Abstract
Generating a functional proteome requires the ribosome to carefully regulate disparate co-translational processes that determine the fate of nascent polypeptides. With protein synthesis being energetically expensive, the ribosome must balance the costs of efficiently making a protein with those of properly folding it. Emerging as a primary means of regulating this trade-off is the nonuniform rate of translation elongation that defines translation kinetics. The varying speeds with which the ribosome progresses along a transcript have been implicated in several aspects of protein biogenesis, including co-translational protein folding and translational fidelity, as well as gene expression by mediating mRNA decay and protein quality control pathways. The optimal translation kinetics required to efficiently execute these processes can be distinct. Thus, the ribosome is tasked with tightly regulating translation kinetics to balance these processes while maintaining adaptability for changing cellular conditions. In this review, we first discuss the regulatory role of translation elongation in protein biogenesis and what factors influence elongation kinetics. We then describe how changes in translation kinetics signal downstream pathways that dictate the fate of nascent polypeptides. By regulating these pathways, the kinetics of translation elongation has emerged as a critical tool for driving gene expression and maintaining proteostasis through varied mechanisms, including nascent chain folding and binding different ribosome-associated machinery. Indeed, a growing number of examples demonstrate the important role of local changes in elongation kinetics in modulating the pathophysiology of human disease.
Keywords: chaperone, translation, protein folding, protein misfolding, ribosome function, cotranslational folding, elongation rate, nascent chain, translational fidelity
Introduction
Fundamental to every cellular process is the accurate conversion of the genetic code into a functional protein. This not only involves generating a polymer of amino acids, but also critically depends on folding this polymer into its native conformation. Productive protein biogenesis requires intricate coordination of several factors in a multistep choreography in which one misstep can disrupt protein homeostasis (proteostasis). Such missteps can drive the pathogenesis of many different diseases (1), either by causing an imbalance in the abundance of native protein or an accumulation of misfolded protein and subsequent cytotoxic protein aggregates.
In this review, we discuss the role that translation elongation kinetics plays in regulating proteostasis by serving as a determinant of accurate synthesis, folding, and targeting of nascent proteins. We will describe the interdependent factors that modulate translation kinetics followed by discussing how such modulation coordinates protein biogenesis. Although many mechanistic details remain unclear, it is now firmly established that the kinetics of translation elongation is a critical factor in generating a healthy proteome.
Proteostasis begins at the ribosome
Pioneering work from Christian Anfinsen demonstrated that proteins can spontaneously adopt their final structure in vitro (2). This led to the proposal that the amino acid sequence carried all of the necessary information for a protein to reach its native conformation in an unassisted fashion. It is now clear, however, that not all proteins can adopt their native conformation through an unassisted folding trajectory (3–5). This is especially true for eukaryotic proteins that tend to be larger with multiple domains as compared with bacterial proteins (3, 6). Larger proteins innately have a greater conformational landscape from which they can sample non-native interactions. Thus, folding these proteins requires strict regulation to ensure that a limited number of productive folding pathways outcompete the many unproductive or misfolding pathways (3, 5, 7).
To overcome this challenge, the cell has evolved the capacity to co-translationally fold nascent polypeptides in a domain-wise fashion (8). Translation is a vectorial process that sequentially produces a nascent polypeptide from its N to C terminus. The benefit of the vectorial nature of translation is that it allows the cell to sequentially partition folding into the distinct structural components of multidomain proteins, thereby limiting the available conformational space of the nascent chain to more productive folding trajectories (9). However, the side effect is that nascent polypeptides remain vulnerable to misfolding and aggregation until they can adopt their final native conformation upon completion of translation (3, 5).
Balancing these competing pathways in vivo to maintain proteostasis requires the constant monitoring of the nascent chain throughout the elongation phase. At the heart of this surveillance is an elaborate proteostasis network, including chaperones, modification enzymes, and targeting factors, that directly binds ribosomes and regulates folding, assembly, targeting, and quality control of nascent polypeptides (3–5, 10). For instance, molecular chaperones and targeting factors are recruited to the ribosome and must coordinate their binding and release in the context of co-translational folding (3). Co-translational folding is also coupled to assembly into large protein complexes (11, 12). Although the interplay between these co-translational processes and the recruitment of proteostasis machinery is not well understood, it is clear that these processes are controlled by the kinetics of processing the nascent chain as translation proceeds. This suggests that the dynamics of translation elongation is coupled with co-translational processes, and translation kinetics has emerged as the driving force underlying such coordination.
Nonuniform rates of translation elongation drive proteostasis
Of the four major phases of translation (initiation, elongation, termination, and ribosome recycling), studies of protein synthesis have mostly focused on translation initiation as the rate-limiting step of translational control. However, increasing evidence, particularly with the advent of ribosome profiling (13), shows that balancing all four phases of translation is important for maintaining proteostasis, with translation elongation being central to determining protein fate.
During elongation, the ribosome acts as a central processing unit. It decodes mRNA and resolves competing signals in the crowded cytoplasmic environment to systematically recruit the machinery necessary to output a properly folded and targeted protein. Elongation proceeds at an average rate of ∼20 amino acids/s in prokaryotes and only ∼4–6 amino acids/s in eukaryotes, although mRNA transcripts have distinct elongation rates (7). By comparison, forming the stable secondary and tertiary structures of folding intermediates happens on the order of milliseconds (7). This indicates that mRNA decoding is rate-limiting as compared with protein folding (14). Therefore, given the length of eukaryotic proteins (median of ∼400 amino acids) (6), the proteostasis machinery must monitor the nascent chain and prevent it from forming non-native contacts for over a minute before the complete polypeptide can acquire its native conformation.
Yet, local elongation rates are not uniform across the length of the transcript, with the ribosome accelerating and slowing down during elongation (15). Variation in these rates can be over an order of magnitude (16). It is now recognized that these local elongation rates, in combination with global rates, create an evolutionarily selected system of trade-offs that modulate the cell's ability to generate a functional proteome (Fig. 1). One set of trade-offs pertains to how the elongation rate regulates gene expression: faster translation efficiently synthesizes a large amount of protein, whereas slow elongation can decrease mRNA stability and initiate mRNA decay and protein degradation pathways (17). For instance, in addition to reducing translation initiation, proteotoxic stress in mammalian cells can attenuate protein biogenesis by causing ribosomes to stall within the first 50 codons of the coding sequence and limiting further elongation (18, 19). However, it should be noted that these findings remain to be validated in light of recent work in yeast demonstrating that using cycloheximide can cause artifacts in ribosome-profiling experiments (20, 21).
Figure 1.
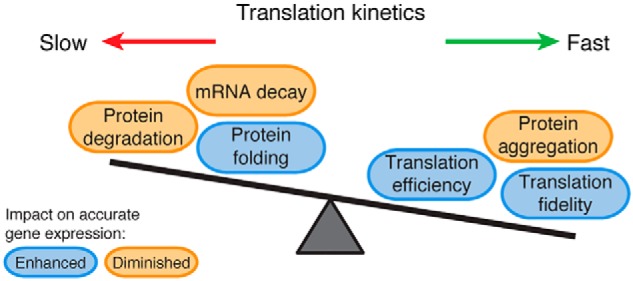
Translation kinetics balances protein production, folding, and quality control pathways. Nonuniform rates of translation elongation along a transcript dictate a set of trade-offs that either enhance (blue) or diminish (orange) accurate gene expression. Slower rates of elongation enhance accurate gene expression by facilitating proper protein folding, but very slow translation (ribosome stalling) diminishes gene expression by causing the turnover of the nascent protein and mRNA. Fast translation also has its trade-offs on accurate gene expression by enhancing the fidelity and efficiency of protein synthesis, while also increasing the likelihood of protein misfolding and aggregation.
The local kinetics of translation elongation also balances a trade-off between protein folding and the accuracy of protein synthesis. Elongation slowdowns generally enhance co-translational protein folding, putatively by giving more time for the protein to properly fold (15, 22). However, slower translation can also decrease translational fidelity, causing frameshifting (23) or amino acid misincorporation that could lead to protein misfolding (5, 24). With such trade-offs, elucidating the underlying mechanisms that regulate the health of the proteome requires greater understanding of the factors that dictate translation kinetics, as well as how translation kinetics subsequently balances these competing determinants of protein fate.
Determinants of translation kinetics
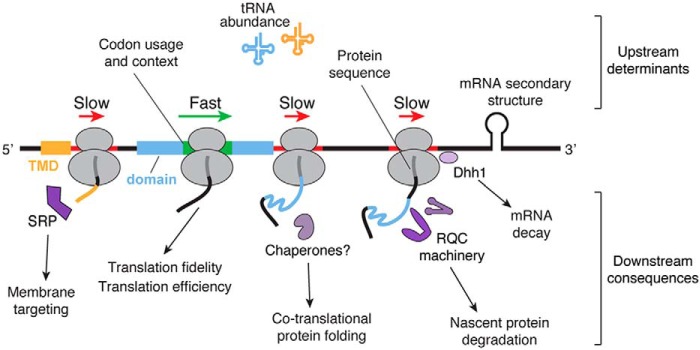
As translation elongation requires the coordination of disparate players, from mRNA and tRNA to ribosomal proteins and proteostasis machinery, several factors influence the rate of translation elongation and its downstream effects (Fig. 2). These include codon usage and codon context, tRNA abundance, protein sequence, and mRNA secondary structure (15, 16, 25). The most commonly studied factor is the frequency of codons in the mRNA transcriptome, referred to as codon usage or bias. The genetic code is degenerate with the same amino acid being encoded by different codons. Synonymous codons are represented in the genome at frequencies that can vary over an order of magnitude between rare and common codons (16). These different frequencies of synonymous codons define an organism's codon usage, which can vary between species. Synonymous codons were long viewed as equivalent, leading to the development of the term “silent” mutations to describe mutations from one synonymous codon to another without a change in amino acid. However, substantial work has shown that the use of “silent” is a misnomer as synonymous mutations are associated with disease (26). For instance, some work has shown that synonymous mutations can drive the development of human cancers (27). By impacting translational processes, as discussed further below, synonymous codons can modulate protein folding and activity (28–36) and even translation fidelity (24, 37, 38).
Figure 2.
Interdependent factors regulate translation kinetics and disparate downstream consequences through the recruitment of trans-acting factors. Several upstream variables dictate the speed at which the ribosome proceeds across an mRNA transcript. These determinants of translation kinetics include codon usage and tRNA abundance, which define codon optimality, along with pairs of codons (codon context), mRNA secondary structure, and protein sequence. Changes in elongation rate then modulate downstream pathways that determine the fate of nascent polypeptides. On the one hand, fast translation of protein structural elements enhances translation fidelity and translation efficiency. On the other hand, transient ribosome pausing facilitates subsequent processes by recruitment of various machinery. This includes membrane targeting of secretory proteins by recruiting the SRP after exposure of targeting signals such as a transmembrane domain (TMD), as well as protein folding possibly by recruiting molecular chaperones. Proper protein folding might then accelerate translation. When a ribosome stalls, RQC machinery is recruited to degrade the nascent polypeptide, and the DEAD-box helicase Dhh1 initiates mRNA decay.
Codon usage dictates the kinetics of translation elongation primarily through the interdependence between codon frequency and tRNA abundance (16, 39). Just as there are 61 sense codons coding for 20 amino acids, the presence of hundreds of tRNA genes in the genome provides an additional level of translation control. There is significant variation in the number of tRNA genes that decode a given codon. Some codons have only a single cognate tRNA gene, whereas other codons are decoded by multiple isoacceptor tRNA genes (same anticodon, but possibly sequence variation elsewhere in the tRNA). For instance, in yeast there is only one genomic copy of the arginine tRNA gene decoding codon CGG but 12 isoacceptor gene copies of the tRNA that decodes the arginine codon AGA. Still other codons do not have a cognate tRNA with perfect Watson-Crick base pairing, such as the arginine codon CGA, and instead require wobble interactions at the third nucleotide of the codon (first nucleotide of the anticodon) for decoding. Such variation in number of gene copies coding for a given tRNA positively correlates with the abundance of that tRNA within the cytoplasm: more abundant tRNAs have a greater number of gene copies (40, 41). This creates a tRNA pool of diverse composition, with tRNAs of different abundance competing for incorporation into the ribosome's peptidyl transferase center (PTC).2 Such competition influences the rate of elongation, particularly by the relative concentration of a tRNA to its near-cognate tRNAs (42). A small cognate/near-cognate tRNA ratio, where a low abundant tRNA must outcompete more abundant near-cognate tRNAs, slows the rate of translation as this competition necessitates more time to accurately incorporate the correct tRNA (15, 43).
In considering tRNA abundance in combination with codon usage, these variables compose the two sides of a supply (tRNA) and demand (mRNA) balance that is necessary for effective mRNA decoding and defines what is termed codon optimality (16, 22). Codon optimality is a measure of differential codon recognition that accounts for the correlated differences in codon usage and the availability of the decoding tRNA. It also accounts for the differences in interactions between the codon and the tRNA anticodon that cause variable decoding rates, such as wobble interactions that slow translation through inefficient codon–anticodon base-pairing (17, 44, 45). Optimal codons generally represent common codons that are efficiently decoded by abundant tRNA. By contrast, nonoptimal codons generally consist of rare codons decoded by wobble interactions or a lower abundant tRNA and are often under-represented in coding sequences (16, 39, 43). The net consequence is that nonoptimal codons are decoded more slowly than their synonymous optimal counterparts, thereby slowing translation (17, 45, 46). Altering codon optimality provides a versatile means to fine-tune the folding efficiency of a protein without altering its protein sequence (16, 33). Modifications of tRNA have also been shown to influence the elongation rate, presumably also by modulating decoding efficiencies (47). Interestingly, the composition of the tRNA pool is dynamic and can change with different cellular conditions or disease (48–50). As altered cellular conditions also influence the transcriptome, such changes will modulate the codon optimality classification and thus translation efficiency. As a result, genes with co-varying expression patterns, such as those in different tissues or pathways, have similar codon optimality (51). Importantly, if the supply and demand balance between the tRNA pool and the transcriptome is not maintained, this can disrupt proteostasis (52).
In addition to the dynamic nature of codon optimality determining the kinetics of translation, nascent protein sequence also plays a role (53). After accurate mRNA decoding, the next step in the elongation cycle involves the peptidyl transferase reaction promoting the addition of the incoming amino acid to the growing nascent chain. The efficiency of this reaction influences the rate of elongation. As proline creates unfavorable positioning for this reaction, both as a donor and acceptor (25, 54), the peptidyl transferase reaction is inefficient when proline is present in the A or P site, which consequently slows translation (55). This is particularly true at proline-rich motifs, thereby necessitating the recruitment of the elongation factor eIF5A (EF-P in bacteria) for progression (56–59). Through a distinct mechanism, regions rich in positively charged amino acids also cause a slowdown in translation (60–63). Such pausing occurs when these residues electrostatically interact with the negatively charged wall of the ribosome exit tunnel (64). These electrostatic interactions occur within 10 amino acids from the PTC where ribosomal proteins constrict the ribosome exit tunnel. Interestingly, it is proposed that these ribosomal proteins might facilitate the sensing of translation slowdowns by trans-acting factors (53).
Additional factors have also been identified as regulating translation kinetics. For instance, throughout the typical elongation cycle, as more than one tRNA is simultaneously incorporated in the ribosome, particular codon pairs have been shown to impact translation rate (55). Referred to as codon context, translation slows just when two nonoptimal codons are adjacent (55, 65). Other determinants of translation kinetics include ribosomal arrest peptides and mRNA secondary structure (66), which are reviewed in depth elsewhere (15).
Elongation speed regulates co-translational protein folding
Substantial work has shown that co-translational protein folding is modulated by the factors discussed above that dictate translation kinetics, with recent work confirming that elongation rate is altered (28, 34, 67, 68). These studies support a model by which slower translation facilitates proper protein folding. Increasing the abundance of tRNAs in prokaryotes, a common strategy in heterologous gene expression, was shown to increase protein aggregation presumably by enhancing elongation rate (69). Similarly, using synonymous codons to alter codon optimality has been shown to dictate folding pathways (28, 29, 33, 34, 68, 70). For instance, increasing the codon optimality of the cystic fibrosis transmembrane receptor or the circadian clock protein FRQ in Neurospora led to faster translation and greater abundance, but also less functionality and increased aggregation (29, 70). By contrast, replacing optimal codons with nonoptimal codons reduced the chance of protein misfolding (68). Still, other work has shown that optimal codons decrease the likelihood of mistranslation (24, 37, 71), such as at structurally sensitive sites like regions buried within the core of a protein where mistranslation would increase the chance of misfolding (71). Collectively, these studies demonstrate that the supply and demand of coding mRNA and decoding tRNA is not necessarily optimized for greatest gene expression. Instead, it is proposed that this balance might have evolved to ensure a properly folded proteome (72). However, the underlying mechanisms by which the rate of translation impacts protein folding remain poorly defined.
One mechanism underlying how slower translation facilitates protein folding is by regulating the synthesis of secondary and tertiary structural elements. Synonymous codons are not uniformly and randomly distributed. There are clusters of optimal and nonoptimal codons that are conserved in equal measure and show position-dependent evolutionary conservation (22). Clusters of rare codons are even present in highly expressed genes that generally have high codon optimality (5). By regulating translation kinetics and protein biogenesis, such conservation demonstrates that these clusters are maintained under positive selection. In particular, optimal codons are often enriched in highly conserved regions of structural domains where accurate translation is required (71, 73). By contrast, the locations of nonoptimal clusters correlate with structural boundaries, such as linker regions downstream of protein structural domains or separating smaller secondary structural motifs within the larger protein domain (16, 22, 74). Slowing translation at these points, when the structural elements have just emerged from the ribosome, is suggested to provide additional time for sub-domains and domains to sequentially fold into a lower energy folding intermediate before more of the protein emerges. Indeed, recent work used nuclear magnetic resonance (NMR) to demonstrate that changes in codon optimality impacted the protein conformational landscape that nascent chains sample (28). However, the details explaining how this additional time facilitates accurate folding remain to be determined.
Some work demonstrates that slowing translation allows the ribosome more time to constrain the folding pathways of the nascent polypeptide (75). The nascent chain traverses an ∼100-Å exit tunnel of the ribosome's large subunit, which encompasses 30–40 amino acids, before emerging and becoming accessible to molecular chaperones and other proteostasis machinery. Within the exit tunnel, the conformation of the nascent chain is largely constrained until it enters a wider part of the tunnel near the exit port (76). At this point just before emerging, the nascent chain can adopt α-helical secondary structure (76–79). Upon emergence from the ribosome, however, the folding energy landscape of the nascent chain greatly expands, necessitating additional mechanisms to help prevent the formation of non-native contacts and facilitate native contacts. It is proposed that the rate of translation helps modulate the folding energy landscape to influence the probability of these differential contacts (15, 80). By restricting this landscape, the ribosome stabilizes folding intermediates (14, 75, 81), and it might also delay folding until a sufficient length of polypeptide has been synthesized to form productive contacts. Moreover, folding exerts force on the nascent chain (82, 83), leading to an attractive hypothesis in which translation rate and folding are in a “tug-of-war” where translation slows to facilitate folding followed by productive folding leading to translation acceleration. Recent work might support this model by demonstrating that the rate of elongation accelerates when the Hsp70 Ssb is bound to the ribosome–nascent chain complex (84).
Another possible mechanism to explain how translation rate facilitates productive protein biogenesis is through the recruitment of machinery that recognizes the nascent chain and assists with its maturation and cellular targeting (Fig. 2). In fact, the ribosome serves as platform for recruiting a wide variety of such factors (5). For instance, a slowdown in translation enhances secretion efficiency by promoting the recruitment of the signal recognition particle (SRP) (85). A rare codon cluster that slows translation is often present 35–40 codons downstream of the targeting signals that are recognized by the SRP and co-translationally target secretory proteins to the endoplasmic reticulum (85, 86). When the nonoptimal codon cluster is in the ribosome's PTC, these targeting signals have just emerged from the ribosome exit tunnel. Hence, although it is not the predominant pathway for SRP recruitment (86, 87), this suggests that a slowdown in translation facilitates SRP recruitment and subsequent membrane targeting. Conversely, enrichment of optimal codons at conserved structural elements can reduce the dependence on chaperones for proper folding of certain nascent proteins (88, 89). This suggests that faster translation of these proteins correlates with decreased molecular chaperone dependences. This provides additional evidence to support the model in which recruitment of protein folding machinery to the ribosome correlates with a local slowdown in translation to fold the nascent chain.
Ribosome stalling dictates gene expression
In addition to the elongation rate regulating co-translational protein folding, the flow of ribosomal traffic also influences gene expression (17). One way is through codon optimality being correlated with gene expression: Highly expressed genes generally have higher codon optimality leading to more efficient translation as compared with lowly expressed genes that often have lower codon optimality (90). This indicates that codon optimality is under evolutionary selection pressure to balance gene expression with proper protein folding (16).
Codon optimality also plays a role at the 5′ end of coding sequences, where nonoptimal codons are enriched. In this context, rather than slowing elongation, the reduced mRNA structure caused by AT-rich nonoptimal codons is suggested to enhance the rate of translation and increase translation efficiency, possibly by optimizing the spacing between ribosomes (40). Yet, these nonoptimal codons might also impact the efficiency of translation initiation, as including more than eight consecutive nonoptimal codons in the 5′ end of coding sequences causes decreased protein expression (91, 92).
Whereas the enhanced translation efficiency of fast translation is at one extreme on the continuum of translation kinetics, ribosomal traffic coming to a standstill is at the other extreme. Such ribosome stalling triggers protein degradation and mRNA decay pathways (93, 94). Although ribosome stalling and ribosome pausing are terms that are often used interchangeably, here we refer to ribosome stalling as a more extreme case of ribosome pausing in which ribosomes undergo elongation arrest as distinguished from the transient slowdowns of ribosome pausing. A growing number of examples demonstrate the involvement of ribosome stalling in gene expression (95). These cases include regulatory functions of ribosome stalling, in which the absence of stalling leads to pathological conditions (e.g. the loss of FMRP function causing Fragile X syndrome), as well as abnormal cases of ribosome stalling where it is the presence of stalling that leads to disease (e.g. coupling mutant tRNA with impaired ribosome recycling causes neurodegeneration (96)).
There are many triggers of ribosome stalling resulting from abnormal translation or aberrant mRNA. These triggers include chemical damage of the mRNA, mRNA cleavage, translation of the poly(A) tail or premature polyadenylation of a new transcript, translation of the 3′ UTR, or even certain amino acid sequences in the nascent polypeptide (94, 97). Such triggers lead to the turnover of the mRNA transcript through nonstop decay or no-go decay pathways (98). Indeed, codon optimality globally influences mRNA decay (99). Decreased optimality slows A-site decoding, and in yeast it ultimately recruits the DEAD-box helicase protein Dhh1 to initiate mRNA decay (100, 101).
Coupled with mRNA decay is the degradation of the nascent chain by the ribosome-associated quality control (RQC) pathway (60, 102), which is reviewed in detail elsewhere (94, 97). Of note, however, protein degradation pathways can also be activated without mRNA decay (103). Upon stalling of a ribosome, it was recently shown in yeast that upstream ribosomes will collide with the stalled ribosome (104). Such ribosome queuing then triggers the RQC cascade involving the systematic recruitment of trans-acting factors. Ribosomal proteins of the 40S small subunit are ubiquitinated by the ubiquitin ligase Hel2 (ZNF598 in mammals), which is facilitated by Asc1/RACK1 (60, 104–108). The 80S ribosome then dissociates from the mRNA after recruitment of ribosome recycling factors, i.e. Dom34/Hbs1/Rli1 in yeast or Pelota/Hbs1L or Gtpbp2/ABCE1 in mammals (109). Some work also suggests that the canonical translation termination factors can alternatively dissociate a stalled ribosome (110). After dissociation, the 60S–nascent chain complex associates with Rqc2/NEMF that facilitates the binding and subsequent ubiquitination of the nascent chain by Ltn1/Listerin, followed by extraction of the nascent chain and degradation (102, 111–114). Highlighting the importance of this pathway in regulating the fate of stalled nascent proteins, disruption of RQC can lead to protein aggregation and neurodegeneration (115–117).
To discuss the impact of translation kinetics on gene expression more generally, consider a model that uses translation kinetics to describe the probability of properly folding nascent proteins (Fig. 3). In the optimal conditions of homeostasis, translation kinetics consists of balanced nonuniform rates of elongation that maximize the chances of a nascent polypeptide acquiring its native conformation. Deviating from this balance by aberrantly slowing elongation at some point along the transcript, as might be the case for a mutant gene, can decrease the likelihood of protein folding. A long enough slowdown will trigger mRNA decay and RQC pathways to down-regulate protein production. By contrast, moving the balance of translation kinetics toward aberrantly fast elongation will enhance translation efficiency and protein production, but with the trade-off of increased protein aggregation, as might be seen with other diseases or in some strategies of heterologous protein expression. Therefore, maintaining balanced translation kinetics remains critical for proteostasis as deviations in either direction of this balance combine possibly aberrant gene expression with a decreased likelihood of proper protein folding.
Figure 3.
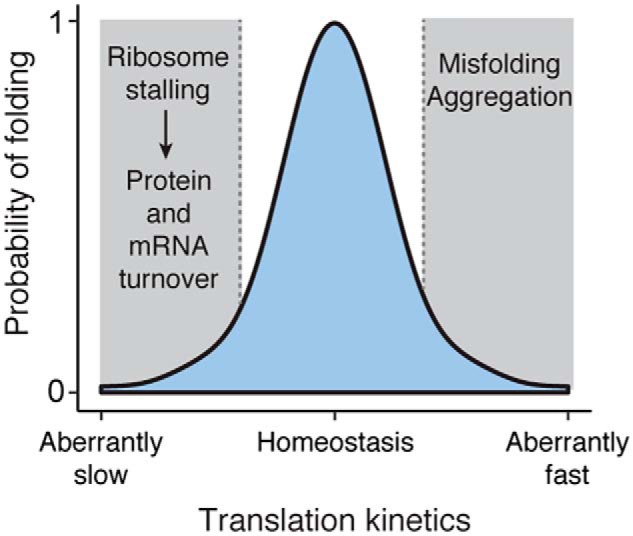
Modeling the impact of regulating translation kinetics. The kinetics of translation along an mRNA transcript is nonuniform and balanced between ribosome pausing and acceleration. In homeostatic conditions in the cell, this balance is optimized for proper folding of the nascent proteome. Deviation from this balance decreases the likelihood of folding and impacts the fate of the protein: Aberrantly fast translation leads to misfolding and aggregation, and aberrantly slow translation leads to protein and mRNA turnover. The details that define the probability distribution of protein folding will change in response to changing cellular environments. For instance, alterations in the tRNA pool will influence what codons are considered optimal versus nonoptimal, which subsequently impacts protein folding.
Conclusions and future perspectives
In this review, we have discussed how the kinetics of translation elongation impacts protein biogenesis. Nonuniform rates of elongation are conserved to balance accurate protein folding with efficient protein production. At one level, there is a trade-off between slower elongation helping to facilitate protein folding and faster elongation helping to prevent mistranslation. The net consequence of disrupting either side of this balance is often the same, both resulting in protein misfolding. At another level, there is a trade-off in translation kinetics regulating gene expression. So as not to sequester cellular resources, faster translation ensures highly abundant and longer proteins are efficiently synthesized. This is balanced with slower translation, and even ribosome stalling events, serving as a means of down-regulating gene expression by initiating mRNA and protein turnover.
The trade-offs represented by each of these two levels reveal both the complexity and importance of translation kinetics. Indeed, it becomes imperative to take context into consideration as changes in cellular conditions can alter balanced kinetics. This is epitomized by analyzing the composition of the tRNA pool, which can change in different environmental conditions or different tissues, and thereby alter codon optimality and its effects on the health of the proteome. Moreover, we discussed the emerging concept of translation kinetics being correlated with the activity of trans-acting factors that are recruited to the ribosome. In light of the number of chaperones and other proteins that associate with the ribosome, future work is needed to dissect what and how trans-acting factors respond to changes in translation kinetics depending on the cellular conditions. The advent of ribosome profiling and the development of additional technology have opened new doors to probe these questions in the context of the cell. This has helped establish the importance of translation kinetics in generating a functional proteome, and it has paved the way for understanding the role that translation kinetics plays in the context of human disease.
Acknowledgments
We apologize to those who have made valuable contributions to our understanding of translation kinetics, but whose work could not be cited due to space limitations. We thank members of the Frydman laboratory for valuable discussions.
This work was supported by National Institutes of Health Grants AG047126 (to K. C. S.) and GM056433 (to J. F.). This is the first article in the JBC Reviews series “Molecular chaperones and protein quality control.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PTC
- peptidyl transferase center
- SRP
- signal recognition particle
- RQC
- ribosome-associated quality control.
References
- 1. Chiti F., and Dobson C. M. (2017) Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 10.1146/annurev-biochem-061516-045115 [DOI] [PubMed] [Google Scholar]
- 2. Anfinsen C. B. (1973) Principles that govern the folding of protein chains. Science 181, 223–230 10.1126/science.181.4096.223 [DOI] [PubMed] [Google Scholar]
- 3. Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M., and Hartl F. U. (2013) Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- 4. Kramer G., Boehringer D., Ban N., and Bukau B. (2009) The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 10.1038/nsmb.1614 [DOI] [PubMed] [Google Scholar]
- 5. Pechmann S., Willmund F., and Frydman J. (2013) The ribosome as a hub for protein quality control. Mol. Cell 49, 411–421 10.1016/j.molcel.2013.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Netzer W. J., and Hartl F. U. (1997) Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature 388, 343–349 10.1038/41024 [DOI] [PubMed] [Google Scholar]
- 7. Kaiser C. M., and Liu K. (2018) Folding up and moving on-nascent protein folding on the ribosome. J. Mol. Biol. 430, 4580–4591 10.1016/j.jmb.2018.06.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frydman J., Nimmesgern E., Ohtsuka K., and Hartl F. U. (1994) Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature 370, 111–117 10.1038/370111a0 [DOI] [PubMed] [Google Scholar]
- 9. Frydman J., Erdjument-Bromage H., Tempst P., and Hartl F. U. (1999) Co-translational domain folding as the structural basis for the rapid de novo folding of firefly luciferase. Nat. Struct. Biol. 6, 697–705 10.1038/10754 [DOI] [PubMed] [Google Scholar]
- 10. Preissler S., and Deuerling E. (2012) Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 37, 274–283 10.1016/j.tibs.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 11. Duncan C. D., and Mata J. (2011) Widespread cotranslational formation of protein complexes. PLoS Genet. 7, e1002398 10.1371/journal.pgen.1002398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shiber A., Döring K., Friedrich U., Klann K., Merker D., Zedan M., Tippmann F., Kramer G., and Bukau B. (2018) Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561, 268–272 10.1038/s41586-018-0462-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ingolia N. T., Ghaemmaghami S., Newman J. R., and Weissman J. S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holtkamp W., Kokic G., Jäger M., Mittelstaet J., Komar A. A., and Rodnina M. V. (2015) Cotranslational protein folding on the ribosome monitored in real time. Science 350, 1104–1107 10.1126/science.aad0344 [DOI] [PubMed] [Google Scholar]
- 15. Rodnina M. V. (2016) The ribosome in action: tuning of translational efficiency and protein folding. Protein Sci. 25, 1390–1406 10.1002/pro.2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chaney J. L., and Clark P. L. (2015) Roles for synonymous codon usage in protein biogenesis. Annu. Rev. Biophys. 44, 143–166 10.1146/annurev-biophys-060414-034333 [DOI] [PubMed] [Google Scholar]
- 17. Brule C. E., and Grayhack E. J. (2017) Synonymous codons: choose wisely for expression. Trends Genet. 33, 283–297 10.1016/j.tig.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu B., Han Y., and Qian S.-B. (2013) Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol. Cell 49, 453–463 10.1016/j.molcel.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shalgi R., Hurt J. A., Krykbaeva I., Taipale M., Lindquist S., and Burge C. B. (2013) Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell 49, 439–452 10.1016/j.molcel.2012.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gerashchenko M. V., and Gladyshev V. N. (2014) Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 42, e134 10.1093/nar/gku671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hussmann J. A., Patchett S., Johnson A., Sawyer S., and Press W. H. (2015) Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet. 11, e1005732 10.1371/journal.pgen.1005732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pechmann S., and Frydman J. (2013) Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 20, 237–243 10.1038/nsmb.2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolf A. S., and Grayhack E. J. (2015) Asc1, homolog of human RACK1, prevents frameshifting in yeast by ribosomes stalled at CGA codon repeats. RNA 21, 935–945 10.1261/rna.049080.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Drummond D. A., and Wilke C. O. (2008) Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134, 341–352 10.1016/j.cell.2008.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Choi J., Grosely R., Prabhakar A., Lapointe C. P., Wang J., and Puglisi J. D. (2018) How messenger RNA and nascent chain sequences regulate translation elongation. Annu. Rev. Biochem. 87, 421–449 10.1146/annurev-biochem-060815-014818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hunt R. C., Simhadri V. L., Iandoli M., Sauna Z. E., and Kimchi-Sarfaty C. (2014) Exposing synonymous mutations. Trends Genet. 30, 308–321 10.1016/j.tig.2014.04.006 [DOI] [PubMed] [Google Scholar]
- 27. Supek F., Miñana B., Valcárcel J., Gabaldón T., and Lehner B. (2014) Synonymous mutations frequently act as driver mutations in human cancers. Cell 156, 1324–1335 10.1016/j.cell.2014.01.051 [DOI] [PubMed] [Google Scholar]
- 28. Buhr F., Jha S., Thommen M., Mittelstaet J., Kutz F., Schwalbe H., Rodnina M. V., and Komar A. A. (2016) Synonymous codons direct cotranslational folding toward different protein conformations. Mol. Cell 61, 341–351 10.1016/j.molcel.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim S. J., Yoon J. S., Shishido H., Yang Z., Rooney L. A., Barral J. M., and Skach W. R. (2015) Protein folding. Translational tuning optimizes nascent protein folding in cells. Science 348, 444–448 10.1126/science.aaa3974 [DOI] [PubMed] [Google Scholar]
- 30. Kimchi-Sarfaty C., Oh J. M., Kim I. W., Sauna Z. E., Calcagno A. M., Ambudkar S. V., and Gottesman M. M. (2007) A “Silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528 10.1126/science.1135308 [DOI] [PubMed] [Google Scholar]
- 31. Kirchner S., Cai Z., Rauscher R., Kastelic N., Anding M., Czech A., Kleizen B., Ostedgaard L. S., Braakman I., Sheppard D. N., and Ignatova Z. (2017) Alteration of protein function by a silent polymorphism linked to tRNA abundance. PLoS Biol. 15, e2000779 10.1371/journal.pbio.2000779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O'Brien E. P., Vendruscolo M., and Dobson C. M. (2012) Prediction of variable translation rate effects on cotranslational protein folding. Nat. Commun. 3, 868 10.1038/ncomms1850 [DOI] [PubMed] [Google Scholar]
- 33. Sander I. M., Chaney J. L., and Clark P. L. (2014) Expanding Anfinsen's principle: contributions of synonymous codon selection to rational protein design. J. Am. Chem. Soc. 136, 858–861 10.1021/ja411302m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu C.-H., Dang Y., Zhou Z., Wu C., Zhao F., Sachs M. S., and Liu Y. (2015) Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell 59, 744–754 10.1016/j.molcel.2015.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang G., Hubalewska M., and Ignatova Z. (2009) Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 16, 274–280 10.1038/nsmb.1554 [DOI] [PubMed] [Google Scholar]
- 36. Komar A. A. (2009) A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 34, 16–24 10.1016/j.tibs.2008.10.002 [DOI] [PubMed] [Google Scholar]
- 37. Kramer E. B., Vallabhaneni H., Mayer L. M., and Farabaugh P. J. (2010) A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA 16, 1797–1808 10.1261/rna.2201210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zaher H. S., and Green R. (2009) Fidelity at the molecular level: lessons from protein synthesis. Cell 136, 746–762 10.1016/j.cell.2009.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ikemura T. (1985) Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2, 13–34 [DOI] [PubMed] [Google Scholar]
- 40. Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., and Pilpel Y. (2010) An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141, 344–354 10.1016/j.cell.2010.03.031 [DOI] [PubMed] [Google Scholar]
- 41. Gingold H., and Pilpel Y. (2011) Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 7, 481 10.1038/msb.2011.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fluitt A., Pienaar E., and Viljoen H. (2007) Ribosome kinetics and aa-tRNA competition determine rate and fidelity of peptide synthesis. Comput. Biol. Chem. 31, 335–346 10.1016/j.compbiolchem.2007.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hanson G., and Coller J. (2018) Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 19, 20–30 10.1038/nrm.2017.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stadler M., and Fire A. (2011) Wobble base-pairing slows in vivo translation elongation in metazoans. RNA 17, 2063–2073 10.1261/rna.02890211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Letzring D. P., Dean K. M., and Grayhack E. J. (2010) Control of translation efficiency in yeast by codon-anticodon interactions. RNA 16, 2516–2528 10.1261/rna.2411710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gardin J., Yeasmin R., Yurovsky A., Cai Y., Skiena S., and Futcher B. (2014) Measurement of average decoding rates of the 61 sense codons in vivo. eLife 2014 3, 10.7554/eLife.03735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nedialkova D. D., and Leidel S. A. (2015) Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161, 1606–1618 10.1016/j.cell.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gingold H., Tehler D., Christoffersen N. R., Nielsen M. M., Asmar F., Kooistra S. M., Christophersen N. S., Christensen L. L., Borre M., Sørensen K. D., Andersen L. D., Andersen C. L., Hulleman E., Wurdinger T., Ralfkiær E., et al. (2014) A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292 10.1016/j.cell.2014.08.011 [DOI] [PubMed] [Google Scholar]
- 49. Sagi D., Rak R., Gingold H., Adir I., Maayan G., Dahan O., Broday L., Pilpel Y., and Rechavi O. (2016) Tissue- and time-specific expression of otherwise identical tRNA genes. PLoS Genet. 12, e1006264 10.1371/journal.pgen.1006264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goodarzi H., Nguyen H. C. B., Zhang S., Dill B. D., Molina H., and Tavazoie S. F. (2016) Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell 165, 1416–1427 10.1016/j.cell.2016.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Man O., and Pilpel Y. (2007) Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat. Genet. 39, 415–421 10.1038/ng1967 [DOI] [PubMed] [Google Scholar]
- 52. Yona A. H., Bloom-Ackermann Z., Frumkin I., Hanson-Smith V., Charpak-Amikam Y., Feng Q., Boeke J. D., Dahan O., and Pilpel Y. (2013) tRNA genes rapidly change in evolution to meet novel translational demands. eLife 2, e01339 10.7554/eLife.01339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wilson D. N., Arenz S., and Beckmann R. (2016) Translation regulation via nascent polypeptide-mediated ribosome stalling. Curr. Opin. Struct. Biol. 37, 123–133 10.1016/j.sbi.2016.01.008 [DOI] [PubMed] [Google Scholar]
- 54. Wohlgemuth I., Brenner S., Beringer M., and Rodnina M. V. (2008) Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J. Biol. Chem. 283, 32229–32235 10.1074/jbc.M805316200 [DOI] [PubMed] [Google Scholar]
- 55. Gamble C. E., Brule C. E., Dean K. M., Fields S., and Grayhack E. J. (2016) Adjacent codons act in concert to modulate translation efficiency in yeast. Cell 166, 679–690 10.1016/j.cell.2016.05.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Woolstenhulme C. J., Guydosh N. R., Green R., and Buskirk A. R. (2015) High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 11, 13–21 10.1016/j.celrep.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Doerfel L. K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., and Rodnina M. V. (2013) EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339, 85–88 10.1126/science.1229017 [DOI] [PubMed] [Google Scholar]
- 58. Gutierrez E., Shin B.-S., Woolstenhulme C. J., Kim J.-R., Saini P., Buskirk A. R., and Dever T. E. (2013) eIF5A promotes translation of polyproline motifs. Mol. Cell 51, 35–45 10.1016/j.molcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schuller A. P., Wu C. C., Dever T. E., Buskirk A. R., and Green R. (2017) eIF5A functions globally in translation elongation and termination. Mol. Cell 66, 194–205.e5 10.1016/j.molcel.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C. C., Li G.-W., Zhou S., King D., Shen P. S., Weibezahn J., Dunn J. G., Rouskin S., Inada T., Frost A., and Weissman J. S. (2012) A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 10.1016/j.cell.2012.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dimitrova L. N., Kuroha K., Tatematsu T., and Inada T. (2009) Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 284, 10343–10352 10.1074/jbc.M808840200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sabi R., and Tuller T. (2015) A comparative genomics study on the effect of individual amino acids on ribosome stalling. BMC Genomics 16, Suppl. 10, S5 10.1186/1471-2164-16-S10-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weinberg D. E., Shah P., Eichhorn S. W., Hussmann J. A., Plotkin J. B., and Bartel D. P. (2016) Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 14, 1787–1799 10.1016/j.celrep.2016.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lu J., and Deutsch C. (2008) Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 384, 73–86 10.1016/j.jmb.2008.08.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Letzring D. P., Wolf A. S., Brule C. E., and Grayhack E. J. (2013) Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA 19, 1208–1217 10.1261/rna.039446.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Keedy H. E., Thomas E. N., and Zaher H. S. (2018) Decoding on the ribosome depends on the structure of the mRNA phosphodiester backbone. Proc. Natl. Acad. Sci. U.S.A. 115, E6731–E6740 10.1073/pnas.1721431115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thommen M., Holtkamp W., and Rodnina M. V. (2017) Co-translational protein folding: progress and methods. Curr. Opin. Struct. Biol. 42, 83–89 10.1016/j.sbi.2016.11.020 [DOI] [PubMed] [Google Scholar]
- 68. Spencer P. S., Siller E., Anderson J. F., and Barral J. M. (2012) Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol. 422, 328–335 10.1016/j.jmb.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fedyunin I., Lehnhardt L., Böhmer N., Kaufmann P., Zhang G., and Ignatova Z. (2012) tRNA concentration fine tunes protein solubility. FEBS Lett. 586, 3336–3340 10.1016/j.febslet.2012.07.012 [DOI] [PubMed] [Google Scholar]
- 70. Zhou M., Guo J., Cha J., Chae M., Chen S., Barral J. M., Sachs M. S., and Liu Y. (2013) Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495, 111–115 10.1038/nature11833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhou T., Weems M., and Wilke C. O. (2009) Translationally optimal codons associate with structurally sensitive sites in proteins. Mol. Biol. Evol. 26, 1571–1580 10.1093/molbev/msp070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dana A., and Tuller T. (2014) The effect of tRNA levels on decoding times of mRNA codons. Nucleic Acids Res. 42, 9171–9181 10.1093/nar/gku646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lee Y., Zhou T., Tartaglia G. G., Vendruscolo M., and Wilke C. O. (2010) Translationally optimal codons associate with aggregation-prone sites in proteins. Proteomics 10, 4163–4171 10.1002/pmic.201000229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chaney J. L., Steele A., Carmichael R., Rodriguez A., Specht A. T., Ngo K., Li J., Emrich S., and Clark P. L. (2017) Widespread position-specific conservation of synonymous rare codons within coding sequences. PLoS Comput. Biol. 13, e1005531 10.1371/journal.pcbi.1005531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kaiser C. M., Goldman D. H., Chodera J. D., Tinoco I. Jr., and Bustamante C. (2011) The ribosome modulates nascent protein folding. Science 334, 1723–1727 10.1126/science.1209740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lu J., and Deutsch C. (2005) Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 12, 1123–1129 10.1038/nsmb1021 [DOI] [PubMed] [Google Scholar]
- 77. Marino J., von Heijne G, Beckmann R. (2016) Small protein domains fold inside the ribosome exit tunnel. FEBS Lett. 590, 655–660 10.1002/1873-3468.12098 [DOI] [PubMed] [Google Scholar]
- 78. Nilsson O. B., Hedman R., Marino J., Wickles S., Bischoff L., Johansson M., Müller-Lucks A., Trovato F., Puglisi J. D., O'Brien E. P., Beckmann R., and von Heijne G. (2015) Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep. 12, 1533–1540 10.1016/j.celrep.2015.07.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kosolapov A., and Deutsch C. (2009) Tertiary interactions within the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 16, 405–411 10.1038/nsmb.1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tsai C.-J., Sauna Z. E., Kimchi-Sarfaty C., Ambudkar S. V., Gottesman M. M., and Nussinov R. (2008) Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J. Mol. Biol. 383, 281–291 10.1016/j.jmb.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cabrita L. D., Cassaignau A. M. E., Launay H. M. M., Waudby C. A., Wlodarski T., Camilloni C., Karyadi M.-E., Robertson A. L., Wang X., Wentink A. S., Goodsell L., Woolhead C. A., Vendruscolo M., Dobson C. M., and Christodoulou J. (2016) A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding. Nat. Struct. Mol. Biol. 23, 278–285 10.1038/nsmb.3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cymer F., and von Heijne G. (2013) Cotranslational folding of membrane proteins probed by arrest-peptide-mediated force measurements. Proc. Natl. Acad. Sci. U.S.A. 110, 14640–14645 10.1073/pnas.1306787110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Goldman D. H., Kaiser C. M., Milin A., Righini M., Tinoco I. Jr., and Bustamante C. (2015) Ribosome. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science 348, 457–460 10.1126/science.1261909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Döring K., Ahmed N., Riemer T., Suresh H. G., Vainshtein Y., Habich M., Riemer J., Mayer M. P., O'Brien E. P., Kramer G., and Bukau B. (2017) Profiling Ssb-nascent chain interactions reveals principles of Hsp70-assisted folding. Cell 170, 298–311.e20 10.1016/j.cell.2017.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pechmann S., Chartron J. W., and Frydman J. (2014) Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat. Struct. Mol. Biol. 21, 1100–1105 10.1038/nsmb.2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chartron J. W., Hunt K. C., and Frydman J. (2016) Cotranslational signal-independent SRP preloading during membrane targeting. Nature 536, 224–228 10.1038/nature19309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schibich D., Gloge F., Pöhner I., Björkholm P., Wade R. C., von Heijne G. Bukau B., and Kramer G. (2016) Global profiling of SRP interaction with nascent polypeptides. Nature 536, 219–223 10.1038/nature19070 [DOI] [PubMed] [Google Scholar]
- 88. Geller R., Pechmann S., Acevedo A., Andino R., and Frydman J. (2018) Hsp90 shapes protein and RNA evolution to balance trade-offs between protein stability and aggregation. Nat. Commun. 9, 1781 10.1038/s41467-018-04203-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Warnecke T., and Hurst L. D. (2010) GroEL dependency affects codon usage–support for a critical role of misfolding in gene evolution. Mol. Syst. Biol. 6, 340 10.1038/msb.2009.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Komar A. A. (2016) The Yin and Yang of codon usage. Hum. Mol. Genet. 25, R77–R85 10.1093/hmg/ddw207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chu D., Kazana E., Bellanger N., Singh T., Tuite M. F., and von der Haar T. (2014) Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 33, 21–34 10.1002/embj.201385651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Arthur L. L., Chung J. J., Jankirama P., Keefer K. M., Kolotilin I., Pavlovic-Djuranovic S., Chalker D. L., Grbic V., Green R., Menassa R., True H. L., Skeath J. B., and Djuranovic S. (2017) Rapid generation of hypomorphic mutations. Nat. Commun. 8, 14112 10.1038/ncomms14112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schuller A. P., and Green R. (2018) Roadblocks and resolutions in eukaryotic translation. Nat. Rev. Mol. Cell Biol. 19, 526–541 10.1038/s41580-018-0011-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Joazeiro C. A. P. (2017) Ribosomal stalling during translation: providing substrates for ribosome-associated protein quality control. Annu. Rev. Cell Dev. Biol. 33, 343–368 10.1146/annurev-cellbio-111315-125249 [DOI] [PubMed] [Google Scholar]
- 95. Richter J. D., and Coller J. (2015) Pausing on polyribosomes: make way for elongation in translational control. Cell 163, 292–300 10.1016/j.cell.2015.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ishimura R., Nagy G., Dotu I., Zhou H., Yang X.-L., Schimmel P., Senju S., Nishimura Y., Chuang J. H., and Ackerman S. L. (2014) RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–459 10.1126/science.1249749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Brandman O., and Hegde R. S. (2016) Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 23, 7–15 10.1038/nsmb.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Shoemaker C. J., and Green R. (2012) Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 19, 594–601 10.1038/nsmb.2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Presnyak V., Alhusaini N., Chen Y.-H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K. E., Graveley B. R., and Coller J. (2015) Codon optimality is a major determinant of mRNA stability. Cell 160, 1111–1124 10.1016/j.cell.2015.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Radhakrishnan A., Chen Y.-H., Martin S., Alhusaini N., Green R., and Coller J. (2016) The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167, 122–132.e9 10.1016/j.cell.2016.08.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hanson G., Alhusaini N., Morris N., Sweet T., and Coller J. (2018) Translation elongation and mRNA stability are coupled through the ribosomal A-site. RNA 24, 1377–1389 10.1261/rna.066787.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bengtson M. H., and Joazeiro C. A. (2010) Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 10.1038/nature09371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Arribere J. A., Cenik E. S., Jain N., Hess G. T., Lee C. H., Bassik M. C., and Fire A. Z. (2016) Translation readthrough mitigation. Nature 534, 719–723 10.1038/nature18308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Simms C. L., Yan L. L., and Zaher H. S. (2017) Ribosome collision is critical for quality control during no-go decay. Mol. Cell 68, 361–373.e5 10.1016/j.molcel.2017.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sundaramoorthy E., Leonard M., Mak R., Liao J., Fulzele A., and Bennett E. J. (2017) ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol. Cell 65, 751–760.e4 10.1016/j.molcel.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Juszkiewicz S., and Hegde R. S. (2017) Initiation of quality control during poly(A) translation requires site-specific ribosome ubiquitination. Mol. Cell 65, 743–750.e4 10.1016/j.molcel.2016.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Juszkiewicz S., Chandrasekaran V., Lin Z., Kraatz S., Ramakrishnan V., and Hegde R. S. (2018) ZNF598 is a quality control sensor of collided ribosomes. Mol. Cell 72, 469–481.e7 10.1016/j.molcel.2018.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Matsuo Y., Ikeuchi K., Saeki Y., Iwasaki S., Schmidt C., Udagawa T., Sato F., Tsuchiya H., Becker T., Tanaka K., Ingolia N. T., Beckmann R., and Inada T. (2017) Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 8, 159 10.1038/s41467-017-00188-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Tsuboi T., Kuroha K., Kudo K., Makino S., Inoue E., Kashima I., and Inada T. (2012) Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell 46, 518–529 10.1016/j.molcel.2012.03.013 [DOI] [PubMed] [Google Scholar]
- 110. Chiabudini M., Tais A., Zhang Y., Hayashi S., Wölfle T., Fitzke E., and Rospert S. (2014) Release factor eRF3 mediates premature translation termination on polylysine-stalled ribosomes in Saccharomyces cerevisiae. Mol. Cell. Biol. 34, 4062–4076 10.1128/MCB.00799-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lyumkis D., Oliveira dos Passos D., Tahara E. B., Webb K., Bennett E. J., Vinterbo S., Potter C. S., Carragher B., and Joazeiro C. A. (2014) Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proc. Natl. Acad. Sci. U.S.A. 111, 15981–15986 10.1073/pnas.1413882111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Shao S., von der Malsburg K, and Hegde R. S. (2013) Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell 50, 637–648 10.1016/j.molcel.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Shao S., Brown A., Santhanam B., and Hegde R. S. (2015) Structure and assembly pathway of the ribosome quality control complex. Mol. Cell 57, 433–444 10.1016/j.molcel.2014.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Shen P. S., Park J., Qin Y., Li X., Parsawar K., Larson M. H., Cox J., Cheng Y., Lambowitz A. M., Weissman J. S., Brandman O., and Frost A. (2015) Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75–78 10.1126/science.1259724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Choe Y.-J., Park S.-H., Hassemer T., Körner R., Vincenz-Donnelly L., Hayer-Hartl M., and Hartl F. U. (2016) Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature 531, 191–195 10.1038/nature16973 [DOI] [PubMed] [Google Scholar]
- 116. Yonashiro R., Tahara E. B., Bengtson M. H., Khokhrina M., Lorenz H., Chen K.-C., Kigoshi-Tansho Y., Savas J. N., Yates J. R. 3rd., Kay S. A., Craig E. A., Mogk A., Bukau B., and Joazeiro C. A. (2016) The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. eLife 5, e11794 10.7554/eLife.11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Chu J., Hong N. A., Masuda C. A., Jenkins B. V., Nelms K. A., Goodnow C. C., Glynne R. J., Wu H., Masliah E., Joazeiro C. A., and Kay S. A. (2009) A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 106, 2097–2103 10.1073/pnas.0812819106 [DOI] [PMC free article] [PubMed] [Google Scholar]