J. F., Shinde V. M., Miura M., et al. , 2012. ER stress is involved in T17M rhodopsin-induced retinal degeneration. Invest. Ophthalmol. Vis. Sci. 53: 3792–3800. 10.1167/iovs.11-9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P. V., Goldman R., Karagiannis K., Narsule T., Simonyan V., et al. , 2013. Structure-based comparative analysis and prediction of N-linked glycosylation sites in evolutionarily distant eukaryotes. Genomics Proteomics Bioinformatics 11: 96–104. 10.1016/j.gpb.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent J. M., Young J. H., Kachroo A. H., Marcotte E. M., 2016. Efforts to make and apply humanized yeast. Brief. Funct. Genomics 15: 155–163. 10.1093/bfgp/elv041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVail M. M., Nishikawa S., Steinberg R. H., Naash M. I., Duncan J. L., et al. , 2018. Phenotypic characterization of P23H and S334ter rhodopsin transgenic rat models of inherited retinal degeneration. Exp. Eye Res. 167: 56–90. 10.1016/j.exer.2017.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., et al. , 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. H., Li H., Yasumura D., Cohen H. R., Zhang C., et al. , 2007. IRE1 signaling affects cell fate during the unfolded protein response. Science 318: 944–949. 10.1126/science.1146361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. Y., Liu J., Mehrotra D., Liu Y., Guo Y., et al. , 2013. Thermal stability of rhodopsin and progression of retinitis pigmentosa: comparison of S186W and D190N rhodopsin mutants. J. Biol. Chem. 288: 17698–17712. 10.1074/jbc.M112.397257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Nahon D., le Roy B., Lenselink E. B., IJzerman A. P., 2015. Scanning mutagenesis in a yeast system delineates the role of the NPxxY(x)(5,6)F motif and helix 8 of the adenosine A(2B) receptor in G protein coupling. Biochem. Pharmacol. 95: 290–300. 10.1016/j.bcp.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Liu R., Wong W., IJzerman A. P., 2016. Human G protein-coupled receptor studies in Saccharomyces cerevisiae. Biochem. Pharmacol. 114: 103–115. 10.1016/j.bcp.2016.02.010 [DOI] [PubMed] [Google Scholar]
- Luo D. G., Yue W. W., Ala-Laurila P., Yau K. W., 2011. Activation of visual pigments by light and heat. Science 332: 1307–1312. 10.1126/science.1200172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke J. P., Davenport C. M., Jacobson S. G., Hennessey J. C., Gonzalez-Fernandez F., et al. , 1993. Identification of novel rhodopsin mutations responsible for retinitis pigmentosa: implications for the structure and function of rhodopsin. Am. J. Hum. Genet. 53: 80–89. [PMC free article] [PubMed] [Google Scholar]
- Maeda S., Sun D., Singhal A., Foggetta M., Schmid G., et al. , 2014. Crystallization scale preparation of a stable GPCR signaling complex between constitutively active rhodopsin and G-protein. PLoS One 9: e98714 10.1371/journal.pone.0098714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsili S., Genini S., Sudharsan R., Gingrich J., Aguirre G. D., et al. , 2015. Exclusion of the unfolded protein response in light-induced retinal degeneration in the canine T4R RHO model of autosomal dominant retinitis pigmentosa. PLoS One 10: e0115723 10.1371/journal.pone.0115723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes H. F., van der Spuy J., Chapple J. P., Cheetham M. E., 2005. Mechanisms of cell death in rhodopsin retinitis pigmentosa: implications for therapy. Trends Mol. Med. 11: 177–185. 10.1016/j.molmed.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Min K. C., Zvyaga T. A., Cypess A. M., Sakmar T. P., 1993. Characterization of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Mutations on the cytoplasmic surface affect transducin activation. J. Biol. Chem. 268: 9400–9404. [PubMed] [Google Scholar]
- Molday R. S., MacKenzie D., 1983. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry 22: 653–660. 10.1021/bi00272a020 [DOI] [PubMed] [Google Scholar]
- Mollaaghababa R., Davidson F. F., Kaiser C., Khorana H. G., 1996. Structure and function in rhodopsin: expression of functional mammalian opsin in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93: 11482–11486. 10.1073/pnas.93.21.11482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz O. L., Tam B. M., Papermaster D. S., Nakayama T., 2001. A functional rhodopsin-green fluorescent protein fusion protein localizes correctly in transgenic Xenopus laevis retinal rods and is expressed in a time-dependent pattern. J. Biol. Chem. 276: 28242–28251. 10.1074/jbc.M101476200 [DOI] [PubMed] [Google Scholar]
- Morrow J. M., Castiglione G. M., Dungan S. Z., Tang P. L., Bhattacharyya N., et al. , 2017. An experimental comparison of human and bovine rhodopsin provides insight into the molecular basis of retinal disease. FEBS Lett. 591: 1720–1731. 10.1002/1873-3468.12637 [DOI] [PubMed] [Google Scholar]
- Moser F., Horwitz A., Chen J., Lim W., Voigt C. A., 2013. Genetic sensor for strong methylating compounds. ACS Synth. Biol. 2: 614–624. 10.1021/sb400086p [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley M. A., Mancini J. D., Young C. L., McCusker E. C., Raden D., et al. , 2009. Progress toward heterologous expression of active G-protein-coupled receptors in Saccharomyces cerevisiae: linking cellular stress response with translocation and trafficking. Protein Sci. 18: 2356–2370. 10.1002/pro.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opefi C. A., South K., Reynolds C. A., Smith S. O., Reeves P. J., 2013. Retinitis pigmentosa mutants provide insight into the role of the N-terminal cap in rhodopsin folding, structure, and function. J. Biol. Chem. 288: 33912–33926. 10.1074/jbc.M113.483032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outeiro T. F., Lindquist S., 2003. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science 302: 1772–1775. 10.1126/science.1090439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pándy-Szekeres G., Munk C., Tsonkov T. M., Mordalski S., Harpsoe K., et al. , 2018. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 46: D440–D446. 10.1093/nar/gkx1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfitt D. A., Aguila M., McCulley C. H., Bevilacqua D., Mendes H. F., et al. , 2014. The heat-shock response co-inducer arimoclomol protects against retinal degeneration in rhodopsin retinitis pigmentosa. Cell Death Dis. 5: e1236 10.1038/cddis.2014.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P. S., 2014. Constitutively active rhodopsin and retinal disease. Adv. Pharmacol. 70: 1–36. 10.1016/B978-0-12-417197-8.00001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perocchi F., Mancera E., Steinmetz L. M., 2008. Systematic screens for human disease genes, from yeast to human and back. Mol. Biosyst. 4: 18–29. 10.1039/B709494A [DOI] [PubMed] [Google Scholar]
- Ploier B., Caro L. N., Morizumi T., Pandey K., Pearring J. N., et al. , 2016. Dimerization deficiency of enigmatic retinitis pigmentosa-linked rhodopsin mutants. Nat. Commun. 7: 12832 10.1038/ncomms12832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoczy E. P., Kiel C., McKeone R., Stricher F., Serrano L., 2011. Analysis of disease-linked rhodopsin mutations based on structure, function, and protein stability calculations. J. Mol. Biol. 405: 584–606. 10.1016/j.jmb.2010.11.003 [DOI] [PubMed] [Google Scholar]
- Ramon E., Cordomi A., Aguila M., Srinivasan S., Dong X., et al. , 2014. Differential light-induced responses in sectorial inherited retinal degeneration. J. Biol. Chem. 289: 35918–35928. 10.1074/jbc.M114.609958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao V. R., Cohen G. B., Oprian D. D., 1994. Rhodopsin mutation G90D and a molecular mechanism for congenital night blindness. Nature 367: 639–642. 10.1038/367639a0 [DOI] [PubMed] [Google Scholar]
- Reeves P. J., Thurmond R. L., Khorana H. G., 1996. Structure and function in rhodopsin: high level expression of a synthetic bovine opsin gene and its mutants in stable mammalian cell lines. Proc. Natl. Acad. Sci. USA 93: 11487–11492. 10.1073/pnas.93.21.11487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovati G. E., Capra V., Neubig R. R., 2007. The highly conserved DRY motif of class A G protein-coupled receptors: beyond the ground state. Mol. Pharmacol. 71: 959–964. 10.1124/mol.106.029470 [DOI] [PubMed] [Google Scholar]
- Sancho-Pelluz J., Tosi J., Hsu C. W., Lee F., Wolpert K., et al. , 2012. Mice with a D190N mutation in the gene encoding rhodopsin: a model for human autosomal-dominant retinitis pigmentosa. Mol. Med. 18: 549–555. 10.2119/molmed.2011.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. O., 2010. Structure and activation of the visual pigment rhodopsin. Annu. Rev. Biophys. 39: 309–328. 10.1146/annurev-biophys-101209-104901 [DOI] [PubMed] [Google Scholar]
- Sokolina K., Kittanakom S., Snider J., Kotlyar M., Maurice P., et al. , 2017. Systematic protein-protein interaction mapping for clinically relevant human GPCRs. Mol. Syst. Biol. 13: 918 10.15252/msb.20167430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita L. M., Young D. L., Islam M., Kitzman J. O., Gullingsrud J., et al. , 2015. Massively parallel functional analysis of BRCA1 RING domain variants. Genetics 200: 413–422. 10.1534/genetics.115.175802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita L. M., Ahituv N., Dunham M. J., Kitzman J. O., Roth F. P., et al. , 2017. Variant interpretation: functional assays to the rescue. Am. J. Hum. Genet. 101: 315–325. 10.1016/j.ajhg.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson P. D., Mort M., Ball E. V., Shaw K., Phillips A., et al. , 2014. The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 133: 1–9. 10.1007/s00439-013-1358-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovic A., Hwang I., Khorana H. G., Hwa J., 2003. Retinitis pigmentosa rhodopsin mutations L125R and A164V perturb critical interhelical interactions: new insights through compensatory mutations and crystal structure analysis. J. Biol. Chem. 278: 39020–39028. 10.1074/jbc.M303625200 [DOI] [PubMed] [Google Scholar]
- Stoy H., Gurevich V. V., 2015. How genetic errors in GPCRs affect their function: possible therapeutic strategies. Genes Dis. 2: 108–132. 10.1016/j.gendis.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Flock T., Deupi X., Maeda S., Matkovic M., et al. , 2015. Probing Galphai1 protein activation at single-amino acid resolution. Nat. Struct. Mol. Biol. 22: 686–694. 10.1038/nsmb.3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Yang F., Tan G., Costanzo M., Oughtred R., et al. , 2016. An extended set of yeast-based functional assays accurately identifies human disease mutations. Genome Res. 26: 670–680. 10.1101/gr.192526.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung C. H., Schneider B. G., Agarwal N., Papermaster D. S., Nathans J., 1991. Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 88: 8840–8844. 10.1073/pnas.88.19.8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung C. H., Davenport C. M., Nathans J., 1993. Rhodopsin mutations responsible for autosomal dominant retinitis pigmentosa. Clustering of functional classes along the polypeptide chain. J. Biol. Chem. 268: 26645–26649. [PubMed] [Google Scholar]
- Sung C. H., Makino C., Baylor D., Nathans J., 1994. A rhodopsin gene mutation responsible for autosomal dominant retinitis pigmentosa results in a protein that is defective in localization to the photoreceptor outer segment. J. Neurosci. 14: 5818–5833. 10.1523/JNEUROSCI.14-10-05818.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam B. M., Moritz O. L., 2009. The role of rhodopsin glycosylation in protein folding, trafficking, and light-sensitive retinal degeneration. J. Neurosci. 29: 15145–15154. 10.1523/JNEUROSCI.4259-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam L. C., Kiang A. S., Campbell M., Keaney J., Farrar G. J., et al. , 2010. Prevention of autosomal dominant retinitis pigmentosa by systemic drug therapy targeting heat shock protein 90 (Hsp90). Hum. Mol. Genet. 19: 4421–4436. 10.1093/hmg/ddq369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui I., Chou C. L., Palmer N., Lin C. S., Tsang S. H., 2008. Phenotype-genotype correlations in autosomal dominant retinitis pigmentosa caused by RHO, D190N. Curr. Eye Res. 33: 1014–1022. 10.1080/02713680802484645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration, 2017 Approval Letter - LUXTURNA. Available at: https://www.fda.gov/downloads/BiologicsBloodVaccines/CellularGeneTherapyProducts/ApprovedProducts/UCM589690.pdf. Accessed: January 30, 2018.
- Wang J., Deretic D., 2014. Molecular complexes that direct rhodopsin transport to primary cilia. Prog. Retin. Eye Res. 38: 1–19. 10.1016/j.preteyeres.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weile J., Sun S., Cote A. G., Knapp J., Verby M., et al. , 2017. A framework for exhaustively mapping functional missense variants. Mol. Syst. Biol. 13: 957 10.15252/msb.20177908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodsmith J., Apelt L., Casado-Medrano V., Ozkan Z., Timmermann B., et al. , 2017. Protein interaction perturbation profiling at amino-acid resolution. Nat. Methods 14: 1213–1221. 10.1038/nmeth.4464 [DOI] [PubMed] [Google Scholar]
- Yang F., Sun S., Tan G., Costanzo M., Hill D. E., et al. , 2017. Identifying pathogenicity of human variants via paralog-based yeast complementation. PLoS Genet. 13: e1006779 10.1371/journal.pgen.1006779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Kolesnikov A. V., Jastrzebska B., Mustafi D., Sawada O., et al. , 2013. Autosomal recessive retinitis pigmentosa E150K opsin mice exhibit photoreceptor disorganization. J. Clin. Invest. 123: 121–137. 10.1172/JCI66176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvyaga T. A., Fahmy K., Siebert F., Sakmar T. P., 1996. Characterization of the mutant visual pigment responsible for congenital night blindness: a biochemical and Fourier-transform infrared spectroscopy study. Biochemistry 35: 7536–7545. 10.1021/bi960391n [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. All supplemental figures, tables, and files have been uploaded to figshare. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7414109.