Abstract
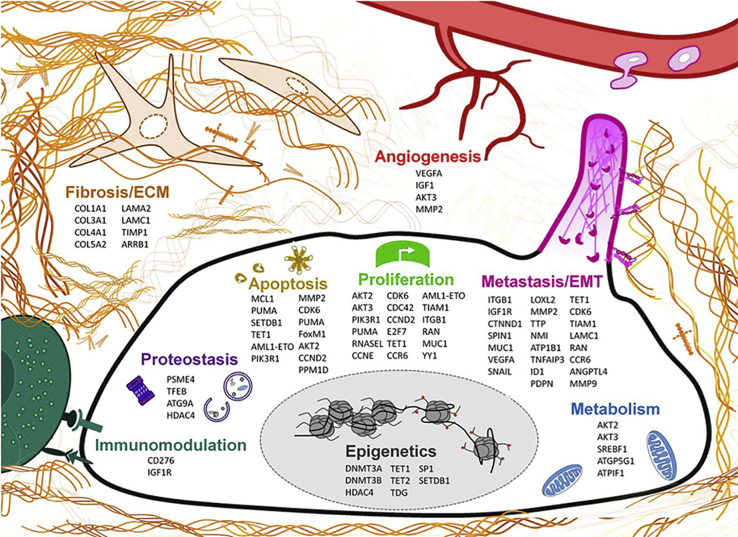
MicroRNAs (miRNA) are small non-coding RNAs (∼22 nt in length) that are known as potent master regulators of eukaryotic gene expression. miRNAs have been shown to play a critical role in cancer pathogenesis, and the misregulation of miRNAs is a well-known feature of cancer. In recent years, miR-29 has emerged as a critical miRNA in various cancers, and it has been shown to regulate multiple oncogenic processes, including epigenetics, proteostasis, metabolism, proliferation, apoptosis, metastasis, fibrosis, angiogenesis, and immunomodulation. Although miR-29 has been thoroughly documented as a tumor suppressor in the majority of studies, some controversy remains with conflicting reports of miR-29 as an oncogene. In this review, we provide a systematic overview of miR-29’s functional role in various mechanisms of cancer and introspection on the contradictory roles of miR-29.
Main Text
MicroRNAs (miRNAs) are small non-coding RNAs, approximately 22 nt in length, that are known as powerful regulators of gene expression in eukaryotes. Since the first miRNA, lin-4, was reported in 1993,1 the field of miRNA biology has exploded in the past quarter century, as revelations were made that these minute RNAs had colossal implications in a multitude of physiological processes and diseases. In the context of cancer pathogenesis, miR-15a/miR-16 was the first miRNA cluster found to be aberrantly regulated, as their encoding genomic region was found to be deleted in chronic lymphocytic leukemia (CLL).2 Since then, dysregulated miRNA signatures have become a well-established feature of various cancers. Among dozens of miRNAs that have been reported to be abnormally expressed in cancer, miR-29 has been recognized as one of the critical miRNAs that play a role in cancer pathogenesis.
miR-29 has been shown to have an important role in a multitude of pathophysiological processes, ranging from cardiovascular3 to retinal functions4 and even Alzheimer disease (AD).5 An increasing number of in vivo and in vitro studies have demonstrated miR-29 to exhibit strong anti-fibrotic activity by negative regulation of mRNAs encoding extracellular matrix (ECM) proteins, such as collagen type I, alpha 1 and 2 (COL1A1, COL1A2), collagen type III alpha 1 (COL3A1), elastin (ELN), and fibrillin 1 (FBN1), which play essential roles in matrix deposition, epithelial-mesenchymal transition (EMT), and the progression of fibrosis.6, 7 Consistently, miR-29 is significantly downregulated in human fibrotic disorders of multiple organs. For example, in cardiac, pulmonary, hepatic, and renal fibroses, miR-29 is found to be downregulated by transforming growth factor β (TGF-β)-SMAD signaling, which in turn results in enhanced expression of the collagen proteins, promoting pathogenesis of the disease.7, 8, 9, 10, 11, 12 In hepatic stellate cells (HSCs), key contributors of collagen production and fibrogenic reactions in liver, miR-29 overexpression markedly inhibits the increased expression of HSC-activating genes α-SMA, DDR2, FN1, ITGB1, and platelet-derived growth factor receptor (PDGFR)-β in vitro.13 Taken together, deregulation of miR-29 is associated with conditions such as myocardial fibrosis, cardiac hypertrophy, congestive heart failure, chronic hepatic injury, hepatitis C virus (HCV) infection and inflammation, hypertensive and diabetic nephropathies, and chronic kidney diseases.3, 12, 14, 15, 16, 17, 18
In addition to fibrogenic disorders, miR-29 is reported to negatively regulate insulin signaling via the inhibition of insulin receptor substrate 1, phosphoinositide 3 kinase, and hexokinase 2, thereby playing a pivotal role in glucose and fatty acid metabolism and type 2 diabetes.19, 20 Furthermore, recent studies have delineated the role of miR-29 in neurodegenerative diseases such as AD.21 In AD, patients exhibit high levels of beta-secretase 1 (BACE1), an enzyme responsible for β-amyloid peptide (Aβ) formation from amyloid precursor protein gene (APP). Loss of miR-29 results in an associated increase in BACE1 expression and Aβ levels, promoting AD pathogenesis.5, 21, 22
In addition to the aforementioned diseases, a large body of literature has demonstrated the significant role of miR-29 in various cancers. The majority of these studies have reported that miR-29 functions as a potent tumor suppressor gene, yet a few other reports have also found oncogenic function of miR-29. To comprehensively summarize the role of miR-29 in cancer, we have curated >150 primary research articles pertaining to miR-29 spanning 13 different cancer types. The search strategy was directed toward English language articles obtained through the PubMed electronic database to identify peer-reviewed articles published and available to date. Here we offer a systematic review synthesizing the current findings of miR-29 and its roles in various mechanisms of cancer.
miRNA Biology and the miR-29 Family
miRNAs are normally transcribed by RNA polymerase II and are commonly embedded in the introns and exons of both coding and non-coding genes.23 Within these transcripts, they form a characteristic hairpin structure and are referred to as primary miRNAs (pri-miRNAs).23 Subsequently, pri-miRNAs are processed by ribonuclease enzyme Drosha in conjunction with a co-factor (DiGeorge syndrome critical region in gene 8 [DGCR8] in humans or Pasha in Drosophila melanogaster and Caenorhabditis elegans), forming the microprocessor complex, which cleaves at the base of the hairpin.24, 25 This liberated 60- to 70-nt hairpin structure is referred to as precursor miRNA (pre-miRNA), which is exported out of the nucleus by the RanGTP-dependent nuclear transport receptor exportin 526, 27 and further processed in the cytoplasm by an endonuclease RNase III enzyme, Dicer, to yield an ∼22-nt long double-stranded mature miRNA composed of a 5 prime (−5p) and a 3 prime (−3p) strand.28, 29 Finally, one of these strands of mature miRNA (passenger strand) is degraded, while the other strand (guide strand or miRNA) is loaded onto the Argonaut proteins (AGOs) to form the RNA-induced silencing complex known as RISC.30, 31 In the case of miR-29, the miR-29-3p arm represents the most abundant and functionally relevant arm of all three family members.32, 33 Following its association within RISC, miRNAs function through a 6-8-mer sequence, known as a seed sequence, that complementarily binds to the 3′ UTR of mRNA transcripts through canonical Watson-Crick base pairing to regulate target expression.23, 27 miRNA biogenesis is a highly complex machinery, and we have provided a very simplified overview of the entire process. A more detailed review of miRNA biogenesis is available for further reading.25, 27, 29, 31
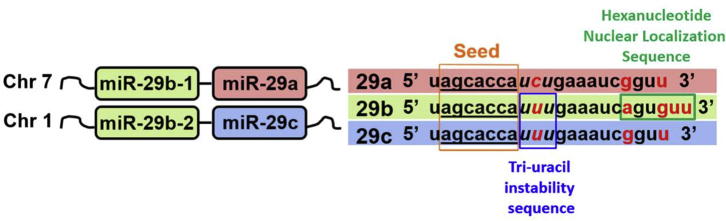
Several miRNAs have common seed sequences and have been known to largely regulate a similar repertoire of target transcripts. Accordingly, evolutionarily conserved miRNAs sharing analogous seed sequences have been organized into 87 distinct miRNA families.34 One of these miRNA families, miR-29, consists of three members, miR-29a, miR-29b, and miR-29c. miR-29a and -29b-1 are encoded on human chromosome 7q32.3, known as the miR-29a/b-1 cluster, and miR-29b-2 and -29c are found on chromosome 1q32.2, designated as the miR-29c/b-2 cluster. While all three family members share a common 7-nt seed sequence, there are unique sequence-functional features of each miR-29 family member that have been reported (Figure 1).
Figure 1.
Schematic Representation of the miR-29 Family Members: miR-29a, -29b, and -29c
miR-29 family members have identical seed sequences (orange box and underlined) along with similar mature miRNA sequences. However, notable differences in nucleotides are indicated in red. Tri-uracil nucleotides at positions 9–11 nt (blue box) present in miR-29b and -29c contribute to instability and shorter half-life, and the hexanucleotide sequence at positions 18–23 nt (green box) is unique to miR-29b, leading to nuclear localization.
One of the notable differences among miR-29 family members resides within a 6-nt segment that is unique to miR-29b (Figure 1). This hexanucleotide sequence present in nucleotide positions 18–23 has been functionally shown to lead to miR-29b nuclear localization.35 In fact, the simple addition of the miR-29b nuclear localization sequence (NLS) to the 3′ end of small interfering RNAs (siRNAs) was found to be sufficient enough to cause nuclear localization.35 Consistently, another report, evaluating the therapeutic efficacy of miR-29b in targeting a subunit of the proteasome, found that the deletion of the hexanucleotide NLS led to cytoplasmic enrichment of miR-29b and further enhanced the downregulation of its target.36 Although miR-29b’s function within the nucleus has yet to be fully elucidated, this initial work has opened the gates to the discovery of various other miRNAs that similarly localize to the nucleus and elicit non-canonical gene regulation beyond binding to the 3′ UTR of mRNA transcripts in the cytoplasm.37, 38 The study of miRNA nuclear function is still active and evolving. A number of reviews are available on this subject matter,39, 40 but, for simplicity, we will focus on the canonical-cytoplasmic function of miR-29 for the remainder of this review.
In addition, miR-29a has a distinct cytosine residue at position 10 (Figure 1). This difference is of particular importance, as uracil residues located in nucleotide positions 9–11 of miRNAs have been found to lead to rapid decay or turnover.41 This tri-uracil sequence is found in both miR-29b and miR-29c, whereas the cytosine residue at nucleotide position 10 of miR-29a lends to its greater stability.41 This is consistent with pulse-chase experiments of synthetic miR-29a and -29b in HeLa cells demonstrating a longer half-life of miR-29a compared to -29b,36 in addition to numerous findings demonstrating that miR-29a is the most abundantly expressed family member.42, 43, 44, 45
In Vivo Function of miR-29
A single miRNA family has been known to have, on average, >400 target transcripts that have matching, evolutionarily conserved 3′ UTR-binding sites.46 Furthermore, >60% of translated genes in the human genome possess at least one miRNA-binding site. Therefore, it is not difficult to imagine that dysregulation of even a single miRNA can profoundly impact normal physiological processes and lead to disease.
In fact, a number of in vivo studies in murine models have demonstrated the essential role of miR-29 in development and general physiology. Whole-body miR-29 knockout (KO) mice have a wide range of developmental defects, such as premature thymic involution,47 muscle wasting,48 growth retardation, and shorter lifespan.49 In addition, liver-specific miR-29 KO mice had a robust increase in hepatic fibrosis and carcinogenesis,16 implicating its physiological relevance.
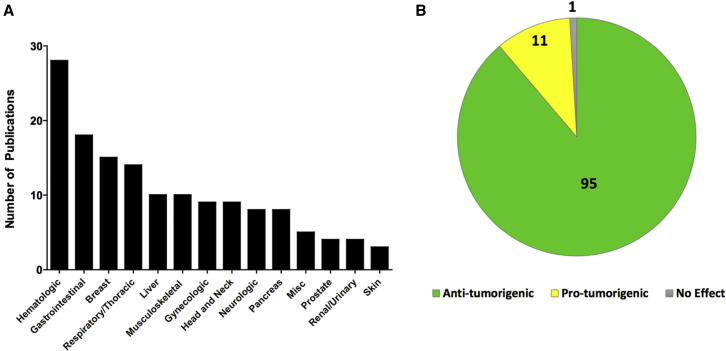
A recent report profiled miR-29a and miR-29c function in the pancreata of whole-body KO mouse models in the context of glucose regulation and diabetes.44 miR-29a KO mice had a defect in insulin secretion, but, interestingly, miR-29c KO mice did not.44 Similarly, miR-29a KO in an insulitis transgenic model (insHEL) resulted in diabetes, whereas wild-type (WT) mice did not.44 Taken together, miR-29a plays a vital role in proper pancreatic function and elicits a protective effect against diabetes in the context of insulitis.44 miR-29 clearly has a critical role in various organs in vivo to help maintain homeostasis and normal function, and its loss leads to developmental problems and disease. Thus, it is not surprising that the misregulation of miR-29 is reported in a variety of different cancer types (Figure 2A) and that it predominantly functions as a tumor suppressor (Figure 2B).
Figure 2.
miR-29 Studies in Cancer Pathogenesis
A systematic curation of English language, primary research articles related to miR-29 in the context of cancer were comprehensively surveyed using the PubMed electronic database in order to gain a better insight into the role of miR-29 in various cancers. (A) Bar graph indicating the number of existing publications based on cancer type, with Miscellaneous (Misc) indicating studies that investigate miR-29 in a broader, multi-cancer context. (B) Pie chart with the numbers of publications demonstrating miR-29 as a tumor suppressor (green), oncogenic (yellow), and having no effect (gray).
Epigenetics
The genome is more than just the simple sequence of its nucleotides. The term epigenetics was originally coined in 1942,50 where the regulatory mechanism of gene expression was considered as a transcending process to explain phenotype. Epigenetics is the study of higher-order regulatory mechanisms influencing the expression of genes without altering the underlying sequence. Since the original conception of the term in 1942, the field of epigenetics has expanded in the past several decades to include a multitude of molecular mechanisms (e.g., histone modifications, DNA methylation, miRNAs, and long non-coding RNAs [lncRNAs]).
DNA methylation is a well-studied mechanism of epigenetic regulation, and it has been shown to be aberrantly regulated in cancer. Methylated DNA effectively silences genes through preventing the association of activator proteins and recruiting methyl-CpG-binding proteins (MBD) to deacetylate neighboring histones. DNA methylation of CpG islands is catalyzed by a class of proteins known as DNA methyltransferases (DNMTs). Furthermore, DNMT family members (DNMT1, DNMT3A, and DNMT3B) have been shown to be elevated in a number of different cancers.51, 52, 53, 54
As one of the earliest implications of miR-29’s role in epigenetic regulation, all three miR-29 family members were found to act as tumor suppressors in lung cancer through the direct targeting of a 3′ UTR-binding site on DNMT3A and DNMT3B transcripts.55 This was of particular interest as DNMT3 was shown to be increased in lung cancer and promote tumor growth by silencing various tumor suppressor genes.51 Further in vivo studies demonstrated miR-29’s ability to effectively repress tumor growth in a xenograft mouse model.55 Various other reports followed, corroborating the tumor-suppressive role of miR-29 through targeting DNMT3s in other cancer contexts (Table S1).
In addition to DNA methylation, the post-translational modification of histones is another major epigenetic regulatory mechanism. Due to the negatively charged phosphate backbone, DNA naturally associates with the positively charged lysine and arginine residues of histone proteins, effectively packaging the DNA into units called nucleosomes.56 In the packed heterochromatic state, the DNA associated with histones is effectively inaccessible, and promoter regions or DNA elements within these regions are turned off. The acetylation of lysine residues on the N-terminal tails of histones can neutralize the positive charge of the side chain group, thus loosening its association with DNA. This leaves the DNA more open to transcriptional regulators. The reaction of acetylating lysine residues of histones is facilitated by a class of enzymes called histone acetyltransferases (HATs). Conversely, the removal of acetyl groups is carried out by histone deacetylases (HDACs). HDACs have been shown to promote oncogenic processes and are considered therapeutic targets in a multitude of cancers.57, 58 In fact, several clinical trials are ongoing in an attempt to implement HDAC inhibitors (HDACi) in combination with other therapeutic agents (ClinicalTrials.gov: NCT01543074 and NCT01486277).
In a study involving multiple myeloma, synthetic miR-29b mimics were shown to effectively inhibit class II HDAC4 expression,59 and the 3′ UTR miR-29-binding site was validated through luciferase reporter assay. Furthermore, miR-29b expression was inversely correlated with HDAC4 expression in various cancer cell lines as well as in patient samples.59 The study went on to demonstrate that miR-29b overexpression caused increased acetylation of histone H4, increased apoptosis, and reduced migratory potential of myeloma cells. Interestingly, HDAC inhibition also led to the upregulation of miR-29b, indicating a feedforward mechanism. Finally, the administration of miR-29b mimics in combination with a pan-HDAC inhibitor, Vorinostat, led to a significant increase in survival and decreased tumor volume in vivo.59
In addition to miR-29 targeting DNMT3 and HDAC4, Thymine DNA glycosylase (TDG) and ten-eleven translocation 1 (TET1) have been indicated as direct targets of miR-29. TDG and TET1 are known to facilitate active demethylation of DNA, where TET1 initiates DNA demethylation, through hydroxylating methylated cytosine,60, 61 and TDG subsequently excises the modified methyl group, effectively demethylating DNA.62
One of the first reports of miR-29 targeting TET1 and TDG was in the human lung adenocarcinoma cell line A549.63 The miR-29-binding sites within the 3′ UTR of TET1 and TDG were validated via luciferase reporter assay.63 Consistently, TET1 and TDG targeting by miR-29, in the context of various cancers through bioinformatics analysis of data, was identified with The Cancer Genome Atlas (TCGA) data.64 Using a statistical method to derive recurrence scores between miRNAs and mRNAs in complement to target prediction, a database of high-confidence miRNA pathway networks for multiple cancer types was created (http://cancerminer.org). As a proof of concept for their analysis, the authors described a potential regulatory role of miR-29 in the DNA demethylation pathway through targeting TET1 as well as TDG.64
Interestingly, TET1 has been shown to act as a potent tumor suppressor in various oncogenic contexts.65, 66 A study in breast cancer cell lines revealed miR-29b overexpression led to increased proliferation, colony formation, and migration through inhibiting TET1 to regulate ZEB2.67 In contrast, thorough analysis of TCGA datasets across a number of cancer types revealed that miR-29a is generally downregulated, and TET1 and TDG are typically upregulated.64 These seemingly contradictory results led to the idea that miR-29’s role in epigenetic regulation as a tumor suppressor versus oncogene may be more subtle and context dependent. However, it is clear from the given studies that TET1 and TDG are bona fide targets of miR-29. More recent work in stem cell differentiation has shown consistent results of miR-29 effectively targeting TET1 in embryonic stem cell differentiation.68, 69 Further work is needed to better understand these paradoxical findings.
Proteostasis
As the cell is challenged with various environmental stresses, maintaining a balance of protein biogenesis and degradation, called proteostasis, is a vital process in preserving cellular homeostasis. The loss of this balance can lead to dysfunction and cell death. Cancerous cells undergo a constant barrage of environmental stresses, including hypoxia and nutrient deprivation, while meeting the demands of oncogenic growth. Therefore, proteostasis is critically important for the survival of transformed cells, and it has been proposed as an appealing therapeutic target. Therapeutic targeting of proteostasis in the context of cancer have been covered in several reviews.70, 71, 72, 73 The proteasome and the process of macroautophagy are essential for protein clearance and turnover, and, in recent years, miR-29 has emerged as a novel regulator of proteostasis by targeting several genes that are pertinent to proteasome and autophagy pathways.
The proteasome is a protein complex that hydrolyzes peptide bonds of polyubiquitinated proteins.72 The ubiquitin proteasome pathway (UPP) is vital for regulating protein half-life and degrading damaged or misfolded proteins.72 In complement to the proteasome, macroautophagy, herein referred to as autophagy, is a key cellular process that degrades large macromolecules for the maintenance of cellular homeostasis and survival under stress conditions.74 Autophagy is the process in which cytosolic components are enveloped by double-membrane vesicles, called autophagosomes, and trafficked to the lysosome for degradation and recycling.75 Subsequently, the hydrolases of the lysosomal compartments degrade cytoplasmic cargo and release the degraded components into the cytosol for reuse.75 Recent studies document that the upregulation of autophagy can serve as a survival mechanism in various malignancies.76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87 As cancer frequently relies upon the UPP and autophagy, the development of proteasome inhibitors and small molecule inhibitors of autophagy for cancer therapy is an active area of research.
The first indication of miR-29’s role in proteostasis was reported in the context of chemosensitizing myeloma to bortezomib, a therapeutic proteasome inhibitor.36 Initially, there was an aim to screen several thousands of miRNAs differentially expressed in bortezomib-resistant cells, and miR-29a, 29b-1, miR-29b-2, and miR-29c were all identified as significantly downregulated miRNAs. Upon the inhibition of miR-29b and miR-29c using antagomirs, the viability of bortezomib-treated myeloma cells was found to be increased. miRNA target prediction algorithms revealed a potential target involved in proteasome activation called PSME4 as a direct target of miR-29b. Finally, utilizing transgenic xenograft murine models, miR-29b replacement was shown to decrease tumor volume and increase overall survival. Although further studies have not investigated miR-29 suppression of PSME4 in the context of cancer, another recent report found similar results in the context of cardiovascular disease, where miR-29b overexpression led to a PSME4 decrease and increased oxidative stress.88
In addition to the proteasome, miR-29 has been shown to play a significant role in regulating autophagy. Initial studies in bovine cells infected with virus revealed endogenous bovine miR-29 had an anti-viral protective effect of the host cell by upregulating miR-29b, which led to the targeting of ATG14 and ATG9A. Bovine diarrhea virus (BVDV) utilized host cell autophagy machinery as a means of promoting viral replication. miR-29b-mediated inhibition of autophagy led to a decrease in BVDV replication.89
Shortly thereafter, miR-29a was shown to have a similar effect inhibiting autophagy in pancreatic cancer cells. Earlier work demonstrated that pancreatic cancer cells heavily upregulated autophagy for survival and that this upregulation also conferred chemoresistance against a nucleoside analog, gemcitabine.87 miR-29a was significantly downregulated in pancreatic cancer cells, and miR-29a overexpression led to increased gemcitabine sensitization in chemoresistant cell lines. Subsequently, miR-29 overexpression led to a late-stage blockage of autophagy, where co-localization of GFP-labeled autophagosomes and lysosomal markers indicated a blockage in autophagosome-lysosome fusion. By utilizing miRNA predictive algorithms, two predicted autophagy-related miR-29 targets were identified, TFEB and ATG9A. TFEB and ATG9A are known to be vital for lysosomal function and autophagosome trafficking, respectively, and they are indispensable for autophagy. Indeed, miR-29 overexpression led to a drastic decrease in expression of both targets. Parallel knockdown of TFEB and ATG9A phenocopied the late-stage blockage of autophagy similar to miR-29 overexpression, but, interestingly, only the knockdown of ATG9A demonstrated a reduction in autophagosome-lysosome fusion.
While initial findings of miR-29b targeting bovine ATG9A were originally reported, we found consistent regulation in human carcinoma. Furthermore, in a study involving the silencing of HDAC4 by miR-29b in multiple myeloma, miR-29 overexpression led to a decrease in TFEB.59 Although postulated that this was an indirect effect of repression through the knockdown of HDAC4, it may be possible they were, in fact, observing a direct targeting of TFEB by miR-29b. However, in subsequent studies, overexpression of miR-29a in myeloma cells had no impact on autophagy flux.59 These results may indicate that miR-29-mediated autophagy inhibition may be context dependent or differ between miR-29a and -29b family members. Further investigation is warranted to determine if miR-29 has an impact on proteasome function in pancreatic cancer.
Oncogenic Metabolism
miR-29 has been implicated in regulating metabolism in numerous tissue types, including β cells, skeletal muscle, and adipocytes,20, 90, 91 and it has been shown to regulate branch-chain amino acid synthesis and insulin secretion.91, 92 Therefore, it is not surprising that miR-29 is also found to have an impact on cancer metabolism. The area of cancer metabolism has been well studied over the past several decades, and many alterations in metabolic processes, including glycolysis, oxidative phosphorylation, and mitochondrial function, have been discovered as a compensatory mechanism of cancer cells to meet the energy demands of oncogenesis.
One of the first studies elucidating the role of miR-29 in cancer metabolism found the downregulation of miR-29b in ovarian cancer, and its restoration led to an inhibition of glycolysis and glucose metabolism in cancer cells by directly targeting AKT2 and AKT3.93 AKT, also known as protein kinase B, is a key hub within the PI3K/AKT/mTOR-signaling pathway. Furthermore, AKT is well known to be activated in tumors and regulates glucose metabolism to promote cancer growth.94, 95 The 3′ UTR miR-29-binding sites of AKT2/3 were validated, and miR-29b overexpression led to decreased tumor formation in vivo.93 Although outside of the metabolism context, various other groups have demonstrated a similar tumor-suppressive function of miR-29 by targeting AKT (Table S1), thus reassuring that AKT is a bona fide target of miR-29.
In addition to facilitating a tumor-suppressive function through regulating glucose metabolism, miR-29 has also been shown to be concertedly regulated in the context of lipid metabolism, and it functions as a tumor suppressor in glioblastoma multiforme (GBM) through an autoregulatory loop involving Sterol regulatory element-binding protein 1 (SREBP-1).96 SREBP-1 is a transcription factor that concertedly regulates genes involved in sterol biosynthesis and glucose and lipid metabolism,97 and it has been known to promote tumorigenesis.98 SREBP-1 was found to suppress miR-29 expression at both loci (miR-29a/b-1 and miR-29c/b-2), and deletion of sterol regulatory element (SRE)-binding sites in the promoters of miR-29 loci derepressed miR-29 expression.96 Interestingly, SREBP1 itself was shown to be a direct target transcript of miR-29, and, ultimately, miR-29 forced expression in GBM led to a significantly increased survival and decreased tumor growth in vivo.
Finally, miR-29 has also been implicated to directly target ATGP5G1 and ATPIF1, two vital subunits of ATP synthase.99 The overexpression of miR-29a/b-1 caused tamoxifen sensitization in tamoxifen-resistant cells.99 Gene ontology analysis of the miR-29a transcriptome in ovarian cancer cells revealed various differentially expressed genes (DEGs) related to oxidative phosphorylation and ATP metabolism.99 Further investigations revealed miR-29a overexpression led to a decreased oxygen consumption rate, mediated by the direct inhibition of ATP5G1 and ATPIF1 expression.99 Although these findings necessitate further in vivo verification, they convey additional evidence of miR-29’s tumor-suppressive role in regulating cancer metabolism.
Proliferation
Uncontrolled proliferation is a hallmark of cancer cells, and several cell cycle regulators are well known to be dysregulated in tumors. Understanding the molecular details of cell cycle regulation and checkpoint abnormalities in cancer have been under intense investigation for the past several years. The targeting of cell cycle control mechanisms has risen as a promising therapeutic strategy.100
The main regulatory proteins that play key roles in controlling cell-cycle progression include cyclins, cyclin-dependent kinases (CDKs), and various cyclin substrates.101 CDK6 plays an important role in the transition of cycling cells into S phase. Studies have reported that cyclin-CDK6 complexes induce cancer cell transition from G1 to S phase by phosphorylation of retinoblastoma (Rb), while CDK6 inhibition blocks the cell cycle and suppresses tumor growth.102, 103 In fact, several CDK6 inhibitors have been developed to target proliferating cancer cells.104 CDK6 has been shown to be a direct target of miR-29 in several malignancies, including mantle cell lymphoma,105 acute myeloid leukemia (AML),106 and cervical cancer.107 For example, CDK6 was shown to be directly targeted by miR-29 in melanoma cell lines. In various melanoma cell lines, miR-29a/b expression inversely correlated to cancer cell proliferation. Previous work had demonstrated an anti-proliferative effect of interferon (IFN)-γ in many cancers.108 Accordingly, IFN-γ as well as miR-29 exhibit anti-proliferative activities in melanoma cells involving downregulation of CDK6.109 Consistently, miR-29 was shown to be involved in Burkitt lymphoma pathogenesis by altering the expression of target genes, including CDK6, DNMT3B, TCL1, and MCL1, which are involved in cell cycle control, DNA methylation, and apoptosis inhibition, respectively.110 Moreover, miR-29 has also been observed to enrich the E7 protein-dependent cell cycle pathway by targeting CDK6, resulting in decreased proliferation of cervical cancer cells.111 Furthermore, cell cycle analysis revealed that miR-29 overexpression caused a significant accumulation of cervical cancer cells at the G1 phase and decreased the number of cells in the S and G2/M phases.112 In schwannoma, miR-29a has been shown to downregulate CDK6 expression and disrupt cell cycle progression by the deactivation of JNK and p38MAPK/ERK pathways. Overexpression of all three members of the miR-29 family in Schwann cells inhibited cell viability, migration, and invasion in vitro.113
Interestingly, a recent study identified a novel oncogenic circular RNA, circRNA_100290, which contained two miR-29-binding sites that acted as a sponge to sequester miR-29 family members in oral squamous cell carcinoma (OSCC).114 The inhibition of miR-29b led to the upregulation of CDK6 expression. Knockdown of circRNA_100290 was found to induce G1/S arrest and inhibit in vitro and in vivo OSCC cell proliferation and reduce CDK6 expression.
Another miR-29 target, cell division cycle 42 (CDC42), is a well-known member of the Ras homolog (Rho) family, and it regulates crucial cellular processes, including cell cycle and cell cytoskeleton organization.115, 116 miR-29a was shown to exert a tumor suppressor role in breast cancer cells by arresting the cell cycle at the G0/G1 phase through the negative regulation of CDC42 expression.117 In corroboration with this study, miR-29 was found to be downregulated in clear cell renal cell carcinoma, and restoration of all three miR-29 family members inhibited cell proliferation along with migration-invasion.117
As covered in the Oncogenic Metabolism section, the AKT kinase family is well known for its role in promoting tumorigenesis. One mechanism by which AKT functions as an oncogene is through promoting cell proliferation by accelerating G2-M phase transition.118 miR-29 has been shown to inhibit myoblast proliferation by targeting AKT3 as well as another proliferation-associated gene, p85α. siRNA-mediated AKT3 knockdown in the C2C12 myoblast cell line was shown to phenocopy miR-29 overexpression, leading to cell-cycle arrest in the G0/G1 stage and a significant decrease in proliferation rate.119 In addition to AKT3, AKT2 and CCND2 have also been shown to be direct targets of miR-29. AKT2 functions as the hub in the PI3K/AKT-signaling pathway,120, 121, 122 and CCND2 is a member of the cyclins, promoting the G1-to-S phase transition through regulating the phosphorylation of Rb.123, 124 The role of miR-29 in significantly inhibiting AML cell proliferation and promotion of apoptosis was attributed to the decrease of these two key signaling molecules.125 Furthermore, in vivo analysis demonstrated that the reintroduction of each miR-29 member could partially correct abnormal cell proliferation and apoptosis repression and mediate myeloid differentiation arrest in AML.125
In addition to CCND2, miR-29 also targets E2F7, as demonstrated in Rhabdomyosarcoma (RMS). E2F7 is essential for cell survival and embryonic development in mice. Furthermore, ectopic expression of E2F7 has been shown to block cell cycle transition, resulting in G1 arrest.126, 127 CCND2 and E2F7 were upregulated in both RMS types, and miR-29 expression was shown to be inversely correlated to these cell cycle genes, suggesting that miR-29 may function as a tumor suppressor in RMS by targeting CCND2 and E2F7. Ultimately, overexpression of miR-29 downregulated the expression of these cell cycle genes and induced partial G1 arrest, leading to decreased cell proliferation.128
Finally, miR-29 also targets FOXM1. FOXM1 is a conservative transcription factor that can mediate cell growth, proliferation, and apoptosis through regulating gene expression.129 A study demonstrated that the overexpression of miR-29 inhibited human leukemia K562 cell growth and proliferation and promoted apoptosis by downregulating FOXM1 expression level.
Apoptosis
The apoptotic pathway is an important pathway in all stages of tumor development and metastasis. Apoptosis is a naturally acquired process that typically plays an important role in the development and life of multicellular organisms, by removal of damaged, aged, or autoimmune cells through a controlled cell death mechanism.130 Apoptosis involves a fine-tuned regulatory mechanism, as it is categorized as a type I form of programmed cell death; therefore, the mechanism of apoptosis is complex and involves numerous signaling pathways. Critical changes may occur at any point along these pathways, leading to tumorigenesis, metastasis, and resistance to anticancer drugs.131, 132 A large body of experimental evidence documents that miRNAs function as important regulators of cancer cell death. In an effort to re-establish miRNAs function in apoptotic pathways, many strategies have been designed to either block the expression of oncomiRs or to boost the expression of tumor suppressor miRNAs.133
miR-29a and -29b have been shown to regulate critical anti-apoptotic genes such as MCL1. MCL1, part of the BCL family, is an anti-apoptotic protein that promotes cancer cell survival and proliferation, and it is frequently overexpressed in cancer, notably in AML.134, 135, 136, 137 miR-29b has been shown to be deregulated in primary AML blasts, and transcriptome analysis after miR-29b overexpression in leukemia cells revealed a tumor suppressor function, in which miR-29 induced apoptosis through directly targeting MCL1, as indicated by Annexin V/propidium iodide (PI) assay and increased caspase activity.138 These results of miR-29b targeting MCL1 to elicit a tumor suppressor effect were consistent in vivo.138 In addition, an inverse relationship of miR-29 and MCL-1 expression in the transformation of myelodysplastic syndromes (MDS) to overt leukemia (OL) was observed.138 Furthermore, miR-29a has been shown to have anti-invasive and anti-proliferative effects on lung cancer cells in vitro, which can be explained in part by the ability of the miR-29 family to downregulate Mcl-1 activity and target RAN, a member of the RAS oncogene family.139
miR-29 also regulates environmental signals like growth factors that have an extrinsic impact on cell cycle control. It was found that the knockdown of miR-29 inhibited proliferation and induced apoptosis in MG-63 osteosarcoma cells by TGF-β1/PUMA signaling. PUMA is a BH3-only member of the Bcl-2 family and a target of p53-mediated apoptosis.140 It activates an apoptotic cascade by facilitating Bax activation, causing cytochrome c release from the mitochondria, caspase-3 activation, and DNA fragmentation.141, 142 It was also found that miR-29 could target pro-apoptotic PUMA protein and protect against ischemia-reperfusion injury, suggesting that PUMA might be negatively regulated by miR-29. Some studies have reported that PUMA was a direct TGF-β target gene in B cells and that TGF-β induces PUMA to aid induction of the intrinsic cell death pathway.143 Furthermore, this study also demonstrated the tumor suppressor effect of miR-29 in vivo.144
miR-29 also affects proliferation through epigenetic control. miR-29 was identified as a negative regulator of SETDB1, thereby reducing hepatocellular carcinoma (HCC) cell proliferation in vitro and suppressing orthotopic tumorigenicity in vivo. Downregulation of miR-29 expression in human HCC contributed to SETDB1 upregulation by relieving its post-transcriptional regulation. The biological reasons behind this global upregulation of epigenetic regulators in human HCC remain to be elucidated. One possible explanation is that cancer cells are actively dividing and undergoing more dynamic epigenetic reprogramming during cell cycles, and, therefore, they may require increasing amounts of epigenetic regulators to sustain their continuous growth and clonal evolution.145 From these studies, it is becoming clear that the miR-29 family plays an anti-apoptotic role in several cancers by targeting key regulators in apoptotic pathways.
Metastasis and EMT
Metastasis occurs when cancer cells undergo EMT, a cellular reprogramming event in which epithelial cells revert to a pseudo-stem cell state that is characterized by a loss of polarity and increased migratory behavior.146 EMT plays a vital role in normal physiology during development as well as in wound healing, but it is also exhibited in cancer metastasis, where a subset of tumor cells disseminates to distant organ sites beyond the site of origination. As a defining signature of stage IV cancer, metastasis is regarded as a major cause of cancer mortality.147 This is largely because, by the time cancer has metastasized, surgical resection is typically no longer a viable option and metastatic tumors are often refractory to conventional therapies. miR-29 has been implicated to inhibit a number of genes that are involved in EMT and metastasis, even causing the reversion of mesenchymal-epithelial transition (MET) in some cases. However, in contradiction, miR-29 itself has also been shown to directly elicit EMT in a minority of cases.
An initial study in gastric cancers revealed δ-catenin (CTNND1) as a target of miR-29.148 δ-catenin plays an important role in cell adhesion between cells and mediates metastatic signal transduction. Mislocalization of δ-catenin in the cytoplasm or nucleus resulted in the advancement of migration and invasion via regulating Rho GTPase activity and growth factor receptor signaling, implying CTNND1’s role as an oncogene.149, 150 A follow-up study in gastric cancer found that the loss of miR-29 contributed to chemoresistance, and overexpression of all three miR-29 family members directly inhibited CTNND1, leading to an increased F-actin and Cofilin phosphorylation via activated RhoA.151 As the F-actin/RhoA pathway is known to influence cell migration, in vitro migration assays demonstrated that miR-29c potently inhibited the migratory potential of gastric cancer cells. Consistently, miR-29c inhibited gastric cancer cell metastasis in vivo as well.151 These results confirm that miR-29 controls gastric cancer cell movement by suppressing the catenin-δ pathway. Similarly, another group also reported that miR-29c/b-2 delivery via recombinant adenovirus decreased gastric cancer cell proliferation and migration in vitro.152
Dysregulation of miR-29 s also affects cancer cell invasion and migration by the activation of several oncogenic pathways, such as the Wnt/β-catenin pathway. For example, miR-29a overexpression led to decreased β-catenin expression and cell proliferation in non-small-cell lung cancer (NSCLC).153 miR-29b was shown to inhibit both Wnt/β-catenin and AKT signaling by downregulating SPIN1 in triple-negative breast cancer (TNBC), and miR-29b overexpression decreased in vitro TNBC cell growth, self-renewal, migration, and invasiveness by inhibiting the aforementioned pathways.154 On the other hand, miR-29 has also been shown as a novel target of β-catenin/Dicer. β-catenin represses Dicer, a key component of the miRNA-processing machinery. Silencing of β-catenin or overexpressing Dicer or miR-29 mimics in highly metastatic ovarian cancer led to significantly reduced migration of cancer cells in vitro.155
A similar anti-metastatic role of miR-29 has been found in HCC, where miR-29a overexpression in HepG2 cells resulted in a significant decrease in migration in vitro. Prior reports have implicated a role for insulin-like growth factor (IGF) and type 1 IGF receptor (IGF-1R) in metastasis.156 The study went on to validate miR-29a’s direct repression of IGF-1R expression.157 Taken together, miR-29a-mediated inhibition of IGF1R exhibited a dual tumor suppressor role in HCC by suppressing HCC cell migration while simultaneously increasing CD8+ T lymphocyte migration.157 This secondary role of increasing immune cell migration is further covered in the Immunomodulation section.
It is important to note that, in addition to internal reprogramming from EMT, the tumor microenvironment can promote and facilitate metastasis. Different cancers tend to spread to specific organ sites, lending to the seed and soil hypothesis of metastatic microenvironments. Overexpression of ECM components, the major stromal proteins in cancer, is observed in several tumor types, and it contributes to cancer cell progression by the dysregulation of cell adhesion, polarity, and structural remodeling through ECM-modifying enzymes.
miR-29 has been extensively shown to target integrin beta-1 (ITGB1).158 ITGB1 is part of the integrin family of cell surface receptors that have been well established to promote cell growth, migration, and tumor metastasis in a majority of cancers.159 Integrins typically heterodimerize in alpha/beta pairs and attach to ECM proteins, which serve as tracks for chemotaxis.160 All three miR-29 family members have been shown to act as potent tumor suppressors in several cancer cell lines, by directly inhibiting ITGB1 and, thereby, inhibiting migration and invasion (Table S1).
The lysyl oxidase (LOX) protein family functions in covalent crosslinking of collagen and/or elastin in the ECM. Dysregulation of LOXL2 has been reported to correlate with disease progression in several diseases and cancers. Furthermore, LOXL2 interacts with SNAIL1 transcription factor and represses E-cadherin along with inducing EMT. miR-29 has been shown to have a tumor-suppressive role by negatively regulating LOXL2 expression and inhibiting cancer cell migration and invasion in renal cell carcinoma in vitro.117 Moreover, LOXL2 was shown to enhance cancer cell invasion in vitro and promote the tumor microenvironment and metastatic niche formation in HCC, where it is activated by HIF-1α/SMAD4 and negatively regulated by miR-26 and miR-29.161
Metallomatrix protease 2 (MMP2) is another ECM-modifying enzyme that has been shown to promote metastasis and was found to be a direct target of miR-29. MMP2 plays a critical role in cancer metastasis, and the effects of MMPs on the ECM are well established.159 Moreover, MMP2 is overexpressed in a variety of primary malignancies, and it is considered an appealing therapeutic target.162 miR-29c has been shown to target MMP2 expression, causing decreased pancreatic cancer cell invasion and metastasis in vitro and in an orthotopic implantation model in nude mice. Moreover, miR-29 expression was also found to inversely correlate with MMP2 expression, metastasis, and survival in pancreatic cancer patients.163 Additionally, the other members of the miR-29 family also regulate MMP2 expression.164 Similarly, miR-29b suppresses invasion and metastasis by downregulating MMP2 in HCC, OSCC, and colorectal cancer (CRC) (Table S1).
Similar to the previous reports on miR-29c, miR-29a overexpression was also found to inhibit migration/invasion in pancreatic cancer cell lines,165, 166 but through different mechanisms. One study found that miR-29a inhibited metastatic potential through targeting membrane-bound mucin (MUC1).166 MUC1 is an epithelial marker of polarized epithelial cells,167, 168 but aberrant overexpression of MUC1 in cancer cells is correlated with an induction of EMT through the Wnt/β-catenin-signaling pathway.169 miR-29a was shown to directly inhibit MUC1 expression, thereby deregulating signaling pathways promoting EMT.166 Further studies on the anti-metastatic effects of miR-29 have been linked to its direct inhibition of genes encoding ECM proteins. These related targets are covered more extensively in the following Fibrosis/ECM section.
Finally, GATA3 is a well-studied transcription factor to have potent tumor-suppressive function in various cancers, and it is known to counteract metastasis by inducing MET.170 In breast cancer cells, GATA3 mediated its tumor-suppressive role through the upregulation of miR-29b.171 Through an extensive series of experiments, the impact of miR-29b in breast cancer was thoroughly examined, and it revealed that miR-29b inhibition led to a drastic decrease in various epithelial markers, along with increases in mesenchymal markers.171 Furthermore, miR-29 inhibition in vivo resulted in a significant increase in lung metastasis.171 Most impressively, the study went on to show that miR-29 potently inhibited 15 different targets, including Angiopoietin-like proteins, integrins, LOXL2, MMP, PDGF, TGFB, and vascular endothelial growth factor A (VEGFA), which are all known to influence metastasis and promote tumor microenvironment progression.171 In summation, miR-29 functions epistatically to GATA3 as a potent master regulator to inhibit metastasis in breast cancer.
In contrast to a large body of experimental evidence lending to the anti-metastatic function of miR-29, aberrant expression of the miR-29 family has been reported to induce migration/invasion in certain malignant contexts as well. In pancreatic cancer, miR-29a was reported to act as an oncogene by downregulating tristetraprolin (TTP), whose activity is known to be regulated by phosphorylation through the p38 MAPK and ERK-MAPK pathways. Downregulation of TTP increased the expression of pro-inflammatory factors and EMT markers,172 and restoration of TTP decreased cell viability and migration in vitro while inhibiting tumor growth and EMT phenotype in vivo.172 Furthermore, miR-29 was also shown to negatively regulate EMT regulator N-myc interactor (NMI) in breast cancer. Increased levels of miR-29 in breast cancer cells inhibited NMI expression, leading to increased cancer cell invasion and promotion of the EMT phenotype.173 With the absence of NMI, inactivation of GSK3β leads to miR-29 upregulation through unrestricted Wnt/β-catenin signaling.173 Moreover, in breast cancer, progestin-regulated miR-29 can control progesterone receptor (PR) action by targeting progestin-responsive genes such as ATP1B1 as well as controlling PGR expression itself.174 Downregulation of miR-29 relieves the repression of ATP1B1, allowing it to adjust the progestin response.174 ATP1B1 limited cell motility, and knockdown of ATP1B1 led to increased breast cancer cell migration and invasion in vitro.174
It is perplexing that miR-29 can have such stark contrasting effects on metastasis reported within the same cancer types. These confounding incongruities warrant further studies to better understand these seemingly contradictory results.
Fibrosis/ECM
The ECM is a vital structure that has a dynamic and complex organization, and it can trigger multiple biological activities that are essential for normal organ development and tissue homeostasis. In mammals, the ECM is composed of about 300 proteins known as the core matrisome, which comprises proteins such as collagens; proteoglycans; and glycoproteins such as laminin, elastin, and fibronectin. Dysregulated ECM remodeling leads to many diseases, namely, fibrosis, which also increases the risk of cancer. Fibrosis is a complex process that involves excessive deposition and reorganization of the ECM, leading to EMT.175 Primarily, fibrosis is mediated by cancer cells that activate fibroblasts, where activated fibroblasts then further perpetuate sustained activation of themselves and other neighboring fibroblasts175 and lead to fibrotic stromal reaction. Stroma is known to impair drug delivery to the tumor core, promoting tumor progression and metastasis. Consequently, many targeted therapies sought to completely ablate the stroma. However, increasing evidence suggests a greater level of complexity to the story, as the stroma was shown to restrain tumor growth in preclinical studies.176, 177 Recent work has demonstrated that the reductionist approach of all or nothing may not be the appropriate means of targeting the stroma, rather a more subtle degree of crosslinking and stromal density may be key to having a therapeutic impact.178 With the failure of current stroma-depleting therapeutic strategies, efforts are now focused on developing novel stroma-targeted therapies that appropriately control the fibrotic stroma.
PDGFs and TGF-β are known to induce fibrosis by causing the proliferation of fibroblasts that differentiate into myofibroblasts and produce fibrotic ECM proteins. TGF-β1 is a well-known proinflammatory growth factor that is associated with the pathogenesis of several cancers. TGF-β1 binds to TGF-β receptors (type I and type II) to form an activated hetero-tetramer with serine-threonine kinase activity, which phosphorylates downstream transcription factors SMAD2 and SMAD3. Upon phosphorylation and activation, pSMAD2/3 form a heterogeneous complex with SMAD4, translocate into the nucleus, and directly regulate target gene expression. Several of these target genes include ECM proteins, such as COL1A1, COL3A1, and TIMP1, as well as about 60 other ECM-related genes.
Several studies have implicated that miRNAs function as potent regulators of the ECM, and, in particular, the miR-29 family has been well established as a potent anti-fibrotic miRNA. Initial studies found that miR-29 restoration quelled fibrosis and ECM protein production in the context of numerous organs, including cardiac,6 liver,179 lung,10 and kidney180 fibroses. Moreover, TGF-β signaling is responsible for the decreased miR-29 expression during fibrosis. Hence, targeting the miR-29 family and TGF-β may be an effective strategy for the treatment and management of fibrosis.181, 182 The interaction between the miR-29 family and TGF-β was revealed in lung and cardiac fibroses.6, 8 After treatment of TGF-β1 on miR-29 knockdown IMR-90 cells,8 certain genes were confirmed to be upregulated by TGF-β, and these genes mainly consisted of miR-29-predicted targets, such as collagens. Furthermore, the stimulation of TGF-β1 and knockdown of miR-29 exhibited similar behavior in lung cells, and they led to the upregulation of specific fibrotic genes, such as COL1A1, COL3A1, and COL1A2. Various laminins, integrins, MMPs, and ADAMs are upregulated with miR-29 downregulation as well, but not with TGF-β1 stimulation.8 Therefore, these observations show that the miR-29 family regulates gene expression via TGF-β-dependent and -independent signaling pathways.
More recent studies revealed how miR-29 mediates TGF-β1 pathways by targeting a variety of ECM network genes. For example, Hsp47, a collagen-binding protein and collagen-specific chaperone, is a hub of the ECM network and controls the tumor microenvironment by the deposition regulation of several ECM proteins. A transcription network analysis showed that Hsp47 expression was activated during breast cancer development and progression, while Hsp47 silencing reprogrammed human breast cancer cells to form growth-arrested and/or non-invasive structures in 3D cultures, and, moreover, it restricted tumor growth in xenograft assays by decreasing the deposition of collagen (COL1A1 and COL4A1) and fibronectin (FN1). Co-expression network analysis showed miR-29b and -29c were inversely correlated with Hsp47 expression and other ECM network genes. In summary, in an in vitro and in vivo breast cancer study, miR-29 mediated TGF-β1-induced Hsp47 expression, promoting breast cancer by increased ECM deposition.183 An interesting study found that knockdown of miR-29 enhanced the ionizing radiation-induced expression of type I collagen through the TGF-β-Smad3-signaling pathway in irradiated cells. Inhibition of TGF-β-Smad3 signaling blocked the significant loss of miR-29, and miR-29 overexpression inhibited the ionizing radiation-induced expression of type I collagen, suggesting that miR-29 may be an important regulator of radiation-induced fibrosis (RIF).184 However, further studies are required to dissect the role of miR-29 in molecular mechanisms associated with radiation.
Moreover, miR-29 downregulation leads to increased ECM protein expression levels of collagens and laminins, as observed in a number of cancers. Based on miRNA-mRNA covariation and sequence-based target predictions, miR-29a/c were identified as novel regulators of LAMA2, an ECM protein associated with poor prognosis in posterior fossa (PF) ependymoma. Decreased miR-29a/c expression correlated with elevated LAMA2 expression are features of PF ependymoma post-transcriptional regulation.185 Moreover, the miR-29 family was identified to have one of the strongest interactions between DEGs in an interaction network derived from gene expression profile data (GEO: GSE12452) of nasopharyngeal carcinoma (NPC) tissue specimens along with an miRNA-sequencing dataset (GEO: GSE14738). COL3A1, COL1A1, COL4A1, and COL5A2 were found to be regulated by several miR-29 family members, suggesting that miR-29 may be related to the development of NPC by the regulation of these genes involved in ECM-receptor interaction.186
Similarly, in a study aiming to explore the regulatory mechanism of CRC and potential novel biomarkers for screening, COL1A1 was predicted to be targeted by miR-29 from examining the protein-protein interaction network (PPI) from microarray data (GEO: GSE44861).187 In 2013, a study found that the reduction of miR-29 expression led to increased cisplatin resistance of ovarian cancer cells, partly through upregulating the expression of ECM components such as COL1A1 in vitro and in vivo and also via increasing the activation of ERK1/2 and inactivation of GSK3β. Therefore, the downregulation of miR-29 in ovarian cancer cells manipulates the surrounding ECM to enhance survival signal transduction upon cisplatin treatment. Ectopic expression of miR-29 alone or in combination with cisplatin effectively reduced tumorigenicity of CP70 ovarian cancer cells in vivo, suggesting that miR-29 overexpression may have therapeutic implications as a potential sensitizer to cisplatin treatment by remodeling the ECM.188 Furthermore, a recent study displayed the role of the miR-29 family in preserving cardiac health in the regulation of age-dependent increases of oxidative stress and cardiac fibrosis. It was found that the miR-29 family is one of the most upregulated miRNAs during aging, and its increase induces the downregulation of known targets such as collagens, DNMTs, and 5′methylcytosine (5mC). In addition, under hypoxic conditions, miR-29a/-29b downregulation was found to be responsible for collagen deposition and fibrosis. Therefore, the miR-29 family upregulation may play a role as an endogenous mechanism against cardiac fibrosis and hypertrophy in age-dependent cardiac damage.189
Pancreatic cancer is notoriously known to be one of the most fibrotic cancers, and its associated stroma has been shown to play an integral role in tumor progression and resistance to therapy.190 Pancreatic ductal adenocarcinoma (PDAC) is unique in that the degree of intratumoral fibrosis is so extreme in many cases that the bulk of the tumor is made up of the stromal compartment.190 Hence, pancreatic cancer frequently serves as a surrogate in studying cancer-stroma interactions. In PDAC tumors, pancreatic stellate cells (PSCs) are mainly responsible for the production of ECM proteins and the dense fibrotic stroma surrounding the tumor bed. Cancer cells secrete a slew of pro-inflammatory growth factors and cytokines, including TGF-β1, which activate PSCs. In turn, activated PSCs and fibroblasts produce pro-inflammatory growth factors and chemokines that act in an autocrine fashion to maintain their sustained activity and produce fibrotic stromal ECM proteins.191 In recent years, we found a significant downregulation of miR-29a and miR-29b in TGF-β1-activated PSCs, fibroblasts, as well as cancer-associated fibroblasts (CAFs).45 Furthermore, we found that miR-29 was significantly downregulated in PSCs of a KRAS mutant genetically engineered mouse model of pancreatic cancer (KC) as well as in PDAC patient biopsies via miRNA in situ hybridization.45 Restoration of miR-29a or -29b in activated PSCs caused a drastic decrease in the expression of various ECM proteins, and it reduced cancer colony growth in co-culture.45 Taken together, our studies indicate that miR-29 may have therapeutic relevance beyond just the cancer cells themselves. Further studies may reveal miR-29 to be a potent anti-tumorigenic agent in other stromal cells as well.
miR-29 has also been observed to target genes outside of the ECM network to provide protective effects against fibrosis. Arrestin beta 1 (ARRB1) is a member of the arrestin and beta-arrestin protein family thought to participate in agonist-mediated desensitization of G-protein-coupled receptors and cause dampening of cellular responses to stimuli, such as hormones, neurotransmitters, or sensory signals. In a 2017 study, miR-29 along with miR-652 were indicated as biomarkers in liver fibrosis. In a luciferase reporter-based target validation assay, ARRB1 was identified as a target gene of these two miRNAs in regulating the onset and progression of liver fibrosis. Decreased expression of ARRB1 in addition to Th17, IL-17A, and IL-22 was observed in miR-29a-transfected CD4+ T cells isolated from the spleen of normal mice. Moreover, miR-29 overexpression led to decreased ALT, AST, Th17, and ARRB1 levels in vivo, suggesting the protective effects of miR-29 overexpression in liver fibrosis.192
The wider scope of these studies demonstrates anti-fibrotic activity of miR-29 and its role in regulating a number of ECM proteins, and it suggests an important role in the homeostasis of the ECM. A considerable amount of experimental evidence has shown that restoring downregulated miR-29 expression with synthetic miRNA mimics may pose a suitable strategy to overcome fibrosis. Though, this strategy still needs to be validated in advanced preclinical studies and in clinical settings.
Angiogenesis
The vascular system provides a vital channel to the delivery of nutrients and oxygen, and it has been shown to be crucial for tumor growth.193, 194 Consistently, cancer cells exploit the process of angiogenesis to generate new blood vessels to redirect nutrients and oxygen for their growth.195 In addition to supplying provisions to cancer cell growth, blood vessels also provide an effective conduit for metastasis.195 Therefore, numerous efforts have been underway to target angiogenesis in various cancers.196
miR-29 has been well implicated in the process of angiogenesis as an anti-angiogenic agent by potently inhibiting VEGFA.197, 198 VEGFA is a functionally dominant member of the VEGF family of growth factor proteins that is typically secreted and acts on endothelial cells to promote their growth, migration, and angiogenic properties.199 As expected, it has been reported that various cancers overexpress VEGF,200 and a number of studies have shown all three miR-29 family members to be potent tumor suppressors through directly targeting VEGFA.171, 201, 202, 203, 204, 205, 206
However, beyond the canonical downregulation of VEGF via 3′ UTR binding, a couple of other studies have indicated new mechanisms of regulation involving miR-29 sponging and a role in cancer-stroma cross-talk. An interesting report on osteosarcoma claimed a novel mechanism by which the miR-29-binding site within the 3′ UTR of IGF1 functions as a competing endogenous RNA for miR-29.207 By effectively sponging miR-29, authors showed an increase in VEGFA mRNA expression.207 This study stands alone in this claim, but this newly proposed mechanism of competitive binding warrants further studies nonetheless.
More recently, miR-29 has been proposed to be involved in a paracrine cross-talk mechanism between cancer-secreted IGF2 on VEGF expression in CAFs.203 Initial findings indicated that there was a concordant elevated expression of IGF2 in cancer cells with an increase in VEGF expression in CAFs, which conferred an unfavorable prognosis in esophageal squamous cell carcinoma.203 Utilizing predictive algorithms, miR-29c and miR-127 were identified to have some of the highest predictive scores for VEGF 3′ UTR-binding sites, and they were also found to be some of the most significantly downregulated miRNAs in IGF2-treated CAFs.203 Follow-up functional studies with both miRNAs revealed that only the overexpression of miR-29c led to a significant and consistent downregulation of VEGF in CAFs.203
Furthermore, miR-29 expression has even been shown to correlate with levels of tumor vasculature in clinical samples. In a study looking at the expression levels of various miRNAs in endometrial patient biopsies, miR-29b was found to be significantly downregulated in tumor samples compared to normal endometria and had a significant inverse correlation with the degree of tumor vascular invasion.208 In addition, low miR-29 expression levels conferred poorer survival.208
Collectively, an insurmountable number of studies demonstrate that miR-29 effectively targets VEGFA, which is an appealing therapeutic target in cancer. In fact, several clinical trials have been conducted or have been ongoing in an attempt to target VEGFA in various cancers (ClinicalTrials.gov: NCT01351415, NCT01863693, and NCT01239732). However, anti-angiogenesis treatment alone does not eliminate cancer completely and requires a combination therapeutic approach.196 Perhaps, miR-29 may also serve as an alternative and effective means of targeting VEGFA in combination with other therapies.
Immunomodulation
miR-29 has been shown to play a role in various immune-related aspects, from adaptive and innate immune responses107 to viral pathogenesis.89, 209 Although miR-29 has not been studied extensively in cancer immunity, a few studies have started to reveal its role in the context of cancer immunity.
B7-H3 is a potent immune checkpoint inhibitor that is known to be overexpressed by cancer cells and causes T cell suppression.210 Unsurprisingly, B7-H3 has become an appealing therapeutic target in cancer.210 In fact, a monoclonal antibody targeting B7-H3 is already undergoing clinical trials (ClinicalTrials.gov: NCT02475213). Moreover, a study investigating various solid tumors found all three miR-29 family members to be significantly downregulated in multiple cancers compared to normal tissues and inversely correlated with B7-H6 expression.211 The B7-H6 miR-29-binding site was validated, and overexpression of miR-29a led to a potent inhibition of B7-H6 expression.211 Although these findings convincingly demonstrate a potential use of miR-29 as a therapeutic inhibitor of B7-H6, further studies are needed to provide evidence of functional significance in vivo.
Another interesting immune-related mechanism of miR-29 was discovered in liver cancer, where miR-29a overexpression in HCC cells downregulated cancer cell migration while simultaneously increasing migration of CD8+ T lymphocytes in vivo.157 miR-29 overexpression downregulated IGF-1R in HCC cells, which led to an increased production of chemokine ligand 5 (CCL5).157 CCL5 is important for chemotactic movement of CD8+ T lymphocytes,212 and this was of particular significance as patients with high CCL5 exhibited better survival rates due to increased immune cell infiltration into the tumor.157 Albeit the mechanism was through indirect effects, these results still provide additional evidence of miR-29’s immunomodulatory function in cancer.
Beyond miR-29’s intrinsic function within cancer cells, miR-29 has also been shown to elicit an inflammatory response extrinsically. A subset of immune cells has been known to detect exogenous RNA through Toll-like receptors (TLRs) and trigger an inflammatory immune response.213, 214 As miRNAs had been reported to be released by cells to function extracellularly, one study sought to determine if secreted miRNAs interact with TLRs in the context of cancer.215 Indeed, miR-29a was shown to be secreted and co-immunoprecipitated with TLR7.215 Furthermore, miR-29a was found to co-localize with macrophage TLR7 and TLR8 in vivo.215 Ultimately, miR-29 binding to TLRs led to nuclear factor κB (NF-κB) activation and subsequent cytokine production that enhanced the metastatic potential of cancer.157 This proved to be a novel means by which miRNAs can function in an extrinsic manner beyond their canonical target mRNA 3′ UTR-binding mechanisms.
Only a handful of reports have demonstrated miR-29’s role in cancer immunomodulation, and, among these few studies, all were within the context of cancer cells. It has yet to be determined whether miR-29 plays an intrinsic role within immune cells themselves in relation to cancer pathogenesis. Certainly, miR-29’s role in immune cells outside of the tumor setting has been studied. For example, systematic screening of >100 individual miRNAs in helper T cells revealed miR-29 as a critical regulator of the IFN-γ pathway through the direct targeting of two critical transcription factors, T-bet and Eomes.216 T cells are known to be involved in the cancer immune response,217 and, therefore, it is not hard to imagine that miR-29 could have a functionally important role in cancer immunity as well. The investigation of miR-29 expression status and function in the context of intratumoral immune cells is of particular interest and remains an open area of research.
Clinical Findings and Prognostic Implications
The majority of accounts have reported miR-29 to have a tumor suppressor function in various cancer types. However, compared to the number of mechanistic studies of miR-29 in cancer, there are noticeably fewer studies interrogating the clinical ramifications of miR-29 expression for patient overall survival and outcomes. Herein, we touch on the current state of miR-29 as a potential prognostic marker.
In lung cancer, a study profiling the miRNA expression levels in tumor biopsies found miR-29b to be one of the most significantly downregulated miRNAs.218 Consistently, another study sought to define molecular features that distinguish lung squamous cell carcinoma versus adenocarcinoma, and the researchers found that all three miR-29 family members were more heavily downregulated in squamous cell carcinoma, with miR-29a as one of the most prominently downregulated miRNAs.219 Even though miR-29 was significantly downregulated in lung tumors, these studies highlighted various other miRNAs as having a more prominent effect in predicting overall survival.218, 219 However, a more recent report specifically looking at the impact of miR-29 expression level on overall survival and relapse-free survival in lung cancer patients found that patients with tumors expressing high miR-29 led to significantly better survival outcomes.220 Oftentimes, all three miR-29 family members were found to negatively correlate with prognostically unfavorable pulmonary neuroendocrine tumor grades. In contrast to miR-29 expression levels in tumors, two independent groups profiled circulating miRNA levels, and they found that higher levels of miR-29 in plasma conferred lower survival rates.221, 222 Although tumor expression and liquid biopsies are not one in the same, these results are still confounding.
These incongruent results are found in various lymphoma-related studies as well. In a single study, miR-29c was found to be upregulated in AML patients and was associated with a higher risk of relapse.223 In stark contrast, miR-29a was found to be significantly decreased in the bone marrow samples of 106 AML patients.224 Furthermore, high miR-29a expression was associated with more favorable prognosis, increased survival, and relapse-free survival.224 Concordant with these results, another study found both miR-29a and miR-29b expression levels to be significantly downregulated in myeloid leukemia and inversely correlated with anti-apoptotic gene BCL2 and MCL1 expression levels.225 The authors noted that lower levels of BCL2 and MCL1 had been reported to confer better overall survival in myeloid leukemia patients. However, the study did not demonstrate that miR-29 expression correlated with survival within their patient samples.225
There are limited reported studies regarding the prognostic value of miR-29 in other cancer types. Nevertheless, many of these studies seem to consistently indicate that high miR-29 expression leads to increased survival. In the case of mantle cell lymphoma, patients with significantly decreased miR-29 levels had shorter survival and had an inverse correlation with CDK6.105 Similar findings were reported in gastric cancer,226 melanoma,227 and serum miR-29 in high-grade glioma,228 where increased miR-29 indicated better survival.
From the existing evidence, the prognostic value of miR-29 expression is not clear and further studies are needed. Yet, most of these biomarker studies simply dichotomize patient cohorts based on high versus low miR-29 expression. Perhaps, further studies with a higher degree of scrutiny in patient stratification will better clarify and rectify these mixed results.
Discussion
Since the discovery of the first miRNA, the area of miRNA research has rapidly expanded and moved beyond basic biology into therapeutic use. Just within the past two decades, >2,000 patent documents related to miRNAs have been published.229 Nonetheless, the development of miRNA therapeutics lags behind many other forms of treatment, including RNAi-based drugs.230 Furthermore, recent setbacks were made when the first miRNA replacement therapy clinical trial, testing the therapeutic effect of miR-34 (MRX34) in cancer, was halted at phase I due to immune-related side effects.231 However, some arguments have been made that the adverse events of the MRX34 clinical trials were not due to miR-34 itself, but rather caused by the method of delivery (e.g., liposomes) or immunostimulation induced by double-stranded RNA.231, 232
Nevertheless, the promise of miRNA-based therapies in their ability to simultaneously target multiple disease-related pathways has remained appealing, as indicated by numerous therapeutic products still actively being pursued within the pipelines of several biotech companies.230 In fact, the miR-29 mimic MRG-201 is currently being tested in phase I clinical trials via intradermal injection (ClinicalTrials.gov: NCT02603224). Recently, a major milestone has been accomplished in the area of small RNA therapeutics, with the advent of the first FDA-approved siRNA treatment for a rare neurological disease, hereditary transthyretin-mediated amyloidosis (hATTR). The siRNA, patisiran, functions by inhibiting abnormal transthyretin (TTR) to reduce amyloid deposits and significantly improves symptoms associated with hATTR, including reflexes and motor strength.233 The delivery method in the clinical study also utilized a lipid nanoparticle to package and deliver the siRNAs, but it is important to note that there were complications associated with patisiran infusion.233 However, with the administration of anti-inflammatory drugs and antihistamines alleviated these issues.233 As new and improved modes of gene delivery are developed and tested,234, 235, 236 it may be just a matter of time before the therapeutic delivery of miR-29 will be tested in cancer.
Although miR-29 may seem to show a high degree of promise as a potent tumor suppressor, the minority of studies reporting miR-29 as an oncogene are still grounds for circumspection and must be strongly taken into consideration (Figure 2B). The opposing effects of miR-29 reported even within the same cancer types, such as breast cancer174, 183, 237, 238 and pancreatic cancer,158, 163, 165, 166, 172 are confounding.
In breast cancer, two studies took a systematic approach of assessing miR-29 status in breast cancer, revealing similar results of miR-29 as a tumor suppressor miRNA.174, 183 While Zhu and colleagues183 conducted a network analysis of gene co-expressions in hundreds of clinical breast cancer samples, Duhachek-Muggy and Zolkiewska237 profiled a panel of 50 breast cancer cell lines. Although overlapping breast cancer cell lines (BT549 and MDA-MB-231) were utilized in validating downstream mechanisms in both studies, only Zhu et al.183 showed functional impact of miR-29b and -29c in reducing colony formation. In two separate studies investigating the role of miR-29 in the context of breast cancer cell response to progesterone receptor activation, Cochrane et al.174 postulated that miR-29a and miR-29b function as tumor promoters by inhibiting ATP1B1 expression. In direct contrast, Cittelly et al.238 demonstrated that miR-29 inhibition in T47D and BT474 cells using antagomirs leads to increased growth in 3D colony formation assays. Further in vivo validation in an orthotopic mouse model demonstrated mir-29 inhibition leads to greater tumor growth and metastases. Although Cittelly and colleagues238 utilized the same breast cancer cell line, T47D, they found contradictory results. However, Cochrane et al.174 only conducted an in vitro viability assay in the T47D cell line, and they did not verify the impact of miR-29 in vivo.
In pancreatic cancer, four studies found that miR-29 exhibited a tumor-suppressive function,158, 163, 165, 166 as opposed to a single study that showed entirely opposite effects.172 Four studies functionally verified the tumor-suppressive properties of miR-29 by transfecting miR-29 mimics in pancreatic cancer cells and subjecting them to various in vitro assays to assess migration/invasion, anchorage-independent growth, and proliferation.158, 165, 166 Two studies went on to test the functional effect of miR-29 in vivo, and they consistently found that miR-29 suppressed tumor growth.158, 163 In direct contrast to these studies, Sun and colleagues172 found that miR-29a functions as a tumor promoter in various in vitro assays as well as in vivo. Further confusion arises when multiple mechanisms are proposed to explain the anti-tumor effects of miR-29 within the same cancer. For example, miR-29 was shown to have anti-metastatic potential in pancreatic cancer, yet all three reports offered alternative hypotheses for which miR-29 target downregulation leads to reduced migration/invasion (MMP2, MUC1, ITGB1).158, 163, 166
Some of the discrepancies of these studies can be explained by subtle differences in methodologies and the utilization of different cell lines. For example, although Cochrane et al.174 and Cittelly et al.238 found opposite results for miR-29 in the same cell line, T47D, Cittelly tested the impact of miR-29 in a particular CD44+ cancer stem cell subpopulation.238 However, there are examples in which using the exact same methodology and cell lines yielded contradictory results. Sun et al.172 found miR-29a to be increased in Panc-1 and BxPC-3 cell lines compared to a normal pancreatic epithelial line. In direct contrast, Tréhoux et al.166 and Kwon et al.165 consistently found its significant downregulation in various pancreatic cancer cell lines, including MIA PaCa-2, Panc-1, and BxPC-3, compared to normal pancreatic epithelial cell lines. Further discrepancies arise whereby Sun et al.172 overexpressed miR-29a mimic similar to both Tréhoux et al.166 and Kwon et al.,165 only to find the exact opposite results of increased cell proliferation and migration. A potential explanation for these confounding results could be explained by recent reports demonstrating heterogeneity between the same cell line from different labs.239 27 strains of the same well-utilized cell line MCF7 demonstrated immense genetic diversity and a large variability in drug response.239 By speculation, the opposing results of these studies could be explained by the diversity that resides within the same cell lines. Nevertheless, these incongruities demand the need for further studies and greater meticulous measures in validating cell lines and reagents to better understand these contradictory results.
Genomic heterogeneity is a well-known feature of cancer exists between tumors of different patients and is even evident within the same tumor.240 In recent years, there has been a paradigm shift toward classifying cancers based on genetic alterations rather than in the context of anatomical location. In fact, Pembrolizumab was the first recently FDA-approved cancer therapy on the basis of the patients’ tumor gene expression profile instead of the cancer’s originating tissue type. As miRNAs function in a context-dependent manner,241, 242, 243, 244 the discrepancies of miR-29 function and alternative mechanisms of action may be due to a lack of contextualizing miR-29 against specific genetic backgrounds. This is particularly relevant given that several different mechanisms of regulating miR-29 expression have been reported. Common mechanisms of miR-29 dysregulation in cancers have been identified, such as MYC directly suppressing miR-29 expression in various cancers.125, 245, 246 However, a few context-specific dysregulations of miR-29 expression have been demonstrated as well.
Given the anti-inflammatory role of miR-29 in fibrosis, it is not surprising that TGF-β1-SMAD signaling has been shown to directly downregulate miR-29 expression.7, 8, 9, 10, 11, 12 Consistently, restoration of miR-29 in activated pancreatic fibroblasts has been shown to reduce cancer cell proliferation in co-culture systems, and miR-29 downregulation in these fibroblasts was found to be SMAD3 dependent.45 These findings coincide with similar effects reported in ovarian cancer, where TGF-β1 has been shown to elicit pro-tumorigenic function by simultaneously downregulating miR-29.247 However, the majority of reports demonstrating TGF-β1-SMAD3/4-mediated miR-29 suppression have been in the context of fibroblasts and fibrosis. Thus, this mechanism of miR-29 dysregulation may be a more fibroblast-specific phenomenon.
A notable example of miR-29 dysregulation involves NF-κB and YY1. Both NF-κB and YY1 have been shown to be vital in the process of regulating myocyte differentiation.248, 249 In rhabdomyosarcoma, miR-29 has been found to be a tumor suppressor, where the molecule downregulates several cell cycle regulatory genes.128 Unsurprisingly, a rhabdomyosarcoma-specific mechanism of miR-29 dysregulation was found to be mediated by an NF-κB-YY1-miR-29-signaling axis, whereby YY1, under the regulation of NF-κB, directly binds to the miR-29 promoter site, and it modulates its expression epigenetically via a negative feedback loop mechanism and by closing the accessibility of chromatin modulation at the miR-29 promoter site.250
Another context-specific example of miR-29 dysregulation involves the transcription factor GATA3. Specifically, GATA3 has been shown to be a tumor suppressor in breast cancer by maintaining luminal cell differentiation and inhibiting metastasis.251, 252 Accordingly, the loss of GATA3 has been shown to be associated with poor prognosis, and it was found to be mutated in several cases of breast cancer.253, 254 In elucidating the mechanism by which GATA3 functions as a tumor suppressor, miR-29 was found to be a critical downstream target of GATA3.171 Specifically, miR-29b was found to be the most differentially upregulated miRNA by GATA3, where GATA3 directly binds to the promoter of miR-29.171 Finally, miR-29 was found to be a dominant effector of GATA3, where the inhibition of miR-29 was found to be sufficient for ablating the GATA3 tumor suppressor function.171
Most of the current miR-29-related literature in cancer frequently generalizes the function of miR-29 within the framework of organ-based stratification, without acknowledging the genetic background of the cells and biopsies. This may be due to utilizing a traditional, reductionist approach in biology. Factoring in many genetic variables is a daunting if not impossible task. However, recent advancements in systems biology and high-throughput technologies, such as proteogenomics, next-generation sequencing, single-cell sequencing, and functional genomics, have allowed the field to gain a more refined insight into global gene regulation, as well as having provided a more systematic approach to testing the function of genes on a large scale. Furthermore, the accumulation of large genomic databases (e.g., Human Cell Atlas, TCGA, The Human Protein Atlas, Project Achilles, DepMap, PRISM, and Cancer Cell Line Encyclopedia) has allowed for a greater degree of resolution in studying the heterogeneity associated with cancer.
Indeed, some systematic approaches have been conducted for miR-29. For example, miRNA expression across 10 cancer types was analyzed in TCGA database to determine commonalities in miRNA-target networks.64 Notably, “miR-29a was generally downregulated…in tumors of most cancer types as compared with representative normal samples.”64 Still, other analyses utilizing comprehensive, unbiased approaches find similarly consistent results of miR-29 as a tumor suppressor.255, 256 With time, further methodical studies employing these techniques and utilizing big data may provide a more auspicious approach to better understanding miR-29’s function in the context of cancer, and they may clarify its disputed role as tumor suppressor versus tumor promoter.
The multifaceted function of miR-29 casts doubts in claiming it as a panacea for cancer. It seems that miR-29’s dichotomous role as a tumor suppressor versus oncogene may be context dependent. However, at the current moment, miR-29 has been predominantly shown to function as a tumor suppressor in the majority of publications, with >85% of all miR-29-cancer-related studies (Figure 2B) demonstrating miR-29 inhibiting various cancer-related targets (Figure 3). Future studies are of interest to see if investigating the function of miR-29 in a more comprehensive and systematic manner will resolve these disputes and elucidate a more refined perspective on miR-29’s role in cancer.
Figure 3.
miR-29 Targets in the Mechanisms of Cancer
Schematic illustration depicts the reported targets of miR-29 involved in epigenetics, proteostasis, metabolism, proliferation, apoptosis, metastasis, fibrosis, angiogenesis, and immunomodulation.
Author Contributions
J.J.K. and J.K. conceived this work. J.J.K. and T.D.F. generated all figures. J.J.K., T.D.F., and S.D. manually curated the literature, organized the data, and wrote the manuscript.
Acknowledgments
This work was supported by grants to J.K. from Research Scholar Grant, RSG-18-105-01-RMC from the American Cancer Society, the Elsa U. Pardee Foundation, and the IU Simon Cancer Center, and J.J.K. was supported by the NCI (F31-CA213731-01A1).
Footnotes
Supplemental Information includes one table and can be found with this article online at https://doi.org/10.1016/j.omto.2018.12.011.
Supplemental Information
References
- 1.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sassi Y., Avramopoulos P., Ramanujam D., Grüter L., Werfel S., Giosele S., Brunner A.D., Esfandyari D., Papadopoulou A.S., De Strooper B. Cardiac myocyte miR-29 promotes pathological remodeling of the heart by activating Wnt signaling. Nat. Commun. 2017;8:1614. doi: 10.1038/s41467-017-01737-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., Shen B., Zhang D., Wang Y., Tang Z., Ni N., Jin X., Luo M., Sun H., Gu P. miR-29a regulates the proliferation and differentiation of retinal progenitors by targeting Rbm8a. Oncotarget. 2017;8:31993–32008. doi: 10.18632/oncotarget.16669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira P.A., Tomás J.F., Queiroz J.A., Figueiras A.R., Sousa F. Recombinant pre-miR-29b for Alzheimer's disease therapeutics. Sci. Rep. 2016;6:19946. doi: 10.1038/srep19946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Rooij E., Sutherland L.B., Thatcher J.E., DiMaio J.M., Naseem R.H., Marshall W.S., Hill J.A., Olson E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cushing L., Kuang P., Lü J. The role of miR-29 in pulmonary fibrosis. Biochem. Cell Biol. 2015;93:109–118. doi: 10.1139/bcb-2014-0095. [DOI] [PubMed] [Google Scholar]
- 8.Cushing L., Kuang P.P., Qian J., Shao F., Wu J., Little F., Thannickal V.J., Cardoso W.V., Lü J. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roderburg C., Urban G.W., Bettermann K., Vucur M., Zimmermann H., Schmidt S., Janssen J., Koppe C., Knolle P., Castoldi M. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 10.Xiao J., Meng X.M., Huang X.R., Chung A.C., Feng Y.L., Hui D.S., Yu C.M., Sung J.J., Lan H.Y. miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol. Ther. 2012;20:1251–1260. doi: 10.1038/mt.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin W., Chung A.C., Huang X.R., Meng X.M., Hui D.S., Yu C.M., Sung J.J., Lan H.Y. TGF-β/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J. Am. Soc. Nephrol. 2011;22:1462–1474. doi: 10.1681/ASN.2010121308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung A.C., Lan H.Y. MicroRNAs in renal fibrosis. Front. Physiol. 2015;6:50. doi: 10.3389/fphys.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekiya Y., Ogawa T., Yoshizato K., Ikeda K., Kawada N. Suppression of hepatic stellate cell activation by microRNA-29b. Biochem. Biophys. Res. Commun. 2011;412:74–79. doi: 10.1016/j.bbrc.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 14.Guo S., Guo X., Wang S., Nie Q., Ni G., Wang C. Role of miR-29 as marker of risk of acute rejection after heart transplant. Br. J. Biomed. Sci. 2017;74:187–192. doi: 10.1080/09674845.2017.1333265. [DOI] [PubMed] [Google Scholar]
- 15.Dawson K., Wakili R., Ordög B., Clauss S., Chen Y., Iwasaki Y., Voigt N., Qi X.Y., Sinner M.F., Dobrev D. MicroRNA29: a mechanistic contributor and potential biomarker in atrial fibrillation. Circulation. 2013;127:1466–1475. doi: 10.1161/CIRCULATIONAHA.112.001207. [DOI] [PubMed] [Google Scholar]
- 16.Kogure T., Costinean S., Yan I., Braconi C., Croce C., Patel T. Hepatic miR-29ab1 expression modulates chronic hepatic injury. J. Cell. Mol. Med. 2012;16:2647–2654. doi: 10.1111/j.1582-4934.2012.01578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy S., Benz F., Luedde T., Roderburg C. The role of miRNAs in the regulation of inflammatory processes during hepatofibrogenesis. Hepatobiliary Surg. Nutr. 2015;4:24–33. doi: 10.3978/j.issn.2304-3881.2015.01.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Q., Mi Q.S., Dong Z. The regulation and function of microRNAs in kidney diseases. IUBMB Life. 2013;65:602–614. doi: 10.1002/iub.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W.M., Jeong H.J., Park S.Y., Lee W. Induction of miR-29a by saturated fatty acids impairs insulin signaling and glucose uptake through translational repression of IRS-1 in myocytes. FEBS Lett. 2014;588:2170–2176. doi: 10.1016/j.febslet.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Massart J., Sjögren R.J.O., Lundell L.S., Mudry J.M., Franck N., O’Gorman D.J., Egan B., Zierath J.R., Krook A. Altered miR-29 Expression in Type 2 Diabetes Influences Glucose and Lipid Metabolism in Skeletal Muscle. Diabetes. 2017;66:1807–1818. doi: 10.2337/db17-0141. [DOI] [PubMed] [Google Scholar]
- 21.Qiu L., Tan E.K., Zeng L. microRNAs and Neurodegenerative Diseases. Adv. Exp. Med. Biol. 2015;888:85–105. doi: 10.1007/978-3-319-22671-2_6. [DOI] [PubMed] [Google Scholar]
- 22.Hébert S.S., Horré K., Nicolaï L., Papadopoulou A.S., Mandemakers W., Silahtaroglu A.N., Kauppinen S., Delacourte A., De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 25.Bråte J., Neumann R.S., Fromm B., Haraldsen A.A.B., Tarver J.E., Suga H., Donoghue P.C.J., Peterson K.J., Ruiz-Trillo I., Grini P.E., Shalchian-Tabrizi K. Unicellular Origin of the Animal MicroRNA Machinery. Curr. Biol. 2018;28:3288–3295.e5. doi: 10.1016/j.cub.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lund E., Güttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 27.Michlewski G., Cáceres J.F. Post-transcriptional control of miRNA biogenesis. RNA. 2019;25:1–16. doi: 10.1261/rna.068692.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 29.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 30.Pratt A.J., MacRae I.J. The RNA-induced silencing complex: a versatile gene-silencing machine. J. Biol. Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koh W., Sheng C.T., Tan B., Lee Q.Y., Kuznetsov V., Kiang L.S., Tanavde V. Analysis of deep sequencing microRNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of hepatic nuclear factor 4 alpha. BMC Genomics. 2010;11(Suppl 1):S6. doi: 10.1186/1471-2164-11-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang H.W., Wentzel E.A., Mendell J.T. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 36.Jagannathan S., Vad N., Vallabhapurapu S., Vallabhapurapu S., Anderson K.C., Driscoll J.J. MiR-29b replacement inhibits proteasomes and disrupts aggresome+autophagosome formation to enhance the antimyeloma benefit of bortezomib. Leukemia. 2015;29:727–738. doi: 10.1038/leu.2014.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeffries C.D., Fried H.M., Perkins D.O. Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA. 2011;17:675–686. doi: 10.1261/rna.2006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khudayberdiev S.A., Zampa F., Rajman M., Schratt G. A comprehensive characterization of the nuclear microRNA repertoire of post-mitotic neurons. Front. Mol. Neurosci. 2013;6:43. doi: 10.3389/fnmol.2013.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts T.C. The MicroRNA Biology of the Mammalian Nucleus. Mol. Ther. Nucleic Acids. 2014;3:e188. doi: 10.1038/mtna.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang H., Zhang J., Zen K., Zhang C.Y., Chen X. Nuclear microRNAs and their unconventional role in regulating non-coding RNAs. Protein Cell. 2013;4:325–330. doi: 10.1007/s13238-013-3001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z., Zou J., Wang G.K., Zhang J.T., Huang S., Qin Y.W., Jing Q. Uracils at nucleotide position 9-11 are required for the rapid turnover of miR-29 family. Nucleic Acids Res. 2011;39:4387–4395. doi: 10.1093/nar/gkr020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tumaneng K., Schlegelmilch K., Russell R.C., Yimlamai D., Basnet H., Mahadevan N., Fitamant J., Bardeesy N., Camargo F.D., Guan K.L. YAP mediates crosstalk between the Hippo and PI(3)K–TOR pathways by suppressing PTEN via miR-29. Nat. Cell Biol. 2012;14:1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y., Taylor N.E., Lu L., Usa K., Cowley A.W., Jr., Ferreri N.R., Yeo N.C., Liang M. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension. 2010;55:974–982. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dooley J., Garcia-Perez J.E., Sreenivasan J., Schlenner S.M., Vangoitsenhoven R., Papadopoulou A.S., Tian L., Schonefeldt S., Serneels L., Deroose C. The microRNA-29 Family Dictates the Balance Between Homeostatic and Pathological Glucose Handling in Diabetes and Obesity. Diabetes. 2016;65:53–61. doi: 10.2337/db15-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon J.J., Nabinger S.C., Vega Z., Sahu S.S., Alluri R.K., Abdul-Sater Z., Yu Z., Gore J., Nalepa G., Saxena R. Pathophysiological role of microRNA-29 in pancreatic cancer stroma. Sci. Rep. 2015;5:11450. doi: 10.1038/srep11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papadopoulou A.S., Dooley J., Linterman M.A., Pierson W., Ucar O., Kyewski B., Zuklys S., Hollander G.A., Matthys P., Gray D.H. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-α receptor. Nat. Immunol. 2011;13:181–187. doi: 10.1038/ni.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cushing L., Costinean S., Xu W., Jiang Z., Madden L., Kuang P., Huang J., Weisman A., Hata A., Croce C.M., Lü J. Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature. PLoS Genet. 2015;11:e1005238. doi: 10.1371/journal.pgen.1005238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith K.M., Guerau-de-Arellano M., Costinean S., Williams J.L., Bottoni A., Mavrikis Cox G., Satoskar A.R., Croce C.M., Racke M.K., Lovett-Racke A.E., Whitacre C.C. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J. Immunol. 2012;189:1567–1576. doi: 10.4049/jimmunol.1103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waddington C.H. The epigenotype. 1942. Int. J. Epidemiol. 2012;41:10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 51.Lin R.K., Hsu H.S., Chang J.W., Chen C.Y., Chen J.T., Wang Y.C. Alteration of DNA methyltransferases contributes to 5'CpG methylation and poor prognosis in lung cancer. Lung Cancer. 2007;55:205–213. doi: 10.1016/j.lungcan.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 52.Girault I., Tozlu S., Lidereau R., Bièche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin. Cancer Res. 2003;9:4415–4422. [PubMed] [Google Scholar]
- 53.Saito Y., Kanai Y., Nakagawa T., Sakamoto M., Saito H., Ishii H., Hirohashi S. Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int. J. Cancer. 2003;105:527–532. doi: 10.1002/ijc.11127. [DOI] [PubMed] [Google Scholar]
- 54.Patra S.K., Patra A., Zhao H., Dahiya R. DNA methyltransferase and demethylase in human prostate cancer. Mol. Carcinog. 2002;33:163–171. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- 55.Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E., Liu S., Alder H., Costinean S., Fernandez-Cymering C. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allis C.D., Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 57.Hideshima T., Cottini F., Ohguchi H., Jakubikova J., Gorgun G., Mimura N., Tai Y.T., Munshi N.C., Richardson P.G., Anderson K.C. Rational combination treatment with histone deacetylase inhibitors and immunomodulatory drugs in multiple myeloma. Blood Cancer J. 2015;5:e312. doi: 10.1038/bcj.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lane A.A., Chabner B.A. Histone deacetylase inhibitors in cancer therapy. J. Clin. Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 59.Amodio N., Stamato M.A., Gullà A.M., Morelli E., Romeo E., Raimondi L., Pitari M.R., Ferrandino I., Misso G., Caraglia M. Therapeutic Targeting of miR-29b/HDAC4 Epigenetic Loop in Multiple Myeloma. Mol. Cancer Ther. 2016;15:1364–1375. doi: 10.1158/1535-7163.MCT-15-0985. [DOI] [PubMed] [Google Scholar]
- 60.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morita S., Horii T., Kimura M., Ochiya T., Tajima S., Hatada I. miR-29 represses the activities of DNA methyltransferases and DNA demethylases. Int. J. Mol. Sci. 2013;14:14647–14658. doi: 10.3390/ijms140714647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacobsen A., Silber J., Harinath G., Huse J.T., Schultz N., Sander C. Analysis of microRNA-target interactions across diverse cancer types. Nat. Struct. Mol. Biol. 2013;20:1325–1332. doi: 10.1038/nsmb.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsai Y.P., Chen H.F., Chen S.Y., Cheng W.C., Wang H.W., Shen Z.J., Song C., Teng S.C., He C., Wu K.J. TET1 regulates hypoxia-induced epithelial-mesenchymal transition by acting as a co-activator. Genome Biol. 2014;15:513. doi: 10.1186/s13059-014-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu B.K., Brenner C. Suppression of TET1-dependent DNA demethylation is essential for KRAS-mediated transformation. Cell Rep. 2014;9:1827–1840. doi: 10.1016/j.celrep.2014.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H., An X., Yu H., Zhang S., Tang B., Zhang X., Li Z. MiR-29b/TET1/ZEB2 signaling axis regulates metastatic properties and epithelial-mesenchymal transition in breast cancer cells. Oncotarget. 2017;8:102119–102133. doi: 10.18632/oncotarget.22183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tu J., Ng S.H., Luk A.C., Liao J., Jiang X., Feng B., Lun Mak K.K., Rennert O.M., Chan W.Y., Lee T.L. MicroRNA-29b/Tet1 regulatory axis epigenetically modulates mesendoderm differentiation in mouse embryonic stem cells. Nucleic Acids Res. 2015;43:7805–7822. doi: 10.1093/nar/gkv653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cui Y., Li T., Yang D., Li S., Le W. miR-29 regulates Tet1 expression and contributes to early differentiation of mouse ESCs. Oncotarget. 2016;7:64932–64941. doi: 10.18632/oncotarget.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oakes S.A. Endoplasmic reticulum proteostasis: a key checkpoint in cancer. Am. J. Physiol. Cell Physiol. 2017;312:C93–C102. doi: 10.1152/ajpcell.00266.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cohen-Kaplan V., Livneh I., Avni N., Cohen-Rosenzweig C., Ciechanover A. The ubiquitin-proteasome system and autophagy: Coordinated and independent activities. Int. J. Biochem. Cell Biol. 2016;79:403–418. doi: 10.1016/j.biocel.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 72.Crawford L.J., Walker B., Irvine A.E. Proteasome inhibitors in cancer therapy. J. Cell Commun. Signal. 2011;5:101–110. doi: 10.1007/s12079-011-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Drie J.H. Protein folding, protein homeostasis, and cancer. Chin. J. Cancer. 2011;30:124–137. doi: 10.5732/cjc.010.10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaur J., Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol. 2015;16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 75.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Apel A., Herr I., Schwarz H., Rodemann H.P., Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 77.Li J., Yang B., Zhou Q., Wu Y., Shang D., Guo Y., Song Z., Zheng Q., Xiong J. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis. 2013;34:1343–1351. doi: 10.1093/carcin/bgt063. [DOI] [PubMed] [Google Scholar]
- 78.Fels D.R., Ye J., Segan A.T., Kridel S.J., Spiotto M., Olson M., Koong A.C., Koumenis C. Preferential cytotoxicity of bortezomib toward hypoxic tumor cells via overactivation of endoplasmic reticulum stress pathways. Cancer Res. 2008;68:9323–9330. doi: 10.1158/0008-5472.CAN-08-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Claerhout S., Verschooten L., Van Kelst S., De Vos R., Proby C., Agostinis P., Garmyn M. Concomitant inhibition of AKT and autophagy is required for efficient cisplatin-induced apoptosis of metastatic skin carcinoma. Int. J. Cancer. 2010;127:2790–2803. doi: 10.1002/ijc.25300. [DOI] [PubMed] [Google Scholar]
- 80.Akalay I., Janji B., Hasmim M., Noman M.Z., André F., De Cremoux P., Bertheau P., Badoual C., Vielh P., Larsen A.K. Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res. 2013;73:2418–2427. doi: 10.1158/0008-5472.CAN-12-2432. [DOI] [PubMed] [Google Scholar]
- 81.Kanzawa T., Germano I.M., Komata T., Ito H., Kondo Y., Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448–457. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 82.Li M., Jiang X., Liu D., Na Y., Gao G.F., Xi Z. Autophagy protects LNCaP cells under androgen deprivation conditions. Autophagy. 2008;4:54–60. doi: 10.4161/auto.5209. [DOI] [PubMed] [Google Scholar]
- 83.Hashimoto D., Bläuer M., Hirota M., Ikonen N.H., Sand J., Laukkarinen J. Autophagy is needed for the growth of pancreatic adenocarcinoma and has a cytoprotective effect against anticancer drugs. Eur. J. Cancer. 2014;50:1382–1390. doi: 10.1016/j.ejca.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 84.Donadelli M., Dando I., Zaniboni T., Costanzo C., Dalla Pozza E., Scupoli M.T., Scarpa A., Zappavigna S., Marra M., Abbruzzese A. Gemcitabine/cannabinoid combination triggers autophagy in pancreatic cancer cells through a ROS-mediated mechanism. Cell Death Dis. 2011;2:e152. doi: 10.1038/cddis.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang M.C., Wang H.C., Hou Y.C., Tung H.L., Chiu T.J., Shan Y.S. Blockade of autophagy reduces pancreatic cancer stem cell activity and potentiates the tumoricidal effect of gemcitabine. Mol. Cancer. 2015;14:179. doi: 10.1186/s12943-015-0449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang A., Rajeshkumar N.V., Wang X., Yabuuchi S., Alexander B.M., Chu G.C., Von Hoff D.D., Maitra A., Kimmelman A.C. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4:905–913. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang S., Wang X., Contino G., Liesa M., Sahin E., Ying H., Bause A., Li Y., Stommel J.M., Dell’antonio G. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang F., Ma H., Liang W.J., Yang J.J., Wang X.Q., Shan M.R., Chen Y., Jia M., Yin Y.L., Sun X.Y. Lovastatin upregulates microRNA-29b to reduce oxidative stress in rats with multiple cardiovascular risk factors. Oncotarget. 2017;8:9021–9034. doi: 10.18632/oncotarget.14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fu Q., Shi H., Ni W., Shi M., Meng L., Zhang H., Ren Y., Guo F., Wang P., Qiao J. Lentivirus-mediated Bos taurus bta-miR-29b overexpression interferes with bovine viral diarrhoea virus replication and viral infection-related autophagy by directly targeting ATG14 and ATG9A in Madin-Darby bovine kidney cells. J. Gen. Virol. 2015;96:85–94. doi: 10.1099/vir.0.067140-0. [DOI] [PubMed] [Google Scholar]
- 90.Song H., Ding L., Zhang S., Wang W. MiR-29 family members interact with SPARC to regulate glucose metabolism. Biochem. Biophys. Res. Commun. 2018;497:667–674. doi: 10.1016/j.bbrc.2018.02.129. [DOI] [PubMed] [Google Scholar]
- 91.Pullen T.J., da Silva Xavier G., Kelsey G., Rutter G.A. miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1) Mol. Cell. Biol. 2011;31:3182–3194. doi: 10.1128/MCB.01433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mersey B.D., Jin P., Danner D.J. Human microRNA (miR29b) expression controls the amount of branched chain alpha-ketoacid dehydrogenase complex in a cell. Hum. Mol. Genet. 2005;14:3371–3377. doi: 10.1093/hmg/ddi368. [DOI] [PubMed] [Google Scholar]
- 93.Teng Y., Zhang Y., Qu K., Yang X., Fu J., Chen W., Li X. MicroRNA-29B (mir-29b) regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3. Oncotarget. 2015;6:40799–40814. doi: 10.18632/oncotarget.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Young C.D., Nolte E.C., Lewis A., Serkova N.J., Anderson S.M. Activated Akt1 accelerates MMTV-c-ErbB2 mammary tumourigenesis in mice without activation of ErbB3. Breast Cancer Res. 2008;10:R70. doi: 10.1186/bcr2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ru P., Hu P., Geng F., Mo X., Cheng C., Yoo J.Y., Cheng X., Wu X., Guo J.Y., Nakano I. Feedback Loop Regulation of SCAP/SREBP-1 by miR-29 Modulates EGFR Signaling-Driven Glioblastoma Growth. Cell Rep. 2016;16:1527–1535. doi: 10.1016/j.celrep.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eberlé D., Hegarty B., Bossard P., Ferré P., Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 98.Cheng C., Ru P., Geng F., Liu J., Yoo J.Y., Wu X., Cheng X., Euthine V., Hu P., Guo J.Y. Glucose-Mediated N-glycosylation of SCAP Is Essential for SREBP-1 Activation and Tumor Growth. Cancer Cell. 2015;28:569–581. doi: 10.1016/j.ccell.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Muluhngwi P., Alizadeh-Rad N., Vittitow S.L., Kalbfleisch T.S., Klinge C.M. The miR-29 transcriptome in endocrine-sensitive and resistant breast cancer cells. Sci. Rep. 2017;7:5205. doi: 10.1038/s41598-017-05727-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Golias C.H., Charalabopoulos A., Charalabopoulos K. Cell proliferation and cell cycle control: a mini review. Int. J. Clin. Pract. 2004;58:1134–1141. doi: 10.1111/j.1742-1241.2004.00284.x. [DOI] [PubMed] [Google Scholar]
- 101.McDonald E.R., 3rd, El-Deiry W.S. Checkpoint genes in cancer. Ann. Med. 2001;33:113–122. doi: 10.3109/07853890109002066. [DOI] [PubMed] [Google Scholar]
- 102.Cole A.M., Myant K., Reed K.R., Ridgway R.A., Athineos D., Van den Brink G.R., Muncan V., Clevers H., Clarke A.R., Sicinski P., Sansom O.J. Cyclin D2-cyclin-dependent kinase 4/6 is required for efficient proliferation and tumorigenesis following Apc loss. Cancer Res. 2010;70:8149–8158. doi: 10.1158/0008-5472.CAN-10-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rivadeneira D.B., Mayhew C.N., Thangavel C., Sotillo E., Reed C.A., Graña X., Knudsen E.S. Proliferative suppression by CDK4/6 inhibition: complex function of the retinoblastoma pathway in liver tissue and hepatoma cells. Gastroenterology. 2010;138:1920–1930. doi: 10.1053/j.gastro.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sherr C.J., Beach D., Shapiro G.I. Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov. 2016;6:353–367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao J.J., Lin J., Lwin T., Yang H., Guo J., Kong W., Dessureault S., Moscinski L.C., Rezania D., Dalton W.S. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115:2630–2639. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garzon R., Liu S., Fabbri M., Liu Z., Heaphy C.E., Callegari E., Schwind S., Pang J., Yu J., Muthusamy N. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma F., Xu S., Liu X., Zhang Q., Xu X., Liu M., Hua M., Li N., Yao H., Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat. Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 108.Kortylewski M., Komyod W., Kauffmann M.E., Bosserhoff A., Heinrich P.C., Behrmann I. Interferon-gamma-mediated growth regulation of melanoma cells: involvement of STAT1-dependent and STAT1-independent signals. J. Invest. Dermatol. 2004;122:414–422. doi: 10.1046/j.0022-202X.2004.22237.x. [DOI] [PubMed] [Google Scholar]
- 109.Schmitt M.J., Philippidou D., Reinsbach S.E., Margue C., Wienecke-Baldacchino A., Nashan D., Behrmann I., Kreis S. Interferon-γ-induced activation of Signal Transducer and Activator of Transcription 1 (STAT1) up-regulates the tumor suppressing microRNA-29 family in melanoma cells. Cell Commun. Signal. 2012;10:41. doi: 10.1186/1478-811X-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mazzoccoli L., Robaina M.C., Apa A.G., Bonamino M., Pinto L.W., Queiroga E., Bacchi C.E., Klumb C.E. MiR-29 silencing modulates the expression of target genes related to proliferation, apoptosis and methylation in Burkitt lymphoma cells. J. Cancer Res. Clin. Oncol. 2018;144:483–497. doi: 10.1007/s00432-017-2575-3. [DOI] [PubMed] [Google Scholar]
- 111.Baldwin A., Li W., Grace M., Pearlberg J., Harlow E., Münger K., Grueneberg D.A. Kinase requirements in human cells: II. Genetic interaction screens identify kinase requirements following HPV16 E7 expression in cancer cells. Proc. Natl. Acad. Sci. USA. 2008;105:16478–16483. doi: 10.1073/pnas.0806195105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li Y., Wang F., Xu J., Ye F., Shen Y., Zhou J., Lu W., Wan X., Ma D., Xie X. Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV-related target genes for miR-29. J. Pathol. 2011;224:484–495. doi: 10.1002/path.2873. [DOI] [PubMed] [Google Scholar]
- 113.Ma J., Li T., Yuan H., Han X., Shui S., Guo D., Yan L. MicroRNA-29a inhibits proliferation and motility of schwannoma cells by targeting CDK6. J. Cell. Biochem. 2018;119:2617–2626. doi: 10.1002/jcb.26426. [DOI] [PubMed] [Google Scholar]
- 114.Chen L., Zhang S., Wu J., Cui J., Zhong L., Zeng L., Ge S. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–4561. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Arias-Romero L.E., Chernoff J. Targeting Cdc42 in cancer. Expert Opin. Ther. Targets. 2013;17:1263–1273. doi: 10.1517/14728222.2013.828037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cerione R.A. Cdc42: new roads to travel. Trends Cell Biol. 2004;14:127–132. doi: 10.1016/j.tcb.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 117.Nishikawa R., Chiyomaru T., Enokida H., Inoguchi S., Ishihara T., Matsushita R., Goto Y., Fukumoto I., Nakagawa M., Seki N. Tumour-suppressive microRNA-29s directly regulate LOXL2 expression and inhibit cancer cell migration and invasion in renal cell carcinoma. FEBS Lett. 2015;589:2136–2145. doi: 10.1016/j.febslet.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 118.Cristiano B.E., Chan J.C., Hannan K.M., Lundie N.A., Marmy-Conus N.J., Campbell I.G., Phillips W.A., Robbie M., Hannan R.D., Pearson R.B. A specific role for AKT3 in the genesis of ovarian cancer through modulation of G(2)-M phase transition. Cancer Res. 2006;66:11718–11725. doi: 10.1158/0008-5472.CAN-06-1968. [DOI] [PubMed] [Google Scholar]
- 119.Wei W., He H.B., Zhang W.Y., Zhang H.X., Bai J.B., Liu H.Z., Cao J.H., Chang K.C., Li X.Y., Zhao S.H. miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 2013;4:e668. doi: 10.1038/cddis.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Francy J.M., Nag A., Conroy E.J., Hengst J.A., Yun J.K. Sphingosine kinase 1 expression is regulated by signaling through PI3K, AKT2, and mTOR in human coronary artery smooth muscle cells. Biochim. Biophys. Acta. 2007;1769:253–265. doi: 10.1016/j.bbaexp.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 121.Li X., Leu S., Cheong A., Zhang H., Baibakov B., Shih C., Birnbaum M.J., Donowitz M. Akt2, phosphatidylinositol 3-kinase, and PTEN are in lipid rafts of intestinal cells: role in absorption and differentiation. Gastroenterology. 2004;126:122–135. doi: 10.1053/j.gastro.2003.10.061. [DOI] [PubMed] [Google Scholar]
- 122.Liu A.X., Testa J.R., Hamilton T.C., Jove R., Nicosia S.V., Cheng J.Q. AKT2, a member of the protein kinase B family, is activated by growth factors, v-Ha-ras, and v-src through phosphatidylinositol 3-kinase in human ovarian epithelial cancer cells. Cancer Res. 1998;58:2973–2977. [PubMed] [Google Scholar]
- 123.Wang L., Wang J., Blaser B.W., Duchemin A.M., Kusewitt D.F., Liu T., Caligiuri M.A., Briesewitz R. Pharmacologic inhibition of CDK4/6: mechanistic evidence for selective activity or acquired resistance in acute myeloid leukemia. Blood. 2007;110:2075–2083. doi: 10.1182/blood-2007-02-071266. [DOI] [PubMed] [Google Scholar]
- 124.Decker T., Schneller F., Hipp S., Miething C., Jahn T., Duyster J., Peschel C. Cell cycle progression of chronic lymphocytic leukemia cells is controlled by cyclin D2, cyclin D3, cyclin-dependent kinase (cdk) 4 and the cdk inhibitor p27. Leukemia. 2002;16:327–334. doi: 10.1038/sj.leu.2402389. [DOI] [PubMed] [Google Scholar]
- 125.Gong J.N., Yu J., Lin H.S., Zhang X.H., Yin X.L., Xiao Z., Wang F., Wang X.S., Su R., Shen C. The role, mechanism and potentially therapeutic application of microRNA-29 family in acute myeloid leukemia. Cell Death Differ. 2014;21:100–112. doi: 10.1038/cdd.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Logan N., Graham A., Zhao X., Fisher R., Maiti B., Leone G., La Thangue N.B. E2F-8: an E2F family member with a similar organization of DNA-binding domains to E2F-7. Oncogene. 2005;24:5000–5004. doi: 10.1038/sj.onc.1208703. [DOI] [PubMed] [Google Scholar]
- 127.Di Stefano L., Jensen M.R., Helin K. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 2003;22:6289–6298. doi: 10.1093/emboj/cdg613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li L., Sarver A.L., Alamgir S., Subramanian S. Downregulation of microRNAs miR-1, -206 and -29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma. Lab. Invest. 2012;92:571–583. doi: 10.1038/labinvest.2012.10. [DOI] [PubMed] [Google Scholar]
- 129.Liu Y., Chen X., Gu Y., Zhu L., Qian Y., Pei D., Zhang W., Shu Y. FOXM1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and mediates sensitivity to cisplatin in A549 cells via the JNK/mitochondrial pathway. Neoplasma. 2015;62:61–71. doi: 10.4149/neo_2015_008. [DOI] [PubMed] [Google Scholar]
- 130.Lima R.T., Busacca S., Almeida G.M., Gaudino G., Fennell D.A., Vasconcelos M.H. MicroRNA regulation of core apoptosis pathways in cancer. Eur. J. Cancer. 2011;47:163–174. doi: 10.1016/j.ejca.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 131.Wong R.S. Apoptosis in cancer: from pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Indran I.R., Tufo G., Pervaiz S., Brenner C. Recent advances in apoptosis, mitochondria and drug resistance in cancer cells. Biochim. Biophys. Acta. 2011;1807:735–745. doi: 10.1016/j.bbabio.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 133.Pileczki V., Cojocneanu-Petric R., Maralani M., Neagoe I.B., Sandulescu R. MicroRNAs as regulators of apoptosis mechanisms in cancer. Clujul Med. 2016;89:50–55. doi: 10.15386/cjmed-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Calin G.A., Cimmino A., Fabbri M., Ferracin M., Wojcik S.E., Shimizu M., Taccioli C., Zanesi N., Garzon R., Aqeilan R.I. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl. Acad. Sci. USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Warr M.R., Shore G.C. Unique biology of Mcl-1: therapeutic opportunities in cancer. Curr. Mol. Med. 2008;8:138–147. doi: 10.2174/156652408783769580. [DOI] [PubMed] [Google Scholar]
- 136.Yagi T., Morimoto A., Eguchi M., Hibi S., Sako M., Ishii E., Mizutani S., Imashuku S., Ohki M., Ichikawa H. Identification of a gene expression signature associated with pediatric AML prognosis. Blood. 2003;102:1849–1856. doi: 10.1182/blood-2003-02-0578. [DOI] [PubMed] [Google Scholar]
- 137.Kaufmann S.H., Karp J.E., Svingen P.A., Krajewski S., Burke P.J., Gore S.D., Reed J.C. Elevated expression of the apoptotic regulator Mcl-1 at the time of leukemic relapse. Blood. 1998;91:991–1000. [PubMed] [Google Scholar]
- 138.Garzon R., Heaphy C.E., Havelange V., Fabbri M., Volinia S., Tsao T., Zanesi N., Kornblau S.M., Marcucci G., Calin G.A. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Muniyappa M.K., Dowling P., Henry M., Meleady P., Doolan P., Gammell P., Clynes M., Barron N. MiRNA-29a regulates the expression of numerous proteins and reduces the invasiveness and proliferation of human carcinoma cell lines. Eur. J. Cancer. 2009;45:3104–3118. doi: 10.1016/j.ejca.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 140.Yu J., Wang Z., Kinzler K.W., Vogelstein B., Zhang L. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:1931–1936. doi: 10.1073/pnas.2627984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ouyang Y.B., Xu L., Lu Y., Sun X., Yue S., Xiong X.X., Giffard R.G. Astrocyte-enriched miR-29a targets PUMA and reduces neuronal vulnerability to forebrain ischemia. Glia. 2013;61:1784–1794. doi: 10.1002/glia.22556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim H., Tu H.C., Ren D., Takeuchi O., Jeffers J.R., Zambetti G.P., Hsieh J.J., Cheng E.H. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Spender L.C., Carter M.J., O’Brien D.I., Clark L.J., Yu J., Michalak E.M., Happo L., Cragg M.S., Inman G.J. Transforming growth factor-β directly induces p53-up-regulated modulator of apoptosis (PUMA) during the rapid induction of apoptosis in myc-driven B-cell lymphomas. J. Biol. Chem. 2013;288:5198–5209. doi: 10.1074/jbc.M112.410274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang C.Y., Ren J.B., Liu M., Yu L. Targeting miR-29 induces apoptosis of osteosarcoma MG-63 cells via regulation of TGF-β1/PUMA signal. Eur. Rev. Med. Pharmacol. Sci. 2016;20:3552–3560. [PubMed] [Google Scholar]
- 145.Wong C.M., Wei L., Law C.T., Ho D.W., Tsang F.H., Au S.L., Sze K.M., Lee J.M., Wong C.C., Ng I.O. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63:474–487. doi: 10.1002/hep.28304. [DOI] [PubMed] [Google Scholar]
- 146.Heerboth S., Housman G., Leary M., Longacre M., Byler S., Lapinska K., Willbanks A., Sarkar S. EMT and tumor metastasis. Clin. Transl. Med. 2015;4:6. doi: 10.1186/s40169-015-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 148.Gong J., Li J., Wang Y., Liu C., Jia H., Jiang C., Wang Y., Luo M., Zhao H., Dong L. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis. 2014;35:497–506. doi: 10.1093/carcin/bgt337. [DOI] [PubMed] [Google Scholar]
- 149.Gravdal K., Halvorsen O.J., Haukaas S.A., Akslen L.A. A switch from E-cadherin to N-cadherin expression indicates epithelial to mesenchymal transition and is of strong and independent importance for the progress of prostate cancer. Clin. Cancer Res. 2007;13:7003–7011. doi: 10.1158/1078-0432.CCR-07-1263. [DOI] [PubMed] [Google Scholar]
- 150.Anastasiadis P.Z. p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochim. Biophys. Acta. 2007;1773:34–46. doi: 10.1016/j.bbamcr.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 151.Wang Y., Liu C., Luo M., Zhang Z., Gong J., Li J., You L., Dong L., Su R., Lin H. Chemotherapy-Induced miRNA-29c/Catenin-δ Signaling Suppresses Metastasis in Gastric Cancer. Cancer Res. 2015;75:1332–1344. doi: 10.1158/0008-5472.CAN-14-0787. [DOI] [PubMed] [Google Scholar]
- 152.Luo A., Du C., Zhao X., Liang J., Chen Y. [Effects of recombinant adenovirus Ad-miR-29b2c on HGC-27 cell proliferation and migration] Sheng Wu Gong Cheng Xue Bao. 2017;33:1136–1144. doi: 10.13345/j.cjb.170003. [DOI] [PubMed] [Google Scholar]
- 153.Tan M., Wu J., Cai Y. Suppression of Wnt signaling by the miR-29 family is mediated by demethylation of WIF-1 in non-small-cell lung cancer. Biochem. Biophys. Res. Commun. 2013;438:673–679. doi: 10.1016/j.bbrc.2013.07.123. [DOI] [PubMed] [Google Scholar]
- 154.Drago-Ferrante R., Pentimalli F., Carlisi D., De Blasio A., Saliba C., Baldacchino S., Degaetano J., Debono J., Caruana-Dingli G., Grech G. Suppressive role exerted by microRNA-29b-1-5p in triple negative breast cancer through SPIN1 regulation. Oncotarget. 2017;8:28939–28958. doi: 10.18632/oncotarget.15960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.To S.K.Y., Mak A.S.C., Eva Fung Y.M., Che C.M., Li S.S., Deng W., Ru B., Zhang J., Wong A.S.T. β-catenin downregulates Dicer to promote ovarian cancer metastasis. Oncogene. 2017;36:5927–5938. doi: 10.1038/onc.2017.185. [DOI] [PubMed] [Google Scholar]
- 156.Bähr C., Groner B. The IGF-1 receptor and its contributions to metastatic tumor growth-novel approaches to the inhibition of IGF-1R function. Growth Factors. 2005;23:1–14. doi: 10.1080/08977190400020229. [DOI] [PubMed] [Google Scholar]
- 157.Wang X., Liu S., Cao L., Zhang T., Yue D., Wang L., Ping Y., He Q., Zhang C., Wang M. miR-29a-3p suppresses cell proliferation and migration by downregulating IGF1R in hepatocellular carcinoma. Oncotarget. 2017;8:86592–86603. doi: 10.18632/oncotarget.21246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lu Y., Hu J., Sun W., Li S., Deng S., Li M. MiR-29c inhibits cell growth, invasion, and migration of pancreatic cancer by targeting ITGB1. OncoTargets Ther. 2015;9:99–109. doi: 10.2147/OTT.S92758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Blandin A.F., Renner G., Lehmann M., Lelong-Rebel I., Martin S., Dontenwill M. β1 Integrins as Therapeutic Targets to Disrupt Hallmarks of Cancer. Front. Pharmacol. 2015;6:279. doi: 10.3389/fphar.2015.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schwartz M.A., Schaller M.D., Ginsberg M.H. Integrins: emerging paradigms of signal transduction. Annu. Rev. Cell Dev. Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- 161.Wong C.C., Tse A.P., Huang Y.P., Zhu Y.T., Chiu D.K., Lai R.K., Au S.L., Kai A.K., Lee J.M., Wei L.L. Lysyl oxidase-like 2 is critical to tumor microenvironment and metastatic niche formation in hepatocellular carcinoma. Hepatology. 2014;60:1645–1658. doi: 10.1002/hep.27320. [DOI] [PubMed] [Google Scholar]
- 162.Zhong Y., Lu Y.T., Sun Y., Shi Z.H., Li N.G., Tang Y.P., Duan J.A. Recent opportunities in matrix metalloproteinase inhibitor drug design for cancer. Expert Opin. Drug Discov. 2018;13:75–87. doi: 10.1080/17460441.2018.1398732. [DOI] [PubMed] [Google Scholar]
- 163.Zou Y., Li J., Chen Z., Li X., Zheng S., Yi D., Zhong A., Chen J. miR-29c suppresses pancreatic cancer liver metastasis in an orthotopic implantation model in nude mice and affects survival in pancreatic cancer patients. Carcinogenesis. 2015;36:676–684. doi: 10.1093/carcin/bgv027. [DOI] [PubMed] [Google Scholar]
- 164.Fang J.H., Zhou H.C., Zeng C., Yang J., Liu Y., Huang X., Zhang J.P., Guan X.Y., Zhuang S.M. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54:1729–1740. doi: 10.1002/hep.24577. [DOI] [PubMed] [Google Scholar]
- 165.Kwon J.J., Willy J.A., Quirin K.A., Wek R.C., Korc M., Yin X.M., Kota J. Novel role of miR-29a in pancreatic cancer autophagy and its therapeutic potential. Oncotarget. 2016;7:71635–71650. doi: 10.18632/oncotarget.11928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tréhoux S., Lahdaoui F., Delpu Y., Renaud F., Leteurtre E., Torrisani J., Jonckheere N., Van Seuningen I. Micro-RNAs miR-29a and miR-330-5p function as tumor suppressors by targeting the MUC1 mucin in pancreatic cancer cells. Biochim. Biophys. Acta. 2015;1853(10 Pt A):2392–2403. doi: 10.1016/j.bbamcr.2015.05.033. [DOI] [PubMed] [Google Scholar]
- 167.Hollingsworth M.A., Swanson B.J. Mucins in cancer: protection and control of the cell surface. Nat. Rev. Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 168.Jonckheere N., Van Seuningen I. The membrane-bound mucins: how large O-glycoproteins play key roles in epithelial cancers and hold promise as biological tools for gene-based and immunotherapies. Crit. Rev. Oncog. 2008;14:177–196. doi: 10.1615/critrevoncog.v14.i2-3.30. [DOI] [PubMed] [Google Scholar]
- 169.Roy L.D., Sahraei M., Subramani D.B., Besmer D., Nath S., Tinder T.L., Bajaj E., Shanmugam K., Lee Y.Y., Hwang S.I. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30:1449–1459. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Takaku M., Grimm S.A., Shimbo T., Perera L., Menafra R., Stunnenberg H.G., Archer T.K., Machida S., Kurumizaka H., Wade P.A. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 2016;17:36. doi: 10.1186/s13059-016-0897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chou J., Lin J.H., Brenot A., Kim J.W., Provot S., Werb Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat. Cell Biol. 2013;15:201–213. doi: 10.1038/ncb2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sun X.J., Liu B.Y., Yan S., Jiang T.H., Cheng H.Q., Jiang H.S., Cao Y., Mao A.W. MicroRNA-29a Promotes Pancreatic Cancer Growth by Inhibiting Tristetraprolin. Cell. Physiol. Biochem. 2015;37:707–718. doi: 10.1159/000430389. [DOI] [PubMed] [Google Scholar]
- 173.Rostas J.W., 3rd, Pruitt H.C., Metge B.J., Mitra A., Bailey S.K., Bae S., Singh K.P., Devine D.J., Dyess D.L., Richards W.O. microRNA-29 negatively regulates EMT regulator N-myc interactor in breast cancer. Mol. Cancer. 2014;13:200. doi: 10.1186/1476-4598-13-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cochrane D.R., Jacobsen B.M., Connaghan K.D., Howe E.N., Bain D.L., Richer J.K. Progestin regulated miRNAs that mediate progesterone receptor action in breast cancer. Mol. Cell. Endocrinol. 2012;355:15–24. doi: 10.1016/j.mce.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bussard K.M., Mutkus L., Stumpf K., Gomez-Manzano C., Marini F.C. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016;18:84. doi: 10.1186/s13058-016-0740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rhim A.D., Oberstein P.E., Thomas D.H., Mirek E.T., Palermo C.F., Sastra S.A., Dekleva E.N., Saunders T., Becerra C.P., Tattersall I.W. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Özdemir B.C., Pentcheva-Hoang T., Carstens J.L., Zheng X., Wu C.C., Simpson T.R., Laklai H., Sugimoto H., Kahlert C., Novitskiy S.V. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Levental K.R., Yu H., Kass L., Lakins J.N., Egeblad M., Erler J.T., Fong S.F., Csiszar K., Giaccia A., Weninger W. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.He Y., Huang C., Zhang S.P., Sun X., Long X.R., Li J. The potential of microRNAs in liver fibrosis. Cell. Signal. 2012;24:2268–2272. doi: 10.1016/j.cellsig.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 180.Wang B., Komers R., Carew R., Winbanks C.E., Xu B., Herman-Edelstein M., Koh P., Thomas M., Jandeleit-Dahm K., Gregorevic P. Suppression of microRNA-29 expression by TGF-β1 promotes collagen expression and renal fibrosis. J. Am. Soc. Nephrol. 2012;23:252–265. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Harmanci D., Erkan E.P., Kocak A., Akdogan G.G. Role of the microRNA-29 family in fibrotic skin diseases. Biomed. Rep. 2017;6:599–604. doi: 10.3892/br.2017.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bonnans C., Chou J., Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhu J., Xiong G., Fu H., Evers B.M., Zhou B.P., Xu R. Chaperone Hsp47 Drives Malignant Growth and Invasion by Modulating an ECM Gene Network. Cancer Res. 2015;75:1580–1591. doi: 10.1158/0008-5472.CAN-14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yano H., Hamanaka R., Nakamura-Ota M., Zhang J.J., Matsuo N., Yoshioka H. Regulation of type I collagen expression by microRNA-29 following ionizing radiation. Radiat. Environ. Biophys. 2018;57:41–54. doi: 10.1007/s00411-017-0723-4. [DOI] [PubMed] [Google Scholar]
- 185.Lourdusamy A., Rahman R., Smith S., Grundy R. microRNA network analysis identifies miR-29 cluster as key regulator of LAMA2 in ependymoma. Acta Neuropathol. Commun. 2015;3:26. doi: 10.1186/s40478-015-0206-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhu L.H., Miao X.T., Wang N.Y. Integrated miRNA-mRNA analysis of Epstein-Barr virus-positive nasopharyngeal carcinoma. Genet. Mol. Res. 2015;14:6028–6036. doi: 10.4238/2015.June.1.20. [DOI] [PubMed] [Google Scholar]
- 187.Wang J., Yu H., Ye L., Jin L., Yu M., Lv Y. Integrated regulatory mechanisms of miRNAs and targeted genes involved in colorectal cancer. Int. J. Clin. Exp. Pathol. 2015;8:517–529. [PMC free article] [PubMed] [Google Scholar]
- 188.Yu P.N., Yan M.D., Lai H.C., Huang R.L., Chou Y.C., Lin W.C., Yeh L.T., Lin Y.W. Downregulation of miR-29 contributes to cisplatin resistance of ovarian cancer cells. Int. J. Cancer. 2014;134:542–551. doi: 10.1002/ijc.28399. [DOI] [PubMed] [Google Scholar]
- 189.Heid J., Cencioni C., Ripa R., Baumgart M., Atlante S., Milano G., Scopece A., Kuenne C., Guenther S., Azzimato V. Age-dependent increase of oxidative stress regulates microRNA-29 family preserving cardiac health. Sci. Rep. 2017;7:16839. doi: 10.1038/s41598-017-16829-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kota J., Hancock J., Kwon J., Korc M. Pancreatic cancer: Stroma and its current and emerging targeted therapies. Cancer Lett. 2017;391:38–49. doi: 10.1016/j.canlet.2016.12.035. [DOI] [PubMed] [Google Scholar]
- 191.Apte M.V., Wilson J.S. Dangerous liaisons: pancreatic stellate cells and pancreatic cancer cells. J. Gastroenterol. Hepatol. 2012;27(Suppl 2):69–74. doi: 10.1111/j.1440-1746.2011.07000.x. [DOI] [PubMed] [Google Scholar]
- 192.Xuan J., Guo S.L., Huang A., Xu H.B., Shao M., Yang Y., Wen W. MiR-29a and miR-652 Attenuate Liver Fibrosis by Inhibiting the Differentiation of CD4+ T Cells. Cell Struct. Funct. 2017;42:95–103. doi: 10.1247/csf.17005. [DOI] [PubMed] [Google Scholar]
- 193.Parangi S., O’Reilly M., Christofori G., Holmgren L., Grosfeld J., Folkman J., Hanahan D. Antiangiogenic therapy of transgenic mice impairs de novo tumor growth. Proc. Natl. Acad. Sci. USA. 1996;93:2002–2007. doi: 10.1073/pnas.93.5.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Holmgren L., O’Reilly M.S., Folkman J. Dormancy of micrometastases: balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat. Med. 1995;1:149–153. doi: 10.1038/nm0295-149. [DOI] [PubMed] [Google Scholar]
- 195.Folkman J. Tumor angiogenesis: therapeutic implications. N. Engl. J. Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 196.Rajabi M., Mousa S.A. The Role of Angiogenesis in Cancer Treatment. Biomedicines. 2017;5:E34. doi: 10.3390/biomedicines5020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yang Z., Wu L., Zhu X., Xu J., Jin R., Li G., Wu F. MiR-29a modulates the angiogenic properties of human endothelial cells. Biochem. Biophys. Res. Commun. 2013;434:143–149. doi: 10.1016/j.bbrc.2013.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Shepard S.B., Broverman S.A., Muskavitch M.A. A tripartite interaction among alleles of Notch, Delta, and Enhancer of split during imaginal development of Drosophila melanogaster. Genetics. 1989;122:429–438. doi: 10.1093/genetics/122.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Simons M., Gordon E., Claesson-Welsh L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 2016;17:611–625. doi: 10.1038/nrm.2016.87. [DOI] [PubMed] [Google Scholar]
- 200.Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69(Suppl 3):4–10. doi: 10.1159/000088478. [DOI] [PubMed] [Google Scholar]
- 201.Li Y., Cai B., Shen L., Dong Y., Lu Q., Sun S., Liu S., Ma S., Ma P.X., Chen J. MiRNA-29b suppresses tumor growth through simultaneously inhibiting angiogenesis and tumorigenesis by targeting Akt3. Cancer Lett. 2017;397:111–119. doi: 10.1016/j.canlet.2017.03.032. [DOI] [PubMed] [Google Scholar]
- 202.Chen H.X., Xu X.X., Tan B.Z., Zhang Z., Zhou X.D. MicroRNA-29b Inhibits Angiogenesis by Targeting VEGFA through the MAPK/ERK and PI3K/Akt Signaling Pathways in Endometrial Carcinoma. Cell. Physiol. Biochem. 2017;41:933–946. doi: 10.1159/000460510. [DOI] [PubMed] [Google Scholar]
- 203.Xu W.W., Li B., Guan X.Y., Chung S.K., Wang Y., Yip Y.L., Law S.Y., Chan K.T., Lee N.P., Chan K.W. Cancer cell-secreted IGF2 instigates fibroblasts and bone marrow-derived vascular progenitor cells to promote cancer progression. Nat. Commun. 2017;8:14399. doi: 10.1038/ncomms14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhang K., Cai H.X., Gao S., Yang G.L., Deng H.T., Xu G.C., Han J., Zhang Q.Z., Li L.Y. TNFSF15 suppresses VEGF production in endothelial cells by stimulating miR-29b expression via activation of JNK-GATA3 signals. Oncotarget. 2016;7:69436–69449. doi: 10.18632/oncotarget.11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chen L., Xiao H., Wang Z.H., Huang Y., Liu Z.P., Ren H., Song H. miR-29a suppresses growth and invasion of gastric cancer cells in vitro by targeting VEGF-A. BMB Rep. 2014;47:39–44. doi: 10.5483/BMBRep.2014.47.1.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Szczyrba J., Nolte E., Hart M., Döll C., Wach S., Taubert H., Keck B., Kremmer E., Stöhr R., Hartmann A. Identification of ZNF217, hnRNP-K, VEGF-A and IPO7 as targets for microRNAs that are downregulated in prostate carcinoma. Int. J. Cancer. 2013;132:775–784. doi: 10.1002/ijc.27731. [DOI] [PubMed] [Google Scholar]
- 207.Gao S., Cheng C., Chen H., Li M., Liu K., Wang G. IGF1 3'UTR functions as a ceRNA in promoting angiogenesis by sponging miR-29 family in osteosarcoma. J. Mol. Histol. 2016;47:135–143. doi: 10.1007/s10735-016-9659-2. [DOI] [PubMed] [Google Scholar]
- 208.Hiroki E., Akahira J., Suzuki F., Nagase S., Ito K., Suzuki T., Sasano H., Yaegashi N. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. 2010;101:241–249. doi: 10.1111/j.1349-7006.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Frattari G., Aagaard L., Denton P.W. The role of miR-29a in HIV-1 replication and latency. J. Virus Erad. 2017;3:185–191. [PMC free article] [PubMed] [Google Scholar]
- 210.Picarda E., Ohaegbulam K.C., Zang X. Molecular Pathways: Targeting B7-H3 (CD276) for Human Cancer Immunotherapy. Clin. Cancer Res. 2016;22:3425–3431. doi: 10.1158/1078-0432.CCR-15-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Xu H., Cheung I.Y., Guo H.F., Cheung N.K. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: potential implications for immune based therapy of human solid tumors. Cancer Res. 2009;69:6275–6281. doi: 10.1158/0008-5472.CAN-08-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Amicarella F., Muraro M.G., Hirt C., Cremonesi E., Padovan E., Mele V., Governa V., Han J., Huber X., Droeser R.A. Dual role of tumour-infiltrating T helper 17 cells in human colorectal cancer. Gut. 2017;66:692–704. doi: 10.1136/gutjnl-2015-310016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Lund J.M., Alexopoulou L., Sato A., Karow M., Adams N.C., Gale N.W., Iwasaki A., Flavell R.A. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., Lipford G., Wagner H., Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 215.Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G.J. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Steiner D.F., Thomas M.F., Hu J.K., Yang Z., Babiarz J.E., Allen C.D., Matloubian M., Blelloch R., Ansel K.M. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.van der Woude L.L., Gorris M.A.J., Halilovic A., Figdor C.G., de Vries I.J.M. Migrating into the Tumor: a Roadmap for T Cells. Trends Cancer. 2017;3:797–808. doi: 10.1016/j.trecan.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 218.Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R.M., Okamoto A., Yokota J., Tanaka T. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 219.Landi M.T., Zhao Y., Rotunno M., Koshiol J., Liu H., Bergen A.W., Rubagotti M., Goldstein A.M., Linnoila I., Marincola F.M. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin. Cancer Res. 2010;16:430–441. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wu D.W., Hsu N.Y., Wang Y.C., Lee M.C., Cheng Y.W., Chen C.Y., Lee H. c-Myc suppresses microRNA-29b to promote tumor aggressiveness and poor outcomes in non-small cell lung cancer by targeting FHIT. Oncogene. 2015;34:2072–2082. doi: 10.1038/onc.2014.152. [DOI] [PubMed] [Google Scholar]
- 221.Zhang L., Ye Y., Tu H., Hildebrandt M.A., Zhao L., Heymach J.V., Roth J.A., Wu X. MicroRNA-related genetic variants in iron regulatory genes, dietary iron intake, microRNAs and lung cancer risk. Ann. Oncol. 2017;28:1124–1129. doi: 10.1093/annonc/mdx046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Joerger M., Baty F., Früh M., Droege C., Stahel R.A., Betticher D.C., von Moos R., Ochsenbein A., Pless M., Gautschi O. Circulating microRNA profiling in patients with advanced non-squamous NSCLC receiving bevacizumab/erlotinib followed by platinum-based chemotherapy at progression (SAKK 19/05) Lung Cancer. 2014;85:306–313. doi: 10.1016/j.lungcan.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 223.Butrym A., Rybka J., Baczyńska D., Poręba R., Kuliczkowski K., Mazur G. Clinical response to azacitidine therapy depends on microRNA-29c (miR-29c) expression in older acute myeloid leukemia (AML) patients. Oncotarget. 2016;7:30250–30257. doi: 10.18632/oncotarget.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhu C., Wang Y., Kuai W., Sun X., Chen H., Hong Z. Prognostic value of miR-29a expression in pediatric acute myeloid leukemia. Clin. Biochem. 2013;46:49–53. doi: 10.1016/j.clinbiochem.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 225.Xu L., Xu Y., Jing Z., Wang X., Zha X., Zeng C., Chen S., Yang L., Luo G., Li B., Li Y. Altered expression pattern of miR-29a, miR-29b and the target genes in myeloid leukemia. Exp. Hematol. Oncol. 2014;3:17. doi: 10.1186/2162-3619-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang D., Fan Z., Liu F., Zuo J. Hsa-miR-21 and Hsa-miR-29 in Tissue as Potential Diagnostic and Prognostic Biomarkers for Gastric Cancer. Cell. Physiol. Biochem. 2015;37:1454–1462. doi: 10.1159/000438514. [DOI] [PubMed] [Google Scholar]
- 227.Nguyen T., Kuo C., Nicholl M.B., Sim M.S., Turner R.R., Morton D.L., Hoon D.S. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics. 2011;6:388–394. doi: 10.4161/epi.6.3.14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Wu J., Li L., Jiang C. Identification and Evaluation of Serum MicroRNA-29 Family for Glioma Screening. Mol. Neurobiol. 2015;52:1540–1546. doi: 10.1007/s12035-014-8937-9. [DOI] [PubMed] [Google Scholar]
- 229.van Rooij E., Purcell A.L., Levin A.A. Developing microRNA therapeutics. Circ. Res. 2012;110:496–507. doi: 10.1161/CIRCRESAHA.111.247916. [DOI] [PubMed] [Google Scholar]
- 230.Chakraborty C., Sharma A.R., Sharma G., Doss C.G.P., Lee S.S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Slabáková E., Culig Z., Remšík J., Souček K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017;8:e3100. doi: 10.1038/cddis.2017.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Robbins M., Judge A., Ambegia E., Choi C., Yaworski E., Palmer L., McClintock K., MacLachlan I. Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation. Hum. Gene Ther. 2008;19:991–999. doi: 10.1089/hum.2008.131. [DOI] [PubMed] [Google Scholar]
- 233.Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 234.Wang H., Jiang Y., Peng H., Chen Y., Zhu P., Huang Y. Recent progress in microRNA delivery for cancer therapy by non-viral synthetic vectors. Adv. Drug Deliv. Rev. 2015;81:142–160. doi: 10.1016/j.addr.2014.10.031. [DOI] [PubMed] [Google Scholar]
- 235.Riley M.K., Vermerris W. Recent Advances in Nanomaterials for Gene Delivery-A Review. Nanomaterials (Basel) 2017;7:E94. doi: 10.3390/nano7050094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Herrera-Carrillo E., Liu Y.P., Berkhout B. Improving miRNA Delivery by Optimizing miRNA Expression Cassettes in Diverse Virus Vectors. Hum. Gene Ther. Methods. 2017;28:177–190. doi: 10.1089/hgtb.2017.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Duhachek-Muggy S., Zolkiewska A. ADAM12-L is a direct target of the miR-29 and miR-200 families in breast cancer. BMC Cancer. 2015;15:93. doi: 10.1186/s12885-015-1108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Cittelly D.M., Finlay-Schultz J., Howe E.N., Spoelstra N.S., Axlund S.D., Hendricks P., Jacobsen B.M., Sartorius C.A., Richer J.K. Progestin suppression of miR-29 potentiates dedifferentiation of breast cancer cells via KLF4. Oncogene. 2013;32:2555–2564. doi: 10.1038/onc.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ben-David U., Siranosian B., Ha G., Tang H., Oren Y., Hinohara K., Strathdee C.A., Dempster J., Lyons N.J., Burns R. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560:325–330. doi: 10.1038/s41586-018-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Dagogo-Jack I., Shaw A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 241.Flynt A.S., Lai E.C. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bartel D.P., Chen C.Z. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 243.Brenner J.L., Jasiewicz K.L., Fahley A.F., Kemp B.J., Abbott A.L. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr. Biol. 2010;20:1321–1325. doi: 10.1016/j.cub.2010.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Erhard F., Haas J., Lieber D., Malterer G., Jaskiewicz L., Zavolan M., Dölken L., Zimmer R. Widespread context dependency of microRNA-mediated regulation. Genome Res. 2014;24:906–919. doi: 10.1101/gr.166702.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Zhang X., Zhao X., Fiskus W., Lin J., Lwin T., Rao R., Zhang Y., Chan J.C., Fu K., Marquez V.E. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell. 2012;22:506–523. doi: 10.1016/j.ccr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 246.Mott J.L., Kurita S., Cazanave S.C., Bronk S.F., Werneburg N.W., Fernandez-Zapico M.E. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J. Cell. Biochem. 2010;110:1155–1164. doi: 10.1002/jcb.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Teng Y., Zhao L., Zhang Y., Chen W., Li X. Id-1, a protein repressed by miR-29b, facilitates the TGFβ1-induced epithelial-mesenchymal transition in human ovarian cancer cells. Cell. Physiol. Biochem. 2014;33:717–730. doi: 10.1159/000358647. [DOI] [PubMed] [Google Scholar]
- 248.Bakkar N., Wang J., Ladner K.J., Wang H., Dahlman J.M., Carathers M., Acharyya S., Rudnicki M.A., Hollenbach A.D., Guttridge D.C. IKK/NF-kappaB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J. Cell Biol. 2008;180:787–802. doi: 10.1083/jcb.200707179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Caretti G., Di Padova M., Micales B., Lyons G.E., Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wang H., Garzon R., Sun H., Ladner K.J., Singh R., Dahlman J., Cheng A., Hall B.M., Qualman S.J., Chandler D.S. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kouros-Mehr H., Slorach E.M., Sternlicht M.D., Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Kouros-Mehr H., Bechis S.K., Slorach E.M., Littlepage L.E., Egeblad M., Ewald A.J., Pai S.Y., Ho I.C., Werb Z. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–152. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Jacquemier J., Charafe-Jauffret E., Monville F., Esterni B., Extra J.M., Houvenaeghel G., Xerri L., Bertucci F., Birnbaum D. Association of GATA3, P53, Ki67 status and vascular peritumoral invasion are strongly prognostic in luminal breast cancer. Breast Cancer Res. 2009;11:R23. doi: 10.1186/bcr2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Usary J., Llaca V., Karaca G., Presswala S., Karaca M., He X., Langerød A., Kåresen R., Oh D.S., Dressler L.G. Mutation of GATA3 in human breast tumors. Oncogene. 2004;23:7669–7678. doi: 10.1038/sj.onc.1207966. [DOI] [PubMed] [Google Scholar]
- 255.Plaisier C.L., Pan M., Baliga N.S. A miRNA-regulatory network explains how dysregulated miRNAs perturb oncogenic processes across diverse cancers. Genome Res. 2012;22:2302–2314. doi: 10.1101/gr.133991.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Park S.Y., Lee J.H., Ha M., Nam J.W., Kim V.N. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat. Struct. Mol. Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.