Abstract
Pattern-recognition receptors (PRRs), which are single transmembrane proteins belonging to the receptor-like kinase (RLK) and receptor-like protein (RLP) super families, sense microbe- and host-derived molecular patterns to activate immune responses in plants. PRRs associate with co-receptors, scaffold proteins and receptor-like cytoplasmic kinases (RLCKs) to form immune receptor complexes at the cell surface, allowing activation of cellular responses upon perception of extracellular ligands. Recent advances have uncovered new mechanisms by which these immune receptor complexes are regulated at the levels of composition, stability and activity. It has become clear that RLCKs are central components directly linking PRRs to multiple downstream signalling modules. Furthermore, new studies have provided important insights into the regulation of reactive oxygen species, mitogen-activated protein (MAP) kinase cascades and heterotrimeric G proteins, which has not only deepened our understanding of immunity, but also expanded our view of transmembrane signalling in general.
This article is part of the theme issue ‘Biotic signalling sheds light on smart pest management’.
Keywords: innate immunity, receptor-like kinases, MAP kinases, heterotrimeric G proteins
1. Introduction
In the absence of an adaptive immune system, plants rely on cell surface-localized and intracellular immune receptors to detect potential pathogens. A large number of proteins belonging to receptor-like kinase (RLK) and receptor-like protein (RLP) super families act as cell surface immune receptors or components of receptor complexes in plant disease resistance. Some of these proteins function as surface-localized pattern-recognition receptors (PRRs) or components of PRR complexes to perceive microbe- and host-derived molecular patterns associated with pathogen attacks and trigger defences in plants [1]. Meanwhile, others recognize apoplastic effectors to trigger plant immunity [2]. On the other hand, cytoplasmic nucleotide binding leucine-rich repeat domain-containing receptors (NLRs) recognize pathogen-secreted cytoplasmic effectors to trigger immune responses in plants [3].
Plant PRRs perceive ligands of diverse biochemical nature through their variable ectodomains (ECDs). PRRs form dynamic receptor complexes with co-receptors, which participate in the perception of ligands and signalling. PRRs also dynamically interact with receptor-like cytoplasmic kinases (RLCKs), which play key roles in transducing signals from PRRs to downstream signalling. Readers are referred to recent reviews for detailed coverage [4–6].
New studies show that the composition of PRR complexes and the regulation of their stability and activity are more complex than previously thought. In addition, we are beginning to unravel how PRRs regulate major downstream signalling events that are crucial for plant defences. In this review, we highlight recent data and discuss how the new findings have advanced our understanding of pattern-triggered immunity.
2. Composition of pattern-recognition receptor complexes
RLCKs have emerged as central players in receptor complexes, linking PRRs to downstream signalling [6]. RLCKs involved in pattern-triggered immunity include family VII, which contains 46 members, and family XII, which contains 12 members [7]. Among these, several RLCKs including BOTRYTIS-INDUCED KINASE1 (BIK1) and PBS1-LIKE1 (PBL1) have been known to play roles in pattern-triggered immunity by directly interacting with PRRs, but whether different RLCK members are differentially recruited to distinct PRRs remains elusive. A recent systematic analysis of higher-order mutants showed that clades 5, 7 and 8 of RLCK VII are genetically linked to multiple PRRs, while clade 4 is specifically required for signalling downstream of chitin receptors [8], indicating that numerous RLCK VII members function as crucial components in multiple PRR complexes and a specific subgroup of RLCK VII proteins are recruited by chitin receptors.
In addition to PRRs and RLCKs, recent studies have uncovered increasingly complex PRR complexes. FLAGELLIN SENSING 2 (FLS2) and ELONGATION FACTOR-TU (EF-Tu) RECEPTOR (EFR), which are receptors recognizing bacterial flagellin epitope flg22 and bacterial elongation factor-TU epitope elf18, respectively, have been shown to associate with FERONIA (FER), a receptor of RAPID ALKALIZATION FACTORs (RALFs) [9,10]. FER also weakly interacts with BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), which is a co-receptor for both FLS2 and EFR, to promote pattern-induced FLS2–BAK1 and EFR–BAK1 interactions [10], suggesting that FER functions as a scaffold protein for ligand-induced dimerization of the receptors and co-receptor. The glycosylphosphatidylinositol (GPI)-anchored protein LLG1, which was first identified as a chaperon and co-receptor of FER, was recently shown to interact constitutively with FLS2 and EFR and form a complex with BAK1 in a ligand-dependent manner [11,12]. LLG1 is required for steady state accumulation of FLS2 in the resting state and ligand-induced degradation of the FLS2 protein [12], indicating a crucial role of LLG1 in the dynamic control of FLS2. It should be noted that a number of RLPs such as lysin motif (LYM)-containing proteins possess GPI [13], raising a possibility that GPI-anchored proteins are broadly involved in the regulation of cell surface receptors. In addition, the leucine-rich repeat (LRR)-RLK FLS2-INTERACTING RECEPTOR (FIR) was recently identified as a component of FLS2 complex by a high-throughput interactome study [14]. FIR interacts with both FLS2 and BAK1 and facilitates flg22-triggered FLS2–BAK1 complex formation [14]. In addition, another small LRR–RLK APEX constitutively associates with both PLANT ELICITOR PEPTIDE RECEPTOR 1 (PEPR1) and PEPR2, which perceive plant elicitor peptides (Peps) [14]. Interestingly, both loss-of-function mutation and overexpression of APEX lead to diminished Pep signalling, indicating that appropriate APEX dosage is required for PEPR complex formation or signalling. Furthermore, the Arabidopsis malectin-like/LRR–RLK IMPAIRED OOMYCETE SUSCEPTIBILITY1 (IOS1) interacts with multiple PRRs and is critical for pattern-triggered immunity [15]. These findings suggest that PRR complexes are composed of multiple components, which allow plants to initiate robust immune response once stimulated by molecular patterns.
Inappropriate and excessive immune signalling is detrimental to plants. Thus, negative regulatory components must be included in PRR complexes. Before pattern perception, PRR complexes must be maintained at a resting state. For example, a LRR–RLK BAK1-INTERACTING RECEPTOR-LIKE KINASE2 (BIR2) interacts with BAK1 and prevents unwanted interactions with PRRs in the absence of patterns [16]. After pattern perception, BIR2 dissociates from BAK1 and results in PRR–BAK1 complex formation. Recently, it was reported that another LRR–RLK BIR3 interacts with not only BAK1 but also PRRs to prevent PRR–BAK1 complex formation and immune activation before pattern perception [17]. As with BIR2, BIR3 is released from BAK1 after ligand-induced activation of the PRR complexes. These findings indicated that PRR complexes are highly regulated in the resting state to avoid abnormal immune activation. In addition, ligand-induced PRR complex formation is also tightly controlled. For example, two malectin-like RKs ANXUR1 (ANX1) and ANX2, which are receptors of RALF4 and RALF19, negatively regulate immune responses triggered by multiple patterns [18,19]. They constitutively associate with both FLS2 and BAK1, but interfere with flg22-induced FLS2–BAK1 complex formation to prevent excessive immune activation [19]. In the future, it will be interesting to test whether these negative regulatory components compete with positive regulatory components in the formation of PRR complexes.
3. Regulation of protein phosphorylation and stability in pattern-recognition receptor complexes
In addition to dynamic regulation of the composition of PRR complexes, the stability and phosphorylation of PRR complex components are also subject to delicate regulation. A most recent study showed that flg22 and elf18 induce BAK1 phosphorylation at Ser602, Thr603, Ser604 and Ser612 in vivo, and these four phosphosites are required for flg22 but not brassinosteroid (BR) signalling [20]. In addition to serine/threonine phosphorylation, an increasing number of RLKs have been shown to undergo tyrosine phosphorylation, and this modification is important for their activation [20–23]. A Tyr residue is present in kinase subdomain VIa (Tyr-VIa) and conserved in about 80% Arabidopsis LRR–RLKs. The Tyr-VIa (Tyr403) phosphorylation of co-receptor BAK1 is required for elf18 but not BR signalling [20]. Interestingly, the Tyr-VIa position is conserved in EFR but not in BR INSENSITIVE 1 (BRI1), the receptor of BR, indicating that the common co-receptor may differentially regulates two class of LRR–RLKs based on the presence or absence of this Tyr residue in the receptors [20]. Indeed, elf18-triggered Tyr-VIa (Tyr836) phosphorylation of EFR is required for EFR activation and elf18-triggered immunity [20,21]. Moreover, the Tyr-VIa (Tyr428) phosphorylation of CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1), a co-receptor for LysM-type receptor kinases, is also indispensable for CERK1 activation upon chitin perception [22,23]. These findings suggest that Tyr-VIa phosphorylation serves as a phospho-code determining specificity of RLKs [20].
In addition to PRRs and co-receptors, RLCKs are also subject to phosphorylation. For example, BIK1 is autophosphorylated and/or transphosphorylated by BAK1 on Tyr243 and Tyr250 in vitro. Genetic analysis indicated that Tyr243 and Tyr250 are required for immune function of BIK1 [24]. A recent study suggested that BIK1 is directly phosphorylated at Ser89 and Thr90 by EFR, and that is required for elf18-triggered growth inhibition [25]. BIK1 has been shown to trans-phosphorylate BAK1 in vitro, but it remains unknown whether BIK1 and other RLCKs in turn phosphorylate PRRs and co-receptors in vivo. The redundancy of various RLCK members makes it difficult to determine whether they are required for the observed phosphorylation in vivo. To this end, a series of rlck vii higher order mutants have been constructed, which may provide important genetic resources for this analysis. For example, the rlck vii-4 sextuple mutant is severely impaired in chitin-triggered immune responses [8]. This mutant will be suitable for analyses of RLCK-mediated CERK1 phosphorylation.
Protein stability of the PRR immune components is also tightly controlled. For example, FLS2 stability is regulated by 26S proteasome pathway, which has been discussed by several excellent reviews [4,26,27]. Recent studies showed that BIK1 is also subject to degradation by 26S proteasome pathway. It was reported that CALCIUM-DEPENDENT PROTEIN KINASE 28 (CPK28) constitutively associates with BIK1 to negatively control BIK1 accumulation in a proteasome-dependent manner [28]. Another study showed that Arabidopsis heterotrimeric G proteins are directly coupled to the FLS2-BIK1 complex and attenuate the proteasome-dependent degradation of BIK1 in the resting state [29]. These findings indicated that BIK1 accumulation is tightly controlled by multiple components and the detailed mechanism has been uncovered by a most recent study [30]. A pair of closely related ubiquitin E3 ligases PLANT U-BOX25 (PUB25) and PUB26 poly-ubiquitinate and promote degradation of non-activated BIK1 [30]. Interestingly, both CPK28 and heterotrimeric G proteins regulate BIK1 stability through PUB25/26. Before ligand perception, heterotrimeric G proteins directly inhibit PUB25/26 E3 activity to stabilize BIK1, ensuring optimum signalling competence. Upon ligand-induced PRR activation, CPK28 directly phosphorylates PUB25/26 to enhance the E3 ligase activity and accelerates degradation of non-activated BIK1, preventing over-accumulation of activated BIK1 and excessive immunity. At the same time, the activated BIK1 is protected from degradation, allowing it to activate downstream signalling [30]. These findings suggested that PUB25/26, CPK28 and heterotrimeric G proteins form a regulatory module to fine-tune the homeostasis of BIK1. Interestingly, a recent study showed that serine/threonine kinase 1 (SIK1) also stabilizes BIK1 in the resting state, indicating that BIK1 stability is tightly regulated by multiple mechanisms [31]. Similarly, the turnover of non-activated OsRLCK176 is also regulated by OsCPK4 in rice [32], indicating that there is a conserved regulatory mechanism for RLCK stability in Arabidopsis and rice.
4. Activation of downstream signalling components
Increasing evidence suggests that RLCKs regulate multiple early signalling events in the vicinity of plasma membrane [33–37]. A recent study reported that a sub-population of BIK1 localizes to the nucleus, suggesting a direct regulation of nuclear events, a possibility requires future attention [25]. Recent advances show that RLCKs directly regulate heterotrimeric G protein-mediated signalling, production of reactive oxygen species (ROS), and mitogen-activated protein kinase (MAPK) activation through phosphorylation-relay.
(a). Calcium influx
Pattern-triggered transient calcium influx is one critical cellular response, which almost participates in all the cellular responses [38]. Recent work showed that BIK1 and PBL1 are required for FLS2-, EFR-, PEPR1- and PEPR2-mediated calcium influx, suggesting that BIK1 and PBL1 may directly or indirectly regulate the unknown calcium channel in calcium influx [36,37]. Although plasma membrane-localized calcium permeable channels such as cyclic nucleotide-gated channels (CNGCs), ionotropic glutamate receptors (GLRs) and reduced hyperosmolality-induced [Ca2+] increase 1 (OSCA1) exist in plants [39], no genetic evidence has supported the involvement of these channels in pattern-triggered calcium influx. It was reported that nuclear membrane-localized CNGC15 and Ca2+-dependent adenosine triphosphatase (Ca2+-ATPase) facilitate symbiotic calcium oscillations in response to symbiotic elicitors [40,41]. Interestingly, flg22 also induces calcium oscillation in the guard cell and the pumps Ca2+-ATPase 8 (ACA8) and ACA10 are required for FLS2-mediated immunity [42,43], suggesting that calcium channels and pumps may be similarly involved in pattern-triggered calcium oscillation. In addition, plasma membrane-localized GLR3.3 and GLR3.6 have been implicated in wound-induced systemic electrical signalling and response to aphid feeding [44,45]. The GLR-mediated calcium influx further activates vacuolar ion channel TWO-PORE CHANNEL1 (TPC1) to release vacuolar Ca2+, activating defences against aphids [45]. It is worth noting that the aphid-elicited calcium influx depends on BAK1, suggesting that unknown PRRs are involved in plant responses to aphids [45]. Consistent with this notion, treatment of plants with inhibitors of iGluR (mammalian GLR homologues) attenuates pattern-triggered calcium influx [46]. Thus, a major task in the future will be to identify calcium channels involved in pattern-triggered calcium influx and to investigate the mechanisms by which PRRs activate these channels.
(b). Heterotrimeric G protein activation
G protein activation is another critical response. Heterotrimeric G proteins are common signalling components in eukaryotes, which are composed of α, β and γ subunits. In animals, the activation of heterotrimeric G protein is controlled through interaction with seven-transmembrane G protein-coupled receptors (GPCRs). In the resting state, a GDP-bound Gα interacts with a Gβγ dimer to form an inactive heterotrimer. Upon activation by ligands, GPCRs promote GDP–GTP exchange in the Gα subunit, leading to Gα activation and the dissociation of Gα from the Gβγ subunits [47,48]. Gα has intrinsic GTPase activity to hydrolyse of GTP, allowing Gα to cycle back to the GDP-bound resting state [49]. A class of regulator of G protein signalling (RGS) proteins act as GTPase accelerating proteins (GAPs) to negatively regulate G protein signalling. Arabidopsis contains four Gα proteins (GPA1, EXTRA-LARGE GTP-BINDING PROTEIN1 (XLG1), XLG2, and XLG3), one Gβ protein (AGB1), and three Gγ proteins (AGG1, AGG2, and AGG3) [50]. Plant Gα proteins also have spontaneous GTP hydrolysis activity, which is similarly enhanced by RGS proteins [51,52], although some plant species appear to use alternative regulatory proteins to enhance Gα GTP hydrolysis [53].
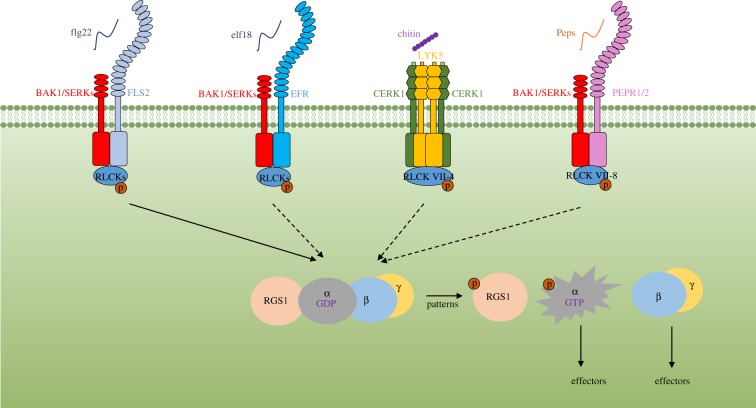
Unlike animals, plants contain no functional GPCR proteins. Instead, plant heterotrimeric G proteins are coupled to receptor kinases, including FLS2 [29,54,55]. In soya bean, Gα proteins interact with RLK Nod factor receptor 1 (NFR1) to control nodule formation, and RGS proteins negatively regulate Gα protein activity during this process [55]. In this case, NFR1 phosphorylates and enhances the GAP activity of RGS proteins, indicating a distinct regulatory mechanism of G protein signalling. A recent study showed that G protein inactivation/activation in the FLS2 receptor complex is directly controlled by RGS1 (figure 1; [56]). In the resting state, RGS1 directly interacts with and maintains Gα proteins in a GDP-bound form by enhancing GTP hydrolysis. RGS1 also directly associates with FLS2–BIK1 complex. As such, RGS1 maintains the G proteins in the resting state. After flg22 perception, BIK1 and its related PBS1-Like (PBL) kinases directly phosphorylate RGS1 at multiple sites including Ser428 and Ser431 to trigger its dissociation from FLS2 and Gα proteins. This leads to the de-repression of Gα proteins and G protein activation (figure 1). In addition to flg22, elf18, chitin, and Pep2 also trigger RGS1 phosphorylation (figure 1; [56]), indicating that the aforementioned mechanism of G protein regulation is used by multiple PRRs.
Figure 1.
A model for G protein activation by pattern-recognition receptors (PRRs). In the resting state, regulator of G protein signalling 1 (RGS1) associates with PRR complexes to maintain the Gα protein in the inactive state. Upon perception of patterns, RLCKs are activated, which in turn phosphorylate RGS1. Phosphorylation on RGS1 leads to its dissociation from the Gα proteins, and the G proteins are auto-activated in the absence of RGS1-mediated inhibition. The activated G proteins then positively regulate downstream immune signalling. SERKs, SOMATIC EMBRYOGENESIS RECEPTOR KINASES; p, phosphorylation.
In addition to phosphorylating RGS1, BIK1 also directly phosphorylates XLG2/3 N-terminus after flg22 treatment, which is required for full activation of ROS production and disease resistance to Pseudomonas syringae. However, the N-terminus of XLG2 is dispensable for the G protein-mediated stabilization of BIK1 and G protein regulation by RGS1 [29,56], suggesting that the phosphorylation of XLG2 independently modulates immune responses through an unknown mechanism (figure 1).
(c). Reactive oxygen species
Pattern-triggered ROS production is a robust immune signalling readout, which plays a crucial role in stomatal closure and callose deposition. In plants, pattern-triggered ROS production is largely produced in apoplast [57]. In some cases, ROS can be produced in chloroplasts, peroxisomes and mitochondria [58]. Pattern-triggered apoplastic ROS production is fast and transient, and can reach peak within 30 min. Pattern-triggered ROS production in the apoplast is mainly mediated by a plasma membrane-localized NADPH oxidase called Respiratory Burst Oxidase Homologue D (RBOHD) and cell wall peroxidases [57,59]. Before activation, RBOHD directly associates with multiple PRR complexes. Upon activation of the PRR complexes, RBOHD is phosphorylated by multiple kinases including BIK1, PBL1, CPK5 and SIK1 [31,35,37,60]. RBOHD activity is further regulated by binding to calcium and phosphatidic acid (PA), and interaction with G proteins [29,61,62]. Recent studies showed that in addition to RLCK VII-8 including BIK1 and PBL1, RLCK II-5 and -7 are genetically required for ROS production triggered by multiple patterns, while RLCK VII-4 members function specifically in CERK1-mediated ROS production [8], although it remains to be tested whether these RLCK VII members directly phosphorylate RBOHD to activate ROS production.
Interestingly, a more recent study showed that in addition to apoplastic ROS production, lipopolysaccharides (LPS) also trigger second long-lasting ROS production [63]. Microscopic observation demonstrated that the LPS-triggered second ROS production is largely associated with chloroplasts, which only partially depends on LPS receptor LORE [63], indicating that the additional PRRs or other components may be involved in LPS recognition and/or response.
(d). Mitogen-activated protein kinase activation
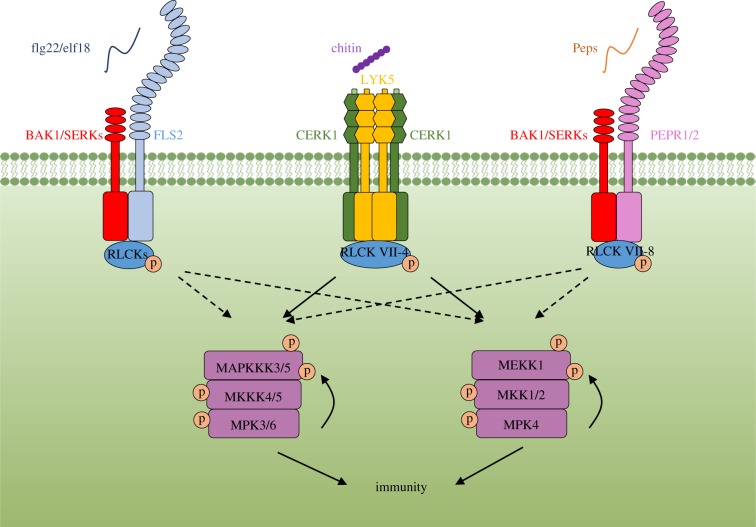
MAPK activation plays vital roles in the establishment of disease resistance [64]. Patterns commonly trigger activation of two MAPK cascades within minutes. One cascade is composed of MAPK kinase kinase (MAPKKK) MEKK1, MAPKKs MKK1 and MKK2, and a MAPK MPK4 [65,66]. The second cascade is composed of two MAPKKs, MKK4 and MKK5, and two MAPKs, MPK3 and MPK6 [67]. The identity of MAPKKKs upstream of MPK3/6 has been debated for more than 16 years. A previous study reported that MAPKKK5 positively regulates chitin-triggered MPK3/6 activation but negatively regulates flg22-triggered MPK3/6 [68], whereas another study reported that MAPKKK5 positively regulates flg22- rather than chitin-triggered MPK3/6 activation [69]. Until recently, two independent groups showed that the closely related MAPKKK3/5 function redundantly to activate MPK3/6 downstream of multiple PRRs (figure 2; [70,71]). In mapkkk3 mapkkk5 double mutants, the activation of MPK3/6 is greatly compromised, but not abolished, indicating that additional MAPKKKs are involved. It is possible that ARABIDOPSIS NUCLEUS- AND PHRAGMOPLAST-LOCALIZED KINASE1 (NPK1)-RELATED PROTEIN KINASEs (ANPs) function redundantly with MAPKKK3/5 in pattern-triggered MPK3/6 activation, as they are reported to be required for MAPK activation triggered by oligogalacturonides [72]. YODA (YDA), a MAPKKK closely related to MAPKKK3/5, is known to form a cascade with MKK4/5 and MPK3/6 to negatively regulate stomatal development [73]. Surprisingly, silencing of YDA resulted in a stronger activation of MAPKs triggered by flg22, and mapkkk3 mapkkk5 mutation suppressed the developmental defects of yda [71], indicating that antagonistic interactions exist between these two MAPK pathways. Interestingly, MAPKKK7 also plays a negative role in flg22-induced MAPK activation [74], providing another example of antagonism among distinct MAPKKKs. It may well be that YODA and MAPKKK7 compete for MKK4/5 binding with the MAPKKK3/5.
Figure 2.
Pattern-recognition receptor (PRR)-mediated activation of mitogen-activated protein kinase (MAPK) cascades. In the chitin-triggered immune pathway, the chitin co-receptor CERK1 phosphorylates RLCK VII-4 members, which then phosphorylate the MAPK kinase kinases MAPKKK3/5 and MEKK1. This phosphorylation positively regulates the activation of two MAPKs, MPK3/6 and MPK4, respectively. The activated MPK3/6 and MPK4 further phosphorylate MAPKKK5 and MEKK1, respectively, further enhancing their activity. Other PRRs such as FLS2 and EFR activate MAPK cascade likely by employing other RLCKs, whose identity remains to be defined. SERKs, SOMATIC EMBRYOGENESIS RECEPTOR KINASES; p, phosphorylation.
Another important endeavour of MAPK studies is to understand how PRRs regulate their activation. A number of studies have implicated a role of RLCKs in connecting PRRs to the activation of MAPK cascades [69,75–77]. However, the subtle phenotypes of rlck single or double mutants are difficult to reproduce owing to their functional redundancy. Systematic analyses of higher order mutants showed that RLCK VII-4 members play a major role in chitin-triggered MAPK activation [8]. These members directly phosphorylate MAPKKK5 at Ser599 and MEKK1 at Ser603 to activate MPK3/6 and MPK4, respectively (figure 2; [70]). MAPKKK3/5 are similarly phosphorylated in response to diverse patterns including flg22, elf18, and Pep2 [70]. Importantly, phospho-dead mutations in MAPKKK5Ser599 and MEKK1Ser603 not only abolish the chitin-triggered MPK activation, but also flg22-triggered MPK activation, indicating that these phosphorylations are similarly regulated by various RLCKs downstream of different PRRs (figure 2; [70]). The complete sets of RLCKs responsible for MAPK activation downstream of FLS2, EFR, and PEPRs, however, remain to be elucidated.
Interestingly, MAPKKK5 is further phosphorylated by the activated MPK3/6 at Ser682 and Ser692, and this enhances pattern-triggered MPK3/6 activation and disease resistance, indicating a positive feedback regulation (figure 2; [70]). Likewise, MEKK1Ser603 is subjected to positive feedback regulation by MPK4 [70]. Moreover, it was reported that BRASSINOSTEROID-SIGNALING KINASE 1 (BSK1) phosphorylates Ser289 in the N-terminal of MAPKKK5 to positively regulate immunity [69], indicating that both N- and C-termini of MAPKKK5 and MEKK1 are subjected to regulation by RLCKs. It remains to be determined in the future whether the aforementioned phosphorylation in MAPKKKs changes their subcellular localization, enhances the kinase activity, or increases protein stability.
5. Conclusion
PRRs associate with co-receptors, scaffold proteins and RLCKs to form PRR complexes at the cell surface in the wake of pathogen attacks. The composition of PRR complexes is highly intricate, and their stability and activity are tightly regulated. Future studies are expected to uncover the full complement of PRR complex components and how they are assembled at the cell surface in response to different patterns. Questions at large include whether different PRR complexes are similarly organized in nanodomains and how the PRR complexes generate signalling specificity.
RLCKs directly link by phospho-relay PRRs to multiple downstream signalling components, including RBOHD, MAPK cascades, and heterotrimeric G proteins. Future efforts are needed to identify additional downstream components such as channels controlling calcium influx and other ions and to understand how these channels are regulated during immune responses.
Acknowledgements
We apologize to all colleagues whose work is not cited owing to space limitations.
Data accessibility
This article has no additional data.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by Chinese Academy of Sciences (Strategic Priority Research Program Grant No. XDB11020200).
References
- 1.Boller T, Felix G. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. ( 10.1146/annurev.arplant.57.032905.105346) [DOI] [PubMed] [Google Scholar]
- 2.Wu Y, Zhou JM. 2013. Receptor-like kinases in plant innate immunity. J Integr. Plant Biol. 55, 1271–1286. ( 10.1111/jipb.12123) [DOI] [PubMed] [Google Scholar]
- 3.Jones JD, Vance RE, Dangl JL. 2016. Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395 ( 10.1126/science.aaf6395) [DOI] [PubMed] [Google Scholar]
- 4.Tang D, Wang G, Zhou JM. 2017. Receptor kinases in plant–pathogen interactions: more than pattern recognition. Plant Cell 29, 618–637. ( 10.1105/tpc.16.00891) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zipfel C, Oldroyd GED. 2017. Plant signalling in symbiosis and immunity. Nature 543, 328–336. ( 10.1038/nature22009) [DOI] [PubMed] [Google Scholar]
- 6.Liang X, Zhou JM. 2018. Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling. Annu. Rev. Plant Biol. 69, 267–299. ( 10.1146/annurev-arplant-042817-040540) [DOI] [PubMed] [Google Scholar]
- 7.Shiu SH, Bleecker AB. 2001. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl Acad. Sci. USA 98, 10 763–10 768. ( 10.1073/pnas.181141598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao S, Zhou Z, Miao P, Bi G, Hu M, Wu Y, Feng F, Zhang X, Zhou JM. 2018. Roles of receptor-like cytoplasmic kinase VII members in pattern-triggered immune signaling. Plant Physiol. 177, 1679–1690. ( 10.1104/pp.18.00486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haruta M, Sabat M, Stecker K, Minkoff BB, Sussman MR. 2014. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. ( 10.1126/science.1244454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C. 2017. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289. ( 10.1126/science.aal2541) [DOI] [PubMed] [Google Scholar]
- 11.Li C, et al. 2015. Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. Elife 4, e06587 ( 10.7554/eLife.06587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Q, Bourdais G, Pan H, Robatzek S, Tang D. 2017. Arabidopsis glycosylphosphatidylinositol-anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity. Proc. Natl Acad. Sci. USA 114, 5749–5754. ( 10.1073/pnas.1614468114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang XC, Cannon SB, Stacey G. 2009. Evolutionary genomics of LysM genes in land plants. BMC Evol. Biol. 9, 183 ( 10.1186/1471-2148-9-183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smakowska-Luzan E, et al. 2018. An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553, 342–346. ( 10.1038/nature25184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh YH, et al. 2016. The Arabidopsis malectin-like/LRR–RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 28, 1701–1721. ( 10.1105/tpc.16.00313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halter T, et al. 2014. The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24, 134–143. ( 10.1016/j.cub.2013.11.047) [DOI] [PubMed] [Google Scholar]
- 17.Imkampe J, et al. 2017. The Arabidopsis leucine-rich repeat receptor kinase BIR3 negatively regulates BAK1 receptor complex formation and stabilizes BAK1. Plant Cell 29, 2285–2303. ( 10.1105/tpc.17.00376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge Z, et al. 2017. Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358, 1596–1600. ( 10.1126/science.aao3642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mang H, et al. 2017. Differential regulation of two-tiered plant immunity and sexual reproduction by ANXUR receptor-like kinases. Plant Cell 29, 3140–3156. ( 10.1105/tpc.17.00464) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perraki A, et al. 2018. Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 561, 248–252. ( 10.1038/s41586-018-0471-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macho AP, et al. 2014. A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 343, 1509–1512. ( 10.1126/science.1248849) [DOI] [PubMed] [Google Scholar]
- 22.Suzuki M, et al. 2016. Autophosphorylation of specific threonine and tyrosine residues in Arabidopsis CERK1 is essential for the activation of chitin-induced immune signaling. Plant Cell Physiol. 57, 2312–2322. ( 10.1093/pcp/pcw150) [DOI] [PubMed] [Google Scholar]
- 23.Liu J, et al. 2018. A tyrosine phosphorylation cycle regulates fungal activation of a plant receptor Ser/Thr kinase. Cell Host Microbe 23, 241–253. ( 10.1016/j.chom.2017.12.005) [DOI] [PubMed] [Google Scholar]
- 24.Lin W, Li B, Lu D, Chen S, Zhu N, He P, Shan L. 2014. Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc. Natl Acad. Sci. USA 111, 3632–3637. ( 10.1073/pnas.1318817111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lal NK, et al. 2018. The receptor-like cytoplasmic kinase BIK1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host Microbe 23, 485–497. ( 10.1016/j.chom.2018.03.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Yu Y, Zhou Z, Zhou JM. 2016. Plant pattern-recognition receptors controlling innate immunity. Sci. China Life Sci. 59, 878–888. ( 10.1007/s11427-016-0115-2) [DOI] [PubMed] [Google Scholar]
- 27.Couto D, Zipfel C. 2016. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. ( 10.1038/nri.2016.77) [DOI] [PubMed] [Google Scholar]
- 28.Monaghan J, et al. 2014. The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16, 605–615. ( 10.1016/j.chom.2014.10.007) [DOI] [PubMed] [Google Scholar]
- 29.Liang X, et al. 2016. Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. Elife 5, e13568 ( 10.7554/eLife.13568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, et al. 2018. A regulatory module controlling homeostasis of a plant immune kinase. Mol. Cell 69, 493–504. ( 10.1016/j.molcel.2017.12.026) [DOI] [PubMed] [Google Scholar]
- 31.Zhang M, et al. 2018. The MAP4 kinase SIK1 ensures robust extracellular ROS burst and antibacterial immunity in plants. Cell Host Microbe 24, 379–391. ( 10.1016/j.chom.2018.08.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, et al. 2018. The kinase OsCPK4 regulates a buffering mechanism that fine-tunes innate immunity. Plant Physiol. 176, 1835–1849. ( 10.1104/pp.17.01024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. 2010. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl Acad. Sci. USA 107, 496–501. ( 10.1073/pnas.0909705107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, et al. 2010. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7, 290–301. ( 10.1016/j.chom.2010.03.007) [DOI] [PubMed] [Google Scholar]
- 35.Kadota Y, et al. 2014. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55. ( 10.1016/j.molcel.2014.02.021) [DOI] [PubMed] [Google Scholar]
- 36.Ranf S, Eschen-Lippold L, Fröhlich K, Westphal L, Scheel D, Lee J. 2014. Microbe-associated molecular pattern-induced calcium signaling requires the receptor-like cytoplasmic kinases, PBL1 and BIK1. BMC Plant Biol. 14, 374 ( 10.1186/s12870-014-0374-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, et al. 2014. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338. ( 10.1016/j.chom.2014.02.009) [DOI] [PubMed] [Google Scholar]
- 38.Lecourieux D, Ranjeva R, Pugin A. 2006. Calcium in plant defence-signalling pathways. New Phytol. 171, 249–269. ( 10.1111/j.1469-8137.2006.01777.x) [DOI] [PubMed] [Google Scholar]
- 39.Demidchik V, Shabala S, Isayenkov S, Cuin TA, Pottosin I. 2018. Calcium transport across plant membranes: mechanisms and functions. New Phytol. 220, 49–69. ( 10.1111/nph.15266) [DOI] [PubMed] [Google Scholar]
- 40.Capoen W, et al. 2011. Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl Acad. Sci. USA 108, 14 348–14 353. ( 10.1073/pnas.1107912108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charpentier M, et al. 2016. Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352, 1102–1105. ( 10.1126/science.aae0109) [DOI] [PubMed] [Google Scholar]
- 42.Thor K, Peiter E. 2014. Cytosolic calcium signals elicited by the pathogen-associated molecular pattern flg22 in stomatal guard cells are of an oscillatory nature. New Phytol. 204, 873–881. ( 10.1111/nph.13064) [DOI] [PubMed] [Google Scholar]
- 43.Frei dit Frey N, et al. 2012. Plasma membrane calcium ATPases are important components of receptor-mediated signaling in plant immune responses and development. Plant Physiol. 159, 798–809. ( 10.1104/pp.111.192575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mousavi SA, Chauvin A, Pascaud F, Kellenberger S, Farmer EE. 2013. GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500, 422–426. ( 10.1038/nature12478) [DOI] [PubMed] [Google Scholar]
- 45.Vincent TR, et al. 2017. Interplay of plasma membrane and vacuolar ion channels, together with BAK1, elicits rapid cytosolic calcium elevations in Arabidopsis during aphid feeding. Plant Cell 29, 1460–1479. ( 10.1105/tpc.17.00136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwaaitaal M, Huisman R, Maintz J, Reinstädler A, Panstruga R. 2011. Ionotropic glutamate receptor (iGluR)-like channels mediate MAMP-induced calcium influx in Arabidopsis thaliana. Biochem. J. 440, 355–365. ( 10.1042/BJ20111112) [DOI] [PubMed] [Google Scholar]
- 47.McCudden CR, Hains MD, Kimple RJ, Siderovski DP, Willard FS. 2005. G-protein signaling: back to the future. Cell. Mol. Life Sci. 62, 551–577. ( 10.1007/s00018-004-4462-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oldham WM, Hamm HE. 2008. Heterotrimeric G protein activation by G-protein coupled receptors. Nat. Rev. Mol. Cell Biol. 9, 60–71. ( 10.1038/nrm2299) [DOI] [PubMed] [Google Scholar]
- 49.Neubig RR, Siderovski DP. 2002. Regulators of G-protein signaling as new central nervous system drug targets. Nat. Rev. Drug Discov. 1, 187–197. ( 10.1038/nrd747) [DOI] [PubMed] [Google Scholar]
- 50.Stateczny D, Oppenheimer J, Bommert P. 2016. G protein signaling in plants: minus times minus equals plus. Curr. Opin. Plant Biol. 34, 127–135. ( 10.1016/j.pbi.2016.11.001) [DOI] [PubMed] [Google Scholar]
- 51.Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. 2003. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301, 1728–1731. ( 10.1126/science.1087790) [DOI] [PubMed] [Google Scholar]
- 52.Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, Siderovski DP, Jones AM, Willard FS. 2007. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc. Natl Acad. Sci. USA 104, 17 317–17 322. ( 10.1073/pnas.0704751104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma Y, et al. 2015. COLD1 confers chilling tolerance in rice. Cell 160, 1209–1221. ( 10.1016/j.cell.2015.01.046) [DOI] [PubMed] [Google Scholar]
- 54.Bommert P, Je BI, Goldshmidt A, Jackson D. 2013. The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502, 555–558. ( 10.1038/nature12583) [DOI] [PubMed] [Google Scholar]
- 55.Choudhury SR, Pandey S. 2015. Phosphorylation-dependent regulation of G-protein cycle during nodule formation in soybean. Plant Cell 27, 3260–3276. ( 10.1105/tpc.15.00517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang X, et al. 2018. Ligand-triggered de-repression of Arabidopsis heterotrimeric G proteins coupled to immune receptor kinases. Cell Res. 28, 529–543. ( 10.1038/s41422-018-0027-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qi J, Wang J, Gong Z, Zhou JM. 2017. Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 38, 92–100. ( 10.1016/j.pbi.2017.04.022) [DOI] [PubMed] [Google Scholar]
- 58.Camejo D, Guzmán-Cedeño Á, Moreno A. 2016. Reactive oxygen species, essential molecules, during plant–pathogen interactions. Plant Physiol. Biochem. 103, 10–23. ( 10.1016/j.plaphy.2016.02.035) [DOI] [PubMed] [Google Scholar]
- 59.Torres MA, Dangl JL, Jones JD. 2002. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl Acad. Sci. USA 99, 517–522. ( 10.1073/pnas.012452499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T. 2013. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl Acad. Sci. USA 110, 8744–8749. ( 10.1073/pnas.1221294110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ogasawara Y, et al. 2008. Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 283, 8885–8892. ( 10.1074/jbc.M708106200) [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, et al. 2009. Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21, 2357–2377. ( 10.1105/tpc.108.062992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shang-Guan K, et al. 2018. Lipopolysaccharides trigger two successive bursts of reactive oxygen species at distinct cellular locations. Plant Physiol. 176, 2543–2556. ( 10.1104/pp.17.01637) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meng X, Zhang S. 2013. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. ( 10.1146/annurev-phyto-082712-102314) [DOI] [PubMed] [Google Scholar]
- 65.Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, Zhang Y. 2008. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18, 1190–1198. ( 10.1038/cr.2008.300) [DOI] [PubMed] [Google Scholar]
- 66.Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF, Krysan PJ. 2007. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143, 661–669. ( 10.1104/pp.106.091389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. ( 10.1038/415977a) [DOI] [PubMed] [Google Scholar]
- 68.Yamada K, et al. 2016. The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 35, 2468–2483. ( 10.15252/embj.201694248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan H, Zhao Y, Shi H, Li J, Wang Y, Tang D. 2018. BRASSINOSTEROID-SIGNALING KINASE1 phosphorylates MAPKKK5 to regulate immunity in Arabidopsis. Plant Physiol. 176, 2991–3002. ( 10.1104/pp.17.01757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bi G, et al. 2018. Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 30, 1543–1561. ( 10.1105/tpc.17.00981) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun T, Nitta Y, Zhang Q, Wu D, Tian H, Lee JS, Zhang Y. 2018. Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep. 19, e45324 ( 10.15252/embr.201745324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Savatin DV, Bisceglia NG, Marti L, Fabbri C, Cervone F, De Lorenzo G.. 2014. The Arabidopsis NUCLEUS- AND PHRAGMOPLAST-LOCALIZED KINASE1-related protein kinases are required for elicitor-induced oxidative burst and immunity. Plant Physiol. 165, 1188–1202. ( 10.1104/pp.114.236901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. 2007. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63–73. ( 10.1105/tpc.106.048298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mithoe SC, et al. 2016. Attenuation of pattern recognition receptor signaling is mediated by a MAP kinase kinase kinase. EMBO Rep. 17, 441–454. ( 10.15252/embr.201540806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, Zhou JM. 2012. A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485, 114–118. ( 10.1038/nature10962) [DOI] [PubMed] [Google Scholar]
- 76.Yamada K, Yamashita-Yamada M, Hirase T, Fujiwara T, Tsuda K, Hiruma K, Saijo Y. 2016. Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK1. EMBO J. 35, 46–61. ( 10.15252/embj.201591807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kong Q, et al. 2016. Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis. Plant Physiol. 171, 1344–1354. ( 10.1104/pp.15.01954) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.