Abstract
Plants have evolved tightly regulated signaling networks to respond and adapt to environmental perturbations, but the nature of the signaling hub(s) involved have remained an enigma. We have previously established that methylerythritol cyclodiphosphate (MEcPP), a precursor of plastidial isoprenoids and a stress-specific retrograde signaling metabolite, enables cellular readjustments for high-order adaptive functions. Here, we specifically show that MEcPP promotes two Brassicaceae-specific traits, namely endoplasmic reticulum (ER) body formation and induction of indole glucosinolate (IGs) metabolism selectively, via transcriptional regulation of key regulators NAI1 for ER body formation and MYB51/122 for IGs biosynthesis). The specificity of MEcPP is further confirmed by the lack of induction of wound-inducible ER body genes as well as IGs by other altered methylerythritol phosphate pathway enzymes. Genetic analyses revealed MEcPP-mediated COI1-dependent induction of these traits. Moreover, MEcPP signaling integrates the biosynthesis and hydrolysis of IGs through induction of nitrile-specifier protein1 and reduction of the suppressor, ESM1, and production of simple nitriles as the bioactive end product. The findings position the plastidial metabolite, MEcPP, as the initiation hub, transducing signals to adjust the activity of hardwired gene circuitry to expand phytochemical diversity and alter the associated subcellular structure required for functionality of the secondary metabolites, thereby tailoring plant stress responses.
Keywords: glucosinolates, ER body, retrograde signaling, MEP pathway, MEcPP, stress
INTRODUCTION
To survive frequent environmental challenges, plants have evolved delicately balanced regulatory mechanisms to reprogram the metabolic output and modify the cellular and subcellular structures to ultimately mount appropriate physiological responses. A key goal of metabolic reprograming is the biosynthesis of secondary metabolites, a remarkable repository of structurally diverse compounds known for their roles in shaping the phylogenetic specificity and tailoring plant responses to the nature of the challenges encountered (Weng et al., 2012). Among the secondary metabolites with proven adaptive roles are the glucosinolates (GSLs), which are nitrogen- and sulfur-containing phytochemicals found mainly in the order Capparales. In Arabidopsis thaliana, GSLs are produced predominantly from tryptophan (indole glucosinolates [IGs]) and chain-elongated methionine (short- or long-chained aliphatic glucosinolates [AGs]) (Sonderby et al., 2010b). The accumulation of GSLs is regulated in response to a range of stresses, including herbivory or wounding and treatment with jasmonates (Kliebenstein et al., 2002; Petersen et al., 2002; Brown et al., 2003; Mikkelsen et al., 2003; Mewis et al., 2006; Kim and Jander, 2007; Bidart-Bouzat and Kliebenstein, 2008). The GSL profile displays both quantitative and qualitative variation depending not only on genetic variation between Arabidopsis accessions but also in response to various developmental and environmental cues (Kerwin et al., 2015). This leads to each tissue having differing GSL patterns such as high concentrations of AGs in seeds, flowers, and siliques, and the IGs being predominantly in the vegetative parts of plants, rosette leaves, and roots (Kliebenstein et al., 2001; Brown et al., 2003). Moreover, in addition to natural selection favoring a given GSLs profile, there is a highly regulated and robust co-expression of the respective genes within plants, as demonstrated by grouped induction of genes in the IGs and the tryptophan biosynthetic pathways in response to stress stimuli (Gachon et al., 2005), and by omics-based identification of transcription factors regulating AGs biosynthesis (Hirai et al., 2007).
Biosynthesis of GSLs occurs in three steps: (1) amino acid chain elongation, (2) core structure synthesis, and (3) secondary modifications, with the biosynthetic genes being regulated by at least two transcription factor families, R2R3-MYB and basic helix-loop-helix (bHLH) MYC (Gachon et al., 2005; Grubb and Abel, 2006; Gigolashvili et al., 2007; Sonderby et al., 2010b; Frerigmann and Gigolashvili, 2014; Li et al., 2014). Transcription factors MYB28, MYb29, and MYB76 predominantly regulate AG synthesis (Hirai et al., 2007; Gigolashvili et al., 2008; Sonderby et al., 2010a), whereas MYB34, MYB51, and MYB122 largely control IG synthesis (Gigolashvili et al., 2007; Frerigmann and Gigolashvili, 2014; Frerigmann et al., 2016). MYC transcription factors mainly MYC2, MYC3, and MYC4 interact with both sets of MYBs to regulate the biosynthesis of both aliphatic and indolic GSLs (Dombrecht et al., 2007; Schweizer et al., 2013). The complexity of the regulatory network controlling GSL is further revealed through post-translational modification by mitogen-activated protein kinase3 (MPK3) and MPK6 via their substrate ethylene response factor 6 (ERF6) on regulation of MYB51 and MYB122 (Xu et al., 2016).
The hydrophilic GSLs are spatially compartmentalized apart from their specific β-glucosidase hydrolyzing enzymes, myrosinases (Wittstock and Burow, 2010). Cellular disruption caused by insect chewing or wounding enables mixing of the intact GSLs with the activating myrosinase, thereby facilitating defense-induced activation of GSLs. Subsequently, myrosinases hydrolyze the thioglucosidic bond to produce unstable aglycones (Bones and Rossiter, 2006). Rapid conversion of aglycones to various bioactive compounds such as isothiocyanates, thiocyanates, and nitriles by specifier proteins and associated modifier proteins, provide direct and indirect defense against a range of invaders, including insects, nematodes, and microorganisms (Lambrix et al., 2001; Burow et al., 2007, 2008, 2009; Pfalz et al., 2009; Fan et al., 2011; Bednarek, 2012; Fan and Doerner, 2012).
Classical myrosinases are expressed in scattered cells termed myrosin cells, allowing cellular level compartmentalization of enzymes from GSLs, which are stored in S cells (Koroleva et al., 2000; Andreasson et al., 2001; Husebye et al., 2002; Ueda et al., 2006; Shroff et al., 2008; Kissen et al., 2009). However, some non-canonical myrosinases are separated from IGs not at the cellular level but at the subcellular level in structures known as ER bodies, derived from and continuous with the ER (Yamada et al., 2009; Nakano et al., 2014, 2017). The ER bodies in Arabidopsis thaliana are induced de novo by wounding or jasmonic acid (JA) treatment, suggestive of their potential function in defense responses such as resistance against pathogens or herbivores (Yamada et al., 2009; Nakano et al., 2014, 2017). In roots, ER bodies are constitutively present and contain large amounts of the putative non-canonical myrosinase proteins, mainly BGLU23/PYK10 (Nakano et al., 2017). In addition to housing β-glucosidase/myrosinases, the root ER bodies also contain two other family proteins, jacalin-related lectin (JAL) and GDSL lipase-like/myrosinase associated protein (GLL) (Nagano et al., 2005, 2008; Ahn et al., 2010). In fact, four of the JALs are nitrile-specifier proteins (NSP1–4) that control production of the end bioactive metabolites, nitriles and isothiocyanates (Lambrix et al., 2001; Burow et al., 2008, 2009; Nagano et al., 2008; Agee et al., 2010). Similarly, several of the GLLs are modifier proteins (ESM1/AGG1 family members include GLL22, 23, and 25) functioning as putative myrosinase chaperones and controlling nitrile and isothiocyanate production (Zhang et al., 2006; Burow et al., 2008). Thus, the composition of the ER bodies may control both the rate of GSL activation and the ultimate bioactive end product, thereby providing a structural platform with key roles in plant defense responses against biotic stresses (Barth and Jander, 2006; Bednarek et al., 2009).
The formation of ER body structure is controlled by the basic helix-loop-helix type transcription factor NAI1 and, by extension, the genes encoding ER-body-localized proteins, including BGLU23, NAI2, MEB1, MEB2, NSP/JALs, and ESM/GLLs, which collectively form activated myrosinase complex regulating the turnover of GSLs end products (Matsushima et al., 2004; Yamada et al., 2008, 2013; Nakano et al., 2014).
GSLs activation and ER body formation have been implicated in tailoring Arabidopsis response to various biotic stimuli, including infection by the beneficial fungus Piriformospora indica (P. indica). Interestingly, the bglu23/pyk10 and the respective transcription factor nai1 mutants do not display beneficial effects of P. indica but instead allow uncontrolled fungal overgrowth. This suggests that the defense response conferred by the ER body enables controlled fungal colonization, thereby establishing a mutualistic interaction between the symbiotic partners (Sherameti et al., 2008). Accordingly, the roots of pen2 mutant, deficient in a BGLU23/PYK10 phylogenetically related BGLU gene, also display enhanced fungus colonization (Jacobs et al., 2011).
We have previously identified methylerythritol cyclodiphosphate (MEcPP), an intermediate of the plastidial methylerythritol phosphate (MEP) pathway, as a stress-specific retrograde signal, using ceh1 (constitutively expressing hydroperoxide lyase1), a mutant line with constitutively high levels of this signaling metabolite (Xiao et al., 2012; Walley et al., 2015; Benn et al., 2016). This mutant is the result of a point mutation in the enzyme, 4-hydroxy-3-methylbut-2-enyl diphosphate synthase (HDS), responsible for conversion of MEcPP to HMBPP (Xiao et al., 2012). In addition, we have established that accumulation of MEcPP results in a range of pleiotropic stress phenotypes, including stunted growth, high salicylic acid (SA) levels, high expression of JA response genes, constitutive general stress response, and potentiation of unfolded protein response in the ER (Xiao et al., 2012; Walley et al., 2015; Benn et al., 2016; Lemos et al., 2016; Bjornson et al., 2017). These phenotypes led us to hypothesize a potential expanded function of MEcPP in the reorganization of subcellular structures, such as the ER body, a storage body for myrosinases, and to explore the profile of the GSLs as the substrate for myrosinases.
Here, we have utilized a combination of genetics and multi-omics approaches to reveal the principles by which MEcPP functions as the initiating signaling hub coordinating the two Brassicaceae-specific traits, through reprogramming the transcriptional output responsible for ER body formation and selective induction of the biosynthesis and hydrolysis of IGs. Collectively, these findings highlight the role of MEcPP, a primary metabolic intermediate, as a master switch for expansion of the phytochemical diversity by enabling selective biosynthesis and degradation of secondary metabolites, in concert with formation of the associated subcellular structures, thus providing a previously unidentified foundation for the regulatory roles of core metabolites in stress biology.
RESULTS
Stress-inducible ER Bodies Are Constitutive in the ceh1 Mutant
It is well established that ER bodies are constitutively present in the epidermal cells of cotyledons, hypocotyls, and roots of young Arabidopsis seedlings, but are absent in rosette leaves (Ogasawara et al., 2009). However, stresses such as wounding not only increase the number of constitutive ER bodies but also induce their formation in rosette leaves (Ogasawara et al., 2009). Formation of the ER bodies is accompanied by induction of at least 17 genes encoding proteins that either regulate, associate, or localize to ER bodies (Supplemental Table 1) (Matsushima et al., 2004; Nagano et al., 2008; Yamada et al., 2008; Agee et al., 2010; Nakano et al., 2012, 2014; Hakenjos et al., 2013; Yamada et al., 2013; Nakano et al., 2017).
We have previously established that MEcPP induces a number of ER stress response genes and their encoded proteins (Walley et al., 2015). Closer examination of these data revealed a statistically significant increase in the expression levels of ~50% of ER-body-associated genes and ~60% of ER body proteins in ceh1, the high MEcPP accumulating mutant (Supplemental Table 2). Using qRT-PCR analyses of selected genes such as NAI2 and BGLU23, whose transcripts are regulated by developmental cues, and those regulated by environmental stress signals such as TSK-associating protein 1 (TSA1) and BGLU18, in various genotypes (wild-type [WT], ceh1, and HDS complemented [CP] lines) further confirmed the earlier transcriptomic data and identified HDS mutation in the ceh1 mutant background as the causative agent responsible for induction of the genes (Figure 1A and 1B).
Figure 1. Stress-lnducible ER Bodies Are Constitutive in the ceh1 Mutant.
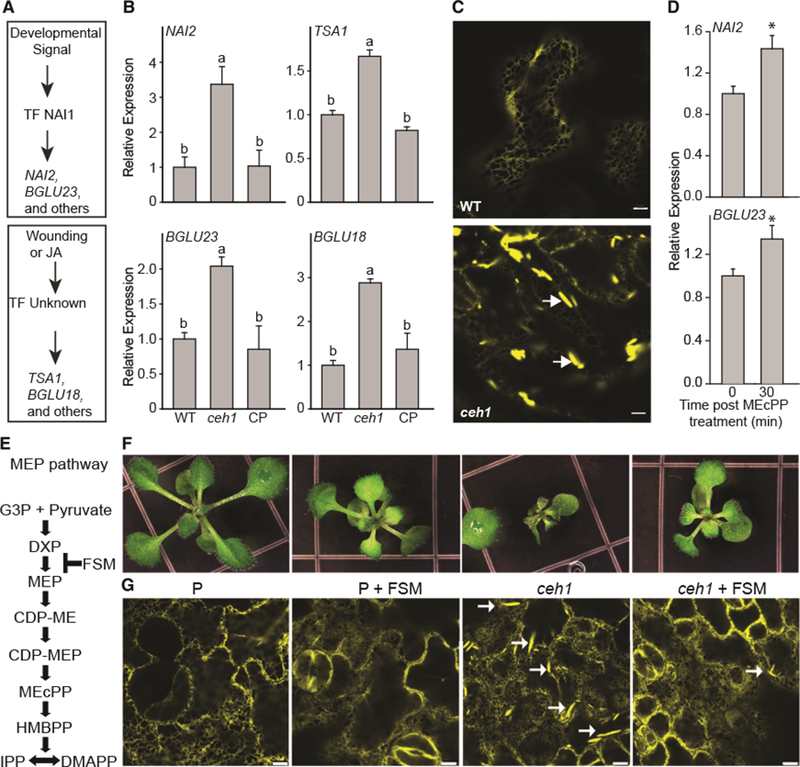
(A) Depiction of ER body marker genes induced by developmental or stress signals.
(B) Relative expression levels of genes regulated by developmental (NAI2 and BGLU23) and stress (TSA1 and BGLU18) signals, in WT, ceh1, and ceh1 complemented (CP) lines. Total RNA extracted from these genotypes was subjected to real-time qPCR analysis. The transcript levels were normalized to At4g26410 (M3E9) measured in the same samples. Data are the mean fold difference ± SD of three biological replicates each with three technical repeats. Different letters represent statistically significant differences (p < 0.05). p values were determined by Student’s t-test.
(C) Representative confocal images of WT and the ceh1 mutant rosette leaves depicting constitutive presence of otherwise wound-inducible ER bodies exclusively in ceh1. White arrows show the ER body. Bars, 5 μm.
(D) Relative expression levels of genes in WT plants treated exogenously with MEcPP. The transcript levels were normalized to At4g26410 (M3E9) measured in the same samples. Data are the mean fold difference ± SD of three biological replicates each with three technical repeats. Asterisks show the statistically significant differences relative to time 0 (p < 0.05). p values were determined by Student’s t-test.
(E) Schematic presentation of the MEP pathway depicting the site of fosmidomycin (FSM) action.
(F and G) Representative images of seedlings (F) together with the confocal images depicting ER body structure selectively shown by white arrows (G), grown in the presence (+) or absence (−) of FSM. Bars, 5 μm.
Next, we questioned whether the induction of the ER body marker genes could have resulted in the constitutive presence of otherwise stress-inducible ER bodies in the ceh1 leaves. To address this, we generated homozygous lines resulting from crosses performed between ER-localized yellow fluorescent protein (ER-YFP) (Nelson et al., 2007) and the ceh1 mutant line. Subsequent confocal imaging of the leaves clearly displayed the presence of ER-derived spindle-shaped structures earlier designated as ER bodies (Matsushima et al., 2003) in epidermal cells of rosette leaves in ceh1, but not in WT (Figure 1C). To confirm the potential role of MEcPP in the ceh1 mutant in ER body formation, we used a competitive inhibitor of DXR fosmidomycin (FSM) previously shown to arrest the flux through the MEP pathway (Gonzalez-Cabanelas et al., 2015) (Figure 1E). These data clearly show notable reduction of ER body formation in FMS-treated ceh1 plants, thereby providing evidence for he specificity of MEcPP in the formation of this subcellular structure (Figure 1F and 1G).
To confirm the specificity of MEcPP in the induction of gene expression, we further examined the relative transcript levels of NAI2 and BGLU23 in WT plants exogenously treated with MEcPP (Figure 1D). The data clearly confirm the induction of these genes 30 min post treatment, thereby providing evidence for the function of MEcPP in transcriptional regulation of these genes. In addition, we examined the expression levels of two wound-inducible ER body genes (TSAI and BGLU18) exploiting various RNAi lines silencing the MEP-pathway genes (Supplemental Figure 1). Exclusive induction of TSA1 and BGLU18 in the ceh1 mutant and none of the other MEP pathway silenced lines establishes the specificity of MEcPP function in the induction of wound-inducible ER-body-forming genes.
Collectively, the data established the constitutive presence of otherwise stress-inducible ER body structures in the ceh1 mutant, supported by the induction of the pertinent transcripts.
Constitutive Presence of ER Bodies in ceh1 Is SA-Independent
The presence of high levels of the defense-induced hormone SA in addition to elevated MEcPP in the ceh1 mutant (Xiao et al., 2012) led us to examine a potential role of SA in the constitutive induction of otherwise stress-inducible ER bodies. Toward this goal, we obtained homozygous ceh1/eds16 lines (Xiao et al., 2012) generated from crosses between the ceh1 harboring ER-YFP and eds16–1, an SA-deficient mutant encoding a dysfunctional Isochorismate synthase 1 (Wildermuth et al., 2001). These plants together with the corresponding controls, namely WT, ceh1, and eds16–1 (herein referred to as eds16), were examined for the presence of ER bodies in their leaves. Representative confocal images clearly showed the constitutive presence of ER bodies in the ceh1 and ceh1/eds16 genotypes but not in the respective controls (WT and eds16), thus establishing SA-independent constitutive formation of otherwise stress-inducible ER body structures (Figure 2A).
Figure 2. Constitutive Presence of ER Bodies in the ceh1 Is SA Independent.
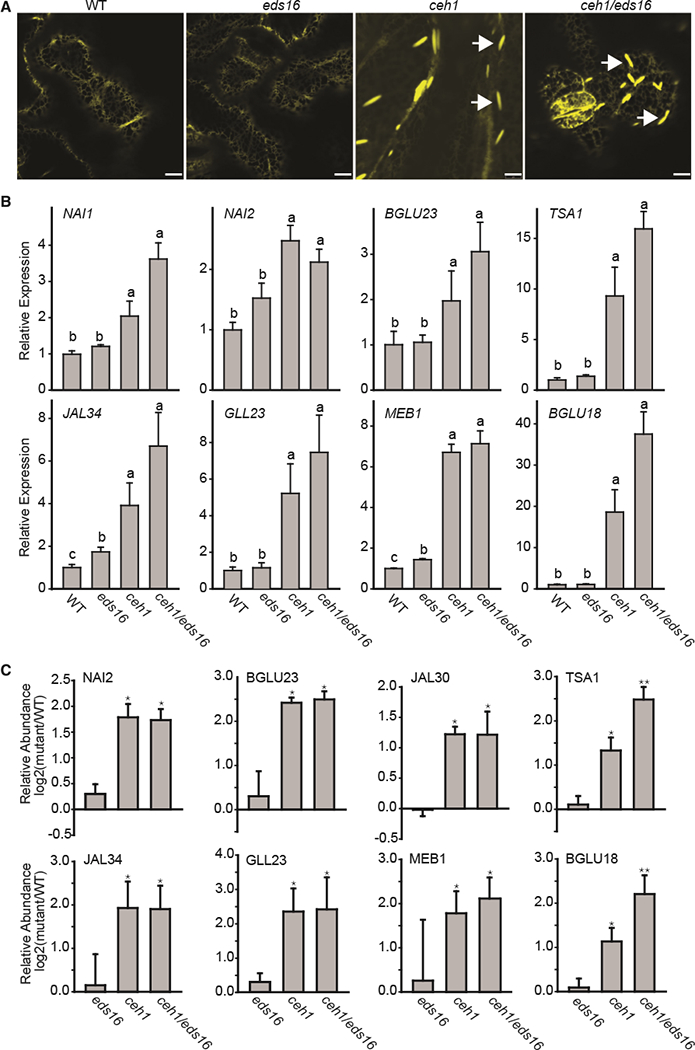
(A) Representative confocal images of WT, the SA-deficient mutant eds16, ceh1, and ceh1/eds16 leaves depicting constitutive presence of otherwise wound-inducible ER bodies independently of SA in ceh1 mutant backgrounds. White arrows show the ER body. Bars, 5 μm.
(B) Relative expression levels of ER marker genes in the aforementioned genotypes. Total RNA extracted from these genotypes was subjected to real-time qPCR analysis. The transcript levels were normalized to At4g26410 (M3E9) measured in the same samples. Data are the mean fold difference ± SD of three biological replicates each with three technical repeats. Different letters represent statistically significant differences (p < 0.05).
(C) Normalized iTRAQ protein abundance ratios of detected ER marker proteins in mutants (eds16, ceh1, and ceh1/eds16) relative to the WT plants. Data are means of n = 3 ± SEM. Single and double asterisks denote a statistically significant difference relative to WT and ceh1, respectively (p < 0.05) as determined by t-tests.
In addition, qRT-PCR-based examination of the expression profile of ER body marker genes in the aforementioned genotypes confirmed higher transcripts levels of all genes examined in the ceh1 and ceh1/eds16 compared with WT and eds16 plants (Figure 2B). Among the most notably induced genes are the two known co-expressed genes, namely TSA1 and BGLU18 (Nakano et al., 2014).
In concordance with the transcriptomic data, the relative abundance of the detectable ER-body-associated proteins, as described by a previous report (Walley et al., 2015) and accessible at (massive.ucsd.edu/ProteoSAFe/static/massive.jsp), is also higher in ceh1 and ceh1/eds16 compared with the controls (Figure 2C). However, one notable discordance between the transcriptomics and proteomic data is the statistically significantly higher relative protein abundance of TSA1 and BGLU18 in ceh1/eds16 compared with ceh1 (Figure 2C). This suggests that while SA is not involved in determining the expression levels of TSA1 and BGLU18, it is involved in enhancement of the stability and or translational capacity of the respective encoded products.
Altogether, these results support the notion that elevated MEcPP causes constitutive formation of typically stress-inducible ER bodies through enhanced expression levels and, by extension, abundance of the respective encoded products, predominantly independently of SA.
ER Body Formation in the ceh1 Mutant Is NAI1-Dependent
The ER body formation in roots is regulated by a basic helix-loop-helix- (bHLH) type transcription factor NAI1 (AtbHLH20) that controls expression of BGLU23, NAI2, MEB1, MD2-related lipid recognition protein 3 (ML3), NSP1 /JALs, and ESM1/GDSL lipase-like myrosinase chaperone genes (GLL23 and GLL25) (Matsushima et al., 2004; Nagano et al., 2005, 2008; Hakenjos et al., 2013). To examine the role of NAI1 in MEcPP-mediated regulation of ER body formation, homozygous ceh1/nai1 double mutants were generated through performing crosses between the ceh1 -harboring ER-YFP line and the knockdown nai1 mutant (nai1–3 allele) line. The similar phenotype between WT and nai1 opposed to enhanced compromised growth of the ceh1/nai1 double mutant relative to ceh1, suggests the role of a functional NAI1 in seedling growth in the ceh1 mutant background (Supplemental Figure 2). The representative confocal images from the leaves of the four genotypes (WT, ceh1, ceh1/nai1, and nai1) showed that in contrast to the abundant presence ER bodies in ceh1, these structures are no longer visible rosette leaves of ceh1/nai1 (Figure 3A).
Figure 3. ER Body Formation in the ceh1 Mutant Is NAI1 Dependent.
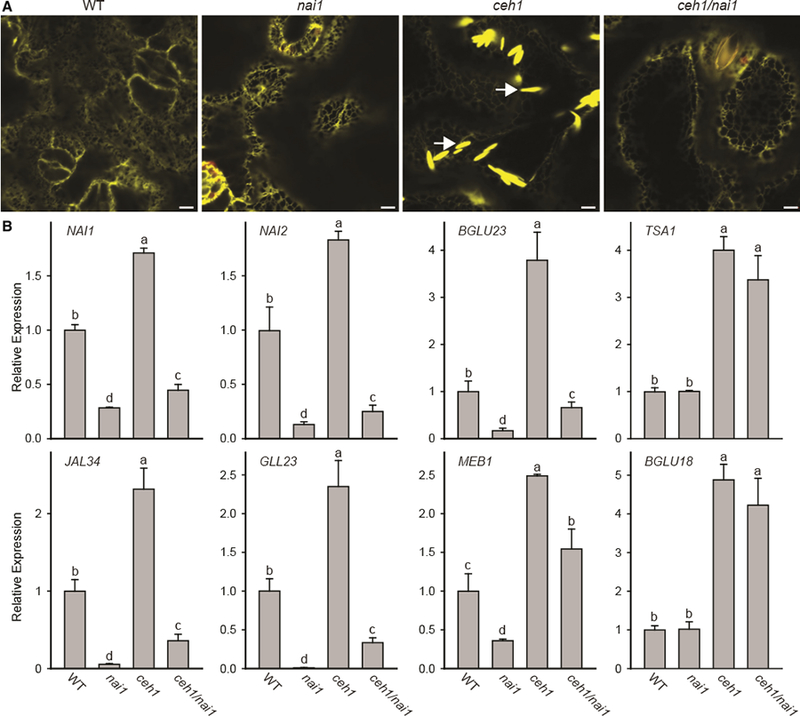
(A) Representative confocal images of WT, nai1, ceh1, and ceh1/nai1 leaves depicting the NAI1-dependent constitutive presence of otherwise wound-inducible ER bodies the in ceh1 mutant background. White arrows show the ER body. Bars, 5 μm.
(B) Relative expression levels of ER marker genes in the aforementioned genotypes. Total RNA extracted from these genotypes was subjected to real-time qPCR analysis. The transcript levels were normalized to At4g26410 (M3E9) measured in the same samples. Data are the mean fold difference ± SD of three biological replicates each with three technical repeats. Different letters represent significant differences (p < 0.05). p values were determined by Student’s t-test.
In concordance with the ER body phenotype, the qRT-PCR-based analyses using the aforementioned genotypes showed reduced expression levels of genes associated with constitutive ER body formation in ceh1/nai1 compared with the ceh1 mutant (Figure 3B). However, the expression levels of TSA1 and BGLU18 in ceh1/nai1 lines remained similar to that of the ceh1 plant (Figure 3B). These results corroborate the earlier report on NAI1-independent wounding induction of BGLU18 (Ogasawara et al., 2009) and further establish the key role of NAH in ER body formation in ceh1.
We also measured the MEcPP levels of these four genotypes to ascertain that the absence of ER body phenotype in ceh1/nai1 mutant lines is not due to alterations in the MEcPP levels (Supplemental Figure 3). These data clearly show similarly high MEcPP levels in ceh1 and ceh1/nai1 mutant lines compared with the controls, thereby negating any possible regulation of MEcPP levels by NAI1. This result supports the notion that the MEcPP-mediated ER body formation is NAI1-dependent.
Constitutive ER Body Formation in ceh1 Is Dependent on the Jasmonate Signaling Pathway
Reports showing JA-dependent de novo formation of ER bodies locally and systemically in wounded leaves (Matsushima et al., 2002; Ogasawara et al., 2009) led us to test the intersection of MEcPP and JA signaling in ER body formation in the ceh1 mutant background. Toward this goal, we generated homozygous double-mutant lines of ceh1/aos and ceh1/coi1 (Lemos et al., 2016) by performing crosses between ceh1 harboring ER-YFP and jasmonate synthesis mutant (Allene oxide synthase [aos]) and the signaling component mutant coi1 (co/7–7) (Xie et al., 1998; Savchenko et al., 2014). The larger plant phenotype in ceh1/coi1 versus ceh1 compared with the similar size of WT and coi1 seedlings suggests that the presence of jasmonate perception in the ceh1 mutant contributes to the compromised growth in this mutant line (Supplemental Figure 4). Furthermore, representative images of the ER structure displayed a significant reduction, albeit not total abolishment, of ER bodies both in ceh1/aos and ceh1/coi1 double mutants compared with the ceh1 mutant background (Figure 4A). In addition, expression analyses of selected ER body genes indicated a reduction in transcript levels of all genes examined in the double=mutant backgrounds compared with ceh1 alone (Figure 4B). Among the genes examined, the expression levels of TSA1 and BGLU18 are most significantly reduced, as their transcript levels in single (aos and coi1) and double mutants (ceh1/aos or/coi1) are below the basal WT levels (Figure 4B). This suggests that MEcPP function is mediated via the jasmonate signaling pathway followed by the consequential NA11-dependent induction of ER-body-associated genes and ultimately formation of the ER body structure.
Figure 4. Constitutive ER Body Formation in ceh1 Is Dependent on the Jasmonate Signaling Pathway.
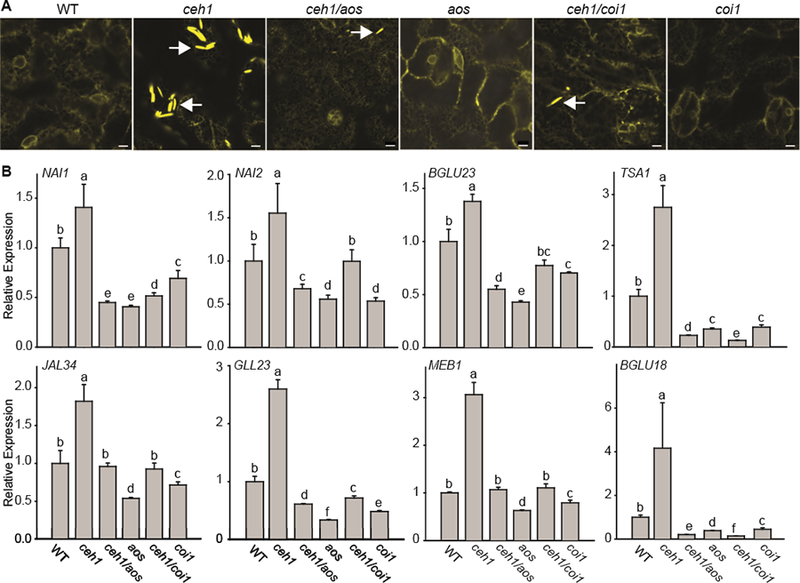
(A) Representative confocal images of WT, ceh1, ceh1/aos, aos, ceh1/coi1, and coi1 leaves depicting the jasmonate-dependent constitutive presence of otherwise wound-inducible ER bodies in the ceh1 mutant background. White arrows show the ER body. Bars, 5 μm.
(B) Relative expression levels of ER marker genes in the aforementioned genotypes. Total RNA extracted from these genotypes was subjected to real-time qPCR analysis. The transcript levels were normalized to At4g26410 (M3E9) measured in the same samples. Data are the mean fold difference ± SD of three biological replicates each with three technical repeats. Different letters represent statistically significant differences (p < 0.05). p values were determined by Student’s t-test.
MEcPP Mediates the Induction of Genes Associated with IG Synthesis
The presence of high levels of β-glucosidases in ER bodies together with the reported remarkable co-expression of ER body and GSLs genes and the genes encoding nitrile specifier proteins that control the profile of the activation products of myrosinase-mediated GSLs metabolism strongly support the notion of integration of the transcriptional network and functional coordination between ER bodies and GSLs (Yamada et al., 2008; Nakano et al., 2014, 2017). Accordingly, we tested the potential role of MEcPP in the induction of GSL accumulation.
Using existing RNA-seq data measuring the transcriptional network response to MEcPP, we investigated the regulation of all known GSL genes divided into three categories: known transcription factors (TFs), core structure, and side-chain modification of the IG and AG biosynthetic pathway (Figure 5A and 5B and Supplemental Figure 5). To distinguish between MEcPP regulation and potential secondary effects from elevated SA in ceh 1, we performed the analyses in four genotypes (WT, ceh1, ceh1/eds16, and eds16).
Figure 5. MEcPP Mediates Induction of Genes Associated with Indole Glucosinolate Synthesis.
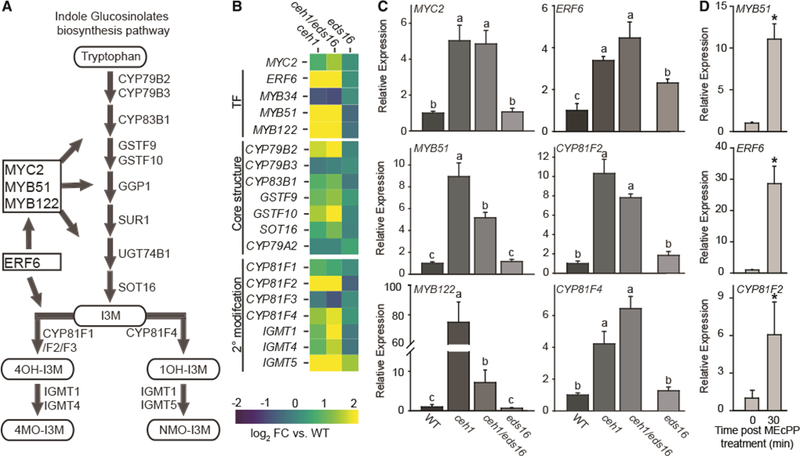
(A) Schematic of the indole glucosinolate (IGs) biosynthesis pathway depicting the TFs in boxes and the enzyme substrates lassoed.
(B) Heatmap of genes within each IG category (TFs, the core structure synthesis, and 2° modification proteins). The log2 fold change for ceh1 (left), ceh1/eds16 (middle), and eds16 (right) versus WT is represented by color, from −2 (blue) to 2 (yellow).
(C) Relative expression levels of selected genes within each of the three aforementioned categories. Total RNA extracted from these genotypes was subjected to real-time qPCR analysis. The transcript levels were normalized to At4g26410 (M3E9) measured in the same samples. Data are the mean fold difference ± SD of three biological replicates each with three technical repeats. Different letters represent significant differences (p < 0.05). p values were determined by Student’s t-test.
(D) Relative expression levels of genes in WT plants treated exogenously with MEcPP. The transcript levels were normalized to At4g26410 (M3E9) measured in the same samples. Data are the mean fold difference ± SD of three biological replicates each with three technical repeats. Asterisks show the statistically significant differences relative to time 0 (p < 0.05). p values were determined by Student’s t-test.
This analysis showed that the IG genes (TFs, core structure synthesis, and side-chain modification) are induced in both ceh1 and ceh1/eds16, as displayed by a high ratio (9/19) of IG biosynthesis-associated genes significantly changed (q value <0.05, log2 fold change >2) in ceh1, with an even higher ratio of genes in ceh1/eds16 (14/19) (Figure 5A and 5B and Supplemental Table 3). This finding enables identification of fully or partially SA-independent genes. Interestingly, however, the ratio for the AG altered genes is rather modest, as there are only 4 and 5/27 genes in ceh1 and ceh1/eds16 mutants, respectively (Supplemental Figure 5B and 5C and Supplemental Table 3). The higher ratio of genes altered in the IG pathway compared with those of the AG pathway strongly suggests a preference for IG biosynthesis.
Guided by the heatmap results, we focused on confirmation of altered transcriptomic data by qRT-PCR analyses. We specifically analyzed the expression levels of selected genes, among them two of the three TFs (MYB51 and MYB122) whose co-functionality in regulating IG biosynthesis is established (Frerigmann and Gigolashvili, 2014). We also included MYC2, a TF-encoding gene with the capacity to regulate the accumulation of both IGs and AGs, as well as two downstream genes encoding IG-specific modifying enzymes, cytochrome P450 monooxygenases (CYP81F2 and CYP81F4). In addition, the expression levels of ERF6, a gene with an established link to the regulation of specific IG structures was also examined (Xu et al., 2016). The results illustrate equal induction of most genes in ceh1 and ceh1/eds16 with the exception of MYB51 and MYB122, whose expression is lower in ceh1/eds16 compared with ceh1, reflecting the synergistic function of MEcPP and SA in the induction of the two (Figure 5C).
To confirm the specificity of MEcPP in the induction of gene expression, we further examined the relative transcript levels of two TFs (MYB51 and ERF6) and one biosynthetic gene (CYP81F2) in WT plants exogenously treated with MEcPP (Figure 5D). The data clearly confirm induction of these genes 30 min post treatment, thereby providing evidence for the function of MEcPP in transcriptional regulation of both TFs and the biosynthetic gene. Together, these analyses suggest that the plastidial metabolite MEcPP selectively mediates the induction of IG’s regulatory and metabolic genes.
Specificity of MEcPP in the Induction of IG Production
The altered transcriptomic profiles of IG regulatory genes prompted us to include metabolomics analyses and to explore the concordance between the two omics approaches in the four genotypes of WT, ceh1, ceh1/eds16, and eds16. Initial analyses performed using LC-MS are consistent with metabolic reprogramming for increased GSL biosynthesis (Bjornson et al., 2017). Moreover, direct measurement of GSLs revealed increased levels of various IGs, including I3M, 4MOI3M, and NMOI3M in ceh1 and ceh1/eds16, albeit at different levels (Figure 6A). Next, we exploited the well-established targeted HPLC methods (Kliebenstein et al., 2001) to more accurately quantitate the levels of these metabolites. These analyses established enhanced levels of various species of IGs at comparable levels between ceh1 and ceh1/eds16 and further illustrate SA-independent but MEcPP-mediated induction of these IGs (Figure 6B). However, these data do not differentiate between the specific function of MEcPP and the potential general impact of MEP-pathway perturbation in the ceh1 mutant backgrounds.
Figure 6. MEcPP Specifically Induces the Production of IGs.
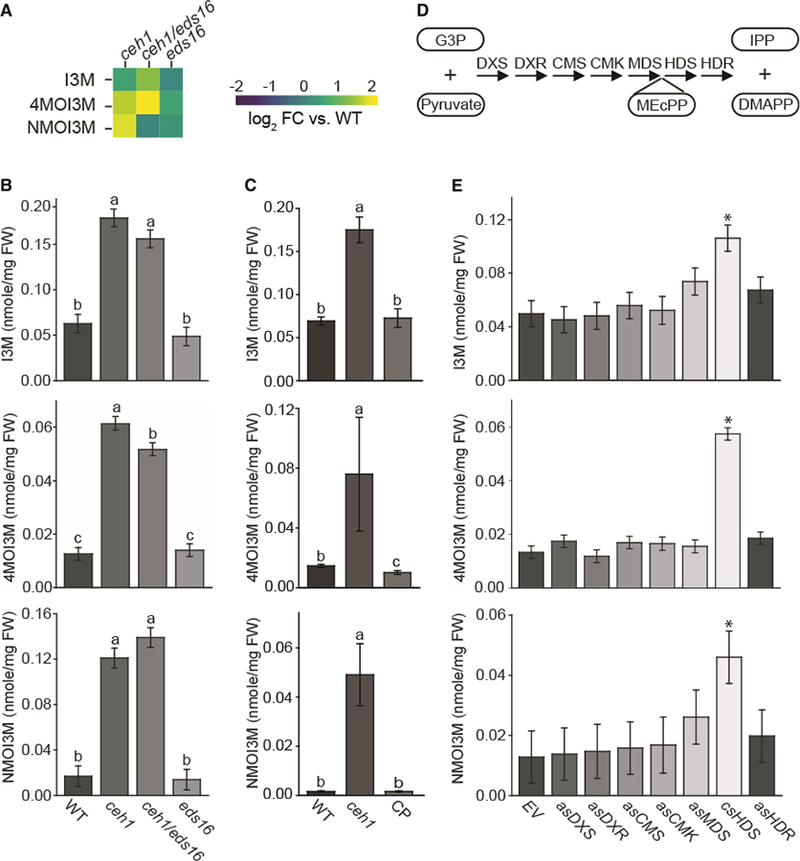
(A) Heatmap of IG levels in ceh1, ceh1/eds16, and eds16 relative to WT extracted from the metabolomics analyses using LC-MS-based methods. The log2 fold change is presented by color, from −2 (blue) to 2 (yellow).
(B and C) HPLC-based targeted measurements of IG levels in WT, ceh1, ceh1/eds16, and eds16 (B), and in WT, ceh1, and HDS complementation line (CP) (C). Data are means ± SEM; n = 3. Different letters represent statistically significant differences (p < 0.05).
(D) Schematic of the MEP-pathway genes and lassoed selected metabolites involved.
(E) Measurement of IG levels in WT transformed with empty vector (EV), RNAi lines silencing MEP-pathway genes individually, and co-suppressed HDS (csHDS). Data are means ± SEM; n = 3. Asterisks show the statistically significant differences relative to EV (p < 0.05). All the statistical analyses were performed using ANOVA.
To differentiate between the role of MEcPP and general MEP-pathway perturbation, we first examined the levels of different IGs in complemented HDS lines and established the reversion of the high IG levels in the HDS complementation line back to the WT levels (Figure 6C). Next, we performed these analyses using all the previously described RNAi lines with reduced levels of the individual MEP-pathway genes (Xiao et al., 2012) (Figure 6D). Specifically, we examined different species of IGs as well as AGs, as the control, in the RNAi and HDS co-suppression (csHDS) lines in concert with the control empty vector transgenic lines (Figure 6C–6E and Supplemental Figure 5). These comparative analyses using lines altered in the levels of all the seven MEP-pathway enzymes displayed enhanced IG levels exclusively in the csHDS line (Figure 6E). Furthermore, these analyses showed no specificity in changing the levels of AG species among various MEP-pathway altered lines (Supplemental Figure 6). Lastly, measurement of NMOI3M levels in ceh1 and the aforementioned RNAi lines 3 days post botrytis (B05) infection further established the function of MEcPP in the induction of IG production (Supplemental Figure 7). Collectively, these data identified MEcPP as the specific signaling core metabolite that selectively induces IGs biosynthesis.
MEcPP-Mediated Selective Induction of IGs Is COI1-Dependent
The induction of JA-responsive genes in spite of high SA levels in the ceh1 mutant (Lemos et al., 2016), together with the tight regulation of IG synthesis by the jasmonate pathway (Brader et al., 2001; Dombrecht et al., 2007), and the dependency of ER body formation on the pathway (Figure 4A and 4B), led us to question the intersection of MEcPP and jasmonate signaling in the induction of IGs. Thus, we examined the accumulation of different IGs and the transcript levels of IG regulatory genes in homozygous ceh1/aos and ceh1/coi1 double mutants in concert with the single mutants and the WT plants (Figure 7A and 7B). These analyses showed significantly decreased levels of the common substrate I3M in jasmonate synthesis and signaling mutant lines (ceh1/aos, aos, ceh1/coi1, and coi1) relative to the levels in ceh1 and WT seedlings (Figure 7A). The I3M downstream metabolites, 4MOI3M and NMOI3M, display a different trend, i.e., modest but statistically significant higher levels in the double (ceh1/aos and ceh1/coi1) than the single mutants, implying an MEcPP-targeted function in mediating the conversion of I3M to 4MOI3M and/or NMOI3M (Figure 7A).
Figure 7. MEcPP-Mediated Selective Induction of IGs Is COI1 Dependent.
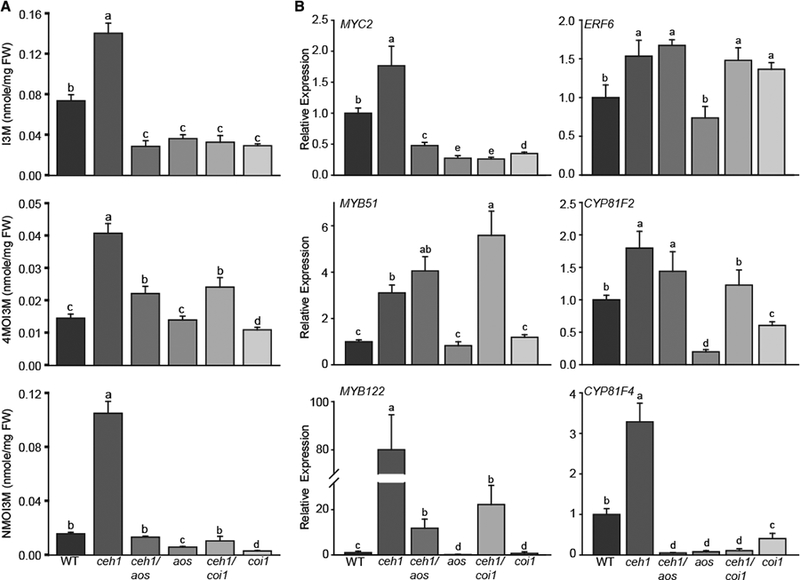
(A) HPLC-based measurement of IG levels in WT, ceh1, ceh1/aos, aos, ceh1/coi1, and coi1. Data are means ± SEM; n = 3. Different letters represent significant differences (p < 0.05). p values were determined by ANOVA.
(B) Relative expression levels of IG-associated genes encoding transcription factors (MYC2, ERF2, MYB51, and MYB122) and biosynthetic enzymes (CYP81F2, CYP81F4) in the aforementioned genotypes by qRT-PCR. Total RNA extracted from these genotypes was subjected to real-time qPCR analysis. The transcript levels were normalized to At4g26410 (M3E9) measured in the same samples. Data are the mean fold difference ± SD of three biological replicates each with three technical repeats. Different letters represent significant differences (p < 0.05). p values were determined by Student’s t-test.
We also examined the expression levels of genes encoding TFs and the modifying enzymes of the IG pathway using qRT-PCR in the aforementioned genotypes (Figure 7B). The analyses show notable reduction of MYC2 expression in the absence of a functional jasmonate signaling pathway in all backgrounds examined. In contrast, the ERF6 expression level is unresponsive to production and perception of jasmonates, specifically in the ceh1 background. This is consistent with the model that MYC2 is a specific component in the JA pathway, whereas ERF6 expression is regulated by multiple hormones and stress cues. Moreover, the expression levels of the direct IG regulator, MYB51, is not reduced in jasmonate signaling pathway mutants in the ceh1 background, whereas the transcript level of MYB122 is reduced in ceh1/aos and ceh1/coi1 compared with the ceh1 mutant, although these levels are still higher than that of the WT. Lastly, while the transcript level of CYP81F2 is only slightly changed in ceh1/coi1 compared with ceh1, the expression of CYP81F4 is notably reduced in the single and double mutants of the jasmonate pathway, consistent with the corresponding profiles of IG metabolites in the respective lines (Figure 7A and 7B). The above data support the predominant but not exclusive COM-dependent MEcPP-mediated selective induction of IGs.
MEcPP-Mediated Induction of Simple Nitriles Is Partially NAH-Dependent
The constitutive presence of normally stress-inducible ER bodies together with the elevated BGLU23 expression levels in the ceh1 mutant prompted us to examine the interplay between ER bodies and the hydrolysis of IGs in the ceh1 mutant (Yamada et al., 2009; Nakano et al., 2014). Toward this goal, we compared ceh1 with the nai1 single and ceh1/nai1 double mutants as they lack ER body structures and show reduced BGLU23 expression levels (Figure 3A and 3B). We first examined the levels of various species of IGs in these genotypes and determined that downregulation of NAI1 has not reduced the levels of any of the IG species examined; if anything the levels are slightly but significantly increased in ceh1/nai1 compared with the ceh1 single mutant (Figure 8A). With the notion that MEcPP regulates ER bodies which harbor the enzymes catalyzing IG hydrolysis, we postulated that the GSL activation products may potentially be altered in ceh1 compared with ceh1/nai1 mutants. To examine this possibility, we employed a targeted approach and analyzed the formation of indole-3-acetonitrile, one of the end products of simple nitrile, which is normally produced during GSL activation through myrosinase-catalyzed glucosinolate hydrolysis (Figure 8B) (Burow et al., 2009). Consistent with the role of ER bodies in storing the myrosinases, ceh1/nai1 indeed displays lower levels of indole-3-acetonitrile compared with the ceh1 mutant background (Figure 8C).
Figure 8. MEcPP-Mediated Induction of Simple Nitriles Is Partially NAI1 Dependent.
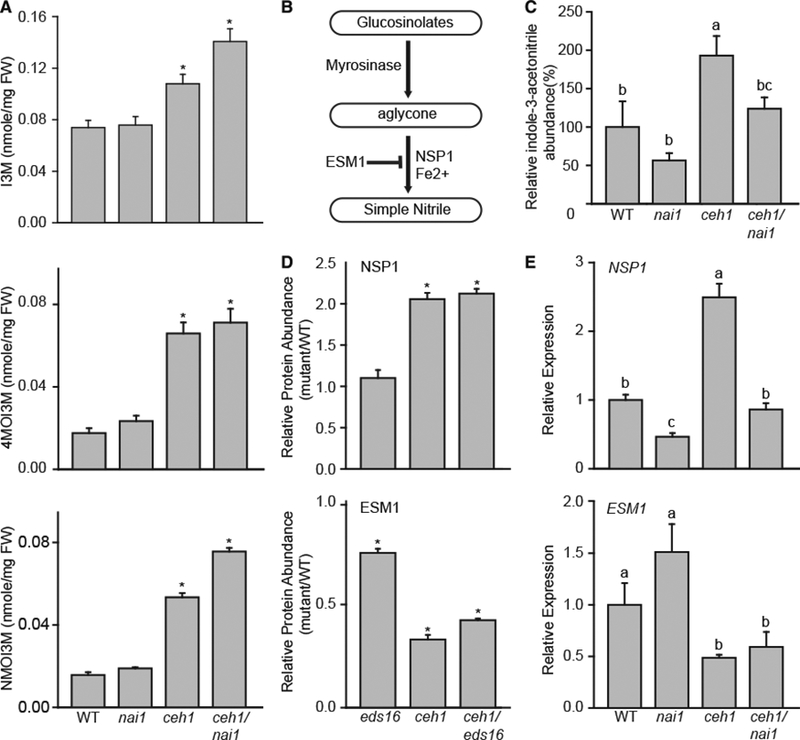
(A) HPLC-based measurement of IG levels in WT, nai1, ceh1, and ceh1/nai1. Data are means ± SEM; n = 3. Asterisks show the statistically significant differences relative to WT (p < 0.05).
(B) Schematic of IG hydrolysis with intermediate and terminal products lassoed and associated enzymes and cofactors.
(C) Relative levels of indole-3-acetonitrile in WT, nai1, ceh1, and ceh1/nai1 measured by LC-MS. Data are means ± SEM; n = 4. Letters represent significant differences if not shared (p < 0.05).
(D) Normalized protein abundance of NSP1 and ESM1 (eds16, ceh1, and ceh1/eds16) relative to the WT plants. Data are means ± SEM, n = 3. Asterisks denote a statistically significant difference relative to WT (p < 0.05).
(E) Relative expression levels of NSP1 and ESM1 in WT, nai1, ceh1, and ceh1/nai1. Data are means ± SEM; n = 4. Letters represent statistically significant differences if not shared (p < 0.05). P values were determined by Student’s t-test for (C-E).
The formation of nitriles from GSLs following tissue disruption is controlled by multiple genes, among them epithiospecifier modifier 1 (ESM1), a GLL in the ER body nomenclature that inhibits the formation of indol-3-acetonitrile from I3M following tissue disruption (Burow et al., 2008). The others are nitrile-specifier proteins (NSPs, JAL in the ER body nomenclature) responsible for the enzymatic formation of simple nitriles. Here, we specifically focused on NSP1 since the expression of this gene is strongly induced by herbivory in the rosette leaves, and leaves of nsp1 knockout lines fail to produce simple nitriles (Burow et al., 2009; Wittstock and Burow, 2010). Interestingly, the search of the proteomic dataset on eds16, ceh1, and ceh1/eds16 lines exhibited enhanced abundance of NSP1 and reduced levels of ESM1 proteins in ceh1 compared with the WT, independently of SA (Figure 8D). These findings led us to examine the transcript levels of NSP1 and ESM1 in single and double ceh1/nai1 mutants. The analyses show the presence of higher levels of NSP1 in ceh1 relative to the WT or nai1 single-mutant background, and the reduced levels in ceh1/nai1 mutant plants (Figure 8E). In contrast, the expression levels of ESM1 in both ceh1 and ceh1/nai1 remained below that of the of WT or nai1 plants. These results collectively establish MEcPP-mediated induction of NSP1 and suppression of ESM1 as a molecular mechanism enabling accumulation of simple nitrile.
DISCUSSION
A central quest of modern biology is to decipher how regulatory functions result in secondary metabolite diversification and alteration of the associated subcellular infrastructure reflecting higher-order adaptive responses. Among such adaptive responses are the evolutionarily associated traits present in a few lineages of the order Brassicales: production of specialized metabolite, IGs, structurally specific activation of the IGs, and formation of ER body structures storing the associated activation enzymes. A transcriptional link between these taxonomically limited traits regulating the adaptive responses within the Brassicaceae family has been suggested (Nelson et al., 2007) but the nature of the regulatory switch that coordinates these processes had remained elusive. In this study, we revealed that MEcPP, a bifunctional metabolite acting as an intermediate of the essential plastidial MEP pathway present in all plants, as well as a stress-specific retrograde signaling metabolite, is an initiator of the signal transduction pathway leading to the induction of ER body formation and selective metabolism of IGs.
We specifically established that MEcPP mediates induction of a number of ER body formation genes and their corresponding translated proteins via the jasmonate signaling component COI1. We further demonstrated that the MEcPP function is independent of the stress-inducible hormone SA. On the contrary, SA reduces the abundance of TSA1 and BGLU18 proteins, the two components of stress-inducible ER body structures. Among the MEcPP-induced genes is NAI1, encoding a central TF required for ER body formation (Matsushima et al., 2004; Yamada et al., 2008, 2013; Nakano et al., 2014). It is noteworthy that the constitutive presence of ER bodies in roots and cotyledons as opposed to their stress-inducible formation in rosette leaves illustrates a different activation mechanism of NAI1. In green tissue such as leaves, general stresses such as wounding or high light result in accumulation of MEcPP, thereby activating the COI1-dependent jasmonate signaling pathway resulting in enhanced expression of NAI1 and ultimately ER body formation. In addition, using various genotypes in the nai1 mutant background confirmed NAI1 as the master transcriptional activator of stress-inducible ER body formation as the presence of high levels of stress-inducible retrograde signal MEcPP or the elevated BGLU18 transcripts in the ceh1/nai1 background cannot substitute for the NAI1 function in the formation of ER bodies. Moreover, expression of BGLU18 in the nai1 mutant background corroborates the earlier report on NAI1-independent wound induction of BGLU18 (Ogasawara et al., 2009), illustrating the complexity of the regulatory components involved and further raising a question about the subcellular localization of BGLU18 described earlier to be exclusively present in the ER bodies formed directly at wound site (Matsushima et al., 2002; Ogasawara et al., 2009). One explanation for this observation is that BGLU18 may also accumulate in the ER, as previously shown by transient expression assays in protoplasts (Lee et al., 2006). The neutral pH in the ER (Shen et al., 2013), similar to ER bodies, renders the enzyme inactive while physically barring it from reaching the substrate in an intact cell. Alternatively, the ER body functions not only as the repository site but also as a stabilizing site for the BGLU18-encoded product.
Our data further support the previously suggested link between the BGLU18 gene cluster and the metabolism of GSLs as it provides evidence for the MEcPP-mediated regulation of ER body formation and selective induction of IGs, thereby expanding the notion of developmental (Brown et al., 2003), to stress-specific differential regulation and potentially distribution of AGs and IGs.
The initiation of MEcPP signaling both for diversification of IGs and the ER body formation is via the jasmonate pathway in a COM-dependent manner. Interestingly, however, our previous data showed that accumulation of MEcPP enhances mainly the levels of the JA precursor, 12-OPDA, a plastidial produced metabolite that upon transfer to the peroxisome is converted to JA (Lemos et al., 2016; Bjornson et al., 2017). The function of 12-OPDA as a signaling molecule, with distinct as well as overlapping responses with JA (Savchenko and Dehesh, 2014; Savchenko et al., 2014), further denotes the role of plastids as the sensory hub for shaping adaptive responses.
Furthermore, high levels of 4MOI3M and the corresponding biosynthetic enzyme CYP81F2 in ceh1/aos and ceh1/coi1 relative to aos or coi1 single mutants suggests the CO11 -independent role of MEcPP in the regulation of this group of IGs. These data, together with the reported role of CYP81F2 in plant innate immune response (Clay et al., 2009) and defense specifically against the green peach aphid (Pfalz et al., 2009), raises a question about the mode of MEcPP function in fine-tuning 4MOI3M production in response to stresses.
In addition to regulation of biosynthesis, MEcPP mediates hydrolysis of IGs by inverse regulation of two functionally opposing genes, namely by reducing the levels of ESM1 expression and inducing NSP1 transcripts. The end result is enhanced levels of the terminal product, simple nitriles in the plants with high MEcPP levels in the absence of an external inducer such as wounding. Collectively, these data establish MEcPP as the regulatory switch for integration of IG biosynthesis/degradation as well as the formation of the ER body structure.
In summary, as depicted in the simplified schematic model (Figure 9), MEcPP initiates a stringent transcriptional regulation, predominantly via COM, enabling diversification of secondary metabolites and the formation of the associated stress-inducible subcellular structures in the ER. The identification of MEcPP, a precursor of plastidial isoprenoids and a retrograde signaling metabolite initiating these adaptive responses, expands the function of the metabolic hub, plastids, to a stress-sensing center providing cues enabling cellular readjustments for high-order adaptive functions. As an example, the mutualistic interaction between plants and P. indica requiring ER body formation and IG metabolism (Sherameti et al., 2008; Jacobs et al., 2011), the two supported traits by MEcPP, highlights plastidial sensory functions. This notion offers an exciting paradigm of strategies for plastidial-derived metabolic control of plant-specialized metabolism and uncovers the intersection between primary and secondary metabolisms and new principles of metabolic regulation in support of biological processes central to tailored adaptive responses.
Figure 9. Simplified Schematic Models of MEcPP-Mediated Induction of IG Biosynthesis/Breakdown and the Associated ER Body Formation.
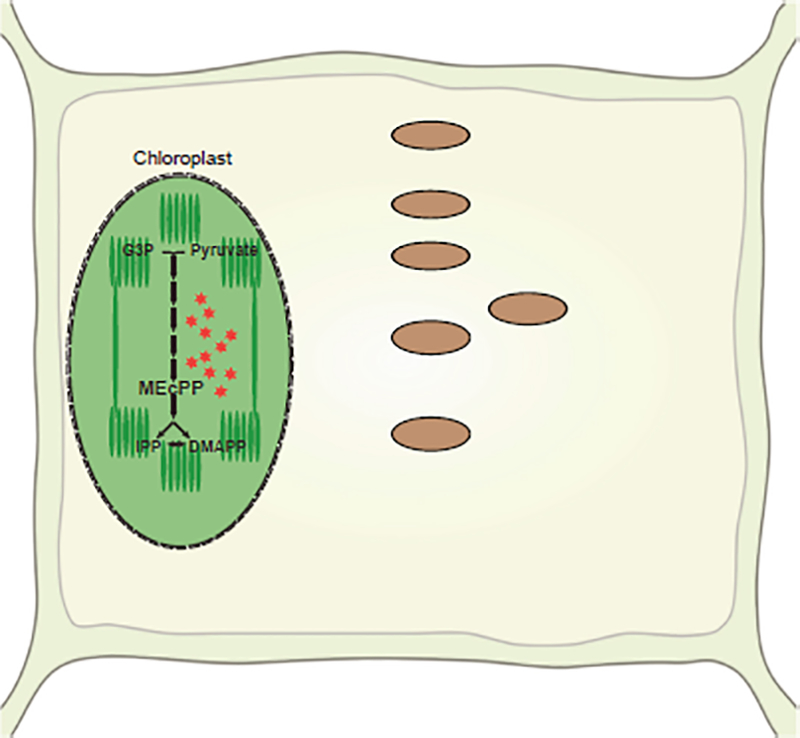
Stress-induced accumulation of MEcPP enables readjustment of the activity of hard-wired gene circuitry (lassoed), resulting in alteration of the levels of IG biosynthesis and hydrolytic products (boxed), and the formation of the associated cellular infrastructure ER bodies.
METHODS
Plant Materials and Growth Conditions
All the experiments were conducted on seedlings grown under 16 h light/8h dark cycles for 2 weeks on half-strength Murashige and Skoog (MS) medium.
Fosmidomycin and Botrytis Treatment
Seedlings were grown for 8 days on half-strength MS containing 10 μM FSM prior to confocal imaging of the ER body structure. Images were from 14-day-old seedlings grown on half-strength MS containing 10 μM FSM.
Botrytis B05 infection was carried out as described previously (Xiao et al., 2012).
RNA-Seq and Quantification of Gene Expression
RNA-seq data were obtained from the previously described report (Bjornson et al., 2017). Gene expression quantification was conducted using quantitative real-time PCR. Real-time PCR and data normalization were performed as described previously (Walley et al., 2015). All primer sequences are listed in Supplementary Table 3.
Glucosinolate Extraction and Quantification
The glucosinolates levels were initially extracted from the metabolomics data described previously (Bjornson et al., 2017) followed by generation of the heatmap using the ggplot2 package in R. Targeted GSL extraction and quantification were performed as described previously (Kliebenstein et al., 2001). The statistical analysis for GSL levels was conducted by Tukey’s method using ANOVA.
Simple Nitrile Extraction and Analyses
Approximately 55 (±7) mg of rosette leaves was placed in 1.5 ml Eppendorf-like tubes with two glass beads (2.5 mm in diameter) and frozen in liquid N2. The material was ground in liquid N2 using a bead beater (2x for 30 s; Mini-Beadbeater; Biospecs Products). Samples were extracted with 400 μl of 9:1 water:methanol (methanol containing 1% acetic acid). Samples were vortexed thoroughly, incubated on ice for 20 min with vortexing every 5 min, followed by 5 min centrifugation at 4°C and 18 000 RCF. After centrifugation, the material was vortexed and centrifuged once more at 4°C and 18 000 RCF. The supernatant was collected into a fresh tube and centrifuged at 4°C and 18 000 RCF to pellet any remaining debris. A suitable aliquot of the supernatant (100 μl) was collected into LC-MS vials for injection.
LC-MS analysis was conducted on a Dionex Ultimate 3000 binary RSLC system coupled to a Thermo Q-Exactive Focus mass spectrometer with a heated electrospray ionization source. Chromatographic separation was done on an Acclaim RSLC 120 C18 column (100 × 2.1 mm, particle size 2.2 μm; Thermo Scientific 068982). Gradient elution was done with acetonitrile containing 0.1 % formic acid (A) and water containing 0.1 % formic acid (B). The separation was conducted using the following gradient profile (f (min), %A, %B): (0, 5, 95), (20, 95, 5), (25, 95, 5), (25.01, 5, 95), (35, 5, 95). The injected volume was 20 μl and the flow rate was kept at 200 μl/min. The column temperature was maintained at 35°C. Mass spectra in negative mode were acquired under the following conditions: spray voltage, 4.50 kV; sheath gas flow rate 45, auxiliary gas flow rate 20, sweep gas flow rate 2, capillary temperature of 250°C, S-lens RF level 50, and auxiliary gas heater temperature 250°C. The compound of interest was identified by accurate mass (MS1), retention time, and mass transition monitoring on purified standards (indole-3-acetonitrile; Sigma-Aldrich 129453) and plant extracts. The area under the peak (MS1; Thermo Trace Finder Software) divided by the initial fresh weight mass was used for relative quantitation and the final data presented are normalized to WT levels.
MEcPP Measurements and Treatment
Analyses of MEcPP levels were carried out as described previously (Lemos et al., 2016). Exogenous MEcPP treatment was performed on 2-week old seedlings sprayed with 100 μM MEcPP.
Confocal Laser Scanning Microscopy
The ER-YFP seeds were obtained from ABRC. The construct was introduced into different backgrounds using crosses. Homozygous lines were employed in confocal laser scanning microscopy analysis. Images were taken with a Zeiss Axio inverted microscope Observer Z1 coupled to an LSM 710 laser scanning confocal system.
Supplementary Material
ACKNOWLEDGMENTS
We thank Professor Claus Schwechheimer for providing nai1–3 seeds. No conflict of interest declared.
FUNDING
This work was supported by National Science Foundation grants IOS-1036491 and IOS-1352478; and NIH R01GM107311 to K.D. This work is dedicated to the memory of Klaus Apel.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information is available at Molecular Plant Online.
REFERENCES
- Agee AE, Surpin M, Sohn EJ, Girke T, Rosado A, Kram BW, Carter C, Wentzell AM, Kliebenstein DJ, Jin HC, et al. (2010). MODIFIED VACUOLE PHENOTYPE1 is an Arabidopsis myrosinase-associated protein involved in endomembrane protein trafficking. Plant Physiol. 152:120–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn YO, Shimizu BI, Sakata K, Gantulga D, Zhou CH, Bevan DR, and Esen A (2010). Scopolin-hydrolyzing beta-glucosidases in roots of Arabidopsis. Plant Cell Physiol. 51:132–143, [DOI] [PubMed] [Google Scholar]
- Andreasson E, Bolt Jorgensen L, Hoglund AS, Rask L, and Meijer J (2001). Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus. Plant Physiol. 127:1750–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C, and Jander G (2006). Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46:549–562. [DOI] [PubMed] [Google Scholar]
- Bednarek P (2012). Chemical warfare or modulators of defence responses - the function of secondary metabolites in plant immunity. Curr. Opin. Plant Biol. 15:407–414. [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323:101–106. [DOI] [PubMed] [Google Scholar]
- Benn G, Bjornson M, Ke H, De Souza A, Baimond EI, Shaw JT, and Dehesh K (2016). Plastidial metabolite MEcPP induces a transcriptionally centered stress-response hub via the transcription factor CAMTA3. Proc. Natl. Acad. Sci. USA 113:8855–8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidart-Bouzat MG, and Kliebenstein DJ (2008). Differential levels of insect herbivory in the field associated with genotypic variation in glucosinolates in Arabidopsis thaliana. J. Chem. Ecol 34:1026–1037. [DOI] [PubMed] [Google Scholar]
- Bjornson M, Balcke GU, Xiao Y, de Souza A, Wang J-Z, Zhabinskaya D, Tagkopoulos I, Tissier A, and Dehesh K (2017). Integrated omics analyses of retrograde signaling mutant delineate interrelated stress response strata. Plant J. 91:70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bones AM, and Rossiter JT (2006). The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry 67:1053–1067. [DOI] [PubMed] [Google Scholar]
- Brader G, Tas E, and Palva ET (2001). Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiol. 126:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, and Gershenzon J (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62:471–481. [DOI] [PubMed] [Google Scholar]
- Burow M, Bergner A, Gershenzon J, and Wittstock U (2007). Glucosinolate hydrolysis in Lepidium sativum–identification of the thiocyanate-forming protein. Plant Mol. Biol 63:49–61. [DOI] [PubMed] [Google Scholar]
- Burow M, Zhang ZY, Ober JA, Lambrix VM, Wittstock U, Gershenzon J, and Kliebenstein DJ (2008). ESP and ESM1 mediate indol-3-acetonitrile production from indol-3-ylmethyl glucosinolate in Arabidopsis. Phytochemistry 69:663–671. [DOI] [PubMed] [Google Scholar]
- Burow M, Losansky A, Muller R, Plock A, Kliebenstein DJ, and Wittstock U (2009). The genetic basis of constitutive and herbivore-induced ESP-independent nitrile formation in Arabidopsis. Plant Physiol. 149:561–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, and Ausubel FM (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323:95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19:2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Crooks C, Creissen G, Hill L, Fairhurst S, Doerner P, and Lamb C (2011). Pseudomonas sax genes overcome aliphatic isothiocyanate-mediated non-host resistance in Arabidopsis. Science 331:1185–1188. [DOI] [PubMed] [Google Scholar]
- Fan J, and Doerner P (2012). Genetic and molecular basis of nonhost disease resistance: complex, yes; silver bullet, no. Curr. Opin. Plant Biol. 15:400–406. [DOI] [PubMed] [Google Scholar]
- Frerigmann H, and Gigolashvili T (2014). MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 7:814–828. [DOI] [PubMed] [Google Scholar]
- Frerigmann H, Pislewska-Bednarek M, Sanchez-Vallet A, Molina A, Glawischnig E, Gigolashvili T, and Bednarek P (2016). Regulation of pathogen-triggered tryptophan metabolism in Arabidopsis thaliana by MYB transcription factors and indole glucosinolate conversion products. Mol. Plant 9:682–695. [DOI] [PubMed] [Google Scholar]
- Gachon CM, Langlois-Meurinne M, Henry Y, and Saindrenan P (2005). Transcriptional co-regulation of secondary metabolism enzymes in Arabidopsis: functional and evolutionary implications. Plant Mol. Biol 58:229–245. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Berger B, Mock HP, Muller C, Weisshaar B, and Flügge U.l. (2007). The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50:886–901. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Engqvist M, Yatusevich R, Muller C, and Flugge U.l. (2008). HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 177:627–642. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cabanelas D, Wright LP, Paetz C, Onkokesung N, Gershenzon J, Rodriguez-Concepcion M, and Phillips MA (2015). The diversion of 2-C-methyl-D-erythritol-2,4-cyclodiphosphate from the 2-C-methyl-D-erythritol 4-phosphate pathway to hemiterpene glycosides mediates stress responses in Arabidopsis thaliana. Plant J. 82:122–137. [DOI] [PubMed] [Google Scholar]
- Grubb CD, and Abel S (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11:89–100. [DOI] [PubMed] [Google Scholar]
- Hakenjos JP, Bejai S, Ranftl Q, Behringer C, Vlot AC, Absmanner B, Hammes U, Heinzlmeir S, Küster B, and Schwechheimer C (2013). ML3 is a NEDD8- and ubiquitin-modified protein. Plant Physiol. 163:135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 104:6478–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye H, Chadchawan S, Winge P, Thangstad OP, and Bones AM (2002). Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 128:1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S, Zechmann B, Molitor A, Trujillo M, Petutschnig E, Lipka V, Kogel KH, and Schafer P (2011). Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 156:726–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerwin R, Feusier J, Corwin J, Rubin M, Lin C, Muok A, Larson B, Li B, Joseph B, Francisco M, et al. (2015). Natural genetic variation in Arabidopsis thaliana defense metabolism genes modulates field fitness. Elife 4 10.7554/eLife.05604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, and Jander G (2007). Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 49:1008–1019. [DOI] [PubMed] [Google Scholar]
- Kissen R, Rossiter JT, and Bones AM (2009). The ‘mustard oil bomb’: not so easy to assemble?! Localization, expression and distribution of the components of the myrosinase enzyme system. Phytochem. Rev 8:69–86. [Google Scholar]
- Kliebenstein DJ, Figuth A, and Mitchell-Olds T (2002). Genetic architecture of plastic methyl jasmonate responses in Arabidopsis thaliana. Genetics 161:1685–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, and Mitchell-Olds T (2001). Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126:811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva OA, Davies A, Deeken R, Thorpe MR, Tomos AD, and Hedrich R (2000). Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol. 124:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrix V, Reichelt M, Mitchell-Olds T, Kliebenstein DJ, and Gershenzon J (2001). The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia ni herbivory. Plant Cell 13:2793–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, and Hwang I (2006). Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126:1109–1120. [DOI] [PubMed] [Google Scholar]
- Lemos M, Xiao Y, Bjornson M, Wang JZ, Hicks D, Souza A, Wang CQ, Yang P, Ma S, Dinesh-Kumar S, et al. (2016). The plastidial retrograde signal methyl erythritol cyclopyrophosphate is a regulator of salicylic acid and jasmonic acid crosstalk. J. Exp. Bot 67:1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gaudinier A, Tang M, Taylor-Tee pies M, Nham NT, Ghaffari C, Benson DS, Steinmann M, Gray JA, Brady SM, et al. (2014). Promoter-based integration in plant defense regulation. Plant Physiol. 166:1803–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Hayashi Y, Kondo M, Shimada T, Nishimura M, and Hara-Nishimura I (2002). An endoplasmic reticulum-derived structure that is induced under stress conditions in Arabidopsis. Plant Physiol. 130:1807–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Hayashi Y, Yamada K, Shimada T, Nishimura M, and Hara-Nishimura I (2003). The ER body, a novel endoplasmic reticulum-derived structure in Arabidopsis. Plant Cell Physiol. 44:661–666. [DOI] [PubMed] [Google Scholar]
- Matsushima R, Fukao Y, Nishimura M, and Hara-Nishimura I (2004). NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. Plant Cell 16:1536–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewis I, Tokuhisa JG, Schultz JC, Appel HM, Ulrichs C, and Gershenzon J (2006). Gene expression and glucosinolate accumulation in Arabidopsis thaliana in response to generalist and specialist herbivores of different feeding guilds and the role of defense signaling pathways. Phytochemistry 67:2450–2462. [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, and Halkier BA (2003). Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 131:298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano AJ, Matsushima R, and Hara-Nishimura I (2005). Activation of an ER-body-localized beta-glucosidase via a cytosolic binding partner in damaged tissues of Arabidopsis thaliana. Plant Cell Physiol. 46:1140–1148. [DOI] [PubMed] [Google Scholar]
- Nagano AJ, Fukao Y, Fujiwara M, Nishimura M, and Hara-Nishimura I (2008). Antagonistic jacalin-related lectins regulate the size of ER body-type beta-glucosidase complexes in Arabidopsis thaliana. Plant Cell Physiol. 49:969–980. [DOI] [PubMed] [Google Scholar]
- Nakano RT, Matsushima R, Nagano AJ, Fukao Y, Fujiwara M, Kondo M, Nishimura M, and Hara-Nishimura I (2012). ERM03/MVP1/G0LD36 is involved in a cell type-specific mechanism for maintaining ER morphology in Arabidopsis thaliana. PLoS One 7:e49103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano RT, Yamada K, Bednarek P, Nishimura M, and Hara-Nishimura I (2014). ER bodies in plants of the Brassicales order: biogenesis and association with innate immunity. Front. Plant Sci. 5:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano RT, Pislewska-Bednarek M, Yamada K, Edger PP, Miyahara M, Kondo M, Böttcher C, Mori M, Nishimura M, Schulze-Lefert P, et al. (2017). PYK10 myrosinase reveals a functional coordination between endoplasmic reticulum bodies and glucosinolates in Arabidopsis thaliana. Plant J. 89:204–220. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, and Nebenfuhr A (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51:1126–1136. [DOI] [PubMed] [Google Scholar]
- Ogasawara K, Yamada K, Christeller JT, Kondo M, Hatsugai N, Hara-Nishimura I, and Nishimura M (2009). Constitutive and inducible ER bodies of Arabidopsis thaliana accumulate distinct beta-glucosidases. Plant Cell Physiol. 50:480–488. [DOI] [PubMed] [Google Scholar]
- Petersen BL, Chen S, Hansen CH, Olsen CE, and Halkier BA (2002). Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 214:562–571. [DOI] [PubMed] [Google Scholar]
- Pfalz M, Vogel H, and Kroymann J (2009). The gene controlling the indole glucosinolate modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21:985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savchenko T, and Dehesh K (2014). Drought stress modulates oxylipin signature by eliciting 12-OPDA as a potent regulator of stomatal aperture. Plant Signal Behav. 9:e28304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savchenko T, Kolia VA, Wang CQ, Nasafi Z, Hicks DR, Phadungchob B, Chehab WE, Brandizzi F, Froehlich J, and Dehesh K (2014). Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol. 164:1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F, Fernandez-Calvo P, Zander M, Diez-Diaz M, Fonseca S, Glauser G, Lewsey MG, Ecker JR, Solano R, and Reymond P (2013). Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25:3117–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zeng Y, Zhuang X, Sun L, Yao X, Pimpl P, and Jiang L (2013). Organelle pH in the Arabidopsis endomembrane system. Mol. Plant 6:1419–1437. [DOI] [PubMed] [Google Scholar]
- Sherameti I, Venus Y, Drzewiecki C, Tripathi S, Dan VM, Nitz I, Varma A, Grundier FM, and Oelmuller R (2008). PYK10, a beta-glucosidase located in the endoplasmatic reticulum, is crucial for the beneficial interaction between Arabidopsis thaliana and the endophytic fungus Piriformospora indica. Plant J. 54:428–439. [DOI] [PubMed] [Google Scholar]
- Shroff R, Vergara F, Muck A, Svatos A, and Gershenzon J (2008). Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. USA 105:6196–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonderby IE, Burow M, Rowe HC, Kliebenstein DJ, and Halkier BA (2010a). A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiol. 153:348–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonderby IE, Geu-Flores F, and Halkier BA (2010b). Biosynthesis of glucosinolates-gene discovery and beyond. Trends Plant Sci. 15:283–290. [DOI] [PubMed] [Google Scholar]
- Ueda H, Nishiyama C, Shimada T, Koumoto Y, Hayashi Y, Kondo M, Takahashi T, Ohtomo I, Nishimura M, and Hara- Nishimura I (2006). AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol. 47:164–175. [DOI] [PubMed] [Google Scholar]
- Walley J, Xiao Y, Wang JZ, Baidoo EE, Keasling JD, Shen Z, Briggs SP, and Dehesh K (2015). Plastid-produced interorgannellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 112:6212–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng JK, Philippe RN, and Noel JP (2012). The rise of chemodiversity in plants. Science 336:1667–1670. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, and Ausubel FM (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414:562–565. [DOI] [PubMed] [Google Scholar]
- Wittstock U, and Burow M (2010). Glucosinolate breakdown in Arabidopsis: mechanism, regulation and biological significance. Arabidopsis Book 8:e0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EE, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, and Dehesh K (2012). Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149:1525–1535. [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, and Turner JG (1998). COM: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280:1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu J, Meng J, Meng X, Zhao Y, Liu J, Sun T, Liu Y, Wang Q, and Zhang S (2016). Pathogen-responsive MPK3 and MPK6 reprogram the biosynthesis of indole glucosinolates and their derivatives in Arabidopsis immunity. Plant Cell 28:1144–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Nagano AJ, Nishina M, Hara-Nishimura I, and Nishimura M (2008). NAI2 is an endoplasmic reticulum body component that enables ER body formation in Arabidopsis thaliana. Plant Cell 20:2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Nagano AJ, Ogasawara K, Hara-Nishimura I, and Nishimura M (2009). The ER body, a new organelle in Arabidopsis thaliana, requires NAI2 for its formation and accumulates specific beta-glucosidases. Plant Signal Behav. 4:849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Nagano AJ, Nishina M, Hara-Nishimura I, and Nishimura M (2013). Identification of two novel endoplasmic reticulum body-specific integral membrane proteins. Plant Physiol. 161:108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ober JA, and Kliebenstein DJ (2006). The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 18:1524–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


