Abstract
Tacrolimus exhibits low and variable drug exposure after oral dosing, but the contributing factors remain unclear. Based on our recent report showing a positive correlation between fecal abundance of Faecalibacterium prausnitzii and oral tacrolimus dose in kidney transplant patients, we tested whether F. prausnitzii and other gut abundant bacteria are capable of metabolizing tacrolimus. Incubation of F. prausnitzii with tacrolimus led to production of two compounds (the major one named M1), which was not observed upon tacrolimus incubation with hepatic microsomes. Isolation, purification, and structure elucidation using mass spectrometry and nuclear magnetic resonance spectroscopy indicated that M1 is a C-9 keto-reduction product of tacrolimus. Pharmacological activity testing using human peripheral blood mononuclear cells demonstrated that M1 is 15-fold less potent than tacrolimus as an immunosuppressant. Screening of 22 gut bacteria species revealed that most Clostridiales bacteria are extensive tacrolimus metabolizers. Tacrolimus conversion to M1 was verified in fresh stool samples from two healthy adults. M1 was also detected in the stool samples from kidney transplant recipients who had been taking tacrolimus orally. Together, this study presents gut bacteria metabolism as a previously unrecognized elimination route of tacrolimus, potentially contributing to the low and variable tacrolimus exposure after oral dosing.
Introduction
Tacrolimus is a commonly used immunosuppressant for kidney transplant recipients as well as patients with glomerular diseases such as membranous nephropathy and focal segmental glomerulosclerosis. However, due to its narrow therapeutic index, underexposure or overexposure to tacrolimus in kidney transplant recipients increases the risks for graft rejection or drug-related toxicity, respectively (Staatz and Tett, 2004). Maintaining therapeutic blood concentrations of tacrolimus has been difficult in part because tacrolimus pharmacokinetics show large interindiviudal and intraindividual variability (Press et al., 2009; Shuker et al., 2015). For example, tacrolimus oral bioavailability in individual patients ranges from 5% to 93% (average ∼25%) (Staatz and Tett, 2004). A better understanding of the factors responsible for the variability is crucial for maintaining target therapeutic concentrations of tacrolimus and improving kidney transplant outcomes.
The human gut is home to trillions of microbes that can influence multiple aspects of host physiology (Schroeder and Bäckhed, 2016). In particular, intestinal bacteria can mediate diverse chemical reactions such as hydrolysis and reduction of orally administered drugs, ultimately affecting the efficacy and/or toxicity of drugs (Wallace et al., 2010; Haiser et al., 2013; Koppel et al., 2017). For example, digoxin is converted to the pharmacologically inactive metabolite, dihydrodigoxin, by the gut bacterium Eggerthella lenta (Haiser et al., 2013). The expression of the enzyme responsible for digoxin metabolism in E. lenta is influenced by dietary protein content (Haiser et al., 2013), indicating that in addition to the abundance of drug-metabolizing bacteria, diet composition may also govern the extent of drug metabolism in the gut and alter systemic drug exposure. For most clinically used drugs, the detailed roles of gut bacteria in their metabolism and/or disposition remain unknown.
Faecalibacterium prausnitzii is one of the most abundant human gut bacteria [108–109 16S ribosomal RNA (rRNA) gene copies per gram of mucosal tissue in ileum and colon], taxonomically belonging to the Clostridiales order (Qin et al., 2010; Arumugam et al., 2011). Because of its anti-inflammatory effects, F. prausnitzii has been investigated as a potential preventative and/or therapeutic agent for dysbiosis (Miquel et al., 2015; Rossi et al., 2016). We have recently shown that in 19 kidney transplant patients, fecal F. prausnitzii abundance positively correlates with oral tacrolimus doses required to maintain therapeutic blood concentrations, independent of gender and body weight (Lee et al., 2015). It remains unknown, however, whether F. prausnitzii is directly involved in tacrolimus elimination in the gut. Herein, we tested a hypothesis that gut bacteria, including F. prausnitzii, metabolize tacrolimus into less potent metabolite(s).
Materials and Methods
Reagents.
Tacrolimus was purchased from AdipoGen (San Diego, CA). Casitone and yeast extract were purchased from HIMEDIA (Nashik, Maharashtra, India) and BD (Sparks, MD), respectively. Other components for media were purchased from Thermo Fisher Scientific (Waltham, MA) or Sigma-Aldrich (St. Louis, MO).
Peripheral blood mononuclear cells (PBMCs) were purchased from PromoCell (Heidelberg, Germany). Phytohemagglutinin and 5-bromo-2′-deoxyuridine were purchased from Sigma-Aldrich. 3,3′,5,5′-Tetramethylbenzidine was purchased from Thermo Fisher Scientific.
Bacterial Strains and Growth.
F. prausnitzii A2-165 was obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). F. prausnitzii VPI C13-20-A (ATCC 27766), and F. prausnitzii VPI C13-51 (ATCC 27768) were obtained from American Type Culture Collection (Manassas, VA). Other gut bacteria were obtained from the Biodefense and Emerging Infections Research Resources Repository (Bethesda, MD) (Supplemental Table 1). Unless stated otherwise, all of the bacterial strains were grown anaerobically (5% H2, 5% CO2, 90% N2) on YCFA agar or broth at 37°C in an anaerobic chamber (Anaerobe Systems, Morgan Hill, CA), and colonies from the agar plate were inoculated into prereduced YCFA broth for preparation of overnight cultures. Optical density at 600 nm (OD600) was measured for estimation of bacterial concentration.
Tacrolimus Metabolism by Gut Bacteria.
To examine tacrolimus metabolism by gut bacteria, cells of a bacterial strain grown as described previously were incubated tacrolimus. Typically, tacrolimus (100 μg/ml) was incubated with bacterial cells in the anaerobic chamber at 37°C for 24–48 hours. Reaction was terminated by adding the same volume of ice-cold acetonitrile. After vortexing for 30 seconds, samples were centrifuged at 16,100g for 10 minutes and the supernatant was collected for high-performance liquid chromatography (HPLC)/UV analysis as described subsequently.
M1 Detection.
The reaction mixture was analyzed by using a 2695 HPLC system (Waters, Milford, MA) coupled with a 2487 UV detector (Waters). Typically, 50 μl of a sample was injected and resolved on a C8 column (Eclipse XDB-C8; 4.6 × 250 nm; 5 μm; Agilent, Santa Clara, CA) using water (0.02 M KH2PO4, pH 3.5; solvent A) and acetonitrile (solvent B) as the mobile phase with the following gradient: 0–12 minutes (50% B), 12–17 minutes (50%–70% B), 17–23 minutes (70% B), 24–30 minutes (90% B), and 30–40 minutes (50% B). Eluates were monitored at 210 nm.
For further verification of M1 production by gut bacteria, the supernatant was also analyzed by HPLC–tandem mass spectrometry (HPLC-MS/MS), an Agilent 1200 HPLC interfaced with an Applied Biosystems (Foster City, CA) Qtrap 3200 using an electrospray ion source. The mobile phase consisted of water with 0.1% formic acid and 0.1% ammonium formate (v/v; solvent A) and methanol (solvent B), and the following gradient was used: 0–2 minutes (40% B), 2–6 minutes (95% B), and 6–12 minutes (40% B). Separation was performed on an Xterra MS C18 column (2.1 × 50 mm, 3.5 μm; Waters) at a flow rate of 0.3 ml/min, and M1 was detected at mass-to-charge (m/z) ratio 828.5/463.5 in the multiple reactions monitoring mode.
Tacrolimus Metabolism by Hepatic Microsomes.
Mouse or human hepatic microsomes (purchased from Corning Life Sciences (Durham, NC); 3 mg microsomal protein/ml) were incubated with tacrolimus (100 μg/ml) in a reaction mixture (1 mM NADP+, 5 mM MgCl2, 0.2 U/l isocitrate dehydrogenase, and 5 mM isocitric acid) at 37°C for 2 hours aerobically. The reaction was terminated by adding the same volume of ice-cold acetonitrile, followed by centrifugation at 16,100g for 10 minutes, and the supernatant was analyzed by HPLC/UV as described previously.
Purification of the Metabolite M1.
F. prausnitzii cells were harvested from 1 l of an overnight culture grown in YCFA media and resuspended in 500 ml phosphate-buffered saline (PBS) containing 50 mg of tacrolimus. After incubation at 37°C for 4 days, cells were removed by centrifugation and supernatant was collected. The supernatant was extracted twice, each with 500 ml of ethyl acetate. The upper organic layer was collected and evaporated using a rotary evaporator. Dried extracts were then dissolved in 800 µl of methanol and the metabolite M1 was purified using a semipreparative 996 HPLC coupled with a photodiode array detector (Waters) and equipped with a Microsorb 60-C8 Dynamax column (Agilent; 250 × 10 mm). The mobile phase consisted of water (solvent A) and acetonitrile (solvent B), and the following gradient was used: 0–12 minutes (60% B), 12–17 minutes (60%–70% B), 17–23 minutes (70% B), 23–25 minutes (70%–100% B), 25–35 minutes (100% B), 35–40 minutes (100%–60% B), and 40–50 minutes (60% B). A peak at 19.5 minutes corresponding to M1 was collected, dried, and subjected to structure determination.
Infrared and Nuclear Magnetic Resonance Spectroscopy.
Infrared spectra were acquired on neat samples using a Thermo-Nicolet (Thermo Electron Corporation, Madison WI) 6700 with the Smart iTRTM accessory. One- and two-dimensional NMR spectra were obtained on a Bruker (Billerica, MA) AVII 900 MHz spectrometer equipped with a 5 mm TCI CryoProbe (Bruker). NMR chemical shifts were referenced to residual solvent peaks (CDCl3 δH 7.26 and δC 77.16). NMR experiments included 1H NMR, distorsionless enhancement by polarization transfer quaternary, homonuclear 1H-1H correlation spectroscopy, heteronuclear single quantum coherence spectroscopy, heteronuclear multiple bonds correlation spectroscopy, and 1H-13C heteronuclear single quantum coherence–total correlated spectroscopy.
Mass Spectrometry for M1 Identification.
Experiments were performed on a Shimadzu (Columbia, MD) ultra-performance liquid chromatography mass spectrometry ion trap-time of flight. Samples were run on a C18 column (Kinetex; 50 × 2.1 mm; 1.7 μm; Phenomenex, Torrance, CA) at a flow rate of 0.5 ml/min with water/0.1% formic acid (solvent A) and acetonitrile/0.1% formic acid (solvent B) as the mobile phase. The gradient program was set from 20% to 100% B for 7 minutes, held at 100% for 1 minute, and returned to initial conditions for re-equilibration. High-resolution mass spectrometry spectra were acquired in both positive and negative modes with a scanning range from 150 to 2000 m/z, detector voltage at 1.7 kV, nebulizing gas (N2) flow at 1.5 l/min, drying gas (N2) pressure at 130 kPa, Curved Desolvation Line temperature at 200°C, and block heater temperature at 200°C. MS/MS fragmentation was performed with collision energy and collision gas set to 50% and frequency set to 45 kHz. Additional MS/MS analyses were performed on an Impact II QTOF (Bruker) with a scanning range from 50 to 1500 m/z, capillary voltage at 4.5 kV, nebulizer gas pressure (N2) at four bars, drying gas flow at 12 l/min, and temperature at 225°C. The three most intense ions per MS1 were selected for MS2, with active exclusion after three spectra. Each spectrum is an average of 65%–100% stepping with the collision energy set at 70 eV.
Immunosuppressant Activity.
The immunosuppressant activity of tacrolimus and M1 was determined by measuring the proliferation of PBMCs as previously described (Messele et al., 2000) with a slight modification. Briefly, cryopreserved PBMCs were stabilized in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA) containing 10% heat-inactivated fetal bovine serum at 37°C and 5% CO2 for 24 hours. Cells were seeded at 1 × 106 cells/ml in 96-well round-bottom plates. After incubation for 24 hours, cells were pretreated with tacrolimus, M1, or vehicle for 1 hour, followed by treatment with phytohemagglutinin (5 µg/ml) and 5-bromo-2′-deoxyuridine (20 µM) for 48 hours. Cells were centrifuged at 1000g for 5 minutes, washed with PBS, and fixed with 4% paraformaldehyde for 15 minutes. The fixed cells were permeabilized with 0.4% Triton X-100 for 5 minutes and incubated with 2 N HCl at 37°C for 30 minutes. After washing with PBS, the cells were incubated with 100 mM borate buffer (pH 8.0) for 10 minutes and washed again with PBS. After blocking with 2% bovine serum albumin for 1 hour, cells were incubated with horseradish peroxidase–conjugated 5-bromo-2′-deoxyuridine antibody for 1 hour at room temperature. Cells were then washed with PBS and incubated with 3,3′,5,5′-tetramethylbenzidine (a horseradish peroxidase substrate) for 30 minutes. The reaction was stopped by adding 2 N HCl. The absorbance was measured at 450 nm on a plate reader (BioTek, Winooski, VT).
Antifungal Assay.
The antifungal activity of tacrolimus and M1 was examined as previously described (Ianiri et al., 2017). Briefly, Malassezia sympodialis M1154/77 (a gift from Dr. Joseph Heitman, Duke University, Durham, NC), grown overnight in modified Dixon medium at 37°C, was plated on modified Dixon agar. After 1 hour incubation, an aliquot (3 µl) of tacrolimus or M1 at different concentrations was spotted on top of the agar, and incubated at 37°C for 2 days. The agar plates were visually inspected, and the images were taken using a camera.
Healthy Volunteers’ Stool Samples.
Fresh stool samples from healthy adults (100 mg wet weight/ml) were incubated with tacrolimus (100 μg/ml) anaerobically for 48 hours at 37°C. As controls, the stool samples were boiled for 10 minutes and then incubated with tacrolimus. The incubation mixtures were analyzed by HPLC/UV as described previously. The study protocol for human stool sample collection was approved by the Institutional Review Board at the University of Illinois at Chicago (protocol number 2018-0810).
Kidney Transplant Recipients’ Stool Samples.
Stool samples were collected from 10 kidney transplant recipients during the first month after transplantation at Weill Cornell Medicine and stored at −80°C until analysis. Tacrolimus dosing in each patient was adjusted to achieve a target therapeutic level of 8–10 ng/ml. The study protocol for kidney transplant stool sample collection was approved by the Institutional Review Board at Weill Cornell Medicine (protocol number 1207012730).
The microbiota composition of the stool samples was determined using 16S rRNA gene deep sequencing as previously described (Lee et al., 2018). In brief, DNA from stool samples was isolated using a phenol chloroform bead-beater extraction method. The V4-V5 hypervariable region was amplified by polymerase chain reaction and the fragments were sequenced on an Illumina (San Diego, CA) MiSEq system (250 × 250 base pairs). The 16S rRNA gene paired-end reads were analyzed using UPARSE (Edgar, 2013) and taxonomic classification was performed using a custom Python script incorporating BLAST (Altschul et al., 1990) with the National Center for Biotechnology Information RefSeq database (Tatusova et al., 2014) as a reference training set.
For the measurement of baseline levels of tacrolimus and M1 in stool samples, an aliquot of stool samples was suspended in PBS (final concentration 20 mg/ml). Also, to measure the capacity of stool samples to produce M1, an aliquot of stool samples was suspended in PBS (10 mg/ml) and incubated with tacrolimus anaerobically for 24 hours at 37°C. These samples were mixed with five volumes of acetonitrile containing ascomycin as an internal standard. An aliquot (10 μl) was injected into an Agilent 1290 UPLC coupled with Applied Biosystems Qtrap 6500. The mobile phase consisted of water with 0.1% formic acid and 10 mM ammonium formate (solvent A) and methanol (solvent B), and the following gradient was used: 0–2 minutes (20% B), 2–5 minutes (90% B), and 5–8 minutes (20% B). Separation was performed on the Xterra MS C18 column (2.1 × 50 mm, 3.5 μm; Waters) at a flow rate of 0.3 ml/min, with the column temperature set at 50°C. M1, tacrolimus, and ascomycin were detected at m/z ratios of 828.5/463.4, 821.6/768.6, and 809.5/756.5, respectively, in the multiple reactions monitoring mode. Standard curves (2–100 ng/ml for both tacrolimus and M1) were prepared by spiking tacrolimus or M1 into the stool samples of healthy volunteers.
Estimation of the Extent of Tacrolimus Metabolism by Intestinal Bacteria.
F. prausnitzii was grown overnight in YCFA medium. The overnight culture typically reaches an OD600 of ∼2, which corresponds to ∼1.6 × 108 F. prausnitzii cells/ml. Cells were harvested by centrifugation at 2000g for 5 minutes, resuspended in PBS, and then serially diluted in PBS (OD600 0.02, 0.2, 0.4, 0.8, 1.6, and 2). To determine the relationship between the number of bacterial cells and the extent of M1 formation, the cell suspensions at different densities were incubated with tacrolimus (10 μg/ml) at 37°C for 2 hours under anaerobic conditions. The reaction was stopped by adding four volumes of ice-cold acetonitrile containing ascomycin as an internal standard. After vortexing (1 minute) and centrifugation at 16,100g (10 minutes), the supernatant (2 μl) of each sample was injected into the HPLC-MS/MS system (Agilent 1200 HPLC interfaced with Applied Biosystems Qtrap 3200) and M1 concentrations were determined as described previously. To examine the relationship between incubation time and M1 formation, F. prausnitzii cells (OD600 0.8, equivalent to 6.3 × 107 cells/ml) in PBS were incubated with tacrolimus (10 μg/ml) for different times (0.5, 1, 2, 4, 8, and 24 hours), and M1 formation was determined as described previously. To examine the relationship between tacrolimus concentrations and M1 formation, tacrolimus at different concentrations (2, 10, 20, 40, and 50 μg/ml) was incubated with F. prausnitzii cells (OD600 0.8) for 1 hour, and M1 formation was determined as described previously. Assuming that the capabilities of bacteria in human small intestine to produce M1 are similar to that of F. prausnitzii cells in PBS, the total amount of M1 formed in the small intestine was estimated as previously reported (McCabe et al., 2015) with modifications:
 |
(1) |
 |
(2) |
The value of 4 × 1010 cells was used as the total number of bacteria in the small intestine (Sender et al., 2016), and 3.3 hours was used as the small intestine transit time (Yu et al., 1996).
Statistical Analysis.
Statistical analyses for comparison between two groups were performed using Wilcoxon rank sum testing. Correlational analysis between two continuous variables was performed using the Spearman correlation. A value of P ≤ 0.05 was considered statistically significant. All statistical analyses were performed using R version 3.3.1 and RStudio version 0.99.902.
Results
F. prausnitzii Potentially Metabolizes Tacrolimus.
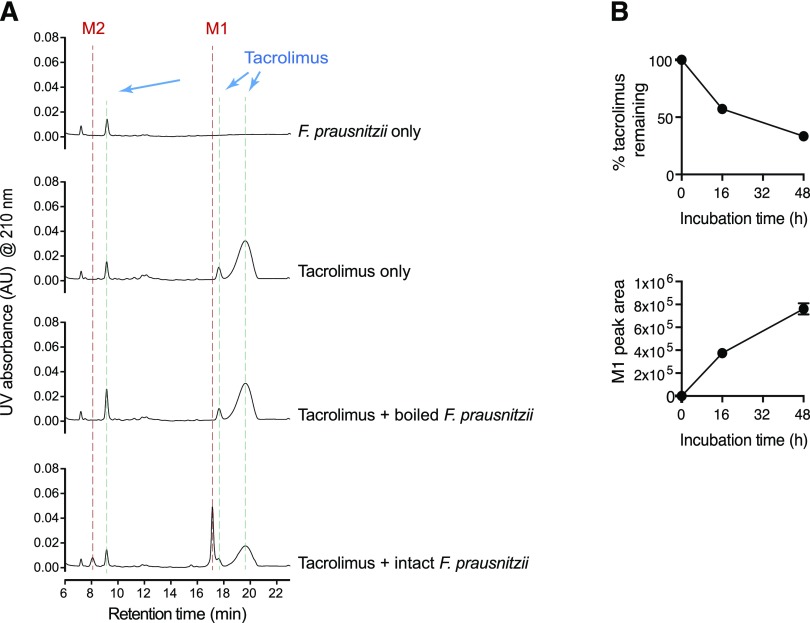
To determine whether F. prausnitzii is capable of metabolizing tacrolimus, cells of F. prausnitzii A2-165 strain grown overnight (in YCFA media) were incubated with tacrolimus (100 μg/ml; 124 μM) anaerobically at 37°C. After 48-hour incubation, the mixture was resolved using HPLC and analyzed by a UV detector. The HPLC chromatogram of intact tacrolimus showed multiple peaks, demonstrating tautomer formation as previously reported (Namiki et al., 1993) (Fig. 1A). For estimation of a concentration of intact tacrolimus, the area of the largest peak at the retention time of 19.7 minutes was used. After 24-hour incubation with F. prausnitzii, the concentration of tacrolimus was decreased by ∼50% (Fig. 1B), which was accompanied by appearance of two new peaks (designated as M1 and M2 in Fig. 1A). The M1 and M2 peaks were not observed when tacrolimus was incubated with boiled F. prausnitzii cells (Fig. 1A), indicating that the production of M1 and M2 requires live bacterial cells. Similar to strain A2-165, two additional strains of F. prausnitzii (ATCC 27766 and ATCC 27768; American Type Culture Collection) were found to produce M1 and M2 (Supplemental Fig. 1), suggesting that this function is likely conserved in different strains of F. prausnitzii.
Fig. 1.
F. prausnitzii metabolizes tacrolimus. (A) F. prausnitzii (OD600 2.6) cultured in YCFA media was incubated with tacrolimus (100 μg/ml) anaerobically at 37°C for 48 hours. The mixture was analyzed by using HPLC/UV. (B) Time profiles of tacrolimus disappearance and M1 appearance upon anaerobic incubation of tacrolimus (100 μg/ml) with F. prausnitzii.
M1 Is a C9 Keto-Reduction Metabolite of Tacrolimus.
To gain insight into the chemical identity of M1 and M2, high-resolution mass spectrometry and HPLC-MS/MS experiments were performed. The m/z values of M1 and M2 were [M + Na]+ 828.4846 and 846.4974, respectively, which are consistent with the formulas C44H71NO12Na (with a calculated mass of 828.4874 Da) for M1 (Supplemental Fig. 2) and C44H73NO13Na (with a calculated mass of 846.4980 Da) for M2. The calculated formulas suggested M1 to be a reduction product of tacrolimus (i.e., addition of 2H to the parent tacrolimus) and M2 to be a tautomer of M1. The fragmentation pattern of M1 compared with that of tacrolimus indicated that M1 is likely a keto-reduction product of tacrolimus (Supplemental Figs. 2 and 3).
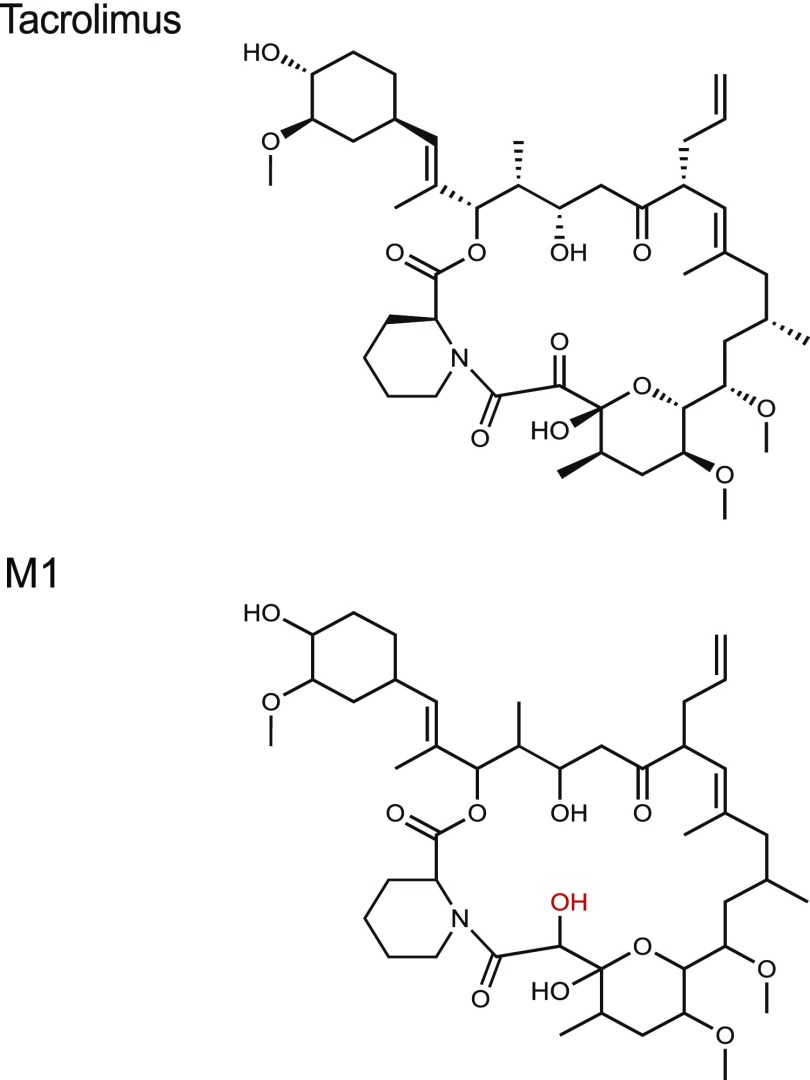
For structural elucidation, we focused on the major product M1. M1 was mass produced by incubating large amounts of tacrolimus with F. prausnitzii, followed by purification using preparative HPLC. The chemical structure of M1 was then determined using various spectroscopic methods. Of note, when the purified M1 was reinjected into HPLC/UV, it resolved into multiple peaks (including one corresponding to M2), indicative of isomerization and/or tautomerization of M1 into M2 (Supplemental Fig. 4). Infrared spectroscopy further supported that M1 is a product of a carbonyl reduction from tacrolimus (Supplemental Fig. 5). Major differences were observed in the C=O and O–H stretch regions of the infrared spectra. NMR spectra showed three major isomers of M1 in CDCl3, for which all resonances were assigned (Supplemental Tables 3–5). Detailed analysis of one- and two-dimensional NMR spectra revealed the site of carbonyl reduction at C-9 and the identity of M1 to be 9-hydroxy-tacrolimus (Supplemental Figs. 6–12). In particular, analysis of the distorsionless enhancement by polarization transfer quaternary spectrum of M1 revealed the absence of the resonances associated with the carbonyl carbon C-9 found in tacrolimus (δC 196.3 for the major isomer; δC 192.7 for the minor isomer) (Supplemental Fig. 13). Instead, three resonances consistent with the reduction of the carbonyl at C-9 to an alcohol were observed at δC 73.0 ppm (isomer I), δC 68.4 ppm (isomer II), and δC 69.7 ppm (isomer III). These resonances were associated with protons at δH 4.02, 4.51, and 4.37 ppm, respectively, in the heteronuclear single quantum coherence spectrum. In turn, the latter resonances showed homonuclear 1H–1H correlation spectroscopy correlations to exchangeable protons (δH 4.23, 3.21, and 3.58, respectively). The heteronuclear multiple bonds correlation spectroscopy correlations from H-9 to C-8 and C-10 were observed (Supplemental Tables 3–5), supporting the assignment of M1 as 9-hydroxy-tacrolimus. These results establish the structure of M1 as the C-9 keto-reduction product of tacrolimus (Fig. 2).
Fig. 2.
Chemical structures of tacrolimus and F. prausnitzii–derived metabolite M1. M1 structure was identified using mass spectrometry and nuclear magnetic resonance spectroscopy.
M1 Is a Less Potent Immunosuppressant than Tacrolimus.
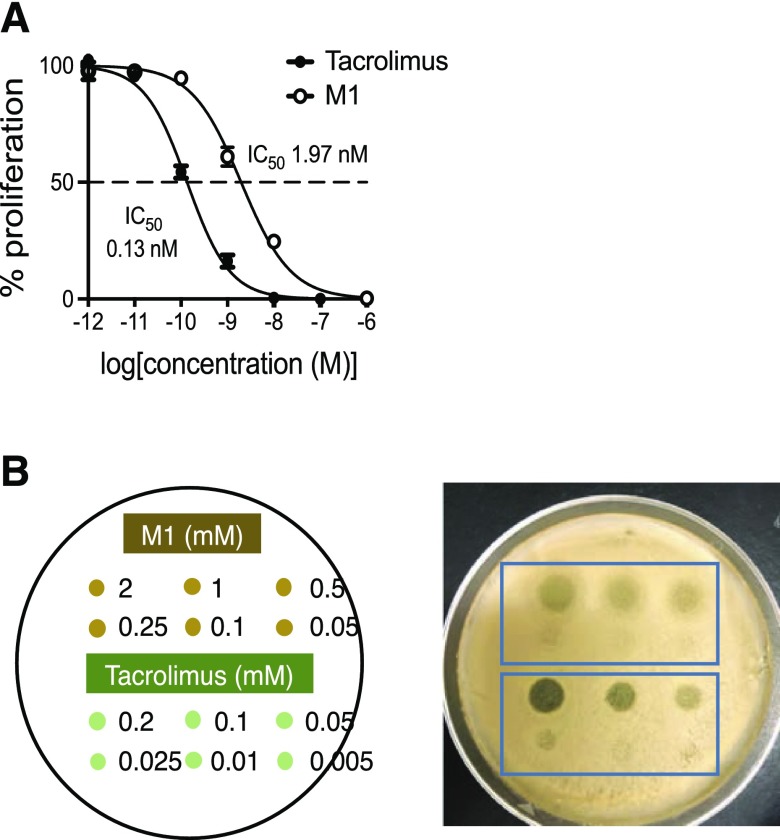
We compared the activities of M1 and tacrolimus by measuring PBMC proliferation after treatment with T-lymphocyte mitogen phytohemagglutinin in the presence of M1 or tacrolimus (Messele et al., 2000). The IC50 value of M1 was 1.97 nM, whereas the IC50 value of tacrolimus was 0.13 nM, demonstrating that M1 was ∼15-fold less potent than the parent tacrolimus in inhibiting T-lymphocyte proliferation (Fig. 3A). Tacrolimus is known to exhibit antifungal activity via the same mechanism for immunosuppression (Steinbach et al., 2007). To further examine the pharmacological activity of M1, an antifungal assay was performed. An aliquot of M1 or tacrolimus was placed onto a lawn of the yeast M. sympodialis, and the antifungal activities were estimated based on the size of the halo formed. M1 was about 10–20-fold less potent than tacrolimus in inhibiting the yeast growth (Fig. 3B), consistent with the results obtained from the PBMC proliferation assay. Taken together, these results demonstrate that M1 is less potent as an immunosuppressant and antifungal agent than the parent drug tacrolimus is.
Fig. 3.
M1 is less potent than tacrolimus as an immunosuppressant and antifungal agent. (A) Immunosuppressant activities of tacrolimus and M1 were examined in PBMCs by measuring cell proliferation after treatment with a T-lymphocyte mitogen in the presence of tacrolimus or M1. (B) Antifungal activities of tacrolimus and M1 were examined using Malassezia sympodialis. The yeast was inoculated on a modified Dixon agar plate. After 1 hour incubation, an aliquot of tacrolimus or M1 at different concentrations was placed on the plate, as shown in the left panel, and incubated at 37°C for 2 days.
Tacrolimus Is Metabolized by a Wide Range of Commensal Gut Bacteria.
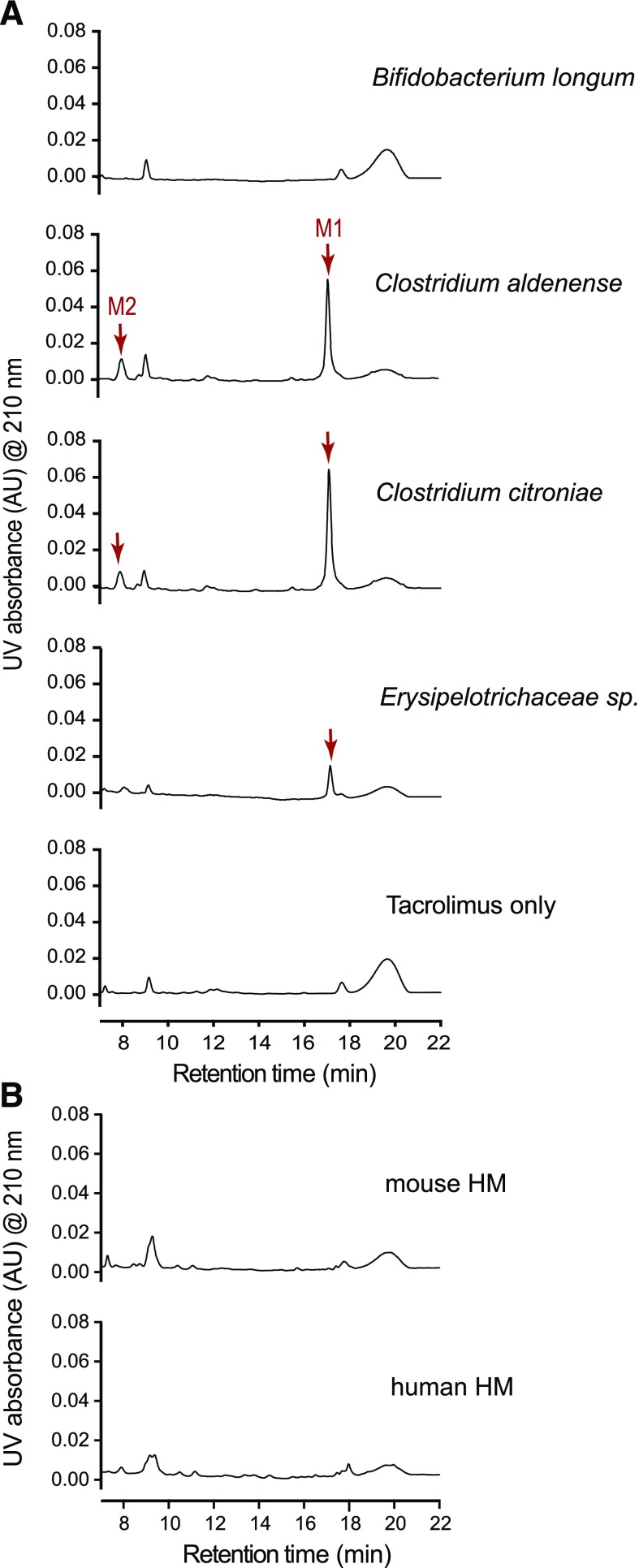
To determine whether other gut bacteria can produce M1/M2 from tacrolimus, we obtained 22 human gut bacteria from the Biodefense and Emerging Infections Research Resources Repository (Supplemental Table 1) and tested them for potential tacrolimus metabolism. The tested bacteria included those belonging to major orders that are known to be highly abundant in the human gut (Qin et al., 2010; Arumugam et al., 2011). Bacteria grown overnight in YCFA media anaerobically were incubated with tacrolimus (100 μg/ml) for 48 hours, and the mixtures were analyzed by HPLC/UV. Apparently, gut bacteria in the orders of Clostridiales and Erysipelotrichales (but not those in Bacteroidales and Bifidobacteriales) produced M1 (Fig. 4A; Table 1). To further verify the results, the mixtures were reanalyzed by HPLC-MS/MS, which exhibits higher sensitivity than HPLC/UV. M1 production by bacteria in Clostridiales was verified (a representative chromatogram of Clostridium citroniae is shown in Supplemental Fig. 14). M1 production by bacteria in Bacteroidales was detectable by HPLC-MS/MS, albeit at ∼100-fold lower levels than that by bacteria in Clostridiales (Supplemental Fig. 14). The M1 peak was not detected upon tacrolimus incubation with Bifidobacterium longum (Supplemental Fig. 14). The formation of M1 was not observed when tacrolimus was incubated with either human or mouse hepatic microsomes (Fig. 4B), also verified by HPLC-MS/MS (data not shown), suggesting that M1 is uniquely produced by gut bacteria.
Fig. 4.
Multiple commensal gut bacteria convert tacrolimus to M1. (A) Representative chromatograms of bacteria incubated with tacrolimus. M1 nonproducer (Bifidobacterium longum) or producer (Clostridium aldenense, Clostridium citroniae, and Erysipelotrichaceae sp.) cultured overnight in YCFA media was incubated with tacrolimus (100 μg/ml) anaerobically at 37°C for 48 hours. The mixture was analyzed by using HPLC/UV at 210 nm. (B) Mouse or human hepatic microsomes [(HMs); 3 mg microsomal protein/ml] were incubated with tacrolimus (100 μg/ml) at 37°C for 2 hours aerobically. The mixture was analyzed by using HPLC/UV.
TABLE 1.
Screening gut bacteria for tacrolimus conversion to M1 in YCFA culture
| Order | Bacterium | OD600 | M1 Production Detected |
|---|---|---|---|
| Bifidobacteriales | Bifidobacterium longum | 1.8 | No |
| Bacteroidales | Bacteroides cellulosilyticus | 0.6 | Yesa |
| Bacteroides finegoldii | 3.4 | Yesa | |
| Bacteroides ovatus | 4.2 | Yesa | |
| Parabacteroides merdae | 2.7 | Yesa | |
| Parabacteroides johnsonii | 3.6 | Yesa | |
| Parabacteroides goldsteinii | 3.3 | Yesa | |
| Clostridiales | Ruminococcaceae sp. | 0.5 | Yes |
| Clostridium innocuum | 3.4 | Yes | |
| Anaerostipes sp. | 2.7 | Yes | |
| Dorea formicigenerans | 2.4 | Yes | |
| Clostridium clostridioforme | 3.0 | Yes | |
| Clostridium hathewayi | 2.6 | Yes | |
| Blautia sp. | 4.7 | Yes | |
| Clostridium aldenense | 1.4 | Yes | |
| Clostridium symbiosum | 2.5 | Yes | |
| Clostridium citroniae | 1.7 | Yes | |
| Coprococcus sp. | 2.4 | Yes | |
| Clostridium bolteae | 3.6 | Yes | |
| Clostridium cadaveris | 1.4 | Yes | |
| Ruminococcus gnavus | 3.4 | Yes | |
| Erysipelotrichales | Erysipelotrichaceae sp. | 3.8 | Yes |
M1 production observed only when using sensitive HPLC-MS/MS for detection.
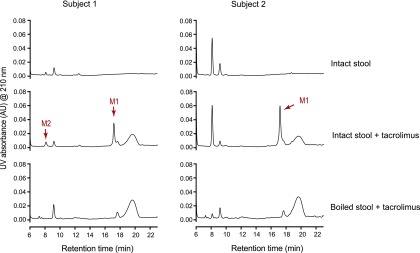
To examine whether tacrolimus metabolism is indeed mediated by human gut microbiota, fresh stool samples from two healthy adults were incubated with tacrolimus, and M1 production was assessed. Both stool samples produced M1, whereas the control stool samples that were boiled prior to tacrolimus incubation did not (Fig. 5). Taken together, these results show that commensal gut bacteria belonging to different genera metabolize tacrolimus into the less potent M1 metabolite.
Fig. 5.
Human gut microbiota convert tacrolimus to M1. Tacrolimus (100 μg/ml) was incubated anaerobically with human stool samples from two different subjects (100 mg wet weight/ml) for 48 hours at 37°C. A separate set of samples was boiled for 10 minutes before incubation with tacrolimus. The incubation mixtures were analyzed by HPLC/UV.
M1 Is Detected in Transplant Patients’ Stool Samples.
F. prausnitzii is one of the most abundant human gut bacteria species (Qin et al., 2010; Arumugam et al., 2011), and its fecal abundance was shown to have a positive correlation with oral tacrolimus dosage (Lee et al., 2015). To explore a potential role of F. prausnitzii in tacrolimus metabolism in kidney transplant recipients, we evaluated 10 stool samples from kidney transplant recipients who were taking oral tacrolimus (demographic information provided in Table 2 and Supplemental Table 2). Based on the sequencing results of the V4-V5 hypervariable region of the 16S rRNA gene in stool samples, we selected five kidney transplant recipients whose stool samples had a relative gut abundance of F. prausnitzii greater than 25% (designated as the high F. prausnitzii group) and five kidney transplant recipients whose stool samples showed no to little (if any) presence of F. prausnitzii (designated as the low F. prausnitzii group). We first determined the baseline levels of tacrolimus and M1 in the stool samples. We were able to measure baseline tacrolimus levels in 8 of the 10 stool samples, but we did not detect a significant difference in the baseline tacrolimus level between the high and low F. prausnitzii groups (median 0.63 vs. 0.29 ng/mg, respectively, P = 0.46). We were also able to measure baseline M1 levels in five of the 10 stool samples, but we did not detect a significant difference in the baseline M1 level between the high and low F. prausnitzii groups (median 0.12 vs. <0.1 ng/mg, respectively, P = 0.48). Next, we tested the stool samples of both high and low F. prausnitzii groups for the capability of M1 production by incubating each of them with tacrolimus (10 μg/ml) for 24 hours. M1 production was detected in all 10 samples, but the amount produced was similar between the high and low F. prausnitzii groups (median 4.5 vs. 7.1 ng/mg, respectively, P = 0.31). The 16S rDNA sequencing analysis revealed that gut bacteria belonging to the Clostridiales order (a main group of bacteria that are expected to produce the majority of M1) were highly abundant in all 10 samples (Table 2). However, the relative abundance of neither F. prausnitzii (ρ = −0.36, P = 0.31) nor Clostridiales (ρ = 0.44, P = 0.20) showed a significant correlation with M1 production. Oral tacrolimus doses (to maintain therapeutic blood concentrations) were similar between the high and low F. prausnitzii groups (median 6 vs. 4 mg/d, respectively, P = 0.34) (Table 2).
TABLE 2.
M1 levels in kidney transplant patients’ stool samples
| Patient |
Age |
Gender |
Post-Transplant Day |
Tacrolimus Oral Dosea |
Fecal Abundance |
Baseline Level in Stool Samples |
M1 Production upon Tacrolimus Incubation |
||
|---|---|---|---|---|---|---|---|---|---|
|
F. prausnitzii |
Clostridiales |
Tacrolimus |
M1 |
||||||
| yr | mg/day | % | % | ng/mg stool | ng/mg stool | ng/mg stool | |||
| 1 | 45 | Female | 31 | 9 | 46 | 86 | 0.88 | 0.38 | 5.1 |
| 2 | 56 | Male | 18 | 3 | 39 | 89 | BQLb | BQLb | 3.5 |
| 3 | 61 | Male | 20 | 5 | 32 | 71 | 0.63 | BQLb | 4.5 |
| 4 | 59 | Female | 12 | 6 | 27 | 76 | 0.71 | 0.12 | 2.9 |
| 5 | 50 | Male | 32 | 10 | 26 | 79 | 0.37 | 0.41 | 6.4 |
| 6 | 52 | Female | 28 | 6 | ND | 15 | 0.29 | BQLb | 3.5 |
| 7 | 57 | Male | 15 | 3 | ND | 44 | 0.85 | BQLb | 4.1 |
| 8 | 71 | Male | 18 | 4 | ND | 95 | BQLb | 0.60 | 7.1 |
| 9 | 25 | Male | 27 | 4 | ND | 74 | 0.49 | BQLb | 12.6 |
| 10 | 52 | Male | 32 | 6 | ND | 95 | 0.14 | BQLb | 11.0 |
BQL, below the quantification limit; ND, not detected.
At the time of stool collection.
Below the quantification limit (i.e., 0.1 ng/mg stool).
Extensive Tacrolimus Metabolism May Occur in Human Small Intestine.
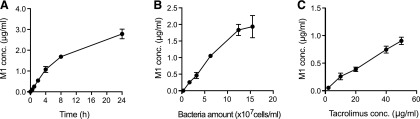
For gross estimation of the extent of tacrolimus metabolism in human small intestine, M1 production kinetic profiles were obtained using F. prausnitzii as a model bacterium. M1 production increased linearly with incubation time up to 4 hours (Fig. 6A) and the amount of F. prausnitzii up to 1.2 × 108 cells/ml (Fig. 6B). M1 production increased with the increasing concentrations of tacrolimus (Fig. 6C) and did not reach a plateau at the highest concentration tested (50 μg/ml; a concentration attained when a typical tacrolimus oral dose of 5 mg is dissolved in 100 ml water). Based on the assumption that bacteria in human small intestine exhibit M1 production capabilities similar to that of F. prausnitzii in PBS, the extent of M1 production in small intestine (at the 50 μg/ml tacrolimus concentration) was estimated to be 1.9 mg.
Fig. 6.
M1 formation by small intestinal bacteria may be extensive. (A) Tacrolimus (10 μg/ml) was incubated anaerobically with F. prausnitzii (6.3 × 107 cells/ml) for varying amounts of time. (B) Tacrolimus (10 μg/ml) was incubated with varying amount of F. parusnitzii in PBS at 37°C for 2 hours. (C) Tacrolimus at varying concentrations was incubated with F. prausnitzii (6.3 × 107 cells/ml) for 1 hour. M1 concentrations in the reaction mixtures were analyzed by liquid chromatography–MS/MS.
Discussion
In this study, we have demonstrated that a wide range of commensal gut bacteria can metabolize tacrolimus into a novel metabolite M1 (9-hydroxy-tacrolimus). To the best of our knowledge, this represents the first experimental evidence for commensal gut bacteria being involved in the metabolism of tacrolimus.
The extent of M1’s contribution to overall immunosuppression by tacrolimus therapy is unclear. M1 is ∼15-fold less potent than tacrolimus in inhibiting both the proliferation of activated T-lymphocytes and the growth of the yeast M. sympodialis. This result is consistent with the currently available structure-activity relationships of tacrolimus analogs; modifications at the C-9 position affect the interaction of tacrolimus with its effector protein (i.e., FK506 binding protein 12) and lead to decreased immunosuppressant activities (Goulet et al., 1994). While the systemic concentrations of M1 after oral tacrolimus dosing remain to be measured, results from previous tacrolimus disposition studies using a radiolabeled compound (Möller et al., 1999) indicate that the blood concentrations of metabolites are likely lower than that of tacrolimus. These results suggest that pharmacological activity originated from circulating M1 is likely less than that from tacrolimus. Of note, certain tacrolimus metabolites (e.g., 13-O-demethyltacrolimus), independent of their immunosuppressive activities, crossreact with the antibodies used in the immunoassays for measurement of tacrolimus blood concentrations, leading to overestimation of tacrolimus concentrations (Staatz and Tett, 2004; Dubbelboer et al., 2012). Interestingly, the extent of such overestimation could not be fully explained by the crossreactivity of currently known tacrolimus metabolites (Dubbelboer et al., 2012). Whether the novel metabolite M1 crossreacts with the antibodies, accounting in part for the overestimation of tacrolimus concentrations, is currently being investigated.
Multiple factors have been reported to contribute to the low and variable bioavailability of orally administered tacrolimus. These include differential expression and/or activity levels of cytochrome P450 enzymes (especially CYP3A4 and CYP3A5 isoforms) and the drug transporter P-glycoprotein (P-gp) in the intestine and liver (Staatz and Tett, 2004). Previous pharmacokinetics studies in healthy volunteers and renal transplant recipients have shown that hepatic extraction of tacrolimus is very low (i.e., 4%–8%) (Floren et al., 1997; Tuteja et al., 2001), suggesting that the low oral bioavailability of tacrolimus is mainly due to drug loss in the gut. P-gp-mediated drug efflux and intestinal CYP3A-mediated metabolism were proposed as major contributors to the loss. However, results from drug-drug interaction studies have shown that oral bioavailability of tacrolimus increases to at most ∼30% when coadministered with ketoconazole, a potent inhibitor of CYP3As and P-gp (Floren et al., 1997; Tuteja et al., 2001); 70% of oral dose is lost (not reaching systemic circulation) even when intestinal CYP3A and P-gp activities are blocked by ketoconazole. Our results suggest that tacrolimus conversion to M1 in the gut may represent a previously unrecognized pathway of tacrolimus elimination in the gut, potentially contributing to tacrolimus loss in the gut.
We attempted to estimate the overall magnitude of tacrolimus metabolism in the human small intestine using F. prausnitzii as a model gut bacterium. F. prausnitzii was chosen because it is one of the most abundant bacterium (at the bacterial species level) in the human gut, including the small intestine (Sokol et al., 2008; Qin et al., 2010; Lopez-Siles et al., 2015). Our estimation indicates that about 1.9 mg of M1 may be produced in the small intestine during drug transit through the organ. Considering that the typical oral dose of tacrolimus ranges from 2 to 5 mg, a significant fraction of the orally administered tacrolimus may be lost by gut bacterial metabolism before absorption. On the other hand, it should be noted that our calculation may grossly overestimate or underestimate the true extent of tacrolimus metabolism in the gut because: 1) bacterial gene expression (and thereby function) in the gut is likely different from that in the laboratory medium used in our study, 2) the capacity of other gut bacteria to metabolize tacrolimus may be widely different compared with that of F. prausnitzii, and 3) a low solubility drug such as tacrolimus may reach the lower gastrointestinal tract (Sousa et al., 2008) and be presented to a large amount of gut bacteria in the colon. Slow absorption of tacrolimus over a prolonged period has been reported clinically (Venkataramanan et al., 1995). Studies are currently ongoing to measure the extent of tacrolimus metabolism by gut bacteria in mice.
Our results revealed that multiple commensal gut bacteria are capable of metabolizing tacrolimus, suggesting that differences in gut bacterial composition may lead to differential tacrolimus exposure in kidney transplant recipients. Gut bacteria that extensively metabolized tacrolimus into M1 (including F. prausnitzii) belong to the Clostridiales order. On the other hand, bacteria in Bacteroidales were found to be weak producers of M1 (i.e., detectable only by sensitive HPLC-MS/MS), and B. longum in Bifidobacteriales did not produce detectable amounts of M1. A previous study has shown that fecal abundance of F. prausnitzii (belonging to the Clostridiales order) was positively correlated with oral tacrolimus dose in 19 kidney transplant patients (Lee et al., 2015). However, we observed no differences in M1 production between high and low F. prausnitzii groups of stool samples. Also, we did not observe a correlation between Clostridiales abundance and M1 production in the stool samples. This may be due to the small number of samples used for this exploratory study and/or the quality of samples being nonoptimal for enzymatic assays. The presence of multiple factors affecting gut bacterial gene expression in vivo such as nutritional status of the gut may further explain why we did not observe a correlation between our in vitro culture-based results and in vivo abundance of gut bacteria. For example, the amino acid arginine was shown to repress the expression of the gene encoding digoxin-metabolizing enzyme in E. lenta, thus reducing digoxin elimination by gut bacteria (Haiser et al., 2013). Obviously, in vitro culture-based systems do not fully reflect the bacterial functions activated in the physiologic gut ecosystem. In this regard, our follow-up study is focused on the identification of the bacterial gene(s) responsible for tacrolimus metabolism. Such information will enable us to examine the prevalence and abundance of tacrolimus-metabolizing enzymes in the gut bacterial community and identify factors such as diet or drugs that alter gut bacterial composition and/or gene expression specific for tacrolimus metabolism.
In summary, we present evidence of tacrolimus metabolism by gut bacteria, providing potential explanations for its low oral bioavailability. Tacrolimus metabolism into M1 may represent a novel elimination pathway that occurs before intestinal absorption of tacrolimus. While the extent of gut metabolism of tacrolimus on variable tacrolimus exposure remains to be determined, our data provide a novel understanding of tacrolimus metabolism and may explain variability in tacrolimus exposures in kidney transplant recipients and patients with glomerular diseases on tacrolimus therapy.
Acknowledgments
We thank Joseph Heitman (Duke University, Durham, NC) for kindly providing the yeast strain and Jennifer C. Chang for proofreading the manuscript.
Abbreviations
- HPLC
high-performance liquid chromatography
- MS/MS
tandem mass spectrometry
- m/z
mass-to-charge ratio
- OD600
optical density at 600 nm
- PBMC
peripheral blood mononuclear cell
- PBS
phosphate-buffered saline
- P-gp
P-glycoprotein
- rRNA
ribosomal RNA
Authorship Contributions
Participated in research design: Guo, Crnkovic, Won, Yang, J. R. Lee, Orjala, H. Lee, Jeong.
Conducted experiments: Guo, Crnkovic, Won, Yang.
Performed data analysis: Guo, Crnkovic, Won, Yang, J. R. Lee, Orjala, H. Lee, Jeong.
Wrote or contributed to the writing of the manuscript: Guo, Crnkovic, Won, Yang, J. R. Lee, Orjala, H. Lee, Jeong.
Footnotes
This work was supported by the National Institutes of Health [Grant K23 AI 124464] (to J.R.L.) and Chicago Biomedical Consortium [Catalyst Award C‐066] (to H.J.).
J.R.L. receives research support from BioFire Diagnostics, LLC. The other authors have no conflicts of interest.
An earlier version of this paper appears in bioRχiv under the doi 10.1101/426197.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. MetaHIT Consortium (2011) Enterotypes of the human gut microbiome. Nature 473:174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubbelboer IR, Pohanka A, Said R, Rosenborg S, Beck O. (2012) Quantification of tacrolimus and three demethylated metabolites in human whole blood using LC-ESI-MS/MS. Ther Drug Monit 34:134–142. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. [DOI] [PubMed] [Google Scholar]
- Floren LC, Bekersky I, Benet LZ, Mekki Q, Dressler D, Lee JW, Roberts JP, Hebert MF. (1997) Tacrolimus oral bioavailability doubles with coadministration of ketoconazole. Clin Pharmacol Ther 62:41–49. [DOI] [PubMed] [Google Scholar]
- Goulet M, Rupprecht K, Sinclar P, Wyvratt M, Parsons W. (1994) The medicinal chemistry of FK-506. Perspect Drug Discov Des 2:145–162. [Google Scholar]
- Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. (2013) Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341:295–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianiri G, Applen Clancey S, Lee SC, Heitman J. (2017) FKBP12-dependent inhibition of calcineurin mediates immunosuppressive antifungal drug action in Malassezia. MBio 8:e01752-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel N, Maini Rekdal V, Balskus EP. (2017) Chemical transformation of xenobiotics by the human gut microbiota. Science 356:eaag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Magruder M, Zhang L, Westblade LF, Satlin MJ, Robertson A, Edusei E, Crawford C, Ling L, Taur Y, et al. (2018) Gut microbiota dysbiosis and diarrhea in kidney transplant recipients. Am J Transplant DOI: 10.1111/ajt.14974 [published ahead of print]. [DOI] [PMC free article] [PubMed]
- Lee JR, Muthukumar T, Dadhania D, Taur Y, Jenq RR, Toussaint NC, Ling L, Pamer E, Suthanthiran M. (2015) Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS One 10:e0122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M, Martinez-Medina M, Abellà C, Busquets D, Sabat-Mir M, Duncan SH, Aldeguer X, Flint HJ, Garcia-Gil LJ. (2015) Mucosa-associated Faecalibacterium prausnitzii phylotype richness is reduced in patients with inflammatory bowel disease. Appl Environ Microbiol 81:7582–7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe M, Sane RS, Keith-Luzzi M, Xu J, King I, Whitcher-Johnstone A, Johnstone N, Tweedie DJ, Li Y. (2015) Defining the role of gut bacteria in the metabolism of deleobuvir: in vitro and in vivo studies. Drug Metab Dispos 43:1612–1618. [DOI] [PubMed] [Google Scholar]
- Messele T, Roos MT, Hamann D, Koot M, Fontanet AL, Miedema F, Schellekens PT, Rinke de Wit TF. (2000) Nonradioactive techniques for measurement of in vitro T-cell proliferation: alternatives to the [3H]thymidine incorporation assay. Clin Diagn Lab Immunol 7:687–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S, Leclerc M, Martin R, Chain F, Lenoir M, Raguideau S, Hudault S, Bridonneau C, Northen T, Bowen B, et al. (2015) Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. MBio 6:e00300-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller A, Iwasaki K, Kawamura A, Teramura Y, Shiraga T, Hata T, Schäfer A, Undre NA. (1999) The disposition of 14C-labeled tacrolimus after intravenous and oral administration in healthy human subjects. Drug Metab Dispos 27:633–636. [PubMed] [Google Scholar]
- Namiki Y, Kihara N, Koda S, Hane K, Yasuda T. (1993) Tautomeric phenomenon of a novel potent immunosuppressant (FK506) in solution. I. Isolation and structure determination of tautomeric compounds. J Antibiot (Tokyo) 46:1149–1155. [DOI] [PubMed] [Google Scholar]
- Press RR, Ploeger BA, den Hartigh J, van der Straaten T, van Pelt J, Danhof M, de Fijter JW, Guchelaar HJ. (2009) Explaining variability in tacrolimus pharmacokinetics to optimize early exposure in adult kidney transplant recipients. Ther Drug Monit 31:187–197. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. MetaHIT Consortium (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi O, van Berkel LA, Chain F, Tanweer Khan M, Taverne N, Sokol H, Duncan SH, Flint HJ, Harmsen HJ, Langella P, et al. (2016) Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci Rep 6:18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BO, Bäckhed F. (2016) Signals from the gut microbiota to distant organs in physiology and disease. Nat Med 22:1079–1089. [DOI] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R. (2016) Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuker N, van Gelder T, Hesselink DA. (2015) Intra-patient variability in tacrolimus exposure: causes, consequences for clinical management. Transplant Rev (Orlando) 29:78–84. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. (2008) Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 105:16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, Basit AW. (2008) The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm 363:1–25. [DOI] [PubMed] [Google Scholar]
- Staatz CE, Tett SE. (2004) Clinical pharmacokinetics and pharmacodynamics of tacrolimus in solid organ transplantation. Clin Pharmacokinet 43:623–653. [DOI] [PubMed] [Google Scholar]
- Steinbach WJ, Reedy JL, Cramer RA, Jr, Perfect JR, Heitman J. (2007) Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat Rev Microbiol 5:418–430. [DOI] [PubMed] [Google Scholar]
- Tatusova T, Ciufo S, Fedorov B, O’Neill K, Tolstoy I. (2014) RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res 42:D553–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja S, Alloway RR, Johnson JA, Gaber AO. (2001) The effect of gut metabolism on tacrolimus bioavailability in renal transplant recipients. Transplantation 71:1303–1307. [DOI] [PubMed] [Google Scholar]
- Venkataramanan R, Swaminathan A, Prasad T, Jain A, Zuckerman S, Warty V, McMichael J, Lever J, Burckart G, Starzl T. (1995) Clinical pharmacokinetics of tacrolimus. Clin Pharmacokinet 29:404–430. [DOI] [PubMed] [Google Scholar]
- Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh LA, Mani S, et al. (2010) Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330:831–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LX, Crison JR, Amidon GL. (1996) Compartmental transit and dispersion model analysis of small intestinal transit flow in humans. Int J Pharm 140:111–118. [Google Scholar]