Abstract
hiHep is a new type of hepatocyte-like cell that is predicted to be a potential unlimited source of hepatocytes for a bioartificial liver. However, hiHep cannot currently be used in clinical settings because serum must be added during the culture process. Thus, a defined medium is required. Because serum is complex, an efficient statistical approach based on the Plackett–Burman design was used. In this manner, an original medium and several significant cell growth factors were identified. These factors include insulin, VH, and VE, and the original medium was optimized based on these significant factors. Additionally, hiHep liver-specific functions and metabolism in the optimized serum-free medium were measured. Results showed that hiHep functions, such as glycogen storage, albumin secretion, and urea production, were well maintained in our optimized serum-free medium. In summary, we created a chemically defined, serum-free medium in which cell growth, proliferation, metabolism, and function were well maintained. This medium has the potential to support the clinical use of hiHep.
Electronic supplementary material
The online version of this article (10.1007/s10616-018-0289-2) contains supplementary material, which is available to authorized users.
Keywords: hiHep, Serum-free, Medium, Bioartificial liver
Introduction
Liver disease has become a prevalent medical condition worldwide, and liver failure may arise from many causes, such as cirrhosis and viral infections (Wang et al. 2010). Liver failure is a severe clinical syndrome in which the liver’s metabolic functions such as detoxification and biotransformation are negatively affected (Carpentier et al. 2009). To date, drug treatment can only relieve patients’ pain or repair minor injuries, but it cannot heal massive injures to the liver. Currently, liver transplantation is the only curative treatment for end-stage liver failure (Raju et al. 2013; Kogiso et al. 2013). However, the clinical application of liver transplantation is limited by the limited availability of donated organs (Vacanti and Kulig 2014; Hu and Li 2015); there were approximately 10,000 patients who did not receive a liver transplant because of the shortage of donated livers in the United States alone in 2012 (Aravindan et al. 2011). Therefore, alternate therapies to liver transplantation or an effective way to prolong the patient’s life until a liver becomes available are required. Bioartificial liver (BAL) as a new in vitro artificial liver support system that has been developed and is becoming increasingly recognized (Carpentier et al. 2009).
The BAL device contains a cell-housing bioreactor and it can temporarily replace the primary and most important functions of the liver (e.g. oxidative detoxification, biotransformation, and synthesis). BAL can improve patients’ symptoms and provide hope for the patient (Carpentier et al. 2009). However, BAL requires a large amount of hepatocytes per treatment (1–3 × 108/kg body weight; Hu and Li 2015). Difficulties in expanding and maintaining primary hepatocytes in culture remain a major obstacle for the use of BAL (Raju et al. 2013). Currently, the only hepatocyte source for BAL is another liver, but hepatocytes readily undergo dedifferentiation and hepatocyte functions decrease rapidly during culture in vitro (Hannoun et al. 2016; Carpentier et al. 2009; Pareja et al. 2010). Thus, BAL is largely limited by the low availability of mature functional hepatocytes and an alternate source of hepatocytes that can overcome these difficulties for BAL is required.
Human induced hepatocyte (hiHep) cells were generated from fibroblasts using lentiviral expression of FOXA3, HNF1A, and HNF4A (Zhu et al. 2016). hiHep as a type of hepatocyte-like cells that is expandable and expresses liver-specific gene programs, such as albumin secretion, glycogen synthesis, and even detoxification (Huang et al. 2014; Ni et al. 2016). Fibroblasts can be easily obtained from patients and converted into hiHep cells. This suggests that hiHep cells may also possess a low risk of immunogenicity. These advantages of hiHep suggest that it may be a solution for the cell source in BAL, and it could support the clinical use of BAL (Carpentier et al. 2009). Additionally, Hui et al. (Shi et al. 2016) showed that hiHep had similar therapeutic effects compared to human primary hepatocytes in vivo and hiHep demonstrated a therapeutic effect by improving survival in pigs with acute liver failure using a hiHep-based BAL (Shi et al. 2016; Huang et al. 2014). Thus, hiHep is a highly desirable cell resource for BAL.
However, hiHep cell growth still strongly depends on fetal bovine serum supplementation in vitro. Serum is a complex mixture containing many ingredients, such as cytokines, proteins, and inorganic salts. To date, there has been no clear standardization for serum because of its complex constituents (Runge et al. 2000). The application of serum or any animal products is banned for clinical use. Therefore, for medical use of hiHep, including BAL and cell transplantation (Raju et al. 2013), serum and any other animal component must be removed from the medium to meet the clinical requirements. Additionally, the medium should also support growth and maintain the function of hiHep for a long period of time. However, similar to hepatocytes, a medium that could maintain cell growth and function simultaneously has not existed until now. Therefore, removing serum from the medium so that hiHep can be used clinically is a significant challenge.
Many kinds of materials have replaced the role of serum in cell culture (Zhao et al. 2017), but most of these materials contain undefined raw materials that were derived from microbes, such as hydrolysates (Gupta et al. 2015). Therefore, we focused on developing a defined medium.
The most common approaches for medium development and optimization are based on successively changing one or more factors to assess their effect on cell growth or function (Richardson et al. 2015), but this method is usually very labor intensive (Knöspel et al. 2010). To overcome this limitation, the statistical design of experiments was used. The approach was based on a two-step strategy, using the Plackett–Burman design for screening for significant factors out of the 20 factors and the original medium. Then, the original medium was optimized after the significant factors were added. The growth, morphology, proliferation, integrity, metabolism, and function of hiHep in the optimized medium was investigated. Additionally, long-term proliferation and liver-specific gene expression of hiHep in the optimized medium were also investigated.
Materials and methods
Cell and culture conditions
hiHep cells were directly reprogramed from fibroblasts using lentiviral expression of FOXA3, HNF1A, and HNF4A in Lijian Hui’s lab, and were then subcultured in rat tail collagen-pre-coated dishes with hepatocyte maintenance medium (HMM; Ni et al. 2016). Cells were digested using trypsin (Gibco, the USA) when their confluence reached 80–90%. Then, the harvested cells were seeded onto collagen-coated plates or dishes to measure cell growth and function, according to the culture medium. hiHep cells were seeded onto plates at a density of 10,000 cells/cm2. The medium was changed every 48 h and frozen in − 80 °C for further analysis. For the proliferation assay, the cells were inoculated onto a collagen-coated (Bioroot biology, Shanghai, China) dish at a density of 1.25 × 104 cells/cm2 and the medium was changed every other day. Cells were routinely passaged every 6 days and cell numbers were counted to determine their proliferation. hiHep cells were cultured under humidified conditions at 37 °C and 5% CO2.
Medium
Basal medium comprised DMEM/F12 (Gibco, the USA) supplemented with 0.544 mg/L ZnCl2 (Sinopharm, Shanghai, China), 0.75 mg/L ZnSO4·7H2O (Sinopharm, Shanghai, China), 0.2 mg/L CuSO4·5H2O (Sinopharm, Shanghai, China), 0.025 mg/L MnSO4 (Sinopharm, Shanghai, China), hormone mixture(cholestenone, Histamine and Estradiol)(Macklin, Shanghai, China), 4 g/L bovine serum albumin (Sigma-Aldrich, USA), 2 g/L galactose (Sigma-Aldrich, USA), 0.1 g/L ornithine, 0.03 g/L proline, 0.61 g/L nicotinamide, 40 ng/mL transforming growth factor (TGF)-α (Peprotech, USA), 40 ng/mL epidermal growth factor (EGF; Peprotech, USA), and 10 µM dexamethasone(Sigma-Aldrich, USA; Huang et al. 2014).
Serum-free medium was prepared according to the Plackett–Burman matrix. Twenty-four types of serum-free media were prepared for each set of experiments.
Analytical method
Determination of metabolic activity
Metabolic activity was tested during the cell culture process by analyzing glucose and lactate in the supernatant. Measurements were performed using an automatic chemistry analyzer (BP400, Nova Biomedical, USA). Amino acid concentrations were also determined in collected samples using high performance liquid chromatography (HPLC, Agilent 1200, USA).
Cell proliferation assay
Cells were seeded in 24-well plates at a density of 1 × 104 cells/cm2. The number of Cell or the absorbance of value of Cell counting kit (CCK-8, DOJINDO, Japan) were measured every 2 days. The number of cells was counted by blood counting chamber. The assay was performed in triplicate for each experimental group.
Periodic acid-Schiff stain
hiHep cultures on the plates were stained using a periodic acid-Schiff (PAS) kit (Solarbio cat: G1281, Beijing, China), according to the manufacturer’s instructions.
Urea and albumin determination
The urea concentration in the medium was measured using a commercially available kit (Nanjing Jiancheng Bioengineering Institute cat: co13-2). The albumin content was determined using the corresponding enzyme-linked immunosorbent assay kit (e80-129).
Pcr
hiHep cells were harvested after termination of the culture. All RNA was extracted from the cells using the TRIzol reagent (Invitrogen) and reverse transcription of 1 μg RNA was performed according to the manufacturer’s instructions. Random sequence primers were used and reverse transcription was performed with SuperScript III reverse transcriptase (Invitrogen). Primers sequences for ALB, AAT, TAT, C5, Transferrin, CK18, TTR, CDH1, and GAPDH were listed in Table 1. GAPDH was used as housekeeping gene.
Table 1.
Primers used for real-time PCR
| Gene name | Forward primers | Reverse primers |
|---|---|---|
| GAPDH | CCACCTTTGACGCTGGG | CATACCAGGAAATGAGCTTGACA |
| ALB | GCACAATGAAGTGGGTAA | TACTGAGCAAAGGCAATC |
| Transferrin | TGTCTACATAGCGGGCAAGT | GTTCCAGCCAGCGGTTCT |
| CK-18 | TCGCAAATACTGTGGACAATGC | GCAGTCGTGTGATATTGGTGT |
| TTR | TGGGAGCCATTTGCCTCTG | AGCCGTGGTGGAATAGGAGTA |
| AAT | TATGATGAAGCGTTTAGGC | CAGTAATGGACAGTTTGGGT |
| C5 | CTGTTGAAGCCCGAGAGAAC | AGGGAAAGAGCATACGCAGA |
| TAT | GCATCCTATGTCGCACCC | TCAGCAACTAACCGCTCC |
| CDH1 | CGAGAGCTACACGTTCACGG | GGCCTTTTGACTGT |
The semi-quantitative PCR experiment was performed in a thermal cycler (ExPress Thermo Hybaid, Middlesex, United Kindom). Amplification of DNA was performed at the following PCR experimental condition: initial denaturation at 95 °C for 5 min, followed by 30 amplification cycles (95°Cfor 45 s, 60°Cfor 45 s, 72 °C for 45 s), and a final extension step at 72 °C for 5 min.
The agarose 1 percent gel was electrophoresed at 120 V for 30 min and on completion, the gel was analyzed in Gel Documentation System (BIO-RAD, USA). PCR products were stained by Gelred and visualized under ultraviolet transilluminator.
The real-time polymerase chain reaction (RT-PCR) experiment was performed using a Real-Time PCR System (Bio-Rad CFX96) and SYBR Green Jumpstart Taq ready mix (Sigma-Aldrich). The PCR experimental conditions were 95 °C for 4 min, followed by 45 cycles of amplification at 95 °C for 10 s and 60 °C for 40 s. Gene expression was analyzed according to the ΔΔCT method and presented relative to human Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Krøyer 2017). Ct values for GAPDH were not different between the samples (supplement, Fig. 1). The specificity of the amplicons was determined by melting curve analysis. It indicated good specificity when the amplicons of melting curve only exist a single peak. Just like the Fig. 2 in the supplement. The primer sequences are shown in Table 1.
LDH release
The lactate dehydrogenase (LDH) level in the medium was measured using a Cytotoxicity LDH Assay kit-WST (DOJINDO cat: CK12, Japan). The LDH concentration in the medium indicates the potential for cellular necrosis.
AST release
The aspartate aminotransferase (AST) concentration in the medium was determined using an aspartate aminotransferase assay kit (Nanjing Jiancheng Bioengineering Institute cat: c010-2), according to the manufacturer’s instructions.
CYP activity
To test cytochrome (CYP) enzyme activity, cells were induced in medium containing the indicated drug (2.5 μM testosterone and 2.5 μM phenacetin) for 0, 0.5, 1, 2, and 4 h. The activity of CYP was reversed by the variation of corresponding substrate concentration. The supernatants were collected for measurement of metabolized compounds using liquid chromatography-tandem mass spectrometry (Huang et al. 2014; LC–MS/MS, Agilent 1200 HPLC and ABI 4000 mass spectrometer).
Statistical methodology
Fetal bovine serum is a complex mixture that contains many components. Based on the published factors summarized in Table 2. A matrix containing all of variables was designed using a Plackett–Burman design. Twenty-three variables (including three dummy variables) were investigated in this study and the matrix is shown in Table 3.
Table 2.
The list of all test substances
| Factors | Factors |
|---|---|
| Biotin (VH) | Putrescine |
| Carnitine | Retinyl acetate |
| Corticosterone | Selenium |
| Galactose | T3 (triodo-1-tyronine) |
| Glutathione | DL-a-tocopherol (VE) |
| Linoleic acid | DL-a-tocopherol acetate |
| Linolenic acid | Bovine serum albumin |
| Progesterone | Catalase |
| Insulin | Superoxide dismutase |
| Transferrin | Ethanolamine |
Table 3.
Placket-Burman matrix for the study of 23 variables using 24 experiments
| Experimental variables | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | ||
| 1 | − | − | − | − | + | − | + | – | − | + | + | − | − | + | + | − | + | − | + | + | − | − | − |
| 2 | − | + | − | + | − | − | + | + | − | − | + | + | – | + | − | + | + | + | + | + | − | + | − |
| 3 | − | + | − | + | + | + | + | + | − | − | − | − | + | − | + | − | − | + | + | − | − | + | − |
| 4 | + | + | + | + | + | − | − | − | − | + | − | + | − | − | + | + | − | − | + | + | + | + | + |
| 5 | + | − | − | + | + | − | + | − | + | + | + | + | + | − | − | − | − | + | − | + | + | − | − |
| 6 | − | − | − | + | − | + | − | − | + | + | − | − | + | + | − | + | − | + | + | + | − | − | − |
| 7 | − | + | + | − | + | − | + | + | + | + | + | − | − | − | − | + | − | + | − | − | − | + | + |
| 8 | + | + | − | − | − | − | + | − | + | − | − | + | + | − | − | + | + | − | + | − | + | + | − |
| 9 | + | + | + | + | − | − | − | − | + | − | + | − | − | + | + | − | − | + | + | − | + | + | + |
| 10 | + | − | − | + | + | − | − | + | + | − | + | − | + | + | + | + | + | − | − | − | + | − | − |
| 11 | − | + | + | − | − | + | + | − | + | − | + | + | + | + | + | − | − | − | − | + | − | + | + |
| 12 | + | − | + | − | − | + | + | − | − | + | + | − | + | − | + | + | + | + | + | − | + | − | + |
| 13 | + | + | − | + | − | + | + | + | + | + | − | − | − | − | + | − | + | − | − | + | + | + | − |
| 14 | + | − | + | + | + | + | + | − | − | − | − | + | − | + | − | − | + | + | − | − | + | − | + |
| 15 | + | + | + | − | − | − | − | + | − | + | − | − | + | + | − | − | + | + | − | + | + | + | + |
| 16 | − | + | − | − | + | + | − | − | + | + | − | + | − | + | + | + | + | + | − | − | − | + | − |
| 17 | − | + | + | + | + | + | − | − | − | − | + | − | + | − | − | + | + | − | − | + | − | + | + |
| 18 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 19 | + | + | − | − | + | + | − | + | − | + | + | + | + | + | − | − | − | − | + | − | + | + | − |
| 20 | − | − | + | − | + | − | − | + | + | − | − | + | + | − | + | − | + | + | + | + | − | − | + |
| 21 | + | − | − | − | − | + | − | + | − | − | + | + | − | − | + | + | − | + | − | + | + | − | − |
| 22 | − | − | + | + | − | − | + | + | − | + | − | + | + | + | + | + | − | − | − | − | − | − | + |
| 23 | + | − | + | − | + | + | + | + | + | − | − | − | − | + | − | + | − | − | + | + | + | − | + |
| 24 | − | − | + | + | − | + | − | + | + | + | + | + | − | − | − | − | + | − | + | − | − | − | + |
+ adding; − no adding
All the experiments were performed in 24-well plates according to the design matrix, which was based on the basal medium. Each row represents one trial and each column represents a single variable, and three variables are dummy variables. The (+) and (−) elements refer to the adding and no adding of each variable that is present within each trial, respectively (Aravindan et al. 2011; Shi and Zhu 2007). The experiment was repeated at least three times independently. Cell growth as the response variables was measured using CCK-8.
All the data are presented as the mean ± standard deviation (SD). Statistical analysis was performed using SPSS 13.0 software and an analysis of variance (ANOVA) test was used. The Student–Newman–Kleuss method was used to identify the level of significance. A value of P < 0.05 was considered statistically significant.
Results
Screening of original medium and significant factors
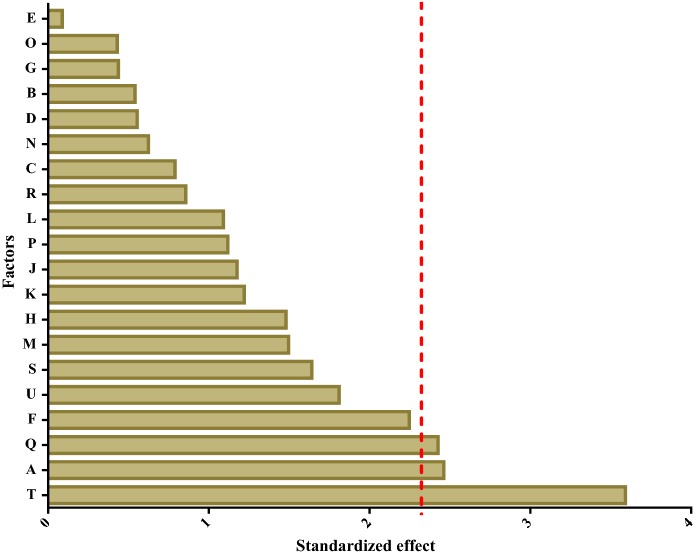
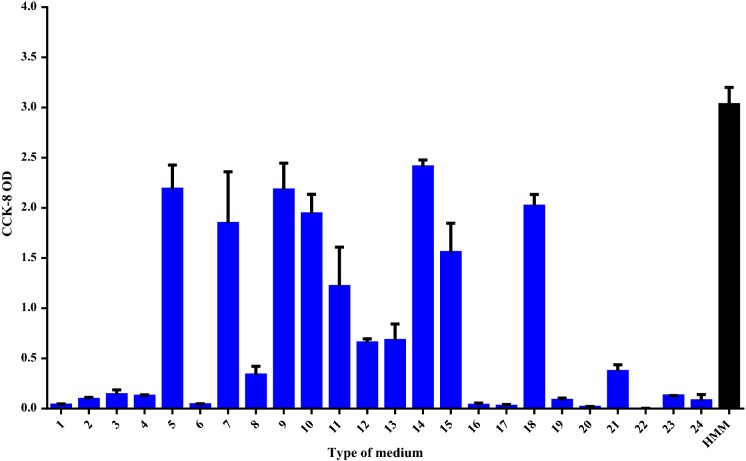
To develop a defined medium, the formulation of the serum replacement needs to be identified. According to previous reports, the important factors for cell growth can be screened using a Plackett–Burman design, so this was used to define the components in our medium that could support hiHep growth in vitro (Vasconcellos et al. 2016; Aravindan et al. 2011). In this study, 20 different variables include insulin, carnitine, VE, VA, VH and so on (Sigma-Aldrich, USA), were completely screened in only 24 experiments and the order of these experiments was randomized according to the matrix (Table 3). Next, the significance level of each factor was determined by Student’s t test. It was accepted only the confidence levels to the 90% level. Data analysis showed that only VE, VH, and insulin exhibited a significant effect on cell growth and the fit of the matrix reached 97.1%, which is considered to be reliable (Fig. 1). After culture in the 24-well plates for 6 days, cell growth was evaluated using CCK-8. Figure 2 shows that cells cultured in media 14, 9, 5, and 18 grew better than those in the other media, indicating that these four types of media can support hiHep cell growth. Furthermore, the cell growth in media 14 superior to others (p < 0.05 vs. 14). Medium 14 was selected as the original medium (ORM). As shown in Fig. 2, the ORM was still insufficient compared with HMM to support hiHep cell growth. Further research and optimization of the medium are required.
Fig. 1.
The effect of factors in the medium. Normal plot of the standardized effects of cell growth factors in a Plackett–Burman design (The significance level of each variable effect was determined using t distribution associated with Student’s t-test,; The red vertical line means the t critical value (α = 0.1). (Color figure online)
Fig. 2.
Screening of the original medium. hiHep was seeded in 24-well plates and cultured in the indicated medium for 8 days. After culturing, cells were subjected to CCK-8 analysis to evaluate their growth. Cells cultured in HMM served as the control
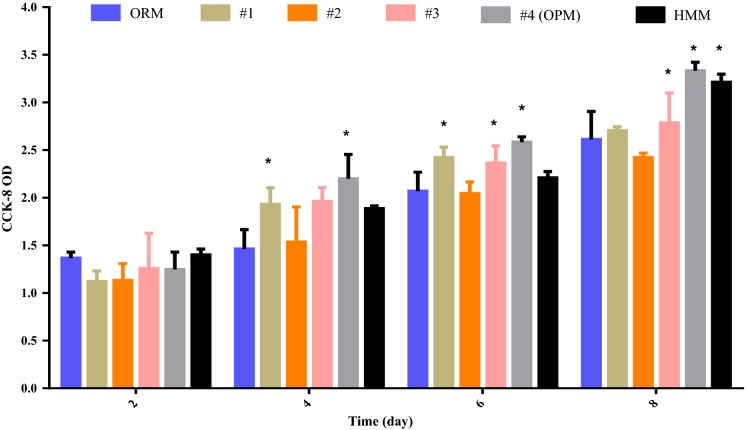
Significant factor supplementation
The significant factors were selectively added to ORM to optimize cell growth and evaluate the effect of significant factors, and the resulting media were called medium 1, 2, and 3, respectively (Fig. 3). The original medium containing all the significant factors was called medium 4. hiHep cell growth in different media was tested using the cell counting kit-8. As shown in Fig. 3, cell growth was better in all media with the significant factors compared with those in the original medium, based on the increased optical density absorption in 450 nm. The three factors that showed significance (VE, VH, and insulin) had positive effects on cell growth. Additionally, either insulin or VH showed a more significant impact than VE, and the most effective culture medium was medium 4, which contained insulin, VE and VH, and which had an optical density absorption that was equivalent to HMM. Our results indicated that supplementation of significant factors is essential for cell growth, and medium 4 was found to be the optimized medium (OPM).
Fig. 3.
The effect of significant factors on cell growth. a The growth of hiHep cells in different media that were supplemented with respective factors. 1: ORM + insulin, 2: ORM + VE, 3: ORM + VH, 4: ORM + VE + VH + insulin (medium 4 was called the optimized medium). Data are present as the mean ± SD of six independent experiments. *p < 0.05 versus ORM
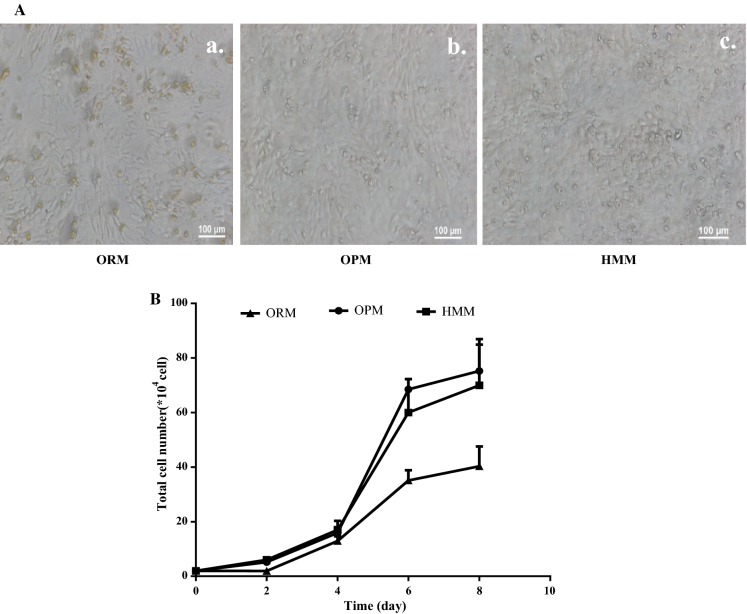
Cell growth and morphology
Morphology of cells cultured in OPM was visualized using light phase contrast microscopy (Fig. 4A). Cells presented a compact polygonal cell shape of mature hepatocytes and binuclear cells were also observed. This result indicated that cells cultured in optimized medium maintain the typical characteristics of hepatocytes. The cell growth curve in the optimized medium was measured (Fig. 4B) and cell numbers in this medium reached 7.5 × 105 within 8 days, which was equivalent to that in HMM. Further, there was no obvious difference between optimized medium and HMM.
Fig. 4.
Cell growth results. A hiHep morphology in ORM, OPM and HMM; (a) ORM; (b)OPM; (c) HMM. B The hiHep growth curves
Cell damage and metabolic activity
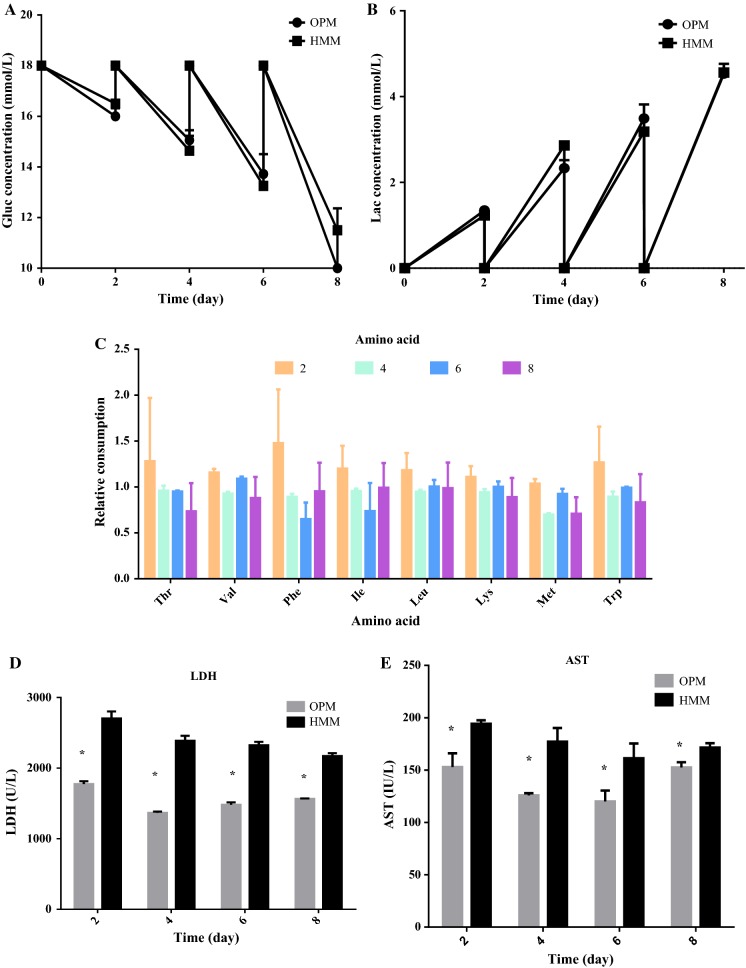
To test hiHep metabolic activity in OPM, glucose, lactate, and amino acid metabolism were tested in the collected supernatant (Fig. 5). Cells cultured with OPM and HMM showed comparable metabolic activities in terms of glucose, lactate, and amino acids. Glucose concentration and lactate production both indicated a similar variable tendency during whole culture process (Fig. 5a, b). Additionally, the relative change in amino acid concentration in OPM and HMM showed no obvious difference throughout the process (Fig. 5c).
Fig. 5.
Characterization of hiHep metabolism and damage. a Glucose concentration; b Lactate production; c relative consumption of eight essential amino acids (The variation of amino acids concentration in OPM versus the corresponding of HMM); d LDH activity; e Transaminase activity. Data are presented as the mean ± SD of three independent experiments. *p < 0.05
To detect the potential for cell damage during culture, the release of intracellular enzymes LDH and AST were tested. The activity of these two enzymes showed a similar trend during the entire culture process and both were lower in the OPM than in HMM (Fig. 5d, e).
Gene analysis
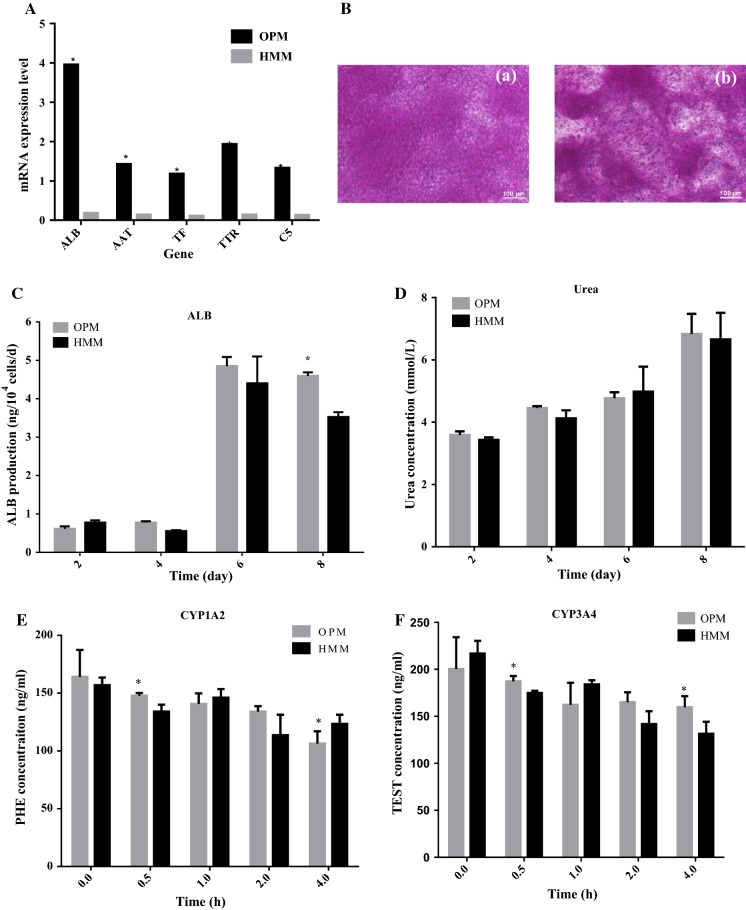
Liver-specific gene analysis showed no marked difference between OPM and HMM, and most genes showed a tendency for higher expression in OPM compared with those in HMM (Fig. 6A).
Fig. 6.
Assessment of hiHep cell functionality. A Liver-specific gene analysis by semi-quantitative PCR; B Cells on day 8 showed the capability to store glycogen, as seen in the areas stained in pink. (a) OPM, (b) HMM; C Albumin secretion during the culture process; D Urea production during the culture process; E CYP1A2 activity; F CYP3A4 activity (The activity of CYP was reflected by the reduction of corresponding substrate concentration, the greater variation means the higher activity). *p < 0.05 versus HMM
Although the gene analysis showed that hiHep cells were well maintained, this cannot be considered a marker for cell function. To further assess the functional status of hiHep cells in OPM, liver-specific functions were measured.
Glycogen storage
Glycogen storage was assayed using PAS staining (Fig. 6B), and the capacity of glycogen storage was shown to be well maintained in cells cultured in the OPM.
Albumin secretion
Albumin secretion is an important functional index of the liver. The speed of albumin secretion gradually increased during the prolonged culture time and the albumin content secreted in the optimized medium was higher than HMM throughout the whole process (Fig. 6C).
Urea production
Urea production is one of the most important characteristics of mature hepatocytes. As shown in Fig. 6D, the accumulation of urea was not different between OPM and HMM.
Cytochrome enzyme viability
To test pharmaceutical metabolism of hiHep cells in serum-free medium, CYP450 3A4 and 1A2 enzymes activities were assessed (Guillouzo et al. 2007). CYP3A4 is the main enzyme that metabolizes several drugs (Mandal et al. 2016; Lu and Li 2001). The cells were cultured with corresponding substrates for 4 h, and the activity of CYP enzymes such as CYP3A4 and CYP1A2 showed almost no loss or decrease in activity compared with cells cultured in HMM (Fig. 6E, F).
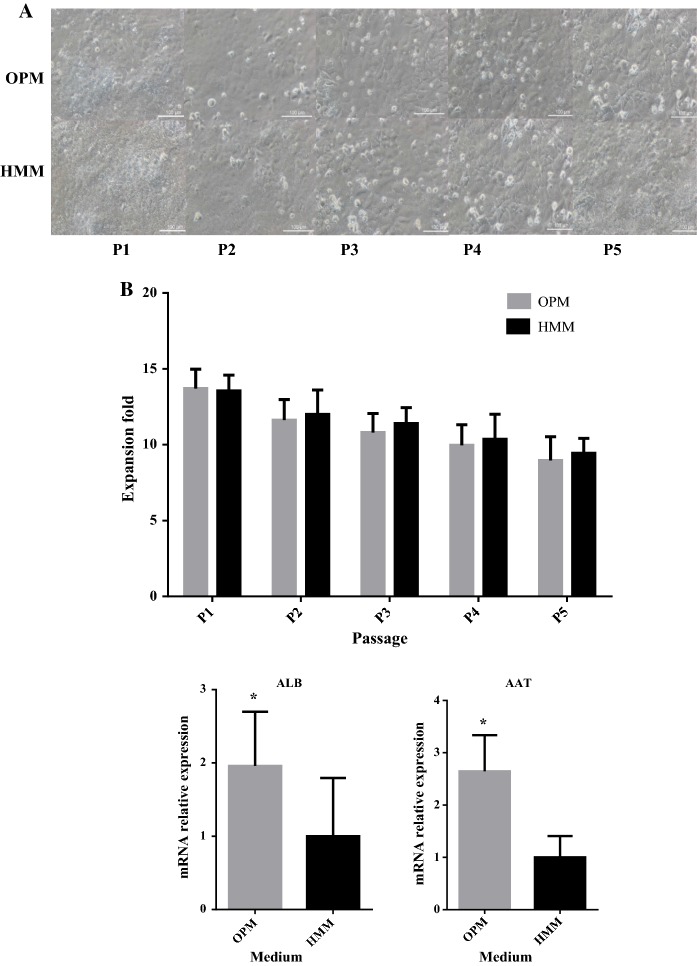
Long-term cell culture
To fulfill the BAL’s requirement for the 1010 cells needed for human treatment, the medium has to continue to support hiHep cell growth, proliferation, and liver-specific function after more than five passages in culture (Ji et al. 2013; Miyajima et al. 2014; Schwartz et al. 2014). As shown in Fig. 7a, b, the hiHep morphology was well maintained in OPM for a long time and cells could continually be passaged in culture, with the cell number remaining equivalent to that of cells in HMM. The results suggested that both OPM and HMM can maintain good cell morphology, growth, and proliferation for a long time.
Fig. 7.
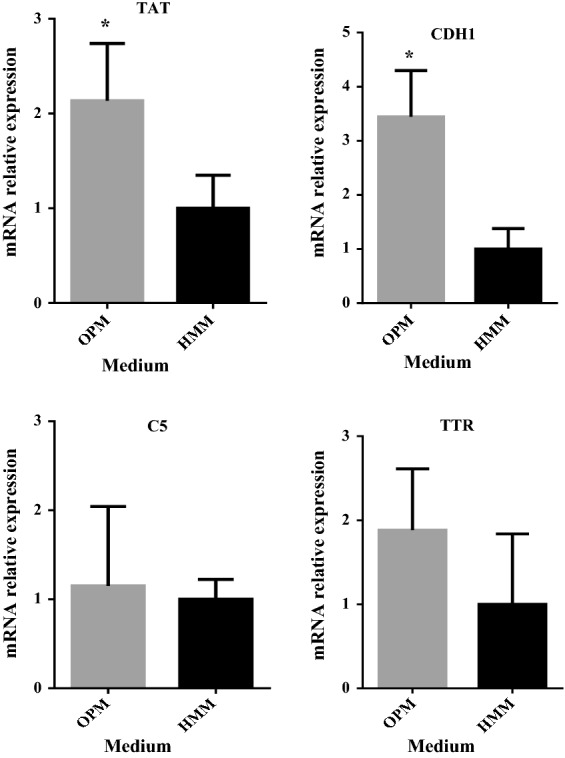
Long-term cell culture. a hiHep cell morphology in each passage (100 μΜ); b hiHep cell proliferation in each passage; c mRNA expression levels in liver-specific functions (i.e., ALB, AAT, TAT, CDH1, C5and TTR) in hiHep cells as tested by qRT-PCR. The data are expressed as the mean ± SD (n = 3). *p < 0.05 versus HMM
Long-term gene array
Expression of liver-specific genes in long-term hiHep culture with OPM was assessed. Cell samples from OPM at the end of the extended culture period showed obvious differences compared with HMM. Most genes showed a significantly higher expression in OPM compared with HMM (Fig. 7c).
Discussion
hiHep is beneficial because it possesses liver-specific function and the potential to be readily expandable, and it could be used as a cost-effective source of cells for a BAL. For future clinical application of hiHep, including BAL and cell transplantation, we used a statistical method to develop a defined medium for hiHep cells that is free of animal components. The medium both supports hiHep growth for a long time and maintains hiHep liver-specific functions.
Original medium screening and optimization
Serum is a complex mixture that includes components such as protein, small amounts of amino acids, and vitamins. To efficiently find a serum replacement, the Plackett–Burman design is a classical method that is used to screen large variables (Ekpenyong et al. 2017). It allows the investigation of up to N-1 variables with N experiments and identifying the most important ones (Zhao et al. 2017; Shi and Zhu 2007). VE, VH, and insulin were identified as important components for cell growth (Fig. 1). However, there was still a gap between ORM and HMM regarding cell growth. The ORM components comprise 12 factors, which is likely because this medium still lacks factors that are essential for cells to grow in ORM. Previous studies have shown that insulin can also stimulate cell metabolism and growth (Han et al. 2016) and promotes glycogen synthesis (Yeom et al. 2015). VE is an efficient antioxidant within the cell and can protect the cell from oxidative damage (Traber and Atkinson 2007; Butt et al. 2017). VH as a member of the B-vitamin family that can stimulate cell metabolism such as fat processing and glucose metabolism (Takechi et al. 2008). This suggests that significant factors maybe have a great impact on hiHep cell growth and function. To improve the supportive capacity on cell growth and explore the material effect of significant factors(Insulin, VE, VH), the significant factors were added into ORM. Figure 3 shows that three positive factors could promote cell growth when supplemented into the original medium. Additionally, cells grew better when all three factors were supplemented compared to other media, and a cell growth curve further confirmed this result (Fig. 4a). Although, some factors which have significant effect on cell growth were found by Plackett–Burman design. The interaction among factors cannot be accurately analyzed. It is mainly used in the experiment which with lots candidate factors and exist no interaction among factors.
The results of metabolic activity and cell integrity were comparable between cells grown in OPM and HMM. Glucose concentration and lactate production best represented cell metabolism (Lubberstedt et al. 2015), and both had the same variation in OPM and HMM, which confirm the consistency of cell metabolism activity in the two groups (Fig. 5a, b; Lubberstedt et al. 2015; Richter et al. 2016). Additionally, relative consumption of eight essential amino acids also showed no obvious difference between the two groups during the culture process (Fig. 5c). Amino acids are the basis for new cellular growth, and the synchronous change in consumption continued to show consistency in hiHep cell growth and metabolism between the two groups. Because the relevant metabolisms were comparable in OPM and HMM, cell metabolic status in OPM was also well maintained (Richter et al. 2016). LDH enzyme activity increased with the number of cells continues to grow in both groups over time. It may be ascribed to the inhibitory effect of gradually growing cellular contact (Zeilinger et al. 2011). However, LDH and AST activity in cells grown in the OPM were lower than that in cells grown in the HMM (Fig. 5d, e). This suggests that less cell damage or death occurs in cells grown in the OPM. Moreover, no drastic peaks in enzyme release, indicating harmful stress to the cells, were observed over the entire culture period (Hoffmann et al. 2015).
Expression of liver function
Gene expression analysis showed that most liver-specific hiHep gene expression in OPM was higher than that in HMM, especially for transport proteins (Fig. 6a). These results suggest that OPM may be more suited to hiHep than HMM. To confirm this suggestion, the liver-specific functions of hiHep in the optimized medium were evaluated. Many mature hepatocyte characteristics of hiHep such as glycogen storage, urea synthesis, albumin secretion, and CYP1A2 and CYP3A4 activity were observed in hiHep cells grown in the OPM (Fig. 6b–f). Most of mature hepatocyte functions were improved or well maintained in OPM. Albumin and urea production are the typical mature function of hepatocyte. Albumin responsible for establishing serum colloid osmotic pressure and transporting fatty acids (Runge et al. 2000).Urea was the final metabolite of nitrogen via the urea cycle for hepatocyte (Hoffmann et al. 2015). In accordance with the previous work (Mueller et al. 2012; Runge et al. 2000), the result indicated both functions were well maintained without decrease or loss in the serum-free medium. In addition, as a further functional parameter, the activity of CYP isoenzyme, which is responsible for hepatic toxicity metabolism were assessed (Montellano 2009; Richter et al. 2016). CYP1A2 and CYP3A4 were the most abundant P450 isoforms in humans (Rogers et al. 2002). And in contrast to Nelson et al. (2013) report about human primary hepatocyte,CYP3A4 and CYP1A2 enzyme activity of hiHep both were well maintained in OPM and HMM. It may imply that the OPM components contain a stimulating component which is enough to trigger the relevant cytochrome P450 isoenzyme activities like serum (Krøyer 2017). Most of these mature hepatocyte functions appeared to improve in OPM. These results supported the previous findings.
Additionally, over 1010 cells are required if a BAL is to be used to treat humans with liver disease. hiHep was evaluated in a long-term culture, and results showed that the OPM and the HMM could maintain hiHep in continuous passage culture (Fig. 6b). In previous study had proved, that the preservation of hepatocyte morphology was closed with the expression of normal in vitro functionality (Nelson et al. 2013, Runge et al. 2000). There was no obvious discrepancy in cell morphology among different passages, and liver-specific genes expressions were also maintained for a long time. It suggested that the relevant hepatic functions of hiHep were well maintained among different passages. Moreover, most genes appeared to be up-regulated in the OPM. This suggests that hiHep cells further matured in OPM. The difference observed between the media may be because of the elimination of serum, which may be a hazardous substance that was removed from the OPM. It’s known that serum is a complex and undefined mixture, and many of its components can inhibit cell growth and proliferation (Klein et al. 2014; Lubberstedt et al. 2015). Over all, these results further confirm that the OPM is better suited for hiHep cells than HMM.
Conclusion
In conclusion, our results demonstrate that we defined a medium that can support hiHep cell growth and proliferation, and maintain their function for a long time. Based on the significant factors that were identified, further investigation is required. Thus, the statistical method can also be as a useful tool to explore hiHep growth and function. Finally, a serum-free medium which can remove the disadvantage of serum for the use of hiHep in clinical settings was developed by this study. There is no doubt that it will speed up the application of hiHep in BAL.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Lijian Hui’s lab for providing hiHep and the cell culturing protocol. We are also grateful to Xuan Ni from Guoyu Pan’s lab for performing CYP activity quantification. This research was supported by the National Natural Science Foundation of China (Grant Nos. 31170951 and 81671841), the Natural Science Foundation of Shanghai (Grant No.16ZR1408700) and Basic Research Key Program Project of Commission of Science and Technology of Shanghai (16JC1400203).
References
- Aravindan V, Gnanaraj J, Madhavi S, Liu HK. Lithium-ion conducting electrolyte salts for lithium batteries. Chemistry. 2011;17:14326–14346. doi: 10.1002/chem.201101486. [DOI] [PubMed] [Google Scholar]
- Butt H, Mehmood A, Ali M, Tasneem S, Anjum MS, Tarar MN, Khan SN, Riazuddin S. Protective role of vitamin E preconditioning of human dermal fibroblasts against thermal stress in vitro. Life Sci. 2017;184:1–9. doi: 10.1016/j.lfs.2017.07.002. [DOI] [PubMed] [Google Scholar]
- Carpentier B, Gautier A, Legallais C. Artificial and bioartificial liver devices: present and future. Gut. 2009;58:1690–1702. doi: 10.1136/gut.2008.175380. [DOI] [PubMed] [Google Scholar]
- Ekpenyong MG, Antai SP, Asitok AD, Ekpo BO. Plackett–Burman design and response surface optimization of medium trace nutrients for glycolipopeptide biosurfactant production. Iran Biomed J. 2017;21:249–260. doi: 10.18869/acadpub.ibj.21.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact. 2007;168:66–73. doi: 10.1016/j.cbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Gupta AJ, Wierenga PA, Gruppen H, Boots JW. Influence of protein and carbohydrate contents of soy protein hydrolysates on cell density and IgG production in animal cell cultures. Biotechnol Prog. 2015;31:1396–1405. doi: 10.1002/btpr.2121. [DOI] [PubMed] [Google Scholar]
- Han C, Wei S, Song Q, He F, Xiong X, Wan H, Liu D, Ye F, Liu H, Li L, Xu H, Du X, Kang B, Zeng X. Insulin stimulates goose liver cell growth by activating PI3 K-AKT-mTOR signal pathway. Cell Physiol Biochem. 2016;38:558–570. doi: 10.1159/000438650. [DOI] [PubMed] [Google Scholar]
- Hannoun Z, Steichen C, Dianat N, Weber A, Dubart-Kupperschmitt A. The potential of induced pluripotent stem cell derived hepatocytes. J Hepatol. 2016;65:182–199. doi: 10.1016/j.jhep.2016.02.025. [DOI] [PubMed] [Google Scholar]
- Hoffmann SA, Müllervieira U, Biemel K, Knobeloch D, Heydel S, Lübberstedt M, Nüssler AK, Andersson TB, Gerlach JC, Zeilinger K. Analysis of drug metabolism activities in a miniaturized liver cell bioreactor for use in pharmacological studies. Biotechnol Bioeng. 2015;109:3172–3181. doi: 10.1002/bit.24573. [DOI] [PubMed] [Google Scholar]
- Hu C, Li L. In vitro culture of isolated primary hepatocytes and stem cell-derived hepatocyte-like cells for liver regeneration. Protein Cell. 2015;6:562–574. doi: 10.1007/s13238-015-0180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Zhang L, Gao Y, He Z, Yao D, Wu Z, Cen J, Chen X, Liu C, Hu Y, Lai D, Hu Z, Chen L, Zhang Y, Cheng X, Ma X, Pan G, Wang X, Hui L. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell. 2014;14:370–384. doi: 10.1016/j.stem.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Ji S, Zhang L, Hui L. Cell fate conversion: direct induction of hepatocyte-like cells from fibroblasts. J Cell Biochem. 2013;114:256–265. doi: 10.1002/jcb.24380. [DOI] [PubMed] [Google Scholar]
- Klein S, Mueller D, Schevchenko V, Noor F. Long-term maintenance of HepaRG cells in serum-free conditions and application in a repeated dose study. J Appl Toxicol. 2014;34:1078–1086. doi: 10.1002/jat.2929. [DOI] [PubMed] [Google Scholar]
- Knöspel F, Schindler RK, Lübberstedt M, Petzolt S, Gerlach JC, Zeilinger K. Optimization of a serum-free culture medium for mouse embryonic stem cells using design of experiments (DoE) methodology. Cytotechnology. 2010;62:557–571. doi: 10.1007/s10616-010-9307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogiso T, Nagahara H, Otsuka M, Shiratori K, Dowdy SF. Transdifferentiation of human fibroblasts into hepatocyte-like cells by defined transcriptional factors. Hepatol Int. 2013;7:937–944. doi: 10.1007/s12072-013-9432-5. [DOI] [PubMed] [Google Scholar]
- Krøyer RM. Induction of Cytochrome P450 mRNA in porcine primary hepatocytes cultured under serum free conditions; comparison of freshly isolated cells and cryopreserved. Exp Cell Res. 2017;360:218. doi: 10.1016/j.yexcr.2017.09.011. [DOI] [PubMed] [Google Scholar]
- Lu C, Li AP. Species comparison in P450 induction: effects of dexamethasone, omeprazole, and rifampin on P450 isoforms 1A and 3A in primary cultured hepatocytes from man, Sprague-Dawley rat, minipig, and beagle dog. Chem BioInterac. 2001;134:271–281. doi: 10.1016/s0009-2797(01)00162-4. [DOI] [PubMed] [Google Scholar]
- Lubberstedt M, Muller-Vieira U, Biemel KM, Darnell M, Hoffmann SA, Knospel F, Wonne EC, Knobeloch D, Nussler AK, Gerlach JC, Andersson TB, Zeilinger K. Serum-free culture of primary human hepatocytes in a miniaturized hollow-fibre membrane bioreactor for pharmacological in vitro studies. J Tissue Eng Regen Med. 2015;9:1017–1026. doi: 10.1002/term.1652. [DOI] [PubMed] [Google Scholar]
- Mandal A, Raju S, Viswanathan C. Long-term culture and cryopreservation does not affect the stability and functionality of human embryonic stem cell-derived hepatocyte-like cells. Vitro Cell Dev Biol Anim. 2016;52:243–251. doi: 10.1007/s11626-015-9956-1. [DOI] [PubMed] [Google Scholar]
- Miyajima A, Tanaka M, Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell. 2014;14:561–574. doi: 10.1016/j.stem.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Montellano PD (2009) The Cytochrome P450 Oxidative System[M].
- Mueller D, Müllervieira U, Biemel KM, Tascher G, Nüssler AK, Noor F. Biotransformation of diclofenac and effects on the metabolome of primary human hepatocytes upon repeated dose exposure. Eur J Pharm Sci. 2012;45:716–724. doi: 10.1016/j.ejps.2012.01.014. [DOI] [PubMed] [Google Scholar]
- Nelson LJ, Treskes P, Howie AF, Walker SW, Hayes PC, Plevris JN. Profiling the impact of medium formulation on morphology and functionality of primary hepatocytes in vitro. Sci Rep. 2013;3:2735. doi: 10.1038/srep02735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X, Gao Y, Wu Z, Ma L, Chen C, Wang L, Lin Y, Hui L, Pan G. Functional human induced hepatocytes (hiHeps) with bile acid synthesis and transport capacities: a novel in vitro cholestatic model. Sci Rep. 2016;6:38694. doi: 10.1038/srep38694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareja E, Cortés M, Martínez A, Vila JJ, López R, Montalvá E, Calzado A, Mir J. Hepatic cell transplantation: a new therapy in liver diseases. Cirugía Espaola. 2010;88:3. doi: 10.1016/j.ciresp.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Raju R, Chau D, Verfaillie CM, Hu WS. The road to regenerative liver therapies: the triumphs, trials and tribulations. Biotechnol Adv. 2013;31:1085–1093. doi: 10.1016/j.biotechadv.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J, Nicklaus M, Shah B, Bondarenko PV, Bhebe P, Kombe MC, Zhang Z. Metabolomics analysis of soy hydrolysates for the identification of productivity markers of mammalian cells for manufacturing therapeutic proteins. Biotechnol Prog. 2015;31:522. doi: 10.1002/btpr.2050. [DOI] [PubMed] [Google Scholar]
- Richter M, Fairhall EA, Hoffmann SA, Tröbs S, Knöspel F, Probert PME, Oakley F, Stroux A, Wright MC, Zeilinger K. Pancreatic progenitor-derived hepatocytes are viable and functional in a 3D high density bioreactor culture system. Toxicol Res. 2016;5:278–290. doi: 10.1039/C5TX00187K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JF, Nafziger AN, Jr JSB. Pharmacogenetics affects dosing, efficacy, and toxicity of cytochrome P450–metabolized drugs. Am J Med. 2002;113:746–750. doi: 10.1016/S0002-9343(02)01363-3. [DOI] [PubMed] [Google Scholar]
- Runge D, Runge DM, Jã¤Ger D, Lubecki KA, Beer SD, Karathanasis S, Kietzmann T, Strom SC, Jungermann K, Fleig WE. Serum-free, long-term cultures of human hepatocytes: maintenance of cell morphology, transcription factors, and liver-specific functions. Biochem Biophys Res Commun. 2000;269:46–53. doi: 10.1006/bbrc.2000.2215. [DOI] [PubMed] [Google Scholar]
- Schwartz RE, Fleming HE, Khetani SR, Bhatia SN. Pluripotent stem cell-derived hepatocyte-like cells. Biotechnol Adv. 2014;32:504–513. doi: 10.1016/j.biotechadv.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Zhu Y. Application of statistically-based experimental designs in medium optimization for spore production of Bacillus subtilis from distillery effluent. Biocontrol. 2007;52:845–853. doi: 10.1007/s10526-006-9055-z. [DOI] [Google Scholar]
- Shi XL, Gao Y, Yan Y, Ma H, Sun L, Huang P, Ni X, Zhang L, Zhao X, Ren H, Hu D, Zhou Y, Tian F, Ji Y, Cheng X, Pan G, Ding YT, Hui L. Improved survival of porcine acute liver failure by a bioartificial liver device implanted with induced human functional hepatocytes. Cell Res. 2016;26:206–216. doi: 10.1038/cr.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takechi R, Taniguchi A, Ebara S, Fukui T, Watanabe T. Biotin deficiency affects the proliferation of human embryonic palatal mesenchymal cells in culture. J Nutr. 2008;138:680. doi: 10.1093/jn/138.4.680. [DOI] [PubMed] [Google Scholar]
- Traber MG, Atkinson J. Vitamin E, antioxidant and nothing more. Free Radical Bio Med. 2007;43:4. doi: 10.1016/j.freeradbiomed.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacanti JP, Kulig KM. Liver cell therapy and tissue engineering for transplantation. Semin Pediatr Surg. 2014;23:150–155. doi: 10.1053/j.sempedsurg.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Vasconcellos R, Alvarenga EC, Parreira RC, Lima SS, Resende RR. Exploring the cell signalling in hepatocyte differentiation. Cell Signal. 2016;28:1773–1788. doi: 10.1016/j.cellsig.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Wang P, Cong M, Liu TH, Yang AT, Cong R, Wu P, Tang SZ, Xu Y, Wang H, Wang BE, Jia JD, You H. Primary isolated hepatic oval cells maintain progenitor cell phenotypes after two-year prolonged cultivation. J Hepatol. 2010;53:863–871. doi: 10.1016/j.jhep.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Yeom CG, Kim DI, Park MJ, Choi JH, Jeong J, Wi A, Park W, Han HJ, Park SH. Insulin-induced CARM1 upregulation facilitates hepatocyte proliferation. Biochem Biophys Res Commun. 2015;461:568–574. doi: 10.1016/j.bbrc.2015.04.099. [DOI] [PubMed] [Google Scholar]
- Zeilinger K, Schreiter T, Darnell M, Söderdahl T, Lübberstedt M, Dillner B, Knobeloch D, Nüssler AK, Gerlach JC, Andersson TB. Scaling down of a clinical three-dimensional perfusion multicompartment hollow fiber liver bioreactor developed for extracorporeal liver support to an analytical scale device useful for hepatic pharmacological in vitro studies. Tissue Eng Part C Methods. 2011;17:549–556. doi: 10.1089/ten.tec.2010.0580. [DOI] [PubMed] [Google Scholar]
- Zhao A, Chen F, Ning C, Wu H, Song H, Wu Y, Chen R, Zhou K, Xu X, Lu Y. Use of real-time cellular analysis and Plackett–Burman design to develop the serum-free media for PC-3 prostate cancer cells. PLoS ONE. 2017;12:e0185470. doi: 10.1371/journal.pone.0185470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XQ, Pan XH, Yao L, Li W, Cui J, Wang G, Mrsny RJ, Hoffman AR, Hu JF. Converting skin fibroblasts into hepatic-like cells by transient programming. J Cell Biochem. 2016;117:589–598. doi: 10.1002/jcb.25355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.