Abstract
Myofibroblast differentiation is characterized by increased expression of cytoskeletal smooth muscle α-actin. In human and murine fibroblasts, the gene encoding smooth muscle α-actin (Acta2) is tightly regulated by a network of transcription factors that either activate or repress the 5′ promoter-enhancer in response to environmental cues signaling tissue repair and remodeling. Purine-rich element binding protein B (Purβ) suppresses the expression of Acta2 by cooperatively interacting with the sense strand of a 5′ polypurine sequence containing an inverted MCAT cis-element required for gene activation. In this study, we evaluated the chemical basis of nucleoprotein complex formation between the Purβ repressor and the purine-rich strand of the MCAT element in the mouse Acta2 promoter. Quantitative single-stranded DNA (ssDNA)-binding assays conducted in the presence of increasing concentrations of monovalent salt or anionic detergent suggested that the assembly of a high-affinity nucleoprotein complex is driven by a combination of electrostatic and hydrophobic interactions. Consistent with the results of pH titration analysis, site-directed mutagenesis revealed several basic amino acid residues in the intermolecular (R267) and intramolecular (K82, R159) subdomains that are essential for Purβ transcriptional repressor function in Acta2 promoter-reporter assays. In keeping with their diminished Acta2 repressor activity in fibroblasts, purified Purβ variants containing an R267A mutation exhibited reduced binding affinity for purine-rich ssDNA. Moreover, certain double and triple point mutants were also defective in binding to the Acta2 corepressor protein, Y-box binding protein 1. Collectively, these findings establish the repertoire of non-covalent interactions that account for the unique structural and functional properties of Purβ.
Graphical Abstract
Myofibroblasts arise from the differentiation of connective tissue fibroblasts and play a key role in granulation tissue formation and wound healing by mediating extracellular matrix deposition and wound closure1–4. Although the smooth muscle cell-like contractile properties of myofibroblasts are beneficial for wound repair, chronic activation and survival of extracellular matrix-producing myofibroblasts provokes aberrant tissue remodeling including hypertrophic scar formation, vascular stiffening, and chronic organ fibrosis particularly of the heart, liver, and lung5–7. To develop better and more effective therapeutic strategies for treating fibrotic diseases, it is necessary to identify and characterize the molecular pathways that govern myofibroblast proliferation and differentiation.
Smooth muscle α-actin (SMαA) is the most widely accepted protein biomarker of fibroblast to myofibroblast conversion and acquisition of a contractile phenotype2, 8–10. Consistent with its phenotype-defining properties, expression of the murine SMαA gene (Acta2) in fibroblasts is tightly regulated by a network of transcriptional activators and repressors11, 12. The 5′-flanking region of Acta2 contains a variety of positive cis-regulatory elements including an inverted muscle-CAT (MCAT) motif, CArG boxes, CAGA elements, and GC-rich sequences that serve as cognate binding sites for certain basal or growth factor-responsive trans-activators, namely transcription enhancer factor 1 (TEF-1), serum response factor (SRF), small mothers against decapentaplegic (Smad) proteins, and specificity proteins 1 and 3 (Sp1/3), respectively13–17. Conversely, repression of Acta2 transcription in fibroblasts is apparently mediated by the coordinate interaction of purine-rich element binding proteins A and B (Purα and Purβ) and Y-box binding protein (YB-1) with the opposing strands of an asymmetric polypurine-polypyrimidine (Pur/Pyr) tract containing the MCAT motif14, 18–20. Although Purα and Purβ both interact with purine-rich single-stranded nucleic acids and possess the capacity to destabilize duplex DNA21–23, Purβ appears to function as a more potent repressor of Acta2 transcription in both fibroblasts and smooth muscle cells24–26.
Purβ is a member of a small but highly conserved family of purine-rich ssDNA/RNA-binding proteins27. Each member of the Pur family (Purα, Purβ, Purγ variant A, Purγ variant B) contains three central sequence elements designated PUR repeats I, II, and III27–29. These elements account for the roughly 70% sequence homology between mammalian Purα and Purβ22. The recently solved x-ray crystal structures of Drosophila melanogaster (Dm) Purα (amino acids 40 to 185) and Borrelia burgdorferi (Bb) Purα (amino acids 8 to 105) revealed that PUR repeat sequences fold to form a 4-stranded anti-parallel β-sheet followed by an α-helix. In the case of Dm Purα, intramolecular association of PUR repeats I and II produces a Whirly-like globular domain (dubbed the PUR domain) capable of ssDNA interaction28. In the case of Bb Purα, intermolecular dimerization of the single encoded PUR repeat forms a ssDNA-binding PUR domain, which is nearly identical to the domain generated by intramolecular association of PUR repeats I and II of Dm Purα29. Because full-length Pur proteins in metazoans contain three PUR repeats, PUR repeat III likely serves to mediate intermolecular interaction of Pur protein monomers to form dimers as first reported in Dm Purα28.
Our efforts to establish the molecular basis for ssDNA-binding and Acta2 repression by Mus musculus (Mm) Purβ have provided additional insight into the similarities and differences in the structural and functional properties of members of the Pur protein family. For example, hydrodynamic analyses revealed that recombinant mouse Purβ reversibly self-associates to form a homodimer in the absence of ssDNA30. Rigorous quantitative analyses of nucleoprotein complex formation between Purβ and the 32 nt sense strand of the MCAT-containing Pur/Pyr element from Acta2 were consistent with a cooperative, multisite binding mechanism leading to formation of a high-affinity 2:1 Purβ:ssDNA complex31. Results of limited tryptic digestion suggested that the presence of all three PUR repeats is required for high affinity binding of Purβ to the purine-rich strand of the Acta2-derived MCAT enhancer element32. To further define the structural elements in Purβ responsible for multisite binding to ssDNA and consequent Acta2 repression in fibroblasts, we recently reported that dimeric Purβ is comprised of three distinct PUR subdomains each capable of interacting with a separate ssDNA binding site33. Consistent with the cooperative nature of Purβ-ssDNA interaction, all three PUR subdomains are required for high affinity interaction of the Purβ homodimer with the Acta2 promoter. Interestingly, when studied in isolation, the central dimerization subdomain of Purβ formed by intermolecular association of two PUR repeat III sequences exhibited markedly higher ssDNA-binding affinity than the comparable subdomain formed by intramolecular association of PUR repeats I and II33.
In this report, the detergent- and salt-sensitivity of the nucleoprotein complex formed between Purβ and its Acta2-derived purine-rich target sequence was initially evaluated to estimate the contribution of hydrophobic and ionic interactions in facilitating the high affinity binding of Purβ to ssDNA. Based on homology modeling of the tertiary and quaternary structure of Purβ in comparison to Purα, several basic amino acid residues in PUR repeats I, II, and III were identified and empirically validated as essential for the Acta2 repressor activity of Purβ in fibroblasts. Biochemical analyses implicated these residues in mediating the high affinity binding of Purβ to purine-rich ssDNA and to the corepressor protein mouse YB-1 (MSY1). Our findings suggest that a combination of hydrophobic and electrostatic interactions are responsible for the unique ssDNA-binding and transcriptional regulatory properties of Purβ.
EXPERIMENTAL PROCEDURES
Computational modeling of Purβ.
Hydrophobic side chains were identified in a Mus musculus Purβ homology model using PyMOL to depict regions of Purβ with high hydrophobic amino acid content33, 34. Additionally, electrostatic surface maps were generated to identify solvent-exposed, charged amino acids. To select positively charged amino acid residues of Purβ that might confer Acta2 repression, a ClustalW multiple pairwise sequence alignment of Mm Purβ and Dm Purα was performed to identify Purβ residues corresponding to the Purα residues (R80, R158, and R229) that had been previously implicated in single-stranded nucleic acid interaction35–37. The lack of conservation of Purα R158 led us to align the crystal structure of Dm Purα (amino acids 40–185) and the homology model of Mm Purβ I-II to identify a positionally conserved arginine in spatial proximity to Dm Purα R158.
Construction of Expression Vectors.
The original mammalian expression plasmid encoding recombinant N-terminally hexahistidine-tagged mouse Purβ (pCI-NHis-Purβ) was described in a previous study14, 20. The QuikChange® Primer Design Program (Stratagene) was used to design complementary oligonucleotide primers (Table S1) for site-directed mutagenesis of the pCI-NHis-Purβ template. Multiple plasmids encoding different combinations of single, double, and triple point mutations of selected lysine or arginine residues in the Purb open reading frame were sequentially produced using the QuikChange® XL site-directed mutagenesis kit as directed by the manufacturer (Agilent). Plasmids encoding Purβ mutants K82A, R159A, R267A, K82A/R159A, K82A/R267A, R159A/R267A, and K82A/R159A/R267A were sequence validated (Vermont Cancer Center DNA Analysis Facility) and purified by double cesium chloride gradient centrifugation. Quantification of isolated plasmids was achieved by measuring the optical density at 260 nm. Plasmid purity was assessed via analytical agarose gel electrophoresis following double restriction enzyme digestion with BamHI and KpnI to validate the size of the cDNA inserts. The cDNAs encoding selected single, double, and triple Purβ mutants (R267A, R159A/R267A, and K82A/R159A/R267A) were excised from pCI with EcoRI and SalI and then subcloned into the bacterial expression plasmid pQE30, which had been treated with the same restriction enzymes. The resulting bacterial expression plasmids were propagated in E. coli JM109 cells and screened for cDNA insertion and sequence fidelity as described above.
Protein Purification.
Wild-type NHis-Purβ and selected deletion and point mutants were expressed in E. coli JM109 cells by induction with isopropyl β-D-1-thiogalactopyranoside (IPTG) as previously described30. The recombinant NHis-tagged proteins expressed included full-length Purβ (Purβ FL), the core ssDNA-binding region (Purβ I-II-III, residues 41–303), the intermolecular subdomain (Purβ III, residues 209–303), the R267A single point mutant, the R159A/R267A double point mutant, and the K82A/R159A/R267A triple point mutant. NHis-Purβ proteins were purified from E. coli cell lysates by a combination of metal chelate affinity chromatography, heparin-agarose ion exchange chromatography, and size exclusion chromatography (SEC)33. Additional details about the purification steps used for each point mutant are provided in Supporting Information. Column fractions were monitored for protein content either by optical density measurement at 280 nm (A280) or by Bradford assay (Thermo Scientific) using bovine serum albumin (BSA) as a standard. The relative size and purity of isolated Purβ proteins were assessed by denaturing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 10% or 12% mini-gels. Resolved proteins were visualized by staining with Coomassie® Brilliant Blue R-250 on gels standardized with the PageRuler™ Broad Range Protein Ladder (Thermo Scientific). The protein concentration of purified Purβ preparations was determined by A280 measurement using theoretical molar extinction coefficients for full-length or truncated Purβ33. Purified protein preparations were assayed to ensure the absence of nuclease contamination30, 33.
Protein Thermostability Assay.
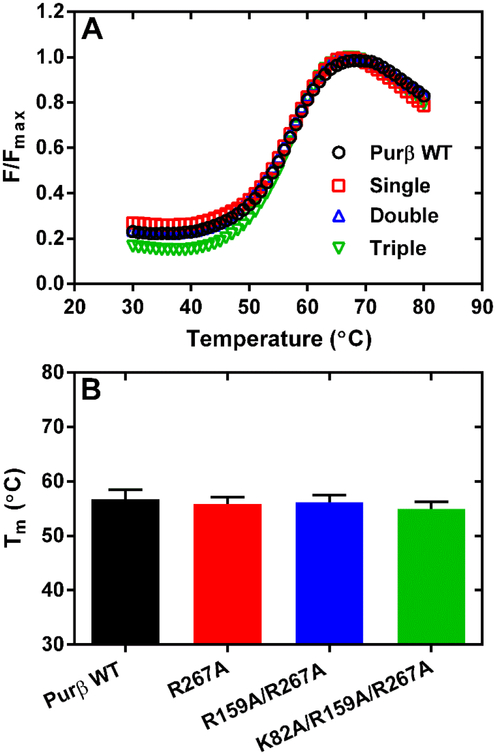
The relative thermostability of wild-type and mutated Purβ proteins was evaluated by thermal shift assay using SYPRO® Orange (Life Technologies) to monitor protein unfolding as a function of temperature38, 39. Assays were conducted at protein concentrations ranging from 0.3–8.5 μM in SEC/storage buffer consisting of 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole pH 8.0 with 10 mM β-mercaptoethanol (BME). In some cases, assays were conducted in buffer composed of 20 mM HEPES, 150 mM NaCl, 10 mM BME adjusted to selected pH values. A 4 μL solution of a 100× stock of SYPRO® Orange pre-diluted in protein storage buffer was combined with solutions of wild-type or mutated Purβ proteins to achieve a final volume of 80 μL. The mixture was then dispensed in 40 μL aliquots into the wells of a 96 well PCR microplate (Axygen Scientific), which was covered with optical quality sealing tape (Applied Biosystems). Fluorescence data points were collected at 1°C increments at temperatures ranging from 25° to 94°C on an AB 7500 Fast Sequence Detection System real-time PCR instrument (Applied Biosystems) with excitation and emission wavelengths set at 480 nm and 568 nm, respectively (Vermont Cancer Center DNA Analysis Facility). The average background fluorescence detected in the buffer only control was subtracted from each incremental fluorescence value obtained in wells containing protein. The data points were then normalized by dividing the background-corrected fluorescence measured at each temperature by the maximum fluorescence (Fmax) obtained in that well. To calculate the temperature midpoint of the protein unfolding transition curve (Tm), data points obtained in the range of 40°C-70°C were fit to the Boltzmann equation using Prism 6 (GraphPad Software, Inc.). Data points acquired below 40°C and after the fluorescence maximum were excluded from the curve fitting analysis due to the well-to-well variability in baseline protein fluorescence seen at low temperature and excessive protein aggregation occurring at high temperature.
Single-Stranded DNA-Binding Assay.
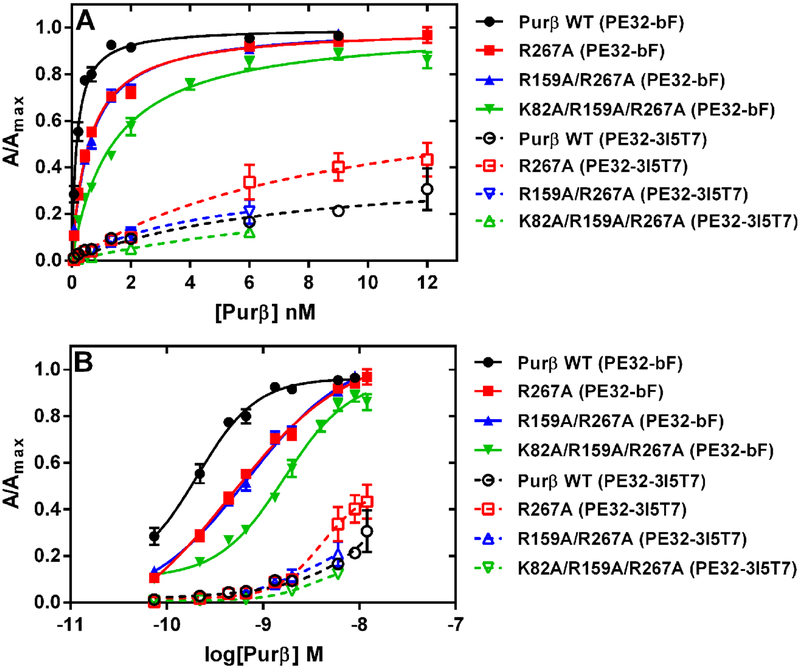
The interaction of purified, recombinant Purβ proteins with a synthetic 3′ biotinylated ssDNA probe corresponding to the purine-rich sense strand of the Acta2 MCAT element (5′-GGGAGCAGAACAGAGGAATGCAGTGGAAGAGA-3′, nt −195 to −164, dubbed PE32-bF) was monitored by enzyme-linked immunorsorbent assay (ELISA) as described previously32, 40. To assess the effects of either detergent or salt on nucleoprotein complex formation, the DNA binding step was carried out by incubating 1.0 nM full-length, wild-type Purβ, 1.0 nM Purβ I-II-III, or 5.0 nM Purβ III with 0.5 nM PE32-bF immobilized on StreptaWells™ (Roche) in binding buffer containing varying concentrations of sodium deoxycholate or NaCl (Sigma). In these experiments, the DNA binding buffer consisted of 20 mM HEPES, 150 mM NaCl, 1.5 mM MgCl2-6H2O pH 7.5 plus 0.2% (w/v) BSA, 0.05% (v/v) Tween 20, 0.5 mM dithiothreitol (DTT), and 100 nM dT32 oligonucleotide. To assess the effects of point mutations on nucleoprotein complex formation, varying concentrations of wild-type Purβ or mutants R267A, R159A/R267A, or K82A/R159A/R267A were incubated with 0.5 nM PE32-bF pre-bound to StreptaWells™ (Roche). After overnight incubation (typically 16 h) at room temperature, wells were washed and solid-phase Purβ:PE32-bF complexes were immunodetected by sequential 1 h incubations with 1.0 μg/ml of an affinity-purified primary rabbit polyclonal antibody directed against mouse Purβ amino acids 210–22920 followed by a 1:8000 dilution of a secondary goat anti-rabbit IgG conjugated to horseradish peroxidase (Santa Cruz Biotechnology). Antibody bound nucleoprotein complexes were detected by the addition of colorimetric substrate solution 2, 2′-AZINO-bis [3-ethylbenziazoline-6-sulfonic acid] (ABTS) (Millipore). After quenching color development with 1% (w/v) sodium dodecyl sulfate, endpoint absorbance measurements were obtained at 405 nm (A405) using a Vmax plate reader (Molecular Devices).
For detergent and salt inhibition assays, data points were fit by nonlinear least squares regression analysis to a four-parameter variable-slope equation to obtain an IC50 value using Prism 6 (Graphpad Software, Inc.). For assessment of the relative ssDNA-binding affinity of wild-type vs. mutant Purβ proteins, A405 readings were corrected for background by subtracting the A405 signal generated in wells without DNA at each concentration of protein tested. Background corrected A405 values were fit by nonlinear least squares regression analysis to the equation for a rectangular hyperbola to estimate the Bmax (i.e. Amax) for each protein using Prism 6 (GraphPad Software, Inc.). The calculated Amax was used to normalize absorbance values at each concentration of protein tested (A/Amax) for the purpose of evaluating the reproducibility of the apparent Kd or EC50 determined in separate trials. In some experiments, the coating concentration of biotinylated ssDNA probe was varied and the amount of fluid-phase Purβ was fixed. To assess ssDNA-binding specificity, titration assays were performed with the mutant probe in which the PUR elements were substituted with thymidylate (PE32-bF-3I5T7)31.
Protein-Protein Interaction Assay.
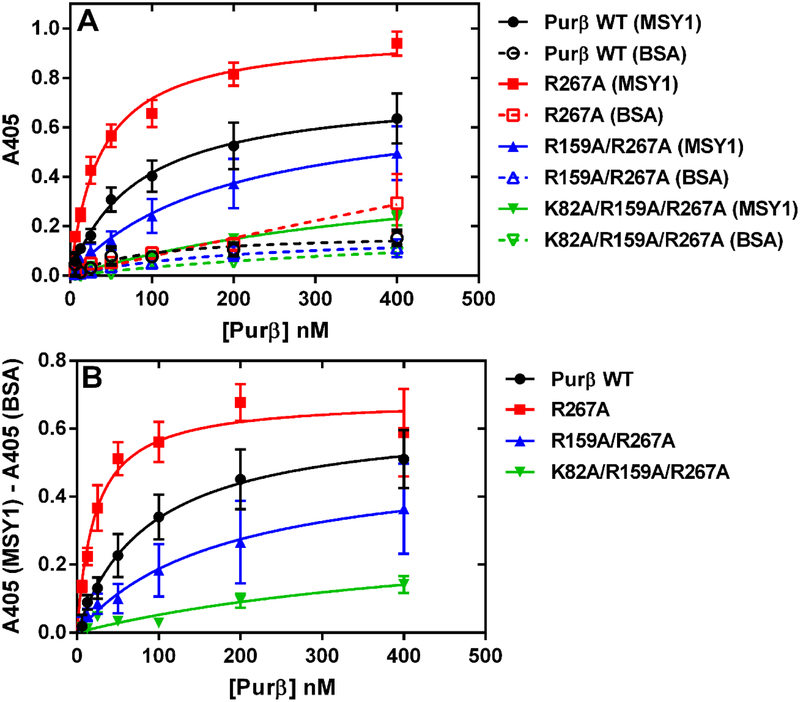
The binding of purified Purβ proteins to purified MSY1 was evaluated by ELISA as previously described40. Briefly, NHis-tagged MSY120 was diluted to 100 nM in coating buffer consisting of 20 mM HEPES, 150 mM NaCl, 1.5 mM MgCl2-6H2O pH 7.5 with 5.0 μg/ml BSA and then applied to microtiter wells (Costar® EIA/RIA plate, 96 Well Easy Wash™, Certified High Binding, Corning, Inc.). After a 3 h incubation at room temperature, the wells were washed, blocked, and solutions of either wild-type or mutant Purβ protein were applied in binding buffer consisting of 20 mM HEPES, 150 mM NaCl, 1.5 mM MgCl2-6H2O pH 7.5 plus 0.2% (w/v) BSA, 0.05% (v/v) Tween 20, and 0.5 mM DTT. After overnight incubation (≥ 16 h) at 4°C, wells were washed and solid-phase Purβ:MSY1 complexes were detected in the same manner as described for the ssDNA-binding ELISA using a primary rabbit antibody recognizing amino acids 210–229 or 302–324 of mouse Purβ20. To estimate the relative affinity of wild-type vs. mutant Purβ proteins for MSY1, A405 readings were corrected for background by subtracting the A405 signal generated in wells coated with BSA only from the MSY1-coated wells at each concentration of Purβ protein tested. Background corrected A405 values were fit by nonlinear least squares regression analysis to the equation for a rectangular hyperbola to estimate the apparent Kd using Prism 6 (GraphPad Software, Inc.).
Transient Transfection Assay.
AKR-2B mouse embryo fibroblasts (MEFs) were seeded at 4.0 × 104 in McCoy’s 5A medium supplemented with 5% heat-inactivated FBS in 6-well plates. Cells were incubated at 37°C in an incubator with 5% CO2 for 24 h to facilitate adherence to the plate. MEFs were transiently transfected with a total of 2 μg/mL of DNA consisting of 0.1 μg/mL of pSV40-βGal, 1 μg/mL of expression plasmid encoding single, double, or triple point mutations, and 0.9 μg/mL of Acta2 promoter-reporter construct, pVSMP8-luciferase33. After 48 h incubation at 37°C, transfected cells were washed with PBS and harvested by lysis in 1× Passive Lysis Buffer (Promega) supplemented with protease inhibitors leupeptin, pepstatin A, and aprotinin at 1.0 μg/mL. Cleared lysates were assayed for total protein content by either Bradford or BCA™ assay (Thermo Scientific) and reporter gene expression by luciferase activity assay (Promega). Datasets were analyzed by performing a one-way analysis of variance and Tukey’s multiple comparison test with significance of p <0.05 using Prism 6 (Graphpad Software, Inc.).
Immunoblotting.
Total soluble protein from cell lysates was precipitated by addition of 5 volumes of cold ethanol and incubation at −20°C for 1 h. Precipitated protein was collected by centrifugation and dissolved in 1× SDS-PAGE loading buffer (20 mM Tris-Cl, pH 6.8, 0.5% w/v SDS, 5% v/v glycerol, 0.005% w/v bromophenol blue, 5% v/v BME), heated for 3–5 minutes at 100°C, and run on a 12% w/v acrylamide/bisacrylamide (29:1) mini-gel with molecular weight standards (BenchMark™ Prestained Protein Ladder, Invitrogen). Protein (10 μg or 20 μg) was transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore) at 125 V for 90 min at 4°C in 25 mM Trizma base, 192 mM glycine, 20% v/v methanol. His-tagged Purβ proteins were detected with a mouse anti-RGS-His monoclonal antibody (Qiagen) at 1.0 μg/ml followed by a goat anti-mouse IgG-HRP antibody. Bands were visualized via chemiluminescent detection (Pierce® ECL Western Blotting Substrate, Thermo Scientific). Blots were reprobed with a mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (clone 6C5, Millipore).
RESULTS
Mapping of polar and nonpolar regions in Mm Purβ.
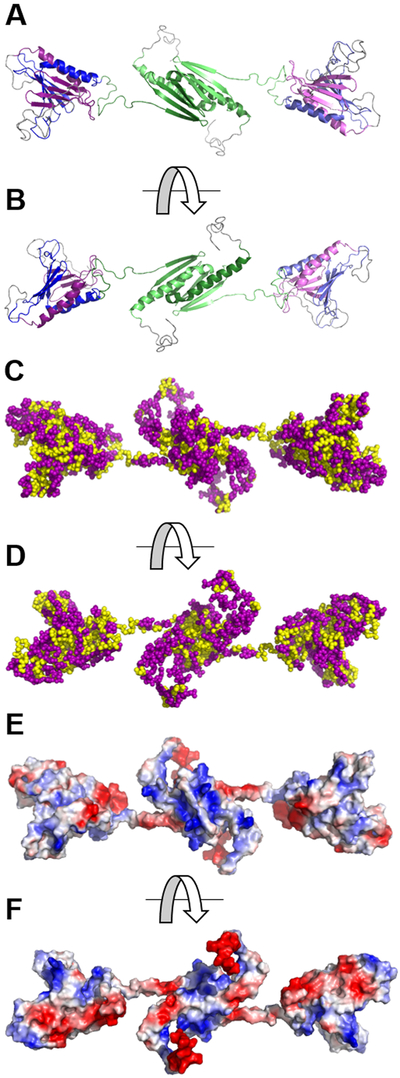
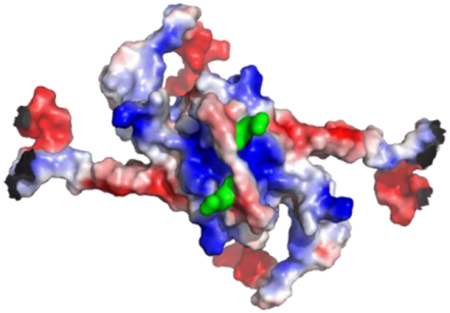
The chemical properties of amino acid residues in the homology model of the homodimeric form of Mm Purβ33 were displayed using hydrophobic and electrostatic maps generated in PyMOL34. As expected, hydrophobic amino acid residues are predominantly located at the interior of both the intramolecular (PUR repeat I-II) and intermolecular subdomains (PUR repeat III) (Figure 1C, D). Hydrophobic side chains in the ββββα structure formed by each PUR repeat protrude away from the surface and in toward the interface of two PUR repeats. This arrangement suggests that a hydrophobic core is located at the center of each PUR subdomain presumably to facilitate protein folding and dimerization.
Figure 1.
Hydrophobic and electrostatic surface maps of the Mm Purβ homodimer generated by computational homology modeling. (A, B) Ribbon model of the Mm Purβ dimer highlighting sequences corresponding to PUR repeats I (violet), II (blue), and III (green). Two intramolecular subdomains are formed by association of PUR repeats I and II. The central dimerization subdomain is formed by intermolecular interaction of PUR repeat III sequences from each monomer. (C, D) Hydrophobic maps of the Mm Purβ dimer show the non-polar core of each subdomain. Yellow spheres represent hydrophobic amino acids while purple spheres represent non-hydrophobic residues. (E, F) Electrostatic surface maps of the Mm Purβ dimer show regions of charged amino acids. Blue areas indicate positively charged residues and red represents negatively charged residues. Images in B, D, and F are rotated 180° around the horizontal axis with respect to the corresponding images in A, C, and E.
An electrostatic map of presumably solvent-exposed residues in the Mm Purβ homodimer model shows regions of localized negative charge around the α-helices of each PUR subdomain and pockets of positively charged residues on the β-sheet surface (Figure 1E, F). Interestingly, the PUR I-II intramolecular domain exhibits one positively charged pocket on the surface of the β-sheets while evidence of two positively charged channels exist at the surface of the β-sheets in the PUR III intermolecular dimerization domain (Figure 1E). These results are consistent with surface charge maps of the Dm Purα I-II crystal structure which exhibited moderately negative charges on the α-helix surface side and positive charges around the β-sheets28, 29. The positively charged channels localized around the β-sheets of the PUR subdomains point to potential binding surfaces for negatively charged nucleic acid.
Effect of detergent or salt on Purβ binding to the Acta2 MCAT element.
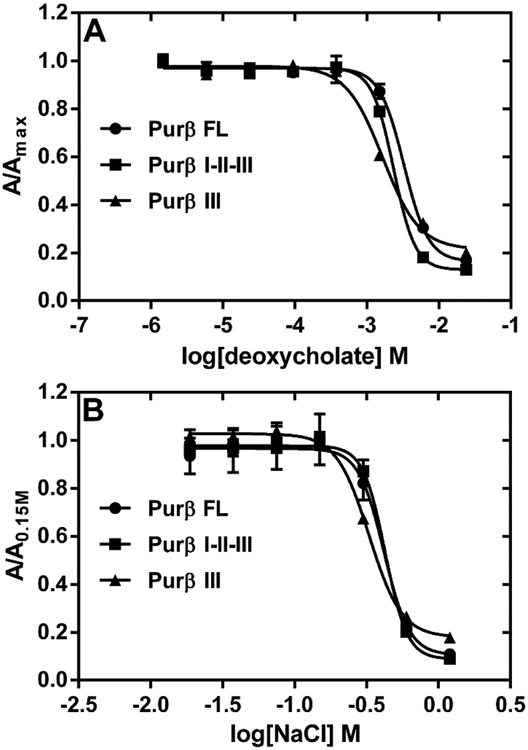
To investigate whether hydrophobic interactions play a role in Purβ nucleoprotein complex formation, ssDNA-binding ELISAs were conducted in binding buffer supplemented with increasing concentrations of selected detergents. In this assay system, a 3’ biotinylated single-stranded oligonucleotide (PE32-bF) corresponding to the purine-rich sense strand of the MCAT enhancer of Acta2 was used as the ssDNA probe. This oligonucleotide has previously been shown to form high-affinity 2:1 Purβ:PE32-F nucleoprotein complex with a macroscopic Kd of approximately 0.3 nM31. Thus, the protein-ssDNA binding step of the assay was conducted with a limiting concentration of PE32-bF (0.5 nM) and a 2–5 fold molar excess of purified recombinant Purβ proteins. Predictably, the non-ionic and non-denaturing detergent, Triton X-100, had no effect on the binding of full-length Purβ to ssDNA, while the anionic and denaturing detergent, sodium dodecyl sulfate, was a much more potent inhibitor than the anionic bile salt detergent sodium deoxycholate (Figure S1). The amphiphilic properties of deoxycholate allow this detergent to interact with hydrophobic surfaces of proteins in a non-cooperative and non-denaturing manner41. Consequently, deoxycholate is useful in disruption of protein-protein interaction42, 43. As shown in Figure 2A, very low concentrations of deoxycholate (< 0.0156% or 0.38 mM) had little to no effect on Purβ binding to purine-rich ssDNA. However, at deoxycholate concentrations exceeding 1 mM, protein binding to PE32-bF was markedly inhibited in the case of full-length Purβ (IC50 = 0.098%, 2.36 mM), the core Purβ I-II-III construct (IC50 = 0.084%, 2.03 mM), and the isolated Purβ III intermolecular subdomain (IC50 = 0.060%, 1.45 mM). The concentration-dependent inhibitory effect of deoxycholate on Purβ-ssDNA interaction is presumably due to detergent-mediated disruption of hydrophobic contacts that dictate the structural stability and/or ssDNA-binding activity of the intermolecular subdomain of the protein. Consistent with this interpretation, thermal shift assay revealed that the isolated Purβ III dimerization domain becomes progressively less thermostable at sodium deoxycholate concentrations between 0.38 and 2.0 mM (Figure S2).
Figure 2.
Effect of anionic detergent and salt on the interaction of Purβ with Acta2 ssDNA. (A) The binding of full-length Purβ (Purβ FL), its core region (Purβ I-II-III), and its intermolecular subdomain (Purβ III) to 0.5 nM PE32-bF ssDNA was assessed in assay buffer containing varying concentrations of sodium deoxycholate. Solid-phase Purβ-PE32-bF complexes were detected by ELISA using rabbit anti-mouse Purβ 210–229 as the primary antibody. (B) The same ELISA format was used to evaluate the effect of varying concentrations of salt on the binding of Purβ FL, Purβ I-II-III, and Purβ III to ssDNA. Data points show absorbance values at 405 nm normalized to the maximum absorbance observed at the lowest concentration of deoxycholate tested (A) or normalized to the absorbance obtained in buffer with 0.15 mM NaCl (B) (mean ± SD, n = 4).
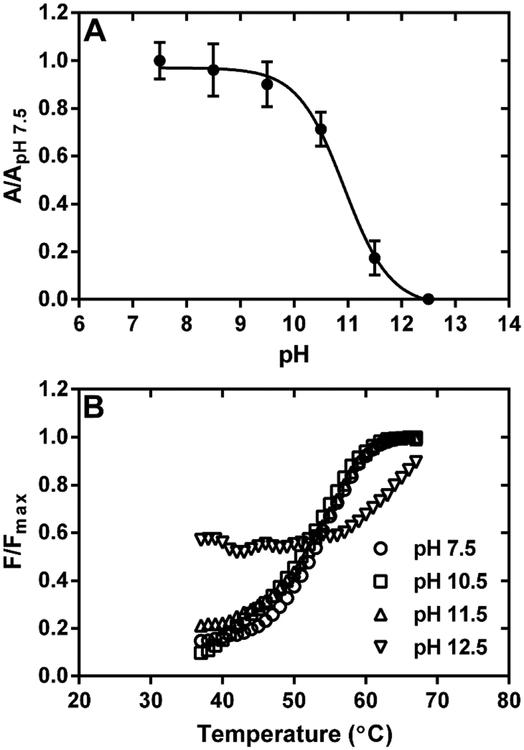
To evaluate the involvement of electrostatic forces in Purβ-ssDNA interaction, the effects of increasing NaCl concentration on the binding of full-length Purβ, the core I-II-III construct, and the isolated Purβ III subdomain to PE32-bF was monitored by ELISA (Figure 2B). Results indicated that salt concentrations greater than ~0.6 M abolished the binding of each Purβ construct to PE32-bF. Given the similarity of the competition curves, these data suggest that each Purβ subdomain possesses charged amino acid residues that enable stable nucleoprotein complex formation between Purβ and ssDNA. In support of this contention, assessment of the effect of pH on the interaction of Purβ with PE32-bF indicated that nucleoprotein complex formation was markedly reduced at solution pH ≥ 10.5 (Fig. 3A). This outcome was not attributable to protein unfolding as thermal shift assay revealed that Purβ retains it native structure at pH 10.5 and 11.5 (Fig. 3B). The thermal shift profiles of full-length Purβ at pH 7.5, 10.5, and 11.5 were virtually identical (Tm = 53.2 ± 0.92 °C) while some degree of structural instability was apparent at pH 12.5. Interestingly, lowering the pH had no substantive effect on the binding of full-length Purβ to PE32-bF, while raising the pH had an analogous inhibitory effect on ssDNA-binding by the core I-II-III construct and the isolated Purβ III subdomain (Fig. S3). Collectively, these results are consistent with the deprotonation of the side chains of solvent-exposed lysine, arginine, and possibly tyrosine residues leading to reduced electrostatic interaction of Purβ with ssDNA.
Figure 3.
Effect of solution pH on the interaction of Purβ with Acta2 ssDNA. (A) The binding of full-length Purβ (1.0 nM) to PE32-bF ssDNA (0.5 nM) was assessed in assay buffer without MgCl2 at pH ranging from 7.5 to 12.5. Solid-phase Purβ-PE32-bF complexes were detected by ELISA using rabbit anti-mouse Purβ 210–229 as the primary antibody. Nonspecific background absorbance at 405 nm in control wells without any DNA was subtracted from the signal generated in PE32-bF-coated wells. Background corrected A405 values measured at each pH were normalized by dividing by the mean A405 value determined at pH 7.5 (mean ± SD, n = 4). (B) Effect of solution pH on the thermostability of Purβ. The unfolding of full-length Purβ was evaulated by thermal shift assay at a protein concentration of 2.8 μM in 20 mM HEPES, 150 mM NaCl, 10 mM β-mercaptoethanol adjusted to pH 7.5, 10.5, 11.5, or 12.5.
Identification of K/R residues in PUR repeats I, II, and III that are essential for Purβ repressor activity.
Based on the reported ability of certain basic amino acid residues to mediate RNA-binding by Dm Purα37, three basic residues located in β-strands of each PUR repeat of Mm Purβ were selected for site-directed mutagenesis. Alignment of human and mouse Purα and Purβ sequences revealed that Dm Purα residues R80 and R229 are positionally conserved in Purα and Purβ orthologs and correspond to residues K82 and R267 located in the fourth β-strand of the PUR I and PUR III repeats in mouse Purβ (Figure S4). The absence of a conserved basic residue in the PUR II repeat led us to align the PUR repeat I-II subdomain of the Mm Purβ homology model with the Dm Purα I-II structure28, 33. This maneuver uncovered a unique R159 residue located in the 3rd β-strand of Purβ repeat II in spatial proximity to Purα R158. Mapping the three residues onto the homology model of the Purβ homodimer shows that each residue is predicted to be surface exposed in the intramolecular (K82, R159) or intermolecular (R267) subdomains (Figure S5).
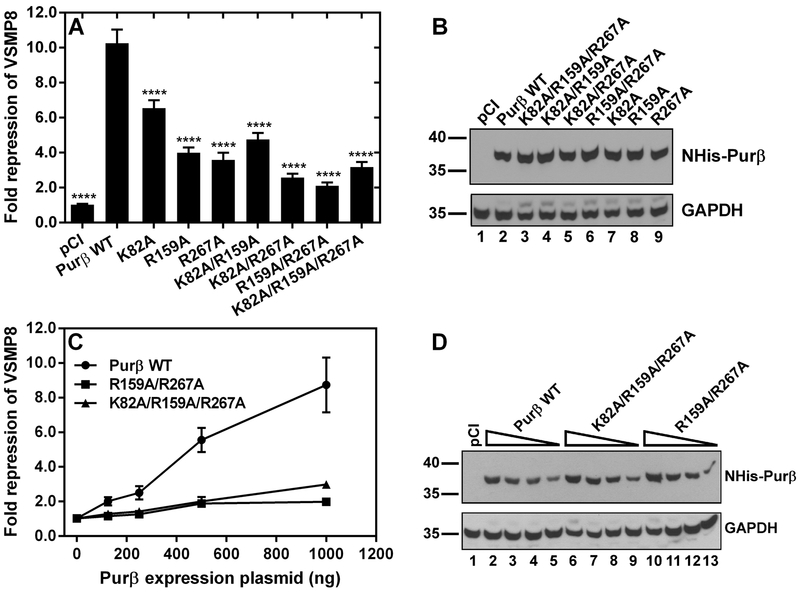
To evaluate the consequence of K82A, R159A, and R267A point mutations on the Acta2 repressor activity of Purβ, AKR-2B MEFs were co-transfected with Purβ expression vectors and a full-length Acta2 promoter-reporter construct (VSMP8-luciferase)33. All single point mutants tested demonstrated reduced repressor activity toward the VSMP8 promoter-reporter in MEFs with R267A being the most influential mutation (Figure 4A). The effects of double and triple point mutations were more pronounced than the single point mutations. Immunoblotting of the cell lysates confirmed that all the Purβ point mutants were stably expressed in MEFs (Figure 4B). Interestingly, all Purβ constructs containing the PUR repeat III R267A mutation exhibited greatly diminished Acta2 repressor activity suggesting that the R267 residue plays a critical role in the physical and functional interaction of Purβ with the Acta2 gene. The K82A/R159A/R267A triple mutant and R159A/R267A double mutant were much weaker repressors than the single R267A mutant alone. The results of transient co-transfection assays comparing the Acta2 repressor activity of Purβ K82A/R159A/R267A and R159A/R267A expressed at increasing concentrations in MEFs confirmed the functional impairment of these mutants relative to wild-type Purβ (Figure 4C, D).
Figure 4.
Evaluation of the Acta2 repressor activity of Purβ point mutants expressed in fibroblasts. (A) Subconfluent AKR-2B MEFs were transiently co-transfected with mammalian expression plasmids encoding either wild-type (WT) Purβ or the indicated point mutants and an Acta2 promoter-luciferase reporter plasmid, VSMP8. After 48 h, transfected MEFs were harvested and whole cell extracts were assayed for both luciferase enzyme activity and total protein concentration. Relative luciferase reporter expression measured in MEFs co-transfected with an empty pCI vector control was defined as 1. Bars show the fold repression of the VSMP8 reporter by each Purβ construct (mean ± SEM, n = 6–15). ****, p < 0.0001 compared to Purβ WT. (B) Immunoblot analysis was performed with a His tag antibody to confirm the expression of single, double, and triple NHis-Purβ point mutants in transfected cells. The anti-His tag blot was reprobed with a GAPDH antibody as a loading control. (C) A titration assay was performed with plasmids encoding the indicated double and triple Purβ point mutants in comparison to the wild-type protein. Symbols show the relative VSMP8 repressor activity of each Purβ construct (mean± SEM, n = 3). (D) Immunoblot analysis was performed to confirm the dose-dependent expression of NHis-Purβ in transfected cells. The anti-His tag blot was reprobed with a GAPDH antibody as a loading control. (B, D) Numbers on the left indicate molecular mass in kilodaltons.
Purification and structural characterization of Purβ K/R mutants.
Results of cell-based promoter-reporter assays suggested that certain basic residues in each Purβ subdomain are likely involved in mediating the functional properties of Purβ. To assess the effect of these mutations on protein structure, mouse Purβ mutants R267A, R159A/R267A, and K82A/R159A/R267A were expressed in and purified from E. coli. Protein preparations obtained after metal chelate and heparin affinity chromatography were analyzed by calibrated SEC. As evidenced by the SEC elution profiles of each protein, the single, double, and triple mutants were each capable of forming homodimers when resolved at protein loading concentrations in the micromolar range (Figure S6). Peaks corresponding to homodimer were collected and the purity and structural integrity of each of the point mutants was evaluated by a combination of SDS-PAGE and thermal shift assay. As shown in Figure 5A, the thermal shift profiles of the Purβ mutants are comparable to the wild-type protein when evaluated under relatively high ionic strength conditions. The calculated midpoint temperatures for the protein unfolding transition were nearly identical when assayed across a range of protein concentrations suggesting that the mutant Purβ proteins are properly folded (Figure 5B). Results of SDS-PAGE revealed that the isolated mutants were of sufficient purity to merit further functional analysis (Figure S7).
Figure 5.
Assessment of the relative thermostability of purified Purβ point mutants. (A) The unfolding of wild-type (WT) Purβ in comparison to point mutants R267A (single), R159A/R267A (double), and K82A/R159A/R267A (triple) was evaulated by thermal shift assay at a protein concentration of 2.8 μM in 50 mM sodium phosphate pH 8.0, 300 mM NaCl, 10 mM imidazole, 10 mM β-mercaptoethanol. (B) Bars show the calculated Tm of each protein determined at multiple protein concentrations ranging from 1.0 to 8.5 μM (mean ± SD, n = 6–10).
Effect of K/R mutations on Purβ binding to ssDNA.
To determine the effect of R267A, R159A/R267A, and K82A/R159A/R267A mutations on Purβ function, the single, double, and triple mutants were quantitatively evaluated for Acta2 ssDNA-binding affinity and specificity. All purified Purβ preparations were checked to ensure that any proteins tested in the ssDNA-binding assay were free of nuclease contamination (Figure S8). Nucleoprotein complex formation between Purβ and the Acta2 probe, PE32-bF, immobilized on streptavidin-coated microtiter wells was measured by ELISA. The primary Purβ antibody used to detect complex formation was initially screened to ensure that the residue 210–229 epitope located between the PUR II and PUR III repeats was recognized equivalently in the wild-type and mutated Purβ proteins (Figure S9). To evaluate the effects of the K/R mutations on ssDNA-binding affinity and specificity, each mutant protein was tested for interaction with PE32-bF and a mutated probe in which each PUR element (3′ end, internal, 5′ end) was substituted with thymidylate (PE32–3I5T7). As shown in Figure 6A and B, the single R267A mutation in PUR repeat III reduced the apparent ssDNA-binding affinity of the protein by ~3-fold (Kd = 0.57 ± 0.04 nM) relative to wild-type Purβ (Kd = 0.17 ± 0.01). The addition of the R159A mutation in PUR repeat II did not result in any further diminishment in ssDNA-binding affinity in the R159A/R267A double mutant (Kd = 0.60 ± 0.04). Importantly, addition of the K82A mutation in PUR repeat I reduced the ssDNA-binding affinity of the triple mutant by ~8-fold (Kd = 1.39 ± 0.12 nM) (Figure 6A and B). Interestingly, the K/R mutations did not affect ssDNA-binding specificity per se as each Purβ mutant tested demonstrated preferential interaction with the wild-type, purine-rich Acta2 probe but only weak binding to the T7 mutated probe (Figure 6). Collectively, these data indicate that specific basic residues in both the intra- and intermolecular subdomains account for the high affinity binding of Purβ to purine-rich ssDNA.
Figure 6.
Effect of R/K point mutations on the interaction of Purβ with Acta2 ssDNA. (A and B) The binding of wild-type (WT) Purβ and the indicated single (R267A), double (R159A/R267A), or triple (K82A/R159A/R267A) point mutants to 0.5 nM PE32-bF ssDNA (filled symbols) or 0.5 nM mutated PE32–3I5T7 ssDNA (open symbols) was evaluated by ELISA. Solid-phase Purβ-ssDNA complexes were detected using rabbit anti-mouse Purβ 210–229 as the primary antibody. (A) Protein concentration ranges were chosen to achieve saturable binding to the PE32-bF probe. Binding isotherms were generated by fitting data points obtained from multiple, independent titration experiments (n = 3–5) to the equation for a rectangular hyperbola. (B) Replot of the same datasets fit to a log(agonist) vs. response (four parameters) equation. The apparent Kd (A) or EC50 (B) of each Purβ protein tested for the PE32-bF probe was extrapolated accordingly.
Effect of K/R mutations on Purβ binding to MSY1.
To evaluate the potential effect of these mutations on heterotypic interaction between Purβ and its Acta2 corepressor partner MSY1, the single, double, and triple mutants were tested for their ability to bind to purified MSY1 by ELISA. As shown in Figure 7, the R267A mutant actually exhibited a somewhat higher binding affinity for MSY1 compared to wild-type Purβ, while the double and triple point mutants demonstrated reduced MSY1 binding affinity consistent with their diminished Acta2 repressor activity. The use of a different Purβ antibody recognizing a C-terminal epitope to detect Purβ-MSY1 complex formation gave essentially identical results (Figure S10) and further validated the distinction in MSY1 binding affinity among the Purβ mutants tested.
Figure 7.
Effect of R/K point mutations on the interaction of Purβ with MSY1. (A) The binding of purified Purβ wild-type (WT) protein or the indicated single (R267A), double (R159A/R267A), or triple (K82A/R159A/R267A) point mutants to immobilized MSY1 (filled symbols, solid lines) or BSA (open symbols, dashed lines) was evaluated by ELISA. Purβ-MSY1 complexes were detected using rabbit anti-Purβ 210–229 as the primary antibody. Binding isotherms were generated by fitting data points obtained from several independent titration experiments (n = 3) to the equation for a rectangular hyperbola. (B) Binding curves generated after subtracting out the nonspecific absorbance measured in BSA only-coated wells from the absorbance obtained in MSY1-coated wells.
DISCUSSION
During normal wound repair, inflammatory cytokine-stimulated trans-differentiation of resident stromal fibroblasts leads to the transient formation of contractile myofibroblasts expressing SMαA3. However, persistent myofibroblast activation resulting from sustained inflammatory signaling can promote excess connective tissue deposition resulting in aberrant tissue remodeling associated with many fibrotic diseases44, 45. A better understanding of the intracellular factors that regulate Acta2 transcription in myofibroblasts is necessary to identify relevant molecular targets for the purpose of developing novel therapies to reverse myofibroblast differentiation and to limit pathological tissue fibrosis. Purβ may represent one such target as this protein functions as a transcriptional repressor of Acta2 by virtue of its ability to interact with the purine-rich strand of a MCAT-containing element in the 5′ flanking region of the gene. Purβ likely operates in conjunction with other ssDNA-binding corepressors, namely MSY1, to disrupt the combinatorial assembly of canonical double-stranded DNA-binding trans-activators on their respective MCAT, CArG, or G/C-rich elements in Acta2 promoter in fibroblasts14, 20, 26, 33.
Previous homology modeling of the tertiary and quaternary structure of Purβ in conjunction with empirical analysis of the physical and functional properties of the intermolecular and intramolecular subdomains indicated the presence of three discrete ssDNA-binding modules in the Purβ homodimer33. In this report, in silico analysis of the distribution of non-polar and polar amino acid residues in each subdomain revealed an asymmetric arrangement with hydrophobic residues predominating in the interior and charged residues on the solvent-exposed surface of each subdomain. While this is not necessarily surprising based on principles of protein folding, there is a clear distinction in the extent of clustering of positively charged residues in the intramolecular subdomain composed of PUR repeats I and II in comparison to the intermolecular dimerization subdomain composed of two PUR III repeats. Notably, the two basic channels evident in the intermolecular dimerization domain may explain why this subdomain demonstrates a higher ssDNA-binding affinity than the isolated intramolecular subdomain33.
Because these computational modeling studies implied the importance of both hydrophobic and electrostatic forces in mediating protein self-association and DNA-binding, we tested this concept by evaluating the effects of anionic detergent, monovalent salt, and pH on the binding of purified Purβ to ssDNA. To ensure conceptual relevance to Acta2 gene regulation, the assembly of nucleoprotein complexes composed of Purβ and the purine-rich strand of the Acta2 promoter element (PE32-bF) containing a functional MCAT motif was monitored by quantitative ELISA. Our results indicated that at concentrations in excess of 1.0 mM, sodium deoxycholate inhibited the binding of full-length Purβ and truncated Purβ proteins containing the PUR repeat III dimerization domain to the PE32-bF probe. The anionic detergent-sensitivity of each of the recombinant Purβ constructs analyzed is consistent with previous findings in which the inhibitory effect of deoxycholate on the interaction of native fibroblast-derived Purα and Purβ with ssDNA was documented by band shift assay in comparison to Triton X-10020. It should be noted that all protein-DNA binding assays presented in this report were conducted in the presence of 0.05% (v/v) Tween 20, another non-ionic detergent which has no apparent effect on Purβ-ssDNA interaction. Consequently, the ability of deoxycholate to hinder Purβ-ssDNA interaction is potentially due to interference with PUR repeat III dimerization and/or obstruction of hydrophobic contacts between Purβ and nucleobases in the ssDNA. Although physical destabilization of the PUR repeat I-II subdomain by deoxycholate cannot be formally excluded, our results do corroborate existing structural data implicating hydrophobic interactions as the principal driver of both intramolecular and intermolecular association of the homologous PUR repeats in Purα29.
With respect to the salt and pH sensitivity of Purβ interaction with ssDNA, protein binding to ssDNA was blocked at concentrations of NaCl exceeding 0.6 M in the case of full-length Purβ, Purβ I-II-III, and the Purβ III intermolecular domain. Purβ-ssDNA complexes were relatively stable across a wide range of solution pH conditions. However, in accordance with a specific role for basic, positively charged residues in mediating nucleoprotein complex formation, assembly of Purβ-ssDNA complexes was abolished at pH ≥ 10.5 where deprotonation of the ε amino and guanidino groups of lysine and arginine would be predicted to occur46. Collectively, our findings suggest that a combination of hydrophobic and electrostatic interactions contribute to the formation and stability of the Purβ-PE32-bF complex. In particular, the hydrophobic core of each subdomain presumably accounts for the structural stability of each ssDNA-binding module while basic residues located on the solvent-exposed surface serve to mediate subdomain contact with nucleic acid. This interpretation is entirely consistent with the subdomain architecture of the Purβ dimer and the cooperative mechanism of nucleoprotein complex assembly31, 33. Furthermore, the recently solved x-ray crystal structure of the Dm Purα I-II subdomain in complex with CGG trinucleotide-repeats substantiates the importance of direct electrostatic and hydrophobic contacts between specific amino acid residues and nucleobases in mediating Purα binding to ssDNA47.
Inspired by the results of the pH and salt sensitivity assays, we investigated the role of several basic residues that are positionally conserved in PUR repeats I (K82), II (R159) and III (R267) of mouse and human Pur proteins and for which data exists indicating their involvement in mediating nucleic acid interaction by Dm Purα28, 37. To evaluate the effect of PUR repeat-specific K82A, R159A, and R267A point mutations on Purβ function, either single, double, or triple mutants were expressed in undifferentiated MEFs and assayed for their ability to repress an Acta2 promoter-luciferase reporter gene. The results showed that Purβ constructs containing the R267A mutation were the most demonstrably defective in Acta2 repressor activity. Of the mutants tested, Purβ R159A/R267A, K82/R159/R267, and K82A/R267A demonstrated the most profound deficiency in Acta2 repressor activity. These findings suggest that residue R267 in PUR repeat III plays a critical role in mediating Purβ repressor activity but that residues K82 and R159 in PUR repeats I and II contribute to Acta2 repression as well. This interpretation was further validated by the finding that the deficiency in Acta2 repressor activity of the R159A/R267A and K82A/R159A/R267A mutants was evident over a broad range of expressed protein concentrations.
Consistent with the stable expression of the Purβ point mutants in fibroblasts, structural analyses performed on the purified R267A, R159A/R267A, and K82/R159A/R267A point mutants indicated that each mutant protein was properly folded and capable of self-association. However, further biochemical analyses of the single, double, and triple point mutants in comparison to wild-type Purβ revealed a 3.5–8.5 fold reduction in binding affinity of the point mutants for the purine-rich strand of the Acta2 MCAT element. Loss of electrostatic contacts between basic, solvent-exposed residues in the protein and potential hydrogen bond acceptors or negatively-charged phosphate groups in the DNA presumably account for this finding. Although we cannot formally exclude the possibility of altered subcellular trafficking of the expressed Purβ point mutants, such a defect in ssDNA-binding function would translate into a deficiency in Acta2 repressor activity. Interestingly, these particular mutations did not impact ssDNA-binding specificity per se as all the mutants demonstrated preferential interaction with purine-rich ssDNA in comparison to a ssDNA probe containing thymidylate substitutions in each of three putative binding sites31. It is important to note that the analogous mutations in the 4th β-strand of PUR repeats I, II, and III of Dm Purα have been reported to abolish RNA-binding without having any effect on Purα protein dimerization37. Moreover, the R80 residue in PUR repeat I of Dm Purα, which is positionally-conserved with the K82 residue in Purβ, has been reported to participate in direct hydrogen bond interaction with guanine in GCGGCGG ssDNA47.
Although the results of ssDNA-binding assays were consistent with changes in ssDNA-binding affinity as the biophysical basis for the impaired Acta2 repressor function of each Purβ mutant, the results of protein-protein interaction assays suggested another possible reason for the diminished repressor activity particularly of the Purβ double and triple point mutants. As previously reported, MSY1 binds to the pyrimidine-rich strand of the Acta2 MCAT element14, 20. Consequently, a deficiency in the ability of Purβ to interact with MSY1 may impair the ability of these corepressor proteins to assemble an inhibitory nucleoprotein complex capable of altering DNA structure and preventing cis-element recognition by transcriptional activators. The fact that the Purβ K82A/R159A/R267A mutant exhibited the most dramatic reduction in MSY1 binding affinity is consistent with the original study suggesting that the entire core ssDNA-binding region of Purα is required to mediate robust interaction of human Purα with human YB-148. In the case of Purβ, we have found that the isolated intermolecular and intramolecular subdomains of the protein interact much more weakly with fibroblast-derived MSY1 than does full-length Purβ or a truncation protein containing all three PUR repeats33. The observation that the single R267A point mutant has apparently enhanced affinity while the R159A/R267A double and K82/R159A/R267A triple mutants have reduced affinity for MSY1 points to a differential role for each subdomain in the modulating the interaction of the assembled Purβ homodimer with MSY1. Consequently, the preponderance of the evidence presented in this report suggests that the loss of Acta2 repressor function, particularly in the triple point mutant, is likely due to decreased interaction of Purβ with both ssDNA and corepressor binding partners.
Supplementary Material
Acknowledgments
Funding: This work was supported by a grant awarded to R.J.K. from the American Heart Association Founders Affiliate (09GRNT2170060). A.E.R. and L.A.F. were supported by an institutional training grant from the National Heart, Lung, and Blood Institute (T32 HL007594). T.R.W. was supported by an Undergraduate Student Summer Fellowship from the American Heart Association Founders Affiliate and by the Cardiovascular Research Institute of Vermont.
Abbreviations:
- Purβ
purine-rich element binding protein B
- SMαA
smooth muscle α-actin
- ssDNA
single-stranded DNA
- MCAT
muscle-CAT box
- Sp(1/3)
specificity protein 1 and 3
- Purα
purine-rich element binding protein A
- Dm
Drosophila melanogaster
- Mm
Mus Musculus
- YB-1
Y-box binding protein 1
- MEF
mouse embryo fibroblast
- Pur/Pyr
polypurine/polypyrimidine
- TGF-β1
transforming growth factor β1
- nt
nucleotide
- SEC
size exclusion chromatography
- BSA
bovine serum albumin
- SDS
sodium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
- BME
β-mercaptoethanol
- DTT
dithiothreitol
- MSY1
mouse YB-1
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
SUPPORTING INFORMATION AVAILABLE
Figures S1 to S10, Table S1, and an expanded methods section. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- [1].Gabbiani G, Hirschel BJ, Ryan GB, Statkov PR, and Majno G (1972) Granulation tissue as a contractile organ. A study of structure and function, J Exp Med 135, 719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gabbiani G, Ryan GB, and Majne G (1971) Presence of modified fibroblasts in granulation tissue and their possible role in wound contraction, Experientia 27, 549–550. [DOI] [PubMed] [Google Scholar]
- [3].Hinz B (2007) Formation and function of the myofibroblast during tissue repair, J Invest Dermatol 127, 526–537. [DOI] [PubMed] [Google Scholar]
- [4].Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, and Brown RA (2002) Myofibroblasts and mechano-regulation of connective tissue remodelling, Nat Rev Mol Cell Biol 3, 349–363. [DOI] [PubMed] [Google Scholar]
- [5].Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, and Gabbiani G (2007) The myofibroblast: one function, multiple origins, Am J Pathol 170, 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zalewski A, Shi Y, and Johnson AG (2002) Diverse origin of intimal cells: smooth muscle cells, myofibroblasts, fibroblasts, and beyond?, Circ Res 91, 652–655. [DOI] [PubMed] [Google Scholar]
- [7].Hao H, Gabbiani G, Camenzind E, Bacchetta M, Virmani R, and Bochaton-Piallat ML (2006) Phenotypic modulation of intima and media smooth muscle cells in fatal cases of coronary artery lesion, Arterioscler Thromb Vasc Biol 26, 326–332. [DOI] [PubMed] [Google Scholar]
- [8].Darby I, Skalli O, and Gabbiani G (1990) Alpha-smooth muscle actin is transiently expressed by myofibroblasts during experimental wound healing, Lab Invest 63, 21–29. [PubMed] [Google Scholar]
- [9].Hinz B, Celetta G, Tomasek JJ, Gabbiani G, and Chaponnier C (2001) Alpha-smooth muscle actin expression upregulates fibroblast contractile activity, Mol Biol Cell 12, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang J, Zohar R, and McCulloch CA (2006) Multiple roles of alpha-smooth muscle actin in mechanotransduction, Exp Cell Res 312, 205–214. [DOI] [PubMed] [Google Scholar]
- [11].Foster DN, Min B, Foster LK, Stoflet ES, Sun S, Getz MJ, and Strauch AR (1992) Positive and negative cis-acting regulatory elements mediate expression of the mouse vascular smooth muscle alpha-actin gene, J Biol Chem 267, 11995–12003. [PubMed] [Google Scholar]
- [12].Stoflet ES, Schmidt LJ, Elder PK, Korf GM, Foster DN, Strauch AR, and Getz MJ (1992) Activation of a muscle-specific actin gene promoter in serum-stimulated fibroblasts, Mol Biol Cell 3, 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gan Q, Yoshida T, Li J, and Owens GK (2007) Smooth muscle cells and myofibroblasts use distinct transcriptional mechanisms for smooth muscle alpha-actin expression, Circ Res 101, 883–892. [DOI] [PubMed] [Google Scholar]
- [14].Carlini LE, Getz MJ, Strauch AR, and Kelm RJ Jr. (2002) Cryptic MCAT enhancer regulation in fibroblasts and smooth muscle cells. Suppression of TEF-1 mediated activation by the single-stranded DNA-binding proteins, Pur alpha, Pur beta, and MSY1, J Biol Chem 277, 8682–8692. [DOI] [PubMed] [Google Scholar]
- [15].Cogan JG, Subramanian SV, Polikandriotis JA, Kelm RJ Jr., and Strauch AR (2002) Vascular smooth muscle alpha-actin gene transcription during myofibroblast differentiation requires Sp1/3 protein binding proximal to the MCAT enhancer, J Biol Chem 277, 36433–36442. [DOI] [PubMed] [Google Scholar]
- [16].Subramanian SV, Polikandriotis JA, Kelm RJ Jr., David JJ, Orosz CG, and Strauch AR (2004) Induction of vascular smooth muscle alpha-actin gene transcription in transforming growth factor beta1-activated myofibroblasts mediated by dynamic interplay between the Pur repressor proteins and Sp1/Smad coactivators, Mol Biol Cell 15, 4532–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sandbo N, Kregel S, Taurin S, Bhorade S, and Dulin NO (2009) Critical role of serum response factor in pulmonary myofibroblast differentiation induced by TGF-beta, Am J Respir Cell Mol Biol 41, 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sun S, Stoflet ES, Cogan JG, Strauch AR, and Getz MJ (1995) Negative regulation of the vascular smooth muscle alpha-actin gene in fibroblasts and myoblasts: disruption of enhancer function by sequence-specific single-stranded-DNA-binding proteins, Mol Cell Biol 15, 2429–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cogan JG, Sun S, Stoflet ES, Schmidt LJ, Getz MJ, and Strauch AR (1995) Plasticity of vascular smooth muscle alpha-actin gene transcription. Characterization of multiple, single-, and double-strand specific DNA-binding proteins in myoblasts and fibroblasts, J Biol Chem 270, 11310–11321. [DOI] [PubMed] [Google Scholar]
- [20].Kelm RJ Jr., Cogan JG, Elder PK, Strauch AR, and Getz MJ (1999) Molecular interactions between single-stranded DNA-binding proteins associated with an essential MCAT element in the mouse smooth muscle alpha-actin promoter, J Biol Chem 274, 14238–14245. [DOI] [PubMed] [Google Scholar]
- [21].Darbinian N, Gallia GL, and Khalili K (2001) Helix-destabilizing properties of the human single-stranded DNA- and RNA-binding protein Puralpha, J Cell Biochem 80, 589–595. [PubMed] [Google Scholar]
- [22].Kelm RJ Jr., Elder PK, Strauch AR, and Getz MJ (1997) Sequence of cDNAs encoding components of vascular actin single-stranded DNA-binding factor 2 establish identity to Puralpha and Purbeta, J Biol Chem 272, 26727–26733. [DOI] [PubMed] [Google Scholar]
- [23].Wortman MJ, Johnson EM, and Bergemann AD (2005) Mechanism of DNA binding and localized strand separation by Pur alpha and comparison with Pur family member, Pur beta, Biochim et Biophys Acta 1743, 64–78. [DOI] [PubMed] [Google Scholar]
- [24].Kelm RJ Jr., Wang SX, Polikandriotis JA, and Strauch AR (2003) Structure/function analysis of mouse Purbeta, a single-stranded DNA-binding repressor of vascular smooth muscle alpha-actin gene transcription, J Biol Chem 278, 38749–38757. [DOI] [PubMed] [Google Scholar]
- [25].Knapp AM, Ramsey JE, Wang SX, Godburn KE, Strauch AR, and Kelm RJ Jr. (2006) Nucleoprotein interactions governing cell type-dependent repression of the mouse smooth muscle alpha-actin promoter by single-stranded DNA-binding proteins Pur alpha and Pur beta, J Biol Chem 281, 7907–7918. [DOI] [PubMed] [Google Scholar]
- [26].Hariharan S, Kelm RJ Jr., and Strauch AR (2014) The Puralpha/Purbeta single-strand DNA-binding proteins attenuate smooth-muscle actin gene transactivation in myofibroblasts, J Cell Physiol 229, 1256–1271. [DOI] [PubMed] [Google Scholar]
- [27].Johnson EM, Daniel DC, and Gordon J (2013) The pur protein family: genetic and structural features in development and disease, J Cell Physiol 228, 930–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Graebsch A, Roche S, and Niessing D (2009) X-ray structure of Pur-alpha reveals a Whirly-like fold and an unusual nucleic-acid binding surface, Proc Natl Acad Sci U S A 106, 18521–18526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Graebsch A, Roche S, Kostrewa D, Soding J, and Niessing D (2010) Of bits and bugs--on the use of bioinformatics and a bacterial crystal structure to solve a eukaryotic repeat-protein structure, PLoS One 5, e13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ramsey JE, Daugherty MA, and Kelm RJ Jr. (2007) Hydrodynamic studies on the quaternary structure of recombinant mouse Purbeta, J Biol Chem 282, 1552–1560. [DOI] [PubMed] [Google Scholar]
- [31].Ramsey JE, and Kelm RJ Jr. (2009) Mechanism of strand-specific smooth muscle alpha-actin enhancer interaction by purine-rich element binding protein B (Purbeta), Biochemistry 48, 6348–6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rumora AE, Steere AN, Ramsey JE, Knapp AM, Ballif BA, and Kelm RJ Jr. (2010) Isolation and characterization of the core single-stranded DNA-binding domain of purine-rich element binding protein B (Purbeta), Biochem Biophys Res Commun 400, 340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rumora AE, Wang SX, Ferris LA, Everse SJ, and Kelm RJ Jr. (2013) Structural basis of multisite single-stranded DNA recognition and ACTA2 repression by purine-rich element binding protein B (Purbeta), Biochemistry 52, 4439–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schrodinger L (2010) The PyMOL Molecular Graphics System, Version 1.3r1.
- [35].Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, and Lopez R (2010) A new bioinformatics analysis tools framework at EMBL-EBI, Nucleic Acids Res 38, W695–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, and Higgins DG (2007) Clustal W and Clustal X version 2.0, Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- [37].Aumiller V, Graebsch A, Kremmer E, Niessing D, and Forstemann K (2012) Drosophila Pur-alpha binds to trinucleotide-repeat containing cellular RNAs and translocates to the early oocyte, RNA Biol 9, 633–643. [DOI] [PubMed] [Google Scholar]
- [38].Ericsson UB, Hallberg BM, Detitta GT, Dekker N, and Nordlund P (2006) Thermofluor-based high-throughput stability optimization of proteins for structural studies, Anal Biochem 357, 289–298. [DOI] [PubMed] [Google Scholar]
- [39].Niesen FH, Berglund H, and Vedadi M (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability, Nat Protoc 2, 2212–2221. [DOI] [PubMed] [Google Scholar]
- [40].Knapp AM, Ramsey JE, Wang SX, Strauch AR, and Kelm RJ Jr. (2007) Structure-function analysis of mouse Pur beta II. Conformation altering mutations disrupt single-stranded DNA and protein interactions crucial to smooth muscle alpha-actin gene repression, J Biol Chem 282, 35899–35909. [DOI] [PubMed] [Google Scholar]
- [41].Makino S, Reynolds JA, and Tanford C (1973) The binding of deoxycholate and Triton X-100 to proteins, J Biol Chem 248, 4926–4932. [PubMed] [Google Scholar]
- [42].Kupfer SR, Marschke KB, Wilson EM, and French FS (1993) Receptor accessory factor enhances specific DNA binding of androgen and glucocorticoid receptors, J Biol Chem 268, 17519–17527. [PubMed] [Google Scholar]
- [43].Meitinger C, Strobl LJ, Marschall G, Bornkamm GW, and Zimber-Strobl U (1994) Crucial sequences within the Epstein-Barr virus TP1 promoter for EBNA2-mediated transactivation and interaction of EBNA2 with its responsive element, J Virol 68, 7497–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wynn TA (2008) Cellular and molecular mechanisms of fibrosis, J Pathol 214, 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ghosh AK, Quaggin SE, and Vaughan DE (2013) Molecular basis of organ fibrosis: potential therapeutic approaches, Exp Biol Med (Maywood) 238, 461–481. [DOI] [PubMed] [Google Scholar]
- [46].Pace CN, Grimsley GR, and Scholtz JM (2009) Protein ionizable groups: pK values and their contribution to protein stability and solubility, J Biol Chem 284, 13285–13289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Weber J, Bao H, Hartlmuller C, Wang Z, Windhager A, Janowski R, Madl T, Jin P, and Niessing D (2016) Structural basis of nucleic-acid recognition and double-strand unwinding by the essential neuronal protein Pur-alpha, eLife 5, e11297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Safak M, Gallia GL, and Khalili K (1999) Reciprocal interaction between two cellular proteins, Puralpha and YB-1, modulates transcriptional activity of JCVCY in glial cells, Mol Cell Biol 19, 2712–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.