Abstract
Daunorubicin (Dnr) is at the forefront of acute myeloid leukemia (AML) therapy, but drug resistance poses a major threat to treatment success. MicroRNA (miR)-9 has been shown to have a pivotal role in AML development. However, little is known about the role of miR-9 in Dnr resistance in AML. We explored the potential role of miR-9 in Dnr resistance in AML cells and its mechanism of action. AML cell lines with high half-maximal inhibitory concentration to Dnr in vivo had significantly low miR-9 expression. miR-9 overexpresssion sensitized AML cells to Dnr, inhibited cell proliferation, and enhanced the ability of Dnr to induce apoptosis; miR-9 knockdown had the opposite effects. Mechanistic studies demonstrated that eukaryotic translation initiation factor 5A-2 (EIF5A2) was a putative target of miR-9, which was inversely correlated with the expression and role of miR-9 in AML cells. miR-9 improved the anti-tumor effects of Dnr by inhibiting myeloid cell leukemia-1 (MCL-1) expression, which was dependent on downregulation of EIF5A2 expression. These results suggest that miR-9 has an essential role in Dnr resistance in AML cells through inhibition of the EIF5A2/MCL-1 axis in AML cells. Our data highlight the potential application of miR-9 in chemotherapy for AML patients.
Keywords: micoRNA-9, EIF5A2, drug resistance, acute myeloid leukemia (AML)
Introduction
“Leukemia” refers to cancer of white blood cells, and can be classified as “acute myeloid leukemia” (AML), “acute lymphoblastic leukemia” or “chronic lymphoblastic leukemia” 1. AML is the most common type of acute leukemia, and ≈80% of adult patients with acute leukemia have AML 2.
The combination of cytarabine and daunorubicin (Dnr) is used as first-line treatment for AML (except for AML-M3) 3. Although 70-85% of patients achieve initial induction remission with this treatment, chemoresistance and therapy-related mortality continue to have a key role in therapeutic failure and poor outcomes 4. Hence, it is essential to explore the potential mechanisms underlying AML chemoresistance and develop effective new strategies for enhancement of the efficacy of AML chemotherapy.
MicroRNAs (mIRs) are a large family of highly conserved endogenous non-coding RNAs. They negatively regulate expression of target genes by binding to the 3′ untranslated region (UTR), thereby degrading or blocking their translation 5, 6. Studies have shown that miRs have vital roles in the initiation, progression, invasion and metastasis of tumor cells as tumor-suppressor genes or proto-oncogenes 7.
Abnormal expression of miRs has been identified in AML chemoresistance 8, 9. For miR-9, tumor suppressor and oncogenic roles have been reported in various types of solid tumor 10-13. In AML, Chen et al. reported that miR-9 was a direct target of mixed-lineage leukemia (MLL) fusion proteins. They also reported that depletion of endogenous miR-9 expression could inhibit the growth and viability of cells and promote apoptosis in human MLL-rearranged AML cells, indicating that miR-9 is a tumor oncogene 14. However, Emmrich et al. reported that miR-9 was a tumor suppressor in pediatric AML with a t (8;21) translocation 15. Nishioka et al. demonstrated that miR-9 has a tumor-suppressor role in AML because it regulates interleukin-10-mediated expression of E-cadherin 16. However, studies focusing on the role of miR-9 in mediating AML chemoresistance are lacking.
We wished to explore the role of miR-9 in Dnr resistance in AML and uncover its potential mechanism of action. Our results demonstrated that miR-9 was expressed at significantly lower levels in Dnr-resistant AML cell lines. We present evidence that miR-9 overexpression can enhance Dnr sensitivity. Then, we identified eukaryotic translation initiation factor 5A-2 (EIF5A2) as a target gene of miR-9. EIF5A2 knockdown increased Dnr sensitivity and downregulated myeloid cell leukemia 1 (MCL-1) expression. We further confirmed that MCL-1 was involved in miR-9-mediated regulation of Dnr sensitivity in AML. Finally, we demonstrated that the effects of miR-9 in AML cells were mediated by EIF5A2. Collectively, we proposed that upregulation of miR-9 expression could improve Dnr sensitivity to AML cells by transcriptional repression of MCL-1 through direct targeting of the 3′UTR region of EIF5A2.
Materials and Methods
Cell culture
The AML cell lines HL-60, KG-1, THP-1 and Kasumi-1 were purchased from the Chinese Academy of Science Cell Bank (Shanghai, China) and maintained in RMPI 1640 medium (Gibco, Billings, MT, USA). All culture media were supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin solution (Gibco) at 37°C in a humidified atmosphere of 5% CO2. Dnr was purchased from Selleck Chemicals (Houston TX, USA) and dissolved in dimethyl sulfoxide.
Small interfering (si)RNA and transfection
Mcl-1, eIF5A2 siRNA, and negative-control siRNA were obtained from Ribobio (Guangzhou, China). miR-9 inhibitors, miR-9 mimics, and their negative control RNA were purchased from Fulengen (Guangzhou, China) (miR-9 mimic, 5ʹ-UCUUUGGUUAUCUAGCUGUAUGA-3ʹ and 5ʹ-AUACAGCUAGAUAACCAAAGAUU-3ʹ; miR-9 inhibitor, 5ʹ-UCAUACAGCUAGAUAACCAAAGA-3ʹ, Negative control: 5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ). Transfection was conducted using Lipofectamine 2000 Reagent following manufacturer (Thermo Scientific, Waltham, MA, USA) instructions.
Cell Counting Kit-8 (CCK-8) assay
AML cells that had undergone various types of transfection were seeded at 10,000 cells/well in 96-well plates. Then, the cells were cultured in different concentrations of Dnr at 37°C in an atmosphere of 5% CO2 in an incubator for 48 h. Cell viability was measured using a CCK-8 kit at the indicated time points according to manufacturer (Dojindo Laboratories, Tokyo, Japan) instructions. Absorbance at 450 nm was measured using a microplate reader (MRX II; Dynex Technologies, Chantilly, VA, USA). The concentration at which each drug produced 50% inhibition of growth (IC50) was estimated using a relative survival curve.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cell lines using TRIzol™ Reagent (Thermo Scientific). To detect miR-9 expression, cDNA was reverse-transcribed using a TaqMan™ miRNA RT kit (Life Technologies, Carlsbad, CA, USA) and U6 was used as an endogenous control. qRT-PCR assays were analyzed by the StepOnePlus™ RT-PCR system (Thermo Scientific) using a SYBR™ PrimeScript RT-PCR kit (TaKaRa Biotechnology, Dalian, China). The primers were as follows:
eIF5A2: Forward: 5'-GCAGACGAAAUUGAUUUCATT-3';
Reverse: 5'-UGAAAUCAAUUUCGUCUGCTT-3';
miR9: 5'-TCTTTGGTTATCTAGCTGTATGA-3'.
Western blotting
Western blotting was carried out according to a standard protocol. Protein concentrations were quantified using a Bicinchoninic Acid Protein Assay kit (Thermo Scientific). Proteins (40 µg) were resolved on 10% sodium dodecyl sulfate-polyacrylamide gels and transferred to polyvinylidene difluoride membranes. Primary antibodies against eIF5A2, MCL-1 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were purchased from Abcam (Cambridge, UK) and used at 1:1000 dilution. Secondary antibodies were also from Abcam and used at 1:2000 dilution.
EdU assay
Proliferation of AML cells was determined using a Click-iT® EdU Imaging kit according to manufacturer (Invitrogen, Carlsbad, CA, USA) protocols. Cell nuclei were stained blue with Hoechst 3344 (Invitrogen) at 5 μg/mL for 30 min. EdU-positive cells were green.
Apoptosis assay
Fluorescein isothiocyanate (FITC)-conjugated annexin-V and a Propidium Iodide (PI) kit were used according to manufacturer (BD Biosciences, San Jose, CA, USA) instructions. Cells (2-3×105) were washed twice with pre-chilled phosphate-buffered saline and resuspended in 100 µL of binding buffer and 5 μL of FITC-conjugated annexin-V, then protected from light for 30 min at room temperature. Next, 100 μL of PI was added followed, 5-min later, with 400 µL of Binding Buffer. Cells were examined with a flow cytometer CANTO™ II (BD Biosciences) and data analyzed by FlowJo (Ashland, OR, USA).
Statistical analyses
SPSS v18.0 (IBM, Armonk, NY, USA) was used for statistical analyses. Experimental data are the mean ± SD and were assessed by two-tailed Student's t-tests. P<0.05 was considered significant.
Results
miR-9 expression was negatively correlated with Dnr sensitivity in AML cells
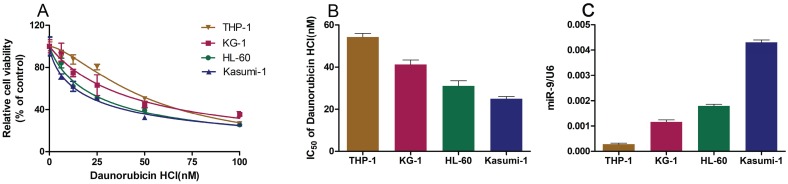
We wished to explore the role of miR-9 in Dnr sensitivity in AML cell lines. First, we employed the CCK-8 assay to study the viability of AML cell lines cultured with different concentrations of Dnr and calculated the IC50 values. The sensitivity of AML cell lines to Dnr varied and was concentration-dependent (Fig. 1A). The IC50 value (in decreasing order) was THP-1, KG-1, HL60 and Kasumi-1 cell lines (Fig. 1B). In general, the AML cell line that was less sensitive to Dnr showed lower expression of miR-9 (Fig. 1C). Thus, high expression of miR-9 could enhance Dnr sensitivity in AML cells.
Fig 1.
AML cell lines resistant to daunorubicin showed decreased miR-9 expression. (A) Viability of AML cell lines under different concentrations of Dnr according to the CCK-8 assay. (B) IC50 of AML cell lines calculated according to the result of the CCK-8 assay. (C) miR-9 expression in AML cell lines examined by qPCR. U6 was used as the internal reference.
miR-9 could improve Dnr sensitivity in AML cell lines
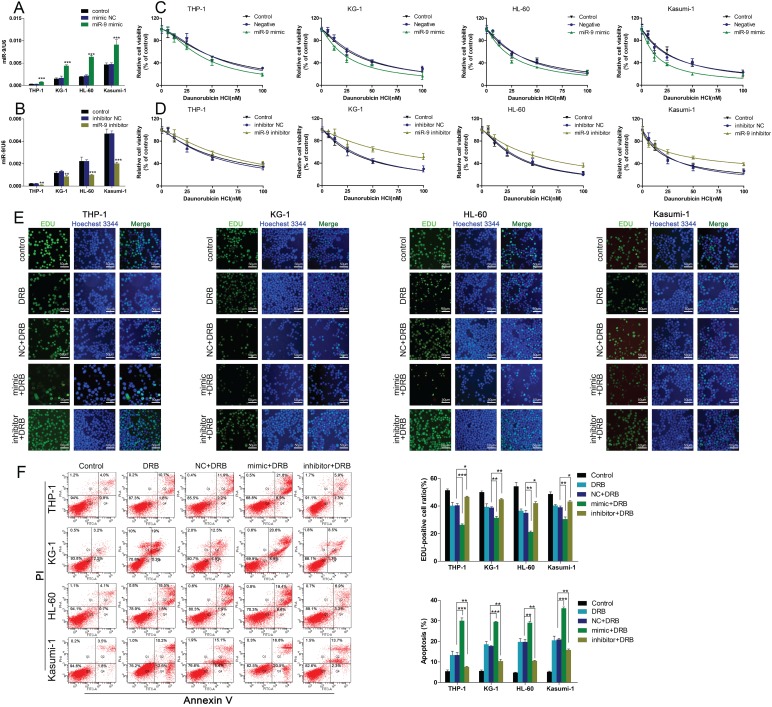
To further confirm the functional association between miR-9 expression and Dnr resistance in AML cells, we transiently transfected AML cell lines with a miR-9 mimic, miR-9 inhibitor, or scrambled control. The efficiency of the miR-9 mimic and miR-9 inhibitor were confirmed by qRT-PCR (Fig. 2A, B). At 48 h after transfection, the CCK-8 assay showed that miR-9 overexpression could lead to enhanced inhibition of cell viability in comparison with controls, and that miR-9 inhibition could lead to Dnr resistance (Fig. 2C, D). EdU assays confirmed that miR-9 overexpression could improve Dnr sensitivity in AML cells (Fig. 2E). Apoptosis assays demonstrated that enforced miR-9 expression increased the proportion of apoptotic cells compared with their respective controls (Fig. 2F). These results suggested that upregulation of miR-9 expression could restore Dnr sensitivity in AML cells.
Fig 2.
miR-134 overexpression enhances the Dnr sensitivity of AML cells. (A-B) Relative expression of miR-9 in AML cells transfected with miR-9 mimics, miR-9 inhibitors or negative control. (C) Effect of miR-9 on viability of AML cells transfected with miR-9 mimics and negative control under different concentrations of Dnr. (D) Effect of miR-9 on the viability of AML cells transfected with a miR-9 inhibitor and negative control under different concentrations of Dnr. (E) Effect of miR-9 on proliferation of AML cells transfected with a miR-9 inhibitor, miR-9 mimics or negative control and treated with Dnr according to the EdU assay. The number of EdU-positive cells was counted. (F) Effect of miR-9 on apoptosis of AML cells transfected with a miR-9 inhibitor, miR-9 mimic or negative control and treated with Dnr according to flow cytometry. *p<0.05, **p<0.01, ***p<0.001.
EIF5A2 was the target gene of miR-9
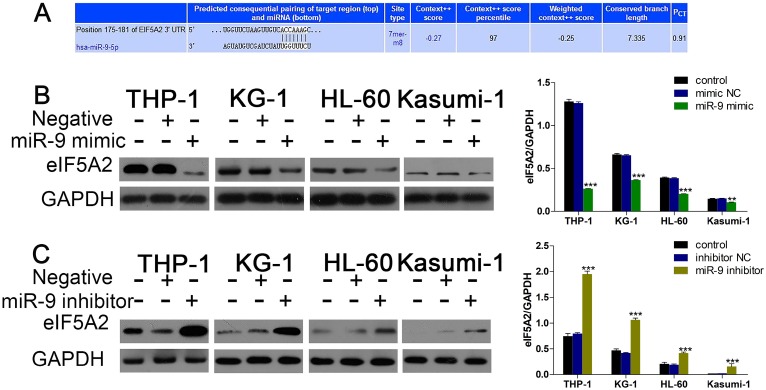
Given that miR-9 could relieve Dnr resistance in AML cells, we sought to uncover the underlying molecular mechanisms of this phenomenon. First, we used bioinformatics methods (TargetScan) to predict the potential targets of miR-9. Among the candidate targets, 3′UTR of EIF5A2 contained regions that matched the seed sequences of miR-9 (Fig. 3A). To further confirm EIF5A2 was a target gene of miR-9, we used western blotting to detect EIF5A2 expression under existing miR-9 mimics and inhibitors. Expression of EIF5A2 protein in AML cell lines was upregulated significantly by miR-9 (Fig. 3B, C). All these data suggested that EIF5A2 was a direct target of miR-9.
Fig 3.
miR-9 negatively regulates EIF5A2. (A) Predicted binding sequences of miR-9 in the 3′UTR of EIF5A2 according to TargetScan. (B) Expression of EIF5A2 protein in AML cells transfected with miR-9 mimics or negative control. **P<0.01,***P<0.001 vs control. (C) Expression of EIF5A2 protein in AML cells transfected with a miR-9 inhibitor or negative control. ***P<0.001 vs control.
EIF5A2 knockdown increased the sensitivity of AML cells to Dnr and inhibited MCL-1 expression
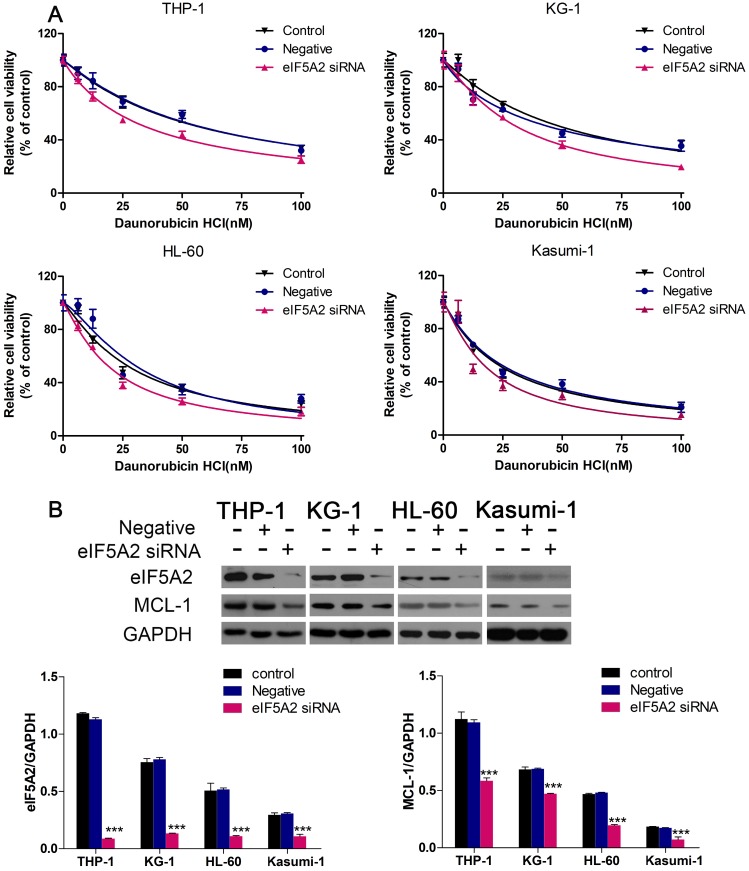
We continued to investigate if EIF5A2 contributed to Dnr resistance in AML cells. We employed EIF5A2 with siRNA to reduce endogenous expression of pleomorphic adenoma gene 1 (PLAG1). Then, 48 h after exposure to different concentrations of Dnr, CCK-8 assays were conducted. AML cells with EIF5A2 knockdown showed much lower viability compared with control groups (Fig. 4A). To further identify the mechanisms implicated in Dnr resistance, we measured protein expression of several classical molecules reported to regulate drug resistance in AML cells. We found that EIF5A2 knockdown could inhibit MCL-1 expression (Fig. 4B), which has been implicated in the drug resistance of AML cells 17-19.
Fig 4.
EIF5A2 knockdown sensitized AML cells to Dnr and decreased MCL-1 expression. (A) Effect of EIF5A2 on the viability of AML cells transfected with EIF5A2 siRNA or negative control under different concentrations of Dnr. (B) Western blots of the expression of EIF5A2 and MCL-1 in AML cell lines transfected with EIF5A2 siRNA or negative control. GAPDH was the control. ***P<0.001 vs control.
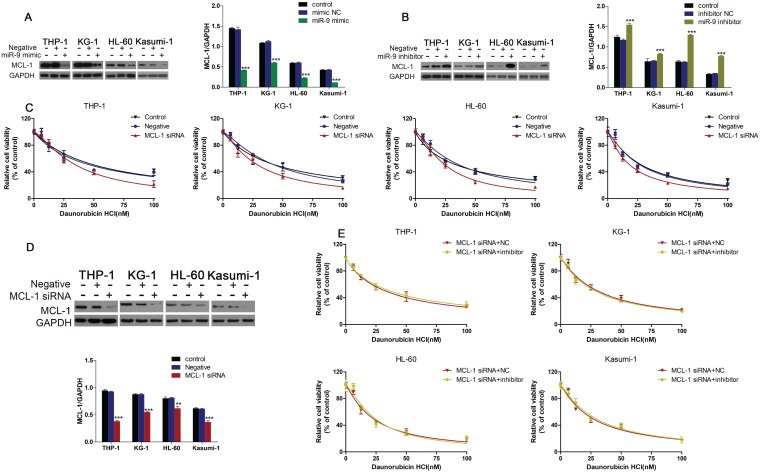
MCL-1 was involved in miR-9-mediated regulation of Dnr sensitivity in AML cells
Western blotting showed that miR-9 mimics could inhibit MCL-1 expression, and that inhibition of miR-9 expression could increase MCL-1 expression (Fig. 5A, B). We wondered if MCL-1 contributed to the Dnr resistance in AML cells. CCK-8 assays demonstrated that AML cells with MCL-1 knockdown showed much lower viability compared with control groups (Fig. 5C). Also, the interference efficiency of MCL-1 siRNA was confirmed by western blotting (Fig. 5D). Finally, we co-transfected AML cells with MCL-1 siRNA plus a miR-9 inhibitor or MCL-1 siRNA plus a negative control and examined cell viability under Dnr. After MCL-1 knockdown, miR-9 inhibitor-induced Dnr resistance vanished, which suggested that MCL-1 participated in miR-9-mediated regulation of Dnr sensitivity.
Fig 5.
miR-9 could inhibit MCL-1 expression and MCL-1 knockdown was involved in miR-9-mediated Dnr sensitivity in AML cells. (A) Expression of MCL-1 protein in AML cells transfected with miR-9 mimics or negative control. ***P<0.001 vs control. (B) Expression of MCL-1 protein in AML cells transfected with a miR-9 inhibitor or negative control. ***P<0.001 vs control. (C) Effect of MCL-1 on the viability of AML cells transfected with MCL-1 siRNA or negative control under different concentrations of Dnr. (D) Interference of MCL-1 siRNA by western blotting. **P<0.01,***P<0.001 vs control. (E) Viability of AML cell lines transfected with MCL-1 siRNA plus a miR-9 inhibitor or CL-1 siRNA plus a negative control under different concentrations of Dnr according to the CCK-8 assay.
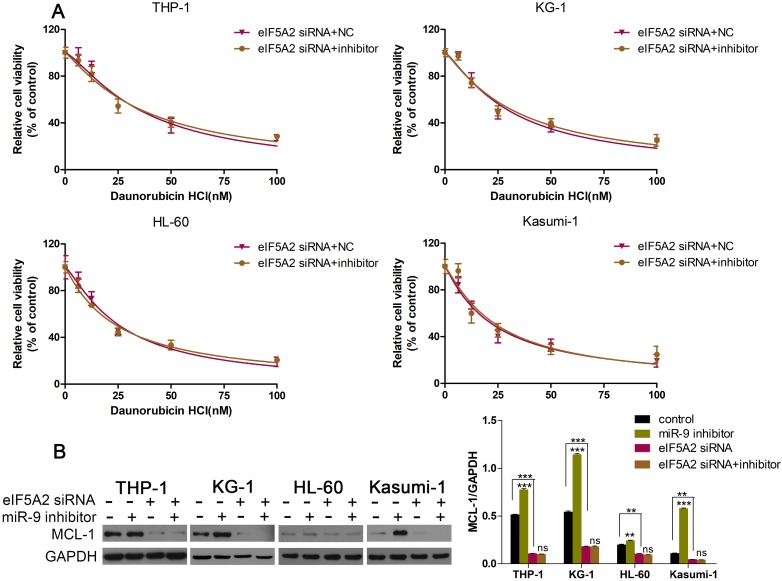
EIF5A2 mediated the effects of miR-9 in AML cells
Given that EIF5A2 was the target gene of miR-9, and that EIF5A2 could regulate MCL-1 expression, we wondered if EIF5A2 was involved in the miR-9-induced effects in AML cells. First, we co-transfected AML cells with EIF5A2 siRNA plus a miR-9 inhibitor or EIF5A2 siRNA plus a negative control, and examined cell viability under Dnr. EIF5A2 siRNA could overcome miR-9 inhibitor-mediated Dnr resistance (Fig. 6A). Additional western blotting showed that EIF5A2 knockdown ablated miR-9 inhibitor-mediated upregulation of MCL-1 expression (Fig. 6B). These data suggested that the effects of miR-9 in AML cells were mediated by EIF5A2, and that a miR-9/EIF5A2/MCL-1 axis regulated Dnr sensitivity in AML cells.
Fig 6.
EIF5A2 was involved in how miR-9 regulated MCL-1 expression and Dnr sensitivity in AML cells. (A) Viability of AML cell lines transfected with EIF5A2 siRNA plus a miR-9 inhibitor or CL-1 siRNA plus a negative control under different concentrations of Dnr according to the CCK-8 assay. (B) Expression of MCL-1 protein in AML cells transfected with EIF5A2 siRNA plus a miR-9 inhibitor or CL-1 siRNA plus a negative control. **P<0.01,***P<0.001.
Discussion
Accumulating evidence suggests that aberrant expression of miRNA is involved in various cancers, including hematologic malignancies 20-22. Here, we identified for the first time, a miR-9/EIF5A2/MCL-1 axis in human AML cells that may be responsible for chemoresistance. We showed that miR-9 overexpression could improve Dnr sensitivity in AML cells by targeting EIF5A2, which led to decreased expression of MCL-1 in AML cells. Collectively, our results demonstrated that miR-9 has a vital role in the chemoresistance of AML cells and provided novel insights into the potential mechanisms underlying AML chemoresistance.
miR-9 has been shown to be involved in the proliferation, autophagy, migration, and apoptosis of cancer cells as an oncogene or tumor suppressor depending on the cancer type 23. For example, miR-9 overexpression has been reported to promote the migration and invasion of ovarian cancer cells 24. miR-9 has been reported to promote the proliferation, migration, and invasion of breast cancer cells by down-regulating expression of forkhead box protein O1 (FOXO1) 25. However, in gastric cancer, miR-9 can inhibit proliferation of gastric cancer cells by targeting tumor necrosis factor-alpha-induced protein 8 (TNFAIP8) 26. miR-9 has been reported to suppress the migration and invasion of colorectal cancer cells by targeting transmembrane 4 L6 family member 1 (TM4SF1) 27.
Studies have shown that miR-9 is involved in the chemoresistance of hematopoietic malignancies. Li et al. reported that miR-9 overexpression was sufficient to reverse cancer-cell resistance to multiple chemotherapeutics by targeting ATP binding cassette subfamily B member 1 (ABCB1) in chronic myeloid leukemia 28. However, in AML, little is known about the role of miR-9 in drug resistance. We have shown here, for the first time, that miR-9 overexpression can enhance Dnr sensitivity in AML.
To uncover the underlying molecular mechanisms by which miR-9 influences Dnr sensitivity in AML cells, we used bioinformatics methods to predict its possible target genes. We found EIF5A2 to be a target gene of miR-9 in AML. EIF5A2 had been shown to play important parts in the proliferation, metastasis and apoptosis of cancer cells, and is considered to be a novel oncogene 29. In hepatocellular carcinoma and non-small-cell lung cancer, EIF5A2 has been identified as the target gene of miR-9 30, 31. However, the role of EIF5A2 and the relationship between miR-9 and EIF5A2 in AMLs are not known. Here we showed, for the first time, that EIF5A2 knockdown could increase Dnr sensitivity and that EIF5A2 mediated the regulatory role of miR-9 in AML cells.
Mcl-1 is a member of the B-cell lymphoma (Bcl)-2 family. Bcl-2 family proteins play vital parts in balancing the decision between cell survival and apoptosis, and escape from apoptosis is a hallmark of cancer 32, 33. Dysregulation of Bcl-2 family proteins has been found in AML, and upregulation of expression of anti-apoptotic family members Bcl-2, Bcl-xL and Mcl-1 have been found to promote AML chemoresistance 34, 35. Here, we demonstrated that MCL-1 knockdown could improve Dnr sensitivity in AML and that MCL-1 expression was regulated by the miR-9/EIF5A2 axis. These novel findings could provide new insights into the regulation of apoptosis in AML.
Conclusions
miR-9 overexpression contributed to high Dnr sensitivity in AML. miR-9 enhanced the chemosensitivity of AML cells to Dnr through an apoptotic pathway mediated by MCL-1. EIF5A2 was identified as a direct target of miR-9 in AML. Identification of the candidate target genes of miR-9 may aid understanding of the potential mechanisms involved in AML. However, additional studies are required to ascertain if miR-9 can be used in AML treatment.
Acknowledgments
This study was supported by grants from the Natural Science Foundation of Henan Province (No. 162300410274, 182300410298), the National Natural Science Foundation of China (No. 81273260).
References
- 1.Meenaghan T, Dowling M, Kelly M. Acute leukaemia: making sense of a complex blood cancer. Br J Nurs. 2012;21:76. doi: 10.12968/bjon.2012.21.2.76. 8-83. [DOI] [PubMed] [Google Scholar]
- 2.Short NJ, Rytting ME, Cortes JE. Acute myeloid leukaemia. Lancet; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy T, Yee KWL. Cytarabine and daunorubicin for the treatment of acute myeloid leukemia. Expert Opin Pharmacother. 2017;18:1765–80. doi: 10.1080/14656566.2017.1391216. [DOI] [PubMed] [Google Scholar]
- 4.Roboz GJ. Current treatment of acute myeloid leukemia. Curr Opin Oncol. 2012;24:711–9. doi: 10.1097/CCO.0b013e328358f62d. [DOI] [PubMed] [Google Scholar]
- 5.Mirzaei H, Masoudifar A, Sahebkar A, Zare N, Sadri Nahand J, Rashidi B, MicroRNA: A novel target of curcumin in cancer therapy. J Cell Physiol; 2017. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–9. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Gabra MM, Salmena L. microRNAs and Acute Myeloid Leukemia Chemoresistance: A Mechanistic Overview. Front Oncol. 2017;7:255. doi: 10.3389/fonc.2017.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zebisch A, Hatzl S, Pichler M, Wolfler A, Sill H. Therapeutic Resistance in Acute Myeloid Leukemia: The Role of Non-Coding RNAs. Int J Mol Sci; 2016. p. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo LM, Pu Y, Han Z, Liu T, Li YX, Liu M. et al. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009;276:5537–46. doi: 10.1111/j.1742-4658.2009.07237.x. [DOI] [PubMed] [Google Scholar]
- 11.Luo H, Zhang H, Zhang Z, Zhang X, Ning B, Guo J. et al. Down-regulated miR-9 and miR-433 in human gastric carcinoma. J Exp Clin Cancer Res. 2009;28:82. doi: 10.1186/1756-9966-28-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D. et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu L, Chen H, Zhou D, Li D, Bai R, Zheng S. et al. MicroRNA-9 up-regulation is involved in colorectal cancer metastasis via promoting cell motility. Med Oncol. 2012;29:1037–43. doi: 10.1007/s12032-011-9975-z. [DOI] [PubMed] [Google Scholar]
- 14.Chen P, Price C, Li Z, Li Y, Cao D, Wiley A. et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc Natl Acad Sci U S A. 2013;110:11511–6. doi: 10.1073/pnas.1310144110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emmrich S, Katsman-Kuipers JE, Henke K, Khatib ME, Jammal R, Engeland F. et al. miR-9 is a tumor suppressor in pediatric AML with t(8;21) Leukemia. 2014;28:1022–32. doi: 10.1038/leu.2013.357. [DOI] [PubMed] [Google Scholar]
- 16.Nishioka C, Ikezoe T, Pan B, Xu K, Yokoyama A. MicroRNA-9 plays a role in interleukin-10-mediated expression of E-cadherin in acute myelogenous leukemia cells. Cancer Sci. 2017;108:685–95. doi: 10.1111/cas.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luedtke DA, Niu X, Pan Y, Zhao J, Liu S, Edwards H. et al. Inhibition of Mcl-1 enhances cell death induced by the Bcl-2-selective inhibitor ABT-199 in acute myeloid leukemia cells. Signal Transduct Target Ther. 2017;2:17012. doi: 10.1038/sigtrans.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin KH, Winter PS, Xie A, Roth C, Martz CA, Stein EM. et al. Targeting MCL-1/BCL-XL Forestalls the Acquisition of Resistance to ABT-199 in Acute Myeloid Leukemia. Sci Rep. 2016;6:27696. doi: 10.1038/srep27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasper S, Breitenbuecher F, Heidel F, Hoffarth S, Markova B, Schuler M. et al. Targeting MCL-1 sensitizes FLT3-ITD-positive leukemias to cytotoxic therapies. Blood Cancer J. 2012;2:e60. doi: 10.1038/bcj.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omer A, Singh P, Yadav NK, Singh RK. microRNAs: role in leukemia and their computational perspective. Wiley Interdiscip Rev RNA. 2015;6:65–78. doi: 10.1002/wrna.1256. [DOI] [PubMed] [Google Scholar]
- 22.Babashah S, Sadeghizadeh M, Tavirani MR, Farivar S, Soleimani M. Aberrant microRNA expression and its implications in the pathogenesis of leukemias. Cell Oncol (Dordr) 2012;35:317–34. doi: 10.1007/s13402-012-0095-3. [DOI] [PubMed] [Google Scholar]
- 23.Yuva-Aydemir Y, Simkin A, Gascon E, Gao FB. MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol. 2011;8:557–64. doi: 10.4161/rna.8.4.16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou B, Xu H, Xia M, Sun C, Li N, Guo E. et al. Overexpressed miR-9 promotes tumor metastasis via targeting E-cadherin in serous ovarian cancer. Front Med. 2017;11:214–22. doi: 10.1007/s11684-017-0518-7. [DOI] [PubMed] [Google Scholar]
- 25.Liu DZ, Chang B, Li XD, Zhang QH, Zou YH. MicroRNA-9 promotes the proliferation, migration, and invasion of breast cancer cells via down-regulating FOXO1. Clin Transl Oncol. 2017;19:1133–40. doi: 10.1007/s12094-017-1650-1. [DOI] [PubMed] [Google Scholar]
- 26.Gao HY, Huo FC, Wang HY, Pei DS. MicroRNA-9 inhibits the gastric cancer cell proliferation by targeting TNFAIP8. Cell Prolif; 2017. p. 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park YR, Lee ST, Kim SL, Liu YC, Lee MR, Shin JH. et al. MicroRNA-9 suppresses cell migration and invasion through downregulation of TM4SF1 in colorectal cancer. Int J Oncol. 2016;48:2135–43. doi: 10.3892/ijo.2016.3430. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Zhao L, Li N, Miao Y, Zhou H, Jia L. miR-9 regulates the multidrug resistance of chronic myelogenous leukemia by targeting ABCB1. Oncol Rep. 2017;37:2193–200. doi: 10.3892/or.2017.5464. [DOI] [PubMed] [Google Scholar]
- 29.Mathews MB, Hershey JW. The translation factor eIF5A and human cancer. Biochim Biophys Acta. 2015;1849:836–44. doi: 10.1016/j.bbagrm.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue F, Liang Y, Li Z, Liu Y, Zhang H, Wen Y. et al. MicroRNA-9 enhances sensitivity to cetuximab in epithelial phenotype hepatocellular carcinoma cells through regulation of the eukaryotic translation initiation factor 5A-2. Oncol Lett. 2018;15:813–20. doi: 10.3892/ol.2017.7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu G, Shao G, Pan Q, Sun L, Zheng D, Li M. et al. MicroRNA-9 regulates non-small cell lung cancer cell invasion and migration by targeting eukaryotic translation initiation factor 5A2. Am J Transl Res. 2017;9:478–88. [PMC free article] [PubMed] [Google Scholar]
- 32.De Blasio A, Vento R, Di Fiore R. Mcl-1 targeting could be an intriguing perspective to cure cancer. J Cell Physiol; 2018. [DOI] [PubMed] [Google Scholar]
- 33.Opferman JT. Attacking cancer's Achilles heel: antagonism of anti-apoptotic BCL-2 family members. FEBS J. 2016;283:2661–75. doi: 10.1111/febs.13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chauhan PS, Bhushan B, Singh LC, Mishra AK, Saluja S, Mittal V. et al. Expression of genes related to multiple drug resistance and apoptosis in acute leukemia: response to induction chemotherapy. Exp Mol Pathol. 2012;92:44–9. doi: 10.1016/j.yexmp.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Doi K, Gowda K, Liu Q, Lin JM, Sung SS, Dower C. et al. Pyoluteorin derivatives induce Mcl-1 degradation and apoptosis in hematological cancer cells. Cancer Biol Ther. 2014;15:1688–99. doi: 10.4161/15384047.2014.972799. [DOI] [PMC free article] [PubMed] [Google Scholar]