Abstract
Replication protein A (RPA) coordinates important DNA metabolic events by stabilizing single-strand DNA (ssDNA) intermediates, activating the DNA damage response and handing off ssDNA to appropriate downstream players. Six DNA binding domains (DBDs) in RPA promote high affinity binding to ssDNA yet also allow RPA displacement by lower affinity proteins. We generated fluorescent versions of Saccharomyces cerevisiae RPA and visualized the conformational dynamics of individual DBDs in the context of the full-length protein. We show that both DBD-A and DBD-D rapidly bind to and dissociate from ssDNA while RPA remains bound to ssDNA. The recombination mediator protein Rad52 selectively modulates the dynamics of DBD-D. These findings reveal how RPA interacting proteins with lower ssDNA binding affinities can access the occluded ssDNA and remodel individual DBDs to replace RPA.
One Sentence Summary:
The choreography of binding and rearrangement of the individual domains of RPA during homologous recombination is revealed.
Introduction
In eukaryotic cells, RPA binds to transiently exposed ssDNA and serves as a hub protein to coordinate DNA replication, recombination, repair, and telomere maintenance1,2. Cellular functions of RPA rely on its high ssDNA binding affinity, ability to physically interact with over two dozen DNA processing enzymes, and to correctly position these enzymes on complex DNA structures. The precise mechanisms by which RPA functions in many contexts and how RPA differentiates between multiple DNA metabolic events is a long-standing puzzle1,3. RPA is heterotrimeric, flexible, and modular in structure. It is composed of three subunits: RPA70, RPA32 and RPA14 (Figs.1a, b), which harbor six oligonucleotide/oligosaccharide binding folds (OB-folds; labeled A through F). We refer to the DNA binding OB-folds as DNA binding domains (DBDs; Fig. 1b). RPA binds to ssDNA with sub-nanomolar affinity, but can be displaced by DNA binding proteins with much lower DNA binding affinity. Recent studies have suggested that the RPA-ssDNA complex is relatively dynamic4,5,6 positing a selective dissociative mechanism where not all DBDs are stably bound to the DNA, whereas, microscopic dissociation of individual DNA binding domains occurs.
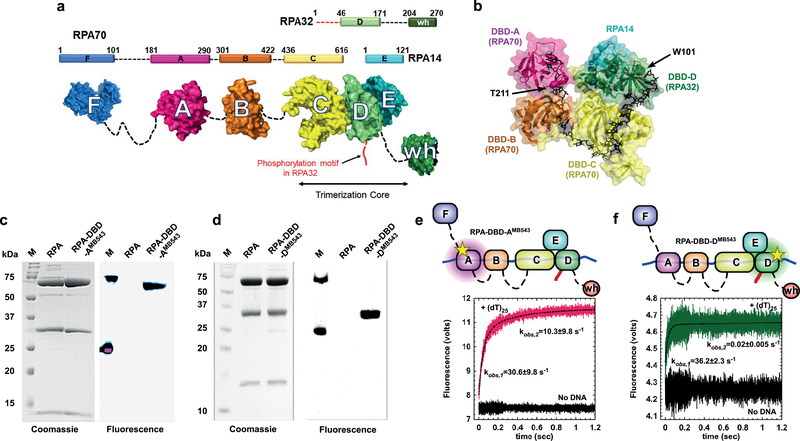
Figure 1. Non-canonical amino acid-based fluorescent RPAs report on individual DBD dynamics.
a) The residue numbers for the three RPA subunits and their respective DNA binding domains (DBDs A-F) are denoted. The winged-helix (wh) domain in RPA32 and DBD-F in RPA70 mediate interactions with RPA-interacting proteins (RIPs). The N-terminus of RPA32 that is phosphorylated is shown in red. Crystal structures of the ordered domains are shown as surface representations with intervening disordered linkers as dotted lines (black). DBD-C, DBD-D and RPA14 interact to form the trimerization core. b) Crystal structure of the DNA binding domains of U. maydis RPA bound to ssDNA (PDB ID:4GNX). Residues T211 in DBD-A and W101 in DBD-D are sites where 4-azidophenylalanine (4AZP) is incorporated (residue numbering in Saccharomyces cerevisiae RPA). The bound ssDNA is shown as sticks (black). c & d) Coomassie and fluorescence imaging of RPA complexes labeled with MB543 at either DBD-A or DBD-D. Only the fluorescently-labeled domains are visualized upon fluorescence imaging suggesting site-specific labeling of each domain, respectively. e & f) RPA-DBD-AMB543 and RPA-DBD-DMB543 binding to ssDNA was analyzed by monitoring the change in MB543 fluorescence. Robust change in fluorescence depicts engagement of specific DBDs onto ssDNA. Data were fit and analyzed as described in Methods. Values depicted in panels e & f represent the mean and s.e. from n=3 independent experiments. Uncropped gel images of panels c & d are shown in Supplementary Dataset 1.
In all existing models for RPA function, DBDs A and B are assigned as high affinity binding domains. Purified DBD-A, DBD-B and DBD-A/DBD-B constructs bind ssDNA with KD values of 2 μM, 20 μM and 50 nM, respectively7–9. The trimerization core made up of DBD-C, DBD-D and DBD-E is considered to have a weaker ssDNA binding affinity (Kd> 5 μM)10. Additionally, mutational analysis of individual aromatic residues that interact with the ssDNA in either DBD-C or DBD-D show minimal perturbations on ssDNA binding affinity6. Paradoxically, in the crystal structure of the RPA-ssDNA complex (Fig. 1b), the ssDNA interactions of all four DBDs are similar, with DBD-C having more contacts with ssDNA bases than DBD-A, DBD-B or DBD-D11. Thus, the exact nature of the contributions from each DBD to RPA function is likely complicated and may be influenced by the dynamics of DBD-ssDNA interaction.
Both the N-terminus of RPA70 and the C-terminus of RPA32 interact with distinct sets of RPA-interacting proteins (RIPs). During DNA processing, RIPs must displace RPA from ssDNA. Displacement may be achieved by modulating the DNA binding activity of specific DBDs within RPA. In such a model, a protein that exchanges for RPA does not dissociate all DBDs at once but displaces individual DBDs after gaining access to DNA that is transiently exposed by dissociation of a DBD. Moreover, if the RPA-ssDNA complex were to be considered as a sequential, linear assembly of DBDs as seen in the crystal structure, then depending on the DBD first displaced, a downstream DNA binding protein could be positioned at the 5′ or 3′ end of the RPA-occluded ssDNA.
The recombination mediator Rad52 is one example of a RIP. It belongs to a group of proteins that orchestrate homologous recombination (HR) and homology directed DNA repair. S. cerevisiae Rad52 regulates recombination by facilitating replacement of RPA on ssDNA with the Rad51 nucleoprotein filament, an active species in homology search and DNA strand exchange12–15. Nucleation of the Rad51 filament is a slow and tightly controlled process as Rad51 fails to compete for binding to ssDNA with RPA16. Thus, Rad52 must physically interact with both RPA and Rad51 to promote Rad51 filament nucleation. The mechanism by which Rad52 loads Rad51 on the ssDNA is unclear except that Rad51 filament formation is simultaneous with displacement of RPA from ssDNA and likely proceeds through a Rad52-RPA-ssDNA intermediate17. Within this complex, Rad52 was shown to stabilize the RPA-ssDNA interaction18, further mystifying its assigned mechanism of action as a recombination mediator to displace RPA18.
To determine how individual DBDs work in the context of the full-length protein and investigate how proteins such as Rad52 modulate RPA binding, we generated fluorescent forms of RPA containing a non-canonical amino acid (ncAA) that is labeled with the fluorescent dye MB543 in either DBD A or D. When positioned near the DNA-binding site, MB543 produces a change in fluorescence intensity upon binding to ssDNA. Using direct measurements of full-length RPA carrying a fluorescently-labeled DBD binding to, and dissociating from ssDNA, we show that both DBD-A and DBD-D are highly dynamic, frequently binding to and dissociating from ssDNA. We also show that RPA-ssDNA complexes exist in at least 4 distinct conformational states offering differential access to the ssDNA within this complex. Rad52 interacts with the RPA-ssDNA complex and selectively modulates the dynamics of DBD-D preventing its full engagement to ssDNA and thereby opening the 3′ end of the RPA-occluded sequence.
Results:
Direct read out of DBD dynamics using non-canonical amino acids and fluorescence.
Directly monitoring the dynamics (binding, dissociation or remodeling) of a single enzyme in multi-protein reactions remains technically challenging. To decipher how the DBDs of RPA function in the context of the heterotrimeric RPA complex, we labeled DBD-A or DBD-D (in RPA70 and RPA32, respectively; Figs.1c,d) of S. cerevisiae RPA19 with MB543, an environmentally sensitive fluorophore. Both fluorescently labeled RPAs are fully active for ssDNA binding with binding parameters and occluded binding site sizes typical of the wild-type RPA protein (Supplementary note 1 and Fig.S1). RPA labeled at domain A (RPA-DBD-AMB543) and domain D (RPA-DBD-DMB543) produce enhanced fluorescence upon binding to ssDNA (Figs.1e,f and Fig.S2)19.
RPA binding to ssDNA is a paradigm for reactions where multiple DNA binding enzymes function together on a single DNA template. Knowledge of where, how, and when each enzyme gains access to the DNA in this multi-enzyme milieu is fundamental to deciphering when and how specific DNA repair/recombination processes are orchestrated. Site-specific labeling with MB543 allows us to monitor the dynamics of individual DBDs in the context of the full-length RPA heterotrimer and in multi-protein reactions. We measured the DNA binding kinetics for RPA-DBD-AMB543 and RPA-DBD-DMB543 providing direct read outs of each domain’s engagement with ssDNA in the context of full-length RPA. RPA-DBD-AMB543or RPA-DBD-DMB543 were rapidly mixed with ssDNA [(dT)25], and the change in fluorescence was measured (Figs.1e, f). Upon binding to ssDNA, both RPA-DBD-AMB543 and RPA-DBD-DMB543 produce a change in fluorescence intensity. The data for RPA-DBD-AMB543 is best described by a two-step model (kobs,1=30.6±9.8 s−1 and kobs,2 = 10.3±9.8 s−1), whereas intensity changes associated with RPA-DBD-DMB543 best fits a single-step DNA binding model (kobs = 36.2±2.3 s−1). The first step in both models is similar and reflects the initial interaction of RPA with ssDNA. The second step for RPA-DBD-AMB543 possibly reflects a rearrangement of DBD-A, as has been observed in structural studies20,21. To probe the nature of these differences further, we performed these binding experiments as a function of increasing DNA concentration using (dT)35, which provides enough space for engagement of all the DBDs of RPA (Fig. 2). While measurements of RPA-ssDNA interaction footprints under our buffer conditions yield occluded site sizes of ~20 nt/RPA (Fig. S1h), the modularity of the DBDs have been shown to produce occluded site-sizes between 18–28 nt22.
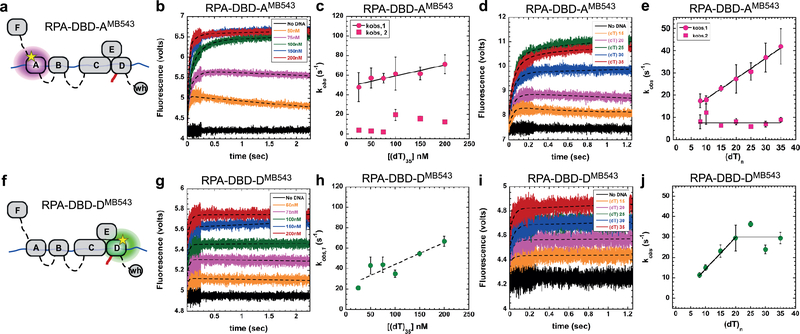
Figure 2. DNA binding dynamics of individual DBDs.
a & f) Cartoons depicting RPA-DBD-AMB543 or RPA-DBD-DMB543 binding to ssDNA and producing a change in fluorescence. Stopped flow experiments done with b & c) increasing concentrations of [(dT)35] ssDNA or d & e) with ssDNA of increasing length, captures the observed rates in fluorescence change for RPA-DBD-AMB543. g – j) Stopped flow analysis of RPA-DBD-DMB543 ssDNA binding dynamics. The data for RPA-DBD-AMB543 are best fit using a two-step model whereas the data for RPA-DBD-DMB543 fit to a one-step process, suggesting distinct DNA context dependent changes in their dynamics. Error bars in panels c, e, h & j represent the mean and s.e. from n=3 independent experiments.
The observed rate for the first association step for both RPA-DBD-AMB543 and RPA-DBD-DMB543 increases as a function of DNA concentration yielding bimolecular kON values (1.1±0.6 ×108M−1s−1 and 2.1±0.4×108M−1s−1, respectively; Figs.2a-c, f-h). The second step, observed only for RPA-DBD-AMB543, is not linear (Fig.2c). This is consistent with a conformational rearrangement of DBD-A after the complex with ssDNA has been established and depends on the protein to DNA ratio in the reaction. Under conditions where RPA is present in excess over ssDNA, we clearly observe biphasic binding and dissociation/rearrangement phases for RPA-DBD-AMB543 (orange and pink traces in Fig.2b), but not for RPA-DBD-DMB543 (Fig.2g). These data suggest that the dynamics of individual DBDs are not identical, possibly reflecting different functional roles; as shown in our single-molecule studies (see below).
FRET analysis confirms primary assessments of DBD-ssDNA dynamics.
In the experiments described above, the change in fluorescence intensity arises from local environmental changes of the fluorophore upon binding to the ssDNA. We suggest that changes in the MB543 fluorescence reflect changes in the electrostatic environment of the dye. (Fig.S2). To reaffirm that the accuracy of the dynamics we measure for each fluorescent DBD reflects ssDNA interactions, we used Förster Resonance Energy Transfer (FRET) to capture DBD-ssDNA binding kinetics. RPA binds to ssDNA with specific polarity where DBD-A is positioned closer to the 5’ end of the ssDNA11,23. Similar to the MB543-labeled proteins, we generated Cy5-labeled RPAs where either DBD-A or DBD-D was labeled with Cy5. We next performed FRET experiments with either 5′ or 3′Cy3-end-labeled DNA [(dT)34]. On 5′Cy3 DNA, a high FRET signal is observed for RPA-DBD-ACy5, and a medium FRET state is captured for RPA-DBD-DCy5 (Fig.S3a,b). In the corollary experiment with 3’Cy3 DNA, a low FRET state for RPA-DBD-ACy5 and a high FRET state for RPA-DBD-DCy5 are observed (Fig.S3c,d). These experiments are consistent with the expected 5′ to 3′ polarity of RPA binding. Strikingly, the observed rate for the appearance of the RPA-DBD-DCy5 high FRET state (36±2s−1; Fig.S3d) agrees with the rate for change in fluorescence intensity of RPA-DBD-DMB543 upon binding to ssDNA (36.2±2s−1; Fig.1f). Similarly, the observed rate of 21±1s−1 for the appearance of the RPA-DBD-ACy5 high FRET state (Fig.S3b) is likely a composite of the two observed phases captured in fluorescence intensity changes of the RPA-DBD-AMB543 ssDNA complex (kobs,1=30s−1 and kobs,2=10s−1; Fig.1e). The FRET data affirm that the ssDNA binding responsive fluorescence intensity enhancements reflect specific DBD-ssDNA interactions.
Differential effects of ssDNA length on DBD conformations.
Since each DBD has varying footprints on ssDNA11 we measured the DBD dynamics as a function of ssDNA length and found that the kobs,1 increases as a function of ssDNA length for RPA-DBD-AMB543 (Fig.2d,e), whereas the same parameter saturated for RPA-DBD-DMB543 at ~20 nt (Fig.2i,j). On shorter DNA lengths, both binding and dissociation phases are clearly observed for RPA-DBD-AMB543 (Fig.2d: (dT)15 and (dT)20 traces); however, only a single binding phase for RPA-DBD-DMB543 is observed with all ssDNA lengths (Fig.2i). Since ssDNA and RPA are in molar equivalents in these experiments, the dissociation from shorter DNA probably occurs from intra-subunit competition between the four DBDs of RPA. DBDs F, A and B are considered the conformationally flexible half24. In contrast, DBDs C, D and E constitutively interact to form the trimerization core and are more conformationally rigid. We considered the possibility that the trimerization core might be outcompeting the more dynamic DBD-A (and possibly DBD-B) under conditions of excess RPA or when the length of the DNA is too short to accommodate all the DBDs. To test this scenario, we generated the RPA-FAB fragment containing DBDs F, A and B and labeled it with MB543 in DBD-A (RPA-FAB-AMB543). Stopped flow measurement of DNA binding kinetics of RPA-FAB-AMB543 yield kon=1.0±0.1×108M−1s−1 (Fig.S4a-d), which is similar to that measured for RPA-DBD-AMB543 (1.1±0.6×108M−1s−1; Fig.2b,c), suggesting that DBD-A has intrinsically distinct DNA binding capacity compared to DBD-D, and possibly other DBDs as well. Interestingly, RPA-FAB-AMB543 binds to ssDNA with monophasic kinetics under both conditions of excess protein or shorter DNA lengths (Fig.S4 b-c and supplementary note 2). This suggests competitive binding and rearrangements between the DBDs within full length RPA when the binding sites on ssDNA are limiting. We propose that when only a short segment of ssDNA ((dT)15 or (dT)20) is available, DBD-A rapidly binds and dissociates, whereas DBD-D (and possibly the trimerization core) forms more stable, longer-lived complexes with ssDNA, thus outcompeting DBD-A from short ssDNA substrates. These data also suggest that the interactions of each DBD and resulting conformations of the RPA-ssDNA complex are sensitive to the context of DNA encountered during various DNA metabolic processes in the cell.
Single-molecule analysis reveals the presence of multiple conformational states involving DBD-D and DBD-A.
Ensemble stopped flow experiments described above suggest that the terminal DBDs of RPA associate with ssDNA with different rates and that upon binding to ssDNA, RPA commences a complex and dynamic rearrangement of its DBDs. Single-molecule total internal reflection microscopy (smTIRFM) was used to directly observe RPA-DBD-AMB543 and RPA-DBD-DMB543 binding to and dissociation from surface-tethered ssDNA in the context of the RPA heterotrimer. In the smTIRFM experiments, biotinylated ssDNA (66 nt) was tethered to the surface of the TIRFM flow cell (see Methods for details). The surface was illuminated with a 532 nm laser to excite the MB543 dye on fluorescent-RPA molecules entering the evanescent field (Fig.3a,b). Binding of a MB543-labeled RPA to surface-tethered ssDNA molecules generates a fluorescence signal at a particular location of the flow cell surface. This signal persists until RPA dissociates, transitions to a dark state and then dissociates, or until the dye photobleaches.
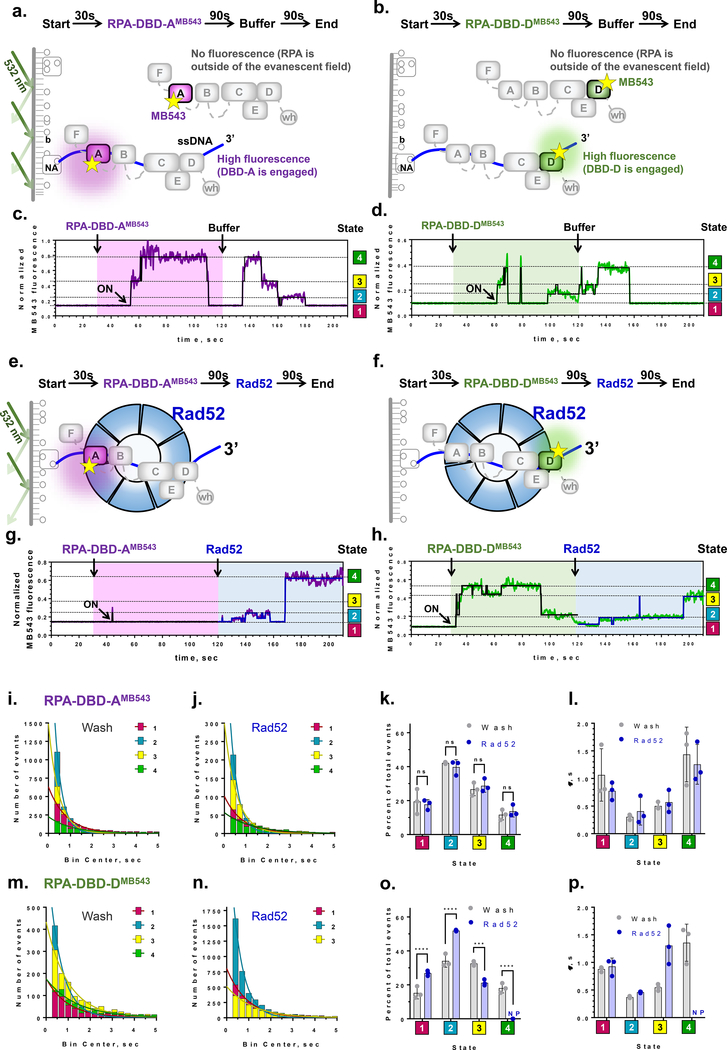
Figure 3. Single-molecule analysis quantifies the conformational dynamics of DBDs and the effect of the recombination mediator Rad52.
a & b) Experimental scheme for visualizing conformational dynamics of DBD-A and DBD-D. Binding of fluorescently labeled RPA to a surface-tethered ssDNA (purple line) brings the MB543 fluorophore within the evanescent field of TIRFM. NA – neutravidin, b – biotin. c & d) Representative fluorescence trajectories for individual RPA-DBD-AMB543and RPA-DBD-DMB543 molecules, (purple and green lines, respectively). Black lines are the results of ebFRET fitting. Additional examples of representative trajectories are presented in Supplementary Datasets 2 & 3. e & f) Experimental scheme for visualizing the effect of Rad52 on the conformational dynamics of DBD-A and DBD-D. g & h) Representative fluorescence trajectories depicting conformational dynamics of the individual RPA-DBD-AMB543and RPA-DBD-DMB543 molecules upon addition of Rad52. Additional trajectories are shown in the Supplementary Datasets 5 & 6. i-j) Dwell time histograms for the four fluorescent states obtained by the ebFRET fitting of RPA-DBD-AMB543 trajectories from three independent experiments after buffer wash (i), and Rad52 wash (j). The trajectories were reduced from 120 sec to 210 sec. Solid lines represent single-exponential fit. The data are summarized in Supplementary Table 1. k) Fractional visitation to each state available to RPA-DBD-AMB543 alone (grey) and in the presence of Rad52 (blue). The 2-way ANOVA analysis suggests no significant differences between the visitation frequencies in the presence and absence of Rad52 (p>0.1). l) Stability of each state available to RPA-DBD-AMB543 alone (grey) and in the presence of Rad52 (blue). The data on Y axis are the lifetimes for the respective dwell time distributions. m - p) The same analysis was carried out for RPA-DBD-DMB543. Only three fluorescence states were detected in the presence of Rad52. NP – not present. Statistical analysis is performed by ANOVA (***, and **** correspond to p=0.0001 and p<0.0001, respectively).
Several hundred molecules are observed in the field of view, each yielding a fluorescence trajectory (i.e. change in fluorescence in a particular location on the slide as a function of time)25. The trajectories allow measurement of binding and dissociation of individual RPA molecules. Moreover, fluorescence changes of the dye in the trajectory can also report on the presence of conformational states in the dye-decorated protein26. The experiments were carried out in three stages: first, the surface was observed for 30 seconds to confirm the absence of the non-protein derived fluorescence spots; second, RPA-DBD-AMB543 or RPA-DBD-DMB543 was injected into the flow cell; finally, at 120 seconds, protein-containing solution was replaced with the buffer (Fig.3a,b). The last step ensured that the observed changes in fluorescence can be attributed to single RPA molecules. Fluorescence trajectories were extracted from the recorded movies and were normalized (see Methods and (Fig.S5)). Resultant trajectories showed dynamics within the RPA-ssDNA complex (Fig.3c,d). Moreover, transitions between different fluorescence states persisted during the last segment of the experiment suggesting that they truly reflect the conformational dynamics of individual RPA-ssDNA complexes. Notably, a surface-tethered RPA displayed essentially invariable, one state fluorescence with only very rare exertions into a dark state (Fig.S6a,b). Global analysis of normalized trajectories for the ssDNA-bound RPA-DBD-AMB543 and RPA-DBD-DMB543 was performed with ebFRET27,28. The number of trajectories and states in each experiment are summarized in the Supplemental Table S1 and Supplemental Dataset 4. This analysis revealed that the fluorescence derived from both proteins best fit a 4-state model, with state 1 corresponding to very low fluorescence and states 2–4 corresponding to increasing fluorescence enhancement (Fig.3c,d). Segments of the trajectories between 120 and 210 seconds, which can be attributed to the dynamics of a single bound RPA, were used in the quantification of the lifetimes and visitation frequencies for all states.
We attribute state 4 in each case to the RPA conformation where the labeled domain is potentially fully engaging the ssDNA; because in bulk experiments, we observed the ssDNA-dependent fluorescence increase in the full-length RPA carrying the fluorophore, and RPA-FAB-AMB543 (containing only DBD-A and DBD-B). To rule out that the lowest fluorescence state (state 1) followed by the reappearance of the fluorescence during the last 90 seconds of the experiment is due to the RPA dissociation and rebinding, we substituted the buffer wash with the buffer supplemented with high concentration of ssDNA. In the absence of additional RPA in the solution, ssDNA competitor cannot strip the bound RPA from the DNA but can sequester all dissociated RPA molecules5. As expected, the addition of ssDNA into the reaction chamber had no effect on the RPA fluorescence states (Fig.S7 and Table S2).
We ruled out photophysical effects as the source of the MB543 fluorescence states by repeating the experiments at three different powers of the excitation laser, as true conformational states should not display any trend in power dependence29–31. The data, summarized in Fig.S6c-d and Table S3, show all four states for RPA-DBD-AMB543 and RPA-DBD-DMB543 thus confirming that this is the case. Additionally, the absence of distinct fluorescence states for RPA alone, with exception of the infrequent visitation of the dimmest state, suggests that the observed states are induced by the interaction with ssDNA (Fig.S6a-b). The four fluorescence states and their dwell times were consistent between independent experiments suggesting that the normalization scheme we developed yields reproducible results (Fig.3k,l,o,p). For both RPA-DBD-AMB543 and RPA-DBD-DMB543, states 1 and 4 were the most stable with average dwell times around 1 second compared to states 2 and 3 whose average dwell times were between 300 and 500 ms. As evident from the representative trajectories (Fig.3c,d, Supplemental Dataset 2,3), RPA spends significant periods of time in states where DBD-A or DBD-D are not fully engaged, providing a window of binding opportunity for lower affinity proteins.
In addition, we found that the collective DNA binding affinities of all DBDs produce stable RPA-ssDNA complexes. DBDs A and B have been canonically assigned as responsible for high affinity DNA binding of the RPA complex. By carrying out single-molecule experiments with the RPA-FAB-AMB543 we found that it forms a less stable complex on ssDNA and readily dissociates (Fig.S4e-h). These findings agree with the results from bulk stopped flow experiments (Fig.S8 and supplementary note 2). Fluorescence trajectories recorded for RPA-FAB-AMB543 were best fit with the three-state model (Fig.S4f). State 1 corresponded to free ssDNA and its lifetime displayed a linear dependence on the RPA-FAB-AMB543 concentration (Fig.S4h). States, 2 and 3 were present in the bound state of RPA-FAB-AMB543 whose two DBDs had been suggested to form a dynamic complex on the ssDNA32. The presence of only two fluorescence states in the RPA-FAB-AMB543 further confirms that the four states observed for the full-length RPA are not photophysical states inherent to the MB543 dye.
Rad52 selectively modulates DBD-D dynamics.
To determine the mechanism by which the recombination mediator Rad52 remodels the RPA-ssDNA complex, RPA-DBD-AMB543 or RPA-DBD-DMB543 bound to the surface-tethered ssDNA in the smTIRFM experiments were challenged with Rad52 (Fig.3e,f, Supplemental Datasets 5,6). The last 90 second portions of the resulting trajectories were normalized and globally analyzed using ebFRET. Dwell times for each state were binned and fit to an exponential decay (Fig.3i,j,m,n, Supplemental Dataset 4 and Table S1). RPA-DBD-AMB543 trajectories after buffer wash or after Rad52 addition fit best to a 4-state model with the same distribution of states and the same dwell times (Fig.3i-l). The trajectories collected for RPA-DBD-DMB543 after Rad52 addition instead best fit a 3-state model (Fig.3m-p). Attempts to fit these trajectories with a 4-state model resulted in overfitting and overlapping states. Intensities of the 3-states of RPA-DBD-DMB543 after Rad52 addition correspond to the 3 lowest states seen after the buffer wash with the highest state absent when Rad52 was present (Fig.3h, blue shaded area). With RPA-DBD-DMB543 occupancy at state 4 lost after Rad52 addition, state 3 occupancy decreases, whereas state 1 and 2 occupancy increases (Fig.3o). This suggests that Rad52 selectively modulates conformational dynamics of the RPA-ssDNA complex: reducing the engagement of DBD-D from ssDNA, providing access to the 3′ end of the occluded ssDNA.
Formation of the RPA-ssDNA-Rad52 complex depends on the physical interaction between RPA and Rad52, which is mediated by ssDNA and is confined to the middle region of the Rad52 C-terminal domain33,34. To test whether the DBD-D modulation by Rad52 depends on the interaction between the two proteins we used human RAD52, which resembles the yeast protein in all its activities, but does not interact with yeast RPA35. We found that human RAD52 does not alter the four states of the RPA-DBD-DMB543 interaction with DNA (Fig.S9a-d). Thus, the modulation of state 4 is specific for yeast Rad52 and therefore requires Rad52-RPA interaction. To highlight the importance of the interaction between Rad52 and ssDNA for modulating the RPA conformational dynamics we performed an experiment in the presence of a Rad52 inhibitor, epigallocatechin (EGC) (Fig.S9e-k). EGC inhibits the DNA binding activity of human RAD5236 and yeast RAD52, but not that of RPA (Fig.S9e-f). We observed the four fluorescence states of RPA-DBD-DMB543 in the presence of Rad52 and EGC (Fig.S9g-k and Table S4). These data suggest that the loss of state 4 also depends on the DNA binding activity of Rad52.
Discussion
Prior RPA binding models have been based on analysis of subcomplexes or mutations. In contrast, we have analyzed the dynamics of individual domains in the context of the full RPA-ssDNA complex. This analysis demonstrates that rather than being composed of “high” and “low” affinity domains, all DNA binding domains transiently engage ssDNA with high affinity, but with differential dynamics. We also observe interplay between the flexible half of RPA (DBDs-F,A and B) and the trimerization core. Thus, the RPA-ssDNA complex consists of an ensemble of domains that dynamically interact with ssDNA. This suggests that integration of these interactions results in overall high affinity to ssDNA while facilitating diverse functions of RPA. Our data also suggest that assembly of DBDs on ssDNA is not sequential but rather the result of dynamic, independent interactions between connected DBDs and DNA.
RPA forms a complex with the recombination mediator Rad5237. RPA is in dynamic equilibrium on ssDNA and Rad52 has been shown to increase the residence time of RPA on ssDNA18. Formation of “early Rad52-bound” RPA and “late Rad52-bound” RPA are proposed to play distinct roles during Rad51 filament formation and second-strand capture during HR18. The ability to observe the individual RPA DBDs binding to, and dissociating from, the ssDNA in real time permits a mechanistic description of RPA-ssDNA-Rad52 interactions. The RPA heterotrimer and the heptameric Rad52 ring have similar ssDNA binding sites. Each Rad52 monomer contains an RPA binding site and RPA has two Rad52 binding sites per heterotrimer. Rad52 is believed to interact with the ssDNA backbone, while the DBDs of RPA, especially within the trimerization core, engage the bases. Our results show that stabilization of RPA by Rad52 is a result of both their physical interactions and their individual interactions with DNA. We therefore envision a ternary complex where Rad52 and RPA interact with one another while both proteins are simultaneously bound to ssDNA. Selective modulation of DBD-D ssDNA engagement by Rad52 provides space for Rad52 to interact with ssDNA and stabilize the ternary complex, which then makes more extensive contacts with the ssDNA than RPA does on its own. By redistributing the ssDNA between RPA and Rad52, and by reducing the contacts between RPA and ssDNA, such selective remodeling by Rad52 may provide Rad51 recombinase access to the 3′ end of RPA-occluded ssDNA, while maintaining its interaction with RPA (Fig.4). Each Rad51 monomer binds to one monomer of Rad52 and to three nucleotides of ssDNA. Six Rad51 monomers are required to achieve a stable nucleation cluster38, which amounts to 18 nucleotides of open ssDNA. This cannot be achieved without the help of a recombination mediator. When Rad52 binds to ssDNA-bound RPA, it modifies the dynamics of the DBD-D engagement. This provides a stretch of ssDNA with sufficient length to initiate Rad51 filament nucleation. We predict that recombination mediators in other species, including human BRCA2 may operate by a similar mechanism. The details of this mechanism, however, will depend on the intrinsic differences in nucleoprotein filament formation by human RAD51, which nucleates on ssDNA through dynamic association of RAD51 dimers39 and grows from heterogeneous nuclei ranging in size from dimers to oligomers40.
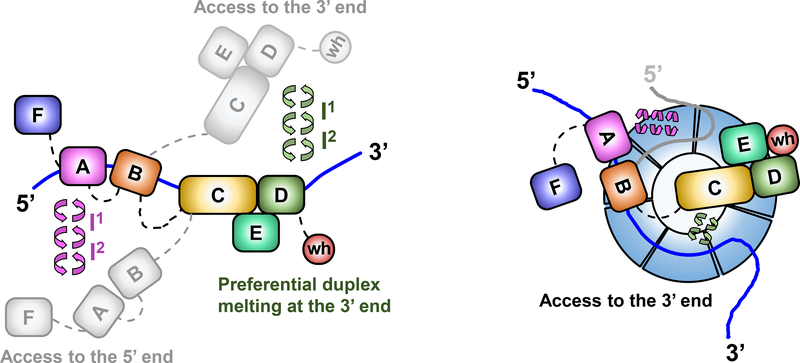
Figure 4. Dynamics of RPA DBDs and modulation by Rad52.
a) Sequential and directional arrangement of the DBDs allows RPA to occlude 20–30 nt of ssDNA (∼20 nt under our experimental conditions; Supplemental Fig 1). When RPA is in a stoichiometric complex with ssDNA, or when the ssDNA is in excess, the individual DBDs of RPA exist in a variety of distinct dynamic conformational DNA bound states. Such conformational flexibility allows access to either the 5′ or the 3′ segment of the DNA to other proteins that function in downstream processes. The circular arrows represent the transitions between multiple fluorescence states we observe in the single molecule experiments and which are implied by the bulk stopped flow experiments. Note that while we illustrate the changes in the conformation of the RPA-ssDNA complex as movement of the DBDs, the same microscopically bound states may arise from ssDNA dissociating and moving away from the respective DBDs. b) The DBDs are also selectively modulated by RPA-interacting proteins (RIPs) such as Rad52. In this case, only the DNA binding dynamics of DBD-D, and possibly the trimerization core, is influenced by Rad52. In the ternary RPA-ssDNA-Rad52 complex, the ssDNA is shared by RPA and Rad52, which also interact with one another. The ability of the DBD-D and other RPA elements contacting the ssDNA near the 3′ end of the occluded sequence is constrained. Such selective DBD modulation may promote loading of Rad51 onto the 3′ end of the ssDNA during homologous recombination.
The myriad roles of RPA in DNA replication, repair, and recombination is also a paradigm for reactions where multiple DNA binding enzymes function together on a single DNA template. Knowledge of where, how, and when each enzyme gains access to DNA in this multi-enzyme milieu is fundamental to deciphering when and how specific DNA repair and recombination processes are established and used. RPA-ssDNA complexes serve as binding targets for recruitment of appropriate enzymes during various DNA metabolic processes, and physical interactions between RPA and more than two dozen enzymes have been identified. Upon recruitment, the bound ssDNA must be handed over from RPA or remodeled in such a way that the DNA is accessible to the incoming enzyme while RPA remains at the site. Microscopic binding and dissociation of the RPA DBDs is likely to enable the persistent residence of RPA at the site of repair as well as its ability to coordinate access to the DNA by helicases and nucleases. Such a mechanism might also be applicable to RPA-like proteins that also carry a multi-OB fold architecture, such as the CST complex associated with telomerase41.
Methods
Chemicals and reagents
4-Azidophenylalanine (4-AZP) was prepared from Fmoc-4-amino-phenylalanine (Angene Inc., Nanjing, China) as described 42. All fluorophores used to generate fluorescently labeled proteins were purchased from Click Chemistry Tools (Scottsdale, AZ, USA). Cy3-labeled, biotinylated and unmodified oligonucleotides were purchased form Integrated DNA Technologies (Coralville, IA).
Plasmids and Protein Expression
Plasmids used in this study are detailed in Supplemental Note 3. S. cerevisiae RPA was purified and fluorescently labeled as described 42. Minor modifications to the published procedure are described in Supplemental Note 3. S. cerevisiae Rad52 was purified as described 43 with the modifications as described in Supplemental Note 3. Site-specific incorporation of 4-AZP was confirmed via MS analysis (Supplemental Dataset 7).
Generation of fluorescently labeled RPA variants
RPA variants carrying 4-AZP were labeled with MB543, Cy3 or Cy5 as previously described 42 with the following modifications. Briefly, ~ 3 ml of RPA4-AZP (10 μM) was incubated on a rocker with 1.5-fold molar excess (15 μM) of either DBCO-MB543, DBCO-Cy3 or DBCO-Cy5 for 2 hours at 4 oC. Labeled RPA variants were separated from excess dye using a Biogel- P4 gel filtration column (65 ml bed volume) using storage buffer (30 mM HEPES, pH 7.8, 100 mM KCl and 10% (v/v) glycerol). Fractions containing labeled RPA were pooled, concentrated using a 30 kDa cut-off spin concentrator and flash frozen using liquid nitrogen. Fluorescent RPA were stored at −80 oC. Labeling efficiency was calculated using absorption values measured at 280 nm with ε280= 98500 M−1cm−1for RPA, at 550 nm with ε550= 105,000 M−1cm−1for DBCO-MB543, at 555 nm with ε555= 150,000 M−1cm−1for DBCO-Cy3, and at 650 nm with ε650= 250,000 M−1cm−1 for DBCO-Cy5 fluorophores. We obtain 45 ± 17 % and 40 ± 25 % labeling efficiencies for the RPA-DBD-AMB543 and RPA-DBD-DMB543 versions, respectively.
Fluorescence measurements
Fluorescence spectra were obtained using a PTI QM40 instrument (Horiba Scientific, Edison, NJ). For RPA-DBD-AMB543, RPA-DBD-ACy5 and RPA-DBD-ACy3 slit widths were set at 1.25 nm for excitation and 3 nm for emission. For RPA-DBD-DCy3 and RPA-DBD-DCy5 slit widths were set at 0.5 nm for excitation and 2 nm for emission. For RPA-DBD-AMB543, 2 ml of 50 nM RPA-AMB543 in reaction buffer (30 mM Hepes, pH 7.8, 100 mM KCl, 5 mM MgCl2, 1 mM β-mercaptoethanol and 6 % v/v glycerol) was added to a quartz cuvette, and the spectra were collected in the absence or presence of 50 nM ssDNA [(dT)45] or ds plasmid DNA (100 nM nucleotides) with constant stirring. Samples were excited at 535 nm and emission spectra (555 – 600 nm) were recorded. A similar experimental setup was used to obtain fluorescence spectra for 50 nM RPA-FABMB543 upon addition of 100 nM (dT)45. For RPA-DBD-DCy3 and RPA-DBD-ACy3, 2 ml of 100 nM protein was used and 100nM of (dT)45 was added with constant stirring. The samples were excited at 559 nm and emission spectra (539 nm- 579 nm) were recorded. Similarly, for RPA-DBD-ACy5 and RPA-DBD-DCy5, 2 ml of 100 nM proteins were used and 100 nM of (dT)45 were added to the reaction. Samples were excited at 690 nm and emission spectra (640– 700 nm) were recorded. All experiments were performed at 25 °C. The total number of nucleotides occupied (site-size) by RPA variants (RPA-wt, RPA-DBD-AMB543 and RPA-DBD-DMB543) in reaction buffer was determined as previously described using poly(dT) ssDNA 44.
DNA Binding
The DNA binding activity of unlabeled and fluorescent RPA was measured using electromobility band shift analysis. 50 nM of 32P-labeled (dT)30 oligonucleotide was incubated with increasing concentrations of RPA-wt or RPA-DBD-AMB543 or RPA-DBD-DMB543 (0 – 1 μM) in reaction buffer for 10 minutes at 4 ºC. 1 ml of DNA loading dye (50 % v/v glycerol and 0.2 % w/v bromophenol blue in 1 X TBE) was added to the samples and resolved using an 8 % TBE-acrylamide gel (110 volts, 25 °C). Gels were exposed overnight onto a phosphorimaging screen and scanned using a STORM scanner (GE Healthcare, Pittsburgh, PA). Bound and unbound DNA signals were quantitated using ImageQuant software and the fraction ssDNA bound to RPA was calculated using the equation:
| (Eq. 1) |
Stopped flow analysis of RPA-DNA interactions
Stopped flow experiments described below to monitor RPA – ssDNA dynamics were performed using an Applied Photophysics SX20 instrument (Surrey, UK) in reaction buffer (30 mM Hepes, pH 7.8, 100 mM KCl, 5 mM MgCl2, 1 mM β-mercaptoethanol and 6 % v/v glycerol) at 25 °C. All the concentrations mentioned in the methods are ‘pre-mixing’ concentrations which are reduced to half upon mixing the samples to provide final ‘post- mixing’ concentrations. RPA labeled with MB543 were excited at 535 nm and emission was monitored using a 555 nm cut-off filter (Newport corp., Irvine, CA). Changes in tryptophan fluorescence were monitored by exciting the samples at 290 nm and emission was measured using a 350 nm cut-off filter. For the FRET experiments, the Cy5-labeled RPA and Cy3-labeled DNA samples were mixed and excited at 555 nm and changes in Cy5 fluorescence was monitored with a 645 nm cut-off emission filter.
RPA-DBD-AMB543, RPA-DBD-DMB543 and RPA-FABMB543 ssDNA binding kinetics
To quantitate RPA-DBD-AMB543, RPA-DBD-DMB543 or RPA binding to ssDNA, 200 nM of the respective protein was rapidly mixed with 200 nm of (dT)25 oligonucleotide, and the change in MB543 fluorescence was captured. All the data were fitted using Kintek Explorer to obtain the appropriate rate constants. For RPA-DBD-AMB543, the data were fit using a phenomenological double exponential plus linear equation:
| (Eq. 2) |
Where, Δf is the change in fluorescence signal, A1 and A2 are the amplitude of fluorescence change, and k1, k2, and k3 are the observed rate constants and t is time. For RPA-DBD-DMB543 and RPA-FABMB543, the data were well described by a single step binding model defined phenomenologically by a single exponential plus linear equation.
| (Eq. 3) |
RPA-DBD-AMB543, RPA-DBD-DMB543, and RPA- FABMB543 association kinetics:
To obtain the rate of association for DBD-A binding to ssDNA, 200 nM RPA-AMB543 in one syringe was rapidly mixed with increasing concentrations (50– 400 nM) of (dT)35 oligonucleotide from the second syringe and the change in fluorescence of RPA-DBD-AMB543 was monitored. All the data obtained were analyzed using Eq.2 to obtain observed rate constants. kobs,1 was plotted against [(dT)35] and a linear fit was used to generate a rate for DBD-A binding to ssDNA. Similarly, to measure DBD-D binding to ssDNA, 200nM RPA-DBD-DMB543 in one syringe was rapidly mixed with increasing concentrations (50– 400 nM) of (dT)35 oligonucleotide from the second syringe and the change in fluorescence of RPA-DMB543 was monitored. Finally, to obtain the rate of association of FAB binding to ssDNA, 300 nM of FABMB543 in one syringe was rapidly mixed with increasing concentration of (dT)30 oligonucleotide (100–1000 nM) from the second syringe and the change in fluorescence of FABMB543 was monitored. For RPA-DBD-DMB543 and RPA-FABMB543, all the data obtained were analyzed using Eq.3 to obtain observed rate constants. kobs,1 was plotted against [(dT)35] and against [(dT)30] for RPA-DBD-DMB543 and RPA-FABMB543 respectively. A linear fit was used to generate a rate for DBD-D and FAB binding to ssDNA.
RPA-AMB543 and RPA-DMB543 length dependent association kinetics:
To measure the oligonucleotide length dependent rate of association for DBD-A, 200 nM RPA-DBD-AMB543 was rapidly mixed with 200 nM (dT)8, (dT)10, (dT)15, (dT)20, (dT)25, (dT)30 or (dT)35 and change in fluorescence was monitored. All the data obtained were analyzed using Eq.2 to obtain the observed rates. kobs,1 was plotted against (dT)n to determine the rate for oligonucleotide length dependent association of DBD-A. Using the same concentration of reactants, similar experiments were performed with RPA-DBD-DMB543 and RPA-FABMB543 to measure the oligonucleotide length dependent rate of association for DBD-D. All the data obtained were analyzed using Eq.3 to obtain the observed rates. kobs,1 was plotted against (dT)n to determine the rate for oligonucleotide length dependent association of FAB and DBD-D.
RPA-DBD-ACy5 and RPA-DBD-DCy5 polarity for DNA binding:
For all FRET experiments, samples (Cy3) were excited at 555 nm and Cy5 fluorescence emission was monitored with a 645 nm long-pass emission filter. 5′ or 3′ Cy3-labeled DNA (100 nM) was mixed with the appropriate Cy5-labeled RPA protein and the change in Cy5 fluorescence was captured. Data were fit to Eq. 3 to obtain observed rate constants for the resulting change in fluorescence.
RPA-DBD-AMB543 and RPA-DBD-DMB543 facilitated exchange kinetics:
200 nM of either RPA-DBD-AMB543 or RPA-DBD-AMB543 was premixed with 200 nM (dT)25 in one syringe to form the RPA-DNA complex and rapidly mixed with increasing concentrations of RPA-wt from the second syringe (100 – 500 nM). The change in MB543 fluorescence was monitored, and the data were analyzed using Eq.3 to obtain observed rate constants. kobs,1 was plotted against [RPA-wt] and a linear fit was used to generate a rate for the facilitated exchange processes.
RPA-FABMB543 facilitated exchange kinetics
To measure the facilitated exchange kinetics of FABMB543, FABMB543- (dT)30 complexes were pre-formed using 375 nM FABMB543 and 100 nM (dT)30 in one syringe. FABMB543- (dT)30 complexes were rapidly mixed with increasing concentrations of RPA-wt from the second syringe (100 – 500 nM) and the change in fluorescence of FABMB543 was monitored. All the data obtained were analyzed using Eq.3 to obtain observed rate constants. kobs,1 was plotted against [RPA-wt] and a linear fit was used to generate a rate for the facilitated exchange processes.
Tryptophan binding kinetics:
Intrinsic tryptophan fluorescence was used to capture the binding of unlabeled and fluorescently labeled RPA variants to ssDNA in reaction buffer. 200 nM RPA (RPA-wt or RPA-DBD-AMB543 or RPA-DBD-DMB543) was rapidly mixed with increasing concentrations (50– 400 nM) of (dT)35 oligonucleotide and the change in Trp fluorescence was monitored by exciting the sample at 290 nm and measuring emission with a 350 nm cut-off filter. Data were fit using Eq.3 and kobs,1 was plotted against [(dT)35]. A linear fit was used to obtain the observed rate constants for RPA-DNA binding.
Single-Molecule Imaging
A custom-built prism-type total internal reflection microscope (TIRFM) was used for all single-molecule experiments. A diode-pumped solid state (DPSS) green laser (532 nm, Coherent, CA, USA) was used to excite the MB543 dye. The laser power output was set to 45 mW at the entrance to the microscope stage, unless otherwise noted. A dual band pass filter (Semrock, NY, USA; FF01–577/690) was used to filter out scattered light in the emission optical path and the MB543 fluorescence was collected using a Chroma ET605/70m filter. Movies were recorded using an electron-multiplying charge-coupled device (EMCCD) camera (Andor, MA, USA; DU-897-E-CSO-#BV) at 100 ms time resolution. Background was set to 400, correction to 1200, and gain to 295 for all movies recorded.
FRET-based Assays for Rad52 Inhibition
Epigallocatechin (EGC, Sigma-Aldrich, MO, USA) has been previously characterized as an inhibitor of hRAD52-ssDNA binding45. In order to verify that EGC also inhibits the ssDNA binding of scRad52 and not scRPA, we carried out FRET based inhibitor assays as described 45. A Cary Eclipse spectrofluorimeter was used to monitor the Cy3 and Cy5 emission simultaneously. Cy3 was excited at 530 nm and emission was collected at 565 nm with slit widths of 10 nm. Cy5 was excited through FRET transfer with Cy3 and emission was collected at 660 nm with a slit width of 10 nm. Experiments were performed at 37°C in buffer containing 50 mM Tris-HCl, pH 7.5, 5 mM MgCl, 100 mM NaCl, 1 mM DTT, and 0.1 mg/ml BSA.
EGC inhibition of Rad52 ssDNA binding was tested by premixing 100 nM Rad52 and 10 nM Cy3-dT30-Cy5 ssDNA to form a stoichiometric complex, which corresponds to the ssDNA fully wrapped around the oligomeric Rad52 ring and, correspondingly, the high FRET state46, and titrating increasing concentrations of EGC. Experiments were carried out in triplicate and FRET signal and IC50 were calculated as previously described 45.
EGC inhibition of RPA ssDNA binding was tested by premixing 10 nM RPA and 10 nM Cy3-dT30-Cy5 ssDNA to form a stoichiometric complex where the DNA is straightened to its contour length 46, and titrating increasing concentrations of EGC. Experiments were carried out in triplicate and FRET signal was calculated as described above.
Single-Molecule Cell Assembly
Slides were washed, coated, and flow cell assembled as described previously 47. Reaction Buffer containing 50 mM Tris-HCl, pH 7.5, 5 mM MgCl, 100 mM NaCl, 1 mM DTT, BSA, 0.8 % glucose in Trolox was used for all single molecule experiments. 12 mM Trolox (6-Hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid, Sigma-Aldrich, MO, USA; 238813–1G) was dissolved in 12 mM NaOH and rotated under a fluorescent light (Sylvaia FM13W/835) for 3 days or until the absorbance at 400 nm is approximately 0.119. Assembled flow cells were first rinsed with T50 (10 mM Tris-HCl, pH 7.5 and 50 mM NaCl), then incubated with 0.2 mg/ml Neutravidin for three minutes and then rinsed with T50 again.
Single Molecule Experiments
In order to tether ssDNA to the slide surface, the flow cell was incubated for three minutes with 100 pM of biotinylated 66 nt oligonucleotide (/5Biosg/ CTC AAG CCA TCC GCA ACG TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT GAA ACA AAG GGC TCC TCA, IDT, IA, USA) in Reaction Buffer, then rinsed with Reaction Buffer. 2100 frame, 210 second movies were recorded with 100 pM RPA-DBD-AMB543 or RPA-DBD-DMB543 added after the first 300 frames. At 1200 frames free RPA was either washed away with Reaction Buffer or replaced with 700 pM Rad52.
Experiments with RPA-FAB-AMB543 were carried out as described above, except 1 nM of RPA-FAB-AMB543 was added at 300 frames with no wash. The experiment with hRAD52 was carried out as described, adding 700 pM hRAD52 instead of scRad52. Epigallocatechin (EGC) inhibitor single molecule experiments were carried out as Rad52 addition to RPA-DBD-DMB543 described above, with the addition of 10 μM EGC premixed with Rad52 in Reaction Buffer. In order to challenge RPA binding to ssDNA, experiments were performed with 1 nM of a 42 nt unlabeled ssDNA oligonucleotide (5’- TTT TTT TTT TTT TTT TTT TTG GAA TTA AGC TCT AAG CCA TCC −3’) added to the Reaction buffer in the wash step.
Except where indicated, the excitation laser power was set to 45mW at the entrance to the microscope stage. Laser power experiments were performed as described above, but at 36 and 27mW and compared to previous experiments carried out at maximum laser power (45mW) of the green laser (532 nm).
Surface-Tethered RPA
RPA-DBD-DMB543 contains a polyhistidine tag on the C-terminus of RPA32, which was used to tether the protein to the slide surface. The slide was first incubated with Neutravidin and rinsed, and was then incubated with Biotin-X-NTA Buffer (50 mM Tris pH 7.5, 50 mM NaCl, 20nM Biotin-X-NTA, Sigma-Aldrich, MO, USA) for ten minutes. The same Reaction Buffer was used, with the addition of 5 μM NiSO4. The chamber was rinsed with Reaction Buffer, followed by addition of 500 pM RPA-DBD-DMB543 which was incubated in the chamber for 5 minutes. The chamber was then rinsed with reaction buffer. A 2100 frame, or 210 second, movie was recorded.
Single-Molecule Data Analysis
An IDL script was used to extract fluorescence intensity trajectories (changes in the fluorescence over time in a particular location on the flow cell) from each movie (available upon request). Only those trajectories that show appearance of the fluorescence signal between 30 and 120 seconds (indicated as ON in Fig. 3c and d) were selected for analysis. Traces were viewed using a Matlab script and selected using the following three criteria: at least 2 transitions, signal-to-noise ratio > 4 (raw signal), no signal prior to addition of a fluorophore. Traces that did not meet these criteria were discarded.
Selected traces were then saved individually and processed for analysis by ebFRET 48,49. Since ebFRET was developed to analyze the two-channel trajectories, where recorded donor and acceptor fluorescence is recalculated into FRET efficiency, which is between 0 and 1, our single-color trajectories required normalization prior to analysis. To prepare fluorescence trajectories for the analysis by ebFRET, we used “emulateFRET”, a program that normalizes intensity in all experimental trajectories to be within the [0,1] range (Fig.S5). Fluorescent trajectories were imported into emulateFRET in batches and analyzed as a group. A percentile calculation determines a threshold for the maximum intensity in the group while excluding outliers. For each trajectory, the fluorescence in each point is then divided by this maximum intensity. This normalizes all traces to the range (0,1). A weighted moving-average algorithm (of width 5) removes a small amount of noise in each trajectory. For each trajectory, a histogram of the values in the trace is produced (a histogram of fluorescence intensity values binned in intervals of 5 units is shown on the right of the raw trajectory in light green color, a brighter green histogram with overlapping Gaussian curve represents the baseline values for the first 30 seconds before addition of fluorescent RPA). This histogram shows a characteristic peak surrounding the values which comprise the baseline of that trajectory. The shape of this peak on the histogram is fit to a Gaussian distribution. From this Gaussian distribution, the new baseline is determined and is placed two standard deviations above the mean of this peak. All values below this baseline are reassigned to surround the new baseline value. This suppresses the ebFRET program’s tendency to assign multiple states to the typically over-represented baseline signal. The edited trajectories are given a dummy “donor” trace, calculated such that the FRET signal of each pair will be equal to the normalized input trace. Each pair of normalized trajectories is then exported to the file format which ebFRET uses to import raw donor and acceptor values. The converted ebFRET compatible traces were then trimmed to exclude portions of the recording prior to the addition of fluorescent material. The trimmed traces were loaded into the ebFRET MatLab program. Analysis was carried out for 2 to 5 states, with 10 repeats and a precision of 10e−06.
After normalization, trajectories from Rad52 addition versus buffer experiments were trimmed to include separate portions of the experiment: 300–1200 frames after RPA addition and 1200–2100 frames after Rad52 or buffer addition. Each portion was analyzed separately using ebFRET. The output from ebFRET was then analyzed by KERA 3.0 to sort dwell times of individual events at each state. The dwell times were binned with the first center at 400 ms with a width of each bin 300 ms. The dwell times were fit to one and two phase exponential decays using GraphPad Prism 7.03. The F-test suggested that single exponential decay was the best fit for dwell time distributions.
Several modifications were introduced to analyze the data from the surface-tethered RPA. Trajectories were selected for analysis only 1) there was no signal in the “red” channel, 2) fluorescence terminated in a single-step photobleaching event at least 30 seconds before the end of the recording, and 3) signal-to-noise ratio > 4. Selected trajectories were normalized using emulateFRET with the baseline selected as the portion occurring after photobleaching (last 30 seconds of the movie). After normalization, the trajectories were trimmed at 10 seconds after photobleaching. Normalized trajectories were analyzed using ebFRET previously described for the surface-tethered ssDNA.
scRad52 vs hRAD52 electrophoretic mobility shift assay (EMSA)
A Cy5-labeled 30nt ssDNA substrate (TGC ATA TCA GAT GCC TCG CGT CCG TCG CCA /Cy5/) was used in the EMSA. Reactions were prepared in KPi buffer (30 mM Kpi (pH 7.4), 5 mM MgCl, 100 mM NaCl, 1 mM DTT, 0.01 mg/ml BSA). Increasing concentrations of scRad52 or hRAD52 (50nM, 150nM, 500nM, 1000nM) were added to 10nM ssDNA coated with scRPA (100nM). Controls with free ssDNA (10nM ssDNA), RPA coated DNA (100nM RPA) and Rad52 on RPA-free DNA (1000nM Rad52) were also prepared. The samples were split in half and one half was crosslinked with 0.01% glutaraldehyde. Samples were incubated at 37° C for 5 minutes. Samples were loaded onto to 1% agarose gel with 10X orange loading dye (TAE, 30% glyceraol, 4mg/ml Orange G). The agarose gel was then run at room temperature for 1 hour at 50V. A Bio-Rad ChemiDoc MP Imaging System was used to image the gel using a Cy5 protocol.
Supplementary Material
Acknowledgements:
We acknowledge the members in our laboratories for their helpful discussions and suggestions. We thank Dr. Theodore Keppel at the Center for Biomedical Mass Spectrometry Research at the Medical College of Wisconsin for MS analysis.
Funding: This work was supported by grants from the National Institutes of Health [7R15GM110671] to E.A. and [R01 GM108617] to M.S. CCC is supported by a NIH T32 Pharmacological Sciences Training Grant [NIH T32 GM067795]. J.T., S.M.A.T. and M.S. acknowledge support from the University of Iowa Carver College of Medicine FUTURE in Biomedicine program. E.A. also acknowledges support from an SFF-RRG grant from Marquette University. S.M.A.T acknowledges the CHAS Faculty Research Activity Grant support from the University of Northern Iowa. E.A. and E.I.C acknowledge salary support from a Department of Energy office of Basic Energy Sciences grant [DE-SC0017866].
Footnotes
Competing Financial Interests Statements:
The authors declare no competing financial interests.
Data and Materials Availability
The data supporting the conclusions of this study are presented in Supplementary Data Sets 1–7. Additional data, plasmids for protein expression, and code for single-molecule data analysis are available from the corresponding authors upon request.
References:
- 1.Chen R & Wold MS Replication protein A: single-stranded DNA’s first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays 36, 1156–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wold MS Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66, 61–92 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Fanning E, Klimovich V & Nager AR A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res 34, 4126–37 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen B et al. Diffusion of human replication protein A along single-stranded DNA. J Mol Biol 426, 3246–3261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibb B et al. Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One 9, e87922 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R, Subramanyam S, Elcock AH, Spies M & Wold MS Dynamic binding of replication protein a is required for DNA repair. Nucleic Acids Res 44, 5758–72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arunkumar AI, Stauffer ME, Bochkareva E, Bochkarev A & Chazin WJ Independent and coordinated functions of replication protein A tandem high affinity single-stranded DNA binding domains. J Biol Chem 278, 41077–82 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Bochkareva E, Korolev S, Lees-Miller SP & Bochkarev A Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J 21, 1855–63 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyka IM, Dhar K, Binz SK & Wold MS Replication protein A interactions with DNA: differential binding of the core domains and analysis of the DNA interaction surface. Biochemistry 42, 12909–18 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Bochkareva E, Frappier L, Edwards AM & Bochkarev A The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J Biol Chem 273, 3932–6 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Fan J & Pavletich NP Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev 26, 2337–47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson FE, Baumann P & West SC Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391, 401–4 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Shinohara A & Ogawa T Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391, 404–7 (1998). [DOI] [PubMed] [Google Scholar]
- 14.New JH, Sugiyama T, Zaitseva E & Kowalczykowski SC Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391, 407–10 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Sung P Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem 272, 28194–7 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Sugiyama T & Kowalczykowski SC Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem 277, 31663–72 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Sugiyama T & Kowalczykowski SC Rad52 Protein Associates with Replication Protein A (RPA)-Single-stranded DNA to Accelerate Rad51-mediated Displacement of RPA and Presynaptic Complex Formation. Journal of Biological Chemistry 277, 31663–31672 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Gibb B et al. Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nat Struct Mol Biol 21, 893–900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pokhrel N et al. Monitoring Replication Protein A (RPA) dynamics in homologous recombination through site-specific incorporation of non-canonical amino acids. Nucleic Acids Res 45, 9413–9426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brosey CA et al. Functional dynamics in replication protein A DNA binding and protein recruitment domains. Structure 23, 1028–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brosey CA et al. A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Res 41, 2313–27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumaran S, Kozlov AG & Lohman TM Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry 45, 11958–73 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolpashchikov DM et al. Polarity of human replication protein A binding to DNA. Nucleic Acids Res 29, 373–9 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brosey CA et al. NMR analysis of the architecture and functional remodeling of a modular multidomain protein, RPA. J Am Chem Soc 131, 6346–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boehm EM, Subramanyam S, Ghoneim M, Washington MT & Spies M Quantifying the Assembly of Multicomponent Molecular Machines by Single-Molecule Total Internal Reflection Fluorescence Microscopy. Methods Enzymol 581, 105–145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghoneim M & Spies M Direct correlation of DNA binding and single protein domain motion via dual illumination fluorescence microscopy. Nano Lett 14, 5920–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van de Meent JW, Bronson JE, Wiggins CH & Gonzalez RL Jr. Empirical Bayes methods enable advanced population-level analyses of single-molecule FRET experiments. Biophys J 106, 1327–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanyam S, Kinz-Thompson CD, Gonzalez RL Jr. & Spies M Observation and Analysis of RAD51 Nucleation Dynamics at Single-Monomer Resolution. Methods Enzymol 600, 201–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu HP & Xie XS Single-molecule spectral fluctuations at room temperature. Nature 385, 143 (1997). [Google Scholar]
- 30.Brender JR et al. Conformational Dynamics of the Isoalloxazine in Substrate-Free p-Hydroxybenzoate Hydroxylase: Single-Molecule Studies. Journal of the American Chemical Society 127, 18171–18178 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Ghoneim M & Spies M Direct Correlation of DNA Binding and Single Protein Domain Motion via Dual Illumination Fluorescence Microscopy. Nano Letters 14, 5920–5931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pretto DI et al. Structural dynamics and single-stranded DNA binding activity of the three N-terminal domains of the large subunit of replication protein A from small angle X-ray scattering. Biochemistry 49, 2880–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plate I et al. Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J Biol Chem 283, 29077–85 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seong C et al. Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J Biol Chem 283, 12166–74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugiyama T, New JH & Kowalczykowski SC DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci U S A 95, 6049–54 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hengel SR et al. Small-molecule inhibitors identify the RAD52-ssDNA interaction as critical for recovery from replication stress and for survival of BRCA2 deficient cells. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park MS, Ludwig DL, Stigger E & Lee SH Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J Biol Chem 271, 18996–9000 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Qiu Y et al. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat Commun 4, 2281 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanyam S, Ismail M, Bhattacharya I & Spies M Tyrosine phosphorylation stimulates activity of human RAD51 recombinase through altered nucleoprotein filament dynamics. Proc Natl Acad Sci U S A 113, E6045–E6054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Candelli A et al. Visualization and quantification of nascent RAD51 filament formation at single-monomer resolution. Proc Natl Acad Sci U S A 111, 15090–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan H, Wang Y & Feigon J Progress in Human and Tetrahymena Telomerase Structure Determination. Annu Rev Biophys 46, 199–225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References.
- 42.Pokhrel N et al. Monitoring Replication Protein A (RPA) dynamics in homologous recombination through site-specific incorporation of non-canonical amino acids. Nucleic Acids Res 45, 9413–9426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibb B et al. Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nat Struct Mol Biol 21, 893–900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumaran S, Kozlov AG & Lohman TM Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry 45, 11958–73 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hengel SR et al. Small-molecule inhibitors identify the RAD52-ssDNA interaction as critical for recovery from replication stress and for survival of BRCA2 deficient cells. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grimme JM & Spies M FRET-based assays to monitor DNA binding and annealing by Rad52 recombination mediator protein. Methods Mol Biol 745, 463–83 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Bain FE, Wu CG & Spies M Single-molecule sorting of DNA helicases. Methods 108, 14–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinz-Thompson CD, Bailey NA & Gonzalez RL Precisely and Accurately Inferring Single-Molecule Rate Constants. Methods in enzymology 581, 187–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van de Meent, Bronson J-W, Jonathan E, Wiggins, Chris H & Gonzalez Jr, Ruben L. Empirical Bayes Methods Enable Advanced Population-Level Analyses of Single-Molecule FRET Experiments. Biophysical Journal 106, 1327–1337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.