Abstract
The knowledge of drug metabolising enzymes (DMEs) in cattle is rather limited. The capability of the bovine foetal hepatocyte-derived cell line BFH12 to serve as model for biotransformation was evaluated. Gene expression analysis of DMEs was performed by reverse transcription PCR (RT-PCR). The presence of efflux transporters was visualised by immunocytochemistry, and functional induction of cytochrome P450 (CYP) 1A was assessed by the ethoxyresorufin-O-deethylase (EROD) assay. The production of bile acids was measured by liquid chromatography-tandem mass spectrometry (LC–MS/MS). RT-PCR revealed the expression of cytochromes 1A1, 1A2, 3A4 and phase II enzymes UGT1A1, UGT1A6 and GSTM1. Immunofluorescence demonstrated efflux transporters ABCG2 and ABCC1. The EROD assay revealed a dose-dependent CYP1A induction after treatment with benzo[a]pyrene (BP). LC–MS/MS analysis of cell culture supernatants showed the production of bile acids including taurocholic acid, tauro-chenodeoxycholic acid, taurodeoxycholic acid and taurolithocholic acid. The results strongly suggest the applicability of the cell line BFH12 for subsequent experiments in the emerging field of bovine biotransformation.
Electronic supplementary material
The online version of this article (10.1007/s10616-018-0279-4) contains supplementary material, which is available to authorized users.
Keywords: Biotransformation, Cytochrome P450, Bovine hepatocyte, Cattle, In vitro model, Bile acid
Introduction
Compared to other species, knowledge on drug metabolising enzymes (DMEs) in cattle is fairly limited, especially with regard to the regulation and induction of these enzymes (Darwish et al. 2010). This is particularly problematic since food-producing animals are frequently exposed to a variety of xenobiotics, including antibiotics, growth hormones, pesticides and toxins (Khaniki 2007). These compounds are enzymatically biotransformed to increase hydrophilicity and facilitate excretion. Dysregulation of this process has been implicated in the development of several diseases (Langmann et al. 2004). The induction or inhibition of DMEs can lead to a reduced or prolonged half-life of a drug, respectively (Leucuta and Vlase 2006). Moreover, DMEs may also contribute to the toxification of certain compounds (Li et al. 2011). Accumulation of these toxic or carcinogenic metabolites in dairy products such as meat or milk may pose a potential risk for human and animal health (Wassermann et al. 2013). Thus, studying drug metabolism in Bos taurus using a well-established and well-characterised model contributes to animal welfare and aids in evaluating residue levels in edible tissues and dairy food products (Dacasto et al. 2005; Ioannides 2006; Zancanella et al. 2010). Only a few models other than animals have been described for investigating cattle biotransformation (Rijk et al. 2012).
To this day, no bovine hepatocyte-derived cell line except BFH12 has been established to our knowledge. Therefore, there is a clear need for further knowledge about CYP enzymes and their regulation in food-producing animals (Fink-Gremmels 2008). The currently most used models for biotransformation are primary hepatocytes, liver microsomes and liver slices. All of them have significant limitations, e.g. low predictive power and experimental robustness, phenotypic changes and large batch variability (Plant 2004). In order to overcome these limitations, we recently established a novel SV40 large T-antigen-transduced hepatocyte cell line (BFH12) derived from foetal cattle liver. BFH12 cells share many characteristics with primary hepatocytes, including an epithelial phenotype, hepatocyte-like metabolism and a stable expression of several phase I, II and III enzymes. These findings suggest that the cells retain important features of primary cells and may be useful for studies on bovine biotransformation (Gleich et al. 2016).
This study was conducted to assess the suitability of BFH12 as in vitro model for biotransformation in cattle. Gene expression of basal and induced levels of DMEs and efflux transporters were analysed. Immunofluorescence staining of several transporters was performed and bile acid production was measured. These studies aimed to show that BFH12 may be used to gain a better understanding of xenobiotic metabolism, how xenobiotics are detoxified and eliminated or why they accumulate in the animal. Furthermore, the carry-over from feed to food, which represents a persistent risk to public health (Anadón 2016), could be studied. BFH12 may be a possible candidate for short- or long-term experiments on bovine biotransformation with a higher predictive power than other in vitro bovine models.
Materials and methods
Materials
All chemicals and reagents were obtained from Sigma-Aldrich (Munich, Germany) unless noted otherwise. Cell culture flasks and multi-well plates were purchased from Greiner Bio-One (Frickenhausen, Germany) and TPP (Trasadingen, Switzerland), respectively. Williams’ Medium E, l-alanyl-l-glutamine, penicillin/streptomycin and standardised FBS were acquired from Biochrom (Berlin, Germany).
Cell culture
BFH12 cells were cultured in Williams’ Medium E containing 5% heat-inactivated FBS, 1% penicillin/streptomycin, 2 mM l-alanyl-l-glutamine, 100 nM dexamethasone and 0.2 U/ml insulin. Cells were seeded in 75 cm2 cell culture flasks at a density of 5 × 103 cells/cm2 (equivalent to 2.5 × 104 cells/ml) and incubated at 37 °C in a humidified atmosphere containing 5% CO2. Medium was changed on day 3. The cells were harvested using Trypsin–EDTA (8 min at 37 °C) and passaged every 7 days. Thus, a passage is defined as the cultivation of BFH12 during the course of 7 days with a routine medium exchange on day 3. Cell number and viability were determined with the Moxi Z cell counter (ORFLO Technologies, Ketchum, USA).
Reverse transcription (RT)-PCR
RNA of biotransformation enzymes and transporters was detected by reverse transcription (RT)-PCR. Genes were chosen based on their significance in drug metabolism and the availability of sequence data.
The expression of hepatocyte-specific genes in BFH12 cells of passage 20, foetal hepatocytes and adult liver tissue was determined by RT-PCR. Total RNA from BFH12 grown in 25 cm2 cell culture flasks was extracted using the ReliaPrep™ RNA Cell MiniPrep System (Promega, Madison, USA). RNA from adult liver tissue was extracted with the ReliaPrep™ RNA Tissue MiniPrep System. Integrity of the RNA extracted from the samples was assessed by gel electrophoresis in an agarose gel stained with ethidium bromide (EtBr). Only intact RNA showing sharp, clear 28S and 18S rRNA bands was used for subsequent cDNA synthesis. Reverse transcription was performed with the Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Scientific, Schwerte, Germany). PCR amplification was carried out on a T1 Thermocycler (Biometra, Göttingen, Germany) using the FastGene® Optima HotStart Ready Mix (Nippon Genetics, Düren, Germany). Initial denaturation was performed at 95 °C for 3 min followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 55 °C for 15 s and extension at 72 °C for 1 min. Genes of interest were cytochrome P450 1A1 (CYP1A1), 1A2 (CYP1A2), 2B6 (CYP2B6), 2C19 (CYP2C19), 2E1 (CYP2E1), 3A4 (CYP3A4), UDP glucuronosyltransferase 1 family polypeptide A1 (UGT1A1) and A6 (UGT1A6), glutathione S-Transferase M1 (GSTM1), P-glycoprotein (P-gp), ATP-binding cassette sub-family G member 2 (ABCG2), ATP-binding cassette sub-family C member 1 (ABCG2), organic cation transporter 1 (OCT1), organic anion-transporting polypeptide 1B3 (OATP1B3), and sodium-taurocholate cotransporting polypeptide (NTCP). β-actin (ACTB) served as positive control. The DME genes are designated by conventional (human) nomenclature. A new nomenclature for bovine DMEs has been proposed (Zancanella et al. 2010) which is not yet standardized or implemented in the National Center for Biotechnology Information (NCBI) database. The gene-specific PCR primers were designed from bovine sequence data using the primer-BLAST tool of the NCBI and were synthesised by Metabion International AG (Planegg, Germany). Primer sequences and PCR conditions are given in Table 1. PCR products were analysed by gel electrophoresis (3.5% agarose) and ethidium bromide staining. Product sizes were determined using a 100 bp DNA ladder (Thermo Scientific GeneRuler 100 bp DNA Ladder; Thermo Scientific, Schwerte, Germany). In order to compare the gene expression of BFH12 and primary cells, experiments were also conducted with adult liver tissue and foetal bovine hepatocytes (cryopreserved cells). The cells were cultivated using the same cell culture protocol as for cultivation of BFH12 before cryopreservation. Gene expression analysis of BFH12 cells and foetal hepatocytes was performed in two independent experiments (n = 2).
Table 1.
Primers and conditions for gene expression analysis by RT-PCR
| Gene | Primer sequences | Accession no. | Amplicon size (bp) |
|---|---|---|---|
| CYP1A1 | F: 5′-CATCCCTGTCCTCCGTTACC-3′ R: 5′-TCCCGGATGTGACCCTTCTC-3′ |
XM_005222018.2 | 132 |
| CYP1A2 | F: 5′-TCATAGGCGCCCTGTTCAAG-3′ R: 5′-GTGATGGTGTCAAACCCAGC-3′ |
XM_010817139.1 | 121 |
| CYP2B6 | F: 5′-CCTTCCTGAGGTTCCAACAGA-3′ R: 5′-GAATGGCCTCTGTCCCACAT-3′ |
XM_005218914.2 | 93 |
| CYP2C19 | F: 5′-TCAGCAGGAAAAAGAGTTTGTG-3′ R: 5′-TCACAGAAGGGTGGAATAGAGA-3′ |
NM_001109792.2 | 167 |
| CYP2E1 | F: 5′-GGCGTTTTCTGCAGGATATGAA-3′ R: 5′-CCTTGCAGGCACAAAGTCAG-3′ |
XM_010820126.1 | 150 |
| CYP3A4 | F: 5′-CGATCCCTTTCTTCTCGCAGT-3′ R: 5′-AGTCCACACGTGGCTTTTGA-3′ |
NM_001099367.1 | 164 |
| UGT1A1 | F: 5′-TTCCCAAGACCCATCATGCC-3′ R: 5′-TCAAATTCCTGAGAGAGTGGCT-3′ |
NM_001105636.1 | 83 |
| UGT1A6 | F: 5′-GAAGGCCTCCATTTGGCTGT-3′ R: 5′-CAAATTCCTGAGACAGGACGC-3′ |
NM_174762.1 | 125 |
| GSTM1 | F: 5′-GAGAAGACAAGTTTCAAGCCCA-3′ R: 5′-CTGTCATAGTCGGGAGCATCT-3′ |
NM_175825.3 | 193 |
| P-gp | F: 5′-AAACTGCCTAATAAATTTGACACCC-3′ R: 5′-GCGATGGCGATTCTCTGCTT-3′ |
AJ829445.1xxx | 83 |
| ABCG2 | F: 5′-GAGCCATAGGTTTCCGCTGT-3′ R: 5′-GCCGTATCGAGGAATGCTCA-3′ |
NM_001037478.3 | 902 |
| ABCC1 | F: 5′-GACCCCGTTTTGTTTTCGGG-3′ R: 5′-CGATCACCCTCGTGTAGTCC-3′ |
NM_174223.1 | 370 |
| OCT1 | F: 5′-CTTCATCCGGAACCTTGGCA-3′ R: 5′-GCATCCTCGATGGTCTCTGG-3′ |
NM_001101098.2 | 207 |
| OATP1B3 | F: 5′-CTCCAGTGGCATCGCATGTA-3′ R: 5′-GGAATTCCTCCTAGCGTTCG-3′ |
NM_205804.2 | 421 |
| NTCP | F: 5′-CTCAACGGACGATGCAAACG-3′ R: 5′-ATGCAAAGGCAGCTTTGGTG-3′ |
NM_001046339.1 | 599 |
| ACTB | F: 5′-CTTCCTGGGCATGGAATCCT-3′ R: 5′-CGTAGAGGTCCTTGCGGATG-3′ |
NM_173979.3 | 89 |
MTT assay
Benzo[a]pyrene (BP) is known to induce CYP1A in bovine hepatocytes. BP was tested in order to exclude any cytotoxic effect on BFH12 in the subsequent EROD assay. Cytotoxicity of BP was determined determined using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay, carried out in 96-well plates according to Riss et al. (2004). Briefly, BFH12 cells were seeded at a density of 0.01 × 106 cells per well, cultivated for 24 h and treated for an additional 24 h with BP at concentrations of 0.01 μM, 0.1 μM, 1 μM and 10 μM. Cyclohexane was used as solvent for the BP stock solution (end concentration: 0.025 μl cyclohexane/μl of assay medium). After cultivation, 20 μl MTT solution (5 mg/ml) were added to each well and the plate was incubated for 2 h. After that, 10 μl SDS (3%) were added and culture medium was completely removed. Formazan crystals were dissolved in DMSO (200 μl/well). Absorption was measured at 595 nm on a Wallac Victor 2 multilabel plate reader (Perkin Elmer, Massachusetts, USA). The MTT assay was conducted in 8 wells containing cells from one cryotube (technical replicates).
Functional activity of cytochrome P450 1A (EROD assay)
Induction of cytochrome P450 1A enzymes by BP was determined by ethoxyresorufin O-deethylase (EROD) assay according to Halwachs et al. (2010). The EROD assay is a well-established method to assess CYP1A activity in bovine liver microsomes (Machala et al. 2003; Giantin et al. 2008; Darwish et al. 2010). BFH12 cells were seeded into 24-well plates (0.05 × 106 cells per well), cultivated under standard culture conditions for 24 h, and induced with BP at concentrations of 0.01 μM, 0.1 μM, 1 μM and 10 μM (in cell culture medium, 200 μl/well) for an additional 24 h. This time period is required for reliable estimates, although maximal induction of enzyme activity may not be achieved within 24 h of incubation. All BP concentrations were prepared independently. Formation of the fluorescent dye resorufin was calculated as pmol per mg protein and minute. Total cell protein was determined using the bicinchoninic acid assay (Smith et al. 1985). The EROD assay was conducted in 12 wells per treatment group containing cells from 2 cryotubes (n = 12).
Immunocytochemistry
Expression and localisation of ATP-binding cassette transporter G2 (ABCG2) and C1 (ABCC1) were analysed by immunocytochemistry. Both transporters are considered to be of great relevance as they exert either protective functions by reducing oral availability/limiting penetration of potential harmful xenobiotics into critical tissues or play a detrimental role by mediating active transport of these compounds into the milk (Platte and Honscha 1996; Halwachs et al. 2010; Wassermann et al. 2013; Mahnke et al. 2016). Furthermore, they are considered as one of the few transporters to be responsible for extruding drugs commonly used in veterinary patients (Mealey 2012). Cells seeded on a 24-well plate at a density of 0.05 × 106 cells per well were cultivated for 48 h. Cells were treated with 10 μM BP for 24 h. Fixation was performed with sodium metaperiodate-lysine-paraformaldehyde solution (McLean and Nakane 1974) for 10 min. Permeabilisation and blocking was done by incubation for 1 h in a PBS solution containing 0.5% Triton X-100 and 4% horse serum (CC pro GmbH, Oberdorla, Germany). Cells were incubated overnight at 4 °C in a dark humidified chamber with suitable primary antibodies against ABCG2 (BXP-21, sc-58222, Santa Cruz Biotech, Dallas, TX; dilution 1:200) and ABCC1 (MRPm6, sc59609, Santa Cruz Biotech, Dallas, TX; dilution 1:200). Subsequently, cells were stained for 2.5 h at 4 °C in a humidified dark chamber with suitable Cy3 conjugated anti-mouse (715-165-161, Dianova; dilution 1:500) and anti-goat (705-165-145, Dianova; dilution 1:500) antibodies. Nuclei were stained with 0.25 µg/ml 4′,6-diamidino-2-phenylindole (DAPI; Roth, Karlsruhe, Germany) for 5 min at room temperature. Labelled cells were imaged on an Olympus IX50 fluorescence microscope and analysed using the cellF software version 2.6 (Olympus, Hamburg, Germany). Secondary antibody controls were included in each experiment. Immunofluorescence analysis was performed in 2 independent experiments.
Bile acid quantification by LC/MS–MS
LC/MS–MS analysis was performed to study the formation and conjugation of bile acids which are important liver-specific metabolic markers. The production of conjugated bile acid is also an indicator for active secondary conjugation (Chiang 2009). BFH12 cells were cultivated in culture medium for 7 days without medium change. During this time period no changes in cell morphology or viability attributable to a lack of nutrients were evident. Concentrations of bile acids were quantified by liquid chromatography-tandem mass spectrometry as previously described (Scherer et al. 2009). In order to calculate the production of metabolites, the amount of bile acids in culture medium before cultivation was quantified and the means of these values were subtracted from values obtained after 7 days. The analysis was performed four times (n = 4). This experiment was conducted using the supernatant of 4 cell culture flasks containing cells from 2 cryotubes (n = 4).
Statistical analysis
All experiments were conducted independently at least two times, except for the MTT assay which was performed once. Data are presented as mean ± standard deviation (SD) unless noted otherwise. D’Agostino-Pearson omnibus test was used to test for normality. In order to determine significant differences between means, one-way analysis of variance (ANOVA) was performed. A p value of less than 0.05 was considered statistically significant. Graphs and statistics were generated using GraphPad Prism 6 (GaphPad Software, La Jolla, USA). Cells from 2 cryobatches were independently cultured in cell culture flasks and then seeded into the experimental flasks or wells, depending on the respective experiment.
Results
Gene expression of drug metabolising enzymes and efflux transporters
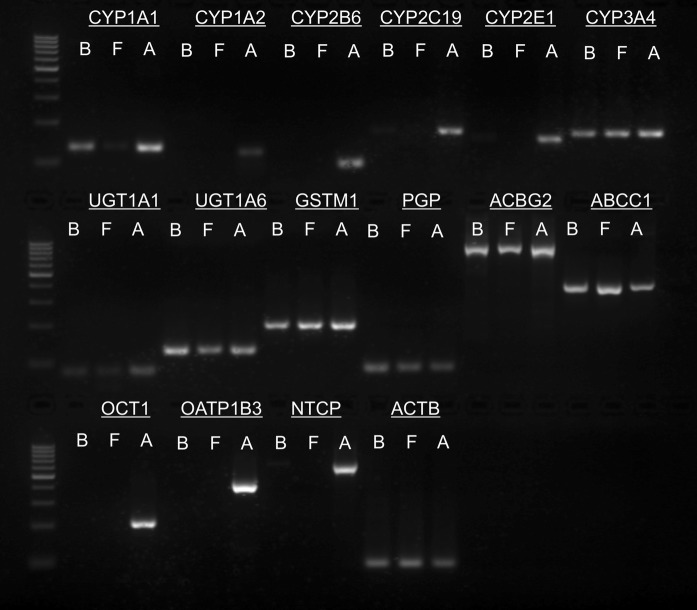
CYP1A1 and CYP3A4 expression was found in BFH12 cells, foetal hepatocytes and adult hepatic tissue (Fig. 1). CYP1A2, 2B6, and 2E1 were not expressed in BFH12 and foetal hepatocytes under standard culture conditions. All three phase II genes (UGT1A1, UGT1A6, GSTM1) could be detected in BFH12 and foetal hepatocytes. Gene expression of the phase III transporters P-gp, ABCG2, and ABCC1 was also observed in BFH12 and foetal hepatocytes. There was no expression of OCT1 and OATP1B3 in BFH12, but a weak band was obtained for CYP2C19 and NTCP. In adult liver tissue, which served as a positive control, all genes were expressed. ACTB served as loading control in the gene expression experiments.
Fig. 1.
Detection of phase I, II and III enzymes/transporters by reverse transcription (RT)-PCR. Representative image of genes expressed in BFH12 cells of passage 20 (“B”), foetal bovine hepatocytes (“F”) and adult liver tissue (“A”). Expression of the following genes was analysed: Cytochrome P450 1A1 (CYP1A1), cytochrome P450 1A2 (CYP1A2), cytochrome P450 2B6 (CYP2B6), cytochrome P450 2C19 (CYP2C19), cytochrome P450 2E1 (CYP2E1), cytochrome P450 3A4 (CYP3A4), UDP glucuronosyltransferase 1 family polypeptide A1 (UGT1A1), UDP glucuronosyltransferase 1 family polypeptide A6 (UGT1A6), glutathione S-transferase M1 (GSTM1), P-glycoprotein (PGP), ATP-binding cassette sub-family G member 2 (ABCG2), ATP-binding cassette sub-family C member 1 (ABCC1), organic cation transporter 1 (OCT1), organic anion-transporting polypeptide 1B3 (OATP1B3), sodium-taurocholate cotransporting polypeptide (NTCP) and β-actin (ACTB). ACTB served as loading control. Product sizes were determined using a 100 bp DNA ladder (n = 2 per gene for BFH12 cells and foetal hepatocytes, n = 1 for adult liver tissue used as positive control)
For purposes of comparison and to better estimate the stability of the system we have included the results of the gene expression analysis of BFH12 cells of passage 13 and 15 as supplementary Fig. 1. Few differences are apparent between the passages. Compared to the late passage (20) in the early passages (13–15) NTCP was expressed whereas UGT1A1 was not. Furthermore, CYP1A1 showed an increase in expression with passaging.
Cell viability is unaffected by benzo[a]pyrene (BP)
Cell viability (%) was unaffected by BP treatment (Fig. 2). Cyclohexane, used as solvent for the BP stock solution, had no effect on cell viability. Cells treated with Triton-X 0.1% served as lysis control. As expected, treatment with Triton-X 0.1% resulted in complete cell death.
Fig. 2.
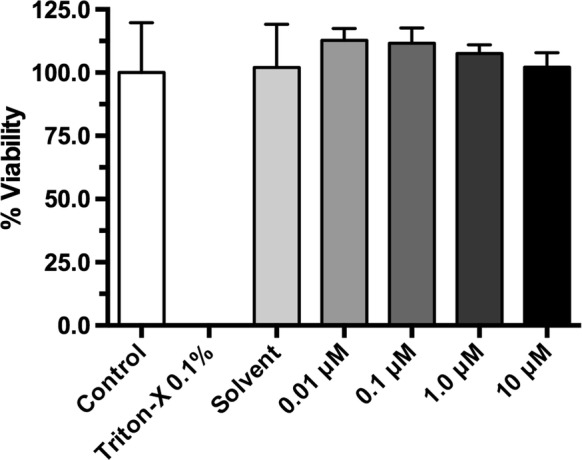
Effect of benzo[a]pyrene on cell viability. The MTT assay was performed to assess the effect of BP on the viability of BFH12 cells. Cell viability was determined measuring the formazan production at a wavelength of 595 nm and calculated as % viability compared to the control. The mean of the absolute absorption value of the control cells was 1.176. Data are shown as mean of eight technical replicates ± SD
Induction of CYP1A1 and CYP1A2 gene expression by BP
The gene expression of CYP1A1 and 1A2 after addition of 10 μM BP was determined by RT-PCR and agarose gel electrophoresis. In untreated cells, only mRNA of CYP1A1 was present (Fig. 3). CYP1A2 expression was not observed. There was a small but visually apparent increase in CYP1A1 expression in cells treated with BP. Furthermore, CYP1A2 gene was expressed at detectable levels after treatment. The bands in Fig. 3 correspond to the expected products CYP1A1 (132 bp) and CYP1A2 (121 bp).
Fig. 3.
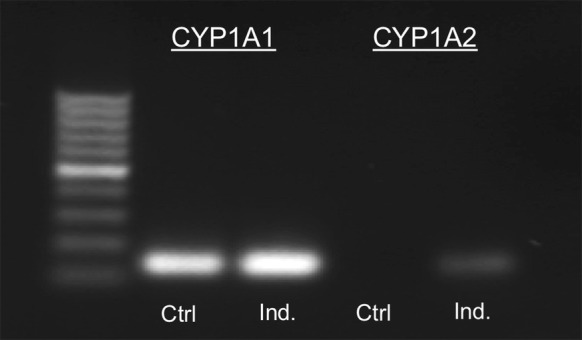
Induction of cytochrome P450 1A1 and 1A2 gene expression by benzo[a]pyrene. Representative image of CYP1A1 and 1A2 gene expression after induction (Ind.) with 10 μM benzo[a]pyrene. Product sizes were validated using a 100 bp DNA ladder (n = 2 per gene)
CYP1A activity is upregulated by benzo[a]pyrene
The EROD assay which is based on the deethylation of 7-ethoxyresorufin to resorufin was used to determine specific CYP1A activity (Fig. 4). BFH12 shows only low constitutive CYP1A activity. However, CYP1A activity was significantly and dose-dependently increased by BP compared to control-treated cells.
Fig. 4.
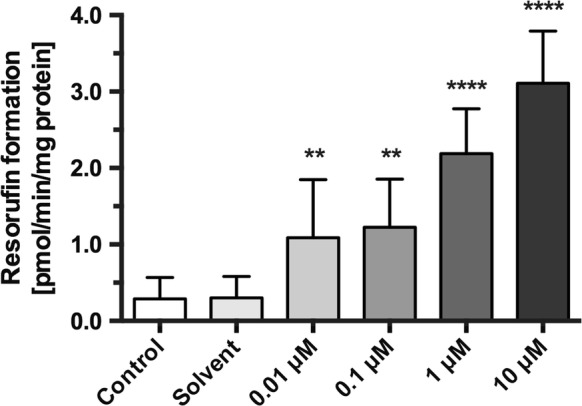
Induction of cytochrome P450 1A activity by benzo[a]pyrene. Production of resorufin from 7-ethoxyresorufin (EROD) was measured as fluorescence intensity and converted to resorufin production per minute and mg protein (pmol/min/mg protein) after an incubation time of 24 h. Asterisks indicate significant differences between cells treated with BP and the solvent control. (**p < 0.01, ****p < 0.0001; n = 12). No significant differences were found between control and solvent
Expression of the efflux transporters ABCG2 and ABCC1
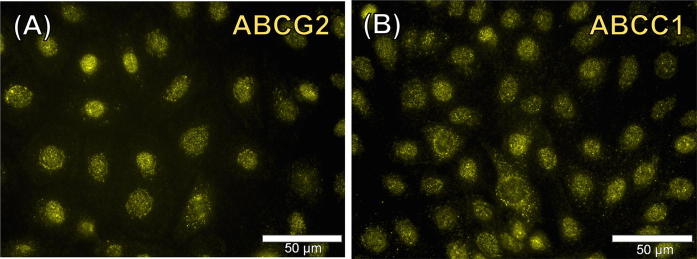
Immunofluorescence analysis revealed the presence of efflux transporters ABCG2 and ABCC1 in control cells (treated with 2.5% cyclohexane). A representative image of the cellular expression of phase III transporter ABCG2 is shown (Fig. 5a). Figure 5b shows the result obtained for the efflux transporter ABCC1. Controls of both Cy3-conjugated antibodies were negative, indicating the specificity of the labelling (see supplementary Fig. 2).
Fig. 5.
Immunofluorescence analysis of the phase III transporters ABCG2 and ABCC1. Representative images of the expression and localisation of ABC transporter G2 (ABCG2) (a) and ATP-binding cassette sub-family C member 1(ABCC1) (b). × 400. Analysis was performed in two independent experiments (n = 2)
Production of taurine-conjugated bile acids
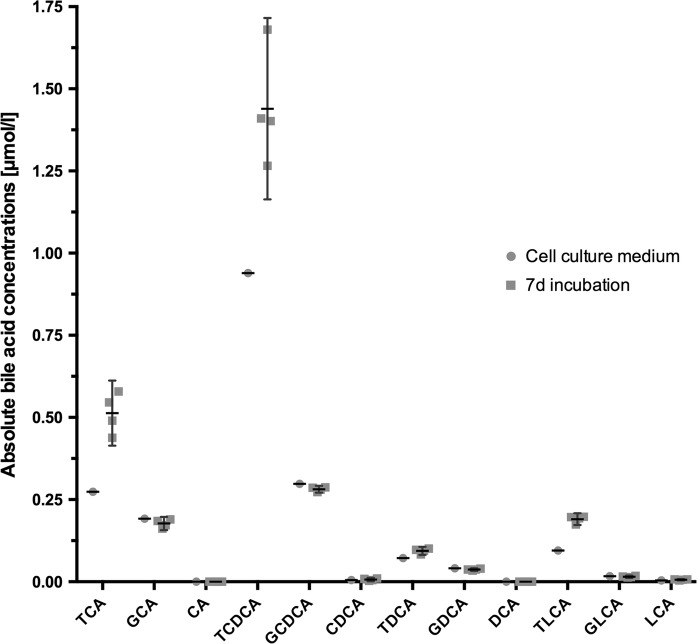
The following taurine-conjugated bile acids were detected in cell culture supernatants by LC–MS/MS (Fig. 6): taurocholic acid (TCA), taurochenodeoxycholic acid (TCDCA), taurodeoxycholic acid (TDCA) and taurolithocholic acid (TLCA). Other bile acids investigated were not detectable in cell culture supernatants. In order to compare bile acid concentrations before and after cultivation data for both groups have been included in Fig. 6. For reasons of clarity, supplementary Fig. 3 presents the relative concentrations (i.e. mean difference).
Fig. 6.
Determination of bile acid production in cell culture supernatants by LC/MS–MS with data of concentrations present in cell culture medium. Concentration of individual bile acids present in cell culture medium supplemented with FBS and amount of each bile acid (in μmol/L) after 7 days of cultivation are shown. The bile acids measured were: Taurocholic acid (TCA), taurochenodeoxycholic acid (TCDCA), taurodeoxycholic acid (TDCA) and taurolithocholic acid (TLCA), glycocholic acid (GCA), cholic acid (CA), glycochen- odeoxycholic acid (GCDCA), chenodeoxycholic acid (CDCA), glycodeoxycholic acid (GDCA), deoxycholic acid (DCA), glycolithocholic acid (GLCA), lithocholic acid (LCA). Data are presented as column scatter plots showing medians and interquartile ranges
Discussion
We evaluated the drug metabolizing capacity of the recently described bovine hepatocyte-derived cell line BFH12. Our findings support the use of BFH12 as a candidate for in vitro studies of bovine drug metabolism. The cell line should be useful for investigations from individual cytochromes and excretion processes to whole biotransformation pathways. Other in vitro models are available, e.g. primary hepatocytes, which are the closest relation to the in vivo situation but they tend to dedifferentiate into a foetal state within few days of cultivation (Guguen-Guillouzo et al. 1980; Nakamura et al. 1988) and also after cryopreservation. Owing to their limited lifespan they are not suitable for long-term studies (Swift et al. 2010). Liver microsomes on the other hand are used to investigate phase I and, at least in part, UGT-mediated metabolism, but a major downside is the artificial character of this model. Liver microsomes lack a plasma membrane, as well as intracellular control mechanisms and metabolic exchange with other subcellular organelles (Shull et al. 1987). Liver slices retain the normal hepatic tissue structure, but not all cells are fully preserved during preparation. The main disadvantages of liver slices include inadequate penetration of medium, short-term viability and the large number of damaged cells with impaired biotransformation (Brandon et al. 2003). As a result of their short longevity, liver slices are not suitable to investigate poorly metabolised drugs (Sivapathasundaram et al. 2004). Therefore, it is helpful and appropriate to establish a new cell line that allows reproducible in vitro analysis and that can be used to investigate different aspects of bovine biotransformation.
In order to gain a deeper understanding of the new established bovine hepatocyte derived cell line BFH12, its gene expression of phase I–II enzymes and phase III efflux transporters was investigated: Transcription of the most common phase I enzymes CYP P450 isoforms 1A1, 1A2, 2B6, 2C19, 2E1 and 3A4 was assessed by RT-PCR. There is only little information available on bovine CYPs compared to respective CYPs of human or murine origin. Therefore, CYPs were selected based on their orthology to human CYPs (Ioannides 2006; Fink-Gremmels 2008; Antonovic and Martinez 2011). A computer-based orthology study found CYP3A28, CYP3A38 and CYP3A48 (original nomenclature: CYP3A4, CYP3A5, CYP3A4 (niphedipine oxidase)) to be the most expressed enzymes (Zancanella et al. 2010). Our gene expression analysis revealed that cytochromes 1A2, 2B6 and 2E1 are not expressed in BFH12. These results are in line with those for primary foetal hepatocytes and provide evidence for the foetal origin of BFH12. Foetal hepatocytes feature a different set of enzymes compared to adult liver tissue, and therefore have their own gene expression pattern (Lazaro et al. 2003; Deurholt et al. 2009; Zhou et al. 2014). CYP3A4 was expressed in BFH12, which is in accordance with the findings for foetal cells. CYP1A1 and CYP2C19 gene levels were weakly expressed in BFH12 but absent in foetal hepatocytes. It is possible that chromosomal aberrations were induced during transformation of the cells with SV40LTAg as described previously (Sack 1981; Woodworth and Isom 1987; Klocke et al. 2002; Priesner et al. 2004; Reid et al. 2009). These alterations may have led to a constitutive expression of both CYPs. Since there is only little data available on other bovine hepatocyte-derived cell lines, we discuss our data in relation to data from other species. For established hepatocyte-derived cell lines from other species, different patterns of gene expression are observed. Cytochromes 3A4 and 2B6 were found in a feeder-cell dependent bovine hepatocyte line, but their expression could not be increased (Talbot et al. 2015). A study investigating the CYP system in the coronary arteries and the liver of cattle found CYP 1A, 2B, 2C, 2D, 2E, 2 J, 3A, 4A subfamilies to be present in the liver (Grasso et al. 2005). Immortalized cells of porcine origin only express CYP1, CYP2 and CYP3 (Pan et al. 2010). However, subfamilies have not been specified. A subline of various transgenic mouse hepatocyte-derived cell lines showed induction of CYP3A2, 1A1, 2B1 and presence of 2E1 (Klocke et al. 2002). Human HepG2 cells express cytochromes 1A1, 2A6, 2D6 and 3A7 while BFH12 express CYPs 1A1, 1A2 and 3A4. Another cell line, HepaRG, features a complete set of cytochromes, phase II enzymes and transporters, but it is of human origin and requires a time consuming and sophisticated cell culture protocol (Kanebratt and Andersson 2008).
It is to be expected that there are significant interspecies differences in CYP isoforms, given that expression and induction of CYPs are species-specific. Moreover, these differences are considered to be a major cause of species differences in xenobiotic metabolism. This implies that data from different species are not directly comparable. Thus, it is important to note that data from different species are given here in order to provide an overview of available information on the CYP profiles. The limited amount of published data on bovine hepatocytes precludes a thorough comparison with other species. In any case, the differences in gene expression profiles should be considered when choosing a cell line for toxicological studies. The gene expression results were performed using cells of passages 13–20 (see Fig. 1 and supplementary Fig. 1). Comparing both figures, it is seen that gene expression is fairly stable over time. However, few differences were observed between passages, which have been described above (see “Gene expression of drug metabolising enzymes and efflux transporters” section). These differences may be related to small genetic differences between early and late passage. However, expression of these genes close to the detection limit of standard RT-PCR could be another explanation. They may be regulated by cell metabolism in our model and thus drop slightly above or below the detection limit, depending on the given replicate used for the RT-PCR. Small changes could also be partly due to the limitations inherent to image acquisition.
Besides the expression of phase I enzymes BFH12 also expresses phase II conjugation enzymes. These included UGT1A1 (expressed in late passage cells) and 1A6 which mediate the glucuronidation (Burchell et al. 1991) and GSTM1 (glutathione conjugation). These results indicate that BFH12 retains an enzymatic conjugation activity. However, this needs to be confirmed using UGT-substrates to measure functional activity. Gene expression analysis further showed that BFH12 expresses phase III efflux transporters, including ABCC1, ABCG2, PGP and, to a lesser extent, NTCP. In contrast, OCT1 and OATP1B3 were not detected. The results are again largely comparable to those of foetal cells. All six transporters were expressed in adult hepatocytes.
The lack of the hepatic uptake transporters OCT1 (SLC22A1) and OATP1B3 (SLCO1B3), which are found in hepatocytes forming the sinusoidal membrane (Jonker and Schinkel 2004; Lee et al. 2008), is consistent with the findings for primary foetal hepatocytes. Further studies will show whether both transporters can be induced or up-regulated using inducers or an alternative culture format. In contrast to the above-mentioned uptake transporters, the efflux transporter PGP was expressed in BFH12. PGP is a transporter for a wide range of structurally diverse compounds, including anticancer agents, immunosuppressants, and steroid hormones. It is primarily found on the canalicular surface of hepatocytes (Amin 2013). NTCP mRNA was also detected in BFH12, although its expression was rather weak. NTCP is a sodium cotransporter translocating taurocholate, bile salts, sulfates and other molecules into the cell (Trauner and Boyer 2003). NTCP might also be involved in the transport of unconjugated bile acids into BFH12 for subsequent conjugation. The similar expression pattern of BFH12 and foetal cells emphasizes the hepatocyte-like character of the cell line. Transporter expression of BFH12 cells is largely in line with findings in other cell lines. The human hepatoma cell line HepG2 also secrets bile acids but does not express OATs or OATPs, only MRPs and MDRs are present (Cui et al. 2003). HPCT-1E3, which is a fusion cell line of primary rat hepatocytes and Fao Reuber hepatoma cells H35, possesses influx transporters and ABCC1 (Halwachs 2005), a transporter also present in BFH12 cells. As expected and reported in the literature, all genes were present in fresh adult liver tissue (Kanebratt and Andersson 2008).
In order to investigate the inducibility of CYP1A1 and 1A2 the cells were treated with BP for 24 h. There was a slightly increased gene expression of CYP1A1 while CYP1A2 was detectable after inducer treatment. Induction of bovine CYP1A was also shown previously (Darwish et al. 2010; Halwachs et al. 2013). The results suggest that, despite the fact that foetal cells have a reduced set of CYPs compared to adult primary hepatocytes, CYP expression can be induced in BFH12. The AhR pathway is likely to be involved in this regulation (Talbot et al. 2015).
Concentration-dependent induction of CYP1A activity, as demonstrated by EROD assay, is another important feature of BFH12. Due to the limited amount of data available, values obtained from different cattle breeds and other species were also discussed. The measured (basal) activity is comparable to those observed for other hepatocyte-derived cell lines such as HepG2 (0.9 ± 0.5) or Mz-Hep-1 (0.4 ± 0.3), but is lower than the value of 4.5 ± 2.0 for human hepatocytes, which was reported in a study (Rodríguez-Antona et al. 2002). Depending on the study and the inducers used, values can range from 5 to 130 (LeCluyse et al. 2000).
CYP1A activities of other species are in the range of 1.9 ± 1.2 for pig hepatocytes to 6.0 ± 4.3 for rat hepatocytes (Donato et al. 1999). Compared to BFH12, bovine liver microsomes show considerably higher specific activities with 337.1 ± 2.4 (Darwish et al. 2010) or 323.5 ± 43.0 to 430.1 ± 59.3 depending on the cattle breed (Giantin et al. 2008). Other studies report values of 400 ± 30 (Pegolo et al. 2010) and 328 ± 28 (Sivapathasundaram et al. 2001), respectively, for Charolais beef cattle liver microsomes. Data from measurements on subcellular fractions obtained from bovine livers show CYP1A activities of 354.1 ± 32.1 (Girolami et al. 2016).
In order to validate the results of the ABCC1 and ABCG2 gene expression analysis, the protein expression levels of both transporters were further analyzed by immunofluorescence staining. Both transporters have a broad and, to some extent, overlapping substrate specificity (Robey et al. 2007). The results show that both efflux transporters were abundantly expressed by BFH12. It has been reported that ABCG2 is mainly located at the apical membrane of polarised cells and can be found in vivo in hepatocytes forming bile canaliculi (Jani et al. 2014). The substrates of ABCG2 comprise anticancer drugs, xenobiotics, toxins, and antibiotics (Mo and Zhang 2012). In contrast, ABCC1 is present in the basolateral membrane of hepatocytes under physiological conditions (Bakos and Homolya 2007). Substances transported by ABCC1 include antivirals, toxicants, metalloids and antibiotics (Deeley and Cole 2006).
Several taurine conjugated bile acids, which are also produced by phase II enzymes, were actively secreted by the cells. The demonstration of basic phase II activity is an important function to study bovine xenobiotic metabolism in vitro (Gusson et al. 2006). Moreover, presence of bile acids reflects hepatic function and emphasizes the hepatocyte-like character of BFH12. The strong predominance of taurine conjugated species is most likely due to a high concentration of taurine in the FBS. In addition, those species were already found in the culture medium before cultivation (see Fig. 6). For HepG2 is has been shown that mainly chenodeoxycholic acid and other primary bile acids are produced by enzymatic conjugation (Javitt 1990), whereas taurine conjugation was found in rat hepatocytes (Rembacz et al. 2010).
Our results suggest that BFH12 might be used for toxicological and metabolic studies which require a stable expression of distinct CYPs. The cell line BFH12 shows some DME capacity and allows the investigation of bovine biotransformation over a prolonged time period although it is clearly more foetal-like in characteristic.
Conclusions
The new bovine hepatocyte cell line, BFH12, expresses a range of phase I and II enzymes and efflux transporters, and is responsive to CYP1A induction by BP. It is also capable of taurine-conjugation of bile acids. Our results support the use of this cell line for in vitro investigations into the metabolism and toxicity of chemicals relevant to bovine exposure.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We gratefully thank the group of Professor Michaela Schulz-Siegmund of the Institute of Pharmaceutical Technology (University of Leipzig) for discussions and feedback. Adult liver tissue samples were obtained from the group of Professor Alexander Starke of the Medizinische Tierklinik (University of Leipzig). We also thank the University Hospital Regensburg for conducting LC/MS–MS measurements of bile acids. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Abbreviations
- ABCC1
ATP-binding cassette sub-family C member 1
- ABCG2
ATP-binding cassette sub-family G member 2
- ACTB
β-actin
- AhR
aryl hydrocarbon receptor
- ANOVA
Analysis of variance
- ARNT
AhR nuclear translocator
- BP
benzo[a]pyrene
- cDNA
complementary DNA
- Cy2
Cyanine
- Cy3
Indocarbocyanine
- CYP
Cytochrome P450
- CYP1A1
Cytochrome P450 1A1
- CYP1A2
Cytochrome P450 1A2
- CYP2B6
Cytochrome P450 2B6
- CYP2C19
Cytochrome P450 2C19
- CYP2C9
Cytochrome P450 2C9
- CYP3A4
DAPI, 4′,6-diamidino-2-phenylindole
- DMEs
drug metabolising enzymes
- EDTA
Ethylenediaminetetraacetic acid
- EROD
7-Ethoxy-resorufin-O-deethylase
- FBS
Foetal bovine serum
- GLUT2
Glucose transporter 2
- GSTM1
Glutathione S-Transferase M1
- LC/MS–MS
Liquid chromatography tandem mass spectroscopy
- MRL
Maximum residue level
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NTCP
Sodium-taurocholate cotransporting polypeptide
- OAT
Organic anion transporter
- OATP
Organic anion-transporting polypeptide
- OATP1B3
Organic anion-transporting polypeptide 1B3
- OCT1
Organic cation transporter 1
- P-gp
P-glycoprotein
- PAH
Polycyclic aromatic hydrocarbons
- PCR
Polymerase chain reaction
- RT-PCR
Reverse transcription polymerase chain reaction
- RT-qPCR
Quantitative reverse transcription PCR
- SD
Standard deviation
- SDS
Sodium dodecyl sulfate
- SLC22A1
Solute Carrier Family 22 Member 1
- SLCO1B3
Solute carrier organic anion transporter family member 1B3
- SV40LTAg
SV40 large-T antigen
- TCA
Taurocholic acid
- TCDCA
Taurochenodeoxycholic acid
- TDCA
Taurodeoxycholic acid
- TLCA
Taurolithocholic acid
- UGT1A1
UDP glucuronosyltransferase 1 family polypeptide A1
- UGT1A6
UDP glucuronosyltransferase 1 family polypeptide A6
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Amin ML. P-glycoprotein inhibition for optimal drug delivery. Drug Target Insights. 2013;2013:27–34. doi: 10.4137/DTI.S12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anadón A. Perspectives in veterinary pharmacology and toxicology. Front Vet Sci. 2016;3:1–12. doi: 10.3389/fvets.2016.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonovic L, Martinez M. Role of the cytochrome P450 enzyme system in veterinary pharmacokinetics: where are we now? Where are we going? Future Med Chem. 2011;3:855–879. doi: 10.4155/fmc.11.37. [DOI] [PubMed] [Google Scholar]
- Bakos E, Homolya L. Portrait of multifaceted transporter, the multidrug resistance-associated protein 1 (MRP1/ABCC1) Pflügers Arch Eur J Physiol. 2007;453:621–641. doi: 10.1007/s00424-006-0160-8. [DOI] [PubMed] [Google Scholar]
- Brandon EFA, Raap CD, Meijerman I, et al. An update on in vitro test methods in human hepatic drug biotransformation research: pros and cons. Toxicol Appl Pharmacol. 2003;189:233–246. doi: 10.1016/S0041-008X(03)00128-5. [DOI] [PubMed] [Google Scholar]
- Burchell B, Nebert DW, Nelson DR, et al. The UDP glucuronosyltransferase gene superfamily: suggested nomenclature based on evolutionary divergence. DNA Cell Biol. 1991;10:487–494. doi: 10.1089/dna.1991.10.487. [DOI] [PubMed] [Google Scholar]
- Chiang JYL. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, König J, Nies AT, et al. Detection of the human organic anion transporters SLC21A6 (OATP2) and SLC21A8 (OATP8) in liver and hepatocellular carcinoma. Lab Invest. 2003;83:527–538. doi: 10.1097/01.LAB.0000065015.02412.48. [DOI] [PubMed] [Google Scholar]
- Dacasto M, Eeckhoutte C, Capolongoa F, et al. Effect of breed and gender on bovine liver cytochrome P450 3A (CYP3A) expression and inter-species comparison with other domestic ruminants. Vet Res. 2005;36:179–190. doi: 10.1051/vetres:2004066. [DOI] [PubMed] [Google Scholar]
- Darwish WS, Ikenaka Y, Eldaly EA, et al. Cytochrome P450 1A-dependent activities in deer, cattle and horses. J Vet Med Sci. 2010;72:561–566. doi: 10.1292/jvms.09-0318. [DOI] [PubMed] [Google Scholar]
- Deeley RG, Cole SPC. Substrate recognition and transport by multidrug resistance protein 1 (ABCC1) FEBS Lett. 2006;580:1103–1111. doi: 10.1016/j.febslet.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Deurholt T, van Til NP, Chhatta AA, et al. Novel immortalized human fetal liver cell line, cBAL111, has the potential to differentiate into functional hepatocytes. BMC Biotechnol. 2009;9:89. doi: 10.1186/1472-6750-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato MT, Castell JV, Gómez-Lechón MJ. Characterization of drug metabolizing activities in pig hepatocytes for use in bioartificial liver devices: Comparison with other hepatic cellular models. J Hepatol. 1999;31:542–549. doi: 10.1016/S0168-8278(99)80049-X. [DOI] [PubMed] [Google Scholar]
- Fink-Gremmels J. Implications of hepatic cytochrome P450-related biotransformation processes in veterinary sciences. Eur J Pharmacol. 2008;585:502–509. doi: 10.1016/j.ejphar.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Giantin M, Giantin M, Carletti M, et al. Effect of breed upon cytochrome P450 s and phase II enzymes expression in cattle liver. Drug Metab Dispos. 2008;11:885–893. doi: 10.1124/dmd.107.019042. [DOI] [PubMed] [Google Scholar]
- Girolami F, Spalenza V, Benedetto A, et al. Comparative liver accumulation of dioxin-like compounds in sheep and cattle: possible role of AhR-mediated xenobiotic metabolizing enzymes. Sci Total Environ. 2016;571:1222–1229. doi: 10.1016/j.scitotenv.2016.07.150. [DOI] [PubMed] [Google Scholar]
- Gleich A, Kaiser B, Schumann J, Fuhrmann H. Establishment and characterisation of a novel bovine SV40 large T-antigen-transduced foetal hepatocyte-derived cell line. In Vitro Cell Dev Biol Anim. 2016;52:662–672. doi: 10.1007/s11626-016-0018-0. [DOI] [PubMed] [Google Scholar]
- Grasso E, Longo V, Coceani F, Giovanni Gervasi P. Cytochrome P450 expression and catalytic activity in coronary arteries and liver of cattle. Biochim Biophys Acta. 2005;1722:116–123. doi: 10.1016/j.bbagen.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Guguen-Guillouzo C, Tichonicky L, Szajnert MF, et al. Maintenance of chromatin and cytoplasmic enzymes of perinatal rat hepatocytes during culture. Ann N Y Acad Sci. 1980;349:393–396. doi: 10.1111/j.1749-6632.1980.tb29546.x. [DOI] [PubMed] [Google Scholar]
- Gusson F, Carletti M, Giuliano Albo A, et al. Comparison of hydrolytic and conjugative biotransformation pathways in horse, cattle, pig, broiler chick, rabbit and rat liver subcellullar fractions. Vet Res Commun. 2006;30:271–283. doi: 10.1007/s11259-006-3247-y. [DOI] [PubMed] [Google Scholar]
- Halwachs S. Endogenous expression of liver-specific drug transporters for organic anions in the rat hepatocytoma fusion cell line HPCT-1E3. Eur J Cell Biol. 2005;84:677–686. doi: 10.1016/j.ejcb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Halwachs S, Lakoma C, Gebhardt R, et al. Dioxin mediates downregulation of the reduced folate carrier transport activity via the arylhydrocarbon receptor signalling pathway. Toxicol Appl Pharmacol. 2010;246:100–106. doi: 10.1016/j.taap.2010.04.020. [DOI] [PubMed] [Google Scholar]
- Halwachs S, Wassermann L, Lindner S, et al. Fungicide prochloraz and environmental pollutant dioxin induce the ABCG2 transporter in bovine mammary epithelial cells by the arylhydrocarbon receptor signaling pathway. Toxicol Sci. 2013;131:491–501. doi: 10.1093/toxsci/kfs304. [DOI] [PubMed] [Google Scholar]
- Ioannides C. Cytochrome p450 expression in the liver of food-producing animals. Curr Drug Metab. 2006;7:335–348. doi: 10.2174/138920006776873544. [DOI] [PubMed] [Google Scholar]
- Jani M, Ambrus C, Magnan R, et al. Structure and function of BCRP, a broad specificity transporter of xenobiotics and endobiotics. Arch Toxicol. 2014;88:1205–1248. doi: 10.1007/s00204-014-1224-8. [DOI] [PubMed] [Google Scholar]
- Javitt NB. Hep G2 cells as a resource for metabolic studies: lipoprotein, cholesterol, and bile acids. FASEB J. 1990;4:161–168. doi: 10.1096/fasebj.4.2.2153592. [DOI] [PubMed] [Google Scholar]
- Jonker JW, Schinkel AH. Pharmacological and physiological functions of the polyspecific organic cation transporters: OCT1, 2, and 3 (SLC22A1-3) J Pharmacol Exp Ther. 2004;308:2–9. doi: 10.1124/jpet.103.053298. [DOI] [PubMed] [Google Scholar]
- Kanebratt K, Andersson T. Evaluation of HepaRG cells as an in vitro model for human drug metabolism studies. Drug Metab Dispos. 2008;36:1444–1452. doi: 10.1124/dmd.107.020016. [DOI] [PubMed] [Google Scholar]
- Khaniki GRJ. Chemical contaminants in milk and public health concerns: a review. Int J Dairy Sci. 2007;2:104–115. doi: 10.3923/ijds.2007.104.115. [DOI] [Google Scholar]
- Klocke R, Gómez-Lechón MJ, Ehrhardt A, et al. Establishment and characterization of immortal hepatocytes derived from various transgenic mouse lines. Biochem Biophys Res Commun. 2002;294:864–871. doi: 10.1016/S0006-291X(02)00579-X. [DOI] [PubMed] [Google Scholar]
- Langmann T, Moehle C, Mauerer R, et al. Loss of detoxification in inflammatory bowel disease: dysregulation of pregnane X receptor target genes. Gastroenterology. 2004;127:26–40. doi: 10.1053/j.gastro.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Lazaro CA, Croager EJ, Mitchell C, et al. Establishment, characterization, and long-term maintenance of cultures of human fetal hepatocytes. Hepatology. 2003;38:1095–1106. doi: 10.1053/jhep.2003.50448. [DOI] [PubMed] [Google Scholar]
- LeCluyse E, Madan A, Hamilton G, et al. Expression and regulation of cytochrome P450 enzymes in primary cultures of human hepatocytes. J Biochem Mol Toxicol. 2000;14:177–188. doi: 10.1002/(SICI)1099-0461(2000)14:4<177::AID-JBT1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Lee W, Belkhiri A, Lockhart AC, et al. Overexpression of OATP1B3 confers apoptotic resistance in colon cancer. Cancer Res. 2008;68:10315–10323. doi: 10.1158/0008-5472.CAN-08-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucuta SE, Vlase L. Pharmacokinetics and metabolic drug interactions. Curr Clin Pharmacol. 2006;1:5–20. doi: 10.2174/157488406775268183. [DOI] [PubMed] [Google Scholar]
- Li N, Xia Q, Ruan J, et al. Hepatotoxicity and tumorigenicity induced by metabolic activation of pyrrolizidine alkaloids in herbs. Curr Drug Metab. 2011;12:823–834. doi: 10.2174/138920011797470119. [DOI] [PubMed] [Google Scholar]
- Machala M, Soucek P, Neca J, et al. Inter-species comparisons of hepatic cytochrome P450 enzyme levels in male ruminants. Arch Toxicol. 2003;77:555–560. doi: 10.1007/s00204-003-0477-4. [DOI] [PubMed] [Google Scholar]
- Mahnke H, Ballent M, Baumann S, et al. The ABCG2 efflux transporter in the mammary gland mediates veterinary drug secretion across the blood-milk barrier into milk of dairy cows. Drug Metab Dispos. 2016;44:700–708. doi: 10.1124/dmd.115.068940. [DOI] [PubMed] [Google Scholar]
- McLean IW, Nakane PK. Periodate-lysine-paraformaldehyd fixative—a new fixative for immunoelectron microscopy. J Histochem Cytochem. 1974;22:1077–1083. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- Mealey KL. ABCG2 transporter: therapeutic and physiologic implications in veterinary species. J Vet Pharmacol Ther. 2012;35:105–112. doi: 10.1111/j.1365-2885.2011.01313.x. [DOI] [PubMed] [Google Scholar]
- Mo W, Zhang JT. Human ABCG2: Structure, function, and its role in multidrug resistance. Int J Biochem Mol Biol. 2012;3:1–27. [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Fujii T, Ichihara A. Autocrine mechanism of growth of neonatal rat hepatocytes in primary culture. J Biochem. 1988;103:700–706. doi: 10.1093/oxfordjournals.jbchem.a122332. [DOI] [PubMed] [Google Scholar]
- Pan X, Du W, Yu X, et al. Establishment and characterization of immortalized porcine hepatocytes for the study of hepatocyte xenotransplantation. Transplant Proc. 2010;42:1899–1906. doi: 10.1016/j.transproceed.2009.11.043. [DOI] [PubMed] [Google Scholar]
- Pegolo S, Merlanti R, Giantin M, et al. High performance liquid chromatography determination of cytochrome P450 1A and 2C activities in bovine liver microsomes. Vet J. 2010;183:81–88. doi: 10.1016/j.tvjl.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Plant N. Strategies for using in vitro screens in drug metabolism. Drug Discov Today. 2004;9:328–336. doi: 10.1016/S1359-6446(03)03019-8. [DOI] [PubMed] [Google Scholar]
- Platte H, Honscha W. Functional characterization of the hepatic sodium-dependent taurocholate transporter stably transfected into an immortalized liver-derived cell line and V79 fibroblasts. Eur J Cell Biol. 1996;70:54–60. [PubMed] [Google Scholar]
- Priesner C, Hesse F, Windgassen D, et al. Liver-specific physiology of immortal, functionally differentiated hepatocytes and of deficient hepatocyte-like variants. Vitro Cell Dev Biol Anim. 2004;40:318–330. doi: 10.1290/0404031.1. [DOI] [PubMed] [Google Scholar]
- Reid Y, Gaddipati JP, Yadav D, Kantor J. Establishment of a human neonatal hepatocyte cell line. In Vitro Cell Dev Biol Anim. 2009;45:535–542. doi: 10.1007/s11626-009-9219-0. [DOI] [PubMed] [Google Scholar]
- Rembacz KP, Woudenberg J, Hoekstra M, et al. Unconjugated bile salts shuttle through hepatocyte peroxisomes for taurine conjugation. Hepatology. 2010;52:2167–2176. doi: 10.1002/hep.23954. [DOI] [PubMed] [Google Scholar]
- Rijk JCW, Bovee TFH, Peijnenburg AACM, et al. Bovine liver slices: a multifunctional in vitro model to study the prohormone dehydroepiandrosterone (DHEA) Toxicol In Vitro. 2012;26:1014–1021. doi: 10.1016/j.tiv.2012.04.012. [DOI] [PubMed] [Google Scholar]
- Riss TL, Niles AL, Minor L. Cell viability assays assay guidance manual. Assay Guid Man. 2004 [Google Scholar]
- Robey RW, Polgar O, Deeken J, et al. ABCG2: Determining its relevance in clinical drug resistance. Cancer Metastasis Rev. 2007;26:39–57. doi: 10.1007/s10555-007-9042-6. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Antona C, Donato MT, Boobis A, et al. Cytochrome P450 expression in human hepatocytes and hepatoma cell lines: molecular mechanisms that determine lower expression in cultured cells. Xenobiotica. 2002;32:505–520. doi: 10.1080/00498250210128675. [DOI] [PubMed] [Google Scholar]
- Sack GH. Human cell transformation by simian virus 40—a review. In Vitro. 1981;17:1–19. doi: 10.1007/BF02618025. [DOI] [PubMed] [Google Scholar]
- Scherer M, Gnewuch C, Schmitz G, Liebisch G. Rapid quantification of bile acids and their conjugates in serum by liquid chromatography-tandem mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2009;877:3920–3925. doi: 10.1016/j.jchromb.2009.09.038. [DOI] [PubMed] [Google Scholar]
- Shull LR, Kirsch DG, Lohse CL, Wisniewski JA. Application of isolated hepatocytes to studies of drug metabolism in large food animals. Xenobiotica. 1987;17:345–363. doi: 10.3109/00498258709043944. [DOI] [PubMed] [Google Scholar]
- Sivapathasundaram S, Magnisali P, Coldham NG, et al. A study of the expression of the xenobiotic-metabolising cytochrome P450 proteins and of testosterone metabolism in bovine liver. Biochem Pharmacol. 2001;62:635–645. doi: 10.1016/S0006-2952(01)00710-9. [DOI] [PubMed] [Google Scholar]
- Sivapathasundaram S, Howells LC, Sauer MJ, Ioannides C. Functional integrity of precision-cut liver slices from deer and cattle. J Vet Pharmacol Ther. 2004;27:79–84. doi: 10.1111/j.1365-2885.2004.00555.x. [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, et al. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Swift B, Pfeifer ND, Brouwer KLR. Sandwich-cultured hepatocytes: an in vitro model to evaluate hepatobiliary transporter-based drug interactions and hepatotoxicity. Drug Metab Rev. 2010;42:446–471. doi: 10.3109/03602530903491881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot NC, Wang L, Garrett WM, et al. Establishment and characterization of feeder cell-dependent bovine fetal liver cell lines. In Vitro Cell Dev Biol Anim. 2015 doi: 10.1007/s11626-015-9982-z. [DOI] [PubMed] [Google Scholar]
- Trauner M, Boyer JL. Bile salt transporters : molecular characterization, function, and regulation. Physiol Rev. 2003;83:633–671. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- Wassermann L, Halwachs S, Baumann D, et al. Assessment of ABCG2-mediated transport of xenobiotics across the blood-milk barrier of dairy animals using a new MDCKII in vitro model. Arch Toxicol. 2013;87:1671–1682. doi: 10.1007/s00204-013-1066-9. [DOI] [PubMed] [Google Scholar]
- Woodworth CD, Isom HC. Regulation of albumin gene expression in a series of rat hepatocyte cell lines immortalized by simian virus 40 and maintained in chemically defined medium. Mol Cell Biol. 1987;7:3740–3748. doi: 10.1128/MCB.7.10.3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancanella V, Giantin M, Lopparelli RM, et al. Proposed new nomenclature for Bos taurus cytochromes P450 involved in xenobiotic drug metabolism. J Vet Pharmacol Ther. 2010;33:528–536. doi: 10.1111/j.1365-2885.2010.01173.x. [DOI] [PubMed] [Google Scholar]
- Zhou M, Zhao F, Li J, et al. Long-term maintenance of human fetal hepatocytes and prolonged susceptibility to HBV infection by co-culture with non-parenchymal cells. J Virol Methods. 2014;195:185–193. doi: 10.1016/j.jviromet.2013.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.