The CRISPR/Cas9 gene editing system continues to push the boundaries of genetic analysis. Here, papers from Farboud, Severson, and Meyer and Poe et al. describe cutting-edge advances for CRISPR use. Farboud, Severson, and Meyer....
Keywords: CRISPR/Cas9, gRNA, Drosophila, tissue-specific, loss-of-function, da neuron, dendrite morphogenesis, Ptp69D, SNAP, NSF
Abstract
Tissue-specific loss-of-function (LOF) analysis is essential for characterizing gene function. Here, we present a simple, yet highly efficient, clustered regularly interspaced short palindromic repeats (CRISPR)-mediated tissue-restricted mutagenesis (CRISPR-TRiM) method for ablating gene function in Drosophila. This binary system consists of a tissue-specific Cas9 and a ubiquitously expressed multi-guide RNA (gRNA) transgene. We describe convenient toolkits for making enhancer-driven Cas9 lines and multi-gRNAs that are optimized for mutagenizing somatic cells. We demonstrate that insertions or deletions in coding sequences more reliably cause somatic mutations than DNA excisions induced by two gRNAs. We further show that enhancer-driven Cas9 is less cytotoxic yet results in more complete LOF than Gal4-driven Cas9 in larval sensory neurons. Finally, CRISPR-TRiM efficiently unmasks redundant soluble N-ethylmaleimide–sensitive factor attachment protein receptor gene functions in neurons and epidermal cells. Importantly, Cas9 transgenes expressed at different times in the neuronal lineage reveal the extent to which gene products persist in cells after tissue-specific gene knockout. These CRISPR tools can be applied to analyze tissue-specific gene function in many biological processes.
TISSUE-SPECIFIC loss-of-function (LOF) analysis is instrumental for elucidating the developmental roles of essential genes, determining cell autonomy, and dissecting cell-to-cell interactions. Conventional methods for studying tissue-specific gene function in Drosophila, such as mosaic analysis with a repressible cell marker (MARCM) (Lee and Luo 1999) and tissue-specific RNA interference (RNAi) (Dietzl et al. 2007; Ni et al. 2011), are powerful approaches for genetic screens and LOF analysis. However, these techniques present several disadvantages. RNAi is prone to off-target effects (Ma et al. 2006), and gene knockdown is rarely complete (Dietzl et al. 2007) because this technique only targets mRNAs for degradation or translational suppression. MARCM produces more reliable LOF of genes of interest, but the process can be labor-intensive and requires multiple components to be combined in the same animal.
The clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system (Jinek et al. 2012) has the potential to surpass the current methods of tissue-specific LOF in Drosophila due to its simplicity and efficiency in creating gene disruption (Bassett et al. 2013; Gratz et al. 2013; Kondo and Ueda 2013; Ren et al. 2013; Yu et al. 2013; Sebo et al. 2014). In this system, Cas9 endonuclease cleaves genomic DNA at a site determined by the protospacer sequence (or target sequence) of a chimeric guide RNA (gRNA) (Jinek et al. 2012). Cas9-mediated double-strand breaks (DSBs) are then repaired through either nonhomologous-end joining (NHEJ) or homology-directed repair (Gaj et al. 2013). Imprecise repair through NHEJ can result in small insertions or deletions (indels) at each target site (Bassett et al. 2013), or deletions of DNA fragments between two target sites (Kondo and Ueda 2013; Ren et al. 2013). CRISPR/Cas9 has been successfully used in Drosophila and other organisms to create heritable mutations (Bassett et al. 2013; Gratz et al. 2013; Yu et al. 2013), to edit genomic sequences precisely (Gratz et al. 2014; Xue et al. 2014a), and to control gene expression (Ghosh et al. 2016; Ewen-Campen et al. 2017).
Tissue-specific mutagenesis has been achieved in Drosophila by combining the CRISPR/Cas9 system with the Gal4/UAS (upstream activating system) system (Port et al. 2014; Xue et al. 2014b; Port and Bullock 2016). In this approach, tissue-specific Gal4 drives UAS-Cas9 expression, while gRNAs are expressed either from ubiquitous promoters (Port et al. 2014; Xue et al. 2014b) or by the UAS (Port and Bullock 2016). Transgenic constructs expressing multiple gRNAs increase mutagenesis efficiency and allow the simultaneous mutagenesis of more than one gene (Port et al. 2014; Xue et al. 2014b; Port and Bullock 2016). Despite these initial successes, Gal4-driven Cas9 and transgenic gRNAs have not been widely used to study tissue-specific gene function, due to uncertainties and limitations associated with this method. For example, gRNAs can vary greatly in their mutagenic efficiency, and it is difficult to know whether a transgenic gRNA reliably causes mutations in the tissue of interest. These concerns worsen when a multiplex gRNA construct is used to knock out two or more genes simultaneously. Gal4-driven Cas9 has several additional potential drawbacks that could limit its applications in developmental studies. First, the intermediate Gal4 expression step can delay Cas9 expression, making it difficult to study early gene functions in specific tissues. Second, the Gal4/UAS system often results in excessive levels of Cas9 expression that can be toxic (Jiang et al. 2014). Finally, using Gal4-driven Cas9 makes the Gal4/UAS system unavailable for other genetic manipulations in the same animal. Thus, a simpler and more robust method of tissue-specific mutagenesis via CRISPR/Cas9 is desirable.
One way to improve mutagenic efficiency is the optimization of transgenic gRNAs. Previous studies in Drosophila exploring choices of the gRNA promoter, the length and sequence composition of the target sequence, and methods of producing multiple gRNAs from a single construct have identified several parameters for making efficient gRNAs (Port et al. 2014; Ren et al. 2014; Xue et al. 2014b; Port and Bullock 2016). However, the goal of most of these studies was to increase the frequency of heritable mutations, leaving room for the optimization of transgenic gRNA design for mutagenesis in somatic cells. In particular, specific modifications of the gRNA scaffold improve Cas9 targeting to DNA in human cells (Chen et al. 2013), but these modifications have not been tested to date in Drosophila. Thus, there is a compelling need for optimized transgenic gRNAs coupled with tissue-specific control of Cas9 efficacy.
Here, we have developed a new CRISPR/Cas9 toolkit that achieves highly efficient knockout of one or multiple Drosophila genes in a tissue-specific manner. Our method of CRISPR-mediated tissue-restricted mutagenesis (CRISPR-TRiM) combines a transgenic Cas9 driven by a tissue-specific enhancer with a transgenic construct that ubiquitously expresses multiple gRNAs. By targeting every gene of interest with two gRNAs, this system mutates all target genes tissue-specifically through indels or large DNA deletions. To build the most efficient reagents, we have generated convenient tools for making and evaluating enhancer-driven Cas9 transgenes, identified a multi-gRNA design that is superior to previous options, and established an in vivo assay for testing gRNA efficiency in causing DSBs. We investigated how the frequency of DNA deletion in individual somatic cells is impacted by the distance between two target sites, and we further found that enhancer-driven Cas9 is more effective in causing LOF and less cytotoxic than Gal4-driven Cas9 in Drosophila sensory neurons. Using genes in the SNARE (soluble N-ethylmaleimide–sensitive factor attachment protein receptor) pathway as examples, we demonstrate that CRISPR-TRiM can efficiently knock out multiple genes in the same cells, and reveal their redundant functions in neurons and epithelial epidermal cells. Our results also underscore the importance of mutagenesis timing for uncovering tissue-specific gene functions: postmitotic knockout of neuronal type-specific genes, such as the receptor protein tyrosine phosphatase Ptp69D, is sufficient and effective for removing gene functions; while housekeeping genes, such as those encoding N-ethylmaleimide–sensitive factor (NSF) and synaptosomal nerve-associated (SNAP) proteins, require mutagenesis earlier in the cell lineage to unmask their LOF phenotypes.
Materials and Methods
Methods including fly stocks, Cas9-LEThAL assay, molecular cloning, Drosophila transgenic lines, western blotting, identification of gRNA target sequence, live imaging, imaginal disc imaging, immunohistochemistry, image analysis and quantification, and statistical analysis are in the Supplemental Material available at Figshare.
Data availability
Supplemental Materials, including supplemental figures, supplemental tables, methods, and supplemental references, are available at Figshare: https://doi.org/10.25386/genetics.7399514. Plasmids are available at Addgene or upon request. Drosophila lines are available at the Bloomington Drosophila Stock Center or upon request.
Results
Generation and evaluation of tissue-specific Cas9 lines
Our CRISPR-TRiM strategy relies on the availability of efficient tissue-specific Cas9 transgenes. To simplify the generation of tissue-specific Cas9 lines, we developed a Cas9 Gateway destination vector pDEST-APIC-Cas9 using the pAPIC (attB P-element insulated CaSpeR) backbone optimized for enhancer-driven transgene expression (Han et al. 2011) (Figure 1A). Tissue-specific enhancers can be conveniently swapped into this vector through the Gateway LR reaction to generate Cas9-expression constructs. This cloning strategy is compatible with over 14,000 FlyLight (Jenett et al. 2012) and VT (Kvon et al. 2014) enhancers, whose expression profiles for multiple developmental stages and tissues in Drosophila are publicly available.
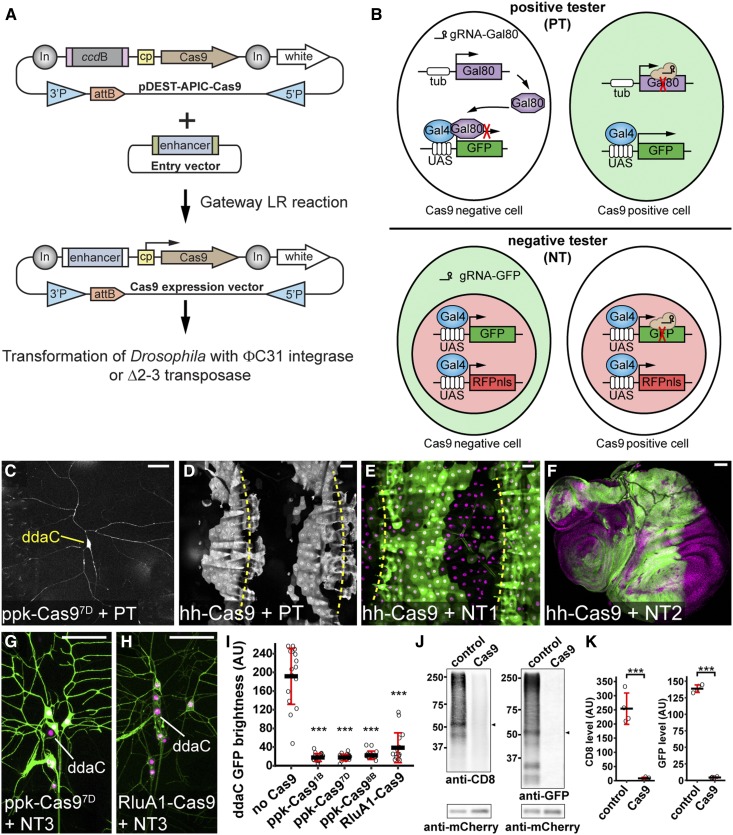
Figure 1.
Generation and evaluation of tissue-specific Cas9 lines, (A) Diagram of Gateway cloning and transgenesis of Cas9 expression vectors. In, Gypsy insulator; cp, core promoter; 3′P and 5′P, P-element sequences. (B) Diagrams of positive tester (PT) and negative tester (NT), illustrating how Cas9-expressing cells are visualized by each type of tester. In PT, two ubiquitous guide RNAs (gRNAs) target Gal80. In NT, two ubiquitous gRNAs target GFP. Full genotypes of Cas9 testers are in Table S1. (C and D) Patterns of Cas9 activity in ppk-Cas97D (C) and hh-Cas9 (D) as visualized by PT. (E and F) Patterns of hh-Cas9 activity in the larval epidermis as visualized by NT1 (E), and in wing, haltere, and leg imaginal discs as visualized by NT2 (F). The positions of body wall segmental borders (muscle attachment sites) are indicated by yellow broken lines in (D and E). ppk-Cas9 is predicted to be active in C4da neurons, including ddaC. hh-Cas9 is predicted to be active in the posterior compartments of epidermal segments and imaginal discs. (G and H) Patterns of Cas9 activity in ppk-Cas97D (G) and RluA1-Cas9 (H) as visualized by NT3. The cell bodies of ddaC neurons are indicated. (I) Quantification of ddaC GFP brightness in NT3 crosses using control (no Cas9) and various da neuron-specific Cas9 lines. *** P ≤ 0.001; one-way ANOVA and Dunnett’s test. n = 16 neurons for each genotype. Black bar, mean; red bars, SD. Bar, 50 μm. AU, arbitrary units. (J) Western blot of CD8 and GFP in larval homogenates of NT2 crossed to w1118 (control) or Act-Cas9 (Cas9). mCherry serves as the loading control. The bands used for quantification are indicated by arrowheads. (K) Quantification of CD8 and GFP levels in control and Cas9 after normalization with mCherry levels. *** P ≤ 0.001; unpaired Student’s t-test. Black bar, mean; red bars, SD. n = 4 biological replicates.
An ideal tissue-specific Cas9 should be consistently and robustly expressed in the tissue of interest but not in unintended tissues. In practice, the insertion site in the genome often modifies the expression pattern, timing, and level of a transgene (Weiler and Wakimoto 1995). This position effect could impact the tissue-specificity and mutagenic efficiency. To evaluate Cas9 transgenes, we developed a series of tester lines, with the positive tester positively labeling Cas9-expressing cells and negative testers negatively labeling Cas9-expressing cells (Supplemental Material, Table S1). The positive tester carries a UAS-GFP and ubiquitously expresses Gal80, Gal4, and two gRNAs targeting Gal80 (Figure 1B). In Cas9-negative cells, Gal80 suppresses Gal4 activity, thereby inhibiting GFP expression. In contrast, in Cas9-expressing cells, the gRNAs induce mutations in Gal80 and thus allow Gal4-driven GFP expression. As examples, we generated random insertions of ppk-Cas9 and hh-Cas9, and evaluated their tissue specificities using the positive tester. The ppk enhancer is specific to class IV dendritic arborization (C4da) sensory neurons on the larval body wall (Grueber et al. 2003), while the R28E04 enhancer of hh drives epidermal expression in the posterior half of every hemisegment (http://flweb.janelia.org). The positive tester allowed us to identify the ppk-Cas9 and hh-Cas9 insertions that most resemble the expected patterns (Figure 1, C and D).
Negative testers help further evaluate the efficiency of Cas9 transgenes for inducing mutations. A negative tester contains a ubiquitous or tissue-specific Gal4, a UAS-driven cytosolic or membrane GFP, a UAS-driven nuclear red fluorescent protein (RFP), and two ubiquitous gRNAs targeting GFP (Figure 1B). With a negative tester, Cas9-negative cells are dually labeled by both GFP and the nuclear RFP, while Cas9-expressing cells are only labeled by the nuclear RFP, due to GFP mutagenesis. When crossed to negative testers ubiquitously expressing Gal4, hh-Cas9, as expected, caused loss of GFP in the posterior compartments of larval epidermal segments (Figure 1E) and the imaginal discs (Figure 1F). A neuronal negative tester expressing the membrane marker CD8-GFP in all da neurons (NT3) showed that ppk-Cas9 specifically knocked out GFP in C4da neurons (Figure 1G). Negative testers are particularly useful for comparing the efficiency of Cas9 lines in mutagenesis; lower persistent GFP signals likely reflect earlier-acting Cas9. Using NT3, we identified two ppk-Cas9 insertions that appeared to be most efficient (ppk-Cas91B and ppk-Cas97D) (Figure 1I). In comparison, Cas9 driven by a pan-da RluA1 enhancer (Figure 1H) led to more variable GFP reductions in C4da neurons (Figure 1I). To understand how Cas9/gRNA-induced mutations affect CD8-GFP proteins, we crossed a negative tester that expresses CD8-GFP in all cells (NT2) to a ubiquitous Act-Cas9 (Port et al. 2014), and examined both the N-terminal CD8 and the C-terminal GFP in the larva by western blot. Surprisingly, targeting GFP alone abolished both CD8 and GFP signals (Figure 1, J and K), indicating that either indel mutations extended into the CD8 coding sequence, or the mutations reduced the mRNA or protein stability.
The Cas9 Gateway cloning vector and the Cas9 tester lines together provide a convenient toolbox for generating and identifying Cas9 transgenes that are most efficient for CRISPR-TRiM.
Optimization of multi-gRNA design for tissue-specific gene knockout in Drosophila
Being able to express multiple gRNAs from a single transgenic construct is desirable for CRISPR-TRiM, as more gRNAs can increase the chance of LOF in a single gene and also enable the simultaneous mutagenesis of multiple genes (Port et al. 2014; Xue et al. 2014b; Port and Bullock 2016). A common strategy for making multiplex gRNA constructs in Drosophila is to use two or three ubiquitous U6 promotors in tandem, each driving a gRNA separately (Port et al. 2014). Alternatively, polycistronic gRNA designs with intervening tRNA sequences have also been reported to be effective in expressing multiple gRNAs in plants and Drosophila (Xie et al. 2015; Port and Bullock 2016). Aiming to optimize the multi-gRNA strategy to achieve the greatest mutagenic efficiency in somatic cells, we compared four dual-gRNA designs that carry the same two targeting sequences for enhanced GFP and GFP in a vector containing P-element/attB sequences and the mini-white selection marker (Figure 2A). Three of them (forward, reverse, and insulated) are variants of a U6:1-gRNA-U6:3-gRNA strategy described previously (Port et al. 2014), with differences in the orientation of the gRNA cassette, and the use of an insulator to separate mini-white and the gRNAs. The fourth design (tgFE) builds upon the tRNA-gRNA strategy (Port and Bullock 2016), and introduces an A–U base pair flip and an extension of the Cas9-binding hairpin (F+E modifications) in the gRNA scaffold (Chen et al. 2013), which have been shown to greatly improve the targeting of Cas9 to the genomic DNA.
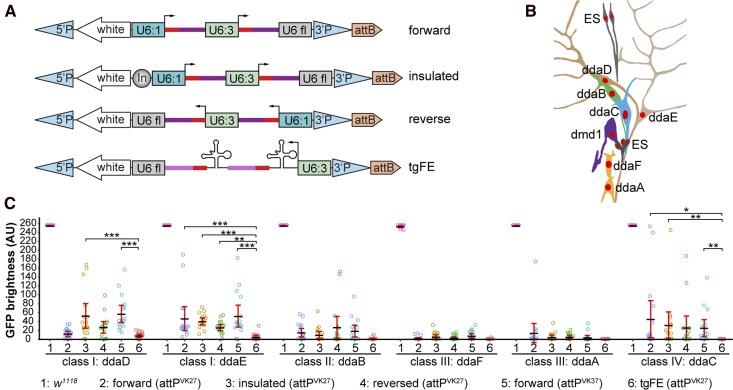
Figure 2.
Optimization of multi-guide RNA (gRNA) design for tissue-specific gene knockout in Drosophila. (A) Four designs of multi-gRNA transgenic vectors. U6:1 and U6:3, U6 promoters; U6 fl, U6 3′ flanking sequence; In, Gypsy insulator. Red bars, gRNA targeting sequence; dark magenta bars, original gRNA scaffold; light magenta bars, E+F gRNA scaffold. (B) Diagram of the dorsal cluster of larval peripheral sensory neurons. (C) Comparison of a control (1) and various gRNA-GFP lines in eliminating GFP signal in each dorsal da neuron using RluA1-Cas9. Da neurons express UAS-CD8-GFP driven by nsyb-Gal4. The integration site for each gRNA line is indicated in parentheses. The GFP signals in most control neurons are saturated under the setting used. Each circle represents an individual neuron (n = 16 for each column). Black bar, mean; red bars, SD. * P ≤ 0.05, ** P ≤ 0.01, ***P ≤ 0.001; one-way ANOVA and Tukey’s honest significant difference test. Only significance levels between #6 and others are indicated. AU, arbitrary units.
We compared these constructs in knocking out GFP in individual neurons of the larval peripheral nervous system (PNS). The dorsal cluster of sensory neurons in every abdominal segment contains six da neurons belonging to four classes (Grueber et al. 2002) (Figure 2B), allowing for the accurate measurement of fluorescence intensity at single-cell resolution. To detect differences in gRNA efficiency, we used the relatively ineffective RluA1-Cas9 to knock out UAS-CD8-GFP driven by pan-neural nsyb-Gal4 (Pfeiffer et al. 2012). The performance of gRNAs based on U6:1-gRNA-U6:3-gRNA varied depending on the neuronal identity and the fact that none of these designs were efficient enough to remove GFP in all neurons (Figure 2C). In contrast, the tgFE design led to near complete elimination of GFP signals in almost all neurons examined (Figure 2C). Therefore, tgFE is a more efficient multiplex gRNA design for tissue-specific mutagenesis. An additional benefit of the tgFE strategy is the convenient cloning of two-to-six gRNAs in a single step (see Table S4 and Cloning of gRNA expression vectors in the Supplemental Material).
Efficiency of dual gRNA-mediated DNA deletion at the single-cell level
When using two gRNAs to target the same gene, large DNA deletions between the two target sites would more likely generate null alleles. To investigate the frequency of large deletions caused by two gRNAs in individual cells, we constructed a reporter nSyb-tdGFP (“td” standing for tandem dimer) (Figure 3A), which labels all 12 neurons in the dorsal cluster of PNS sensory neurons (Figure 2B and Figure 3B). In addition, we designed seven gRNAs (0–6) targeting different sites in the noncoding sequence of this reporter, with site 0 located before the nSyb enhancer, site 1 immediately after the enhancer, and the remaining sites at various distances downstream of the tdGFP coding sequence (Figure 3A). We reasoned that small indels at any of these target sites would probably not abolish GFP expression, but large deletions between site 0 and any of the other targeting sites would (Figure 3C). As a control, we included a gRNA pair that targets two sites in the tdGFP coding sequence (gRNA-GFP), and therefore is predicted to remove GFP expression by either indels or large deletions.
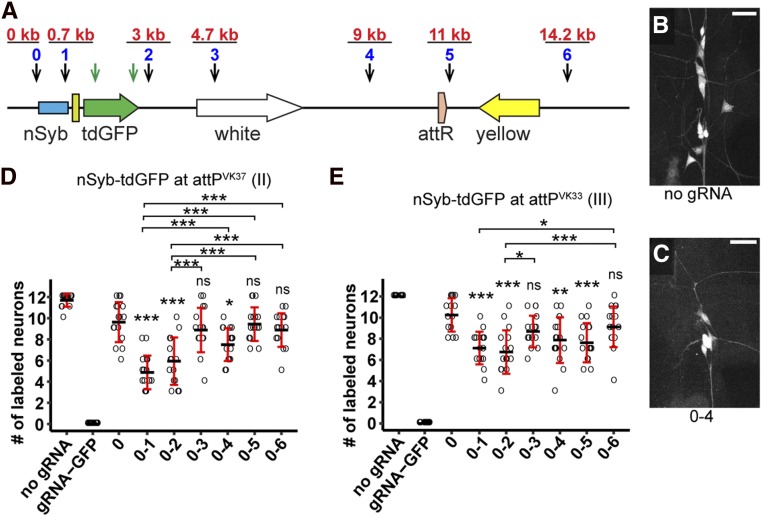
Figure 3.
Efficiency of dual guide RNA (gRNA)-mediated DNA deletion at the single-cell level. (A) Diagram showing the nSyb-tdGFP reporter integrated in the genome and gRNA target sites. Each blue number and the black arrow below it indicate a gRNA-targeting noncoding sequence of the reporter. The distance of each gRNA from gRNA 0 is indicated in red above the gRNA. The two green arrows indicate two gRNAs targeting the coding sequence of tdGFP. (B and C) Dorsal clusters of PNS neurons labeled by the reporter in a control animal (B) and an animal expressing gRNAs 0 and 4 (C). Bar, 25 μm. (D and E) Quantification of the number of GFP-positive neurons for each gRNA pair using nSyb-tdGFP inserted at attPVK37 (D) and attPVK33 (E) sites. Each circle represents an individual neuron (n = 16 neurons for each genotype). Black bar, mean; red bars, SD. * P ≤ 0.05, *** P ≤ 0.001; one-way ANOVA and Tukey’s honest significant difference test.
Using Act-Cas9, we tested the efficiencies of these gRNA pairs in eliminating GFP expression in individual neurons with two different nSyb-tdGFP insertions. In all animals examined, gRNA-GFP completely abolished GFP expression (Figure 3, D and E). Unexpectedly, gRNA 0 alone reduced numbers of labeled neurons in some animals (Figure 3, D and E), likely due to deletions extending into regulatory elements in the nSyb enhancer. Pairing gRNA 0 with gRNAs 1–6 further reduced the number of labeled neurons in some combinations, with a tendency for gRNA pairs positioned closer more often generating fewer GFP-positive neurons (Figure 3, C–E). Although the gRNAs are unlikely to target the genomic DNA with the same efficiency and the nature of mutations could not be confirmed in individual neurons, our data indicate that large deletions occur in random somatic cells, and that an inverse correlation appears to exist between deletion frequency and gRNA distance. Importantly, our results suggest that large deletions do not occur frequently enough to remove gene function in every cell, such that indels in the coding region are more reliable for causing LOF.
Enhancer-driven Cas9 is advantageous over Gal4-driven Cas9 for studying neural development
Conditional mutagenesis can be achieved in Drosophila somatic cells using Gal4-driven Cas9 (Port et al. 2014; Xue et al. 2014b; Port and Bullock 2016), but this method requires an intermediate transcription step that could potentially delay Cas9 expression. Consistent with this assumption, ppk-CD4-tdGFP is expressed at least 8-hr earlier than UAS-CD8-GFP driven by ppk-Gal4 in the embryo (Han et al. 2011). Thus, we predict that enhancer-driven Cas9 will result in earlier Cas9 action, thereby reducing perdurance of wild-type mRNA or protein products of the target gene made prior to mutation induction. We tested this hypothesis by comparing the effectiveness of enhancer-driven Cas9 and Gal4-driven Cas9 in knocking out CD4-tdGFP expression in C4da neurons (Figure 4A). We observed more consistent and stronger reduction of GFP with ppk-Cas9 insertions compared to ppk-Gal4 UAS-Cas9 (ppk > Cas9) (Figure 4B). To ask whether even earlier Cas9 expression could lead to further GFP reduction, we made a Cas9 that is expressed in sensory organ precursors (SOPs), the progenitor cells of da neurons (Powell et al. 2004). Indeed, SOP-Cas9 resulted in complete loss of GFP fluorescence in most animals (Figure 4B). Using ppk-Cas91B, we also found that adding two irrelevant gRNAs targeting Gal80 did not reduce the efficiency of gRNA-GFP in removing CD4-tdGFP signals (Figure 4C).
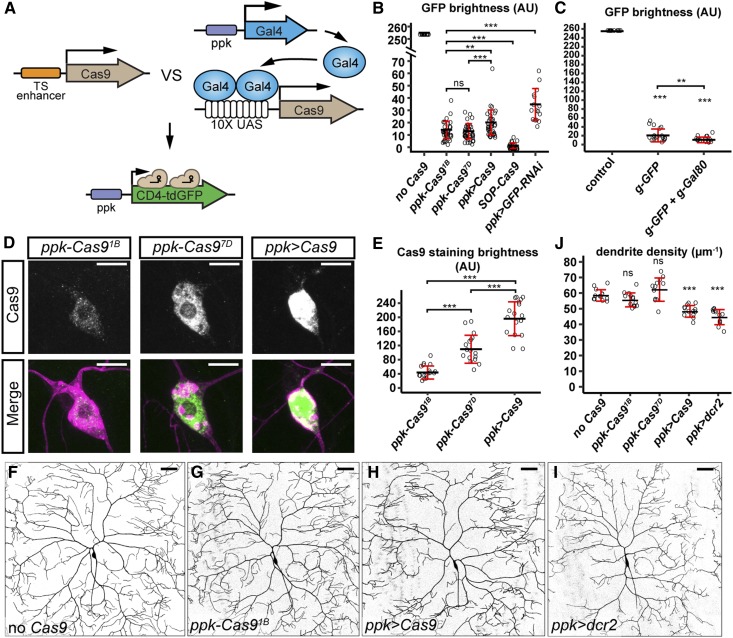
Figure 4.
Enhancer-driven Cas9 is advantageous over Gal4-driven Cas9 in studying neural development. (A) Diagram showing the comparison of tissue-specific (TS) enhancer-driven Cas9 and Gal4-driven Cas9 in knocking out ppk-CD4-tdGFP. (B) Quantification of GFP brightness in C4da neurons in the control, Cas9-expressing animals, and GFP knockdown animals. GFP signals in the control (no Cas9) are saturated. *** P ≤ 0.001; ns, not significant; one-way ANOVA and Tukey’s honest significant difference (HSD) test. (C) Quantification of GFP brightness in C4da neurons in the control (no Cas9), and animals expressing ppk-Cas9 gRNA-GFP and ppk-Cas9 gRNA-GFP gRNA-Gal80. Control values are from (B). *** P ≤ 0.001, ** P ≤ 0.01; one-way ANOVA and Tukey’s HSD test. (D) Cas9 staining in the genotypes indicated. Upper panels show Cas9 staining. Lower panels show Cas9 staining (green) and C4da neurons labeled by ppk > CD4-tdTom (magenta). (E) Quantification of nuclear Cas9 levels in C4da neurons in the genotypes indicated. *** P ≤ 0.001; one-way ANOVA and Tukey’s HSD test. (F–I) DdaC neurons in the control (F), ppk-Cas91B (G), ppk-Gal4 UAS-Cas9 (H), and ppk-Gal4 UAS-dcr2 (I). (J) Quantification of dendrite density in genotypes indicated. *** P ≤ 0.001; ns, not significant; one-way ANOVA and Dunnett’s test. Each circle represents an individual neuron. For (B), n = 16 for the control and ppk > GFP-RNAi; n = 40 for SOP-Cas9 and ppk > Cas9; n = 39 for ppk-Cas91B; and n = 43 for ppk-Cas97D. For (C), n = 16. For (E), n = 17. For (J), n = 13. Black bar, mean; red bars, SD. Bar, 10 μm in (D) and 50 μm in (F–I). AU, arbitrary units.
High levels of Cas9 have been reported to be cytotoxic (Jiang et al. 2014). We found that Gal4-driven Cas9 resulted in higher levels of nuclear Cas9 protein than the enhancer–fusion versions (Figure 4, D and E). Correspondingly, ppk > Cas9 caused obvious dendrite reduction in C4da neurons even in the absence of gRNAs, while ppk-Cas9 lines had much weaker impacts on dendrite morphology (Figure 4, F–H and J). These data suggest that high levels of Cas9 in postmitotic neurons are not desirable for studying neuronal morphogenesis and that enhancer-driven Cas9 could alleviate this concern.
We also compared the effects of RNAi-mediated suppression of GFP expression and CRISPR/Cas9-induced GFP mutagenesis. CD4-tdGFP was knocked down with a publicly available UAS-GFP-RNAi line (Pastor-Pareja and Xu 2011) driven by ppk-Gal4. We also coexpressed Dicer-2 (Dcr-2) in C4da neurons to enhance double-strand RNA (dsRNA)-mediated knockdown (Dietzl et al. 2007). RNAi was found to be less efficient in eliminating GFP than CRISPR-mediated mutagenesis by either enhancer-driven Cas9 or Gal4-driven Cas9 (Figure 4B). In addition, we found that Dcr-2 overexpression, which is commonly employed in Drosophila RNAi experiments, resulted in a strong dendrite reduction (Figure 4, I and J), indicating that Dcr-2 causes cytotoxicity in neurons.
Our results suggest that, at least in larval sensory neurons, enhancer-driven Cas9 outperforms Gal4-driven Cas9 in tissue-specific mutagenesis, and that the CRISPR-TRiM method is more effective than RNAi in LOF studies.
Postmitotic knockout of Ptp69D reveals its function in C4da neurons
To validate the effectiveness of CRISPR-TRiM in studying neuronal morphogenesis, we knocked out the receptor protein tyrosine phosphatase Ptp69D in C4da neurons. Using hemizygous Ptp69D mutants and MARCM, we previously found that loss of Ptp69D in C4da neurons caused dendritic reduction with shortened terminal dendrites (Poe et al. 2017). As hemizygous mutants completely lack zygotic transcription and MARCM removes the Ptp69D gene before the birth of neurons, it is unclear from our previous results whether mutagenesis after the birth of neurons (postmitotic mutagenesis) would be sufficient to remove Ptp69D function. Thus, we made a gRNA-Ptp69D line expressing three gRNAs, each targeting a distinct site in the Ptp69D coding sequence (Table S3).
To validate the efficiency of gRNA-Ptp69D, we established a “Cas9-LEThAL” (for Cas9-induced lethal effect through the absence of Lig4) assay (Figure S2) that was adapted from a previously described method for assessing injection-based gRNA efficiency (Lee et al. 2015). Efficient gRNAs for nonessential genes, such as a published gRNA for e (Port et al. 2014) (Table S2), cause male-specific lethality in pupal stages when males carrying gRNAs are crossed to Act-Cas9 lig4 homozygous females. But if the target gene is essential, in the same cross, efficient gRNAs should cause lethality of both males and females similar to homozygous mutants. gRNA-Ptp69D caused all animals to die at the late pupal stages in this assay, indicating that this gRNA line is efficient (Table S2).
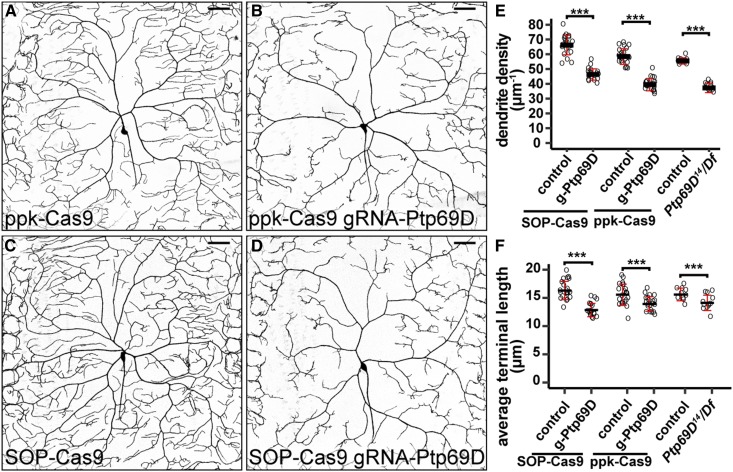
We knocked out Ptp69D in C4da neurons using both ppk-Cas9 and SOP-Cas9. As the ppk enhancer only becomes active in stage-16 embryos after the birth of C4da neurons (Grueber et al. 2003), ppk-Cas9 would only induce mutations postmitotically. In contrast, the SOP enhancer turns on in SOPs (Culi and Modolell 1998) that divide twice to give rise to da neurons (Lai and Orgogozo 2004), enabling SOP-Cas9 to act before the neuronal birth. We found that both ppk-Cas9 and SOP-Cas9 caused consistent and similar degrees of dendritic reduction in C4da neurons in late third-instar larvae (Figure 5, A–D). In both cases, the extent of the dendrite reductions caused by CRISPR-TRiM were also similar to that in Ptp69D14/Df(3L)8ex34 hemizygous null mutant larvae (Poe et al. 2017) (Figure 5, E and F). These data suggest that postmitotic mutagenesis is sufficient to remove Ptp69D gene function, which is consistent with Ptp69D being a neuronal type-specific gene (Desai et al. 1994; Poe et al. 2017).
Figure 5.
Postmitotic knockout of Ptp69D is sufficient to reveal its function in C4da neurons. (A–D) DdaC neurons in ppk-Cas9 control (A), ppk-Cas9 gRNA-Ptp69D (B), SOP-Cas9 control (C), and SOP-Cas9 gRNA-Ptp69D (D). (E and F) Quantification of total dendrite density (E) and average terminal dendrite length (F) in the genotypes indicated. Each circle represents an individual neuron: ppk-Cas9 (n = 22); ppk-Cas9 gRNA-Ptp69D (n = 21); SOP-Cas9 (n = 22); and SOP-Cas9 gRNA-Ptp69D (n = 20). Data for Ptp69D14/Df(3L)8ex34 (n = 12) and its control (n = 10) are cited from Poe et al. (2017) for comparison. *** P ≤ 0.001; unpaired Student’s t-test. Black bar, mean; red bars, SD. Bar, 50 μm.
CRISPR-TRiM reveals the redundancy and perdurance of NSF and SNAP genes in dendrite morphogenesis
CRISPR/Cas9 can simultaneously mutate multiple genes in Drosophila somatic cells (Port and Bullock 2016). Such an application would be very useful for studying the roles of redundant genes during development. To test whether CRISPR-TRiM can efficiently knock out multiple genes that may exhibit redundant functions, we targeted SNARE complex components in C4da neurons. Because SNAREs are required for all vesicle fusions (Wickner and Schekman 2008), interference with the complex should severely hamper C4da dendrite growth. Drosophila contains two NSF genes (comt/Nsf1 and Nsf2), which are necessary for the recycling of the SNARE complex after membrane fusion (Golby et al. 2001). Drosophila also has three SNAP-25 paralogs (Snap24, Snap25, and Snap29) that encode the SNAP (or Qbc.IV) group of SNARE proteins thought to be involved in secretion (Kloepper et al. 2007). The potential functional redundancy of the NSF and SNAP genes has not been examined during neuronal morphogenesis.
To conduct CRISPR-TRiM analyses, we used the tgFE design to generate dual-gRNA constructs for every NSF and SNAP gene (Table S3). Also using the tgFE design, we made four-gRNA constructs to knock out Nsf1/Nsf2 simultaneously and Snap24/Snap25 simultaneously, and a six-gRNA construct to knock out all three SNAP genes (Table S3). The efficiencies of these gRNA lines were first validated with the Cas9-LEThAL assay (Table S2). The lethal phase induced by each single-gene gRNA line was consistent with published results for null mutants of the corresponding gene, indicating that the gRNAs are efficient in mutagenesis. We found that gRNA-Nsf1-Nsf2 was as effective as gRNA-Nsf2 in causing lethality in first-instar larvae, while gRNA-Nsf1 caused lethality in late pupae, suggesting that increasing the number of gRNAs from two to four in one construct may not reduce the efficiency of gRNAs. Interestingly, compared to gRNA-Snap24 or gRNA-Snap25 alone, which produced animals surviving to the late pupal stage, gRNA-Snap24-Snap25 caused lethality in the first-instar larvae, demonstrating that Snap24 and Snap25 are redundantly required for the larval development. gRNA-Snap24-Snap25-Snap29 further advanced the lethal phase to late embryos, suggesting that Snap29 is also redundant with Snap24 and Snap25.
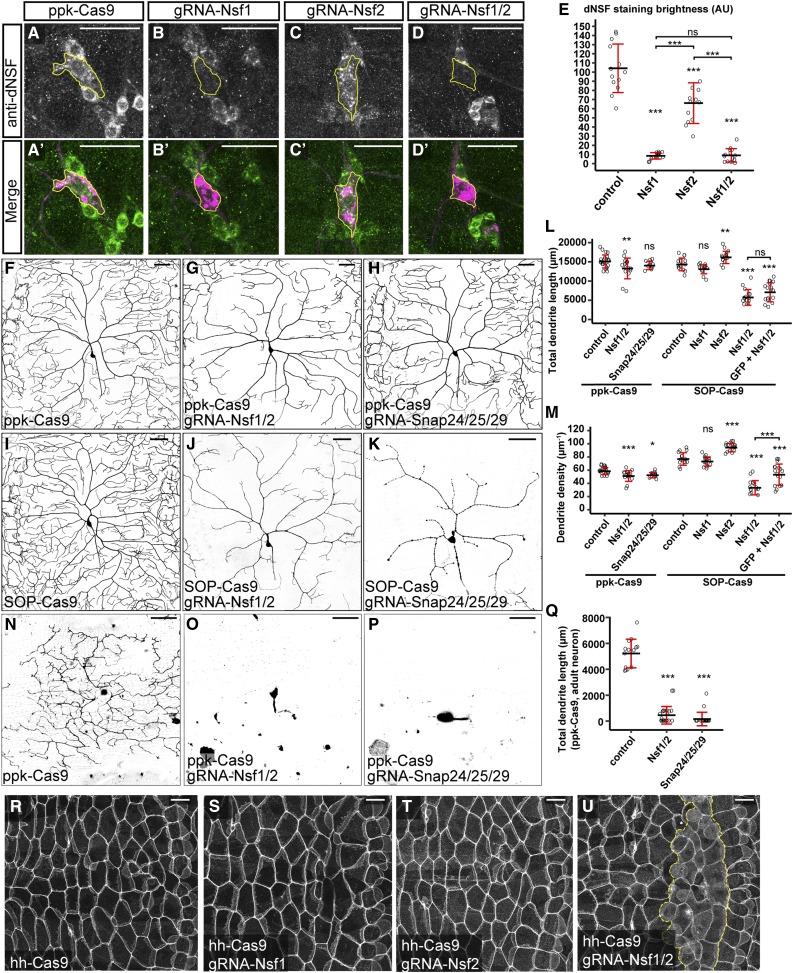
We further validated the efficiency of NSF gRNAs in knocking out Nsf1 by immunostaining. An anti-dNSF antibody that recognizes Nsf1 (Yu et al. 2011) strongly labels the cell bodies of da neurons (Figure 6, A and A’). When combined with ppk-Cas9, both gRNA-Nsf1 and gRNA-Nsf1-Nsf2, but not gRNA-Nsf2, effectively eliminated the staining specifically in C4da neurons (Figure 6, B–E). The gRNA-Nsf2 mildly reduced the staining (Figure 6, C–C’ and E), indicating that Nsf2 may stabilize Nsf1 in neurons.
Figure 6.
Clustered regularly interspaced short palindromic repeats-mediated tissue-restricted mutagenesis analyses of NSF and SNAP genes in C4da dendrite morphogenesis, (A–D’) Nsf1 staining revealed by anti-dNSF in ppk-Cas9 (A and A’), ppk-Cas9 gRNA-Nsf1 (B and B’), ppk-Cas9 gRNA-Nsf2 (C and C’), and ppk-Cas9 gRNA-Nsf1-Nsf2 (D and D’). Upper panels show Nsf1 staining. Lower panels show the Nsf1 staining (green) and C4da neurons labeled by ppk > CD4-tdTom (magenta). Yellow outlines indicate the location of the C4da cell body. (E) Quantification of Nsf1 staining in C4da neurons in the genotypes indicated. *** P ≤ 0.001; ns, not significant; one-way ANOVA and Tukey’s honest significant difference (HSD) test. (F–H) DdaC neurons in ppk-Cas9 (F), ppk-Cas9 gRNA-Nsf1-Nsf2 (G), and ppk-Cas9 gRNA-Snap24-Snap25-Snap29 (H). (I–K) DdaC neurons in SOP-Cas9 (I), SOP-Cas9 gRNA-Nsf1-Nsf2 (J), and SOP-Cas9 gRNA-Snap24-Snap25-Snap29 (K). (L and M) Quantification of total dendrite length (L) and dendrite density (M) in the genotypes indicated. Each circle represents an individual neuron: ppk-Cas9 (n = 22); ppk-Cas9 gRNA-Nsf1-Nsf2 (n = 16); ppk-Cas9 gRNA-Snap24-Snap25-Snap29 (n = 11); SOP-Cas9 (n = 15); SOP-Cas9 gRNA-Nsf1 (n = 15); SOP-Cas9 gRNA-Nsf2 (n = 15); and SOP-Cas9 gRNA-Nsf1-Nsf2 (n = 15). * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001; ns, not significant; one-way ANOVA and Tukey’s HSD test. (N–P) V’ada neurons in day 0 adults of ppk-Cas9 (N), ppk-Cas9 gRNA-Nsf1-Nsf2 (O), and ppk-Cas9 gRNA-Snap24-Snap25-Snap29 (P). (Q) Quantification of total dendrite length of adult v’ada neurons expressing ppk-Cas9 and the gRNAs indicated. Each circle represents an individual neuron: ppk-Cas9 (n = 14), ppk-Cas9 gRNA-Nsf1-Nsf2 (n = 23), or ppk-Cas9 gRNA-Snap24-Snap25-Snap29 (n = 20). *** P ≤ 0.001; one-way ANOVA and Dunnett’s test. (R–U) Epidermal cell morphology revealed by Nrg-GFP in hh-Cas9 (R), hh-Cas9 gRNA-Nsf1 (S), hh-Cas9 gRNA-Nsf2 (T), and hh-Cas9 gRNA-Nsf1-Nsf2 (U) at late second-instar larval stage. Yellow dotted lines in (U) indicate the presumed region of hh-Cas9-expressing cells. Black bar, mean; red bars, SD. Bar, 25 μm for (A–D’), and 50 μm for (F–P) and (R–U). AU, arbitrary units.
We next examined the functional consequences of knocking out the NSF and SNAP genes in C4da neurons using both ppk-Cas9 and SOP-Cas9. Removing individual NSF genes did not cause obvious dendritic reductions (Figure S1, A–C and H–J), but SOP-Cas9/gRNA-Nsf2 neurons instead showed a mild increase in dendrite length and density (Figure 6, L and M). Surprisingly, knocking out both NSF genes using ppk-Cas9 only produced weak and variable C4da dendrite reduction (Figure 6, F, G, L, and M). In contrast, SOP-Cas9/gRNA-Nsf1-Nsf2 animals showed consistent and much stronger dendrite reductions (Figure 6, I, J, L, and M). Adding an irrelevant gRNA transgene, gRNA-GFP, slightly alleviated the dendrite defects in NSF knockout induced by SOP-Cas9 (Figure 6, L and M). These data suggest that Nsf1 and Nsf2 act redundantly to promote dendrite growth. Furthermore, the observation that ppk-Cas9 caused a weaker phenotype than SOP-Cas9 suggests that NSF gene products made before postmitotic mutagenesis allow neurons to grow a significant amount of dendrites. Lastly, expressing more gRNA species could potentially reduce the mutagenic efficiency of each individual gRNA, possibly by competition for Cas9 proteins.
Tissue-specific knockout of individual SNAP genes using ppk-Cas9 or SOP-Cas9 produced either no obvious phenotypes (for Snap24 and Snap25) or weak dendrite reductions (for Snap29) (Figure S1, D–F and K–M). Knocking out both Snap24 and Snap25 similarly did not cause obvious dendrite defects (Figure S1, G and N). We next knocked out all three SNAP genes using gRNA-Snap24-Snap25-Snap29. While ppk-Cas9-mediated knockout only slightly reduced dendrite density (Figure 6, H, L, and M), SOP-Cas9-mediated knockout caused strong C4da dendrite reduction and degeneration (n = 16/19 neurons) in second-instar larvae (Figure 6K), and late larval lethality. Although SOP-Cas9 is highly efficient in da neurons, as shown by the NT3 negative tester (Figure S1O), this lethality might be independent of neuronal defects, because SOP-Cas9 also labeled a small number of random larval epidermal cells with the positive tester (Figure S1P). Nevertheless, our results suggest that, like NSF genes, all three SNAP genes are redundantly required in C4da neurons and that mutagenesis before the neuronal birth is required to unmask the LOF phenotype of SNAP genes.
As the SNARE machinery is required for all vesicle trafficking in the cell, we were curious to know why knocking out all NSF or all SNAP genes in neurons with SOP-Cas9 was not sufficient to suppress all dendritic growth. One possibility is that membrane trafficking-independent mechanisms exist that allow neurons to elaborate dendrites. Alternatively, NSF and SNAP gene products that are contributed maternally or made before SOP-Cas9 activity persist long enough to support a small degree of larval dendrite growth. To distinguish between these possibilities, we turned to adult C4da neurons. C4da neurons ddaC and v’ada prune all their dendrites during metamorphosis, and regrow new dendritic arbors in late pupae (Shimono et al. 2009). Because dendritic pruning removes all existing gene products except for the residual amounts left in the cell body, dendrite regrowth must rely on new transcription. If NSF and SNAP genes are required for all dendrite growth, knocking out all NSF or SNAP genes during larval stages should prevent dendrite regrowth. Indeed, adult v’ada neurons lacking Nsf1 Nsf2 or Snap24 Snap25 Snap29 via ppk-Cas9-mediated knockout either did not regrow primary branches or showed severe reduction in total dendrite length (Figure 6, N–Q). These data suggest that ppk-Cas9 can effectively remove redundant genes in postmitotic neurons and that neuronal dendrite growth absolutely requires SNARE function.
Lastly, we validated that CRISPR-TRiM can also be applied to remove gene functions in the larval epidermis. Using hh-Cas9, we knocked out NSF genes in posterior epidermal cells of each larval segment. Knocking out Nsf1 or Nsf2 individually did not cause obvious changes in epidermal cell morphology (Figure 6, R–T). However, knocking out both NSF genes caused epidermal cells to delaminate and become rounded (Figure 6U), and the animals died in the early third-instar larval stage, suggesting that NSF genes are redundant but together are essential in larval epidermal cells.
Discussion
In this study, we describe an optimized strategy that we call CRISPR-TRiM for tissue-specific gene mutagenesis using CRISPR/Cas9 in Drosophila. To implement this method, we developed a toolkit for generating and evaluating enhancer-driven Cas9 lines, created convenient cloning vectors for making efficient multi-gRNA transgenes, and established an assay for assessing the mutagenic efficiency of transgenic gRNAs. Using our CRISPR-TRiM tools, we demonstrate that postmitotic knockout of Ptp69D is sufficient to cause LOF in neurons, while SNARE complex components are strongly redundant and perdurant in supporting neuronal dendrite development.
Comparison of CRISPR-TRiM with other tissue-specific LOF methods
Flp/FRT-based mosaic analyses have been widely used to investigate the tissue-specific roles of genes in Drosophila (Griffin et al. 2014). Among these techniques, MARCM and its variants are considered gold standards for neuronal studies, due to the positive labeling of homozygous mutant cells and single-cell resolution (Lee and Luo 1999; Yu et al. 2009). However, MARCM and other Flp/FRT-based mosaic analyses also have some obvious limitations. First, they require preexisting mutations in the gene of interest recombined with FRT on the appropriate chromosome arm. Second, because these techniques rely on mitotic chromosome crossovers, which would result in wild-type “twin spots,” it is impossible to remove gene function in every cell of the tissue of interest. Third, these techniques require at least five genetic components in the final genotype, making it harder to introduce additional components. Lastly, generating cells mutant for multiple genes located on different chromosome arms is extremely difficult, if not impossible. In contrast, the bipartite CRISPR-TRiM system requires only transgenic components that are independent of all existing binary expression systems. Using efficient Cas9 and gRNA reagents, LOF in all cells of the target tissue can be expected. These features make CRISPR-TRiM much more convenient than traditional mosaic-based methods.
Compared to UAS-Cas9 driven by tissue-specific Gal4s, at least with the ppk enhancer, the CRISPR-TRiM system has the advantages of faster Cas9 expression (and therefore more complete LOF) and decreased cytotoxicity due to lower Cas9 expression levels. These advantages of enhancer-driven Cas9 are likely more important for studying early gene function in neuronal morphogenesis. An additional benefit of using enhancer-driven Cas9 is that the Gal4/UAS system is available for other genetic manipulations in the same experiment.
Over the last decade, several genome-wide UAS-RNAi resources have greatly accelerated gene identification and characterization in Drosophila (Dietzl et al. 2007; Ni et al. 2011). However, RNAi results in incomplete LOF and suffers from off-target effects (Ma et al. 2006). In comparison, CRISPR methods can generate true gene knockout, and ever-improving gRNA-selection algorithms have mostly mitigated the off-target effects (Chari et al. 2015; Doench et al. 2016; Haeussler et al. 2016). In addition, UAS-Dcr-2 overexpression, which is often necessary for maximizing the knockdown efficiency of dsRNAs, can also cause deleterious effects in the expressing cells. The CRISPR-TRiM method can avoid most of these concerns.
Caveats of CRISPR-TRiM and potential solutions
Due to the nature of CRISPR/Cas9-induced mutagenesis, CRISPR-TRiM will generate tissues composed of heterogenous cells carrying different mutations. This mosaicism could complicate phenotypic analysis, given that different mutations could impact gene function in diverse ways. Although immunostaining could alleviate this problem by revealing whether individual cells make the final protein product, antibodies are not always available nor are all assays compatible with immunostaining. For this reason, we recommend the use of at least two gRNAs for each target gene to enhance the chance of mutagenesis.
Nonetheless, even with multiple efficient gRNAs we observed that CRISPR-TRiM sometimes produced variable phenotypes among cells (e.g., Nsf1 Nsf2 knockout by ppk-Cas9, Figure 6, G and L), likely due to differences in the timing of mutagenesis and/or the nature of the mutations induced in different cells. This variability could actually be beneficial for the analysis of tissues like da neurons where each cell can be evaluated separately, as it could reveal a fuller spectrum of phenotypes associated with different strengths of LOF.
Designing efficient gRNA constructs and assessing gRNA efficiency
Our comparison of several dual-gRNA designs using the same targeting sequences revealed that the tgFE design is particularly efficient for mutagenesis in larval sensory neurons. The same design also performs well in other somatic tissues such as the larval epidermis. Because previous studies indicated that the use of tRNA in polycistronic gRNAs does not seem by itself to enhance mutagenesis (Port and Bullock 2016), the tgFE design’s high efficiency is likely due to the F+E gRNA scaffold. Whether this design also works well in the germ line for creating heritable mutations remains to be determined.
Although large deletions induced by two gRNAs would be more effective in causing LOF, we found that the frequency of large deletions in somatic cells is too low to be reliable. Therefore, to maximize the chance of LOF mutagenesis, we recommend selecting targeting sites in coding sequences shared by all protein isoforms, preferably in conserved protein domains. In our experience, choosing common top hits by using multiple experimentally validated gRNA selection algorithms (Moreno-Mateos et al. 2015; Doench et al. 2016; Chari et al. 2017) usually yields very efficient gRNAs.
We also recommend evaluating the in vivo efficiency of gRNA lines using the Cas9-LEThAL assay before conducting CRISPR-TRiM analyses. In our hands, the lethal phase of male progeny in this assay reliably indicates gRNA efficiency for our CRISPR-TRiM experiments.
CRISPR-TRiM reveals gene functions in neuronal morphogenesis
Our results of CRISPR-TRiM analysis in the dendrite morphogenesis of C4da neurons show that the timing of mutagenesis and the perdurance of gene products influence the extent of LOF; therefore, these parameters must be considered when choosing the most appropriate Cas9 line. The CRISPR-TRiM analysis of Ptp69D shows that postmitotic mutagenesis is sufficient to cause its LOF, because Ptp69D either is expressed late in neuronal development or turns over quickly. In contrast, SNAP and NSF proteins are likely contributed maternally and made throughout the neuronal lineage. The early acting SOP-Cas9 is therefore required to reveal SNARE LOF phenotypes in neurons. Moreover, dendrite regrowth of adult C4da neurons provides an opportunity to unmask fully the requirements of SNARE components for dendrite morphogenesis. This technique should be useful to circumvent potential perdurance because gene products are removed by dendrite pruning prior to the regrowth. Our results imply that perdurance could be an underappreciated concern for studying the developmental roles of housekeeping genes in any mutation-based LOF analysis.
The potential redundancies of SNARE components in neuronal morphogenesis have been mostly elusive. The roles of Drosophila Snap24 and Snap25 in the dendrite growth of da neurons have been investigated by RNAi, but knockdown of each gene only resulted in minor defects (Peng et al. 2015). At the neuromuscular junction, Snap25 mutations were found to cause defective synaptic transmission only in pharate adults, prompting the hypothesis that Snap24 and Snap25 play redundant roles in larval neurotransmission (Vilinsky et al. 2002). However, due to an inability to remove multiple SNAP genes, these previous studies were unable to determine whether Drosophila SNAP genes are redundant in neural development. Using CRISPR-TRiM to mutate SNAP genes in combination, we show that SNAP genes are highly redundant in C4da neurons and that the removal of all three genes is required to block all dendrite branching morphogenesis. Our NSF LOF data demonstrate that Nsf1 and Nsf2 also play redundant roles in C4da neurons, which is consistent with the previous finding that these two genes can substitute for each other in the nervous system (Golby et al. 2001). Interestingly, we observed a distinction between NSF and SNAP LOF phenotypes. Both with SOP-Cas9 in the larva and ppk-Cas9 in the adult, SNAP LOF appears to produce a more severe dendritic reduction than NSF LOF. This distinction likely reflects the different roles of these proteins in the SNARE machinery. Because NSFs are responsible for recycling the SNARE complex after membrane fusion, newly synthesized SNARE components can still mediate vesicle fusion in the absence of NSFs. In contrast, SNAP LOF causes secretion to stop completely, thereby generating a stronger phenotype.
The Drosophila genome contains a large number of paralogous genes that may carry redundant functions. The lack of efficient ways to remove multiple genes simultaneously in specific tissues has hampered the characterization of these genes in development. The CRISPR-TRiM tools that we present here offer an efficient and convenient way for investigating not only the developmental roles of individual genes, but also those of potential redundant gene groups. These tools can be applied to address a broad range of developmental, cell biological, and physiological questions in Drosophila.
Acknowledgments
We thank Ying Peng, Yi Guo, and the Bloomington Drosophila Stock Center for fly stocks; Norbert Perrimon and Addgene for plasmids; Leo Pallanck for antibodies; Richard Ordway for personal communications; Lu Zhu for technical help; and Michael Goldberg, Mariana Wolfner, David Deitcher, Dion Dickman, and Quan Yuan for critical reading and suggestions on the manuscript. This work was supported by a Cornell Fellowship awarded to H.J., and a Cornell start-up fund and National Institutes of Health grants (R01 NS-099125 and R21 OD-023824) awarded to C.H. The authors declare no competing financial interests.
Author contributions: C.H. and A.R.P. designed the experiments. B.W. and Y.H. conducted molecular cloning. A.R.P. performed imaging and quantification. C.H., A.R.P., M.L.S., and H.J. built genetic reagents used in this study. K.L., T.O., R.F., M.G., and Y.Q. screened Cas9 transgenic lines. C.H. and A.R.P. wrote the manuscript.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7399514.
Communicating editor: L. Luo
Literature Cited
- Bassett A. R., Tibbit C., Ponting C. P., Liu J. L., 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4: 220–228. 10.1016/j.celrep.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari R., Mali P., Moosburner M., Church G. M., 2015. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods 12: 823–826. 10.1038/nmeth.3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari R., Yeo N. C., Chavez A., Church G. M., 2017. sgRNA scorer 2.0: a species-independent model to predict CRISPR/Cas9 activity. ACS Synth. Biol. 6: 902–904. 10.1021/acssynbio.6b00343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Gilbert L. A., Cimini B. A., Schnitzbauer J., Zhang W., et al. , 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491 [corrigenda: Cell 156: 373 (2014)] 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culi J., Modolell J., 1998. Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 12: 2036–2047. 10.1101/gad.12.13.2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai C. J., Popova E., Zinn K., 1994. A Drosophila receptor tyrosine phosphatase expressed in the embryonic CNS and larval optic lobes is a member of the set of proteins bearing the “HRP” carbohydrate epitope. J. Neurosci. 14: 7272–7283. 10.1523/JNEUROSCI.14-12-07272.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Doench J. G., Fusi N., Sullender M., Hegde M., Vaimberg E. W., et al. , 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34: 184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen-Campen B., Yang-Zhou D., Fernandes V. R., Gonzalez D. P., Liu L. P., et al. , 2017. Optimized strategy for in vivo Cas9-activation in Drosophila. Proc. Natl. Acad. Sci. USA 114: 9409–9414. 10.1073/pnas.1707635114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F., III, 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31: 397–405. 10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Tibbit C., Liu J. L., 2016. Effective knockdown of Drosophila long non-coding RNAs by CRISPR interference. Nucleic Acids Res. 44: e84 10.1093/nar/gkw063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golby J. A., Tolar L. A., Pallanck L., 2001. Partitioning of N-ethylmaleimide-sensitive fusion (NSF) protein function in Drosophila melanogaster: dNSF1 is required in the nervous system, and dNSF2 is required in mesoderm. Genetics 158: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. 10.1534/genetics.113.152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., et al. , 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961–971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin R., Binari R., Perrimon N., 2014. Genetic odyssey to generate marked clones in Drosophila mosaics. Proc. Natl. Acad. Sci. USA 111: 4756–4763. 10.1073/pnas.1403218111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber W. B., Jan L. Y., Jan Y. N., 2002. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development 129: 2867–2878. [DOI] [PubMed] [Google Scholar]
- Grueber W. B., Ye B., Moore A. W., Jan L. Y., Jan Y. N., 2003. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr. Biol. 13: 618–626. 10.1016/S0960-9822(03)00207-0 [DOI] [PubMed] [Google Scholar]
- Haeussler M., Schonig K., Eckert H., Eschstruth A., Mianne J., et al. , 2016. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17: 148 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Jan L. Y., Jan Y. N., 2011. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc. Natl. Acad. Sci. USA 108: 9673–9678. 10.1073/pnas.1106386108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A., Rubin G. M., Ngo T. T., Shepherd D., Murphy C., et al. , 2012. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2: 991–1001. 10.1016/j.celrep.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Brueggeman A. J., Horken K. M., Plucinak T. M., Weeks D. P., 2014. Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot. Cell 13: 1465–1469. 10.1128/EC.00213-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepper T. H., Kienle C. N., Fasshauer D., 2007. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol. Biol. Cell 18: 3463–3471. 10.1091/mbc.e07-03-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Ueda R., 2013. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195: 715–721. 10.1534/genetics.113.156737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon E. Z., Kazmar T., Stampfel G., Yanez-Cuna J. O., Pagani M., et al. , 2014. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512: 91–95. 10.1038/nature13395 [DOI] [PubMed] [Google Scholar]
- Lai E. C., Orgogozo V., 2004. A hidden program in Drosophila peripheral neurogenesis revealed: fundamental principles underlying sensory organ diversity. Dev. Biol. 269: 1–17. 10.1016/j.ydbio.2004.01.032 [DOI] [PubMed] [Google Scholar]
- Lee H. B., Sebo Z. L., Peng Y., Guo Y., 2015. An optimized TALEN application for mutagenesis and screening in Drosophila melanogaster. Cell. Logist. 5: e1023423 10.1080/21592799.2015.1023423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L., 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Ma Y., Creanga A., Lum L., Beachy P. A., 2006. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443: 359–363. 10.1038/nature05179 [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos M. A., Vejnar C. E., Beaudoin J. D., Fernandez J. P., Mis E. K., et al. , 2015. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12: 982–988. 10.1038/nmeth.3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Q., Zhou R., Czech B., Liu L. P., Holderbaum L., et al. , 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8: 405–407. 10.1038/nmeth.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Pareja J. C., Xu T., 2011. Shaping cells and organs in Drosophila by opposing roles of fat body-secreted Collagen IV and perlecan. Dev. Cell 21: 245–256. 10.1016/j.devcel.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Lee J., Rowland K., Wen Y., Hua H., et al. , 2015. Regulation of dendrite growth and maintenance by exocytosis. J. Cell Sci. 128: 4279–4292. 10.1242/jcs.174771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D., Truman J. W., Rubin G. M., 2012. Using translational enhancers to increase transgene expression in Drosophila. Proc. Natl. Acad. Sci. USA 109: 6626–6631. 10.1073/pnas.1204520109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poe A. R., Tang L., Wang B., Li Y., Sapar M. L., et al. , 2017. Dendritic space-filling requires a neuronal type-specific extracellular permissive signal in Drosophila. Proc. Natl. Acad. Sci. USA 114: E8062–E8071. 10.1073/pnas.1707467114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Bullock S. L., 2016. Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods 13: 852–854. 10.1038/nmeth.3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H. M., Lee T., Bullock S. L., 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111: E2967–E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L. M., Zur Lage P. I., Prentice D. R., Senthinathan B., Jarman A. P., 2004. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol. Cell. Biol. 24: 9517–9526. 10.1128/MCB.24.21.9517-9526.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Sun J., Housden B. E., Hu Y., Roesel C., et al. , 2013. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. USA 110: 19012–19017. 10.1073/pnas.1318481110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Yang Z., Xu J., Sun J., Mao D., et al. , 2014. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 9: 1151–1162. 10.1016/j.celrep.2014.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebo Z. L., Lee H. B., Peng Y., Guo Y., 2014. A simplified and efficient germline-specific CRISPR/Cas9 system for Drosophila genomic engineering. Fly (Austin) 8: 52–57. 10.4161/fly.26828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono K., Fujimoto A., Tsuyama T., Yamamoto-Kochi M., Sato M., et al. , 2009. Multidendritic sensory neurons in the adult Drosophila abdomen: origins, dendritic morphology, and segment- and age-dependent programmed cell death. Neural Dev. 4: 37 10.1186/1749-8104-4-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilinsky I., Stewart B. A., Drummond J., Robinson I., Deitcher D. L., 2002. A Drosophila SNAP-25 null mutant reveals context-dependent redundancy with SNAP-24 in neurotransmission. Genetics 162: 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler K. S., Wakimoto B. T., 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29: 577–605. 10.1146/annurev.ge.29.120195.003045 [DOI] [PubMed] [Google Scholar]
- Wickner W., Schekman R., 2008. Membrane fusion. Nat. Struct. Mol. Biol. 15: 658–664. 10.1038/nsmb.1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Minkenberg B., Yang Y., 2015. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 112: 3570–3575. 10.1073/pnas.1420294112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z., Ren M., Wu M., Dai J., Rong Y. S., et al. , 2014a. Efficient gene knock-out and knock-in with transgenic Cas9 in Drosophila. G3 (Bethesda) 4: 925–929. 10.1534/g3.114.010496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z., Wu M., Wen K., Ren M., Long L., et al. , 2014b. CRISPR/Cas9 mediates efficient conditional mutagenesis in Drosophila. G3 (Bethesda) 4: 2167–2173. 10.1534/g3.114.014159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. H., Chen C. H., Shi L., Huang Y., Lee T., 2009. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nat. Neurosci. 12: 947–953. 10.1038/nn.2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Kawasaki F., Ordway R. W., 2011. Activity-dependent interactions of NSF and SNAP at living synapses. Mol. Cell. Neurosci. 47: 19–27. 10.1016/j.mcn.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Ren M., Wang Z., Zhang B., Rong Y. S., et al. , 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 195: 289–291. 10.1534/genetics.113.153825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplemental Materials, including supplemental figures, supplemental tables, methods, and supplemental references, are available at Figshare: https://doi.org/10.25386/genetics.7399514. Plasmids are available at Addgene or upon request. Drosophila lines are available at the Bloomington Drosophila Stock Center or upon request.